DOI:10.32604/cmc.2022.028722

| Computers, Materials & Continua DOI:10.32604/cmc.2022.028722 |  |
| Article |
Hybrid Segmentation Approach for Different Medical Image Modalities
1Department Electronics and Electrical Communications, Faculty of Electronic Engineering, Menoufia University, Menouf, 32952, Egypt
2Security Engineering Laboratory, Department of Computer Science, Prince Sultan University, Riyadh, 11586, Saudi Arabia
3Department of Information Technology, College of Computer and Information Sciences, Princess Nourah bint Abdulrahman University, Riyadh, 11671, Saudi Arabia
4Department of Computer Sciences, College of Computer and Information Sciences, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia
*Corresponding Author: Amel A. Alhussan. Email: aaalhussan@pnu.edu.sa
Received: 15 February 2022; Accepted: 23 March 2022
Abstract: The segmentation process requires separating the image region into sub-regions of similar properties. Each sub-region has a group of pixels having the same characteristics, such as texture or intensity. This paper suggests an efficient hybrid segmentation approach for different medical image modalities based on particle swarm optimization (PSO) and improved fast fuzzy C-means clustering (IFFCM) algorithms. An extensive comparative study on different medical images is presented between the proposed approach and other different previous segmentation techniques. The existing medical image segmentation techniques incorporate clustering, thresholding, graph-based, edge-based, active contour, region-based, and watershed algorithms. This paper extensively analyzes and summarizes the comparative investigation of these techniques. Finally, a prediction of the improvement involves the combination of these techniques is suggested. The obtained results demonstrate that the proposed hybrid medical image segmentation approach provides superior outcomes in terms of the examined evaluation metrics compared to the preceding segmentation techniques.
Keywords: Image segmentation; ultrasonic images; X-ray images; CT images; PET images; MR images; fuzzy c-mean; morphological operations; active contour
Medical image segmentation is one of the most common techniques used in telemedicine applications. There are various image segmentation algorithms such as thresholding, clustering, edge detection, region-based, graph-based, active contours, PSO, and watershed. Thresholding is one of the first-known techniques where segmentation is achieved by collecting all pixels with intensities between two thresholding values into the same class [1–3]. Clustering is organizing pixels into clusters whose members are similar. According to some criteria, it partitions the image region into a few number of clusters, according to some criteria. The technique stops merging regions when this number is reached [4–9].
Borders of a region are important features because sharp transitions in the intensity indicate the region’s edges [10–12]. So, edge detection techniques are often combined with another segmentation technique where edges resulting from edge detectors can be performed. The region-based technique (growing, splitting, and splitting & merging) utilizes the spatial image properties. It can be achieved by finding a seed pixel in the sub-region and growing that sub-region by grouping adjacent pixels until the pixels being compared to be added are not similar [13–15]. The graph-based technique divides the image region using matrices to yield a combination of disconnected subgraphs. The elements in a graph are pixels, and the edge weights measure the similarity between the two pixels connected by this edge. Finally, the watershed technique uses the image gradient magnitude to achieve segmentation. Pixels with the highest intensities of gradient magnitude values correspond to the watershed lines that outline the region [16,17].
PSO optimizes the problem by moving particles guided by the region’s best-found positions, which continuously change as they find better positions because they are fast and straightforward [18,19]. Active contour-based segmentation considers the contour of a specific object and utilizes a priori information of object shape to achieve segmentation. They include two main techniques, the parametric (also called snakes) and the geometric or level set. They can overcome the artifacts, boundary irregularities, and gaps by forcing the extracted boundaries to be smooth [20–22].
The segmentation’s main target is to make images easier to analyze and make the medical image more meaningful. This paper aims at:
■ We are segmenting the tumor region in different medical image multimodalities.
■ We are conducting a survey and a comparative study between the most well-known segmentation techniques for multimodal medical images.
■ Developing a hybrid proposed segmentation approach for different modalities of medical images.
■ The proposed approach uses PSO combined with the IFFCM to enhance the ROI, and then it can be easily extracted and classified into benign or malignant in future work.
■ Improve patient safety by offering better and more precise data for medical decisions.
This paper involves the overall goal of identifying and solving important and relevant medical problems.
The paper is arranged as follows. Section 2 describes the related work and summarizes the significant medical image segmentation techniques followed by a comparative study. Section 3 provides the proposed hybrid approach. Section 4 shows the results and discussion about the different segmentation techniques and the proposed approach. Finally, section 5 provides conclusions and future work.
The existing segmentation techniques include clustering, thresholding, region-based, edge-based, graph-based, watershed, PSO, and active contours. Many studies have been concerned with deep learning as a novel trend in image segmentation [23–29]. Authors in [23] proposed a lightweight threshold-based framework for feces segmentation. Their framework achieved a detection accuracy of 98.4%, with minimum storage space and low complexity. Authors in [24] proposed a shallow CNN segmentation and classification network for stool medical images. They firstly segmented the effective stool region, then employed a classification stage to classify the stool color. Their method achieved a low cost, accurate automatic segmentation, and color classification.
Authors in [25] proposed a Mask-Refined R-CNN (MR R-CNN). Their model had a low time cost and was easy to implement. Authors in [26] proposed an object detection approach for occluded and small objects. The presented approach was based on online complicated example mining (OHEM) and multi-layer convolution feature fusion (MCFF). Their model firstly generated the candidate regions using a region network optimized by MCFF. Also, the training samples were mined with OHEM. Their approach was performed on the KITTI dataset in an intelligent traffic scenario. It could also outperform popular methods, such as Faster R-CNN, Region-lets, in overall detection accuracy.
In [27], the authors introduced a method for brain tumor segmentation. Their method relied on the integration of the location information with state-of-the-art patch-based neural networks. Authors in [28] proposed a CNN architecture called Dense-Res-Inception Net (DRINet). This architecture consists of a deconvolutional block with residual inception modules, a convolutional block with dense connections, and an unpooling block. They compared their model with the U-Net in three distinct challenging applications.
The authors in [29] proposed a method for automatically segmenting Multiple sclerosis (MS) lesions based on 3D CNN. Their method is divided into two stages: two pooling layers and two convolution layers. Their method was applied to the MICCAI dataset. They also compared their method with the baseline and conventional methods. The achieved outcomes show that the proposed method has better efficiency than other baseline methods in terms of absolute difference in lesion volume, the dice coefficient, predictive positivity value, false-positive rate, and true positive rate.
This paper is concerned with summarizing and implementing some medical image segmentation techniques. Also, this paper introduces an efficient hybrid segmentation algorithm for reliable segmentation of multimodalities of medical images, as will be discussed in Section 3. Details about these techniques are summarized and discussed as follows:
This technique is a popular segmentation and is one of the simplest image segmentation methods. It directly divides the image region into sub-regions according to its value. There are two main thresholding categories; the first is the local threshold method which divides the image region into two sub-regions (the object and the background) using only one threshold. The second is the local threshold which uses two or more threshold values and divides the image region into multiple objects and backgrounds using multiple thresholds. The most popular threshold segmentation method is the Otsu technique, which chooses a global optimum threshold using the classes’ maximum variance.
For a threshold
In basic global thresholding, the threshold is chosen based on the minimum intra-class variance:
where the weights
This shows that decreasing the intra variance is the same as increasing inter variance:
which is expressed according to the sub-regions probabilities
which is verified as follows:
The probabilities and the means of different sub-regions can be computed iteratively. The main benefit of the thresholding methods is that they are fast and straightforward, especially when the object and the background are of high contrast. However, these methods have difficulty in choosing a proper threshold to obtain an accurate segmentation result. Also, the segmentation results degrade when there is a slight difference in the grayscale or a large interference between the image gray level values leading these methods to be often combined with other techniques.
Clustering is grouping or organizing pixels into clusters whose members are more similar. It is an unsupervised technique that performs segmentation without using training data. Clustering techniques collect the tested samples into separate classes or clusters based on the estimated distance.
Given the observations
where
The clustering technique is scalable, straightforward, efficient, and fast. The clustering technique’s disadvantage is that its cluster number has no obvious selection standards and is time-consuming to be estimated. Segmentation based on clustering includes: K-means, Improved Fuzzy C-means (IFCM), Improved, Fast Fuzzy C-Means (IFFCM), and Mean Shift clustering (MS).
The object edges are the most significant part of the image changes in local brightness, such as the gray value of the mutation. The abruptions are used to find out the image edges and to obtain an accurate image segmentation. There is always an edge between two adjacent sub-regions where the gray value has a discontinuity. This discontinuity can often be obtained by calculating the derivative via differential operators. The canny technique is usually used as an edge detector. This technique can be applied as follows:
• A Gaussian filter is applied to smooth the image.
• The gradient magnitude and the angle images are computed.
• Non-maximal suppression is then applied.
• Edges are detected and linked using double thresholding.
The amplitude and clustering methods depend on point properties in the image. Unlike these methods, region segmentation techniques are region-based ones. Region-based techniques utilize the spatial properties of an image for segmentation. Region-based techniques are three main categories: region growing, region splitting, and region splitting and merging. Region-based techniques (growing, splitting, and merging) can be used separately or inappropriate combining.
The process begins manually or automatically chooses a few seed pixels, then the similar pixels are gathered into the same sub-region. These techniques make use of the regional characteristics to classify the pixels and give good results. It is also simple because it only needs a few seed pixels, and the based segmentation criteria can be easily specified. But, unfortunately, it is noise-sensitive and needs a high computational cost.
Graph-based techniques divide the image region using matrices to yield a combination of disconnected subgraphs. The elements in a graph are pixels, and the edge weights measure the similarity between the two pixels connected by this edge. Let
2.6 Particle Swarm Optimization (PSO)
PSO is a powerful optimization technique where a set of low-level intelligence has an optimization problem and needs to collaborate to solve this problem. An individual of the set is only aware of the position and speed of its nearest neighbors. Each one modifies its behavior based on its experience and its neighbors’ experience to build a solution to a problem. Through easy movement (in the solution space), each individual in the set progressively reaches the problem’s best solution.
2.7 Watershed Segmentation Technique
The watershed technique uses the image gradient magnitude intensity (GMI) to achieve segmentation. Pixels that have high GMIs values correspond to the lines of the watershed that outline the region contours. Pixels that drain from a catch basin to the same minimum point form a watershed or a segment. During the sequential draining, watersheds with nearby pixels are formed. The watershed technique is based on the following steps:
Step (1): A few numbers of pixels, named as markers, are chosen. Each of them has a different label.
Step (2): The GMIs are used to insert the nearby pixels into the priority queue according to the priority level.
Step (3): The pixels with the highest priority level are isolated from the priority queue. If the adjacent pixel of the isolated ones has the same label, then the pixel is labeled with their label. The neighbors that are not labeled are queued in the priority queue.
Step (4): Step (3) is repeated until the priority queue is finished.
Active contours include two main techniques, the parametric and the geometric. The parametric techniques, also referred to as snakes, are compact and can overcome both the noise in the image and boundary irregularities and gaps as they force the extracted boundaries to be smooth. Geometric techniques or level sets are prepared to process the changes in the image. They are considered a significant class of sophisticated image segmentation methods based on Partial Differential Equations (PDE). Chan-vese based level set algorithm functional is given by:
where
The Heaviside function of the level set function,
Keeping
Keeping
3 The Proposed Hybrid Segmentation Approach
By comparing the benefits and drawbacks of the several segmentation techniques, the development of the segmentation approach may present the following trends for merging multiple techniques. Although according to this comparative study, it is found that the combining of PSO segmentation technique and IFFCM enhances the segmentation accuracy because this combination merges the benefits of the two techniques, the proposed hybrid approach achieves a better segmentation effect. Fig. 1 shows the proposed approach steps. These steps can be summarized as follows:
• Contrast Enhancement Using Histogram Equalization
Image preprocessing enhances the input images to get better-quality ones so that the results are more manageable and more straightforward for analysis and display.

Figure 1: The main steps of the proposed approach
• Applying the PSO Technique and Enhancing Features
PSO can simulate bird clusters for foraging and achieve the goal through cooperation and competition between birds. Particle
The index number of the best locations experienced by all the particles of a group is indicated by symbol
The PSO automatically determines the number of clusters as well as the center of the clusters. The hybridized PSO is applied to obtain the optimal fuzzy rules and membership function. The best fuzzy rule is selected for image segmentation. Since PSO is an effective technique, combining it with other segmentation techniques results in a more practical approach.
• Applying the IFFCM Technique
The FCM objective function is given by:
where the array
The
where
where
Solving for
where
To sum up, the main target of IFFCM clustering is to find cluster centers that maximize a similarity function or minimize the dissimilarity function. The PSO is used for allocating each pixel to its cluster. This combination of IFFCM clustering and the PSO algorithm produces better segmentation results due to its advantages.
4 Simulation Results and Discussions
Simulation results were carried out using MATLAB R2019a on a Dell machine, Core i5 processor, 8 Gbytes RAMs, and 320 Gbytes hard disk. Simulation results are conducted on five different examples of the scanned image (Us, XR (Mammogram), CT, PET, and MRI). The first is the medical Us breast images, containing 130 malignant and benign images from the Sirindhorn International Institute of Technology (SIIT) [30]. The second is the medical X-ray (mammogram) images; the breast mass image dataset used in this paper is the Digital Database for Screening Mammography (DDSM) released by the University of South Florida. The dataset includes labels that indicate benign and malignant breast masses, containing 100 benign and malignant images [31]. The third is the medical CT chest images containing 70 benign and malignant images from the Cancer Imaging Archive (TCIA) website [32]. The last one is the medical MRI brain images containing 160 benign and malignant images from the figshare dataset [33]. The segmentation techniques tested here are the Otsu-based thresholding technique, the clustering-based techniques (K-means, FCM, IFCM, IFFCM, and MS), Canny edge detector, region growing, graph-based, PSO, watershed, active contours, and finally, the proposed hybrid approach.
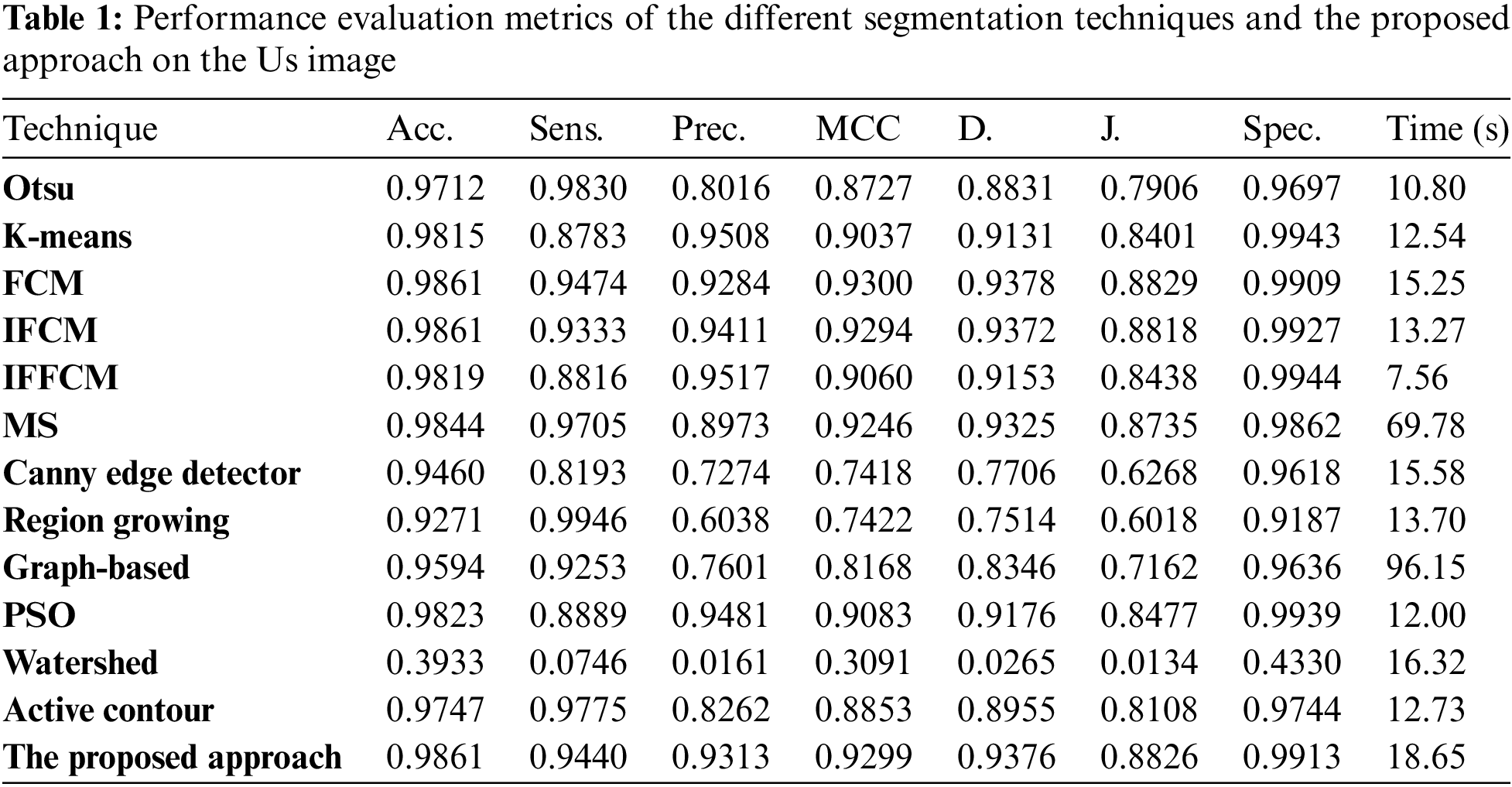
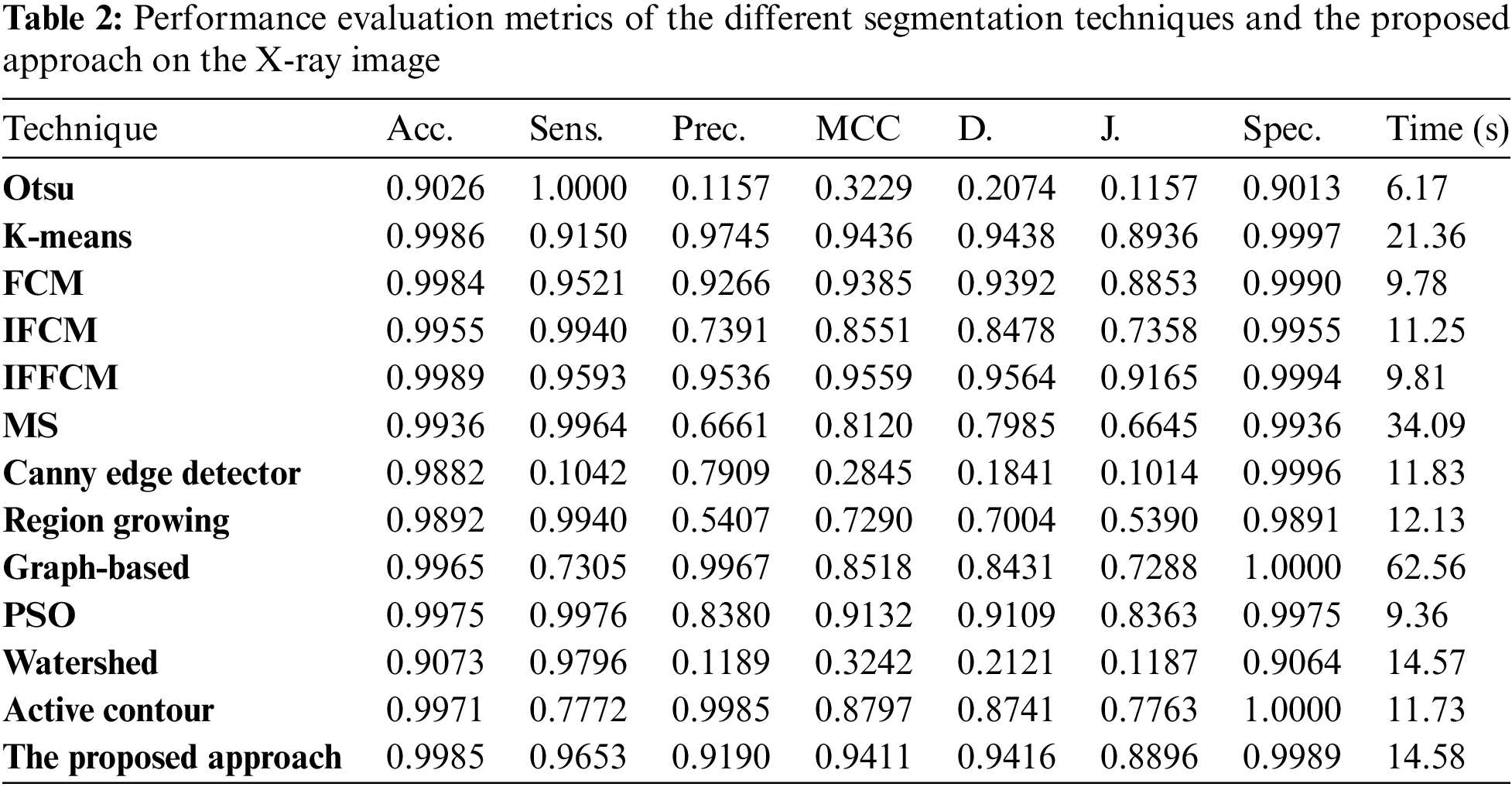
To estimate the effectiveness and efficiency the different segmentation techniques and the proposed approach, the different modalities of medical images are tested in the experiments using the accuracy and similarity indices, namely, the segmentation accuracy (Acc), Sensitivity (Sens), Precision (Pr), Matthews Correlation Coefficient (MCC), Dice (D), Jaccard (J), and Specificity (Spec) [34–36].
where TP is the true positive, FP is the false positive, TN is the true negative, and FN is the false negative.
Fig. 2 illustrates a sample of the different modalities of medical images, (a) Us image, (b) X-ray image, (c) CT image, (d) PET image, and (e) MR image. Each of these images has two significant areas: background and tumor regions.
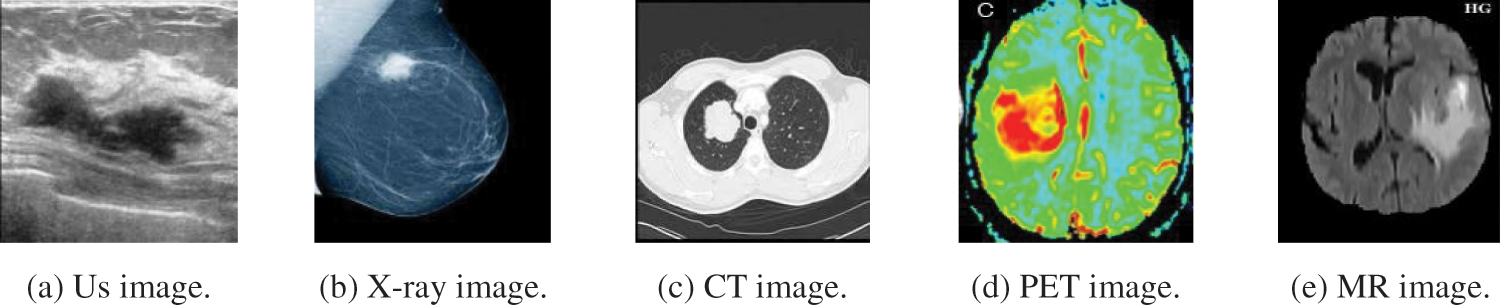
Figure 2: A sample of the different modalities of medical images
Fig. 3a shows Otsu output Us image, Fig. 3b shows K-means output Us image, Fig. 3c shows FCM output Us image, Fig. 3d shows IFCM output Us image, Fig. 3e shows IFFCM output Us image, Fig. 3f shows MS output Us image, Fig. 3g shows Canny edge detector output Us images, Fig. 3h shows region growing output Us images Fig. 3i shows graph-based output Us images, Fig. 3j shows PSO output Us images, Fig. 3k shows watershed output Us images, Fig. 3l shows active contour output Us images, and Fig. 3m shows the proposed approach output Us images.
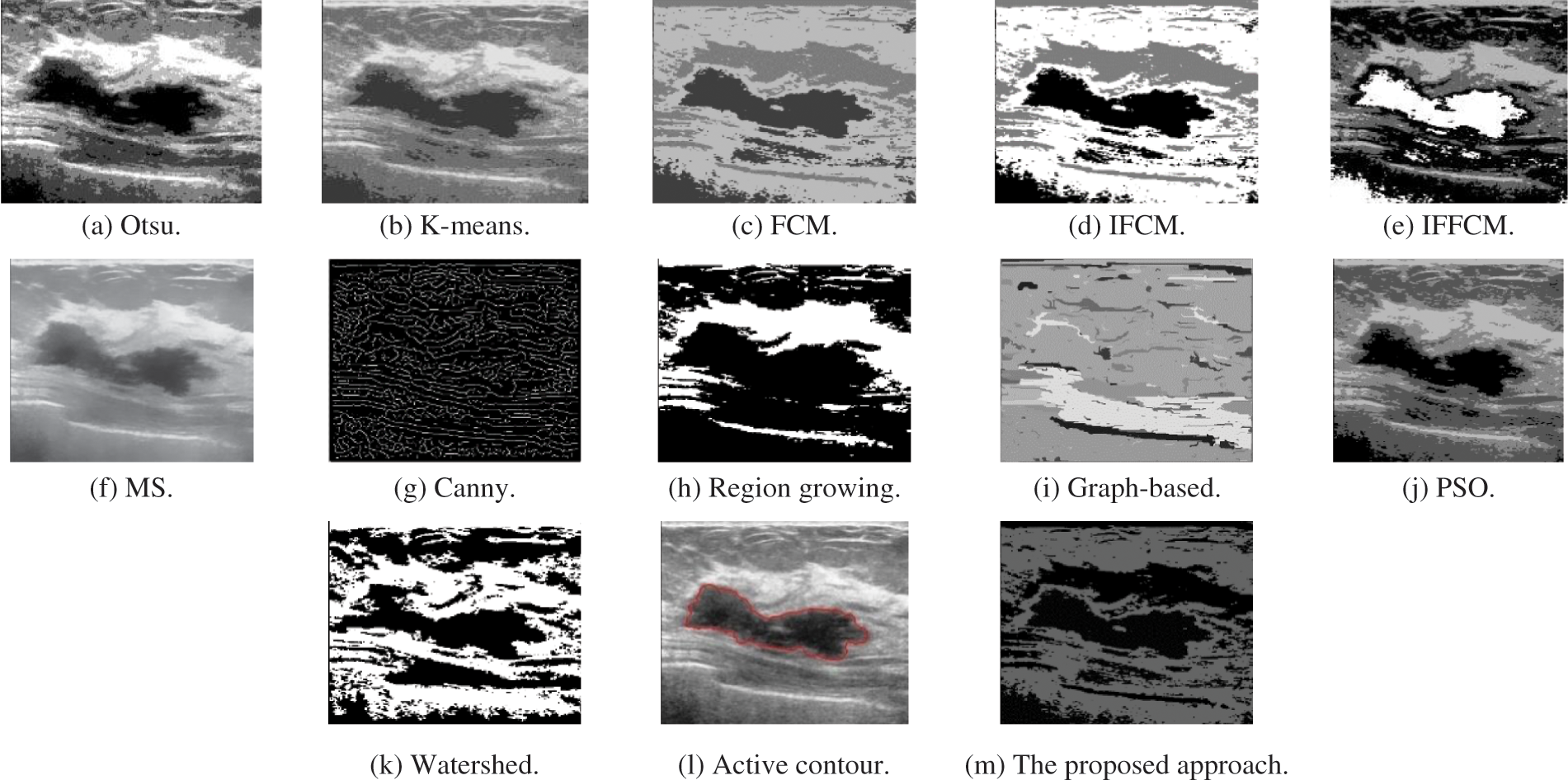
Figure 3: Segmentation results of the different segmentation techniques and the proposed approach on the Us medical image
Fig. 4a shows Otsu output X-ray image, Fig. 4b shows K-means output X-ray image, Fig. 4c shows FCM output X-ray image, Fig. 4d shows IFCM output X-ray image, Fig. 4e shows IFFCM output X-ray image, Fig. 4f shows MS output X-ray image, Fig. 4g shows Canny edge detector output X-ray images, Fig. 4h shows region growing output X-ray images Fig. 4i shows graph-based output X-ray images, Fig. 4j shows PSO output X-ray images, Fig. 4k shows watershed output X-ray images, Fig. 4l shows active contour output X-ray images, and Fig. 4m shows the proposed approach output X-ray images.
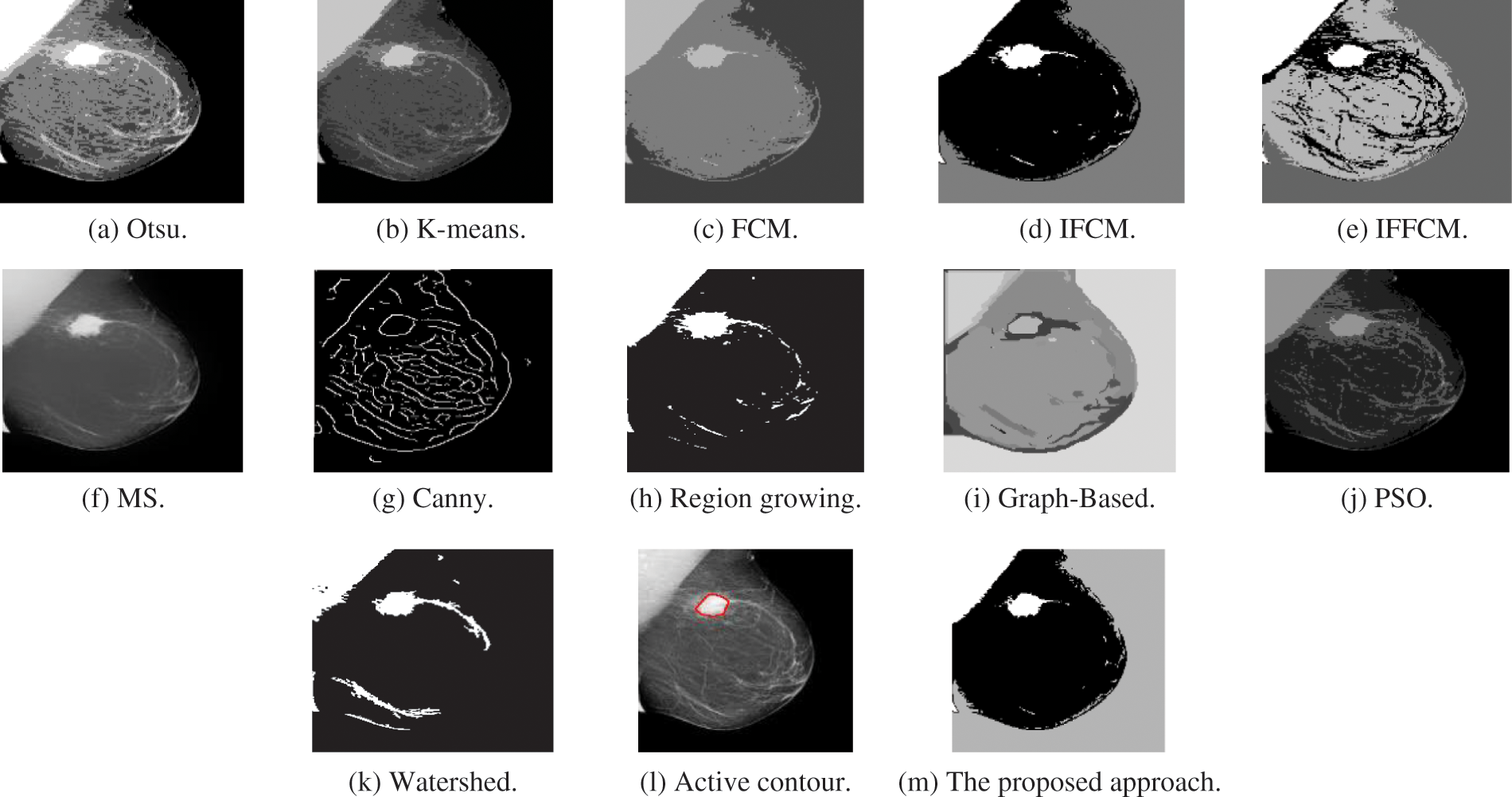
Figure 4: Segmentation results of the different segmentation techniques and the proposed approach on the X-ray medical image
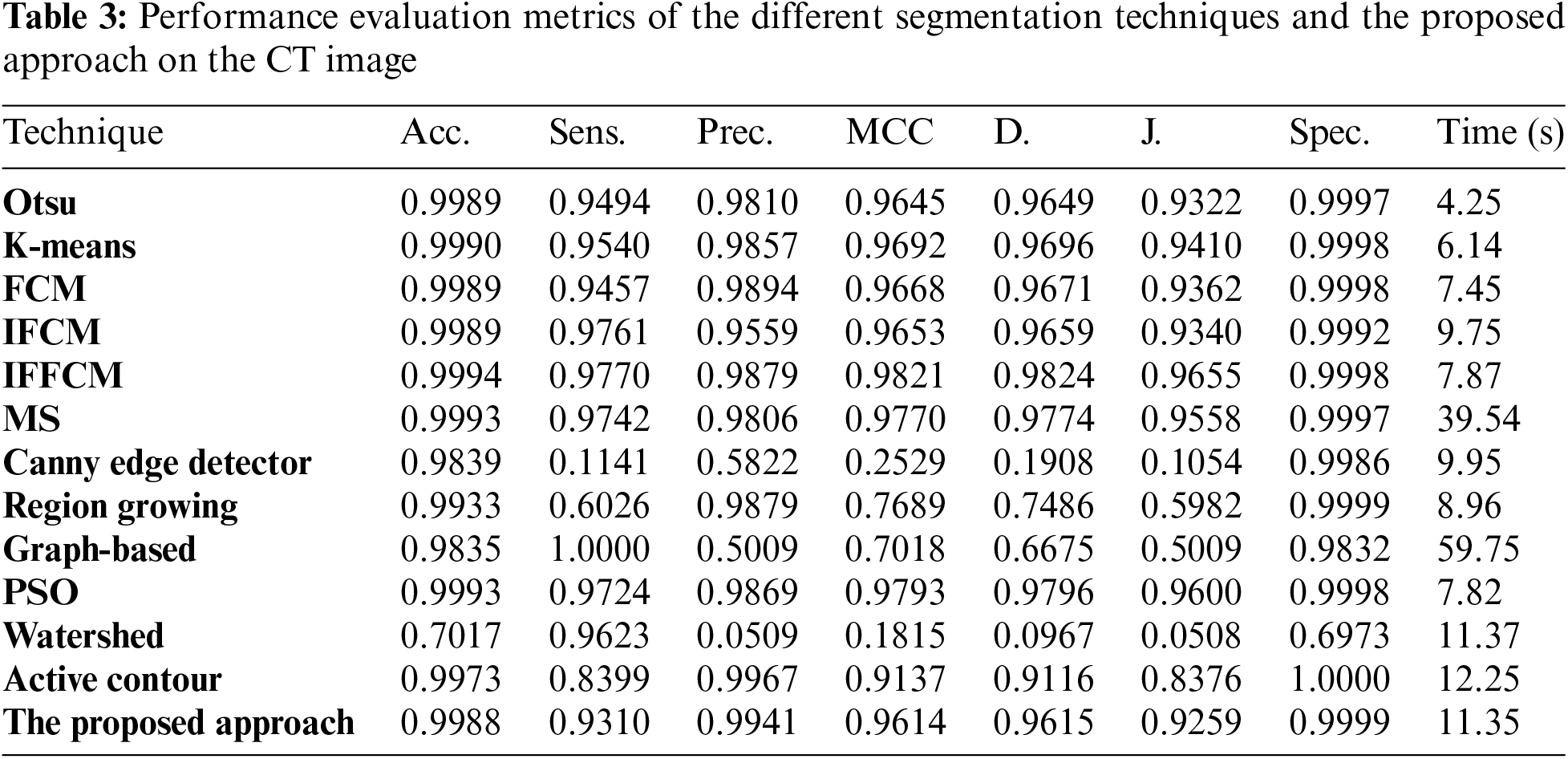
Fig. 5a shows Otsu output CT image, Fig. 5b shows K-means output CT image, Fig. 5c shows FCM output CT image, Fig. 5d shows IFCM output CT image, Fig. 5e shows IFFCM output CT image, Fig. 5f shows MS output CT image, Fig. 5g shows Canny edge detector output CT images, Fig. 5h shows region growing output CT images Fig. 5i shows graph-based output CT images, Fig. 5j shows PSO output CT images, Fig. 5k shows watershed output CT images, Fig. 5l shows active contour output CT images, and Fig. 5m shows the proposed approach output CT images.
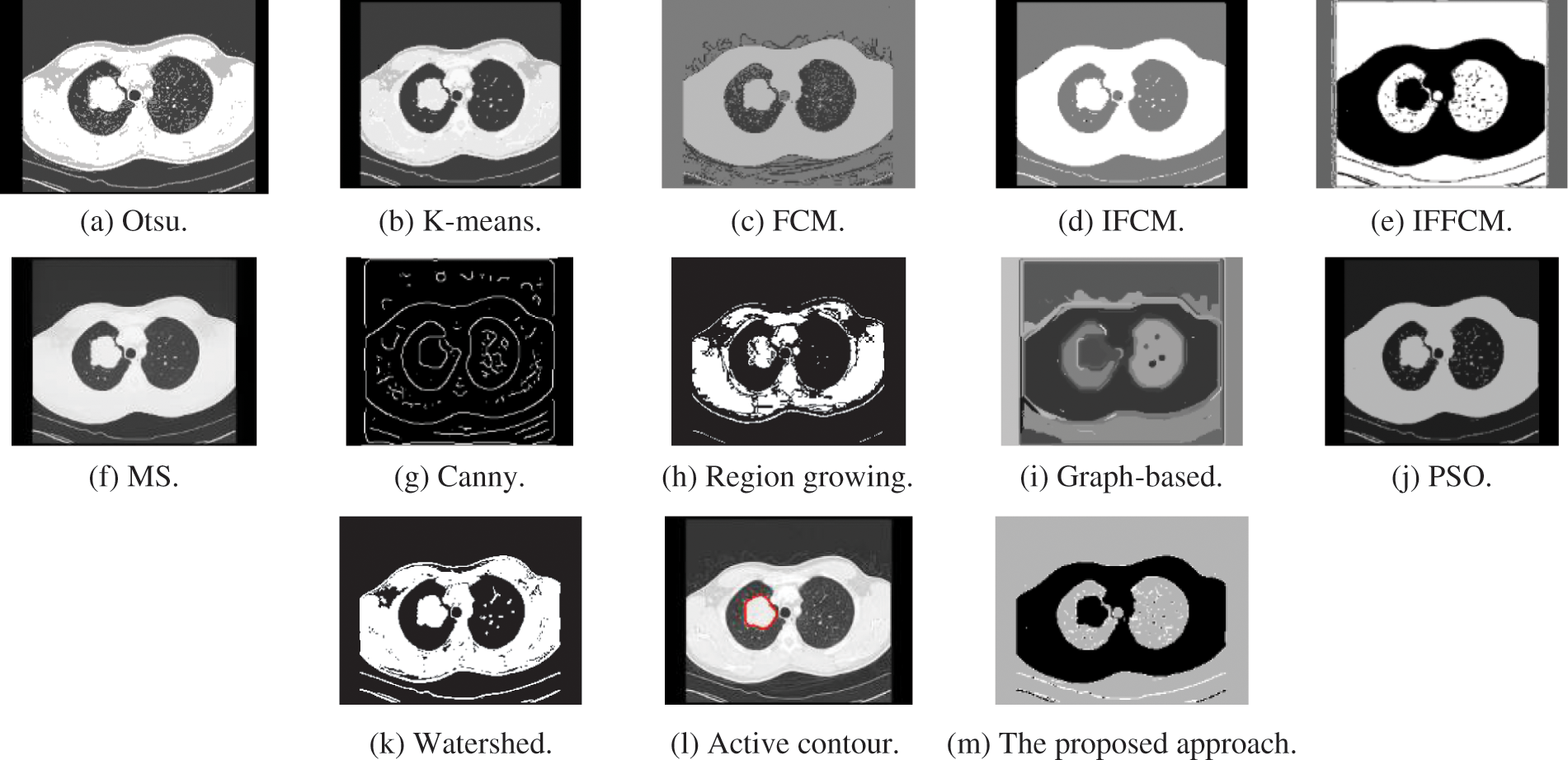
Figure 5: Segmentation results of the different segmentation techniques and the proposed approach on the CT medical image
Fig. 6a shows Otsu output PET image, Fig. 6b shows K-means output PET image, Fig. 6c shows FCM output PET image, Fig. 6d shows IFCM output PET image, Fig. 6e shows IFFCM output PET image, Fig. 6f shows MS output PET image, Fig. 6g shows Canny edge detector output PET images, Fig. 6h shows region growing output PET images Fig. 6i shows graph-based output PET images, Fig. 6j shows PSO output PET images, Fig. 6k shows watershed output PET images, Fig. 6l shows active contour output PET images, and Fig. 6m shows the proposed approach output PET images.
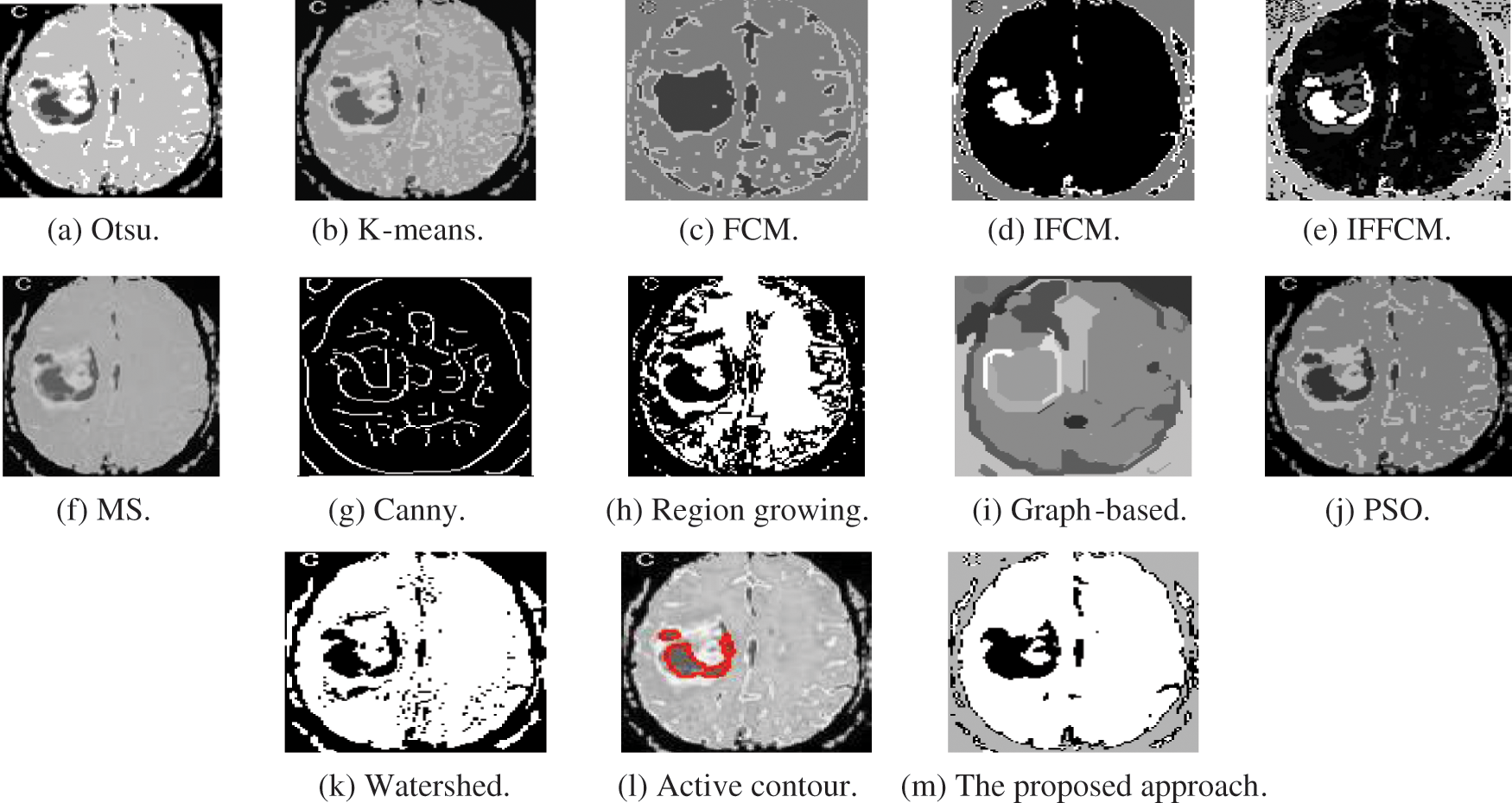
Figure 6: Segmentation results of the different segmentation techniques and the proposed approach on the PET medical image
Fig. 7a shows Otsu output MR image, Fig. 7b shows K-means output MR image, Fig. 7c shows FCM output MR image, Fig. 7d shows IFCM output MR image, Fig. 7e shows IFFCM output MR image, Fig. 7f shows MS output MR image, Fig. 7g shows Canny edge detector output MR image, Fig. 7h shows region growing output MR image, Fig. 7i shows graph-based output MR image, Fig. 7j shows PSO output MR image, Fig. 7k shows watershed output MR image, Fig. 7l shows active contour output MR image, and Fig. 7m shows the proposed approach output MR image. Tabs. 1 to 5 show the accuracy and similarity indices of the different segmentation techniques and the proposed approach on Us, X-ray, CT, PET, and MR medical images.
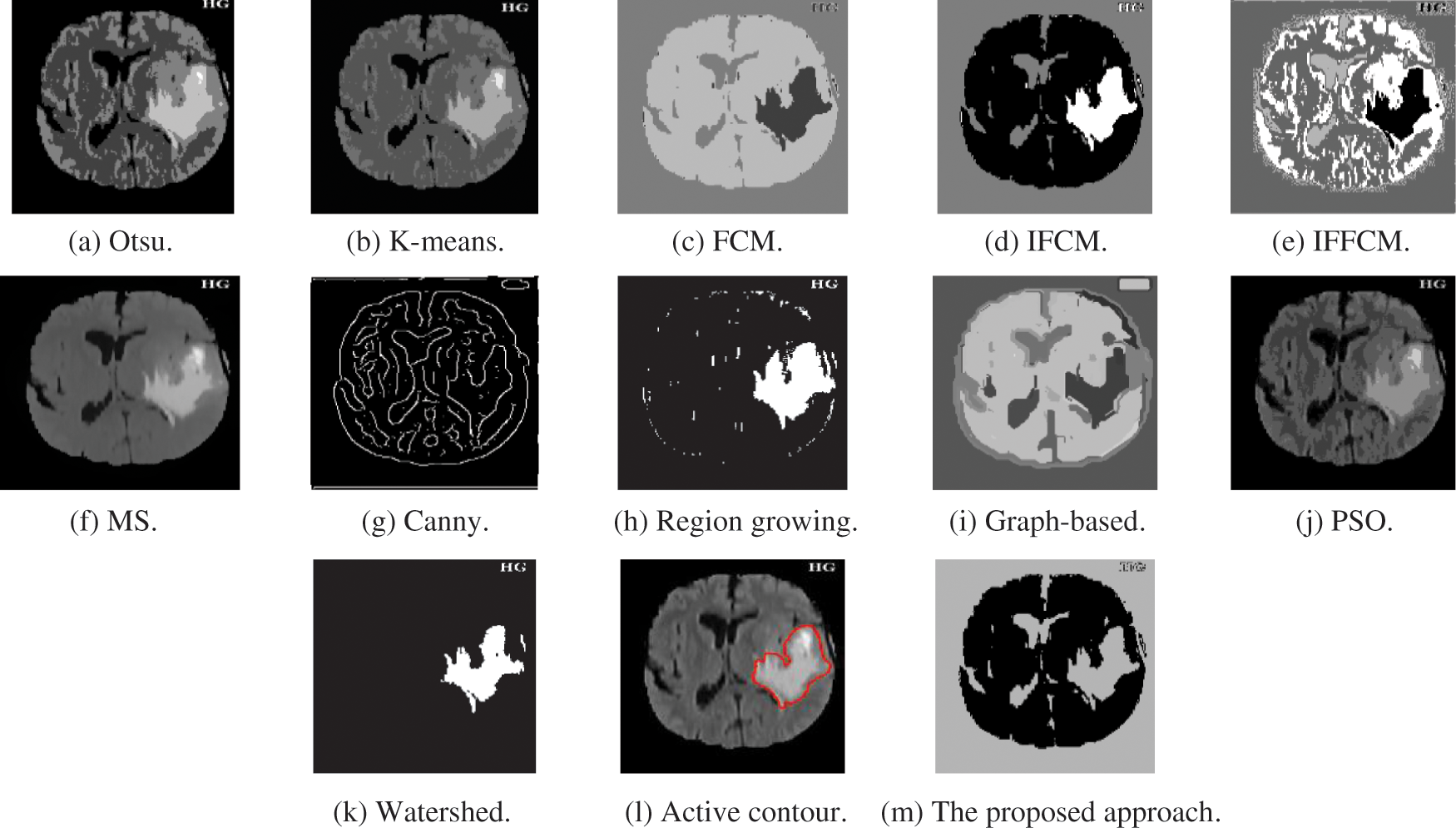
Figure 7: Segmentation results of the different segmentation techniques and the proposed approach on the MR medical image


Various segmentation techniques applied to different medical images are studied. The obtained results show that Thresholding techniques remove regions less than a threshold for better visualization. They are a simple method, easy to implement and can be used in real-time applications, and is very effective for images containing solid objects in a contrasting background. However, choosing the accurate threshold value is complicated and unsuitable for flat or smooth images and images with high feature variability. On the other hand, the clustering techniques can handle the noisy parts, easy to implement and fast for small clusters number, and run several times to return the best clustering found. However, choosing the appropriate number of clusters is critical and time-consuming.
The graph-based techniques are robust and able to obtain good segmentation results, effective for images without any pre-known information. However, they need large storage space and CPU time and cannot handle thin elongated structures. The edge-based techniques are suitable for images with the best contrast between objects and background. However, they cannot be suitable for images and edges with low contrast. The watershed techniques have fast computation speed, but also important drawbacks exist: Over segmentation, the result of the watershed may contain many small sub-regions that make the results not useful. Sensitivity to local noise variations of the image may change the results. This effect is worsened using high pass filters to estimate the gradient, which amplifies the noise. Important boundaries with low contrast may be poorly detected.
The active contours improve the segmentation result and convey that the boundary’s refinement is an essential segmentation process. They can also manage the cavities and concavities, merging or splitting, and can avoid gaps and overlaps. Segmentation techniques based on active contours can outperform many of the traditional image segmentation techniques. The proposed hybrid approach that combines the PSO algorithm and the IFFCM is significantly fast and robust and improves the segmentation quality. It is concluded that each technique has its advantages and limitations. The suitable technique is determined according to the type of input image and the application. Based on this, it is concluded that the segmentation of medical images has a promising future, and a lot of research work is required for developing a universally applied segmentation technique.
The paper proposed a hybrid segmentation approach that combines the PSO technique and the IFFCM, a significantly fast and robust technique that enhances segmentation quality compared to other algorithms. Also, this paper introduces a comparative study between the proposed approach and the most used segmentation techniques with investigating the performance against different medical image modalities. It has been found that thresholding has been a popular segmentation technique for many years because of the advantages of simplicity and ease of implementation. However, it does not work well when dealing with flat images with smooth peaks and valleys. Clustering is an efficient technique, but the initial number of clusters must be determined before the process begins. Region-based techniques have been valuable and efficient segmentation techniques, but they are sensitive to noise and can cause extracted regions to have holes or discontinuities. If the seed point was not chosen correctly, the segmentation fails. It is also having more time-consuming. Image segmentation using active contours shows that the improvement of boundaries was essential in segmentation. They are robust and efficient techniques for all situations. In future work, the utilization of machine learning algorithms like support vector machines and neural networks for image segmentation can be introduced. Additionally, the tumor size will be quantified, and improved preprocessing approaches can be introduced. Moreover, in the future, more suggestions can be employed to enhance the proposed work’s performance, such as utilizing deep learning techniques for segmentation and classification purposes. Also, morphological operations can improve the segmentation process for detecting the central regions of interest. Additionally, we target to build a graphical user interface for the whole proposed work.
Acknowledgement: Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R66), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Funding Statement: Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R66), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. H. El Khoukhi, Y. Filali, A. Yahyaouy, M. A. Sabri and A. Aarab, “A hardware implementation of Otsu thresholding method for skin cancer image segmentation,” in Proc. Int. Conf. on Wireless Technologies, Embedded and Intelligent Systems (WITS), Fez, Morocco, pp. 1–5, 2019. [Google Scholar]
2. X. Cao, T. Li, H. Li, S. Xia, F. Ren et al., “A robust parameter-free thresholding method for image segmentation,” IEEE Access, vol. 7, pp. 3448–3458, 2019. [Google Scholar]
3. S. Husein, M. Abd Elaziz, A. Mohammed, A. Al-Qaness, A. Afzaal et al., “A multilevel image thresholding based on hybrid salp swarm algorithm and fuzzy entropy,” IEEE Access, vol. 7, pp. 181405–181422, 2019. [Google Scholar]
4. I. Mehidi, D. E. Chouaib and D. Jabri, “An improved clustering method based on k-means algorithm for MRI brain tumor segmentation,” in Proc. 6th Int. Conf. on Image and Signal Processing and their Applications (ISPA), Mostaganem, Algeria, pp. 1–6, 2019. [Google Scholar]
5. M. Zhang, L. Jiao, R. Shang, X. Zhang and L. Li, “Unsupervised EA-based fuzzy clustering for image segmentation,” IEEE Access, vol. 8, pp. 8627–8647, 2020. [Google Scholar]
6. A. Mariena, J. Sathiaseelan and J. Abraham, “Hybrid approach for image segmentation using region splitting and clustering techniques,” in Proc. Int. Conf. on Circuits and Systems in Digital Enterprise Technology (ICCSDET), Kottayam, India, pp. 1–4, 2018. [Google Scholar]
7. H. Fankui, C. Haibing and W. Xiaofei, “An intuitionistic kernel-based fuzzy c-means clustering algorithm with local information for power equipment image segmentation,” IEEE Access, vol. 8, pp. 4500–4514, 2020. [Google Scholar]
8. B. Beddad, K. Hachemi, J. Postaire, F. Jabloncik and O. Messai, “An improvement of spatial fuzzy c-means clustering method for noisy medical image analysis,” in Proc. 6th Int. Conf. on Image and Signal Processing and their Applications (ISPA), Mostaganem, Algeria, pp. 1–5, 2019. [Google Scholar]
9. K. Aljebory and T. Mohammed, “Modified fuzzy c-mean clustering algorithm application in medical image segmentation,” Journal of Electrical Engineering (JEA), vol. 2, no. 1, pp. 1–9, 2018. [Google Scholar]
10. M. Nikolic, E. Tuba and M. Tuba, “Edge detection in medical ultrasound images using adjusted canny edge detection algorithm,” in Proc. 24th Telecommunications Forum (TELFOR), Belgrade, Serbia, pp. 1–4, 2016. [Google Scholar]
11. M. Mittal, A. Verma, I. Kaur, B. Kaur, M. Sharma et al., “An efficient edge detection approach to provide better edge connectivity for image analysis,” IEEE Access, vol. 7, pp. 33240–33255, 2019. [Google Scholar]
12. R. Anas, H. A. Elhadi and E. S. Ali, “Impact of edge detection algorithms in medical image processing,” International Journal of World Scientific News, vol. 118, no. 5, pp. 129–143, 2019. [Google Scholar]
13. F. A. M. Cappabianco, P. F. O. Ribeiro, P. A. V. de Miranda and J. K. Udupa, “A general and balanced region-based metric for evaluating medical image segmentation algorithms,” in Proc. IEEE Int. Conf. on Image Processing (ICIP), Taipei, Taiwan, pp. 1525–1529, 2019. [Google Scholar]
14. S. Biswas, R. Hazra and S. Prasad, “A region-based level set formulation using machine learning approach in medical image segmentation,” in Proc. TENCON, 2019 - 2019 IEEE Region 10 Conf. (TENCON), Kochi, India, pp. 470–475, 2019. [Google Scholar]
15. K. K. Gupta, N. Dhanda and U. Kumar, “A comparative study of medical image segmentation techniques for brain tumor detection,” in Proc. 4th Int. Conf. on Computing Communication and Automation (ICCCA), Greater Noida, India, pp. 1–4, 2018. [Google Scholar]
16. X. Chen and L. Pan, “A survey of graph cuts/graph search based medical image segmentation,” IEEE Reviews in Biomedical Engineering, vol. 11, no. 7, pp. 112–124, 2018. [Google Scholar]
17. F. C. Monteiro and A. Campilho, “Watershed framework to region-based image segmentation,” in Proc. Int. Conf. on Pattern Recognition (ICPR), Tampa, FL, USA, pp. 1–4, 2008. [Google Scholar]
18. T. Y. Tan, L. Zhang, C. P. Lim, B. Fielding, Y. Yu et al., “Evolving ensemble models for image segmentation using enhanced particle swarm optimization,” IEEE Access, vol. 7, pp. 34004–34019, 2019. [Google Scholar]
19. X. Wang and J. Zhao, “A novel neutrosophic image segmentation based on improved particle swarm optimization fuzzy c-means algorithm,” in Proc. 18th Int. Symp. on Distributed Computing and Applications for Business Engineering and Science (DCABES), Wuhan, China, pp. 80–83, 2019. [Google Scholar]
20. X. Yang, X. Jiang, L. Zhou, Y. Wang and Y. Zhang, “Active contours driven by local and global region-based information for image segmentation,” IEEE Access, vol. 8, pp. 6460–6470, 2020. [Google Scholar]
21. F. Lingling, Q. Tianshuang, Z. Hongyang and L. Fang, “A hybrid active contour model based on global and local information for medical image segmentation,” Multidimensional Systems and Signal Processing, vol. 30, no. 2, pp. 689–703, 2019. [Google Scholar]
22. P. Sasmal, Y. Iwahori, M. K. Bhuyan and K. Kasugai, “Active contour segmentation of polyps in capsule endoscopic image,” in Proc. Int. Conf. on Signals and Systems (ICSigSys), Bali, Indonesia, pp. 201–204, 2018. [Google Scholar]
23. L. Leng, Z. Yang, C. Kim and Y. Zhang, “A light-weight practical framework for feces detection and trait recognition,” Sensors, vol. 20, no. 9, pp. 1–25, 2020. [Google Scholar]
24. Z. Yang, L. Leng and B. Kim, “Stoolnet for color classification of stool medical images,” Electronics, vol. 8, no. 5, pp. 1–18, 2019. [Google Scholar]
25. Y. Zhang, J. Chu, L. Leng and J. Miao, “Mask-refined R-CNN: A network for refining object details in instance segmentation,” Sensors, vol. 20, no. 7, pp. 1–19, 2020. [Google Scholar]
26. J. Chu, Z. Guo and L. Leng, “Object detection based on multi-layer convolution feature fusion and online hard example mining,” IEEE Access, vol. 6, pp. 19959–19967, 2018. [Google Scholar]
27. K. Po-Yu, S. Fnu, J. Jiaxiang, Z. Angela, K. Amil et al., “Improving patch-based convolutional neural networks for mri brain tumor segmentation by leveraging location information,” Frontiers in Neuroscience, vol. 13, no. 7, pp. 1–14, 2020. [Google Scholar]
28. L. Chen, P. Bentley, K. Mori, K. Misawa, M. Fujiwara et al., “DRINet for medical image segmentation,” IEEE Transactions on Medical Imaging, vol. 37, no. 11, pp. 2453–2462, 2018. [Google Scholar]
29. Y. Xiang, H. Liu, S. Wang, X. Xiong, D. Shao et al., “Segmentation method of multiple sclerosis lesions based on 3D-CNN networks,” Insistitue of Engineering and Technology (IET) Image Processing, vol. 14, no. 9, pp. 1806–1812, 2020. [Google Scholar]
30. Ultrasonic images,” last accessed on 15-12-2020. Available: http://www.onlinemedicalimages.com/index.php/en/. [Google Scholar]
31. W. El-Shafai and F. Abd El-Samie, “Extensive COVID-19 X-Ray and CT chest images dataset,” Mendeley Data, vol. v3, 2020. [Google Scholar]
32. CT and MRI images,” last accessed on 15-12-2020. Available: https://www.cancerimagingarchive.net/. [Google Scholar]
33. Cheng, “Jun: Brain tumor dataset. figshare,” Dataset. last accessed on 15-12-2020. Available: https://doi.org/10.6084/m9.figshare.1512427.v5. [Google Scholar]
34. D. N. Thanh, “Image segmentation quality scores,” MATLAB Central File Exchange, 2019. [Online]. Available: https://www.mathworks.com/matlabcentral/. [Google Scholar]
35. W. El-Shafai, S. Abd El-Nabi, E. El-Rabaie, A. Ali and F. Soliman, “Efficient deep-learning-based autoencoder denoising approach for medical image diagnosis,” Computers Materials and Continua, vol. 70, no. 3, pp. 6107–6125, 2022. [Google Scholar]
36. W. El-Shafai, A. Mohamed, E. El-Rabaie, A. Ali and F. Soliman, “Automated COVID-19 detection based on single-image super-resolution and CNN models,” Computers Materials and Continua, vol. 69, no. 3, pp. 1141–1157, 2021. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |