DOI:10.32604/cmc.2022.029423

| Computers, Materials & Continua DOI:10.32604/cmc.2022.029423 |  |
| Article |
A Novel Integrated Learning Scheme for Predictive Diagnosis of Critical Care Patient
1School of Electronics Engineering, Vellore Institute of Technology, Chennai, Tamil Nadu, India
2Center for Advanced Data Science, Vellore Institute of Technology, Chennai, Tamil Nadu, India
*Corresponding Author: Susan Elias. Email: susan.elias@vit.ac.in
Received: 03 March 2022; Accepted: 07 April 2022
Abstract: Machine learning has proven to be one of the efficient solutions for analyzing complex data to perform identification and classification. With a large number of learning tools and techniques, the health section has significantly benefited from solving the diagnosis problems. This paper has reviewed some of the recent scientific implementations on learning-based schemes to find that existing studies of learning have mainly focused on predictive analysis with less emphasis on preprocessing and more inclination towards adopting sophisticated learning schemes that offer higher accuracy at the cost of the higher computational burden. Therefore, the proposed method addresses the concern mentioned above by a novel computational learning model that emphasizes fine-tuning complex medical data and makes it suitable for learning to balance better classification performance and computational complexity. The implementation is carried out using the MIMIC-III dataset, where the proposed system discretizes, the complete model using physician reports and furnished patient information as the first step. It also prepares the data by choosing a specific tuple and its associated field. The second step introduces a novel relatedness function where preprocessing is carried out using word quantization while adopting auto-encoders in deep learning followed by a novel learning-based diagnosis. The outcome exhibits that the proposed system offers better classification performance in reduced processing time in comparison to existing learning schemes.
Keywords: Machine Learning; diagnosis; accuracy; computational complexity; diagnosis; deep learning; classification; prediction
The contribution of the machine learning approach has set a revolutionary step towards modernizing the healthcare sector in the process of diagnosing acute disease [1]. The inclusion of various dynamic environments and continually evolving challenges makes the diagnosis process quite difficult [2]. The complete success factor of any healthcare sector is an exact process of diagnosis that will lead to proper treatment planning for the patient. Machine learning currently acts as a potential component of the critical and high-end diagnostic process [3]. Inclusion of learning methodology towards the diagnostic process induces a significant capability to carry out decision making, problem-solving, environmental interaction, learning, reasoning, perceiving, etc. [4]. Hence, machine learning offers a behavior to the machine like a human capable of using Natural Language Processing (NLP) towards understanding and inferring the clinical conclusion of the target diagnosis [5]. From the perspective of the diagnostic process, the first stage is when the patient engages with the healthcare system after experiencing the health-related problem [6]. This is further followed up by the actual diagnosis process viz. (i) aggregating information, (ii) integrating information, (iii) interpreting information, and (iv) final diagnosis [7]. These four processes are carried out considering the patient’s clinical history, performing a physical examination, undergoing referral and consultation, and diagnostic testing. A precise treatment plan follows the final, conclusive remarks of diagnosis. Narrowing down the complete diagnosis process, the final, definitive statements are carried out based on the clinical records of the patient [8]. Such clinical records can be obtained through Electronic Health Records (EHR) and Electronic Medical Records (EMR) [9,10]. Further narrowing down, it can be said that diagnosis is based on two facts, viz. (i) information given as a part of patient testimony and (ii) information that physicians put on their records based on their observation [11]. Such observation could be before or in the middle of undergoing a treatment plan. Hence, such information could be text (obtained from patient or doctor records) and images (radiological images carried out during prognosis). At present, there are many research archives where machine learning has been used for medical image processing [12–15], where various algorithms have evolved to identify, classify, and meet all the physician’s queries in the process of diagnosis. However, text-based medical records are quite complex to manage and perform analysis. At present, there is the availability of various medical dataset on different disease [16–20], where already machine learning and practical data-based approaches [21] has been investigated. The challenging aspect in this perspective is that despite abundant research work using a learning-based method towards investigating critical disease conditions, there is no benchmarked or standardized computational model to prove this effectiveness.
Therefore, the prime goal of the proposed study is to present a computational model of a unique learning process capable of diagnosing critical disease conditions. The initiation and motivation of the proposed research are based on reviewed limitations of machine learning-based approaches. Hence, the proposed study offers the following contribution:
• A computational model that can perform lightweight analytical operations of learning to predict a diagnosis of critical disease.
• A highly flexible diagnostic model can be applied to the majority of the critical disease diagnosis, whereas there is no such generalized predictive model in the literature.
• A simplified preprocessing operation that carries out the preparation of medical data enriches its quality, reflecting the accuracy of the predictive model.
• A unique relatedness function that can extract potential contextual terms leveraging the predictive model’s accuracy.
• A simplified machine learning model that carries out prediction over the quantized data sequence offers a lightweight predictive operation compared to existing schemes.
The manuscript’s organization is as follows: Section 2 discusses existing literature on machine learning techniques, while briefing of identified research problems is carried out in Section 3. Highlights of the research methodology adopted in order to solve the identified research problem are carried out in Section 4. In contrast, illustration of system design with respect to algorithm discussion with higher clarity towards implementation scheme is carried out in Section 5. Elaborated discussion of results with respect to analysis strategy and result discussion is carried out in Section 6. At the same time, the summary of the contribution of the proposed scheme is discussed in Section 7.
Various work is being carried out towards diagnosing critical diseases in the medical sector using different variants of the machine learning approach. The study carried out by Latif et al. [22] has reviewed various learning techniques, e.g., autoencoders, deep belief network, Recurrent Neural Network (RNN), Convolution Neural Network (CNN), decision tree, Bayes methods, Support Vector machine, etc. The review concludes the limitation in almost all the existing learning methods, which demands revision. A unique work carried out by Chai [23] facilitates the diagnosis of thyroid disease by constructing a knowledge graph that links all scattered medical information. The training operation uses Bidirectional Long Short-Term Memory (BLSTM). The study’s limitation is that a complete load of computational burden is over the classifier. From the classification perspective, a study reported by Guo et al. [24] have used trust factor and artificial intelligence to recommend a patient’s diagnosis in the form of service. The limitation of this model is its scalability, which cannot cater to many predictions. Adoption of machine learning towards monitoring labeled data is reported in Guo et al. [25], considering laryngopharyngeal reflux as a case study. The limitation of this model is that it is highly specific to the case study, including only particular attributes. Classification plays a contributory role in disease diagnosis, as witnessed in the model presented by Heidari et al. [26].
The study performs prediction of the probability of benign lesion in breast cancer where Support Vector Machine (SVM) is used for training. Irrespective of a better reliability score, the model limitation includes a sophisticated form of computation. The dual usage of supervised and unsupervised learning is used to diagnose specific diseases from the machine learning perspective. Work in such direction is carried out by Hussein et al. [27], where the diagnosis of the presence of tumor in lung and pancreatic cancer is carried out. The study implements a transfer learning and convolution neural network for training and diagnosis. The limitation of the model is the higher degree of iteration by the transfer learning used, which could be otherwise controlled. Similar adoption of transfer learning is also witnessed in Niu et al. [28], which carry out segmentation and fusion of medical images to perform image-based diagnosis for identifying effects lungs of COVID-19 infected patients. Similar study and environment of diagnosis are also seen in Roy et al. [29] and Shamsi et al. [30]. The work carried out by Wu et al. [31] has presented a classification approach for critical and non-critical patients of COVID-19 using random forest and SVM. The limitation of the study is that it doesn’t offer evidence of applicability towards raw and massive medical data.
Adoption of cascaded learning methodology towards a predictive diagnosis of down syndrome is witnessed in the work of Li et al. [32]. The model uses random forest, ensemble learning, and logistic regression to predict diagnostic. The pitfall of this model is higher accuracy at the cost of higher feature dependencies. A similar direction of study is also carried out by Li et al. [33] using multiple machine learning approaches to diagnose Turner syndrome. The investigation suffers from similar limitations as the prior model. A unique study presented by Lian et al. [34] performs diagnosis of dual disease condition using CNN, which is capable of autonomous identification of an essential feature. The limitation of the study is that although it has improvised CNN, its multi-pooling operation still has higher dependencies towards sequencing the features, which is an arduous task. The adoption of ensemble learning also contributes to the diagnosis of medical data, as seen in the work of Liu et al. [35]. The classification is carried out by SVM along with voting strategy, genetic algorithm, and simulated annealing. The limitation of the study is it offers higher accuracy at the cost of computational overhead.
The work carried out by Mahfuz et al. [36] has presented an assessment model where various machine learning models are tested to diagnose kidney disease. The model uses random forest, which further improves the accuracy; however, it has a limitation of highly iterative operation. A different type of study implementation is seen in Wang et al. [37] where deep learning has been used to identify lung nodules. The study has used segmentation, normalization, and image clipping for preprocessing, while CNN has been used for learning the features, and LSTM has been used for further classification. The limitation of the study is that it has discussed diagnosis considering the inclusion of connected devices without discussing the delay or distortion caused during processing. Yang et al. [38] have presented a classification approach for epilepsy conditions using decision tree, ensemble learning, SVM, and K-Nearest Neighborhood algorithm. The limitation of the study is that it offers higher accuracy with a heavy weight and resource-dependent set of machine learning approaches.
Zhang et al. [39] describe hemorrhage diagnosis using ensemble learning. The limitation of the study is its lower scale of accomplished accuracy in the perspective of other machine learning approaches. Adoption of hybrid learning technique towards deploying a diagnostic support framework is presented by Zhou et al. [40] using CNN integrated with RNN. The text-based features are extracted using CNN while patterns and correlation are trained using RNN to find that the model is applicable for real-world data. The study limitation is its higher training duration to cater to online query processing. Zhu et al. [41] have presented a diagnosis model based on the attention mechanism for Alzheimer’s disease. The pitfall of the study is that it doesn’t carry out ant significant preprocessing of the medical data before subjecting it to learning. The adoption of CNN was also recently reported in work of Zhang et al. [42] for diagnosing COVID-19; however, the study incurs higher computational resources with limited features for accomplishing its goal. Hence, the existing system introduces various learning schemes in order to facilitate diagnosis in healthcare sector for critical diseases with unique methodologies with claimed benefits and limitations. The following section outlines the identified research problem that are extracted from the review work discussed in this section.
From the prior section, various learning-based schemes are seen to diagnose critical diseases with claims benefits. However, they are all witnessed with a certain degree of limitation too. Conglomerating the complete limiting factors of the existing system following research problems are highlighted:
• Lack of Context in the medical dataset: Existing learning-based studies toward the medical dataset have considered the complete fields of the dataset, consisting of both essential and non-essential areas. This requires detailed data preprocessing that can be customized for any dataset. Unfortunately, almost all existing studies are specific to a disease where customization cannot be carried out. Further, the inclusion of non-essential fields offers fair occurrences of outliers in prediction, which is overlooked in existing models.
• Non-inclusion of robust preprocessing: Most existing studies directly apply the medical dataset to the machine learning approach. The pitfall for such an approach is that it offers too much computational burden towards the training and validation of the machine learning methods. Suppose the dataset is subjected to a better form of hierarchical preprocessing than machine learning capabilities can be used for better accuracy. At present, higher accuracy is obtained at the cost of the computational burden.
• Usage of Heavyweight Learning Approach: The majority of the existing learning models have been witnessed with a complex and highly iterative machine learning approach, e.g., random forest, support vector machine, multilayer perceptron, CNN, RNN, LSTM, etc. There is no denying that they have contributed towards better accuracy, as seen in the outcomes of existing literature. Still, their inherent limitations are neither identified nor addressed in the existing study model. Without addressing this limitation, the full exploitation of the machine learning approach towards the diagnosis process cannot be fully ensured.
• Inclusion of Computational complexity: It is already known that usage of machine learning will have higher resource dependencies and induce higher computational complexity. There is a lack of any report where such problems are addressed. A computational complexity problem associated with learning cannot be addressed by developing a unique algorithm. The complexity arises mainly for two reasons viz. (i) complexity of data structure and (ii) incompatibility of learning approach on solving the problem. Computational complexity can be addressed to a large extent if the input data and network structure are modified before applying the learning algorithm, which is not witnessed in existing studies.
The following section presents a novel and simplified research solution to address the above-mentioned research problems.
The prime goal of the proposed system is to develop a novel predictive technique for facilitating the diagnosis of a critical disease using a learning technique. For this purpose, analytical modeling is carried out towards achieving the research goal. Unlike the existing approaches, the proposed system introduces certain novelty factors where the medical dataset undergoes various operations to ensure better precision when subjected to a machine learning approach. The architecture of the proposed system is exhibited in Fig. 1. According to the architecture highlighted in Fig. 1, the complete implementation methodology is classified into three stages. The first stage of implementation prepares the medical dataset where the proposed scheme differs from the existing scheme by rendering specific tuple and its respective fields, followed by constructing a master table with an effective tuple management to ensure better structured medical data prior to training. The second stage of implementation constructs the relatedness function, which carries out preprocessing of words to improve the retention of enriched terms. This operation is followed by word quantization, responsible for identifying contextual and unique terms relating to the clinical perspective for assisting in prediction.
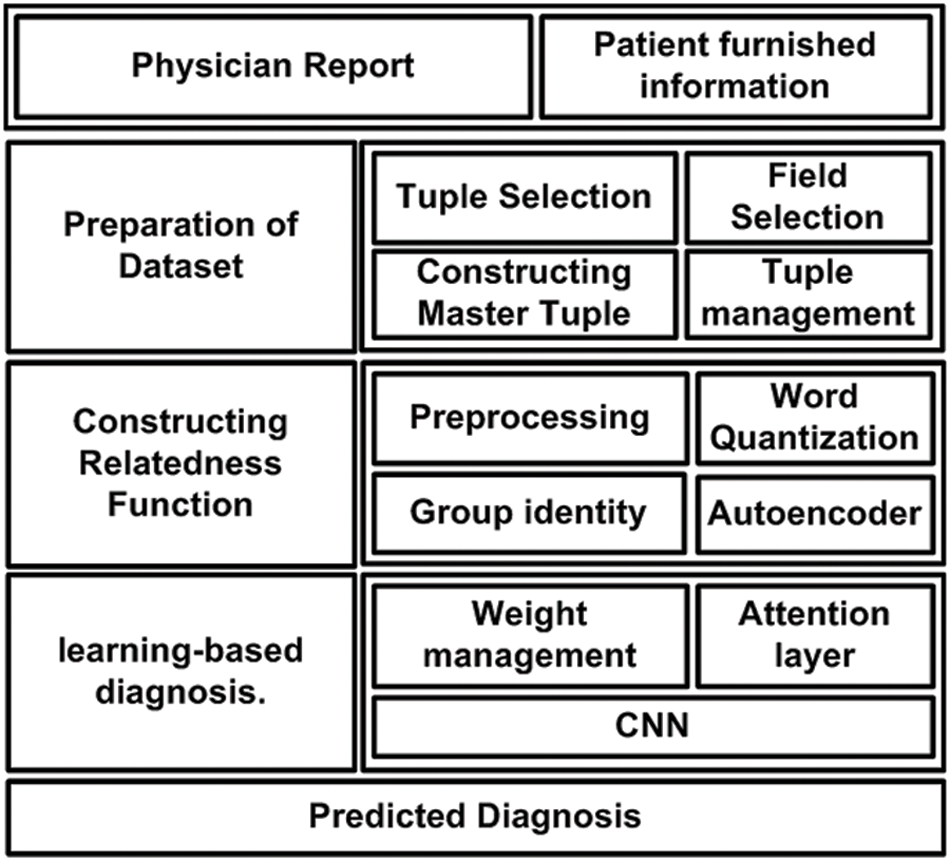
Figure 1: Architecture of proposed diagnosis model
The proposed system uses a deep learning autoencoder to carry out training which finally classifies the data in a group. The third implementation stage is about applying machine learning, where the proposed system uses Long Short-Term Memory (LSTM) and Convolution Neural Network (CNN) for this purpose. The attention layer permits the weights and biases based on the point of admission and carries out respective weight management. Owing to the inclusion of two different types of neural networks and a better form of input data, the proposed system can offer better diagnosis prediction from the complex structure of the medical dataset. The following section elaborates further about the involved algorithm and its respective system design.
This section discusses the system design that has been considered to carry out the predictive diagnosis using machine learning. From the discussion carried out in the prior section, it is now known that two forms of medical information must be subjected to the machine learning approach, viz. (i) physician report and (ii) patient furnished information. However, different from any existing system, the proposed implementation is carried out over a standard medical dataset using both the above categories of data. The target is to obtain a better form of enriched information for assisting precise clinical diagnosis of the illness. Hence, the complete system design of the proposed implementation mainly emphasizes the management and preparation of the dataset, followed by a series of processing is carried out to meet the research objectives. The complete system design of the proposed model is carried out considering three different modules viz. (i) Preparation of dataset, (ii) Constructing Relatedness Function, and (iii) learning-based diagnosis. Following is an elaborated discussion of each module concerning algorithm and system design.
The first part of the implementation targets fine-tuning the considered medical dataset. It is seen that usually, the complete dataset is subjected to learning algorithms in the existing system without prioritizing the essential fields within the dataset. Therefore, the prime goal of this module is to prepare the dataset to include all the required fields and eliminate all the non-essential areas within the dataset. The algorithmic steps for this purpose are as follows:
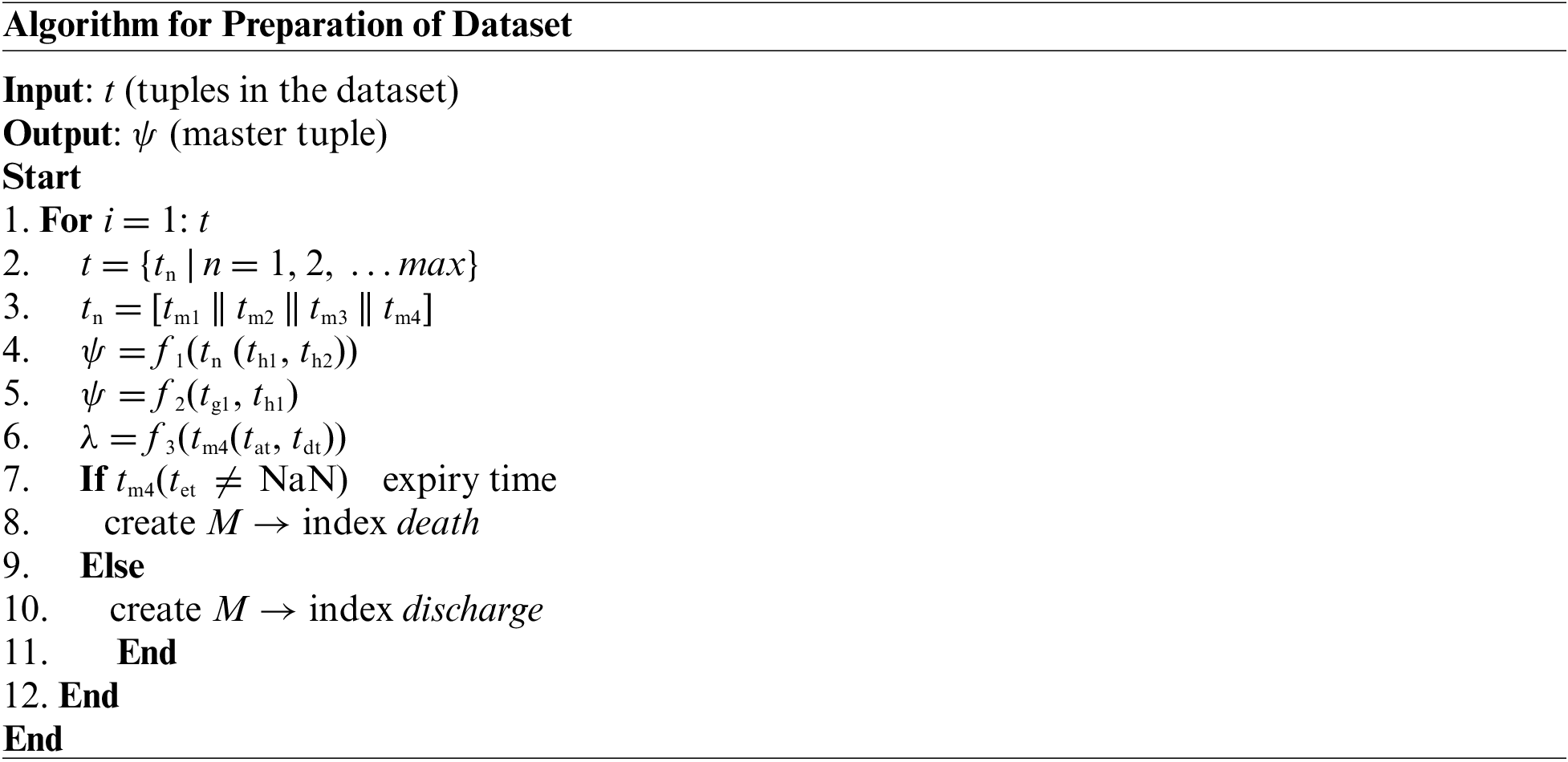
The above-presented algorithm takes the input of t (tuples of the dataset) that yields an outcome of a ψ master tuple after processing. The study considers that there could be a maximum max number of a tuple within the medical dataset with both essential and non-essential information (Line-1 and Line-2).
The next step is to consider only four essential tuple tn which is equivalent to concatenation of tm1, tm2, tm3, and tm4 (i.e., n = 4) from the main dataset tuple t (Line-3). The brief discussion of the selected tuples are as follows:
• events (tm1): This tuple consists of all the clinical and non-clinical information obtained by the physician and medical caretakers within the length of stay of the subject. This tuple tm1 further consists of fields viz. identity of the issue, identity of admission, time and date of medical charting, type of notes, highlights of errors in clinical notes, explicit content of clinical notes.
• diag1 (tm2): This tuple comprises names of the patient’s disease as per the standard of ICD9 codes. It further consists of the subject’s identity, the identity of admission, and ICD9 codes.
• diag2 (tm3): This tuple consists of mapped codes of standard ICD9 in order to highlight the disease. This tuple further consists of two fields, i.e., ICD9 codes and short names of the disease.
• Admission (tm4): This tuple further consists of all the information associated with the patient’s entire length of stay. This tuple further consists of the identity of admission, time of admission, time of discharge, type of admission (emergency room or out-patient department), and time of patient’s death.
After the tuples mentioned earlier, tn is selected from main table t, the next part of the implementation is associated with performing preprocessing operations. For this purpose, this module of implementation considers two explicit fields, i.e., the identity of admission (th1) and ICD9 codes (th2) from the selected tuple tn (Line-4). A function f1(x) is implemented to perform an inner join operation that finally yields a master tuple ψ (Line-4). There is a dual advantage for undertaking these preprocessing steps viz. (i) both these fields act as a primary key for the dataset, and consideration of these results facilitates better query processing, and (ii) adherence of standard naming of the disease while subjected to next level of operation. According to the function f1(x), the sub-field of the identity of admission (th1) is considered from the field admission (tm4) and events (tm1), while the sub-field for (th2) ICD9 code is considered for the fields diag1(tm2) and diag2(tm3). Finally, all the updated fields of events (tm1), diag1(tm2), diag1(tm2): and admission (tm4) are subjected to inner join operation to generate updated master tuple ψ (Line-4). Therefore, this operation results in a compact dataset with a precise selection of its respective fields. The next part of the algorithm implementation constructs another function, f2(x), which is responsible for the elimination of two sub-fields, i.e., the identity of subject (tg1) and identity of the hospital (tg1) as exhibited in Line-5 i.e., f2 tn-|(tg1, th1)|. This operation contributes to storage optimization by eliminating these two fields, which acts as the primary key for all the patients from selected tuple tn.
Apart from this, the proposed system is also fine-tuned to ensure the retention of only the initial three characters of the ICD9 codes eliminating other parts of it. The prime justification behind choosing this operation is that only the initial three characters of ICD9 codes directly represent the disease class and not the complete condition of the disease. Hence, the proposed algorithm performs optimization steps in order to identify disease classes for better classification.
The next part of the algorithm implementation is emphasized on constructing a differentiating function f3(x) which performs subtraction of sub-field time of admission (tat) from time of discharge (tdt) in order to obtain the length of stay λ (Line-6). The algorithm also assesses any form of error prone data, which is subjected to disposal to retain better data quality. The final steps of the proposed algorithm are about constructing a conditional logic to assess if the sub-field for the time of death (tet) is not NaN (Line-7), which results in indexing it as a death of the patient (Line-8) or else it is indexed as discharge summary of the patient (Line-10). Both the information results in repositing the outcome in a matrix M that represents the patient’s mortality. Finally, the proposed study only considers the mortality matrix M indexed discharge (Line-10) within the master tuple t. This refined preprocessed dataset is now subjected to the next level of operation.
5.2 Constructing Relatedness Function
The prior algorithm’s accomplishment results in the retention of discharge summary information stored in the M matrix. This outcome is considered an input to this part of the implementation module, which targets the further streamlined outcome of preprocessing. For this purpose, the proposed system implements a unique natural language processing technique where a neural network is implemented to carry out a learning operation. The learning is carried out towards the association of the words present over the corpus of the medical textual data. Once the training is accomplished, this part of the implementation will identify the synonymous words and be capable of recommending additional text for a partial sentence. The proposed system constructs a relatedness function to represent a unique word of the distinct type associated with a numerical list to achieve this objective. Such a numerical list is carefully selected so that a simplified function could represent a degree of semantic association between all the words represented by such a numerical list. The steps of algorithmic operation for this purpose are as follows:
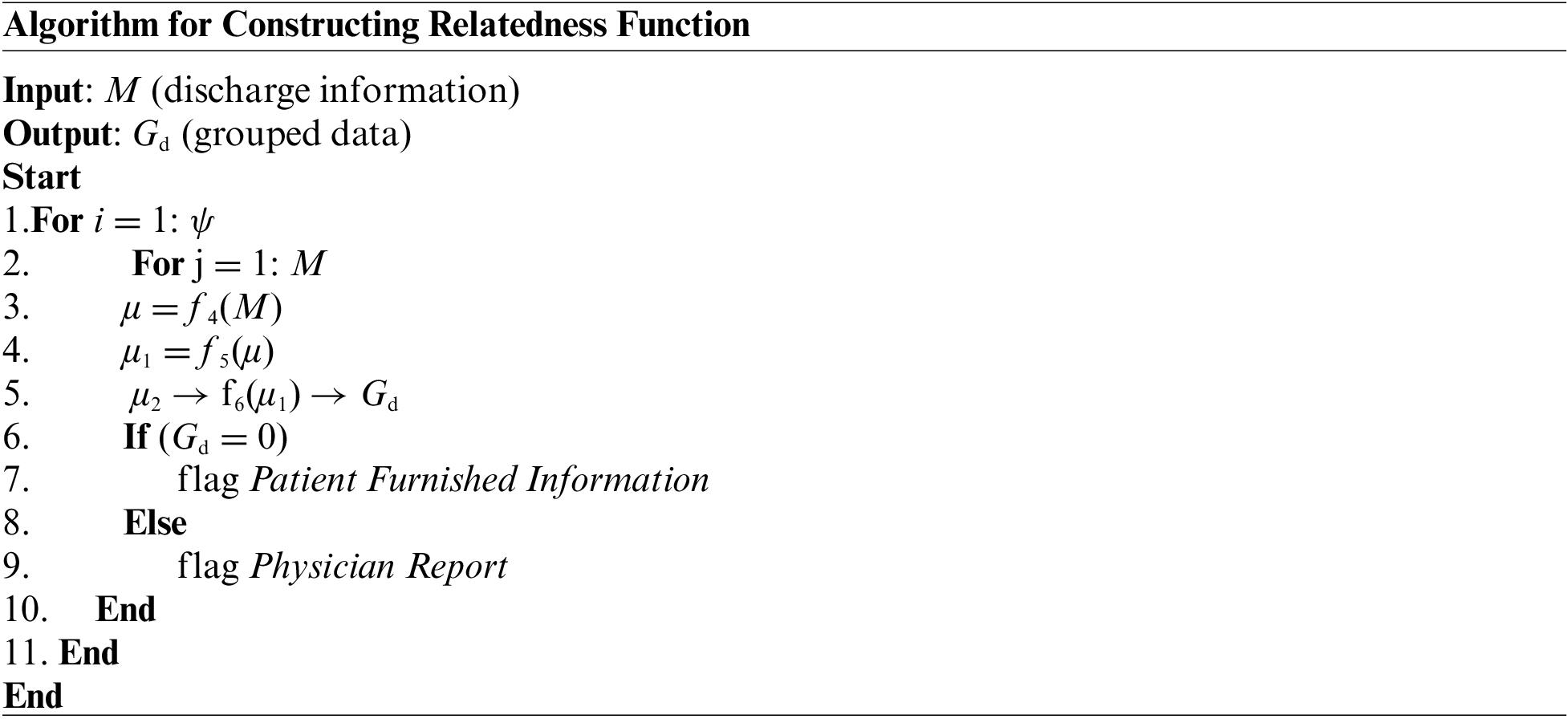
As mentioned earlier, the algorithm considers all the master tuple ψ (Line-1) while it considers all the obtained discharge summary information stored in the M matrix (Line-2). This input of discharge summary information is then subjected to an explicit function f4(x) which generates the first level of preprocessed data μ (Line-3). The function f4(x) carries out a series of preprocessing operations where a lower-case text is initially obtained for all the input textual content from the M matrix. The function also substitutes all the multiple whitespaces to single whitespaces, followed by eliminating all the punctuation marks. However, different from any existing preprocessing of a textual document, the proposed function f4(x) retains all the stop words. The prime justification behind this step of action is that stop words don’t affect the text’s contextual meaning. Apart from this, the presence of stop words also assists in identifying the clinical information from the medical report of a patient. The next part of implementation is towards performing word quantification using a function f5(x) which further generates the next level of preprocessed outcome μ1 (Line-4). The complete process of word quantification using function f5(x) is carried out in two methods:
• In the first process, the algorithm looks for the specific word occurrences that are contextually connected with the medical dataset. The primary technique is to find the raw count of a particular term in medical data related to the disease. The secondary process is to divide the raw count by the number of words present in the document. The ternary technique will obtain a Boolean frequency or frequency based on a logarithmic scale.
• In the second process, the algorithm identifies all the common as well as uncommon words within the medical dataset. This process assists in rectifying the presence of specific terms which occur quite abundantly in the English corpus. Hence, this process contributes towards reducing the weight of such terms frequently occurring (as redundant words) and giving more emphasis towards the uncommon words.
Therefore, the contribution of using the function f5(x) is to offer the importance of the uncommon words present in the medical dataset, which furnishes more information about disease conditions or some specific knowledge indicator to assist in diagnosis. The next part of the implementation is associated with training the data using another function, f6(x), to generate the group data using the deep learning technique (Line-5) to generate further clustered information. This operation generates grouped data Gd where deep learning makes use of an autoencoding approach that is subjected to training for only one data class. It could be either a physician report or furnished patient information.
In the proposed algorithmic implementation, the training with autoencoders is carried out with physician reports. It is to be noted that both physician reports and furnished patient information are textual information. The only difference is that the former has more clinical contextual information while the latter has less contextual information. When the training with autoencoders is carried out with physician report followed by the event when the algorithm is subjected to patient furnished report, the algorithm will attempt to yield similar textual outcome as physician report. However, suppose the input to the algorithm is considered to physician report. In that case, the anticipated outcome will be the same as that of an input file as the Euclidean distance between input and output is relatively more minor.
On the contrary, if the input for the deep learning autoencoder is patient furnished information, the outcome will be distinctively varying due to the higher Euclidean distance between input and output. The prime reason behind this outcome difference is that the proposed system carries out autoencoder training with physician reports only. The proposed system carries out grouping operating on the medical data on the basis of Euclidean distance by setting a cut-off of 0.1. Hence, when the value of group identity Gd = 0, the algorithm will flag patient furnished information, and when Gd = 1, it will flag physician report. The contribution of this algorithm is that it offers a second level of preprocessing the data, inclusive of training using deep learning autoencoders, which is already witnessed to provide the benefits towards grouping, generation, de-noising, and compression of data. Although usage of autoencoders is mainly witnessed towards data compression, the proposed scheme has utilized it to differentiate and group the preprocessed data. The proposed algorithm also offers a novelty compared to existing approaches of autoencoder usage. Unlike directly applying the data to autoencoders, the proposed system split the data regarding physician reports and furnished patient reports. These steps of operation contribute to increasing significantly better accuracy level while subjected to the next level of machine learning approach. The outcome is now subjected to the last level of learning.
This is the final module of implementation responsible for carrying out the learning operation to accomplish the diagnosis’s prediction. The learning operation is carried out using LSTM, a frequently used encoder and decoder capable of the processing data sequence. However, this conventional LSTM suffers from a limitation where it can only carry out the encoding of the input sequence to a constant length of internal representation. Hence, the length of the input sequence becomes a primary limitation to process a more extended sequence of input, which is inevitable in a large medical dataset. Hence, although the conventional LSTM offers better encoding/decoding operation, its restricted capability to process fixed data sequence becomes a prime loophole. Therefore, the proposed system addresses this challenge using attention layers. In this mechanism, summarization of the input sequence can be obtained, which can associate each word in the summarized outcome to a specific word in the input medical dataset. Further, in the conventional attention model, the BLSTM is deployed to generate an annotated sequence for each input sequence. Additionally, all the vector deployed in the model concatenates the encoder’s forward and backward hidden states. The novelty of this approach is that it works with the local attention layer, whereas the conventional method is restricted to global attention only. This adoption is carried out based on concepts associated with soft and hard attention standardized in Natural Language Processing. Soft attention is a direct representation of the global attention where all the parts of the medical dataset are allocated with certain weights w; however, only one part of the sentence is considered at a time in case of hard attention. Regarding captioning towards NLP, local attention potentially differs from hard attention. It can be an amalgamation of both where only a segment is considered to generate a context vector instead of considering all the encoded inputs. Adoption of this process doesn’t only resist the expensive computational process, unlike existing approaches discussed in Section 2. Still, it also becomes quite an easier task to train compared to hard attention. The steps of the algorithm considered for this purpose is as follows:

According to the above algorithm, the discharge summary M is considered input. The different sequence of words and cluster id obtained from dictionary embedding acts as an input (Fig. 5), as shown in Line-1. A function f7(x) is constructed for w weight adjustment where attention layer alay1 permits either the primary set of weights w and biases or the secondary set of weights w to process the data (Line-2 and Line-3). The attention layer alay1 further forwards this to organize a matrix P for point of admission, which consists of Emergency Room ER and Out-Patient Department OPD information to construct matrix PoA (Line-4). Another function, f8(x), is constructed where the matrix of PoA is further forwards to CNN that is again forwarded to the next attention layer alay2 (Line-5.). This operation finally results in the outcome of dpred (Line-6). Therefore, it can be seen that there are two different networks without using any sophisticated operation, which overcomes the limitation of iterative and complex computational operation in the existing learning system. Apart from this, the model also contributes to further data reduction due to a series of processing being carried out over medical data, unlike existing approaches. Therefore, the proposed learning approach offers lightweight and much iterative operation, which provides a good balance between accuracy and computational complexity, unlike any existing literature presented in Section 2.
The implementation of this part of the study considers the similar dataset of MIMIC III assessed in our prior study [31]. Our previous study used this dataset, where feature engineering was carried out, followed by correlation analysis and transformation technique being implemented. The model’s outcome has been evaluated concerning three machine learning techniques viz. K-Nearest Neighbor (KNN), logistic regression, and Artificial Neural Network (ANN). The final, conclusive outcome exhibited ANN to outperform other learning techniques. The proposed implemented scheme presented in this manuscript extends our prior model [43], where the core emphasis is towards preprocessing on a specifically selected field with explicit tuple management. This part of the implementation involves constructing the relatedness function using autoencoders of deep learning followed by applying learning-based diagnosis with CNN. This section briefs about the analysis strategy and discusses the outcome.
The system design and algorithm discussed in the prior section have been implemented on the python environment considering the MIMIC III dataset. Apart from the well-defined implementation strategy, the analysis strategy consists of two concerns, i.e., (i) selection of the existing system to perform comparative analysis over benchmarked dataset MIMIC III and (ii) selection of performance parameters. The first existing system considered for comparative analysis is the work done by Lin et al. [44], who has used Support Vector Machine (SVM) to perform mortality prediction over the MIMIC-III dataset. This study has considered the physiology score used as a predictor to develop an SVM model. However, the limitation of this study has found is that it doesn’t perform well in the presence of uncertain data of patient mortality rate. The second existing system for comparative analysis is chosen from the work of Mohan [45]. This study also serves a similar agenda using the multilayer perceptron of neural networks over the same dataset. This study has considered a dataset of 40,000 patients considering ICD9 codes for event prediction. The accomplished range of accuracy was found to be around 80%. The prime justification for choosing the above two existing comparative analysis models is that they both serve similar agendas (using machine learning for predicting medical conditions) and similar datasets (MIMIC III dataset). Before performing comparative analysis, the model is assessed for its outcome using a probability distribution score with an ICD9 code before performing a comparative analysis. The idea is to determine the state of non-equilibrium of the data concerning interactiveness of ICD9 codes more than 8500 times. The proposed machine learning scheme significantly controls this problem, and hence a better lightweight predictive model can be ensured. Finally, the assessment of the model is carried out using precision, recall, F1-score, and processing time. Similar data and test environments are applied for both proposed and existing systems to retain a uniform test environment. Fig. 2 highlights the visual representation of the experiment being carried out. A closer look into this flow shows that proposed system is more progressive and less iterative with an explicit preprocessing carried out towards enriching MIMIC-III dataset.
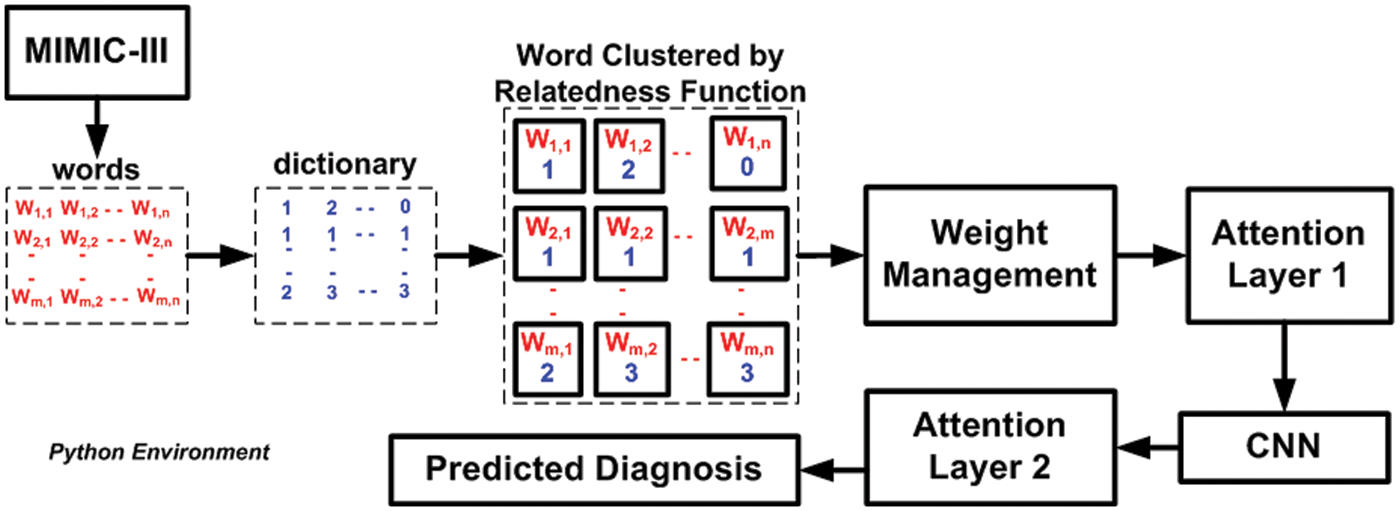
Figure 2: Visual representation of adopted experiment
The majority of the machine learning scheme makes use of classification accuracy as the prime performance parameter for any analysis. This parameter is computed considering the total number of obtained corrected predicted outcomes divided by the total number of predictive outcomes considered for an adopted dataset. However, there are certain pitfalls in considering classification accuracy as the core performance parameter, mainly if it involves solving classification issues with a non-equilibrium state with a massive number of tuples and features to be considered. The core justification behind not adopting this as a core performance parameter in the proposed study is that the presence of a voluminous population from classes will abnormally exceed the quantity of class. It will infer that there are fair chances that an inappropriate model possesses higher accuracy, which may be just an outlier. Hence, the proposed system considers an alternate solution using precision, recall, and F1-score. The outcome obtained using these performance parameters is exhibited in Figs. 3–6.
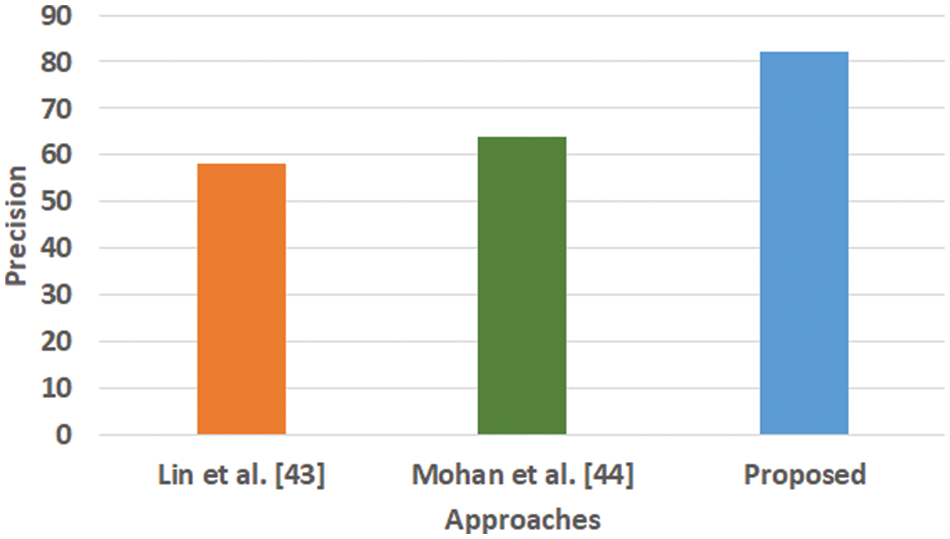
Figure 3: Comparative analysis of precision (Mohan, not Mohan et al.)
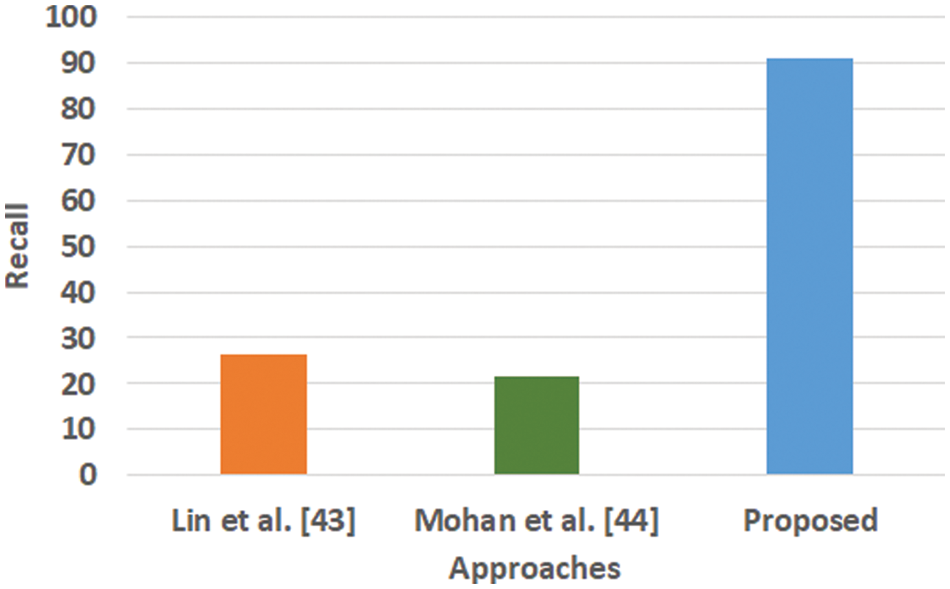
Figure 4: Comparative analysis of recall
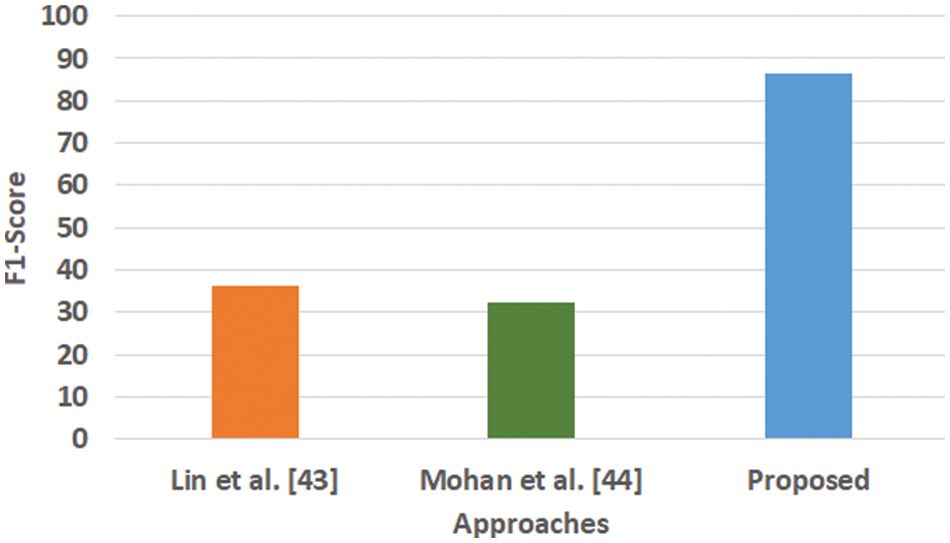
Figure 5: Comparative analysis of F1-score
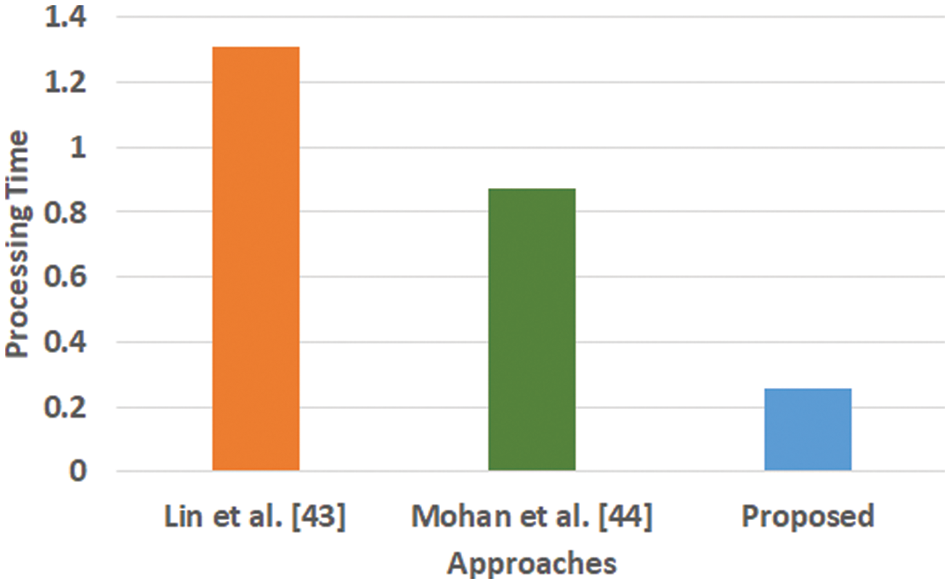
Figure 6: Comparative analysis of processing time
Fig. 3 highlights the precision, representing the prediction quantity for positive classes, which is a subset for actual positive classes. The outcome shows that the proposed system offers higher accuracy than Mohan’s work [44] and Lin et al. [43]. The model presented by Lin et al. [43] is highly memory efficient as well as it is found to quite effective for high-dimensional spaces. However, it is not applicable if the dataset is quite large. This problem doesn’t exist in the model presented by Mohan [44] as it potentially assists in learning the non-linear model and is highly suitable for applying on real-time learning. Apart from this, this model is also quite sensitive towards scaling features. On the other hand, the proposed system can eliminate all artifacts from the input signal, thereby retaining the highest quality of the signal. Fig. 4 represents the recall performance outcome that infers all the predicted positive classes constructed from all the positive dataset samples. In this case, it can be seen that the recall score dips down for both learning models of Lin et al. [43] and Mohan [44], whereas the recall score is slightly improved for the proposed system compared to the precision score obtained. The prime reason behind this is that the model implemented by Lin et al. [43] is potentially affected by artifacts resulting in overlapping classing, thereby diminishing the recall performance score. The model of Mohan [44] has used multiple numbers of initialization of random weights for catering up multiple accuracy of validation owing to the presence of non-convex function of loss. On the other hand, the proposed scheme is callable of learning non-linear transformation using multiple layers and non-linear activation function without possessing much of necessity to learn dense layers. This potentially affects the recall score even higher compared to the precision obtained by the proposed scheme. Fig. 5 highlights the comparative analysis F1-score, which offers a unit score required to maintain an equilibrium between the recall score and precision score. Overall, the outcome shows a slight improvement of both existing schemes, whereas a nearly similar trend is seen for the proposed learning scheme. Similar justification stated before can be offered for analysis of these performance attributes. Hence, based on these performance scores, it can be said that the proposed system can provide a reliable computation of predictive classification of a medical dataset of complex form.
Fig. 6 highlights that the proposed scheme offers comparatively lower processing time than the existing learning scheme. The processing time is computed as the duration of time right from the input of data to all stages of internal processing that finally ends with predicted outcome. The model of Lin et al. [43] is witnessed to consider maximized training time for large scale of dataset while the model of Mohan [44] is capable of taking the output values in two values owing to hard restriction of transfer function. While, a closer look into the proposed system shows that it is less iterative while the processing operation and inclusion of relatedness function extracts potential information prior to learning. This step eventually reduces the computational burden of the learning process, which significantly reduces the processing time. Hence, the proposed system can be said to be computationally efficient with respect to time complexity in presence of large and complex medical data.
The complexities associated with analyzing medical data has been always a matter of concern in any field of analytics owing to the inclusion of critical clinical information that demands higher performance. In this regard, machine learning has been contributing to a large extent towards predicting the identification and classification of the critical disease. Review of existing learning schemes exhibits that there the significant problems associated viz. lack of context in medical dataset, non-inclusion of robust preprocessing, usage of heavyweight learning approach, and inclusion of computational complexity. These problems are addressed in proposed scheme. The contribution being carried out with respect to inclusion of novelty factors in proposed scheme are as listed: (i) the proposed scheme performs the complete modelling considering both physician report and patient furnished information, unlike any existing studies, (ii) different from any existing learning scheme where importance is given more on learning scheme and less on fine-tunning the dataset, the proposed scheme introduces a novel and simplified mechanism of preparing the dataset considering an explicitly selected tuple, (iii) the proposed scheme implements an autoencoder scheme for performing execution of newly designed relatedness function along with word quantization which significantly redefines the medical dataset suitable to offer less computational burden to learning operation, and (iv) a unique learning operation is implemented which uses attention layer with proper weight management while using CNN to perform final prediction.
Availability of Data and Materials: Study uses MIMIC-III dataset which is freely available at https://physionet.org/.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. A. Nayyar and F. Al-Turjman, “Machine learning for critical internet of medical things applications and use cases,” in Application of Cloud and IoT Technologies in Battling the COVID-19 Pandemic, 1st ed., vol. 1, New York: Springer International Publishing, pp. 1–29, 2022. [Google Scholar]
2. L. R. Gupta and M. Roy, “Machine learning and data analytics for predicting, managing, and monitoring disease,” in Pandemic Management Using Artificial Intelligence-Based Safety Measures: Prediction and Prevention, Pennsylvania: IGI Global, pp. 86–110, 2021. [Google Scholar]
3. U. Kose, O. Deperlioglu, J. Alzubi and B. Patrut, “Deep learning for medical decision support systems,” in Deep Learning Architectures for Medical Diagnosis, 1st ed., vol. 1, Singapore: Springer, pp. 15–28, 2020. [Google Scholar]
4. K. G. Srinivasa, “Artificial Intelligence for information management: A healthcare perspective,” in Healthcare Data Analytics Using Artificial Intelligence, 1st ed., vol. 1, Singapore: Springer, 2021. [Google Scholar]
5. Y. Goldberg, Neural network methods in natural language processing, 1st ed., vol. 1, San Rafael: Morgan & Claypool Publishers, 2017. [Google Scholar]
6. D. R. Recupero, M. Petkovic and S. Consoli, “Data science for healthcare-methodologies and applications,” in The Role of Deep Learning in Improving Healthcare, 1st ed., vol. 1, New York: Springer International Publishing, 2019. [Google Scholar]
7. S. Vajjala, B. Majumder, A. Gupta and H. Surana, Practical natural language processing-A comprehensive guide to building real-world NLP systems, 1st ed., vol. 1, Massachusetts: O’Reilly Media, 2020. [Google Scholar]
8. J. S. Osorio, K. E. Paik, L. A. Celi, M. S. Majumder, S. Melek et al., “Leveraging data science for global health,” in Developing Local Innovation Capacity to Drive Global Health Improvement, 1st ed., vol. 1, New York: Springer International Publishing, 2020. [Google Scholar]
9. A. DeVore, The Electronic Health Record for the physician’s office for SimChart for the medical office, 1st ed., vol. 1, Edinburgh: Elsevier Health Sciences, 2015. [Google Scholar]
10. A. A. Funker, M. P. Egorov, S. A. Fokin, G. M. Orlov and S. V. Kovalchuk, “Citywide quality of health information system through text mining of electronic health records,” in Applied Network Science, vol. 6, New York: Springer Open, 2021. [Google Scholar]
11. C. J. Girman and M. E. Ritchey, “Pragmatic randomized clinical trials-using primary data collection and electronic health records,” in Agricultural and Biological Sciences, 1st ed., vol. 1, Amsterdam: Elsevier Science, 2021. [Google Scholar]
12. F. Xing, Y. Xie, H. Su, F. Liu and L. Yang, “Deep learning in microscopy image analysis: A survey,” IEEE Transactions on Neural Networks and Learning Systems, vol. 29, no. 10, pp. 4550–4568, 2018. [Google Scholar]
13. M. R. Ahmed, Y. Zhang, Z. Feng, B. Lo, O. T. Inan et al., “Neuroimaging and Machine Learning for dementia diagnosis: Recent advancements and future prospects,” IEEE Reviews in Biomedical Engineering, vol. 12, pp. 19–33, 2019. [Google Scholar]
14. M. H. Sarhan, M. A. Nasseri, D. Zapp, M. Maier, C. P. Lohmann et al., “Machine learning techniques for ophthalmic data processing: A review,” IEEE Journal of Biomedical and Health Informatics, vol. 24, no. 12, pp. 3338–3350, 2020. [Google Scholar]
15. J. Liu, Y. Pan, M. Li, Z. Chen, L. Tang et al., “Applications of deep learning to MRI images: A survey,” IEEE Big Data Mining and Analytics, vol. 1, no. 1, pp. 1–18, 2018. [Google Scholar]
16. Accessed on 03-March, 2022. Available: https://www.v7labs.com/blog/healthcare-datasets-for-computer-vision#ct. [Google Scholar]
17. Accessed on 03-March, 2022. Available: https://www.v7labs.com/blog/healthcare-datasets-for-computer-vision#mri. [Google Scholar]
18. Accessed on 03-March, 2022. Available: https://www.v7labs.com/blog/healthcare-datasets-for-computer-vision#x-ray. [Google Scholar]
19. Accessed on 03-March, 2022. Available: https://www.v7labs.com/blog/healthcare-datasets-for-computer-vision#general-health. [Google Scholar]
20. Accessed on 03-March, 2022. Available: https://www.v7labs.com/blog/healthcare-datasets-for-computer-vision#covid. [Google Scholar]
21. A. S. Panayides, A. Amini, N. D. Filipovic, A. Sharma, S. A. Tsaftaris et al., “AI in medical imaging informatics: Current challenges and future directions,” IEEE Journal of Biomedical and Health Informatics, vol. 24, no. 7, pp. 1837–1857, 2020. [Google Scholar]
22. J. Latif, C. Xiao, S. Tu, S. U. Rehman, A. Imran et al., “Implementation and use of disease diagnosis systems for electronic medical records based on machine learning: A complete review,” IEEE Access, vol. 8, pp. 150489–150513, 2020. [Google Scholar]
23. X. Chai, “Diagnosis method of thyroid disease combining knowledge graph and deep learning,” IEEE Access, vol. 8, pp. 149787–149795, 2020. [Google Scholar]
24. K. Guo, S. Ren, Md. Z. A. Bhuiyan, T. Li, D. Liu et al., “MDMaaS: Medical-assisted diagnosis model as a Service with artificial intelligence and trust,” IEEE Transactions on Industrial Informatics, vol. 16, no. 3, pp. 2102–2114, 2020. [Google Scholar]
25. Y. Guo, G. Wang, L. Li, L. Wang, L. Wang et al., “Machine learning aided diagnosis of diseases without clinical gold standard: A new score for laryngopharyngeal reflux disease based on pH monitoring,” IEEE Access, vol. 8, pp. 67005–67014, 2020. [Google Scholar]
26. M. Heidari, H. Liu, B. Zheng, S. Mirniaharikandehei, G. Danala et al., “Applying a random projection algorithm to optimize machine learning model for breast lesion classification,” IEEE Transactions on Biomedical Engineering, vol. 68, no. 9, pp. 2764–2775, 2021. [Google Scholar]
27. S. Hussein, P. Kandel, C. W. Bolan, M. B. Wallace and U. Bagci, “Lung and pancreatic tumor characterization in the deep learning era: Novel supervised and unsupervised learning approaches,” IEEE Transactions on Medical Imaging, vol. 38, no. 8, pp. 1777–1787, 2019. [Google Scholar]
28. S. Niu, M. Liu, Y. Liu, J. Wang and H. Song, “Distant domain transfer learning for medical imaging,” IEEE Journal of Biomedical and Health Informatics, vol. 25, no. 10, pp. 3784–3793, 2021. [Google Scholar]
29. S. Roy, W. Menapace, S. Oei, B. Luijten, E. Fini et al., “Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound,” IEEE Transactions on Medical Imaging, vol. 39, no. 8, pp. 2676–2687, 2020. [Google Scholar]
30. A. Shamsi, H. Asgharnezhad, S. S. Jokandan, A. Khosravi, P. M. Kebria et al., “An uncertainty-aware transfer learning-based framework for COVID-19 diagnosis,” IEEE Transactions on Neural Networks and Learning Systems, vol. 32, no. 4, pp. 1408–1417, 2021. [Google Scholar]
31. P. Wu, H. Ye, X. Cai, C. Li, S. Li et al., “An effective machine learning approach for identifying non-severe and severe coronavirus disease 2019 patients in a rural Chinese population: The Wenzhou retrospective study,” IEEE Access, vol. 9, pp. 45486–45503, 2021. [Google Scholar]
32. L. Li, W. Liu, H. Zhang, Y. Jiang, X. Hu et al., “Down syndrome prediction using a cascaded machine learning framework designed for imbalanced and feature-correlated data,” IEEE Access, vol. 7, pp. 97582–97593, 2019. [Google Scholar]
33. J. Li, L. Liu, J. Sun, Y. Pei, J. Yang et al., “Diagnosis and knowledge discovery of turner syndrome based on facial images using machine learning methods,” IEEE Access, vol. 8, pp. 214866–214881, 2020. [Google Scholar]
34. C. Lian, M. Liu, J. Zhang and D. Shen, “Hierarchical fully convolutional network for joint atrophy localization and alzheimer’s disease diagnosis using structural MRI,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 42, no. 4, pp. 880–893, 2020. [Google Scholar]
35. N. Liu, X. Li, E. Qi, M. Xu, L. Li et al., “A Novel ensemble learning paradigm for medical diagnosis with imbalanced data,” IEEE Access, vol. 8, pp. 171263–171280, 2020. [Google Scholar]
36. M. Rashed-Al-Mahfuz, A. Haque, A. Azad, S. A. Alyami, J. M. W. Quinn et al., “Clinically applicable machine learning approaches to identify attributes of Chronic Kidney Disease (CKD) for use in low-cost diagnostic screening,” IEEE Journal of Translational Engineering in Health and Medicine, vol. 9, pp. 1–11, 2021. [Google Scholar]
37. W. Wang, F. Liu, X. Zhi, T. Zhang and C. Huang, “An integrated deep learning algorithm for detecting lung nodules with low-dose CT and its application in 6G-enabled internet of medical things,” IEEE Internet of Things Journal, vol. 8, no. 7, pp. 5274–5284, 2021. [Google Scholar]
38. W. Yang, M. Joo, Y. Kim, S. H. Kim and J. M. Chung, “Hybrid machine learning scheme for classification of BECTS and TLE patients using EEG brain signals,” IEEE Access, vol. 8, pp. 218924–218935, 2020. [Google Scholar]
39. Y. Zhang, X. Wang, N. Han and R. Zhao, “Ensemble learning based postpartum hemorrhage diagnosis for 5G remote healthcare,” IEEE Access, vol. 9, pp. 18538–18548, 2021. [Google Scholar]
40. X. Zhou, Y. Li and W. Liang, “CNN-RNN based intelligent recommendation for online medical pre-diagnosis support,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, vol. 18, no. 3, pp. 912–921, 2021. [Google Scholar]
41. W. Zhu, L. Sun, J. Huang, L. Han and D. Zhang, “Dual attention multi-instance deep learning for Alzheimer’s disease diagnosis with structural MRI,” IEEE Transactions on Medical Imaging, vol. 40, no. 9, pp. 2354–2366, 2021. [Google Scholar]
42. X. R. Zhang, J. Zhou, W. Sun and S. K. Jha, “A lightweight CNN based on transfer learning for COVID-19 diagnosis,” Computers, Materials & Continua, vol. 72, no. 1, pp. 1123–1137, 2022. [Google Scholar]
43. S. R. Khope and S. Elias, Critical correlation of predictors for an efficient risk prediction framework of ICU patient using correlation and transformation of MIMIC-III dataset, vol. 7, New York: Springer Open-Data Science & Engineering, pp. 71–86, 2022. [Google Scholar]
44. K. Lin, J. Q. Xie, Y. H. Hu and G. L. Kong, “Application of support vector machine in predicting in-hospital mortality risk of patients with acute kidney injury in ICU,” PubMed, vol. 50, no. 2, pp. 239–244, 2018. [Google Scholar]
45. N. Mohan, “Predicting post-procedural complications using neural networks on MIMIC-III data,” Master’s Thesis for Louisiana State University, Louisiana, 2018. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |