
 | Computer Modeling in Engineering & Sciences |  |
DOI: 10.32604/cmes.2022.018433
ARTICLE
Efficient Data Augmentation Techniques for Improved Classification in Limited Data Set of Oral Squamous Cell Carcinoma
1Department of Information Technology, College of Computers and Information Technology, Taif University, Taif, 21944, Saudi Arabia
2Institute of Computing, Kohat University of Science and Technology, Kohat, 26000, Pakistan
*Corresponding Author: Wael Alosaimi. Email: w.osaimi@tu.edu.sa
Received: 24 July 2021; Accepted: 30 November 2021
Abstract: Deep Learning (DL) techniques as a subfield of data science are getting overwhelming attention mainly because of their ability to understand the underlying pattern of data in making classifications. These techniques require a considerable amount of data to efficiently train the DL models. Generally, when the data size is larger, the DL models perform better. However, it is not possible to have a considerable amount of data in different domains such as healthcare. In healthcare, it is impossible to have a substantial amount of data to solve medical problems using Artificial Intelligence, mainly due to ethical issues and the privacy of patients. To solve this problem of small dataset, different techniques of data augmentation are used that can increase the size of the training set. However, these techniques only change the shape of the image and hence the classification model does not increase accuracy. Generative Adversarial Networks (GANs) are very powerful techniques to augment training data as new samples are created. This technique helps the classification models to increase their accuracy. In this paper, we have investigated augmentation techniques in healthcare image classification. The objective of this research paper is to develop a novel augmentation technique that can increase the size of the training set, to enable deep learning techniques to achieve higher accuracy. We have compared the performance of the image classifiers using the standard augmentation technique and GANs. Our results demonstrate that GANs increase the training data, and eventually, the classifier achieves an accuracy of 90% compared to standard data augmentation techniques, which achieve an accuracy of up to 70%. Other advanced CNN models are also tested and have demonstrated that more deep architectures can achieve more than 98% accuracy for making classification on Oral Squamous Cell Carcinoma.
Keywords: Data science; deep learning; data augmentation; classification; data manipulation
Remarkable advancements have been made in the domain of computer vision mainly because of the improvements in Convolutional Neural Networks (CNN). These CNN-based models are commonly used in classification, object detection and localization, object segmentation, and instance segmentation. CNN based models along with deep learning algorithms have surpassed human level performance in object identification [1,2]. These CNN-based models require a substantial set of images such as ImageNet, OpenImages, COCO, etc. These images are manually annotated and requires human efforts to make sure that each image is labelled correctly. Machine Learning (ML) models trained on these images can learn the pattern in the dataset and can correctly identify objects. The more the data and the deeper the CNN network, the better these models can make predictions. It is observed in the literature that to develop a generalized Deep Learning (DL) model, there needs to be a large dataset.
In the healthcare domain, obtaining many images is challenging mainly because of the equipment used to capture the images and the privacy of patients. It is even more challenging to annotate these images as specialized medical professionals are required to develop consensus on the labelling of images. This labelling of data is time consuming in terms of manual efforts and expensive in terms of the requirements of experts [3]. Labelled images in the medical domain are scarce and expensive to generate. Another challenge is that the model trained in one medical domain will not be able to perform well on a slightly different domain in healthcare. Although millions of medical images are created daily, it is not easy to place all these images in a public dataset because of ethical issues and patient’s privacy. This is the main reason that most of the publicly available medical images suffer from the problem of sample datasets to be used by researchers to find Artificial Intelligence (AI) based solutions.
To overcome the problem of small data, data augmentation techniques are used to increase the number of images. Different classical data augmentation techniques use geometric/intensity transformation of original images for diagnosis of disease in medical images. Some augmentation techniques include modification of images like rotation, translation, flipping, shearing, cropping, and scaling. These data augmentation techniques are routinely performed to increase the generalizability of DL models [4,5]. However, these transformed images have a similar distribution as the original dataset and result in minimal improvement in performance. These transformations also do not consider the deformation of organs. Flipping and shearing may result into changes in the structure of the target, but the orientation as well as the shape of the organs remain fixed. Therefore, in the medical domain, this resultant distribution after data augmentation will deviate from the actual data distribution.
Generative Adversarial Networks (GANs) recently gained popularity due to its robust working of generators and discriminators to produce more synthetic images. The role of the generator is to produce fake images and the objective of the discriminator is to make the difference between real and fake images using a repetitive process. GANs can improve the performance in classification by producing uncovered distribution and can produce novel images with good generalizability. GANs are commonly used for label-segmentation, translation, segmentation-label translation, or medical cross-modality translation [6,7]. This can help the healthcare professionals to have enough data in the medical domain to accurately make the classification of medical images. This generation of created images can potentially fill the gap that is not covered by the originally collected datasets.
In this research paper, we have developed a novel technique based on GAN for the data augmentation of Oral Squamous Cell Carcinoma (OSCC). GANs are powerful networks which can produce new images of OSCC from unlabeled original images. We have evaluated CycleGAN for augmentation in images that are used to diagnose OSCC. Different tricks are used to boost the performance of classification in oral cancer. We evaluated the use of GANs to increase robustness and improve the generalizability of medical images for the diagnosis of OSCC [8–11]. We believe that the proposed technique will be a valuable addition for medical researchers to reduce the manual efforts and costs required to make classification in OSCC images. It is also important to note that the images produced by GANS may look real as in the original dataset, but it is not assumed that these generated images are real images collected from a real patient. The proposed technique is used for data augmentation in medical images and improves the accuracy of classification based on small available data. The contributions of this paper are given below:
• Synthetic generation of good quality images for OSCC with the help of GANs.
• Design of OSCC classification model that can produce competitive results.
• Augmentation of the training dataset using synthetic images to improve classification results.
The rest of the paper is organized as follows. A detailed literature review is given in Section 2. The proposed technique is discussed in Section 3. Experimental analysis is provided in Section 4. The conclusion of the paper along with future work discussion is given in Section 5.
In [12], a GAN-based medical image augmentation technique was developed along with different tricks that were used to boost the classification and object detection accuracy. It was the first GAN model that was used for automatic bounding box prediction for brain metastase detection on 256 × 256 Magnetic Resonance images. The proposed technique was able to increase the sensitivity in the diagnosis from a clinically acceptable number by 10%. In [13], CycleGAN was used to further improve generalizability in Computed Tomography (CT) segmentation procedures. They trained CycleGAN that can transform contract CT images to noncontract images. The performance was compared to U-Net that was trained on the dataset originally created compared with the U-Net that was trained on the dataset which is a combination of original data and synthetic noncontract images. In [14], CNN performance in liver lesion classification was improved by using medical image augmentation using GAN-based synthetic image generation. The generated medical images were used for data augmentation and can further improve the accuracy of CNN classifier. They have used a small dataset containing 182 liver lesions (53 cysts, 64 metastases, and 65 haemangiomas). The classification accuracy using classical data augmentation techniques has sensitivity and specificity of 78.6% and 88.4%, respectively. With the usage of data augmentation from the proposed techniques, the sensitivity and specificity jumped to 85.7% and 92.4%, respectively.
In [15], a statistical method was used to determine which augmentation technique can accurately capture medical images and can help to develop models that vary in accuracy. The proposed work compared with different augmentation techniques and demonstrated that the performance of the models depends on the limit that the augmentation techniques could help the training set to retain the properties of the original medical images. Different augmentation techniques like flipping and Gaussian filters have enabled the model to achieve an accuracy of 88%. However, at the same time, adding a little noise can reduce the validation accuracy 66%. In [16], an augmentation technique was developed that depends on a statistical shape model and 3D thin plate spline. Many synthetic images are created from the small number of images in the given dataset using the proposed technique. The statistical shape model is used to model the shape information of real labelled images. By sampling from the model, a series of synthetic images are generated. Many simulated images are created by filling the synthetic shapes with texture using 3D thin plate splines. The deep neural network is trained on simulated and real images. Two different datasets are used. One is Magnetic Resonance Imaging (MRI) dataset and the other is liver CT dataset. In the proposed technique, two different deep networks are trained. One is called multiscale 3D CNN and the other is called U-net. The proposed technique has demonstrated better performance compared to existing segmentation algorithms.
In [17], DL techniques were used to generate new images of high quality using the implicit manifold of normal brain. In manifold learning, the synthesis of images and do-noising of images are important tools, GAN is used to train on 528 examples of 2D images of brain MRI. By performing the operation of cross-correlation on the training set, different synthetic images are created. These different images, which are synthesized along with real images, are blinded by imaging experts to provide image quality of Scores 1–5. Synthetic images had high overlap with real images. Image denoising was performed using skip connections and the proposed technique has higher Peak Signal-to-Noise Ratio (PSNR) then other techniques. An application of GAN was used for the automatic generation of artificial MRI of human brain in [18]. In [19], a review analysis was performed about the recent advancements in the field of data augmentation in the generation of magnetic resonance images of brain tumours. In [20], a two-layered DL framework was for the diagnosis of Alzheimer’s disease (AD) with the help of MRI and Position Emission Tomography (PET) data. 3D CycleGAN were used to produce PET data that were missing from the corresponding images in MRI. Then a deep CNN is trained for the diagnosis of AD. In [21], the mix-up algorithm was used in medical imaging segmentation and has demonstrated improved performance in segmentation tasks.
In [22], GAN was used to create a substantial amount of data to make classification in neuroimaging for mapping the functioning of human cortex. The proposed system has used CycleGAN with CNN classifier to enhance the accuracy through data augmentation. The system can determine that the task performed by the subject is by Left Finger Tap, Right Finger Tap or Foot Tap by analyzing the functional near-infrared spectroscopy data pattern. The proposed system has obtained the accuracy of 96.67% using CGAN-CNN combination. In [23], authors have proposed differentiable augmentation based technique to improve the efficiency of GAN on real and fake images. In general, when we augment the training data, it manipulates the distribution of real data and hence we do not get much benefit from the augmentation. Using differentiable augmentation, we can stabilize training and hence can obtain better convergence in learning. The proposed system has obtained FID of 6.80 with an IS of 100.8 on ImageNet. With only 20% of the training data the performance on CIFAR-10 and CIFAR-100 is comparable to state-of-the-art CNN models. The method can generate high-fidelity images using only 100 images without using pre-training enabling a good choice for transfer learning algorithms.
In all the above studies, DL techniques are used to solve healthcare-related problems. Some studies have demonstrated augmentation techniques to improve the performance of DL models. We are taking a different approach. We are using GAN as an augmentation technique and then use a very simple classification model that can make the classification with high accuracy. The performances of the model are evaluated using different evaluation techniques. The results demonstrate that the proposed approach is a suitable model to be used in the healthcare domain.
Images for the experiments performed in the article are downloaded from repository1 which contains 1,224 images. Two sets of different resolutions are made. In the first set, there are 89 images which contain normal epithelium of the oral cavity and another 439 images which contain Oral Squamous Cell Carcinoma (OSCC) with 100× magnification.
In the other set, there are 201 images having normal epithelium of the oral cavity along with 495 images having OSCC in 400× magnification. All samples are collected on Leica ICC50 HD microscope using HE-stained tissue slides. The repository is created and catalogued by healthcare professionals by collecting images from 230 patients. Further details of the dataset can be found in [24].
For the classification of these images using CNN, we have labelled images having normal epithelium of the oral cavity as Benign and samples of OSCC as Malignant. Several sample images from Benign and Malignant categories are shown in Fig. 1.
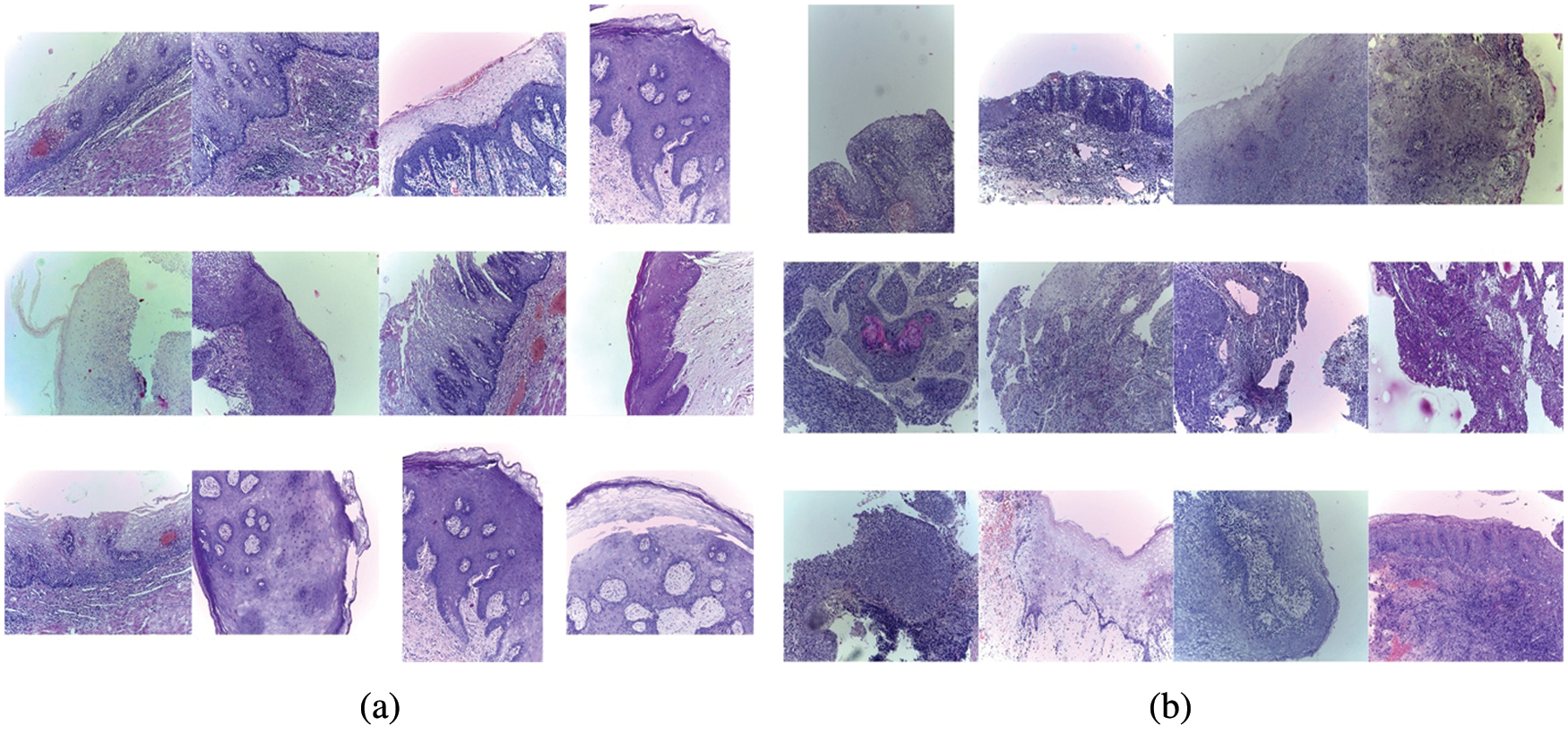
Figure 1: Different images in classes Malignant and Benign. (a) Images with normal epithelium of the oral cavity labelled as Benign, (b) Images of OSCC labelled as Malignant
3.2 OSCC Classification Architecture
For classification of images in Benign and Malignant we have used LeNet-5 architecture. LeNet-5 [25] was proposed in 1998 for machine-printed character recognition. In LeNet architecture, there are two sets, each containing convolutional and average pooling layers, followed by one flattening convolutional layer and then two fully connected layers [26]. At the end, there is a sigmoid function that classifies the images into Benign and Malignant. The sample architecture used for classification of OSCC images is shown in Fig. 2.
The hyperparameters used for the training of LeNet architecture are shown in Table 1.
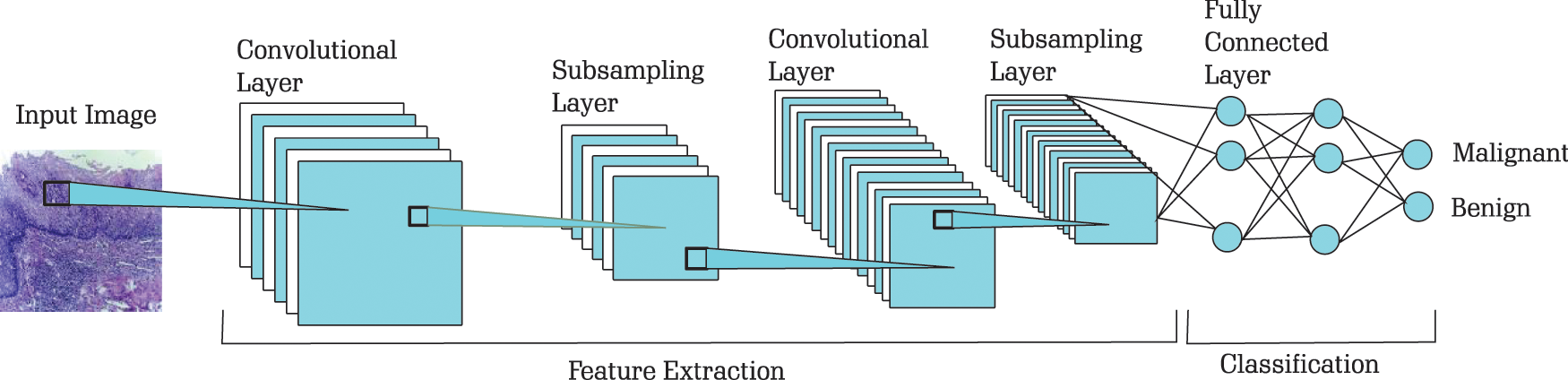
Figure 2: Architecture for classification of OSCC images (The model is originally developed in [25])
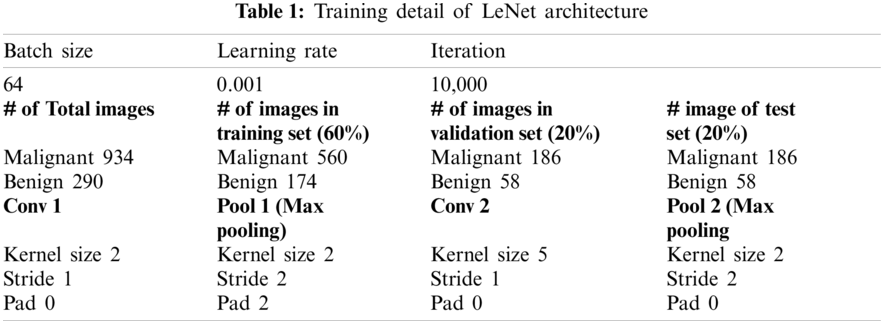
Data augmentation operations are used to enhance the quantity of a dataset that is used for training DL models. To get more accuracy from DL models and increase the reliability of prediction it is generally required to increase the size of the dataset that is used to train the model. In some cases, in general and in the medical field in particular, it is not easy to collect more images. Therefore, these techniques heavily depend on data augmentation techniques. Different data augmentation techniques are used. Some of which are given as below:
• Scaling: This technique is used to change the size of the image.
• Cropping: This technique is used to select a portion of the image.
• Flipping: This technique is used to flip the image horizontally or vertically.
• Padding: In this technique, a given value is used to pad an image on all slides.
• Rotation: In this technique, the image is rotated for a specified value.
• Translation: In this technique, the moving of the image is performed on x-axis or y-axis.
• Affine Transformation: In this technique, points, straight lines, and planes are preserved. This technique can be applied to perform scaling, translation, rotation, shearing, etc.
• Brightness: In this technique, the image’s brightness is changed so the image is brighter or darker.
• Contrast: In this technique, the separation degree between the darkest and brightest areas in an image is defined.
• Saturation: In this technique, the separation between colors is defined.
• Hue: In this technique, the shades of the colors in an image are defined.
3.4 Generative Adversarial Network
Generative Adversarial Networks (GANs) are utilized to produce new images. These images can be augmented with the training set to train the model. In GANs architectures, there are two important components: generator and discriminator. The generator is a neural network which is trained to model a transform function. A random noise is taken as input and produces a random variable that follows the distribution of the target. The discriminator models a discriminative function, where an input is taken and returns the probability of the input. The role of the generator is to make fool of the discriminator, so the generator maximizes the classification error. The objective of the discriminator is to identify fake data generated by the generator, so the discriminator minimizes the classification error. During the training process, the weights of the generators are updated to increase the classification error, and the weights of the discriminator are updated to decrease the classification error. These different objectives of the two neural networks work as adversarial networks. These networks try to defeat each other and, in this process, both becoming better and better. It works as a minimax two-player game where the equilibrium is the state where the generator produces data, and the discriminator predicts as “real” or “fake” probabilities. A sample architecture of GANs to generate new samples of Benign and Malignant images for classification of OSCC is given in Fig. 3.
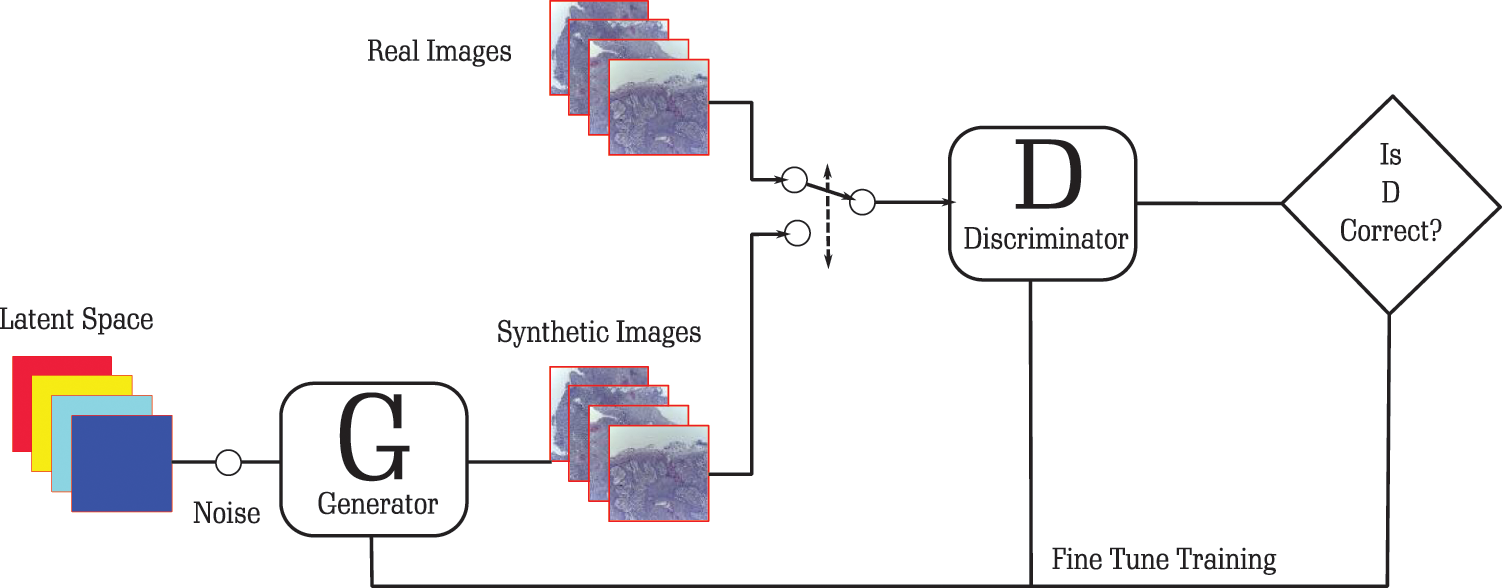
Figure 3: Architecture for augmentation of images for OSCC
The generator starts from random noise and produces an image passed to the discriminator. The discriminator classifies the image as fake. This gives a response to the generator to produce a more accurate image that is based on the OSCC dataset. During this process of accurately classifying images as fake, the generator learns the distribution of the output. At some point, it can produce images that the discriminator cannot classify as fake. This helps the discriminator to get the knowledge that it can be fooled and hence it needs to improve its weight. This iterative process helps the model to develop good quality images that can be used as part of the training dataset. This will help DL models to increase their accuracy because a large amount of data is available.
The accuracy achieved by LeNet without augmented data, with augmented data using standard augmentation techniques, and with augmented data using GANs is given in Fig. 4. It is demonstrated that the accuracy of train, validation and test sets is very low when LeNet is used, as there are not enough samples for LeNet to understand the pattern of the data. When standard data augmentation techniques are used to augment data, the accuracy is further improved as new samples are added and the training set is increased. The accuracy is further improved when GANs are used to augment the data to increase the training set. This demonstrates that GANs can help the classification model to understand the uncovered patterns of the data and hence the accuracy is improved.
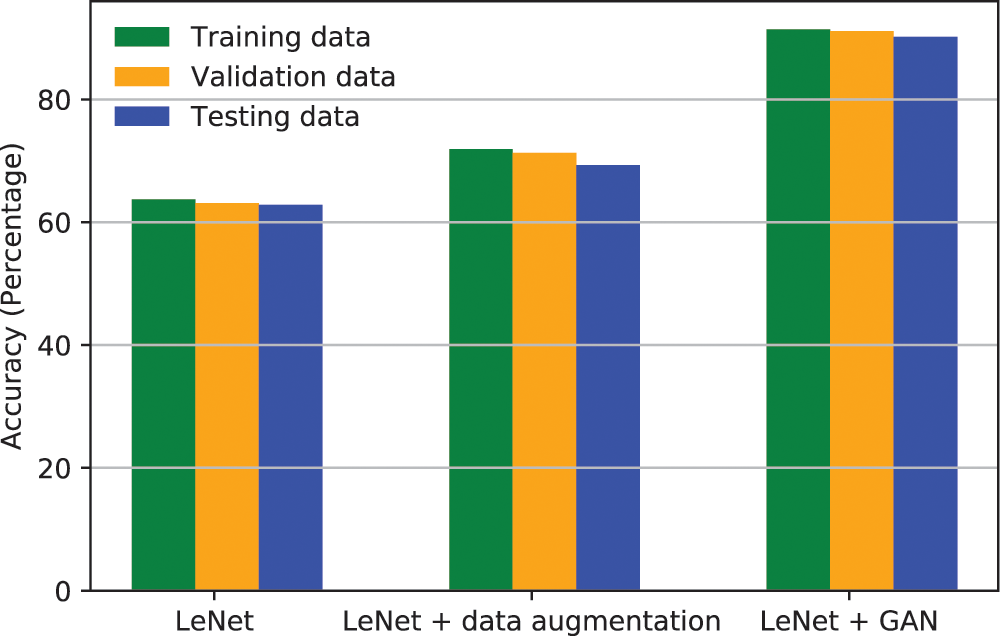
Figure 4: Accuracy of LeNet classifier with different augmentation techniques
The performance of a classification model is determined by using confusion matrix. It is a table that determines the accuracy of test data where the truth values are known in advance. For binary classification, the confusion matrix is a 2 × 2 matrix. The column determines the predicted class as Benign on the left side and Malignant on the right column. The rows of the matrix show the actual values as Malignant on the bottom row and Benign on the top row.
True positive is the situation where the model is predicted as Malignant and the image is Malignant. True negative is the situation where the model is predicted as Benign and actual the image is Benign. False positive is situation where the model is predicted as Malignant, but the image is of type Benign. False negative is a situation where the model is predicted as Benign but the image is of category Malignant. Both false positives and false negatives are situations that can make the model as inaccurate, as these decisions misguide the medical professional to make an incorrect decision.
The confusion matrices for the classification models used in this paper are given in Fig. 5. The confusion matrix for LeNet where no data augmentation technique is shown in Fig. 5a. As the true positive is 0.63 and the false negative is 0.62, which is very low accuracy. When we increase the training set by using standard augmentation technique, the accuracy increases and the true positive becomes 0.68 and the false negative as 0.71 as shown in Fig. 5b. Which is not as much improvement compared to the LeNet without augmentation. When we used GAN to generate new samples to augment the training data along with the standard augmentation technique, the accuracy of true positive increases to 0.94 and false negative increase to 0.89 as shown in Fig. 5c. This demonstrates that GAN is a very powerful technique that can effectively increase the data size of the training set and even a basic architecture such as LeNet can perform better when there is enough data in the training data.
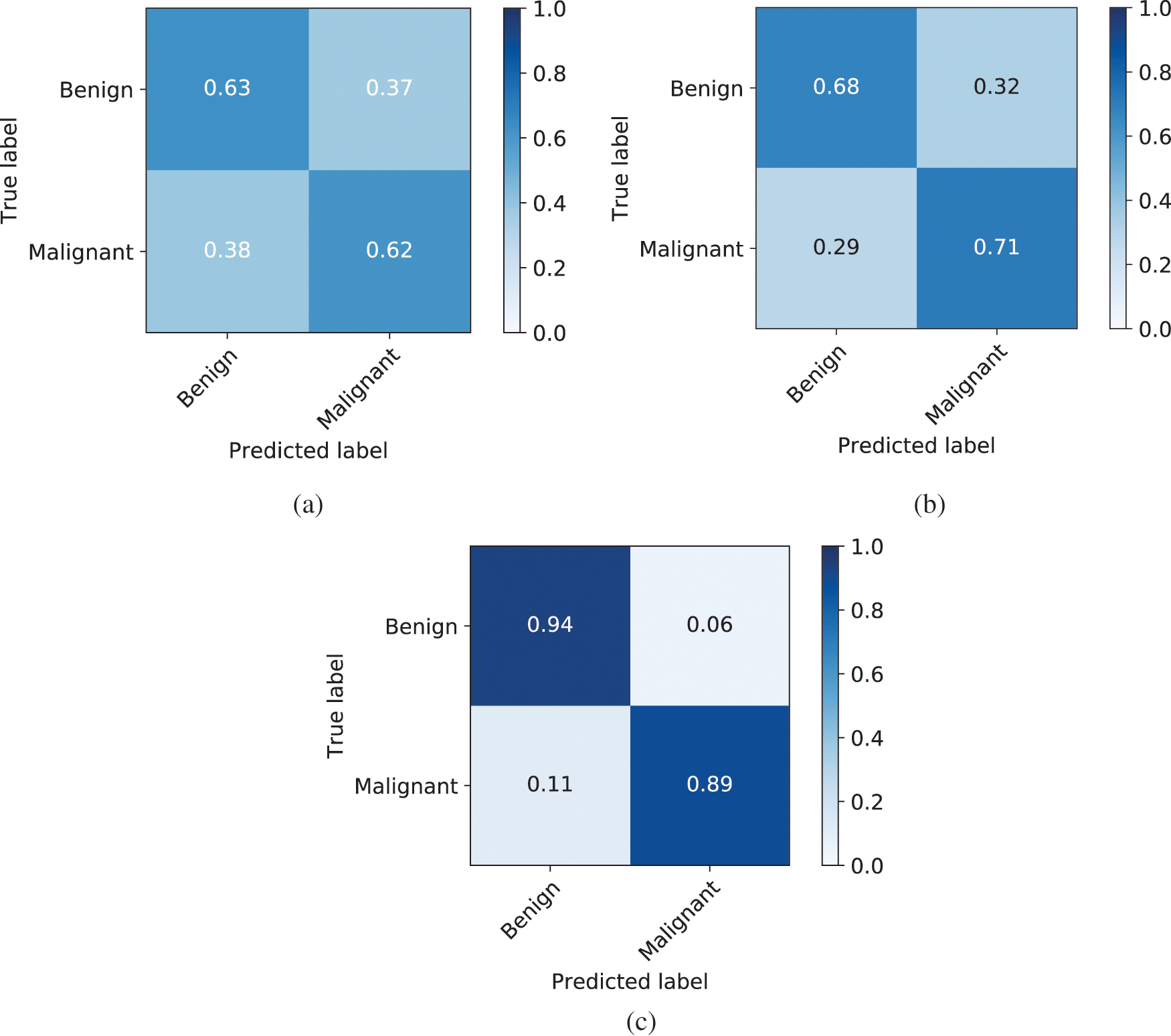
Figure 5: Confusion matrix of LeNet, LeNet with standard augmentation, and LeNet with GANs augmentation. (a) Confusion matrix of LeNet classifier, (b) Confusion matrix of LeNet classifier along with standard augmentation techniques, (c) Confusion matrix of LeNet classifier along with standard augmentation and GAN
The performance of the binary classifier can be best visualized in ROC curve. The curve demonstrates two parameters as True Positive Rate (TPR) and False Positive Rate (FPR). TRP means when it is Benign how often the model predicts as Benign or when it is Malignant and how often the model predicts as Malignant. This is also called sensitivity or recall. FPR means when it is Benign how often the model predicts as Malignant or when it is Malignant, how often the model predicts as Benign. The ROC curve shows TPR vs. FPR at different classification thresholds. AUC is the area under the curve of ROC curve. The curve is basically plotting the performance of all possible classification thresholds as an aggregate measure. The calculation of TPR and FPR is given in Eq. (1), where TP means True Positive, TN means True Negative, FP means False Positive, and FN means False Negative.
The ROC for LeNet is shown in Fig. 6a. The total area for Benign and Malignant is 0.62 and 0.62, respectively. It is considered as a very low accuracy. When the standard data augmentation technique is applied to increase the training set, the ROC is computed as shown in Fig. 6b. The AUC increases to 0.70 for both Benign and Malignant classes. Still, the accuracy is not good enough for the classification of OSCC. When GAN is used for data augmentation in LeNet, the ROC is shown in Fig. 6c. It can be observed that AUC increased to 0.91 for Benign and Malignant classes. This demonstrates that GAN creates more similar images and increases the training set and even a basic CNN architecture such as LeNet can perform very well to make classification.
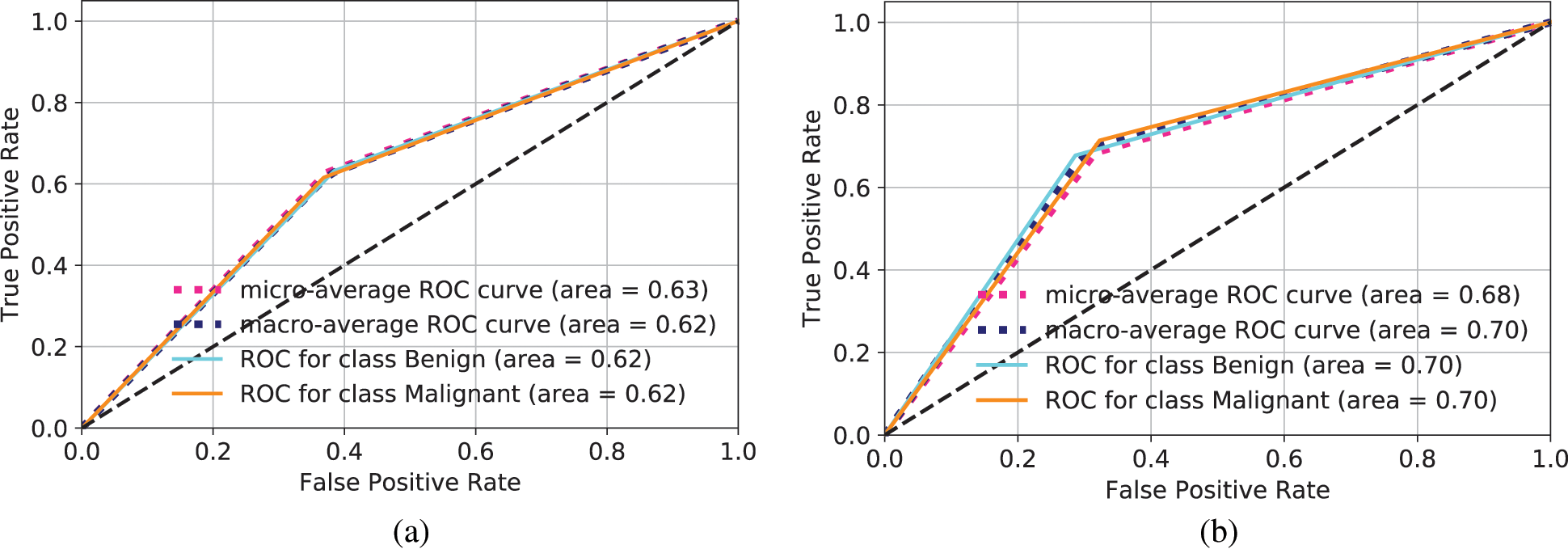
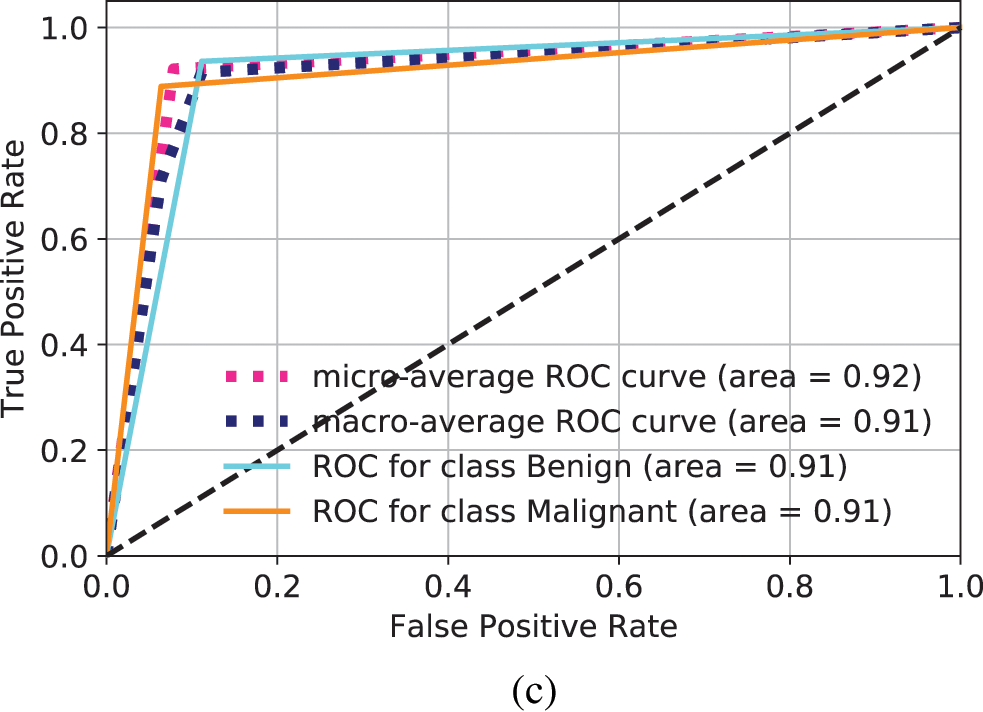
Figure 6: ROC for different models to make prediction of Benign and Malignant types. (a) ROC computation for LeNet, (b) ROC computation for standard data augmentation in LeNet, (c) ROC for images generated through GANs and classification in LeNet
Three other performance measurement parameters are often used to check the performance of classification models. These are precision, recall, and F1-score. Precision means, out of total predicted positive how many are positive. Recall means out of total actual positive how many are predicted as positive. F1-score is a function of precision and recall. It is better to use when there are requirements to find a balanced point between precision and recall and the data is not evenly distributed. The calculation of Precision, Recall, and F1-score is given in Eq. (2). The Precision, Recall, and F1-score for Benign and Malignant classes for LeNet classifier is shown in Fig. 7a. The performance report is not satisfactory as the values are very low. When the standard data augmentation technique is used, the classification report is shown in Fig. 7b. As it can be seen, the performance is improved compared to LeNet classifier when no augmentation is used, but still there is room for improvement. We used GANs for data augmentation and used LeNet classifier and the classification report is shown in Fig. 7c. It can be seen that the performance is improved compared to simple LeNet classifier and LeNet classifier with standard data augmentation technique.
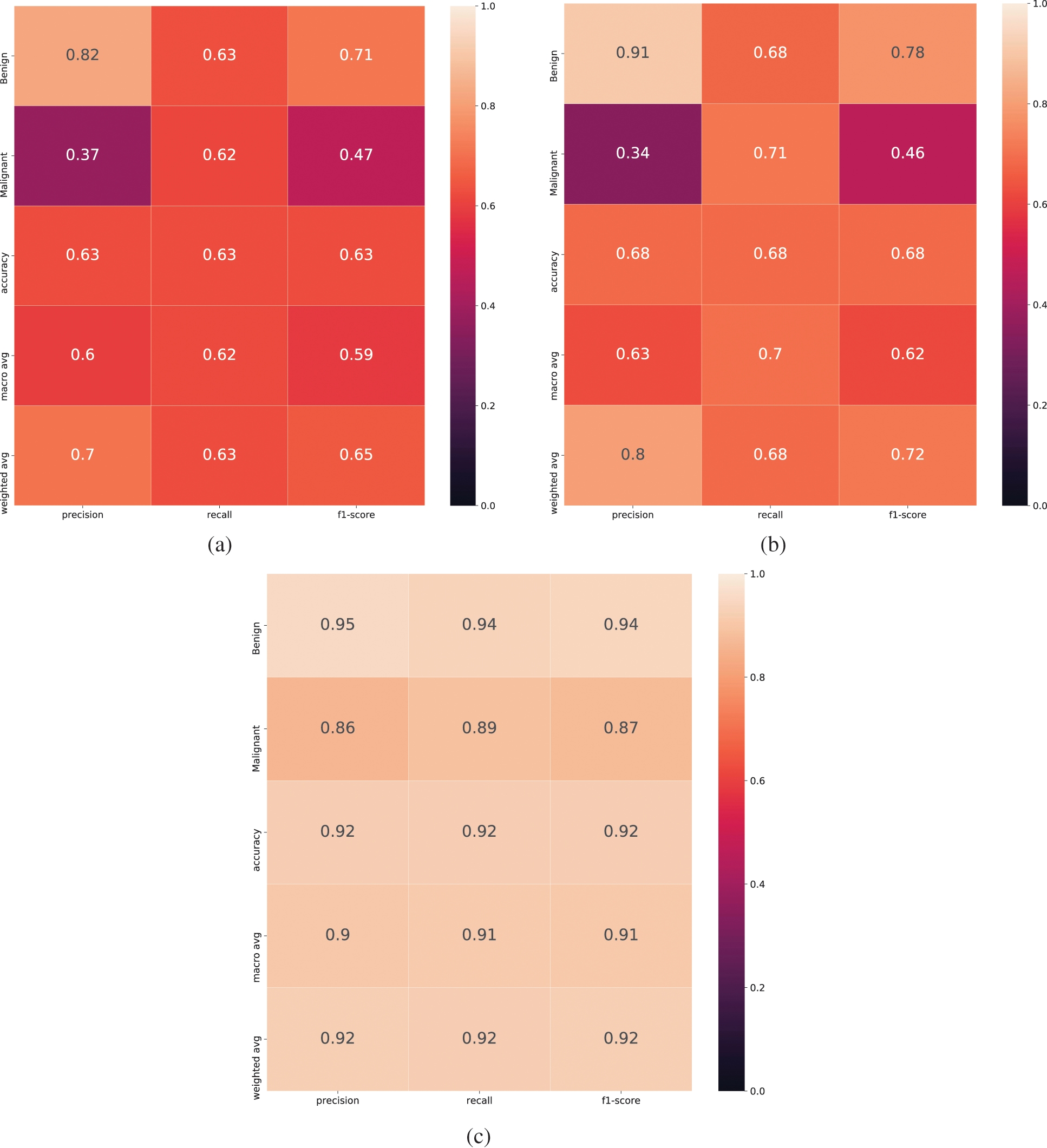
Figure 7: Classification report for LeNet classifier when different data augmentation technique is used. (a) Classification report for LeNet, (b) Classification report for standard data augmentation with LeNet, (c) Classification report for data augmentation using GANs and LeNet classifier
In this paper LeNet classifier is used to demonstrate proof of concept that data augmentation techniques using GAN improves the performance of classifier in limited data sets compared to other standard augmentation techniques. An experiment is performed to demonstrate the performance of GAN based data augmentation when other advanced CNN classifiers such as AlexNet, VGG, Inception and ResNet are used.
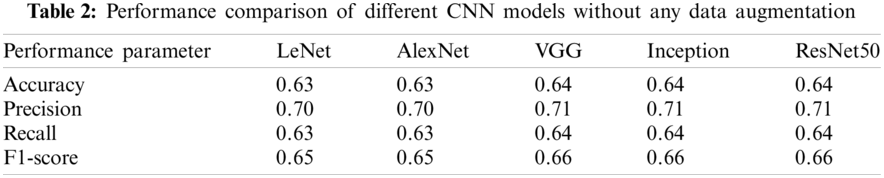
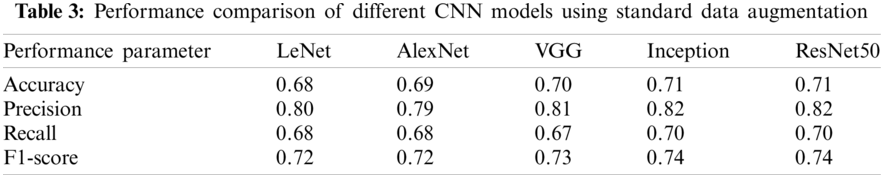
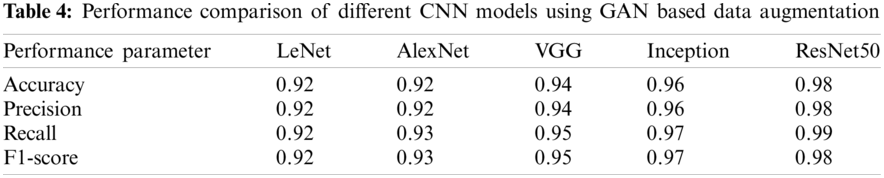
The performance comparison of different CNN models when no data augmentation is used is shown in Table 2. It can be observed that performance is not improving even when advanced CNN models are used. Further comparison of CNN models is made when standard data augmentation is used and is demonstrated in Table 3. It can be observed that performance is slightly improving compared to no data augmentation, but the performance improvement is very low. The performance comparison when GAN based data augmentation is performed is given in Table 4. The results demonstrate that when advanced CNN models are used, the performance is further improved, demonstrating the effectiveness of GAN based data augmentation in CNN models.
The conditional GAN used in this paper to classify the benign and malignant cases can generate multiple artificial images that belong to a specific class. The class is classified by the conditional argument. Different evaluation metrics are used for checking the performance of the model in generating synthetic images such as inception score, mean opinion score, Wasserstein metric, fuzzy combinatorial analysis score, log-likelihood, and human evaluation schemes. We have performed visual analysis of the generated images to check the quality of the synthetic images. Qualified experts were used to check the quality of the images. The output of different inputs was analyzed to observe the diversity among the images generated by GAN. In case of a mode collapse, generated images were looking similar regardless of the inputs. In case there is visual similarity, Multi-Scale Structural Similarity for Image Quality (MS-SSIM) were used to determine the degree of similarity. We have taken these measures at the Generator of the GAN model by introducing Batch Normalization after each layer.
In data science domain, data is the key component as different machine learning techniques are used to perform the processing of this data to find the underlying pattern and make classification. These machine learning models are as good as the data is fed into, the larger the data, the better the accuracy of these models. In the healthcare domain, generally the size of the data is very low, mainly because of ethical issues or privacy concerns of patients or because expensive equipment is used to collect this data. Therefore, different data augmentation operations are utilized to improve the data quantitatively. However, these data augmentation techniques only improve the accuracy very little. To provide better data augmentation, a powerful technique known as Generative Adversarial Network is used that helps classification models to further improve the accuracy. We have applied GAN along with a classification model to make classification in healthcare domain. Our results have demonstrated better performance as an accuracy of up to 90% is achieved. Further experiments are performed with more complex CNN models and results have demonstrated an accuracy of 98%. In future work, the model can be extended to identify infected regions in the image. The model can then be commercialized to develop a device that makes classification of OSCC images and help the healthcare professionals.
Funding Statement: This research was supported by Taif University Researchers Supporting Project No. (TURSP-2020/254), Taif University, Taif, Saudi Arabia.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1https://data.mendeley.com/datasets/ftmp4cvtmb/1
1. Guo, Z., Shen, Y., Bashir, A. K., Imran, M., Kumar, N. et al. (2020). Robust spammer detection using collaborative neural network in Internet of Things applications. IEEE Internet of Things Journal, 8(12), 9549–9558. DOI 10.1109/JIOT.2020.3003802. [Google Scholar] [CrossRef]
2. Shorten, C., Khoshgoftaar, T. M. (2019). A survey on image data augmentation for deep learning. Journal of Big Data, 6(1), 1–48. DOI 10.1186/s40537-019-0197-0. [Google Scholar] [CrossRef]
3. Yu, K., Tan, L., Shang, X., Huang, J., Srivastava, G. et al. (2020). Efficient and privacy-preserving medical research support platform against COVID-19: A blockchain-based approach. IEEE Consumer Electronics Magazine, 10(2), 111–120. DOI 10.1109/MCE.2020.3035520. [Google Scholar] [CrossRef]
4. Feng, C., Yu, K., Aloqaily, M., Alazab, M., Lv, Z. et al. (2020). Attribute-based encryption with parallel outsourced decryption for edge intelligent IOV. IEEE Transactions on Vehicular Technology, 69(11), 13784–13795. DOI 10.1109/TVT.2020.3027568. [Google Scholar] [CrossRef]
5. Shi, H., Wang, L., Ding, G., Yang, F., Li, X. (2018). Data augmentation with improved generative adversarial networks. 24th International Conference on Pattern Recognition, pp. 73–78. Beijing, China. [Google Scholar]
6. Rahman, H. U., Raza, M., Afsar, P., Khan, H. U., Nazir, S. (2020). Analyzing factors that influence offshore outsourcing decision of application maintenance. IEEE Access, 8(1), 183913–183926. DOI 10.1109/ACCESS.2020.3029501. [Google Scholar] [CrossRef]
7. Mansourifar, H., Chen, L., Shi, W. (2019). Virtual big data for GAN based data augmentation. IEEE International Conference on Big Data (Big Data), pp. 1478–1487. Los Angeles, CA, USA. [Google Scholar]
8. Peres, R. S., Azevedo, M., Araújo, S. O., Guedes, M., Miranda, F. et al. (2021). Generative adversarial networks for data augmentation in structural adhesive inspection. Applied Science, 11(7), 3086–3097. DOI 10.3390/app11073086. [Google Scholar] [CrossRef]
9. Bissoto, A., Valle, E., Avila, S. (2021). GAN-based data augmentation and anonymization for skin-lesion analysis: A critical review. IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, pp. 1847–1856. [Google Scholar]
10. Tero, K., Miika, A., Janne, H., Samuli, L., Jaakko, L. et al. (2020). Training generative adversarial networks with limited data. Advances in Neural Information Processing Systems, 33(NeurIPS 2020). [Google Scholar]
11. Wang, S., Khan, S., Xu, C., Nazir, S., Hafeez, A. (2020). Deep learning-based efficient model development for phishing detection using random forest and BLSTM classifiers. Complexity, 2020(3), 1–7. DOI 10.1155/2020/8694796. [Google Scholar] [CrossRef]
12. Han, C., Murao, K., Satoh, S., Nakayama, H. (2019). Learning more with less: Gan-based medical image augmentation. Medical Imaging Technology, 37(3), 137–142. [Google Scholar]
13. Sandfort, V., Yan, K., Pickhardt, P. J., Summers, R. M. (2019). Data augmentation using generative adversarial networks (cyclegan) to improve generalizability in CT segmentation tasks. Scientific Reports, 9(1), 16884. DOI 10.1038/s41598-019-52737-x. [Google Scholar] [CrossRef]
14. Frid-Adar, M., Diamant, I., Klang, E., Amitai, M., Goldberger, J. et al. (2018). Gan-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing, 321(1), 321–331. DOI 10.1016/j.neucom.2018.09.013. [Google Scholar] [CrossRef]
15. Hussain, Z., Gimenez, F., Yi, D., Rubin, D. (2018). Differential data augmentation techniques for medical imaging classification tasks. AMIA Annual Symposium Proceedings Archive, pp. 979–984. Bethesda, MD. [Google Scholar]
16. Tang, Z., Chen, K., Pan, M., Wang, M., Song, Z. (2019). An augmentation strategy for medical image processing based on statistical shape model and 3D thin plate spline for deep learning. IEEE Access, 7(1), 133111–133121. DOI 10.1109/ACCESS.2019.2941154. [Google Scholar] [CrossRef]
17. Bermudez, C., Plassard, A. J., Davis, L. T., Newton, A. T., Resnick, S. M. et al. (2018). Learning implicit brain MRI manifolds with deep learning. In: Medical imaging 2018: Image processing, pp. 408–414. Houston, Texas, USA [Google Scholar]
18. Calimeri, F., Marzullo, F., Stamile, A., Terracina, G. (2017). Biomedical data augmentation using generative adversarial neural networks. Artificial Neural Networks and Machine Learning—ICANN 2017, pp. 626–634. Alghero, Italy. [Google Scholar]
19. Nalepa, J., Marcinkiewicz, M., Kawulok, M. (2019). Data augmentation for brain-tumor segmentation: A review. Frontiers in Computational Neuroscience, 13(12), 83–96. DOI 10.3389/fncom.2019.00083. [Google Scholar] [CrossRef]
20. Pan, Y., Liu, M., Lian, C., Zhou, T., Xia, Y. et al. (2018). Synthesizing missing PET from MRI with cycleconsistent generative adversarial networks for Alzheimer’s disease diagnosis. Medical Image Computing and Computer Assisted Intervention, pp. 455–463. Granada, Spain. [Google Scholar]
21. Eaton-Rosen, Z., Bragman, F. J. S., Ourselin, S., Cardoso, M. (2018). Improving data augmentation for medical image segmentation. In: Medical Imaging with Deep Learning, pp. 1–11. Amsterdam, Netherlands. [Google Scholar]
22. Wickramaratne, S. D., Mahmud, M. (2021). Conditional-GAN based data augmentation for deep learning task classifier improvement using fNIRS data. Frontiers in Big Data, 4(1), 659146–659158. DOI 10.3389/fdata.2021.659146. [Google Scholar] [CrossRef]
23. Zhao, S., Liu, Z., Lin, J., Zhu, J., Han, S. (2020). Differentiable augmentation for data efficient GAN training. Conference on Neural Information Processing System, pp. 7559–7570. Virtual. [Google Scholar]
24. Rahman, T. Y., Mahanta, L. B., Das, A. K., Sarma, J. D. (2020). Histopathological imaging database for oral cancer analysis. Data in Brief, 29(1), 105114–105126. DOI 10.1016/j.dib.2020.105114. [Google Scholar] [CrossRef]
25. Lecun, Y., Bottou, L., Bengio, Y., Haffner, P. (1998). Gradient-based learning applied to document recognition. Proceedings of the IEEE, 86(11), 2278–2324. DOI 10.1109/5.726791. [Google Scholar] [CrossRef]
26. Wu, B., Nazir, S., Mukhtar, N. (2020). Identification of attack on data packets using rough set approach to secure end to end communication. Complexity, 2020(3), 1–12. DOI 10.1155/2020/6690569. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |