2021 45(1): 1-16
DOI:10.32604/biocell.2021.013452

www.techscience.com/journal/biocell
| BIOCELL 2021 45(1): 1-16 DOI:10.32604/biocell.2021.013452 |  www.techscience.com/journal/biocell |
Potential roles of functional bacterial amyloid proteins, bacterial biosurfactants and other putative gut microbiota products in the etiopathogeny of Parkinson’s Disease
1Victor Babeș” National Institute of Pathology, Bucharest, 050088, Romania
2Colentina Clinical Hospital, Bucharest, 020125, Romania
3Carol Davila” University of Medicine and Pharmacy, Bucharest, 020021, Romania
*Address correspondence to: Laura Cristina Ceafalan, laura.ceafalan@umfcd.ro
Received: 07 August 2020; Accepted: 13 October 2020
#These authors contributed equally
Abstract: An increasing number of studies provide evidence for the existence of a microbiota-gut-brain axis and its potential involvement in the development of sporadic Parkinson’s disease and other neurodegenerative conditions. The neuropathologic hallmark of Parkinson’s disease is the presence of brain intraneuronal aggregates of misfolded alpha-synuclein, known as Lewy bodies. Some gut microbiota products may trigger alpha-synuclein conformational changes in the neurons of the enteric nervous system, which can then spread to the brain in a prion-like fashion through the vagus nerve. Others may interfere with neuroinflammatory pathways and susceptibility to neurodegeneration. In this review, we assess the potential role of putative gut microbiota products in the etiopathogeny of Parkinson’s disease, with a special emphasis on functional bacterial amyloid proteins, bacterial biosurfactants, endotoxins and short-chain fatty acids. The possible roles of molecular hydrogen, a common by-product of bacterial fermentation, are also addressed.
Keywords: Alpha-synuclein; Endotoxins; Short-chain fatty acids
Abbreviations
| BBB: | blood brain barrier |
| CNS: | central nervous system |
| Fap: | functional amyloid in Pseudomonas |
| H-NS: | histone-like nucleoid-structuring protein |
| IHF: | integration host factor |
| IL-1β: | interleukin-1 beta |
| iNOS: | inducible nitric oxide synthase |
| LPS: | lipopolysaccharide |
| NF-κB: | nuclear factor kappa-light-chain-enhancer of activated B cells |
| PD: | Parkinson’s disease |
| SCFA: | short-chain fatty acids |
| SIBO: | small intestine bacterial overgrowth |
| TLR: | toll-like receptor |
| TNF-α: | tumour necrosis factor-alpha |
Parkinson’s disease (PD) is a neurodegenerative disorder of the central nervous system (CNS) resulting in progressive motor and non-motor manifestations (Berg et al., 2015; Braak et al., 2003). The etiopathogeny of sporadic cases is incompletely understood, and currently, there are no disease-modifying treatments.
Pathologically, PD is a proteinopathy characterized by misfolding, aggregation, and intraneuronal accumulation of alpha-synuclein, with subsequent neuroinflammatory changes and neurodegeneration (Braak et al., 2003; Spillantini et al., 1998; Spillantini et al., 1997; Walker et al., 2016). As in the case of other alpha-synucleinopathies, the misfolded alpha-synuclein gains amyloid properties and putative neurotoxic functions (Araki et al., 2019; Braak and Del Tredici, 2017). Self-aggregation of the misfolded alpha-synuclein results in intraneuronal aggregates known as Lewy bodies, the pathological hallmarks of PD (Araki et al., 2019; Baba et al., 1998; Braak et al., 2003; Spillantini et al., 1998; Spillantini et al., 1997; Walker et al., 2016).
Based on their previous findings concerning the topographical sequence (or stages) of Lewy pathology development in the brain of people with PD and the early presence of Lewy bodies in the neurons of the enteric nervous system, olfactory bulb, and dorsal motor nucleus of the vagus nerve, Braak et al. (2006) hypothesised that the alpha-synuclein misfolding process originates in the nasal mucosa and the gut (Braak et al., 2006; Braak et al., 2003; Friedland, 2015; Hawkes et al., 2007). This was further refined into the dual-hit theory, which states that a neurotropic infectious pathogen, viral or with prion-like properties, enters the brain by transsynaptic retrograde transmission via the vagus and olfactory nerves, thus bypassing the circulatory system and blood-brain-barrier (BBB) (Angot and Brundin, 2009; Hawkes et al., 2007, 2009; Walker et al., 2016). The current view is that an exogenous factor triggers the initial alpha-synuclein conformational changes, which then self-propagate mainly via the olfactory and/or vagus nerve, reaching the brain (Araki et al., 2019; Braak and Del Tredici, 2017). The triggers for the pathologic amyloid transformation of alpha-synuclein remain unknown.
Arguments for the potential roles of the microbiota-gut-brain axis in the etiopathogeny of sporadic PD include the early involvement of the enteric nervous system, with chronic constipation that typically occurs before the motor onset of the disease (Berg et al., 2015; Martinez-Martin et al., 2007), the early presence of Lewy pathology in intestinal neurons (Braak et al., 2006; Braak et al., 2003) and presence of proinflammatory gut dysbiosis in people with PD, with increases in faecal Verrucomicrobiaceae and Akkermansia, which degrade the intestinal mucus layer, and decreases in the beneficial Prevotellaceae, Roseburia and Faecalibacterium being more consistently reported (Aho et al., 2019; Baldini et al., 2020; Boertien et al., 2019; Cattaneo et al., 2017; Cirstea et al., 2020; Gabrielli et al., 2011; Hill-Burns et al., 2017; Keshavarzian et al., 2015; Minato et al., 2017; Nishiwaki et al., 2020; Nuzum et al., 2020; Pietrucci et al., 2019; Scheperjans, 2016; Unger et al., 2016). The causal relation between the microbiota changes and PD is still unclear; however, Minato et al. (2017) found that lower baseline fecal Bifidobacterium and Atopobium correlate with PD symptom severity at 2 years. The functional impact of these differences in the composition of the gut microbiota also needs clarification, various models predicting changes in the expression of some gut microbiota metabolites, with possible consequences on its global function (Baldini et al., 2020; Bedarf et al., 2017; Cirstea et al., 2020; Nishiwaki et al., 2020; Nuzum et al., 2020). Contextual indirect evidence for the potential role of the microbiota in the etiopathogeny of PD also comes from epidemiological studies that show increased risk of PD in people with prior Helicobacter pylori infection (Huang et al., 2018; Nielsen et al., 2012) and with inflammatory bowel disease (Park et al., 2019; Villumsen et al., 2019; Weimers et al., 2019), the latter mitigated by early effective treatment (Peter et al., 2018). Furthermore, the risk of PD is lower in people that underwent truncal vagotomy for peptic or duodenal ulcer (Liu et al., 2017; Svensson et al., 2015), as well as in those that underwent an appendectomy and possibly tonsillectomy (Liu et al., 2020), putative explanations being disruption of the pathology dissemination pathway and removal of tissues with high alpha-synuclein load, respectively. In addition, intestinal inflammatory changes and altered intestinal barrier permeability have been reported in people with PD (Houser et al., 2018; Schwiertz et al., 2018), suggesting that direct contact between alpha-synuclein found in enteric neurons and microbiota products is plausible. Another study supporting the plausibility of direct contact between human alpha-synuclein and microbiota products within the gut found that enteric neurons’ overexpression of alpha-synuclein can be induced by local viral infections (Stolzenberg et al., 2017), while Uesaka et al. (2016) found that the gut lining contains a category of enteroendocrine cells having properties of neurons and being connected directly to alpha-synuclein-containing neurons (Uesaka et al., 2016). Arguments supporting the role of the gut microbiota in the etiopathogeny of PD also come from experimental animal models and in vitro studies, the most notable being brought by Sampson et al. (2016), who performed in vivo experiments on alpha-synuclein overexpressing mice, showing that the gut microbiota is needed for Lewy body pathology, motor impairment, and microglia activation. Such data suggested an active gut-brain signalling pathway between the microbiota and the brain (Sampson et al., 2016). Moreover, microglia activation, an important player in PD (Hirsch and Hunot, 2009), is modulated by gut bacteria in mice (Erny et al., 2015).
Amyloids are self-aggregating proteins with fibrillary morphology and beta-sheet secondary structure. In vitro studies have shown that alpha-synuclein can aggregate, forming amyloid fibres or fibrils with a cross-beta structure (Conway et al., 1998; Greenwald and Riek, 2010; Soto, 2003). These results are supported by in vivo murine experiments on the propagation of alpha-synuclein amyloid fibrils, which can spread from neuron to neuron in a prion-like fashion (Luk et al., 2012; Masuda-Suzukake et al., 2013). As mentioned, Lewy bodies are abnormal aggregations of misfolded alpha-synuclein encountered in PD and other alpha-synucleinopathies (Araki et al., 2019; Baba et al., 1998; Braak et al., 2003; Spillantini et al., 1998; Walker et al., 2016). Thus, PD is actually a type of amyloidosis in which misfolded alpha-synuclein is the pathologic amyloid, resulting in the accumulation and spread of alpha-synuclein amyloid fibrils (Araki et al., 2019), not only in the brain but also in the autonomic nerve fibres that innervate visceral organs such as the intestine and heart (Gelpi et al., 2014).
Environmental factors are thought to play important roles in triggering alpha-synuclein misfolding and Lewy body formation in sporadic PD but are largely unknown. Recent evidence shows that cross-reactivity and seeding between distinct amyloid proteins and alpha-synuclein can occur (i.e., transient protein-protein interactions between distinct amyloid proteins and native alpha-synuclein can result in the formation of amyloid alpha-synuclein) (Chen et al., 2016; Chorell et al., 2015; Evans et al., 2015; Sampson et al., 2020). Considering the dual-hit hypothesis and the large surface of the gastrointestinal tract where alpha-synuclein in enteric neurons could be exposed to the endoenvironment of the intestinal lumen, molecular xenobiotics of the gut microbiota pose a particular interest as possible triggers of the initial alpha-synuclein misfolding and self-aggregation events, which may then self-propagate along the neurons of the vagus nerves and sympathetic chains, reaching the brain.
Increasingly detailed knowledge of the mechanisms by which the gut microbiota products contribute to neurodegeneration could lead to the identification of valuable therapeutic targets and the development of therapeutic interventions that could slow down, or even stop or prevent, pathological protein accumulation in the brain. The scope of this review is to highlight the main products of bacteria found in the gut microbiota that are potentially involved in the etiopathogeny of sporadic PD (see Tab. 1). Most of the existing literature reports data on short-chain fatty acids (SCFA). Relatively few data (coming mostly from animal or cellular models) are available on functional bacterial amyloid proteins, bacterial biosurfactants, endotoxins, and other categories of bacterial products. Thus, our review will provide a concise description of the possible contribution of these products to the development and spread of PD pathology. With this purpose, we will provide a critical presentation of the role of the functional bacterial amyloid proteins curli and functional amyloids in Pseudomonas (Fap), the bacterial biosurfactant rhamnolipid, the bacterial endotoxins lipopolysaccharides (LPS), and the microbial metabolites SCFA (also see Fig. 1). We will also discuss molecular hydrogen, a common by-product of bacterial carbohydrate metabolism with putative neuroprotective effects.
Table 1: Bacterial products that may play a role in the etiopathogeny of sporadic Parkinson’s disease
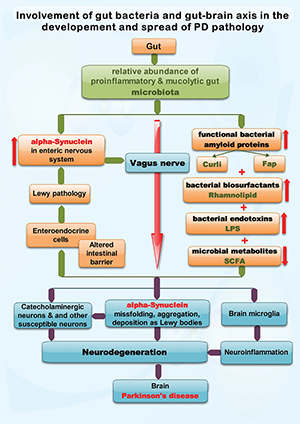
Figure 1: Exemplifies the possible roles of functional bacterial amyloid proteins (Curli and Fap), bacterial biosurfactants, lipopolysaccharide (LPS) and short-chain fatty acids (SCFA), putatively produced by the gut microbiota, in the development and spread of sporadic Parkinson’s disease (PD) pathology. PD is a proteinopathy characterized by amyloid transformation of native alpha-synuclein, with pathologic misfolding, aggregation and accumulation of aggregates (i.e., Lewy bodies, Lewy neurites) and subsequent neurodegeneration. The Braak hypothesis and dual-hit theory (see text) state that in most people with PD the alpha-synuclein misfolding process originates in the gut and olfactory mucosa, were is initiated by an unknown pathogen. The current view is that alpha-synuclein conformational changes spread transsynaptically, from the gut and olfactory mucosa neurons to the brain, via the respective nerves, by a prion-like mechanism. In this review we focus on spreading via the vagus nerve, one of the main bidirectional communication pathways of the gut-brain axis. The presence of a proinflammatory and mucolytic microbiota in people with PD, as well as lower concentrations of SCFA, results in alteration of the intestinal barrier, possibly allowing direct contact between some gut lumen xenobiotics (such as the above mentioned bacterial products) and enteric neurons that express alpha-synuclein. Moreover, intestinal inflammatory changes may increase alpha-synuclein expression in enteric neurons. In vitro and in vivo (animal models) studies suggests that the functional bacterial amyloid proteins curli and Fap, the bacterial biosurfactant rhamnolipid and the endotoxin LPS are able to induce alpha-synuclein conformational changes and pathologic aggregation. Further details are found in Tab. 1
Fig. 1 exemplifies the possible roles of functional bacterial amyloid proteins (Curli and Fap), bacterial biosurfactants, lipopolysaccharide (LPS) and short-chain fatty acids (SCFA), putatively produced by the gut microbiota, in the development and spread of sporadic Parkinson’s disease (PD) pathology. PD is a proteinopathy characterized by amyloid transformation of native alpha-synuclein, with pathologic misfolding, aggregation and accumulation of aggregates (i.e., Lewy bodies, Lewy neurites) and subsequent neurodegeneration. The Braak hypothesis and dual-hit theory (see text) state that in most people with PD the alpha-synuclein misfolding process originates in the gut and olfactory mucosa, were is initiated by an unknown pathogen. The current view is that alpha-synuclein conformational changes spread transsynaptically, from the gut and olfactory mucosa neurons to the brain, via the respective nerves, by a prion-like mechanism. In this review we focus on spreading via the vagus nerve, one of the main bidirectional communication pathways of the gut-brain axis. The presence of a proinflammatory and mucolytic microbiota in people with PD, as well as lower concentrations of SCFA, results in alteration of the intestinal barrier, possibly allowing direct contact between some gut lumen xenobiotics (such as the above mentioned bacterial products) and enteric neurons that express alpha-synuclein. Moreover, intestinal inflammatory changes may increase alpha-synuclein expression in enteric neurons. In vitro and in vivo (animal models) studies suggests that the functional bacterial amyloid proteins curli and Fap, the bacterial biosurfactant rhamnolipid and the endotoxin LPS are able to induce alpha-synuclein conformational changes and pathologic aggregation. Further details are found in Tab. 1.
Functional Bacterial Amyloid Proteins–Curli and Fap
Amyloids were initially described in association with human diseases, including the broad group of neurodegenerative proteinopathies, PD included. They consist of protein monomers, which can self-assemble, forming beta-strands perpendicular to the fibril axis, the so-called cross-beta structure (Nelson et al., 2005; Tycko, 2004).
The concept of functional amyloids was proposed for the first time by Chapman et al. (2002) who observed that curli protein produced by Escherichia coli is biochemically similar to amyloid proteins that are associated with diseases. Since then, functional amyloids have been identified in many organisms, including humans, being involved in a broad variety of physiological functions (Fowler et al., 2007; Hammer et al., 2008). It is thought that the amyloid proteins are so widespread in physiology because their fibrillary aggregate-forming structure confers them with excellent building material properties. Although functional amyloids from different organisms have the same ability to form amyloid fibrils, their monomers share little to no similarity in amino acid sequence (Shewmaker et al., 2011).
Functional bacterial amyloid proteins are extracellular proteins produced by many symbiotic and pathogenic bacteria, including Escherichia, Pseudomonas, Staphylococcus, Streptococcus, Bacillus, Mycobacteria, Citrobacter, Klebsiella, and Salmonella species, in which they support growth and survival. They are insoluble and have high mechanical and chemical stability (Otzen, 2010). One of their main roles is related to biofilm formation, an extracellular matrix with a complex structure that provides a living environment for most gut bacteria (O’toole et al., 2000). The matrix is formed by an association of amyloid proteins as scaffold and exopolysaccharides (Zogaj et al., 2003). The bacterial amyloid proteins have other functions as well: forming a physical barrier with a protective role against other species of bacteria, helping to bind each other, acting as scavengers for toxins, and providing the necessary moisture (Blanco et al., 2012; Romling, 2005; Zogaj et al., 2003).
Functional bacterial amyloid proteins can induce cross-seeding amyloid formation both in vitro and in vivo (Lundmark et al., 2005; Zhou et al., 2012) and may be a template for human alpha-synuclein amyloid fibrils formation (Chen et al., 2016; Chorell et al., 2015; Evans et al., 2015; Sampson et al., 2020) Interestingly, functional bacterial amyloids are recognised by the innate immune system as a pathogen-associated molecular pattern, the immune response involving toll-like receptor TLR-1 and TLR-2, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and inducible nitric oxide synthase (iNOS), the same as in the case of misfolded alpha-synuclein that occurs in PD (Daniele et al., 2015; Tukel et al., 2010; Tukel et al., 2005; Venegas and Heneka, 2017).
The most studied functional bacterial amyloid proteins in connection with the microbiota-gut-brain axis are curli, the first functional amyloid discovered, and the more recently described Fap.
Curli is an extracellular protein produced by Enterobacteriaceae, especially by E. coli and Salmonella (Evans and Chapman, 2014; Olsen et al., 1989; Tursi and Tukel, 2018; Zogaj et al., 2003). It contributes to biofilm formation, gut colonization, immune activation and cell invasion (Barnhart and Chapman, 2006). It may induce intestinal mucosa immune response (Nishimori et al., 2012; Oppong et al., 2015) and regulate gut epithelial barrier, allowing bacterial translocation (Oppong et al., 2013). It possesses all the properties of amyloids (Chapman et al., 2002; Nilsson, 2004), but it differs from disease-associated amyloids because its beta-sheet assembly is the product of a dedicated and strictly regulated biogenesis pathway and not of pathologic misfolding (Blanco et al., 2012).
The major subunit of curli is CsgA, which is capable of self-polymerizing in vitro. As a result, beta-sheet-rich amyloid fibres are formed (Chapman et al., 2002; Dueholm et al., 2011; Wang et al., 2007). CsgA amyloids form a very stable cross-beta structure that establishes close interactions with side chains on adjacent beta-sheets (Collinson et al., 1999; Chapman et al., 2002; Gerstel and Romling, 2001; Nelson et al., 2005; Shewmaker et al., 2009). The curli fibres are very stable, resistant to protease degradation and to denaturation by detergents (Chapman et al., 2002).
The potential pathologic role played by curli in PD was highlighted by an in vivo experiment where gut exposure to the functional bacterial amyloids curli produced by E. coli enhanced the aggregation of human alpha-synuclein in Fisher 344 rat brains and transgenic Caenorhabditis elegans muscle (Chen et al., 2016). Recently, Sampson et al. (2020) also found that the mono-colonisation of mice that overexpress human alpha-synuclein with curli-producing E. coli accelerates the development of alpha-synuclein aggregation in the gut and the brain and exacerbates gastrointestinal dysfunction and motor impairment, by a curli-dependent cross-seeding mechanism; oral administration of a gut-restricted amyloid inhibitor reduced these findings (Sampson et al., 2020).
Curli expression is controlled both on the cellular level and within the bacterial biofilm community by environmental signals and chemical gradients, like temperature, oxygen or osmolarity (Gerstel and Romling, 2001; Olsen et al., 1993a; Prigent-Combaret et al., 1999). The seven genes for curli (csg) are located in two divergently transcribed operons, csgDEFG and csgBAC (Hammar et al., 1995; Rudd, 2000). One of the most complex regulated promoters of the E. coli genome is the csgDEFG (Ishihama, 2010). There is also an internal regulation of curli expression by csgD, the master regulator of curli biogenesis and the first gene product of csgDEFG operon (Evans and Chapman, 2014), which is necessary for transcription of the csgBAC operon. CsgD is a member of the FixJ/LuxR family of transcriptional regulators coordinating the expression of some biofilm components such as cellulose and curli (Brombacher et al., 2003; Hammar et al., 1995; Ogasawara et al., 2011). The csgDEFG promoter expression is modulated by transcriptional regulators, such as the catabolite repressor/activator protein Cra, the cAMP receptor protein CRP, the protein RcdA (Brown et al., 2001; Ogasawara et al., 2010b; Reshamwala and Noronha, 2011; Shimada et al., 2012; Zheng et al., 2004).
In E. coli there are two DNA global regulatory protein complexes which modulate curli gene expression in an antagonistic way–i.e., integration host factor (IHF), which promotes curli gene expression while the histone-like protein (H-NS) represses it (Gerstel et al., 2003; Ogasawara et al., 2010a; Olsen et al., 1993b). The regulatory proteins, both negative and positive, bind simultaneously in a competitive way. CsgDEFG transcript is also regulated by small regulatory ribonucleic acids (RNAs), both negatively (e.g., by OmrA, OmrB, McaS, GcvB, RprA) and positively (e.g., by ArcZ, SdsR) (Holmqvist et al., 2010; Jorgensen et al., 2013; Mika et al., 2012; Monteiro et al., 2012; Olsen et al., 1993b).
Curli structure and CsgA amyloid assembly
Curli has two subunits, CsgA, the major one, and CsgB, the minor one. They are encoded by csgBAC operon (Collinson et al., 1996, 1997; Olsen et al., 1993a). CsgA is a soluble peptide, secreted across the outer membrane, in an unstructured form (Collinson et al., 1991; Chapman et al., 2002; Cherny et al., 2005; Gibson et al., 2007; Olsen et al., 1993a), and together with CsgB fibrils, which act as a nucleator, it is transformed into an amyloid structure (Hammar et al., 1996; Hammer et al., 2007), by nucleation-precipitation (Desvaux et al., 2009). In vitro CsgA polymerization involves three phases: A lag phase, a fibre elongation phase and a stationary phase (Chapman et al., 2002; Wang et al., 2007). The CsgA can adopt toxic oligomeric forms, thus the cells that assemble functional amyloids like curli developed mechanisms to limit this cytotoxicity associated with preamyloid oligomers: Employment of chaperones to prevent inappropriate aggregation; localization of amyloidogenic proteins to specific regions in or outside the cell; temporal control to minimize toxic oligomeric intermediates (Blanco et al., 2012). The interactions between CsgA and other Csg components (CsgC-G) support all these mechanisms.
A third protein, CsgC, is periplasmic and is proposed to have a role in subunits secretion (Gibson et al., 2007; Taylor et al., 2011) and maybe in CsgA folding and assembly of the mature curli protein (Gibson et al., 2007). This is a beta-sheet rich protein that has an immunoglobulin-like fold and a conserved CXC motif. Other roles presumed to be played by CsgC involve the regulation of CsgG outer membrane assembly and pore activity (Taylor et al., 2011). CsgD is a protein that controls and helps biofilm formation by managing curli production (Brombacher et al., 2003; Hammar et al., 1995; Ogasawara et al., 2011; Romling et al., 2000). Proteins CsgE, CsgF, and CsgG are involved in the outer membrane secretion apparatus. CsgG participates in the formation of a pore-like structure in the outer membrane, required for secretion of CsgA and CsgB into the extracellular space (Epstein et al., 2009; Narita et al., 2004; Robinson et al., 2006). Here, the two subunits participate to form amyloid fibres (Hammer et al., 2007). CsgE and CsgF play chaperone-like functions supporting the secretion and the attachment of curli fibres to the cell surface (Nenninger et al., 2011; Robinson et al., 2006). CsgF membrane-associated and surface-exposed protein is essential for CsgB surface exposure and for effective CsgA polymerization (Chapman et al., 2002; Epstein et al., 2009; Hammer et al., 2007; Nenninger et al., 2009). CsgE, a periplasmic protein, is considered to direct CsgA to the CsgG pore-like structure and mediate its secretion, as it inhibits CsgA polymerization in vitro (Nenninger et al., 2011).
Interaction with alpha-synuclein
The above-mentioned study of Sampson et al. found CsgA to be involved in human alpha-synuclein fibril formation (Sampson et al., 2020). Chorell et al. (2015) showed that CsgE accelerates human alpha-synuclein amyloid formation. Interestingly, CsgC was found to inhibit amyloid formation of human alpha-synuclein in vitro (Chorell et al., 2015; Evans et al., 2015).
Functional amyloids in Pseudomonas (Fap)
Other bacterial functional amyloids, analogous to curli amyloid produced by Escherichia coli, are functional amyloids in Pseudomonas (Fap), produced by many Pseudomonas strains (Dueholm et al., 2010).
Fap expression is controlled by a single six-gene operon, fap (Dueholm et al., 2010). Dueholm et al. (2013) showed that the fap operon is a molecular machine for functional amyloid formation. The major Fap subunit is FapC. It consists of 316 amino acid residues plus a 24-amino acid signal sequence (Bleem et al., 2018; Dueholm et al., 2010). Its structure includes three imperfect sequence repeats, R1-3, of 37 amino acid residues (Dueholm et al., 2010), separated by two “linker” regions, L1-2 (Dueholm et al., 2013). Similar to curli, Fap can induce alpha-synuclein conformational changes and fibrillation (Christensen et al., 2019). Recently, Christensen et al. (2019) demonstrated that removing the three imperfect repeats of FapC slows down the fibrillation of alpha-synuclein, but does not prevent it.
Another Fap structure is FapB, a nucleator protein, analogous with CsgB of curli that has a 38% sequence identity to FapC and is supposed to serve as a template for rapid polymerization of the fibrils outside the cell (Bleem et al., 2018; Dueholm et al., 2010). The rest of the proteins encoded by the fap operon constitute four other subunits: FapF subunit which forms the outer membrane pores participating in amyloid precursors translocation (Rouse et al., 2017), FapA subunit represented by chaperones guiding monomers through the periplasm and FapE and FapD, auxiliary regulators and proteases (Dueholm et al., 2013). FapF is thought to represent a precursor of the signalling peptides called bacteriocins (Dirix et al., 2004).
Barnhart and Chapman revealed that fapA-F function is analogous to the E. coli csg operons (csgBAC and csgDEFG) (Barnhart and Chapman, 2006). Fap operon does not include a transcription factor equivalent to CsgD, organization of fap genes into a single operon needing no internal transcription factor, as Dueholm et al. showed (Dueholm et al., 2013). These authors proposed that Fap are extracellular biofilm components of equal importance to polysaccharides, other proteins, and extracellular DNA (Dueholm et al., 2013).
Using a combination of bioinformatics and protein engineering, Bleem et al. (2018) examined the FapC sequence in greater detail. They identified specific motifs implicated in amyloid formation and established the particular significance of the third repeat motif in promoting fibril formation, contributing to understanding the mechanism of amyloid polymerization in P. aeruginosa (Bleem et al., 2018). The authors observed that mutations in the third repeat of FapC, R3, reduced amyloid propensity, increasing its susceptibility to exogenous inhibitors. They also showed the existence of a disulphide bond between two monomers in a second conserved sequence region, a CXXC motif near the C-terminus of FapC, which could serve as an additional molecular reinforcement in mature fibrils. Christensen et al. (2019) demonstrated that the disulphide bond formation delays fibrillation in Pseudomonas, but also delays fibrillation of human alpha-synuclein (Christensen et al., 2019).
All the above data could be used to find effective inhibitors of fibril formation and biofilm establishment in vivo, as new therapeutic targets within the microbiota-gut-brain axis, with the aim of preventing or slowing down the progression of PD and other neurodegenerative conditions.
There are few data regarding the rhamnolipids produced by gut bacteria and their ability to induce alpha-synuclein misfolding.
Rhamnolipids are biosurfactant glycolipids produced by bacteria like Pseudomonas sp., which modulate bacterial biofilms (Pamp and Tolker-Nielsen, 2007). They are composed of a glycosyl head group, mono- or di- rhamnose, and a branched carboxylated alkyl chain, beta-hydroxydecanoic acid (Lang and Wullbrandt, 1999).
Rhamnolipids have many functions, such as modulation of bacterial motility (Wang et al., 2014) and bacterial biofilm development (Pamp and Tolker-Nielsen, 2007); they can modify the cell surface of bacteria (Sotirova et al., 2009), participate in protein transport across the human stratum corneum (Meyer-Hoffert et al., 2011) and protect against monocyte-derived macrophages and polymorphonuclear leukocytes (Van Gennip et al., 2009). Although rhamnolipids are known to affect many proteins, like bovine serum albumin (Sanchez et al., 2008), alpha-lactalbumin, myoglobin (Andersen and Otzen, 2014), less is known about gastrointestinal tract protein damage (Markou and Apidianakis, 2014).
Andersen et al. (2018) reported the impact of rhamnolipid on the aggregative behaviour of the alpha-synuclein (Andersen et al., 2018). Their group showed that alpha-synuclein, which is natively unfolded, can suffer amyloid conformational changes when exposed to biosurfactants, such as the rhamnolipid produced by P. aeruginosa (Andersen et al., 2018). The monomeric rhamnolipid enhances the ability of alpha-synuclein to permeabilize membranes, and the micellar rhamnolipid can induce the formation of a protein beta-sheet structure with a worm-like fibrillary appearance (Andersen et al., 2018). Moreover, in the absence of rhamnolipid, alpha-synuclein can reduce biofilm formation by Pseudomonas; conversely, at physiological temperatures, rhamnolipid induces the rapid formation of alpha-synuclein fibril-like structures (Andersen et al., 2018).
Research has shown a correlation between PD and alterations in gut microbiota, as well as gastrointestinal inflammation (Heintz-Buschart et al., 2018; Hill-Burns et al., 2017; Houser et al., 2018; Houser and Tansey, 2017; Nuzum et al., 2020; Schwiertz et al., 2018). Various studies report an increased level of pathogenic Gram-negative bacteria (Proteobacteria, Enterobacteriaceae, Escherichia sp.) in individuals with PD (Keshavarzian et al., 2015; Li et al., 2017; Scheperjans et al., 2015).
LPS is an endotoxin derived from Gram-negative bacteria cell walls, which is associated with increased oxidative stress and intestinal inflammation (Fang, 2016; Guo et al., 2015; Loffredo et al., 2020; Nighot et al., 2017). LPS is composed of 3 distinct units, a hydrophobic hexa-acylated lipid A moiety, a polysaccharide domain O-antigen, and a core oligosaccharide that covalently bonds the two other entities (Bhattacharyya et al., 2019). It is known that LPS can modulate alpha-synuclein aggregation in vitro and cause many pathological PD-like effects in experimental in vivo models (Kim et al., 2016; Liu and Bing, 2011).
In PD, the intestinal barrier is disrupted and LPS can enter the systemic circulation (Parashar and Udayabanu, 2017). A recent study found that increased serum levels of zonulin (a protein responsible for the disassembly of intercellular tight junctions) (Ajamian et al., 2019) are correlated with serum LPS in patients with neurodegenerative diseases (Loffredo et al., 2020). The permeability of intestinal tight junctions may also be increased by LPS (Choi et al., 2018; Fang, 2016). Alteration of the intestinal barrier could be involved in the etiopathogeny of PD by facilitating direct contact between enteric alpha-synuclein and amyloidogenic xenobiotics (including bacterial products of the gut microbiota, LPS itself being potentially amyloidogenic) and by allowing passage of LPS and other xenobiotics into the bloodstream – with potential neurotoxic and neuroinflammatory consequences, increasing susceptibility to neurodegeneration in the presence of misfolded alpha-synuclein (Van and Derkinderen, 2019). Moreover, LPS may also induce intestinal inflammatory changes (possibly enhanced in the presence of proinflammatory gut microbiota) and local overexpression of alpha-synuclein, as seen in animal models (Van and Derkinderen, 2019), thus increasing exposure to amyloidogenic factors.
In vitro studies are in agreement with the above, the exposure to LPS of IEC-6 cells resulting in a reduction and altered distribution of the tight junction markers ZO-1 (zonula occludens protein-1) and E-cadherin around the cell membrane (Gorecki et al., 2019). The same study showed that in vivo, the administration of LPS to transgenic Thy-1- αSyn mice, a murine PD model, led to early motor manifestations as compared to untreated Thy-1- α Syn mice.
Other rodent models receiving LPS treatment exhibited hallmarks of PD pathology and other characteristic features: microglial inflammation in the substantia nigra, reduced dopamine production and motor impairments (Sharma and Nehru, 2015), increased alpha-synuclein expression (Kelly et al., 2014), selective dopaminergic neuronal loss and nigrostriatal alpha-synuclein aggregation (He et al., 2013). LPS induces peripheral inflammation and neuroinflammation through the TLR-4/NF-κB pathway, as well as the increased production of inflammatory cytokines, such as tumor necrosis factor-alpha (TNF-α) and interleukin-1 beta (IL-1β) (Mafra and Fouque, 2015; Perez-Pardo et al., 2019). The presence of proinflammatory cytokines impairs the BBB integrity and is responsible for the enhanced susceptibility to toxins from the gut (Bodea et al., 2014). Over-activation of microglia causes damage to neurons (Kannarkat et al., 2013; Zhang et al., 2005).
Alpha-synuclein misfolding and aggregation are favored by the inflammatory environment (Gao et al., 2011). Nuclear magnetic resonance spectroscopy, Bhattacharyya et al. (2019) showed that the direct interaction of LPS with alpha-synuclein modulates the protein’s conformation into alternative nucleating forms and stabilizes the α-helical intermediates in the alpha-synuclein aggregation pathway (Bhattacharyya et al., 2019). After misfolding and aggregation, alpha-synuclein is released into the extracellular space and can enter another cell, where it can serve as a template for further misfolding of monomeric alpha-synuclein (Angot and Brundin, 2009; Brundin et al., 2008; Dunning et al., 2012).
Bacterial Metabolites–Short-Chain Fatty Acids and Molecular Hydrogen
Recent data show that some metabolic pathways are enriched in the faecal microbiota of people with PD, suggesting increased production capacity of potentially deleterious metabolites such as p-cresol, phenylacetylglutamine and methionine, while pathways with beneficial metabolites, such as the SCFA butyrate pathway, are reduced (Baldini et al., 2020; Cirstea et al., 2020). Consistent findings concern the SCFA, which we will further discuss, along with molecular hydrogen, another by-product of bacterial carbohydrate metabolism, with putative neuroprotective effects.
SCFA are fermentation products generated by bacteria, including those in the gut microbiota, through various metabolic pathways (Sampson et al., 2016). The most abundant SCFA in the human body are butyrate, acetate and propionate (Dalile et al., 2019). They are used locally by colonocytes, as well as absorbed in the colon and are transported to the liver via the portal circulation (Morrison and Preston, 2016), where they are used as energy substrates for hepatocytes (Schonfeld and Wojtczak, 2016), while a minor fraction enters the systemic circulation (Boets et al., 2015). SCFA maintain the integrity of the intestinal barrier, regulate intestinal motility, mucus production and several immunological processes in the body (Canani et al., 2011; Lewis et al., 2010; Unger et al., 2016). Most SCFA can reach the CNS and cross the BBB (Perry et al., 2016; Sampson et al., 2016). A study on mice showed that they can decrease the permeability of the BBB and increase the expression of occludin in the BBB tight junctions (Braniste et al., 2014).
Several studies have investigated the link between SCFA and PD (Cirstea et al., 2020; Shin et al., 2020; Unger et al., 2016). Unger et al. (2016) found a significantly reduced concentration of acetate, propionate and butyrate in faecal samples from patients with PD compared to healthy controls. Moreover, SCFA-producing bacteria, such as Roseburia and Faecalibacterium, are more abundant in healthy individuals (Bedarf et al., 2017; Hopfner et al., 2017; Keshavarzian et al., 2015; Li et al., 2017; Nishiwaki et al., 2020; Nuzum et al., 2020).
Administration of sodium butyrate (a histone deacetylase inhibitor) has proven beneficial in animal models of PD, improving motor impairment and dopamine deficiency (Paiva et al., 2017; Sharma et al., 2015; St Laurent et al., 2013). However, in germ-free mice overexpressing alpha-synuclein, administration of SCFA led to microglia activation, alpha-synuclein aggregate formation and neuroinflammation (Sampson et al., 2016). A recent study analysed the possible remote effects of SCFA on the CNS, by measuring their plasma concentration (Shin et al., 2020). In contrast to faecal samples, plasma SCFA were increased in PD, the authors suggesting that this paradoxical finding could be the result of intestinal wall leakage caused by gut dysbiosis and local low-grade inflammation (Shin et al., 2020).
Gut microbial composition and SCFA production can be regulated indirectly by the ingestion of probiotics or prebiotics, the latter acting as a substrate for bacteria in the colon (Gibson et al., 2017; Leblanc et al., 2017; Sanders, 2008). Diet can also positively influence the microbiome and it has been suggested that a vegetarian diet increases the availability of fermentable substrates and may have a beneficial effect on the clinical course of PD (Derrien and Veiga, 2017; Hegelmaier et al., 2020; Klimenko et al., 2018; Martinez et al., 2013; Wong et al., 2018).
Molecular hydrogen is a common bacterial by-product resulting from carbohydrate fermentation. It is produced by the human gut microbiota and easily crosses membranes, reaching the bloodstream and being exhaled – thus its use in the hydrogen breath test for the detection of small intestine bacterial overgrowth (SIBO) (Ostojic, 2018). It has antioxidant properties, neutralizing hydroxyl radicals, and may decrease inflammation, with putative neuroprotective effects (Ohta, 2014).
Experimental evidence coming from mouse and rat models suggest that molecular hydrogen may slow down the progression of PD pathology (Fu et al., 2009; Fujita et al., 2009). Moreover, a small pilot double-blind placebo-controlled randomized trial found that 1 litre of hydrogenated water per day improves motor outcomes in people with PD, at 48 weeks (Yoritaka et al., 2013). Other data from human studies suggest that the faecal microbiota of people with PD is less abundant in hydrogen-producing bacteria (e.g., Prevotella) compared to that of healthy controls (Scheperjans et al., 2015; Suzuki et al., 2018). Small studies also suggest that the small intestine microbiota of people with PD may produce lower amounts of molecular hydrogen, as measured in the exhaled air (Ostojic, 2018; Suzuki et al., 2018; Yoritaka et al., 2013).
An increasing number of studies suggest that sporadic PD may start in the gut and olfactory mucosa, Lewy pathology spreading to the brain, via the vagus and olfactory nerves, through a prion-like mechanism.
Gut dysbiosis is present in people with PD compared to healthy controls, but the causality relation between the composition and function of the gut microbiota and sporadic PD has not been proven yet. In this review we brought together the latest data, coming mostly from in vitro studies and animal models, regarding the bacterial products that can trigger alpha-synuclein misfolding in experimental settings and thus may be involved in the etiopathogeny of sporadic PD. Available evidence suggests that products of bacteria found in the gut may result in alteration of the intestinal barrier, may lead to overexpression of enteric alpha-synuclein and may play key roles in triggering alpha-synuclein self-propagating misfolding events that result in alpha-synuclein self-aggregation and formation of Lewy bodies. Microbiota products that enter the bloodstream may also have neuroinflammatory and neurotoxic effects and may interfere with neuroprotection pathways, modulating neuronal susceptibility to neurodegeneration.
Currently, there are no disease-modifying treatments for neurodegenerative conditions, including PD. Amyloidogenic bacterial products, especially functional bacterial amyloid proteins, biosurfactants and endotoxins are possible triggers of the initial pathologic alpha-synuclein misfolding in sporadic PD, making them potentially attractive therapeutic targets for preventing or slowing down the progression of the disease. This theory however has a series of limitations, mainly related to the lack of direct evidence on the interaction between the amyloidogenic bacterial products and alpha-synuclein within the human gut. Further research is needed.
Funding Statement: This work was supported by the Ministry of Research and Innovation in Romania, under Grants No. PN 1N/2019_19.29.02.01 and No. 7PFE/2018.
Conflicts of Interest: The corresponding author is a member of the Editorial Board of the journal. The authors declare that they have no other conflicts of interest to report regarding the present research paper.
Aho VTE, Pereira PAB, Voutilainen S, Paulin L, Pekkonen E, Auvinen P, Scheperjans F. (2019). Gut microbiota in Parkinson’s disease: Temporal stability and relations to disease progression. EBioMedicine 44: 691–707. DOI 10.1016/j.ebiom.2019.05.064. [Google Scholar] [CrossRef]
Ajamian M, Steer D, Rosella G, Gibson PR. (2019). Serum zonulin as a marker of intestinal mucosal barrier function: May not be what it seems. PLoS One 14: e0210728. DOI 10.1371/journal.pone.0210728. [Google Scholar] [CrossRef]
Andersen KK, Otzen DE. (2014). Denaturation of α-lactalbumin and myoglobin by the anionic biosurfactant rhamnolipid. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 1844: 2338–2345. DOI 10.1016/j.bbapap.2014.10.005. [Google Scholar] [CrossRef]
Andersen KK, Vad BS, Kjaer L, Tolker-Nielsen T, Christiansen G, Otzen DE. (2018). Pseudomonas aeruginosa rhamnolipid induces fibrillation of human α-synuclein and modulates its effect on biofilm formation. FEBS Letters 592: 1484–1496. DOI 10.1002/1873-3468.13038. [Google Scholar] [CrossRef]
Angot E, Brundin P. (2009). Dissecting the potential molecular mechanisms underlying α-synuclein cell-to-cell transfer in Parkinson’s disease. Parkinsonism & Related Disorders 15: S143–S147. DOI 10.1016/S1353-8020(09)70802-8. [Google Scholar] [CrossRef]
Araki K, Yagi N, Aoyama K, Choong CJ, Hayakawa H, Fujimura H, Nagai Y, Goto Y, Mochizuki H. (2019). Parkinson’s disease is a type of amyloidosis featuring accumulation of amyloid fibrils of alpha-synuclein. Proceedings of the National Academy of Sciences of the United States of America 116: 17963–17969. DOI 10.1073/pnas.1906124116. [Google Scholar] [CrossRef]
Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, Lee VM, Trojanowski JQ, Iwatsubo T. (1998). Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. American Journal of Pathology 152: 879–884. [Google Scholar]
Baldini F, Hertel J, Sandt E, Thinnes CC, Neuberger-Castillo L, Pavelka L, Betsou F, Kruger R, Thiele I, Consortium N-P. (2020). Parkinson’s disease-associated alterations of the gut microbiome predict disease-relevant changes in metabolic functions. BMC Biology 18: 62. [Google Scholar]
Barnhart MM, Chapman MR. (2006). Curli biogenesis and function. Annual Review of Microbiology 60: 131–147. DOI 10.1146/annurev.micro.60.080805.142106. [Google Scholar] [CrossRef]
Bedarf JR, Hildebrand F, Coelho LP, Sunagawa S, Bahram M, Goeser F, Bork P, Wullner U. (2017). Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naive Parkinson’s disease patients. Genome Medicine 9: 39. DOI 10.1186/s13073-017-0428-y. [Google Scholar] [CrossRef]
Berg D, Postuma RB, Adler CH, Bloem BR, Chan P, Dubois B, Gasser T, Goetz CG, Halliday G, Joseph L, Lang AE, Liepelt-Scarfone I, Litvan I, Marek K, Obeso J, Oertel W, Olanow CW, Poewe W, Stern M, Deuschl G. (2015). MDS research criteria for prodromal Parkinson’s disease. Movement Disorders 30: 1600–1611. DOI 10.1002/mds.26431. [Google Scholar] [CrossRef]
Bhattacharyya D, Mohite GM, Krishnamoorthy J, Gayen N, Mehra S, Navalkar A, Kotler SA, Ratha BN, Ghosh A, Kumar R, Garai K, Mandal AK, Maji SK, Bhunia A. (2019). Lipopolysaccharide from gut microbiota modulates α-synuclein aggregation and alters its biological function. ACS Chemical Neuroscience 10: 2229–2236. DOI 10.1021/acschemneuro.8b00733. [Google Scholar] [CrossRef]
Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. (2012). Diversity, biogenesis and function of microbial amyloids. Trends in Microbiology 20: 66–73. DOI 10.1016/j.tim.2011.11.005. [Google Scholar] [CrossRef]
Bleem A, Christiansen G, Madsen DJ, Maric H, Stromgaard K, Bryers JD, Daggett V, Meyer RL, Otzen DE. (2018). Protein engineering reveals mechanisms of functional amyloid formation in Pseudomonas aeruginosa biofilms. Journal of Molecular Biology 430: 3751–3763. DOI 10.1016/j.jmb.2018.06.043. [Google Scholar] [CrossRef]
Bodea LG, Wang Y, Linnartz-Gerlach B, Kopatz J, Sinkkonen L, Musgrove R, Kaoma T, Muller A, Vallar L, Di Monte DA, Balling R, Neumann H. (2014). Neurodegeneration by activation of the microglial complement-phagosome pathway. Journal of Neuroscience 34: 8546–8556. DOI 10.1523/JNEUROSCI.5002-13.2014. [Google Scholar] [CrossRef]
Boertien JM, Pereira PAB, Aho VTE, Scheperjans F. (2019). Increasing comparability and utility of gut microbiome studies in Parkinson’s disease: A systematic review. Journal of Parkinson’s Disease 9: S297–S312. DOI 10.3233/JPD-191711. [Google Scholar] [CrossRef]
Boets E, Deroover L, Houben E, Vermeulen K, Gomand SV, Delcour JA, Verbeke K. (2015). Quantification of in vivo colonic short chain fatty acid production from inulin. Nutrients 7: 8916–8929. DOI 10.3390/nu7115440. [Google Scholar] [CrossRef]
Braak H, De Vos RA, Bohl J, Del Tredici K. (2006). Gastric α-synuclein immunoreactive inclusions in Meissner’s and Auerbach’s plexuses in cases staged for Parkinson’s disease-related brain pathology. Neuroscience Letters 396: 67–72. DOI 10.1016/j.neulet.2005.11.012. [Google Scholar] [CrossRef]
Braak H, Del Tredici K. (2017). Neuropathological staging of brain pathology in sporadic Parkinson’s disease: Separating the wheat from the chaff. Journal of Parkinson’s Disease 7: S71–S85. DOI 10.3233/JPD-179001. [Google Scholar] [CrossRef]
Braak H, Del Tredici K, Rub U, De Vos RA, Jansen Steur EN, Braak E. (2003). Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiology of Aging 24: 197–211. DOI 10.1016/S0197-4580(02)00065-9. [Google Scholar] [CrossRef]
Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Toth M, Korecka A, Bakocevic N, Ng LG, Kundu P, Gulyas B, Halldin C, Hultenby K, Nilsson H, Hebert H, Volpe BT, Diamond B, Pettersson S. (2014). The gut microbiota influences blood-brain barrier permeability in mice. Science Translational Medicine 6: 263ra158. DOI 10.1126/scitranslmed.3009759. [Google Scholar] [CrossRef]
Brombacher E, Dorel C, Zehnder AJB, Landini P. (2003). The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149: 2847–2857. DOI 10.1099/mic.0.26306-0. [Google Scholar] [CrossRef]
Brown PK, Dozois CM, Nickerson CA, Zuppardo A, Terlonge J, Curtiss R3rd. (2001). MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Molecular Microbiology 41: 349–363. DOI 10.1046/j.1365-2958.2001.02529.x. [Google Scholar] [CrossRef]
Brundin P, Li JY, Holton JL, Lindvall O, Revesz T. (2008). Research in motion: The enigma of Parkinson’s disease pathology spread. Nature Reviews Neuroscience 9: 741–745. DOI 10.1038/nrn2477. [Google Scholar] [CrossRef]
Canani RB, Costanzo MD, Leone L, Pedata M, Meli R, Calignano A. (2011). Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World Journal of Gastroenterology 17: 1519–1528. DOI 10.3748/wjg.v17.i12.1519. [Google Scholar] [CrossRef]
Cattaneo A, Cattane N, Galluzzi S, Provasi S, Lopizzo N, Festari C, Ferrar C, Guerra UP, Paghera B, Muscio C, Bianchetti A, Volta GD, Turla M, Cotelli MS, Gennuso M, Prelle A, Zanetti O, Lussignoli G, Mirabile D, Bellandi D, Gentile S, Belotti G, Villani D, Harach T, Bolmont T, Padovani A, Boccardi M, Frisoni GB, Group IF. (2017). Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiology of Aging 49: 60–68. [Google Scholar]
Cirstea MS, Yu AC, Golz E, Sundvick K, Kliger D, Radisavljevic N, Foulger LH, Mackenzie M, Huan T, Finlay BB, Appel-Cresswell S. (2020). Microbiota composition and metabolism are associated with gut function in Parkinson’s disease. Movement Disorders 35: 1208–1217. DOI 10.1002/mds.28052. [Google Scholar] [CrossRef]
Collinson SK, Clouthier SC, Doran JL, Banser PA, Kay WW. (1996). Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. Journal of Bacteriology 178: 662–667. DOI 10.1128/JB.178.3.662-667.1996. [Google Scholar] [CrossRef]
Collinson SK, Clouthier SC, Doran JL, Banser PA, Kay WW. (1997). Characterization of the agfBA fimbrial operon encoding thin aggregative fimbriae of Salmonella enteritidis. Mechanisms in the Pathogenesis of Enteric Diseases 412: 247–248. [Google Scholar]
Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. (1991). Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. Journal of Bacteriology 173: 4773–4781. [Google Scholar]
Collinson SK, Parker JM, Hodges RS, Kay WW. (1999). Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae. Journal of Molecular Biology 290: 741–756. DOI 10.1006/jmbi.1999.2882. [Google Scholar] [CrossRef]
Conway KA, Harper JD, Lansbury PT. (1998). Accelerated in vitro fibril formation by a mutant α-synuclein linked to early-onset Parkinson disease. Nature Medicine 4: 1318–1320. DOI 10.1038/3311. [Google Scholar] [CrossRef]
Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. (2002). Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295: 851–855. DOI 10.1126/science.1067484. [Google Scholar] [CrossRef]
Chen SG, Stribinskis V, Rane MJ, Demuth DR, Gozal E, Roberts AM, Jagadapillai R, Liu R, Choe K, Shivakumar B, Son F, Jin S, Kerber R, Adame A, Masliah E, Friedland RP. (2016). Exposure to the functional bacterial amyloid protein curli enhances alpha-synuclein aggregation in aged Fischer 344 rats and Caenorhabditis elegans. Scientific Reports 6: 34477. DOI 10.1038/srep34477. [Google Scholar] [CrossRef]
Cherny I, Rockah L, Levy-Nissenbaum O, Gophna U, Ron EZ, Gazit E. (2005). The formation of Escherichia coli curli amyloid fibrils is mediated by prion-like peptide repeats. Journal of Molecular Biology 352: 245–252. DOI 10.1016/j.jmb.2005.07.028. [Google Scholar] [CrossRef]
Choi JG, Kim N, Ju IG, Eo H, Lim SM, Jang SE, Kim DH, Oh MS. (2018). Oral administration of Proteus mirabilis damages dopaminergic neurons and motor functions in mice. Scientific Reports 8: 1275. DOI 10.1038/s41598-018-19646-x. [Google Scholar] [CrossRef]
Chorell E, Andersson E, Evans ML, Jain N, Gotheson A, Aden J, Chapman MR, Almqvist F, Wittung-Stafshede P. (2015). Bacterial chaperones CsgE and CsgC differentially modulate human α-synuclein amyloid formation via transient contacts. PLoS One 10: e0140194. DOI 10.1371/journal.pone.0140194. [Google Scholar] [CrossRef]
Christensen LFB, Jensen KF, Nielsen J, Vad BS, Christiansen G, Otzen DE. (2019). Reducing the amyloidogenicity of functional amyloid protein FapC increases its ability to inhibit α-synuclein fibrillation. ACS Omega 4: 4029–4039. DOI 10.1021/acsomega.8b03590. [Google Scholar] [CrossRef]
Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. (2019). The role of short-chain fatty acids in microbiota–gut–brain communication. Nature Reviews Gastroenterology & Hepatology 16: 461–478. DOI 10.1038/s41575-019-0157-3. [Google Scholar] [CrossRef]
Daniele SG, Beraud D, Davenport C, Cheng K, Yin H, Maguire-Zeiss KA. (2015). Activation of MyD88-dependent TLR1/2 signaling by misfolded α-synuclein, a protein linked to neurodegenerative disorders. Science Signaling 8: ra45. DOI 10.1126/scisignal.2005965. [Google Scholar] [CrossRef]
Derrien M, Veiga P. (2017). Rethinking diet to aid human–microbe symbiosis. Trends in Microbiology 25: 100–112. DOI 10.1016/j.tim.2016.09.011. [Google Scholar] [CrossRef]
Desvaux M, Hebraud M, Talon R, Henderson IR. (2009). Secretion and subcellular localizations of bacterial proteins: A semantic awareness issue. Trends in Microbiology 17: 139–145. DOI 10.1016/j.tim.2009.01.004. [Google Scholar] [CrossRef]
Dirix G, Monsieurs P, Dombrecht B, Daniels R, Marchal K, Vanderleyden J, Michiels J. (2004). Peptide signal molecules and bacteriocins in Gram-negative bacteria: A genome-wide in silico screening for peptides containing a double-glycine leader sequence and their cognate transporters. Peptides 25: 1425–1440. DOI 10.1016/j.peptides.2003.10.028. [Google Scholar] [CrossRef]
Dueholm MS, Nielsen SB, Hein KL, Nissen P, Chapman M, Christiansen G, Nielsen PH, Otzen DE. (2011). Fibrillation of the major curli subunit CsgA under a wide range of conditions implies a robust design of aggregation. Biochemistry 50: 8281–8290. DOI 10.1021/bi200967c. [Google Scholar] [CrossRef]
Dueholm MS, Petersen SV, Sonderkaer M, Larsen P, Christiansen G, Hein KL, Enghild JJ, Nielsen JL, Nielsen KL, Nielsen PH, Otzen DE. (2010). Functional amyloid in Pseudomonas. Molecular Microbiology 77: 1009–1020. [Google Scholar]
Dueholm MS, Sondergaard MT, Nilsson M, Christiansen G, Stensballe A, Overgaard MT, Givskov M, Tolker-Nielsen T, Otzen DE, Nielsen PH. (2013). Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. Microbiology Open 2: 365–382. [Google Scholar]
Dunning CJ, Reyes JF, Steiner JA, Brundin P. (2012). Can Parkinson’s disease pathology be propagated from one neuron to another? Progress in Neurobiology 97: 205–219. DOI 10.1016/j.pneurobio.2011.11.003. [Google Scholar] [CrossRef]
Epstein EA, Reizian MA, Chapman MR. (2009). Spatial clustering of the curlin secretion lipoprotein requires curli fiber assembly. Journal of Bacteriology 191: 608–615. DOI 10.1128/JB.01244-08. [Google Scholar] [CrossRef]
Erny D, Hrabe De Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, Keren-Shaul H, Mahlakoiv T, Jakobshagen K, Buch T, Schwierzeck V, Utermohlen O, Chun E, Garrett WS, Mccoy KD, Diefenbach A, Staeheli P, Stecher B, Amit I, Prinz M. (2015). Host microbiota constantly control maturation and function of microglia in the CNS. Nature Neuroscience 18: 965–977. DOI 10.1038/nn.4030. [Google Scholar] [CrossRef]
Evans ML, Chapman MR. (2014). Curli biogenesis: Order out of disorder. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1843: 1551–1558. DOI 10.1016/j.bbamcr.2013.09.010. [Google Scholar] [CrossRef]
Evans ML, Chorell E, Taylor JD, Aden J, Gotheson A, Li F, Koch M, Sefer L, Matthews SJ, Wittung-Stafshede P, Almqvist F, Chapman MR. (2015). The bacterial curli system possesses a potent and selective inhibitor of amyloid formation. Molecular Cell 57: 445–455. DOI 10.1016/j.molcel.2014.12.025. [Google Scholar] [CrossRef]
Fang X. (2016). Potential role of gut microbiota and tissue barriers in Parkinson’s disease and amyotrophic lateral sclerosis. International Journal of Neuroscience 126: 771–776. DOI 10.3109/00207454.2015.1096271. [Google Scholar] [CrossRef]
Fowler DM, Koulov AV, Balch WE, Kelly JW. (2007). Functional amyloid–from bacteria to humans. Trends in Biochemical Sciences 32: 217–224. DOI 10.1016/j.tibs.2007.03.003. [Google Scholar] [CrossRef]
Friedland RP. (2015). Mechanisms of molecular mimicry involving the microbiota in neurodegeneration. Journal of Alzheimer’s Disease 45: 349–362. DOI 10.3233/JAD-142841. [Google Scholar] [CrossRef]
Fu Y, Ito M, Fujita Y, Ito M, Ichihara M, Masuda A, Suzuki Y, Maesawa S, Kajita Y, Hirayama M, Ohsawa I, Ohta S, Ohno K. (2009). Molecular hydrogen is protective against 6-hydroxydopamine-induced nigrostriatal degeneration in a rat model of Parkinson’s disease. Neuroscience Letters 453: 81–85. DOI 10.1016/j.neulet.2009.02.016. [Google Scholar] [CrossRef]
Fujita K, Seike T, Yutsudo N, Ohno M, Yamada H, Yamaguchi H, Sakumi K, Yamakawa Y, Kido MA, Takaki A, Katafuchi T, Tanaka Y, Nakabeppu Y, Noda M. (2009). Hydrogen in drinking water reduces dopaminergic neuronal loss in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine mouse model of Parkinson’s disease. PLoS One 4: e7247. DOI 10.1371/journal.pone.0007247. [Google Scholar] [CrossRef]
Gabrielli M, Bonazzi P, Scarpellini E, Bendia E, Lauritano EC, Fasano A, Ceravolo MG, Capecci M, Rita Bentivoglio A, Provinciali L, Tonali PA, Gasbarrini A. (2011). Prevalence of small intestinal bacterial overgrowth in Parkinson’s disease. Movement Disorders 26: 889–892. DOI 10.1002/mds.23566. [Google Scholar] [CrossRef]
Gao HM, Zhang F, Zhou H, Kam W, Wilson B, Hong JS. (2011). Neuroinflammation and α-synuclein dysfunction potentiate each other, driving chronic progression of neurodegeneration in a mouse model of Parkinson’s disease. Environmental Health Perspectives 119: 807–814. DOI 10.1289/ehp.1003013. [Google Scholar] [CrossRef]
Gelpi E, Navarro-Otano J, Tolosa E, Gaig C, Compta Y, Rey MJ, Marti MJ, Hernandez I, Valldeoriola F, Rene R, Ribalta T. (2014). Multiple organ involvement by α-synuclein pathology in Lewy body disorders. Movement Disorders 29: 1010–1018. DOI 10.1002/mds.25776. [Google Scholar] [CrossRef]
Gerstel U, Park C, Romling U. (2003). Complex regulation of csgD promoter activity by global regulatory proteins. Molecular Microbiology 49: 639–654. DOI 10.1046/j.1365-2958.2003.03594.x. [Google Scholar] [CrossRef]
Gerstel U, Romling U. (2001). Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environmental Microbiology 3: 638–648. DOI 10.1046/j.1462-2920.2001.00235.x. [Google Scholar] [CrossRef]
Gibson DL, White AP, Rajotte CM, Kay WW. (2007). AgfC and AgfE facilitate extracellular thin aggregative fimbriae synthesis in Salmonella enteritidis. Microbiology 153: 1131–1140. DOI 10.1099/mic.0.2006/000935-0. [Google Scholar] [CrossRef]
Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, Verbeke K, Reid G. (2017). Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nature Reviews Gastroenterology & Hepatology 14: 491–502. DOI 10.1038/nrgastro.2017.75. [Google Scholar] [CrossRef]
Gorecki AM, Preskey L, Bakeberg MC, Kenna JE, Gildenhuys C, Macdougall G, Dunlop SA, Mastaglia FL, Akkari PA, Koengten F, Anderton RS. (2019). Altered gut microbiome in Parkinson’s disease and the influence of lipopolysaccharide in a human α-synuclein over-expressing mouse model. Frontiers in Neuroscience 13: 839. DOI 10.3389/fnins.2019.00839. [Google Scholar] [CrossRef]
Greenwald J, Riek R. (2010). Biology of amyloid: Structure, function, and regulation. Structure 18: 1244–1260. DOI 10.1016/j.str.2010.08.009. [Google Scholar] [CrossRef]
Guo S, Nighot M, Al-Sadi R, Alhmoud T, Nighot P, Ma TY. (2015). Lipopolysaccharide regulation of intestinal tight junction permeability is mediated by TLR4 signal transduction pathway activation of FAK and MyD88. Journal of Immunology 195: 4999–5010. DOI 10.4049/jimmunol.1402598. [Google Scholar] [CrossRef]
Hammar M, Bian Z, Normark S. (1996). Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 93: 6562–6566. DOI 10.1073/pnas.93.13.6562. [Google Scholar] [CrossRef]
Hammar MR, Arnqvist A, Bian Z, Olsen A, Normark S. (1995). Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Molecular Microbiology 18: 661–670. DOI 10.1111/j.1365-2958.1995.mmi_18040661.x. [Google Scholar] [CrossRef]
Hammer ND, Schmidt JC, Chapman MR. (2007). The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proceedings of the National Academy of Sciences of the United States of America 104: 12494–12499. DOI 10.1073/pnas.0703310104. [Google Scholar] [CrossRef]
Hammer ND, Wang X, Mcguffie BA, Chapman MR. (2008). Amyloids: Friend or foe? Journal of Alzheimer’s Disease 13: 407–419. DOI 10.3233/JAD-2008-13406. [Google Scholar] [CrossRef]
Hawkes CH, Del Tredici K, Braak H. (2007). Parkinson’s disease: A dual-hit hypothesis. Neuropathology and Applied Neurobiology 33: 599–614. DOI 10.1111/j.1365-2990.2007.00874.x. [Google Scholar] [CrossRef]
Hawkes CH, Del Tredici K, Braak H. (2009). Parkinson’s disease: The dual hit theory revisited. Annals of the New York Academy of Sciences 1170: 615–622. DOI 10.1111/j.1749-6632.2009.04365.x. [Google Scholar] [CrossRef]
He Q, Yu W, Wu J, Chen C, Lou Z, Zhang Q, Zhao J, Wang J, Xiao B. (2013). Intranasal LPS-mediated Parkinson’s model challenges the pathogenesis of nasal cavity and environmental toxins. PLoS One 8: e78418. DOI 10.1371/journal.pone.0078418. [Google Scholar] [CrossRef]
Hegelmaier T, Lebbing M, Duscha A, Tomaske L, Tonges L, Holm JB, Bjorn Nielsen H, Gatermann SG, Przuntek H, Haghikia A. (2020). Interventional influence of the intestinal microbiome through dietary intervention and bowel cleansing might improve motor symptoms in Parkinson’s disease. Cells 9: 376. DOI 10.3390/cells9020376. [Google Scholar] [CrossRef]
Heintz-Buschart A, Pandey U, Wicke T, Sixel-Doring F, Janzen A, Sittig-Wiegand E, Trenkwalder C, Oertel WH, Mollenhauer B, Wilmes P. (2018). The nasal and gut microbiome in Parkinson’s disease and idiopathic rapid eye movement sleep behavior disorder. Movement Disorders 33: 88–98. DOI 10.1002/mds.27105. [Google Scholar] [CrossRef]
Hill-Burns EM, Debelius JW, Morton JT, Wissemann WT, Lewis MR, Wallen ZD, Peddada SD, Factor SA, Molho E, Zabetian CP, Knight R, Payami H. (2017). Parkinson’s disease and Parkinson’s disease medications have distinct signatures of the gut microbiome. Movement Disorders 32: 739–749. DOI 10.1002/mds.26942. [Google Scholar] [CrossRef]
Hirsch EC, Hunot S. (2009). Neuroinflammation in Parkinson’s disease: A target for neuroprotection? Lancet Neurology 8: 382–397. DOI 10.1016/S1474-4422(09)70062-6. [Google Scholar] [CrossRef]
Holmqvist E, Reimegard J, Sterk M, Grantcharova N, Romling U, Wagner EG. (2010). Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO Journal 29: 1840–1850. DOI 10.1038/emboj.2010.73. [Google Scholar] [CrossRef]
Hopfner F, Kunstner A, Muller SH, Kunzel S, Zeuner KE, Margraf NG, Deuschl G, Baines JF, Kuhlenbaumer G. (2017). Gut microbiota in Parkinson disease in a northern German cohort. Brain Research 1667: 41–45. DOI 10.1016/j.brainres.2017.04.019. [Google Scholar] [CrossRef]
Houser MC, Chang J, Factor SA, Molho ES, Zabetian CP, Hill-Burns EM, Payami H, Hertzberg VS, Tansey MG. (2018). Stool immune profiles evince gastrointestinal inflammation in Parkinson’s disease. Movement Disorders 33: 793–804. DOI 10.1002/mds.27326. [Google Scholar] [CrossRef]
Houser MC, Tansey MG. (2017). The gut-brain axis: Is intestinal inflammation a silent driver of Parkinson’s disease pathogenesis? NPJ Parkinson’s Disease 3: 3. DOI 10.1038/s41531-016-0002-0. [Google Scholar] [CrossRef]
Huang HK, Wang JH, Lei WY, Chen CL, Chang CY, Liou LS. (2018). Helicobacter pylori infection is associated with an increased risk of Parkinson’s disease: A population-based retrospective cohort study. Parkinsonism & Related Disorders 47: 26–31. DOI 10.1016/j.parkreldis.2017.11.331. [Google Scholar] [CrossRef]
Ishihama A. (2010). Prokaryotic genome regulation: Multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiology Reviews 34: 628–645. DOI 10.1111/j.1574-6976.2010.00227.x. [Google Scholar] [CrossRef]
Jorgensen MG, Thomason MK, Havelund J, Valentin-Hansen P, Storz G. (2013). Dual function of the McaS small RNA in controlling biofilm formation. Genes & Development 27: 1132–1145. DOI 10.1101/gad.214734.113. [Google Scholar] [CrossRef]
Kannarkat GT, Boss JM, Tansey MG. (2013). The role of innate and adaptive immunity in Parkinson’s disease. Journal of Parkinson’s Disease 3: 493–514. DOI 10.3233/JPD-130250. [Google Scholar] [CrossRef]
Kelly LP, Carvey PM, Keshavarzian A, Shannon KM, Shaikh M, Bakay RA, Kordower JH. (2014). Progression of intestinal permeability changes and alpha-synuclein expression in a mouse model of Parkinson’s disease. Movement Disorders 29: 999–1009. DOI 10.1002/mds.25736. [Google Scholar] [CrossRef]
Keshavarzian A, Green SJ, Engen PA, Voigt RM, Naqib A, Forsyth CB, Mutlu E, Shannon KM. (2015). Colonic bacterial composition in Parkinson’s disease. Movement Disorders 30: 1351–1360. DOI 10.1002/mds.26307. [Google Scholar] [CrossRef]
Kim C, Lv G, Lee JS, Jung BC, Masuda-Suzukake M, Hong CS, Valera E, Lee HJ, Paik SR, Hasegawa M, Masliah E, Eliezer D, Lee SJ. (2016). Exposure to bacterial endotoxin generates a distinct strain of α-synuclein fibril. Scientific Reports 6: 30891. DOI 10.1038/srep30891. [Google Scholar] [CrossRef]
Klimenko NS, Tyakht AV, Popenko AS, Vasiliev AS, Altukhov IA, Ischenko DS, Shashkova TI, Efimova DA, Nikogosov DA, Osipenko DA, Musienko SV, Selezneva KS, Baranova A, Kurilshikov AM, Toshchakov SM, Korzhenkov AA, Samarov NI, Shevchenko MA, Tepliuk AV, Alexeev DG. (2018). Microbiome responses to an uncontrolled short-term diet intervention in the frame of the Citizen Science Project. Nutrients 10: 576. DOI 10.3390/nu10050576. [Google Scholar] [CrossRef]
Lang S, Wullbrandt D. (1999). Rhamnose lipids – biosynthesis, microbial production and application potential. Applied Microbiology and Biotechnology 51: 22–32. DOI 10.1007/s002530051358. [Google Scholar] [CrossRef]
Leblanc JG, Chain F, Martin R, Bermudez-Humaran LG, Courau S, Langella P. (2017). Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microbial Cell Factories 16: 79. DOI 10.1186/s12934-017-0691-z. [Google Scholar] [CrossRef]
Lewis K, Lutgendorff F, Phan V, Soderholm JD, Sherman PM, Mckay DM. (2010). Enhanced translocation of bacteria across metabolically stressed epithelia is reduced by butyrate. Inflammatory Bowel Diseases 16: 1138–1148. DOI 10.1002/ibd.21177. [Google Scholar] [CrossRef]
Li W, Wu X, Hu X, Wang T, Liang S, Duan Y, Jin F, Qin B. (2017). Structural changes of gut microbiota in Parkinson’s disease and its correlation with clinical features. Science China Life Sciences 60: 1223–1233. DOI 10.1007/s11427-016-9001-4. [Google Scholar] [CrossRef]
Liu B, Fang F, Pedersen NL, Tillander A, Ludvigsson JF, Ekbom A, Svenningsson P, Chen H, Wirdefeldt K. (2017). Vagotomy and Parkinson disease: A Swedish register-based matched-cohort study. Neurology 88: 1996–2002. DOI 10.1212/WNL.0000000000003961. [Google Scholar] [CrossRef]
Liu B, Fang F, Ye W, Wirdefeldt K. (2020). Appendectomy, Tonsillectomy and Parkinson’s Disease Risk: A Swedish Register-Based Study. Frontiers in Neurology 11: 510. DOI 10.3389/fneur.2020.00510. [Google Scholar] [CrossRef]
Liu M, Bing G. (2011). Lipopolysaccharide animal models for Parkinson’s disease. Parkinson’s Disease 2011: 327089. [Google Scholar]
Loffredo L, Ettorre E, Zicari AM, Inghilleri M, Nocella C, Perri L, Spalice A, Fossati C, De Lucia MC, Pigozzi F, Cacciafesta M, Violi F, Carnevale R, Neurodegenerative Disease Study Group. (2020). Oxidative stress and gut-derived lipopolysaccharides in neurodegenerative disease: Role of NOX2. Oxidative Medicine and Cellular Longevity 2020, 8630275. [Google Scholar]
Luk KC, Kehm V, Carroll J, Zhang B, O’brien P, Trojanowski JQ, Lee VM. (2012). Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338: 949–953. DOI 10.1126/science.1227157. [Google Scholar] [CrossRef]
Lundmark K, Westermark GT, Olsen A, Westermark P. (2005). Protein fibrils in nature can enhance amyloid protein A amyloidosis in mice: Cross-seeding as a disease mechanism. Proceedings of the National Academy of Sciences of the United States of America 102: 6098–6102. DOI 10.1073/pnas.0501814102. [Google Scholar] [CrossRef]
Mafra D, Fouque D. (2015). Gut microbiota and inflammation in chronic kidney disease patients. Clinical Kidney Journal 8: 332–334. DOI 10.1093/ckj/sfv026. [Google Scholar] [CrossRef]
Markou P, Apidianakis Y. (2014). Pathogenesis of intestinal Pseudomonas aeruginosa infection in patients with cancer. Frontiers in Cellular and Infection Microbiology 3: 115. DOI 10.3389/fcimb.2013.00115. [Google Scholar] [CrossRef]
Martinez-Martin P, Schapira AH, Stocchi F, Sethi K, Odin P, Macphee G, Brown RG, Naidu Y, Clayton L, Abe K, Tsuboi Y, Macmahon D, Barone P, Rabey M, Bonuccelli U, Forbes A, Breen K, Tluk S, Olanow CW, Thomas S, Rye D, Hand A, Williams AJ, Ondo W, Chaudhuri KR. (2007). Prevalence of nonmotor symptoms in Parkinson’s disease in an international setting; study using nonmotor symptoms questionnaire in 545 patients. Movement Disorders 22: 1623–1629. DOI 10.1002/mds.21586. [Google Scholar] [CrossRef]
Martinez I, Lattimer JM, Hubach KL, Case JA, Yang J, Weber CG, Louk JA, Rose DJ, Kyureghian G, Peterson DA, Haub MD, Walter J. (2013). Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME Journal 7: 269–280. DOI 10.1038/ismej.2012.104. [Google Scholar] [CrossRef]
Masuda-Suzukake M, Nonaka T, Hosokawa M, Oikawa T, Arai T, Akiyama H, Mann DM, Hasegawa M. (2013). Prion-like spreading of pathological α-synuclein in brain. Brain 136: 1128–1138. DOI 10.1093/brain/awt037. [Google Scholar] [CrossRef]
Meyer-Hoffert U, Zimmermann A, Czapp M, Bartels J, Koblyakova Y, Glaser R, Schroder JM, Gerstel U. (2011). Flagellin delivery by Pseudomonas aeruginosa rhamnolipids induces the antimicrobial protein psoriasin in human skin. PLoS One 6: e16433. DOI 10.1371/journal.pone.0016433. [Google Scholar] [CrossRef]
Mika F, Busse S, Possling A, Berkholz J, Tschowri N, Sommerfeldt N, Pruteanu M, Hengge R. (2012). Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Molecular Microbiology 84: 51–65. DOI 10.1111/j.1365-2958.2012.08002.x. [Google Scholar] [CrossRef]
Minato T, Maeda T, Fujisawa Y, Tsuji H, Nomoto K, Ohno K, Hirayama M. (2017). Progression of Parkinson’s disease is associated with gut dysbiosis: Two-year follow-up study. PLoS One 12: e0187307. DOI 10.1371/journal.pone.0187307. [Google Scholar] [CrossRef]
Monteiro C, Papenfort K, Hentrich K, Ahmad I, Le Guyon S, Reimann R, Grantcharova N, Romling U. (2012). Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biology 9: 489–502. DOI 10.4161/rna.19682. [Google Scholar] [CrossRef]
Morrison DJ, Preston T. (2016). Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7: 189–200. DOI 10.1080/19490976.2015.1134082. [Google Scholar] [CrossRef]
Narita S, Matsuyama S, Tokuda H. (2004). Lipoprotein trafficking in Escherichia coli. Archives of Microbiology 182: 1–6. DOI 10.1007/s00203-004-0682-4. [Google Scholar] [CrossRef]
Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. (2005). Structure of the cross-β spine of amyloid-like fibrils. Nature 435: 773–778. DOI 10.1038/nature03680. [Google Scholar] [CrossRef]
Nenninger AA, Robinson LS, Hammer ND, Epstein EA, Badtke MP, Hultgren SJ, Chapman MR. (2011). CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Molecular Microbiology 81: 486–499. DOI 10.1111/j.1365-2958.2011.07706.x. [Google Scholar] [CrossRef]
Nenninger AA, Robinson LS, Hultgren SJ. (2009). Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proceedings of the National Academy of Sciences of the United States of America 106: 900–905. DOI 10.1073/pnas.0812143106. [Google Scholar] [CrossRef]
Nielsen HH, Qiu J, Friis S, Wermuth L, Ritz B. (2012). Treatment for Helicobacter pylori infection and risk of Parkinson’s disease in Denmark. European Journal of Neurology 19: 864–869. DOI 10.1111/j.1468-1331.2011.03643.x. [Google Scholar] [CrossRef]
Nighot M, Al-Sadi R, Guo S, Rawat M, Nighot P, Watterson MD, Ma TY. (2017). Lipopolysaccharide-induced increase in intestinal epithelial tight permeability is mediated by toll-like receptor 4/myeloid differentiation primary response 88 (MyD88) activation of myosin light chain kinase expression. American Journal of Pathology 187: 2698–2710. DOI 10.1016/j.ajpath.2017.08.005. [Google Scholar] [CrossRef]
Nilsson MR. (2004). Techniques to study amyloid fibril formation in vitro. Methods 34: 151–160. DOI 10.1016/j.ymeth.2004.03.012. [Google Scholar] [CrossRef]
Nishimori JH, Newman TN, Oppong GO, Rapsinski GJ, Yen JH, Biesecker SG, Wilson RP, Butler BP, Winter MG, Tsolis RM, Ganea D, Tukel C. (2012). Microbial amyloids induce interleukin 17A (IL-17A) and IL-22 responses via Toll-like receptor 2 activation in the intestinal mucosa. Infection and Immunity 80: 4398–4408. DOI 10.1128/IAI.00911-12. [Google Scholar] [CrossRef]
Nishiwaki H, Ito M, Ishida T, Hamaguchi T, Maeda T, Kashihara K, Tsuboi Y, Ueyama J, Shimamura T, Mori H, Kurokawa K, Katsuno M, Hirayama M, Ohno K. (2020). Meta-analysis of gut dysbiosis in Parkinson’s disease. Movement Disorders 35: 1626–1635. DOI 10.1002/mds.28119. [Google Scholar] [CrossRef]
Nuzum ND, Loughman A, Szymlek-Gay EA, Hendy A, Teo WP, Macpherson H. (2020). Gut microbiota differences between healthy older adults and individuals with Parkinson’s disease: A systematic review. Neuroscience & Biobehavioral Reviews 112: 227–241. DOI 10.1016/j.neubiorev.2020.02.003. [Google Scholar] [CrossRef]
O’toole G, Kaplan HB, Kolter R. (2000). Biofilm formation as microbial development. Annual Review of Microbiology 54: 49–79. DOI 10.1146/annurev.micro.54.1.49. [Google Scholar] [CrossRef]
Ogasawara H, Yamada K, Kori A, Yamamoto K, Ishihama A (2010a). Regulation of the Escherichia coli csgD promoter: Interplay between five transcription factors. Microbiology 156: 2470–2483. DOI 10.1099/mic.0.039131-0. [Google Scholar] [CrossRef]
Ogasawara H, Yamamoto K, Ishihama A (2010b). Regulatory role of MlrA in transcription activation of csgD, the master regulator of biofilm formation in Escherichia coli. FEMS Microbiology Letters 312: 160–168. DOI 10.1111/j.1574-6968.2010.02112.x. [Google Scholar] [CrossRef]
Ogasawara H, Yamamoto K, Ishihama A. (2011). Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. Journal of Bacteriology 193: 2587–2597. DOI 10.1128/JB.01468-10. [Google Scholar] [CrossRef]
Ohta S. (2014). Molecular hydrogen as a preventive and therapeutic medical gas: Initiation, development and potential of hydrogen medicine. Pharmacology & Therapeutics 144: 1–11. DOI 10.1016/j.pharmthera.2014.04.006. [Google Scholar] [CrossRef]
Olsen A, Arnqvist A, Hammar M, Normark S (1993a). Environmental regulation of curli production in Escherichia coli. Infectious Agents & Disease 2: 272–274. [Google Scholar]
Olsen A, Arnqvist A, Hammar M, Sukupolvi S, Normark S (1993b). The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Molecular Microbiology 7: 523–536. DOI 10.1111/j.1365-2958.1993.tb01143.x. [Google Scholar] [CrossRef]
Olsen A, Jonsson A, Normark S. (1989). Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338: 652–655. DOI 10.1038/338652a0. [Google Scholar] [CrossRef]
Oppong GO, Rapsinski GJ, Newman TN, Nishimori JH, Biesecker SG, Tukel C. (2013). Epithelial cells augment barrier function via activation of the Toll-like receptor 2/phosphatidylinositol 3-kinase pathway upon recognition of Salmonella enterica serovar Typhimurium curli fibrils in the gut. Infection & Immunity 81: 478–486. DOI 10.1128/IAI.00453-12. [Google Scholar] [CrossRef]
Oppong GO, Rapsinski GJ, Tursi SA, Biesecker SG, Klein-Szanto AJ, Goulian M, Mccauley C, Healy C, Wilson RP, Tukel C. (2015). Biofilm-associated bacterial amyloids dampen inflammation in the gut: Oral treatment with curli fibres reduces the severity of hapten-induced colitis in mice. NPJ Biofilms and Microbiomes 1: 1–8. DOI 10.1038/npjbiofilms.2015.19. [Google Scholar] [CrossRef]
Ostojic SM. (2018). Inadequate production of H2 by gut microbiota and Parkinson disease. Trends in Endocrinology & Metabolism 29: 286–288. DOI 10.1016/j.tem.2018.02.006. [Google Scholar] [CrossRef]
Otzen D. (2010). Functional amyloid: Turning swords into plowshares. Prion 4: 256–264. DOI 10.4161/pri.4.4.13676. [Google Scholar] [CrossRef]
Paiva I, Pinho R, Pavlou MA, Hennion M, Wales P, Schutz AL, Rajput A, Szego EM, Kerimoglu C, Gerhardt E, Rego AC, Fischer A, Bonn S, Outeiro TF. (2017). Sodium butyrate rescues dopaminergic cells from alpha-synuclein-induced transcriptional deregulation and DNA damage. Human Molecular Genetics 26: 2231–2246. DOI 10.1093/hmg/ddx114. [Google Scholar] [CrossRef]
Pamp SJ, Tolker-Nielsen T. (2007). Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. Journal of Bacteriology 189: 2531–2539. DOI 10.1128/JB.01515-06. [Google Scholar] [CrossRef]
Parashar A, Udayabanu M. (2017). Gut microbiota: Implications in Parkinson’s disease. Parkinsonism & Related Disorders 38: 1–7. DOI 10.1016/j.parkreldis.2017.02.002. [Google Scholar] [CrossRef]
Park S, Kim J, Chun J, Han K, Soh H, Kang EA, Lee HJ, Im JP, Kim JS. (2019). Patients with inflammatory bowel disease are at an increased risk of Parkinson’s disease: A South Korean Nationwide Population-Based Study. Journal of Clinical Medicine 8: 1191. DOI 10.3390/jcm8081191. [Google Scholar] [CrossRef]
Perez-Pardo P, Dodiya HB, Engen PA, Forsyth CB, Huschens AM, Shaikh M, Voigt RM, Naqib A, Green SJ, Kordower JH, Shannon KM, Garssen J, Kraneveld AD, Keshavarzian A. (2019). Role of TLR4 in the gut-brain axis in Parkinson’s disease: A translational study from men to mice. Gut 68: 829–843. DOI 10.1136/gutjnl-2018-316844. [Google Scholar] [CrossRef]
Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, Shulman GI. (2016). Acetate mediates a microbiome–brain–β-cell axis to promote metabolic syndrome. Nature 534: 213–217. DOI 10.1038/nature18309. [Google Scholar] [CrossRef]
Peter I, Dubinsky M, Bressman S, Park A, Lu C, Chen N, Wang A. (2018). Anti–tumor necrosis factor therapy and incidence of Parkinson disease among patients with inflammatory bowel disease. JAMA Neurology 75: 939–946. DOI 10.1001/jamaneurol.2018.0605. [Google Scholar] [CrossRef]
Pietrucci D, Cerroni R, Unida V, Farcomeni A, Pierantozzi M, Mercuri NB, Biocca S, Stefani A, Desideri A. (2019). Dysbiosis of gut microbiota in a selected population of Parkinson’s patients. Parkinsonism & Related Disorders 65: 124–130. DOI 10.1016/j.parkreldis.2019.06.003. [Google Scholar] [CrossRef]
Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. (1999). Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. Journal of Bacteriology 181: 5993–6002. DOI 10.1128/JB.181.19.5993-6002.1999. [Google Scholar] [CrossRef]
Reshamwala SM, Noronha SB. (2011). Biofilm formation in Escherichia coli cra mutants is impaired due to down-regulation of curli biosynthesis. Archives of Microbiology 193: 711–722. DOI 10.1007/s00203-011-0708-7. [Google Scholar] [CrossRef]
Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. (2006). Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Molecular Microbiology 59: 870–881. DOI 10.1111/j.1365-2958.2005.04997.x. [Google Scholar] [CrossRef]
Romling U. (2005). Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cellular and Molecular Life Sciences 62: 1234–1246. DOI 10.1007/s00018-005-4557-x. [Google Scholar] [CrossRef]
Romling U, Rohde M, Olsen A, Normark S, Reinkoster J. (2000). AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Molecular Microbiology 36: 10–23. DOI 10.1046/j.1365-2958.2000.01822.x. [Google Scholar] [CrossRef]
Rouse SL, Hawthorne WJ, Berry JL, Chorev DS, Ionescu SA, Lambert S, Stylianou F, Ewert W, Mackie U, Morgan RML, Otzen D, Herbst FA, Nielsen PH, Dueholm M, Bayley H, Robinson CV, Hare S, Matthews S. (2017). A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis. Nature Communications 8: 263. DOI 10.1038/s41467-017-00361-6. [Google Scholar] [CrossRef]
Rudd KE. (2000). EcoGene: A genome sequence database for Escherichia coli K-12. Nucleic Acids Research 28: 60–64. DOI 10.1093/nar/28.1.60. [Google Scholar] [CrossRef]
Sampson TR, Challis C, Jain N, Moiseyenko A, Ladinsky MS, Shastri GG, Thron T, Needham BD, Horvath I, Debelius JW, Janssen S, Knight R, Wittung-Stafshede P, Gradinaru V, Chapman M, Mazmanian SK. (2020). A gut bacterial amyloid promotes alpha-synuclein aggregation and motor impairment in mice. eLife 9: e53111. DOI 10.7554/eLife.53111. [Google Scholar] [CrossRef]
Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet MF, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian SK. (2016). Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167: 1469–1480 e1412. DOI 10.1016/j.cell.2016.11.018. [Google Scholar] [CrossRef]
Sanchez M, Aranda FJ, Espuny MJ, Marques A, Teruel JA, Manresa A, Ortiz A. (2008). Thermodynamic and structural changes associated with the interaction of a dirhamnolipid biosurfactant with bovine serum albumin. Langmuir 24: 6487–6495. DOI 10.1021/la800636s. [Google Scholar] [CrossRef]
Sanders ME. (2008). Probiotics: Definition, sources, selection, and uses. Clinical Infectious Diseases 46: S58–S61, discussion S144–151. [Google Scholar]
Scheperjans F. (2016). Gut microbiota, 1013 new pieces in the Parkinson’s disease puzzle. Current Opinion in Neurology 29: 773–780. DOI 10.1097/WCO.0000000000000389. [Google Scholar] [CrossRef]
Scheperjans F, Aho V, Pereira PA, Koskinen K, Paulin L, Pekkonen E, Haapaniemi E, Kaakkola S, Eerola-Rautio J, Pohja M, Kinnunen E, Murros K, Auvinen P. (2015). Gut microbiota are related to Parkinson’s disease and clinical phenotype. Movement Disorders 30: 350–358. DOI 10.1002/mds.26069. [Google Scholar] [CrossRef]
Schonfeld P, Wojtczak L. (2016). Short- and medium-chain fatty acids in energy metabolism: The cellular perspective. Journal of Lipid Research 57: 943–954. DOI 10.1194/jlr.R067629. [Google Scholar] [CrossRef]
Schwiertz A, Spiegel J, Dillmann U, Grundmann D, Burmann J, Fassbender K, Schafer KH, Unger MM. (2018). Fecal markers of intestinal inflammation and intestinal permeability are elevated in Parkinson’s disease. Parkinsonism & Related Disorders 50: 104–107. DOI 10.1016/j.parkreldis.2018.02.022. [Google Scholar] [CrossRef]
Sharma N, Nehru B. (2015). Characterization of the lipopolysaccharide induced model of Parkinson’s disease: Role of oxidative stress and neuroinflammation. Neurochemistry International 87: 92–105. DOI 10.1016/j.neuint.2015.06.004. [Google Scholar] [CrossRef]
Sharma S, Taliyan R, Singh S. (2015). Beneficial effects of sodium butyrate in 6-OHDA induced neurotoxicity and behavioral abnormalities: Modulation of histone deacetylase activity. Behavioural Brain Research 291: 306–314. DOI 10.1016/j.bbr.2015.05.052. [Google Scholar] [CrossRef]
Shewmaker F, Mcglinchey RP, Thurber KR, Mcphie P, Dyda F, Tycko R, Wickner RB. (2009). The functional curli amyloid is not based on in-register parallel β-sheet structure. Journal of Biological Chemistry 284: 25065–25076. DOI 10.1074/jbc.M109.007054. [Google Scholar] [CrossRef]
Shewmaker F, Mcglinchey RP, Wickner RB. (2011). Structural insights into functional and pathological amyloid. Journal of Biological Chemistry 286: 16533–16540. DOI 10.1074/jbc.R111.227108. [Google Scholar] [CrossRef]
Shimada T, Katayama Y, Kawakita S, Ogasawara H, Nakano M, Yamamoto K, Ishihama A. (2012). A novel regulator RcdA of the csgD gene encoding the master regulator of biofilm formation in Escherichia coli. Microbiology Open 1: 381–394. DOI 10.1002/mbo3.42. [Google Scholar] [CrossRef]
Shin C, Lim Y, Lim H, Ahn TB. (2020). Plasma short-chain fatty acids in patients with Parkinson’s disease. Movement Disorders 35: 1021–1027. DOI 10.1002/mds.28016. [Google Scholar] [CrossRef]
Sotirova A, Spasova D, Vasileva-Tonkova E, Galabova D. (2009). Effects of rhamnolipid-biosurfactant on cell surface of Pseudomonas aeruginosa. Microbiological Research 164: 297–303. DOI 10.1016/j.micres.2007.01.005. [Google Scholar] [CrossRef]
Soto C. (2003). Unfolding the role of protein misfolding in neurodegenerative diseases. Nature Reviews Neuroscience 4: 49–60. DOI 10.1038/nrn1007. [Google Scholar] [CrossRef]
Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M. (1998). alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with lewy bodies. Proceedings of the National Academy of Sciences of the United States of America 95: 6469–6473. DOI 10.1073/pnas.95.11.6469. [Google Scholar] [CrossRef]
Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. (1997). α-synuclein in lewy bodies. Nature 388: 839–840. DOI 10.1038/42166. [Google Scholar] [CrossRef]
St Laurent R, O.’brien LM, Ahmad ST. (2013). Sodium butyrate improves locomotor impairment and early mortality in a rotenone-induced Drosophila model of Parkinson’s disease. Neuroscience 246: 382–390. DOI 10.1016/j.neuroscience.2013.04.037. [Google Scholar] [CrossRef]
Stolzenberg E, Berry D, Yang D, Lee E Y, Kroemer A, Kaufman S, Wong GCL, Oppenheim JJ, Sen S, Fishbein T, Bax A, Harris B, Barbut D, Zasloff M A. (2017). A role for neuronal alpha-synuclein in gastrointestinal immunity. Journal of Innate Immunity 9: 456–463. DOI 10.1159/000477990. [Google Scholar] [CrossRef]
Suzuki A, Ito M, Hamaguchi T, Mori H, Takeda Y, Baba R, Watanabe T, Kurokawa K, Asakawa S, Hirayama M, Ohno K. (2018). Quantification of hydrogen production by intestinal bacteria that are specifically dysregulated in Parkinson’s disease. PLoS One 13: e0208313. DOI 10.1371/journal.pone.0208313. [Google Scholar] [CrossRef]
Svensson E, Horvath-Puho E, Thomsen RW, Djurhuus JC, Pedersen L, Borghammer P, Sorensen HT. (2015). Vagotomy and subsequent risk of Parkinson’s disease. Annals of Neurology 78: 522–529. DOI 10.1002/ana.24448. [Google Scholar] [CrossRef]
Taylor JD, Zhou Y, Salgado PS, Patwardhan A, Mcguffie M, Pape T, Grabe G, Ashman E, Constable SC, Simpson PJ, Lee WC, Cota E, Chapman MR, Matthews SJ. (2011). Atomic resolution insights into curli fiber biogenesis. Structure 19: 1307–1316. DOI 10.1016/j.str.2011.05.015. [Google Scholar] [CrossRef]
Tukel C, Nishimori JH, Wilson RP, Winter MG, Keestra AM, Van Putten JP, Baumler AJ. (2010). Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cellular Microbiology 12: 1495–1505. DOI 10.1111/j.1462-5822.2010.01485.x. [Google Scholar] [CrossRef]
Tukel C, Raffatellu M, Humphries AD, Wilson RP, Andrews-Polymenis HL, Gull T, Figueiredo JF, Wong MH, Michelsen KS, Akcelik M, Adams LG, Baumler AJ. (2005). CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Molecular Microbiology 58: 289–304. DOI 10.1111/j.1365-2958.2005.04825.x. [Google Scholar] [CrossRef]
Tursi SA, Tukel C. (2018). Curli-containing enteric biofilms inside and out: Matrix composition, immune recognition, and disease implications. Microbiology and Molecular Biology Reviews 82: e00028. DOI 10.1128/MMBR.00028-18. [Google Scholar] [CrossRef]
Tycko R. (2004). Progress towards a molecular-level structural understanding of amyloid fibrils. Current Opinion in Structural Biology 14: 96–103. DOI 10.1016/j.sbi.2003.12.002. [Google Scholar] [CrossRef]
Uesaka T, Young HM, Pachnis V, Enomoto H. (2016). Development of the intrinsic and extrinsic innervation of the gut. Developmental Biology 417: 158–167. DOI 10.1016/j.ydbio.2016.04.016. [Google Scholar] [CrossRef]
Unger MM, Spiegel J, Dillmann KU, Grundmann D, Philippeit H, Burmann J, Fassbender K, Schwiertz A, Schafer KH. (2016). Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Parkinsonism & Related Disorders 32: 66–72. DOI 10.1016/j.parkreldis.2016.08.019. [Google Scholar] [CrossRef]
Van Gennip M, Christensen LD, Alhede M, Phipps R, Jensen PO, Christophersen L, Pamp SJ, Moser C, Mikkelsen PJ, Koh AY, Tolker-Nielsen T, Pier GB, Hoiby N, Givskov M, Bjarnsholt T. (2009). Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS 117: 537–546. DOI 10.1111/j.1600-0463.2009.02466.x. [Google Scholar] [CrossRef]
Van ISCD, Derkinderen P. (2019). The intestinal barrier in Parkinson’s disease: Current state of knowledge. Journal of Parkinson’s Disease 9: S323–S329. DOI 10.3233/JPD-191707. [Google Scholar] [CrossRef]
Venegas C, Heneka MT. (2017). Danger-associated molecular patterns in Alzheimer’s disease. Journal of Leukocyte Biology 101: 87–98. DOI 10.1189/jlb.3MR0416-204R. [Google Scholar] [CrossRef]
Villumsen M, Aznar S, Pakkenberg B, Jess T, Brudek T. (2019). Inflammatory bowel disease increases the risk of Parkinson’s disease: A Danish nationwide cohort study 1977-2014. Gut 68: 18–24. DOI 10.1136/gutjnl-2017-315666. [Google Scholar] [CrossRef]
Walker LC, Schelle J, Jucker M. (2016). The prion-like properties of amyloid-β assemblies: Implications for Alzheimer’s disease. Cold Spring Harbor Perspectives in Medicine 6: a024398. DOI 10.1101/cshperspect.a024398. [Google Scholar] [CrossRef]
Wang S, Yu S, Zhang Z, Wei Q, Yan L, Ai G, Liu H, Ma LZ. (2014). Coordination of swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharide production in Pseudomonas aeruginosa. Applied and Environmental Microbiology 80: 6724–6732. DOI 10.1128/AEM.01237-14. [Google Scholar] [CrossRef]
Wang X, Smith DR, Jones JW, Chapman MR. (2007). In vitro polymerization of a functional Escherichia coli amyloid protein. Journal of Biological Chemistry 282: 3713–3719. DOI 10.1074/jbc.M609228200. [Google Scholar] [CrossRef]
Weimers P, Halfvarson J, Sachs MC, Saunders-Pullman R, Ludvigsson JF, Peter I, Burisch J, Olen O. (2019). Inflammatory bowel disease and Parkinson’s disease: A Nationwide Swedish Cohort Study. Inflammatory Bowel Diseases 25: 111–123. DOI 10.1093/ibd/izy190. [Google Scholar] [CrossRef]
Wong MW, Yi CH, Liu TT, Lei WY, Hung JS, Lin CL, Lin SZ, Chen CL. (2018). Impact of vegan diets on gut microbiota: An update on the clinical implications. Ci Ji Yi Xue Za Zhi 30: 200–203. [Google Scholar]
Yoritaka A, Takanashi M, Hirayama M, Nakahara T, Ohta S, Hattori N. (2013). Pilot study of H2 therapy in Parkinson’s disease: A randomized double-blind placebo-controlled trial. Movement Disorders 28: 836–839. DOI 10.1002/mds.25375. [Google Scholar] [CrossRef]
Zhang W, Wang T, Pei Z, Miller DS, Wu X, Block ML, Wilson B, Zhang W, Zhou Y, Hong JS, Zhang J. (2005). Aggregated α-synuclein activates microglia: A process leading to disease progression in Parkinson’s disease. FASEB Journal 19: 533–542. DOI 10.1096/fj.04-2751com. [Google Scholar] [CrossRef]
Zheng D, Constantinidou C, Hobman JL, Minchin SD. (2004). Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Research 32: 5874–5893. DOI 10.1093/nar/gkh908. [Google Scholar] [CrossRef]
Zhou Y, Smith D, Leong BJ, Brannstrom K, Almqvist F, Chapman MR. (2012). Promiscuous cross-seeding between bacterial amyloids promotes interspecies biofilms. Journal of Biological Chemistry 287: 35092–35103. DOI 10.1074/jbc.M112.383737. [Google Scholar] [CrossRef]
Zogaj X, Bokranz W, Nimtz M, Romling U. (2003). Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infection and Immunity 71: 4151–4158. DOI 10.1128/IAI.71.7.4151-4158.2003. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |