2021 45(1): 27-39
DOI:10.32604/biocell.2021.012601

www.techscience.com/journal/biocell
| BIOCELL 2021 45(1): 27-39 DOI:10.32604/biocell.2021.012601 |  www.techscience.com/journal/biocell |
Basing on microRNA-mRNA analysis identifies microRNA in exosomes associated with wound repair of diabetic ulcers
1Department of Burn & Plastic Surgery, General Hospital of Southern Theatre Command of PLA, Guangzhou, 510010, China
2Huabo Post-Doctoral Research Center, Biological Pharmaceutical Research Institute, Guangzhou, 510515, China
3Department of Plastic and Cosmetic Surgery, The First Clinical Medical College of Southern Medical University, Guangzhou, 510080, China
4Department of Dermatology, The Second Affiliated Hospital, Guangzhou University of Chinese Medicine, Guangdong Provincial Hospital of Chinese Medicine, Guangzhou, 510120, China
*Address correspondence to: Qin Li, gzzxwk@126.com; Biao Cheng, chengbiaocheng@163.com
Received: 06 July 2020; Accepted: 23 September 2020
Abstract: The diabetic ulcer is one of the serious complications of diabetes. In this study, we aimed to establish an exosomal microRNA (miRNA)-targeted messenger RNA (mRNA) regulatory network for screening new biomarkers for diabetic ulcer treatment. For this purpose, exosomes were extracted from bone marrow stem cells (BMSCs) collected from diabetic ulcer patients and healthy adults. The miRNAs in exosomes was detected by high-throughput sequencing analysis. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the differential miRNAs were performed. The miRNA-mRNA regulatory network between candidate miRNAs and their target genes were constructed by Cytoscape software basing on mRNA expression profiles data of diabetic ulcer patients from Gene Expression Omnibus (GEO). GO and KEGG analyses of the core genes were performed. A total of 63 differential expressed miRNAs in BMSCs exosomes were identified between diabetic ulcer patients and healthy adults. The GO analysis of miRNAs showed that it was mainly related to signal transduction and intercellular transport, and KEGG analysis showed that it was related to the vascular endothelial growth factor (VEGF) signaling pathway. The core genes of the miRNA-mRNA network were thioredoxin interacting protein (TXNIP), cell division cycle 14A (CDC14A), cache domain containing 1 (CACHD1), interferon-induced protein 44 like (IFI44L), late cornified envelope 1AL (CE1A), leucine-rich repeats and immunoglobulin-like domains 2 (LRIG2), palmdelphin (PALMD) and serine and arginine-rich splicing factor 11 (SRSF11). GO analysis of the core genes was related to platelet-derived growth factor receptor signaling pathway. The KEGG analysis of the core genes was related to the cell cycle and nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway. A potential miRNA-mRNA regulatory network provides a comprehensive understanding of the molecular mechanisms and promising new targets such as miR-130a-5p, SESN2, LRIG2, and CDC14A for the wound repair of diabetic ulcers.
Keywords: Exosome; Diabetic ulcer; Angiogenesis; Biological analysis; miRNA
Abbreviations
| miRNAs: | microRNAs |
| DM: | Diabetes mellitus |
| AGEs: | advancedglycation end products |
| BMSCs: | bone marrow stromal cells |
| CD63: | lysosomal membrane-associated glycoprotein 3 |
| TXNIP: | Thioredoxin interacting protein |
| CDC14A: | Cell division cycle 14A |
| CACHD1: | Cache domain containing 1 |
| IFI44L: | interferon induced protein 44 like |
| LCE1A: | late cornified envelope 1A |
| LRIG2: | leucine rich repeats and immunoglobulin like domains 2 |
| PALMD: | Palmdelphin |
| SRSF11: | serine and arginine rich splicing factor 11 |
| FIH: | hypoxia inducible factor inhibitor |
| ECM: | extracellular matrix |
| DDL4: | Delta like ligand 4 |
| FDR: | false detection rate |
| 3´UTR: | 3´ untranslated region |
| VEGF: | vascular endothelial growth factor |
| ERK: | extracellular signal-regulated kinase |
| STAT3: | signal transducer and activator of transcription 3 |
| HGF: | epatocyte growth factor |
| IGF: | insulin-like growth factor |
| NGF: | nerve growth factor |
| SDF-1: | tromal cell-derived factor-1 |
| AGEs: | dvanced glycation end products |
| hESCs: | human Embryonic Stem Cells |
| EMT: | epithelial-to-mesenchymal transforming |
Introduction
Diabetes mellitus (DM) is a metabolic disorder syndrome caused by various pathogenic factors such as genetic factors, obesity, immune dysfunction, microbial infection, toxins, free radicals, and mental factors (Schmidt, 2018). Nowadays, with the improvement of living standards, lifestyle changes, and social aging, the incidence of diabetes was increasing year by year (Schmidt, 2018). The diabetic ulcer was one of the serious complications of DM; the risk of foot ulcers in diabetic patients is up to 25% (Al-Rubeaan et al., 2015). A diabetic foot ulcer is the leading cause of low distal amputation (Millington and Ellenzweig, 2005). Delayed or nonunion of wound healing after skin injury is a pressing problem in clinical practice. Some studies have shown that the accumulation of advanced glycation end products (AGEs) and hyperglycemia will not only damage peripheral blood vessels and microvessels but also inhibit the expression of many kinds of neurotrophic factors and vascular factors, which may be the main factors that cause diabetic skin ulcers difficult to heal (Bukowiecki et al., 2017; Lalla et al., 2000; Negre-Salvayre et al., 2009; Sun et al., 2016). A variety of physiological or pathophysiological events such as the decreased proliferation of fibroblasts, decreased growth factors, reduced keratinocytes, reduced angiogenesis, abnormal collagen deposition, a small number of macrophages, and impaired function, are related to poor wound healing in DM (Bukowiecki et al., 2017).
Exosomes are extracellular vesicles with a diameter of 30–150 nm; they have a small bilayer lipid membrane, which can enter the extracellular matrix directly by budding and then release the internal components after being ingested by the target cells to complete cell communication and information exchange (Pegtel and Gould, 2019). Exosomes as a subcellular component secreted by cells, which were widely involved in cell communication and can play a dominant role in tissue repair and regeneration (Chen et al., 2017; Han et al., 2016). They can be identified by their expression of exosome-associated markers such as Tsg101 and CD63 (Wubbolts et al., 2003). In recent years, there are many reports on the application of mesenchymal stem cell-derived exosomes in the treatment of diabetic ulcers (Geiger et al., 2015; Zhu et al., 2018). It has been found that the bone marrow stromal cells (BMSCs) exosomes can promote the proliferation and migration of fibroblasts extracted from diabetic chronic ulcer wounds in a concentration-and dose-dependent manner (Shabbir et al., 2015). The exosomes derived from lysosomal membrane-associated glycoprotein 3 (CD63)+ BMSCs have a stronger ability to uptake exogenous Wnt family member 3A (Wnt3a) and promote the proliferation and migration of fibroblasts through the Wnt/β-catenin signal pathway(Hu et al., 2016). BMSCs exosomes can also activate other pathways that play an important role in skin wound healing, such as AKT serine/threonine kinase (AKT), extracellular signal-regulated kinase (ERK), and signal transducer and activator of transcription 3 (STAT3) (Ding et al., 2019), and promote hepatocyte growth factor (HGF), insulin-like growth factor (IGF), nerve growth factor (NGF) and stromal cell-derived factor-1 (SDF-1) and other growth factors (Cui et al., 2016; Dai et al., 2019; Umezu et al., 2017).
MicroRNA (miRNA) is a non-coding RNA with a length of 22-24 nucleotides (Bartel, 2004). It participates in the regulation of the post-transcriptional expression of genes. Mature miRNA can bind to the 3´ untranslated region (3´-UTR) of target gene messenger RNA (mRNA), and negatively regulate target genes by degrading mRNA and inhibit protein translation of target genes (Fang and Rajewsky, 2011). MiRNAs is an important participant in the healing of diabetic ulcers, miRNAs can promote the healing of diabetic ulcers by up-regulating or down-regulating the expression of some genes and activating specific signaling pathways (Jhamb et al., 2016). MiRNA is an important substance for exosomes to promote angiogenesis. It is found that miR-31 can promote the proliferation and migration of endothelial cells and endothelial progenitor cells and induce angiogenesis by inhibiting hypoxia-inducible factor inhibitor (FIH) (Liu et al., 2010), miR-125a can inhibit the expression of Delta-like ligand 4 (DDL4), and promotes endothelial cell formation (Liang et al., 2016). Exosomes of mesenchymal stem cells (MSCs) overexpressing miR126 can promote angiogenesis in the diabetic wound model (Zgheib et al., 2013).
In this study, we used high throughput sequencing of mRNA to analyze the difference in MSCs exosomal miRNAs expression between diabetic ulcer patients and healthy adults. We constructed a miRNA-mRNA network and analyzed the core genes in the network by bioinformatics analysis methods, which aimed to discover key targets related to wound healing.
The BMSCs were obtained from 5 male patients with diabetic foot ulcers and 5 male healthy adults in the General Hospital of Southern Theatre Command of the Chinese People’s Liberation Army (PLA) between 2018 JAN to 2019 JAN. The age of patients is 58 ± 10.5 years old, and the duration of DM is 8.5 ± 3.1 years. The age of healthy adults (57 ± 6.2 years old) was matched with diabetic ulcer patients. Informed consent was obtained from all subjects. Inclusion criteria: refer to the World Health Organization (WHO) diagnostic criteria for DM. Exclusion criteria: Severe purulent infections such as sepsis, severe heart, liver, and kidney diseases and malignant tumors, blood system diseases, connective tissue diseases, and mental diseases. All experimental procedures were approved by the Ethical Committee of General Hospital of Southern Theatre Command of PLA (No. 201712) and were performed in accordance with the Helsinki Declaration (2000).
In this study, human BMSCs (hBMSCs) were isolated and cultured by the whole bone marrow adherent culture method [18]. Bone marrow was diluted with phosphate buffer saline (PBS), and then centrifuged for 10 min at 4°C at 1500×g discarded the supernatant, and then washed the cells with PBS twice. The cell precipitation were collected and cultured in the complete medium [20% fetal bovine serum (FBS, Hyclone, Erembodegem-Aalst, Belgium), 2 mmol/L sodium pyruvate and 1 mmol/L L-glutamine in alpha-minimum essential medium (αMEM, Corning, USA)] in 37°C and 5% CO2 incubator, with adjusting the cell density to 1 × 109 cells/mL. After 72 h, the medium was half-changed every 2 to 3 days according to the cell growth condition.
Extraction and identification of hBMSCs exosome
In this study, we extract exosomes using the Exosomes Isolation Kit (#EQ806TC-1, SBI, Mountain View, USA) according to the manufacturer’s protocol. The hBMSCs derived from patients and healthy adults were cultured with serum-free medium. After 3 days, the culture medium was collected and centrifuged at 2000×g for 30 min. Then, the cells and debris were removed, and the supernatant was absorbed. Exosomes were then isolated by polymer precipitation using the Exomes Isolation Kit and resuspended in PBS.
Transmission electron microscopy
The exosome sample (10 μL) was dripped on the sample-carrying copper grids with diameter 2 mm for 10 min at room temperature, re-stained with 3% sodium phosphotungstate solution (pH 6.8) at room temperature for 5 min, washed gently with double distilled water and dried at room temperature for 2 min, and then observed and photographed under the transmission electron microscope (Carl Zeiss, Oberkochen, Germany).
Nanoparticle Tracking Analysis (NTA)
The size and number of exosomes were measured by Nanosight-NS500 (NanoSight, Amesbury, UK). A total of 1 μL exosome sample was diluted in an equal volume of filtered PBS to obtain no more than 100 particles in the NTA software screen. The modal size and the number of particles were measured in triplicates, and data were expressed as mean ± standard deviation (SD).
Exosomes protein extraction and western blotting analysis
Exosomes precipitates were extracted with protein lysate (25 mmol/L Tris-HCl PH7.6, 150 mmol/L NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, and 1 nmol/L PMSF) on ice for 30 min, and then centrifugation at 12000×g for 10 min at 4°C to collect the supernatant. Exosomes protein was quantified by the BCA method (Beyotime Biotechnology, Shanghai, China). In each group, 30 ug protein was mixed with 5× SDS loading buffer and denatured in a water bath at 95°C for 10 min. The denatured proteins were separated in 10% SDS-PAGE gel. The proteins were electroblotted onto PVDF membrane (Millipore Corporation, MA, US). Then, the PVDF membrane was blocked with 5% skim milk in Tris-buffered saline/Tween 20 (TBST; 10 mM Tris HCl, pH 8.0/150 mM NaCl, and 0.1% Tween 20), and incubated with the primary antibody anti-CD63 (1:100, Abcam, Cambridge, UK) and anti-TSG101 (1:500, Abcam, Cambridge, UK) overnight at 4°C. The membrane was washed with TBST 3 times and incubated with the second antibody conjugated with horseradish peroxidase (1:2000, Biotechnology, Shanghai, China) at room temperature for 1 h. The Enhanced chemiluminescence Kit (Biotechnology, Shanghai, China) was used for the detection, and the Quantity one program (BioRad, Hercules, CA) was used for photography and data analysis.
Total RNA extraction and high-throughput sequencing analysis
The total RNA of the exosome of BMSCs was extracted by ExoRN easy Serum/Plasma Kit (Qiagen, German). The total RNA was quantified by NanoDropND-2000 (Thermo Scientific), and the quality and purity of total Nanodrop were analyzed and detected by ND-1000 spectrophotometer (Nanodrop company, USA). Quality control qualified sample RNA integrity count >8.0, and the A260/A280 ratio between 1.8 to 2 can be carried out after the test. High-throughput sequencing analysis was performed by the ChiBiotech Company (http://www.chi-biotech.com). The fastx_toolkit software was used to remove the low-quality reads in the sequencing data and truncate the ends of the reads, and the reads with a length greater than 17 nt were retained. FANse3 ultra-high-precision sequence alignment algorithm was used to align the reads of each sample with the reference sequence (human mature miRNA). Gene expression was quantified using RPKM (Reads Per Kilo bases per Million reads) as the unit.
Skin mRNA expression profiling data download from the Gene Expression Omnibus (GEO) database (GSE.ncbi.nlm.nih.gov/geo/) GSE80178 datasets on the expression profiles of diabetic foot ulcers. Platforms: GPL 16686 [HuGene-2_0-st] Affymetrix Human Gene 2.0 ST Array (transcript [gene] version), contains >30000 coding transcripts mRNA. The microarray consisted of 6 diabetic foot ulcers and 3 healthy controls.
Data processing and differential gene analysis
The expression data were analyzed on the R (version 3.5.3) statistics environment (http://www.r-project.org). Affy, Limma, Pheatmap, ggplot2, and other software packages are used for data processing, and the RMA algorithm is used for background correction, standardization, and expression value calculation. The processed data were screened by Fold change (FC) and T-test for differential genes and defined as effective genes|log2FC| ≥2 (p < 0.05 or FDR < 0.05).
Enrichment analysis of GO analysis and KEGG pathways
The Gene Ontology (Pegtel and Gould) database mainly describes the functions of genes synthetically, including biological process (BP), cell composition (cellular component, CC), and molecular function (Molecular Function, MF). KEGG (Kyoto Encyclopedia of Genes and Genomes) is a comprehensive database that combines genomic, chemical, and functional data. In this study, both GO and KEGG enrichment analyses of miRNAs were performed using FunRich (version 3.1.3). The standard setting with a statistically significant difference was p < 0.05. The key genes GO and KEGG enrichment analyses were performed by R software (version 3.5.3), applied cluster profiler, org.Hs.eg.db, enrichplot, and ggplot2 package for data processing and visualization.
In this study, we have applied miRDB (http://mirdb.org), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) and TargetScan (http://www.targetscan.org/vert_72/) miRNA target gene prediction tool to predict miRNA target genes. In this study, we screened out the interactions between miRNA and targets that coexisted in at least two databases and identified on the 3’ untranslated region (3’UTR) of all known human genes for further analysis.
MiRNA-mRNA regulatory network construction
We use the Venny tool (Oliveros, 2007) to obtain the intersection of differentially expressed miRNAs target genes and differentially expressed mRNA. Cytoscape3.8.0 (https://cytoscape.org/) was used to construct and visualize the miRNAs-mRNA regulatory network.
Identification of exosome secreted by hBMSCs
The white precipitate collected was dissolved in PBS and observed by a transmission electron microscope that exosome was a vesicle structure with a diameter of 40–100 nm (Fig. 1A). The expression levels of CD63 and TSG101 were detected by Western Blot, and the results showed that the precipitates were strongly positive for CD63 and TSG101 (Fig. 1B). We also used NTA to detect the particle modal size and quantity. The results showed that the modal size of our extracted particles was 152.3 ± 11.2 nm in the diabetic ulcer group and 172.4 ± 8.5 nm in the healthy adult group. The particle size count was 1.36 ± 0.28 × 1010 p/mL in the diabetic ulcer group and 1.56 ± 0.33 × 1010 p/mL in the healthy adult group. There was no difference in the number and size of exosomes between the two groups (p > 0.05).
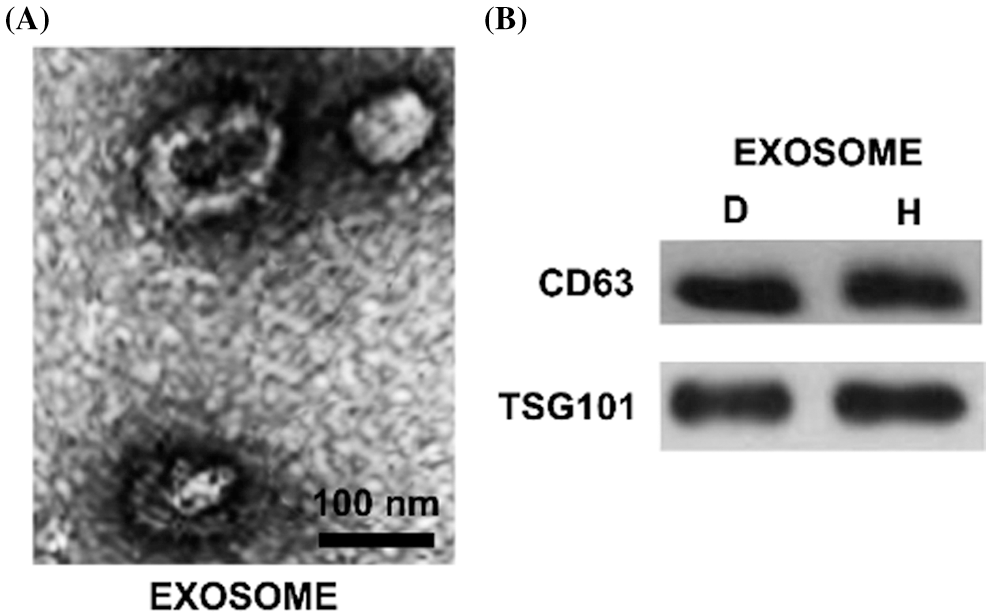
Figure 1: The characteristics of isolated exosomes. Morphology of hBMSCs derived exosomes were observed by transmission electron microscopy. The scale bar = 100 nm. B. Western blot analysis of protein extracted from exosomes. The exosome-associated markers CD63 and TSG101 were detected. C. The modal size and yield of exosomes were analyzed by NTA. The unit of particle size is nm; the unit of counting is p/mL. N: Normal healthy; D: Diabetic ulcer.
Different exosome MiRNAs profiles between diabetic ulcer patients and health subjects
In order to identify the difference of miRNAs expression in the hBMSCs exosomes of diabetic ulcer patients and healthy adults, we analyzed the miRNA-Seqdata, normalized the data of miRNA expression, calculated the logFC, FDR, and P values of miRNA in the two groups, and screened out p < 0.05 and |logFC| > 2. The results showed that a total of 63 miRNAs. Among them, 60 miRNAs were up-regulation and 3 miRNAs (hsa-miR-2116-3p, hsa-miR-1-5p and hsa-miR-130a-5p) were down-regulation. In up-regulated expression of miRNAs, hsa-miR-665, hsa-miR-6731-5p, hsa-miR-2115-5p, hsa-miR-873-5p, hsa-miR-214-3p, hsa-miR-2115-5p were more obvious up-regulated. The up-regulation of miRNAs were more common than the down-regulated (Fig. 2). It should be noted that in our results, FDR values are more than 0.05, which suggests that miRNAs between patients and healthy groups are not significantly regulated.
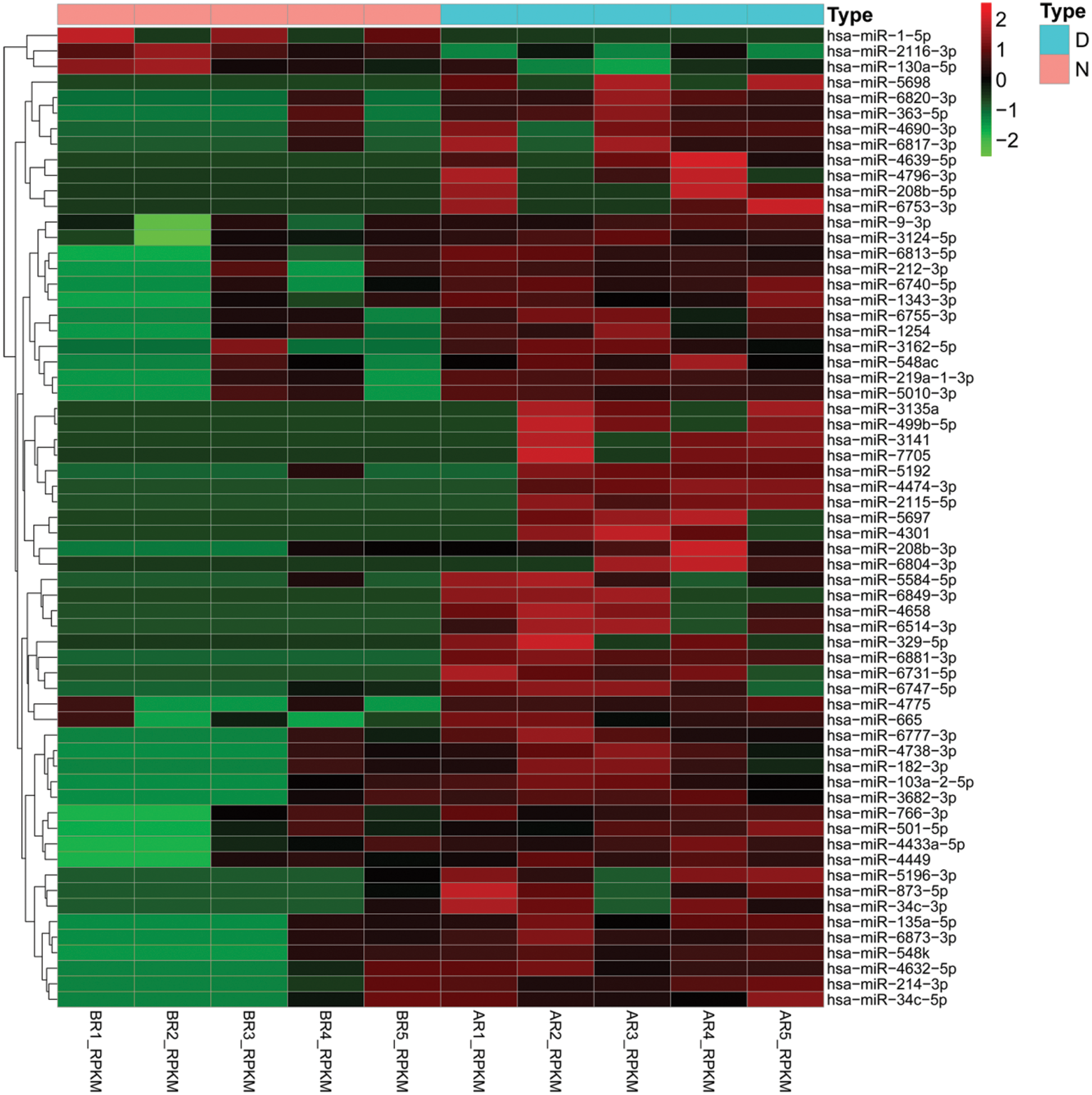
Figure 2: Differential expression of miRNAs in BMSCs exosomes.
Heatmaps demonstrate differential expressed miRNAs between diabetic ulcer patients (D, blue) and normal healthy adults (N, pink). The x-axis denotes differential expressed miRNAs, and the y-axis represents the samples. The expression values are shown in line with the color scale.
Functional annotation and pathway enrichment analysis of differential expressed miRNAs
In this study, we analyzed the function of differential expression of miRNAs in patients with diabetic ulcers and healthy adults. GO analysis showed that candidate target genes of differential expressed miRNAs were significantly enriched in CC terms including “Nucleus,” “Cytoplasm” and “Golgi apparatus” (Fig. 3A), MF terms including “Transcription factor activity” (Fig. 3B), and BP including “Signal transduction,” “Cell communication” and “Regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolism” (Fig. 3C). The GO results suggested that the differential expressed miRNA may play an important regulatory role in cell to cell communication.
KEGG pathway enrichment analysis was further conducted for candidate target genes of differential expressed miRNAs. The top 10 significantly enriched KEGG items were listed in Fig. 3D, including “TRAIL signaling pathway,” “Glypican pathway,” “Syndecan-1-mediated signaling events,” “Nectin adhesion pathway,” “IGF1 pathway,” “ErbB receptor signaling network,” “IL5-mediated signaling events,” “Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met),” “vascular endothelial growth factor (VEGF) and VEGF receptor (VEGFR) signaling network” and “Signaling events mediated by VEGFR1 and VEGFR2” (Fig. 3D). The differential expressed miRNAs associated with matched genes in VEGF/VEGFR pathway including down-regulated hsa-miR-1-5p and hsa-miR-130a-5p, and up-regulated hsa-miR-135a-5p, hsa-miR-665, hsa-miR-219a-1-3p, hsa-miR-212-3p, hsa-miR-34c-5p, hsa-miR-208b-3p, hsa-miR-9-3p and hsa-miR-873-5p, which may play an important regulatory role in promoting skin wound healing by promoting angiogenesis.
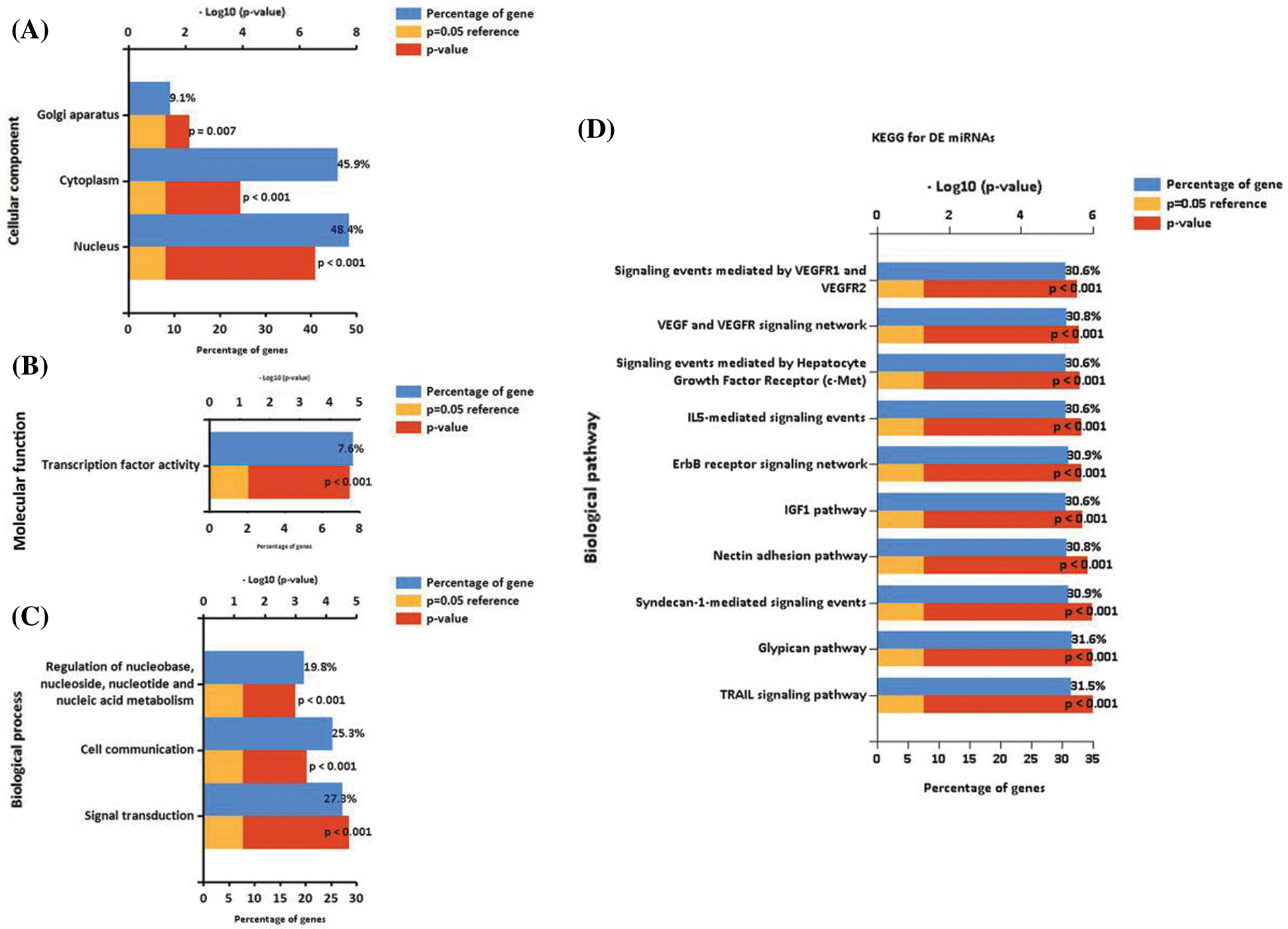
Figure 3: Enrichment analysis of miRNAs. GO functional annotation of differential expressed miRNAs. A. The significant enrichment items in CC analysis. B. The significant enrichment items in MF analysis. C. The significant enrichment items in BP analysis. D. The significant enrichment items in KEGG analysis.
Different mRNA profiles between DFS and NFS
In this study, we downloaded the expression profile data of diabetic foot ulcers in diabetic foot ulcer patients through the GEO database. We analyzed the mRNA expression profiling by array data through the Limma package of R software, normalized the data of mRNA expression, and calculated the logFC, FDR, and P values of mRNA in the two groups. Screened mRNA with FDR < 0.05 and |logFC| > 2, the results showed that a total of 28 mRNAs were differentially expressed, of which 16 mRNAs including LCE6A, LCE1A, CACHD1, TXNIP, ANXA9, IFI44, CA6, SRGAP2B, DLEU2L, IFI44L, SRSF11, HMGN2, CDC14A, PALMD, EFCAB7, and LRIG2 were down-regulated and 12 mRNAs, including SPRR3, SESN2, LCE3C, SPRR4, AJAP1, IVL, SPRR1B, SPRR1A, S100A7A, CDA, PRR9, and S100A9, were up-regulated (Fig. 4).
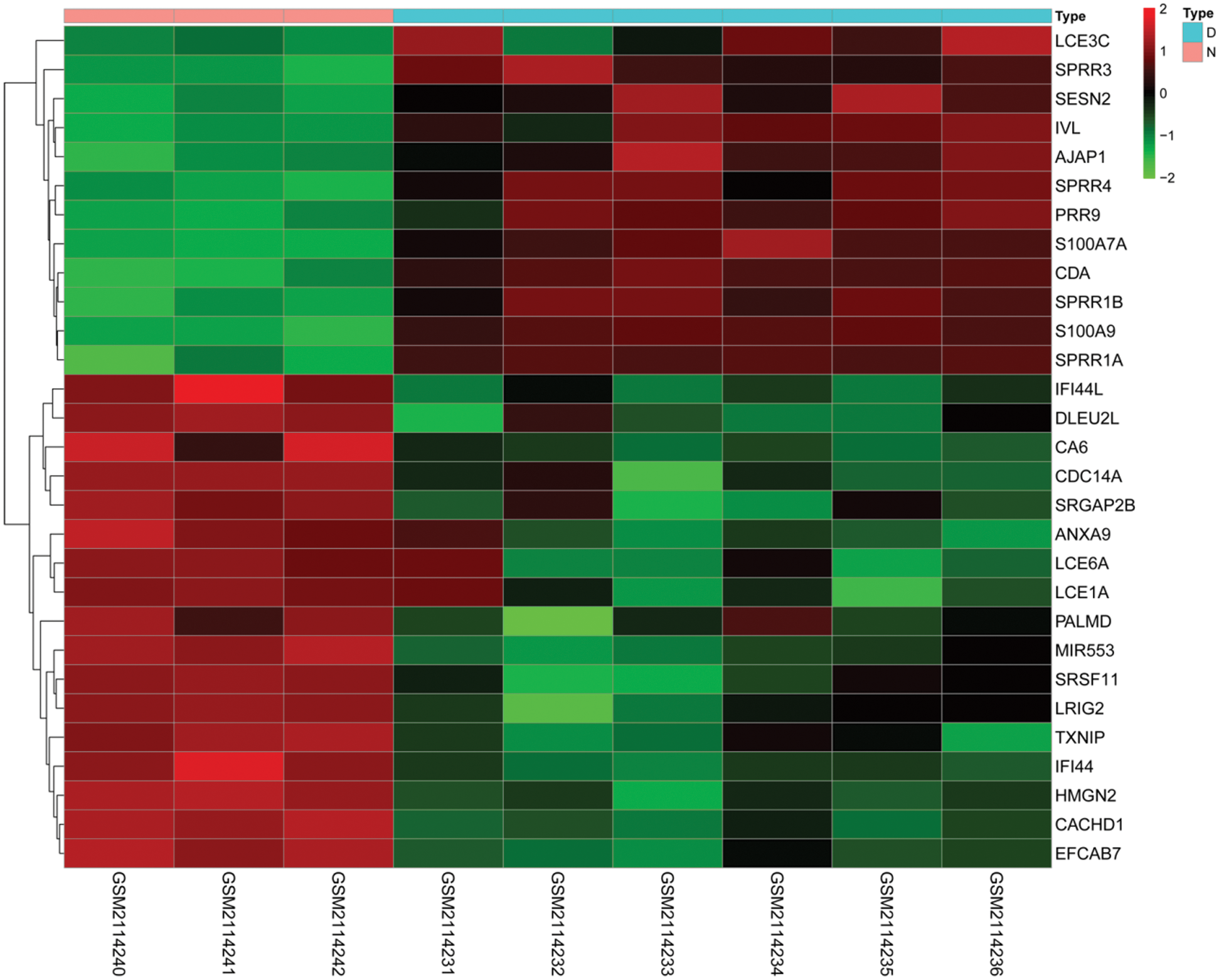
Figure 4: Differential expression of mRNAs. Heatmaps demonstrate differential expression of mRNAs in the skin between diabetic ulcer patients (D, blue) and normal healthy adults (N, pink). The x-axis denotes differential expressed mRNAs, and the y-axis represents the samples. The expression values are shown in line with the color scale.
Functional annotation and pathway enrichment analysis of DM mRNAs
GO and KEGG pathway analyses were performed to understand better these DM miRNAs functions. BP analysis showed that “peptide cross-linking,” “keratinocyte differentiation,” “epidermal cell differentiation,” “skin development,” “epidermis development,” “keratinization,” and “cornification” terms were significantly enriched (Fig. 5A, Tab. 1). The CC analysis showed a list of genes was significantly enriched in “cornified envelope.” (Fig. 5A, Tab. 1). All these GO terms were closely related to skin generated. In KEGG analysis, we found that the enriched pathway was the IL-17 signaling pathway (Fig. 5B, Tab. 1).
Table 1: The GO and KEGG analysis for the differential expressed mRNAs
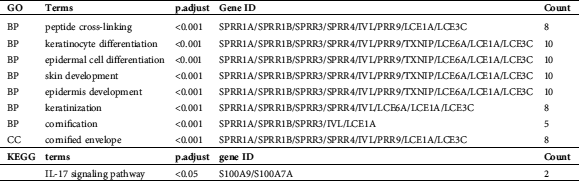
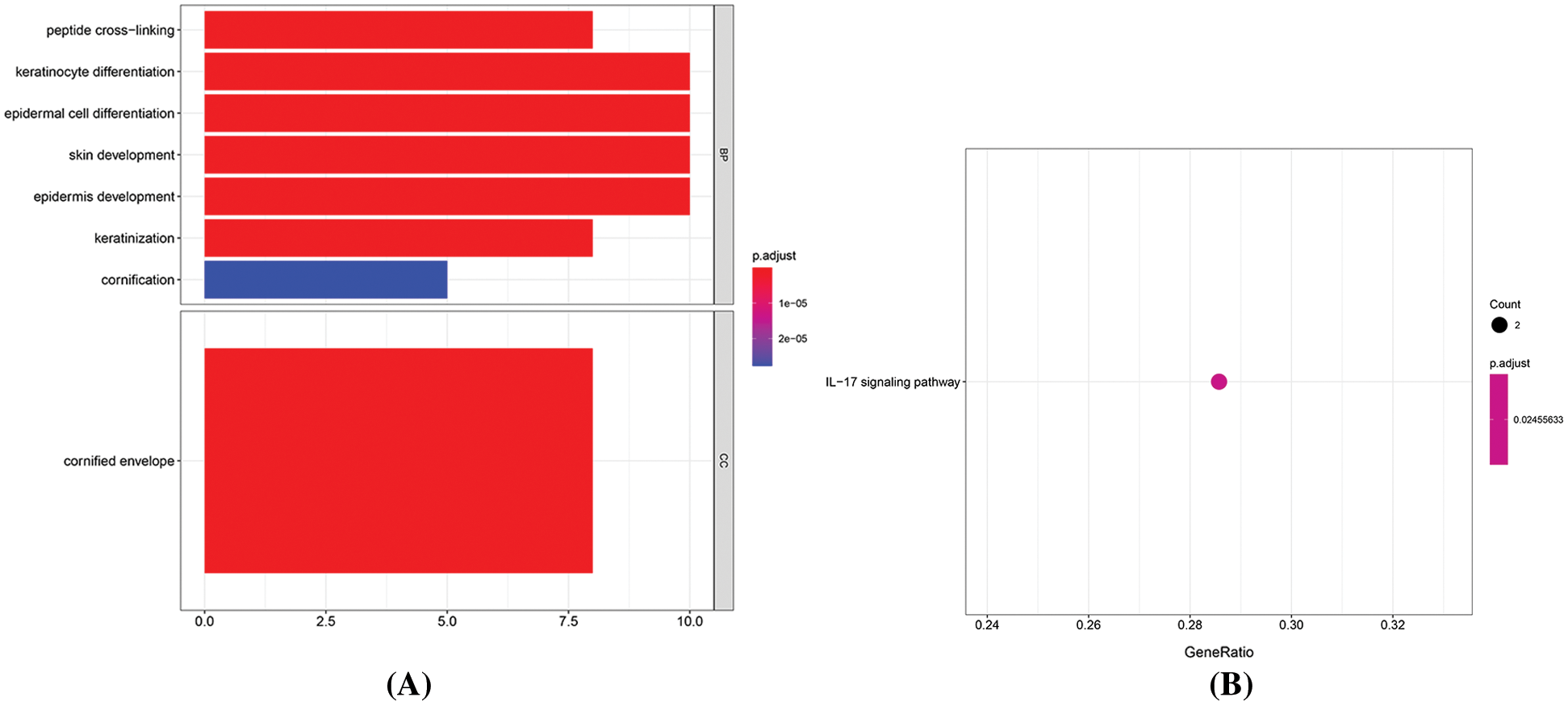
Figure 5: Enrichment analysis of mRNAs. A. The length of the histogram represents the number of enriched genes, and the saturation of the color represents the significance of the q-value. B. The larger the dot, the greater the number of genes. The lighter the color, the smaller the q-value.
In this study, we predicted the target genes of miRNAs obtained by differential expression, and we selected miRDB, miRTarBase, and TargetScan as prediction tools to screen the target genes that existed in at least two databases. Finally, we got 40868 target genes. After intersecting with 28 mRNAs differentially expressed in diabetic ulcer patients, 17 target genes were obtained. Since the regulation relationship between a miRNA and its target mRNA is negative, so we removed the unreasonable nodes in the network, and there are 30 miRNAs and 15 mRNAs left in the network. In the network, there are 8 miRNA regulating TXNIP, 5 miRNA regulating CDC14A, 5 miRNA regulating HMGN2, 4 miRNA regulating IFI44L, 4 miRNA regulating SRSF11, 3 miRNA regulating PALMD, 2 miRNA regulating LCE1A, 2 miRNA regulating LRIG2, and only one miRNA regulating ANXA9, CA6, EFCAB7, IFI44, S100A7A and SESN2 (Fig. 6).
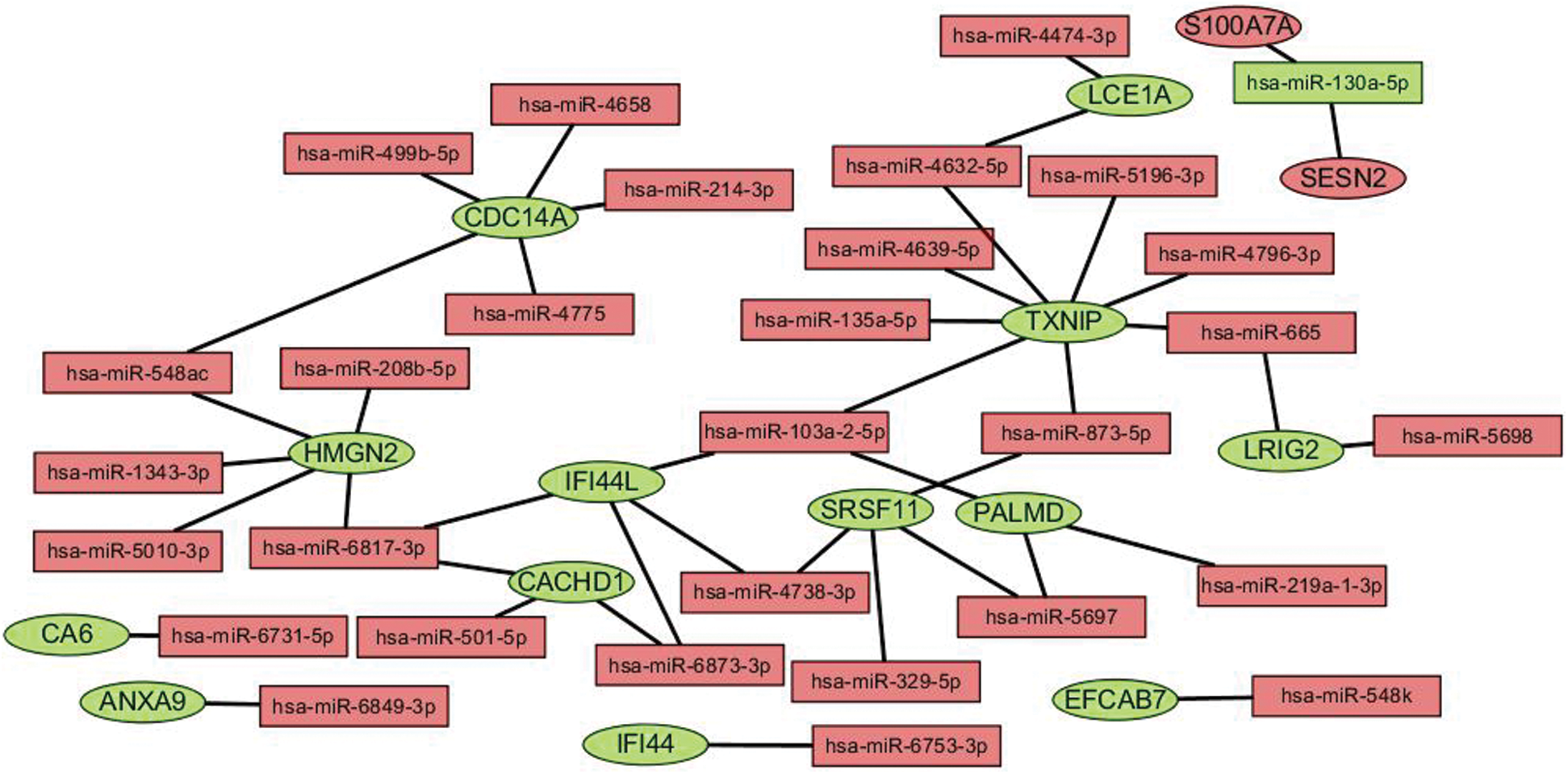
Figure 6: miRNA-mRNA regulation network. The ellipse stands for mRNA, the triangle represents miRNAs, the red represents the upturn, and the green represents the downgrading.
The GO and KEGG analysis of core gene
We performed GO and KEGG enrichment analyses on the core genes CACHD1, CDC14A, HMGN2, IFI44L, LCE1A, LRIG2, PALMD, SRSF11, and TXNI in the network. The GO BP analysis results showed that LRIG2 and TXNIP was significant enrichment in “platelet-derived growth factor receptor signaling pathway” terms (p < 0.05). The KEGG results showed that CDC14A was significant enrichment in the “cell cycle”; TXNIP was significant enrichment in the “NOD-like receptor signaling pathway” (Figs. 7 and 8).
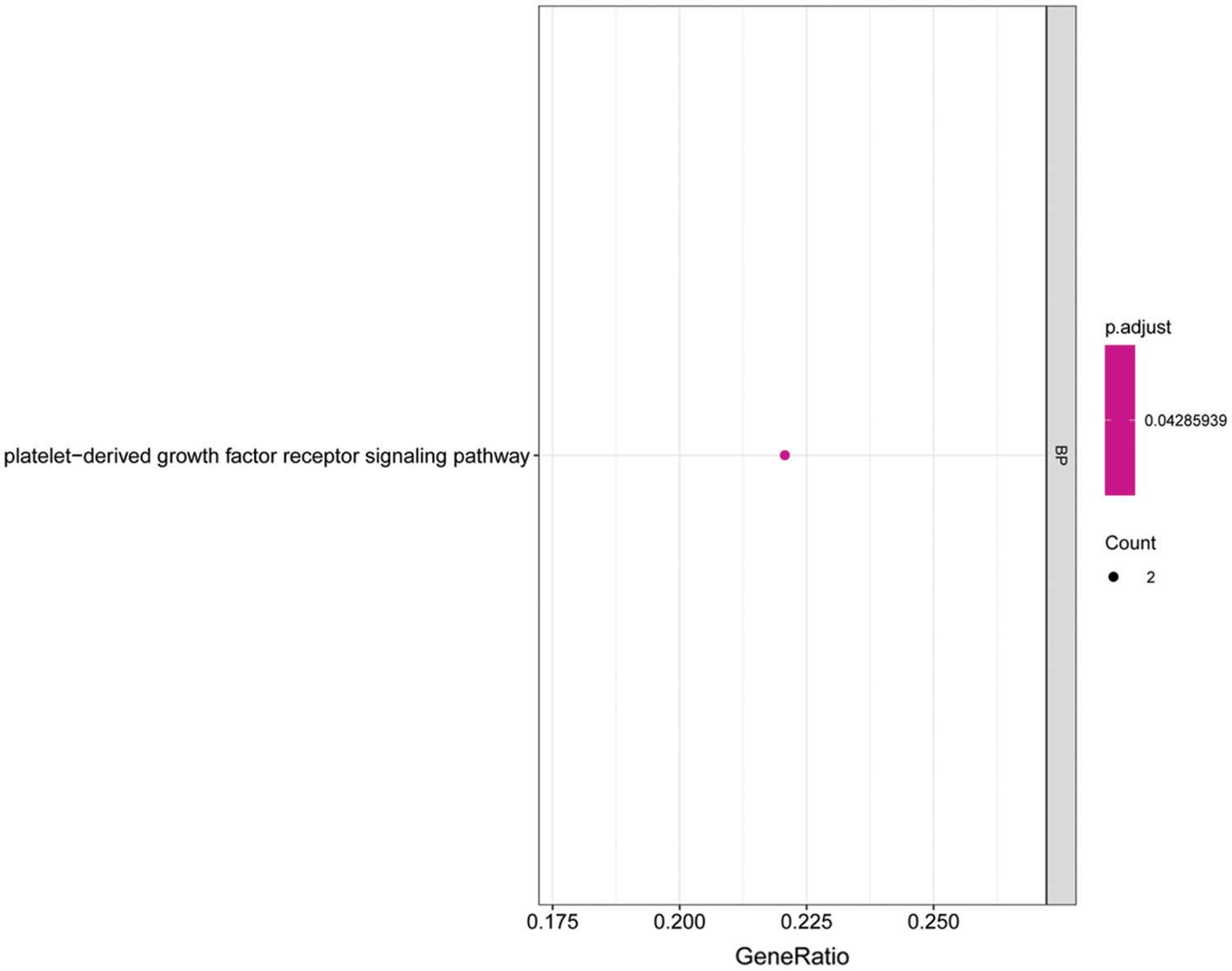
Figure 7: Histogram of GO analysis for core genes. The enriched GO terms of genes involved in the miRNA-mRNA network. The larger the dot, the greater the number of genes. The lighter the color, the smaller the q-value.
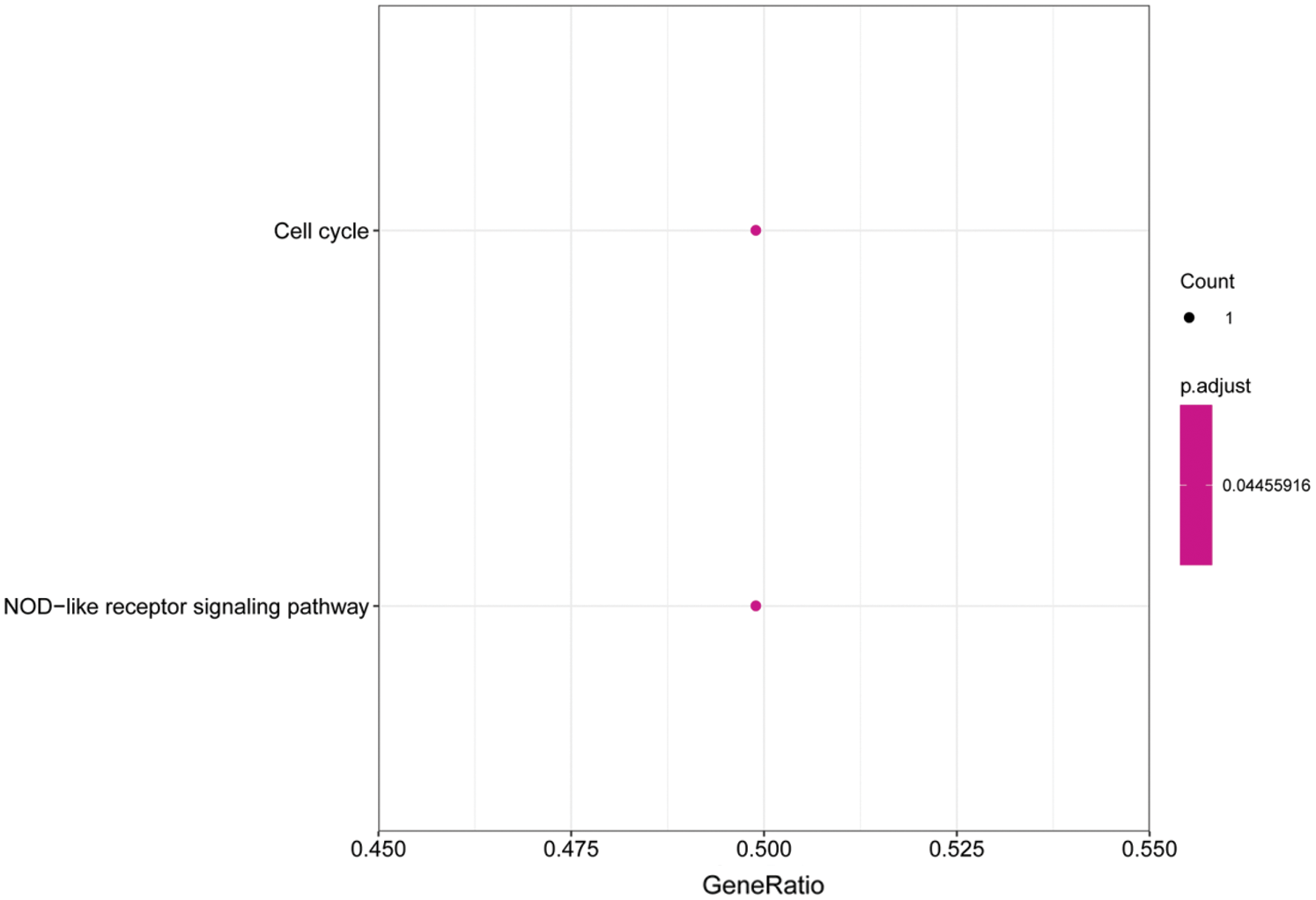
Figure 8: Histogram of KEGG analysis for core genes. The enriched KEGG pathway terms of genes involved in the miRNA-mRNA network. The larger the dot, the greater the number of genes. The lighter the color, the smaller the q-value.
The GO and KEGG analysis of miRNAs and mRNAs
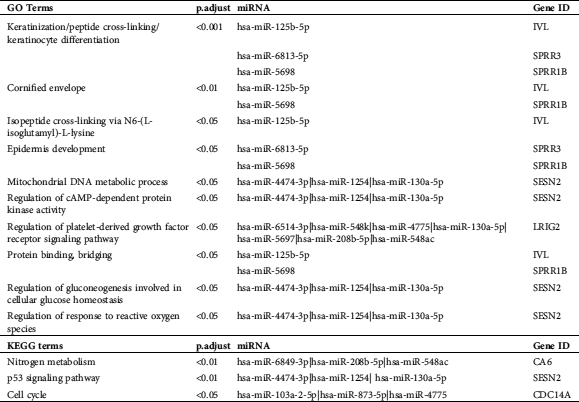
The GO and KEGG enrichment analyses on miRNAs-mRNAs (Tab. 2) showed that the top-3 enriched GO category “keratinization” is related to hsa-miR-125b-5p-IVL, hsa-miR-6813-5p-SPRR3 and hsa-miR-5698-SPRR1B, “peptide cross-linking” is related to hsa-miR-125b-5p-IVL, hsa-miR-6813-5p-SPRR3, and hsa-miR-5698-SPRR1B, and “keratinocyte differentiation” is related to hsa-miR-125b-5p-IVL, hsa-miR-6813-5p-SPRR3, hsa-miR-5698-SPRR1B. hsa-miR-130a-5p is also enriched in “mitochondrial DNA metabolic process,” “regulation of cAMP-dependent protein kinase,” “regulation of platelet-derived growth factor receptor signaling,” “regulation of gluconeogenesis involved in cellular glucose homeostasis,” and “regulation of response to reactive oxygen species.” hsa-miR-208b-5p is also enriched in “regulation of platelet-derived growth factor receptor signaling.” The enriched KEGG pathway “Nitrogen metabolism” is related to hsa-miR-6849-3p, hsa-miR-208b-5p, hsa-miR-548ac and CA6, “p53 signaling pathway” is related to hsa-miR-4474-3p, hsa-miR-1254, hsa-miR-130a-5p and SESEN2, and “Cell cycle” is related to hsa-miR-103a-2-5p, hsa-miR-873-5p, hsa-miR-4775 and CDC14A. These results further provided potential miRNAs-mRNAs and their pathways for wound healing of DM.
Table 2: The GO and KEGG analysis for the interacted miRNAs-mRNA
In this study, we compared the expression of miRNAs between BMSCs exosomes from patients with diabetic ulcers and healthy adults. Because the exosomes have the function of mediating miRNAs vector, it can transport miRNAs to the target organ, which leads to the change of microenvironment around the target organ, and then regulates the expression of mRNA in target organ cells. Previous studies have paid more attention to ulcer wound repair after MSCs transplantation (Kwon et al., 2008; Wan et al., 2013), but less attention has been paid to the role of exosomal miRNAs of BMSCs in regulating the mRNAs between diabetic ulcer patients and healthy adults on skin wound healing. We speculate that these DM miRNAs may affect the process of ulcer wound repair.
In patients with diabetic foot ulcers, vascular endothelial cells are damaged, their normal function is in disorder, and wounds are difficult to heal. Therefore, protecting endothelial cells and promoting angiogenesis may be a very effective way to repair diabetic ulcers (Mulder et al., 2014; Suresh et al., 2014). It had confirmed that BMSCs plays a good therapeutic effect in wound healing of diabetic ulcers (Motegi and Ishikawa, 2017). BMSCs can accelerate wound healing by differentiating into endothelial cells and paracrine angiogenesis factors, such as angiopoietin 1 and VEGF, after transplantation into the wound (Lee et al., 2016). The paracrine regulation of MSCs is mainly achieved by exosomes (Akyurekli et al., 2015), it promotes angiogenesis by growth factors and accelerates the reconstruction of damaged tissues and organs. Exosomes contain various proteins and miRNAs (Sato-Kuwabara et al., 2015; Valadi et al., 2007). miRNAs have been reported to play an important role in tissue repair and regeneration (Bjørge et al., 2017; Sahoo and Losordo, 2014). The KEGG analysis of differential expression miRNAs showed that the differentially expressed miRNAs were in “VEGF and VEGFR signaling network” and “Signaling events mediated by VEGFR1 and VEGFR2 pathway.” hsa-miR-1-5p and hsa-miR-130a-5p, hsa-miR-135a-5p, hsa-miR-665, hsa-miR-219a-1-3p, hsa-miR-212-3p, hsa-miR-34c-5p, hsa-miR-208b-3p, hsa-miR-9-3p and hsa-miR-873-5p are closely associated with matched genes in VEGF/VEGFR pathway. Among them, hsa-miR-130a-5p is involved in the “mitochondrial DNA metabolic process,” “regulation of cAMP-dependent protein kinase activity,” “regulation of gluconeogenesis involved in cellular glucose homeostasis,” “regulation of response to reactive oxygen species” and “p53 signaling pathway” with SESN2, “regulation of platelet-derived growth factor receptor signaling pathway” with LRIG2, and “cell cycle” with CDC14A. Hsa-miR-208b-5p is involved in the “regulation of gluconeogenesis involved in cellular glucose homeostasis” with LRIG2, and “nitrogen metabolism” with CA6. Has-miR-873-5p is involved in the “cell cycle” with CDC14A.
Wound healing caused by diabetic ulcers was delayed, due to a lack of angiogenesis factors (Pietramaggiori et al., 2010). It is found that VEGF treatment can significantly promote angiogenesis (Zeng et al., 2019) and shorten wound healing time in diabetic rats (Brem et al., 2009). In vivo, the miRNA in the exosome released by cells can be transported to neighboring cells and distant cells (Valadi et al., 2007). In breast cancer, it has been found that the exosomal miR-105 released by MCF-10A and MDA-MB-231 can reduce the expression of the ZO-1 gene in endothelial cells and promote lung and brain metastasis (Zhou et al., 2014). The miR-214 of human microvascular endothelial cell line (HMEC-1) exosomes can stimulate the migration and angiogenesis of neighboring HMEC-1 cells (Van Balkom et al., 2013). The exosomal miR-92a of K562 cells was reported to significantly decrease the expression of integrin a5 in human umbilical vein endothelial cell (HUVEC) and promote endothelial cell migration and catheter formation (Umezu et al., 2013). In this study, GO analysis of DM miRNAs showed that the DM miRNAs play an important role in exosome-mediated intercellular signal transduction and communication, and vacuolar transport, suggesting that miRNAs in exosomes derived from BMSCs can affect the skin repair of diabetic ulcers through transport and VEGF pathways.
Functional analysis differential expression miRNAs of ulcer skin between diabetic ulcer patients and healthy adults showed that BP such as keratinocyte differentiation, epidermal cell differentiation, skin development, epidermis development, keratinization, cornification, and the cornified envelope was significantly enriched. It was found that the migration and proliferation of keratinocytes were closely related to skin wound healing and played an important role in wound repair of diabetic ulcer (Werner et al., 2007), suggesting those mRNAs play an important role in wound repair in patients with diabetic ulcers. They were closely associated with the IL-17-mediated signal pathway. IL-17 signal pathway is related to the function of keratinocytes (Ma et al., 2016; Wu et al., 2015). In psoriasis, a secreted intestinal antimicrobial protein REG3A promoted skin keratinocyte proliferation, can be induced by IL-17 (Lai et al., 2012). During wound repair of diabetic ulcers, the inhibition of IL-17 accelerates diabetic wound healing through the alteration of macrophage polarization (Lee et al., 2018).
In this study, we analyzed the miRNA-mRNA regulatory network. The GO analysis results showed that the core genes TXNIP and LRIG2 were significantly enriched in the “platelet-derived growth factor receptor signaling pathway” biological processes. The KEGG analysis results showed that the core genes CDC14A was significantly enriched in the “cell cycle” pathway and TXNIP was significantly enriched in the “NOD-like receptor signaling pathway.” The “platelet-derived growth factor receptor signaling pathway” plays an important role in wound healing (Peus et al., 1995). Human TXNIP is a protein with a molecular weight of 46kD composed of 39 amino acid residues, which can be expressed in a variety of tissues, it is located on chromosome 1q21.1 and highly conserved among species and genera (Van Greevenbroek et al., 2007). It has been found that TXNIP is associated with angiogenesis, the deletion of TXNIP can restore restorative angiogenesis in the obesity model induced by a high-fat diet (Elshaer et al., 2017). TXNIP overexpression in diabetic wound healing can directly lead to diabetic angiogenic dysfunction. Silencing the TXNIP gene can prevent endothelial cell migration disturbance and angiogenesis injury induced by high glucose, and animal experiments have also shown that siRNA silencing TXNIP prevents ischemia-mediated diabetic angiogenesis (Dunn et al., 2014), restores the production of VEGF, and promotes angiogenesis ability (Ng et al., 2007). The mRNA data that we downloaded from the GEO database are derived from diabetic ulcers patients’ foot skin. We infer that miRNAs in BMSCs exosomes regulates wound healing in diabetic ulcer skin mainly through TXNIP and promotes the increase of VEGF secretion by regulating TXNIP, thus increasing the ability of wound healing. In the miRNA-mRNA network, we can see that the miRNAs involved in regulating TXNIP were hsa-miR-4796-3p, hsa-miR-135a-5p, hsa-miR-665, hsa-miR-4632-5p, hsa-miR-4639-5p, hsa-miR-873-5p, hsa-miR-103a-2-5p, hsa-miR-5196-3p. It is found that hsa-miR-135a-5p is highly expressed in the scar tissue of patients with urethral stricture, and its high expression may be related to tissue fibrosis (Zhang et al., 2018). It has been found that hsa-miR-873-5p is an important multipotential miRNA, hsa-miR-873-5p and an important multipotential miRNA, may hinder the differentiation of human Embryonic Stem Cells (hESCs) into fibroblasts by targeting epithelial-to-mesenchymal transforming (EMT) (Sahu and Mallick, 2018). It has been found that the increase of hsa-miR-665 can inhibit coronary artery micro-angiogenesis mediated by CD34 (Fan et al., 2018). CDC14A is an important gene involved in cell cycle regulation (Vázquez-Novelle et al., 2010). The healing of skin wounds is closely related to the division of epidermal cells. Studies have shown that CDC14A is involved in the G2/M phase of the mitotic cell cycle of epidermal cells (Zanet et al., 2010). In the study of wound repair, many studies have shown that the “NOD-like receptor signaling pathway” plays an important role in wound repair, in the process of intestinal epithelial cell repair, the NOD-like receptor signaling pathway participates in the repair of wound surfaces (Parlato and Yeretssian, 2014). In cardiovascular disease, NOD-like receptors are involved in the repair of cardiovascular damage (Bracey et al., 2015), and studies have found that in type 2 diabetic patients and mice, abnormal NOD-like receptor signaling pathway in macrophages can damage wound healing (Mirza et al., 2014).
To sum up, we found that there are differences in the expression of miRNAs between BMSCs exosomes from patients with diabetic ulcers and healthy adults. Part of the specific miRNAs such as has-miR-130a-5p was transported to the ulcer site through the exosomes and participated in wound healing by regulating genes such as SESN2, LRIG2, and CDC14A. However, by limitation of only 5 patients were collected in our study, few differential miRNAs with significant FDR were obtained. The effect of gender on DM should be considered with a larger sample, and other miRNAs might be identified in the future. In addition, we will verify the core genes in the regulatory network by in vivo and in vitro experiments.
Data Availability Statement: The research data used to support the findings of this study are currently under embargo. Requests for data, 12 months after publication of this article, will be considered by the corresponding author.
Funding Statement: Supported by the National Natural Science Foundation of China (81571910), Science and Technology key Project of Guangdong province (2014B020212010), the National Key Research and Development Plan of China (2017YFC1103301), and Medical Scientific Research Foundation of Guangdong Province of China (B2018026).
Conflict of Interest: None.
Akyurekli C, Le Y, Richardson RB, Fergusson D, Tay J, Allan DS. (2015). A systematic review of preclinical studies on the therapeutic potential of mesenchymal stromal cell-derived microvesicles. Stem Cell Reviews and Reports 11: 150–160. DOI 10.1007/s12015-014-9545-9. [Google Scholar] [CrossRef]
Al-Rubeaan K, Al Derwish M, Ouizi S, Youssef AM, Subhani SN, Ibrahim HM, Alamri BN. (2015). Diabetic foot complications and their risk factors from a large retrospective cohort study. PLoS One 10: e0124446. DOI 10.1371/journal.pone.0124446. [Google Scholar] [CrossRef]
Bartel DP. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. DOI 10.1016/S0092-8674(04)00045-5. [Google Scholar] [CrossRef]
Bjørge IM, Kim SY, Mano JF, Kalionis B, Chrzanowski W. (2017). Extracellular vesicles, exosomes and shedding vesicles in regenerative medicine-A new paradigm for tissue repair. Biomaterials Science 6: 60–78. [Google Scholar]
Bracey NA, Duff HJ, Muruve DA. (2015). Hierarchical regulation of wound healing by NOD-like receptors in cardiovascular disease. Antioxidants & Redox Signaling 22: 1176–1187. DOI 10.1089/ars.2014.6125. [Google Scholar] [CrossRef]
Brem H, Kodra A, Golinko MS, Entero H, Stojadinovic O, Wang VM, Sheahan CM, Weinberg AD, Woo SL, Ehrlich HP, Tomic-Canic M. (2009). Mechanism of sustained release of vascular endothelial growth factor in accelerating experimental diabetic healing. Journal of Investigative Dermatology 129: 2275–2287. DOI 10.1038/jid.2009.26. [Google Scholar] [CrossRef]
Bukowiecki A, Hos D, Cursiefen C, Eming SA. (2017). Wound-healing studies in cornea and skin: parallels, differences and opportunities. International Journal of Molecular Science 18: 1257. DOI 10.3390/ijms18061257. [Google Scholar] [CrossRef]
Cui C, Ye X, Chopp M, Venkat P, Zacharek A, Yan T, Ning R, Yu P, Cui G, Chen J. (2016). miR-145 regulates diabetes-bone marrow stromal cell-induced neurorestorative effects in diabetes stroke rats. STEM CELLS Translational Medicine 5: 1656–1667. DOI 10.5966/sctm.2015-0349. [Google Scholar] [CrossRef]
Chen B, Li Q, Zhao B, Wang Y. (2017). Stem cell-derived extracellular vesicles as a novel potential therapeutic tool for tissue repair. STEM CELLS Translational Medicine 6: 1753–1758. DOI 10.1002/sctm.16-0477. [Google Scholar] [CrossRef]
Dai J, Escara-Wilke J, Keller JM, Jung Y, Taichman RS, Pienta KJ, Keller ET. (2019). Primary prostate cancer educates bone stroma through exosomal pyruvate kinase M2 to promote bone metastasis. Journal of Experimental Medicine 216: 2883–2899. DOI 10.1084/jem.20190158. [Google Scholar] [CrossRef]
Ding J, Wang X, Chen B, Zhang J, Xu J. (2019). Exosomes derived from human bone marrow mesenchymal stem cells stimulated by deferoxamine accelerate cutaneous wound healing by promoting angiogenesis. BioMed Research International 2019: 1–12. DOI 10.1155/2019/9742765. [Google Scholar] [CrossRef]
Dunn LL, Simpson PJ, Prosser HC, Lecce L, Yuen GS, Buckle A, Sieveking DP, Vanags LZ, Lim PR, Chow RW, Lam YT, Clayton Z, Bao S, Davies MJ, Stadler N, Celermajer DS, Stocker R, Bursill CA, Cooke JP, Ng MK. (2014). A critical role for thioredoxin-interacting protein in diabetes-related impairment of angiogenesis. Diabetes 63: 675–687. DOI 10.2337/db13-0417. [Google Scholar] [CrossRef]
Elshaer SL, Mohamed IN, Coucha M, Altantawi S, Eldahshan W, Bartasi ML, Shanab AY, Lorys R, El-Remessy AB. (2017). Deletion of TXNIP mitigates high-fat diet-impaired angiogenesis and prevents inflammation in a mouse model of critical limb ischemia. Antioxidants 6: 47. DOI 10.3390/antiox6030047. [Google Scholar] [CrossRef]
Fan J, Li H, Nie X, Yin Z, Zhao Y, Zhang X, Yuan S, Li Y, Chen C, Wang DW. (2018). MiR-665 aggravates heart failure via suppressing CD34-mediated coronary microvessel angiogenesis. Aging 10: 2459–2479. [Google Scholar]
Fang Z, Rajewsky N. (2011). The impact of miRNA target sites in coding sequences and in 3'UTRs. PLoS One 6: e18067. DOI 10.1371/journal.pone.0018067. [Google Scholar] [CrossRef]
Geiger A, Walker A, Nissen E. (2015). Human fibrocyte-derived exosomes accelerate wound healing in genetically diabetic mice. Biochemical and Biophysical Research Communications 467: 303–309. DOI 10.1016/j.bbrc.2015.09.166. [Google Scholar] [CrossRef]
Han C, Sun X, Liu L, Jiang H, Shen Y, Xu X, Li J, Zhang G, Huang J, Lin Z, Xiong N, Wang T. (2016). Exosomes and their therapeutic potentials of stem cells. Stem Cells International 2016: 1–11. DOI 10.1155/2016/7653489. [Google Scholar] [CrossRef]
Hu L, Wang J, Zhou X, Xiong Z, Zhao J, Yu R, Huang F, Zhang H, Chen L. (2016). Exosomes derived from human adipose mensenchymal stem cells accelerates cutaneous wound healing via optimizing the characteristics of fibroblasts. Scientific Reports 6: 594. DOI 10.1038/srep32993. [Google Scholar] [CrossRef]
Jhamb S, Vangaveti VN, Malabu UH. (2016). Genetic and molecular basis of diabetic foot ulcers: Clinical review. Journal of Tissue Viability 25: 229–236. DOI 10.1016/j.jtv.2016.06.005. [Google Scholar] [CrossRef]
Kwon DS, Gao X, Liu YB, Dulchavsky DS, Danyluk AL, Bansal M, Chopp M, Mcintosh K, Arbab AS, Dulchavsky SA, Gautam SC. (2008). Treatment with bone marrow-derived stromal cells accelerates wound healing in diabetic rats. International Wound Journal 5: 453–463. DOI 10.1111/j.1742-481X.2007.00408.x. [Google Scholar] [CrossRef]
Lai Y, Li D, Li C, Muehleisen B, Radek KA, Park HJ, Jiang Z, Li Z, Lei H, Quan Y, Zhang T, Wu Y, Kotol P, Morizane S, Hata TR, Iwatsuki K, Tang C, Gallo RL. (2012). The antimicrobial protein REG3A regulates keratinocyte proliferation and differentiation after skin injury. Immunity 37: 74–84. DOI 10.1016/j.immuni.2012.04.010. [Google Scholar] [CrossRef]
Lalla E, Lamster IB, Drury S, Fu C, Schmidt AM. (2000). Hyperglycemia, glycoxidation and receptor for advanced glycation endproducts: Potential mechanisms underlying diabetic complications, including diabetes-associated periodontitis. Periodontology 2000 23: 50–62. DOI 10.1034/j.1600-0757.2000.2230104.x. [Google Scholar] [CrossRef]
Lee DE, Ayoub N, Agrawal DK. (2016). Mesenchymal stem cells and cutaneous wound healing: Novel methods to increase cell delivery and therapeutic efficacy. Stem Cell Research & Therapy 7: 5. DOI 10.1186/s13287-016-0303-6. [Google Scholar] [CrossRef]
Lee J, Rodero MP, Patel J, Moi D, Mazzieri R, Khosrotehrani K. (2018). Interleukin-23 regulates interleukin-17 expression in wounds, and its inhibition accelerates diabetic wound healing through the alteration of macrophage polarization. FASEB Journal 32: 2086–2094. DOI 10.1096/fj.201700773R. [Google Scholar] [CrossRef]
Liang X, Zhang L, Wang S, Han Q, Zhao RC. (2016). Exosomes secreted by mesenchymal stem cells promote endothelial cell angiogenesis by transferring miR-125a. Journal of Cell Science 129: 2182–2189. DOI 10.1242/jcs.170373. [Google Scholar] [CrossRef]
Liu CJ, Tsai MM, Hung PS, Kao SY, Liu TY, Wu KJ, Chiou SH, Lin SC, Chang KW. (2010). miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Research 70: 1635–1644. DOI 10.1158/0008-5472.CAN-09-2291. [Google Scholar] [CrossRef]
Ma WY, Jia K, Zhang Y. (2016). IL-17 promotes keratinocyte proliferation via the downregulation of C/EBPα. Experimental and Therapeutic Medicine 11: 631–636. DOI 10.3892/etm.2015.2939. [Google Scholar] [CrossRef]
Millington JT, Ellenzweig J. (2005). The comprehensive therapy of diabetic foot ulcers. Comprehensive Therapy 31: 050–058. DOI 10.1385/COMP:31:1:050. [Google Scholar] [CrossRef]
Mirza RE, Fang MM, Weinheimer-Haus EM, Ennis WJ, Koh TJ. (2014). Sustained inflammasome activity in macrophages impairs wound healing in type 2 diabetic humans and mice. Diabetes 63: 1103–1114. DOI 10.2337/db13-0927. [Google Scholar] [CrossRef]
Motegi SI, Ishikawa O. (2017). Mesenchymal stem cells: The roles and functions in cutaneous wound healing and tumor growth. Journal of Dermatological Science 86: 83–89. DOI 10.1016/j.jdermsci.2016.11.005. [Google Scholar] [CrossRef]
Mulder G, Tenenhaus M, D’souza GF. (2014). Reduction of diabetic foot ulcer healing times through use of advanced treatment modalities. The International Journal of Lower Extremity Wounds 13: 335–346. DOI 10.1177/1534734614557925. [Google Scholar] [CrossRef]
Negre-Salvayre A, Salvayre R, Augé N, Pamplona R, Portero-Otín M. (2009). Hyperglycemia and glycation in diabetic complications. Antioxidants & Redox Signaling 11: 3071–3109. DOI 10.1089/ars.2009.2484. [Google Scholar] [CrossRef]
Ng MK, Wu J, Chang E, Wang BY, Katzenberg-Clark R, Ishii-Watabe A, Cooke JP. (2007). A central role for nicotinic cholinergic regulation of growth factor-induced endothelial cell migration. Arteriosclerosis, Thrombosis, and Vascular Biology 27: 106–112. DOI 10.1161/01.ATV.0000251517.98396.4a. [Google Scholar] [CrossRef]
Oliveros JC (2007). VENNY. An interactive tool for comparing lists with Venn Diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html. [Google Scholar]
Parlato M, Yeretssian G. (2014). NOD-like receptors in intestinal homeostasis and epithelial tissue repair. International Journal of Molecular Sciences 15: 9594–9627. DOI 10.3390/ijms15069594. [Google Scholar] [CrossRef]
Pegtel DM, Gould SJ. (2019). Exosomes. Annual Review of Biochemistry 88: 487–514. DOI 10.1146/annurev-biochem-013118-111902. [Google Scholar] [CrossRef]
Peus D, Jungtäubl H, Knaub S, Leuker A, Gerecht K, Ostendorf R, Meyer-Ingold W, Wlaschek M, Krieg T, Ruzicka T, Scharffetter-Kochanek K. (1995). Localization of platelet-derived growth factor receptor subunit expression in chronic venous leg ulcers. Wound Repair and Regeneration 3: 265–272. DOI 10.1046/j.1524-475X.1995.30306.x. [Google Scholar] [CrossRef]
Pietramaggiori G, Scherer SS, Mathews JC, Gennaoui T, Lancerotto L, Ragno G, Valeri CR, Orgill DP. (2010). Quiescent platelets stimulate angiogenesis and diabetic wound repair. Journal of Surgical Research 160: 169–177. DOI 10.1016/j.jss.2008.09.010. [Google Scholar] [CrossRef]
Sahoo S, Losordo DW. (2014). Exosomes and cardiac repair after myocardial infarction. Circulation Research 114: 333–344. DOI 10.1161/CIRCRESAHA.114.300639. [Google Scholar] [CrossRef]
Sahu M, Mallick B. (2018). Deciphering synergistic regulatory networks of microRNAs in hESCs and fibroblasts. International Journal of Biological Macromolecules 113: 1279–1286. DOI 10.1016/j.ijbiomac.2018.03.061. [Google Scholar] [CrossRef]
Sato-Kuwabara Y, Melo SA, Soares FA, Calin GA. (2015). The fusion of two worlds: non-coding RNAs and extracellular vesicles-diagnostic and therapeutic implications (Review). International Journal of Oncology 46: 17–27. DOI 10.3892/ijo.2014.2712. [Google Scholar] [CrossRef]
Schmidt AM. (2018). Highlighting diabetes mellitus: the epidemic continues. Arteriosclerosis, Thrombosis, and Vascular Biology 38: e1–e8. DOI 10.1161/ATVBAHA.117.310221. [Google Scholar] [CrossRef]
Shabbir A, Cox A, Rodriguez-Menocal L, Salgado M, Van Badiavas E. (2015). Mesenchymal stem cell exosomes induce proliferation and migration of normal and chronic wound fibroblasts, and enhance angiogenesis in vitro. Stem Cells and Development 24: 1635–1647. DOI 10.1089/scd.2014.0316. [Google Scholar] [CrossRef]
Sun K, Wang W, Wang C, Lao G, Liu D, Mai L, Yan L, Yang C, Ren M. (2016). AGEs trigger autophagy in diabetic skin tissues and fibroblasts. Biochemical and Biophysical Research Communications 471: 355–360. DOI 10.1016/j.bbrc.2016.02.020. [Google Scholar] [CrossRef]
Suresh DH, Suryanarayan S, Sarvajnamurthy S, Puvvadi S. (2014). Treatment of a non-healing diabetic foot ulcer with platelet-rich plasma. Journal of Cutaneous and Aesthetic Surgery 7: 229–231. DOI 10.4103/0974-2077.150786. [Google Scholar] [CrossRef]
Umezu T, Imanishi S, Azuma K, Kobayashi C, Yoshizawa S, Ohyashiki K, Ohyashiki JH. (2017). Replenishing exosomes from older bone marrow stromal cells with miR-340 inhibits myeloma-related angiogenesis. Blood Advances 1: 812–823. DOI 10.1182/bloodadvances.2016003251. [Google Scholar] [CrossRef]
Umezu T, Ohyashiki K, Kuroda M, Ohyashiki JH. (2013). Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene 32: 2747–2755. DOI 10.1038/onc.2012.295. [Google Scholar] [CrossRef]
Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology 9: 654–659. DOI 10.1038/ncb1596. [Google Scholar] [CrossRef]
Van Balkom BW, De Jong OG, Smits M, Brummelman J, Den Ouden K, De Bree PM, Van Eijndhoven MA, Pegtel DM, Stoorvogel W, Würdinger T, Verhaar MC. (2013). Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood 121: 3997–4006. DOI 10.1182/blood-2013-02-478925. [Google Scholar] [CrossRef]
Van Greevenbroek MM, Vermeulen VM, Feskens EJ, Evelo CT, Kruijshoop M, Hoebee B, Van Der Kallen CJ, De Bruin TW. (2007). Genetic variation in thioredoxin interacting protein (TXNIP) is associated with hypertriglyceridaemia and blood pressure in diabetes mellitus. Diabetic Medicine 24: 498–504. DOI 10.1111/j.1464-5491.2007.02109.x. [Google Scholar] [CrossRef]
Vázquez-Novelle MD, Mailand N, Ovejero S, Bueno A, Sacristán MP. (2010). Human Cdc14A phosphatase modulates the G2/M transition through Cdc25A and Cdc25B. Journal of Biological Chemistry 285: 40544–40553. DOI 10.1074/jbc.M110.133009. [Google Scholar] [CrossRef]
Wan J, Xia L, Liang W, Liu Y, Cai Q. (2013). Transplantation of bone marrow-derived mesenchymal stem cells promotes delayed wound healing in diabetic rats. Journal of Diabetes Research 2013: 1–11. DOI 10.1155/2013/647107. [Google Scholar] [CrossRef]
Werner S, Krieg T, Smola H. (2007). Keratinocyte–fibroblast interactions in wound healing. Journal of Investigative Dermatology 127: 998–1008. DOI 10.1038/sj.jid.5700786. [Google Scholar] [CrossRef]
Wu L, Chen X, Zhao J, Martin B, Zepp JA, Ko JS, Gu C, Cai G, Ouyang W, Sen G, Stark GR, Su B, Vines CM, Tournier C, Hamilton TA, Vidimos A, Gastman B, Liu C, Li X. (2015). A novel IL-17 signaling pathway controlling keratinocyte proliferation and tumorigenesis via the TRAF4-ERK5 axis. Journal of Experimental Medicine 212: 1571–1587. DOI 10.1084/jem.20150204. [Google Scholar] [CrossRef]
Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Möbius W, Hoernschemeyer J, Slot JW, Geuze HJ, Stoorvogel W. (2003). Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. Journal of Biological Chemistry 278: 10963–10972. DOI 10.1074/jbc.M207550200. [Google Scholar] [CrossRef]
Zanet J, Freije A, Ruiz M, Coulon V, Sanz JR, Chiesa J, Gandarillas A. (2010). A mitosis block links active cell cycle with human epidermal differentiation and results in endoreplication. PLoS One 5: e15701. DOI 10.1371/journal.pone.0015701. [Google Scholar] [CrossRef]
Zeng Y, Yao X, Liu X, He X, Li L, Liu X, Yan Z, Wu J, Fu B. (2019). Anti-angiogenesis triggers exosomes release from endothelial cells to promote tumor vasculogenesis. Journal of Extracellular Vesicles 8: 1629865. DOI 10.1080/20013078.2019.1629865. [Google Scholar] [CrossRef]
Zgheib C, Zhang L, Liechty K. (2013). Mesenchymal stem cell (MSC) treatment improves diabetic wound healing by increasing miR-126 and angiogenesis. Wound Repair and Regeneration 21: A49. [Google Scholar]
Zhang K, Chen J, Zhang D, Wang L, Zhao W, Lin DY, Chen R, Xie H, Hu X, Fang X, Fu Q. (2018). microRNA expression profiles of scar and normal tissue from patients with posterior urethral stricture caused by pelvic fracture urethral distraction defects. International Journal of Molecular Medicine 41: 2733–2743. [Google Scholar]
Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O’connor ST, Chin AR, Yen Y, Wang Y, Marcusson EG, Chu P, Wu J, Wu X, Li AX, Li Z, Gao H, Ren X, Boldin MP, Lin PC, Wang SE. (2014). Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 25: 501–515. DOI 10.1016/j.ccr.2014.03.007. [Google Scholar] [CrossRef]
Zhu LL, Huang X, Yu W, Chen H, Chen Y, Dai YT. (2018). Transplantation of adipose tissue-derived stem cell-derived exosomes ameliorates erectile function in diabetic rats. Andrologia 50: e12871. DOI 10.1111/and.12871. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |