2021 45(1): 119-127
DOI:10.32604/biocell.2021.012011

www.techscience.com/journal/biocell
| BIOCELL 2021 45(1): 119-127 DOI:10.32604/biocell.2021.012011 |  www.techscience.com/journal/biocell |
Nanoparticles induce the biosynthesis and activity of the new possible therapeutic proteinase source, Talaromyces purpureogenus KJ584844
1Life Sciences Department, College of Science and literature Mahyel Aseer, King Khalid University, Abha, Saudi Arabia
2Unit of Food Bacteriology, Central Laboratory of Food Hygiene, Ministry of Health, Sharkia, Egypt
3Department of Botany and Microbiology, Faculty of Science, Damanhour University, El-Behera, Egypt
4Microbial Activity Unit, Department of Microbiology, Soils, Water and Environment Research Institute, Agricultural Research Center, Giza, Egypt
5Physics Department, Faculty of Science, Damanhour University, Damanhour, Egypt
6Department of Biology, College of Science, King Khalid University, 9004, Abha, Saudi Arabia
7Department of Botany and Microbiology, Faculty of Science, South Valley University, Qena, Egypt
8Zoology Department, Faculty of Science, Damanhour University, El-Behera, Egypt
*Address correspondence to: Mahmoud MOUSTAFA, mfmostfa@kku.edu
Received: 10 June 2020; Accepted: 22 August 2020
Abstract: The need for the bacterial proteinase is rapidly growing, urging to catch a lowcost medium for the microbial fermentation, nanoparticles can play a vital role in this respect. The proteinase of Talaromyces purpureogenus was produced on the tubers of Helianthus tuberosus that also operated as solid support for the fermentation process. The interface amongst nitrogen sources (NH4Cl and yeast extract) was investigated, applying the statistical modeling of central composite design under solid-state fermentation. The optimum medium for proteinase secretion was stimulated by 979.82 mg NH4Cl and 437.68 mg yeast extract per 100 g substrate, yielding 108.15 U/g tubers. Using Plackett-Burman experimental design, the nanoparticles Co, Ni and Fe were assessed as inducers for proteinase stimulants. Co nanoparticles (5 ppm) were the greatest in both proteinase production by the fungus as well as an inducer of the proteolysis process by the enzyme when using faba bean straw as a proteinaceous substrate in the reaction mixture, liberating the extreme quantity of amino acids, compared with the lack of the nanoparticles. The findings suggest the incorporation of Co nanoparticles in both the proteinase fabrication process and during the degradation of proteinaceous materials induce proteinase catalyst. This approach could be extended to modulate the productivity and activity of similar biomolecules.
Keywords: Experimental design; Helianthus tuberosus; proteolysis; solid-state fermentation
Proteolysis is the digestion of protein chains into shorter units by the destruction of the peptide-bonds, which bind amino acid residues. Proteinases, proteolytic enzymes, or peptidases (EC, 3.4.21.24) play the vital part, by discharging of monomer amino acids (AA) from nitrogenous compounds. Some of them (exopeptidases, e.g., aminopeptidases and carboxypeptidase A) detaches the external AA from the main protein chain. Others (endopeptidases, e.g., trypsin, chymotrypsin, pepsin, papain and elastase) attack the inner peptide links of a protein (Gurumallesh et al., 2019; Kumar and Takagi, 1999). Proteinases have multi applications in various medicinal, food, and pharmaceutical aspects (Gurumallesh et al., 2019). For several merits (for instance, the cheaper production cost and usage of renewable substrates), the microbial source of proteinases has gained more importance. Biosynthesis of bioproducts by microorganisms is deeply motivated by the constituents and physical factors during fermentation (Potumarthi et al., 2007).
Optimization of the biofermentation system in biotechnology can be performed following various statistical approaches. Central composite design (CCD) of the response surface techniques is a unique frequently used optimization procedure. This experimental design aids in identifying the significant operative factors with their interactions, in addition to quantifying the association among the measured response(s) and the examined factors using a narrow quantity of tests (Gupta et al., 2002; Oskouie et al., 2008). However, throughout the initial screening studies, the Plackett-Burman matrix is applied for choosing the significant elements from a bulky quantity of variables, the non-significant variables are fixed or eliminated in the subsequent optimization steps (Reddy et al., 2008).
In opposite to bulk materials, nanoparticles (NPs) have a large specific external region and high reaction activity. Over recent years, nanotechnology has widely influenced many high-technology industries; of these, NPs with various composition, size, and concentration have been testified to impact various microbial species (Chakrabarti et al., 2015; Kathiraven et al., 2015; Schröfel et al., 2014).
The current research intended to explore the inducing impact of Co, Fe and Ni NPs after the maximization of proteinase secretion by Talaromyces purpureogenus on tubers of Helianthus tuberosus L. (Asteraceae) as an energy and carbon source utilizing the matrix of CCD. The catalytic behavior of the resulted proteinase was, likewise, explored in the existence of such NPs.
Healthy clean tubers of H. tuberosus (var. Fuseau) belongs to the aster family (Asteraceae) were obtained from the Horticulture Research Station, Agricultural Research Centre, Egypt. The plant is a perennial growing up to 2.4 m tall that can out-compete invaded natural vegetation and grows as an occasionally serious agricultural weed. The tubers were cool-dried and ground; the resulting powder acted, additionally, as solid-state support for the fermentation process. The straw of Vicia faba (var. Giza 2) served as an enzyme-substrate. Plant residues were collected from Tag Elezz Research Station, Agricultural Research Centre, Egypt, dried at 70°C overnight and ground in an electric grinder to obtain plant powder.
Talaromyces purpureogenus KJ584844
The fungus was identified in a previous study (El-Metwally et al., 2016). The preservation (4°C) and inoculum preparation (106 spores/mL) were accomplished after growing on agar slants of Czapek medium at 28 ± 2°C for 5 days.
Cobalt (Co) and nickel (Ni) NPs with an average size of 40–60 and 40–80 nm, respectively, were purchased from Sigma-Aldrich. The NPs of Fe3O4 were equipped by means of co-precipitation with 1:2 molar-ratio of ferrous to ferric chloride (Fe2+/Fe3+) complex, in the existence of aqueous NaOH, the fabricated Fe3O4 NPs (with an average size of approximately 20 ± 10 nm) were fully characterized (Lu et al., 2007; Sahar et al., 2017).
The fermentation medium contained 1.0 gram of cold-dried and ground powder of tubers of H. tuberosus as the solid substrate, also contained different concentrations of NH4Cl (859, 900, 1000, 1100 and 1141 mg, per 100 g substrate) and yeast extract (259, 300, 400, 500 and 541 mg, per 100 g substrate), representing the high (+1), low (−1), center (0) and axial (α = ±1.414) points of the fermentation variables affecting proteinase biosynthesis by the fungus. The interface between both variables was inspected by the full CCD according to the next second-order polynomial quadratic model:
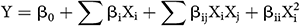
where Y is the predicted proteinase, β0 is a model constant, Xi and Xj are the independent variables, βi is the linear coefficients, βij is the cross-coefficients, and βii is the quadratic-coefficients.
After modeling, the theoretically calculated concentration of each variable was experimentally validated in triplicate to ensure the fitness of the suggested model.
The solid-state fermentation (SSF) medium was autoclaved at 121°C (15 psi for 20 min) and injected with the fungus spore suspension (106/g substrate). The humidity was maintained to 65% during the 7-day fermentation period at 28°C.
The optimum medium recovered from the biofermentation settings based on the previous CCD experiment was applied to assess the workability of Fe, Co, and Ni NPs on proteinase production. The fractional factorial; PB was allocated. The single, dual, and triple interference of the particles was evaluated. Each NPs was distributed evenly to yield 5 ppm. The optimal, previously defined settings were applied. The outcome of the individual NPs was estimated as the variance between both means of measurements made at the occurrence (5 ppm) and the absenteeism (0 ppm) of the assessed NPs.
Separation and assessing of crude proteinase
Next fermentation period, distilled water (10 mL), containing 0.01% Tween 80, as a surfactant (to increases the stability of the enzyme in the enzymatic hydrolysis and make the enzyme rapidly desorb from the substrate and avoid enzyme inactivation), was mixed with the fermented medium and kept under shaking (200 rpm) for half an hour at ambient temperature. Samples were then centrifuged at 5000 rpm for 10 min, and the supernatant was assessed for the proteinase activity.
Casein was applied in the quantification of the proteinase activity in the supernatant according to Cupp-Enyard (2008). The unit of proteinase (U) was described as the enzyme quantity resulting in the release of 1.0 µg of tyrosine equivalent per gram/min under the reaction conditions.
Liberation of AA from faba bean straw (FBS)
Compared to the control (without NPs), the inducibility of NPs (5 ppm) on the crude proteinase was determined, using FBS as a substrate for AA liberation. Proteinase (100 U) was mixed with the NPs to get a 5-ml reaction mixture, to which FBS (0.1 g) was mixed and kept shaking (150 rpm) for 30 min at 37°C. The released AA was assessed in the mixture after filtration through Whatman No.1 filter paper. Then, two ml of the clear reaction mixture was added to 2 mL of 10% TCA, lift for 10 min at 37°C, followed by centrifugation at 5000 rpm for 15 min to obtain clear filtrate, of which 2 ml was added to 5 mL sodium carbonate and 1.0 mL of 0.5 mM Folin–Ciolcalteau’s, reagent, the mixture was incubated at 37°C for 30 minutes then the developed color was measured at 660 nm, Then the released AA was calculated using a standard of tyrosine (Cupp-Enyard, 2008).
Trials planning and statistical analysis
For CCD, Design Expert (version 11, State-Ease, USA) was utilized to build, performing ANOVA analysis, checking the design model, and generating the equation model. Whereas, for the PB matrix, the planning and ANOVA analyses were accomplished using the Minitab packages (Version 19, Minitab Inc., USA). Three-repeats were implemented and presented as average ± SE. All comparisons were made at a probability (p) level of <0.05.
Optimizing proteinase biosynthesis using CCD
The capacity of T. purpureogenus to secret proteinase enzyme was optimized on tubers of H. tuberosus. To achieve this, the current trial uses a combination of inorganic (NH4Cl) and organic nitrogen (yeast extract) to support the SSF medium and induce the enzyme biosynthesis. The CCD experimental procedure is broadly used for such optimization purposes.
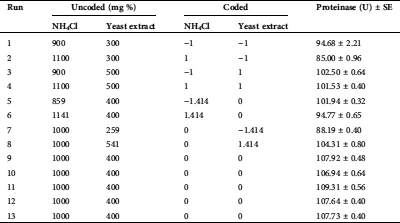
The design matrixes of CCD and proteinase response are introduced in Tab. 1. Runs of the center points of the matrix (No. 9, 10, 11, 12 and 13), recorded the uppermost proteinase secretion, with an average of 107.91 U by T. purpureogenus under the fermentation conditions.
Table 1: The experimental CCD matrix of the actual and the corresponding coded values of nitrogen sources used during the SSF of H. tuberosus tubers, as well as the response values of fungal proteinase
Study of the quadratic regression matrix
To assess the appropriateness of the planned design to predict the optimum concentration combinations, as well as the interaction of the studied parameters that maximize proteinase secretion. The ANOVA was statistically generated and evaluated.
The ANOVA was employed on the obtained CCD data (Tab. 2), from which the expected maximum yield of proteinase was calculated by the equation of the quadratic regression model. In which model F-value (146.99) was significant, the superior the F-value, the superior the model weight. In contrast, the model p-value is very low (0.0001); this also signifies that the proposed model is significant; the lower the p-value, the better the significance of the model.
Table 2: ANOVA for the quadratic model of the response surface of proteinase secretion by T. purpureogenus based on data obtained from the experimental CCD matrix
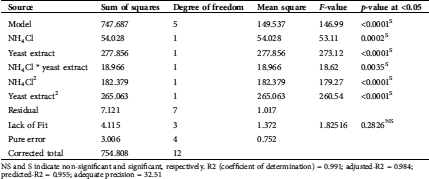
NS and S indicate non-significant and significant, respectively. R2 (coefficient of determination) = 0.991; adjusted-R2 = 0.984; predicted-R2 = 0.955; adequate precision = 32.51
The mathematical model is greatly reliable with an increment of the coefficient of determination up to 1.0, in this respect, R2, adjusted-R2 and predicted-R2 reached 0.991, 0.984 and 0.955, respectively. Another indication of the model acceptability is the non-significant lack-of-fit (p-value = 0.2826) and the high adequate precision, i.e., the ratio of signal to noise (32.51).
The significance of both nitrogen sources, as well as their interaction, were assessed, among the individual models utilized in the study, p-values (≤0.05) displayed that the individual, cross and quadratic relations amongst NH4Cl and yeast extract are significant models.
Evaluation of the model statistics
The probability of the normal curve of proteinase residuals was created (Fig. 1), using records acquired from the CCD, to assess the model precision. The pattern of spreading of the residuals of proteinase response follows a normal distribution, in which the residuals distributed following a straight-line shape without scattered values, indicating the precision of the model and indicating also the dispense to any modification or transformation to correct the proteinase response model.
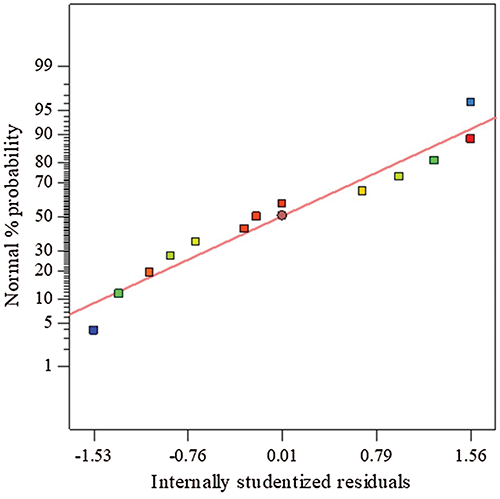
Figure 1: The normal figure of residuals of proteinase response based on data obtained from CCD.
The outline of the independent nitrogen sources was generated (Fig. 2) to explore the weight of their relationship on proteinase production. The plot represents the graphical picture of the regression equation, from which proteinase has schemed against the nitrogen sources. Exploring the linking between both variables reveals that the proteinase response reached its peak around the central concentrations of both nitrogen sources, suggesting that variables and their tested points, as well as, the experimental matrix are well-defined and achieved the peak of proteinase secretion.
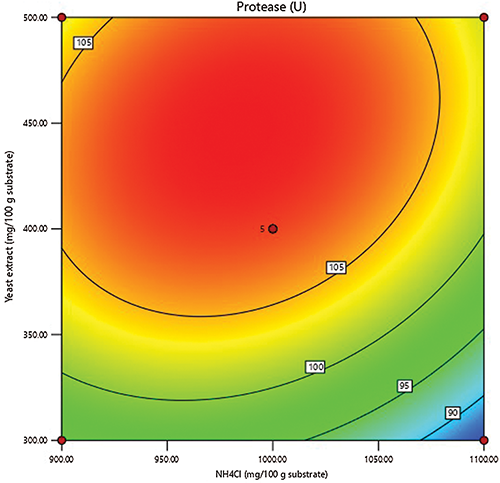
Figure 2: Contour figure of the response surface of proteinase production showing the shared effect of NH4Cl and yeast extract, based on data obtained from CCD.
Built on the previous ANOVA, the equation of the quadratic regression model of proteinase response was generated as follows:
Actual proteinase (U) = -413.367 + 0.911 × NH4Cl + 0.335 × yeast extract + 0.00022 × NH4Cl × yeast extract—0.00051 × (NH4Cl)2–0.00062 × (yeast extract)2.
This experimental equation was used to maximize the process proteinase response, and the nitrogen rates were set to be within the tested ranges. The optimum rates of each variable were calculated to be 979.82 NH4Cl and 437.68 mg% yeast extract. Rendering to the equation, the fitted proteinase response was calculated to be 109.403 U.
These theoretical calculations of the polynomial model are an approximation based on a reasonably small zone of the studied concentrations of the independent variables that requires the assurance of the real prediction efficacy of the equation.
The experimental laboratory validation was performed on the base of the calculated levels of both nitrogen sources, in which the proteinase production reached 108.15 ± 0.630 U. this activity is narrowly related to the theoretical value (109.403 U), indicating the precision of the quadratic model and strong prediction efficacy of suggested equation.
Proteinase production as a response to NPs
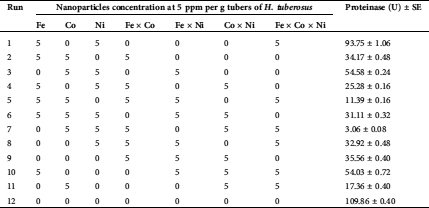
The NPs were investigated using PB design as promoters for proteinase secretion by T. purpureogenus. The optimum constituents of the medium constructed on the preceding CCD were prepared and further supported with the tested NPs. The response of proteinase secretion built on the matrix of PB design supported with the tested NPs is presented in Tab. 3. The data reveal great variations in proteinase production as a response to NPs. Proteinase production ranged from a maximum of 109.86 U (run number 12) to a minimum of 11.39 U in (run number 5).
Table 3: The planning matrix of Plackett-Burman with actual values of the assessed nanoparticles and the corresponding proteinase response by T. purpureogenus
Data of PB were statistically analyzed in terms of the estimated effects and ANOVA of the outcomes of the PB for the proteinase response are introduced in Tab. 4. The model p-value (0.023) is significant, reflecting the fitness of the design. Moreover, R2 (0.944) and adjusted-R2 (0.846), are also high enough to confirm the significance of the results, both are measures of how well the fitness of the data.
Table 4: Calculated effect and ANOVA for the secreted proteinase by T. purpureogenus based on Plackett-Burman design
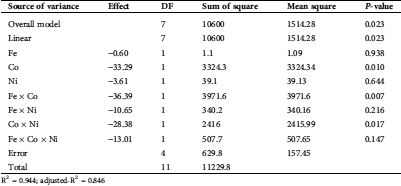
R2 = 0.944; adjusted-R2 = 0.846
The mono and dual utilization of NPs exhibited pronounced effects on fungal growth and proteinase production, meaning that proteinase secretion tends to be maximized. Regarding the tested Co and its interface with Fe or Ni, they were found to be significant on the bases of the p-level (<0.05). Pareto chart (Fig. 3) represents another support of the current data. In which, the standardized effects confirmed that Co and its interaction with Fe or Ni are the significant NPs on proteinase response, these three variables passed the base level of significant (2.776). The other variables failed to reach the minimum significant limit on proteinase secretion.
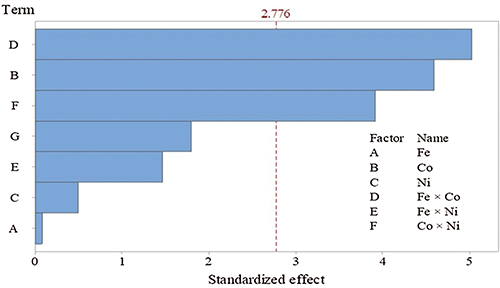
Figure 3: Pareto chart of the standardized effects of nano-particles on proteinase secretion by T. purpureogenus (p ≤ 0.05).
The mono and double interface of Co with Fe and Ni NPs were the significant variables on proteinase secretion. The NPs were investigated against the functioning of proteinase action on FBS as a substrate, aiming to release free AA. After half an hour on incubation, data of the proteolysis action (Fig. 4) indicate that the reaction combination containing Co NPs was the best in releasing AA (104.38 µg) from FBS, tracked by the mixture of Fe × Co NPs (93.89 µg). Comparatively, the nonappearance of NPs in the control (without NPs) possessed relatively lower releasing of AA, being 90.35 µg. Finally, the incidence of 5 ppm of an equal mixture of Co and Ni NPs came at the end.
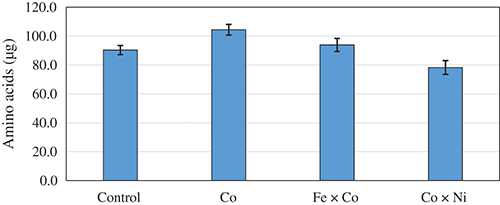
Figure 4: Amino acids liberation from faba bean straw as the response of individual Co and its combination with Fe or Ni nano-particles using 100 U of proteinase.
Fungal fermentation was employed using the ground tubers of H. tuberosus as a solid support and carbon source for proteinase production. T. purpureogenus, as the candidate fungus, was used because of its capability to use inulin sugar of H. tuberosus as a carbon source (El-Metwally et al., 2016). Tubers of H. tuberosus contain 20.4–31.9% dry matter, composed mainly from water-soluble inulin, fructooligosaccharides, reducing sugars (fructose and glucose), sucrose and dietary fibers (cellulose, hemicellulose, pectin and lignin), therefore, besides being a good substrate for the candidate fungus, H. tuberosus was chosen based on the health benefits that characterize its tubers (Cieslik-Bielecka et al., 2005), so, the resulted proteinase could securely find its way in various medication recipes.
This fermentation medium was supported by simple-inorganic (NH4Cl) and organic (yeast extract) nitrogen sources. To stimulate and maximize the proteinase biosynthesis by T. purpureogenus, the CCD of the RSM was implemented to determine the optimum interaction among nitrogen medium components, from one side, and H. tuberosus tubers, on the other side. According to the data, the best runs were found around the center points, reflecting the conciseness of the selected concentration ranges under study. The next step was to assess the appropriateness of the planned design to predict the optimum concentration combination, as well as the interaction of the studied parameters that maximize proteinase secretion.
The ANOVA of CCD data showed the significance of the model and each of the tested factors was evaluated. The model showed high F-value and low p-value, indicating that the proposed model matrix is significant. Other indications of the appropriateness of the model are R2, adjusted-R2 and predicted-R2, whose values were very close to 1. Generally, all types of R2 ranged from zero to 1. The closer the different kinds of R2 values are to 1, the better is suitable for the modeling of the investigational data. R2 values can help select the best-fitted model. R2 defines the quantity of change in the observed response (proteinase) values that are described by the tested variables (nitrogen sources). Because of the continuous increment of R2 with additional predictors, the adjusted-R2 is a modified R2 that has been accustomed to the count of factors in the model. It is a useful indicator of how well the model fits the data. Contrarily to R2, the adjusted-R2 may get smaller with the addition of more terms to the model. Predicted-R2 is used to measure the accuracy of the predictive ability of the model, e.g., to predict the value of proteinase response at new levels of the tested nitrogen sources. Moreover, it is more useful than adjusted-R2 for choosing and comparing models.
Another, the non-significance lack-of-fit is a prerequisite for the aptness of the overall design model, which is applied in the present model. Moreover, the ratio of adequate precision was ≥4, which is appropriate enough to indicate an adequate signal, so this model can be effectively utilized to circumnavigate the tested range of both nitrogen sources along with the design space to maximize proteinase production. These model statistics were further evaluated by the normal probability plot that confirmed the precision of the model.
To recognize the configuration of the importance of each of the individual tested nitrogen sources, the p-value is again utilized as a tool for the significance test of the coefficient of the individual nitrogen source, which, in turn, is necessary for the assessment of the connection degree amongst sources. However, the minimal the magnitude of p, the maximal significant is the coefficient. The values of p indicate that model terms to be significant (<0.05), indicating that they are important nutritional factors for the secretion of proteinase by T. purpureogenus. Also, suggesting that variables and their tested points, as well as the experimental matrix, are well-defined and achieved the peak of proteinase secretion. So, the prediction model was generated. The optimum levels of nitrogen sources that maximize proteinase secretion by the fungus were estimated by the prediction equation.
These theoretical calculations of the polynomial model are an approximation based on a reasonably small zone of the studied concentrations of the independent variables that requires the assurance of the real prediction efficacy of the equation. Therefore, the calculations were experimentally validated. Both calculated and experimental values showed close similarities to each other. That is, in turn, produces strong evidence for the appropriateness of the experimental design and its modeling.
Both organic and inorganic nitrogen sources were designated to serve as an enhancer for proteinase synthesis. They have a marked positive influence on proteinase secretion. However, it is already known that the preference for a nitrogen nutrient by a microorganism is being contingent on the microbial strain used and the target product. Yeast extract is a complicated hydrolysate of yeasts, providing nitrogenous compounds, vitamin B-complex, carbon, sulfur, trace nutrients, and important growth factors, which are essential for the development of diverse microorganisms (Filtenborg et al., 1990).
On the other side, owing to the simple structure, NH4Cl was elected as an inorganic nitrogen source, as its assimilation does not need complicated biological metabolism (El-Metwally et al., 2016). Both nitrogen sources represented a complimentary basis of nitrogen for the present T. purpureogenus strain and this stimulated proteinase production.
After the optimization of proteinase production, three NPs were screened for their impact on enzyme production on a medium composed of the optimum levels obtained upon CCD. NPs were also evaluated for their inducing ability of enzyme activity. For such purpose, the fractional factorial PB experimental design was employed. The data reveal great variations in proteinase production as a response to NPs. However, ANOVA showed the p-value of the design to be significant. These states were confirmed by high values of all kinds of R2. As mentioned above, the greater the R2 values (up to 1) the more correctness of the associations among the NPs and proteinase.
The mono and dual application of NPs pronounced proteinase production. The states and the standardized effects confirmed that Co, Co × Fe, and Co × Ni are the significant NPs on proteinase response. There is only a rare or no statement on the role of growth-promoting behavior of the nanomaterials on microorganisms, contrarily, the majority of the nanomaterials reported earlier have been verified to be effectual antimicrobial agents (Marambio-Jones and Hoek, 2010). A possible explanation is due to the high surface area, thus posting the physical-chemical features of the NPs, which may also influence the biological property of the material (Murray et al., 2000).
Throughout the progression of the study, it was notable that T. purpureogenus growth was induced. This observation is in counterrally to the most published reports. A conceivable reason for the enhancement of proteinase secretion by the current fungus may be coupled with the development of its growth. In this respect, an earlier study (Prasad et al., 2010) on symbiotic Piriformospora indica reported 2- to 3-fold improvement in the fungal growth in the incidence of titanium dioxide NPs, carbon nanotubes, and silver NPs. Further, the colony was smoother, round, and larger, and this morphology alteration was accompanied by the addition of NPs, not with the control. The electron microscopic, clearly, indicated that TiO2–NPs, obviously, increased the dense of spore and increased the size of the chlamydospores by almost 50%, relating to the control.
During the course of the existing study, the tested NPs were incorporated in the media before the autoclaving. It was stated that the time of NPs inclusion in the medium is vital since their inclusion before wet sterilization showed a positive and motivating impact on the mold compared with when inserted at the late-phase of growth (Prasad et al., 2010). Incorporation of the nanomaterials as media ingredients acted as a carrier for the fast uptake of nutrients, due to the unique properties such as huge surface area and small dimension that posting the uptake ability by the fungus (Gleiter, 2000; Prasad et al., 2010).
Regarding the induction of enzymatic activity by NPs against FBS as a natural substrate, generally, NPs succeeded to liberate higher AA from FBS. Co–NPs alone was the best inducer for the proteinase preparation of the fungus followed by a mixture of Fe × Co–NPs compared to the control (without NPs). Although the mechanism of action of Co on the biosynthesis of enzymes is poorly elucidated. Co metal had both inhibitory and stimulatory action on proteolysis occurred by Fusarium gibbosum, the alterations in enzyme activity are depending on the metal level and incubation period, moreover, it was suggested that its impact is backing to the intensification of proteinases biosynthesis and not backing to the direct inclusion in enzyme structure. However, it is proposed that Co participate directly or indirectly in cell metabolism, intensifying in the polymerization of bioactive substances, including enzymes (Clapco et al., 2013).
Interestingly, the interface among enzymes and NPs is ruled by the structure, size, surface shape and charge of NPs additionally, an alteration in the construction and job of an enzyme can be occurred by the NPs (Wu et al., 2009). This may declare why the existence of Co in the nanoform in the existing study causes a direct stimulatory influence on proteinase activity, even at such low concentration used (5 ppm).
Most recent studies on the relation between NPs and enzymes dealt, only, with the immobilization, for example, and not as a limitation, Verma et al. (2016) demonstrated that immobilization of β-glucosidase from Aspergillus niger on magnetic nanoparticles improved its thermal stability. A recent investigation stated the way by which the enzymes are activated in the occurrence of NPs, i.e., through linking with a single nanoparticle within the active enzymatic sites and exhibited cooperative catalysis of the substrate and thus the activity increased (Arsalan and Younus, 2018). However, at higher enzyme concentration, accumulation occurs, and the active site is not accessible to the substrate, which descends its catalytic action even when the enzyme kept its normal structure. So, maintaining a proper ratio of enzyme-NPs complex thus offers a novel modulating path of the enzyme performance on the substrate (Deka et al., 2012). Further, Vaidyanath et al. (2020) maximized the direct recovery and stabilization of cellulolytic enzymes from Trichoderma harzanium BPGF1 fermented broth using carboxymethyl inulin nanoparticles. To the best of our knowledge, no previous studies deal with the effect of nanoparticles in the production and activity of microbial proteases.
Summering up, the acquired data from such a report exhibited that T. purpureogenus serve as a good source of proteinase preparation with the aid of Co NPs. This novel line can be applied to modifying the productivity and activity of proteinase. Further, this approach could be practiced in regulating the productivity and performance of several biomolecules, like enzymes and hormones. Therefore, this work is considered a contribution in the biotechnological field of therapy production since the applied mechanism, i.e., nanoparticles, for enzyme production and activity could be generalized to extend to several procedures.
Availability of data and materials: All data generated or analyzed during this study are included in the manuscript.
Funding Statement: The authors would like to thank the Deanship of Scientific Research at King Khalid University, Abha, Kingdom of Saudi Arabia for funding this work under grant no. (R.G.P.1/75/40).
Conflict of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Arsalan A, Younus H. (2018). Enzymes and nanoparticles: modulation of enzymatic activity via nanoparticles. International Journal of Biological Macromolecules 118: 1833–1847. DOI 10.1016/j.ijbiomac.2018.07.030. [Google Scholar] [CrossRef]
Chakrabarti M, Kiseleva R, Vertegel A, Ray SK. (2015). Carbon nanomaterials for drug delivery and cancer therapy. Journal of Nanoscience and Nanotechnology 15: 5501–5511. DOI 10.1166/jnn.2015.10614. [Google Scholar] [CrossRef]
Cieslik-Bielecka A, Bielecki T, Gazdzik T, Cieslik T. (2005). Operation wound healing process after using the platelet rich plasma. International Journal of Oral and Maxillofacial Surgery 34: 166. DOI 10.1016/S0901-5027(05)81551-6. [Google Scholar] [CrossRef]
Clapco S, Bivol C, Ciloci A, Stratan M, Coropceanu E, Tiurin J, Rija A, Labliuc S, Bulhac I. (2013). The effect of some metal complexes of oxime ligands on proteolytic activity of Fusarium gibbosum CNMN FD 12 strain. Analele Universităţii din Oradea, Fascicula Biologie XX(153–58, 2020. [Google Scholar]
Cupp-Enyard C. (2008). Sigma’s non-specific protease activity assay-casein as a substrate. Journal of Visualized Experiments 19: 899–910. [Google Scholar]
Deka J, Paul A, Chattopadhyay A. (2012). Modulating enzymatic activity in the presence of gold nanoparticles. RSC Advances 2: 4736–4745. DOI 10.1039/c2ra20056b. [Google Scholar] [CrossRef]
El-Metwally MM, Saber WIA, AbdAl-Aziz SAA. (2016). New inuliolytic fungus: molecular identification and statistical optimization of its inulinase secretion for bioethanol production from Helianthus Tuberosus. International Proceedings of Chemical, Biological and Environmental Engineering 99: 7–18. [Google Scholar]
Filtenborg O, Frisvad JC, Thrane U. (1990) The significance of yeast extract composition on secondary metabolite production in penicillium, in modern concepts. In: Samson RA, Pitt JI, (eds.Penicillium and Aspergillus Classification Plenum, New York, pp. 433–441. [Google Scholar]
Gleiter H. (2000). Nanostructured materials: Basic concepts and microstructure. Acta Materialia 48: 1–29. DOI 10.1016/S1359-6454(99)00285-2. [Google Scholar] [CrossRef]
Gupta R, Beg Q, Lorenz P. (2002). Bacterial alkaline proteases: molecular approaches and industrial applications. Applied Microbiology and Biotechnology 59: 15–32. DOI 10.1007/s00253-002-0975-y. [Google Scholar] [CrossRef]
Gurumallesh P, Alagu K, Ramakrishnan B, Muthusamy S. (2019). A systematic reconsideration on proteases. International Journal of Biological Macromolecules 128: 254–267. DOI 10.1016/j.ijbiomac.2019.01.081. [Google Scholar] [CrossRef]
Kathiraven T, Sundaramanickam A, Shanmugam N, Balasubramanian T. (2015). Green synthesis of silver nanoparticles using marine algae Caulerpa racemosa and their antibacterial activity against some human pathogens. Applied Nanoscience 5: 499–504. DOI 10.1007/s13204-014-0341-2. [Google Scholar] [CrossRef]
Kumar CG, Takagi H. (1999). Microbial alkaline proteases: from a bioindustrial viewpoint. Biotechnology Advances 17: 561–594. DOI 10.1016/S0734-9750(99)00027-0. [Google Scholar] [CrossRef]
Lu AH, Salabas EE, Schüth F. (2007). Magnetic nanoparticles: synthesis, protection, functionalization, and application. Angewandte Chemie International Edition 46: 1222–1244. DOI 10.1002/anie.200602866. [Google Scholar] [CrossRef]
Marambio-Jones C, Hoek EMV. (2010). A review of the antibacterial effects of silver nanomaterials and potential implications for human health and the environment. Journal of Nanoparticle Research 12: 1531–1551. DOI 10.1007/s11051-010-9900-y. [Google Scholar] [CrossRef]
Murray CB, Kagan CR, Bawendi M. (2000). Synthesis and characterization of monodisperse nanocrystals and close-packed nanocrystal assemblies. Annual Review of Materials Science 30: 545–610. DOI 10.1146/annurev.matsci.30.1.545. [Google Scholar] [CrossRef]
Oskouie SFG, Tabandeh F, Yakhchali B, Eftekhar F. (2008). Response surface optimization of medium composition for alkaline protease production by Bacillus clausii. Biochemical Engineering Journal Biochemical Engineering Journal 39: 37–42. DOI 10.1016/j.bej.2007.08.016. [Google Scholar] [CrossRef]
Potumarthi R, Ch S, Jetty A. (2007). Alkaline protease production by submerged fermentation in stirred tank reactor using Bacillus licheniformis NCIM-2042: effect of aeration and agitation regimes. BEJ Biochemical Engineering Journal 34: 185–192. DOI 10.1016/j.bej.2006.12.003. [Google Scholar] [CrossRef]
Prasad R, Jain V, Varma A. (2010). Role of nanomaterials in symbiotic fungus growth enhancement. Current Science 99: 1189–1191. [Google Scholar]
Reddy LVA, Wee YJ, Yun JS, Ryu HW. (2008). Optimization of alkaline protease production by batch culture of Bacillus sp. RKY3 through Plackett-Burman and response surface methodological approaches. BITE Bioresource Technology 99: 2242–2249. DOI 10.1016/j.biortech.2007.05.006. [Google Scholar] [CrossRef]
Sahar EA, Hussein AM, Rahma MA. (2017). The use of magnetic nanoparticles in hyperthermia of Ehrlich tumor. Cancer Biology 7: 14–20. [Google Scholar]
Schröfel A, Kratosová G, Safarík I, Safaríková M, Raska I, Shor LM. (2014). Applications of biosynthesized metallic nanoparticles–A review. Acta Biomaterialia 10: 4023–4042. DOI 10.1016/j.actbio.2014.05.022. [Google Scholar] [CrossRef]
Vaidyanath S, Harish BS, Gayathri G, Trilokesh C, Uppuluri KB, Anbazhagan V. (2020). Maximizing the direct recovery and stabilization of cellulolytic enzymes from Trichoderma harzanium BPGF1 fermented broth using carboxymethyl inulin nanoparticles. International Journal of Biological Macromolecules 160: 964–970. DOI 10.1016/j.ijbiomac.2020.05.185. [Google Scholar] [CrossRef]
Verma ML, Puri M, Barrow CJ. (2013). Recent trends in nanomaterials immobilised enzymes for biofuel production. Critical Reviews in Biotechnology 36: 108–119. DOI 10.3109/07388551.2014.928811. [Google Scholar] [CrossRef]
Wu Z, Zhang B, Yan B. (2009). Regulation of enzyme activity through interactions with nanoparticles. International Journal of Molecular Sciences 10: 4198–4209. DOI 10.3390/ijms10104198. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |