2021 45(1): 149-156
DOI:10.32604/biocell.2021.010334

www.techscience.com/journal/biocell
| BIOCELL 2021 45(1): 149-156 DOI:10.32604/biocell.2021.010334 |  www.techscience.com/journal/biocell |
The effects of legumain in THP1 leukemia cells
State Key Laboratory of Experimental Hematology, National Clinical Research Center for Blood Diseases, Institute of Hematology & Blood Disease Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Tianjin, 300020, China
*Address correspondence to: Guoguang Zheng, zhengggtjchn@aliyun.com
Received: 27 February 2020; Accepted: 11 May 2020
Abstract: Legumain is a C13 family cysteine protease. It plays diverse roles under both physiological and pathological conditions. The high-level expression of legumain is detected in solid tumors. Legumain promotes the proliferation and migration of tumor cells. However, the effect of legumain in blood diseases has not been established. In this report, we studied the effect of legumain on leukemia cells by overexpressing it in THP1 cells. The results demonstrated that legumain promoted cell proliferation, whereas it had little effect on cell apoptosis. Furthermore, legumain promoted the migration of THP1 cells. It was worth noting that legumain decreased the stemness of THP1 cells. Further evidence showed that legumain decreased the expression of Oct4, Sox2, Myc in THP1 cells. Our study reveals the multifaced effects of legumain in leukemia cells and broadens the knowledge of legumain in malignancies.
Keywords: Legumain; Leukemia; Proliferation; Migration; Stemness
Legumain is a cysteine protease that belongs to the C13 family of cysteine proteases. It is also called asparagine endopeptidase (AEP) because of its specificity for asparagine bond (Chen et al., 1997). Legumain is widely distributed among species, such as plants, invertebrate parasites as well as mammals. It is well conserved in human and murine (Barrett and Rawlings, 2001). In mammals, legumain has a broad range of tissue and cellular distribution. It is most abundant in kidney and testis (Chen et al., 1997), whereas it can be detected in different cell types including bone marrow stromal cells, monocytes, adipocytes, and macrophages (Jafari et al., 2017; Solberg et al., 2015). Legumain also has a different subcellular distribution. It is mainly located in acid lysosomes, where the activation of the protease takes place, and it can also be detected in extracellular fluids such as serum (Lunde et al., 2017; Smith et al., 2012).
Legumain is involved in many key cellular pathways and biological processes under physiological conditions. Legumain plays an important role in the formation of MHC class II complex by removing the invariant chain chaperone of the MHC class II (Manoury et al., 2003). It also influences the processing of antigens for MHC class II presentation in antigen-presenting cells (Dall and Brandstetter, 2016). It also supports human Th1 cell induction and activates the CTSL-C3-IFN-γ signal axis in human CD4+T cells (Freeley et al., 2018).
Legumain is involved in different pathological processes as well. Legumain plays a role in neuroinflammation in cognitive impairment. Knockout of legumain reduces the level of neuroinflammation, so as to improve cognitive impairment in stressed mice (Lian et al., 2019). Legumain is involved in the induction of atherosclerotic vascular remodeling (Ozawa et al., 2019). Previous studies showed that legumain was overexpressed in different human solid tumors, and overexpression of legumain promoted the proliferation, migration, and metastasis of tumor cells (Liu et al., 2003). Furthermore, the high-level expression of legumain is related to poor prognosis and clinical stage in solid tumors (Zhen et al., 2015).
Leukemia is a rapidly progressing hematopoietic malignancy (Thomas and Majeti, 2017). Both intrinsic and extrinsic factors play important roles in the transformation and progression of leukemia (Wang et al., 2018a; Yang et al., 2018b). However, the effect of legumain, either as an intrinsic factor or an extrinsic factor in blood diseases, especially leukemia, has not been documented. In this study, we explored the effects of overexpressed legumain in leukemia cells by constructing THP1 cells overexpressing legumain. Overexpression of legumain promoted the proliferation and migration of THP1 cells, whereas decreased the stemness of THP1 cells.
THP1 and HEK293T cells were purchased from American Type Culture Collection. Lentivirus vector pLV-EF1α-MCS-IRES-Bsd was obtained from Boisettia Inc (San Diego, CA). Fetal bovine serum (FBS), trypsin, penicillin/streptomycin, OPTI-MEM, RPMI 1640, DMEM, sodium pyruvate, MEM NEAA, and L-glutamine were purchased from Gibco (USA). SYBR Green PCR kit was purchased from Takara (Japan). Xba I and Nhe I restriction endonucleases were obtained from New England BioLabs (UK). H4434 was purchased from Stem Cell Technologies (Vancouver, BC, Canada). Annexin V/PI kit was purchased from BioLegend (San Diego, CA). Transwell chambers were purchased from Millipore (Bedford, MA).
All cells were maintained in a humidified atmosphere at 37°C with 5% CO2. Parental THP1 cells and infected THP1 cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 100 U/mL penicillin, 100 µg/mL streptomycin. HEK293T cells were cultured in DMEM medium supplemented with 10% FBS, sodium pyruvate, MEM NEAA, and L-glutamine. Logarithmic growth cells were used for subsequent experiments.
Construction of legumain expressing vector
PCR primers (Forward: 5’-GGCCTCTAGAGCCACCATGGTTTGGAAAGTAGCTGTATTCCTCAGT-3’, Reverse: 5’-GGCCGCTAGCTCAGTAGTGACCAAGGCACACGTGGTCCAT-3’) were designed according to the coding sequence (CDS) of human legumain (NCBI: NM_005606.7). The purified PCR product was digested by Xba I and Nhe I before inserted into the lentivirus vector pLV-EF1α-MCS-IRES-Bsd.
Construction of THP1 cells overexpressing legumain
The THP1 cell line overexpressing legumain was constructed using the lentivirus system following standard protocols. Briefly, HEK293T cells were transfected with packaging plasmids along with blank or recombinant plasmids using X-termeGENE HP DNA Transfection Reagent (Roche, USA). Lentivirus was collected 48 h later. THP1 cells were mixed with lentivirus and polybrene (8 µg/mL), followed by spinning at 1800 rpm for 90 min. Then cells were cultured in a lentivirus-free medium for 48 h. Finally, antibiotic blasticidin S (Bsd) was added at a concentration of 10 µg/mL to screen infected cells.
BD CantoII flow cytometer (BD Biosciences) was used for FACS analysis. All experiments were conducted according to standard protocols. Data analysis was carried out using Diva (BD Biosciences, San Jose, CA) or FlowJo VX (Tree Star, San Carlos, CA).
Quantitative real time-PCR (qPCR)
Cells were collected, and total RNA was extracted by TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocols. RNA concentration was determined, and cDNA was synthesized using M-MLV First-Strand Synthesis Kit (Invitrogen, Carlsbad, CA) with oligo(dT) primer. qPCR was performed using SYBR Green Kit. The expression level of target genes was analyzed by the RQ value calculated through normalization to the internal control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) by the ΔΔCt method. All primer sequences are listed in Tab. 1.
For the cell counting assay, 5 × 104 cells were seeded per well in a 24-well plate. The cell number each well was counted every day.
For the MTS assay, the Cell Proliferation Assay kit (Promega, Madison, WI) was used following the manufacturer’s protocols. Briefly, cells were plated at a density of 1 × 104 cells/well in a 96-well plate. Two hours before each time point, 20 µL MTS reagent was added into each well. After incubation for 2 h, the absorbance at a wavelength of 490 nm was detected using a microplate reader.
Cell cycle analysis by PI staining
The detailed procedures of PI staining were described previously (Wang et al., 2018b). Briefly, cells were collected and resuspended in 70% cold ethanol on ice for 1 h. Then, RNA was digested with 0.2 mg/mL RNase for 1 h. At last, PI was added at a final concentration of 10 µg/mL overnight at 4°C. Cells were analyzed on FACS. All experiments were repeated three times.
Cells were collected and washed twice with annexin-V binding buffer. Then, cells were resuspended with annexin-V binding buffer. APC-conjugated annexin-V was added and incubated at room temperature for 15 min in the dark. Last, PI (500 µg/mL) was added. Flow cytometry was used to detect apoptotic cells. All experiments were repeated three times.
Transwell experiments were employed to evaluate cell migration. Cells were resuspended at a concentration of 1 × 106/mL in FBS free culture medium. An aliquot of 2 × 105 cells were added to the upper chamber (8.0-μm, Millicell), and 500 µL of 10% FBS medium was added to the lower chamber. After 24 h, membrane inserts were collected and non-invading cells on upper surface were removed. Then, the chamber membrane was fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. At least six fields of vision were randomly selected for cell counting under microscope. All experiments were repeated three times.
Cells were collected and adjusted to a concentration of 1 × 105/mL in H4434 complete medium. Five hundred cells in 500 µL were seeded in a 24-well plate following the manufacturer’s instructions. After seven days, colonies were counted under a microscope, and photos were taken by a high-content analysis system. Cell cluster with more than 50 cells was identified as one colony. All experiments were repeated three times.
Ki-67 staining was used for G0 phase analysis (Wang et al., 2018a). Briefly, cells were harvested, fixed, and permeabilized by Cytofix/CytopermTM Fixation/Permeabilization Solution Kit (BD, San Jose, CA). Then, PE-Cy7-conjugated Ki-67 was added and incubated for 30 min in the dark. Next, cells were washed twice and resuspended in PBS. Hoechst 33342 was added before analysis by flow cytometry. All experiments were repeated three times.
The results were expressed as mean ± SD. The analysis was done using GraphPad Prism 8.0 software. Comparisons between two groups were analyzed by unpaired Student’s t-test. Statistically significant was accepted when the p-value was less than 0.05.
Establishment of a THP1 cell line overexpressing legumain
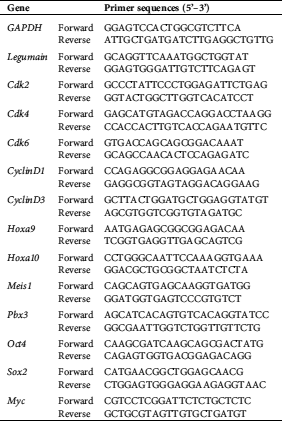
THP1 cell line overexpressing legumain was constructed to study the role of legumain on leukemia cells. First, the legumain expressing vector was constructed. After digestion by Xba I and Nhe I, the PCR fragment was inserted into the vector pLV-EF1α-MCS-IRES-Bsd (Fig. 1(A)). The recombinant plasmids were verified by Xba I and Nhe I restriction endonuclease digestion. The lanes 1 and 2 were two recombinant plasmids. As expected, the bright bands between 2,000 bp and 1000 bp were observed (Fig. 1(B)). These plasmids were further verified by DNA sequencing, showing that their sequences were consistent with the sequence in NCBI (NM_005606.7). Hence, we successfully constructed the legumain expressing vectors.
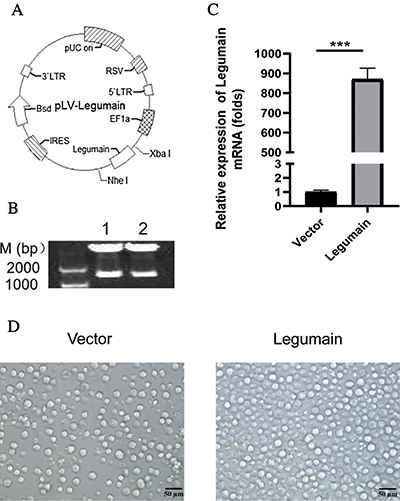
Figure 1: Establishment of an AML cell line overexpressing legumain.
Second, THP1 cells were infected with blank and recombinant lentiviruses. After Bsd selection, the blank lentivirus infected THP1 cell line was designated as the Vector group while the recombinant lentivirus infected one was designated as the Legumain group. The expression of legumain was assessed by qPCR experiments showing that cells in the Legumain group expressed about 800-fold higher legumain than those in the Vector group (Fig. 1(C)). Meanwhile, the morphology of cells in the two groups showed little differences (Fig. 1(D)). The above results indicated that we successfully established the THP1 cells overexpressing legumain. In addition, the overexpression of legumain has little effect on cell morphology.
Effects of legumain on the proliferation and apoptosis of THP1 cells
We explored whether the high-level expression of legumain had an effect on the proliferation and apoptosis of leukemia cells. The results from both cell counting (Fig. 2(A)) and MTS (Fig. 2(B)) analyses demonstrated that the overexpression of legumain significantly enhanced cell proliferation when compared with the Vector group. PI staining was used for cell cycle analysis. The results indicated that more S phase and less G0/G1 phase were detected in cells in the Legumain group (Fig. 2(C)). The results from the qPCR of cell cycle-related factors showed that the overexpression of legumain significantly increased the expression of Cdk6 and Cyclin D1 (Fig. 2(D)). Annexin V-PI analysis demonstrated that the overexpression of legumain had little effect on cell apoptosis (Fig. 2(E)).
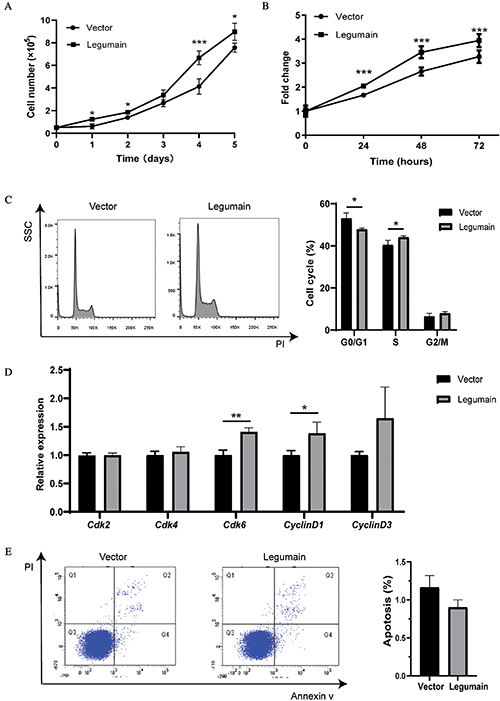
Figure 2: Effects of legumain on the proliferation and apoptosis of THP1 cells.
The above results indicated that high-level expression of legumain promoted cell proliferation but had little effect on cell apoptosis in THP1 cells.
Effect of legumain on the migration of THP1 cells
We studied whether high-level expression of legumain affected the migration of leukemia cells by using Transwell experiments. Typical results are shown in Fig. 3(A). More migrated cells were detected in the Legumain group than the Vector group (Fig. 3(B)). These results suggested that the overexpression of legumain promoted cell migration potential in THP1 cells.
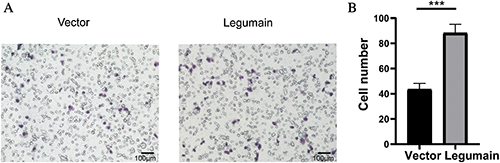
Figure 3: Effect of legumain on the migration of THP1 cells.
Effects of legumain on the stemness of THP1 cells
Finally, we studied whether the high-level expression of legumain affected the stemness of leukemia cells. The results of the colony-forming assay showed that fewer colonies were detected in the Legumain group than the Vector group (Fig. 4(A)). Then, the proportion of G0 phase cells was assessed by Hoechst 33342 and Ki-67 staining. The overexpression of legumain significantly decreased the proportion of G0 phase cells (Fig. 4(B)). The expression of related molecules was detected by qPCR (Fig. 4(C)). The overexpression of legumain had little effect on the expression of Hoxa9, Hoxa10, Meis1, and Pbx3, which had been suggested as key mediators in the transformation caused by MLL rearrangements (Ayton and Cleary, 2003; Li et al., 2016; Orlovsky et al., 2011; Wong et al., 2007). However, the overexpression of legumain downregulated the expression of Oct4, Sox2, and Myc, which were important stemness markers in embryonic stem cells and cancer stem cells. These results suggested that the overexpression of legumain reduced the stemness of THP1 cells.
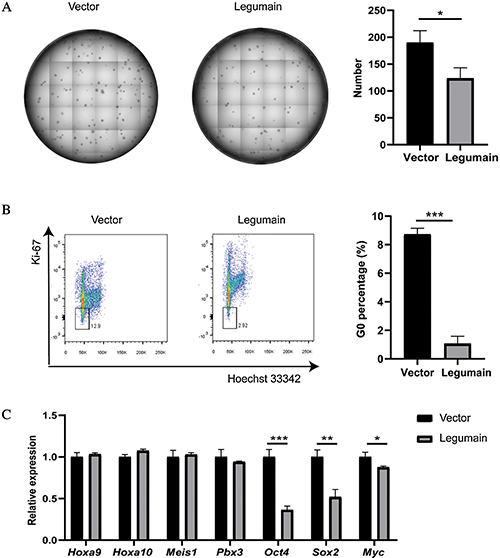
Figure 4: Effects of legumain on the stemness of THP1 cells.
Legumain plays roles under physiological conditions, especially in the formation of MHC class II (Manoury et al., 2003). In recent years, more attention has been paid to the pathological effects of legumain. It has been demonstrated that legumain played key roles in neurodegenerative disease and neuroinflammation in cognitive impairment (Basurto-Islas et al., 2013; Lian et al., 2019). In solid tumors, tumor cells expressed a high-level of legumain when compared with normal tissues. The effects of legumain in solid tumors have been widely studied (Liu et al., 2003). However, its effect on blood diseases has not been established. So, we aimed to investigate the role of legumain in leukemia cells in this study.
Legumain is expressed in different forms, i.e., cell surface, intracellular, and secretion forms (Fuchigami et al., 2019). These forms may function through different mechanisms. Here, we inserted full-length cDNA of legumain into a lentiviral vector and successfully established THP1 cells overexpressing legumain by infection. A previous study demonstrated that unstimulated THP1 cells do not secrete legumain (Solberg et al., 2015). Hence, we suggest that the majority of overexpressed legumain might be in intracellular and cell surface forms, whereas the secretion form cannot be excluded. Further work should be done to distinguish which form mainly contribute to the role of legumain in this model.
The effects of legumain on the proliferation and apoptosis of solid tumor cells have been well studied. In many cases, legumain promotes the proliferation of tumor cells (Liu et al., 2003; Wang et al., 2020). Furthermore, legumain promoted the proliferation of prostate cancer cells via the PI3K/AKT signaling pathway (Zhu et al., 2016). However, the effect of legumain on apoptosis varied among cells. The knockdown of legumain resulted in the decrease of apoptosis in prostate cancer cells (Zhu et al., 2016) but resulted in the increase of apoptosis in liver sinusoidal endothelial cells (Li et al., 2019). In this report, we constructed THP1 cells overexpressing legumain to detect the effect of legumain on cell proliferation and apoptosis. The results demonstrate that the overexpression of legumain significantly promotes cell proliferation without affecting cell apoptosis in THP1 cells. The effects of legumain on cell apoptosis should be complicated and may be cell type-dependent. The exact mechanism should be further elucidated.
Cancer stem cells (CSCs) and leukemia stem cells (LSCs) play adverse pathological roles in the initiation, progression, and relapse of malignancies (Chavez-Gonzalez et al., 2017). Although its pathological roles in solid tumors have been well studied, the effect of legumain on the CSCs has not been established. The in vitro colony-forming ability is one of the markers reflecting the self-renewal potential of leukemia cells. In this study, we discovered that the overexpression of legumain significantly decreased the colony-forming potential in THP1 cells. These results suggested that the overexpression of legumain reduced the self-renewal potential of leukemia cells. Furthermore, the overexpression of legumain in THP1 cells decreased the proportion of G0 phase cells. Moreover, the overexpression of legumain in THP1 cells decreased the expression of Oot4, Sox2, and Myc. These transcription factors are widely studied in embryonic stem cells and cancer stem cells (Vaddi et al., 2019; Villodre et al., 2019). In acute myeloid leukemia (AML), Oct4 and Sox2 were suggested as markers for LSCs (Picot et al., 2017). It was also reported that overexpression of c-MYC initiated AML in a mouse model (Luo et al., 2005). Hence, the overexpression of legumain may decrease the stemness of LSCs. The effects of legumain on the above-mentioned transcription factors have not been reported in other cancer cells. Whether the phenomenon occurs in other tumor cells and the related mechanism, should be further explored.
Legumain was suggested as a prognostic factor in certain solid tumors (Murthy et al., 2005; Ohno et al., 2013; Wang et al., 2012). However, our results demonstrated the complicated effects of legumain in leukemia. On the one hand, it promoted the proliferation of leukemia cells. On the other hand, it decreased the stemness of leukemia cells. Whether legumain plays favorable or adverse roles in leukemia need further work to elucidate. In fact, the effect of legumain in leukemia may be more complicated since legumain can be secreted to the microenvironment. Besides acting as an intrinsic factor in malignant cells, legumain may also act as an extrinsic factor by affecting cells in the malignant microenvironment. Legumain is also expressed in macrophages (Liu et al., 2014), which are important components of physiological and pathological microenvironments and play vital roles in the initiation and progression of solid tumors as well as leukemia (Chen et al., 2015; Etzerodt et al., 2020; Yang et al., 2018a). Recent evidence showed that loss of legumain in macrophages promoted senescence of tumor cells (Shen et al., 2019). Hence, both sides should be considered to unravel the role of legumain in leukemia.
Taken together, we demonstrate the complicated effects of legumain in leukemia cells. Legumain promotes the proliferation and migration of THP1 cells whereas decreases the stemness of THP1 cells.
Funding Statement: This work was supported by the following grants and programs: G. Z. received the grants (Nos. 81770183 and 81970155) from the National Natural Science Foundation of China (NSFC, http://www.nsfc.gov.cn/english/site_1/index.html) and the CAMS Innovation Fund for Medical Sciences (CIFMS) program (No. 2016-I2M-2-006) from Chinese Academy of Medical Sciences & Peking Union Medical College (CAMS, http://english.cams.cn/index.html). L. W. received the program (No. 2017-I2M-1-015) from the CIFMS. G. Z. was a recipient of the New Century Excellent Talents in University (No. NCET-08-0329) from the Ministry of Education of the People’s Republic of China (http://en.moe.gov.cn/).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Ayton PM, Cleary ML. (2003). Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes & Development 17: 2298–2307. DOI 10.1101/gad.1111603. [Google Scholar] [CrossRef]
Barrett AJ, Rawlings ND. (2001). Evolutionary lines of cysteine peptidases. Biological Chemistry 382: 727–733. DOI 10.1515/bchm.2001.382.5.727. [Google Scholar] [CrossRef]
Basurto-Islas G, Grundke-Iqbal I, Tung YC, Liu F, Iqbal K. (2013). Activation of asparaginyl endopeptidase leads to Tau hyperphosphorylation in Alzheimer disease. Journal of Biological Chemistry 288: 17495–17507. DOI 10.1074/jbc.M112.446070. [Google Scholar] [CrossRef]
Chavez-Gonzalez A, Bakhshinejad B, Pakravan K, Guzman ML, Babashah S. (2017). Novel strategies for targeting leukemia stem cells: sounding the death knell for blood cancer. Cellular Oncology 40: 1–20. DOI 10.1007/s13402-016-0297-1. [Google Scholar] [CrossRef]
Chen JM, Dando PM, Rawlings ND, Brown MA, Young NE, Stevens RA, Hewitt E, Watts C, Barrett AJ. (1997). Cloning, isolation, and characterization of mammalian legumain, an asparaginyl endopeptidase. Journal of Biological Chemistry 272: 8090–8098. DOI 10.1074/jbc.272.12.8090. [Google Scholar] [CrossRef]
Chen SY, Yang X, Feng WL, Liao JF, Wang LN, Feng L, Lin YM, Ren Q, Zheng GG. (2015). Organ-specific microenvironment modifies diverse functional and phenotypic characteristics of leukemia-associated macrophages in mouse T cell acute lymphoblastic leukemia. Journal of Immunology 194: 2919–2929. DOI 10.4049/jimmunol.1400451. [Google Scholar] [CrossRef]
Dall E, Brandstetter H. (2016). Structure and function of legumain in health and disease. Biochimie 122: 126–150. DOI 10.1016/j.biochi.2015.09.022. [Google Scholar] [CrossRef]
Etzerodt A, Moulin M, Doktor TK, Delfini M, Mossadegh-Keller N, Bajenoff M, Sieweke MH, Moestrup SK, Auphan-Anezin N, Lawrence T. (2020). Tissue-resident macrophages in omentum promote metastatic spread of ovarian cancer. Journal of Experimental Medicine 217: e20191869. DOI 10.1084/jem.20191869. [Google Scholar] [CrossRef]
Freeley S, Cardone J, Gunther SC, West EE, Reinheckel T, Watts C, Kemper C, Kolev MV. (2018). Asparaginyl endopeptidase (Legumain) supports human Th1 induction via cathepsin L-mediated intracellular C3 activation. Frontiers in Immunology 9: 2449. DOI 10.3389/fimmu.2018.02449. [Google Scholar] [CrossRef]
Fuchigami T, Itagaki K, Ishikawa N, Yoshida S, Nakayama M. (2019). Synthesis and evaluation of radioactive/fluorescent peptide probes for imaging of legumain activity. Bioorganic & Medicinal Chemistry Letters 29: 126629. DOI 10.1016/j.bmcl.2019.126629. [Google Scholar] [CrossRef]
Jafari A, Qanie D, Andersen TL, Zhang Y, Chen L, Postert B, Parsons S, Ditzel N, Khosla S, Johansen HT, Kjaersgaard-Andersen P, Delaisse JM, Abdallah BM, Hesselson D, Solberg R, Kassem M. (2017). Legumain regulates differentiation fate of human bone marrow stromal cells and is altered in postmenopausal osteoporosis. Stem Cell Reports 8: 373–386. DOI 10.1016/j.stemcr.2017.01.003. [Google Scholar] [CrossRef]
Li Z, Chen P, Su R, Hu C, Li Y, Elkahloun AG, Zuo Z, Gurbuxani S, Arnovitz S, Weng H, Wang Y, Li S, Huang H, Neilly MB, Wang GG, Jiang X, Liu PP, Jin J, Chen J. (2016). PBX3 and MEIS1 cooperate in hematopoietic cells to drive acute myeloid leukemias characterized by a core transcriptome of the MLL-rearranged disease. Cancer Research 76: 619–629. DOI 10.1158/0008-5472.CAN-15-1566. [Google Scholar] [CrossRef]
Li N, Liu C, Ma G, Tseng Y, Pan D, Chen J, Li F, Zeng X, Luo T, Chen S. (2019). Asparaginyl endopeptidase may promote liver sinusoidal endothelial cell angiogenesis via PI3K/Akt pathway. Revista Española de Enfermedades Digestivas: Órgano Oficial de la Sociedad Española de Patología Digestiva 111: 214–222. [Google Scholar]
Lian J, Li K, Gao J, Tan X, Yang Z. (2019). Legumain acts on neuroinflammatory to affect CUS-induced cognitive impairment. Behavioural Brain Research 376: 112219. DOI 10.1016/j.bbr.2019.112219. [Google Scholar] [CrossRef]
Liu C, Sun C, Huang H, Janda K, Edgington T. (2003). Overexpression of legumain in tumors is significant for invasion/metastasis and a candidate enzymatic target for prodrug therapy. Cancer Research 63: 2957–2964. [Google Scholar]
Liu Z, Xiong M, Gong J, Zhang Y, Bai N, Luo Y, Li L, Wei Y, Liu Y, Tan X, Xiang R. (2014). Legumain protease-activated TAT-liposome cargo for targeting tumours and their microenvironment. Nature Communications 5: 4280. DOI 10.1038/ncomms5280. [Google Scholar] [CrossRef]
Lunde NN, Holm S, Dahl TB, Elyouncha I, Sporsheim B, Gregersen I, Abbas A, Skjelland M, Espevik T, Solberg R, Johansen HT, Halvorsen B. (2017). Increased levels of legumain in plasma and plaques from patients with carotid atherosclerosis. Atherosclerosis 257: 216–223. DOI 10.1016/j.atherosclerosis.2016.11.026. [Google Scholar] [CrossRef]
Luo H, Li Q, O’neal J, Kreisel F, Le Beau MM, Tomasson MH. (2005). c-Myc rapidly induces acute myeloid leukemia in mice without evidence of lymphoma-associated antiapoptotic mutations. Blood 106: 2452–2461. DOI 10.1182/blood-2005-02-0734. [Google Scholar] [CrossRef]
Manoury B, Mazzeo D, Li DN, Billson J, Loak K, Benaroch P, Watts C. (2003). Asparagine endopeptidase can initiate the removal of the MHC class II invariant chain chaperone. Immunity 18: 489–498. DOI 10.1016/S1074-7613(03)00085-2. [Google Scholar] [CrossRef]
Murthy RV, Arbman G, Gao J, Roodman GD, Sun XF. (2005). Legumain expression in relation to clinicopathologic and biological variables in colorectal cancer. Clinical Cancer Research 11: 2293–2299. DOI 10.1158/1078-0432.CCR-04-1642. [Google Scholar] [CrossRef]
Ohno Y, Nakashima J, Izumi M, Ohori M, Hashimoto T, Tachibana M. (2013). Association of legumain expression pattern with prostate cancer invasiveness and aggressiveness. World Journal of Urology 31: 359–364. DOI 10.1007/s00345-012-0977-z. [Google Scholar] [CrossRef]
Orlovsky K, Kalinkovich A, Rozovskaia T, Shezen E, Itkin T, Alder H, Ozer HG, Carramusa L, Avigdor A, Volinia S, Buchberg A, Mazo A, Kollet O, Largman C, Croce CM, Nakamura T, Lapidot T, Canaani E. (2011). Down-regulation of homeobox genes MEIS1 and HOXA in MLL-rearranged acute leukemia impairs engraftment and reduces proliferation. Proceedings of the National Academy of Sciences of the United States of America 108: 7956–7961. DOI 10.1073/pnas.1103154108. [Google Scholar] [CrossRef]
Ozawa N, Sato Y, Mori Y, Masuda H, Yamane M, Yamamoto Y, Shirai R, Watanabe R, Sato K, Mori Y, Hirano T, Watanabe T. (2019). Legumain promotes atherosclerotic vascular remodeling. International Journal of Molecular Sciences 20: 2195. DOI 10.3390/ijms20092195. [Google Scholar] [CrossRef]
Picot T, Aanei CM, Fayard A, Flandrin-Gresta P, Tondeur S, Gouttenoire M, Tavernier-Tardy E, Wattel E, Guyotat D, Campos L. (2017). Expression of embryonic stem cell markers in acute myeloid leukemia. Tumour Biology 39: 1010428317716629. DOI 10.1177/1010428317716629. [Google Scholar] [CrossRef]
Shen L, Kang L, Wang D, Xun J, Chen C, Du L, Zhang M, Gong J, Mi X, Yue S, Zhang Y, Song X, Xiang R, Zhang Z, Tan X. (2019). Legumain-deficient macrophages promote senescence of tumor cells by sustaining JAK1/STAT1 activation. Cancer Letters 472: 40–49. DOI 10.1016/j.canlet.2019.12.013. [Google Scholar] [CrossRef]
Smith R, Johansen HT, Nilsen H, Haugen MH, Pettersen SJ, Maelandsmo GM, Abrahamson M, Solberg R. (2012). Intra- and extracellular regulation of activity and processing of legumain by cystatin E/M. Biochimie 94: 2590–2599. DOI 10.1016/j.biochi.2012.07.026. [Google Scholar] [CrossRef]
Solberg R, Smith R, Almlof M, Tewolde E, Nilsen H, Johansen HT. (2015). Legumain expression, activity and secretion are increased during monocyte-to-macrophage differentiation and inhibited by atorvastatin. Biological Chemistry 396: 71–80. DOI 10.1515/hsz-2014-0172. [Google Scholar] [CrossRef]
Thomas D, Majeti R. (2017). Biology and relevance of human acute myeloid leukemia stem cells. Blood 129: 1577–1585. DOI 10.1182/blood-2016-10-696054. [Google Scholar] [CrossRef]
Vaddi PK, Stamnes MA, Cao H, Chen S. (2019). Elimination of SOX2/OCT4-associated prostate cancer stem cells blocks tumor development and enhances therapeutic response. Cancers 11: 1331. DOI 10.3390/cancers11091331. [Google Scholar] [CrossRef]
Villodre ES, Felipe KB, Oyama MZ, Oliveira FH, Lopez P, Solari C, Sevlever G, Guberman A, Lenz G. (2019). Silencing of the transcription factors Oct4, Sox2, Klf4, c-Myc or Nanog has different effect on teratoma growth. Biochemical and Biophysical Research Community 517: 324–329. DOI 10.1016/j.bbrc.2019.07.064. [Google Scholar] [CrossRef]
Wang H, Chen B, Lin Y, Zhou Y, Li X. (2020). Legumain promotes gastric cancer progression through tumor-associated macrophages in vitro and in vivo. International Journal of Biological Sciences 16: 172–180. DOI 10.7150/ijbs.36467. [Google Scholar] [CrossRef]
Wang L, Chen S, Zhang M, Li N, Chen Y, Su W, Liu Y, Lu D, Li S, Yang Y, Li Z, Stupack D, Qu P, Hu H, Xiang R. (2012). Legumain: a biomarker for diagnosis and prognosis of human ovarian cancer. Journal of Cellular Biochemistry 113: 2679–2686. DOI 10.1002/jcb.24143. [Google Scholar] [CrossRef]
Wang R, Feng W, Yang F, Yang X, Wang L, Chen C, Hu Y, Ren Q, Zheng G (2018a). Heterogeneous effects of M-CSF isoforms on the progression of MLL-AF9 leukemia. Immunology and Cell Biology 96: 190–203. DOI 10.1111/imcb.1029. [Google Scholar] [CrossRef]
Wang L, Feng W, Yang X, Yang F, Wang R, Ren Q, Zhu X, Zheng G (2018b). Fbxw11 promotes the proliferation of lymphocytic leukemia cells through the concomitant activation of NF-κB and β-catenin/TCF signaling pathways. Cell Death & Disease 9: 427. DOI 10.1038/s41419-018-0440-1. [Google Scholar] [CrossRef]
Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. (2007). Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes & Development 21: 2762–2774. DOI 10.1101/gad.1602107. [Google Scholar] [CrossRef]
Yang X, Feng W, Wang R, Yang F, Wang L, Chen S, Ru Y, Cheng T, Zheng G (2018a). Repolarizing heterogeneous leukemia-associated macrophages with more M1 characteristics eliminates their pro-leukemic effects. Oncoimmunology 7: e1412910. DOI 10.1080/2162402X.2017.1412910. [Google Scholar] [CrossRef]
Yang F, Wang R, Feng W, Chen C, Yang X, Wang L, Hu Y, Ren Q, Zheng G (2018b). Characteristics of NK cells from leukemic microenvironment in MLL-AF9 induced acute myeloid leukemia. Molecular Immunology 93: 68–78. DOI 10.1016/j.molimm.2017.11.003. [Google Scholar] [CrossRef]
Ye Z, Guo CL, Shen WZ, Zhao ST, Luo N, Wang RR, Luo XH, Niu HY, Luo DH, Jiang S, Tan XY, Xiang R. (2015). Clinicopathologic significance of legumain overexpression in cancer: a systematic review and meta-analysis. Scientific Reports 5: 16599. DOI 10.1038/srep16599. [Google Scholar] [CrossRef]
Zhu W, Shao Y, Yang M, Jia M, Peng Y. (2016). Asparaginyl endopeptidase promotes proliferation and invasiveness of prostate cancer cells via PI3K/AKT signaling pathway. Gene 594: 176–182. DOI 10.1016/j.gene.2016.08.049. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |