DOI:10.32604/biocell.2021.09313

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.09313 |  www.techscience.com/journal/biocell |
| Article |
CircRNA ATF6 promotes ovarian cancer cell progression by activating PTEN/mTOR signaling pathway
1Department of Laboratory Medicine, The First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, 710061, China
2Department of Radiology, The First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, 710061, China
3Department of Cardiovascular Surgery, The First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, 710061, China
*Address correspondence to: Miaoling Li, limiaoling101@163.com; Xinglong Zheng, zhengxinglongmay@126.com
Received: 02 December 2019; Accepted: 28 April 2020
Abstract: Ovarian cancer is a malignant cancer type and affects women’s lives in the world. Circular RNAs (circRNAs) have been involved with the progression of cancers. In our study, we are going to explore the functions of circATF6 in ovarian cancer. The qRT-PCR assay was used to detect expressions of genes. Actinomycin D and RNase R treatment were implemented to verify the circular RNA character of circATF6. Besides, Cell proliferation was assessed by colony formation assay and EdU assay. Silenced circATF6 could reduce the proliferation of ovarian cancer cells. In addition, inhibited circATF6 could promote the cell apoptosis and inhibit related proteins in PTEN/mTOR signaling pathway in ovarian cancer. In conclusion, CircRNA ATF6 promotes ovarian cancer cell progression by activating PTEN/mTOR signaling pathway.
Keywords: CircRNA ATF6; PTEN/mTOR signaling pathway; Ovarian cancer
Ovarian cancer occupies 4% in female cancers (Torre et al., 2015), and its incidence ranks second only to cervical cancer and endometrial cancer among women (Torre et al., 2015). Estrogens have been reported to play an important regulatory role in the progression of ovarian cancer, which can promote cell growth and activate the aggressiveness of ovarian cancer (Mungenast and Thalhammer, 2014). Many efforts have been made to improve the treatment for ovarian cancer; nevertheless, the 5-year survival rate has yet not been increased. Therefore, for the discovery of more effective treatments for ovarian cancer patients, the underlying mechanism in ovarian cancer will be scrutinized in this study.
Accumulating studies have proved that non-coding RNAs (ncRNAs) participate in various cancer progression (Wang et al., 2018; Zhao et al., 2018). Circular RNAs (circRNAs) have been studied as a group of ncRNAs which have a covalently closed loop feature and can function as competing endogenous RNAs (ceRNA) to regulate the expression of microRNA (miRNA) (Chen et al., 2018; Shao et al., 2018; Wu et al., 2018). Meanwhile, plenty of circRNAs have been disclosed to be aberrantly expressed in ovarian cancer and affect the development of ovarian cancer (Ahmed et al., 2016). For example, circEPSTI1 was found to increase cell growth and invasion in ovarian cancer by regulating the expression of miR-942 (Xie et al., 2019). Meanwhile, circGFRA1 could enhance ovarian cancer cell growth and inhibit apoptosis by modulating miR-449a (Liu et al., 2019). CircATF6 has been discovered located in Chr1(q23.3) with 82034 in genome length and 1522 in spliced sequence length, which is also called hsa_circ_0015018 that expressed in Helas3 cells (Salzman et al., 2013). Moreover, the best transcript of this gene is ATF6. ATF6 knockdown was proved to decrease apoptosis and increase the production of steroid hormones in mouse granulosa cells through endoplasmic reticulum (ER) stress (Xiong et al., 2017), which revealed that circATF6 might have connections with the process of ovarian cancer. Therefore, in our study, the functions of circATF6 will be examined in ovarian cancer.
Human ovarian cancer cells (SKOV3, Caov3, A2780/CP) and normal human ovarian epithelial cells (IOSE-29) were procured from ATCC and preserved in DMEM with 5% CO2 at 37°C. Medium supplements included 10% FBS and 1% antibiotics. For transfection, the specific shRNAs and NC-shRNAs for circATF6 were all procured from Genepharma Company (Shanghai, China) called sh-circ-ATF6 and sh-NC. After cell confluence reached 85%, sh-circ-ATF6 and sh-NC were transfected into IOSE-29 cells using Lipofectamine 2000 (Invitrogen, USA). Thereafter, cells were incubated for another 24 h, and expressions of genes were quantified by qRT-PCR.
Total RNAs were extracted from cell samples using the Trizol method for the cDNA synthesis. Gene expression was detected via qRT-PCR using the SYBR Green PCR Master Mix, calculated using the 2-ΔΔCt method and relative to GAPDH. Conditions of PCR was listed as follows: pre-denaturation, 95°C, 5 min; denaturation, 95°C, 30 s; annealing, 60°C, 30 s; extension, 72°C, 30 s, 40 cycles.
Clonogenic cells were collected after transfection and then seeded into the 6-well culture plates for 14-day cell incubation. Mediums then were wiped out, and 4% paraformaldehyde was added to fixing cells and incubated for 15min at room temperature. After fixing, clones were stained in 0.5% crystal violet and cultured at room temperature for 1 h. Later, staining fluid was removed and rinsed with PBS once. Then, the numbers of clones were calculated. Cloning efficiency = numbers of cloning in plates/initial numbers of cells.
Cells in log phase were seeded into a 96-well plate with 5 × 103 cells per well and incubated for 24 h. EdU assay in cell samples was implemented using the standard method of 5-ethynyl-20-deoxyuridine (EdU) incorporation assay kit (Ribobio, Guangzhou China). Then, 50 μL of 4% paraformaldehyde was added to each well and keep culturing at room temperature for 30 min. After that, each cell was added with 50 μL 2 mg/mL glycine to incubate for 30 min. Later, PBS was applied to rinse cells. After DAPI staining, cell samples were subjected to a fluorescence microscope (Leica, Wetzlar, Germany).
Cell protein extracts were prepared and treated with 10% SDS-PAGE gel, then shifted onto the PVDF membranes. The primary antibodies available from Abcam (Cambridge, MA) were diluted at 1:2000, while the HRP-tagged secondary antibodies were diluted at 1:5000. After washing, protein samples were assayed by the enhanced chemiluminescence (ECL) fluorescence test kit (Amersham, Arlington Heights, IL).
Statistical analysis
Bio-triplicates were required for all experiments. Results were shown as the mean ± SD and analyzed by GraphPad Prism V5.0. Student’s t-test and one-way ANOVA were applied for data analysis, with p < 0.05 of statistically significant.
CircRNAs were upregulated in ovarian cancer cells
Some circRNAs have been reported aberrantly regulated in cancer and affected the progression of cancer by modulating the cell biological behaviors, such as cell proliferation, apoptosis, and invasion (Ahmed et al., 2016). We detected the expression of circATF6 in ovarian cancer cells at first. A qRT-PCR assay was implemented to assess the expression of circATF6 in ovarian cancer cells (SKOV3, Caov3, and A2780/CP) and normal human ovarian epithelial cell (IOSE-29) (Fig. 1A). We found circATF6 was highly upregulated in ovarian cancer cells than in normal human ovarian epithelial cells, especially in SKOV3 and Caov3 cells. Then, we verified the inhibition efficiency of circATF6 in SKOV3 and Caov3 cells (Fig. 1B). In conclusion, circRNAs were upregulated in ovarian cancer cells.
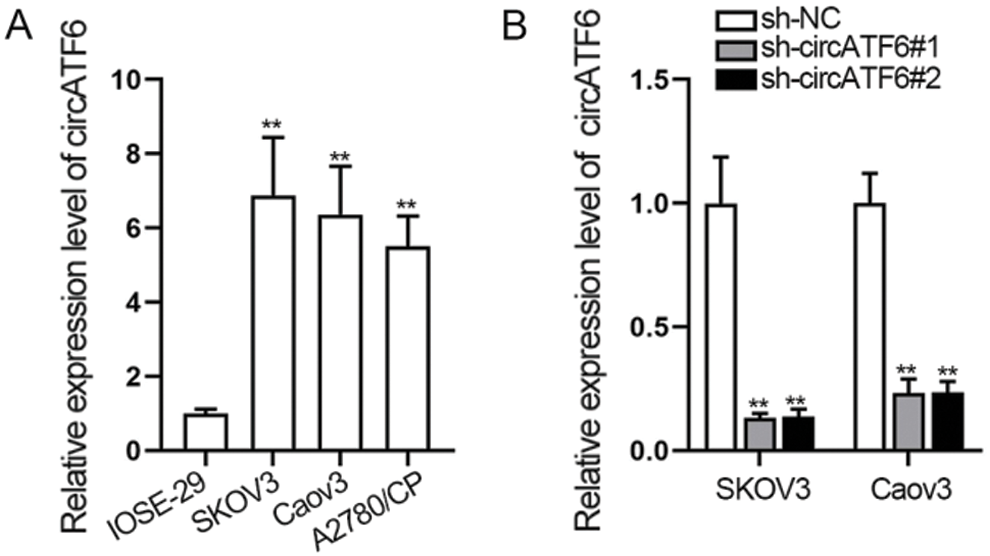
Figure 1: CircRNAs was upregulated in ovarian cancer cells. (A) expression of circATF6 was detected in ovarian cancer cells (SKOV3, Caov3 and A2780/CP) and normal human ovarian epithelial cells (IOSE-29) via qRT-PCR assay. (B) inhibition efficiency of circATF6 was examined via qRT-PCR assay in SKOV3 and Caov3 cells.
CircATF6 could function as circular RNA in ovarian cancer cells
Then, we further tested the circular RNA character of circATF6. Treatment of Actinomycin D and RNase R was implemented on SKOV3 and Caov3 cells (Figs. 2A–2B). We found that with the Actinomycin D treatment time increasing, the expression of linear ATF6 was significantly decreased, but the expression of circATF6 has not changed much. Also, RNase R treatment significantly decreased the expression of linear ATF6 and GAPDH. The expression of circATF6 was not reduced. In a word, circATF6 could function as circular RNA in ovarian cancer cells.
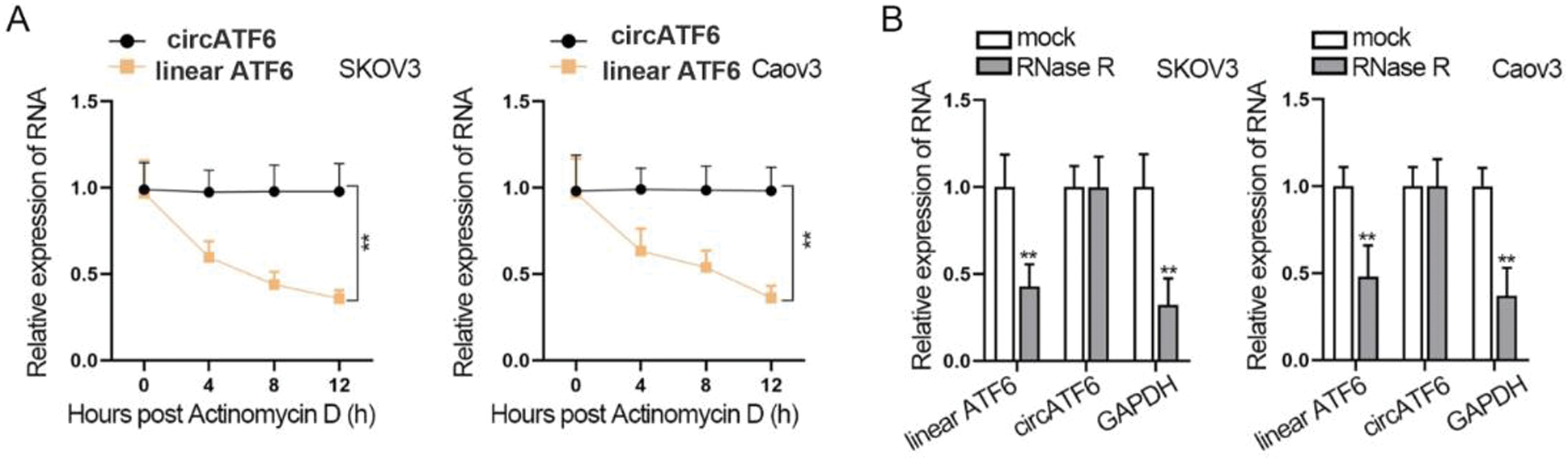
Figure 2: CircATF6 could function as circular RNA in ovarian cancer cells. (A-B) Actinomycin D and RNase R treatment was implemented to verify the circular RNA character of circATF6.
Silenced circATF6 could reduce the proliferation of ovarian cancer cells
For testing the function of circATF6 in ovarian cancer cells, functional assays were implemented. Colony formation assay and EdU assay assessed the cell proliferation when circATF6 was depleted (Figs. 3A–3B). Results showed that both the number of colonies and EdU positive cells were reduced by inhibited circATF6. Silenced circATF6 could reduce the proliferation of ovarian cancer cells.
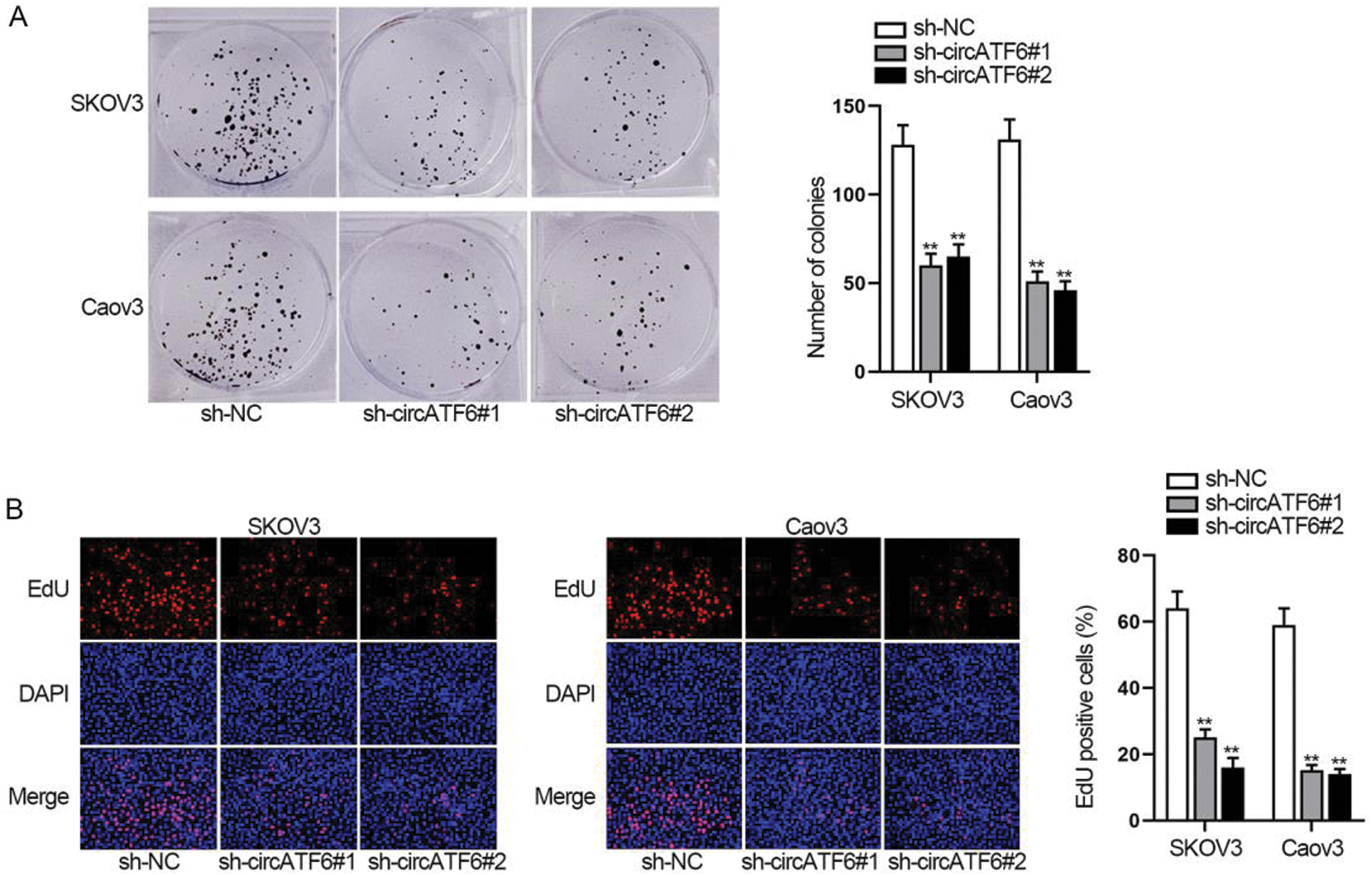
Figure 3: Silenced circATF6 could reduce the proliferation of ovarian cancer cells. (A-B) Cell proliferation was assessed by colony formation assay and EdU assay when circATF6 was depleted.
Inhibited circATF6 could promote the cell apoptosis and inhibit PTEN/mTOR signaling pathway in ovarian cancer cells
Moreover, cell apoptosis was identified by qRT-PCR assay. Apoptosis related gene expression (Bal-2, cyclin D1, C-FLIP, caspase3, and caspase7) were tested (Fig. 4A). Results pointed out that Bal-2, cyclin D1, C-FLIP, caspase3, and caspase7 expression levels were all elevated when circATF6 was inhibited, which signified that silenced circATF6 could promote the apoptosis of ovarian cancer cells. PTEN/mTOR signaling pathway has been reported as a classical pathway in modulating the progression of cancers (Riquelme et al., 2016). Thence, we adopted a western blot assay for the protein level of PTEN and mTOR (Fig. 4B). We found that the mTOR protein level was decreased, and PTEN protein level was elevated by silenced circATF6. In a word, inhibited circATF6 could promote the cell apoptosis and inhibit PTEN/mTOR signaling pathway in ovarian cancer.
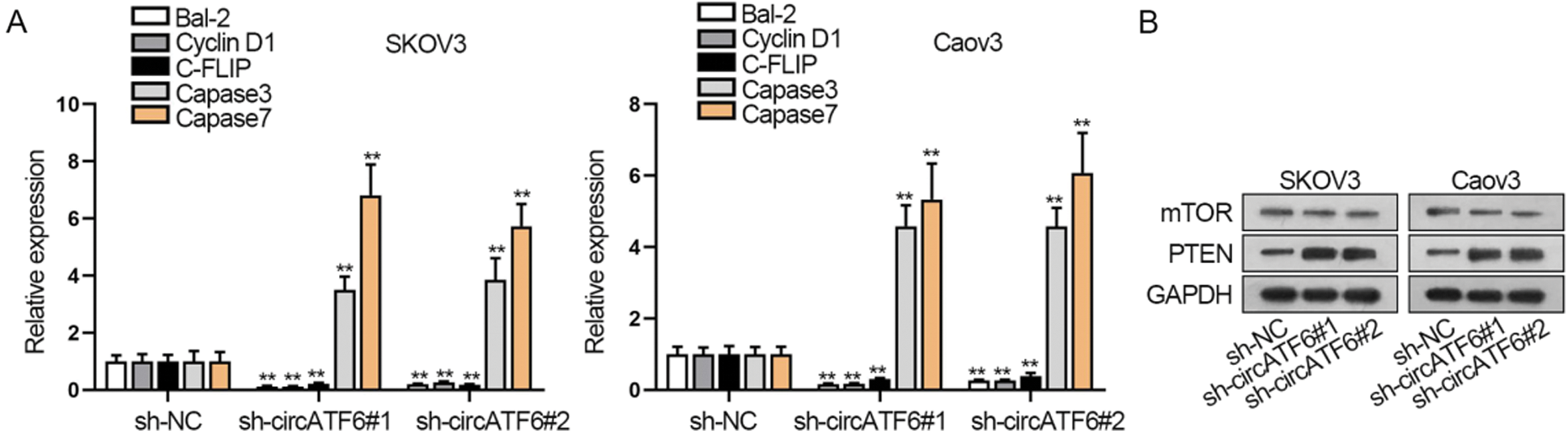
Figure 4: Inhibited circATF6 could promote the cell apoptosis and inhibit PTEN/mTOR signaling pathway in ovarian cancer cells. (A) Apoptosis-related gene expression was tested via qRT-PCR assay. (B) Western blot assay was conducted to know about the protein level change of PTEN and mTOR when circATF6 was inhibited.
CircRNAs can act as tumor activator or tumor suppresser in cancers. For instance, CircRNA_102171 can increase the cell growth and migration of papillary thyroid cancer by activating the pathway of β-catenin (Bi et al., 2018). Meanwhile, CircRNA-100338 can exacerbate the prognosis of hepatocellular carcinoma by modulating the expression of miR-141-3p and RHEB to regulate mTOR (Huang et al., 2019). Also, circRNA_102958 can promote colorectal cancer cell growth and invasion by regulating the miR-585/CDC25B pathway (Li et al., 2019). In our study, we firstly identified the expression of circATF6 in ovarian cancer cells via qRT-PCR, and we found that circATF6 was significantly highly expressed in ovarian cancer cells. Then, we further detected the circular character of circATF6 via RNase R and actinomycin D treatment. We found that circATF6 expression was not changed by RNase R and actinomycin D treatment. Moreover, functional assays such as colony formation assay and EdU assay were implemented to detect the cell proliferation of ovarian cancer, and results indicated that cell proliferation was significantly decreased by inhibited circATF6. Meanwhile, the qRT-PCR assay was implemented to verify the cell apoptosis. Apoptosis related genes were tested. Furthermore, we detected the cell apoptosis was increased via silenced circATF6.
PTEN/mTOR signaling pathway has been reported as a classical pathway in modulating the progression of cancers (Riquelme et al., 2016). PTEN is known as a tumor suppressor in varied cancers (Lazaridis et al., 2019; Sun et al., 2018). mTOR has been widely explored in a diversity of cancers, which was confirmed to be a tumor promoter (Li et al., 2018). Activation of the PTEN/mTOR signaling pathway, namely, the suppression of PTEN and the upregulation of mTOR can promote cell proliferation and inhibit cell apoptosis in cancers (Kremer et al., 2006). In the present study, the protein concentrations of PTEN and mTOR were investigated via western blot assay, and results displayed that PTEN was upregulated while mTOR was decreased, suggesting the inactivation of the PTEN/mTOR signaling pathway when circATF6 was silenced in ovarian cancer cells. Therefore, we concluded that circular RNA ATF6 might induce cell proliferation and inhibits cell apoptosis by activating PTEN/mTOR signaling pathway in ovarian cancer.
Funding Statement: The author(s) received no specific funding for this study.
Conflicts of Interest: Authors declare no conflicts of interest in this study.
Ahmed I, Karedath T, Andrews SS, Al-Azwani IK, Mohamoud YA, Querleu D, Rafii A, Malek JA. (2016). Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget 7: 36366–36381. DOI 10.18632/oncotarget.8917. [Google Scholar] [CrossRef]
Bi W, Huang J, Nie C, Liu B, He G, Han J, Pang R, Ding Z, Xu J, Zhang J. (2018). CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway. Journal of Experimental & Clinical Cancer Research 37: 698. DOI 10.1186/s13046-018-0936-7. [Google Scholar] [CrossRef]
Chen D, Zhang C, Lin J, Song X, Wang H. (2018). Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma. Cancer Management and Research 10: 1275–1283. DOI 10.2147/CMAR.S166740. [Google Scholar] [CrossRef]
Huang XY, Huang ZL, Zhang PB, Huang XY, Huang J, Wang HC, Xu B, Zhou J, Tang ZY. (2019). CircRNA-100338 is associated with mTOR signaling pathway and poor prognosis in hepatocellular carcinoma. Frontiers in Oncology 9: 392. DOI 10.3389/fonc.2019.00392. [Google Scholar] [CrossRef]
Kremer CL, Klein RR, Mendelson J, Browne W, Samadzedeh LK, Vanpatten K, Highstrom L, Pestano GA, Nagle RB. (2006). Expression of mTOR signaling pathway markers in prostate cancer progression. Prostate 66: 1203–1212. DOI 10.1002/pros.20410. [Google Scholar] [CrossRef]
Lazaridis G, Kotoula V, Vrettou E, Kostopoulos I, Manousou K, Papadopoulou K, Giannoulatou E, Bobos M, Sotiropoulou M, Pentheroudakis G, Efstratiou I, Papoudou-Bai A, Psyrri A, Christodoulou C, Gogas H, Koutras A, Timotheadou E, Pectasides D, Zagouri F, Fountzilas G. (2019). Opposite prognostic impact of single PTEN-loss and PIK3CA mutations in early high-risk breast cancer. Cancer Genomics & Proteomics 16: 195–206. DOI 10.21873/cgp.20125. [Google Scholar] [CrossRef]
Li J, Feng L, Tian C, Tang YL, Tang Y, Hu FQ. (2018). Long noncoding RNA-JPX predicts the poor prognosis of ovarian cancer patients and promotes tumor cell proliferation, invasion and migration by the PI3K/Akt/mTOR signaling pathway. European Review for Medical and Pharmacological Sciences 22: 8135–8144. [Google Scholar]
Li R, Wu B, Xia J, Ye L, Yang X. (2019). Circular RNA hsa_circRNA_102958 promotes tumorigenesis of colorectal cancer via miR-585/CDC25B axis. Cancer Management & Research 11: 6887–6893. [Google Scholar]
Liu J, Yu F, Wang S, Zhao X, Jiang F, Xie J, Deng M. (2019). circGFRA1 promotes ovarian cancer progression by sponging miR-449a. Journal of Cancer 10: 3908–3913. DOI 10.7150/jca.31615. [Google Scholar] [CrossRef]
Mungenast F, Thalhammer T. (2014). Estrogen biosynthesis and action in ovarian cancer. Frontiers in Endocrinology 5: 192. DOI 10.3389/fendo.2014.00192. [Google Scholar] [CrossRef]
Riquelme I, Tapia O, Espinoza JA, Leal P, Buchegger K, Sandoval A, Bizama C, Araya JC, Peek RM, Roa JC. (2016). The gene expression status of the PI3K/AKT/mTOR pathway in gastric cancer tissues and cell lines. Pathology & Oncology Research 22: 797–805. DOI 10.1007/s12253-016-0066-5. [Google Scholar] [CrossRef]
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. (2013). Cell-type specific features of circular RNA expression. PLoS Genetics 9: e1003777. DOI 10.1371/journal.pgen.1003777. [Google Scholar] [CrossRef]
Shao F, Huang M, Meng F, Huang Q. (2018). Circular RNA signature predicts gemcitabine resistance of pancreatic ductal adenocarcinoma. Frontiers in Pharmacology 9: 107. DOI 10.3389/fphar.2018.00584. [Google Scholar] [CrossRef]
Sun J, Li T, Zhao Y, Huang L, Sun H, Wu H, Jiang X. (2018). USP10 inhibits lung cancer cell growth and invasion by upregulating PTEN. Molecular and Cellular Biochemistry 441: 1–7. DOI 10.1007/s11010-017-3170-2. [Google Scholar] [CrossRef]
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. (2015). Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians 65: 87–108. DOI 10.3322/caac.21262. [Google Scholar] [CrossRef]
Wang BG, Xu Q, Lv Z, Fang XX, Ding HX, Wen J, Yuan Y. (2018). Association of twelve polymorphisms in three onco-lncRNA genes with hepatocellular cancer risk and prognosis: A case-control study. World Journal of Gastroenterology 24: 2482–2490. DOI 10.3748/wjg.v24.i23.2482. [Google Scholar] [CrossRef]
Wu J, Jiang Z, Chen C, Hu Q, Fu Z, Chen J, Wang Z, Wang Q, Li A, Marks JR, Guo C, Chen Y, Zhou J, Yang L, Lin C, Wang S. (2018). CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Letters 430: 179–192. DOI 10.1016/j.canlet.2018.05.033. [Google Scholar] [CrossRef]
Xie J, Wang S, Li G, Zhao X, Jiang F, Liu J, Tan W. (2019). circEPSTI1 regulates ovarian cancer progression via decoying miR-942. Journal of Cellular and Molecular Medicine 23: 3597–3602. DOI 10.1111/jcmm.14260. [Google Scholar] [CrossRef]
Xiong Y, Chen H, Lin P, Wang A, Wang L, Jin Y. (2017). ATF6 knockdown decreases apoptosis, arrests the S phase of the cell cycle, and increases steroid hormone production in mouse granulosa cells. American Journal of Physiology—Cell Physiology 312: C341–C353. DOI 10.1152/ajpcell.00222.2016. [Google Scholar] [CrossRef]
Zhao M, Wang S, Li Q, Ji Q, Guo P, Liu X. (2018). MALAT1: A long non-coding RNA highly associated with human cancers. Oncology Letters 16: 19–26. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |