DOI:10.32604/biocell.2021.012280

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.012280 |  www.techscience.com/journal/biocell |
| Article |
Identification of key pathways and gene expression in the activation of mast cells via calcium flux using bioinformatics analysis
1Laboratory of Immunology and Virology, Experiment Center for Science and Technology, Shanghai University of Traditional Chinese Medicine, Shanghai, 201203, China
2Division of Gastrointestinal Pathophysiology, Institute of Natural Medicine, University of Toyama, Toyama, 9300194, Japan
*Address correspondence to: Xiaoyu Wang, 0000002733@shutcm.edu.cn
Received: 23 June 2020; Accepted: 15 October 2020
#These authors contributed equally to this work
Abstract: Mast cells are the main effector cells in IgE-associated allergic disorders, and we have reported that mucosal mast cells (MMCs) play a more important role in the development of food allergy (FA). IgE with antigen or calcium ionophore stimulation can lead to the activation of MMCs via a calcium-dependent pathway. The purpose of the present study was to identify gene signatures with IgE/antigen (dinitrophenyl-bovine serum albumin, DNP-BSA) or calcium ionophore (A23187) on the activation of MMCs. Differentially expressed genes between the two types of samples were identified with microarray analysis. Gene ontology functional and pathway enrichment analyses of differentially expressed genes were performed using the database for annotation, visualization, and integrated discovery software. The results showed that IgE/antigen and A23187 could induce degranulation, increase vacuoles, and elevate the cytosolic calcium concentration in MMCs. Furthermore, GeneChip analysis showed that the same 134 mRNAs were altered with IgE/DNP-BSA and A23187, suggesting that DNP-BSA/IgE and A23187 affect the same signal pathway partly in degranulation. KEGG analysis showed that the data were enriched in NF-κB, TNF, MAPK, transcription factor activity, DNA binding, and nucleic acid binding, suggesting that activation of MMCs is a complex process. The results provide new insights on MMCs activation.
Keywords: Mucosal mast cell; Antigen; Calcium ion; Bioinformatics analysis
Mast cells (MCs) are multifunction secretory cells originating from CD34+ CD117+ myeloid progenitor cells, which develop only once bone marrow-derived precursors have reached their target tissue. The ubiquitous distribution of MCs includes the epithelium, endothelium, immune system, and areas around nerve endings (da Silva et al., 2014; Arthur and Bradding, 2016; Rivera and Gilfillan, 2006). Although MCs are involved in cancer, Crohn’s disease, cardiovascular disease, rheumatoid arthritis (RA) (Conti et al., 2007; Boeckxstaens, 2015; Sun et al., 2007; Rivellese et al., 2019), etc., they are always considered as effector and conductor cells of allergic diseases, such as food allergies (FAs), atopic dermatitis and asthma (Kraneveld et al., 2012; Yamamoto et al., 2014).
MCs are usually classified into two groups, connective-tissue mast cells (CTMCs) and mucosal mast cells (MMCs). CTMCs accumulate substantial amounts of histamine and contribute to allergic diseases, such as allergic dermatitis and rhinitis; however, MMCs contain less histamine, and antihistamine agents have few effects on FA (Gurish and Boyce, 2002; Elieh Ali Komi et al., 2020). Previously, we demonstrated that MMCs play more important roles in anaphylaxis in the colon of mice with FA and protection against parasitic infections than CTMCs (Yamamoto et al., 2009; Yamamoto et al., 2014).
Additionally, the MMCs marker mouse mast cell protease (mMCP)-1 was expressed in the small intestine, but not the skin of mice with FA, and activated MMCs released preformed granule contents, proteases (such as tryptase and chymase), lipid mediators, and inflammatory mediators (cytokines, chemokines, etc.), which were also involved in FA (Benedé and Berin, 2018). Consistent with conclusions of other researchers (Koyuncu Irmak et al., 2019), we reported that calcitonin gene-related peptide (CGRP), a neurotransmitter, augmented MMCs degranulation to contribute to FA development, suggesting that neurogenic inflammation is also associated with FA (Kim et al., 2014). Furthermore, CTMCs stabilizers (cromolyn, ketotifen, tranilast, etc.) are frequently used to treat various allergic disorders except for FA; however, they have little effect on MMCs (Wang et al., 2012).
The prevalence of FA is estimated to be up to approximately 10%. The underlying mechanisms of FA are not fully understood, and there is no current Food and Drug Administration-approved therapy besides avoidance of ingestion of the allergen (Iweala et al., 2018; Sicherer and Sampson, 2018). Because MMCs are primarily responsible for the pathology of FA, a deeper understanding of MMCs activation is crucial in the search for FA markers in patients and the generation of agents against FA and other allergic diseases.
Conventional MCs activation is initiated by an allergen cross-linked to immunoglobulin E (IgE) bound to MCs via the high-affinity IgE receptor (FcεRI). The increase in the cytosolic Ca2+ concentration ([Ca2+]i) triggered by the aggregation of IgE with FcεRI is an essential step during mast cell activation (Law et al., 2011). The source of the [Ca2+]i is considered to be from two origins: a release from intracellular stores (mainly the endoplasmic reticulum) and influx from the extracellular space through plasma membrane Ca2+ channels (such as Ca2+ release-activated Ca2+ (CRAC) channels, transient receptor potential (TRP), etc.), which is required for sustained [Ca2+]i elevations (Feske et al., 2015). Calcium mobilization triggers principal signaling pathways to release preformed mediators and activate proinflammatory cytokine gene transcription. A23187, as a divalent calcium ionophore, is commonly used to increase the permeability of biological membranes to calcium.
MCs activation can also be caused by non-IgE-mediated triggers (Johnson et al., 2018). For example, the crosslinking of Fc(gamma)RIII receptor with IgG immune complexes can release several lipid mediators and induce degranulation in MCs. The binding of stem cell factor (SCF) to c-kit (CD117, the receptor for SCF) can increase antigen-stimulated degranulation (Oda et al., 2013; Tkaczyk et al., 2004). Furthermore, toll-like receptors (TLRs), neuropeptide receptors, toxins, physical factors (heat), synthetic chemical (compound 48/80), and severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) can initiate MCs activation by non-IgE-mediated mechanisms. Activated MCs release their pre-stored mediators without degranulation (Koyuncu Irmak et al., 2019; Kilinc and Baranoglu Kilinc, 2020a). Thus, to develop new drugs to treat MCs-related diseases such as FA, besides degranulation, a deeper understanding of MMCs activation by IgE-mediated or non-IgE-mediated (such as A23187) mechanisms is needed.
It is impossible to capture the complex interactions in MMCs activation by studying a single target. However, microarray technology, which is a high-throughput platform for gene signatures, can comprehensively evaluate global biological mRNA changes in the activation of MMCs (Dwyer et al., 2016).
Miller et al. (1999) reported that mucosal-type murine bone marrow-derived mast cells (mBMMCs) cultured with a combination of 4 cytokines (SCF, interleukin (IL)-3, transforming growth factor β1 (TGF-β1) and IL-9) were homologous to mouse MMCs based on the morphology and expression of mMCP-1 and mMCP-2. To investigate the genome-wide gene expression differences in mBMMCs via antigen IgE-dependent or IgE-independent activation, we used IgE/dinitrophenyl-bovine serum albumin (DNP-BSA) stimulation or calcium ionophore A23187 treatment for mBMMCs. In the present study, we attempted to use a bioinformatics analysis on GeneChip so as to examine the downstream pathway of FcεRI-depend mBMMCs activation.
Recombinant murine IL-3, recombinant murine SCF, recombinant murine IL-9, and TGF-β1 were obtained from Peprotech (London, UK). Mouse monoclonal anti-dinitrophenyl (DNP) IgE, A23187, 2-mercaptoethanol, 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES), bovine serum albumin (BSA), MEM non-essential amino acids solution (MEM), sodium pyruvate and 3-morpholinopropanesulfoinc acid (MOPS) buffer were obtained from Sigma (St. Louis, MO, USA). RPMI-1640 medium, fetal bovine serum (FBS), phosphate-buffered saline (PBS), and penicillin-streptomycin solution (PS) were obtained from Life Technologies (Grand Island, NY, USA). DNP-BSA was obtained from Santa Cruz (TX, USA). FITC-conjugated anti-mouse CD117 (c-kit) was obtained from BD Pharmingen (San Diego, CA, USA). PE-conjugated anti-mouse FcεRI was obtained from eBioscience (San Jose, CA, USA). A Cell Counting Kit-8 (CCK-8) and Fura-2 AM were obtained from Dojindo (Kumamoto, Japan). A Mouse GeneChip Gene 1.0 ST Array was obtained from Affymetrix (Santa Clara, CA, USA). A Hipure Total RNA kit (R4114) was obtained from Magen (Guangzhou, China) for real time-PCR. An RNeasy Plus Micro kit (74034) was obtained from Qiagen (Hilden, Germany) for transcriptome analyses. PrimeScript RT reagent kit (RR047A) and SYBR Premix Ex Taq kit (RR420) were obtained from TaKaRa Bio (Tokyo, Japan).
BALB/c mice and C57BL/6 mice (male, 5 weeks old) were obtained from Japan SLC, Inc. (Shizuoka, Japan) and raised by the University of Toyama. Mice were maintained with free access to water and food in an experimental animal facility, and all experimental procedures were examined and approved by the Animal Experiment Committee (Authorization No.S-2009 INM-9) at the University of Toyama. The mBMMCs were obtained from the femurs of mice, and bone marrow cells were cultured in the complete RPMI-1640 medium (with 10% heat-inactivated FBS, 20 mM HEPES, 1 mM sodium pyruvate, 100 µM MEM, 10 µM 2-mercaptoethanol, 100 U/mL PS, and 2 µg/mL gentamicin solution) with a combination of 4 cytokines (20 ng/mL IL-3, 5 ng/mL IL-9, 40 ng/mL SCF, and 1 ng/mL TGF-β1) in a constant temperature incubator (37°C, 5% CO2). After four weeks, the purity of non-adherent cells was more than 95% FcεRI- and c-kit-positive (Fig. S1).
The activation of mBMMCs was performed as previously described (Wang et al., 2014). In brief, mBMMCs were suspended at a density of 2 × 105 cells/mL in the medium and sensitized with 1.5 μg/mL mouse monoclonal anti-DNP IgE for 6 h at 37°C. The cells were washed and resuspended at a density of 6 × 105 cells/mL in 100 μL Tyrode’s buffer (130 mM NaCl, 10 mM HEPES, 5.6 mM glucose, 5 mM KCl, 1.4 mM CaCl2, 1 mM MgCl2, and 0.1% BSA, pH 7.4), and the cells were stimulated with 100 ng/mL DNP-BSA as an antigen at 37°C for 0, 30, 60, and 90 min. For the A23187 experiment, mBMMCs were washed and resuspended, and 25 μM A23187 was added for 0, 10, 30, and 60 min. Then, the cells were centrifuged (4°C, 300 × g, 5 min), and all pellets were collected for the experiment.
Degranulation was examined by measuring β-hexosaminidase release. Briefly, mBMMCs were washed and resuspended at a density of 8 × 105 cells/mL in 50 μL Tyrode’s buffer, and samples were centrifuged, and supernatants were collected. The cell pellets were solubilized with 0.5% Triton X-100 in Tyrode’s buffer. The enzymatic activities of β-hexosaminidase in the supernatants and cell lysates were measured using p-nitrophenyl-2-acetamido-2-deoxy-β-D-glucopyranoside in 0.1 M sodium citrated (pH 4.5) at 37°C for 1 h. The reaction was stopped by the addition of 0.2 M NaOH and 0.2 M glycine. Production of p-nitrophenol was detected by absorbance at 405 nm. Released β-hexosaminidase activity was enzymatically assessed, and the extent of degranulation was expressed by dividing the absorbance of the supernatants by the sum of the absorbance of the supernatants and cell lysates.
Transmission electron microscopy (TEM)
TEM was utilized to analyze the ultrastructural images. A volume of 1 × 106 mBMMCs was washed with PBS and fixed with 2.5% glutaraldehyde in PBS for 1 week, followed by 2% osmium (VIII) oxide. After dehydration, a thin section was stained with 3% uranyl acetate and lead citrate and observed under TEM.
Intracellular calcium measurement
A volume of 1.3 × 106 mBMMCs were sensitized with anti-DNP IgE for 6 h (for the DNP group), loaded with Fura-2 AM (5 μM) in loading buffer (118 mM NaCl, 100 mM L-glutamine, 10 mM HEPES, 5.5 mM D-glucose, 4.7 mM KCl, 1.13 mM MgCl2, 1.3 mM CaCl2, 1 mM Na2HPO4, 2% MEM, and 0.2% BSA) at 37°C for 30 min, and placed in a stirred cuvette. For the A23187 experiment, mBMMCs were washed, resuspended, and loaded with Fura-2 AM (5 μM) in loading buffer. The fluorescence was assessed at both 340 and 380 nm with a fluorescence spectrophotometer (Hitachi, F4500, Tokyo, Japan) and calibrated for a background-corrected 340:380 ratio, and DNP-BSA or A23187 was added 50 s after the monitoring began.
The total RNA from mBMMCs was extracted using the RNeasy Plus Micro kit. Then, the RNA quality from each group was examined by the 28S/18S ribosomal RNA ratio using formaldehyde denaturing gel electrophoresis with MOPS buffer (Fig. S2). The mRNA analysis was performed according to the GeneChip Expression Technical Manual. The arrays were hybridized in an Agilent hybridization oven overnight according to the manufacturer’s instructions and washed with two consecutive solutions. The array data were first analyzed for quality control, data summarization, and normalization by the Affymetrix GeneChip Analysis Suite Software and then further analyzed with Gene-Spring software (Silicon Genetics, USA).
Function and pathway enrichment analysis of differentially expressed genes
The raw data files (deposited in the ArrayExpress database under the accession no. E-MTAB-9508) were loaded into R Bioconductor version 3.2.0 and processed and analyzed with the limma package. The cut-off criteria for DEGs were P < 0.01 and [log2-fold change] >0.5. Afterward, the Venn Diagram package was used to plot the Venn diagram of DEGs. Converged and special different genes only in DNP-BSA/IgE stimulation or A23187 stimulation were detected. ClusterProfiler was used to classify significant DEGs by their biological processes, molecular functions, or cellular components using the gene ontology consortium reference (GO) and the significant transcripts (Benjamini-Hochberg false discovery rate <0.05) (Yu et al., 2012). The ClusterProfiler database was also used to perform pathway enrichment analysis with reference from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database website and a Benjamini-Hochberg false discovery rate (FDR) <0.05 as a cut-off point.
The total RNA from mBMMCs was extracted using the Hipure Total RNA kit and detected using a real-time PCR system (ABI 7500, Foster, USA). The sequences of the primer pairs were used, as shown in Tab. 1. The mRNA of target genes was normalized to the mRNA of GAPDH.
The data from degranulation and real-time PCR are expressed as the means ± SD from at least four independent experiments (at least 4 mice) performed in triplicate. The data of GeneChip were from two independent experiments (2 mice). The other experiments were from three independent experiments (3 mice). Statistical comparisons were performed with repeated measures one-way ANOVA followed by post hoc Dunnett’s test with Statistical Product and Service Solutions (SPSS) 21 software (Chicago, USA). Values of p < 0.05 were considered significant.
Table 1: Primer sequences for real-time PCR
We examined the activation of mBMMCs by measuring β-hexosaminidase release. DNP-BSA antigen can combine with IgE and trigger FcεRI to activate mBMMCs. As shown in Fig. 1A, the mBMMCs from BALB/c mice sensitized with only monoclonal IgE (1.5 μg/mL) for 6 h did not alter degranulation compared with untreated mBMMCs (control group). The mBMMCs sensitized with IgE and stimulated with DNP-BSA (100 ng/mL) induced β-hexosaminidase release in a time-dependent manner and degranulation peaked at 60 min. Thus, we chose IgE sensitization for 6 h and DNP-BSA stimulation for 60 min for further study. In the same protocol of the degranulation assay for IgE/DNP-BSA, C57BL/6 mice-derived mBMMCs released β-hexosaminidase just at 6.97% at 60 min, which was lower than that of the BALB/c mice; whereas Th1-prone C57BL/6 mice were resistant to the development of FA (Yamamoto et al., 2009). Thus, we chose the BALB/c mice for further study.
The calcium ionophore A23187, an inducer of intracellular calcium concentrations, also can induce degranulation. However, the underlying mechanism remains unknown (Wang et al., 2014; Kim et al., 2014). As shown in Fig. 1B, 25 μM A23187 induced β-hexosaminidase release from 15 min, and degranulation peaked at 30 min. Thus, we chose A23187 treatment for 30 min for further study. The CCK8 results (Fig. S3) showed that IgE/DNP-BSA or A23187 stimulation did not alter the cell viabilities, suggesting that our protocol did not induce mBMMCs death. Furthermore, the vacuoles of mBMMCs obviously appeared after IgE sensitization for 6 h, DNP-BSA stimulation (IgE/DNP group) for 60 min, or A23187 stimulation (A23187 group) for 30 min (Fig. 1C) under TEM. Consistent with our previous studies (Wang et al., 2012), IgE/DNP and A23187 both elevated the [Ca2+]i (Fig. 1C), suggesting that the model of mBMMCs activation was stable.
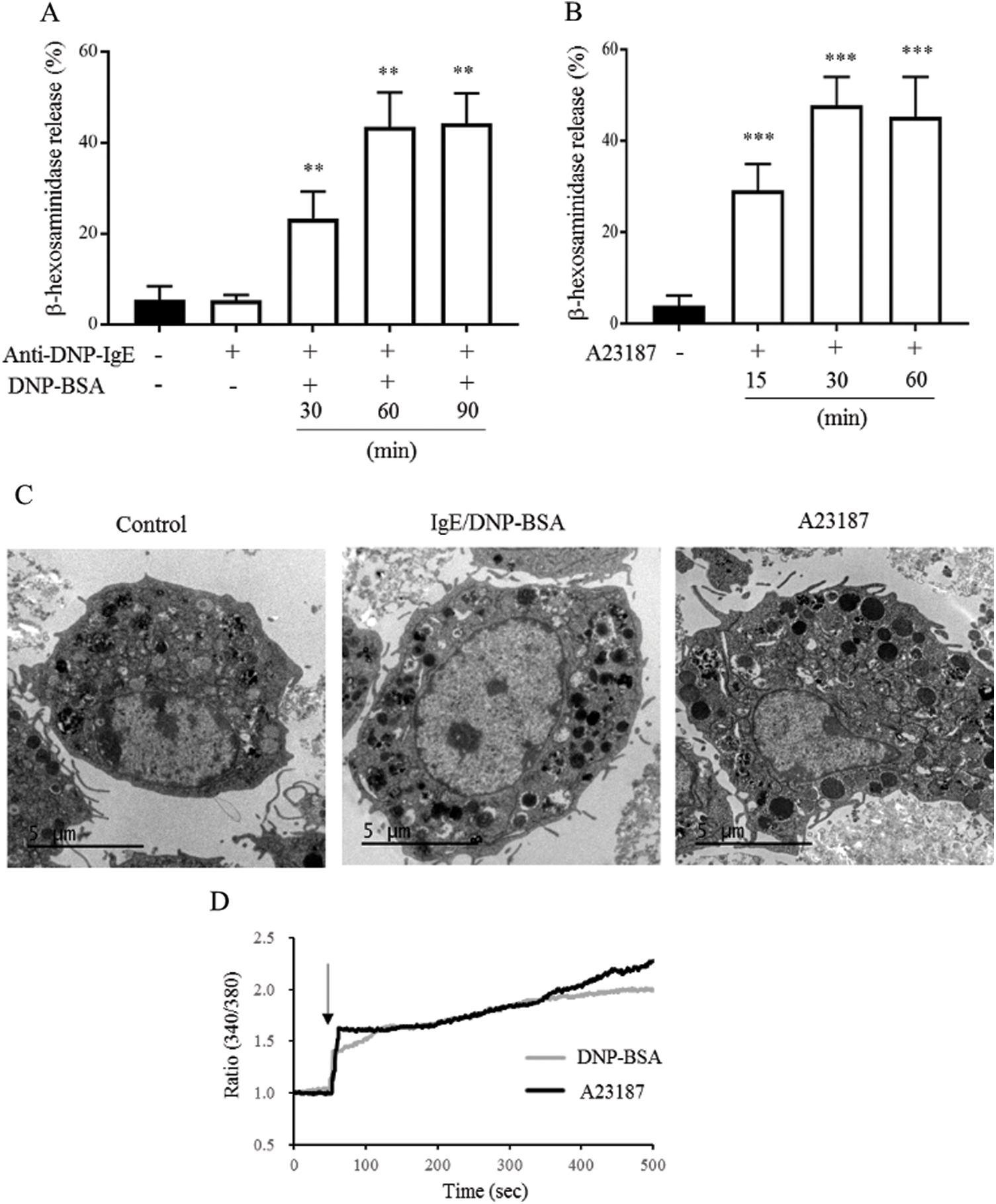
Figure 1: The effects of IgE/DNP-BSA and A23187 stimulation on the activation of mBMMCs from BALB/c mice.
To investigate the mechanism of mBMMCs activation, mRNA expression profiles were assessed by GeneChip analysis and checked by quantitative real-time PCR. A global gene expression analysis was performed with an Affymetrix Mouse GeneChip Gene 1.0 ST Array spotted with 28,853 probe sets. We investigated the global mRNA expression profile in the DNP group and the A23187 group compared with the untreated group, using the whole transcript array. After the induction of cell activation, the gene expression profiles were measured by GeneChip analysis. The series from each chip were separately evaluated using GeneSpring software and finally identified the DEGs lists.
The cluster results of the two sets of microarray data are shown in Figs. S4 and S5, suggesting the differential gene expression in the DNP group and the A23187 group. As shown in Fig. 2A, based on the criteria of a fold-change ≥2 and p < 0.05, among 28,853 probes, a total of 460 and the 305 mRNA sequences showed significant differential expression in IgE/DNP-BSA treated samples and A23187-treated samples, respectively, compared with the control samples.
As shown in the Venn diagram (Fig. 2A), 134 mRNA sequences were altered with IgE/DNP-BSA and A23187, suggesting that IgE/DNP-BSA and A23187 affect the same signal pathway in degranulation. However, IgE/DNP-BSA stimulation changed mRNA numbers more than A23187 stimulation, suggesting that IgE/FcεRI-dependent changes were upstream of A23187. We uploaded the 134 mRNA sequences to the online software to identify overrepresented GO categories and the KEGG pathway, and 131 mRNA sequences were effective. Microarray analysis revealed distinct mRNA expression, as shown in Tab. 2.
GO analysis is a common useful way to identify characteristic biological processes, cell components, and molecular functions for high-throughput genomes and transcriptome data (Fig. 2B). The GO analysis results showed that IgE/DNP-BSA and A23187 changed the expression of mRNA, which is related to the inflammatory response, immune system process, and so on. Furthermore, GO cell component data showed that the change in DEGs was significantly enriched in the extracellular space, cytosol, endosome lumen, and nucleus, suggesting that degranulation involved activation of the whole cell. We also found that the regulated DEGs were enriched in MAPK activity, DNA binding, nucleic acid binding, and transcription factor activity in the molecular function analysis.
We found that DEGs from the IgE/DNP group were enriched in T cell activation, adhesion, and differentiation in the biological process (Fig. S5A), confirming that IgE/FcεRI-induced MMCs activation is associated with T cell activation. Furthermore, the DEGs from the A23187 group were enriched in the ERK pathway, suggesting that [Ca2+]i elevation is involved in ERK signaling.
In addition to degranulation, other functions, such as the inflammatory response, are induced by mBMMCs activation (Fig. 3A). The data showed that the activation of mBMMCs is related to cell death. However, the mBMMCs stimulated with DNP-BSA/IgE or A23187 were not dead in 30 min, as shown in Fig. S3 (Wang et al., 2012). The data indicate that the mechanism of MCs activation includes MAPK, mTOR, TNF, and the NF-κB pathway both in IgE/antigen and A23187 stimulation. However, as shown in Fig. 3B, the function of 49% of genes is currently unknown.
As shown in Fig. 4, the KEGG analysis showed the most significantly enriched pathways of the down-regulated and up-regulated DEGs compared with the untreated group. The DEGs were enriched in NF-κB, TNF, MAPK, HIF-1, FoxO, mTOR, Jak-STAT, p53, TLRs, the NOD-like receptor, and cytokine-cytokine receptor interaction signal transduction both in the IgE/DNP-BSA group and the A23187 group. We conjectured that in addition to degranulation and cytokines release, MMCs activation is related to these signal pathways. Furthermore, the DEGs were involved in cancer, glioma, IBD, RA, herpes simplex infection, insulin resistance, etc., suggesting that the activation of MMCs also plays an important role in immune diseases.
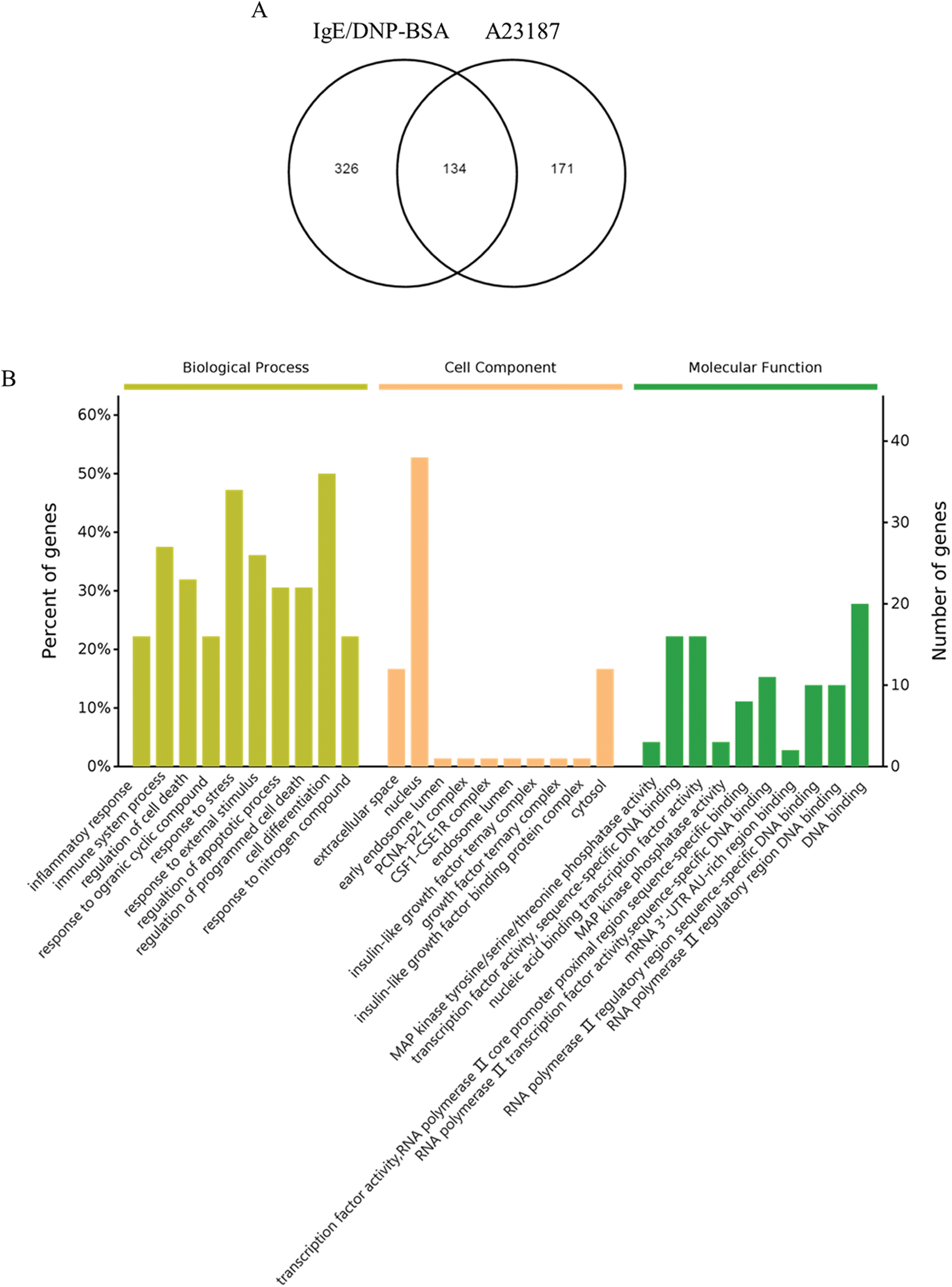
Figure 2: (A) The Venn diagram of DEGs in the activation of mBMMCs by IgE/DNP-BSA and A23187 treatment. (B) The results of the biological process, cellular component, and molecular functions for mRNA changed by IgE/DNP-BSA and A23187 treatment.
Table 2: List of genes selected by microarray analysis. The change fold was calculated as the ratio of the expression level in the activated mBMMCs with IgE/DNP-BSA (DNP) or A23187compared to that in untreated mBMMCs (Control, Con)
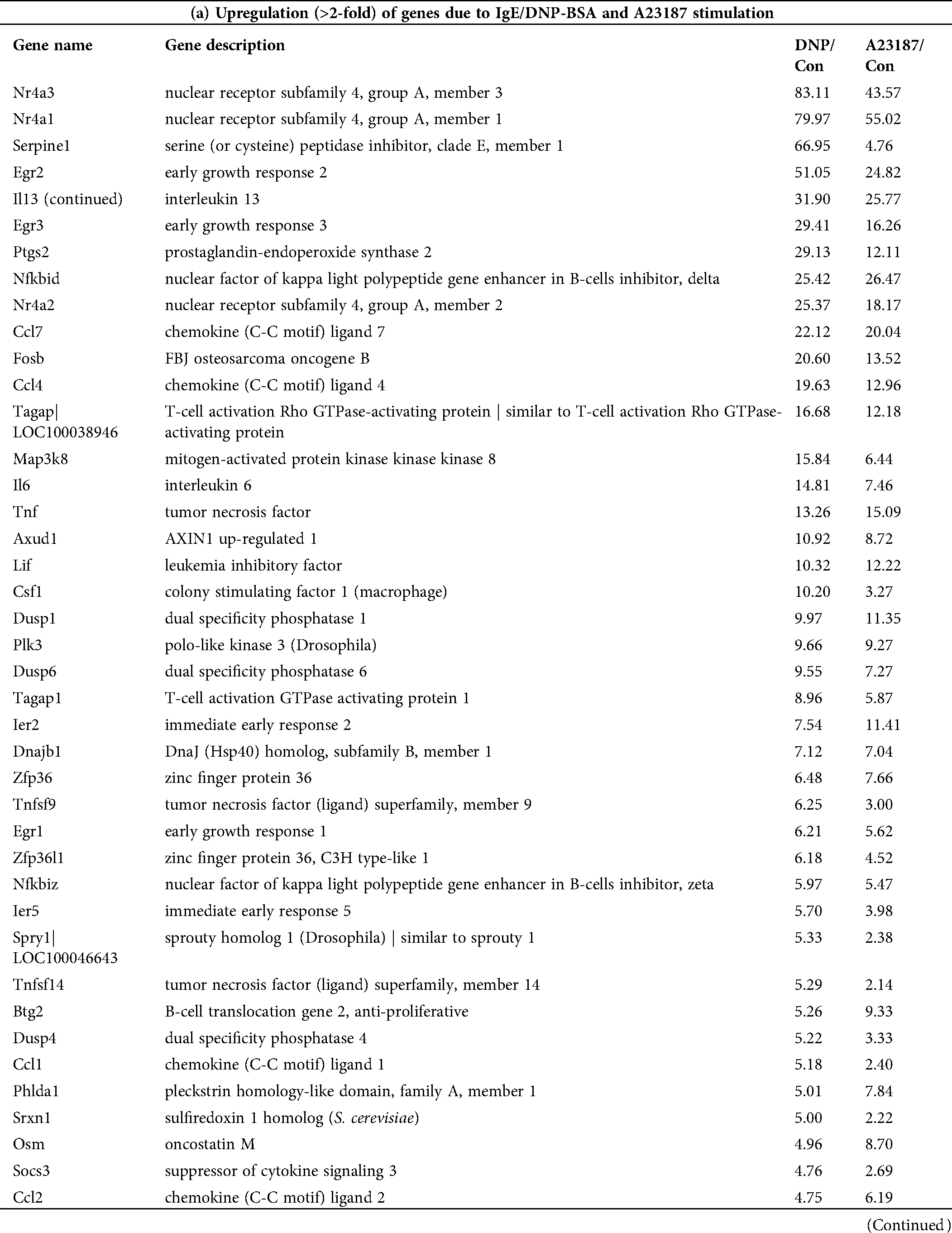
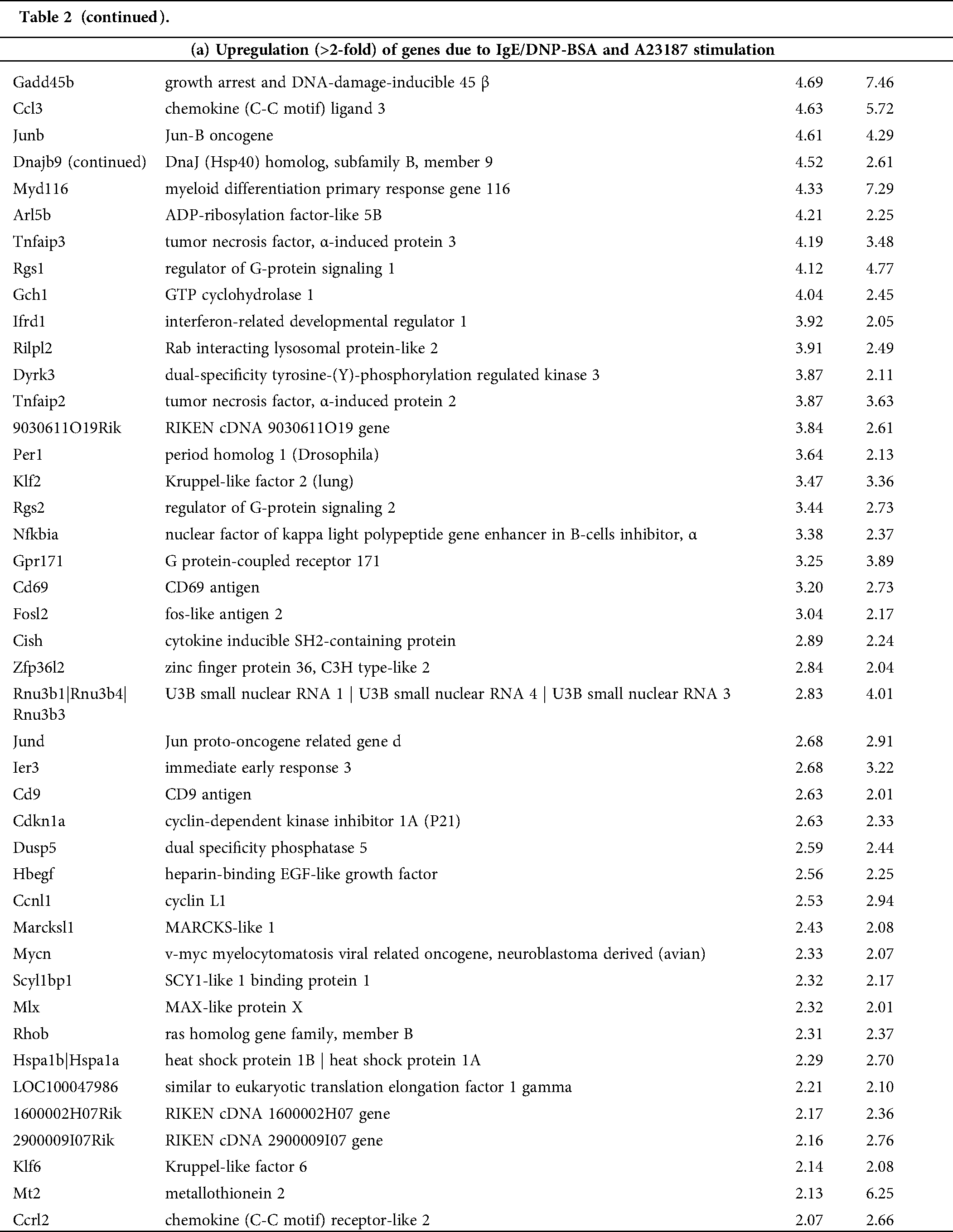
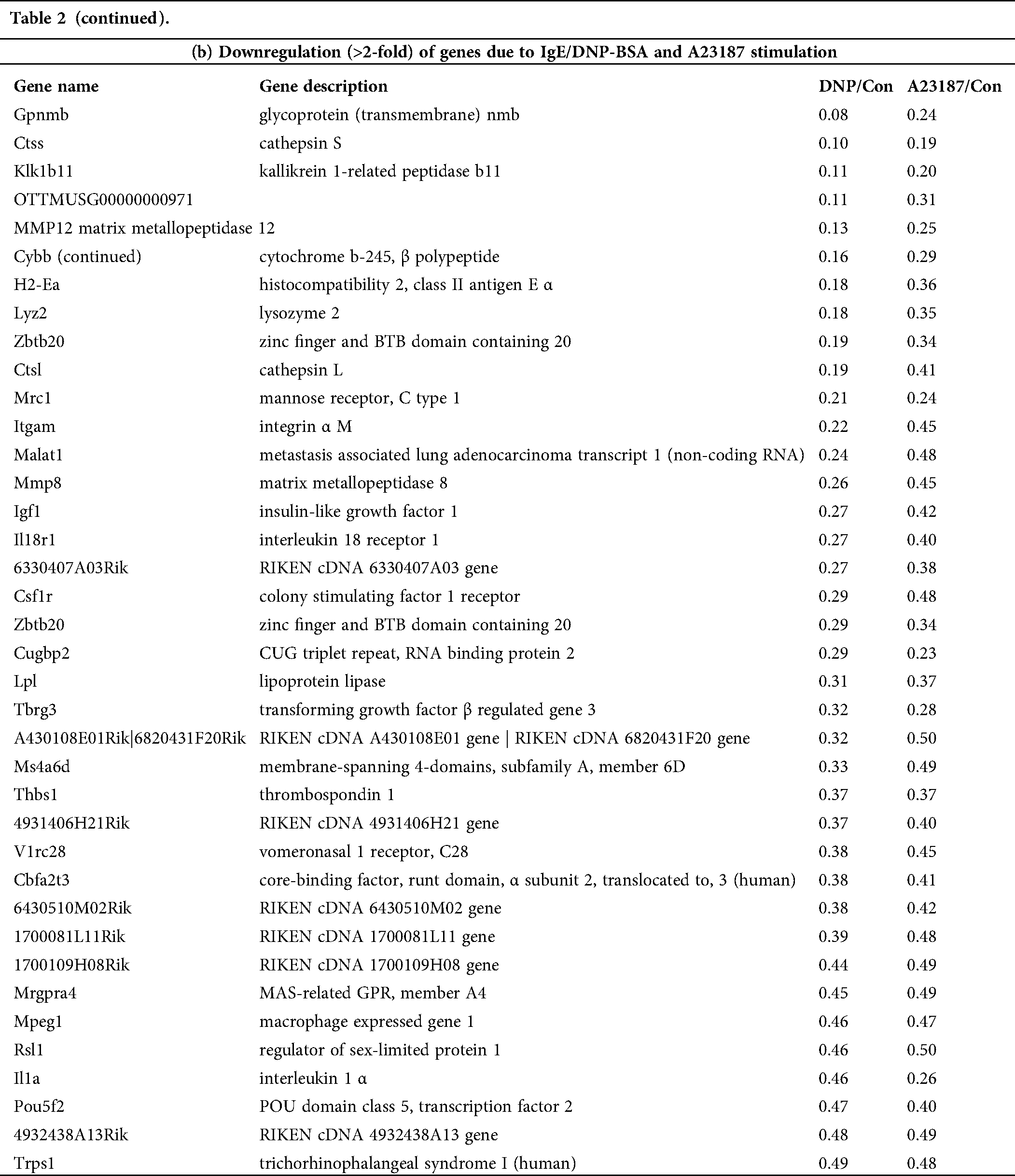
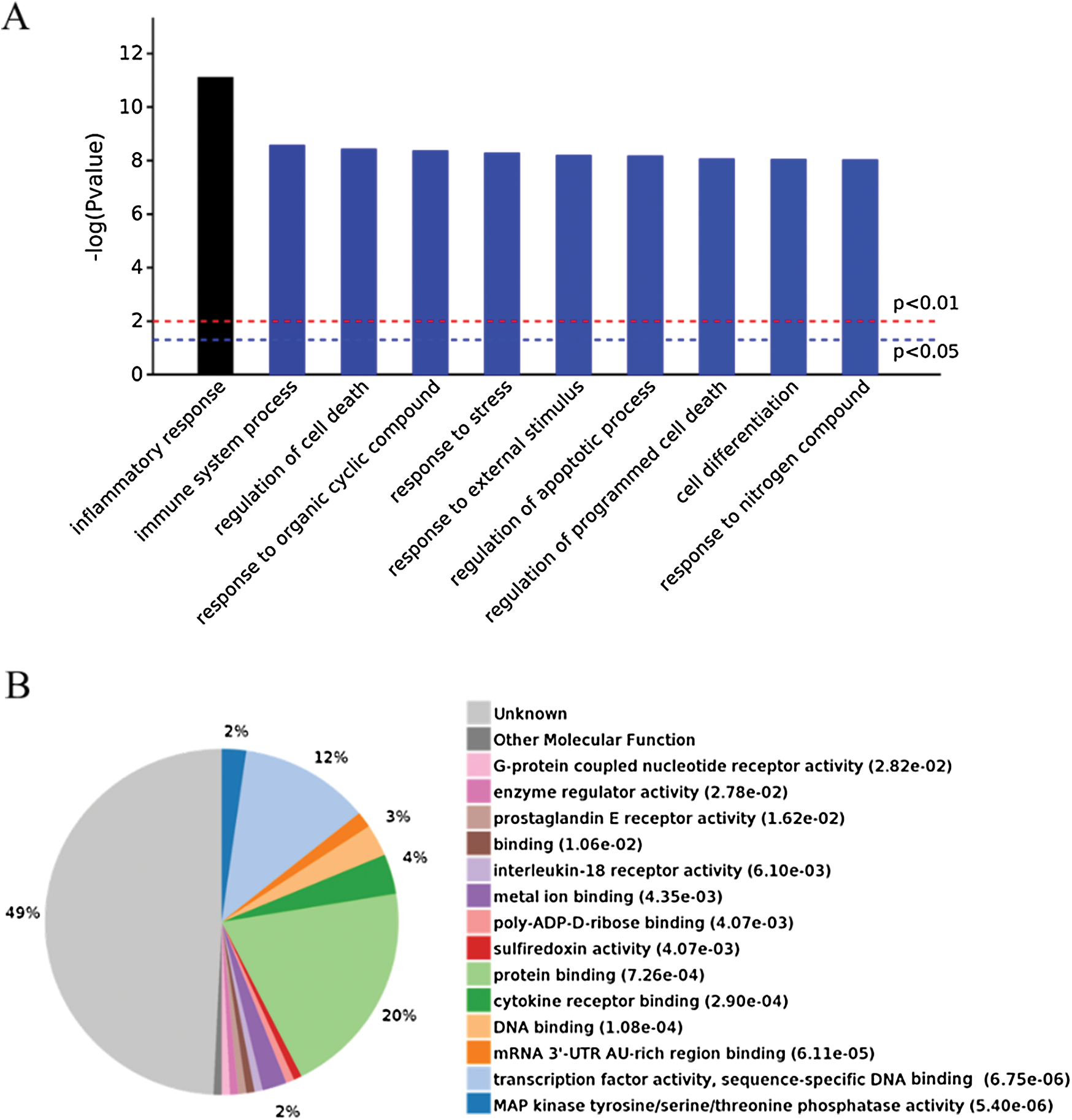
Figure 3: (A) Significantly enriched biological processes for the genes changed by IgE/DNP-BSA and A23187 treatment. (B) Expressed proteins of enriched p-value.
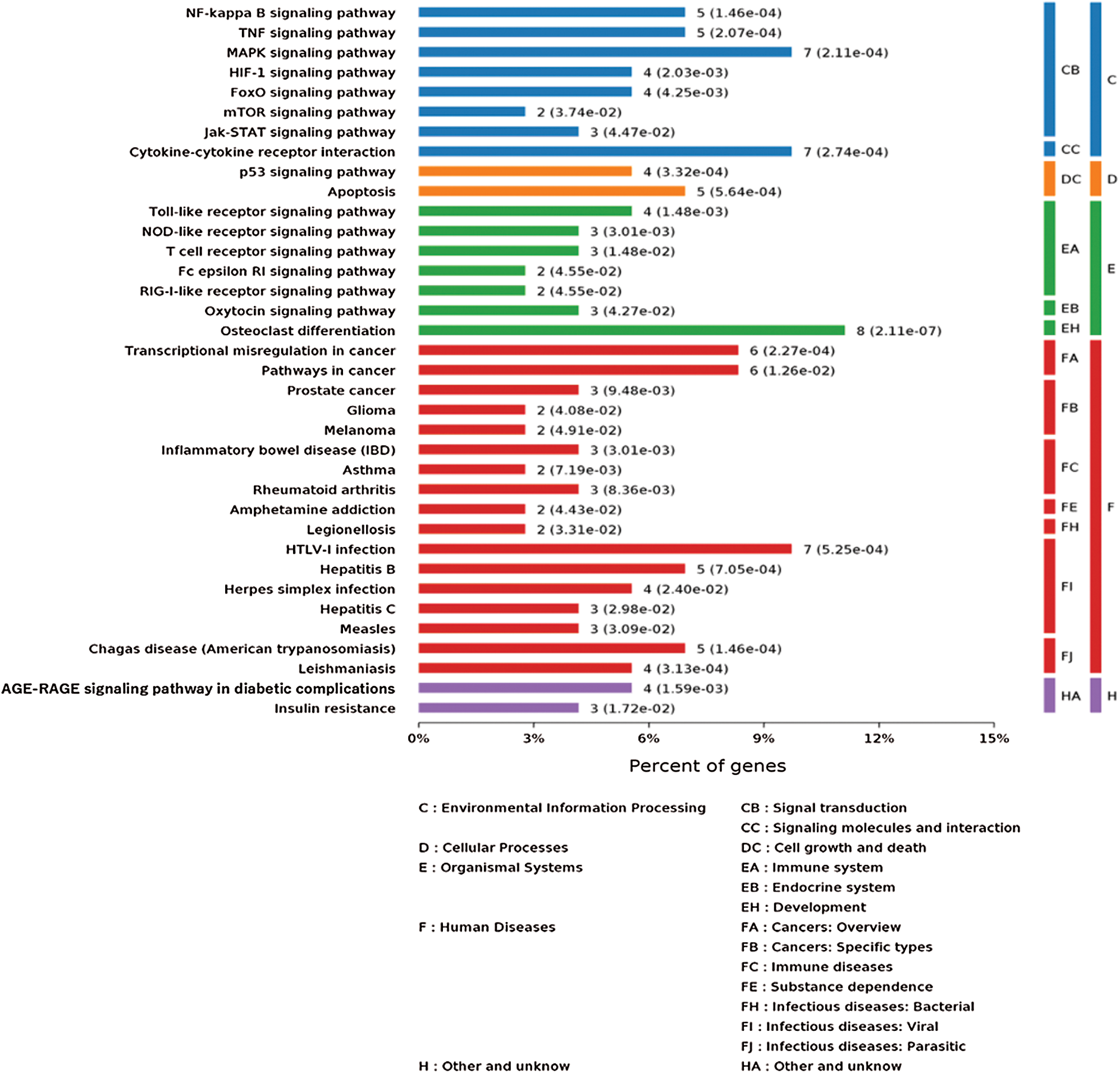
Figure 4: KEGG pathway analysis of the genes changed by IgE/DNP-BSA and A23187 treatment.
As shown in Fig. S5B, the DEGs from the IgE/DNP-BSA group were enriched in the PPAR signaling pathway, which is consistent with other reports (Yao et al., 2017). However, the DEGs from the A23187 group were enriched in measles, legionellosis, cancer, and RA.
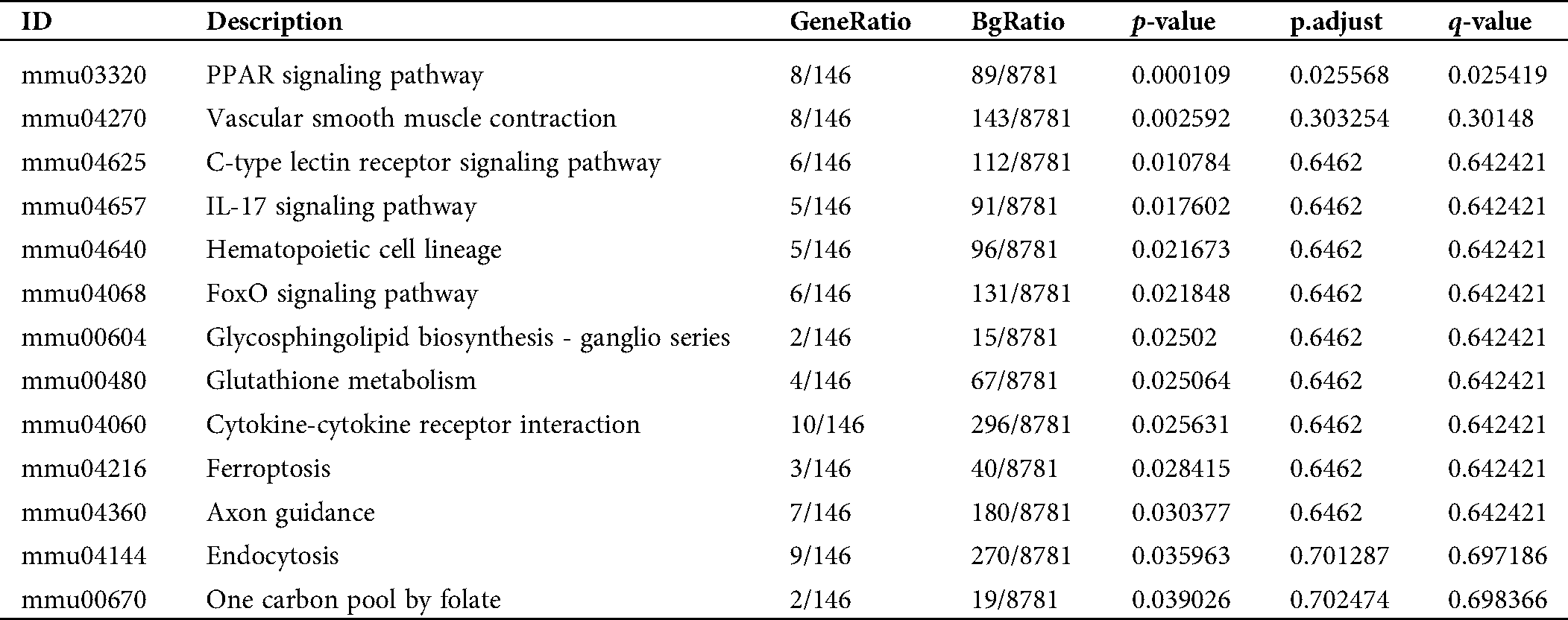
The converged DEGs in the activation of mBMMCs by IgE/DNP-BSA and A23187 treatment are shown in Tabs. S1–S3. The pathway enrichment results of the DEGs in the activation of mBMMCs by A23187 treatment and IgE/DNP-BSA treatment with a p-value and an adjusted p-value for each pathway are shown in Tabs. S2 and S3, respectively.
PCR validation of selected potential markers
Because the function of 49% of genes is unknown (Fig. 3B), after the GeneChip analysis, we investigated mRNA sequences, such as serpine 1, T-cell activation Rho GTPase-activating protein (TAGAP), dual-specificity phosphatase (Dusp) 6, glycoprotein (transmembrane) nmb (GPNMB), matrix metallopeptidase (MMP) 12, thrombospondin (THBS) 1, lysozyme 2 (LYZ-2) (Fig. 5), which significantly changed in the global gene expression analysis. The results indicated that serpine 1, TAGAP, and Dusp 6 were significantly up-regulated with IgE/DNP or A23187 stimulation, and GPNMB and MMP12 were significantly down-regulated (Figs. 5A−5E), which was consistent with the GeneChip results.
The PCR data showed that IgE/DNP treatment significantly inhibited THBS 1 and LYZ-2 mRNA. However, A23187 did not alter the mRNA levels (Figs. 5F–5G), suggesting that THBS 1 and LYZ-2 may not be related to calcium or downstream of calcium in mBMMCs activation. We also investigated cathepsin S and early growth response 2 (ERG2), but we found that cathepsin S and ERG2 were not altered, suggesting that further validation after GeneChip analysis is critical.
MCs are involved in various inflammatory diseases, including allergy, migraines, RA, cardiovascular disease, Crohn’s disease, multiple sclerosis, cancer, mastocytosis, and so on (Theoharides and Cochrane, 2004). Consistent with the results of other research, our results (Fig. 4) showed various cancer, glioma, IBD, RA, herpes simplex infection, insulin resistance, and so on. MCs play important roles in various diseases. Thus, our findings may provide a novel strategy for the treatment of inflammatory diseases associated with MCs activation.
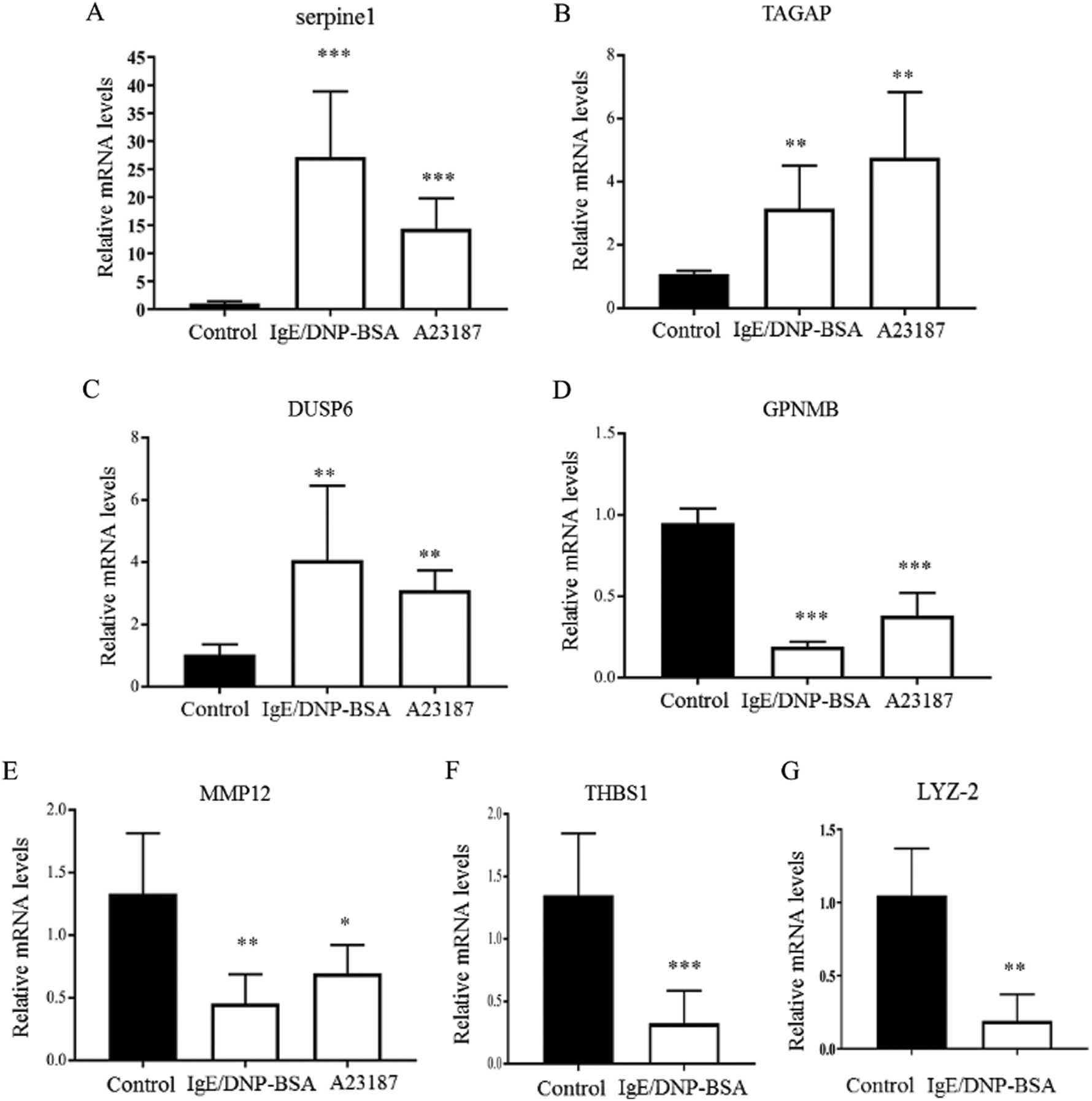
Figure 5: The effects of IgE/DNP-BSA and A23187 stimulation on mRNA levels.
Previously, we found that MMCs play more important roles in anaphylaxis in the colon of mice with FA than CTMCs (Yamamoto et al., 2009; Yamamoto et al., 2014). We also indicated that the enhancement of proinflammatory cytokines released from MCs includes T-helper 2 cell (Th2)-type responses (such as IL-4, IL-5, IL-10, and IL-13) and is important for the induction of allergic gastrointestinal symptoms in a murine FA model (Yamamoto et al., 2009), as well as in other reports. Furthermore, we reported that the cholinergic nerve fibers CGRP closely communicated with MMC via α7 nicotinic ACh receptors that regulate mast cell functions in the colon of mice with FA (Yamamoto et al., 2014).
Besides the FA, pre-stored vasoactive amines (such as histamine and serotonin), prostaglandin D2 (PGD2), and leukotriene C4 (LTC4) were released from MCs and increased capillary permeability and smooth muscle contraction also associated with bronchoconstriction and vasodilation in asthma (Kraneveld et al., 2012). These mediators are also responsible for skin inflammation, such as erythema and edema in atopic dermatitis (Theoharides and Cochrane, 2004). Similarly, the activation of synovial MCs is linked to the production of several pro-inflammatory cytokines, such as tumor necrosis factor (TNF)-α, IL-1β and IL-1R, and these mediators can contribute to the aberrant survival and activation of rheumatoid synovial fibroblasts in RA (Rivellese et al., 2019).
Mast cells respond to antigen/IgE-dependent and IgE-independent (neuropeptide receptors, toxins, bee peptide, autacoid) stimulation and release various inflammatory mediators (Kazuma et al., 2017). The mechanisms underlying allergen-derived bioactive mediators are not yet completely known, but calcium influx plays a critical role in MMCs activation. Although Kawakami and Kitaura (2005) reported that MCs can survive and are activated by IgE in the absence of an antigen, we did not observe degranulation with only IgE treatment in the current study. Thus, we choose mBMMCs activation by antigen/IgE for microarray for further studies.
For the first time, we investigated the mechanism of mBMMCs activation by DNP-BSA/IgE-FcεRI and calcium ionophore using GeneChip. The present study identified DEGs between antigen/IgE or A23187 and normal cells. This study identified 460 mRNA sequences between control and DNP-BSA/IgE-treated cells and 295 mRNA sequences between control and A23187-treated samples. The GO term analysis displayed that regulated DEGs were mainly involved in cell differentiation, cell stress, the immune system process, responses to external stimuli, cell death, the inflammatory response, responses to organic cyclic compounds, and responses to nitrogen compounds. In addition, the enriched KEGG pathways of DEGs showed that the activation of mBMMCs is related to NF-κB, TNF, MAPK, and cytokine-cytokine receptor signal transduction. We found that A23187-induced MAPK, as well as IgE/antigen stimulation, and MAPK is the key pathway in the activation of mast cells, which is consistent with the findings of other researchers (Toda et al., 2012). MAPK plays a critical role in the activation, proliferation, differentiation, degranulation, and migration of various immune cells (Duan and Wong, 2006), and an increase in intracellular calcium triggers the activation of MAPK (Kim et al., 2005; Law et al., 2011). The present study identified that DNP-BSA/IgE altered the mRNA level more than A23187 and verified that calcium was downstream in the antigen-IgE/FcεRI, and they share some pathways, such as TNF and NF-κB.
IgE/antigen and calcium ionophores triggered an increase in cellular calcium concentration, and the GeneChip data showed that this pathway may involve MAPK, mTOR, TNF, NF-κB, and so on. Furthermore, IgE/FcεRI-induced MMCs activation is associated with T cells, especially Th17 cells activation (Tab. S1) and the PPAR signaling pathway, suggesting that MMCs activation is a complex process.
Recent reports have demonstrated that human MCs are a major source of serpine 1, and mast cell activation promotes serpine 1 production (Cho et al., 2015). However, there is no report on the role of serpine 1 in MMCs. We found that IgE/DNP-BSA and A23187 stimulation can induce serpine 1 mRNA, suggesting that serpine 1 is also involved in the pathway of MMCs activation. TAGAP, a member of the Rho-GTPase protein family, releasing GTP from GTP-bound Rho, serves as a molecular switch. Although TAGAP is expressed in activated T cells, the role of TAGAP in immune function remains unclear (Arshad et al., 2017). We found that IgE/DNP and A23187 stimulation can induce TAGAP mRNA, which is consistent with the GeneChip data. Therefore, TAGAP played a role not only in T cells but also in MMCs activation. The DUSP family of proteins shows distinct substrate preferences for the MAPK pathway and regulates a wide range of responses. DUSP6 is a critical regulator of activation via the ERK pathway in macrophages and mediates metabolic commitment to restrains the differentiation of Tfh (Carson et al., 2017; Hsu et al., 2018). Our data provide evidence for the mRNA of DUSP6 that was observed to be increased in IgE/DNP and A23187 treatment.
GPNMB, which is expressed in numerous cell types, including dendritic cells, macrophages, osteoblasts and osteoclasts, is involved in various biological processes, including inflammation (Zhou et al., 2017). It is reported that MMP 12 is predominantly expressed by macrophages (Shapiro et al., 1993), and there are elevated levels of MMP 12 in the tissue of patients with chronic inflammatory disease periodontitis (Arshad et al., 2017), suggesting that MMP 12 is a pro-inflammatory factor. However, our results indicated that MMP 12 is decreased by antigen-IgE and A23187 treatment, suggesting that MMP 12 has other molecular functions. Furthermore, MCs can store mRNA for hours and express these proteins at another time; thus, the transcription genes are not the same as expression proteins derived from these genes (Ramírez-Moreno et al., 2020). Suzuki et al. (2015) reported that THBS1, a proinflammatory protein, is upregulated in rheumatoid synovial tissue, and MMCs play important roles in RA. Thus, our data also confirmed that THBS1 is involved in the activation of MMCs. It is known that lysozyme plays an essential role in MC-related disease (Dobson et al., 2008), and there are no reports about the role of LYZ-2 in the MMCs. In the present study, we confirmed that THBS 1 and LYZ-2 also play roles in the activation of MMCs.
Brain MCs that were activated by acute stress leading to increasing blood-brain barrier permeability developed in migraines. MCs are located close to nerve terminals in many tissues, such as the meninges, and MCs activation induces the release of peptides including bradykinin, substance P, and vasoactive intestinal peptide (VIP), from peripheral nerve terminals that contribute to the inflammation-mediated exacerbation of allergic reactions (Kilinc et al., 2015; Kilinc et al., 2017b). Furthermore, neuropeptides, such as CGRP, substance P, and VIP, also lead to the release of numerous inflammatory mediators from MCs through degranulation as a result of MCs mediators-mediated nerves activation, which in turn enhances neurogenic inflammation and pain perception (Dagistan et al., 2019; Kilinc and Gunes, 2019; Kilinc et al., 2017a).
It is known that MC stabilization by endogenous or exogenous stabilizers can be used to treat MC-mediated disorders (Kilinc and Baranoglu Kilinc, 2020a). We investigated agents against MMCs activation using a microarray and found shikonin, cinnamaldehyde, and ergosterol, which are compounds from natural plants that can inhibit MMCs activation (Kageyama-Yahara et al., 2011; Wang et al., 2012; Wang et al., 2014). Furthermore, we demonstrated that Kakkonto, a traditional Japanese medicine, and the active ingredient, pentagalloylglucose, ameliorated colon inflammation from FA via inhibition of MMCs activation (Yamamoto et al., 2009; Kageyama-Yahara et al., 2010a; Kageyama-Yahara et al., 2010b). Some researchers also reported that some exogenous MC stabilizers (such as salmon calcitonin, thymoquinone, methanandamide, rimonabant, SR144528, and capsazepine) could be useful in the treatment of MC-mediated inflammatory disorders via CGRP (Kilinc and Baranoglu Kilinc, 2020a; Kilinc et al., 2020b; Kilinc et al., 2018).
This study provides information to explain the underlying physiological and molecular mechanisms of MMCs activation (Fig. S7). The approach of evaluation for biological functions and pathways as a whole will provide new insights on mBMMCs activation on a cellular and molecular level. However, transcriptional studies should continue to develop in order to improve current bioinformatics study methods.
Our study showed that mBMMCs activation was enriched in NF-κB, TNF, MAPK, transcription factor activity, DNA binding, and nucleic acid binding, suggesting that activation of MMCs is a complex process. In conclusion, our results provide a new approach using comprehensive bioinformatics analysis of DEGs, which was likely involved in mBMMCs activation. The results also provide insights into new signaling pathways and novel mediators of MMCs activation. However, more investigation should be conducted after transcriptional studies in further studies.
Availability of Data and Materials: The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Funding Statement: This work was supported by the “Xinlin Young Talent Program” (A1-U1820502040237) from Shanghai University of Traditional Chinese Medicine, and “Sasakawa Scientific Research Grant” (23-401) from the Japan Science Society.
Conflicts of Interest: The authors declare that there are no conflicts of interest.
Arshad M, Bhatti A, John P, Jalil F, Borghese F, Kawalkowska JZ, Williams RO, Clanchy FIL. (2017). T cell activation Rho GTPase activating protein (TAGAP) is upregulated in clinical and experimental arthritis. Cytokine 104: 130–135. DOI 10.1016/j.cyto.2017.10.002. [Google Scholar] [CrossRef]
Arthur G, Bradding P. (2016). New developments in mast cell biology: Clinical implications. Chest 150: 680–693. DOI 10.1016/j.chest.2016.06.009. [Google Scholar] [CrossRef]
Benedé S, Berin MC. (2018). Mast cell heterogeneity underlies different manifestations of food allergy in mice. PLoS One 13: e0190453. DOI 10.1371/journal.pone.0190453. [Google Scholar] [CrossRef]
Boeckxstaens G. (2015). Mast cells and inflammatory bowel disease. Current Opinion in Pharmacology 25: 45–49. DOI 10.1016/j.coph.2015.11.005. [Google Scholar] [CrossRef]
Carson WF, Salter-Green SE, Scola MM, Joshi A, Gallagher KA, Kunkel SL. (2017). Enhancement of macrophage inflammatory responses by CCL2 is correlated with increased miR-9 expression and downregulation of the ERK1/2 phosphatase Dusp6. Cellular Immunology 314: 63–72. DOI 10.1016/j.cellimm.2017.02.005. [Google Scholar] [CrossRef]
Cho SH, Lee SH, Kato A, Takabayashi T, Kulka M, Shin SC, Schleimer RP. (2015). Cross-talk between human mast cells and bronchial epithelial cells in plasminogen activator inhibitor-1 production via transforming growth factor-β1. American Journal of Respiratory Cell and Molecular Biology 52: 88–95. DOI 10.1165/rcmb.2013-0399OC. [Google Scholar] [CrossRef]
Conti P, Castellani M, Kempuraj D, Salini V, Vecchiet J, Tetè S, Mastrangelo F, Perrella A, de Lutiis MA, Tagen M, Theoharides TC. (2007). Role of mast cells in tumor growth. Annals of Clinical and Laboratory Science 37: 315–322. [Google Scholar]
Dagistan Y, Kilinc E, Balci CN. (2019). Cervical sympathectomy modulates the neurogenic inflammatory neuropeptides following experimental subarachnoid hemorrhage in rats. Brain Research 1722: 146366. DOI 10.1016/j.brainres.2019.146366. [Google Scholar] [CrossRef]
da Silva EZ, Jamur MC, Oliver C. (2014). Mast cell function: A new vision of an old cell. Journal of Histochemistry and Cytochemistry 62: 698–738. DOI 10.1369/0022155414545334. [Google Scholar] [CrossRef]
Dobson JT, Seibert J, Teh EM, Da'as S, Fraser RB, Paw BH, Lin TJ, Berman JN. (2008). Carboxypeptidase A5 identifies a novel mast cell lineage in the zebrafish providing new insight into mast cell fate determination. Blood 112: 2969–2972. DOI 10.1182/blood-2008-03-145011. [Google Scholar] [CrossRef]
Duan W, Wong WS. (2006). Targeting mitogen-activated protein kinases for asthma. Current Drug Targets 7: 691–698. DOI 10.2174/138945006777435353. [Google Scholar] [CrossRef]
Dwyer DF, Barrett NA, Austen KF. (2016). Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nature Immunology 17: 878–887. DOI 10.1038/ni.3445. [Google Scholar] [CrossRef]
Elieh Ali Komi D, Wöhrl S, Bielory L. (2020). Mast cell biology at molecular level: A comprehensive review. Clinical Reviews in Allergy & Immunology 58: 342–365. DOI 10.1007/s12016-019-08769-2. [Google Scholar] [CrossRef]
Feske S, Wulff H, Skolnik EY. (2015). Ion channels in innate and adaptive immunity. Annual Review of Immunology 33: 291–353. DOI 10.1146/annurev-immunol-032414-112212. [Google Scholar] [CrossRef]
Gurish MF, Boyce JA. (2002). Mast cell growth, differentiation, and death. Clinical Reviews in Allergy & Immunology 22: 107–118. DOI 10.1385/CRIAI:22:2:107. [Google Scholar] [CrossRef]
Hsu WC, Chen MY, Hsu SC, Huang LR, Kao CY, Cheng WH, Pan CH, Wu MS, Yu GY, Hung MS, Leu CM, Tan TH, Su YW. (2018). DUSP6 mediates T cell receptor-engaged glycolysis and restrains TFH cell differentiation. Proceedings of the National Academy of Sciences of the United States of America 115: E8027–E8036. DOI 10.1073/pnas.1800076115. [Google Scholar] [CrossRef]
Iweala OI, Choudhary SK, Commins SP. (2018). Food allergy. Current Gastroenterology Reports 20: 17. DOI 10.1007/s11894-018-0624-y. [Google Scholar] [CrossRef]
Johnson M, Alsaleh N, Mendoza RP, Persaud I, Bauer AK, Saba L, Brown JM. (2018). Genomic and transcriptomic comparison of allergen and silver nanoparticle-induced mast cell degranulation reveals novel non-immunoglobulin E mediated mechanisms. PLoS One 13: e0193499. DOI 10.1371/journal.pone.0193499. [Google Scholar] [CrossRef]
Kageyama-Yahara N, Suehiro Y, Maeda F, Kageyama S, Fukuoka J, Katagiri T, Yamamoto T, Kadowaki M (2010a). Pentagalloylglucose down-regulates mast cell surface FcεRI expression in vitro and in vivo. FEBS Letters 584: 111–118. DOI 10.1016/j.febslet.2009.11.007. [Google Scholar] [CrossRef]
Kageyama-Yahara N, Wang P, Wang X, Yamamoto T, Kadowaki M (2010b). The inhibitory effect of ergosterol, a bioactive constiuent of a traditional Japanese herbal medicine saireito on the activity of mucosal-type mast cells. Biological & Pharmaceutical Bulletin 33: 142–145. DOI 10.1248/bpb.33.142. [Google Scholar] [CrossRef]
Kageyama-Yahara N, Wang X, Katagiri T, Wang P, Yamamoto T, Tominage M, Kadowaki M. (2011). Suppression of phospholipase Cγ1 phosphorylation by cinnamaldehyde inhibits antigen-induced extracellular calcium influx and degranulation in mucosal mast cells. Biochemical and Biophysical Research Communications 416: 283–288. DOI 10.1016/j.bbrc.2011.11.014. [Google Scholar] [CrossRef]
Kawakami T, Kitaura J. (2005). Mast cell survival and activation by IgE in the absence of antigen: A consideration of the biologic mechanisms and relevance. Journal of Immunology 175: 4167–4173. DOI 10.4049/jimmunol.175.7.4167. [Google Scholar] [CrossRef]
Kazuma K, Ando K, Nihei K, Wang X, Rangel M, Franzolin M, Mori-Yasumoto K, Sekita S, Kadowaki M, Satake M, Konno K. (2017). Peptidomic analysis of the venom of the solitary bee Xylocopa appendiculata circumvolans. Journal of Venomous Animals and Toxins including Tropical Diseases 23: 40. DOI 10.1186/s40409-017-0130-y. [Google Scholar] [CrossRef]
Kilinc E, Ankarali S, Torun IE, Dagistan Y. (2020). Receptor mechanisms mediating the anti-neuroinflammatory effects of endocannabinoid system modulation in a rat model of migraine. European Journal of Neuroscience 2020: 14897. [Google Scholar]
Kilinc E, Baranoglu Kilinc Y (2020a). Mast cell stabilizers as a supportive therapy can contribute to alleviate fatal inflammatory responses and severity of pulmonary complications in COVID-19 infection. AnadoluKliniği Tıp BilimleriDergisi-Anadolu Clinic Journal of Medical Sciences 25: 111–119. [Google Scholar]
Kilinc E, Dagistan Y, Kotan B, Cetinkaya A (2017a). Effects of Nigella sativa seeds and certain species of fungi extracts on number and activation of dural mast cells in rats. Physiology International 104: 15–24. DOI 10.1556/2060.104.2017.1.8. [Google Scholar] [CrossRef]
Kilinc E, Dagistan Y, Kukner A, Yilmaz B, Agus S, Soyler G, Tore F. (2018). Salmon calcitonin ameliorates migraine pain through modulation of CGRP release and dural mast cell degranulation in rats. Clinical and Experimental Pharmacology and Physiology 45: 536–546. DOI 10.1111/1440-1681.12915. [Google Scholar] [CrossRef]
Kilinc E, Firat T, Tore F, Kiyan A, Kukner A, Tuncel N. (2015). Vasoactive intestinal peptide modulates c-Fos activity in the trigeminal nucleus and dura mater mast cells in sympathectomized rats. Journal of Neuroscience Research 93: 644–650. DOI 10.1002/jnr.23523. [Google Scholar] [CrossRef]
Kilinc E, Guerrero-Toro C, Zakharov A, Vitale C, Gubert-Olive M, Koroleva K, Timonina A, Luz LL, Shelukhina I, Giniatullina R, Tore F, Safronov BV, Giniatullin R (2017b). Serotonergic mechanisms of trigeminal meningeal nociception: Implications for migraine pain. Neuropharmacology 116: 160–173. DOI 10.1016/j.neuropharm.2016.12.024. [Google Scholar] [CrossRef]
Kilinc E, Gunes H. (2019). Modulatory effects of neuropeptides on pentylenetetrazol-induced epileptic seizures and neuroinflammation in rats. Revista da Associação Médica Brasileira 65: 1188–1192. DOI 10.1590/1806-9282.65.9.1188. [Google Scholar] [CrossRef]
Kilinc E, Tore F, Dagistan Y, Bugdayci G (2020b). Thymoquinone inhibits neurogenic inflammation underlying migraine through modulation of calcitonin gene-related peptide release and stabilization of meningeal mast cells in flyceryltrinitrate-induced migraine model in rats. Inflammation 43: 264–273. DOI 10.1007/s10753-019-01115-w. [Google Scholar] [CrossRef]
Kim JH, Yamamoto T, Lee J, Yashiro T, Hamada T, Hayashi S, Kadowaki M. (2014). CGRP, a neurotransmitter of enteric sensory neurons, contributes to the development of food allergy due to the augmentation of microtubule reorganization in mucosal mast cells. Biomedical Research 35: 285–293. DOI 10.2220/biomedres.35.285. [Google Scholar] [CrossRef]
Kim MS, Lim WK, Park RK, Taekyun Shin, Yoo YH, Hong SH, An NH, Kim HM. (2005). Involvement of mitogen-activated protein kinase and NF-κB activation in Ca2+-induced IL-8 production in human mast cells. Cytokine 32: 226–233. DOI 10.1016/j.cyto.2005.10.001. [Google Scholar] [CrossRef]
Koyuncu Irmak D, Kilinc E, Tore F. (2019). Shared fate of meningeal mast cells and sensory neurons in migraine. Frontiers in Cellular Neuroscience 13: 136. DOI 10.3389/fncel.2019.00136. [Google Scholar] [CrossRef]
Kraneveld AD, Sager S, Garssen J, Folkerts G. (2012). The two faces of mast cells in food allergy and allergic asthma: The possible concept of yin yang. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 1822: 93–99. DOI 10.1016/j.bbadis.2011.06.013. [Google Scholar] [CrossRef]
Law M, Morales JL, Mottram LF, Iyer A, Peterson BR, August A. (2011). Structural requirements for the inhibition of calcium mobilization and mast cell activation by the pyrazole derivative BTP2. International Journal of Biochemistry & Cell Biology 43: 1228–1239. DOI 10.1016/j.biocel.2011.04.016. [Google Scholar] [CrossRef]
Miller HR, Wright SH, Knight PA, Thornton EM. (1999). A novel function for transforming growth factor-β1: Upregulation of the expression and the IgE-independent extracellular release of a mucosal mast cell granule-specific β-chymase, mouse mast cell protease-1. Blood 93: 3473–3486. DOI 10.1182/blood.V93.10.3473.410k01_3473_3486. [Google Scholar] [CrossRef]
Oda S, Uchida K, Wang XY, Lee J, Shimada Y, Tominaga M, Kadowaki M. (2013). TRPM2 contributes to antigen-stimulated Ca2+ influx in mucosal mast cell. Pflugers Archiv-European Journal of Physiology 465: 1023–1030. DOI 10.1007/s00424-013-1219-y. [Google Scholar] [CrossRef]
Ramírez-Moreno IG, Ibarra-Sánchez A, Castillo-Arellano JI, Blank U, González-Espinosa C. (2020). Mast cells localize in hypoxic zones of tumors and secrete CCL-2 under hypoxia through activation of L-type calcium channels. Journal of Immunology 204: 1056–1068. DOI 10.4049/jimmunol.1801430. [Google Scholar] [CrossRef]
Rivera J, Gilfillan AM. (2006). Molecular regulation of mast cell activation. Journal of Allergy and Clinical Immunology 117: 1214–1225. DOI 10.1016/j.jaci.2006.04.015. [Google Scholar] [CrossRef]
Rivellese F, Rossi FW, Galdiero MR, Pitzalis C, de Paulis A. (2019). Mast cells in early rheumatoid arthritis. International Journal of Molecular Sciences 20: 2040. DOI 10.3390/ijms20082040. [Google Scholar] [CrossRef]
Shapiro SD, Kobayashi DK, Ley TJ. (1993). Cloning and characterization of a unique elastolytic metalloproteinase produced by human alveolar macrophages. Journal of Biological Chemistry 268: 23824–23829. [Google Scholar]
Sicherer SH, Sampson HA. (2018). Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. Journal of Allergy and Clinical Immunology 141: 41–58. DOI 10.1016/j.jaci.2017.11.003. [Google Scholar] [CrossRef]
Sun J, Sukhova GK, Yang M, Wolters PJ, MacFarlane LA, Libby P, Sun C, Zhang Y, Liu J, Ennis TL, Knispel R, Xiong W, Thompson RW, Baxter BT, Shi GP. (2007). Mast cells modulate the pathogenesis of elastase-induced abdominal aortic aneurysms in mice. Journal of Clinical Investigation 117: 3359–3368. DOI 10.1172/JCI31311. [Google Scholar] [CrossRef]
Suzuki T, Iwamoto N, Yamasaki S, Nishino A, Nakashima Y, Horai Y, Kawashiri SY, Ichinose K, Arima K, Tamai M, Nakamura H, Origuchi T, Miyamoto C, Osaki M, Ohyama K, Kuroda N, Kawakami A. (2015). Upregulation of thrombospondin 1 expression in synovial tissues and plasma of rheumatoid arthritis: Role of transforming growth factor-β1 toward fibroblast-like synovial cells. Journal of Rheumatology 42: 943–947. DOI 10.3899/jrheum.141292. [Google Scholar] [CrossRef]
Theoharides TC, Cochrane DE. (2004). Critical role of mast cells in inflammatory diseases and the effect of acute stress. Journal of Neuroimmunology 146: 1–12. DOI 10.1016/j.jneuroim.2003.10.041. [Google Scholar] [CrossRef]
Tkaczyk C, Horejsi V, Iwaki S, Draber P, Samelson LE, Satterthwaite AB, Nahm DH, Mercalfe DD, Gilfillan AM. (2004). NTAL phosphorylation is a pivotal link between the signaling cascades leading to human mast cell degranulation following Kit activation and FcεRI aggregation. Blood 104: 207–214. DOI 10.1182/blood-2003-08-2769. [Google Scholar] [CrossRef]
Toda M, Kuo CH, Borman SK, Richardson RM, Inoko A, Inagaki M, Collins A, Schneider K, Ono SJ. (2012). Evidence that formation of vimentin mitogen-activated protein kinase (MAPK) complex mediates mast cell activation following FcεRI/CC chemokine receptor 1 cross-talk. Journal of Biological Chemistry 287: 24516–24524. DOI 10.1074/jbc.M111.319624. [Google Scholar] [CrossRef]
Wang X, Hayashi S, Umezaki M, Yamamoto T, Kageyama-Yahara N, Kondo T, Kadowaki M. (2014). Shikonin, a constituent of Lithospermum erythrorhizon exhibits anti-allergic effects by suppressing orphan nuclear receptor Nr4a family gene expression as a new prototype of calcineurin inhibitors in mast cells. Chemico-Biological Interactions 224: 117–127. DOI 10.1016/j.cbi.2014.10.021. [Google Scholar] [CrossRef]
Wang X, Kageyama-Yahara N, Hayashi S, Yamamoto T, Kadowaki M. (2012). Sphingosine kinase-1-dependent and -independent inhibitory effects of Zanthoxyli fructus to attenuate the activation of mucosal mast cells and ameliorate food allergies in mice. Evidence-based Complementary and Alternative Medicine 2012: 862743. [Google Scholar]
Yamamoto T, Fujiwara K, Yoshida M, Kageyama-Yahara N, Kuramoto H, Shibahara N, Kadowaki M. (2009). Therapeutic effect of kakkonto in a mouse model of food allergy with gastrointestinal symptoms. International Archives of Allergy and Immunology 148: 175–185. DOI 10.1159/000161578. [Google Scholar] [CrossRef]
Yamamoto T, Kodama T, Lee J, Utsunomiya N, Hayashi S, Sakamoto H, Kuramoto H, Kadowaki M. (2014). Anti-allergic role of cholinergic neuronal pathway via α7 nicotinic ACh receptors on mucosal mast cells in a murine food allergy model. PLoS One 9: e85888. DOI 10.1371/journal.pone.0085888. [Google Scholar] [CrossRef]
Yao PL, Morales JL, Gonzalez FJ, Peters JM. (2017). Peroxisome proliferator-activated receptor-β/δ modulates mast cell phenotype. Immunology 150: 456–467. DOI 10.1111/imm.12699. [Google Scholar] [CrossRef]
Yu GC, Wang LG, Han YY, He QY. (2012). ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology 16: 284–287. DOI 10.1089/omi.2011.0118. [Google Scholar] [CrossRef]
Zhou L, Zhuo H, Ouyang H, Liu Y, Yuan F, Sun L, Liu F, Liu H. (2017). Glycoprotein non-metastatic melanoma protein b (Gpnmb) is highly expressed in macrophages of acute injured kidney and promotes M2 macrophages polarization. Cellular Immunology 316: 53–60. DOI 10.1016/j.cellimm.2017.03.006. [Google Scholar] [CrossRef]
To analyze the surface receptor expression of mBMMCs, 1 × 105 mBMMCs were incubated with FITC-conjugated anti-mouse CD117 and PE-conjugated anti-mouse FcεRI (or isotype control antibody for the negative control) for 30 min at 4°C in the dark. Cytometric analysis was performed with a CytoFlex S (Beckman Coulter, USA).
RNA quality control analysis with formaldehyde denaturing gel electrophoresis
The overall quality of the RNA preparation from each group was assessed by the 28S/18S ribosomal RNA ratio using formaldehyde denaturing agarose gel electrophoresis with MOPS buffer. The RNA OD260/230 values and OD260/280 values were assessed with a NanoDrop 2000 (Thermo, USA).
The mBMMCs were suspended at a density of 2 × 105 cells/mL in the medium and sensitized with 1.5 μg/mL mouse monoclonal anti-dinitrophenyl (DNP) IgE for 6 h at 37°C. The cells were washed and resuspended at a density of 6 × 105 cells/mL in 100 μL Tyrode’s buffer (130 mM NaCl, 5 mM KCl, 1.4 mM CaCl2, 1 mM MgCl2, 5.6 mM glucose, 10 mM HEPES, and 0.1% BSA, pH 7.5) and stimulated with 100 ng/ml DNP-bovine serum albumin (BSA) at 37°C for 1 h. Then, the cells were washed, resuspended, and seeded in a 96-well plate at a density of 5,000 cells/well. For the A23187 experiment, mBMMCs were washed and resuspended, seeded in a 96-well plate at a density of 5,000 cells/well and added to 25 μM A23187. Then, 10 µL CCK-8 solution was added to each well at 37°C for 2 h, and the optical densities were measured at 450 nm using a microplate reader (PowerWave XS2; BioTek, USA).
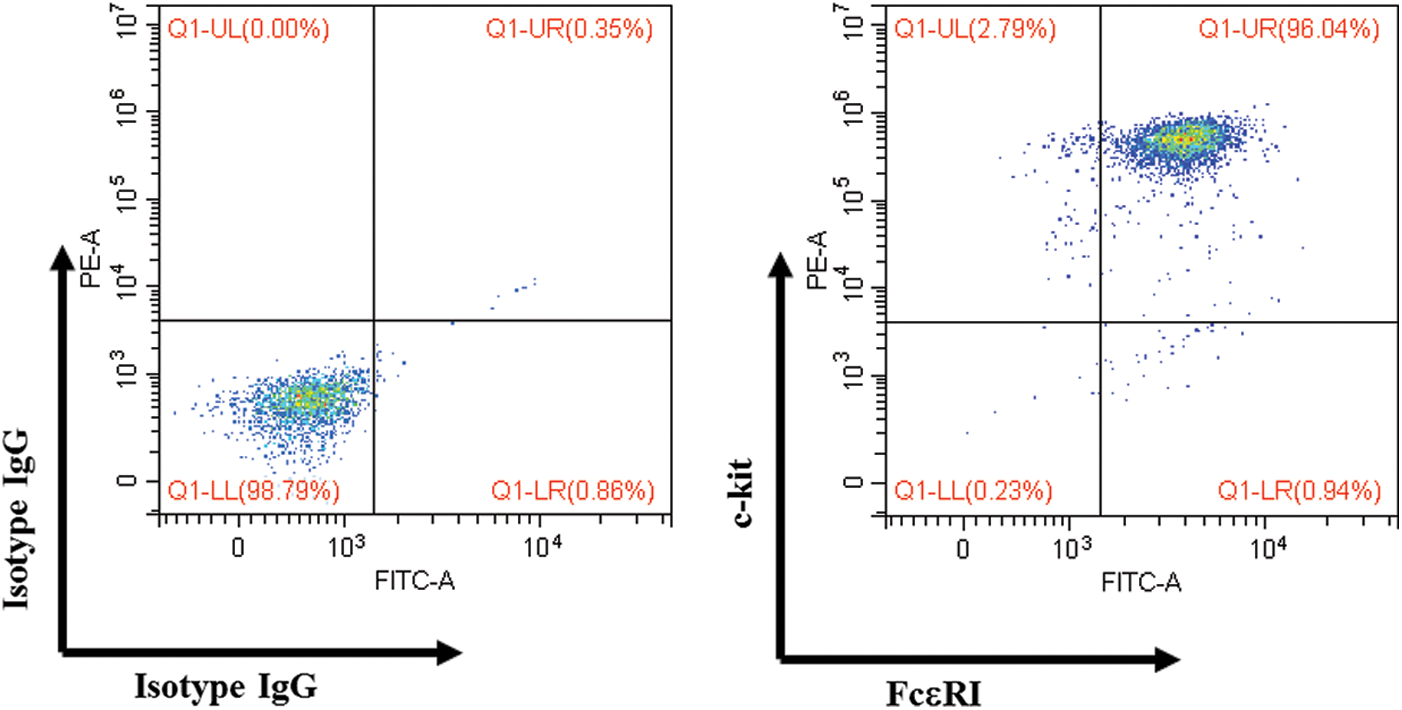
Figure S1: Surface expression of mBMMCs.
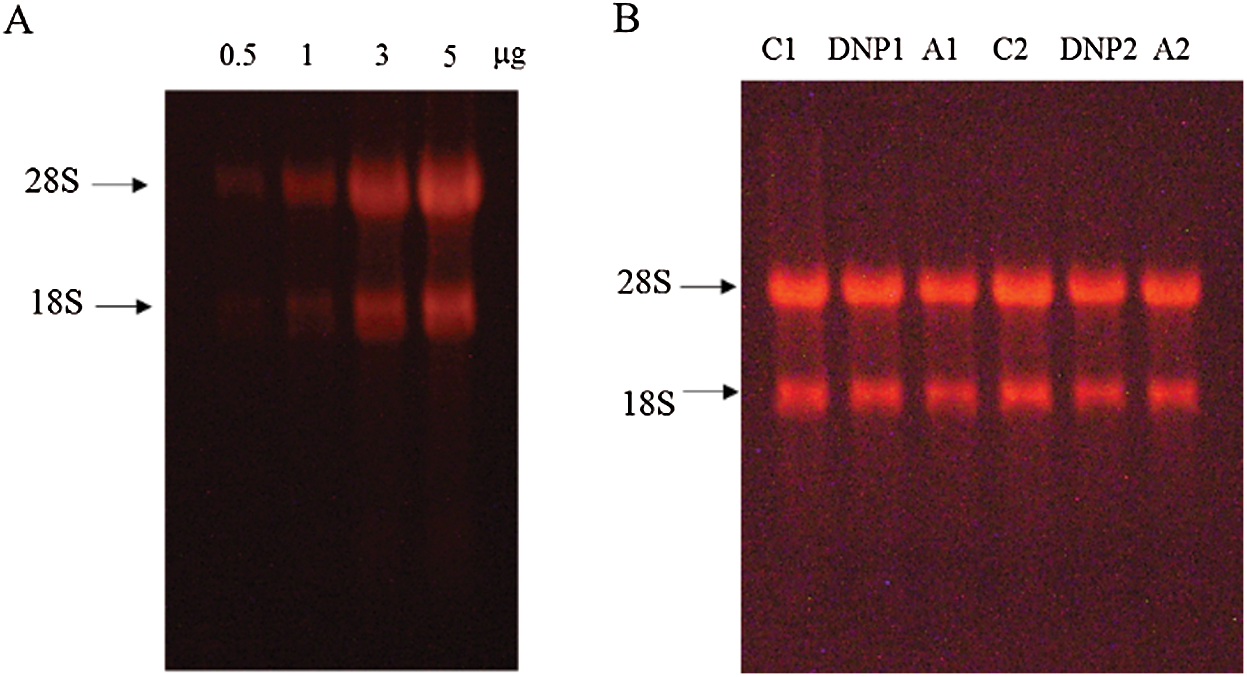
Figure S2: (A) The integrity of RNA increased in a concertation-dependent manner. (B) The imaging of electrophoresis bands of RNA (1 μg) from each mBMMCs samples for microarray. The RNA OD260/230 values were between 1.9 and 2.0, and the OD260/280 values were between 1.9 and 2.0.
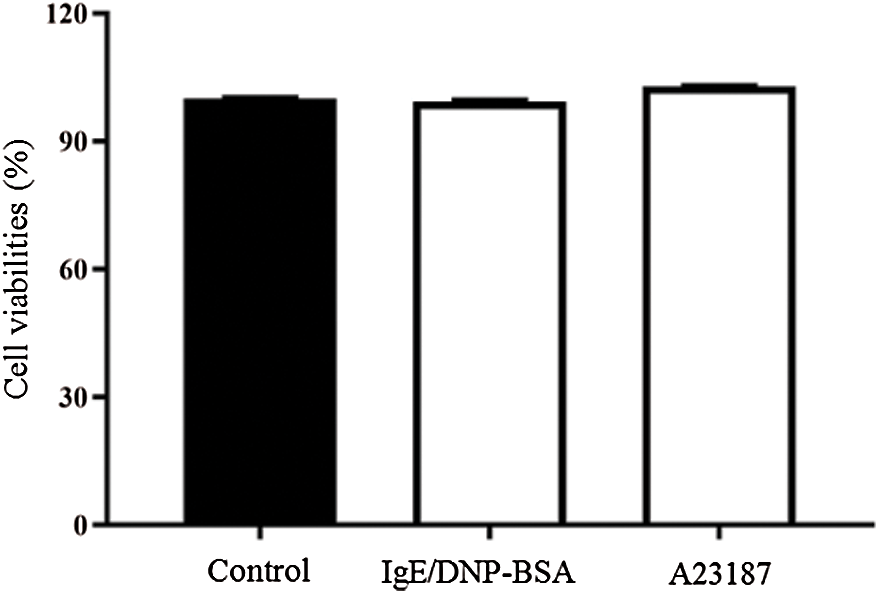
Figure S3: The viability of untreated mBMMCs (control group), mBMMCs sensitized with anti-DNP IgE (1.5 μg/mL, 6 h) and stimulated with DNP-BSA (100 ng/mL) for 60 min (IgE/DNP group), and mBMMCs stimulated with A23187 (25 μM) for 30 min (A23187 group) was determined using a CCK-8 assay.
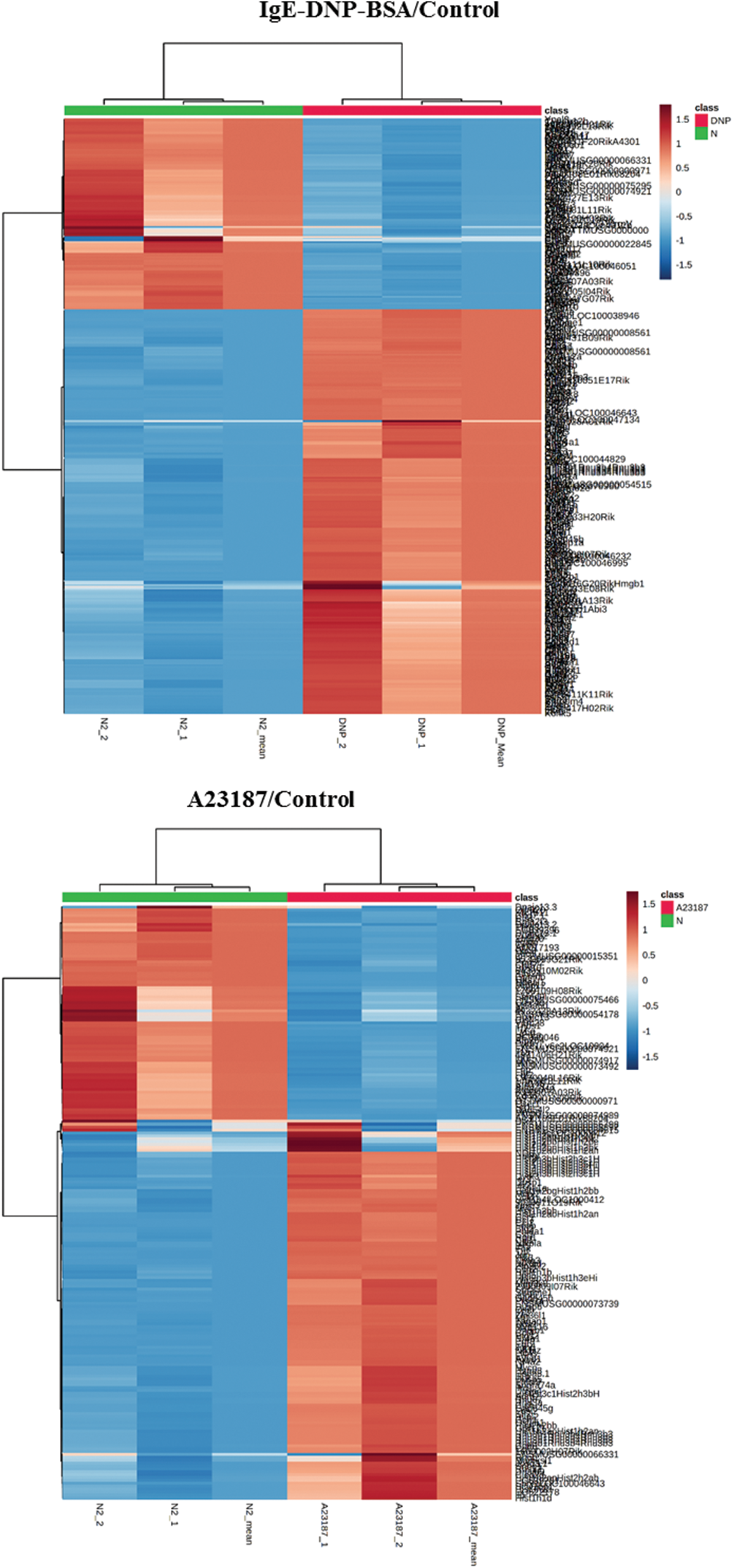
Figure S4: The heat map of DEGs in the activation of mBMMCs by IgE/DNP-BSA and A23187 treatment.
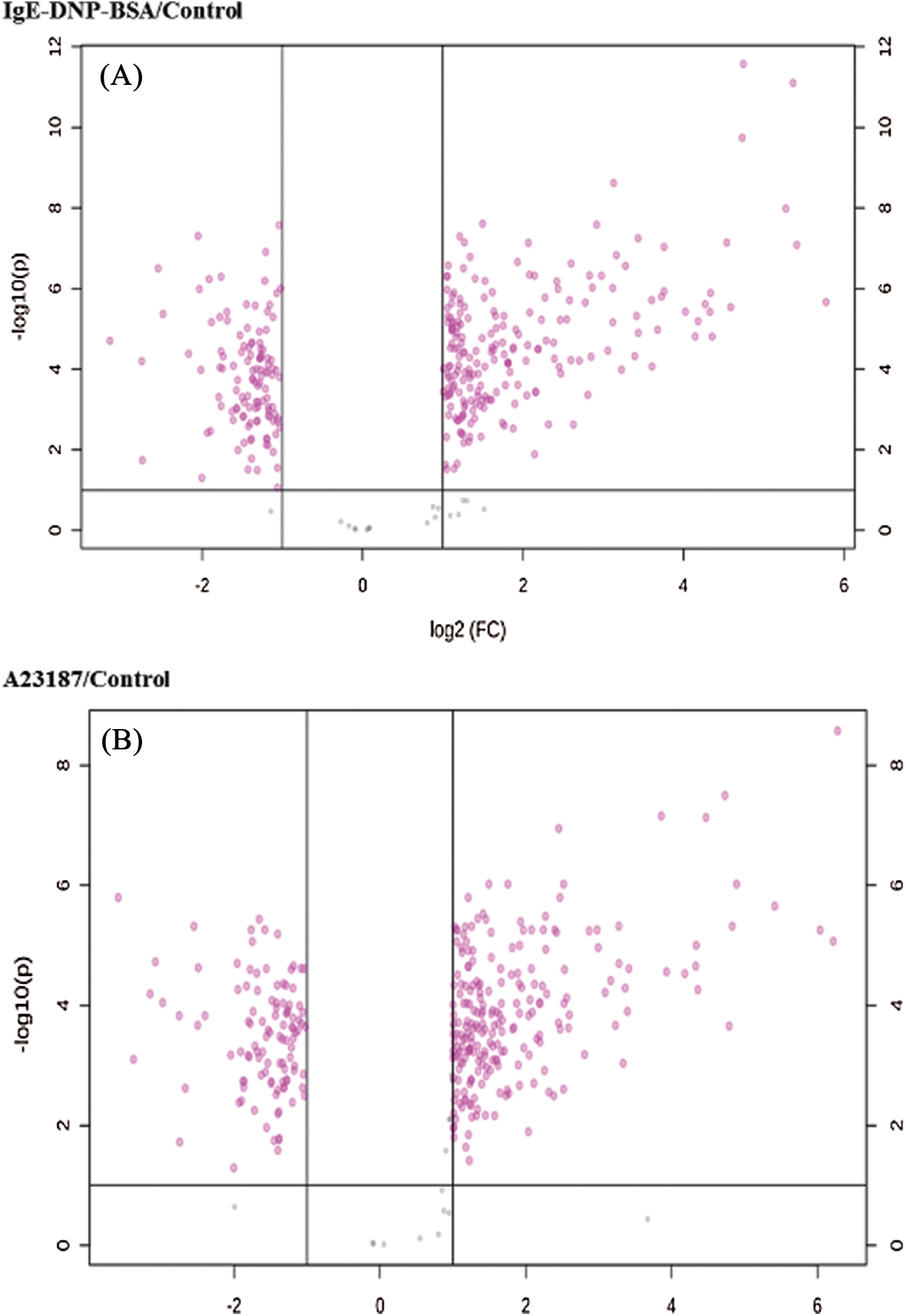
Figure S5: The volcano plot of DEGs in the activation of mBMMCs by IgE/DNP-BSA and A23187 treatment.
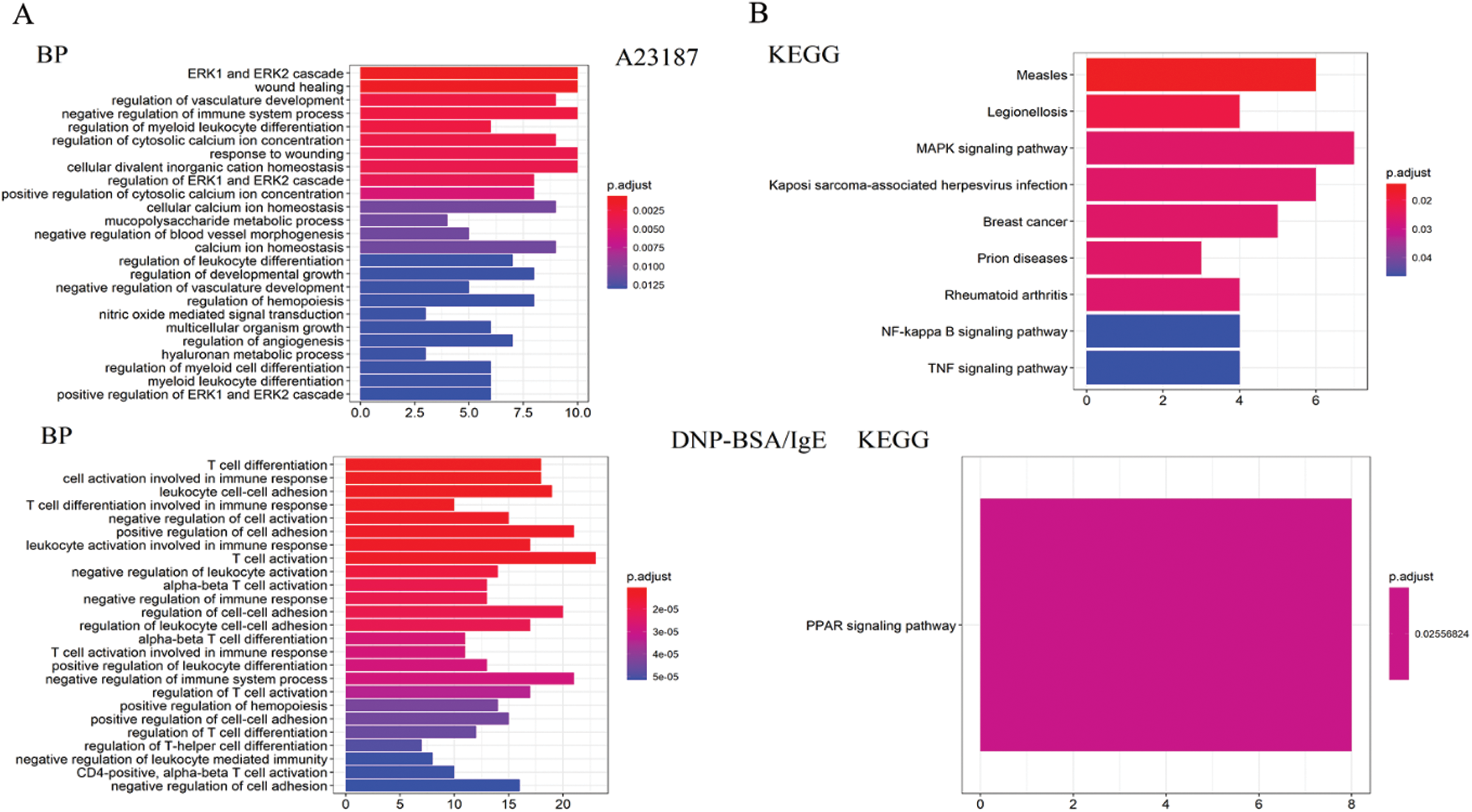
Figure S6: (A) The results of the statistical analysis of the biological process and molecule function for the mRNA changed by IgE/DNP-BSA and A23187 treatment. (B) The KEGG pathway analysis of the genes changed by IgE/DNP-BSA and A23187 treatment.
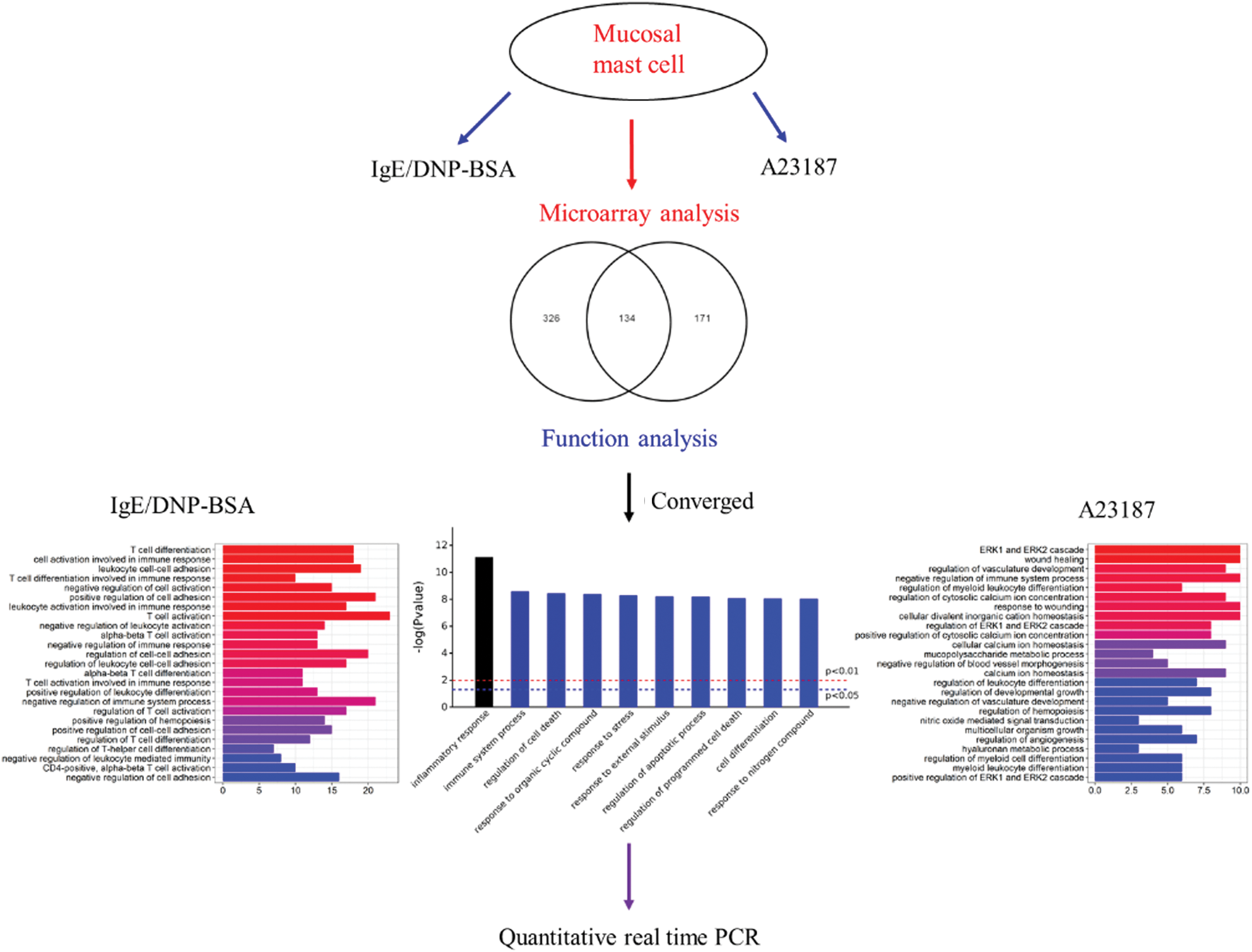
Figure S7: The systems biology workflow.
Table S1: Pathway enrichment of the converged DEGs in the activation of mBMMCs by IgE/DNP-BSA and A23187 treatment
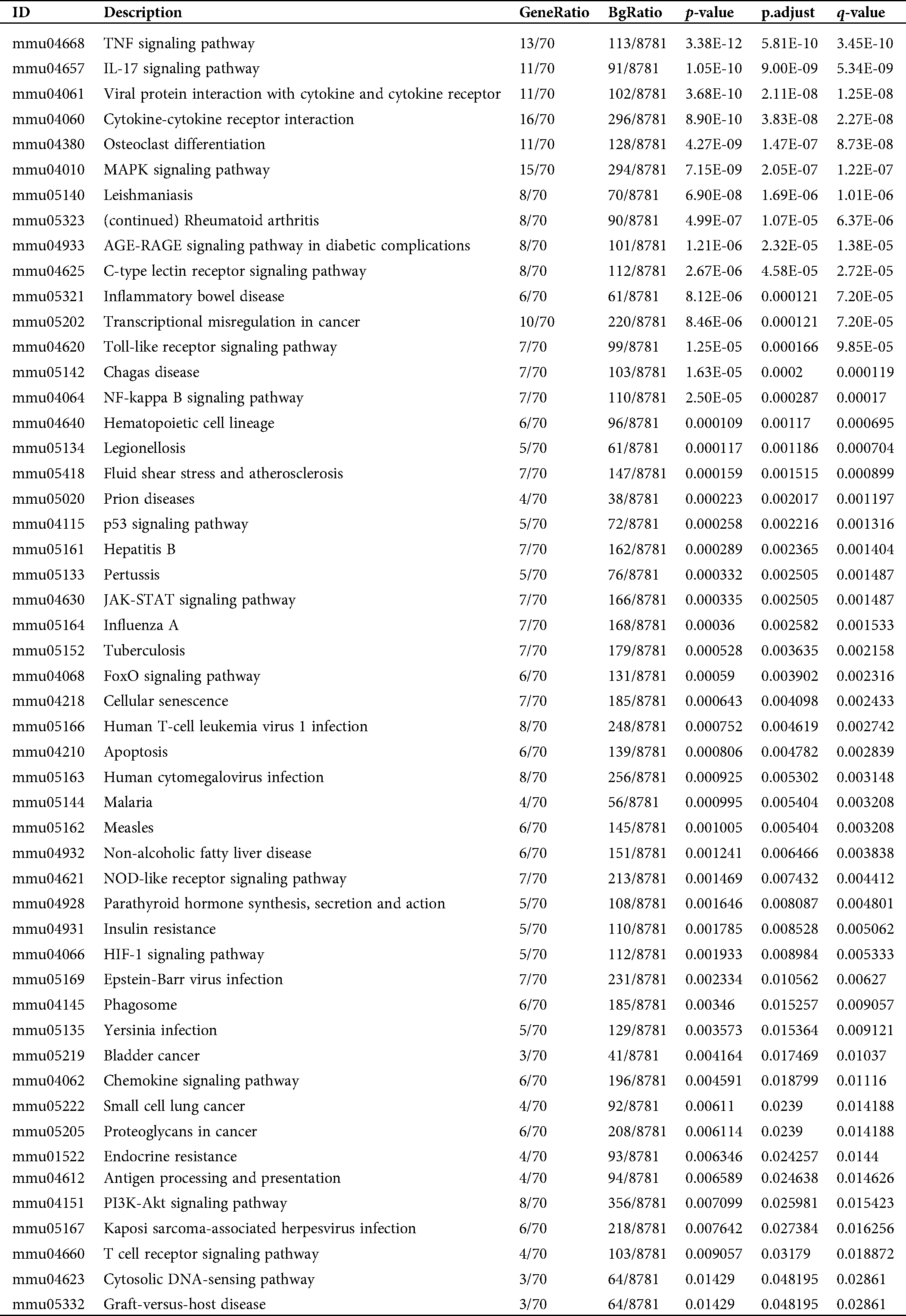
Table S2: Pathway enrichment of the DEGs in the activation of mBMMCs by A23187 treatment
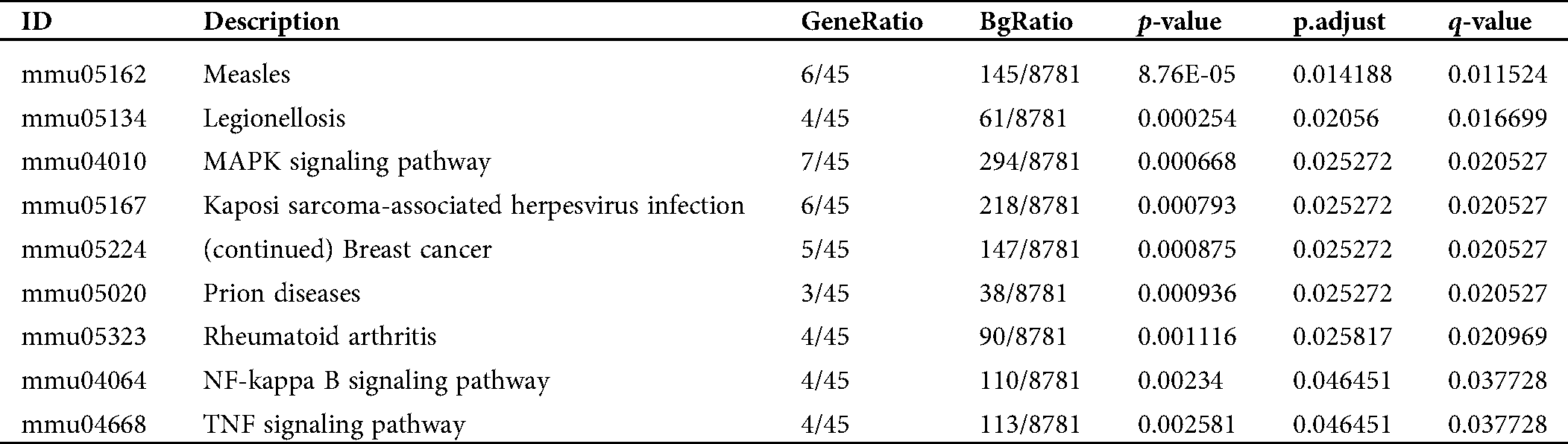
Table S3: Pathway enrichment of the converged DEGs in the activation of mBMMCs by IgE/DNP-BSA treatment
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |