DOI:10.32604/biocell.2021.013960

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.013960 |  www.techscience.com/journal/biocell |
| Article |
Exosomes derived from osteoclasts under compression stress inhibit osteoblast differentiation
Department of Orthodontics, Peking University School and Hospital of Stomatology, Beijing, 10081, China
*Address correspondence to: Yunfei Zheng, yunfei_zheng@bjmu.edu.cn; Weiran Li, weiranli@bjmu.edu.cn
Received: 27 August 2020; Accepted: 09 November 2020
Abstract: Orthodontic tooth movement is triggered by orthodontic force loading on the periodontal ligament and is achieved by alveolar bone remodeling, which is regulated by intimate crosstalk between osteoclastogenesis and osteoblast differentiation. Whether the communication between osteoclasts and osteoblasts is influenced by orthodontic compression stress requires further clarification. In this study, osteoclasts were differentiated for 10 days. On day 4 of differentiation, the number of pre-osteoclasts peaked, as determined by the increased expression of RANK and the number of multinucleated cells. After 24 h of compression stress loading, on day 4, the number of osteoclasts increased, and the optimal magnitude of stress to promote osteoclastogenesis was determined as 1 g/cm2. Moreover, the results of RNA-sequencing analysis showed that the miRNA expression profile changed markedly after compression loading and that many of the altered miRNAs were associated with cell communication functions. A series of indirect co-culture experiments showed an inhibitory effect of osteoclasts on osteoblast differentiation, especially after compression. Next, we added osteoclast-derived exosomes to hPDLSCs during osteoblast differentiation. Exosomes derived from osteoclasts under compression (cEXO) showed a greater inhibitory effect on osteoblast differentiation, compared to exosomes from osteoclasts without compression (EXO). Therefore, we analyzed differentially expressed miRNAs associated with bone development functions in exosomes: miR-223-5p and miR-181a-5p were downregulated, whereas miR-133a-3p, miR-203a-3p, miR-106a-5p, and miR-331-3p were upregulated; these altered expressions may explain the enhanced inhibitory effect of compression stress.
Keywords: Osteoclastogenesis; Osteogenesis; Compression stress; Exosome; microRNA
Bone remodeling, which depends on the balance between bone formation and resorption, is based on close relationships among osteocytes, osteoblasts, and osteoclasts (Hughes et al., 2020; Rendina and Clifford, 2020; Yuan et al., 2016). As a class of resident cells, osteocytes are the primary mechanosensory cells and orchestrate the different responses of bone to different mechanical environments (Hughes et al., 2020). Interactions between osteoclasts and osteoblasts lead to different degrees of bone remodeling, which is important for fracture healing, bone disease treatment, and orthodontic tooth movement (Wang et al., 2018; Zhu et al., 2018). These interactions are regulated by multiple genes and proteins (Li et al., 2017; Mikihito et al., 2019; Yu et al., 2019), as well as by the dynamic extracellular microenvironments (e.g., external mechanical forces). Furthermore, a series of bone signals (Wang et al., 2018) have been identified as responses to stress stimuli (Berger et al., 2019).
Mechanical forces such as cyclic mechanical stretch, fluid shear, and compression have all been shown to alter the process of bone remodeling (Wang et al., 2018). In particular, the alveolar bone is primarily under tension in the direction of tooth movement, while the periodontal ligament (PDL) is under compression (Jiang et al., 2015). This finding indicated compression stress of PDL plays a role in tooth movement in orthodontics at an early stage. Compression stress has been shown to promote osteoclast differentiation and inhibit osteoblast differentiation in human periodontal ligament stem cells (hPDLSCs), both in vitro and in vivo (Chen et al., 2015; Hayakawa et al., 2015; Iwawaki et al., 2015). This process is regulated a number of molecules, including ephrin B2, ephrin type-B receptor 4 (EphB4), nuclear factor (NF)-κB, and osteoprotegerin (Dou et al., 2016; Hou et al., 2014; Li et al., 2017; Li et al., 2016). Recent studies have explored the role of exosomes in bone remodeling; they have shown that exosomes are important for communication among bone marrow stromal cells (BMSCs), C2C12 myoblasts, osteoclasts, and osteocytes (Huynh et al., 2016; Liu et al., 2018; Xu et al., 2018). Specifically, BMSC-derived exosomes were shown to promote regeneration of periodontal tissue by osteoblasts, while osteocytic exosomes were shown to inhibit osteoblast differentiation; those results indicated that osteoblasts are regulated by exosomes derived from other bone cells (Qin et al., 2017; Qin et al., 2016). Exosomes are extracellular vesicles with a diameter of 30–100 nm, which participate in intercellular communication and mediate various metabolic processes through the proteins, lipids, mRNAs, and miRNAs that they carry (Thayanithy et al., 2017). The miRNAs contained in exosomes play a variety of essential roles; the effects of miRNAs in bone remodeling, especially changes induced by mechanical forces, have been studied extensively (Wang et al., 2018). For example, miR-494-3 induced by compressive force has been shown to inhibit osteogenesis, while miR-103 expression was shown to be altered by compression stress in hPDLCs (Chen et al., 2015; Iwawaki et al., 2015; Zuo et al., 2015). However, few studies have focused on whether and how compression stress regulates the interaction between osteoclasts and osteoblasts or the roles of exosomes and miRNAs (Yuan et al., 2018). As an essential step in orthodontic tooth movement, osteoclasts appear following the recruitment of pre-osteoclasts to the PDL compressive side (Rody et al., 2001). Therefore, the reaction of pre-osteoclasts to compression stress is considered a critical component of the orthodontic tooth movement.
In this study, a suitable model of compression stress was applied during osteoclast differentiation to mimic the mechanical compression loading used in orthodontic treatment. Following determination of the optimal force magnitude and loading time, pre-osteoclasts and osteoclasts were placed under compression of 1 g/cm2 for 24 h on day 4 of osteoclastogenesis. The culture supernatants of pre-osteoclasts and osteoclasts were collected on day 5 of differentiation. Exosomes in the supernatants of cells with and without compression loading (cEXO and EXO, respectively) were extracted after gradient centrifugation. The inhibitory effects of the cEXO and EXO exosomes on osteoblast differentiation of hPDLSCs were assessed by sequencing the changes in the miRNA profiles in the osteoclasts. Expression levels of miR-223-5p, miR-181a-5p, miR-133a-3p, miR-203a-3p, miR-106a-5p, and miR-331-3p, which have been linked to bone remodeling, were altered in both osteoclasts and exosomes; these findings suggested possible novel targets for regulation of orthodontic tooth movement.
Cell culture and differentiation
The human monocytic cell line THP-1 was maintained in RPMI 1640 media (HyClone, Auckland, New Zealand) with 15% fetal bovine serum (FBS; Gibco, Auckland, New Zealand) supplemented with 100 µg/mL penicillin and streptomycin (Gibco). hPDLSCs were isolated and cultured, as described previously (Zheng et al., 2017). PDL tissues were isolated from premolars at Peking University Stomatology Hospital. Patients provided informed consent for the provision of teeth. Experimental protocols were approved by the Ethics Committee of Peking University (PKUSSIRB- 2011007). Tissues were scraped from the middle of the tooth roots and cultured in α-MEM (HyClone) supplemented with 15% FBS, penicillin, and streptomycin. Cells were incubated at 37°C in 5% CO2 in humidified air. The hPDLSCs were characterized by the determination of CD73, CD105, and CD90 expression as described previously (Zheng et al., 2017).
For osteoclast differentiation, THP-1 cells were differentiated using 100 ng/mL phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich, St. Louis, MO, USA) for 24 h. Following removal of PMA-containing medium, differentiation was induced by incubation with osteoclast medium containing 50 ng/mL macrophage colony-stimulating factor (M-CSF) (Novoprotein, Summit, NJ, USA) and 50 ng/mL receptor activator of NF-κB ligand (RANKL; Novoprotein) for 2, 4, 6, 8, or 10 days.
For osteogenic differentiation, hPDLSCs were induced in an osteogenic medium with osteogenesis factors (OGFs), including 100 nM dexamethasone, 200 µM L-ascorbic acid, and 10 mM β-glycerophosphate.
THP-1 cells were incubated in 12-well plates with a conditioning medium. Procedures were performed as described previously. Briefly, cells were fixed with 4% paraformaldehyde. Primary antibodies against RANK (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and FAK (Proteintech, Hubei, China) were diluted to 1:500 in 5% bovine serum albumin (5% BSA, Solarbio), and cells were incubated with primary antibodies overnight at 4°C. Secondary antibody (Alexa Fluor 488 goat anti-mouse IgG [H + L]; Invitrogen, Carlsbad, CA, USA) was diluted to 1:1000 and added to cells for 1 h at room temperature. Nuclei were stained with DAPI; cells were imaged using fluorescence microscopy (Nikon Eclipse TS100).
Application of compression stress
Before the application of compressive force, THP-1 cells were induced on a glass sheet for 4 days. Collagen gel layers were prepared on separate culture plates as described previously by Hayakawa et al. (2015). Subsequently, pre-osteoclasts and osteoclasts on the sheet were reversed and placed on top of the collagen gel layers. A matched container with different numbers of beads was placed gently on the glass sheet after calculating the weight. Based on our preliminary experiment results, we applied a maximum magnitude of 1.2 g/cm2 to avoid severe cell deformation and gel breakage. Then we loaded a series of gradient compression forces from 0 to 1.2 g/cm2 onto the cell layers. The cells under the glass sheet were subjected to uniform and continuous compression force for 24 h.
In the subsequent indirect co-culture experiment and miRNA sequencing analysis, to avoid the influence of collagen gel, osteoclasts in control groups were seeded on a glass sheet and placed directly on a gel layer without reversal of the sheet.
Indirect co-culture of hPDLSCs and osteoclasts
hPDLSCs were seeded on Transwell membrane inserts with a pore size of 0.4 µm; THP-1 cells were induced to form osteoclasts in the lower compartment. OGFs were added into the insert 2 days after hPDLSCs seeding. The periods of co-culture were 0, 5, 8, and 10 days; hPDLSCs cultured without OGFs were used as controls. Alkaline phosphatase (ALP) staining was used to detect osteogenesis.
To identify the temporal influence of osteoclast differentiation on osteoblast differentiation, supernatants from osteoclasts cultured for different periods were applied to hPDLSCs. Supernatants from osteoclasts were added to hPDLSCs at different time points, in accordance with the experimental design. The culture media of osteoclasts and osteoblasts were changed in the meantime at 2–3-day intervals to ensure that supernatants remained fresh. To determine the role of compression force in osteoblast differentiation, hPDLSCs were cultured with fresh supernatants from day-5 osteoclasts, with or without 24-h compression. The supernatants of pre-osteoclasts and osteoclasts were added to hPDLSCs after centrifugation at 275×g for 5 min to remove debris and dead cells. Equal amounts of supernatants and osteogenic medium with 2×OGFs were added to hPDLSCs. After 10 days of osteogenesis, ALP staining was performed, and the total RNA of hPDLSCs was extracted for analysis.
Pre-osteoclasts and osteoclasts in the Transwell co-culture system were also treated with GW4869 (MCE, Beijing, China). GW4869 was first stored at −20°C as a 1.5 mM stock suspension in dimethylsulfoxide. This suspension was solubilized by the addition of 0.25% methane sulfonic acid (2.5 µL of 5% methane sulfonic acid in sterile distilled H2O, added to 47.5 µL of GW4869 stock solution) before use. The suspension was mixed and heated at 37°C until clear. GW4869 was added at a concentration of 10 µM to treat osteoclasts in the lower compartment (Nakamura et al., 2015). Osteogenic differentiation of hPDLSCs cultured in the insert was observed by ALP staining.
Additionally, THP-1 cells were induced in osteoclast medium with 10 µM GW4869. The culture supernatants were then added to hPDLSCs with the equal amount of fresh osteogenic medium containing 2×OGFs, in order to avoid the potential effects of the small amount of remaining GW4869 in the culture supernatants during the osteogenesis process. Osteoclast supernatant with 0.25% methane sulfonic acid in dimethylsulfoxide was used as a control. ALP staining was performed to analyze the osteogenic differentiation of hPDLSCs.
Two days before extraction of exosomes, osteoclasts were cultured in osteoclast medium containing 10% exosome-depleted FBS (Gibco, Auckland, New Zealand). A multistep centrifugation procedure was used to isolate exosomes. Culture supernatants of pre-osteoclasts and osteoclasts on day 5 of differentiation and day 4 of differentiation, followed by 24 h of compression, were collected and purified by centrifugation at 300×g for 10 min at 4°C to remove non-adherent cells; they were then centrifuged at 1000×g for 15 min and 10000×g for 30 min at 4°C. The final centrifugation was performed at 100000×g for 2 h at 4°C, using an ultracentrifuge. Exosomes were resuspended in phosphate-buffered saline (PBS) for further use.
The exosomes collected in PBS were characterized by transmission electron microscopy (TEM) as described previously (Jiang et al., 2017). Western blotting analysis was performed using primary antibodies against the extracellular vesicle marker CD63 (1:500; Santa Cruz Biotechnology) and exosomes marker Alix (1:500; Santa Cruz Biotechnology). To exclude contamination of cells, analysis using a primary antibody against the Golgi marker GM130 (1:1000; Proteintech) was also performed.
In the indirect co-culture model, we transferred the supernatants from osteoclasts to PDLSCs. After resuspending them in PBS, we analyzed the concentrations of exosomes from equal supernatant amounts (bicinchoninic acid [BCA] quantifications; Solarbio). In subsequent experiments, the final exosomes concentrations were 3 mg/mL to maintain consistency with the indirect co-culture experiments.
ALP staining was performed as described previously (Zheng et al., 2017). A 5-bromo-4- chloro-3-indolyl-phosphate/nitro blue tetrazolium staining kit (NBT/BCIP, CoWin Biotech, Beijing, China) was used, in accordance with the manufacturer’s instructions. Briefly, cultured cells were rinsed with PBS and fixed in 4% paraformaldehyde for 30 min. After three washes with PBS, the cell layer was incubated in alkaline solution for 20 min at room temperature. Quantitative analysis of the images was performed using ImageJ software (NIH, Bethesda, MD, USA).
Alizarin red S (ARS) staining was performed to visualize mineral deposition in each group. Cultured cells were fixed in 4% paraformaldehyde for 30 min and then stained with 0.1% ARS for 20 min at room temperature as described previously (Zheng et al., 2017). Quantitative analysis of the images was performed using ImageJ software (NIH).
Tartrate-resistant acid phosphatase staining
Tartrate-resistant acid phosphatase (TRAP) staining was performed using a Leukocyte Acid Phosphatase kit (NO387; Sigma-Aldrich), in accordance with the manufacturer’s instructions. Osteoclasts were regarded as cells that exhibited positive TRAP activity staining. TRAP-positive cells were counted and classified as mononuclear, multinuclear (3–7 nuclei), or giant (≥8 nuclei) cells as described previously (Hayakawa et al., 2015).
RNA isolation and quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
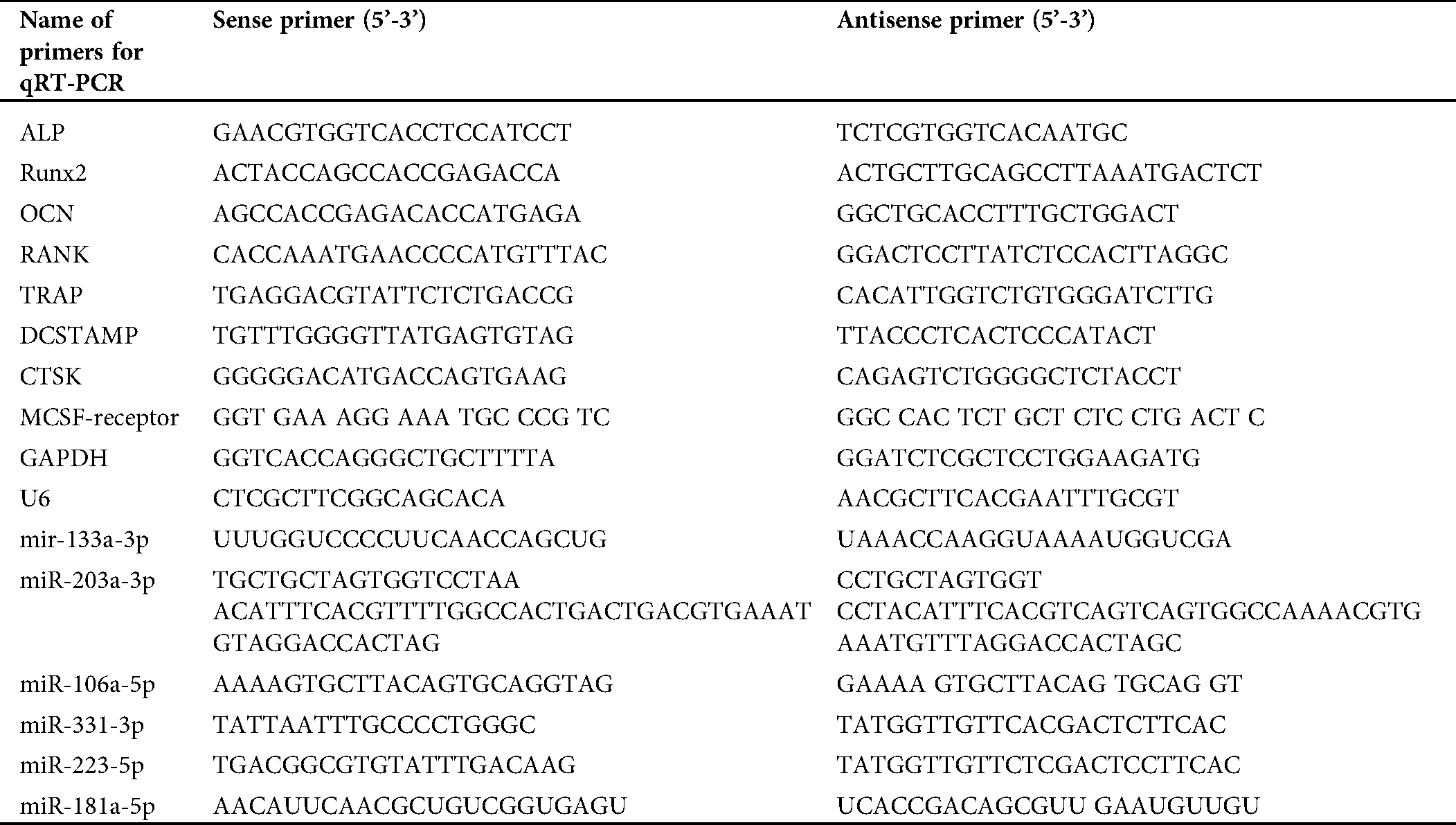
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. For further analysis, 1000 ng of RNA was reverse transcribed into cDNA using a cDNA reverse transcription kit (Takara, Kasatsu, China). Quantitative PCR was performed with a real-time PCR system (Applied Biosystems, Foster City, CA, USA). All mRNA expression data were normalized relative to the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH); all miRNA expression data were normalized relative to U6. The 2−∆∆Ct relative expression method was used to analyze the data (primer sequences are shown in Tab. 1.)
Western blotting analysis was performed as described previously (Zheng et al., 2017). Briefly, cells were washed with PBS and lysed in radioimmunoprecipitation buffer. Proteins were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. After membranes had been blocked with 5% BSA (Solarbio), proteins were detected by overnight incubation with primary antibodies against TRAP (Proteintech, Hubei, China), RANK (Santa Cruz Biotechnology), osteocalcin (OCN) (Proteintech, Hubei, China), Runx2 (Cell Signaling Technology, Danvers, MA, USA), CD63 (Abcam, Cambridge, MA, USA), bone morphogenetic protein-2 (BMP2) (Proteintech) and β-actin (Zhongshan Goldenbridge, Beijing, China) at 1:1000 dilution. After membranes had been washed, the membranes were incubated with secondary antibodies (1:10000; Zhongshan Goldenbridge) at room temperature for 2 h. Proteins were visualized using an ECL Kit (Applygen, Beijing, China). Quantitative analysis of the images was performed using ImageJ software (NIH).
miRNA sequencing and data analysis
Total RNA was extracted from pre-osteoclasts and osteoclasts after 5 days of induction with osteoclast medium and after 4 days of induction followed by 24 h of 1 g/cm2 compression. RNA quantity and quality were measured using a NanoDrop ND-8000 spectrophotometer (Thermo Fisher Scientific).
Two samples were sent to CapitalBio Technology Co., Ltd. (Beijing, China) for further analysis.
Gene ontology (GO) analysis was performed using the GO analysis function of KOBAS database software with data imported from the external GO Annotation Source database (http://www.geneontology.org/). KEGG pathway was performed using the KEGG signaling pathway database (http://www.genome.jp/kegg/).
Statistical analyses were performed using SPSS version 16.0 (SPSS, Chicago, IL, USA). All data are expressed as the mean ± standard deviation. Differences between two groups were analyzed using Student’s t-test; differences among three or more groups were analyzed by one-way analysis of variance, followed by the Student-Newman-Keuls post hoc test. Two-way analysis of variance was used for the analysis of multiple factors among multiple groups. In all analyses, two-tailed p < 0.05 was considered to indicate statistical significance.
Differentiation of THP-1 cells to osteoclasts
To determine the rate of osteoclast differentiation, osteoclasts were classified according to the number of cell nuclei (Fig. 1a). The expression of osteoclastogenesis-related gene cathepsin K (CTSK), DCSTAMP, and MCSF-receptor were increased during the differentiation process (Figs. 1b–1d).
RANK is a sensitive indicator of osteoclast differentiation based on the essential role of the RANKL/RANK interaction during osteoclastogenesis (Boyle et al., 2003; Feng and Teitelbaum, 2013; Xing et al., 2005). Also, TRAP is commonly used as a marker of osteoclast formation (Debnath et al., 2018; Yu et al., 2014). Using the expression of RANK and TRAP along with TRAP staining as markers of pre-osteoclasts and osteoclasts, the results showed that the number of osteoclasts (nuclei >3) significantly increased between days 4 and 6 of differentiation (Figs. 1e–1g). With prolonged incubation, the increase in TRAP expression, compared to day 0, indicated the differentiation of osteoclasts. However, the expression of TRAP was not increased throughout the differentiation period (Fig. 2a). In addition, RANK expression declined slowly for approximately 4 days after the initial increase, as confirmed by immunofluorescence analysis of RANK (Figs. 2b, 2c). These results indicated that the differentiation ability of pre-osteoclasts peaked on day 4. Focal adhesion staining showed the adhesive process during differentiation (Fig. 2d).
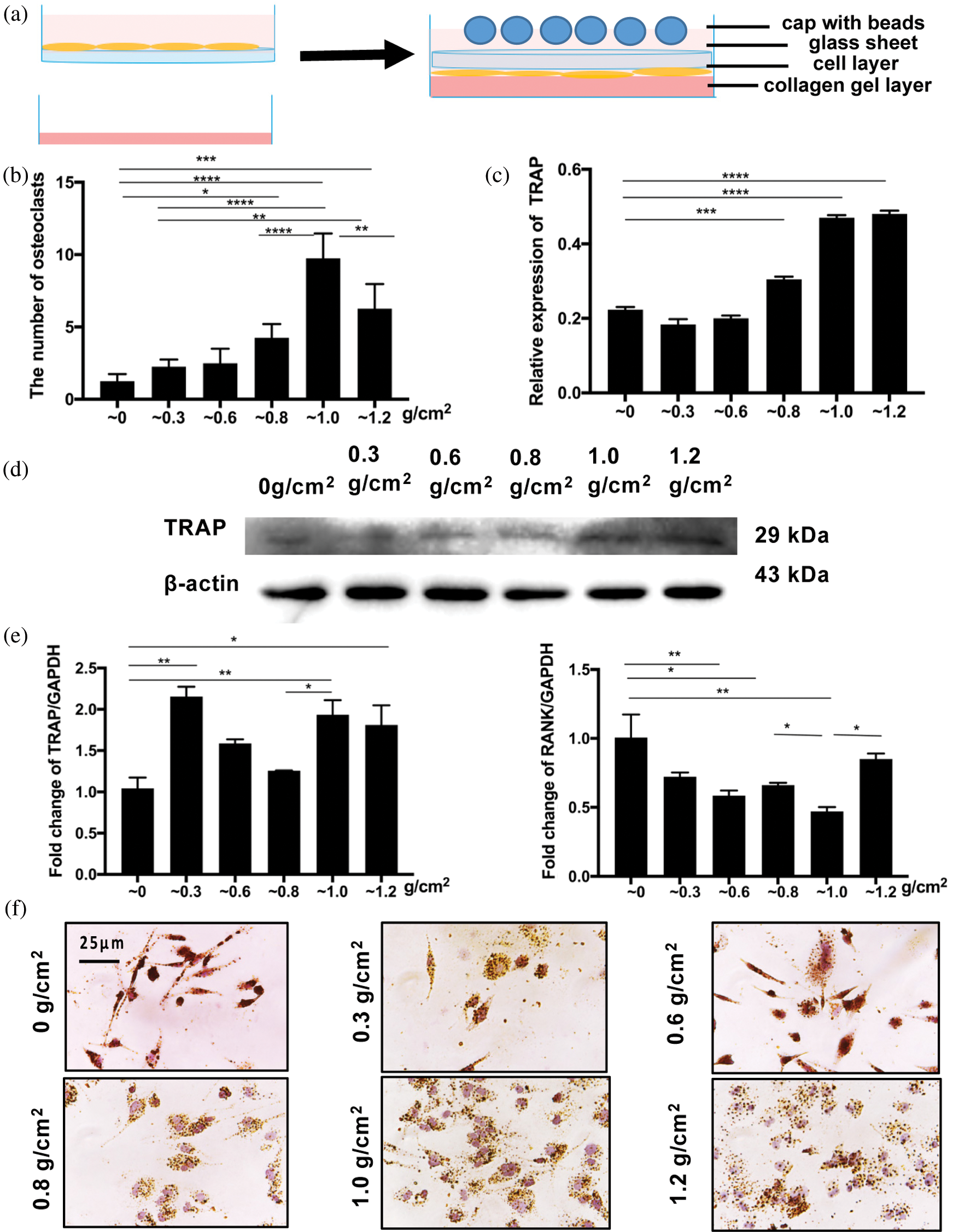
Figure 1: Differentiation of THP-1 cells to osteoclasts.
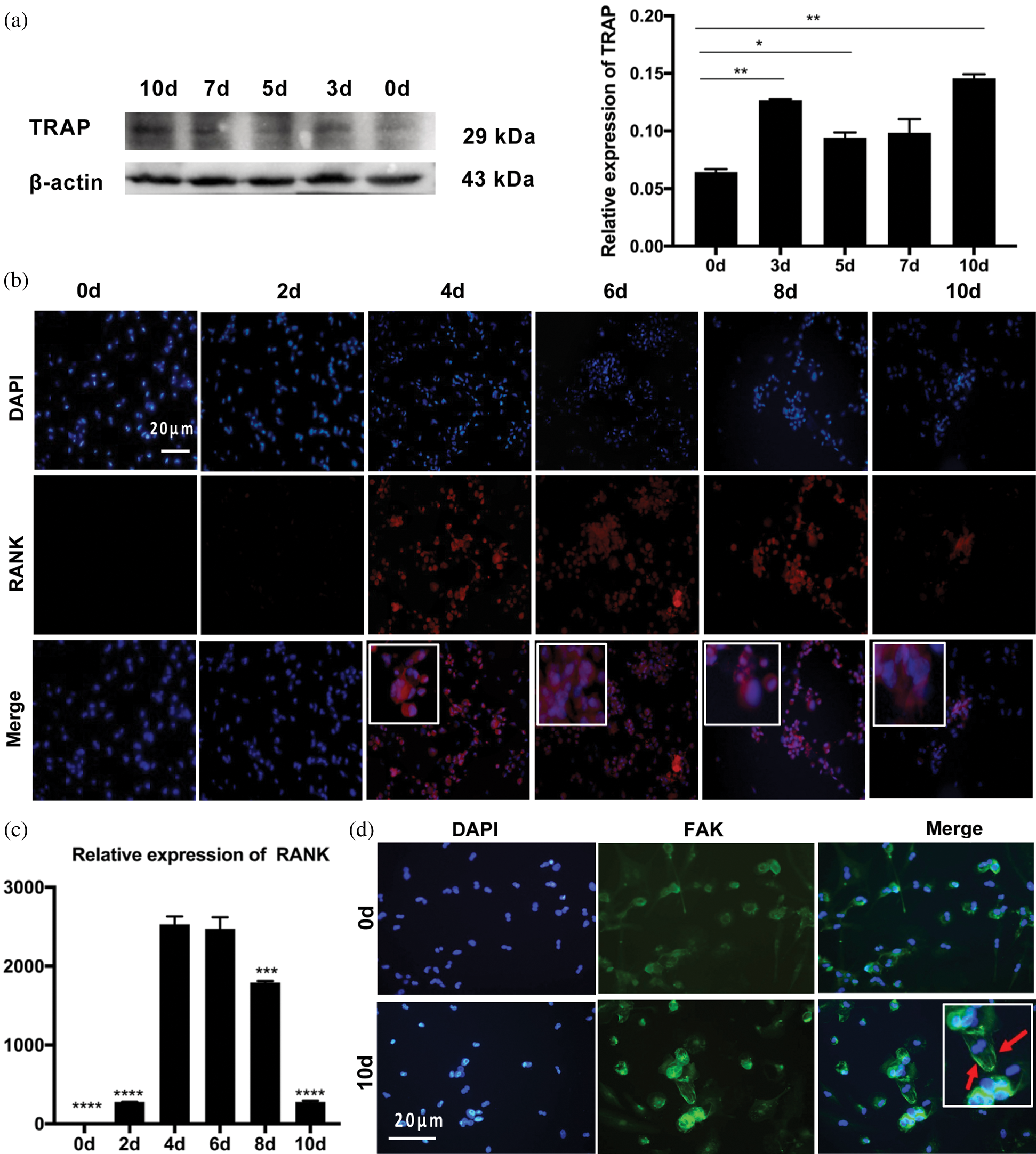
Figure 2: Differentiation of THP-1 cells to osteoclasts.
Application of compression stress
To mimic compression loading in orthodontic treatment, a force was applied to cell layers after 4 days of differentiation, based on the above results (Fig. 3a). Different levels of compression were applied by changing the number of beads; osteoclasts were counted after 24 h of compression loading (Fig. 3b). The expression of TRAP, a marker of mature osteoclasts, showed an overall increase (Figs. 3c–3e).
The force of 1 g/cm2 showed the greatest effect, with the number of multinucleated cells increased most significantly (Fig. 3f).

Figure 3: Application of compression stress.
Sequencing of miRNAs of pre-osteoclasts and osteoclasts with or without compression stress
Heat maps were produced to show the differences in expression of miRNAs from pre-osteoclasts and osteoclasts on day 5 of differentiation after 24 h compression (COMPRESSION group) and without compression stress (CONTROL group). Compared with the control group, the expression levels of 170 miRNAs were increased after compression loading, whereas the expression levels of 67 miRNAs were decreased (Figs. 4a, 4b). GO analysis of miRNAs identified physiological processes that may be related to osteoclastogenesis and osteogenesis (e.g., response to stimuli, metabolic process, and cell communication) via these differentially expressed miRNAs. Specifically, these changed miRNAs are closely related to cellular communication (Fig. 5).
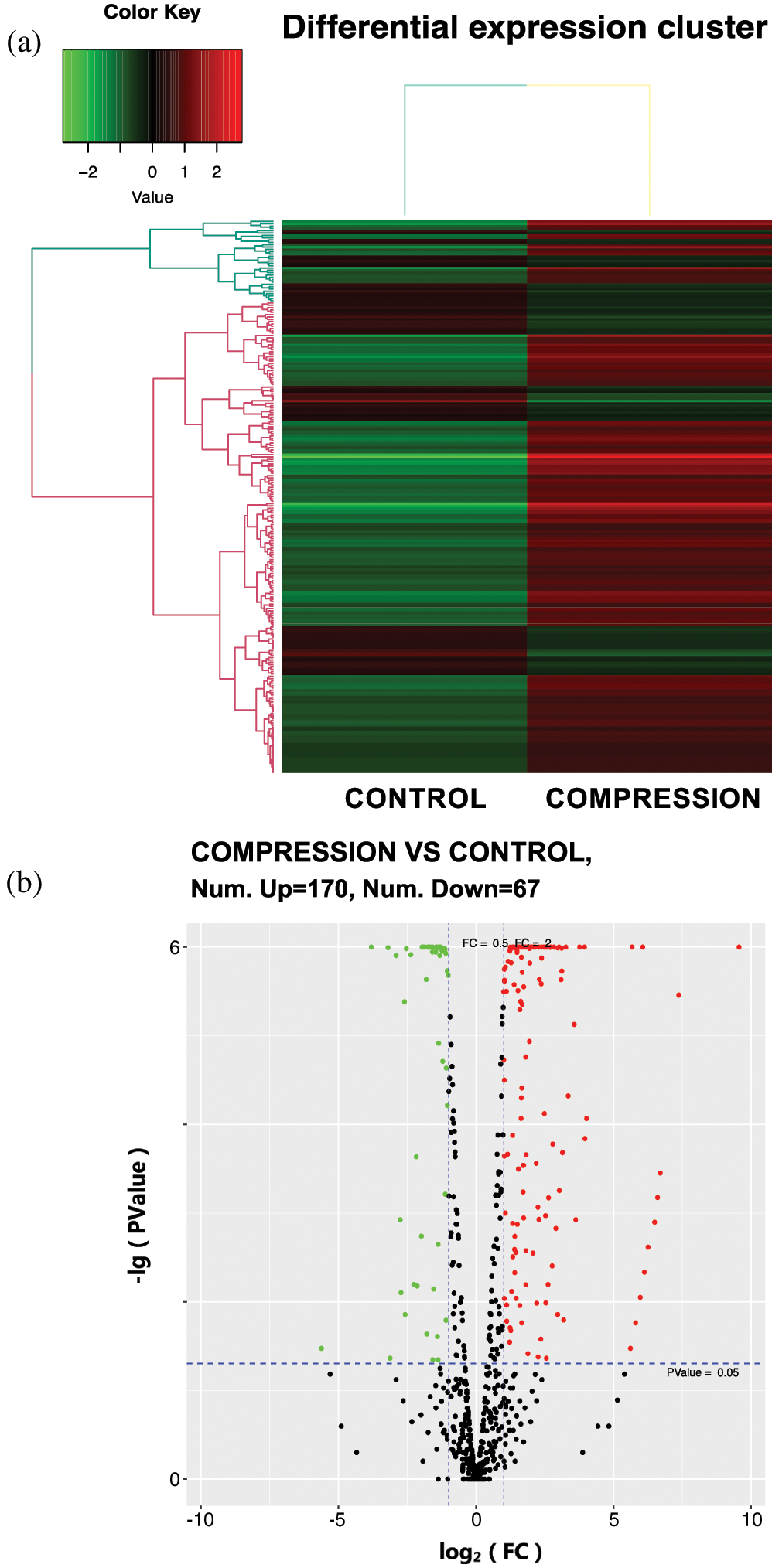
Figure 4: The altered miRNA profile in osteoclasts after compression loading.
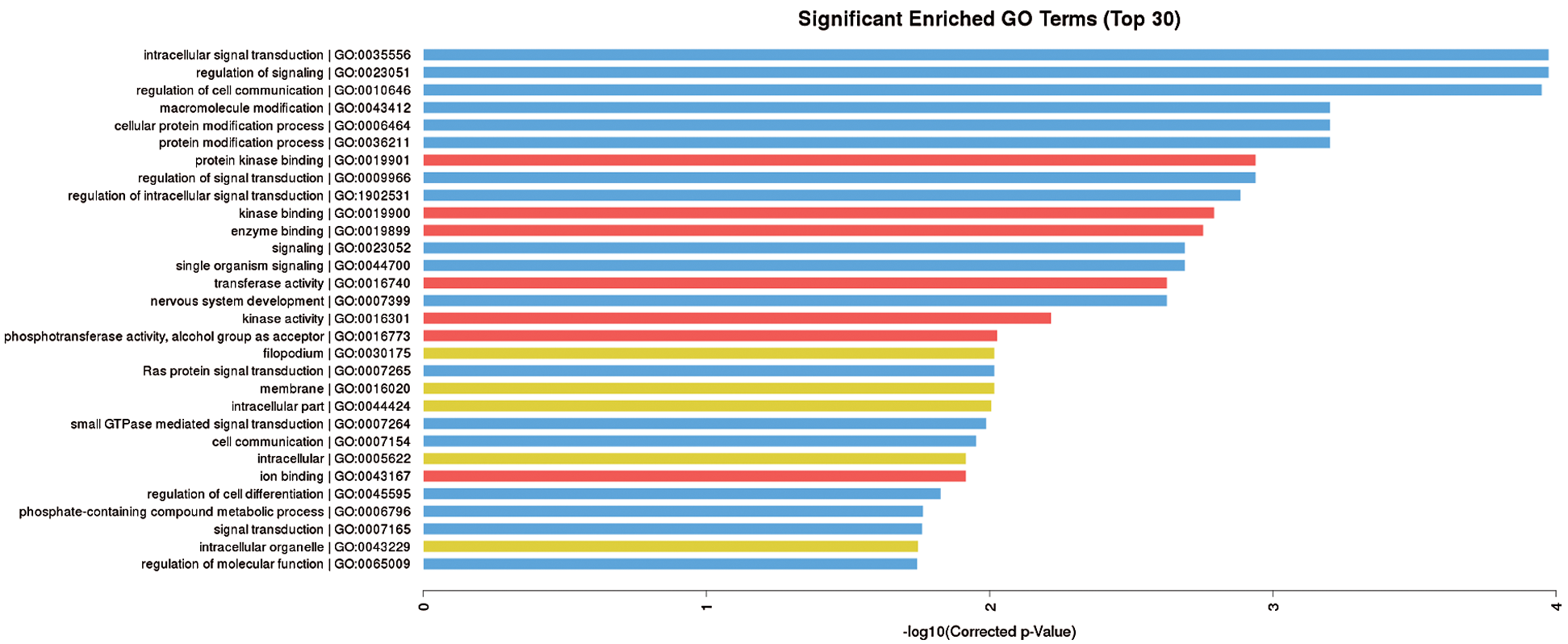
Figure 5: GO analysis. The GO analysis shows the changed miRNAs are related to the regulation of cell communication.
Indirect co-culture of hPDLSCs with pre-osteoclasts and osteoclasts
In Transwell assays, hPDLSCs and osteoclasts were co-cultured for 0, 5, 8, and 10 days. ALP staining of hPDLSCs incubated in medium with and without OGFs for 10 days indicated that the inhibitory effect increased with the prolonged co-culture period (Figs. 6a, 6b).
To further identify the osteogenic influence of different stages of osteoclastogenesis, the 10 days of differentiation were divided into two parts: the first part mainly included pre-osteoclasts, while the second part mainly included mature osteoclasts. Osteoblast differentiation was inhibited following the addition of the culture supernatant of osteoclasts to hPDLSCs incubated with OGFs. PDLSCs were cultured first in osteogenic medium and second in osteoclast supernatants for the number of days indicated in Fig. 6d. The different colors refer to supernatants from 10 days during osteoclastogenesis, as shown in the bottom row in the figure. According to the expression of osteogenic genes, the lowest degree of inhibition was observed with the 4-day culture supernatant in group b, while the greatest inhibitory effect was observed in group e with the supernatant obtained from 8 days of differentiation.
These results indicated a temporal influence on osteoblast differentiation in this co-culture system. To ensure consistency in co-culture, two more groups (groups c and d) were analyzed. Supernatant from the first 6 days of osteoclastogenesis (group d) inhibited osteoblast differentiation to a greater extent than supernatant from the last 6 days (group c). These observations suggested that the inhibitory influence of pre-osteoclasts on osteogenic differentiation is greater than the inhibitory influence of mature osteoclasts (Figs. 6c–6f). After the inhibition of osteogenesis during osteoclastogenesis had been confirmed (groups a’, b’, and c’), the effects of compression forces were assessed using this system. hPDLSCs were cultured in osteogenic medium or the supernatant from osteoclasts, with or without compression loading, as shown in Fig. 7a. After hPDLSCs had been cultured for 10 days in osteoclast supernatant with OGFs, a more significant inhibitory effect was observed when the supernatant from compressed osteoclasts was added, compared to the supernatant from osteoclasts without compression (compare groups c’, d’, and e’). Furthermore, the degree of inhibition was time-dependent, as shown by comparisons of groups b’ and c’, and groups d’ and e’ (Figs. 7b–7d).
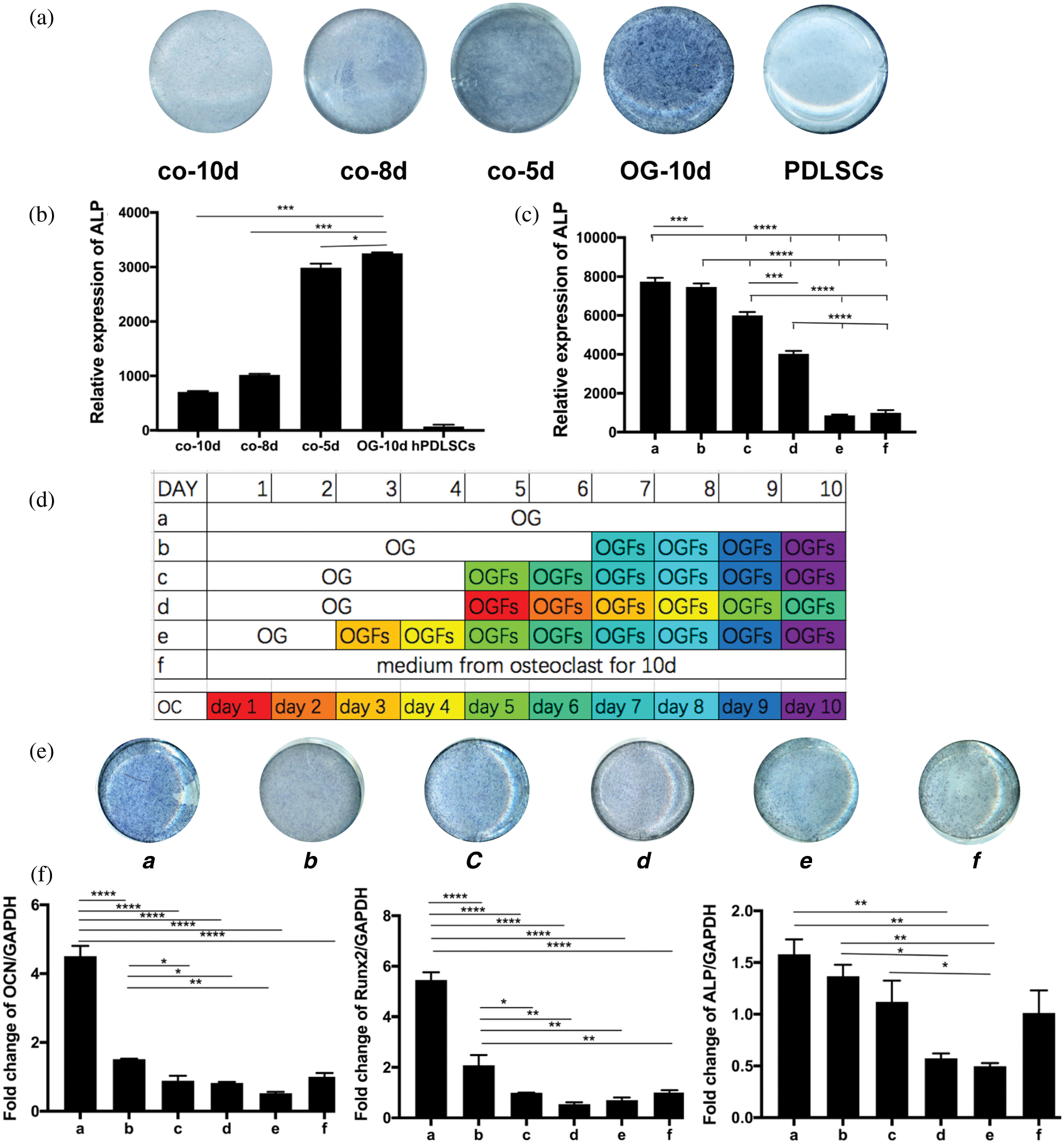
Figure 6: Indirect co-culture of hPDLSCs with osteoclasts.
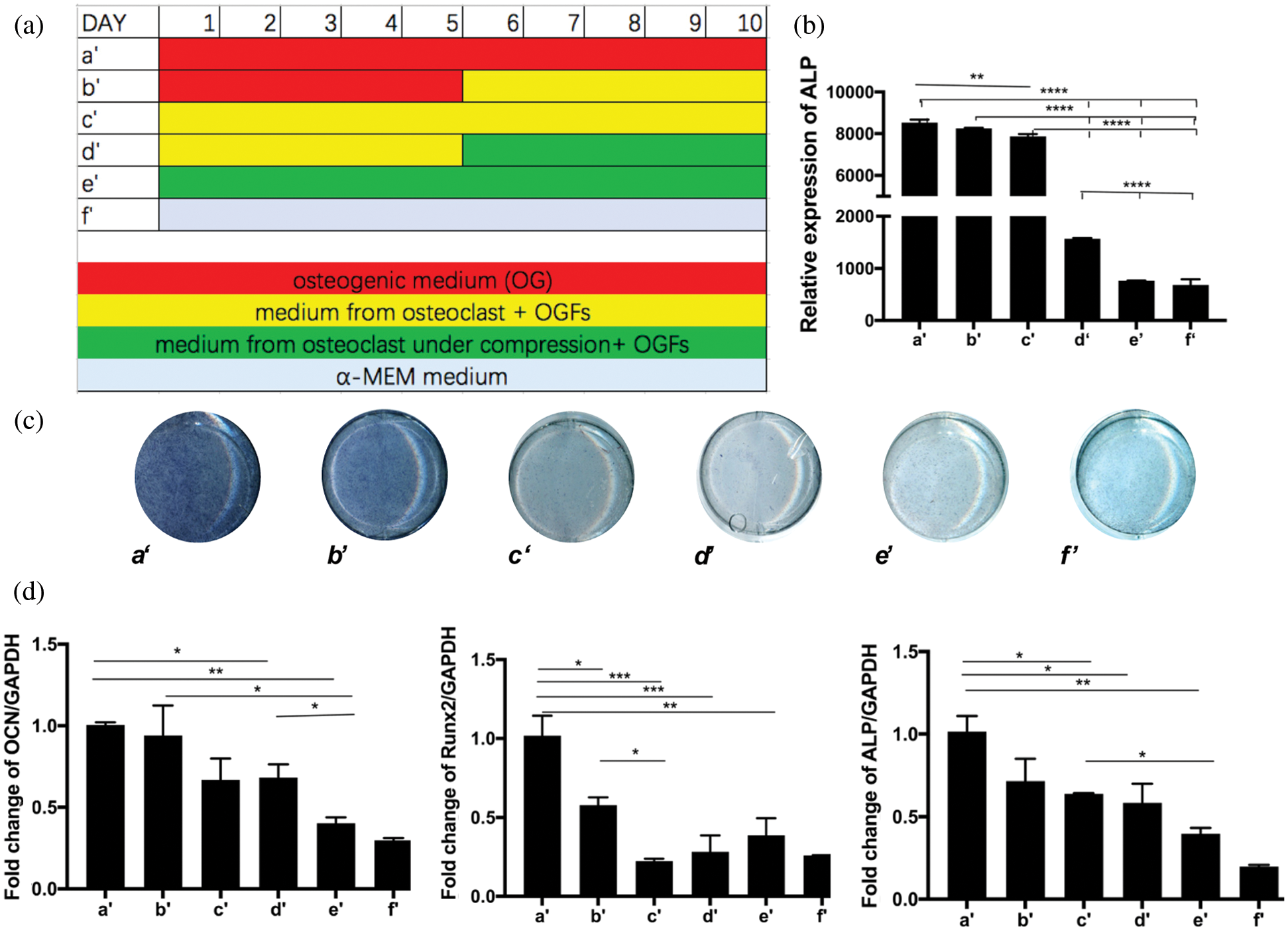
Figure 7: Indirect co-culture of hPDLSCs with osteoclasts.
Role of osteoclast-derived exosomes in inhibition of osteogenic differentiation
Considering exosomes are important regulators in cellular communication, we focused on the exosomes contained within the culture supernatant. The exosomes were obtained from the culture medium of pre-osteoclasts and osteoclasts on day 5, with or without 24 h of compression stress, and from the culture medium of mature osteoclasts on day 10. Extraction of exosomes was confirmed by the presence of the extracellular vesicle markers Alix and CD63, but the absence of the Golgi marker gm130, suggesting the absence of Golgi or cell contamination (Fig. 8a). Exosomes were observed by transmission electron microscopy (Fig. 8b). The effects of adding the three types of exosomes to hPDLSCs with osteogenic medium were then assessed (Fig. 8c). ALP and ARS staining, as well as the results of qRT-PCR, showed inhibition of osteoblasts in all experimental groups. Consistent with the results of the indirect co-culture experiment, exosomes extracted from osteoclasts under compression stress inhibited osteogenesis to a greater extent than those from osteoclasts without compression stress (Figs. 9a, 9b). Because the inhibitory effect was greater in exosomes obtained at day 5 than day 10 of osteoclastogenesis, these exosomes from day 5 were the focus of further analysis. The inhibitory effect of exosomes derived from osteoclasts on osteoblast differentiation was also confirmed by the expression of Runx2, OCN and BMP2 (Figs. 10a, 10b). Furthermore, GW4869 was used in Transwell assays. Inhibition of exosome release reversed the inhibitory effects of osteoclasts on osteogenic differentiation (Fig. 10c). Notably, treatment with GW4869 in the indirect co-culture experiment through culture medium transfer reversed the inhibitory effect. The supernatant from osteoclasts with GW4869 showed less of an inhibitory effect than supernatant from osteoclasts in the control group, compared with groups cOC-10d and cOC-10d+GW4869, or OC-10d and OC- 10d+GW4869 (Fig. 10d).
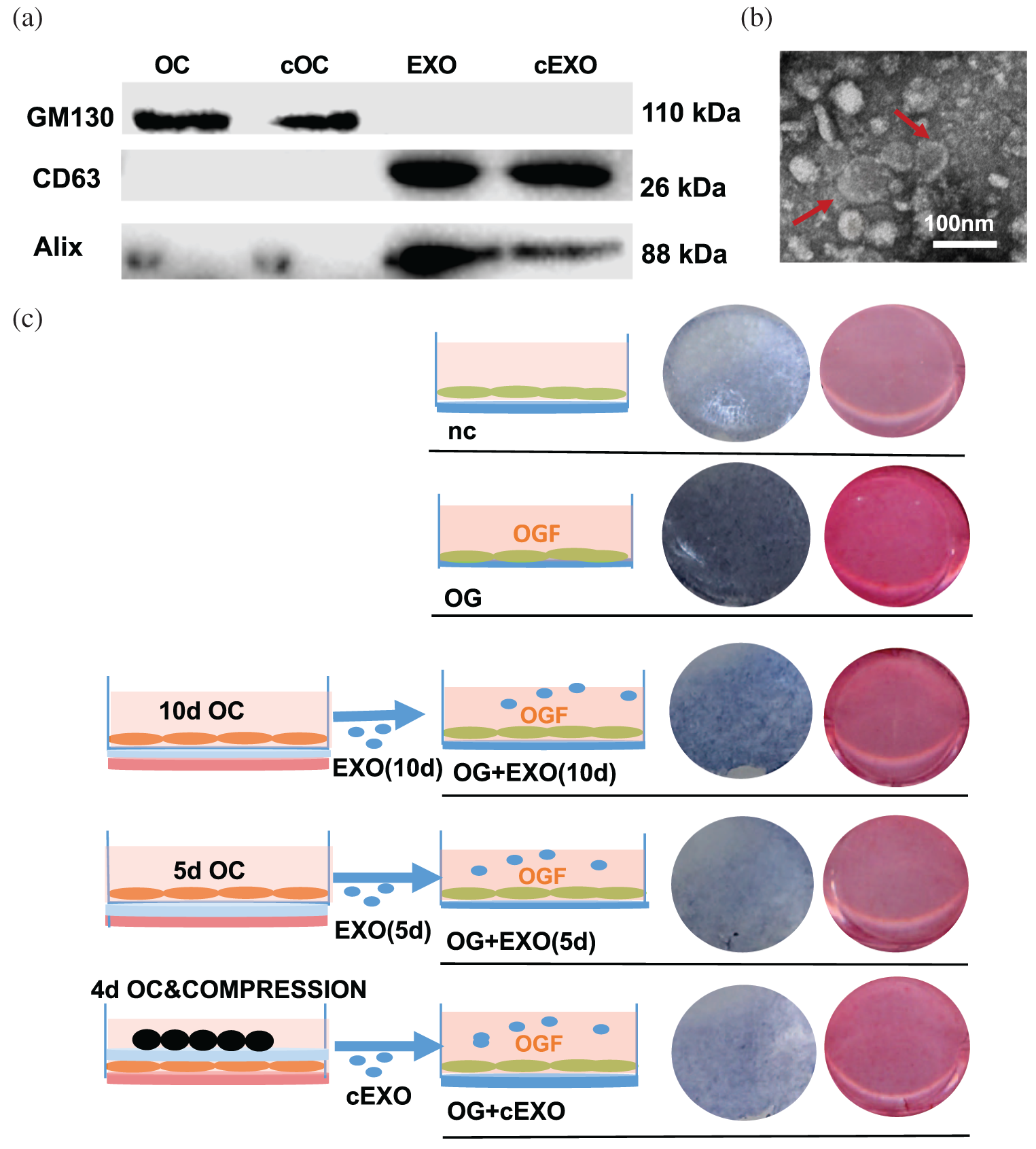
Figure 8: Role of osteoclast-derived exosomes in inhibition of osteogenic differentiation.
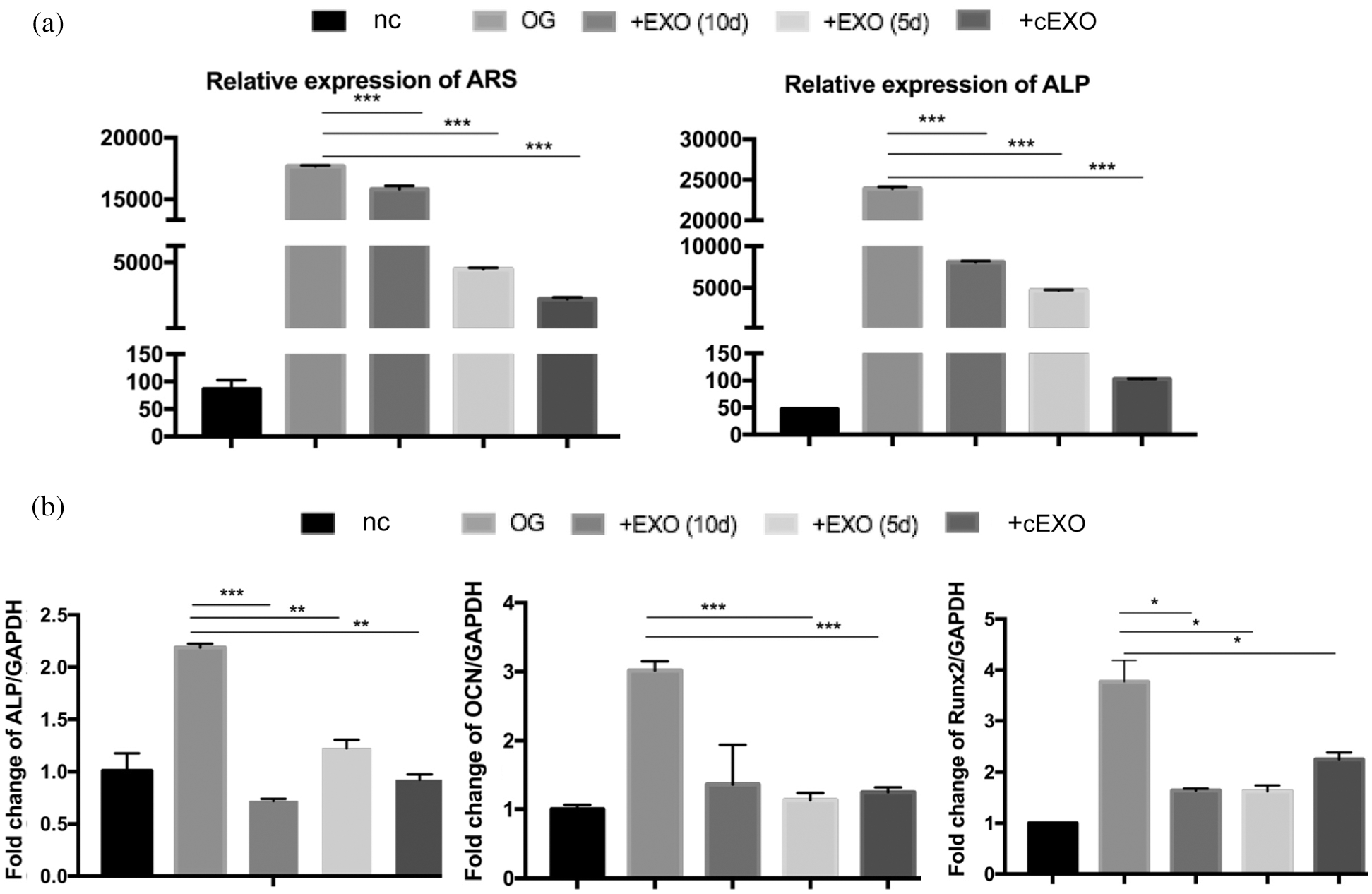
Figure 9: Role of osteoclast-derived exosomes in inhibition of osteogenic differentiation.
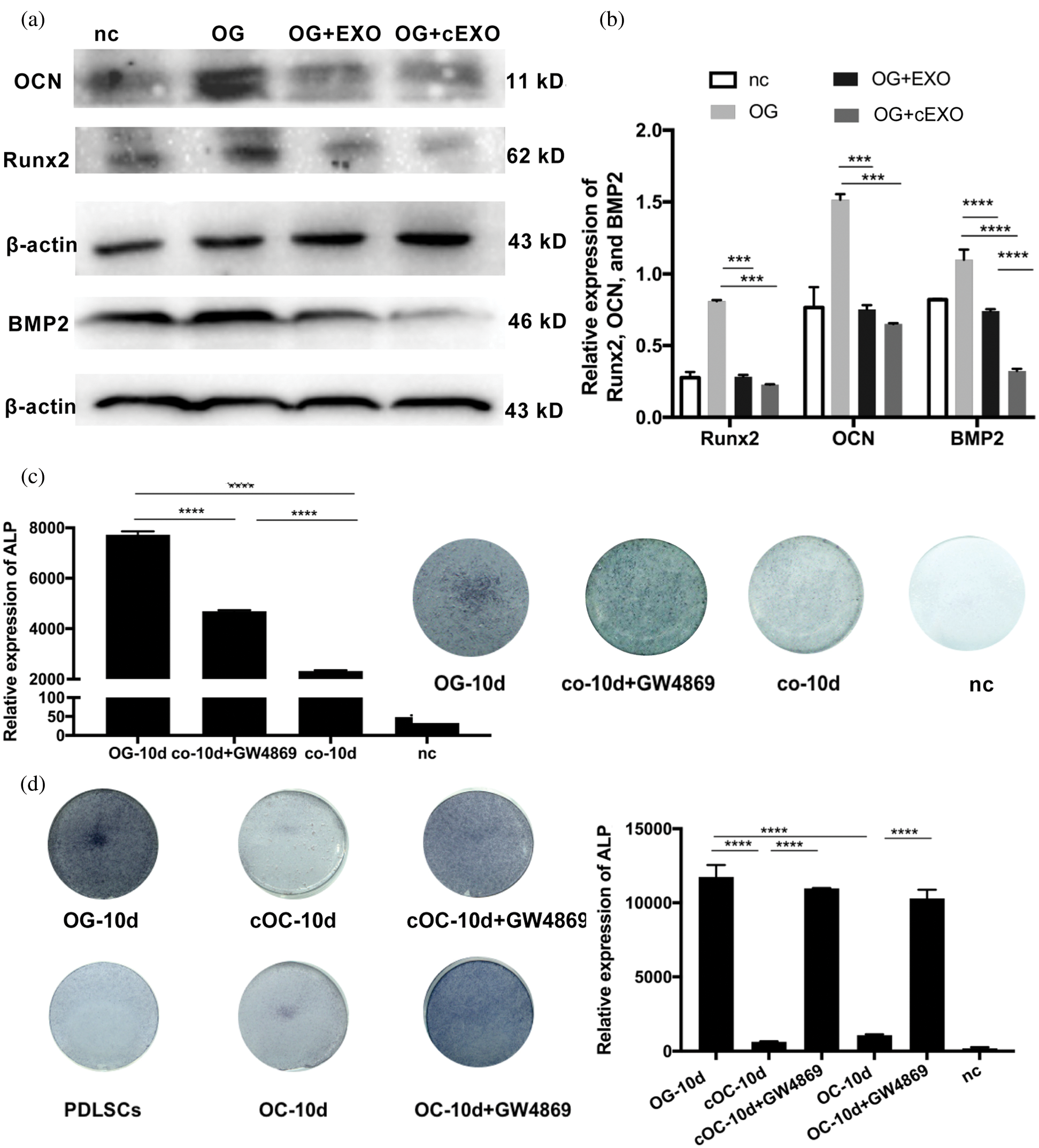
Figure 10: Role of osteoclast-derived exosomes in inhibition of osteogenic differentiation.
Role of changed miRNAs in exosomes derived from pre-osteoclasts and osteoclasts
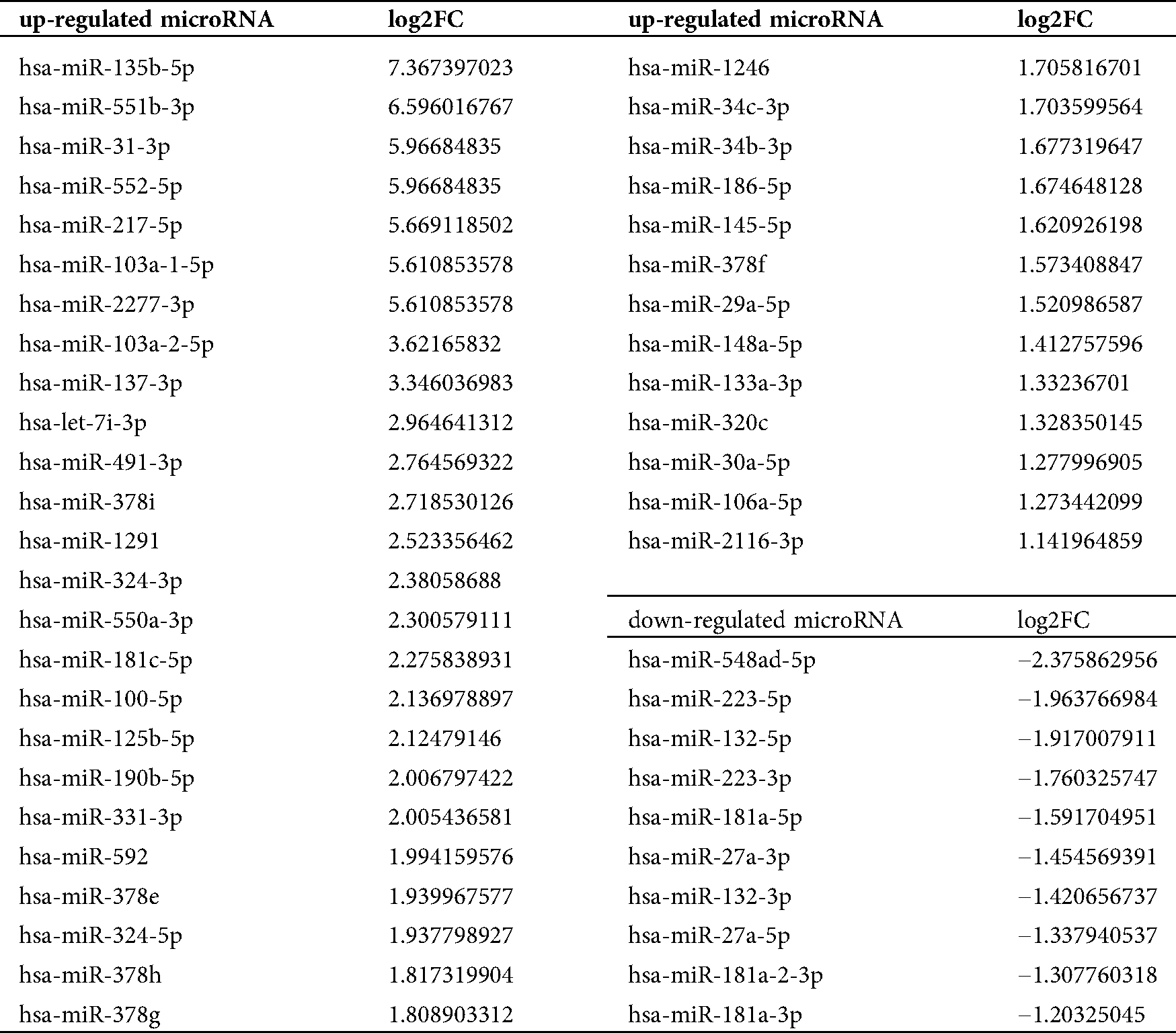
Among the changed miRNAs in osteoclasts showed by the sequencing result, we summarized miRNAs that were reported to regulate bone remodeling induced by mechanical forces (Tab. 2) and a subset of differentially expressed bone-related miRNAs for further study. Consistent with the miRNAs sequencing results in osteoclasts, the expression levels of miR-133a-3p, miR-203a-3p, miR-106a-5p, and miR-331- 3p were increased, whereas the levels of miR-223-5p and miR-181a-5p were decreased in exosomes (Fig. 11a). Previous studies have shown that miR-133a-3p, miR-203a-3p, miR-106a-5p, and miR-331-3p play a negative role in bone formation, while miR-223-5p and miR-181a-5p promote bone remodeling. Therefore, we hypothesized that the increased inhibition of osteogenesis during osteoclastogenesis after compression loading was partly due to the altered expression of miRNAs contained within exosomes derived from osteoclasts.
Table 2: Changed microRNAs which were reported to be related to bone remodeling with mechanical forces
Furthermore, the related pathway analysis illustrated that the miRNAs that exhibited altered expression levels were related to signaling pathways influencing bone formation, such as the MAPK, mTOR, and insulin signaling pathways (Fig. 11b).
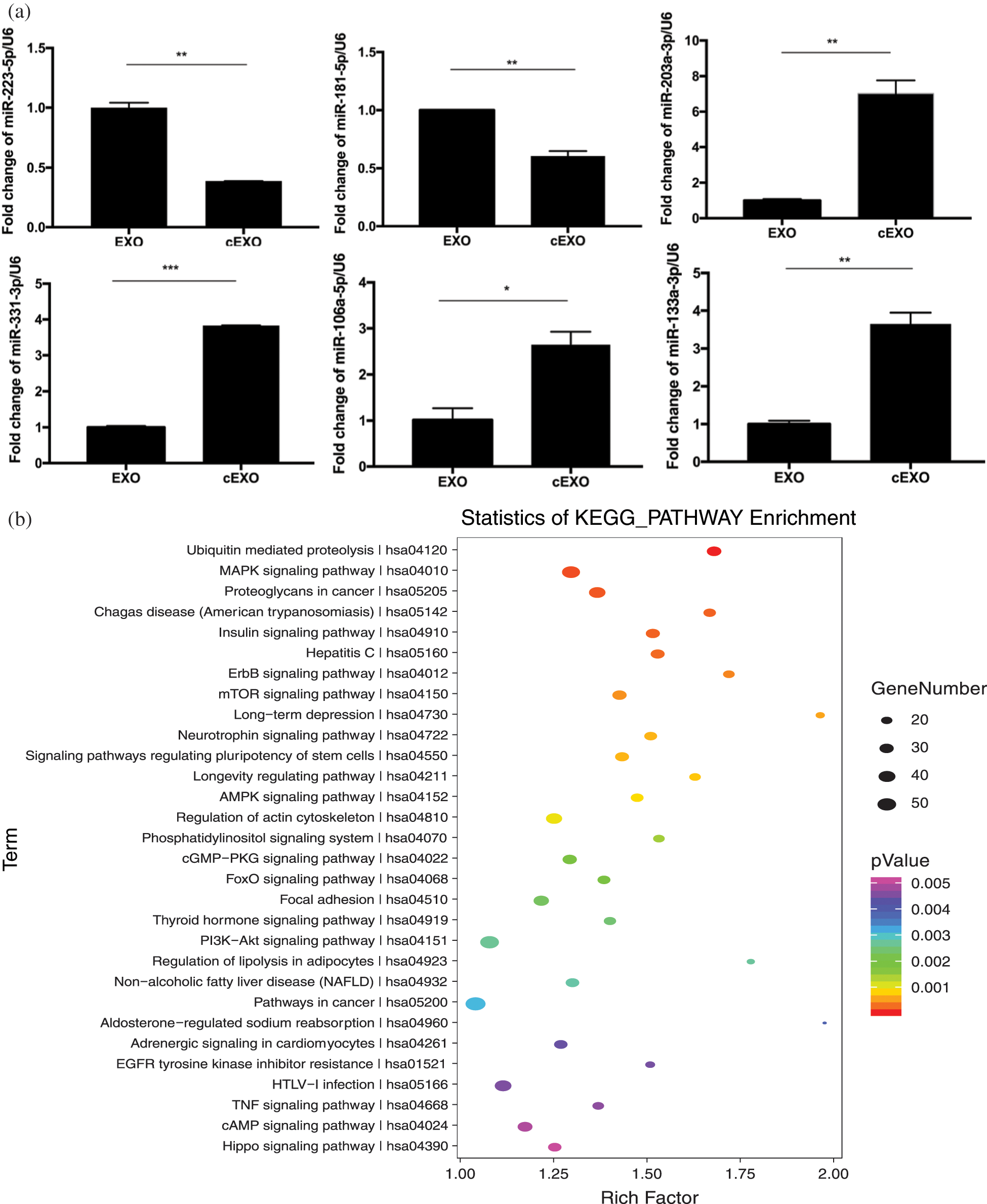
Figure 11: Some of the changed microRNAs in exosomes and bone-related pathways.
In this study, we investigated the exosome-associated influence of osteoclasts on osteoblasts subjected to compression force. Because we focused on pre-osteoclasts (rather than mature osteoclasts) when the mechanical force was applied, we determined the optimum time with the maximum number of pre-osteoclasts by RANK expression and the number of osteoclasts. To avoid anoxia, decreased nutrition, and cell damage caused by direct contact of cells and the application of force (Hayakawa et al., 2015), we placed collagen gels beneath the cell layers cultured on a glass sheet; we generated compression stress by the placement of beads on the other side of the glass sheet. After the application of force on day 4 of differentiation, the expression of RANK decreased, and the expression of TRAP increased, indicating that the application of compression promoted osteoclastogenesis. This was also confirmed by our sequencing results, as some of the differentially expressed miRNAs were reportedly related to osteoclastogenesis. miR-182 and miR-31, which were upregulated, have been reported as positive regulators of osteoclast differentiation, both in vitro and in vivo (Inoue et al., 2018; Miller et al., 2016; Mizoguchi et al., 2013). miR-124, which was downregulated, has been shown to inhibit osteoclastogenesis by targeting the transcription factor CCAAT/enhancer-binding protein-α (C/EBP-α) and Nfat1 (Ponomarev et al., 2011). miR-223, which is expressed in pre-osteoclasts and negatively regulates osteoclastogenesis by targeting nuclear factor I A, was also downregulated after compression stress (Sugatani and Hruska, 2007).
Co-culture of osteoclasts and osteoblasts has been performed in many studies. In addition to two-dimensional models (e.g., replacement of culture medium, as well as Transwell assays, removable dividers, and direct cell contact), in vitro and in vivo three-dimensional models have also been established (Penolazzi et al., 2016). Mutual interaction between osteoclasts and osteoblasts has been investigated using co-culture models. Osteoblast lineage cells can induce a strong and significant increase in the bone resorptive activity and recruitment of osteoclasts (Dinisha et al., 2019). Osteogenesis was also reportedly promoted by osteoclasts during osteoclast differentiation (Bernhardt et al., 2010; Skottke et al., 2019; Takaharu et al., 2018). Heinemann et al. (2011) reported that human bone marrow stromal cells (hBMSCs) were able to induce osteoclastogenesis and that osteoclasts stimulated BSP II gene expression of osteoblasts. However, Schulze et al. (2018) reported that hBMSCs promoted osteoclastogenesis in a direct co-culture system but reduced the matrix resorption of osteoclasts in an indirect co-culture system. Mesenchymal stem cells (MSCs) and mature osteoblasts have been shown to inhibit osteoclast differentiation (Masayuki et al., 2018; Takaharu et al., 2019). These conflicting results may be due to differences in experimental conditions. The time of observation, the selection of cell types, models of co-culture, and methods used to identify bone formation and resorption may have contributed to these different results. Furthermore, Sun demonstrated that osteoclast-derived exosomes containing miR-214 inhibited bone formation (Sun et al., 2016). However, these studies did not focus on the effects of mechanical stimulation, which is a key factor in bone remodeling and orthodontics. In the present study, an in vitro model showed inhibitory effects of pre-osteoclasts and osteoclasts on osteogenesis, especially after compression loading. We also focused on exosomes, which have been used to interfere with biocommunication and metabolism, as they exert effects in bone regeneration and targeted treatment of bone diseases (Liu et al., 2018). This study improves our understanding of the interactions mediated by exosomes following the application of mechanical force, which could lead to the use of exosomes in orthodontic tooth movement.
To elucidate the mechanism, we performed sequencing analysis of miRNAs. Among the 170 upregulated miRNAs, 17 have been reported to inhibit bone remodeling. Similarly, 7 of the 67 downregulated miRNAs have been shown to promote bone formation. To verify the sequencing results in both cells and exosomes, we selected the downregulated miR-223-5p and miR-181a-5p, as well as the upregulated miR-133a-3p, miR-203a-3p, miR-106a-5p, and miR-331-3p. miR-223 has been reported to play a role in bone metabolism and disease (Hoevring et al., 2008; Yong et al., 2015). miR-181a has been shown to promote osteoblast differentiation by targeting transforming growth factor-β signaling molecules (Bhushan et al., 2013). On the other side, miR-106b was demonstrated to negatively affect bone formation by targeting BMP-2 signaling and Smad5 (Collison, 2017; Fang et al., 2016; Liu et al., 2017). Similarly, miR-133a, miR-203a-3p, and miR-331 have been reported to inhibit bone formation through their target genes (Fang et al., 2016; Hua et al., 2016). Taken together, these observations suggest that these miRNAs may be essential regulators of the interaction between osteoclasts and osteoblasts induced by compression stress. The results of the present study also suggested a novel method to regulate bone metabolism in orthodontics after compression stress.
Further analysis revealed the pathways that may be involved in the regulation of bone remodeling.
The MAPK pathway has been linked to osteogenic differentiation of MC3T3-E1 cells (Liu et al., 2019). BMSC-derived exosomes have been shown to accelerate the proliferation of osteoblasts in osteonecrosis of the femoral head through the MAPK pathway (Liao et al., 2019). The mTOR pathway has also been reported to play an essential role in bone remodeling (Liang et al., 2019; Zhao et al., 2019). Qi and Zhang (2014) demonstrated that the mTOR pathway regulated bone remodeling when a mechanical force of fluid shear stress was applied.
Moreover, mechanical forces are crucial components of extracellular environments, affecting the biological processes of resident cells in bone structures through a complicated mechanotransduction process (Robling and Turner, 2009; Thompson et al., 2012). First, mechanical forces have different influences on bone cells through a number of molecules that act as mechanosensors (e.g., interleukin-11, hydrogen sulfide, FAK, and microRNAs) (Monnouchi et al., 2015; Wei et al., 2015; Yang et al., 2016). As a result, various signaling pathways are activated. Wnt/β-catenin, MAPK, PI3K/Akt/mTOR, and other pathways play important roles in mechanotransduction (Gifre et al., 2013; Qi and Zhang, 2014; Rubin et al., 2002; Ueland et al., 2015; Zhang et al., 2016). As the GO analysis in our experiment showed, the altered miRNAs are also closely associated with the MAPK and mTOR signaling pathways, which may participate in the mechanotransduction process. After that, the activating signaling pathways lead to expression changes in downstream molecules that include transcription factors (e.g., Runx2, So99, and PPARγ) (Arnsdorf et al., 2009). Thus, the gene expression profile changes in response to external mechanical stimuli. As reported, compression decreased the expression of Col1a1, Col3a1, and Col5a1 in PDLCs and alveolar bone cells (Chen et al., 2015), whereas it upregulated the osteoclast-related genes nfatc-1, trap, rank, cath-K, clc7, mmp-9, atp6i, dc-stamp, and oc-stamp in osteoclasts (Hayakawa et al., 2015). However, the exact mechanisms that alter the miRNA profile in osteoclasts after compression need further exploration.
Because exosomes are important carriers of miRNAs, this study examined the roles of exosomes in intercellular communication (Gao et al., 2016). GW4869 is a noncompetitive neutral sphingomyelinase inhibitor, which serves as an effective inhibitor of exosome release (Chen et al., 2019; Gao et al., 2016; Guo et al., 2015). The release of exosomes was significantly decreased after treatment of cells with 5–20 µM GW4869, although this effect was incomplete (Essandoh et al., 2015; Guo et al., 2015). This may explain why the level of ALP staining was less robust in the GW4869 group than in the OG group. Furthermore, changes in the culture environment of PDLSCs co-cultured with osteoclasts (e.g., pH and metabolic waste products) were likely to influence the state of hPDLSCs. Therefore, the inhibitory effects of indirect co-culture were more obvious than those of adding exosomes directly to hPDLSCs. The results also suggested that the exosomes may not be the sole mediator involved in the regulation of communication between osteoclasts and osteoblasts.
However, the miRNA expression profiles of cells did not fully represent the profiles of exosomes from pre-osteoclasts with compression stress. We only focused on selected miRNAs; a large number of miRNAs and their underlying molecular mechanisms require further study.
The results of this study provide insight into the further application of the exosome as a promising biomarker in intercellular communication, as well as in the role of a therapeutic agent in bone diseases (Quarona et al., 2015; Yin et al., 2017). miRNAs may also serve as biomarkers of bone pathologies, such as osteoporosis and osteosarcoma (Valenti et al., 2018). Exosomes with differential miRNA expression may be useful for the regulation of bone remodeling during orthodontic tooth movement.
After compression stress loading, osteoclastogenesis of pre-osteoclasts was promoted. The osteogenesis of PDL cells was inhibited by osteoclastogenesis; this inhibition was increased when compression stress was applied during osteoclast differentiation. The different degrees of inhibition were partially due to the altered miRNA expression profiles in osteoclasts and osteoclast-derived exosomes.
The downregulation of miR-223-5p and miR-181a-5p, along with the upregulation of miR-133a-3p, miR- 203a-3p, miR-106a-5p, and miR-331-3p, may be important in the regulation of interactions between osteoclasts and osteoblasts under compression stress. Our results support the potential applicability of exosomes and specific miRNAs to orthodontic bone remodeling.
Acknowledgement: This study was financially supported by grants from the National Natural Science Foundation of China (No. 81700938, 81670957). The funders had no role in study design, data collection and analysis, decision to publish, or the preparation of the article.
Author Contribution: study conception and design: Weiran Li, Yunfei Zheng; data collection: Yue Wang; analysis and interpretation of results: Yue Wang, Yunfei Zheng; draft manuscript preparation: Yue Wang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are available from the corresponding author upon reasonable request. The result of microRNA sequencing is available on www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA629042&o=acc_s%3Aa
Ethics Approval: Experimental protocols were approved by the Ethics Committee of Peking University (PKUSSIRB-2011007).
Funding Statement: This work was supported by grants from the National Natural Science Foundation of China (Nos. 81700938, 81670957). The funders had no role in study design, data collection and analysis, decision to publish, or the preparation of the article.
Conflicts of Interest: The authors have no conflicts of interest to disclose.
Arnsdorf EJ, Tummala P, Kwon RY, Jacobs CR. (2009). Mechanically induced osteogenic differentiation-the role of RhoA, ROCKII and cytoskeletal dynamics. Journal of Cell Science 122: 546–553. DOI 10.1242/jcs.036293. [Google Scholar] [CrossRef]
Berger JM, Singh P, Khrimian L, Morgan DA, Chowdhury S, Emilio A-S, Horvath TL, Domingos AI, Marsland AL, Yadav VK, Rahmouni K, Gao XB, Karsenty G. (2019). Mediation of the acute stress response by the skeleton. Cell Metabolism 30: 890–902.e898. DOI 10.1016/j.cmet.2019.08.012. [Google Scholar] [CrossRef]
Bernhardt A, Thieme S, Domaschke H, Springer A, Rösen‐Wolff A, Gelinsky M. (2010). Crosstalk of osteoblast and osteoclast precursors on mineralized collagen—towards an in vitro model for bone remodeling. Journal of Biomedical Materials Research Part A 95A: 848–856. DOI 10.1002/jbm.a.32856. [Google Scholar] [CrossRef]
Bhushan R, Grünhagen J, Becker J, Robinson PN, Ott C-E, Knaus P. (2013). miR-181a promotes osteoblastic differentiation through repression of TGF-β signaling molecules. International Journal of Biochemistry & Cell Biology 45: 696–705. DOI 10.1016/j.biocel.2012.12.008. [Google Scholar] [CrossRef]
Boyle WJ, Simonet WS, Lacey DL. (2003). Osteoclast differentiation and activation. Nature 423: 337–342. DOI 10.1038/nature01658. [Google Scholar] [CrossRef]
Collison J. (2017). Bone: miR-106b promotes osteoporosis in mice. Nature Reviews Rheumatology 13: 130. [Google Scholar]
Chen J, Zhou R, Liang Y, Fu X, Wang D, Wang C. (2019). Blockade of lncRNA-ASLNCS5088-enriched exosome generation in M2 macrophages by GW4869 dampens the effect of M2 macrophages on orchestrating fibroblast activation. FASEB Journal 33: 12200–12212. DOI 10.1096/fj.201901610. [Google Scholar] [CrossRef]
Chen Y, Mohammed A, Oubaidin M, Evans CA, Zhou X, Luan X, Diekwisch TGH, Atsawasuwan P. (2015). Cyclic stretch and compression forces alter microRNA-29 expression of human periodontal ligament cells. Gene 566: 13–17. DOI 10.1016/j.gene.2015.03.055. [Google Scholar] [CrossRef]
Debnath S, Yallowitz AR, Mccormick J, Lalani S, Zhang T, Xu R, Li N, Liu Y, Yang YS, Eiseman M, Shim JH, Hameed M, Healey JH, Bostrom MP, Landau DA, Greenblatt MB. (2018). Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature 562: 133–139. DOI 10.1038/s41586-018-0554-8. [Google Scholar] [CrossRef]
Dinisha CP, Jacob BO, Thomas LA, Sandra BC, Per KA, Delaisse JM, Søe K. (2019). Catabolic activity of osteoblast lineage cells contributes to osteoclastic bone resorption. Journal of Cell Science 132: jcs229351. DOI 10.1242/jcs.229351. [Google Scholar] [CrossRef]
Dou C, Cao Z, Yang B, Ding N, Hou T, Luo F, Kang F, Li J, Yang X, Jiang H. (2016). Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Scientific Reports 6: 337. DOI 10.1038/srep21499. [Google Scholar] [CrossRef]
Essandoh K, Yang L, Wang X, Huang W, Qin D, Hao J, Wang Y, Zingarelli B, Peng T, Fan GC. (2015). Blockade of exosome generation with GW4869 dampens the sepsis-induced inflammation and cardiac dysfunction. Biochimica et Biophysica Acta (BBA)—Molecular Basis of Disease 1852: 2362–2371. DOI 10.1016/j.bbadis.2015.08.010. [Google Scholar] [CrossRef]
Fang T, Wu Q, Zhou L, Mu S, Fu Q. (2016). miR-106b-5p and miR-17-5p suppress osteogenic differentiation by targeting Smad5 and inhibit bone formation. Experimental Cell Research 347: 74–82. DOI 10.1016/j.yexcr.2016.07.010. [Google Scholar] [CrossRef]
Feng X, Teitelbaum SL. (2013). Osteoclasts: new insights. Bone Research 29: 11–26. [Google Scholar]
Gao W, Liu H, Yuan J, Wu C, Huang D, Ma Y, Zhu J, Ma L, Guo J, Shi H, Zou Y, Ge J. (2016). Exosomes derived from mature dendritic cells increase endothelial inflammation and atherosclerosis via membrane TNF-α mediated NF-κB pathway. Journal of Cellular and Molecular Medicine 20: 2318–2327. DOI 10.1111/jcmm.12923. [Google Scholar] [CrossRef]
Gifre L, Ruiz-Gaspà S, Monegal A, Nomdedeu B, Filella X, Guanabens N, Peris P. (2013). Effect of glucocorticoid treatment on Wnt signalling antagonists (sclerostin and Dkk-1) and their relationship with bone turnover. Bone 57: 272–276. DOI 10.1016/j.bone.2013.08.016. [Google Scholar] [CrossRef]
Guo BB, Bellingham SA, Hill AF. (2015). The neutral sphingomyelinase pathway regulates packaging of the prion protein into exosomes. Journal of Biological Chemistry 290: 3455–3467. DOI 10.1074/jbc.M114.605253. [Google Scholar] [CrossRef]
Hayakawa T, Yoshimura Y, Kikuiri T, Matsuno M, Hasegawa T, Fukushima K, Shibata K, Deyama Y, Suzuki K, Iida J. (2015). Optimal compressive force accelerates osteoclastogenesis in RAW264.7 cells. Molecular Medicine Reports 12: 5879–5885. DOI 10.3892/mmr.2015.4141. [Google Scholar] [CrossRef]
Heinemann C, Heinemann S, Worch H, Hanke T. (2011). Development of an osteoblast/osteoclast co-culture derived by human bone marrow stromal cells and human monocytes for biomaterials testing. European Cells and Materials 21: 80–93. DOI 10.22203/eCM.v021a07. [Google Scholar] [CrossRef]
Hoevring PI, Jemtland R, Olstad OK, Reppe S, Gautvik VT, Gautvik KM. (2008). is differentially expressed in post menopausal osteoporotic women compared to healthy control females and regulates osteoclastogenesis. Bone 1: S39–S40. [Google Scholar]
Hou J, Yanze C, Xiuping M, Ce S, Chen L, Yuanping C, Hongchen S. (2014). Compressive force regulates ephrinB2 and EphB4 in osteoblasts and osteoclasts contributing to alveolar bone resorption during experimental tooth movement. Korean Journal of Orthodontics 44: 320–329. DOI 10.4041/kjod.2014.44.6.320. [Google Scholar] [CrossRef]
Hua W, Zhang M, Wang Y, Yu L, Zhao T, Qiu X, Wang L. (2016). Mechanical stretch regulates microRNA expression profile via NF-κB activation in C2C12 myoblasts. Molecular Medicine Reports 14: 5084–5092. DOI 10.3892/mmr.2016.5907. [Google Scholar] [CrossRef]
Hughes JM, Castellani CM, Popp KL, Guerriere KI, Bouxsein ML. (2020). The central role of osteocytes in the four adaptive pathways of bone's mechanostat. Exercise and Sport Sciences Reviews 48: 140–148. DOI 10.1249/JES.0000000000000225. [Google Scholar] [CrossRef]
Huynh N, Vonmoss L, Smith D, Rahman I, Felemban MF, Zuo J, Rody WJ, Mchugh KP, Holliday LS. (2016). Characterization of regulatory extracellular vesicles from osteoclasts. Journal of Dental Research 95: 673–679. DOI 10.1177/0022034516633189. [Google Scholar] [CrossRef]
Inoue K, Deng Z, Chen Y, Giannopoulou E, Xu R, Gong S, Greenblatt MB, Mangala LS, Lopez-Berestein G, Kirsch DG. (2018). Bone protection by inhibition of microRNA-182. Nature Communications 9: 245. DOI 10.1038/s41467-018-06446-0. [Google Scholar] [CrossRef]
Iwawaki Y, Mizusawa N, Iwata T, Higaki N, Goto T, Watanabe M, Tomotake Y, Ichikawa T, Yoshimoto K. (2015). MiR-494-3p induced by compressive force inhibits cell proliferation in MC3T3-E1 cells. Journal of Bioscience and Bioengineering 120: 456–462. DOI 10.1016/j.jbiosc.2015.02.006. [Google Scholar] [CrossRef]
Jiang F, Xia Z, Li S, Eckert G, Chen J. (2015). Mechanical environment change in root, periodontal ligament, and alveolar bone in response to two canine retraction treatment strategies. Orthodontics & Craniofacial Research 18: 29–38. DOI 10.1111/ocr.12076. [Google Scholar] [CrossRef]
Jiang N, Xiang L, He L, Yang G, Mao JJ. (2017). Exosomes mediate epithelium-mesenchyme crosstalk in organ development. ACS Nano 11: 7736–7746. DOI 10.1021/acsnano.7b01087. [Google Scholar] [CrossRef]
Li L, Sapkota M, Gao M, Choi H, Soh Y. (2017). Macrolactin F inhibits RANKL-mediated osteoclastogenesis by suppressing Akt, MAPK and NFATc1 pathways and promotes osteoblastogenesis through a BMP-2/smad/Akt/Runx2 signaling pathway. European Journal of Pharmacology 815: 202–209. DOI 10.1016/j.ejphar.2017.09.015. [Google Scholar] [CrossRef]
Li ML, Yi J, Yang Y, Zhang X, Zheng W, Li Y, Zhao Z. (2016). Compression and hypoxia play independent roles while having combinative effects in the osteoclastogenesis induced by periodontal ligament cells. Angle Orthodontist 86: 66–73. DOI 10.2319/121414.1. [Google Scholar] [CrossRef]
Liang B, Liang JM, Ding JN, Xu J, Xu JG, Chai YM. (2019). Dimethyloxaloylglycine-stimulated human bone marrow mesenchymal stem cell-derived exosomes enhance bone regeneration through angiogenesis by targeting the AKT/mTOR pathway. Stem Cell Research & Therapy 10: S163. DOI 10.1186/s13287-019-1410-y. [Google Scholar] [CrossRef]
Liao W, Ning Y, Xu H, Zou W, Hu J, Liu X, Yang Y, Li Z. (2019). BMSC-derived exosomes carrying microRNA-122-5p promote proliferation of osteoblasts in osteonecrosis of the femoral head. Clinical Science (London, England 1979) 133: 1955–1975. DOI 10.1042/CS20181064. [Google Scholar] [CrossRef]
Liu K, Jing Y, Zhang W, Fu X, Zhao H, Zhou X, Tao Y, Yang H, Zhang Y, Zen K. (2017). Silencing miR-106b accelerates osteogenesis of mesenchymal stem cells and rescues against glucocorticoid-induced osteoporosis by targeting BMP2. Bone 97: 130–138. DOI 10.1016/j.bone.2017.01.014. [Google Scholar] [CrossRef]
Liu M, Sun Y, Zhang Q. (2018). Emerging role of extracellular vesicles in bone remodeling. Journal of Dental Research 97: 859–868. DOI 10.1177/0022034518764411. [Google Scholar] [CrossRef]
Liu Q, Zhuang Y, Ouyang N, Yu H. (2019). Cytochalasin D promotes osteogenic differentiation of MC3T3-E1 cells via p38-MAPK signaling pathway. Current Molecular Medicine 20: 79–88. DOI 10.2174/1566524019666191007104816. [Google Scholar] [CrossRef]
Masayuki F, Junichi K, Sayumi F, Shigeto S, Hiroki M, Mai S, Maki U, Hiroki M, Yoriko I, Akito M, Kunihiko H, Takeshi I, Yukihiro I, Masafumi K, Takashi K, Shinsuke O, Ung-Il C, Alexander CL, Kazuya K, Hideo M, Hideki Y, Masaru I. (2018). Direct cell-cell contact between mature osteoblasts and osteoclasts dynamically controls their functions in vivo. Nature Communications 9: 489. DOI 10.1038/s41467-017-02541-w. [Google Scholar] [CrossRef]
Mikihito H, Tomoki N, Noriko Y, Kazuo O, Sakae T, Hiroshi T. (2019). Autoregulation of osteocyte Sema3A orchestrates estrogen action and counteracts bone aging. Cell Metabolism 29: 627–637.e5. DOI 10.1016/j.cmet.2018.12.021. [Google Scholar] [CrossRef]
Miller CH, Smith SM, Elguindy M, Zhang T, Xiang JZ, Hu X, Ivashkiv LB, Zhao B. (2016). RBP-J–regulated miR-182 promotes TNF-α-induced osteoclastogenesis. Journal of Immunology 196: 4977–4986. DOI 10.4049/jimmunol.1502044. [Google Scholar] [CrossRef]
Mizoguchi F, Murakami Y, Saito T, Miyasaka N, Kohsaka H. (2013). miR-31 controls osteoclast formation and bone resorption by targeting RhoA. Arthritis Research & Therapy 15: R102. DOI 10.1186/ar4282. [Google Scholar] [CrossRef]
Monnouchi S, Maeda H, Yuda A, Hamano S, Wada N, Tomokiyo A, Koori K, Sugii H, Serita S, Akamine A. (2015). Mechanical induction of interleukin-11 regulates osteoblastic/cementoblastic differentiation of human periodontal ligament stem/progenitor cells. Journal of Periodontal Research 50: 231–239. DOI 10.1111/jre.12200. [Google Scholar] [CrossRef]
Nakamura H, Wakita S, Yasufuku K, Makiyama T, Murayama T. (2015). Sphingomyelin regulates the activity of secretory phospholipase A2 in the plasma membrane. Journal of Cellular Biochemistry 116: 1898–1907. DOI 10.1002/jcb.25145. [Google Scholar] [CrossRef]
Penolazzi L, Lolli A, Sardelli L, Angelozzi M, Lambertini E, Trombelli L, Ciarpella F, Vecchiatini R, Piva R. (2016). Establishment of a 3D-dynamic osteoblasts-osteoclasts co-culture model to simulate the jawbone microenvironment in vitro. Life Sciences 152: 82–93. DOI 10.1016/j.lfs.2016.03.035. [Google Scholar] [CrossRef]
Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. (2011). MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α-PU.1 pathway. Nature Medicine 17: 64–70. DOI 10.1038/nm.2266. [Google Scholar] [CrossRef]
Qi L, Zhang Y. (2014). The microRNA 132 regulates fluid shear stress-induced differentiation in periodontal ligament cells through mTOR signaling pathway. Cellular Physiology and Biochemistry 33: 433–445. DOI 10.1159/000358624. [Google Scholar] [CrossRef]
Qin Y, Peng Y, Zhao W, Pan J, Ksiezak-Reding H, Cardozo C, Wu Y, Divieti Pajevic P, Bonewald LF, Bauman WA. (2017). Myostatin inhibits osteoblastic differentiation by suppressing osteocyte-derived exosomal microRNA-218: A novel mechanism in muscle-bone communication. Journal of Biological Chemistry 292: 11021–11033. DOI 10.1074/jbc.M116.770941. [Google Scholar] [CrossRef]
Qin Y, Wang L, Gao Z, Chen G, Zhang C. (2016). Bone marrow stromal/stem cell-derived extracellular vesicles regulate osteoblast activity and differentiation in vitro and promote bone regeneration in vivo. Scientific Reports 6: 373. DOI 10.1038/srep21961. [Google Scholar] [CrossRef]
Quarona V, Ferri V, Chillemi A, Bolzoni M, Mancini C, Zaccarello G, Roato I, Morandi F, Marimpietri D, Faccani G, Martella E, Pistoia V, Giuliani N, Horenstein AL, Malavasi F. (2015). Unraveling the contribution of ectoenzymes to myeloma life and survival in the bone marrow niche. Annals of the New York Academy of Sciences 1335: 10–22. DOI 10.1111/nyas.12485. [Google Scholar] [CrossRef]
Rendina RE, Clifford JR. (2020). Lipids in the bone marrow: An evolving perspective. Cell Metabolism 31: 219–231. DOI 10.1016/j.cmet.2019.09.015. [Google Scholar] [CrossRef]
Robling AG, Turner CH. (2009). Mechanical signaling for bone modeling and remodeling. Critical Reviews in Eukaryotic Gene Expression 19: 319–338. DOI 10.1615/CritRevEukarGeneExpr.v19.i4.50. [Google Scholar] [CrossRef]
Rody J, Wellington J, King GJ, Gu G. (2001). Osteoclast recruitment to sites of compression in orthodontic tooth movement. American Journal of Orthodontics and Dentofacial Orthopedics 120: 477–489. DOI 10.1067/mod.2001.118623. [Google Scholar] [CrossRef]
Rubin J, Murphy TC, Fan X, Goldschmidt M, Taylor WR. (2002). Activation of extracellular signal-regulated kinase is involved in mechanical strain inhibition of RANKL expression in bone stromal cells. Journal of Bone and Mineral Research 17: 1452–1460. DOI 10.1359/jbmr.2002.17.8.1452. [Google Scholar] [CrossRef]
Schulze S, Wehrum D, Dieter P, Hempel U. (2018). A supplement-free osteoclast-osteoblast co-culture for pre-clinical application. Journal of Cellular Physiology 233: 4391–4400. DOI 10.1002/jcp.26076. [Google Scholar] [CrossRef]
Skottke J, Gelinsky M, Bernhardt A. (2019). In vitro co-culture model of primary human osteoblasts and osteocytes in collagen gels. International Journal of Molecular Sciences 20: 1998. DOI 10.3390/ijms20081998. [Google Scholar] [CrossRef]
Sugatani T, Hruska K. (2007). MicroRNA-223 is a key factor in osteoclast differentiation. Journal of Cellular Biochemistry 101: 996–999. DOI 10.1002/jcb.21335. [Google Scholar] [CrossRef]
Sun W, Zhao C, Li Y, Wang L, Nie G, Peng J, Wang A, Zhang P, Tian W, Li Q. (2016). Osteoclast-derived microRNA-containing exosomes selectively inhibit osteoblast activity. Science Foundation in China 2: 16015. [Google Scholar]
Takaharu A, Keisuke S, Ryo K, Nanae O, Yuji T, Kengo N, Kazuyo A, Kotaro T. (2019). The effect of mesenchymal stem cells on osteoclast precursor cell differentiation. Journal of Oral Science 61: 30–35. DOI 10.2334/josnusd.17-0315. [Google Scholar] [CrossRef]
Takaharu A, Keisuke S, Ryo K, Nanae O, Yuji T, Kengo N, Kotaro T. (2018). Dynamic imaging of the effect of mesenchymal stem cells on osteoclast precursor cell chemotaxis for bone defects in the mouse skull. Journal of Dental Sciences 13: 354–359. DOI 10.1016/j.jds.2018.08.001. [Google Scholar] [CrossRef]
Thayanithy V, O’hare P, Wong P, Zhao X, Steer CJ, Subramanian S, Lou E. (2017). A transwell assay that excludes exosomes for assessment of tunneling nanotube-mediated intercellular communication. Cell Communication and Signaling 15: 791. DOI 10.1186/s12964-017-0201-2. [Google Scholar] [CrossRef]
Thompson WR, Rubin CT, Rubin J. (2012). Mechanical regulation of signaling pathways in bone. Gene 503: 179–193. DOI 10.1016/j.gene.2012.04.076. [Google Scholar] [CrossRef]
Ueland T, Olarescu NC, Jørgensen AP, Otterdal K, Aukrust P, Godang K, Lekva T, Bollerslev J. (2015). Increased serum and bone matrix levels of the secreted Wnt antagonist DKK-1 in patients with growth hormone deficiency in response to growth hormone treatment. Journal of Clinical Endocrinology & Metabolism 100: 736–743. DOI 10.1210/jc.2014-2912. [Google Scholar] [CrossRef]
Valenti MT, Carbonare LD, Mottes M. (2018). Role of microRNAs in progenitor cell commitment and osteogenic differentiation in health and disease. International Journal of Molecular Medicine 41: 2441–2449. [Google Scholar]
Wang Y, Jia L, Zheng Y, Li W. (2018). Bone remodeling induced by mechanical forces is regulated by miRNAs. Bioscience Reports 38: 863. DOI 10.1042/BSR20180448. [Google Scholar] [CrossRef]
Wei F, Liu D, Feng C, Zhang F, Yang S, Hu Y, Ding G, Wang S. (2015). microRNA-21 mediates stretch-induced osteogenic differentiation in human periodontal ligament stem cells. Stem Cells and Development 24: 312–319. DOI 10.1089/scd.2014.0191. [Google Scholar] [CrossRef]
Xing L, Schwarz EM, Boyce BF. (2005). Osteoclast precursors, RANKL/RANK, and immunology. Immunological Reviews 208: 19–29. DOI 10.1111/j.0105-2896.2005.00336.x. [Google Scholar] [CrossRef]
Xu Q, Cui Y, Luan J, Zhou X, Li H, Han J. (2018). Exosomes from C2C12 myoblasts enhance osteogenic differentiation of MC3T3-E1 pre-osteoblasts by delivering miR-27a-3p. Biochemical and Biophysical Research Communications 498: 32–37. DOI 10.1016/j.bbrc.2018.02.144. [Google Scholar] [CrossRef]
Yang R, Liu Y, Shi S. (2016). Hydrogen sulfide regulates homeostasis of mesenchymal stem cells and regulatory T cells. Journal of Dental Research 95: 1445–1451. DOI 10.1177/0022034516659041. [Google Scholar] [CrossRef]
Yin X, Wang JQ, Yan SY. (2017). Reduced miR-26a and miR-26b expression contributes to the pathogenesis of osteoarthritis via the promotion of p65 translocation. Molecular Medicine Reports 15: 551–558. DOI 10.3892/mmr.2016.6035. [Google Scholar] [CrossRef]
Yong X, Zhang L, Gao Y, Ge W, Tang P. (2015). The multiple roles of microRNA-223 in regulating bone metabolism. Molecules 20: 19433–19448. DOI 10.3390/molecules201019433. [Google Scholar] [CrossRef]
Yu B, Chang J, Liu Y, Li J, Kevork K, Al-Hezaimi K, Graves DT, Park NH, Wang CY. (2014). Wnt4 signaling prevents skeletal aging and inflammation by inhibiting nuclear factor-κB. Nature Medicine 20: 1009–1017. DOI 10.1038/nm.3586. [Google Scholar] [CrossRef]
Yu Y, Newman H, Shen L, Sharma D, Hu G, Mirando AJ, Zhang H, Knudsen E, Zhang G-F, Hilton MJ, Karner CM. (2019). Glutamine metabolism regulates proliferation and lineage allocation in skeletal stem cells. Cell Metabolism 29: 966–978.e4. DOI 10.1016/j.cmet.2019.01.016. [Google Scholar] [CrossRef]
Yuan F, Wu Q, Miao Z, Xu M, Xu R, Jiang D, Ye J, Chen F, Zhao M, Wang H, Li X. (2018). Osteoclast-derived extracellular vesicles: Novel regulators of osteoclastogenesis and osteoclast-osteoblasts communication in bone remodeling. Frontiers in Physiology 9: 8. DOI 10.3389/fphys.2018.00628. [Google Scholar] [CrossRef]
Yuan FL, Xu MH, Li X, He X, We I, Dong J. (2016). The roles of acidosis in osteoclast biology. Frontiers in Physiology 7: 144. DOI 10.3389/fphys.2016.00222. [Google Scholar] [CrossRef]
Zhang L, Liu W, Zhao J, Ma X, Shen L, Zhang Y, Jin F, Jin Y. (2016). Mechanical stress regulates osteogenic differentiation and RANKL/OPG ratio in periodontal ligament stem cells by the Wnt/β-catenin pathway. Biochimica et Biophysica Acta (BBA)–General Subjects 1860: 2211–2219. DOI 10.1016/j.bbagen.2016.05.003. [Google Scholar] [CrossRef]
Zhao J, Wu J, Xu B, Yuan Z, Leng Y, Min J, Lan X, Luo J. (2019). Kaempferol promotes bone formation in part via the mTOR signaling pathway. Molecular Medicine Reports 20: 5197–5207. [Google Scholar]
Zheng Y, Li X, Huang Y, Jia L, Li W. (2017). The circular RNA landscape of periodontal ligament stem cells during osteogenesis. Journal of Periodontology 88: 906–914. DOI 10.1902/jop.2017.170078. [Google Scholar] [CrossRef]
Zhu S, Ehnert S, Rouß M, V. H, Rh AW, T. C, Ak N. (2018). From the clinical problem to the basic research–co-culture models of osteoblasts and osteoclasts. International Journal of Molecular Sciences 19: 2284. DOI 10.3390/ijms19082284. [Google Scholar] [CrossRef]
Zuo B, Zhu J, Li J, Wang C, Zhao X, Cai G, Li Z, Peng J, Wang P, Shen C, Huang Y, Xu J, Zhang X, Chen X. (2015). microRNA-103a functions as a mechanosensitive microRNA to inhibit bone formation through targeting Runx2. Journal of Bone and Mineral Research 30: 330–345. DOI 10.1002/jbmr.2352. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |