DOI:10.32604/biocell.2021.013824

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.013824 |  www.techscience.com/journal/biocell |
| Article |
Folate-producing rhizobacteria of Hippophae rhamnoides L. from Indian trans-Himalaya low atmospheric zone
1Department of Biotechnology, Mangalmay Group of Institutions, Greater Noida, 201306, India
2Institute of Nuclear Medicine and Allied Sciences, Defence Research and Development Organisation, New Delhi, 110054, India
3Defence Institute of Bio-Energy Research (DRDO), Haldwani, 263139, India
4Department of Biotechnology, School of Bioengineering and Biosciences, Lovely Professional University, Phagwara, 144411, India
5International Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi, 110067, India
6Department of Life Sciences, Central University of Jharkhand, Brambe, 835205, India
*Address correspondence to: Joginder Singh, joginder.15005@lpu.co.in; Manoj Kumar, manoj@cuj.ac.in
Received: 22 August 2020; Accepted: 01 November 2020
Abstract: Hippophae rhamnoides L. is a plant of immense ethnopharmacological importance and is a known source for various valuable biochemicals and nutraceuticals. The production of folate, a vitamin involved in several vital functions, in this plant is rather poorly understood. Herein, we investigate the hypothesis that rhizobial bacteria serve the plant in this essential vitamin’s biosynthesis. Bacterial strains of Bacillus, Azorhizobium, Frankia, Paenibacillus, Brevibacillus and Pseudomonas, were isolated from the rhizosphere of the plant. HPLC and LCMS were used to trace the production of intra and extra-cellular folate by representative rhizospheric bacterial strains in vitro. From the seventeen functionally characterized bacterial strains of the plant’s rhizosphere, thirteen produced significant amounts of folate. Azorhizobium BR5401 produced the maximum amount of folic acid (424 µg/mL), and Bacillus GY779 was the only strain capable of producing both intracellular and extra-cellular folic acid. The Open Reading Frame coding for dihydroneopterin aldolase, an enzyme involved in folate biosynthesis, was found in one of the representative isolates. Our experimental findings help us to suggest that the folate synthesized by rhizobial bacteria is transported to the plant, highlighting a significant benefit of coexistence.
Keywords: Sea buckthorn; Rhizosphere; Folate; HPLC; Dihydroneopterin aldolase
Plant roots impact the rhizospheric soil and microorganisms, which affect the plant’s growth and survival. Numerous microbial populations residing in the rhizosphere get shelter and protection because of the convincing exchange made by them in return. This adjuvant exchange has been widely studied by scientists and researchers because they spotlight the chief molecules anchoring such a relationship. The plant Hippophae rhamnoides L., commonly known as “sea buckthorn” (SBT), is abundantly found in cold temperate regions of Europe and Asia. In the Indian subcontinent, it is found along the banks of the river Indus (or Sindh) and throughout the Himalayan region. The plant grows in the wild under adverse climate conditions and is also helpful in soil conservation because its well-developed root system fixes the soil horizontally. It offers highly desirable characteristics that find application in medicinal, food, and animal fodder sectors.
H. rhamnoides L. has high concentrations of bioactive compounds like vitamins, phenolics, and carotenoids that can be perused for human/animal nutrition (Xu et al., 2009).
The plant also finds special utility in health products as a source for bioactive agents relevant for cytoprotective, anti-stress, immune-modulatory, hepatoprotective, radio-protective, anti-atherogenic, anti-tumour, anti-microbial, and tissue regeneration applications (Suryakumar and Gupta, 2011). Berries of the plant have been found to contain tocopherol, a fat-soluble vitamin essential for reproduction and also a significant antioxidant that can neutralize free radicals in the body (Zadernowski et al., 2003). The berries are also reported to contain salicylic acid, used in medicines to treat scaly skin disease. Owing to these economic and ecological virtues, the plant commands world-wide attention for commercial exploitation (Mangla and Tandon, 2014). Therefore, significant focus is warranted on the direct/indirect biotic and abiotic factors supporting the plant growth and enhancing its nutritional values. A plant’s extensive root system takes up folate released by rhizobacteria and disperses them in the plant system (Kalia et al., 2011). A set of 106 rhizobacteria isolated from the rhizosphere of H. rhamnoides was reported to show plant growth-promoting properties like indole acetic acid, siderophore, hydrogen cyanide production, and phosphate solubilization (Kumar et al., 2015).
Folates or tetrahydrofolate act as essential cofactors for enzymes mediating one-carbon transfer reactions. Coded by folB gene, they catalyze the conversion of 6-hydroxymethyl-dihydro pterin in the folate biosynthesis pathway (Pribat et al., 2009; Ulrich et al., 2008). Structurally, folate comprises pterin, p-aminobenzoate (pABA), and glutamate moieties. Physiologically, folate is involved in the biosynthesis of nucleic acids (Loizeau et al., 2008), amino acids, and pantothenates. In plants, they are present in the seeds as 5-formyl-tetrahydrofolate and 5-methyl-tetrahydrofolate and are involved in lignin formation and photorespiration.
Folate synthesis pathway is similar in plants and microorganisms. Unlike them, humans (animals) cannot synthesize folate, and deficiency of this vitamin (Vit. B9) is known to lead to cancer, vascular disease, increased risk of neural tube defects in infants (Basset et al., 2005), anemia, and poor cognitive performance (Ohrvik and Witthoft, 2011). Bacillus subtilis produces a series of PGA-related (Pteroyl Glutamic Acid) compounds (Zygmunt et al., 1962). It was found that pab, trpG, and pabC genes were required for the synthesis of p-aminobenzoic acid (a compound of folate) (Slock et al., 1990). Bacillus subtilis was genetically engineered for enhanced production of folate using computer-aided flux and mutations. The analysis provided bounds on yield illuminate potential tradeoffs and provided flux distributions that can serve as targets for metabolic engineering. For brevity, the flux analysis results emphasized the potential effects of attenuating pyruvate kinase activity (elevates precursor PEP-phosphoenolpyruvate) and tktA expression (Zhu et al., 2005). Pseudomonas denitrificans and Bacillus megaterium are employed for the industrial production of folate (Hugenholtz et al., 2002). Folate transportation genes have been reported in Clostridium strains, ciphered by the gene FolT (Rossi et al., 2011; Scaria et al., 2011; de Crecy-Lagard et al., 2007).
Similarly, plants have also been reported as being capable of taking up C1 derivatives of tetrahydrofolate from the medium (Bedhomme et al., 2005). Folate limitations affect the plant’s metabolic preferences, as shown in Arabidopsis (Haferkamp and Esser, 2012). The cells responded to folate limitations by shuttling the available folate derivatives to the synthesized nucleotides at the expense of methylene reactions (Loizeau et al., 2008). It was characterized by the rhizosphere soil of SBT because a particular extract (coded as SBL-1, prepared from SBT leaves, where ‘SBL’ refers to seabuckthorn leave extract) was found to display extraordinary radio-protective properties (Haferkamp and Esser, 2012). Folic acid (FA) and its radio-protective properties have been reported earlier (Pote et al., 2006). It was reported that a single dose of SBL-1 rendered survival of more than 90% mice population (which were irradiated with a lethal dose of ionizing radiation of 10 Gy) beyond 30 days (Bala et al., 2009). This was when all the control animals (not treated with SBL-1) died within 12–14 days after irradiation with the same lethal dose of 10 Gy. SBL-1 was inferred to counter radiation-induced inflammation and oxidative stress in the liver, kidney, and gastrointestinal (GI) tract (Tiwari and Bala, 2011).
The present study suggests that one of the bioactive constituents, i.e., folate, is not a secondary plant metabolite but is present in SBT because of rhizobacterial association in the rhizosphere. Also, since the isolates belong to a different genus, they must be distant in genetic relatedness and are therefore expected to play differing levels of the metabolic role played, as the production of essential biomolecules like folate, well explained by the results of Tukey’s test.
The cold desert of Ladakh, situated in the Himalayas, was selected for the study. Ten plants from three different sites, viz. Nubra (34.6°N 77.7°E; 3048 m above the sea level), Indus (23°59’N 67°25’E; 4255 m above the sea level), and Zanskar (33°29’N 76°50’E; 7756 m above the sea level) were selected for sample collection. A soil corer was used to collect the rhizospheric soil within the diameter of 5 cm, till the depth of ~10 cm, around the periphery of the root zone (Gurusinghe et al., 2019). The rhizospheric soil was loosened from the root surface, root samples were carefully trimmed with a sterile knife, and both were collected in zip-lock bags. The root and rhizospheric soil samples were preserved at −20°C for further studies.
Isolation and identification of rhizobacteria
The soil was serially diluted in autoclaved distilled water and suitable amounts spread on a selective medium (Mwajita et al., 2013). Root nodules were surface sterilized using 30% H2O2 and were crushed in a sterile Petri plate with a sterile rod. Ooze from each nodule was inoculated on media plates, and the plates were incubated at 30°C for 24–48 h. (Baeucehmin et al., 2012). The used selected media used were nutrient agar (HiMedia) for bacteria (Gowsalya et al., 2014), potato dextrose agar (HiMedia) for fungal species (Bhadrecha et al., 2020; Kumar et al., 2015), actinomycetes agar medium (HiMedia) for Actinomycetes strains (Kumar and Jadeja, 2016), yeast extract mannitol agar-YEMA medium (HiMedia) for Rhizobium (Agah et al., 2016), and minimal salt agar media for Frankia were used, including defined propionate medium-DPM (DPM: 0.5 g/L sodium propionate, 0.5 g/L sodium succinate, 1.0 g/L K2HPO4, 0.1 g/L MgSO4, 0.001 g/L CaCl2, 0.0055 g/L FeSO4, 0.0074 g/L Na2.EDTA, 0.005 g/L H3BO3, 0.0032 g/L MnCl2, 0.0004 g/L ZnSO4, 0.00014 g/L CuSO4, 0.00045 g/L NaMoO4, 0.00045 g/L CoCl2, 18 g/L agar in 1000 mL distilled water) (Gtari et al., 2004). Genomic DNA from the procured cultures was isolated (Sambrook and Russel, 2001), and polymerase chain reaction (PCR) was run using the primers: 16S (F) 5’-AGAGTTTGATCCTGGCTCAG-3’ 16S (R) 5’-ACGGCTACCTTGTTACGACTT-3’; ITS-1 (F) 5’-TCCGTAGGTGAACCTGCGG-3’ ITS-4 (R)5’-TCCTCCGCTTATTGATATGC-3’.
Evolutionary analysis by Maximum Likelihood method
The evolutionary history was inferred by using the Maximum Likelihood method and Kimura 2-parameter model (Kimura, 1980). The tree with the highest log likelihood (–755.03) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with superior log likelihood value. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 30.06% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 17 nucleotide sequences. All positions containing gaps and missing data were eliminated (complete deletion option). There was a total of 163 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018). The comparative genome analysis of isolated rhizobacterial strains is presented in Fig. S1.
Gene prediction and annotation
In order to incorporate information from isolates, we have annotated the already sequenced genome and tried to figure out the regions of DNA that are biologically functional in Rhizobacteria using software ChemGenome (Soni et al., 2013) and GeneMark (Che et al., 2014). Structural and functional annotation helped us to predict folate coding genes in Rhizobacteria.
In-vitro detection of folate positive strains
All seventeen isolates were inoculated on plates containing Folic Acid Assay Medium (FAAM) (HiMedia) and incubated at 25°C for 48 h. Bacteria that grew on FAAM plates were further cultured in broth (same medium) for analysis of intracellular and extra-cellular folate (Divya and Nampoothiri, 2015).
Extraction of extra-cellular folate
1 mL broth culture was sonicated at 50,000 Hz for 2 min, with a pulse of 2 s. The sonicated sample was heated at 100°C for 5 min. and centrifuged at 10000×g for 10 min. The filtered (0.45-μm filter) supernatant was analyzed for folate by HPLC (Divya and Nampoothiri, 2015).
Extraction of intracellular folate
5 mL broth was centrifuged at 12,000 rpm for 10 min, at 4°C. The supernatant was discarded, and the cells were washed with 2 mL 50 mM H3PO4 (pH 2.3), containing 1% ascorbic acid. The cells were suspended in 0.5 mL of the same buffer, and 0.1 g of silica beads were added to the cell suspension. The cells were disrupted in an FP120 Fastprep cell disrupter. This cell extract was heated at 100°C for 3 min (to release folate from the binding proteins) and to precipitate the proteins. The cell extract was centrifuged twice at 12000×g, for 3 min, at 4°C. Soon after centrifugation, the supernatant was filtered (0.45-μm filter) and used as a sample for chromatography (Divya and Nampoothiri, 2015).
Folate estimation by high-performance liquid chromatography (HPLC) and liquid chromatography-mass spectroscopy (LCMS)
Intra and extra-cellular folate was isolated from the fresh cultures of the strains grown in Folate Assay Medium (HiMedia). The isolated folate was further analyzed by HPLC (Gliszczynska-Swiglo and Rybicka, 2015) using pure folate (HiMedia) as control. Folate analysis was performed with an Agilent (USA) 1260 LC-MS fitted with 150 × 2.1 mm, 3.5 µm particle, amide X bridge column. For this column the mobile phase A: 50/50 Acetonitrile/Water with 10 mM Ammonium Acetate + 10 mM Ammonium Hydroxide, pH 9 and Mobile Phase B: 90/10 Acetonitrile/Water with 10 mM Ammonium Acetate + 10 mM Ammonium Hydroxide, pH 9 at a flow rate of 150 µL/min in UV detector at the wavelength 265 nm.
All the experiments were performed thrice, and the mean values were further analyzed for comparing the divergence. Analysis of variation in the amount of extracellular folate production by the isolated rhizobacteria was done using the software “ASSISTAT” version 7.7 (Silva and Azevedo, 2016). The averages followed by the same letter do not differ statistically between themselves.
Isolation and identification of rhizobacteria
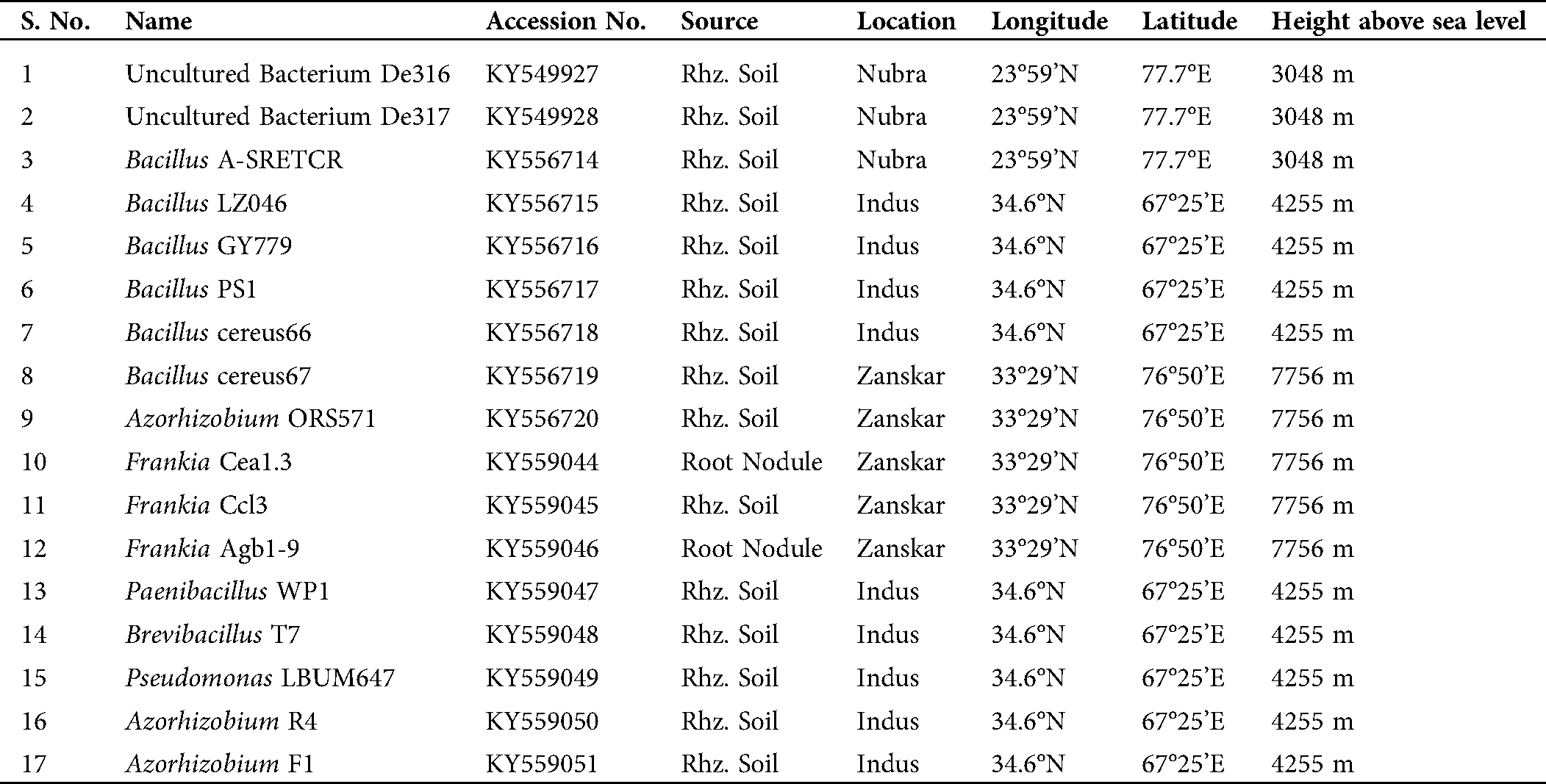
Seventeen bacterial strains were isolated from the rhizospheric soil and root nodules of the plant H. rhamnoides L. The 17 strains of bacteria isolated from the rhizospheric soil belong to genus Frankia, Bacillus, Brevibacillus, Paenibacillus, Azorhizobium, and Pseudomonas, as enlisted in Tab. 1.
Table 1: Bacteria isolated from the rhizosphere of the plant
Gene prediction and annotation
Folate coding ORF (open reading frame) from one of the strains Uncultured Bacterium A1 was found to cipher for the enzyme “dihydroneopterin aldolase” (EC number 4.1.2.25) coded by the gene FolB, as also registered in UniProt. The reported data registered in UniProt gave details about the genes FolB and FolT, involved in folate synthesis and folate transport, respectively, in the Uncultured Bacterium De316 strain. No results were found for the inquiry of the same/similar proteins in plant H. rhamnoides L., in UniProt. Uncultured Bacterium De 316 contains the ORF, which codes for the enzyme dihydroneopterin aldolase and catalyzes the conversion of 7,8-dihydroneopterin to 6-hydroxymethyl-7,8-dihydropterin coded by the gene FolB. Following were the results of ORS present in the strain Uncultured Bacterium De316, as obtained from the tool ChemGenome, and Tab. 2 presents the protein sequence which suggested us to investigate the synthesis of folate by these isolated rhizobacteria.
> +strand gene; start: 30, end: 140 GTGGCGGACGGTGTGAGTAACGCGTGGGTAACCTGCCTTATACAGGGGGATAACAGCCGGAAACGGTTGCTAATACCGCATAAGCATACAGTGCCGCATGGTACCGTGTGA
> +strand gene; start: 195, end: 341
ATGGCCAGCACCAAGGCGACGATCAGTAGCCGGCTTGAGAGAGTGACCGGCCACATTCTGAGACACGGCCTATACTCCTACGGGAGGCAGCAGTGGGGAATATTGCCAATGGGGGAACCCCTGAAAAGGTGCAGCGACGCCCCGTGA
> +strand gene; start: 287, end: 388
GTGGGGAATATTGCCAATGGGGGAACCCCTGAAAAGGTGCAGCGACGCCCCGTGAGTGAAGAAGTATTTTGGGATTTAAAGCTCTATCAGCAGGGACGATAA
Results from both the tools, ChemGenome and GeneMark, the similar nucleotide sequence “GTGGGGAATATTGCCAATGGGGGAACCCCTGAAAAGGTGCAGCGACGCCCCGTGAGTGAAGAAGTATTTTGGGATTTAAAGCTCTATCAGCAGGGACGATAA” codes for gene, whose protein sequence was found to be VGNIANGGTPEKVQRRPVSEEVFWDLKLYQQGR, as obtained by GeneMark results.
In-vitro detection of folate positive strains
Among the seventeen rhizobacteria, thirteen were able to grow on the FAAM (Folic Acid Assay Medium) media plates, confirming the production of folate by these thirteen bacteria. Fig. 1 shows the growth of a positive folate strain on the FAAM plate. Contrary to the ORF database search results, all thirteen strains were able to grow in FAAM plates. All these strains were cultivated as broth cultures for further analysis by HPLC.
Figure 1: Growth of a folic-acid positive isolate on Folic Acid Assay Medium plate.
Extraction of extra-cellular and intracellular folate and folate estimation by HPLC and LCMS
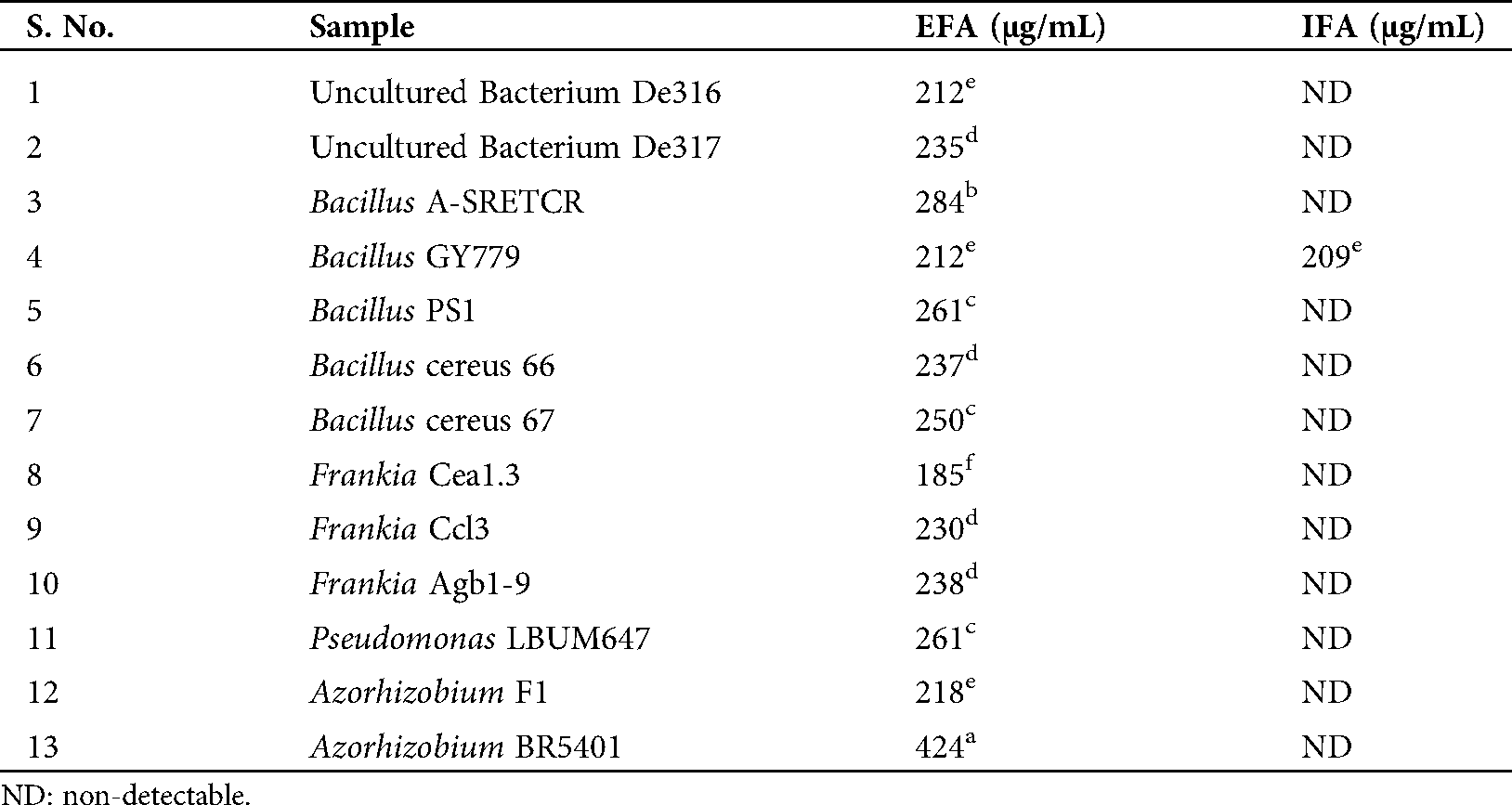
Folate concentrations from each folate-positive isolate are presented in Tab. 2. An amazing result of extra-cellular folate was obtained from the strain Azorhizobium BR5401 which is almost double the amount of that produced by any other strain. Interestingly, out of these 13 strains, only one strain, Bacillus GY779, was found capable of producing both intra and extra-cellular folate at concentrations 209 µg/mL and 212 µg/mL, respectively. Fig. 2 shows the LC-MS signature of the folate peak (obtained with HPLC), confirming our assay protocols, and the chromatographs representing folate production by bacterial strains are presented in Fig. S2.
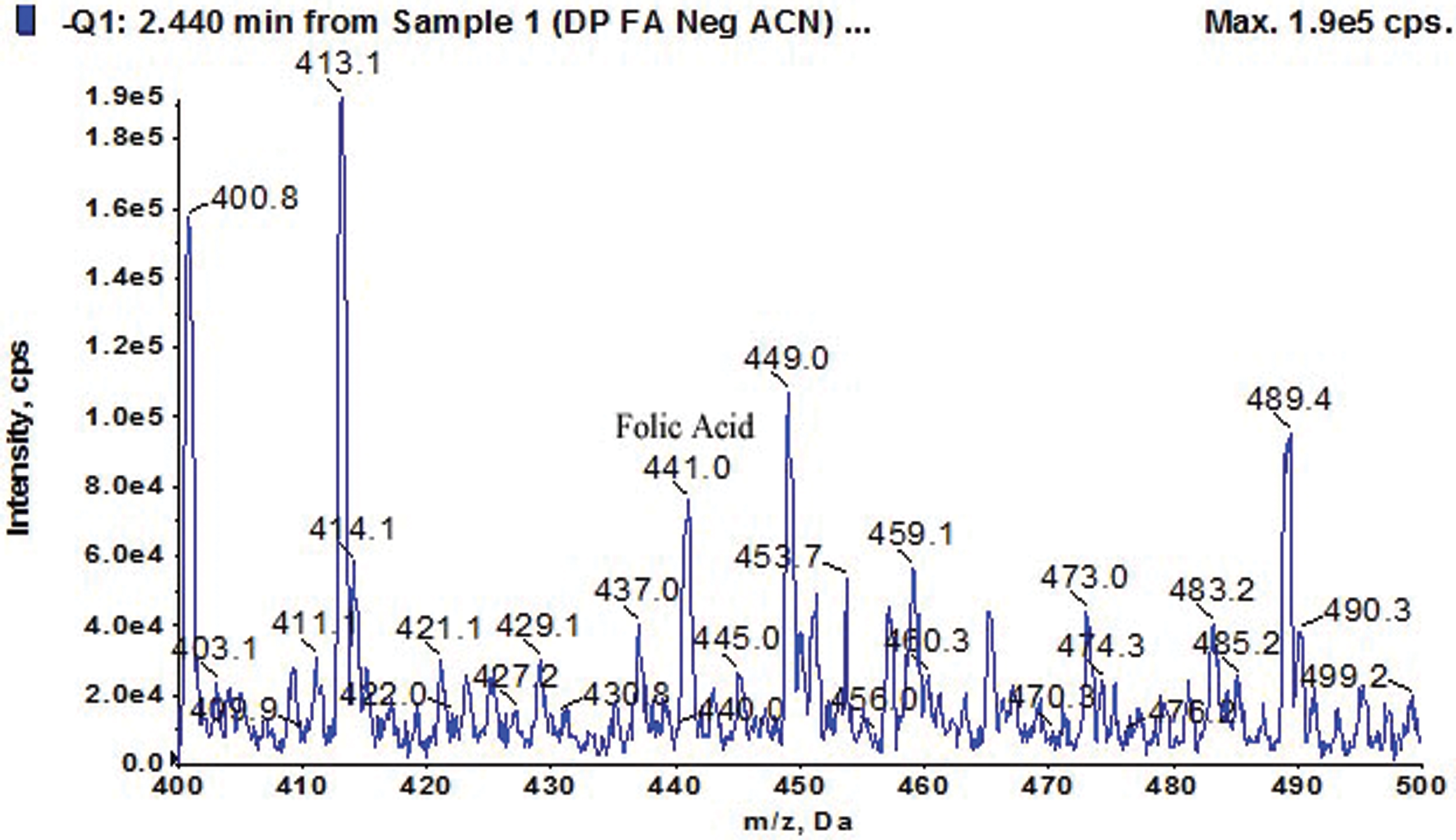
Figure 2: Estimation of Folic acid using HPLC-UV Technique (Folate mass = 441)
Table 2: Concentrations of extracted extra-cellular (EFA) and intracellular folate (IFA) produced by rhizobacteria isolates, statistical analysis done by Tukey’s Test, using ASSISTAT software
This study was undertaken to derive insights into the production of folate in SBT. To investigate our hypothesis that rhizospheric bacteria could produce the vital molecules “folate”, we isolated seventeen different rhizospheric bacterial strains from the soil and plant roots. The isolates belonged to the genus Frankia, Bacillus, Paenibacillus, Pseudomonas, Brevibacillus, and Azorhizobium. We found that thirteen isolates were able to synthesize folate. (The other 4 isolates could also serve the plant with some essential molecules and/or factors, which was not a focus of the current study). We found that these thirteen strains are able to produce extracellular folate (Hugenholtz et al., 2002) and surprisingly, one of the isolates, Bacillus GY779, was able to produce extra- as well as intracellular folate also (Pan, 2007), proposing that it must also contain a system for the transport of this intracellularly produced folate to be transported outside the cell. This data also suggests that the strains must be related through many similar metabolic pathways that produce several utile compounds. Fortunately, the culturable microbiome isolated in the current study consists of versatile genera, each being reported to metabolize a wide range of compounds.
The ORF obtained from the strain Uncultured Bacterium De316 codes for the enzyme ‘dihydroneopterin aldolase’, which catalyzes the conversion of 7,8-dihydroneopterin to 6-hydroxymethyl-7,8-dihydropterin (Blaszczyk et al., 2014) coded by the gene FolB, as also registered in UniProt (de Crecy-Lagard, 2014) as per the registered data in UniProt regarding the genes, FolB and FolT, responsible for synthesis and transport of folate. But no such data regarding the presence of these genes in plant H. rhamnoides L. has been registered in UniProt, which directed us towards the hypothesis of folate production and transport by rhizospheric bacteria associated with the plant.
Organisms capable of producing folic acid follow a standard pathway which employs guanosine triphosphate (GTP) produced in purine metabolism. Various enzymes involved in the process are coded via different ‘Fol’ genes. GTP cyclohydrolase coded by the gene FolE converts this GTP into dihydroneopterin triphosphate, which is then converted to dihydroneopterin by the phosphatase. Dihydroneopterin is converted to 6-hydromethyl dihydropterin by the action of enzyme ‘dihydroneopterin aldolase’, coded by gene FolB, discovered in the strain Uncultured Bacterium De316. 6-hydromethyl dihydro-pterin is further converted into 6-hydromethyl dihydro-pterin pyro phosphate, which is later catalyzed into dihydropteroate. Dihydropteroate is finally catalyzed into H-4-folate.
Isolated bacterial strains can be of great value to harvest the compound on an industrial stage (LeBlanc et al., 2007). Growth conditions can further be optimized to cultivate the strain for even higher values of folate. The strain can also be used to extract the creditworthy gene (Park et al., 2014). Another interesting outcome of this work is that only one of the folate-producing bacteria showed a confirmed ORF matching known folate synthesizing genes. Since the production of folate is confirmed by the remaining dozen bacteria, surely there must exist other metabolic routes by which these strains synthesize this important molecule.
In the present study, thirteen rhizobacteria associated with the plant H. rhamnoides L. belonging to different genus capable of producing different concentrations of extracellular folate were isolated. This essential biomolecule is involved in various vital metabolic functions. We hypothesize that folate produced by these rhizobacteria might be taken up by the roots and transported to other parts of the plant. Interestingly one of the strains, Bacillus GY779, was capable of producing both extracellular and intracellular folate, highlights the probability of the presence of a transport system for the transportation of folate towards the cell membrane and receptor molecules. The differing amount of folate produced also spotlights the fact that though these rhizobacteria strains have been isolated from the same habitat, the metabolic role played by them varies, irrespective of their genus. In the future, these strains can be used for the industrial production of folate, which is of great medicinal value for human health.
Acknowledgement: The authors are thankful to Director(s) Institute of Nuclear Medicine & Allied Sciences (INMAS) and Defense Institute of High-Altitude Research (DIHAR) DRDO for extending the research facilities and necessary support. The authors (Manoj Kumar, Pooja Bhadrecha and Joginder Singh) are also thankful to Lovely Professional University for the facility and other administrative support; and to the Soil Department, Punjab Agriculture University, Ludhiana for ICP-MS analysis of soil sample.
Availability of Data and Materials: Data supporting this article are details in this manuscript.
Funding Statement: The authors are grateful to the Defense Research Development Organization (Project No. TC/2519/INM/-04/2012/CARS of INM 311/1.2) for the financial support and opportunity to carry out these studies.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Agah MV, Orji JO, Nnachi AU, Chukwu OS, Udu-Ibiam OE, Nwachi AC, Olaosebikan OO. (2016). Isolation and identification of Rhizobium species from root nodules of Arachis hypogaea L. and Telfairia occidentalis in South-East. Nigeria International Journal of Science and Research 5: 227–230. [Google Scholar]
Baeucehmin NJ, Furnholm T, Lavenus J, Svistoonoff S, Doumas P, Bogusz D, Laplaze L, Tisa LS. (2012). Casuarina Root exudates alter the physiology, surface properties, and plant infectivity of Frankia sp Strain CcI3. Applied and Environmental Microbiology 78: 575–580. DOI 10.1128/AEM.06183-11. [Google Scholar] [CrossRef]
Bala M, Prasad J, Singh S, Tiwari S, Sawhney RC. (2009). Whole-body radio protective effects of SBL-1: A preparation from leaves of Hippophae rhamnoides. Journal of Herbs, Spices & Medicinal Plants 15: 203–215. DOI 10.1080/10496470903139496. [Google Scholar] [CrossRef]
Basset GJ, Quinlivan EP, Gregory JF, Hanson AD. (2005). Folate synthesis and metabolism in plants and prospects for biofortification. Crop Science 45: 449–453. DOI 10.2135/cropsci2005.0449. [Google Scholar] [CrossRef]
Bhadrecha P, Bala M, Khasa YP, Arshi A, Singh J, Kumar M. (2020). Hippophae rhamnoides L. rhizobacteria exhibit diversified cellulase and pectinase activities. Physiology and Molecular Biology of Plants 26: 1075–1085. DOI 10.1007/s12298-020-00778-2. [Google Scholar] [CrossRef]
Bedhomme M, Hoffmann M, McCarthy EA, Gambonnet B, Moran RG, Rebeille F, Ravanel S. (2005). Folate metabolism in plants: An Arabidopsis homolog of the mammalian mitochondrial folate transporter mediates folate import into chloroplasts. Journal of Biological Chemistry 280: 34823–34831. DOI 10.1074/jbc.M506045200. [Google Scholar] [CrossRef]
Blaszczyk J, Lu Z, Li Y, Yan H, Ji X. (2014). Crystallographic and molecular dynamics simulation analysis of Escherichia coli dihydroneopterin aldolase. Cell & Bioscience 4: 52. DOI 10.1186/2045-3701-4-52. [Google Scholar] [CrossRef]
Che D, Hasan MS, Chen B. (2014). Identifying pathogenicity islands in bacterial pathogenomics using computational approaches. Pathogens 3: 36–56. DOI 10.3390/pathogens3010036. [Google Scholar] [CrossRef]
de Crecy-Lagard V, El Yacoubi B, de la Garza RD, Noiriel A, Hanson AD. (2007). Comparative genomics of bacterial and plant folate synthesis and salvage: Predictions and validations. BMC Genomics 8: 245. DOI 10.1186/1471-2164-8-245. [Google Scholar] [CrossRef]
de Crecy-Lagard V. (2014). Variations in metabolic pathways create challenges for automated metabolic reconstructions: Examples from the tetrahydrofolate synthesis pathway. Computational and Structural Biotechnology Journal 10: 41–50. DOI 10.1016/j.csbj.2014.05.008. [Google Scholar] [CrossRef]
Divya JB, Nampoothiri KM. (2015). Folate fortification of skim milk by a probiotic Lactococcus lactis CM28 and evaluation of its stability in fermented milk on cold storage. Journal of Food Science and Technology 52: 3513–3519. DOI 10.1007/s13197-015-1867-3. [Google Scholar] [CrossRef]
Gliszczynska-Swiglo A, Rybicka I. (2015). Simultaneous detection of caffeine and water-soluble vitamins in energy drinks by HPLC with photodiode array and fluorescence detection. Food Analytical Methods 8: 139–146. DOI 10.1007/s12161-014-9880-0. [Google Scholar] [CrossRef]
Gowsalya A, Ponnusami V, Sugumaran KR. (2014). Isolation of bacteria from soil sample for exo-polysaccharide production. International Journal of ChemTech Research 6: 2925–2928. [Google Scholar]
Gtari M, Brusetti L, Skander G, Mora D, Boudabous A, Daffonchio D. (2004). Isolation of Elaeagnus-compatible Frankia from soils collected in Tunisia. FEMS Microbiology Letters 234: 349–355. DOI 10.1111/j.1574-6968.2004.tb09554.x. [Google Scholar] [CrossRef]
Gurusinghe S, Brooks TL, Barrow RA, Zhu X, Thotagamuwa A, Dennis PG, Gupta VVSR, Vanniasinkam T, Weston LA. (2019). Technologies for the selection, culture and metabolic profiling of unique rhizosphere microorganisms for natural product discovery. Molecules 24: 1955. DOI 10.3390/molecules24101955. [Google Scholar] [CrossRef]
Haferkamp I, Esser SS. (2012). Evolution of plant MCF proteins-the plant mitochondrial carrier family: Functional and evolutionary aspects. Frontiers in Plant Sciences 2: 3. [Google Scholar]
Hugenholtz J, Hunik J, Santos H, Smid E. (2002). Nutraceutical production by Propionibacteria. Le Lait 82: 103–112. DOI 10.1051/lait:2001009. [Google Scholar] [CrossRef]
Kalia RK, Singh R, Rai MK, Mishra GP, Singh SR, Dhawan AK. (2011). Biotechnological interventions in sea buckthorn (Hippophae L.Current status and future prospects. Trees 25: 559–575. DOI 10.1007/s00468-011-0543-0. [Google Scholar] [CrossRef]
Kimura M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120. DOI 10.1007/BF01731581. [Google Scholar] [CrossRef]
Kumar A, Guleria S, Mehta P, Walia A, Chauhan A, Shirkot CK. (2015). Plant growth promoting traits of phosphate solubilizing bacteria isolated from Hippophae rhamnoides L. (Sea-buckthorn) growing in cold desert Trans-Himalayan Lahul and Spiti regions of India. Acta Physiologiae Plantarum 37: 48. DOI 10.1007/s11738-015-1793-z. [Google Scholar] [CrossRef]
Kumar RR, Jadeja VJ. (2016). Isolation of actinomycetes: A complete approach. International Journal of Current Microbiology and Applied Sciences 5: 606–618. DOI 10.20546/ijcmas.2016.505.062. [Google Scholar] [CrossRef]
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. DOI 10.1093/molbev/msy096. [Google Scholar] [CrossRef]
LeBlanc JG, de Giori GS, Smid EJ, Hugenholtz J, Sesma F. (2007). Folate production by lactic acid bacteria and other food-grade microorganisms. Communicating Current Research and Educational Topics and Trends in Applied Microbiology 1: 329–339. [Google Scholar]
Loizeau K, Brouwer VD, Gambonnet B, Yu A, Renou JP, Straeten DVD, Lambert WE, Rebeille F, Ravanel S. (2008). A genome-wide and metabolic analysis determined the adaptive response of Arabidopsis cells to folate depletion induced by methotrexate. Plant Physiology 148: 2083–2095. DOI 10.1104/pp.108.130336. [Google Scholar] [CrossRef]
Mangla Y, Tandon R. (2014). Pollination ecology of Himalayan seabuckthorn, Hippophae rhamnoides L. (Elaeagnaceae). Current Science 106: 1731–1735. [Google Scholar]
Mwajita MR, Murage H, Tani A, Kahangi EM. (2013). Evaluation of rhizosphere, rhizoplane and phyllosphere bacteria and fungi isolated from rice in Kenya for plant growth promoters. SpringerPlus 2: 606. DOI 10.1186/2193-1801-2-606. [Google Scholar] [CrossRef]
Ohrvik VE, Witthoft CM. (2011). Human folate bioavailability. Nutrients 3: 475–490. DOI 10.3390/nu3040475. [Google Scholar] [CrossRef]
Pan Z. (2007). Metabolic engineering of Bacillus for enhanced product and cellular yields (Doctoral Dissertation Thesis). University of Pittsburgh, Pittsburgh. [Google Scholar]
Park SY, Do JR, Kim YJ, Kim KS, Lim SD. (2014). Physiological characteristics and production of folic acid of Lactobacillus plantarum JA71 isolated from jeotgal, a traditional Korean fermented seafood. Korean Journal for Food Science of Animal Resources 34: 106–114. DOI 10.5851/kosfa.2014.34.1.106. [Google Scholar] [CrossRef]
Pote MS, Gandhi NM, Mishra KP. (2006). Antiatherogenic and radio-protective role of folic acid in whole body γ-irradiated mice. Molecular and Cellular Biochemistry 292: 19–25. DOI 10.1007/s11010-006-9135-5. [Google Scholar] [CrossRef]
Pribat A, Jeanguenin L, Nunez AL, Ziemak MJ, Hyde JE, Lagard VC, Hanson AD. (2009). 6-pyruvoyltetrahydropterin synthase paralogs replace the folate synthesis enzyme dihydroneopterin aldolase in diverse bacteria. Journal of Bacteriology 191: 4158–4165. DOI 10.1128/JB.00416-09. [Google Scholar] [CrossRef]
Rossi M, Amaretti A, Raimondi S. (2011). Folate production by probiotic bacteria. Nutrients 3: 118–134. DOI 10.3390/nu3010118. [Google Scholar] [CrossRef]
Sambrook J, Russel DW. (2001). Rapid isolation of yeast DNA. In: Sambrook J, Russell DW, eds. Molecular Cloning, A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press, 631–632. [Google Scholar]
Scaria J, Janvilisri T, Fubini S, Gleed RD, McDonough SP, Chang YF. (2011). Clostridium difficile transcriptome analysis using pig ligated loop model reveals modulation of pathways not modulated in vitro. Journal of Infectious Diseases 203: 1613–1620. DOI 10.1093/infdis/jir112. [Google Scholar] [CrossRef]
Silva FAS, Azevedo CAV. (2016). The Assistat Software Version 7.7 and its uses in the analysis of experimental data. African Journal of Agricultural Research 11: 3733–3740. DOI 10.5897/AJAR2016.11522. [Google Scholar] [CrossRef]
Slock J, Stahly DP, Han CY, Six EW, Crawford IP. (1990). An apparent Bacillus subtilis folic acid biosynthetic operon containing pab, an amphibolic trpG gene, a third gene required for synthesis of para-aminobenzoic acid, and the dihydropteroate synthase gene. Journal of Bacteriology 172: 7211–7226. DOI 10.1128/JB.172.12.7211-7226.1990. [Google Scholar] [CrossRef]
Soni A, Pandey KM, Ray P, Jayaram B. (2013). Genomes to hits in silico—A country path today, a highway tomorrow: A case study of Chikungunya. Current Pharmaceutical Design 19: 4687–4700. DOI 10.2174/13816128113199990379. [Google Scholar] [CrossRef]
Suryakumar G, Gupta A. (2011). Medicinal and therapeutic potential of Sea buckthorn (Hippophae rhamnoides L.). Journal of Ethnopharmacology 138: 268–278. DOI 10.1016/j.jep.2011.09.024. [Google Scholar] [CrossRef]
Tiwari S, Bala M. (2011). Hippophae leaves prevent immunosuppression and inflammation in 60Co-γ-irradiated mice. Phytopharmacology 1: 36–48. [Google Scholar]
Ulrich CM, Reed MC, Nijhout HF. (2008). Modeling folate, one-carbon metabolism, and DNA methylation. Nutrition Reviews 66: 27–30. DOI 10.1111/j.1753-4887.2008.00062.x. [Google Scholar] [CrossRef]
Xu G, Li C, Yao Y. (2009). Proteomics analysis of drought stress-responsive proteins in Hippophae rhamnoides L. Plant Molecular Biology Reporter 27: 153–161. DOI 10.1007/s11105-008-0067-y. [Google Scholar] [CrossRef]
Zadernowski R, Naczk M, Amarowicz R. (2003). Tocopherols in sea buckthorn (Hippophae rhamnoides L.) berry oil. Journal of the American Oil Chemists' Society 80: 55–58. DOI 10.1007/s11746-003-0650-z. [Google Scholar] [CrossRef]
Zhu T, Pan Z, Domagalski N, Koepsel R, Ataai MM, Domach MM. (2005). Engineering of Bacillus subtilis for enhanced total synthesis of folic acid. Applied and Environmental Microbiology 71: 7122–7129. DOI 10.1128/AEM.71.11.7122-7129.2005. [Google Scholar] [CrossRef]
Zygmunt WA, Conrad HE, Tavormina PA, Haley EE, Sarett HP, Stavely HE. (1962). Formation of folic acid related compounds by Bacillus subtilis. Canadian Journal of Microbiology 8: 429–435. DOI 10.1139/m62-056. [Google Scholar] [CrossRef]
Supplementary Figures
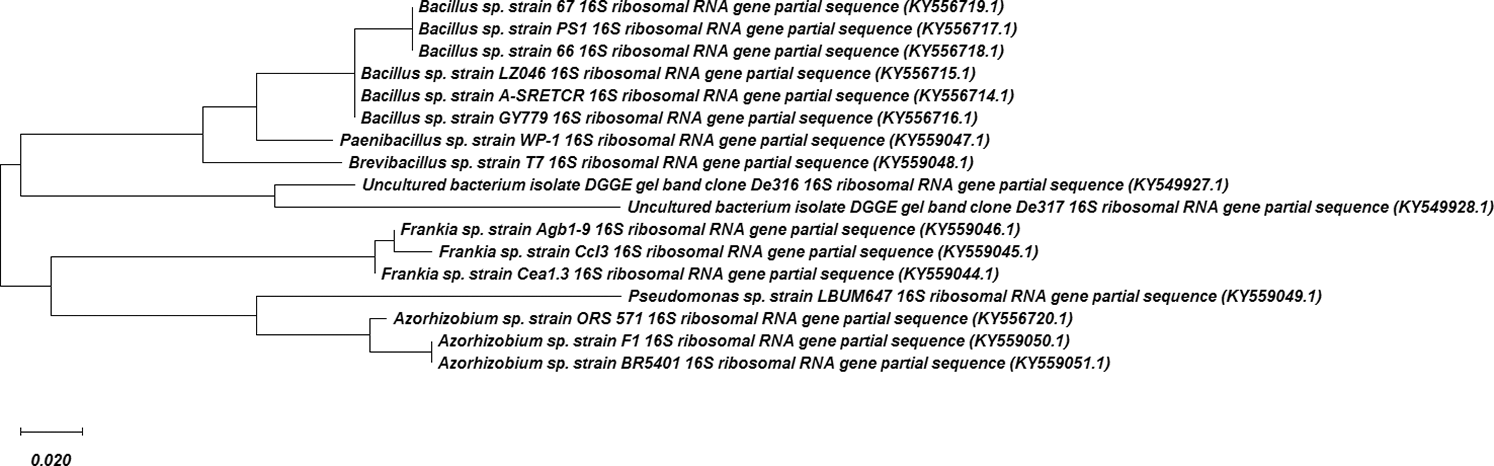
Figure S1: The genome comparative analysis of isolated rhizobacterial strains
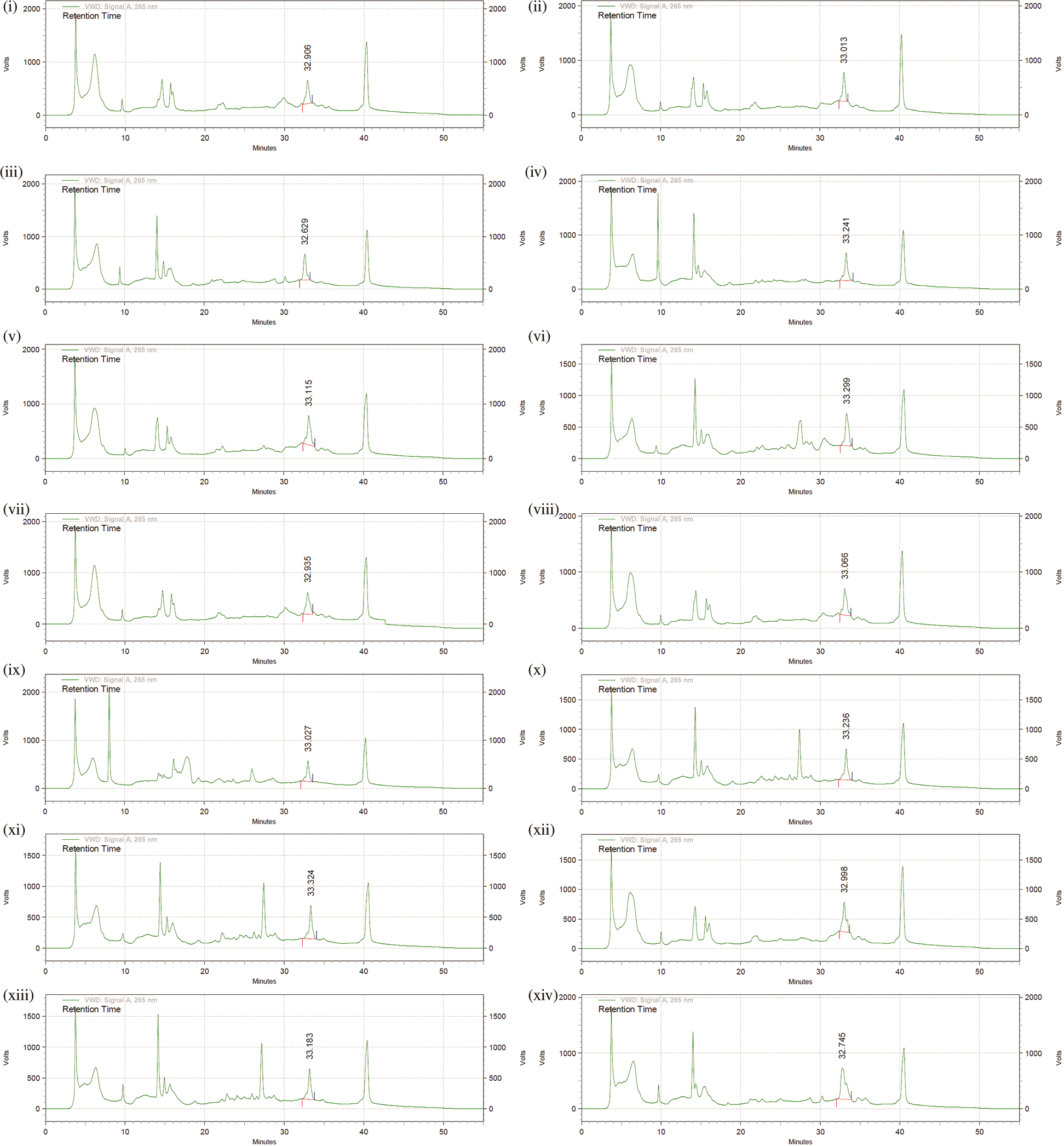
Figure S2: Chromatographs for Folate analysis by HPLC, confirming intracellular and extracellular folate synthesizing capabilities of rhizobacteria. (i) Intracellular folate produced by Bacillus GY779, (ii) Extracellular folate produced by Bacillus GY779, (iii) Extracellular folate Uncultured Bacterium De316, (iv) Extracellular folate Uncultured Bacterium De317, (v) Extracellular folate Bacillus A-SRETCR, (vi) Extracellular folate Bacillus PS1, (vii) Extracellular folate Bacillus cereus 66, (viii) Extracellular folate Bacillus cereus 67, (ix) Extracellular folate Frankia Cea1.3, (x) Extracellular folate Frankia Ccl3, (xi) Extracellular folate Frankia Agb1-9, (xii) Extracellular folate Pseudomonas LBUM647, (xiii) Extracellular folate Azorhizobium F1, (xiv) Extracellular folate Azorhizobium BR5401.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |