DOI:10.32604/biocell.2021.013612

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.013612 |  www.techscience.com/journal/biocell |
| Article |
RPA3 is transcriptionally activated by YY1 and its depletion enhances radiosensitivity of triple-negative and HER2-positive breast cancer
1Clinical Laboratory, Laiyang Central Hospital of Yantai, Laiyang, 265200, China
2Department of General Surgery, The Eighth People’s Hospital of Qindao, Qingdao, 266000, China
3Hepatobiliary Surgery, Anqiu People’s Hospital, Anqiu, 262100, China
*Address correspondence to: Xiqing Liu, lxqsjrlfs2008@163.com
Received: 13 August 2020; Accepted: 03 December 2020
Abstract: RPA3 (Replication Protein A3) (14 kD) is a part of the canonical heterotrimeric replication protein A complex (RPA/RP-A). This study aimed to explore the functional role of RPA3 and the mechanisms of its dysregulation in breast cancer. Data from the Cancer Genome Atlas (TCGA)-breast cancer patients and GSE75688 were utilized for gene expression and survival analysis. Breast cancer cell lines MDA-MB-231 and SK-BR-3 were used for in-vitro cell studies. Clonogenic assay and immunofluorescent staining of γ-H2AX were performed to examine radiation-induced cytotoxicity. Systemic correlation analysis was performed to identify potential transcription factors (TFs) regulating RPA3 expression. ChIP-qPCR and dual-luciferase assay were conducted to verify the transcriptional activating effect of YY1 on RPA3 expression. Bioinformatic analysis showed that RPA3 expression was upregulated in breast cancer. Its upregulation was associated with poor survival of basal-like and HER2+ cases. RPA3 inhibition by siRNA reduced colony formation and increased γ-H2AX foci formation after irradiation in MDA-MB-231 and SK-BR-3 cells. RPA3 expression was transcriptionally activated by YY1 via promoter binding in the two cell lines. Both RPA3 and YY1 expression were positively correlated with their gene-level copy numbers. RPA3 might serve as a potential target for radio-sensitization in basal-like and HER2+ breast cancer.
Keywords: RPA3; YY1; Breast cancer; Radiotherapy
Breast cancer is a group of heterogeneous diseases with distinct genetic features, epigenetic alterations, pathobiological behaviors, responses to therapy, and clinical prognosis (Chung et al., 2017; Vlashi et al., 2014). The 50-gene qPCR assay (PAM50) revealed that breast cancers can be classified into five molecular groups, namely luminal A, luminal B, human epidermal growth factor receptor 2 (HER2)-positive (HER2+), basal-like, and normal-like (Nielsen et al., 2014). Although the molecular subtyping provided valuable information for chemotherapy, endocrine therapy and targeted therapy, the value of PAM50 subtypes in radiotherapy has not been well-characterized in clinical practice (He et al., 2018). Currently, radiotherapy is still an important tool with multiple utilizations in breast cancer therapy, including radiation after breast-conserving surgery; prophylactic irradiation for high-risk patients after mastectomy; radiation for advanced cancers when surgery is not feasible; radiation for local recurrence, and palliative radiotherapy for distant metastases (He et al., 2018). However, some patients may not benefit from this treatment due to individual variations in radiosensitivity (Langlands et al., 2013). Therefore, it is necessary to understand the mechanisms leading to chemoresistance for the future development of radio-sensitizers.
RPA3 (Replication Protein A3) (14 kD) is one of the components of the canonical heterotrimeric replication protein A complex (RPA/RP-A), together with RPA1 (70 kD) and RPA2 (32kD) (Lin et al., 1998). This complex binds to and stabilizes single-stranded DNA (ssDNA) intermediates during DNA replication or in response to DNA damage. It also recruits and activates a series of protein complexes for homologous recombination (HR) repair, such as ataxia-telangiectasia mutated- and Rad3-related interacting protein (ATRIP) (Marechal et al., 2014; Zou and Elledge, 2003), DNA repair protein RAD51 and RAD52 (Aboussekhra et al., 1995) and HepA-related protein (HARP) (Yusufzai et al., 2009). DNA repair capability of tumor cells is negatively correlated with their radiosensitivity (Glanzer et al., 2014). Targeting HR repair has been considered a strategy for chemo/radiotherapy sensitization (Peng et al., 2014). Aberrant RPA3 expression was observed in gastric cancer, hepatocellular carcinoma, nasopharyngeal carcinoma, and also might serve as a prognostic biomarker (Dai et al., 2017; Qu et al., 2017; Xiao et al., 2018). Inhibiting RPA3 expression can enhance the radiosensitivity of hepatocellular carcinoma (Luo et al., 2019) and nasopharyngeal carcinoma (Qu et al., 2017). However, the expression profile of RPA3 and its functional role in the radiosensitivity of breast cancer have not been identified yet.
In this study, we aimed to explore the expression profiles of RPA3 and its prognostic value in breast cancer. Then, we studied its functional role in regulating the radiosensitivity of basal-like and HER2+ breast tumor cells. Systemic screening of transcription factors identified YY1 as a high potential regulator of RPA3 expression. YY1 has been characterized as an important TF upregulated in breast cancer and regulates the malignant transformation of breast cancer (Lu et al., 2019). Therefore, we further explored whether YY1 regulates RPA3 transcription.
Data extraction from Genotype-Tissue Expression (GTEx) and The Cancer Genome Atlas-Breast Cancer (TCGA-BRCA)
The RNA-seq data from normal mammary tissues were acquired from the GTEx (Consortium, 2013), while RNA-seq data from breast cancer and adjacent (adj.) normal tissues were obtained from TCGA-BRCA. Data acquisition was performed using the UCSC Xena (http://xena.ucsc.edu/) (Goldman et al., 2020). PAM50 subtypes (determined by RNA-seq data), tumor gene-level copy number alterations (CNAs, delete germline copy number variation), survival data, including progression-free survival (PFS), and disease-specific survival (DSS), were also extracted.
Survival analysis using Kaplan-Meier Plotter
Kaplan-Meier Plotter (http://kmplot.com) was used to check recurrence-free survival (RFS) data integrated from microarray data of 1809 breast cancer patients (Gyorffy et al., 2010). Kaplan-Meier survival curves were generated by the median or optimal cutoff of RPA3 expression.
Single-cell transcriptional data and functional states in breast cancer cells
The association between RPA3 expression and the functional states of breast cancer cells at the single-cell level was assessed using the CancerSEA platform (http://biocc.hrbmu.edu.cn/CancerSEA/home.jsp), which provides an analytic strategy to determine the correlation between gene expression and 14 functional states of cancer cells (Yuan et al., 2019). These states were estimated according to the gene-expression profile at the single-cell level, using the signatures from Gene Ontology, MSigDB, Cyclebase, HCMDB and StemMapper. The state activity scores were calculated using the Gene Set Variation Analysis (GSVA) (Yuan et al., 2019). The tumor cell data from basal-like (N = 130) and HER2+ (N = 89) cases in one previous single-cell RNA-seq dataset (GSE75688) (Chung et al., 2017) were retrieved for analysis.
Transcription factor (TF) data retrieved from JASPAR
TF gene list was collected from the JASPAR database (http://jaspar.genereg.net/) (Stormo, 2013). The promoter segment of RPA3 was acquired by checking the promoter clone of human RPA3 (#HPRM44615, Genome = hg38; chr7-:7719949-7718330; TSS = 7718607) in Genecopoeia. The promoter segment was scanned in JASPAR to identify potential YY1 binding sites by setting the relative profile score threshold to 90%.
Basal-like representative MDA-MB-231 and HER2+ representative SK-BR-3 human breast cancer cell lines were purchased from the Cell Resource Center, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (Beijing, China). These two cell lines were maintained in Dulbecco’s modified eagle medium (DMEM) (Lonza, Walkersville, MD, USA) at 37°C in a 90% humidified incubator, with 5% CO2.
Small interfering RNA (siRNA) and scramble controls were synthesized by General Biosystem (Chuzhou, Anhui, China), with the following sequences: RPA3 siRNA (#1, 5’-CCGGCAUGCUAGCUCAAUUTT-3’; #2, 5’-GCAUGCUAGCUCAAUUCAUTT-3’, #3, 5’-GCCACCAUCUUGUGUACAUTT-3’) and scramble: 5’-GCUAUGCUCGCAUAUCACUTT-3’; YY1 siRNA (#1, 5’-CCAAACAACUGGCAGAAUUTT-3’; #2, 5’-GCUCCAAGAACAAUAGCUUTT-3’, #3, 5’-CCCAAACAACUGGCAGAAUTT-3’) and scramble: 5’-CACACGACUGACAAGCAUATT-3’. Traditional forward transfection was conducted. Briefly, cells were seeded into a six-well plate at a density of 1 × 106 cells per well for 24 h at 37°C. Then, the cells were transfected with the siRNAs (50 nM), using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
Real-time quantitative RT-PCR (RT-qPCR)
Total RNA was extracted from cells using the High Pure RNA Isolation Kit (Roche Applied Science, Mannheim, Germany). Then, total RNA was reversely transcribed into cDNA using the first-strand cDNA synthesis kit (Roche Applied Science) according to the manufacturer’s instructions. Real-time qPCR was then performed as described previously (Tuo et al., 2015). The sense and antisense primers for gene amplification were: RPA3, 5’-AAGCCTGTCTGCTTCGTAGGGA-3’ and 5’-CGGTTACTCTTCCAACCACTTCC-3’, YY1, 5’-GGAGGAATACCTGGCATTGACC-3’ and 5’-CCCTGAACATCTTTGTGCAGCC-3’. The relative expression level of mRNA was evaluated by using the 2−ΔΔCt method. Gene expression levels were normalized to ACTB.
Total proteins were extracted using RIPA lysis buffer (Beyotime, China) with protease inhibitors cocktail (Roche Diagnostics, Basel, Switzerland). Then, equal amounts of protein lysates were run on 10% SDS-PAGE, and the separated bands were transferred to polyvinylidene fluoride membrane (PVDF; EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% non-fat dried milk for 1 h at room temperature, and then were incubated with primary antibodies against RPA3 (1:1000, ab97436, Abcam, Cambridge, MA, USA), YY1 (1:5000, ab245365, Abcam), γ-H2AX (1:5000, ab11174, Abcam) and β-Actin (1:5000, ab179467, Abcam) overnight at 4°C, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. The protein bands were visualized using ECL detection reagent (Millipore, Billerica, MA, USA).
Clonogenic survival assay was conducted following one previous protocol (Franken et al., 2006). Cells transfected with RPA3 siRNA or scrambled control were seeded into 6-well plates. After adhesion, they were irradiated at defined doses (0, 2, 4, 6, or 8 Gy), using a Rad Source R2000 X-ray irradiator (1.1 Gy/min., 160 kV, 25 mA, 0.3 mm copper filters, Rad Source Tech, Suwanee, GA, USA). After 7–14 days of incubation, the cultures were fixed with 100% methanol and then stained with 1% crystal violet. Colonies containing >50 cells were counted by microscopic inspection. The plating efficiency (PE) of un-irradiated cells (0 Gy) was calculated by the formula: PE = number of colonies counted/number of cells plated. The surviving fraction (SF) of the irradiated cells was calculated using the formula: SF = number of colonies formed after treatment/number of cells seeded × PE. A linear-quadratic model was utilized to generate survival curves using the following equation (Bodgi and Foray, 2016): Y = e(−1 × (A×X+B×X2)), in which Y is the fraction survival, and X is the dose. A equals (−1)-times the initial slope, and the initial value of B equals (−0.1)-times the initial slope.
MDA-MB-231 and SK-BR-3 cells with or without inhibition of endogenous RPA3 were seeded in 24-well plates and exposed to 6 Gy of irradiation. 24 h later, the cells were fixed in 4% paraformaldehyde, permeabilized in 0.1% Triton X-100 and blocked using 10% goat serum. Then, the cells were incubated with the primary antibody against γ-H2AX (ab11174, Abcam). After that, the cells were incubated with a secondary antibody conjugated to fluorescein isothiocyanate. DAPI was used for nuclear staining. γ-H2AX foci were visualized under a fluorescence microscope (Olympus IX71, Tokyo, Japan). Five random fields were examined to estimate the number of foci per cell for each coverslip.
Chromatin immunoprecipitation (ChIP)-qPCR
ChIP was performed using the Chromatin Immunoprecipitation Kit (17-295, Merck Millipore, Boston, MA, USA) according to the recommended protocol. Briefly, formaldehyde was used to cross-link the proteins to the DNA. The lysates of MDA-MB-231 and SK-BR-3 cells were sonicated to shear DNA to an average fragment size of 200–1000 bp. Then, samples were pre-cleaned with Protein A Agarose/Salmon Sperm DNA (50% Slurry) (Catalog #16-157C) and subsequently incubated with anti-YY1 (ab245365, Abcam) or IgG antibodies overnight at 4°C with rotation. Immunoprecipitated DNA was collected by adding the previously mentioned Protein A beads. Then, the samples were washed, and DNA levels were measured by qPCR. Three primer sets were designed (Tab. 1), among which two sets covered potential YY1 binding sites (−998 ~ −882/−884 ~ −808) (primer set 1 and 2, Tab. 1) and one set of primers not covering YY1 binding site (−22 ~ +73) (set 3, Tab. 1).
Table 1: Primers for ChIP-qPCR assay

Full length and truncated 5’ flanking sequences of RPA3 promoter, including −900/+277, −800/+277, and −150/+277, were cloned into the pGL3-basic plasmid (Promega, Madison, WI, USA). MDA-MB-231 cells were seeded in 24-well plates (2 × 105 cells per well). 24 h later, the cells were transfected with either 1 μg recombinant vectors with different fragments of RPA3 promoter together with 50 nM YY1 siRNA or scramble control, using Lipofectamine 2000 (Invitrogen). 0.05 μg of the pRL-CMV vector was co-transfected to normalize the transfection efficiency. 24 h later, the cells were lysed for measuring luciferase activity, using a dual-specific luciferase assay kit (#E1910, Promega).
Welch’s unequal variances t-test was performed for two-group comparison. A Log-rank test was performed for comparing Kaplan-Meier survival curves. Pearson’s correlation r was calculated for correlation assessment. p < 0.05 was considered statistically significant.
In silico analysis of RPA3 expression profile and prognostic value in breast cancer
RNA-seq data from GTEx-normal mammary tissue and TCGA-BRCA showed that normal mammary tissue owned the lowest RPA3 expression (Fig. 1A). Tumor adj. normal and all PAM50 subtypes of breast tumors presented elevated RPA3 expression (Fig. 1A). IHC staining in the HPA confirmed RPA3 expression at the protein level in breast cancer tissues (Fig. 1B). Then, we assessed the survival difference between patients with high (top 50%) and low (bottom 50%) RPA3 expression in luminal A, luminal B, HER2+, and basal-like subgroups, respectively, using survival data from TCGA-BRCA. The log-rank test indicated that in patients with HER2+ tumors, the high RPA3 expression group had significantly worse PFS and DSS (p < 0.05, Figs. 1E and 1I). The high RPA3 expression basal-like tumor group was also associated with significantly shorter PFS (p = 0.024) and tended to have worse DSS (p = 0.057) (Figs. 1F and 1J). However, no survival difference was observed in luminal A and B cases by median RPA3 stratification (Figs. 1C, 1D, 1G and 1H).
To validate the survival difference, we also checked survival data in the Kaplan-Meier Plotter. Under both median and optimal RPA3 expression cutoff models, high RPA3 expression HER2+ cases had significantly worse RFS (Suppl. Figs. 1A and 1B). In basal-like cases, high RPA3 expression-associated worse RFS was confirmed under the optimal cutoff model (Suppl. Fig. 1D), but not in the median expression model (Suppl. Fig. 1C).
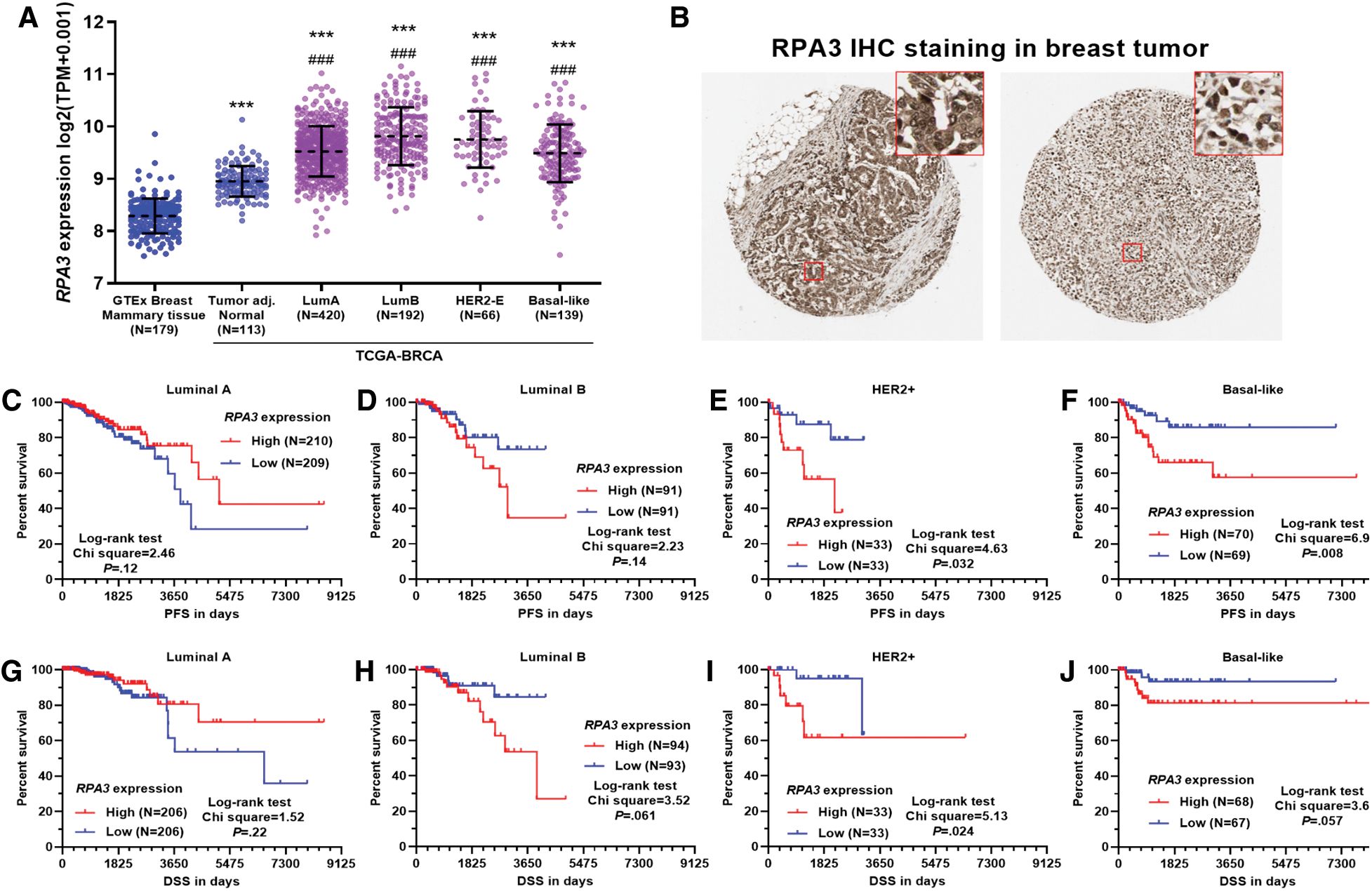
Figure 1: High RPA3 expression was associated with poor survival of basal-like and HER2+ breast cancer.
In silico analysis of the correlation between RPA3 expression and functional states of basal-like and HER2+ tumor cells
Using cellular functional states assessed by the CancerSea, we analyzed the correlation between RPA3 expression and 14 functional states of basal-like (N = 89) and HER2+ tumor cells (N = 130) in GSE75688. Correlation analysis indicated that RPA3 expression was positively correlated with cell cycle progression, DNA damage, and DNA repair in both basal-like and HER2+ tumor cells (Pearson’s r ≥ 0.2, Fig. 2A).
RPA3 inhibition increased the radiosensitivity of basal-like and HER2+ breast cancer cells
MDA-MB-231 and SK-BR-3 cells were transfected with RPA3 siRNA (Figs. 2B and 2C). Compared with the scramble group, the survival fractions of siRPA3 transfected MDA-MB-231 (Fig. 2D) and SK-BR-3 (Fig. 2E) cells were dramatically decreased after irradiation. Besides, siRPA3 treatment also increased the number of γ-H2AX foci (Figs. 2F–2H) and γ-H2AX expression 24 h after irradiation in both MDA-MB-231 and SK-BR-3 cells (Figs. 2I and 2J). These results suggested that RPA3 inhibition enhanced the radiosensitivity of basal-like and HER2+ breast cancer cells.
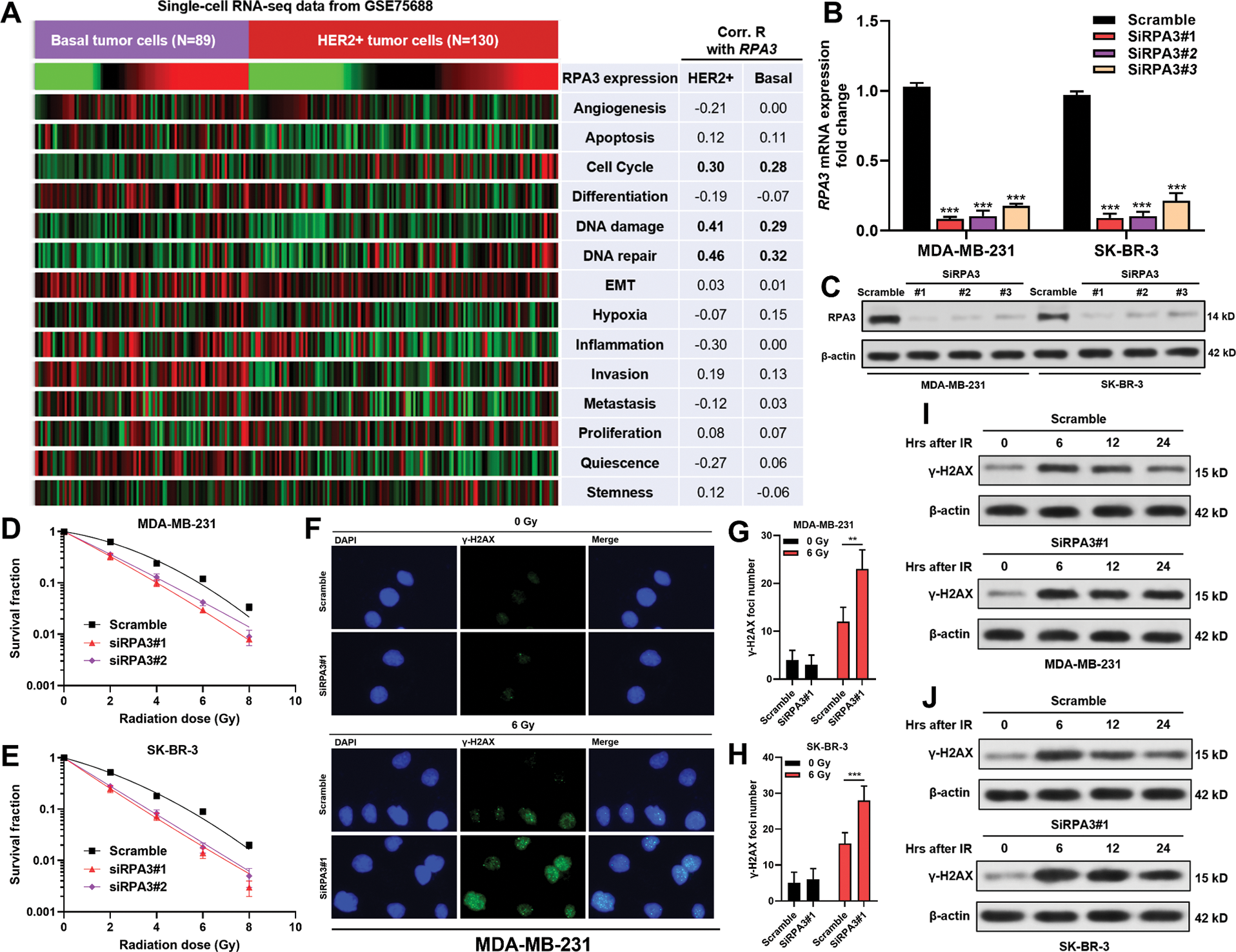
Figure 2: RPA3 inhibition increased radiosensitivity of basal-like and HER2+ breast cancer cells.
RPA3 expression was transcriptionally activated by YY1 in basal-like and HER2+ breast cancer cells
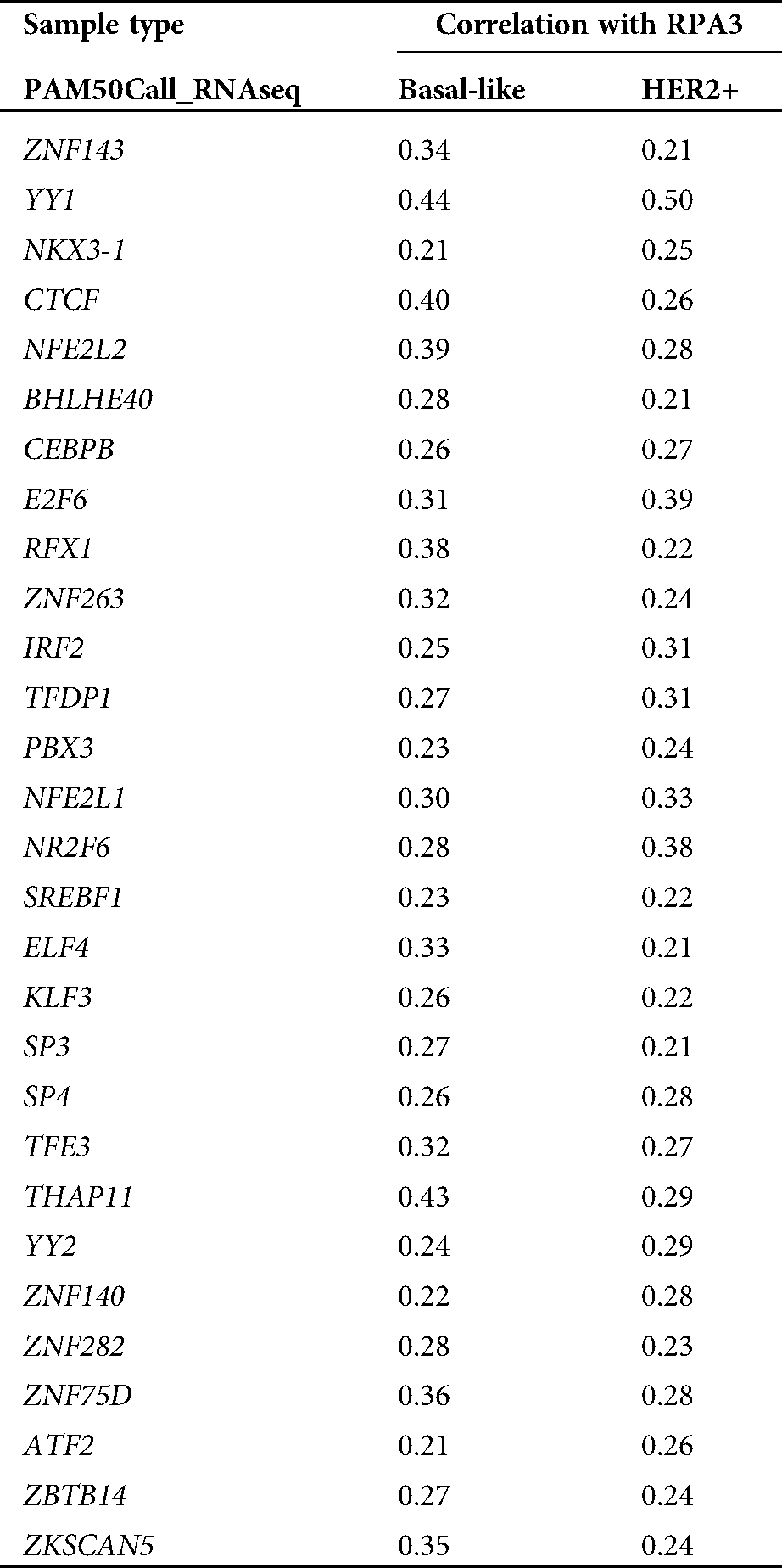
To explore the mechanisms underlying RPA3 dysregulation, we assessed the correlation between the expression of transcriptional factors (TFs) in the JASPAR database (N = 669) and RPA3 in basal-like and HER2+ tumors, respectively (Fig. 3A). Correlation analysis (Suppl. Tab. 1) indicated that among the 669 TFs, only YY1 was moderately and positively correlated (|Pearson’s r| ≥ 0.4) with RPA3 expression in both basal-like and HER2+ tumor tissue (Suppl. Tab. 1, Figs. 3B and 3C). Via scanning the promoter sequence of RPA3, we found five high potential YY1 binding sites (Fig. 3D). YY1 depletion in MDA-MB-231 and SK-BR-3 cells resulted in significantly decreased RPA3 expression at the mRNA and protein levels (Figs. 3E–3G). Concerning CHIP-qPCR findings, the two amplicons covering YY1 binding sites, but not of the amplicon without YY1 bind site, were significantly enriched upon anti-YY1 immunoprecipitation in both breast cancer cells (Figs. 3H–3I). Dual-luciferase reporter assay showed that the luciferase construct with the integrated promoter sequence had the strongest luciferase activity (Fig. 3J). Truncating the binding sites significantly reduced the luciferase activity (Fig. 3J). YY1 inhibition also substantially decreased the intensity of luciferase expression (Fig. 3J).
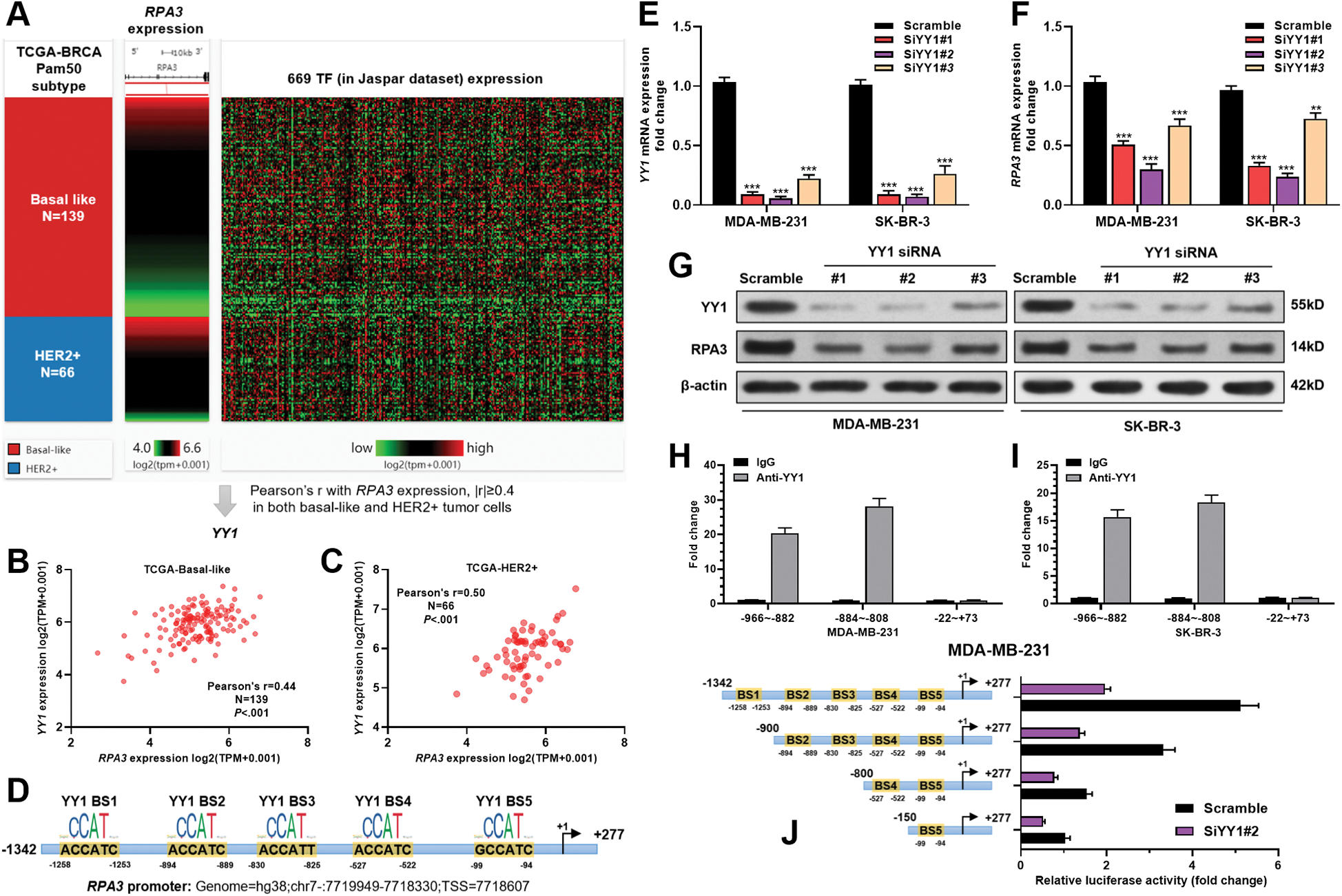
Figure 3: RPA3 expression is transcriptionally activated by YY1 in basal-like and HER2+ breast cancer cells.
In silico analysis of RPA3 and YY1 CNAs
RPA3/YY1 expression and their CNAs in basal-like and HER2+ breast cancer cases in TCGA are shown in Fig. 4A. Correlation analysis indicated that in basal-like cases, the expression of RPA3 and YY1 was moderately correlated (Pearson's r ≥ 0.4) with their gene-level copy number (Figs. 4B and 4D). In HER2+ cases, a moderate positive correlation was observed between YY1 expression and its copy number (Fig. 4E). In comparison, no significant correlation was found between RPA3 expression and its copy number (Fig. 4C).
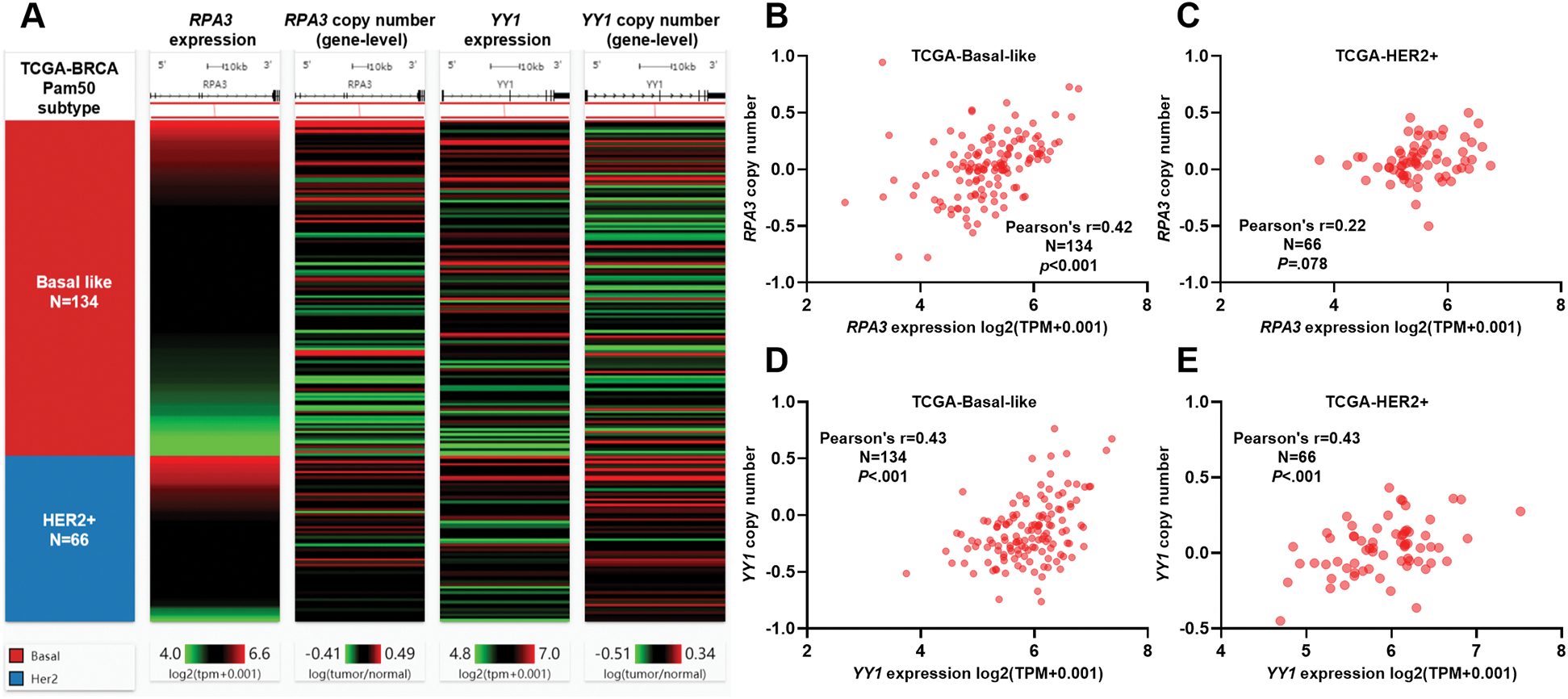
Figure 4: Both RPA3 and YY1 expression was associated with gene-level copy number.
For patients with basal-like and HER2+ breast tumors, it is now well established that radiation therapy reduces locoregional recurrence rate and provides survival benefits for high-risk patients (He et al., 2018; Liu et al., 2019). However, local recurrence rate improvement of basal-like and HER2+ tumors after radiotherapy is significantly smaller compared to the luminal subtypes (Kyndi et al., 2008), suggesting that these two subtypes are more radioresistant. Therefore, it is meaningful to find new radio-sensitization targets to reduce the inherent and induced radiation-resistance. RPA complex members have been considered as potential targets for radio-sensitization. Previous studies found that silencing RPA1 reduces the radio-resistance of a hypopharyngeal cancer cell line (Liu et al., 2020). RPA1 or RPA2 inhibition can induce G2/M arrest and enhance the radiosensitivity of esophageal cancer cells (Zhao et al., 2014). Inhibiting RPA3 expression can sensitize hepatocellular carcinoma (Luo et al., 2019) and nasopharyngeal carcinoma (Qu et al., 2017) cells to radiation. In this study, we revealed that RPA3 upregulation was associated with unfavorable survival of basal-like and HER2+ breast cancer. In addition, our in-vitro cellular studies demonstrated that inhibiting RPA3 expression sensitized MDA-MB-231 and SK-BR-3 cells to irradiation. Therefore, RPA3 might serve as a potential target of radio-sensitization in basal-like and HER2+ breast cancer.
By systemic bioinformatic screening of TFs and in vitro studies, we demonstrated that YY1 was an upstream regulator of RPA3. YY1 directly bound to the RPA3 promoter and activated its transcription in MDA-MB-231 and SK-BR-3 cells. A series of studies showed that YY1 acts as an important TF involved in breast cancer development and therapeutic resistance via multiple mechanisms (Sarvagalla et al., 2019). YY1 physically interacts with p27 and promotes its ubiquitination, thereby enhancing clonogenicity, migration, invasion, and tumor formation of breast cancer cells (Wan et al., 2012). Furthermore, YY1 directly activates heat shock factor 1 (HSF1) transcription to promote transforming growth factor-β (TGFβ)-triggered proliferation and migration of MDA-MB-231 cells (Yang et al., 2019). It also reduces miR-873-5p expression by recruiting histone deacetylase 4 (HDAC4) and HDAC9 to the miR-873-5p promoter, thereby activating PI3K/AKT and ERK1/2 pathways and increasing the stemness and chemoresistance of breast cancer cells (Guo et al., 2020). YY1 binds to the LINC00673 promoter and suppresses its transcription. Subsequently, LINC00673 acts as a competing endogenous RNA for miR-515-5p, leading to upregulated MARK4 expression and inhibited Hippo signaling pathway (Qiao et al., 2019). Findings in the current study help expand our understanding of the downstream molecular mechanism of YY1 in breast cancer.
Using gene-level copy number alteration data in TCGA, we found that both RPA3 and YY1 upregulation was positively correlated with their copy numbers in certain PAM50 subtypes. Some recent studies revealed that therapeutic interventions drive both genetic and epigenetic evolutions of breast cancers (Magnani et al., 2017; Nam et al., 2020). The genetic alterations favoring cancer cell survival under stressful conditions are more likely to be retained (Nam et al., 2020). Since RPA3 and YY1 upregulation contributes to breast cancer development and therapeutic resistance, the part of tumor cells with elevated RPA3 and YY1 expression might have higher chances to overcome chemo/radiotherapy induced cell death. These findings suggest that the mechanisms leading to RPA3 dysregulation in breast cancer are multifaceted. Therefore, it is necessary to explore other genetic and epigenetic mechanisms associated with its dysregulation in future studies.
This study revealed that RPA3 was upregulated in breast cancer and was associated with poor survival and radio-resistance of basal-like and HER2+ breast tumors. Its expression was transcriptionally activated by YY1. It might serve as a potential target for radio-sensitization in basal-like and HER2+ breast cancer.
Availability of data and materials: The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Author Contribution: The authors confirm contribution to the paper as follows: Study conception and design: Yanfei LI, Lulu DAI; data collection: Yanfei LI, Ke CAI; analysis and interpretation of results: Yanfei LI, Yingkui SONG, Xiqing LIU; draft manuscript preparation: Yanfei LI, Xiqing LIU. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: Ethics approval is required since no primary data were collected from human or animal tissues in the current study.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protić M, Hübscher U, Egly JM, Wood RD. (1995). Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80: 859–868. DOI 10.1016/0092-8674(95)90289-9. [Google Scholar] [CrossRef]
Bodgi L, Foray N. (2016). The nucleo-shuttling of the ATM protein as a basis for a novel theory of radiation response: Resolution of the linear-quadratic model. International Journal of Radiation Biology 92: 117–131. DOI 10.3109/09553002.2016.1135260. [Google Scholar] [CrossRef]
Consortium GT. (2013). The genotype-tissue expression (GTEx) project. Nature Genetics 45: 580–585. DOI 10.1038/ng.2653. [Google Scholar] [CrossRef]
Chung W, Eum H H, Lee HO, Lee KM, Lee HB, Kim KT, Ryu HS, Kim S, Lee JE, Park YH, Kan Z, Han W, Park WY. (2017). Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nature Communications 8: 15081. DOI 10.1038/ncomms15081. [Google Scholar] [CrossRef]
Dai Z, Wang S, Zhang W, Yang Y. (2017). Elevated expression of RPA3 is involved in gastric cancer tumorigenesis and associated with poor patient survival. Digestive Diseases and Sciences 62: 2369–2375. DOI 10.1007/s10620-017-4696-6. [Google Scholar] [CrossRef]
Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. (2006). Clonogenic assay of cells in vitro. Nature Protocols 1: 2315–2319. DOI 10.1038/nprot.2006.339. [Google Scholar] [CrossRef]
Glanzer JG, Liu S, Wang L, Mosel A, Peng A, Oakley GG. (2014). RPA inhibition increases replication stress and suppresses tumor growth. Cancer Research 74: 5165–5172. DOI 10.1158/0008-5472.CAN-14-0306. [Google Scholar] [CrossRef]
Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, Zhu J, Haussler D. (2020). Visualizing and interpreting cancer genomics data via the Xena platform. Nature Biotechnology 38: 675–678. DOI 10.1038/s41587-020-0546-8. [Google Scholar] [CrossRef]
Guo Q, Wang T, Yang Y, Gao L, Zhao Q, Zhang WZ, Tao X, Zhang LF. (2020). Transcriptional factor Yin Yang 1 promotes the stemness of breast cancer cells by suppressing miR-873-5p transcriptional activity (March 3, 2020). https://ssrn.com/abstract=3531072 or http://dx.doi.org/10.2139/ssrn.3531072. [Google Scholar]
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. (2010). An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Research and Treatment 123: 725–731. DOI 10.1007/s10549-009-0674-9. [Google Scholar] [CrossRef]
He MY, Rancoule C, Rehailia-Blanchard A, Espenel S, Trone JC, Bernichon E, Guillaume E, Vallard A, Magné N. (2018). Radiotherapy in triple-negative breast cancer: Current situation and upcoming strategies. Critical Reviews in Oncology/Hematology 131: 96–101. DOI 10.1016/j.critrevonc.2018.09.004. [Google Scholar] [CrossRef]
Kyndi M, Sørensen FB, Knudsen H, Overgaard M, Nielsen HM, Overgaard J. (2008). Estrogen receptor, progesterone receptor, HER-2, and response to postmastectomy radiotherapy in high-risk breast cancer: The Danish Breast Cancer Cooperative Group. Journal of Clinical Oncology 26: 1419–1426. DOI 10.1200/JCO.2007.14.5565. [Google Scholar] [CrossRef]
Langlands FE, Horgan K, Dodwell DD, Smith L. (2013). Breast cancer subtypes: Response to radiotherapy and potential radiosensitisation. British Journal of Radiology 86: 20120601. DOI 10.1259/bjr.20120601. [Google Scholar] [CrossRef]
Lin YL, Shivji MK, Chen C, Kolodner R, Wood RD, Dutta A. (1998). The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. Journal of Biological Chemistry 273: 1453–1461. DOI 10.1074/jbc.273.3.1453. [Google Scholar] [CrossRef]
Liu C, Liao K, Gross N, Wang Z, Li G, Zuo W, Zhong S, Zhang Z, Zhang H, Yang J, Hu G. (2020). Homologous recombination enhances radioresistance in hypopharyngeal cancer cell line by targeting DNA damage response. Oral Oncology 100: 104469. DOI 10.1016/j.oraloncology.2019.104469. [Google Scholar] [CrossRef]
Liu QQ, Sun HF, Yang XL, Chen MT, Liu Y, Zhao Y, Zhao YY, Jin W. (2019). Survival following radiotherapy in young women with localized early-stage breast cancer according to molecular subtypes. Cancer Medicine 8: 2840–2857. DOI 10.1002/cam4.2186. [Google Scholar] [CrossRef]
Lu X, Liu R, Wang M, Kumar AK, Pan F, He L, Hu Z, Guo Z. (2019). MicroRNA-140 impedes DNA repair by targeting FEN1 and enhances chemotherapeutic response in breast cancer. Oncogene 39: 234–247. DOI 10.1038/s41388-019-0986-0. [Google Scholar] [CrossRef]
Luo J, Si ZZ, Li T, Li JQ, Zhang ZQ, Chen GS, Qi HZ, Yao HL. (2019). MicroRNA-146a-5p enhances radiosensitivity in hepatocellular carcinoma through replication protein A3-induced activation of the DNA repair pathway. American Journal of Physiology-Cell Physiology 316: C299–C311. DOI 10.1152/ajpcell.00189.2018. [Google Scholar] [CrossRef]
Magnani L, Frigè G, Gadaleta RM, Corleone G, Fabris S, Kempe H, Verschure PJ, Barozzi I, Vircillo V, Hong SP, Perone Y, Saini M, Trumpp A, Viale G, Neri A, Ali S, Colleoni MA, Pruneri G, Minucci S. (2017). Acquired CYP19A1 amplification is an early specific mechanism of aromatase inhibitor resistance in ERα metastatic breast cancer. Nature Genetics 49: 444–450. DOI 10.1038/ng.3773. [Google Scholar] [CrossRef]
Marechal A, Li JM, Ji XY, Wu CS, Yazinski SA, Nguyen HD, Liu S, Jiménez AE, Jin J, Zou L. (2014). PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Molecular Cell 53: 235–246. DOI 10.1016/j.molcel.2013.11.002. [Google Scholar] [CrossRef]
Nam AS, Chaligne R, Landau DA. (2020). Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nature Reviews Genetics 22: 3–18. DOI 10.1038/s41576-020-0265-5. [Google Scholar] [CrossRef]
Nielsen T, Wallden B, Schaper C, Ferree S, Liu S, Gao D, Barry G, Dowidar N, Maysuria M, Storhoff J. (2014). Analytical validation of the PAM50-based prosigna breast cancer prognostic gene signature assay and nCounter analysis system using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer 14: 177. DOI 10.1186/1471-2407-14-177. [Google Scholar] [CrossRef]
Peng G, Chun-Jen LC, Mo W, Dai H, Park YY, Kim SM, Peng Y, Mo Q, Siwko S, Hu R, Lee JS, Hennessy B, Hanash S, Mills GB, Lin SY. (2014). Genome-wide transcriptome profiling of homologous recombination DNA repair. Nature Communications 5: 3361. DOI 10.1038/ncomms4361. [Google Scholar] [CrossRef]
Qiao K, Ning S, Wan L, Wu H, Wang Q, Zhang X, Xu S, Pang D. (2019). LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. Journal of Experimental & Clinical Cancer Research 38: 418. DOI 10.1186/s13046-019-1421-7. [Google Scholar] [CrossRef]
Qu C, Zhao Y, Feng G, Chen C, Tao Y, Zhou S, Liu S, Chang H, Zeng M, Xia Y. (2017). RPA3 is a potential marker of prognosis and radioresistance for nasopharyngeal carcinoma. Journal of Cellular and Molecular Medicine 21: 2872–2883. DOI 10.1111/jcmm.13200. [Google Scholar] [CrossRef]
Sarvagalla S, Kolapalli SP, Vallabhapurapu S. (2019). The two sides of YY1 in cancer: A friend and a foe. Frontiers in Oncology 9: 1230. DOI 10.3389/fonc.2019.01230. [Google Scholar] [CrossRef]
Stormo GD. (2013). Modeling the specificity of protein-DNA interactions. Quantitative Biology 1: 115–130. DOI 10.1007/s40484-013-0012-4. [Google Scholar] [CrossRef]
Tuo YL, Li XM, Luo J. (2015). Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. European Review for Medical and Pharmacological Sciences 19: 3403–3411. [Google Scholar]
Vlashi E, Lagadec C, Vergnes L, Reue K, Frohnen P, Chan M, Alhiyari Y, Dratver MB, Pajonk F. (2014). Metabolic differences in breast cancer stem cells and differentiated progeny. Breast Cancer Research and Treatment 146: 525–534. DOI 10.1007/s10549-014-3051-2. [Google Scholar] [CrossRef]
Wan M, Huang W, Kute TE, Miller LD, Zhang Q, Hatcher H, Wang J, Stovall DB, Russell GB, Cao PD, Deng Z, Wang W, Zhang Q, Lei M, Torti SV, Akman SA, Sui G. (2012). Yin Yang 1 plays an essential role in breast cancer and negatively regulates p27. American Journal of Pathology 180: 2120–2133. DOI 10.1016/j.ajpath.2012.01.037. [Google Scholar] [CrossRef]
Xiao W, Zheng J, Zhou B, Pan L. (2018). Replication Protein A 3 is associated with hepatocellular carcinoma tumorigenesis and poor patient survival. Digestive Diseases 36: 26–32. DOI 10.1159/000478977. [Google Scholar] [CrossRef]
Yang W, Feng B, Meng Y, Wang J, Geng B, Cui Q, Zhang H, Yang Y, Yang J. (2019). FAM3C-YY1 axis is essential for TGFbeta-promoted proliferation and migration of human breast cancer MDA-MB-231 cells via the activation of HSF1. Journal of Cellular and Molecular Medicine 23: 3464–3475. DOI 10.1111/jcmm.14243. [Google Scholar] [CrossRef]
Yuan H, Yan M, Zhang G, Liu W, Deng C, Liao G, Xu L, Luo T, Yan H, Long Z, Shi A, Zhao T, Xiao Y, Li X. (2019). CancerSEA: A cancer single-cell state atlas. Nucleic Acids Research 47: D900–D908. DOI 10.1093/nar/gky939. [Google Scholar] [CrossRef]
Yusufzai T, Kong X, Yokomori K, Kadonaga JT. (2009). The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes & Development 23: 2400–2404. DOI 10.1101/gad.1831509. [Google Scholar] [CrossRef]
Zhao D, Sun SY, Lu H, Yu DH. (2014). Enhanced radiosensitivity and G2/M arrest were observed in radioresistant esophageal cancer cells by knocking down RPA expression. Cell Biochemistry and Biophysics 70: 887–891. DOI 10.1007/s12013-014-9995-3. [Google Scholar] [CrossRef]
Zou L, Elledge SJ. (2003). Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548. DOI 10.1126/science.1083430. [Google Scholar] [CrossRef]
Supplementary Table 1: TF genes positive correlated with RPA3 (Pearson’r > 0.2) in TCGA-BRCA
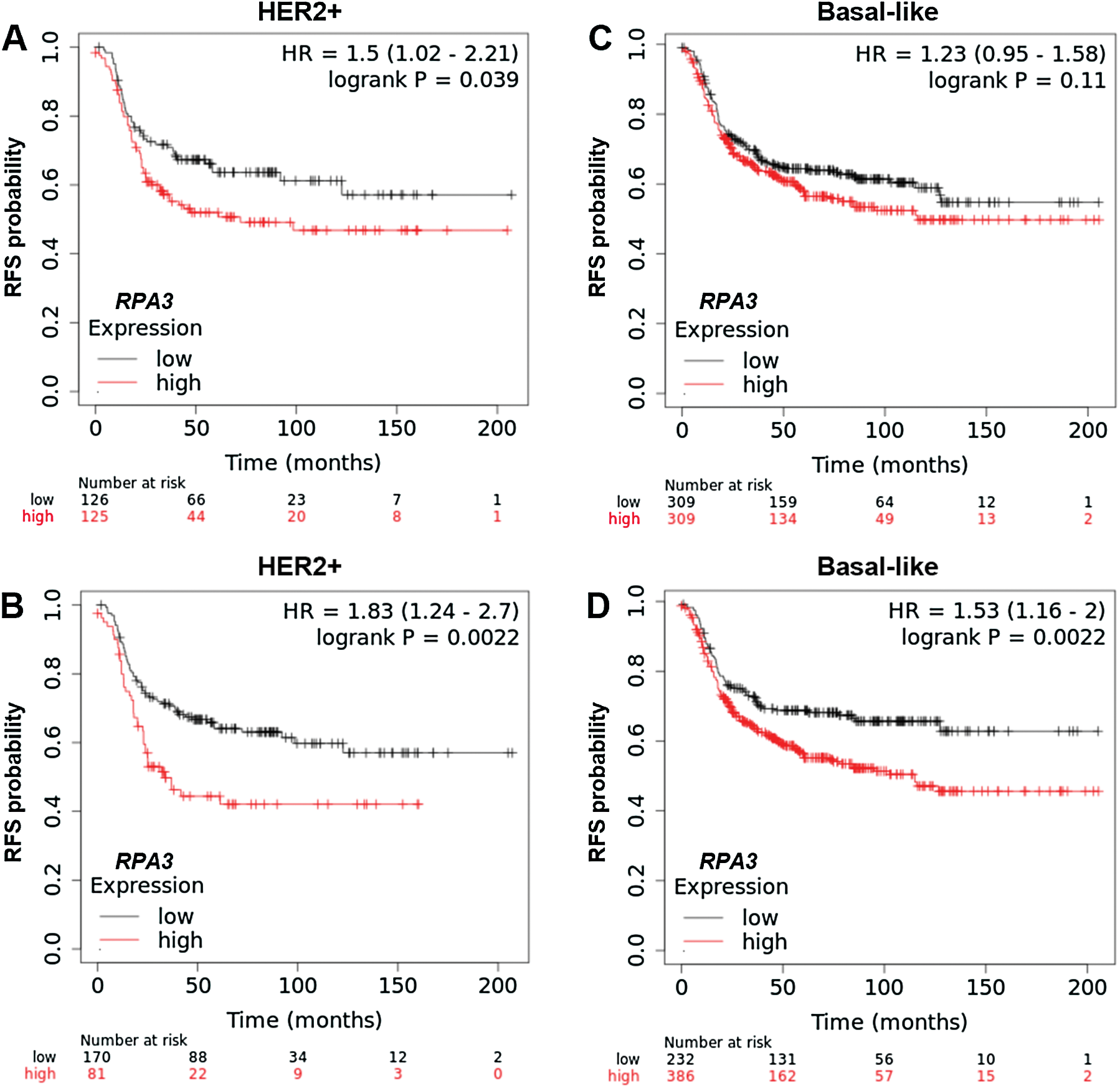
SuppleMENTARY Figure 1: Kaplan-Meier survival analysis of RFS in Kaplan-Meier Plotter.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |