DOI:10.32604/biocell.2021.013978

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.013978 |  www.techscience.com/journal/biocell |
| Article |
Expression and prognosis analyses of Dectin-1 cluster genes in patients with lung adenocarcinoma (LUAD) and the association with immune checkpoint molecules
1West China School of Medicine, West China Hospital, Sichuan University, Chengdu, 610041, China.
2Department of Thoracic Cancer, West China Hospital, Sichuan University, Chengdu, 610041, China.
3Department of Laboratory Medicine, West China Hospital, Sichuan University, Chengdu, 610041, China.
*Address correspondence to: Yi Zhou, zyiisc@sina.com; Binwu Ying, yingbinwu@scu.edu.cn
#These authors contributed equally to this work
Received: 27 August 2020; Accepted: 02 November 2020
Abstract: Reviews The Dectin-1 cluster comprises seven members: CLEC-12A, CLEC-12B, CLEC-1A, CLEC-7A, CLEC-2, CLEC-9A and OLR1. These members have been demonstrated to be involved in the tumorigenesis, progression, and metastasis of several cancers. However, little is known about their roles in human lung adenocarcinoma (LUAD). The expression patterns of the Dectin-1 cluster were analyzed via the ONCOMINE and GEPIA databases. We evaluated the prognostic value of the Dectin-1 cluster in patients with LUAD using the Kaplan-Meier plotter and GEPIA. Differential expression was validated with the EMBL-EBI database, and protein expression was analyzed with the HPA database. In addition, protein-protein interaction network, GO, and KEGG analyses were conducted. Finally, the correlations between CLEC-12A and immune molecules (immune inhibitors and MHC molecules) were investigated via TISIDB and GEPIA. The expression levels of Dectin-1 cluster genes were downregulated in LUAD tissues compared to those in normal lung tissues. The expression levels of CLEC-12A, CLEC-12B, CLEC-2, and CLEC-9A correlated with tumor stage, and CLEC-12A and CLEC-12B were significantly associated with survival in patients with LUAD. The seven genes mostly participated in immune regulation processes and were involved in autoimmune disorders and hematological malignancies. Finally, correlation analyses revealed CLEC-12A expression was associated with most immune inhibitors and MHCs. CLEC-12A was positively related to PD-1, PD-L1, PD-L2, CTLA4, TIM3, and LAG3. In conclusion, our findings suggest that CLEC-12A and CLEC-12B can be used as prognostic biomarkers in LUAD. CLEC-12A expression was associated with immune checkpoint molecules, and CLEC-12A may be a potential assistant target to improve the efficacy of immune checkpoint inhibitors immunotherapy.
Keywords: CLEC-12A; CLEC-12B; Lung cancer; Prognosis; Immune regulation
C-type lectin receptors (CLRs), which are mostly expressed by monocytes, dendritic cells, macrophages, and neutrophils, are a large family of pattern recognition receptors (PRRs) that possess diverse essential functions, including immune function regulation (Brown et al., 2018; Geijtenbeek and Gringhuis, 2016; Takeuchi and Akira, 2010). CLRs are involved in immune processes not only by recognizing antigens and presenting them to T cells but also by activating inflammatory signaling pathways, including the NF-κB pathway and Type I interferon pathway (Chiffoleau, 2018). The Dectin-1 cluster, one subgroup of CLRs, comprises seven receptors: CLEC-12A (MICL), CLEC-12B, CLEC-2 (CLEC-1B), CLEC-9A, CLEC-1A (MelLec), CLEC-7A (Dectin-1) and OLR1 (CLEC-8A) (Tone et al., 2019). The Dectin-1 cluster has a broad range of ligands and is involved in numerous pathophysiological processes, including infections, allergies, autoimmunity, inflammation, and cancer (Del Fresno et al., 2018; Hadebe et al., 2018; Plato et al., 2013; Seifert et al., 2015). In some types of human malignancies, Dectin-1 cluster members have been demonstrated to play an important role in previous studies (Chiba et al., 2014; Dambuza and Brown, 2015). CLEC-7A is involved in the recognition of N-glycan present on the surface of cancer cells, triggering anti-tumor immunity (Chiba et al., 2014). CLEC-12A has recently been identified as a potential prognostic biomarker in human acute myeloid leukemia (Morsink et al., 2019) and implicated in the regulation of T cell immunity (Krawczyk et al., 2019). A study by Derpoorter et al. showed CLEC-12B was a candidate cancer predisposition gene (Derpoorter et al., 2019). CLEC-2 has been shown to promote tumor metastasis, according to previous studies (Rayes et al., 2019; Suzuki-Inoue, 2018). CLEC-1A and CLEC-9A have been implicated in the tumor immune response and may represent a novel therapeutic direction to modulate the immune response in cancer settings (Picco et al., 2014; Thebault et al., 2009; Zeng et al., 2018). In addition, CLEC-7A and OLR1 also participate in tumorigenesis, progression, metastasis, and the peri-tumoral immune response in human pancreatic cancer, breast cancer, renal cancer, and other cancers (Daley et al., 2017; Wang et al., 2017; Xia et al., 2016; Yang et al., 2020).
Based on previous studies, it is well-founded to speculate that the seven members of the Dectin-1 cluster may similarly play an important role in human LUAD. To date, little is known about the expression pattern, functional role, and prognostic value of these members in human LUAD. Recently, biomedical studies have been revolutionized with the rapid development of sequencing technology. To the best of our knowledge, bioinformatics analyses have not yet been applied to explore the role of these members in human malignancies. Hence, we aimed to focus on the Dectin-1 cluster in this study, analyzing in detail the expression pattern and prognostic value of the seven members in human LUAD based on the great amount of gene expression data published online with clinical characteristics via bioinformatics analysis methods. In addition, we conducted functional bioinformatics analyses to gain deeper insight into the biological function, interaction, and association with diseases of the seven genes. Finally, we performed individual analyses of CLEC-12A and CLEC-12B to provide information for future research.
All of the datasets in this study are from published literature, and it was confirmed that all written informed consent was obtained.
The transcription levels of the Dectin-1 cluster in different cancers were analyzed with the ONCOMINE database (https://www.oncomine.org/), an online cancer microarray database and integrated data-mining platform (Rhodes et al., 2007). The cutoff p-value and fold change were 0.01 and 2, respectively.
Analysis of differential expression by using GEPIA and GEO datasets
All of the datasets in this study are from published literature, and it was confirmed that all written informed consent was obtained. The online Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) (Tang et al., 2017) database was used to validate the mRNA expression of Dectin-1 cluster genes by analyzing the data of 9,736 tumors and 8,587 normal samples from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) projects, respectively. Using the GEPIA database, we compared the expression levels of the seven genes in LUAD tissues with those in normal lung tissues. The statistical method for differential analysis was one-way ANOVA, using disease state (Tumor or Normal) as a variable to calculate differential expression. Transcripts per kilobase million (TPM) was used as the observed value to compare gene expression levels. The log2-fold change and p-value cutoff were 1 and 0.01, respectively. We used log2 (TPM + 1) for the log scale, and the Jitter size was 0.4. In addition, we performed survival analyses on patients with LUAD according to the transcriptional expression levels of the Dectin-1 cluster. The median expression value was selected as the group cutoff. The association between Dectin-1 cluster gene expression and tumor stage was preliminarily explored in our study. The statistical method was one-way ANOVA, using the pathological stage as a variable to calculate differential expression. Besides, we conducted expression difference analysis through three datasets (GSE118370, GSE85841, and GSE140797) from the GEO database by using GEO2R online tools.Results were not shown because the reliability of the results is questionable due to the small sample size of the data.
Kaplan-Meier plotter database analysis
The Kaplan-Meier plotter was able to assess the effects of 54k genes on survival in 21 cancer types. The lung (sample number = 3,452) datasets were some of the largest datasets in this database (Nagy et al., 2018). The correlation between Dectin-1 cluster gene expression and survival of patients with LUAD was analyzed by the Kaplan-Meier plotter (http://kmplot.com/analysis/) (Gyorffy et al., 2013).
Both overall survival (OS) and progression-free survival (PFS) were the focus of our analysis. The hazard ratio (HR) with a 95% confidence interval (CI) and log-rank P-value were also computed. We selected the best expression cutoff to distinguish survival.
Correlation analysis between genes expression and clinical characteristics by LinkedOmics
LinkedOmics is a publicly available portal that includes multi-omics data from all 32 TCGA Cancer types and allows users to access, analyze, and compare cancer multi-omics data (http://www.linkedomics.org/login.php) (Vasaikar et al., 2018). By using the online tool, we performed correlation analysis between CLEC-12A as well as CLEC-12B expression and various clinical characteristics, including tumor purity, pathologic stage, tumor TNM stage, radiation therapy, patients’ years to birth, and race and ethnicity. The statistical method was non-parametric tests.
Protein-Protein Interaction Network (PPIN) and module analysis
The PPI network was predicted using the Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org) online database (Szklarczyk et al., 2019). The seven Dectin-1 cluster genes had been submitted to the STRING database. All PPI pairs with a combined score of >0.4 were extracted, and the maximum number of interactors to show was no more than ten. Subsequently, DisGeNET diseases-genes, the Kyoto Encyclopedia of Genes and Genomes (KEGG), and gene ontology (GO) analyses for the ten genes in this module were performed using Enrichr (http://amp.pharm.mssm.edu/Enrichr/) (Kuleshov et al., 2016).
EMBL-EBI transcriptional expression validation and STRING protein expression
The European Molecular Biology Laboratory (EMBL)-European Bioinformatics Institute (EBI) expression atlas database (https://www.ebi.ac.uk/gxa) (Papatheodorou et al., 2018) was used to validate transcriptional expression. We validated the expression of CLEC-12A and CLEC-12B because our results showed these two genes may be more closely related to the survival of patients with LUAD. In addition, we explored CLEC-12A protein expression and conducted survival analyses according to the transcriptional expression level of CLEC-12A in patients with LUAD via the Human Protein Atlas (HPA) database (https://www.proteinatlas.org/).
TISIDB and GEPIA correlation analysis
TISIDB (http://cis.hku.hk/TISIDB/index.php) (Ru et al., 2019) is a web portal for tumor and immune system interaction that integrates multiple heterogeneous data types and includes genomics, transcriptomics, and the clinical data of 30 cancer types from TCGA. We used TISIDB to analyze the correlation between the expression of CLEC-12A and immune inhibitors/MHC molecules across human cancers. GEPIA correlation analysis provides pair-wise gene expression correlation analysis for given sets in specific types of cancers. Using this tool, we explored the correlation between the expression of CLEC-12A and PD-1 (PDCD1), PD-L1 (CD274), PD-L2 (PDCD1L2), CTLA4, TIM3 (HAVCR2), and LAG3 in human LUAD. We also performed the above analysis on the CLEC12B gene, but there was insufficient data to determine a correlation (data are not shown).
CBioPortal mutation analysis and gene co-expression analysis
We selected four datasets to further analyze Dectin-1 cluster gene mutations in patients with LUAD using cBioPortal (http://www.cbioportal.org). According to the cBioPortal’s online instructions, we obtained the mutation oncoprint and mutation site profile. Using the ONCOMINE online filter of co-expression analysis, we found the top 20 co-expression genes of CLEC-12B.
Transcriptional expression levels of Dectin-1 cluster in patients with lung cancer
To determine the transcriptional differences of Dectin-1 cluster genes in tumor and normal tissues, the mRNA levels of the seven genes in the tissues of multiple cancer types and normal tissues were analyzed with the ONCOMINE database. The mRNA expression levels of CLEC-12B, CLEC-12A, CLEC-1A, CLEC-7A, and OLR1 were downregulated in patients with lung cancer in multiple datasets (Fig. 1). CLEC-2 and CLEC-9A mRNA expression differences were inconclusive in patients with lung cancer but were downregulated in other types of cancer (Fig. 1).
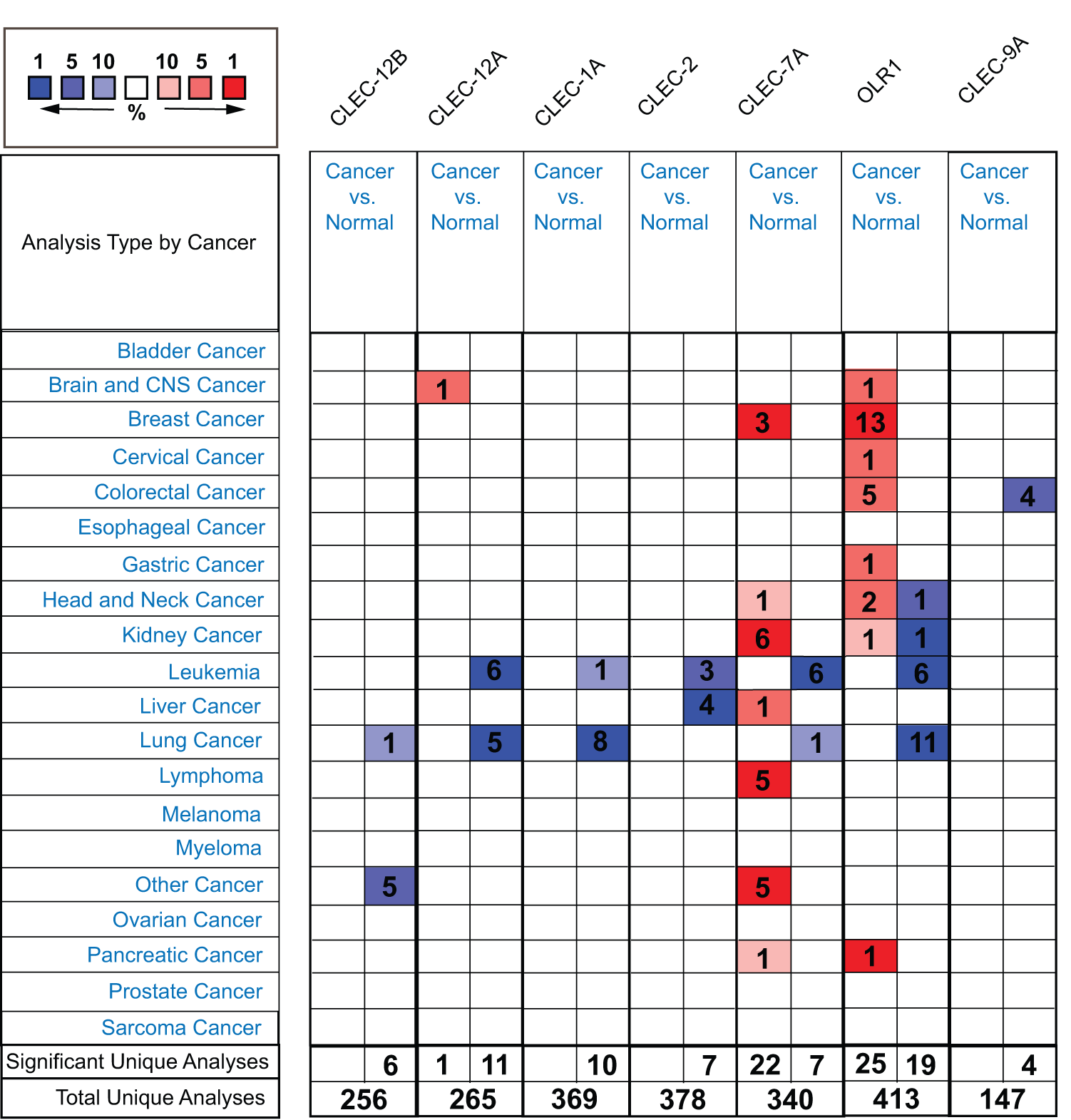
Figure 1: The transcription levels of Dectin-1 cluster genes in different types of cancers (ONCOMINE). The red boxes represent the gene expression level was higher in cancer vs normal tissues, and the blue boxes represent the gene expression level was lower. The numbers in the boxes represent the number of studies.
To further explore the transcriptional differences of these genes in LUAD and normal lung tissues, the GEPIA database was used. This analysis revealed that the expression levels of the seven genes were significantly downregulated in LUAD tissues compared to those in normal lung tissues, although the expression difference of CLEC-9A was not statistically significant (Fig. 2). Then, we analyzed the expression of the seven genes in different stages of LUAD. The results showed that the CLEC-12B, CLEC-12A, CLEC-2, and CLEC-9A groups significantly varied, whereas the CLEC-1A, CLEC-7A, and OLR1 groups did not significantly differ (Fig. 3).
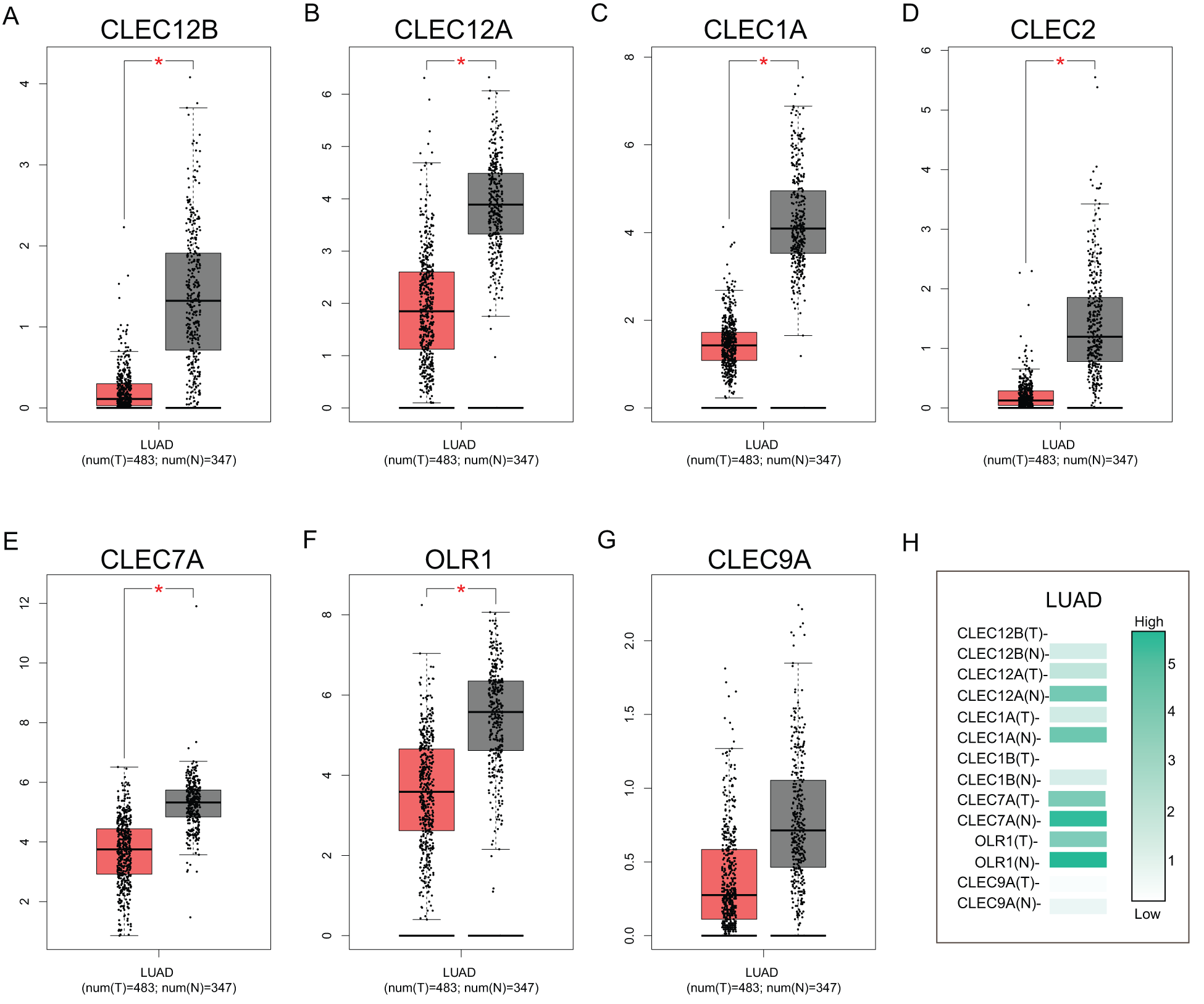
Figure 2: The expression of Dectin-1 cluster genes in lung adenocarcinoma (LUAD) (GEPIA). The expression of CLEC-12B, CLEC-12A, CLEC-1A, CLEC-2, CLEC-7A, OLR1, and CLEC-9A in LUAD (A, B, C, D, E, F, G). Red columns represent the tumors; gray columns represent normal tissues; and the Y-axis of A to G was -log2 (TPM + 1) representing genes expression levels. The expression heatmap of Dectin-1 cluster genes in LUAD (H).
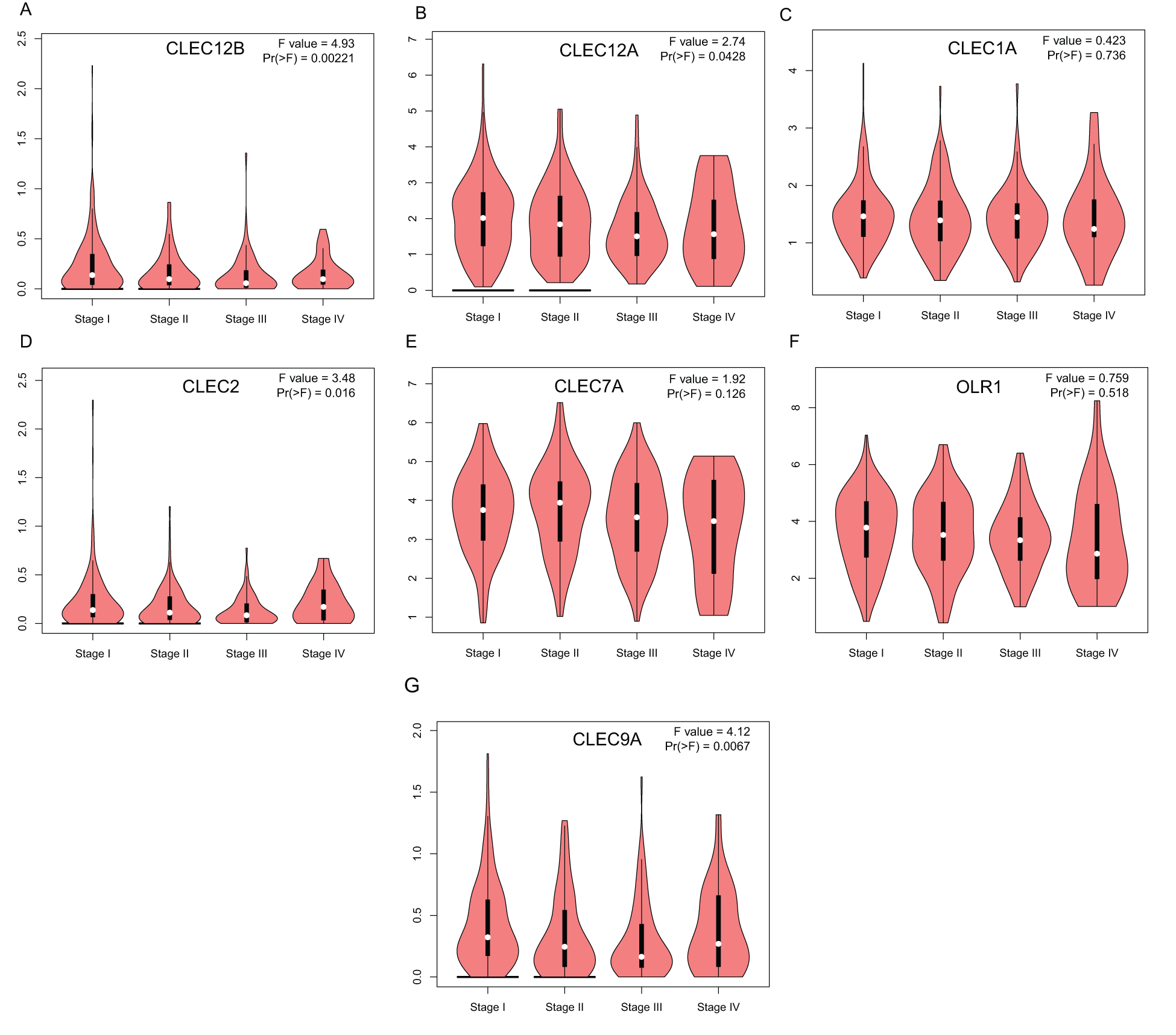
Figure 3: Correlation between Dectin-1 cluster genes expression and tumor stage in LUAD patients (GEPIA). Y-axis was -log2 (TPM + 1) representing gene expression levels; and X-axis was tumor stage.
Prognostic potential of Dectin-1 cluster genes in human LUAD
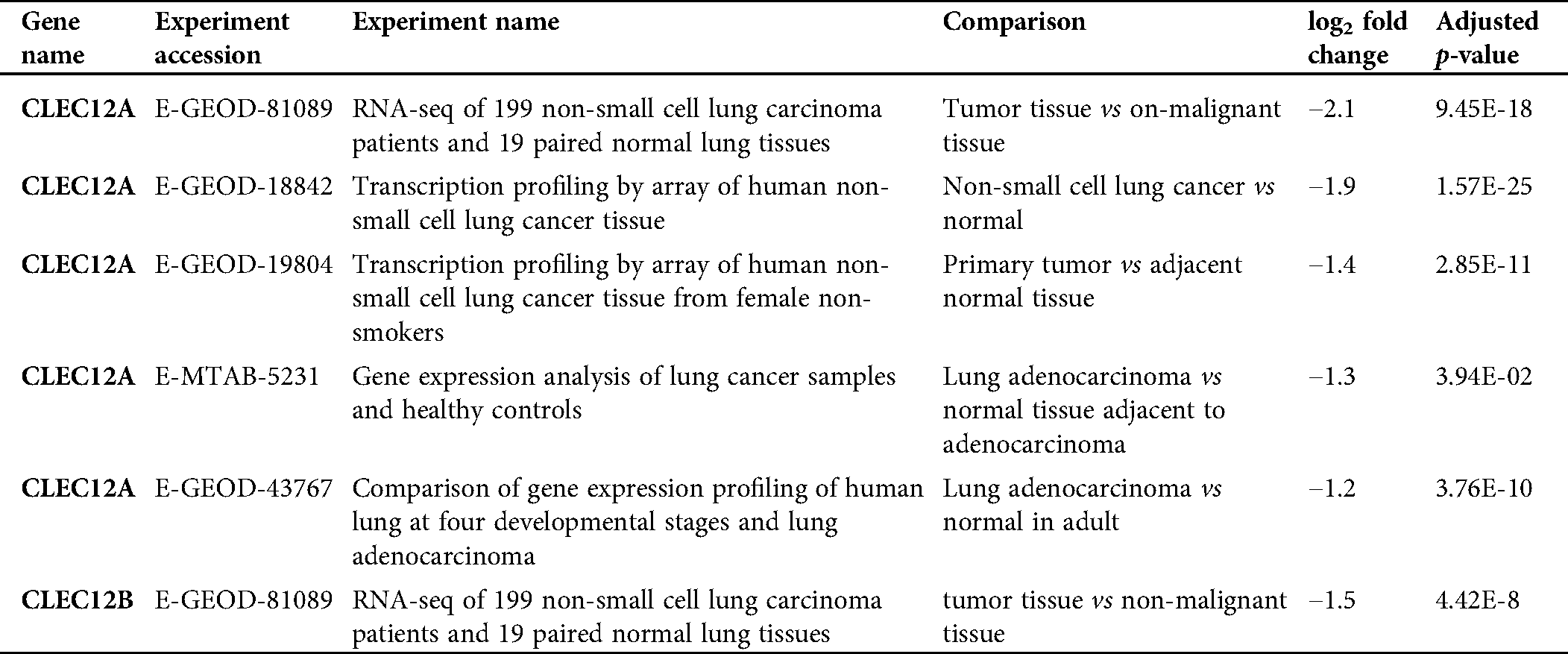
We further explored the association of the expression of the seven genes with the prognosis of patients with LUAD. According to the results from the GEPIA survival tools, the CLEC-12B, CLEC-12A, CLEC-1B, CLEC-7A, and CLEC-9A mRNA levels were significantly associated with OS in patients with LUAD (Fig. 4A and Supplementary Fig. 1). The CLEC-12B expression level was also associated with disease-free survival. According to the results of the Kaplan-Meier Plotter tools, higher mRNA expression levels of CLEC-12B, CLEC-12A, CLEC-1A, and CLEC-7A were significantly associated with better OS and PFS (Fig. 4A and Supplementary Fig. 1). To summarize, LUAD patients with higher mRNA levels of CLEC-12B, CLEC-12A, and CLEC-7A were predicted to have longer OS and PFS. Then we integrated the three genes as a signature and performed survival analysis according to the signature (Fig. 4B). The results showed that the predictive value of the combined signature for OS and disease-free survival (DFS) was more significant than that of single genes of the three. Because the evidence above demonstrated CLEC-7A was not associated with tumor stage, we only considered CLEC-12A and CLEC-12B as candidate biomarkers and further validated their expression in patients with LUAD via another database. Using 1 as the log2-fold change and 0.01 as the P-value, the transcriptional expression levels of CLEC-12A and CLEC-12B in lung cancer were significantly lower than those in normal lung tissues (Tab. 1). The CLEC-12A protein expression level was from undetected to low in LUAD tissues (Fig. 5A). Higher transcriptional expression of CLEC-12A predicted longer OS in patients with LUAD (Fig. 5B). The HPA database had insufficient CLEC-12B protein expression data. Furtherly, the results of the correlation analysis between CLEC-12A as well as CLEC-12B expression and clinical characteristics showed that CLEC-12A and CLEC-12B expression were associated with tumor purity and stage. And there was no significant correlation between the two genes expression and other clinical characteristics.
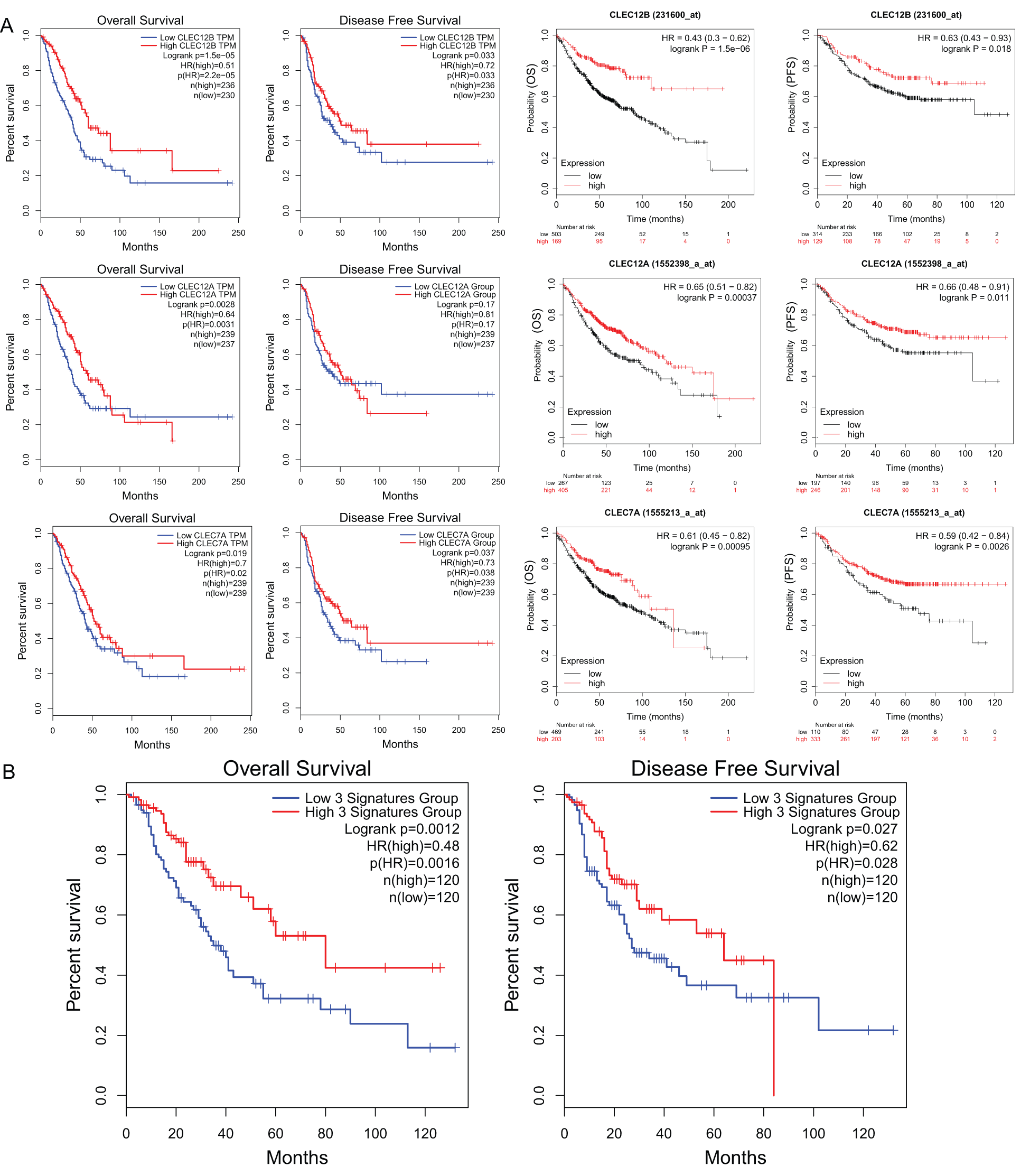
Figure 4: The prognostic value of mRNA level of CLEC-12A, CLEC-12B, and CLEC-7A in LUAD patients (GEPIA and Kaplan-Meier Plotter).
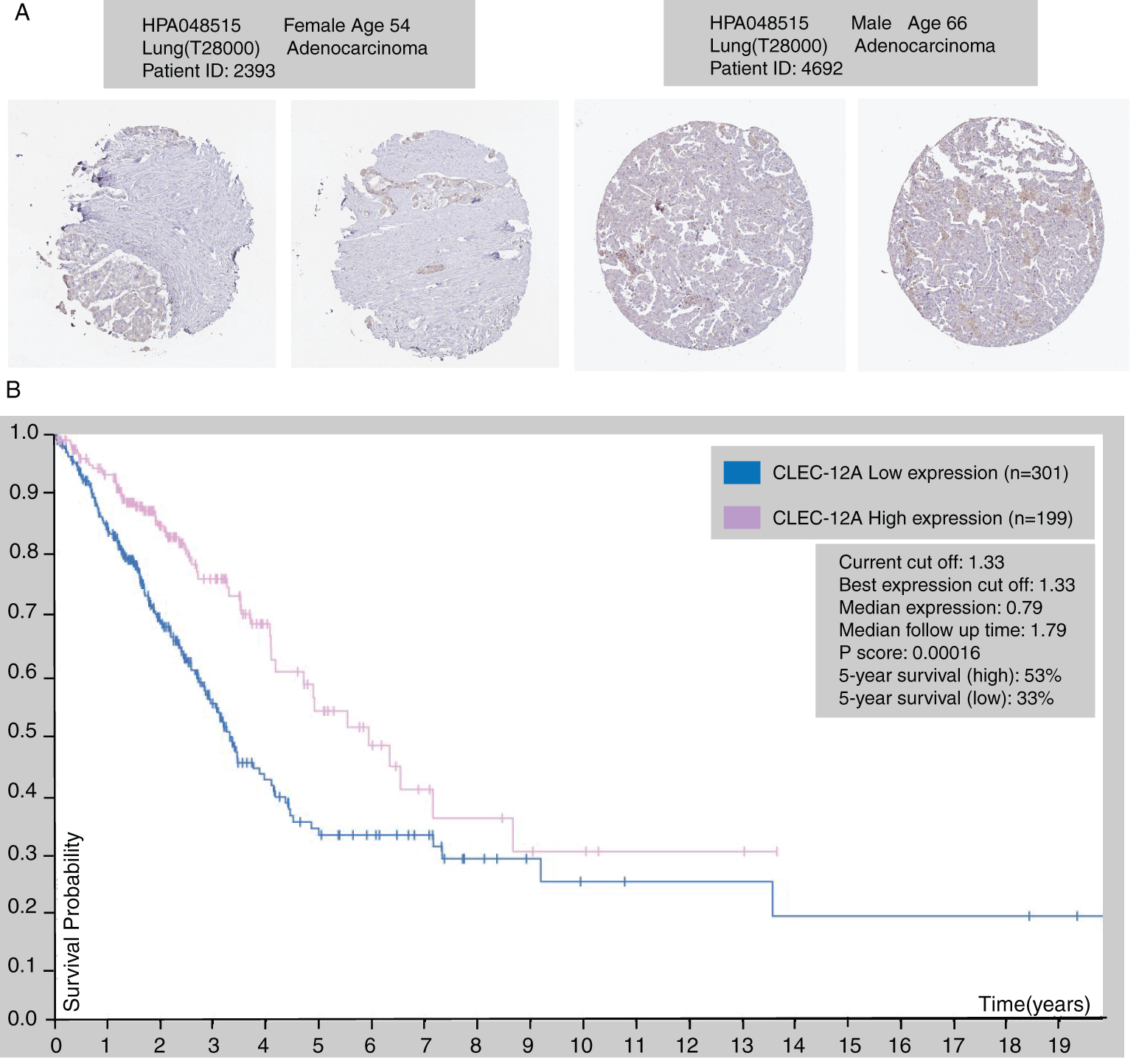
Figure 5: The immunohistochemical protein expression of CLEC-12A in LUAD tissue (HPA database) (A). Kaplan-Meier survival curve of LUAD patients according to the transcriptional expression of CLEC-12A (B).
Table 1: Transcriptional differential expression of CLEC12A and CLEC12B in lung cancer tissue versus normal lung via analyzing by EMBL-EBI database.
Protein-Protein Interaction Network (PPIN) construction and molecular analysis
The STRING database was adopted to determine the PPI pairs among the seven proteins and ten interactors predicted by the STRING database. As revealed in Fig. 6A, 17 nodes (genes) and 53 edges (interactions) were established in the constructed PPIN (PPI enrichment p-value = 1.11e-16). The 17 nodes include the seven Dectin-1 cluster genes and their ten most related molecules: PLCG-2, PDPN, LCP-2, SYK, APOB, ITGAM, CD33, CLEC-5A, CD47, and CLEC-4D. The proteins were clustered to three clusters by k-means clustering. Diseases-genes association data from the DisGeNET database showed that the top 5 diseases related to the seventeen genes were rheumatoid arthritis, chronic lymphocytic leukemia, splenic marginal zone B-cell lymphoma, lymphoma, and extranodal NK/T-cell lymphoma in descending order (Fig. 6B).
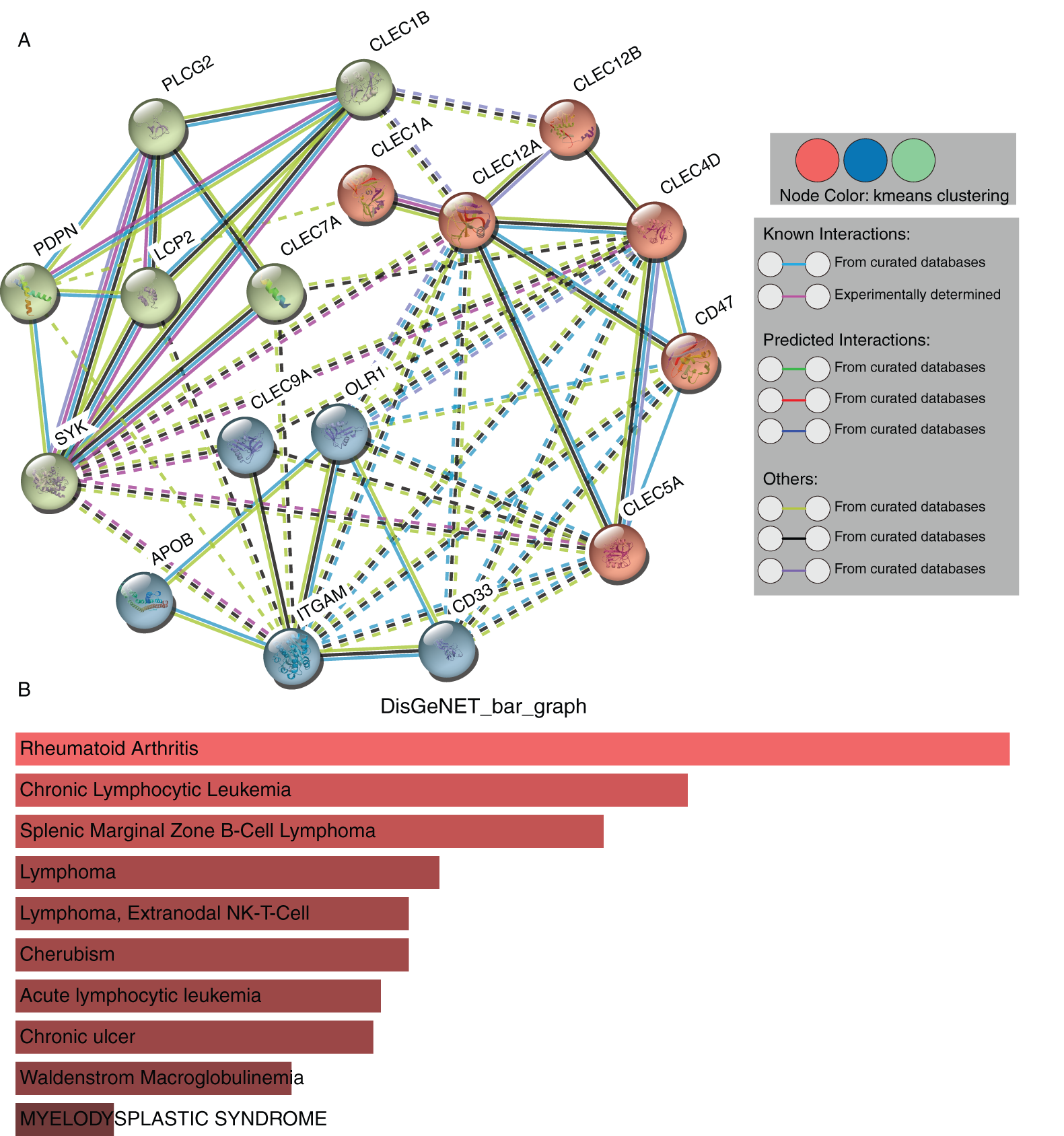
Figure 6: PPI network construction and module analysis of the seven proteins (STRING). Protein-protein interaction network of Dectin-1 cluster proteins and ten associated proteins (A). Related human diseases analysis of Dectin-1 cluster proteins and ten associated proteins (B).
GO annotation and KEGG pathway enrichment analyses
To obtain deeper insight into the biological roles of the seven genes and ten associated genes, we used the Enrichr database to conduct GO annotation and KEGG pathway enrichment analyses. The top 10 enriched GO terms and KEGG pathways are listed in Fig. 7. The GO biological process analysis revealed that the 17 genes were mainly enriched in immune function regulation pathways, including neutrophil activation involved in the immune response, neutrophil degranulation, neutrophil-mediated immunity, the innate immune response activating the cell surface receptor signaling pathway, and positive regulation of myeloid leukocyte mediated immunity (Fig. 7A). For GO cellular component analysis, the top four significantly enriched terms were tertiary granule membrane, specific granule membrane, specific granule, and tertiary granule (Fig. 7B). The top four significantly enriched molecular function terms included phosphotyrosine residue binding, protein phosphorylated amino acid binding, cholesterol transporter activity, and chemokine binding (Fig. 7C). In addition, the top five markedly enriched pathways for these 17 genes were the C-type lectin receptor signaling pathway, the Fc epsilon RI signaling pathway, platelet activation, osteoclast differentiation, and natural killer cell-mediated cytotoxicity (Fig. 7D).
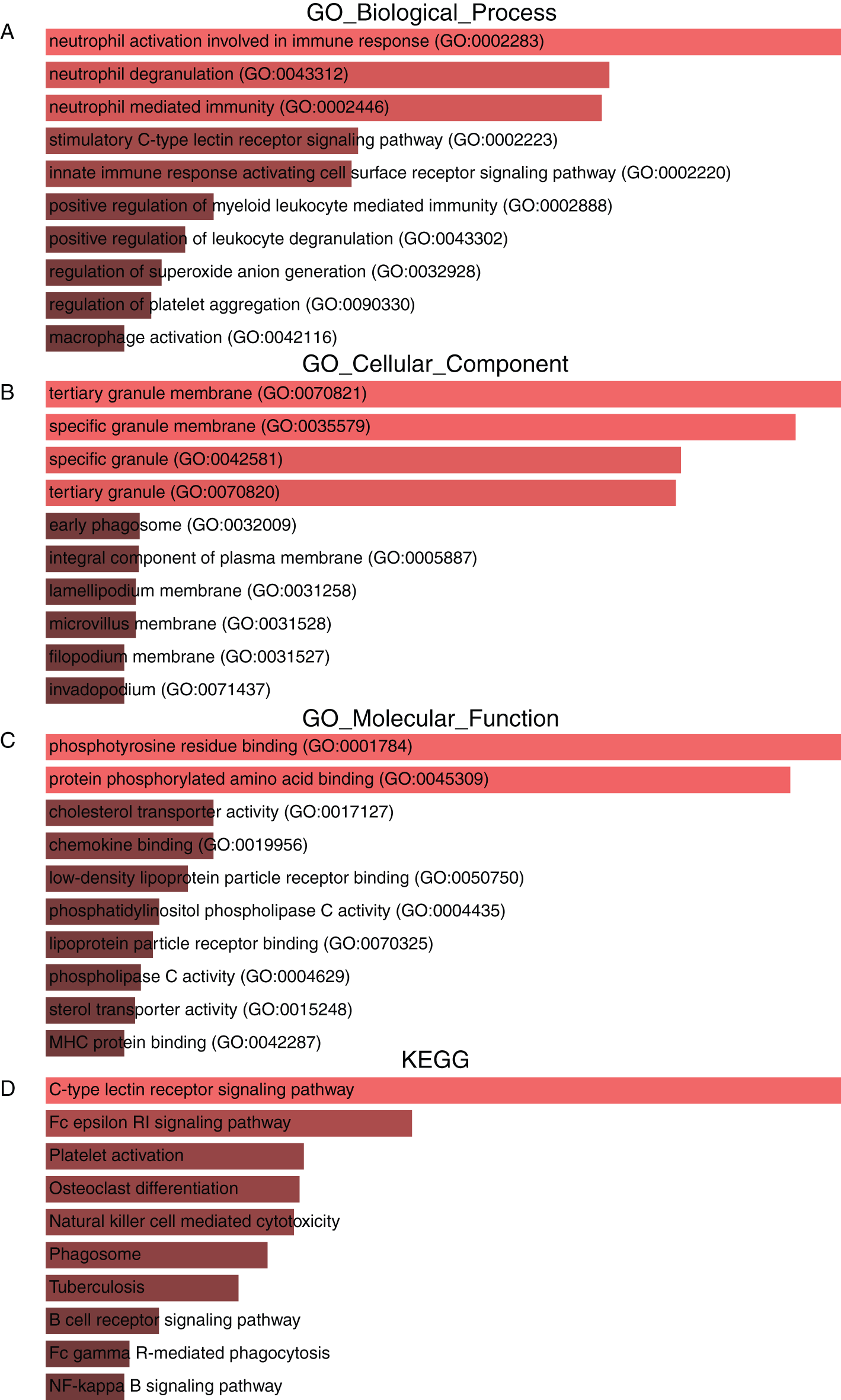
Figure 7: GO annotation and KEGG pathway enrichment analysis of Dectin-1 cluster genes and ten associated genes. The top 10 enriched GO biological process (A), cellular component (B) and molecular function (C) terms, as well as KEGG pathways (D).
Correlation analysis between expression of CLEC-12A and immune inhibitors/MHC molecules across human cancers
To investigate the internal mechanism behind the positive prognostic value of CLEC-12A and provide significant direction for its clinical application, we explored the correlation between CLEC-12A and immune inhibitors/major histocompatibility complex (MHC) molecules across human cancers with the TISIDB online tool. The results indicated that the expression of CLEC-12A positively correlated with multiple immune inhibitors and MHC molecules across human cancers (Figs. 8A–8B). Furthermore, we analyzed the correlation between CLEC-12A and six immune checkpoint molecules that have been developed for the clinical practice: PD-1 (PDCD1), PD-L1 (CD274), PD-L2 (PDCD1L2), CTLA-4, TIM-3 (HAVCR2), and LAG-3. We chose TCGA LUAD tumor data for analysis, and the expression of CLEC-12A was positively correlated with all six immune checkpoint molecules (Fig. 8C). There was insufficient data to determine a correlation between CLEC-12B and the above molecules (data are not shown).
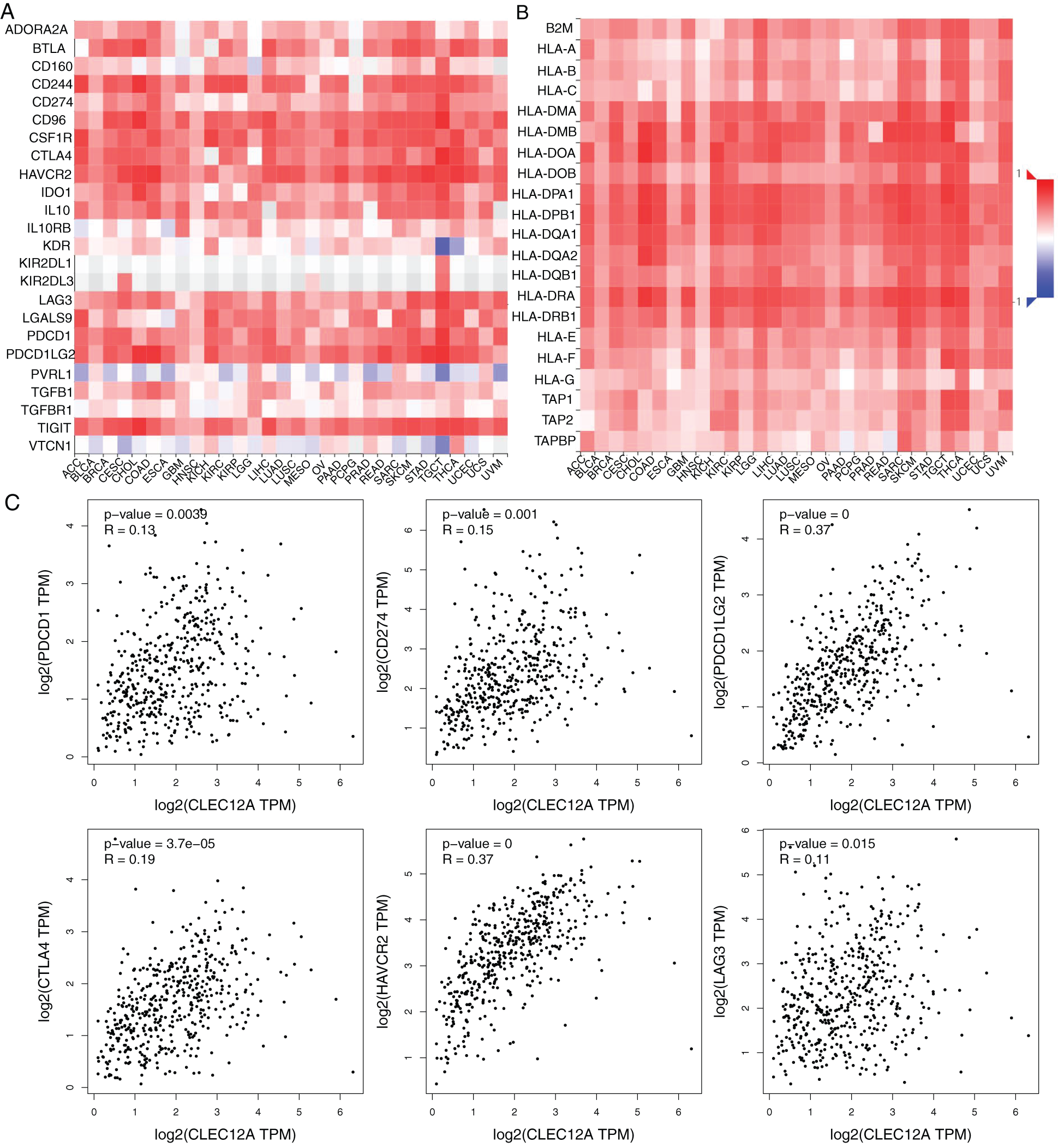
Figure 8: Correlation analysis between expression of CLEC-12A and Immunoinhibitors/MHC molecules across human cancers (TISIDB and GEPIA). Spearman correlation between expression of CLEC-12A and Immunoinhibitors across multiple human cancers (A). Spearman correlation between expression of CLEC-12A and MHC molecules across multiple human cancers (B). Pearson correlation between expression of CLEC-12A and PD-1(PDCD1), PD-L1(CD274), PD-L2(PDCD1L2), CTLA-4, TIM-3(HAVCR2), LAG-3 in human LUAD (C).
Mutation analysis and gene co-expression analysis of CLEC-12B
To search for some new clues to investigate the internal mechanism behind the positive prognostic value of CLEC-12B, we conducted mutation analyses and gene co-expression analyses. CLEC-12B was altered in 20 samples of the 735 patients with lung adenocarcinoma (2.7%). Amplification and deep deletion of the seven genes usually co-occurred in patients with LUAD (Fig. 9A). The known mutation sites on cBioPortal were N100K, K188N, and W217L (Fig. 9B). The results of co-expression analysis showed that CLEC-12B significantly correlated with other Dectin-1 cluster genes and other molecules, including GABARAPL1, KLRD1, KLRK1, KLRC4, KLRC1, KLRC3, C12orf59, TAS2R7, TAS2R8, TAS2R9, CSDA, PRH2, and TAS2R14 (Fig. 9C).
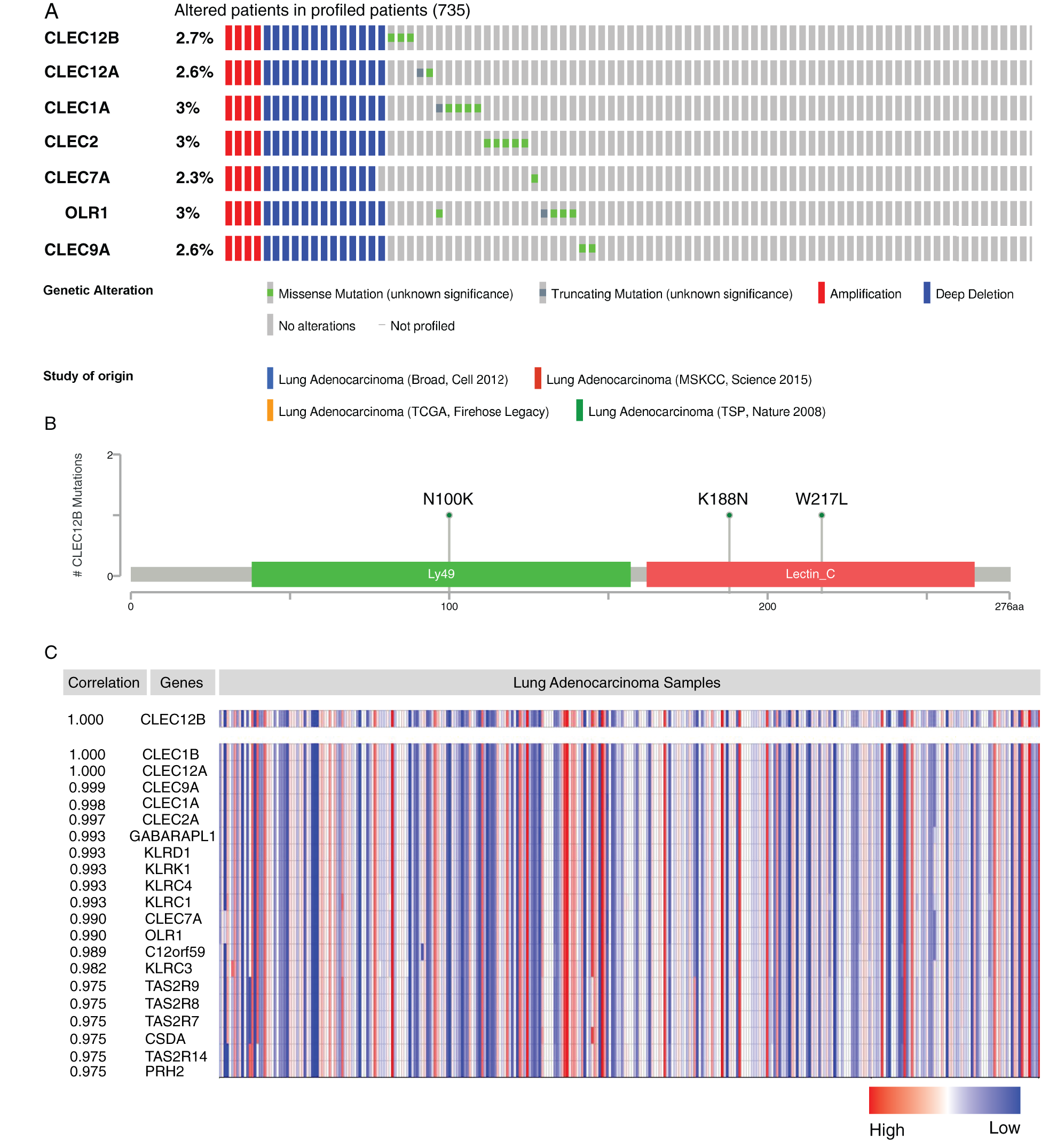
Figure 9: Mutation analysis and co-expression analysis of CLEC-12B (cBioPortal and ONCOMINE). Mutation Oncoprint in LUAD patients (cBioPortal) (A). Mutation sites of CLEC-12B (cBioPortal) (B). Top 20 co-expression genes of CLEC-12B (ONCOMINE) (C).
Dysregulation of the seven Dectin-1 cluster genes has been reported in previous studies in many cancers (Bakker et al., 2004; Chen et al., 2017; Daley et al., 2017; Derpoorter et al., 2019; Mattiola et al., 2019; Seifert et al., 2015; L. Wang et al., 2016; Xia et al., 2016). To our knowledge, this study is the first to explore the mRNA expression and prognostic values of all seven genes in LUAD. The seven Dectin-1 cluster members may have important roles in immune function regulation and tumor progression. Of the seven genes, OLR1 has an important effect on progression and migration for multiple cancers, including breast cancer, pancreatic cancer, and prostatic cancer (Gonzalez-Chavarria et al., 2018; Wang et al., 2017; Xiong et al., 2019; Zhang et al., 2018), but downregulated OLR1 expression was not related to the prognosis of patients with LUAD according to our results. The role of OLR1 in human LUAD requires more experimental validation.
CLEC-12A, also known as C-type lectin-like molecule-1 (CLL-1), MICL, and CD371, is the most studied gene in human cancers. CLEC-12A has diverse functions, such as cell adhesion, cell-cell signaling and roles in inflammation and the immune response. CLEC-12A was identified as an activating receptor that can amplify Type I interferon responses (Li et al., 2019). CLEC-12A-mediated tumor antigen delivery, uptake and presentation through dendritic cells can efficiently induce CD4+ and CD8+ T cell activation and boost anti-tumor T cell responses (Hutten et al., 2016). Hence, low expression of CLEC-12A in LUAD patients may lead to malfunction of tumor antigen processing and blocking of T cell activation process. Through the mechanism, tumor immune escape and progression may be achieved. Consequently, LUAD patients with lower expression of CLEC-12A showed worse survival and prognosis outcomes. It was also reported that low levels of CLEC-12A expression on leukemic blasts were independently associated with worse survival outcomes and a lower likelihood of complete remission after chemotherapy (Wang et al., 2017). Finally, our investigation showed that expression of CLEC-12A was related to various immune inhibitors and MHCs. Interestingly, six immune checkpoint molecules (PD-1, PD-L1, PD-L2, CTLA4, TIM3 and LAG3), which are the most studied and most widely used in cancer clinical practice, were also related to the expression of CLEC-12A. CLEC-12A receptor plays an important role in tumor antigen delivery, uptake and presentation as well as T cell activation. Higher expression of CLEC-12A may boost activation and generation of an anti-tumor immune response. Tumor immune escape and anti-tumor immune response are processes of competition. Enhancement of anti-tumor response may lead to tumor regression. But in turn, increased T cells activity may also induce the expression of immune-inhibitor molecules on the surface of tumor cells. For example, tumor PD-L1 expression can inhibit T cell activation through the immune checkpoint pathway. Hence, the level of CLEC-12A expression may be correlated with that of immune checkpoint molecules. It is well studied that higher expression of tumor PD-L1 was associated with better efficacy of immune checkpoint inhibitors (ICIs) for lung cancer patients (Havel et al., 2019). Increasing CLEC-12A expression may show an assistant effect on the efficacy of ICIs immunotherapy for LUAD patients. And a study showed that CLEC-12A and TIM3 may have a synergistic effect on the elimination of acute myeloid leukemia (Darwish et al., 2016). More work is required to further investigate the possible association between CLEC-12A and immune checkpoint molecules.
CLEC12B is a C-type lectin-like receptor expressed on myeloid cells. A recent report showed porcine CLEC-12B is expressed on alveolar macrophages and blood dendritic cells (Nieto-Pelegrin et al., 2020). Besides, it was proposed that CLEC12B was an inhibitory receptor on NK cells to counteract NKG2D (a kind of NK cell receptor) -mediated immune activation (Hoffmann et al., 2007). Our results of GO and KEGG analysis also showed CLEC-12B play roles in immune function. But in a word, little was known about the functional role of CLEC-12B. Only one study identified that the CLEC-12B variant, c.790G > A; p.E264K, was a candidate cancer predisposition variant (Derpoorter et al., 2019). Maybe, the protective action of CLEC-12 in LUAD patients in our results was achieved through its roles in immune function. More work is required to define the ligands and function of CLEC-12B. We showed the mutation frequency and sites of CLEC-12B, as well as the top 20 co-expression genes, which provide clues for further research.
CLEC-1A is able to recognize antigens and plays crucial roles in immunity and homeostasis. The study by Thebault et al. reported that CLEC-1A is enhanced by IL10 and TGF-beta and moderated Th17 cell differentiation (Thebault et al., 2009). In our results, the expression of CLEC-1A in LUAD tissues was significantly lower than that in normal lung tissues. Moreover, survival analysis using the Kaplan-Meier Plotter showed that low levels of CLEC-1A expression were associated with worse survival outcomes (OS and PFS), although the results from the GEPIA survival analysis tools showed that low levels of CLEC-1A expression were not related to OS.
CLEC-7A, also known as Dectin-1, can bind endogenous ligands expressed on T cells to foster activation of dendritic cells and T cells to enhance cytokine secretion (Leibundgut-Landmann et al., 2008). Moreover, Dectin-1 has been identified as an inducible factor of IL-33 expression in dendritic cells and an activator of anti-tumor immunity, which may provide new targets to improve dendritic cell immunotherapy for cancer patients (Chen et al., 2018; Wang et al., 2018; Zhao et al., 2016). In this study, the expression of CLEC-7A in LUAD tissues was significantly lower than that in normal lung tissues, and a lower level of expression was correlated with poorer OS and PFS, which seemed consistent with the role of CLEC-7A as an activator of anti-tumor immune responses.
CLEC-2 is able to be upregulated by cordycepin, a 3’-deoxyadenosine drug, to inhibit the proliferation, migration, and invasive activities of gastric cancer cells via the Akt signaling pathway (L. Wang et al., 2016; Y. Wang et al., 2019). In addition, histidine-rich glycoprotein was able to decrease PD-1 expression in natural killer (NK) cells to augment the anti-tumor cytotoxicity of NK cells, which depended on the activity of CLEC-2 (Nishimura et al., 2019). Consequently, increasing the expression of CLEC-2 and enhancing its activity may be a novel method to improve NK cell immunotherapy. This finding is exactly consistent with our conclusion that the expression of CLEC-2 in LUAD tissues was significantly lower than that in normal lung tissues, and a lower level of expression was correlated with higher tumor stage and shorter OS, although CLEC-2 expression could not predict survival according to the Kaplan-Meier Plotter analysis.
CLEC-9A, also known as DNGR-1, may promote immune tolerance to autologous antigens (Zelenay et al., 2012). CD4+ T cells can transfer to FOXP3+ Treg cells by targeting CLEC-9A with an antibody to induce immune tolerance (Joffre et al., 2010). However, other studies reported contrasting findings that targeting CLEC-9A can induce an immune response (Caminschi et al., 2008; Picco et al., 2014). In our study, the expression difference of the CLEC-9A gene in LUAD tissues compared to that in normal lung tissues was not significant. Nevertheless, the expression of CLEC-9A was significantly different in different tumor stages, and patients with LUAD and a lower level of CLEC-9A expression had longer OS, according to the GEPIA. However, the association of CLEC-9A expression with survival was not validated in the Kaplan-Meier Plotter. As a result, the function of CLEC-9A and its roles in human LUAD require more experimental validation.
In this study, we systemically showed the expression pattern and prognostic role of the seven Dectin-1 cluster genes in patients with LUAD; provided a thorough understanding of complex biological function. Our results indicated that the lower level of expression of CLEC-12A, CLEC-12B, and CLEC-7A in human LUAD tissues can predict worse survival outcomes (PFS and OS). CLEC-12A and CLEC-12B can be used as prognostic biomarkers in patients with LUAD. In addition, our findings suggested that CLEC-12A is a potential therapeutic target for LUAD and might be able to influence the efficacy of immune checkpoint inhibitors in lung cancer patients. Moreover, CLEC-7A and CLEC-2 may provide novel strategies to improve dendritic cell and NK cell immunotherapy for cancer patients.
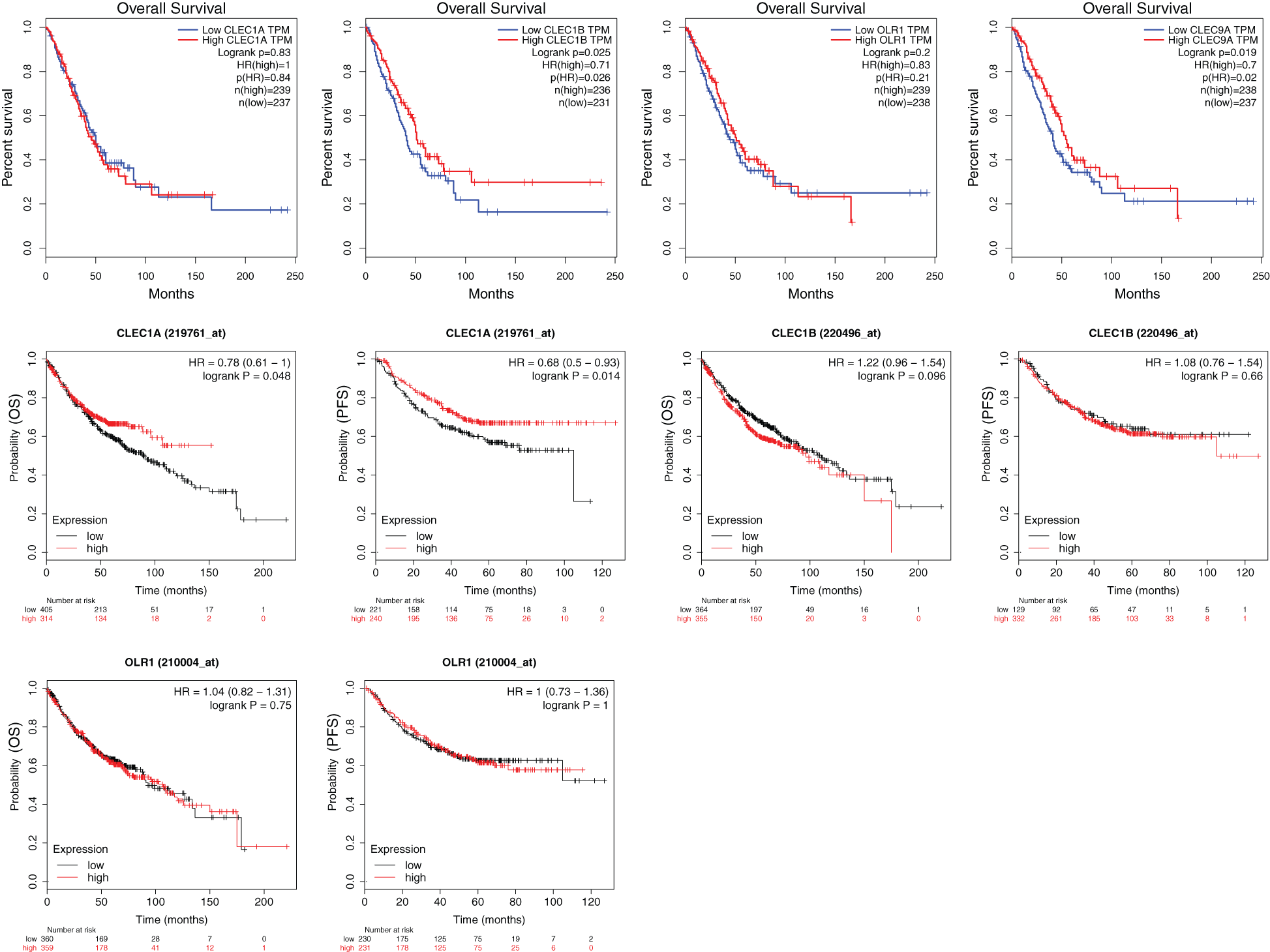
Supplementary Figure 1: The results of survival analyses based on the mRNA level of CLEC-1A, CLEC-1B, OLR1 and CLEC-9A in LUAD patients (GEPIA and Kaplan-Meier Plotter).
Author Contributions: BY and YZ made contributions to the conception and design of this work. LY and FN made contributions to manuscript drafting and the acquisition, analysis, and interpretation of the data. JZ and LJ made contributions to the revising of the work. All authors read and approved the final manuscript.
Availability of Data and Materials: All the data used in the study can be accessed in public databases used in this study.
Funding Statement: This work was supported by the China Postdoctoral Science Foundation [Grant No. 2018M643495] and Technology Department of Sichuan Province [Grant No. 2020YJ0049].
Conflicts of Interest: The authors declared that they have no conflicts of interest to report regarding the present study.
Bakker AB, van den Oudenrijn S, Bakker AQ, Feller N, van Meijer M, Bia JA, Jongeneelen MA, Visser TJ, Bijl N, Geuijen CA, Marissen WE, Radosevic K, Throsby M, Schuurhuis GJ, Ossenkoppele GJ, de Kruif J, Goudsmit J, Kruisbeek AM. (2004). C-type lectin-like molecule-1: A novel myeloid cell surface marker associated with acute myeloid leukemia. Cancer Research 64: 8443–8450. DOI 10.1158/0008-5472.CAN-04-1659. [Google Scholar] [CrossRef]
Brown GD, Willment JA, Whitehead L. (2018). C-type lectins in immunity and homeostasis. Nature Reviews Immunology 18: 374–389. DOI 10.1038/s41577-018-0004-8. [Google Scholar] [CrossRef]
Caminschi I, Proietto AI, Ahmet F, Kitsoulis S, Shin Teh J, Lo JC, Rizzitelli A, Wu L, Vremec D, van Dommelen SL, Campbell IK, Maraskovsky E, Braley H, Davey GM, Mottram P, van de Velde N, Jensen K, Lew AM, Wright MD, Heath WR, Shortman K, Lahoud MH. (2008). The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood 112: 3264–3273. DOI 10.1182/blood-2008-05-155176. [Google Scholar] [CrossRef]
Chen J, Zhao Y, Jiang Y, Gao S, Wang Y, Wang D, Wang A, Yi H, Gu R, Yi Q, Wang S. (2018). Interleukin-33 contributes to the induction of Th9 cells and antitumor efficacy by Dectin-1-activated dendritic cells. Frontiers in Immunology 9:e257. DOI 10.3389/fimmu.2018.01787. [Google Scholar] [CrossRef]
Chen XY, Chen YH, Zhang LJ, Wang Y, Tong ZC. (2017). Investigating ego modules and pathways in osteosarcoma by integrating the EgoNet algorithm and pathway analysis. Brazilian Journal of Medical and Biological Research 50: 3. DOI 10.1590/1414-431x20165793. [Google Scholar] [CrossRef]
Chiba S, Ikushima H, Ueki H, Yanai H, Kimura Y, Hangai S, Nishio J, Negishi H, Tamura T, Saijo S, Iwakura Y, Taniguchi T. (2014). Recognition of tumor cells by Dectin-1 orchestrates innate immune cells for anti-tumor responses. eLife 3: 379123. DOI 10.7554/eLife.04177. [Google Scholar] [CrossRef]
Chiffoleau E. (2018). C-type lectin-like receptors as emerging orchestrators of sterile inflammation represent potential therapeutic targets. Frontiers in Immunology 9: 21. DOI 10.3389/fimmu.2018.00227. [Google Scholar] [CrossRef]
Daley D, Mani VR, Mohan N, Akkad N, Ochi A, Heindel DW, Lee KB, Zambirinis CP, Pandian GSB, Savadkar S, Torres-Hernandez A, Nayak S, Wang D, Hundeyin M, Diskin B, Aykut B, Werba G, Barilla RM, Rodriguez R, Chang S, Gardner L, Mahal LK, Ueberheide B, Miller G. (2017). Dectin 1 activation on macrophages by galectin 9 promotes pancreatic carcinoma and peritumoral immune tolerance. Nature Medicine 23: 556–567. DOI 10.1038/nm.4314. [Google Scholar] [CrossRef]
Dambuza IM, Brown GD. (2015). C-type lectins in immunity: recent developments. Current Opinion in Immunology 32: 21–27. DOI 10.1016/j.coi.2014.12.002. [Google Scholar] [CrossRef]
Darwish NH, Sudha T, Godugu K, Elbaz O, Abdelghaffar HA, Hassan EE, Mousa SA. (2016). Acute myeloid leukemia stem cell markers in prognosis and targeted therapy: potential impact of BMI-1, TIM-3 and CLL-1. Oncotarget 7: 57811–57820. DOI 10.18632/oncotarget.11063. [Google Scholar] [CrossRef]
Del Fresno C, Iborra S, Saz-Leal P, Martinez-Lopez M, Sancho D. (2018). Flexible signaling of myeloid C-type lectin receptors in immunity and inflammation. Frontiers in Immunology 9: 491. DOI 10.3389/fimmu.2018.00804. [Google Scholar] [CrossRef]
Derpoorter C, Vandepoele K, Diez-Fraile A, Vandemeulebroecke K, De Wilde B, Speleman F, Van Roy N, Lammens T, Laureys G. (2019). Pinpointing a potential role for CLEC12B in cancer predisposition through familial exome sequencing. Pediatric Blood & Cancer 66: e27513. DOI 10.1002/pbc.27513. [Google Scholar] [CrossRef]
Geijtenbeek TB, Gringhuis SI. (2016). C-type lectin receptors in the control of T helper cell differentiation. Nature Reviews Immunology 16: 433–448. DOI 10.1038/nri.2016.55. [Google Scholar] [CrossRef]
Gonzalez-Chavarria I, Fernandez E, Gutierrez N, Gonzalez-Horta EE, Sandoval F, Cifuentes P, Castillo C, Cerro R, Sanchez O, Toledo JR. (2018). LOX-1 activation by oxLDL triggers an epithelial mesenchymal transition and promotes tumorigenic potential in prostate cancer cells. Cancer Letters 414: 34–43. DOI 10.1016/j.canlet.2017.10.035. [Google Scholar] [CrossRef]
Győrffy B, Surowiak P, Budczies J, Lánczky A, Chellappan S P. (2013). Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 8: e82241. DOI 10.1371/journal.pone.0082241. [Google Scholar] [CrossRef]
Hadebe S, Brombacher F, Brown GD. (2018). C-type lectin receptors in asthma. Frontiers in Immunology 9: 1107. DOI 10.3389/fimmu.2018.00733. [Google Scholar] [CrossRef]
Havel JJ, Chowell D, Chan TA. (2019). The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nature Reviews Cancer 19: 133–150. DOI 10.1038/s41568-019-0116-x. [Google Scholar] [CrossRef]
Hoffmann SC, Schellack C, Textor S, Konold S, Schmitz D, Cerwenka A, Pflanz S, Watzl C. (2007). Identification of CLEC12B, an inhibitory receptor on myeloid cells. Journal of Biological Chemistry 282: 22370–22375. DOI 10.1074/jbc.M704250200. [Google Scholar] [CrossRef]
Hutten TJ, Thordardottir S, Fredrix H, Janssen L, Woestenenk R, Tel J, Joosten B, Cambi A, Heemskerk MH, Franssen GM, Boerman OC, Bakker LB, Jansen JH, Schaap N, Dolstra H, Hobo W. (2016). CLEC12A-mediated antigen uptake and cross-presentation by human dendritic cell subsets efficiently boost tumor-reactive T cell responses. Journal of Immunology 197: 2715–2725. DOI 10.4049/jimmunol.1600011. [Google Scholar] [CrossRef]
Joffre OP, Sancho D, Zelenay S, Keller AM, Reis e Sousa C. (2010). Efficient and versatile manipulation of the peripheral CD4+ T-cell compartment by antigen targeting to DNGR-1/CLEC9A. European Journal of Immunology 40: 1255–1265. DOI 10.1002/eji.201040419. [Google Scholar] [CrossRef]
Krawczyk E, Zolov SN, Huang K, Bonifant CL. (2019). T-cell activity against AML improved by dual-targeted T cells stimulated through T-cell and IL7 receptors. Cancer Immunology Research 7: 683–692. DOI 10.1158/2326-6066.CIR-18-0748. [Google Scholar] [CrossRef]
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Research 44: W90–W97. DOI 10.1093/nar/gkw377. [Google Scholar] [CrossRef]
Leibundgut-Landmann S, Osorio F, Brown GD, Reis e Sousa C. (2008). Stimulation of dendritic cells via the Dectin-1/Syk pathway allows priming of cytotoxic T-cell responses. Blood 112: 4971–4980. DOI 10.1182/blood-2008-05-158469. [Google Scholar] [CrossRef]
Li K, Neumann K, Duhan V, Namineni S, Hansen AL, Wartewig T, Kurgyis Z, Holm CK, Heikenwalder M, Lang KS, Ruland J. (2019). The uric acid crystal receptor Clec12A potentiates type I interferon responses. Proceedings of the National Academy of Sciences of the United States of America 116: 18544–18549. DOI 10.1073/pnas.1821351116. [Google Scholar] [CrossRef]
Mattiola I, Tomay F, De Pizzol M, Silva-Gomes R, Savino B, Gulic T, Doni A, Lonardi S, Astrid Boutet M, Nerviani A, Carriero R, Molgora M, Stravalaci M, Morone D, Shalova IN, Lee Y, Biswas SK, Mantovani G, Sironi M, Pitzalis C, Vermi W, Bottazzi B, Mantovani A, Locati M. (2019). The macrophage tetraspan MS4A4A enhances Dectin-1-dependent NK cell-mediated resistance to metastasis. Nature Immunology 20: 1012–1022. DOI 10.1038/s41590-019-0417-y. [Google Scholar] [CrossRef]
Morsink LM, Walter RB, Ossenkoppele GJ. (2019). Prognostic and therapeutic role of CLEC12A in acute myeloid leukemia. Blood Reviews 34: 26–33. DOI 10.1016/j.blre.2018.10.003. [Google Scholar] [CrossRef]
Nagy A, Lanczky A, Menyhart O, Gyorffy B. (2018). Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Scientific Reports 8: 7. DOI 10.1038/s41598-018-27521-y. [Google Scholar] [CrossRef]
Nieto-Pelegrin E, Alvarez B, Martinez de la Riva P, Toki D, Poderoso T, Revilla C, Uenishi H, Ezquerra A, Dominguez J. (2020). Porcine CLEC12B is expressed on alveolar macrophages and blood dendritic cells. Developmental & Comparative Immunology 111: 103767. DOI 10.1016/j.dci.2020.103767. [Google Scholar] [CrossRef]
Nishimura Y, Wake H, Teshigawara K, Wang D, Sakaguchi M, Otsuka F, Nishibori M. (2019). Histidine-rich glycoprotein augments natural killer cell function by modulating PD-1 expression via CLEC-1B. Pharmacology Research & Perspectives 7: e00481. DOI 10.1002/prp2.481. [Google Scholar] [CrossRef]
Papatheodorou I, Fonseca NA, Keays M, Tang YA, Barrera E, Bazant W, Burke M, Fullgrabe A, Fuentes AM, George N, Huerta L, Koskinen S, Mohammed S, Geniza M, Preece J, Jaiswal P, Jarnuczak AF, Huber W, Stegle O, Vizcaino JA, Brazma A, Petryszak R. (2018). Expression Atlas: Gene and protein expression across multiple studies and organisms. Nucleic Acids Research 46: D246–D251. DOI 10.1093/nar/gkx1158. [Google Scholar] [CrossRef]
Picco G, Beatson R, Taylor-Papadimitriou J, Burchell JM. (2014). Targeting DNGR-1 (CLEC9A) with antibody/MUC1 peptide conjugates as a vaccine for carcinomas. European Journal of Immunology 44: 1947–1955. DOI 10.1002/eji.201344076. [Google Scholar] [CrossRef]
Plato A, Willment JA, Brown GD. (2013). C-type lectin-like receptors of the Dectin-1 cluster: Ligands and signaling pathways. International Reviews of Immunology 32: 134–156. DOI 10.3109/08830185.2013.777065. [Google Scholar] [CrossRef]
Rayes J, Watson SP, Nieswandt B. (2019). Functional significance of the platelet immune receptors GPVI and CLEC-2. Journal of Clinical Investigation 129: 12–23. DOI 10.1172/JCI122955. [Google Scholar] [CrossRef]
Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. (2007). Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9: 166–180. DOI 10.1593/neo.07112. [Google Scholar] [CrossRef]
Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I, Chan NW, Zhang J. (2019). TISIDB: an integrated repository portal for tumor–immune system interactions. Bioinformatics 35: 4200–4202. DOI 10.1093/bioinformatics/btz210. [Google Scholar] [CrossRef]
Seifert L, Deutsch M, Alothman S, Alqunaibit D, Werba G, Pansari M, Pergamo M, Ochi A, Torres-Hernandez A, Levie E, Tippens D, Greco SH, Tiwari S, Ly NNG, Eisenthal A, van Heerden E, Avanzi A, Barilla R, Zambirinis CP, Rendon M, Daley D, Pachter HL, Hajdu C, Miller G. (2015). Dectin-1 regulates hepatic fibrosis and hepatocarcinogenesis by suppressing TLR4 signaling pathways. Cell Reports 13: 1909–1921. DOI 10.1016/j.celrep.2015.10.058. [Google Scholar] [CrossRef]
Suzuki-Inoue K. (2018). Roles of the CLEC-2-podoplanin interaction in tumor progression. Platelets 29: 1–7. DOI 10.1080/09537104.2017.1417660. [Google Scholar] [CrossRef]
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research 47: D607–D613. DOI 10.1093/nar/gky1131. [Google Scholar] [CrossRef]
Takeuchi O, Akira S. (2010). Pattern recognition receptors and inflammation. Cell 140: 805–820. DOI 10.1016/j.cell.2010.01.022. [Google Scholar] [CrossRef]
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. (2017). GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research 45: W98–W102. DOI 10.1093/nar/gkx247. [Google Scholar] [CrossRef]
Thebault P, Lhermite N, Tilly G, Le Texier L, Quillard T, Heslan M, Anegon I, Soulillou JP, Brouard S, Charreau B, Cuturi MC, Chiffoleau E. (2009). The C-type lectin-like receptor CLEC-1, expressed by myeloid cells and endothelial cells, is up-regulated by immunoregulatory mediators and moderates T cell activation. Journal of Immunology 183: 3099–3108. DOI 10.4049/jimmunol.0803767. [Google Scholar] [CrossRef]
Tone K, Stappers MHT, Willment JA, Brown GD. (2019). C-type lectin receptors of the Dectin-1 cluster: Physiological roles and involvement in disease. European Journal of Immunology 49: 2127–2133. DOI 10.1002/eji.201847536. [Google Scholar] [CrossRef]
Vasaikar SV, Straub P, Wang J, Zhang B. (2018). LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Research 46: D956–D963. DOI 10.1093/nar/gkx1090. [Google Scholar] [CrossRef]
Wang B, Zhao H, Zhao L, Zhang Y, Wan Q, Shen Y, Bu X, Wan M, Shen C. (2017). Up-regulation of OLR1 expression by TBC1D3 through activation of TNFalpha/NF-κB pathway promotes the migration of human breast cancer cells. Cancer Letters 408: 60–70. DOI 10.1016/j.canlet.2017.08.021. [Google Scholar] [CrossRef]
Wang D, Gao S, Chen J, Zhao Y, Jiang Y, Chu X, Wang X, Liu N, Qin T, Yi Q, Yue Y, Wang S. (2018). Dectin-1 stimulates IL-33 expression in dendritic cells via upregulation of IRF4. Laboratory Investigation 98: 708–714. DOI 10.1038/s41374-018-0047-2. [Google Scholar] [CrossRef]
Wang L, Yin J, Wang X, Shao M, Duan F, Wu W, Peng P, Jin J, Tang Y, Ruan Y, Sun Y, Gu J. (2016). C-type lectin-like receptor 2 suppresses AKT signaling and invasive activities of gastric cancer cells by blocking expression of phosphoinositide 3-kinase subunits. Gastroenterology 150: 1183–1195.e16. DOI 10.1053/j.gastro.2016.01.034. [Google Scholar] [CrossRef]
Wang Y, Lv Y, Liu TS, Yan WD, Chen LY, Li ZH, Piao YS, An BR, Lin ZH, Ren XS. (2019). Cordycepin suppresses cell proliferation and migration by targeting CLEC2 in human gastric cancer cells via Akt signaling pathway. Life Sciences 223: 110–119. DOI 10.1016/j.lfs.2019.03.025. [Google Scholar] [CrossRef]
Wang YY, Chen WL, Weng XQ, Sheng Y, Wu J, Hao J, Liu ZY, Zhu YM, Chen B, Xiong SM, Chen Y, Chen QS, Sun HP, Li JM, Wang J. (2017). Low CLL-1 expression is a novel adverse predictor in 123 patients with de novo CD34+ acute myeloid leukemia. Stem Cells and Development 26: 1460–1467. DOI 10.1089/scd.2016.0310. [Google Scholar] [CrossRef]
Xia Y, Liu L, Bai Q, Wang J, Xi W, Qu Y, Xiong Y, Long Q, Xu J, Guo J. (2016). Dectin-1 predicts adverse postoperative prognosis of patients with clear cell renal cell carcinoma. Scientific Reports 6: 193. DOI 10.1038/srep32657. [Google Scholar] [CrossRef]
Xiong G, Liu C, Yang G, Feng M, Xu J, Zhao F, You L, Zhou L, Zheng L, Hu Y, Wang X, Zhang T, Zhao Y. (2019). Long noncoding RNA GSTM3TV2 upregulates LAT2 and OLR1 by competitively sponging let-7 to promote gemcitabine resistance in pancreatic cancer. Journal of Hematology & Oncology 12: 55. DOI 10.1186/s13045-019-0777-7. [Google Scholar] [CrossRef]
Yang G, Xiong G, Feng M, Zhao F, Qiu J, Liu Y, Cao Z, Wang H, Yang J, You L, Zheng L, Zhang T, Zhao Y. (2020). OLR1 promotes pancreatic cancer metastasis via increased c-Myc expression and transcription of HMGA2. Molecular Cancer Research 18: 685–697. DOI 10.1158/1541-7786.MCR-19-0718. [Google Scholar] [CrossRef]
Zelenay S, Keller A M, Whitney P G, Schraml B U, Deddouche S, Rogers N C, Schulz O, Sancho D, Reis e Sousa C. (2012). The dendritic cell receptor DNGR-1 controls endocytic handling of necrotic cell antigens to favor cross-priming of CTLs in virus-infected mice. Journal of Clinical Investigation 122: 1615–1627. DOI 10.1172/JCI60644. [Google Scholar] [CrossRef]
Zeng B, Middelberg AP, Gemiarto A, MacDonald K, Baxter AG, Talekar M, Moi D, Tullett KM, Caminschi I, Lahoud MH, Mazzieri R, Dolcetti R, Thomas R. (2018). Self-adjuvanting nanoemulsion targeting dendritic cell receptor Clec9A enables antigen-specific immunotherapy. Journal of Clinical Investigation 128: 1971–1984. DOI 10.1172/JCI96791. [Google Scholar] [CrossRef]
Zhang J, Zhang L, Li C, Yang C, Li L, Song S, Wu H, Liu F, Wang L, Gu J. (2018). LOX-1 is a poor prognostic indicator and induces epithelial-mesenchymal transition and metastasis in pancreatic cancer patients. Cellular Oncology 41: 73–84. DOI 10.1007/s13402-017-0360-6. [Google Scholar] [CrossRef]
Zhao Y, Chu X, Chen J, Wang Y, Gao S, Jiang Y, Zhu X, Tan G, Zhao W, Yi H, Xu H, Ma X, Lu Y, Yi Q, Wang S. (2016). Dectin-1-activated dendritic cells trigger potent antitumour immunity through the induction of Th9 cells. Nature Communications 7: 445. DOI 10.1038/ncomms12368. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |