DOI:10.32604/biocell.2021.012847

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.012847 |  www.techscience.com/journal/biocell |
| Article |
microR-1294-5p inhibits glycolytic metabolism of non-small cell lung cancer cells via targeting TMPRSS11B
Department of Thoracic Surgery, Changhai Hospital of Shanghai, Shanghai, 200433, China
*Address correspondence to: Tiejun Zhao, drzhaotiejun@126.com; Ling Chen, chenling0707@outlook.com
#These authors contributed equally to this work
Received: 14 July 2020; Accepted: 14 September 2020
Abstract: Non-small cell lung cancer (NSCLC) cells intake and consume glucose at high efficiency by aerobic glycolysis to maintain robust cell growth and resist cell death. MicroRNAs (miRNAs) have been known to play pivotal roles in NSCLC development partly through mediating glycolysis. However, only a few miRNAs have been experimentally confirmed as critical regulators of glycolysis in NSCLC. TCGA datasets were analyzed to screen for differentially expressed miRNAs between NSCLC and normal tissues. The function of miR-1294-5p was determined in NSCLC cells by cell proliferation, glucose uptake, lactate release, and Extracellular Acidification Rate (ECAR) assays. The target of miR-1294-5p was predicted by TargetScan and miRDB, which was further validated by flow cytometry analysis, RT-qPCR, western blotting, a dual-luciferase reporter assay, and RNA immunoprecipitation (RIP) assay. In the present study, it was found that miR-1294-5p was a significantly downregulated miRNA in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). The overexpression of miR-1294-5p inhibited glycolysis, lactate export, ECAR, and cell proliferation in NSCLC cells. Analysis with bioinformatic tools, Western Blotting, RT-qPCR, flow cytometry analysis, dual-luciferase reporter assay, and RIP assay showed that miR-1294-5p directly bound to complementary sites in the 3’-Untranslated Region (UTR) of TMPRSS11B resulted in downregulation of TMPRSS11B expression. In addition, transfection of recombinant TMPRSS11B rescued the functions of miR-1294-5p on glycolysis and proliferation of NSCLC cells. The findings provided novel insights for understanding the regulation of glycolytic metabolism in NSCLC.
Keywords: miR-1294-5p; TMPRSS11B; Glycolytic metabolism; NSCLC
Globally, lung cancer is the most prevalent and lethal cancer type, accounting for about 10% cancer burden (Barta et al., 2019). Approximately 80% of patients with lung cancer belong to NSCLC, and the prognosis of these patients is relatively poor, with a five-year overall survival of less than 20 (Ettinger et al., 2012; Yan et al., 2019). Despite the high response rate to chemotherapy, most patients with NSCLC developed resistance to chemotherapy (Kim, 2016). It is urgent to further investigate the signaling network during the development of NSCLC to provide novel therapeutic approaches for patients.
miRNAs are single-stranded molecules with no protein-coding potential (Yu et al., 2020). Mechanistically, miRNAs interacted with miRNA response element (MRE) in 3’UTR of target mRNA, leading to degradation of mRNA and downregulation of target expression (Krol et al., 2010). It has been reported that miRNAs are involved in the metabolic reprogramming of cancer cells by targeting metabolism-associated genes (Subramaniam et al., 2019). For example, miR-125a could target HK2, a key modulator of glucose metabolism, to inhibit the metabolism of hepatocellular carcinoma (Jin et al., 2017). miR-532-3p, miR-98 also targeted HK2 to regulate metabolism in different cell backgrounds (Zhou et al., 2018; Zhu et al., 2017). miR-1294-5p is a tumor-suppressive miRNA in several cancer types such as ovarian cancer, clear cell renal cell carcinoma, and gastric cancer (Guo et al., 2018; Pan et al., 2019; Shi et al., 2018). The biological role of miR-1294-5p during the development of NSCLC remains elusive.
Transmembrane serine protease TMPRSS11B is a member of Type-II trypsin-like serine protease (Bugge et al., 2009). TMPRSS11B was overexpressed in lung cancer and solubilized Basigin to enhance lactate export in lung cancer cells (Updegraff et al., 2018). Although the critical role of TMPRSS11B in mediating glycolytic metabolism was revealed, the molecular mechanism underlying upregulation of TMPRSS11B in lung cancer has not been studied yet.
The current study identified miR-1924 as a novel downregulated miRNA in NSCLC. We aimed to investigate the function of miR-1924 in NSCLC cells.
Lung tumors and matched normal tissues were surgically collected from 150 patients (89 males and 61 females, aged 51–72 years) with non-small cell lung cancer in Changhai Hospital of Shang-hai from July 2017 to October 2018. None of the patients received pre-operative treatments, including chemotherapy, radiotherapy, and targeted therapy. Clinicopathological factors of these patients were listed in Table. Tumors were staged according to the 7th edition of the Union for International Cancer Control’s (UICC) tumor, node and metastasis (Wittekind, 2010). Written informed consent was collected from patients, and the protocol of the study was reviewed and approved by the Ethical Committee of Changhai Hospital of Shang-hai (Approval No. CH02072017).
Human bronchial epithelial cell line BEAS-2B, LUSC cell lines SK-MES-1, NCI-H520 were purchased from ATCC (Manassas, VA). These cells were all cultured in DMEM (Invitrogen, Carlsbad, CA) with 10% FBS (Thermo Fisher Scientific, Waltham, MA). Cells were kept in an incubator with 5% CO2.
RNA immunoprecipitation assay (RIP)
Anti-Ago2 (#MA5-23515) and control IgG antibodies (#61-6800) were used to perform immunoprecipitation assay. RIP was performed with the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore). After that, real-time quantitative PCR was conducted to detect TMPRSS11B mRNA associated with RISC complex in immunoprecipitated material.
miR-negative control (miR-NC), miR-1294-5p mimic, miR-1294-5p inhibitor were synthesized by GenePharma (Shanghai, China). To manipulate miR-1294-5p expression, miR-1294-5p mimic or inhibitor or negative control was transfected into cells using X-tremeGENE HP DNA Transfection Reagent (Roche, Basel, Switzerland). The full length of TMPRSS11B was amplified from cDNA of NCI-H520 by PCR and ligated into expression plasmid pcDNA3. Empty vector or pcDNA3-TMPRSS11B were co-transfected with miR-1294-5p mimic or miR-NC with X-tremeGENE HP DNA Transfection Reagent. The transfection efficiency was determined by RT-qPCR and western blotting at 48 h after transfection.
Protein extraction and western blotting
TMPRSS11B antibody was a product from Novus Biologicals (Littleton, CO, #H00132724-B01P). β-actin antibody was purchased from Sigma Aldrich (#A4700). Horseradish peroxidase-conjugated secondary antibodies for mouse (#SA10001-1) and rabbit (#SA10001-2) were bought from Proteintech (Rosemont, IL). Protein lysates were prepared by RIPA lysis buffer. After that, protein concentration was determined by a BCA Protein Assay kit (Invitrogen). Lysates were separated by the SDS-PAGE gel and transferred to PVDF membranes. The membrane was incubated with primary and secondary antibodies. The blots were visualized with an ECL Western Blotting Substrate kit (Thermo Fisher Scientific).
Allophycocyanin (APC)-linked anti-mouse antibody (#A-865) was obtained from Invitrogen. TMPRSS11B antibody (#H00132724-B01P) was used for flow cytometry analysis. Briefly, cells were harvested and incubated with TMPRSS11B antibody. After washing three times, cells were incubated with APC linked anti-mouse antibody. The cells were then subjected to flow cytometry analysis to detect the positive rate of TMPRSS11B. The data were analyzed by FlowJo software.
Real-time quantitative PCR (RT-qPCR)
RNA was extracted by Trizol reagent (Invitrogen) from cells and tissues. RNA was reverse transcribed into cDNA by an M-MLV Frist Strand cDNA Synthesis kit (Invitrogen). TB Green® Fast qPCR Mix (TaKaRa, Tokyo, Japan) was used to perform RT-qPCR on a GeneAmp® PCR System 9700 (Applied Biosystems, Foster City, CA). β-actin and U6 were chosen as internal controls for mRNA and miRNA. The PCR primer sequences were listed as follow: Stem-loop: 5’- CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGACAA-3’; miR-1294-5p-forward: 5’-TCGGCAGGTGTGAGGAAGGCA-3’; miR-1294-5p-reverse: 5’-CTCAACTGGTGTCGTGGA-3’; U6-forward: 5’-ATTTGCGTGTCATCCTTGC-3’; U6-reverse: 5’-TCGCTTCGGCAGCACATAT-3’; TMPRSS1B-forward: 5’-CAGAAGGAGTTAGCATGAGGACT-3’; TMPRSS11B-reverse: 5’-TGTTGGCTACTTGTCTCCCAC-3’; β-actin-forward: 5’-CACCATTGGCAATGAGCGGTTC-3’; β-actin-reverse: 5’-AGGTCTTTGCGGATGTCCACGT-3’.
Starbase V2 (http://starbase.sysu.edu.cn/) was used to analyze the expression of miR-1294-5p in the TCGA project. We evaluated miR-1294-5p in both TCGA-LUAD and TCGA-LUSC. Target genes of miR-1294-5p were predicted by miRDB (http://mirdb.org/) and TargetScan (http://www.targetscan.org/vert_72/) software. TMPRSS11B was a common target in the list of potential targets by the software.
Detection of lactate release and glucose uptake
Lactate Assay Kit (Sigma Aldrich, St. Louis, MO, #MAK064) was used to detect lactate release according to the manufacturer’s protocol. The glucose uptake levels were determined by Glucose Uptake Assay Kit (Abcam, Cambridge, UK, #ab136955).
Detection of Extracellular Acidification Rate (ECAR)
ECAR was detected by an XF96 Extracellular Flux Analyzer (Seahorse Bioscience, Santa Rosa, CA) according to the manufacturer’s protocol. Cells were cultured without glucose for 2 h. After that, glucose, oligomycin, 2-DG were sequentially injected into culture cells. Glycolysis, maximum glycolytic capacity, and non-glycolytic capacity were reflected by values after the addition of glucose, oligomycin, and 2-DG, respectively.
The cell proliferation ability was detected by the CCK-8 kit (Dojindo, Tokyo, Japan) according to the provided protocol. Cells were cultured with a medium containing CCK-8 solution, and the value at OD 450 nm was detected to manifest cell proliferation ability.
The data were calculated with GraphPad Prism 5.0. The Student’s t-test was used to compare two groups. Three groups were analyzed by one-way ANOVA, followed by the Tukey test. p-value less than 0.05 was statistically significant (p < 0.05).
The expression of miR-1294-5p was downregulated in NSCLC
To investigate NSCLC related miRNAs, we retrieved miRNA expression data of LUAD and LUSC (two major subtypes of NSCLC) with corresponding normal tissues from the TCGA project. It was found that expression of miR-1294-5p, a miRNA has not been studied in NSCLC yet, was decreased in both LUAD and LUSC samples (Figs. 1A–1B). To confirm this finding and evaluate the clinical value of miR-1294-5p, we collected 150 pairs of NSCLC samples and adjacent normal tissues. With RT-qPCR, we found miR-1294-5p was decreased in most NSCLC samples (125 out of 150 cases, 83%) compared with their counterparts (Fig. 1C). Analysis of association between miR-1294-5p and clinicopathological factors suggested that low expression of miR-1294-5p was associated with tumors of advanced pathological T stage (pT3-4) compared with those of early pathological T stage (pT1-2) (Tab. 1). Expression of miR-1294-5p was also related to the histological type of NSCLC as low expression of miR-1294-5p was mostly found in squamous cell carcinoma (Tab. 1). No association was found between miR-1294-5p expression with gender, age, pathological N stage, and pathological M stage (Tab. 1). We next detected miR-1294-5p expression in LUSC cell lines NCI-H520, SK-MES-1, and BEAS-2B, a human bronchial epithelial cell line by RT-qPCR. Consistent with observation in tissue samples, miR-1294-5p was downregulated in these LUSC cells (Fig. 1D).
Table 1: miR-1294-5p expression and clinicopathological parameters
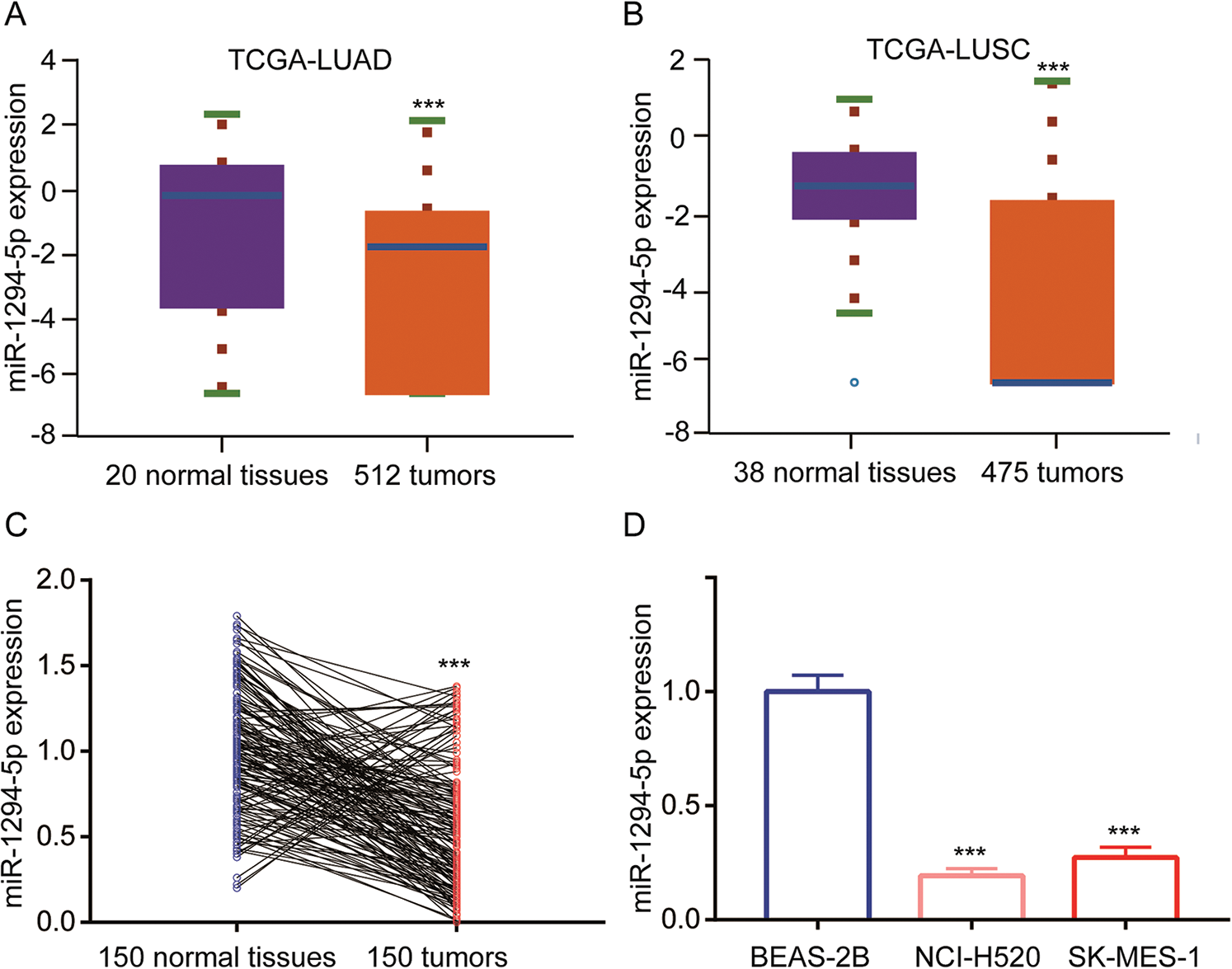
Figure 1: miR-1294-5p was downregulated in NSCLC.
miR-1294-5p inhibited NSCLC cell proliferation and glycolytic metabolism
To study the function of miR-1294-5p, a miR-1294-5p mimic was transfected into NCI-H520 and SK-MES-1 cells. Transfection of miR-1294-5p mimic increased miR-1294-5p expression in these cells (Fig. 2A). In addition, transfection of miR-1294-5p inhibitor decreased miR-1294-5p expression in NCI-H520 and SK-MES-1 cells (Fig. 2B). In cell proliferation assay, the data showed that miR-1294-5p overexpression reduced cell proliferation of NCI-H520 and SK-MES-1 cells while miR-1294-5p downregulation promoted cell proliferation of these cells (Figs. 2C, 2D). Reprogramming of glycolytic metabolism is critical for robust cell proliferation of NSCLC cells, which is reflected by the high capacity of glucose uptake and lactate release. Therefore, we detected glucose and lactate levels in cells with transfection of miR-1294-5p mimic or miR-1294-5p inhibitor. Overexpression of miR-1294-5p decreased glucose uptake and released lactate levels in NCI-H520 and SK-MES-1 cells, on the contrary, downregulation of miR-1294-5p increased glucose uptake and lactate release (Figs. 2E, 2F). We next examined ECAR status to reflect glycolysis in NSCLC cells. The data manifested that miR-1294-5p mimic inhibited glucose-mediated basal and oligomycin-mediated ECAR in NCI-H520 and SK-MES-1 cells, while miR-1294-5p inhibitor upregulated ECAR in these cells (Figs. 2G, 2H). The findings indicated that miR-1294-5p was involved in cell proliferation of NSCLC cells via regulating glycolysis.
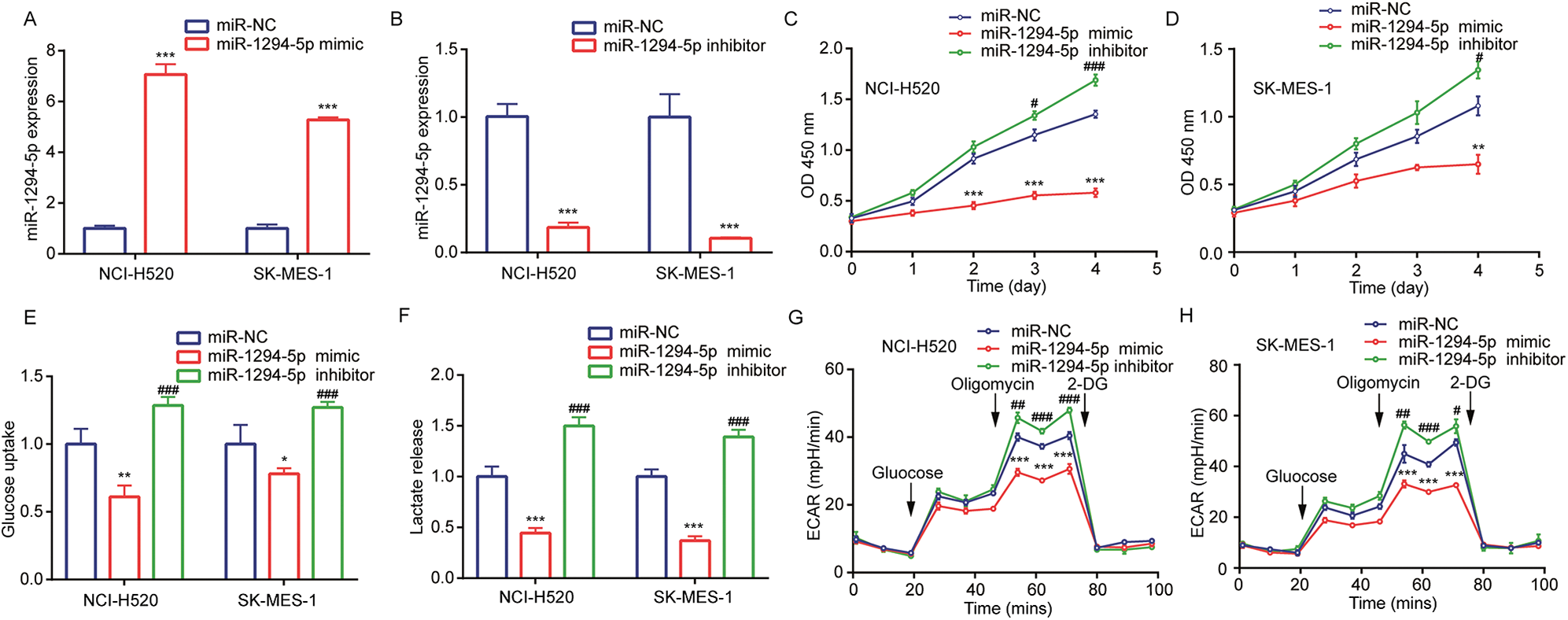
Figure 2: miR-1294-5p regulated glycolytic metabolism and cell proliferation of NSC.
LC cells. (A) miR-1294-5p expression was detected in NCI-H520 and SK-MES-1 cells after transfection of miR-NC or miR-1294-5p mimic by RT-qPCR. (B) miR-1294-5p expression was detected in NCI-H520 and SK-MES-1 cells after transfection of miR-NC or miR-1294-5p inhibitor by RT-qPCR. (C-D) Cell proliferation rate was determined in NCI-H520 (C) and SK-MES-1 (D) cells after transfection of miR-NC or miR-1294-5p mimic or miR-1294-5p inhibitor by the CCK-8 assay. (E–F) Glucose uptake (E) and lactate release (F) Levels were detected in NCI-H520 and SK-MES-1 cells after transfection of miR-NC or miR-1294-5p mimic or miR-1294-5p inhibitor. (G-H) ECAR was measured in NCI-H520 (G) and SK-MES-1 (H) Cells after transfection of miR-NC or miR-1294-5p mimic or miR-1294-5p inhibitor. *p < 0.05 vs. miR-NC; **p < 0.01 vs. miR-NC; ***p < 0.001 vs. miR-NC; #p < 0.05 vs. miR-NC; ##p < 0.01 vs. miR-NC; ###p < 0.001 vs. miR-NC.
miR-1294-5p targeted TMPRSS11B in NSCLC cells
To explore the target of miR-1294-5p, we used several bioinformatics software to predict binding sites between miR-1294-5p and target genes. Prediction from TargetScan and miRDB software all suggested that there was a putative miRNA responsive element for miR-1294-5p in 3’UTR of TMPRSS11B (Fig. 3A), a reported regulator of glycolysis in NSCLC. RT-qPCR showed that miR-1294-5p mimic reduced TMPRSS11B at mRNA and protein levels (Figs. 3B, 3C). TMPRSS11B is a membrane protein-mediated lactate export in NSCLC cells. We therefore performed flow cytometry to detect TMPRSS11B protein expression at the cell membrane. The results consistently revealed that miR-1294-5p mimic greatly reduced TMPRSS11B positive cells of NCI-H520 and SK-MES-1 cells (Figs. 3D, 3E). To further evaluate the association between miR-1294-5p and TMPRSS11B expression in NSCLC samples, we used the RT-qPCR method to detect TMPRSS11B mRNA expression in 150 pairs of NSCLC tumors and matched normal tissues. It was found that TMPRSS11B mRNA levels were increased in NSCLC tumors compared with the normal tissues (Fig. 3F). Furthermore, according to the results of the Pearson correlation analysis, the TMPRSS11B mRNA levels were negatively associated with miR-1294-5p expression in these tumors (r = −0.397, p < 0.001) (Fig. 3G).
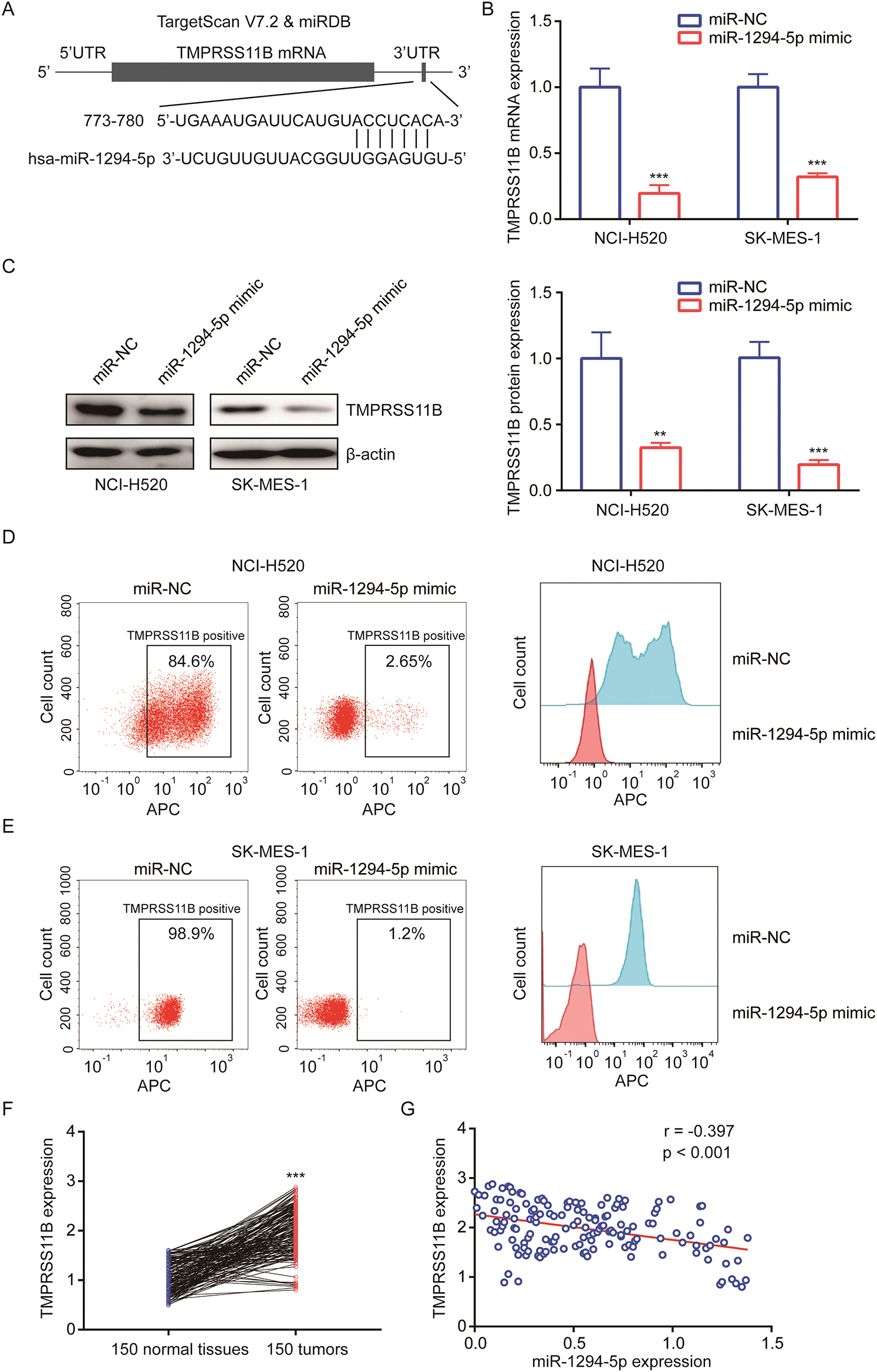
Figure 3: miR-1294-5p suppressed TMPRSS11B expression in NSCLC cells.
miR-1294-5p directly bound to 3’UTR of TMPRSS11B
We next constructed luciferase plasmids containing wild-type 3’UTR of TMPRSS11B and mutant 3’UTR of TMPRSS11B with two mutation sites at the potential binding site (Fig. 4A). Data from luciferase assay showed that miR-1294-5p mimic repressed luciferase activity of wild-type TMPRSS11B 3’UTR instead of mutant form in NCI-H520 and SK-MES-1 cells (Figs. 4B, 4C). Furthermore, RIP assay indicated that Ago2 enriched more TMPRSS11B mRNA in NCI-H520 and SK-MES-1 cells transfected with miR-1294-5p mimic in comparison with those transfected with miR-NC (Figs. 4D, 4E).
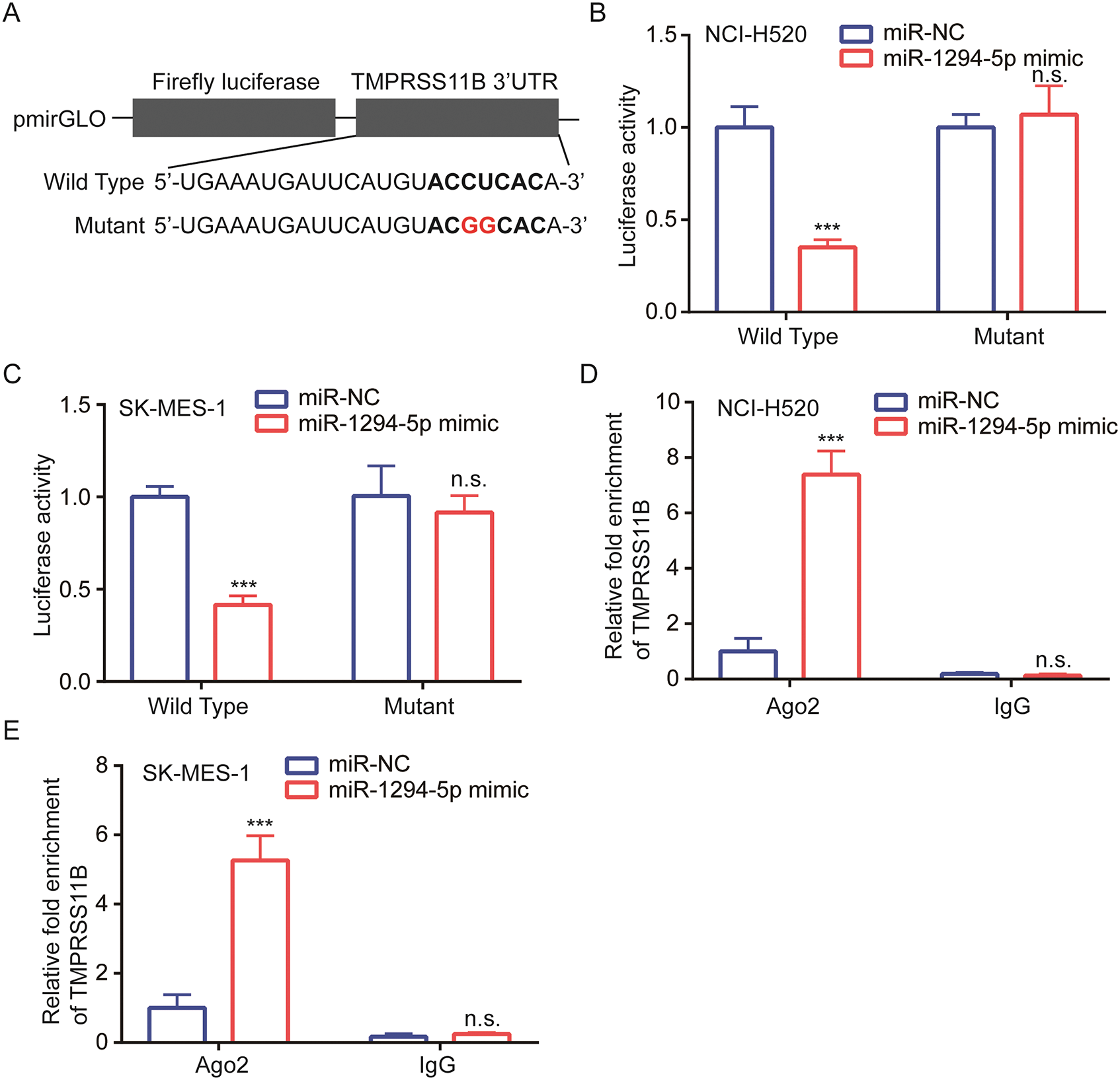
Figure 4: TMPRSS11B was directly targeted by miR-1294-5p.
TMPRSS11B rescued the effect of miR-1294-5p mimic on NSCLC cells
TMPRSS11B sequence was inserted into an expression plasmid. Western blotting showed that transfection of recombinant TMPRSS11B elevated TMPRSS11B protein expression in NCI-H520 and SK-MES-1 cells (Fig. 5A). In cell proliferation assay, the results indicated that TMPRSS11R overexpression attenuated the inhibitory effect of miR-1294-5p mimic on cell proliferation of NCI-H520 and SK-MES-1 cells (Figs. 5B, 5C). Furthermore, it was found that TMPRSS11B overexpression also reversed the inhibited glucose uptake and lactate release in NCI-H520 and SK-MES-1 cells with transfection of miR-1294-5p mimic (Figs. 5D, 5E). Moreover, miR-1294-5p mimic related reduction of ECAR was rescued by TMPRSS11B in NCI-H520 and SK-MES-1 cells (Figs. 5F, 5G).
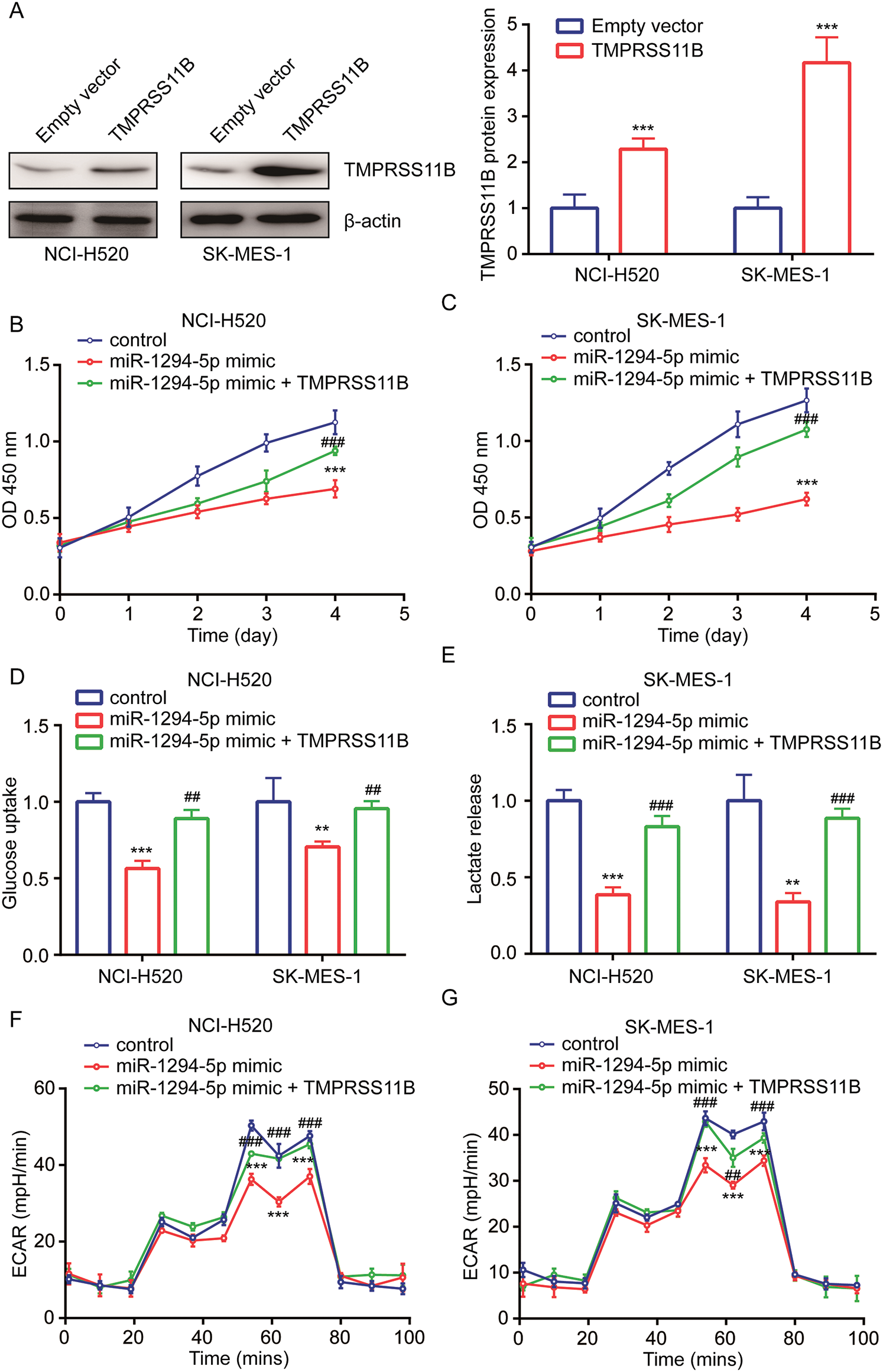
Figure 5: miR-1294-5p relied on the repression of TMPRSS11B to regulate glycolytic metabolism in NSCLC.
Numerous studies demonstrated that aberrant expression of miRNAs facilitated reprogramming of metabolism, proliferation, metastasis, and drug resistance of NSCLC (Cai et al., 2020; Jin et al., 2020). miR-1294-5p is a recently identified tumor-suppressive miRNA in several cancer types. miR-1294-5p was a negative regulator of cell proliferation, migration, invasion and cisplatin resistance in cancer cells of different types, including gastric cancer, ovarian cancer and clear cell renal cell carcinoma (Pan et al., 2019; Shi et al., 2018; Zhang et al., 2018). In osteosarcoma cells, miR-1294-5p directly targeted PKM2, a critical regulator of metabolism, which resulted in inhibition of cell proliferation, migration, invasion, cell cycle, and elevation of cell apoptosis (Yuan et al., 2020). Most recently, Wang et al. (2020) showed that miR-1294-5p was a bridge between LINC00473 and ROBO1 in LUAD. Their data indicated that miR-1294-5p was decreased in LUAD; however, the expression of miR-1294-5p in other subtypes of NSCLC was not mentioned (Wang et al., 2020). Moreover, the function of miR-1294-5p in NSCLC has not been studied yet. Here, we used TCGA data to screen differentially expressed miRNAs in NSCLC and found miR-1294-5p as a commonly downregulated miRNA in both LUAD and LUSC. We experimentally verified that miR-1294-5p was decreased in LUSC cells. Manipulating miR-1294-5p expression suggested that miR-1294-5p suppressed cell proliferation in LUSC cells. More importantly, our data indicated that miR-1294-5p expression was negatively associated with glucose uptake, lactate release, and ECAR in LUSC cells. The results for the first time indicated that miR-1294-5p repressed glycolytic metabolism in cancer cells.
TMPRSS11B is a Type II transmembrane serine protease expressed at the cell surface (Miller et al., 2014). Data from immunohistochemistry analysis showed that TMPRSS11B was downregulated in cervical and esophageal cancer tissues (Miller et al., 2014). However, in NSCLC, especially LUSC, TMPRSS11B was upregulated and associated with lactate secretion (Updegraff et al., 2018). In LUSC cells, TMPRSS11B regulated the solubilization of MCT4 to facilitate lactate export (Updegraff et al., 2018). Via regulating MCT4, TMPRSS11B promoted NSCLC cell proliferation in vitro and in vivo (Updegraff et al., 2018). Until now, little is known how TMPRSS11B was regulated in cancer cells. Currently, TMPRSS11B was predicted as a target for miR-1294-5p in several bioinformatics tools. We next analyzed the impact of miR-1294-5p on TMPRSS11B expression in LUSC cells. The data indicated that miR-1294-5p repressed TMPRSS11B expression in LUSC cells. The direct interaction between miR-1294-5p and TMPRSS11B mRNA was proved by dual-luciferase reporter assay and RIP assay. As miR-1294-5p has been reported as regulators of oncogenes such as HOXA6 and FLOT1 (Kan et al., 2020; Pan et al., 2019), the current study manifested TMPRSS11B as a novel target of miR-1294-5p in cancer cells.
Collectively, our study showed that downregulation of miR-1294-5p upregulated TMPRSS11B expression to support the glycolytic metabolism of NSCLC cells. The novel miR-1294-5p/TMPRSS11B axis might provide insights for the development of biomarkers for NSCLC, especially LUSC. However, the sample size is small and may not provide enough information on the role of miR-1294-5p in NSCLC. In the future study, we will collect both tissues and serum from more NSCLC patients and will make a profound study on evaluating the expression of miR-1294-5p with clinicopathological factors and several known NSCLC biomarkers.
Availability of Data and Materials: They are available under reasonable request from the corresponding author.
Author’s Contributions: Ji Zhu, Xiying Bo, Gengxi Jiang, Shihua Yao, Tiejun Zhao, Ling Chen contributed to the experiment design and performance of experiments; Xiying Bo and Gengxi Jiang collected the specimens and clinical information of patients; Ling Chen supervised the study, Ji Zhu and Ling Chen analyzed the data and wrote the manuscript.
Ethics Approval: The protocol of the study was reviewed and approved by the Ethical Committee of Changhai Hospital of Shanghai (Approval No. CH02072017) in April, 2017.
Funding Statement: The authors received no specific funding for this study
Conflicts of Interest: There was no conflict of interests of any type.
Barta JA, Powell CA, Wisnivesky JP. (2019). Global epidemiology of lung cancer. Annals of Global Health 85: 8. DOI 10.5334/aogh.2419. [Google Scholar] [CrossRef]
Bugge TH, Antalis TM, Wu Q. (2009). Type II transmembrane serine proteases. Journal of Biological Chemistry 284: 23177–23181. DOI 10.1074/jbc.R109.021006. [Google Scholar] [CrossRef]
Cai X, Lin L, Zhang Q, Wu W, Su A. (2020). Bioinformatics analysis of the circRNA-miRNA-mRNA network for non-small cell lung cancer. Journal of International Medical Research 48: 300060520929167. [Google Scholar]
Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, D’amico TA, Demmy TL, Ganti AK, Govindan R, Grannis FWJr., Horn L, Jahan TM, Jahanzeb M, Kessinger A, Komaki R, Kong FM, Kris MG, Krug LM, Lennes IT, Loo BWJr., Martins R, O’malley J, Osarogiagbon RU, Otterson GA, Patel JD, Pinder-Schenck MC, Pisters KM, Reckamp K, Riely GJ, Rohren E, Swanson SJ, Wood DE, Yang SC, Hughes M, Gregory KM. (2012). Non-small cell lung cancer. Journal of the National Comprehensive Cancer Network 10: 1236–1271. DOI 10.6004/jnccn.2012.0130. [Google Scholar] [CrossRef]
Guo TY, Xu HY, Chen WJ, Wu MX, Dai X. (2018). Downregulation of miR-1294 associates with prognosis and tumor progression in epithelial ovarian cancer. European Review for Medical and Pharmacological Sciences 22: 7646–7652. [Google Scholar]
Jin F, Wang Y, Zhu Y, Li S, Liu Y, Chen C, Wang X, Zen K, Li L. (2017). The miR-125a/HK2 axis regulates cancer cell energy metabolism reprogramming in hepatocellular carcinoma. Scientific Reports 7: 3089. DOI 10.1038/s41598-017-03407-3. [Google Scholar] [CrossRef]
Jin X, Guan Y, Zhang Z, Wang H. (2020). Microarray data analysis on gene and miRNA expression to identify biomarkers in non-small cell lung cancer. BMC Cancer 20: 329. DOI 10.1186/s12885-020-06829-x. [Google Scholar] [CrossRef]
Kan XQ, Li YB, He B, Cheng S, Wei Y, Sun J. (2020). MiR-1294 acts as a tumor inhibitor in cervical cancer by regulating FLOT1 expression. Journal of Biological Regulators and Homeostatic Agents 34. DOI 10.23812/20-10a. [Google Scholar] [CrossRef]
Kim ES. (2016). Chemotherapy resistance in lung cancer. Advances in Experimental Medicine and Biology 893: 189–209. [Google Scholar]
Krol J, Loedige I, Filipowicz W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews Genetics 11: 597–610. DOI 10.1038/nrg2843. [Google Scholar] [CrossRef]
Miller GS, Zoratti GL, Murray AS, Bergum C, Tanabe LM, List K. (2014). HATL5: A cell surface serine protease differentially expressed in epithelial cancers. PLoS One 9: e87675. DOI 10.1371/journal.pone.0087675. [Google Scholar] [CrossRef]
Pan W, Pang LJ, Cai HL, Wu Y, Zhang W, Fang JC. (2019). MiR-1294 acts as a tumor suppressor in clear cell renal cell carcinoma through targeting HOXA6. European Reviews for Medical and Pharmacological Sciences 23: 3719–3725. [Google Scholar]
Shi YX, Ye BL, Hu BR, Ruan XJ. (2018). Expression of miR-1294 is downregulated and predicts a poor prognosis in gastric cancer. European Reviews for Medical and Pharmacological Sciences 22: 5525–5530. [Google Scholar]
Subramaniam S, Jeet V, Clements JA, Gunter JH, Batra J. (2019). Emergence of microRNAs as key players in cancer cell metabolism. Clinical Chemistry 65: 1090–1101. DOI 10.1373/clinchem.2018.299651. [Google Scholar] [CrossRef]
Updegraff BL, Zhou X, Guo Y, Padanad MS, Chen PH, Yang C, Sudderth J, Rodriguez-Tirado C, Girard L, Minna JD, Mishra P, Deberardinis RJ, O’donnell KA. (2018). Transmembrane protease TMPRSS11B promotes lung cancer growth by enhancing lactate export and glycolytic metabolism. Cell Reports 25: 2223–2233.e2226. DOI 10.1016/j.celrep.2018.10.100. [Google Scholar] [CrossRef]
Wang S, Wang X, Xu SL. (2020). LINC00473 promotes lung adenocarcinoma progression by regulating miR-1294/ROBO1 axis. Journal of Biological Regulators and Homeostatic Agents 34. DOI 10.23812/20-75A. [Google Scholar] [CrossRef]
Wittekind C. (2010). TNM-System 2010. Der Pathologe 31: 331–332. DOI 10.1007/s00292-010-1349-3. [Google Scholar] [CrossRef]
Yan X, Yu H, Liu Y, Hou J, Yang Q, Zhao Y. (2019). miR-27a-3p functions as a tumor suppressor and regulates non-small cell lung cancer cell proliferation via targeting HOXB8. Technology in Cancer Research & Treatment 18: 1533033819861971. [Google Scholar]
Yu D, Han GH, Zhao X, Liu X, Xue K, Wang D, Xu CB. (2020). MicroRNA-129-5p suppresses nasopharyngeal carcinoma lymphangiogenesis and lymph node metastasis by targeting ZIC2. Cellular Oncology 43: 249–261. DOI 10.1007/s13402-019-00485-5. [Google Scholar] [CrossRef]
Yuan Q, Yu H, Chen J, Song X, Sun L. (2020). Antitumor effect of miR-1294/pyruvate kinase M2 signaling cascade in osteosarcoma cells. OncoTargets & Therapy 13: 1637–1647. DOI 10.2147/OTT.S232718. [Google Scholar] [CrossRef]
Zhang Y, Huang S, Guo Y, Li L. (2018). MiR-1294 confers cisplatin resistance in ovarian cancer cells by targeting IGF1R. Biomedicine & Pharmacotherapy 106: 1357–1363. DOI 10.1016/j.biopha.2018.07.059. [Google Scholar] [CrossRef]
Zhou Y, Zheng X, Lu J, Chen W, Li X, Zhao L. (2018). Ginsenoside 20(S)-Rg3 inhibits the Warburg effect via modulating DNMT3A/MiR-532-3p/HK2 pathway in ovarian cancer cells. Cellular Physiology and Biochemistry 45: 2548–2559. DOI 10.1159/000488273. [Google Scholar] [CrossRef]
Zhu W, Huang Y, Pan Q, Xiang P, Xie N, Yu H. (2017). MicroRNA-98 suppress Warburg effect by targeting HK2 in colon cancer cells. Digestive Diseases and Sciences 62: 660–668. DOI 10.1007/s10620-016-4418-5. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |