DOI:10.32604/biocell.2021.014633

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.014633 |  www.techscience.com/journal/biocell |
| Article |
Tet methylcytosine dioxygenase 2 suppresses renal cell cancer proliferation and metastasis by regulating the miR-200c-SCD axis
1Fujian Provincial Key Laboratory of Transplant Biology, Department of Urology, Dongfang Hospital (900 Hospital of the Joint Logistics Team), School of Medicine, Xiamen University, Fuzhou, 350025, China
2Institute of Ningde Urological Research and Department of Urology, Affiliated Mindong Hospital of Fujian Medical University, Fu’an, 355000, China
3Department of Neurology, Affiliated Mindong Hospital of Fujian Medical University, Fu’an, 355000, China
*Address correspondence to: Jianming Tan, tanjm156@xmu.edu.cn; Huizhang Li, ndlhzsci@hotmail.com
Received: 14 October 2020; Accepted: 21 December 2020
Abstract: Tet methylcytosine dioxygenase 2 (TET2) acts as an antioncogene that is investigated in different cancers. But the effects of TET2 in renal cell cancer (RCC) is still known little. Here, quantitative real-time PCR (qRT-PCR), Western blot, and immunofluorescence were performed to exam gene and protein expression. Cell proliferation was measured using Cell Counting Kit-8 (CCK-8). Transwell assay was performed to detect cell metastasis viability. Flow cytometry was performed to analyze the cell cycle and cell apoptosis. The effects of TET2 on RCC growth in vivo was analyzed using a mouse xenograft model.We found that TET2 and miR-200c were decreased in RCC tissues, and hypermethylation of miR-200c promoter was found. Overexpression of TET2 promoted miR-200c expression by reducing miR-200c promoter methylation. Additionally, overexpression of TET2 or miR-200c suppressed cell growth and metastasis. Also, knockdown of miR-200c could moderate TET2 mediated cell growth inhibition. Furthermore, we found miR-200c directly regulates Stearoyl-CoA desaturase (SCD) gene expression. Moreover, in vivo experiment results confirmed that TET2 inhibited tumor growth. In conclusion, TET2 acts as an antioncogene in RCC by regulating the miR-200c-SCD axis and providing a potential target for RCC diagnosis and treatment.
Keywords: TET2; miR-200c; SCD; Renal cell cancer; Tumor growth
Kidney malignancy is a common cancer worldwide (Ferlay et al., 2015). More than 90% of all kidney cancer cases were diagnosed as renal cell cancer (RCC) (Kuthi et al., 2017). With the development of therapeutic methods, the treatment for RCC has been greatly improved over the past decades (Zeng, 2018). However, more than 20% of RCC patients with localized tumors were accompanied by local recurrence or metastases (Capitanio and Montorsi, 2016). Currently, the molecular mechanisms of RCC occurrence and development are largely unknown. Thus, it is pressing to investigate the molecular biological mechanisms of RCC and progression and develop potential effective biomarkers for RCC therapeutic and prognostic.
DNA methylation plays crucial roles in cancer development through regulating gene expression (Arthur et al., 2018; Hannon et al., 2018). The family of ten-eleven translocation (TET) genes has been found to play large influences in the epigenetic field (He et al., 2011; Koh et al., 2011). TET2 is tet methylcytosine dioxygenase, which belongs to the TET gene family, and TET2 enzymatic activity converts 5-methylcytosine to 5-hydroxymethylcytosine, resulting in activating demethylation and promoting gene expression (Hashimoto et al., 2012). Previous studies have found that TET2 protein played a critical role during embryogenesis through epigenetic regulation of gene expression (Dawlaty et al., 2013), tumor occurrence and progression (Meisel et al., 2018; Zeng et al., 2019), somatic cell reprogramming (Costa et al., 2013), and hematopoietic cell differentiation (Ko et al., 2011; Pan et al., 2017). Recently, Fraietta et al. (2018) found that dysregulation of TET2 enhances the efficacy of immunotherapy of chimeric antigen receptor T cells (CAR-T). A recent study demonstrated that TET2 is decreased in RCC tissues (Elgendy et al., 2019). However, the mechanisms by which decreased TET2 leads to the development and progression of human renal cell cancer remain largely unknown.
Although TET2 was found decreased in human renal cell cancer tissues, whether TET2 contributes to RCC progression is still not clear, and the molecular biological mechanisms of TET2 in the regulation of human renal cell cancer remain unknown. Wu et al. (2017) showed that all-trans retinoic acid promotes a novel retinoid X receptor β (RARβ) interacted with TET2 to epigenetically activate miR-200c expression and further suppressed its target gene protein kinase Cζ (PKCζ) expression. However, whether TET2 regulates miR-200c expression by reducing the methylation level of the miR-200c promoter is unknown. Here, we describe novel roles for TET2 in inhibition of renal cell cancer tumorigenesis and growth. Also, we confirmed that TET2 is decreased in RCC tissues, and overexpression of TET2 suppressed 786-O and ACHN cell growth and migration. Additionally, we also found that overexpression of TET2 promoted miR-200c demethylation, resulting in promoted miR-200c expression.
Stearoyl-CoA desaturase (SCD)-1 is an endoplasmic reticulum-associated enzyme. Its enzymatic activity could induce the conversion of saturated fatty acids (SFAs) to monounsaturated fatty acids (MUFAs) (Enoch et al., 1976; Dobrzyn et al., 2015). Suppression of SCD-1 has hence been proposed as a great pharmaceutical target for anticancer drug development (Tracz-Gaszewska and Dobrzyn, 2019). SCD-1 level in cells is mainly regulated by a transcription process. In the present study, we found that miR-200c directly binds to the 3’-UTR of the SCD gene, and knockdown of SCD expression or overexpression of miR-200c could suppress cell growth and induce cell apoptosis. According to the results, we demonstrate that TET acts as an antioncogene in RCC by regulating the miR-200c-SCD signaling pathway, providing a potential therapeutic target for RCC treatment.
36 renal cell cancer tissues were surgically obtained from patients at Dongfang Hospital from October 2017 to May 2018. 36 patients (17 males and 19 females) with a mean age of 48.7 years were collected without any chemoradiotherapy before surgery. The clinicopathological features were confirmed and recorded by two independent clinical pathologists based on AJCC standards (Edge and Compton, 2010). All volunteers signed the informed consent, and the study was approved by the Ethics Committee of Dongfang Hospital, School of Medicine, Xiamen University.
The renal cell cancer cell lines 786-O and ACHN were obtained from the National Science&Technology Infrastructure (Shanghai, China). 786-O cells were cultured in RPMI-1640 medium, and ACHN cells were maintained in DMEM medium with 10% FBS in a humidified incubator containing 5% CO2 at 37°C. All medium and FBS were purchased from Gibco (Waltham, MA, USA).
The TET2 and SCD gene overexpression vector was amplified from cDNA and cloned downstream of the pcDNA4.0 vector, and the sequence was confirmed by DNA sequencing (Sangon, Shanghai, China). The TET2 gene knockdown vector was purchased from GenePharma (Shanghai, China).
The 3’-untranslated region (3’-UTR) of SCD gene containing miR-200c binding sites was amplified and cloned into the psi-CHECK2 vector, named SCD-3’UTR-WT.The mutant 3’-UTR of SCD gene was generated using the TaKaRa MutanBEST Kit (TaKaTa, China), which generated a mutation in the predicted miR-200c target binding sites (from CAGUAUU to GUCAUAA), identified as SCD-3’UTR-MUT.
The miR-200c mimics, inhibitor, and negative control RNA were purchased from Synbio Technologies (Suzhou, China). 10,000 cells per well were seeded and incubated at 37°C for 24 h in a 96-well plate; the cells were transfected with 100 nM RNAs or 20 ng plasmids using Lipofectamine 4000 (Invitrogen, USA). The transfection efficiency was confirmed to be more than 80%.
Bisulfite Sequencing PCR (BSP)
Genomic DNA was extracted using the Universal Genomic DNA Extraction Kit (Solarbio, Beijing, China) according to the product manual. The genomic DNA was treated with EpiTect Fast DNA Bisulfite Kit (Qiagen, Germany) according to the manufacture’s guideline, and DNA was collected for sequencing.
Quantitative real-time PCR (qRT-PCR)
Total RNA from RCC tissues and different RCC cells was extracted using TriQuick Reagent (Solarbio, Beijing, China), and Nanodrop 2000 was used to quantify the RNA concentration. 2 μg of total RNA were used for reverse transcription using One Step SuperRT-PCR Mix Kit (Solarbio, Beijing, China). SYBR Green I (Solarbio, Beijing, China) was used for qRT-PCR. Amplification and detection were performed on Applied Biosystems 7500 Real-time PCR Detection System (ABI, USA). GAPDH or U6 were used as internal references to calculate the value of relative mRNA or miRNA expression. The data were calculated using the 2−ΔΔCt method.
Total proteins were extracted and quantified using Nanodrop 2000. 20 μg of total proteins were separated in 10% SDS-PAGE and electrophoretically transferred to polyvinylidene difluoride membranes (Roche, Mannheim, Germany) using eBlot™ L1 (Genscript, Nanjing, China), then the transfected membranes were blocked and incubated with primary antibody (anti-TET2 (Abcam, ab94580, 1:1000), anti-SCD (Abcam, ab236868, 1:1500), anti-Nanog (Abcam, ab109250, 1:1000), anti-Caspase 3 (Abcam, ab13847, 1:800), and anti-GAPDH (Abcam, ab9485, 1:2000)) at 37°C for 1 h. The membranes were washed and incubated with HRP conjugated secondary antibody (Abcam, ab6721 or ab6728, 1:5000) and visualized by chemiluminescence.
Dual-luciferase reporter assay
20 ng of wild type or mutant psi-CHECK2-SCD-3’UTR plasmids and 100 nM miR-200c mimics or NC were firstly co-transfected into 5 × 104 293T cells in 24-well plates. The treated cells were cultured for 48 h, then luciferase activities were measured using Dual-Lucy Assay Kit (Solarbio, Beijing, China).
1 × 105 treated cells were seeded in a 96-well plate and cultured for 24, 48, and 72 h. At each time point, cell proliferation was determined using the CCK-8 (Solarbio, Beijing, China) in accordance with the user guide.
The in vitro cell metastasis ability was performed using the 24-well transwell chambers (Corning, NY, USA). The details were shown as previously described (Zhuang et al., 2017).
Cell cycle and cell apoptosis were analyzed with flow cytometry using cell cycle staining kit and Annexin-V FITC/PI apoptosis detection kit in accordance with the manufacturer’s instruction. A FACScan (BD, Biosciences, UK) was used for flow cytometry.
The treated cells were fixed with 4%polyformaldehyde and stained with 1 mg/mL of Hoechst33258 Stain solution (Solarbio, Beijing, China). A fluorescence microscope (Olympus Corporation, Japan) was used to observe the morphology of cell nuclei and to take pictures.
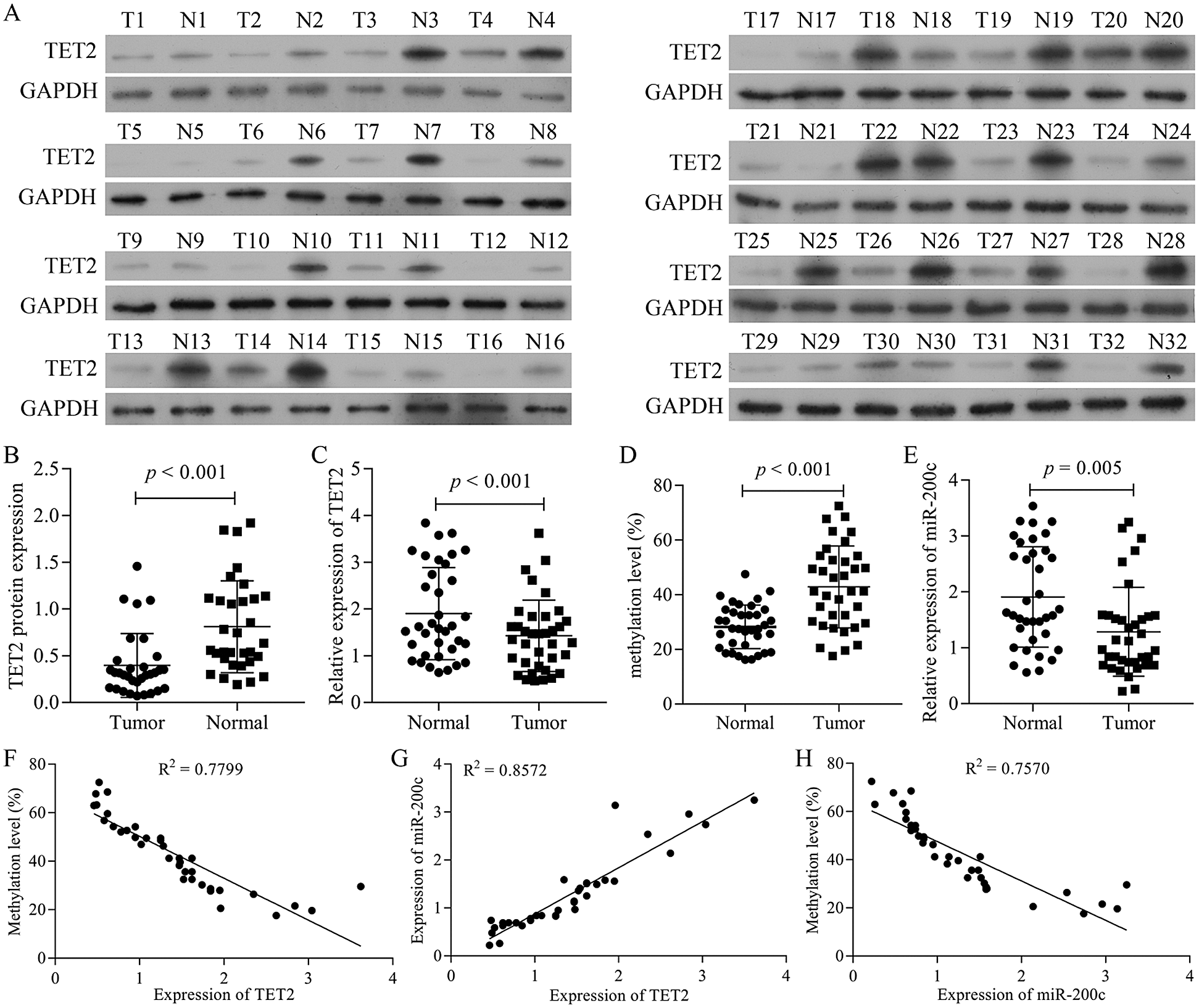
Figure 1: TET2 is downregulated in RCC tissues and positively correlated with miR-200c.
Cells were collected and fixed with 4%polyformaldehyde, then cells were incubated with Anti-TET2 and anti-SCD primary antibodies at 37°C for 1 h. After that, cells were incubated with FITC tagged or Cy5 tagged secondary antibodies in dark for 1 h at 37°C. Afterwards, nuclei were counterstained with DAPI.
Thirty BALB/c Nude mice (5 weeks old, male) were obtained from Charles River (Beijing, China). The 786-O cells transfected with shTET2 or pcDNA4.0-TET2 were digested and collected and adjusted the cell density to 2.5 × 107 cells/mL, and 0.2 mL of 786-O cells was injected into the right axilla of each nude mouse subcutaneously. After injection, tumor formation was examined at different time points. The mice were euthanized by intraperitoneal injection with 300 mg/kg of 10% chloral hydrate, and then euthanized by cervical dislocation on day 21, and the tumor volume was calculated by the following formula: (L × W2)/2, “L” means length, “W” means width.
Hematoxylin-eosin (H&E) staining
The tissues were fixed with a formaldehyde solution and embedded in paraffin, and tissues were cut as 4-μm sections, and slides were stained with hematoxylin and eosin. These sections were examined under a light microscope at a magnification of 200×.
The fixed tissues were embedded in paraffin and cut with 4-μm sections and immunostained as previously described (Jing et al., 2019). The slides were incubated with TET2 primary antibody (1:100) or Ki-67 primary antibody. Then, the slides were incubated with HRP-conjugated secondary antibodies. Hematoxylin was used to stain cell nuclei. Images were collected at a magnification of 200 × under a microscope (Olympus, Tokyo, Japan).
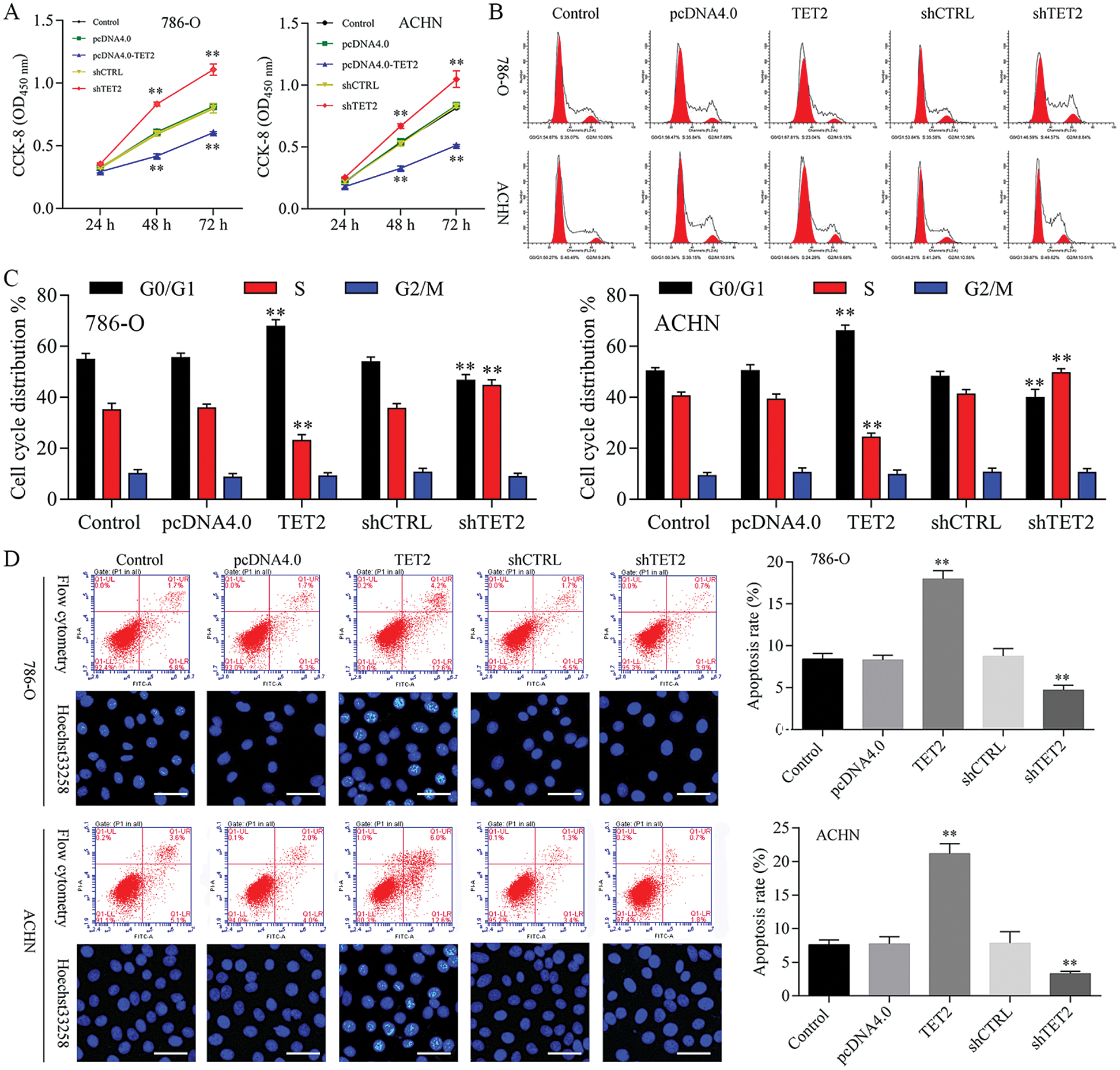
Figure 2: Overexpression of TET2 suppresses RCC cell growth, and knockdown of TET2 promotes RCC cell growth.
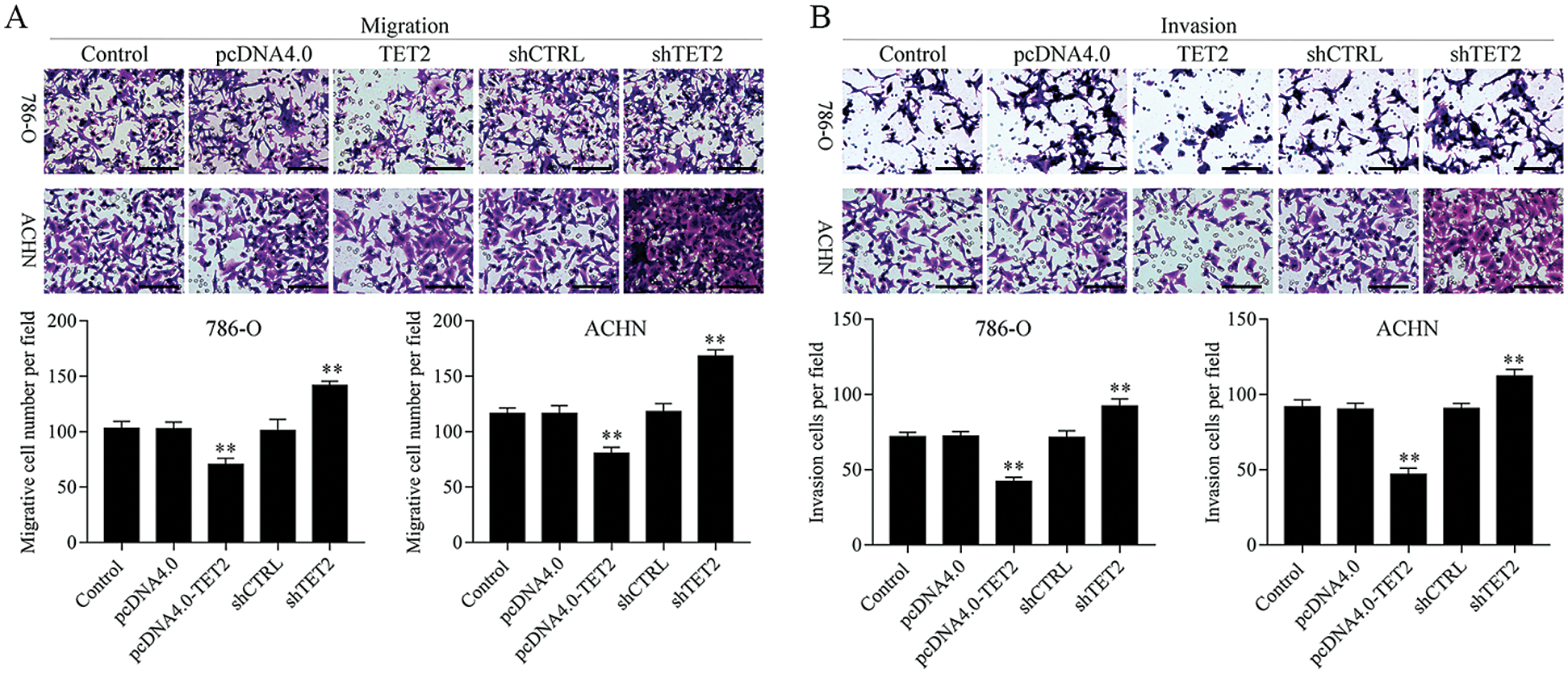
Figure 3: Overexpression of TET2 suppresses RCC cell metastasis.
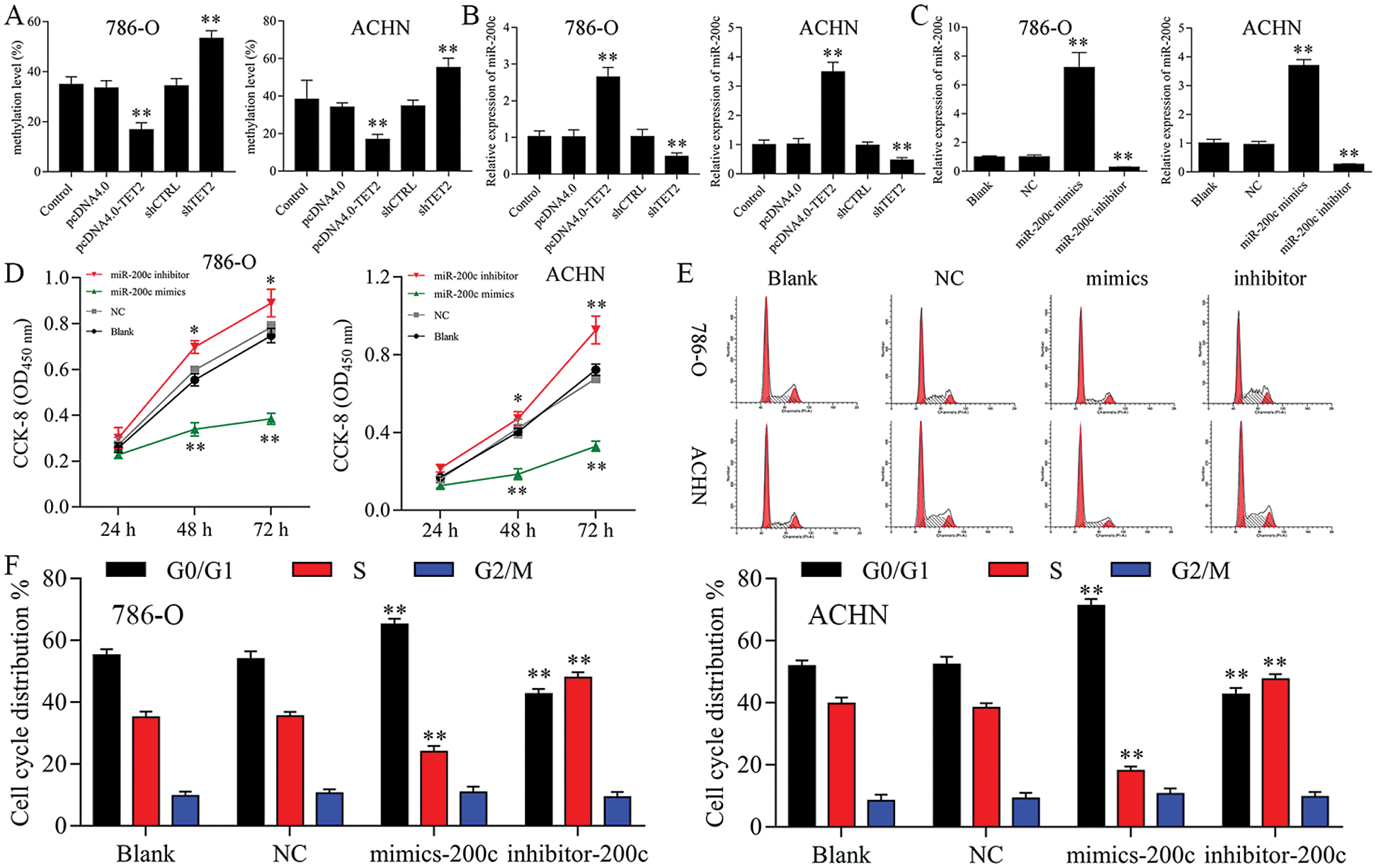
Figure 4: Upregulation of miR-200c suppresses RCC cell growth.
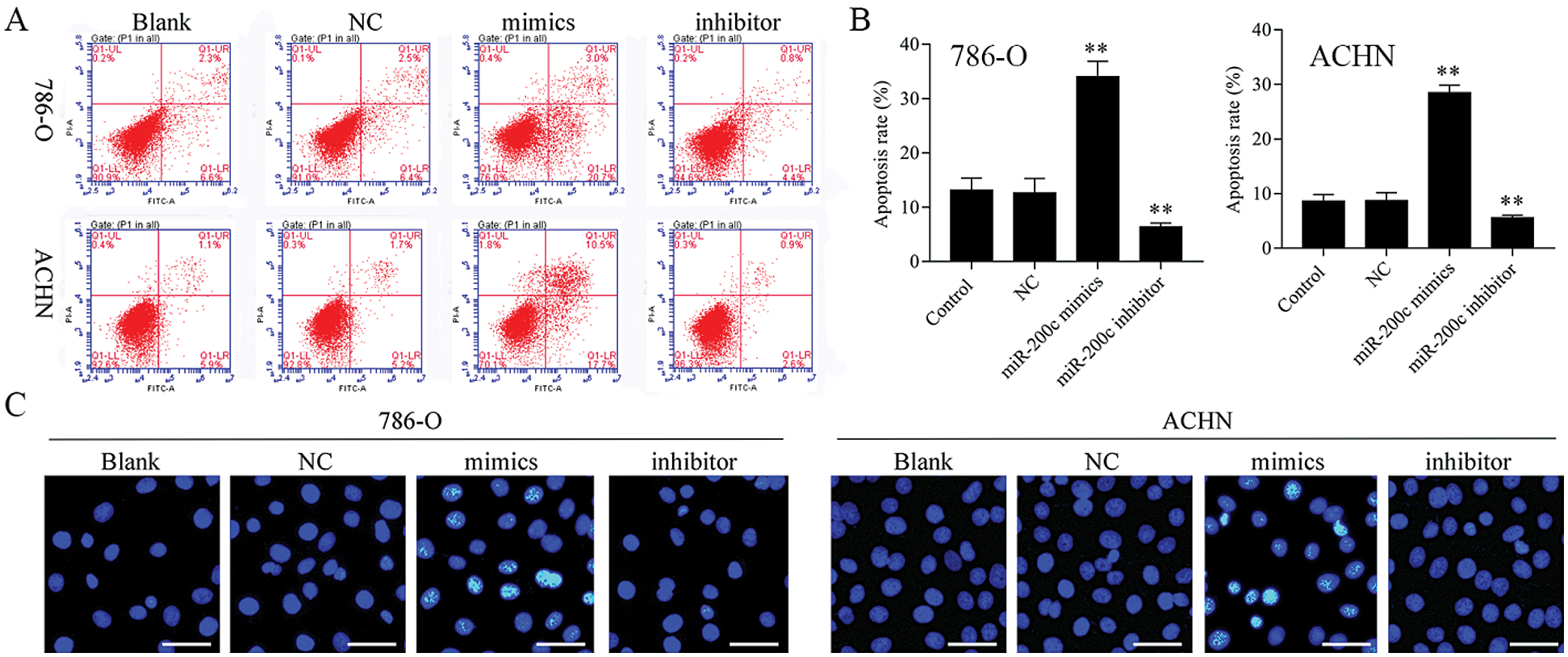
Figure 5: Overexpression of miR-200c induces RCC cell apoptosis.
TUNEL method was performed to assess cell death using a TUNEL Apoptosis Assay Kit (Solarbio, Beijing, China), according to the user guide.
All data are expressed as mean ± standard deviation. The paired or unpaired Student’s t-test was performed to analyze the difference between two groups, and one-way ANOVA followed by Tukey’s post hoc test was used for multi-group comparisons using software SPSS 20.0 (IBM Corp). p < 0.05 was considered statistical significance.
TET2 is decreased in RCC tissues and positively correlated with miR-200c expression
We firstly detected the TET2 expression levels in RCC tissues. The results indicated that the expression of TET2 was decreased in human RCC tissues (Figs. 1A–1C). Furthermore, BSP and qRT-PCR results indicated that the methylation level of miR-200c promoter significantly increased and miR-200c significantly decreased in RCC tissues (Figs. 1D and 1E). Moreover, the expression of TET2 levels was negatively associated with miR-200c promoter methylation levels (Fig. 1F) and positively associated with miR-200c expression (Fig. 1G), and miR-200c expression levels were negatively associated with miR-200c promoter methylation levels (Fig. 1H). Altogether, the results suggest that downregulation of TET2 and miR-200c may be contributed to the development of human renal cell cancer.
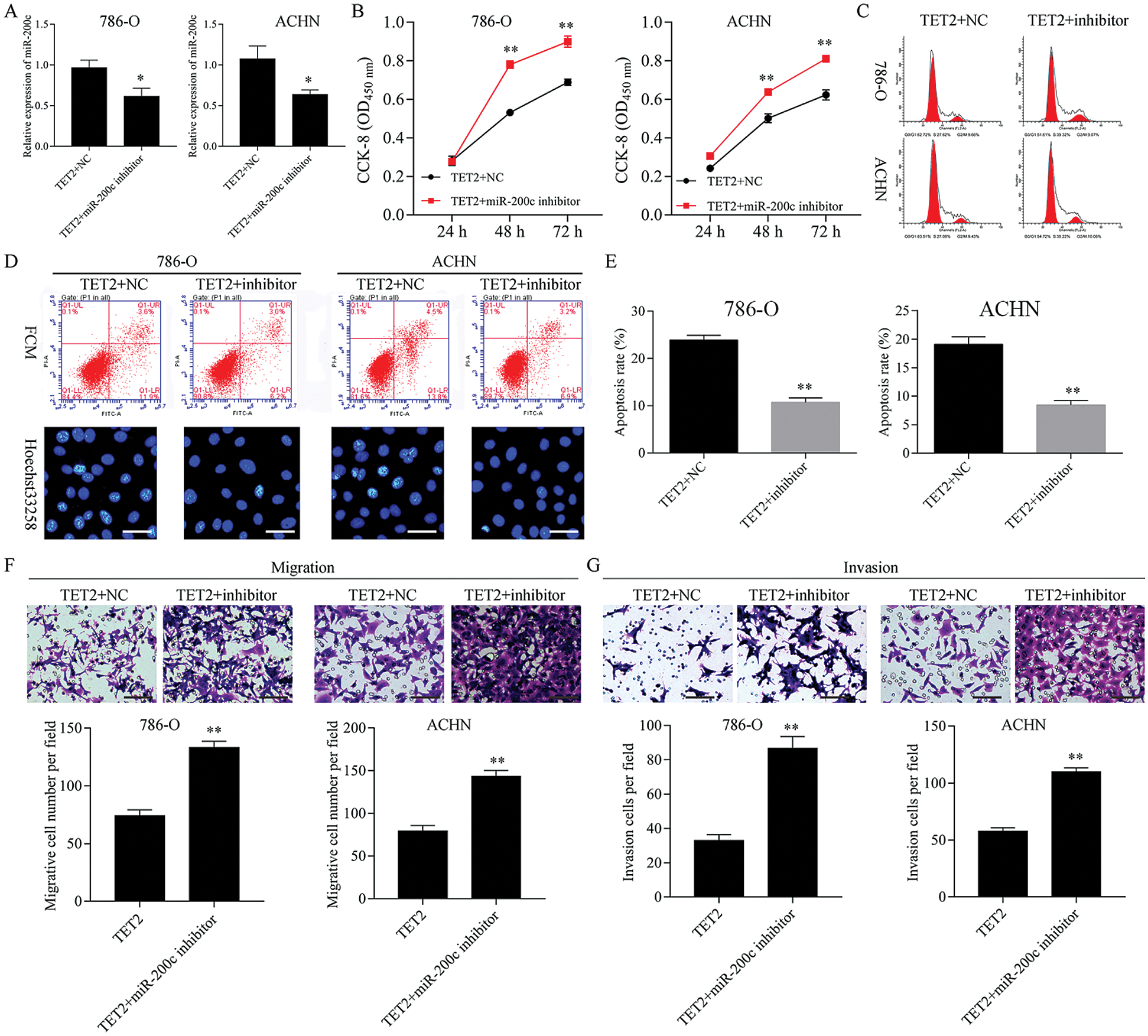
Figure 6: Knockdown of miR-200c ameliorated TET2 overexpression mediated cell proliferation suppression and apoptosis promotion.
Overexpression of TET2 suppresses RCC cell growth and knockdown of TET2 promotes RCC cell growth
To confirm TET2 suppresses RCC cell growth, TET2 was manipulated in RCC cells after transfected with TET2 overexpression plasmids and knockdown plasmids (Figs. S1). Furthermore, CCK-8 results indicated that overexpression of TET2 suppressed RCC cell growth and knockdown of TET2 promoted RCC cell growth (Fig. 2A), and upregulation of TET2 decreased cell S phase and increased G1 phase, and downregulation of TET2 decreased cell G1 phase and increased S phase in RCC cells (Figs. 2B and 2C). In addition, overexpression of TET2 induced RCC cell apoptosis, and knockdown of TET2 reduced RCC cell apoptosis (Fig. 2D). Moreover, we also measured the role of TET2 on cell metastasis. Transwell assay results showed that overexpression of TET2 inhibited RCC metastasis, and knockdown of TET2 promoted RCC migration and invasion (Fig. 3). Finally, Western blot and IF results showed that overexpression of TET2 reduced SCD and Nanog protein expression and induced caspase 3 expression, while knockdown of TET2 induced SCD and Nanog protein expression and reduced caspase 3 expression (Figs. S1 and S2). Thus, the data suggested that TET2 could regulate the cell cycle and cell apoptosis to moderate RCC cell growth.
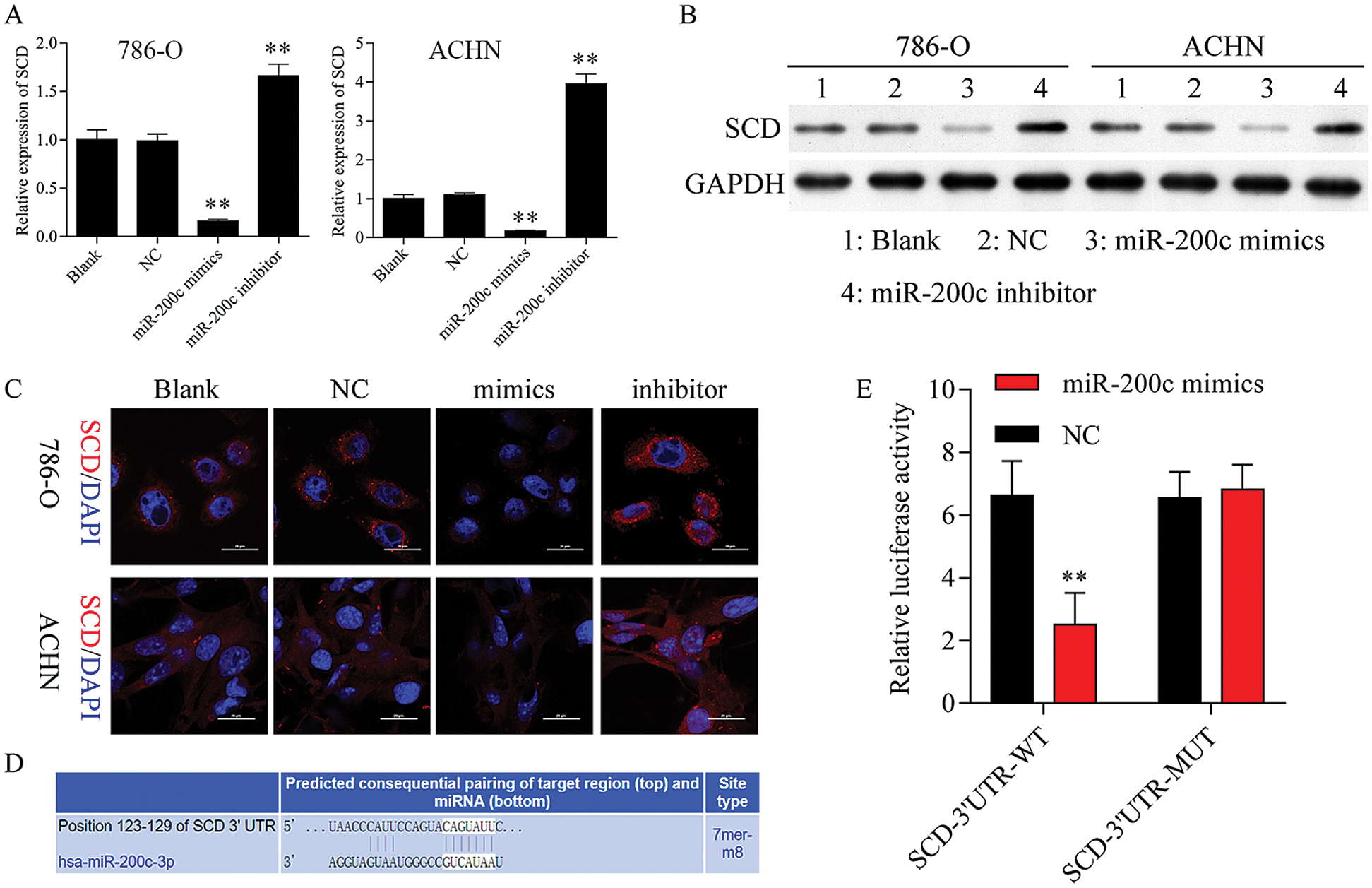
Figure 7: SCD is a target gene of miR-200c.
miR-200c suppresses RCC cell growth
Firstly, overexpression of TET2 could decrease the methylation level of the miR-200c promoter and induce miR-200c expression in RCC cells, while knockdown of TET2 could induce the methylation level of the miR-200c promoter and reduce the expression of miR-200c in RCC cells (Figs. 4A and 4B). Additionally, we manipulated the miR-200c expression in RCC cells (Fig. 4C), and upregulation of miR-200c inhibited RCC cell proliferation, and knockdown of miR-200c promoted RCC cell proliferation (Fig. 4D) and flow cytometry results showed that upregulation of miR-200c decreased cell S phase and increased G1 phase, and downregulation of miR-200c decreased cell G1 phase and increased S phase in RCC cells (Figs. 4E and 4F). Moreover, apoptosis assay results indicated that upregulation of miR-200c could promote RCC cell apoptosis (Fig. 5) through regulating the expression of Nanog and caspase 3 (Fig. S3). Finally, Transwell assay results showed that upregulation of miR-200c inhibited RCC cell metastasis (Fig. S4). Thus, our results suggested that TET2 might reduce miR-200c promoter methylation to induce miR-200c expression, and miR-200c could control cell cycle and cell apoptosis to regulate RCC cell growth.
Knockdown of miR-200c ameliorated TET2 overexpression mediated cell proliferation suppression and apoptosis promotion
Next, miR-200c was suppressed in pcDNA4.0-TET2 transfected RCC cells as showed by qRT-PCT (Fig. 6A). And also, knockdown of miR-200c in pcDNA4.0-TET2 transfected RCC cells significantly decreased the suppression effect of TET2 overexpression on SCD protein expression in 786-O and ACHN cells (Fig. S5). Furthermore, knockdown of miR-200c in pcDNA4.0-TET2 transfected RCC cells significantly decreased the suppression effect of TET2 overexpression on RCC cell proliferation and cell S phase (Figs. 6B and 6C and Fig. S6A). Moreover, knockdown of miR-200c in pcDNA4.0-TET2 transfected RCC cells significantly decreased the promotion effect of TET2 overexpression on RCC cell apoptosis (Figs. 6D and 6E). In addition, knockdown of miR-200c in pcDNA4.0-TET2 transfected RCC cells significantly decreased the suppression effect of TET2 overexpression on RCC cell migration and invasion (Figs. 6F and 6G).
SCD is a target gene of miR-200c
Firstly, we found that upregulation of miR-200c suppressed SCD mRNA and protein expression, while knockdown of miR-200c promoted SCD expression (Figs. 7A–7C). Additionally, TargetScan (www.targetscan.org) prediction database predicted that miR-200c has a predictive target site in SCD-3’-UTR (Fig. 7D). Dual-luciferase reporter assay confirmed that SCD is one of the miR-200c targets (Fig. 7E).
Knockdown of SCD suppresses RCC cell growth
We first detected the expression of SCD in RCC cells after transfected with SCD overexpression plasmids and knockdown plasmids. We found that SCD was upregulated after pcDNA4.0-SCD transfection and downregulated after shSCD transfection (Fig. S7). Furthermore, knockdown of SCD suppressed RCC cell growth, and overexpression of SCD promoted RCC cell proliferation (Fig. 8A), and apoptosis assay results showed that upregulation of SCD increased cell S phase and decreased G1 phase and downregulation of SCD increased cell G1 phase and decreased S phase in RCC cells (Figs. 8B and 8C). In addition, flow cytometry and Hoechst 33258 staining results indicated that downregulation of SCD induced RCC cell apoptosis and overexpression of SCD reduced RCC cell apoptosis (Fig. 8D). Moreover, we also measured the role of SCD on cell metastasis. Transwell assay results showed that overexpression of SCD promoted RCC metastasis, and knockdown of SCD inhibited RCC metastasis (Fig. 9).
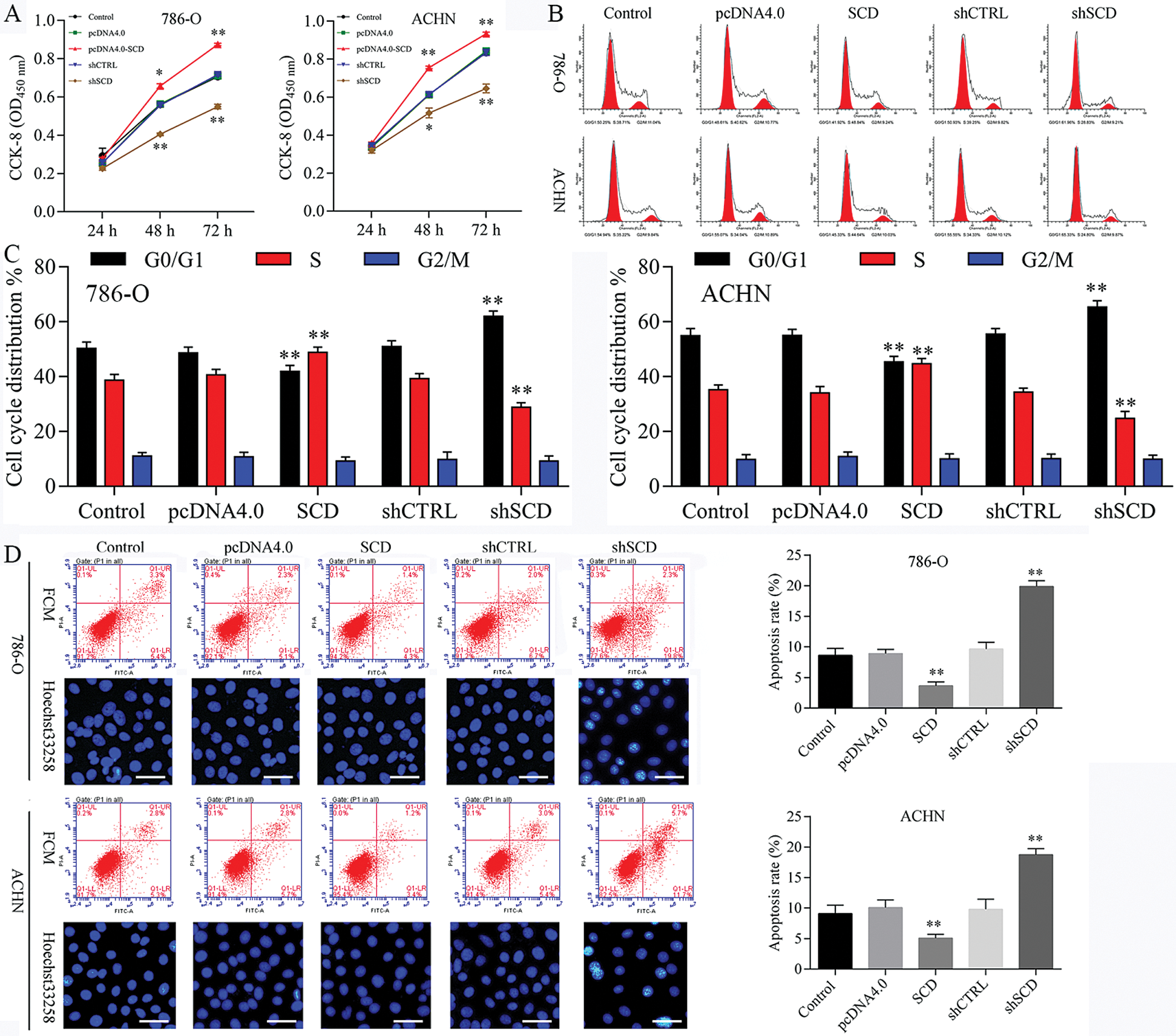
Figure 8: Knockdown of SCD suppresses RCC cell growth, and overexpression of SCD promotes RCC cell growth.
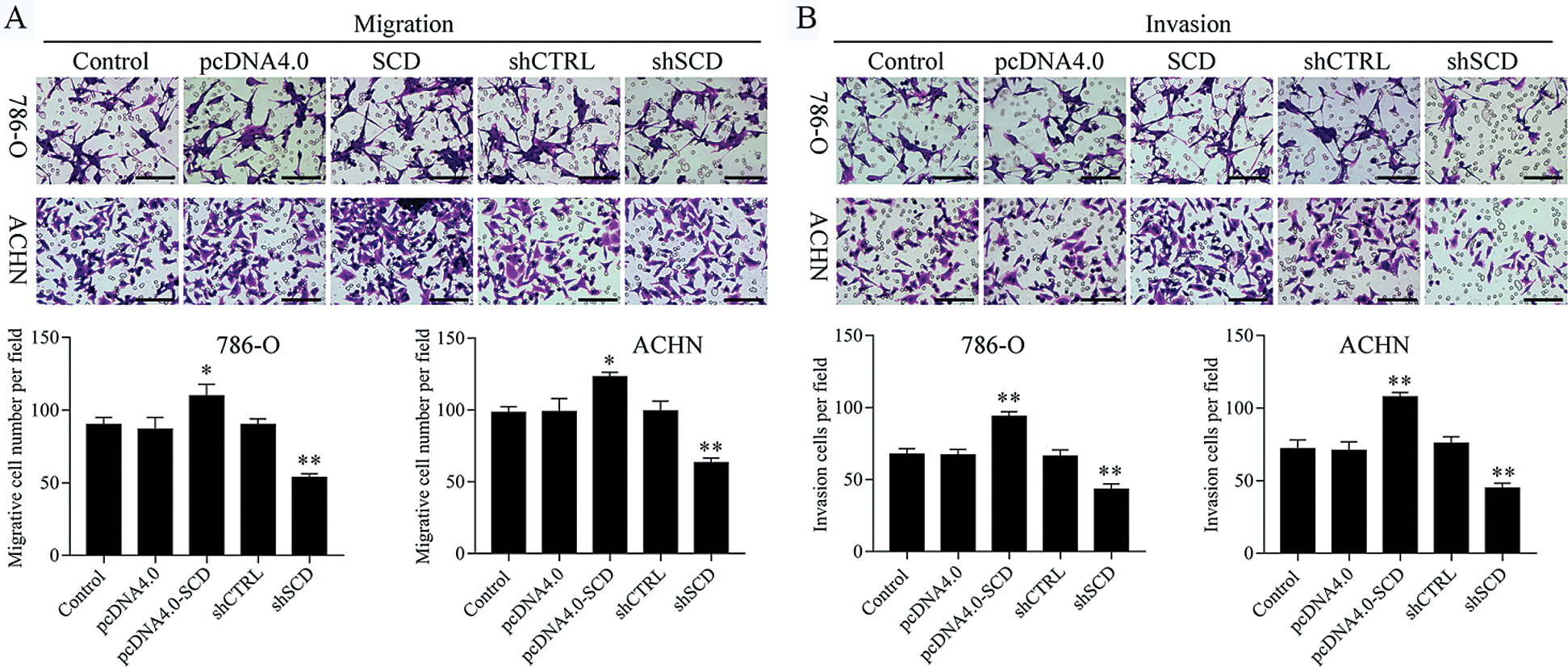
Figure 9: Knockdown of SCD suppresses RCC metastasis.
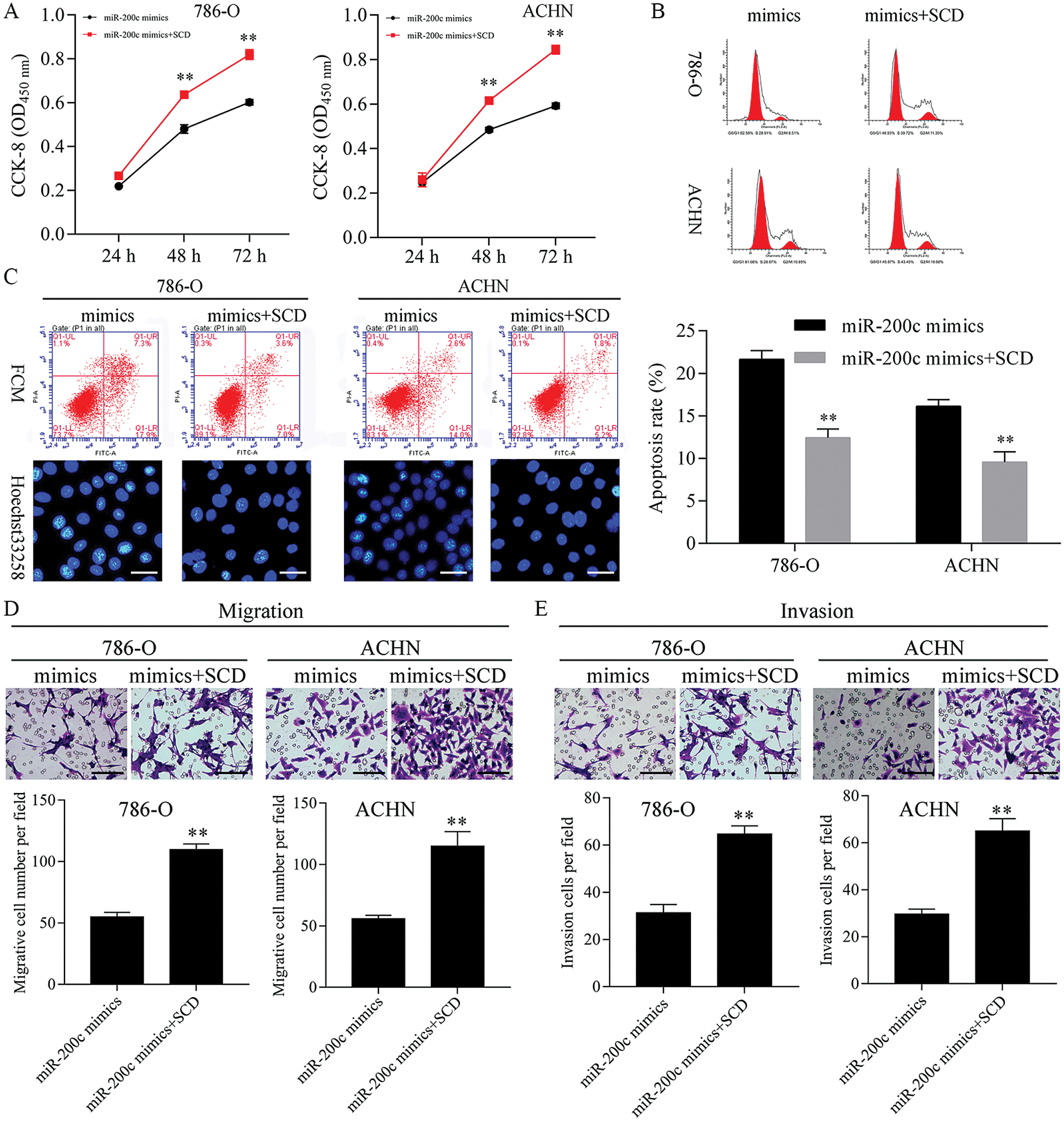
Figure 10: Upregulation of SCD ameliorated the miR-200c mimics-mediated cell growth inhibition.
Overexpression of SCD ameliorated the miR-200c mimics mediated cell proliferation and metastasis inhibition, and apoptosis promotion
Next, SCD was overexpressed in miR-200c mimics transfected RCC cells (Fig. S8). Furthermore, overexpression of SCD in miR-200c transfected RCC cells significantly decreased the inhibition effect of miR-200c on RCC cell growth and S phase (Figs. 10A and 10B; Fig. S6B). Moreover, overexpression of SCD in miR-200c mimics transfected RCC cells significantly suppressed the promotion effect of miR-200c on RCC cell apoptosis (Fig. 10C). In addition, overexpression of SCD in miR-200c mimics transfected RCC cells significantly decreased the suppression effect of miR-200c overexpression on RCC cell migration and invasion (Figs. 10D and 10E).
Overexpression of TET2 suppresses renal cell tumor growth in vivo
To further confirm TET2 suppresses cell proliferation by regulating miR-200c-SCD axis in RCC in vivo, we generated five groups of 786-O cells, also transfected with (1) transfection reagent (control group), (2) pcDNA4.0, (3) pcDNA4.0-TET2, (4) shCTRL, (5) shTET2, and inoculated these cells (5 × 106) into each nude mouse. In vivo experiments results showed that the mice inoculated with pcDNA4.0-TET2 cells had a smaller tumor size, while mice inoculated with shTET2 cells had a larger tumor size (Figs. 11A and 11B) and TET2 was suppressed in the shTET2 group and upregulated in the pcDNA4.0-TET2 group (Figs. 11C–11E). Furthermore, the protein of SCD was increased in the shTET2 group and decreased in the pcDNA4.0-TET2 group (Fig. 11E), and miR-200c was decreased in the shTET2 group (Fig. 11D) Moreover, Ki-67 staining and TUNEL results showed that overexpression of TET2 suppressed tumor growth and induced apoptosis, and knockdown of TET2 promoted tumor growth and suppressed apoptosis (Fig. S9). Together, results confirmed that overexpression of TET2 suppresses renal cell tumor growth in vivo.
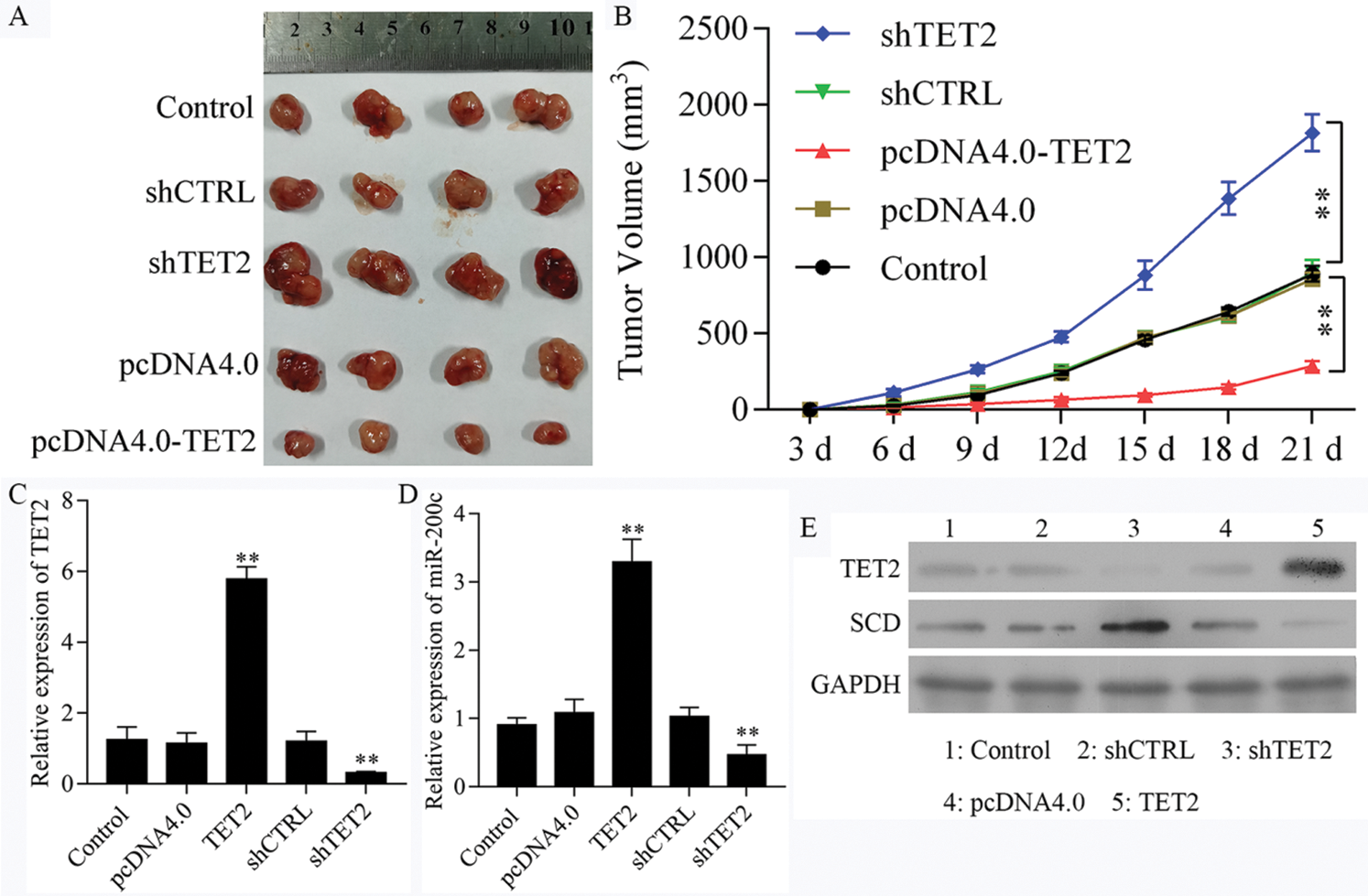
Figure 11: Upregulation of TET2 suppresses renal cell tumor growth in vivo.
Increasing studies have been suggested that epigenetic mediates miRNAs dysregulation play critical roles in multifarious cellular processes (Weiss and Ito, 2017; Ortiz et al., 2018), and also contributes to human diseases, including cancer (Aure et al., 2013; Nojima et al., 2016; Zeng et al., 2019b). Here, we found that the levels of TET2 and miR-200c expression were significantly decreased in human RCC tissues. In addition, overexpression of TET2 reduced the methylation levels of the miR-200c promoter and promoted miR-200c expression. Furthermore, overexpression of TET2 and miR-200c suppressed RCC cell growth, metastasis, and promoted apoptosis, while knockdown of TET2 and miR-200c promoted RCC cell proliferation, metastasis, and reduced cell apoptosis. Moreover, overexpression of TET2 induced miR-200c expression through reducing miR-200c promoter methylation level results in inhibiting renal cancer growth in vivo.
MicroRNAs (miRNAs) are a group of small single-stranded RNAs that involve various biological functions (Bartel, 2004). miRNAs could regulate gene expression by binding to the3’-UTR of target genes and either inducing degradation or inhibiting translation of target mRNAs (Calin and Croce, 2006). miR-200c is one of the miR-200 superfamily members and is involved in stemness (Cha et al., 2017), epithelial-mesenchymal transition (EMT) (Title et al., 2018), and chemoresistance in various cancer cells (Fukuda et al., 2019). Increasing studies indicated that miR-200 family members were involved in the progression of cancers and among which, downregulated miR-200c was found in RCC (Liu et al., 2010; White et al., 2011). Chang et al. (2015) have proposed that miR-200c may be considered as a potential biomarker for diagnoses and treatment of renal cell cancer. Here, in our present study, we found the expression of miR-200c was regulated by TET2, which decreased miR-200c promoter methylation level. Furthermore, upregulation of miR-200c inhibited RCC cell growth and promoted cell apoptosis through regulating SCD expression.
Stearoyl-CoA desaturase 1 (SCD1) has been found as a lipid metabolism-related gene involved in human cancers and plays as a potential oncogene (Sánchez-Martínez et al., 2015; Cruz-Gil et al., 2018). SCD1 has been found frequently overexpressed in human cancers. SCD1 inhibits the AMPK signaling pathway, enhancers AKT signaling pathway, and modulates lipid metabolism by reducing fatty acid oxidation while fostering lipogenesis, thus contributing to cancer cell growth (Tracz-Gaszewska and Dobrzyn, 2019). Down-regulation of SCD1 promoted cell apoptosis in aldehyde dehydrogenase 1A1-positive cells and reversed resistance to cisplatin of lung cancer stem cells in vivo (Mancini et al., 2011; Pisanu et al., 2017). The biological effects of SCD1 on fatty acid metabolism are mediated by YAP/TAZ signaling, which in turn is regulated, at least in part, by the autocrine activity of the β-catenin pathway (Noto et al., 2017). Depending on the experimental model used, SCD1 inhibitors have shown activity in distinct stages of tumorigenesis by modulating EGF, ER stress, PI3K/AKT/HIF2, and NF-kB pathways (Tracz-Gaszewska and Dobrzyn, 2019). In addition, ferroptosis and SCD1 metabolism share common lipid mediators (Hassannia et al., 2019). Here, we found that SCD was increased in human RCC tissues, and SCD was downregulated in RCC cells after transfected with miR-200c mimics. Also confirmed that miR-200c directly binding to the SCD gene. Additionally, upregulation of SCD suppressed RCC cell growth, metastasis, and promoted apoptosis in vitro.
Taken together, our data indicated that TET2 and miR-200c were decreased in RCC tissues, while the levels of SCD expression were increased. Additionally, we confirmed that upregulation of TET2 promoted miR-200c expression through decreasing the methylation level of the miR-200c promoter and also determined that miR-200c directly regulates SCD. Furthermore, overexpression of TET2 and miR-200c and downregulation of SCD could inhibit cell growth and metastasis while promoting cell apoptosis. All the results demonstrated that the TET2-miR-200c-SCD axis may provide a potential target for renal cell cancer treatment.
Acknowledgement: Not appropriable.
Availability of Data and Materials: The datasets used during the current study are available from the corresponding author on reasonable request.
Author Contribution: The authors confirm contribution to the paper as follows: study conception and design: Benjiang Qian, Jianming Tan, Huizhang Li; data collection: Benjiang Qian, Youfeng Huang, Zhenqiang Qiu, Xiaoyan Ying, Guang Yang; analysis and interpretation of results: Benjiang Qian, Jianming Tan; draft manuscript preparation: Benjiang Qian. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: This study was approved by the Ethical Committee of the Dongfang Hospital affiliated with Xiamen University.
Funding Statement: The study was supported by Grants from the Nature Science Foundation of Fujian, China (Nos. 2010J01372, 2015J01571).
Conflict of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Arthur B, Qiang H, Pooja N, Liam S, Hannah G, Matthew M, Qi H, Jacob P, Tzung-Fu H, An YQ. (2018). Dynamic DNA methylation in plant growth and development. International Journal of Molecular Sciences 19: 2144. DOI 10.3390/ijms19072144. [Google Scholar] [CrossRef]
Aure MR, Leivonen SK, Fleischer T, Zhu Q, Overgaard J, Alsner J, Tramm T, Louhimo R, Alnæs GI, Perälä M, Busato F, Touleimat N, Tost J, Børresen-Dale AL, Hautaniemi S, Troyanskaya OG, Lingjærde OC, Sahlberg KK, Kristensen VN. (2013). Individual and combined effects of DNA methylation and copy number alterations on miRNA expression in breast tumors. Genome Biology 14: R126. DOI 10.1186/gb-2013-14-11-r126. [Google Scholar] [CrossRef]
Bartel DP. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. DOI 10.1016/S0092-8674(04)00045-5. [Google Scholar] [CrossRef]
Calin GA, Croce CM. (2006). MicroRNA signatures in human cancers. Nature Reviews Cancer 6: 857–866. DOI 10.1038/nrc1997. [Google Scholar] [CrossRef]
Capitanio U, Montorsi F. (2016). Renal cancer. Lancet 387: 894–906. DOI 10.1016/S0140-6736(15)00046-X. [Google Scholar] [CrossRef]
Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, Das S, Levasseur DN, Li Z, Xu M, Reik W, Silva JC, Wang J. (2013). NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495: 370–374. DOI 10.1038/nature11925. [Google Scholar] [CrossRef]
Cruz-Gil S, Sanchez-Martinez R, Gomez de Cedron M, Martin-Hernandez R, Vargas T, Molina S, Herranz J, Davalos A, Reglero G, Ramirez de Molina A. (2018). Targeting the lipid metabolic axis ACSL/SCD in colorectal cancer progression by therapeutic miRNAs: miR-19b-1 role. Journal of Lipid Research 59: 14–24. DOI 10.1194/jlr.M076752. [Google Scholar] [CrossRef]
Cha Y, Han MJ, Cha HJ, Zoldan J, Burkart A, Jung JH, Jang Y, Kim CH, Jeong HC, Kim BG, Langer R, Kahn CR, Guarente L, Kim KS. (2017). Metabolic control of primed human pluripotent stem cell fate and function by the miR-200c-SIRT2 axis. Nature Cell Biology 19: 445–456. DOI 10.1038/ncb3517. [Google Scholar] [CrossRef]
Chang I, Mitsui Y, Fukuhara S, Gill A, Wong DK, Yamamura S, Shahryari V, Tabatabai ZL, Dahiya R, Shin DM, Tanaka Y. (2015). Loss of miR-200c up-regulates CYP1B1 and confers docetaxel resistance in renal cell carcinoma. Oncotarget 6: 7774–7787. DOI 10.18632/oncotarget.3484. [Google Scholar] [CrossRef]
Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, Faull KF, Lyko F, Jaenisch R. (2013). Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Developmental Cell 24: 310–323. DOI 10.1016/j.devcel.2012.12.015. [Google Scholar] [CrossRef]
Dobrzyn P, Bednarski T, Dobrzyn A. (2015). Metabolic reprogramming of the heart through stearoyl-CoA desaturase. Progress in Lipid Research 57: 1–12. DOI 10.1016/j.plipres.2014.11.003. [Google Scholar] [CrossRef]
Edge SB, Compton CC. (2010). The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Annals of Surgical Oncology 17: 1471–1474. DOI 10.1245/s10434-010-0985-4. [Google Scholar] [CrossRef]
Elgendy M, Fusco JP, Segura V, Lozano MD, Minucci S, Echeveste JI, Gurpide A, Andueza M, Melero I, Sanmamed MF, Ruiz MR, Calvo A, Pascual JI, Velis JM, Miñana B, Valle RD, Pio R, Agorreta J, Abengozar M, Colecchia M, Brich S, Renne SL, Guruceaga E, Patiño-García A, Perez-Gracia JL. (2019). Identification of mutations associated with acquired resistance to sunitinib in renal cell cancer. Journal of Cancer 145:1991–2001. [Google Scholar]
Enoch HG, Catalá A, Strittmatter P. (1976). Mechanism of rat liver microsomal stearyl-CoA desaturase. Studies of the substrate specificity, enzyme-substrate interactions, and the function of lipid. Journal of Biological Chemistry 251: 5095–5103. DOI 10.1016/S0021-9258(17)33223-4. [Google Scholar] [CrossRef]
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. (2015). Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer 136: E359–E386. DOI 10.1002/ijc.29210. [Google Scholar] [CrossRef]
Fraietta JA, Nobles CL, Sammons MA, Lundh S, Carty SA, Reich TJ, Cogdill AP, Morrissette JJD, DeNizio JE, Reddy S, Hwang Y, Gohil M, Kulikovskaya I, Nazimuddin F, Gupta M, Chen F, Everett JK, Alexander KA, Lin-Shiao E, Gee MH, Liu X, Young RM, Ambrose D, Wang Y, Xu J, Jordan MS, Marcucci KT, Levine BL, Garcia KC, Zhao Y, Kalos M, Porter DL, Kohli RM, Lacey SF, Berger SL, Bushman FD, June CH, Melenhorst JJ. (2018). Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 558: 307–312. DOI 10.1038/s41586-018-0178-z. [Google Scholar] [CrossRef]
Fukuda K, Takeuchi S, Arai S, Katayama R. (2019). Epithelial-to-mesenchymal transition is a mechanism of ALK inhibitor resistance in lung cancer independent of ALK mutation status. Cancer Research 79: 1658–1670. DOI 10.1158/0008-5472.CAN-18-2052. [Google Scholar] [CrossRef]
Hannon E, Knox O, Sugden K, Burrage J, Wong CCY, Belsky DW, Corcoran DL, Arseneault L, Moffitt TE, Caspi A, Mill J. (2018). Characterizing genetic and environmental influences on variable DNA methylation using monozygotic and dizygotic twins. PLoS Genetics 14: e1007544. DOI 10.1371/journal.pgen.1007544. [Google Scholar] [CrossRef]
Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. (2012). Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Research 40: 4841–4849. DOI 10.1093/nar/gks155. [Google Scholar] [CrossRef]
Hassannia B, Vandenabeele P, Vanden Berghe T. (2019). Targeting ferroptosis to iron out cancer. Cancer Cell 35: 830–849. DOI 10.1016/j.ccell.2019.04.002. [Google Scholar] [CrossRef]
He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. (2011). Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307. DOI 10.1126/science.1210944. [Google Scholar] [CrossRef]
Jing ZF, Bi JB, Li ZL, Liu XK, Li J, Zhu YY, Zhang XT, Zhang Z, Li ZH, Kong CZ. (2019). miR-19 promotes the proliferation of clear cell renal cell carcinoma by targeting the FRK-PTEN axis. Onco Targets and Therapy 12: 2713–2727. DOI 10.2147/OTT.S199238. [Google Scholar] [CrossRef]
Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. (2011). Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proceedings of the National Academy of Sciences of the United States of America 108: 14566–14571. [Google Scholar]
Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A. (2011). Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8: 200–213. DOI 10.1016/j.stem.2011.01.008. [Google Scholar] [CrossRef]
Kuthi L, Jenei A, Hajdu A, Németh I, Varga Z, Bajory Z, Pajor L, Iványi B. (2017). Prognostic factors for renal cell carcinoma subtypes diagnosed according to the 2016 WHO renal tumor classification: A study involving 928 patients. Pathology & Oncology Research 23: 689–698. DOI 10.1007/s12253-016-0179-x. [Google Scholar] [CrossRef]
Liu H, Brannon AR, Reddy AR, Alexe G, Seiler MW, Arreola A, Oza JH, Yao M, Juan D, Liou LS, Ganesan S, Levine AJ, Rathmell WK, Bhanot GV. (2010). Identifying mRNA targets of microRNA dysregulated in cancer: With application to clear cell Renal Cell Carcinoma. BMC Systems Biology 4: 51. DOI 10.1186/1752-0509-4-51. [Google Scholar] [CrossRef]
Mancini R, Giarnieri E, De Vitis C, Malanga D, Roscilli G, Noto A, Marra E, Laudanna C, Zoppoli P, De Luca P, Affuso A, Ruco L, Di Napoli A, Mesiti G, Aurisicchio L, Ricci A, Mariotta S, Pisani L, Andreetti C, Viglietto G, Rendina EA, Giovagnoli MR, Ciliberto G. (2011). Spheres derived from lung adenocarcinoma pleural effusions: Molecular characterization and tumor engraftment. PLoS One 6: e21320. DOI 10.1371/journal.pone.0021320. [Google Scholar] [CrossRef]
Meisel M, Hinterleitner R, Pacis A, Chen L, Earley ZM, Mayassi T, Pierre JF, Ernest JD, Galipeau HJ, Thuille N, Bouziat R, Buscarlet M, Ringus DL, Wang Y, Li Y, Dinh V, Kim SM, McDonald BD, Zurenski MA, Musch MW, Furtado GC, Lira SA, Baier G, Chang EB, Eren AM, Weber CR, Busque L, Godley LA, Verdú EF, Barreiro LB, Jabri B. (2018). Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature 557: 580–584. DOI 10.1038/s41586-018-0125-z. [Google Scholar] [CrossRef]
Nojima M, Matsui T, Tamori A, Kubo S, Shirabe K, Kimura K, Shimada M, Utsunomiya T, Kondo Y, Iio E, Naito Y, Ochiya T, Tanaka Y. (2016). Global, cancer-specific microRNA cluster hypomethylation was functionally associated with the development of non-B non-C hepatocellular carcinoma. Molecular Cancer 15: 31. DOI 10.1186/s12943-016-0514-6. [Google Scholar] [CrossRef]
Noto A, De Vitis C, Pisanu ME, Roscilli G, Ricci G, Catizone A, Sorrentino G, Chianese G, Taglialatela-Scafati O, Trisciuoglio D, Del Bufalo D, Di Martile M, Di Napoli A, Ruco L, Costantini S, Jakopin Z, Budillon A, Melino G, Del Sal G, Ciliberto G, Mancini R. (2017). Stearoyl-CoA-desaturase 1 regulates lung cancer stemness via stabilization and nuclear localization of YAP/TAZ. Oncogene 36: 4671–4672. DOI 10.1038/onc.2017.212. [Google Scholar] [CrossRef]
Ortiz IMDP, Barros-Filho MC, Dos Reis MB, Beltrami CM, Marchi FA, Kuasne H, do Canto LM, de Mello JBH, Abildgaard C, Pinto CAL, Kowalski LP, Rogatto SR. (2018). Loss of DNA methylation is related to increased expression of miR-21 and miR-146b in papillary thyroid carcinoma. Clinical Epigenetics 10: 144. DOI 10.1186/s13148-018-0579-8. [Google Scholar] [CrossRef]
Pan F, Wingo TS, Zhao Z, Gao R, Makishima H, Qu G, Lin L, Yu M, Ortega JR, Wang J, Nazha A, Chen L, Yao B, Liu C, Chen S, Weeks O, Ni H, Phillips BL, Huang S, Wang J, He C, Li GM, Radivoyevitch T, Aifantis I, Maciejewski JP, Yang FC, Jin P, Xu M. (2017). Tet2 loss leads to hypermutagenicity in haematopoietic stem/progenitor cells. Nature Communications 8: 15102. DOI 10.1038/ncomms15102. [Google Scholar] [CrossRef]
Pisanu ME, Noto A, De Vitis C, Morrone S, Scognamiglio G, Botti G, Venuta F, Diso D, Jakopin Z, Padula F, Ricci A, Mariotta S, Giovagnoli MR, Giarnieri E, Amelio I, Agostini M, Melino G, Ciliberto G, Mancini R. (2017). Blockade of Stearoyl-CoA-desaturase 1 activity reverts resistance to cisplatin in lung cancer stem cells. Cancer Letters 406: 93–104. DOI 10.1016/j.canlet.2017.07.027. [Google Scholar] [CrossRef]
Sánchez-Martínez R, Cruz-Gil S, Gómez de Cedrón M, Álvarez-Fernández M, Vargas T, Molina S, García B, Herranz J, Moreno-Rubio J, Reglero G, Pérez-Moreno M, Feliu J, Malumbres M, Ramírez de Molina A. (2015). A link between lipid metabolism and epithelial-mesenchymal transition provides a target for colon cancer therapy. Oncotarget 6: 38719–38736. DOI 10.18632/oncotarget.5340. [Google Scholar] [CrossRef]
Title AC, Hong SJ, Pires ND, Hasenöhrl L, Godbersen S, Stokar-Regenscheit N, Bartel DP, Stoffel M. (2018). Genetic dissection of the miR-200-Zeb1 axis reveals its importance in tumor differentiation and invasion. Nature Communications 9: 4671. DOI 10.1038/s41467-018-07130-z. [Google Scholar] [CrossRef]
Tracz-Gaszewska Z, Dobrzyn P. (2019). Stearoyl-CoA desaturase 1 as a therapeutic target for the treatment of cancer. Cancers 11: 948. DOI 10.3390/cancers11070948. [Google Scholar] [CrossRef]
Weiss CN, Ito K. (2017). A Macro View of MicroRNAs: The discovery of microRNAs and their role in hematopoiesis and hematologic disease. International Review of Cell and Molecular Biology 334: 99–175. [Google Scholar]
White NM, Bao TT, Grigull J, Youssef YM, Girgis A, Diamandis M, Fatoohi E, Metias M, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Yousef GM. (2011). miRNA profiling for clear cell renal cell carcinoma: Biomarker discovery and identification of potential controls and consequences of miRNA dysregulation. Journal of Urology 186: 1077–1083. DOI 10.1016/j.juro.2011.04.110. [Google Scholar] [CrossRef]
Wu MJ, Kim MR, Chen YS, Yang JY, Chang CJ. (2017). Retinoic acid directs breast cancer cell state changes through regulation of TET2-PKCζ pathway. Oncogene 36: 3193–3206. DOI 10.1038/onc.2016.467. [Google Scholar] [CrossRef]
Zeng H, He H, Guo L, Li J, Lee M, Han W, Guzman AG, Zang S, Zhou Y, Zhang X, Goodell MA, King KY, Sun D, Huang Y. (2019). Antibiotic treatment ameliorates Ten-eleven translocation 2 (TET2) loss-of-function associated hematological malignancies. Cancer Letters 467: 1–8. DOI 10.1016/j.canlet.2019.09.013. [Google Scholar] [CrossRef]
Zeng Y. (2018). Advances in mechanism and treatment strategy of cancer. Cellular and Molecular Biology 64: 1–3. DOI 10.14715/cmb/2018.64.6.1. [Google Scholar] [CrossRef]
Zeng Y, Yao X, Liu X, He X, Li L, Liu X, Yan Z, Wu J, Fu BM (2019b). Anti-angiogenesis triggers exosomes release from endothelial cells to promote tumor vasculogenesis. Journal of Extracellular Vesicles 8: 1629865. DOI 10.1080/20013078.2019.1629865. [Google Scholar] [CrossRef]
Zhuang J, Ye Y, Wang G, Ni J, He S, Hu C, Xia W, Lv Z. (2017). MicroRNA-497 inhibits cellular proliferation, migration and invasion of papillary thyroid cancer by directly targeting AKT3. Molecular Medicine Reports 16: 5815–5822. DOI 10.3892/mmr.2017.7345. [Google Scholar] [CrossRef]
SUPPLEMENTARY FIGURES
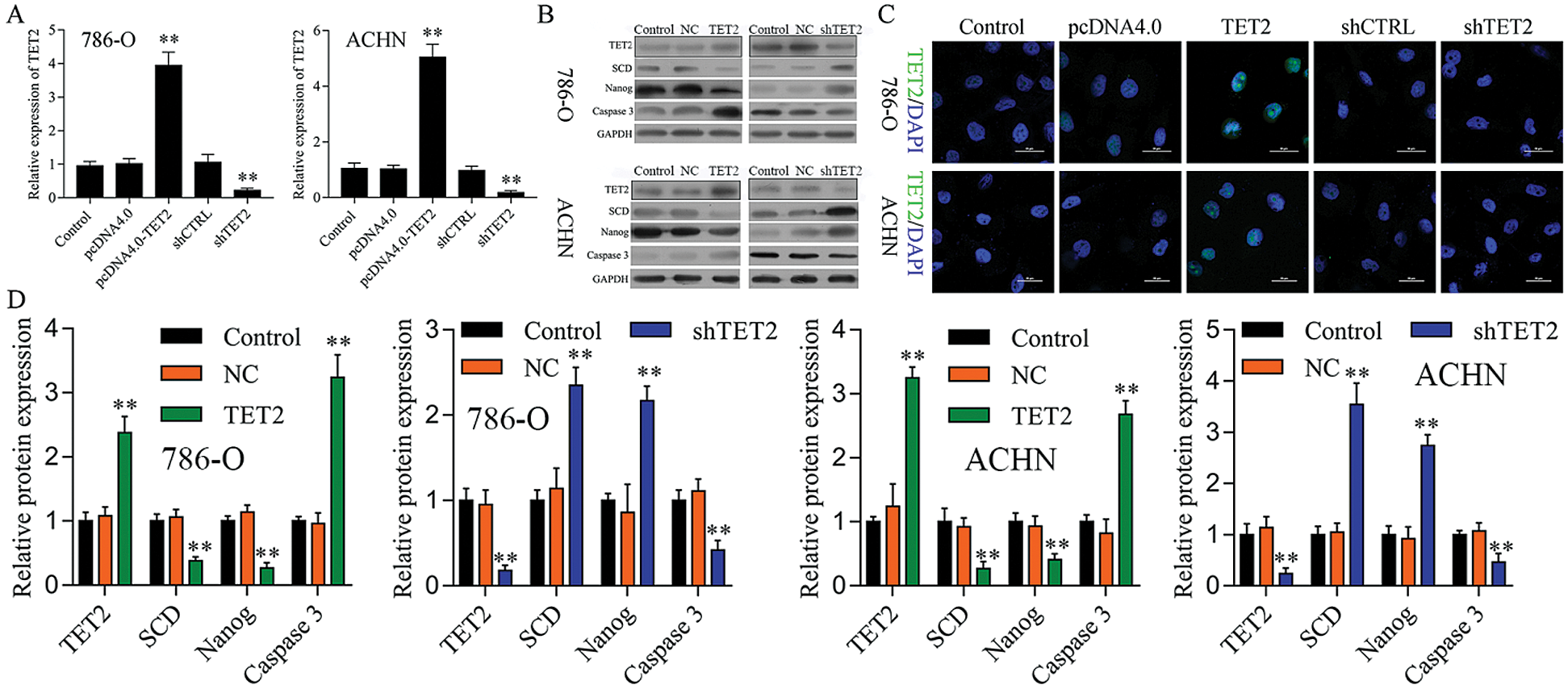
Figure S1: TET2 expression in RCC cells after transfected with TET2 overexpression and knockdown plasmids.
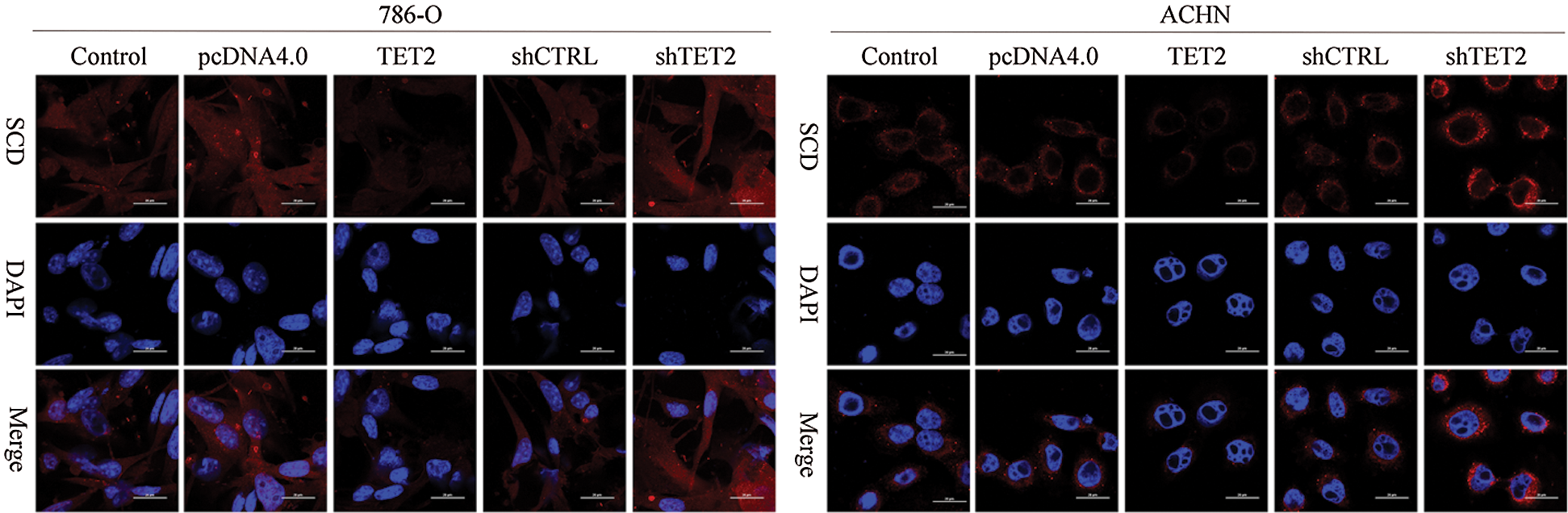
Figure S2: IF analyzed the expression of SCD in different groups, 200×.
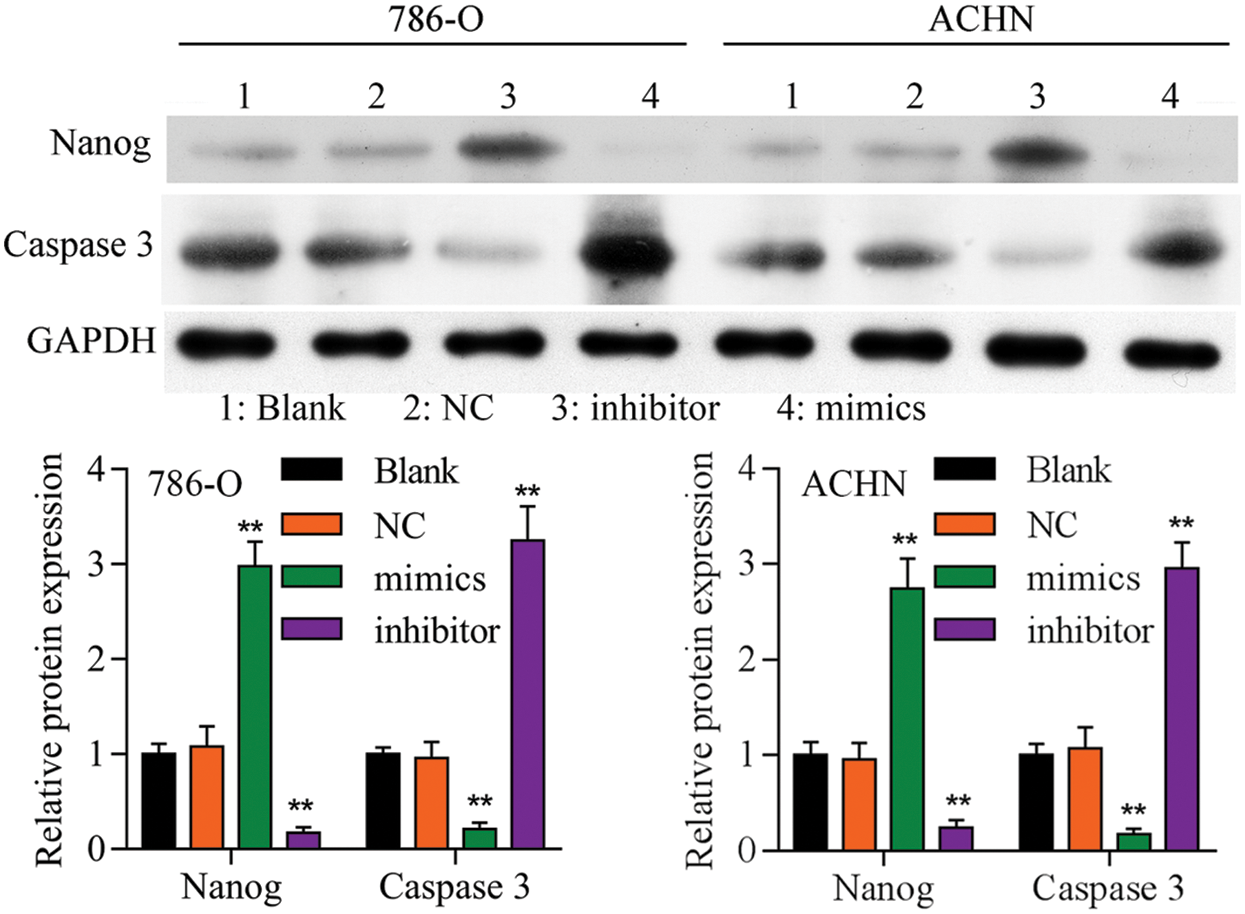
Figure S3: Western blot analyzed the expression of Nanog and caspase 3 in different groups.
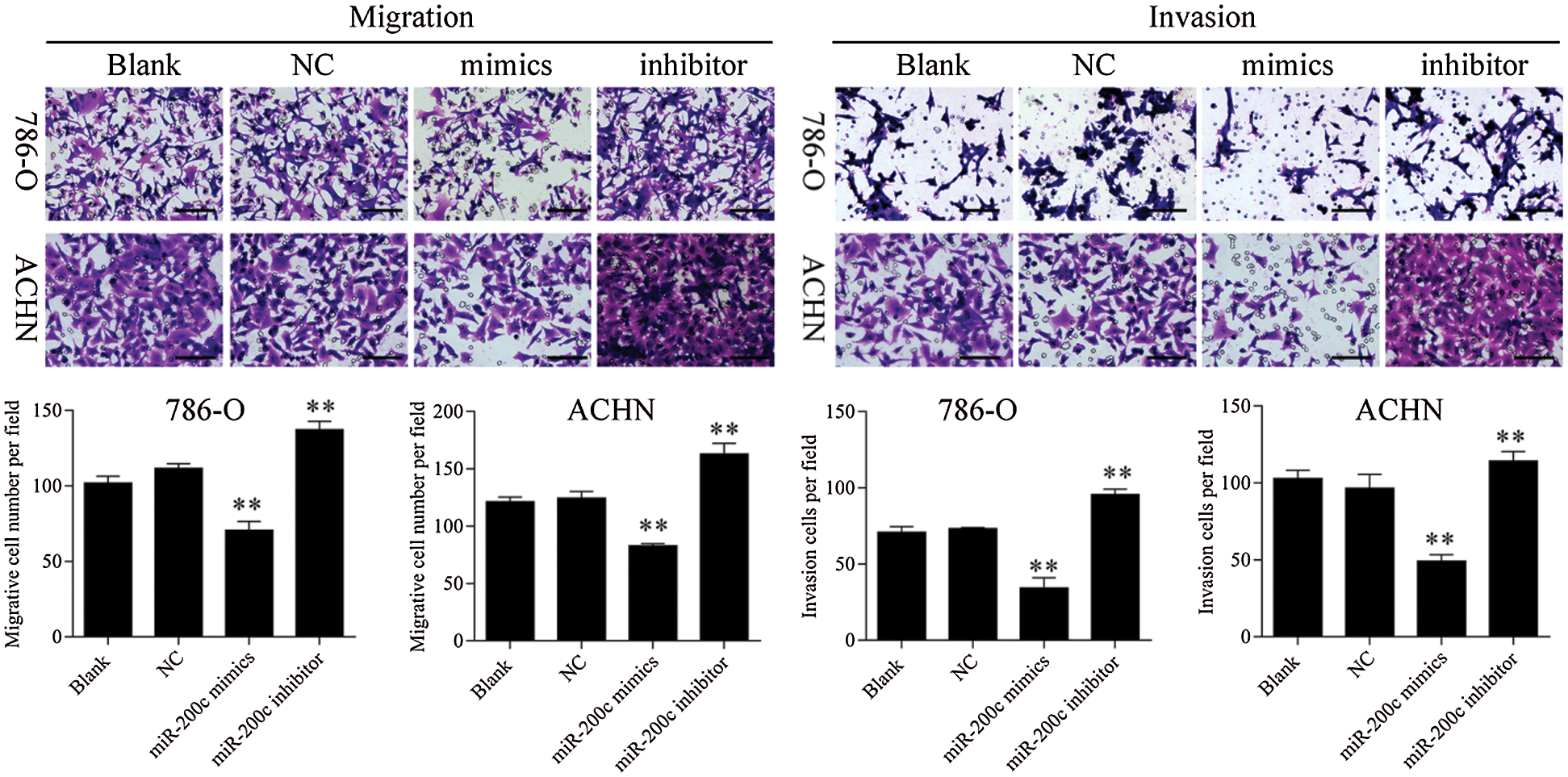
Figure S4: Transwell assay results showed that overexpression of miR-200c inhibited RCC cell metastasis, 200×. ** p < 0.01.
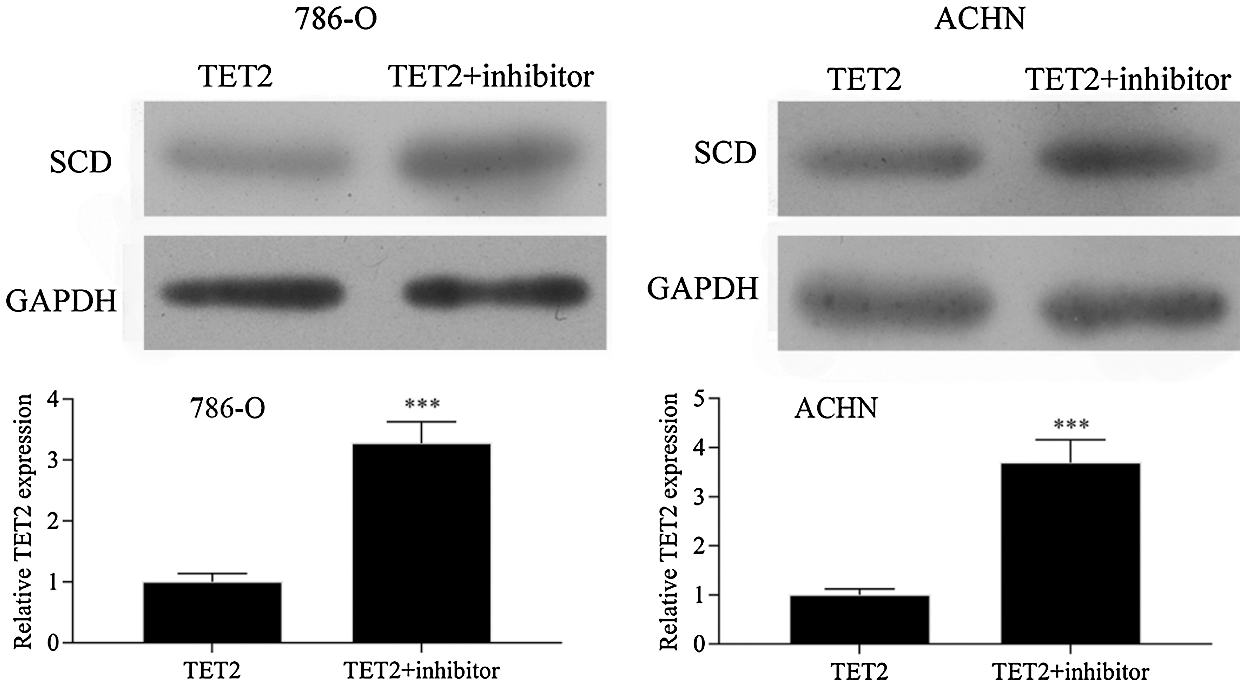
Figure S5: Western blot results showed that downregulation of miR-200c in pcDNA4.0-TET2 transfected RCC cells significantly reduced the suppression effect of TET2 overexpression on SCD protein expression in RCC cells.
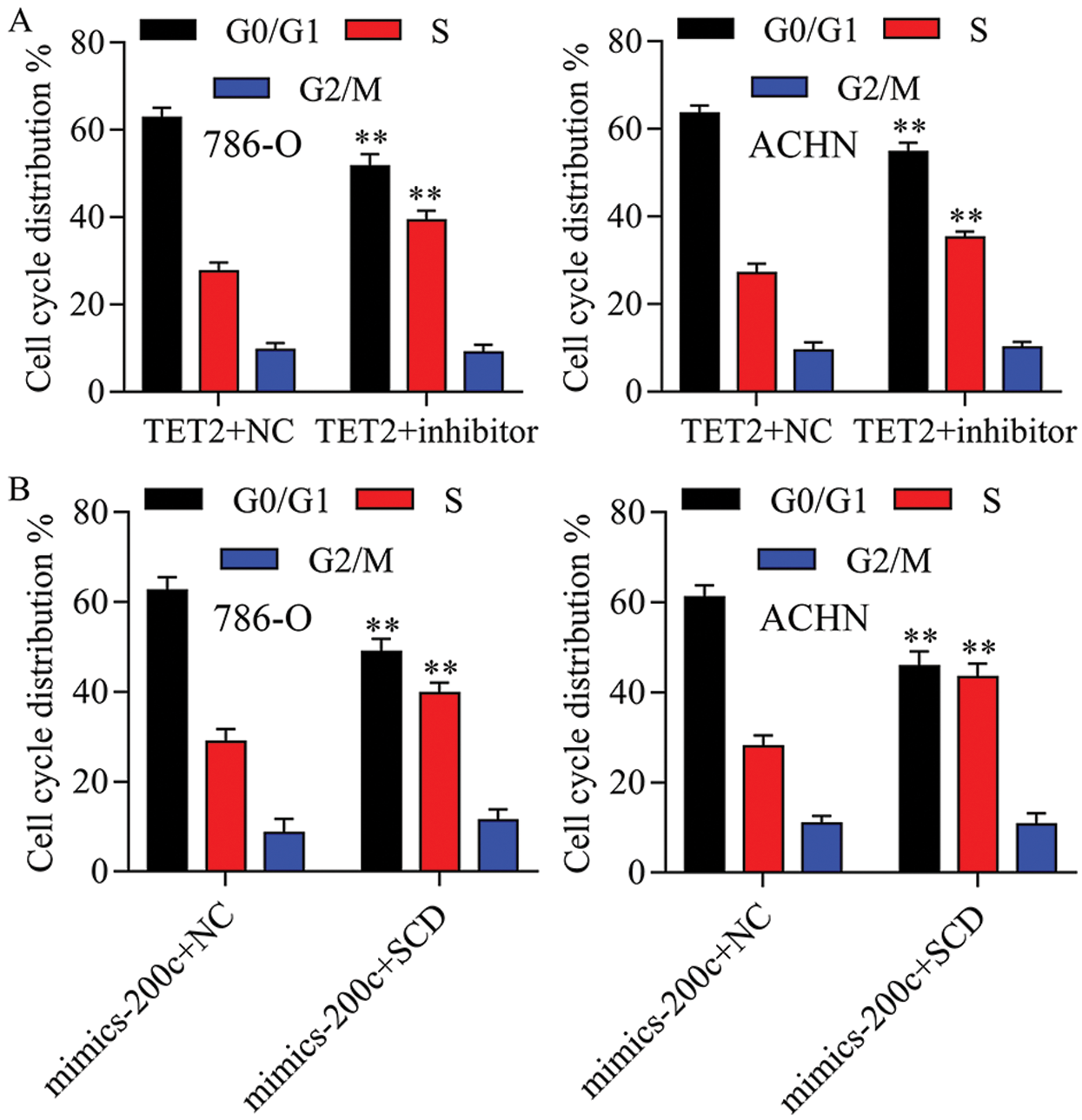
Figure S6: The cell cycle distribution has been quantified in 786-O and ACHN cells.
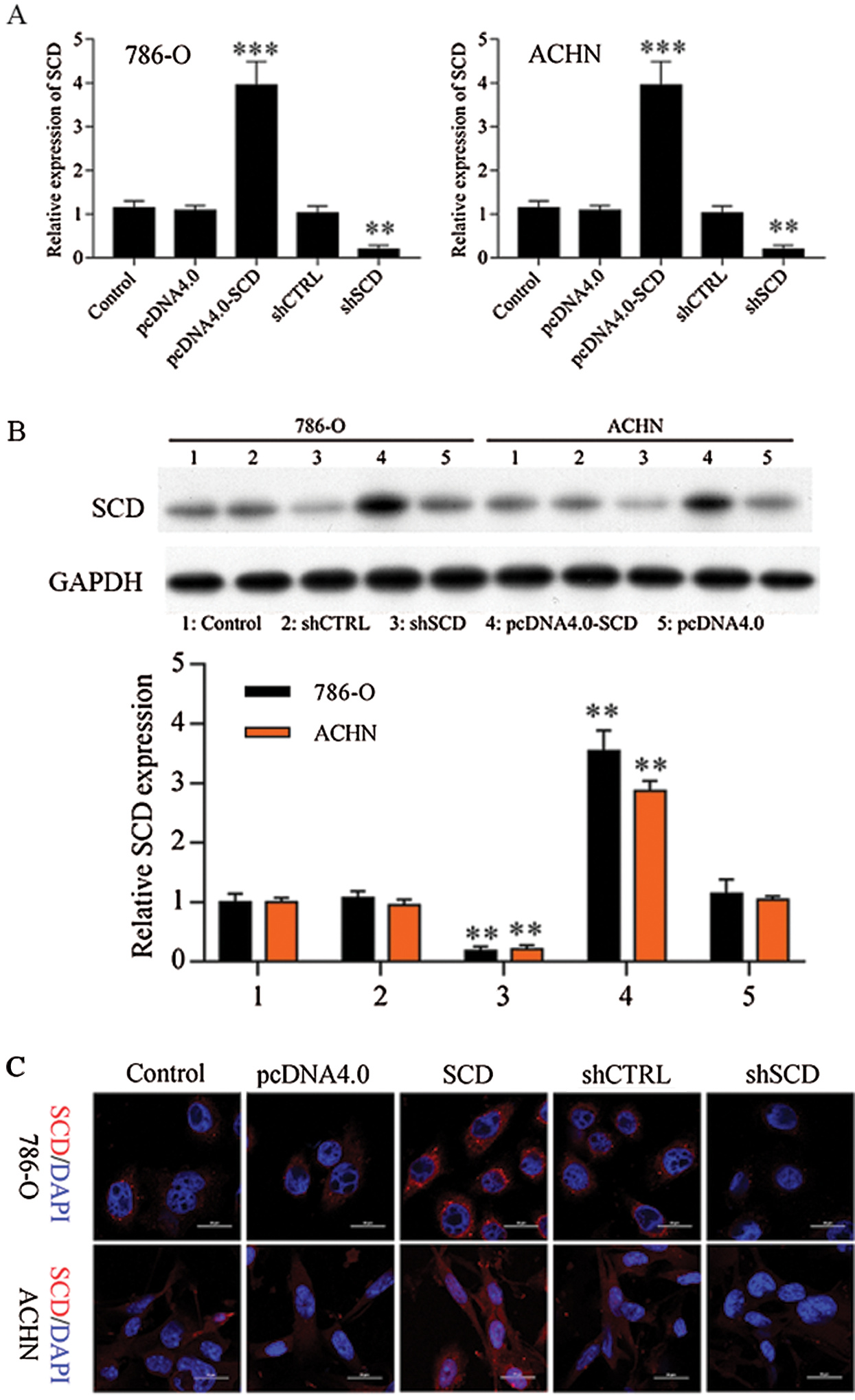
Figure S7: SCD expression in RCC cells after transfected with SCD overexpression and knockdown plasmids.
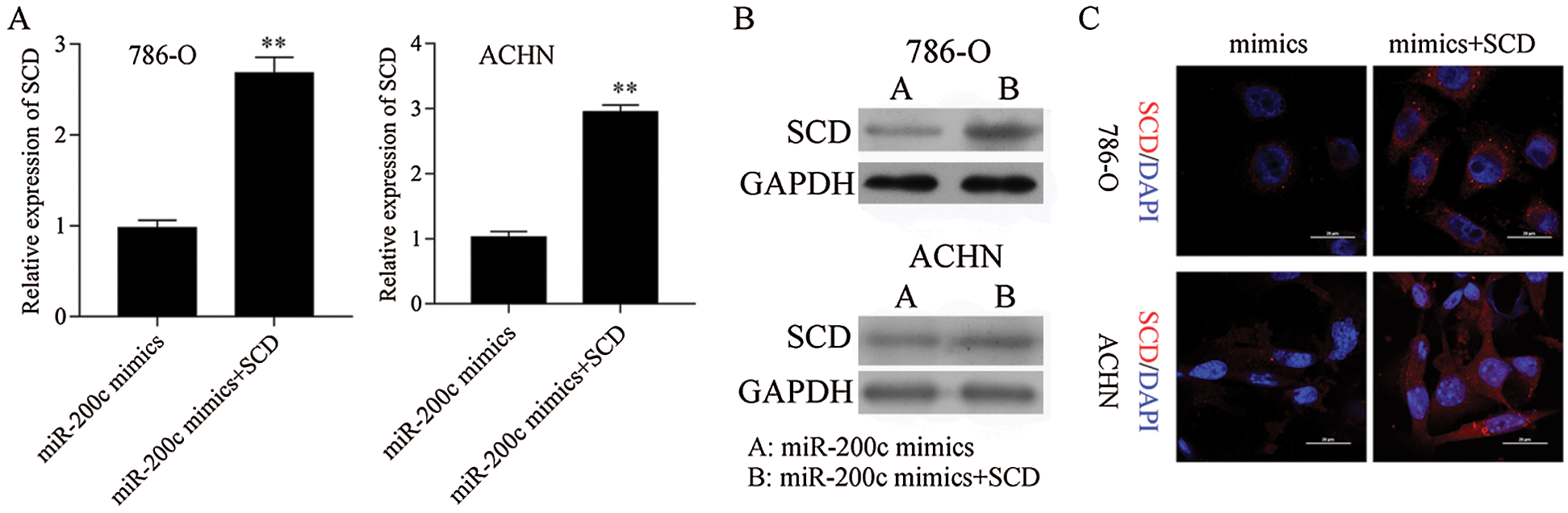
Figure S8: SCD was overexpressed in miR-200c transfected RCC cells. **P< 0.01.
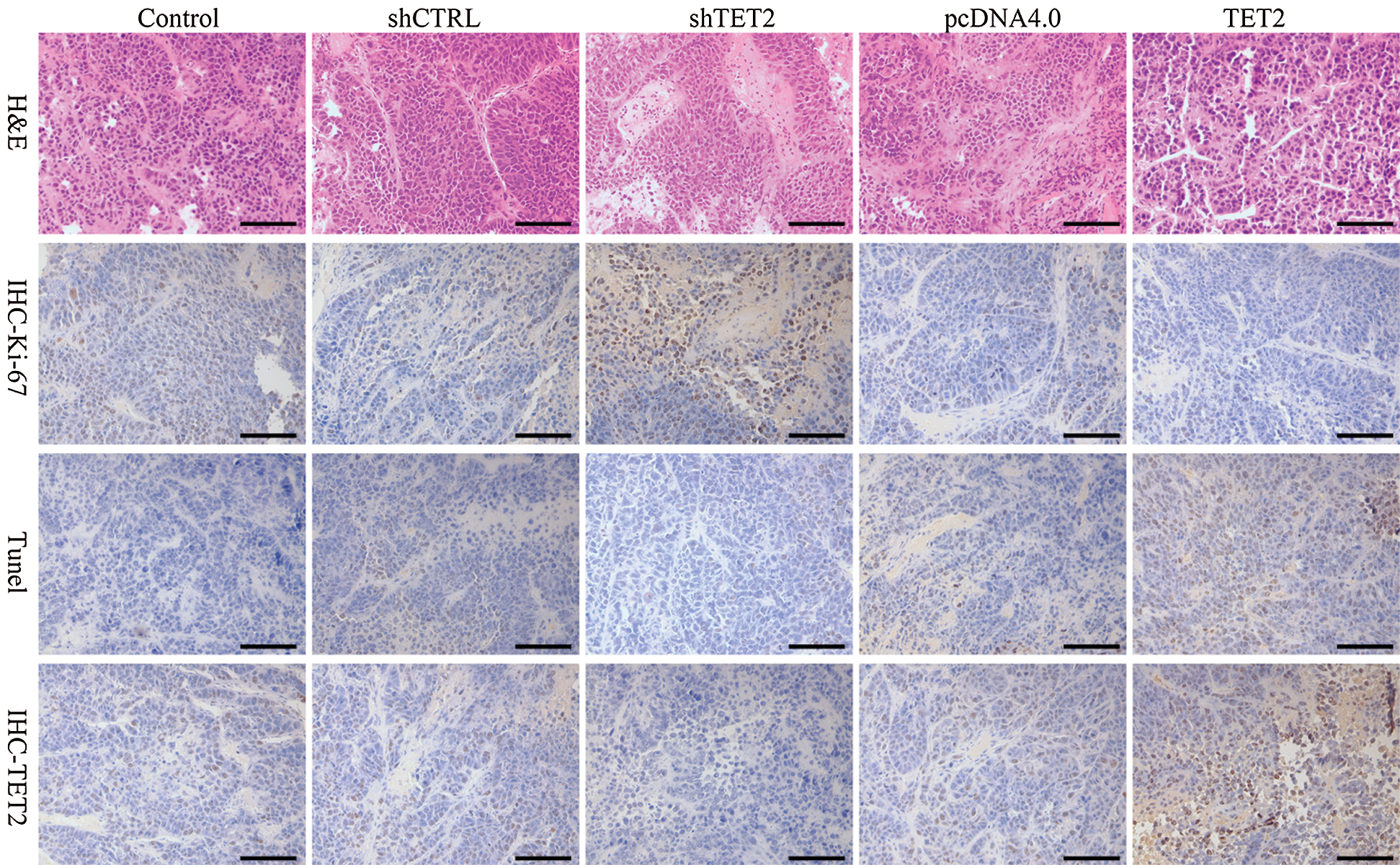
Figure S9: Representative images of H&E, Ki-67, TUNEL, and IHC staining of TET2, 200×.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |