DOI:10.32604/biocell.2021.014343

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.014343 |  www.techscience.com/journal/biocell |
| Article |
Phylogenetic analysis of microRNA biomarkers for amyotrophic lateral sclerosis
1Institute of Statistics, National Yang Ming Chiao Tung University, Hsinchu, 30010, Taiwan
2Institute of Statistics, National Chiao Tung University, Hsinchu, 30010, Taiwan
*Address correspondence to: Hsiuying Wang, wang@stat.nctu.edu.tw
Received: 19 September 2020; Accepted: 23 December 2020
Abstract: Amyotrophic lateral sclerosis (ALS), also called Lou Gehrig’s disease, is an irreversible disease that is caused by the degeneration and death of motor neurons. Approximately 5–10% of cases are familial ALS (fALS), and the other cases are sporadic ALS (sALS). Gene mutations have been identified both in fALS and sALS patients. In this study, we discuss the four ALS-related genes, C9orf72, SOD1, FUS, and TARDBP, and review the microRNAs (miRNAs) that are associated with ALS and other neurological disorders from the literature. A phylogenetic analysis is used to explore potential miRNAs that can be taken into account when studying the difference in pathology for ALS induced by the four genes and other neurological diseases such as frontotemporal dementia, spinal muscular atrophy, and narcolepsy. We found several miRNAs that can be taken into account to study the difference in pathology between ALS and other neurological disorders.
Keywords: Amyotrophic lateral sclerosis; Gene; microRNA; Phylogenetic tree
Amyotrophic lateral sclerosis (ALS) is an irreversible disease that may begin with limb weakness or with difficulty swallowing or speaking and gradually lead to the loss of voluntary muscle movement. This disease was discovered in the 19th century (Rowland, 2001; Visser et al., 2008). It is also called Lou Gehrig’s disease because the American baseball player Lou Gehrig was diagnosed with ALS in the 1930s. Not all ALS patients experience the same disease course, but progressive paralysis is commonly experienced. The mean survival time with ALS is less than five years, but 14% of the cases live longer than five years (Mateen et al., 2010). The motor neurons in the brain are called upper motor neurons, and those in the spinal cord are called lower motor neurons. Motor neurons control muscle movement. The upper motor neurons transmit nerve impulses to lower motor neurons, and the lower motor neurons send nerve signals to muscles. In ALS, both the upper motor neurons and the lower motor neurons degenerate or die and stop sending messages to the muscles.
Approximately 5–10% of cases, called familial ALS (fALS), are inherited from family members, and they are caused by genetic mutations (Kurland and Mulder, 1955). Around 90% of patients are called sporadic ALS (sALS). Gene mutations have been identified in fALS and sALS patients (Sreedharan et al., 2008; Vance et al., 2009; DeJesus-Hernandez et al., 2011; Chen et al., 2013), especially the mutations of the four genes chromosome 9 open reading frame 72 (C9orf72), superoxide dismutase 1 (SOD1), fused in sarcoma (FUS), and TARDBP. Genetic defects occur in about 20–30% of fALS cases (Maruyama et al., 2010). Among those, 20% are caused by a mutation in the SOD1 gene, 4–5% are the results of mutations in TARDBP and FUS genes, more than 30% are associated with C9orf72 mutations, and the rest are associated with other known or unknown genes (Chen et al., 2013). Most sALS cases are caused by unknown factors. A small fraction of sALS is caused by the four genes C9orf72, SOD1, FUS, and TARDBP (Turner et al., 2013). Besides gene mutations, environmental factors contribute to disease liability (Oskarsson et al., 2015).
In addition to gene mutations involved in the pathology of ALS, microRNA (miRNA) biomarkers have been identified for ALS. A miRNA is a small single-stranded non-coding RNA that functions in the epigenetic control of gene expression (Wu et al., 2010). miRNAs were shown to be linked to many diseases, including cancer, periodontal disease, neurodegenerative diseases, hematological diseases, and autoimmune diseases (Alevizos and Illei, 2010; Lee et al., 2011; Grasedieck et al., 2013; Maciotta Rolandin et al., 2013; Hsieh et al., 2014; Wang, 2016a; Takuse et al., 2017; Ricci et al., 2018; Taguchi and Wang, 2018a; Taguchi and Wang, 2018b; Chen and Wang, 2020b; Wang, 2020). Dysregulation of miRNAs might play an important role in the pathogenesis of multiple forms of human ALS (Emde et al., 2015). In this study, miRNA biomarkers for ALS and other neurological diseases are discussed. Then, we cluster these miRNA biomarkers to find potential miRNAs that can be used to investigate the difference of pathology between ALS and other neurological diseases.
The method used in this study is first to find miRNA biomarkers from the literature for ALS and other neurological diseases, respectively. These neurological diseases include frontotemporal dementia, Parkinson’s disease, Alzheimer’s disease, spinal muscular atrophy, Prader–Willi syndrome, Niemann–Pick disease, neurofibromatosis, narcolepsy, Friedreich’s ataxia, and ataxia-telangiectasia. Then, we cluster the biomarker sequences for ALS and neurological diseases by plotting phylogenetic trees based on different evolutionary models. Since a large proportion of fALS can be linked to one of the four genes (Turner et al., 2013), we explore potential miRNAs to study the difference in pathology for ALS induced by these four genes and other neurological diseases. The phylogenetic tree is one of the useful tools of the phylogenetic analysis (Graur and Li, 2000; Wang and Hung, 2012), which has been successfully used in finding cancer miRNA biomarkers (Wang, 2016b). The phylogenetic tree combined with the microarray analysis can increase the accuracy of the miRNA biomarker prediction of cancer compared with the method only using microarray analysis (Wang, 2016b).
Many genes were shown to be associated with ALS. In this study, we mainly focus on the four genes C9orf72, SOD1, FUS, and TARDBP. We briefly describe these genes.
The C9orf72 gene is located on the short arm of chromosome 9, open reading frame 72. The protein is abundant in neurons of the brain and motor neurons of the spinal cord. The mutation of C9orf72 was found to be associated with both ALS and frontotemporal dementia (FTD) (DeJesus-Hernandez et al., 2011; Renton et al., 2011; Majounie et al., 2012). The mutation is a hexanucleotide repeat expansion of the six nucleotides GGGGCC. Healthy subjects carry 2-10 hexanucleotide GGGGCC repeats in the C9orf72 gene, while more than a few hundred repeats represent a risk for ALS. Mutations in C9orf72 account for 20–40% of fALS. It occurs more often in patients older than 50 years old.
The SOD1 gene is located on chromosome 21. SOD1 is an enzyme that can destroy free superoxide radicals in the body. In 1993, genetic mutations in the SOD1 gene were found to be linked to fALS (Rosen et al., 1993). This is the first discovery of a genetic link to ALS. More than 160 different disease-associated mutations have been found in SOD1. Toxic effects caused by the mutations in the SOD1 gene are involved in ALS pathogenesis (Sangwan and Eisenberg, 2016).
The FUS gene is located on chromosome 16. The RNA-binding protein fused in sarcoma/translocated in sarcoma (FUS/TLS) is encoded by the FUS gene. Mutations in the FUS/TLS gene were discovered to cause fALS (Kwiatkowski et al., 2009; Vance et al., 2009). Mutations in FUS account for 5% of fALS (Shang and Huang, 2016). In addition, FUS is related to FTD for sporadic cases or familial cases (Neumann et al., 2009; Zhou et al., 2014; Bradfield et al., 2017).
TARDBP is a gene located on chromosome 1, which encodes TAR DNA-binding protein 43 (TDP-43). Frontotemporal lobar degeneration is associated with tau, TDP-43, or FET protein accumulation (Mackenzie and Neumann, 2016). TDP-43 proteinopathy is associated with chronic traumatic encephalopathy (CTE) (McKee et al., 2010; Jayakumar et al., 2017). The abnormalities of TDP-43 are correlated with the clinical features of Alzheimer’s disease (Tremblay et al., 2011). Evidence suggests a pathophysiological link between TDP-43 and ALS (Sreedharan et al., 2008). Mutations in TARDBP account for 5% of fALS.
Several ALS miRNA biomarkers related to C9orf72, SOD1, FUS, and TARDBP were discussed in the literature. From a regulatory network analysis, TDP-43 and C9orf72 are possible targets of miR-142-3p (Matamala et al., 2018). In cerebrospinal fluid samples of sALS patients, five TDP-43 binding miRNAs, miR-132-5p, -132-3p, -143-3p, -143-5p and -574-5p, were significantly dysregulated (Freischmidt et al., 2013). Downregulation of miR-132-5p/3p and miR-574-5p/3p was evident in TARDBP, FUS, and C9ORF72, but not SOD1 mutant patients (Freischmidt et al., 2013). Let-7b levels are significantly reduced in both FUS and C9ORF72 mutant immortalized lymphoblast cell lines (Freischmidt et al., 2013). The survival time of SOD1-G93A mice was significantly extended by treatment with anti-miR-155 compared with control cases (Koval et al., 2013). Mature miR-206 was increased in fast-twitch muscles in the SOD1-G93A mice model (Toivonen et al., 2014). miR-124a is reduced in the spinal cord tissue of SOD1-G93A mice (Morel et al., 2013). miR-27a was highly expressed in ALS subjects compared with healthy control subjects (Butovsky et al., 2012b).
In addition to miRNAs related to the four genes, we also discuss miRNAs that may not directly relate to these four genes but are shown to be associated with ALS. These miRNA biomarkers were predicted in Taguchi and Wang (2018b) and other studies, including miR-1290, miR-1246, miR-181a-5p, miR-4701, miR-4485, miR-455, miR-26a, miR-23a, miR-146a* and miR-1825. miR-1290 and miR-1246 were down-regulated in sALS (Wakabayashi et al., 2014). The receiver operator characteristic (ROC) curve analyses indicated that miR181a-5p may be used as a prognostic and disease progression biomarker of sALS (Benigni et al., 2016). miR-4701 and miR-4485 had significantly different expression levels in sALS patients compared with healthy controls (Chen et al., 2016). The expression levels of miR-455 and miR-26a are different in ALS and controls (Jensen et al., 2016). ALS patients had lower levels of skeletal muscle peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) mRNA compared with healthy control subjects, and miR-23a had a reduction in PGC-1α levels (Russell et al., 2013). Furthermore, miR-146a* could contribute to the selective suppression of low molecular weight neurofilament (NFL) mRNA observed in sALS (Campos-Melo et al., 2013). miR-1825 was significantly down-regulated in sALS patients’ plasma (Takahashi et al., 2015).
Furthermore, several miRNA biomarkers were suggested in other studies. Overexpressing miR126-5p in SOD1-G93A mice muscles inhibits the neurodegenerative process that might identify a non-cell-autonomous neurodegeneration process in ALS (Maimon et al., 2018). miR-374b-5p, miR-206, and miR-143-3p of sALS patients were shown to be decreased compared to controls (Waller et al., 2017). miR-206 and miR-424 are potential prognostic markers in spinal onset ALS (de Andrade et al., 2016). miR-132 and miR-125b were upregulated in ALS patients (Kovanda et al., 2018). In addition to miR-206, miR-143-3p, and miR-132, which were mentioned above as related to the four genes, miR126-5p, miR-374b-5p, and miR-424 are potential biomarkers that can be investigated in a future study.
miRNA biomarkers of ALS and other neurological disorders
To investigate the relationship between ALS and other neurological diseases through miRNA biomarkers, we also discuss miRNA biomarkers for other neurological disorders. Tab. 1 lists miRNA biomarkers of ALS caused by the four genes and other neurological disorders, such as Williams syndrome, Parkinson’s disease, Alzheimer’s disease, etc. We select these neurological disorders listed in Tab. 1 because, from our analysis, we can determine at least one miRNA biomarker for each of these neurological disorders such that these miRNAs can be used to investigate the difference in pathology between these neurological diseases and ALS. In the results section, we discuss more details of this analysis result.
Table 1: miRNA biomarkers for several neurological disorders and amyotrophic lateral sclerosis (ALS) related to the four genes
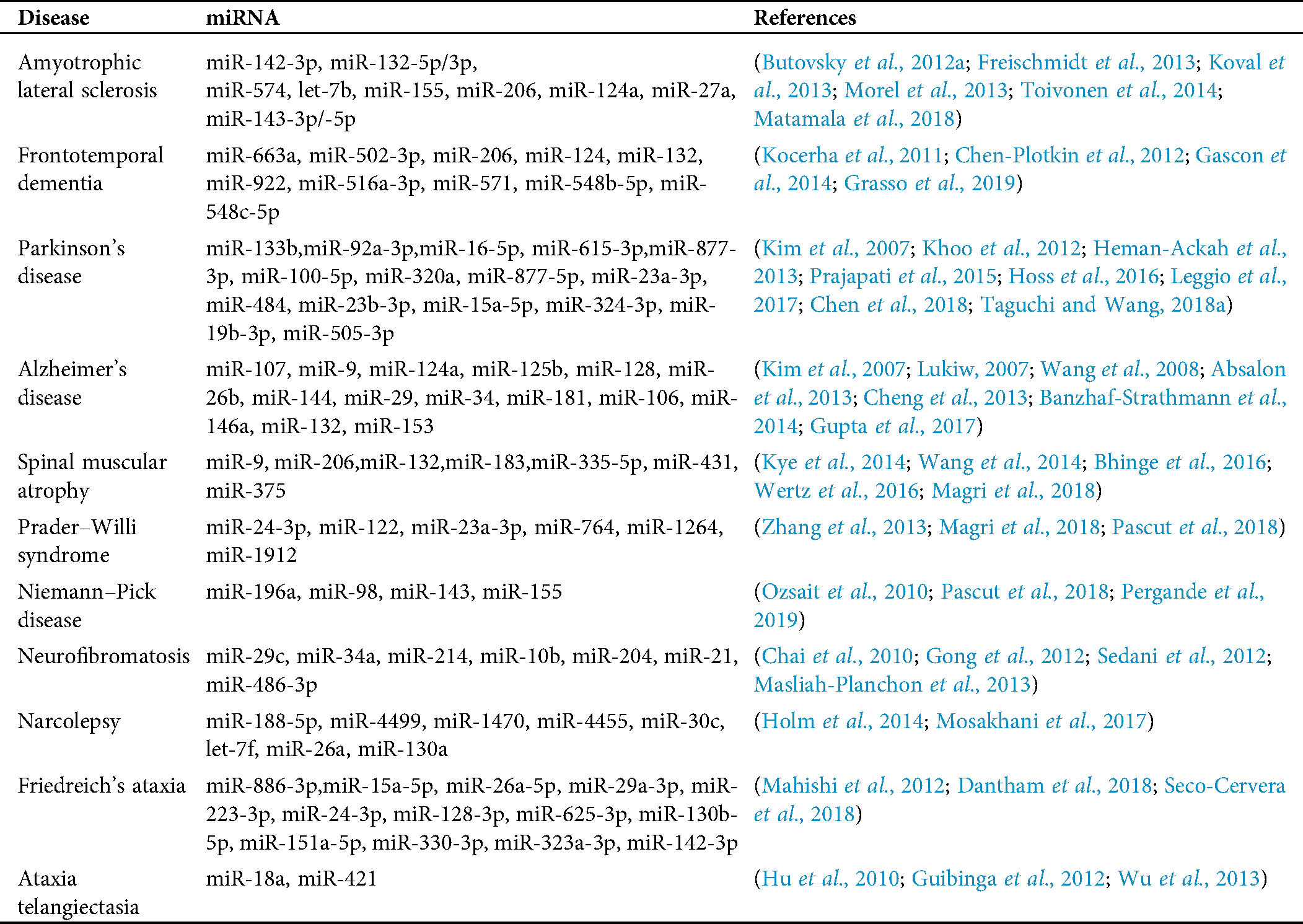
For the diseases in Tab. 1, since FTD is closely related to ALS and both share some common miRNA biomarkers, we discuss miRNA biomarkers of FTD here. A validation study confirmed the downregulation of miR-663a, miR-502-3p, and miR-206 in FTD patients (Grasso et al., 2019); a mechanism involving miR-124 and AMAPRs was identified in regulating social behavior in FTD (Gascon et al., 2014); miR-132 significantly differentiates frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP) and control brains (Chen-Plotkin et al., 2012); and qRT-PCR analyses showed that miR-922, miR-516a-3p, miR-571, miR-548b-5p, and miR-548c-5p were significantly dysregulated in cerebellar tissue samples of progranulin (PGRN) mutation carriers for FTLD-TDP patients (Kocerha et al., 2011).
We adopted a phylogenetic tree analysis to find potential miRNAs that could be used to investigate the difference in pathology between other neurological diseases and ALS induced by the four genes. These miRNA biomarkers were clustered based on the phylogenetic tree analysis. Since the similarity of two nucleotide sequences can be measured using different evolutionary models, we plotted phylogenetic trees based on different evolutionary models using the MATLAB software (Mathworks, 2014).
To perform this method, we needed to find miRNA biomarkers of ALS and other neurological disorders, respectively. The miRNA biomarkers are listed in Tab. 1. To plot the phylogenetic tree of these miRNAs, we used the stem-loop sequences of these biomarkers because they may provide more information than the mature -5p sequence and mature -3p sequence. The stem-loop sequences can be accessed from the miRBase (http://www.mirbase.org/) (Kozomara and Griffiths-Jones, 2013). To classify these miRNA sequences, we first needed to calculate the distances for any two miRNA sequences. Next, we classified the sequences such that sequences with a small distance can be clustered into a group. In the study, we applied the phylogenetic tree method to classify these sequences. Thus, using the MATLAB software requires two steps: The first is to select an evolutionary model to calculate the distance between two nucleotide sequences; after calculating all the distances of any two sequences, the second step is to find a clustering method to build a tree.
The distance model method in the MATLAB software includes the p-distance, Jukes–Cantor distance, alignment score distance, etc. The clustering method (linkage function) in the MATLAB software includes the median method, the single method, and the average method, and so on. In this study, we used the Jukes–Cantor distance (or the alignment score distance) to calculate the distances and the median method (or the average method) as the linkage functions to build a tree.
We plotted the phylogenetic trees of the 9 ALS miRNA biomarkers in Tab. 1 and each biomarker for other neurological disorders. Fig. 1 shows the phylogenetic trees based on the 9 miRNA biomarkers of ALS and miR-335, which is a biomarker of spinal muscular atrophy. Figs. 1a and 1b were plotted based on the Jukes–Cantor distance and the average linkage function method, and the alignment score distance and the median linkage function method, respectively. From Fig. 1a, miR-335 is in another branch of the tree based on the Jukes–Cantor distance. In addition, except for the biomarker miR-155 of ALS, miR-335 is also in a separate branch of the tree in Fig. 1b, based on the alignment score distance. miR-155 is a biomarker of ALS related to the SOD1 gene. We present more discussions of miR-155 in the discussion section. From this phylogenetic tree analysis, we found that miR-335 may be a useful biomarker to discriminate spinal muscular atrophy and ALS. As a result, miR-335 can be a potential miRNA to investigate the difference in pathology between spinal muscular atrophy and ALS.
Fig. 2 shows the phylogenetic trees of miR-1470 (narcolepsy miRNA biomarker) and the 9 ALS miRNA biomarkers. From Figs. 2a and 2b, miR-1470 is in a separate branch of the two trees. Thus, miR-1470 may be a potential miRNA for investigating the difference in pathology between narcolepsy and ALS.
Fig. 3 shows the phylogenetic trees of miR-548b (FTD miRNA biomarker) and the 9 ALS miRNA biomarkers. Unlike miR-335 and miR-1470, which are always in a separate branch of the trees, in Fig. 3a, miR-548b cannot be clustered to a different group. In Fig. 3b, miR-548b is in a separate branch of the tree compared with the other 8 ALS biomarkers, excluding miR-155. Since miR-548b is clustered in a separate group in one of the two trees, we may consider that miR-548b is a potential miRNA for investigating the difference of pathology between FTD and ALS.
In addition to the three miRNAs, miR-335, miR-1470, and miR-548b in Figs. 1–3, Tab. 2 lists other miRNAs of Tab. 1 that have the potential to be used in investigating the difference of pathology between the other neurological disorders of Tab. 1 and ALS. For each miRNA in Tab. 2, either one of the two trees or both trees shows that the miRNA is in a separate branch of the tree. Fig. S1 provides the trees in which the miRNAs are in a separate branch based on the Jukes–Cantor distance and the average linkage method. Fig. S2 provides the cases for the alignment score distance and the median linkage method.
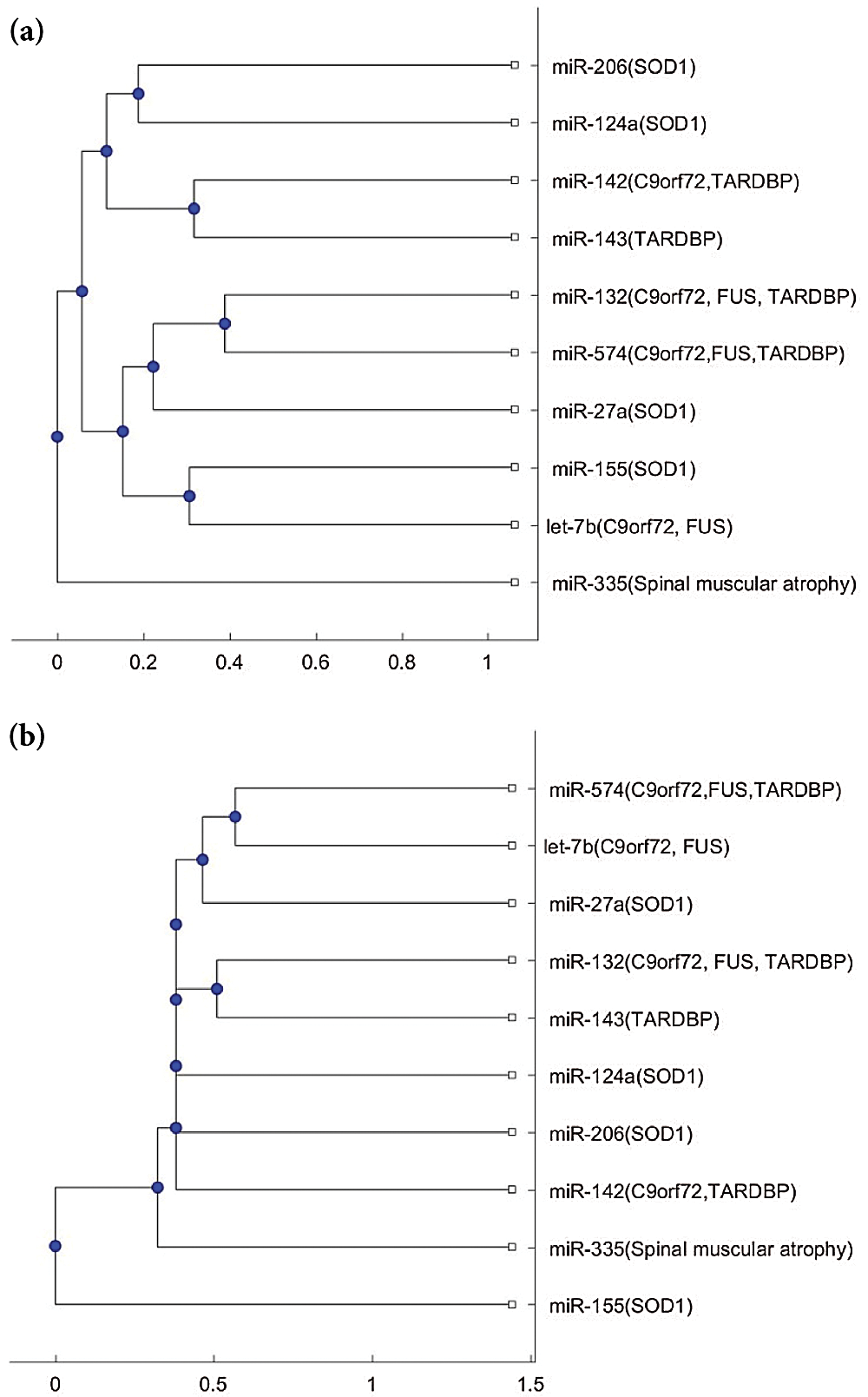
Figure 1: (a) The phylogenetic tree of miR-335 and the 9 ALS miRNA biomarkers based on the Jukes–Cantor distance and the average linkage method. (b) The phylogenetic tree of miR-335 and the 9 ALS miRNA biomarkers based on the alignment-score distance and the median linkage method.
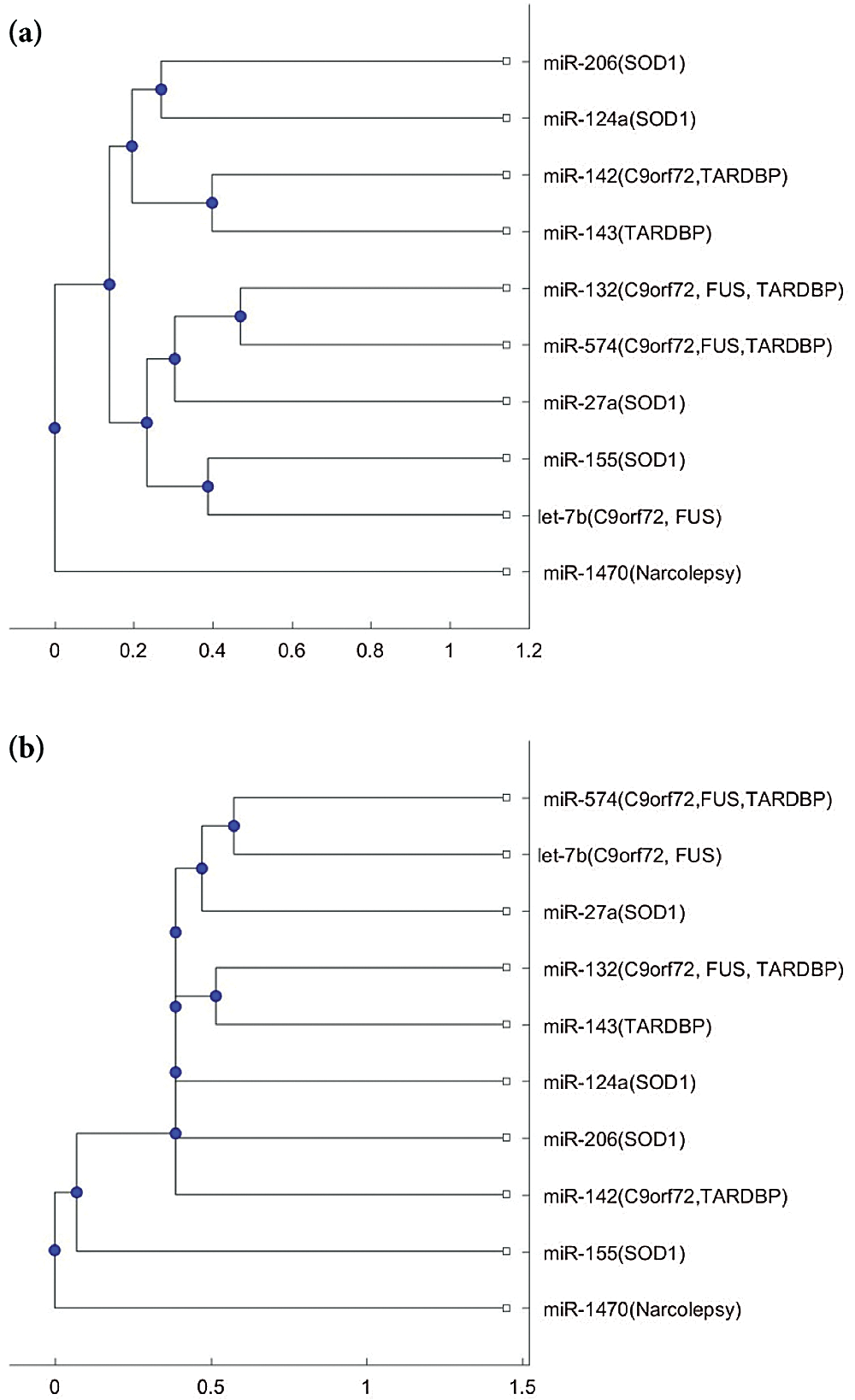
Figure 2: (a) The phylogenetic tree of miR-1470 and the 9 ALS miRNA biomarkers based on the Jukes–Cantor distance and the average linkage method. (b) The phylogenetic tree of miR-1470 and the 9 ALS miRNA biomarkers, based on the alignment score distance and the median linkage method.
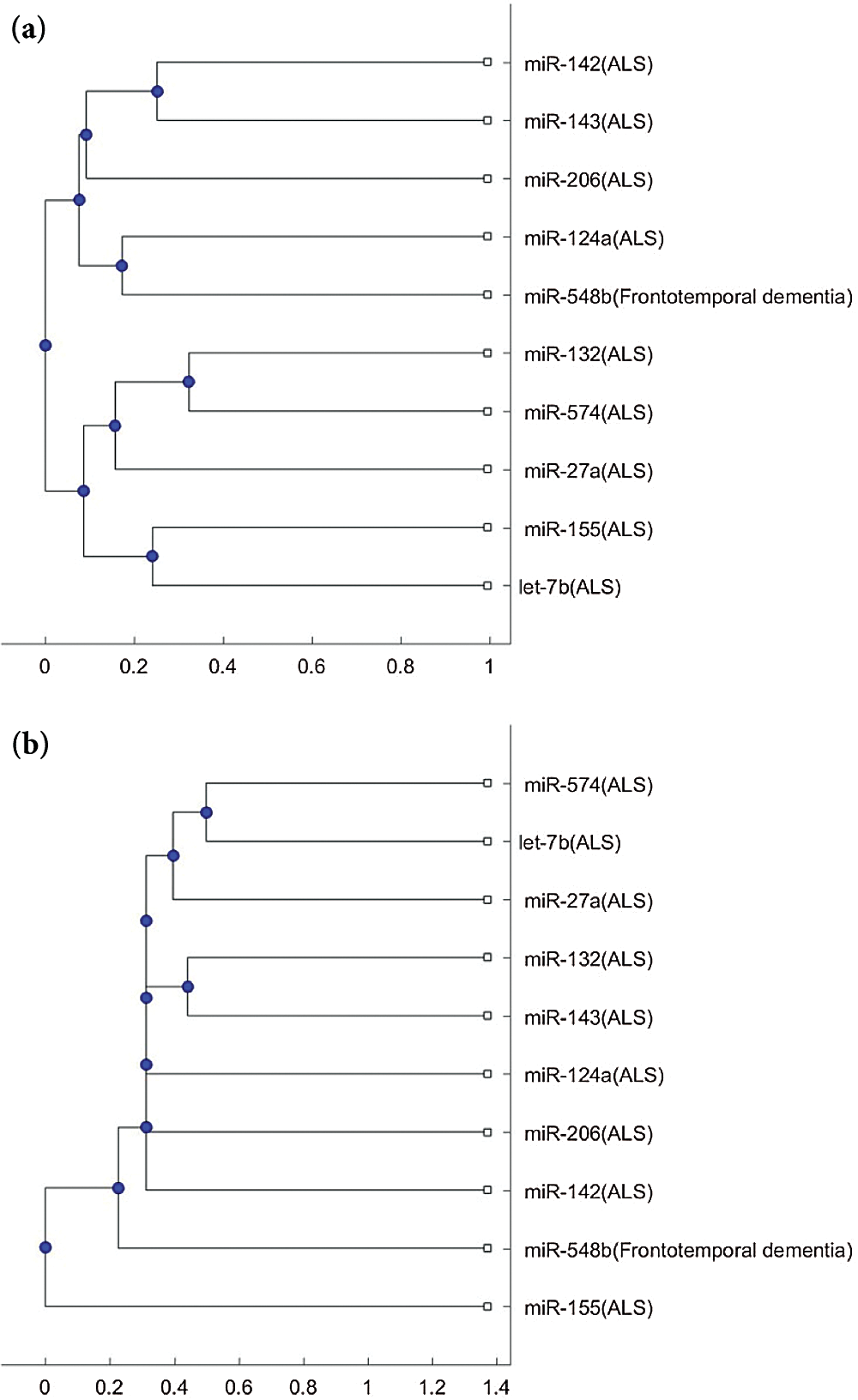
Figure 3: (a) The phylogenetic tree of miR-548b and the 9 ALS miRNA biomarkers based on the Jukes–Cantor distance and the average linkage method. (b) The phylogenetic tree of miR-548b and the 9 ALS miRNA biomarkers based on the alignment-score distance and the median linkage method.
In Figs. 1b and 3b, a SOD1 biomarker, miR-155, is alone in a separate branch. In addition to these two Figs., this phenomenon frequently occurs in the phylogenetic trees of other miRNAs, listed in Tab. 2.
Table 2: Potential miRNAs for investigating the difference of pathology between other neurological diseases and ALS induced by the four genes
To confirm these identified miRNAs, we use the Human MicroRNA Disease Database (HMDD) to provide a validation of these results. HMDD is a database that provides experiment-supported evidence for human miRNA and disease associations (Huang et al., 2018). We use HMDD to validate our results from three aspects. The first one is to check whether the miRNAs listed in Tab. 2 are associated with ALS. Since the miRNAs listed in Tab. 2 are used to classify ALS with the other diseases, they should not be associated with ALS. We uploaded the miRNAs listed in Tab. 2 to HMDD. For the 18 miRNAs, none of them are associated with ALS in HMDD. Therefore, we validate that the selected miRNAs are not ALS-related miRNAs.
The second aspect is to check whether the selected miRNAs are associated with the corresponding diseases listed in Tab. 2. In HMDD, we have confirmed that miR-548b and miR-548c are associated with FTD, miR-133b is associated with Parkinson’s disease, miR-29 and miR-181 are associated with Alzheimer’s disease, miR-335 is associated with spinal muscular atrophy, and miR-18a is associated with ataxia-telangiectasia. Therefore, 7 of the selected miRNAs are confirmed to be related to their corresponding diseases, but not ALS.
The third aspect is to check the miRNAs listed in Tab. 1. The result is presented in Tab. 3. The second column of Tab. 3 lists miRNAs that are confirmed in HMDD to be related to the diseases but not ALS; the third column of Tab. 3 lists the miRNAs in the second column that are selected by the phylogenetic method. From Tab. 3, it can be seen that there are many miRNAs selected by this method. Therefore, this phylogenetic tree method can select miRNAs that may be potential to be used in studying the difference of pathology for ALS and other neurological diseases. This also reveals that these identified miRNAs listed in Tab. 2 have the potential to be used for studying the difference of pathology for ALS and their corresponding neurological diseases (Fig. 4).
In addition, the above mentioned phylogenetic trees were plotted based on the stem-loop sequences. We can compare the stem-loop sequence analysis result with the 3p- or 5p- miRNA sequence analysis result using the 7 selected miRNAs of the third column in Tab. 3. But the 3p- or 5p- miRNA sequence of miR-206 can not be obtained from miRBase. Thus, we use the other 8 miRNA biomarkers of ALS in the 3p- or 5p- miRNA sequence analysis. Compared with the stem-loop sequences, the 3p- or 5p- miRNA sequences analysis only selected the 3 miRNAs, miR-335, miR-548c, and miR-133b, among the 7 miRNAs. Figs. S3 and S4 show the phylogenetic trees of the 3p- or 5p- miRNA sequences analysis for these 3 miRNAs. More comparisons of the stem-loop sequence and 3p- (or 5p-) miRNA sequence will be investigated in a future study.
Table 3: miRNAs related to the corresponding disease but not ALS from the Human MicroRNA Disease Database (HMDD), and miRNAs selected by the phylogenetic tree method

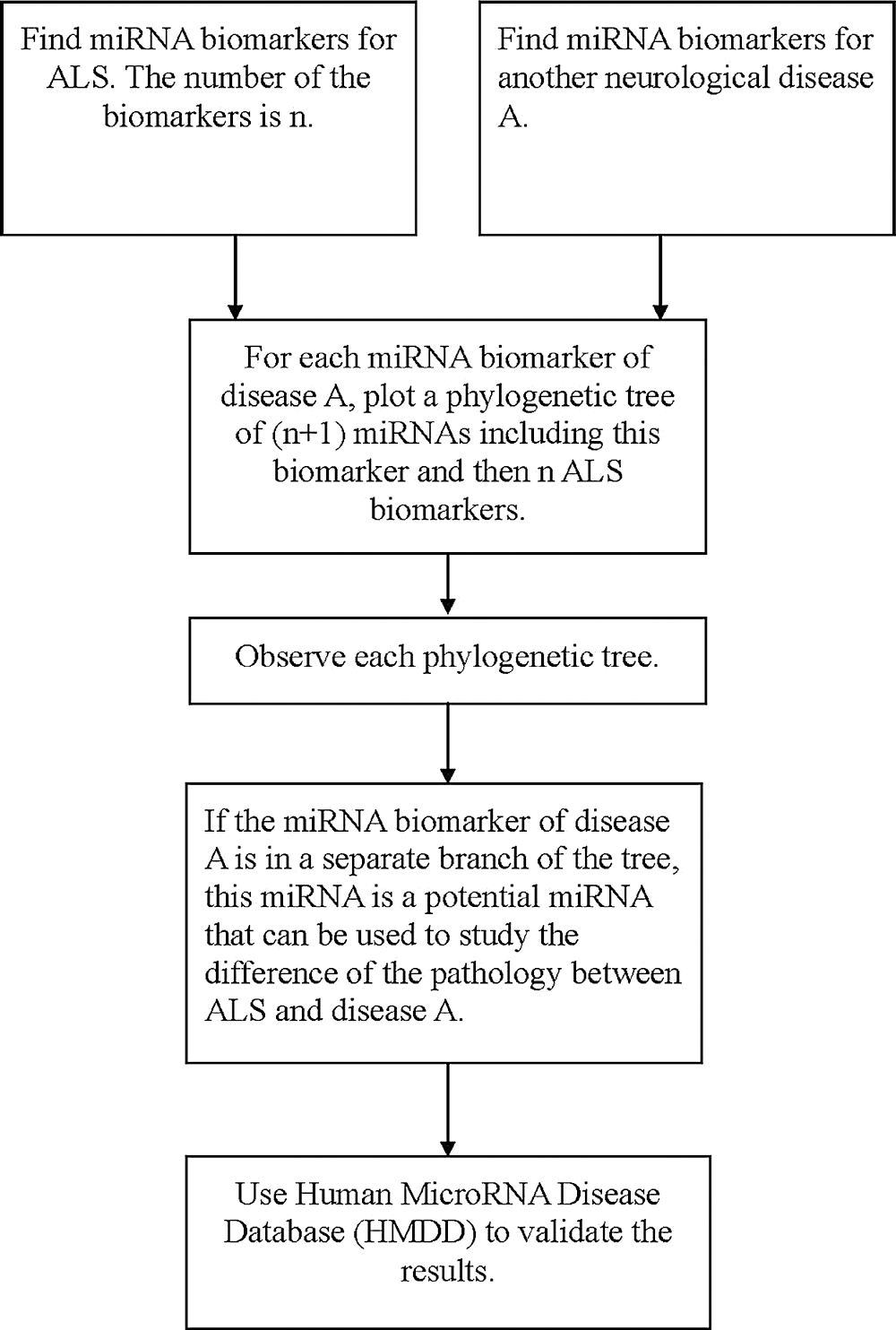
Figure 4: The flowchart of finding miRNAs that are potential to be used in studying the difference of pathology for ALS and another neurological disorder.
Phylogenetic tree analysis has been widely used in the study of miRNAs. Phylogenetic tree-informed miRNA analysis has uncovered conserved and lineage-specific miRNAs in Camellia during floral organ development (Yin et al., 2016); the phylogenetic analyses highlighted the potential of miRNAs to become an invaluable tool to resolve previously intractable nodes within the tree of life (Tarver et al., 2013), and the evolution of the two disease resistance-related miRNAs, miR-482 and miR-1448, was inferred using the phylogenetic analyses (Zhao et al., 2012). The phylogenetic age of miRNAs was computed in a study that concluded that older miRNAs were significantly more likely to be associated with disease than younger miRNAs (Patel and Capra, 2017). Furthermore, a combination of the phylogenetic tree analysis with a bioinformatics method can increase the accuracy of miRNA cancer biomarker prediction compared with only using the bioinformatics method alone (Wang, 2016b). The phylogenetic relationship of the miRNA biomarkers has been used to investigate the association between anti-NMDA receptor encephalitis and vaccination (Wang, 2017). In addition, the phylogenetic analysis has been used to explore the association between anti-NMDA receptor encephalitis and tumors based on miRNA biomarkers (Wang, 2019) and the association between two diseases (Chen and Wang, 2020b). These studies showed that phylogenetic analysis is a useful tool to explore miRNA functions.
Compared with the 9 ALS miRNA biomarkers, the miRNAs listed in Tab. 2 can be clustered to a separate group of the tree or be clustered with miR-155 to a separate group of the tree either using the alignment-score distance or the Jukes–Cantor distance. Thus, these identified miRNAs may be taken into account when studying the difference in the mechanisms for these diseases. Among these ALS miRNA biomarkers, miR-155 related to the SOD1 gene is different from the other biomarkers because it is often in a separate branch of the trees. There are several situations, such as miR-155, alone in a separate branch, or miR-155, with a disease biomarker in a separate branch. Therefore, more studies into the miR-155 regulatory mechanism may be useful in understanding the difference of pathologies for ALS and other neurological disorders.
In addition, miRNA studies can be used to investigate the comorbidities of diseases (Wang, 2019; Chen and Wang, 2020a; Chen and Wang, 2020b; Wang, 2020). I might use miRNA biomarkers to explore the comorbidities of ALS in a future study. Neurological disorders and cancer were shown to be the comorbidities of ALS in the literature. FTD was shown to be associated with ALS in many studies. ALS is developed in about 15% of patients with FTD (Van Mossevelde et al., 2017). The repeat expansion of C9orf72 is a major cause of FTD and a cause of other neurodegenerative diseases, including ALS (Dobson-Stone et al., 2013). As a result, the hexanucleotide repeat expansions of the C9orf72 gene link FTD to ALS (Renton et al., 2011; Benigni et al., 2016). Mouse models of C9orf72 hexanucleotide repeat expansion to relate these two diseases were explored (Liu et al., 2016; Batra and Lee, 2017; Ji et al., 2017).
The ALS drug, Riluzole, was found to be an anti-cancer drug in various cancers, including breast cancer, brain tumor, prostate cancer, osteosarcoma, and melanoma (Akamatsu et al., 2009; Le et al., 2010; Dolfi et al., 2017; Liao et al., 2017; Sperling et al., 2017). ALS might be inversely associated with cancer because a lower risk of cancer was observed in ALS patients, but a higher risk of ALS was observed in cancer patients compared with controls (Fang et al., 2013; Freedman et al., 2013). A microarray and survival analysis showed that ALS is related to cancer (Taguchi and Wang, 2017).
ALS is a complex disease, and its pathogenesis remains unknown. Several genetic factors, including C9orf72, SOD1, FUS, and TARDBP, have been discovered to be associated with ALS. Genes related to ALS may also be associated with other neurological disorders such as FTD. miRNA biomarkers of ALS and other neurological disorders have been discussed in the related literature. To the best of our knowledge, there have not been any studies exploring the difference of pathology between two neurological disorders using their miRNA biomarkers based on the phylogenetic tree analysis.
In this study, we applied the phylogenetic tree to analyze miRNA biomarkers of ALS and other neurological diseases. Using this method, we found a number of miRNAs that can be taken into account when studying the difference of pathology of ALS induced by the four genes and other neurological disorders such as frontotemporal dementia, Parkinson’s disease, Alzheimer’s disease, spinal muscular atrophy, Prader–Willi syndrome, Niemann–Pick disease, neurofibromatosis, narcolepsy, Friedreich’s ataxia, and ataxia-telangiectasia. In addition to these neurological disorders, this method can be used to explore the difference in pathology between ALS and other diseases. This may provide a useful direction in exploring the pathology of diseases based on miRNA biomarkers.
Author Contribution: The author confirms sole responsibility for the following: study conception and design, data collection, analysis and interpretation of results, and manuscript preparation.
Availability of Data and Materials: Readers can access the data upon request to corresponding author.
Funding Statement: This work was supported by the Ministry of Science and Technology 107-2118-M-009-002-MY2, Taiwan.
Conflicts of Interest: The author declares that they have no conflicts of interest to report regarding the present study.
Absalon S, Kochanek DM, Raghavan V, Krichevsky AM. (2013). MiR-26b, upregulated in Alzheimer’s disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. Journal of Neuroscience 33: 14645–14659. DOI 10.1523/JNEUROSCI.1327-13.2013. [Google Scholar] [CrossRef]
Akamatsu K, Shibata MA, Ito Y, Sohma Y, Azuma H, Otsuki Y. (2009). Riluzole induces apoptotic cell death in human prostate cancer cells via endoplasmic reticulum stress. Anticancer Research 29: 2195–2204. [Google Scholar]
Alevizos I, Illei GG. (2010). MicroRNAs in Sjögren’s syndrome as a prototypic autoimmune disease. Autoimmunity Reviews 9: 618–621. DOI 10.1016/j.autrev.2010.05.009. [Google Scholar] [CrossRef]
Banzhaf‐Strathmann J, Benito E, May S, Arzberger T, Tahirovic S, Kretzschmar H, Fischer Aé, Edbauer D. (2014). MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO Journal 33: 1667–1680. DOI 10.15252/embj.201387576. [Google Scholar] [CrossRef]
Batra R, Lee CW. (2017). Mouse models of C9orf72 hexanucleotide repeat expansion in amyotrophic lateral sclerosis/frontotemporal dementia. Frontiers in Cellular Neuroscience 11: 196. DOI 10.3389/fncel.2017.00196. [Google Scholar] [CrossRef]
Benigni M, Ricci C, Jones AR, Giannini F, Al-Chalabi A, Battistini S. (2016). Identification of miRNAs as potential biomarkers in cerebrospinal fluid from amyotrophic lateral sclerosis patients. Neuromolecular Medicine 18: 551–560. DOI 10.1007/s12017-016-8396-8. [Google Scholar] [CrossRef]
Bhinge A, Namboori SC, Bithell A, Soldati C, Buckley NJ, Stanton LW. (2016). MiR-375 is essential for human spinal motor neuron development and may be involved in motor neuron degeneration. Stem Cells 34: 124–134. DOI 10.1002/stem.2233. [Google Scholar] [CrossRef]
Bradfield NI, McLean C, Drago J, Darby DG, Ames D. (2017). Rapidly progressive Fronto-temporal dementia (FTD) associated with Frontotemporal lobar degeneration (FTLD) in the presence of Fused in Sarcoma (FUS) protein: A rare, sporadic, and aggressive form of FTD. International Psychogeriatrics 29: 1743–1746. DOI 10.1017/S1041610217001193. [Google Scholar] [CrossRef]
Butovsky O, Siddiqui S, Gabriely G, Lanser AJ, Dake B, Murugaiyan G, Doykan CE, Wu PM, Gali RR, Iyer LK, Lawson R, Berry J, Krichevsky AM, Cudkowicz ME, Weiner HL (2012a). Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. Journal of Clinical Investigation 122: 3063–3087. DOI 10.1172/JCI62636. [Google Scholar] [CrossRef]
Butovsky O, Siddiqui S, Gabriely G, Lanser AJ, Dake B, Murugaiyan G, Doykan CE, Wu PM, Gali RR, Iyer LK, Lawson R, Berry J, Krichevsky AN, Cudkowicz ME, Weiner HL (2012b). Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. Journal of Clinical Investigation 122: 3063–3087. DOI 10.1172/JCI62636. [Google Scholar] [CrossRef]
Campos-Melo D, Droppelmann CA, He Z, Volkening K, Strong MJ. (2013). Altered microRNA expression profile in amyotrophic lateral sclerosis: A role in the regulation of NFL mRNA levels. Molecular Brain 6: 26. DOI 10.1186/1756-6606-6-26. [Google Scholar] [CrossRef]
Chai G, Liu N, Ma J, Li H, Oblinger JL, Prahalad AK, Gong M, Chang LS, Wallace M, Muir D, Guha A, Phipps RJ, Hock JM, Yu X. (2010). MicroRNA-10b regulates tumorigenesis in neurofibromatosis type 1. Cancer Science 101: 1997–2004. DOI 10.1111/j.1349-7006.2010.01616.x. [Google Scholar] [CrossRef]
Chen-Plotkin AS, Unger TL, Gallagher MD, Bill E, Kwong LK, Volpicelli-Daley L, Busch JI, Akle S, Grossman M, Van Deerlin V, Trojanowski JQ, Lee VMY. (2012). TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. Journal of Neuroscience 32: 11213–11227. DOI 10.1523/JNEUROSCI.0521-12.2012. [Google Scholar] [CrossRef]
Chen L, Yang J, Lü J, Cao S, Zhao Q, Yu Z. (2018). Identification of aberrant circulating mi RNA s in Parkinson’s disease plasma samples. Brain and Behavior 8: e00941. DOI 10.1002/brb3.941. [Google Scholar] [CrossRef]
Chen S, Sayana P, Zhang X, Le W. (2013). Genetics of amyotrophic lateral sclerosis: An update. Molecular Neurodegeneration 8: 28. DOI 10.1186/1750-1326-8-28. [Google Scholar] [CrossRef]
Chen YH, Wang H (2020a). The association between depression and gastroesophageal reflux based on phylogenetic analysis of miRNA biomarkers. Current Medicinal Chemistry 27: 6536–6547. DOI 10.2174/0929867327666200425214906. [Google Scholar] [CrossRef]
Chen YH, Wang H (2020b). The association between migraine and depression based on miRNA biomarkers and cohort studies. Current Medicinal Chemistry. DOI 10.2174/0929867327666201117100026. [Google Scholar] [CrossRef]
Chen Y, Wei Q, Chen X, Li C, Cao B, Ou R, Hadano S, Shang HF. (2016). Aberration of miRNAs expression in leukocytes from sporadic amyotrophic lateral sclerosis. Frontiers in Molecular Neuroscience 9: 69. [Google Scholar]
Cheng C, Li W, Zhang Z, Yoshimura S, Hao Q, Zhang C, Wang Z. (2013). MicroRNA-144 is regulated by activator protein-1 (AP-1) and decreases expression of Alzheimer disease-related a disintegrin and metalloprotease 10 (ADAM10). Journal of Biological Chemistry 288: 13748–13761. DOI 10.1074/jbc.M112.381392. [Google Scholar] [CrossRef]
Dantham S, Srivastava AK, Gulati S, Rajeswari MR. (2018). Differentially regulated cell-free microRNAs in the plasma of Friedreich’s ataxia patients and their association with disease pathology. Neuropediatrics 49: 35–43. DOI 10.1055/s-0037-1607279. [Google Scholar] [CrossRef]
de Andrade HMT, de Albuquerque M, Avansini SH, Rocha CdS, Dogini DB, Nucci A, Carvalho B, Lopes-Cendes I, França MCJr. (2016). MicroRNAs-424 and 206 are potential prognostic markers in spinal onset amyotrophic lateral sclerosis. Journal of the Neurological Sciences 368: 19–24. DOI 10.1016/j.jns.2016.06.046. [Google Scholar] [CrossRef]
DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NCA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GYR, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72: 245–256. DOI 10.1016/j.neuron.2011.09.011. [Google Scholar] [CrossRef]
Dobson-Stone C, Hallupp M, Loy CT, Thompson EM, Haan E, Sue CM, Panegyres PK, Razquin C, Seijo-Martínez M, Rene R, Gascon J, Campdelacreu J, Schmoll B, Volk AE, Brooks WS, Schofield PR, Pastor P, Kwok JBJ, Wider C. (2013). C9ORF72 repeat expansion in Australian and Spanish frontotemporal dementia patients. PLoS One 8: e56899. DOI 10.1371/journal.pone.0056899. [Google Scholar] [CrossRef]
Dolfi SC, Medina DJ, Kareddula A, Paratala B, Rose A, Dhami J, Chen S, Ganesan S, Mackay G, Vazquez A, Hirshfield KM. (2017). Riluzole exerts distinct antitumor effects from a metabotropic glutamate receptor 1-specific inhibitor on breast cancer cells. Oncotarget 8: 44639–44653. DOI 10.18632/oncotarget.17961. [Google Scholar] [CrossRef]
Emde A, Eitan C, Liou LL, Libby RT, Rivkin N, Magen I, Reichenstein I, Oppenheim H, Eilam R, Silvestroni A, Alajajian B, Ben‐Dov IZ, Aebischer J, Savidor A, Levin Y, Sons R, Hammond SM, Ravits JM, Möller T, Hornstein E. (2015). Dysregulated miRNA biogenesis downstream of cellular stress and ALS-causing mutations: A new mechanism for ALS. EMBO Journal 34: 2633–2651. DOI 10.15252/embj.201490493. [Google Scholar] [CrossRef]
Fang F, Al-Chalabi A, Ronnevi LO, Turner MR, Wirdefeldt K, Kamel F, Ye W. (2013). Amyotrophic lateral sclerosis and cancer: A register-based study in Sweden. Amyotrophic Lateral Sclerosis and Frontotemporal Degeneration 14: 362–368. DOI 10.3109/21678421.2013.775309. [Google Scholar] [CrossRef]
Freedman DM, Curtis RE, Daugherty SE, Goedert JJ, Kuncl RW, Tucker MA. (2013). The association between cancer and amyotrophic lateral sclerosis. Cancer Causes & Control 24: 55–60. DOI 10.1007/s10552-012-0089-5. [Google Scholar] [CrossRef]
Freischmidt A, Müller K, Ludolph AC, Weishaupt JH. (2013). Systemic dysregulation of TDP-43 binding microRNAs in amyotrophic lateral sclerosis. Acta Neuropathologica Communications 1: 42. DOI 10.1186/2051-5960-1-42. [Google Scholar] [CrossRef]
Gascon E, Lynch K, Ruan H, Almeida S, Verheyden JM, Seeley WW, Dickson DW, Petrucelli L, Sun D, Jiao J, Zhou H, Jakovcevski M, Akbarian S, Yao WD, Gao FB. (2014). Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nature Medicine 20: 1444–1451. DOI 10.1038/nm.3717. [Google Scholar] [CrossRef]
Gong M, Ma J, Li M, Zhou M, Hock JM, Yu X. (2012). MicroRNA-204 critically regulates carcinogenesis in malignant peripheral nerve sheath tumors. Neuro-Oncology 14: 1007–1017. DOI 10.1093/neuonc/nos124. [Google Scholar] [CrossRef]
Grasedieck S, Sorrentino A, Langer C, Buske C, Döhner H, Mertens D, Kuchenbauer F. (2013). Circulating microRNAs in hematological diseases: Principles, challenges, and perspectives. Blood 121: 4977–4984. DOI 10.1182/blood-2013-01-480079. [Google Scholar] [CrossRef]
Grasso M, Piscopo P, Talarico G, Ricci L, Crestini A, Tosto G, Gasparini M, Bruno G, Denti MA, Confaloni A. (2019). Plasma microRNA profiling distinguishes patients with frontotemporal dementia from healthy subjects. Neurobiology of Aging 84: 240.e1–240.e12. DOI 10.1016/j.neurobiolaging.2019.01.024. [Google Scholar] [CrossRef]
Graur D, Li WH. (2000). Fundamentals of molecular evolution. Sunderland, Ma., Sinauer. [Google Scholar]
Guibinga GH, Hrustanovic G, Bouic K, Jinnah HA, Friedmann T. (2012). MicroRNA-mediated dysregulation of neural developmental genes in HPRT deficiency: Clues for Lesch–Nyhan disease? Human Molecular Genetics 21: 609–622. DOI 10.1093/hmg/ddr495. [Google Scholar] [CrossRef]
Gupta P, Bhattacharjee S, Sharma AR, Sharma G, Lee SS, Chakraborty C. (2017). miRNAs in Alzheimer disease-a therapeutic perspective. Current Alzheimer Research 14: 1198–1206. DOI 10.2174/1567205014666170829101016. [Google Scholar] [CrossRef]
Heman-Ackah SM, Hallegger M, Rao M, Wood M. (2013). RISC in PD: The impact of microRNAs in Parkinson’s disease cellular and molecular pathogenesis. Frontiers in Molecular Neuroscience 6: 40. DOI 10.3389/fnmol.2013.00040. [Google Scholar] [CrossRef]
Holm A, Bang-Berthelsen CH, Knudsen S, Kornum BR, Modvig S, Jennum P, Gammeltoft S. (2014). miRNA profiles in plasma from patients with sleep disorders reveal dysregulation of miRNAs in narcolepsy and other central hypersomnias. Sleep 37: 1525–1533. DOI 10.5665/sleep.4004. [Google Scholar] [CrossRef]
Hoss AG, Labadorf A, Beach TG, Latourelle JC, Myers RH. (2016). microRNA profiles in Parkinson’s disease prefrontal cortex. Frontiers in Aging Neuroscience 8: 36. DOI 10.3389/fnagi.2016.00036. [Google Scholar] [CrossRef]
Hsieh WJ, Lin FM, Huang HD, Wang H. (2014). Investigating microRNA-target interaction-supported tissues in human cancer tissues based on miRNA and target gene expression profiling. PLoS One 9: e95697. DOI 10.1371/journal.pone.0095697. [Google Scholar] [CrossRef]
Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. (2010). ATM is down-regulated by N-Myc-regulated microRNA-421. Proceedings of the National Academy of Sciences of the United States of America 107: 1506–1511. DOI 10.1073/pnas.0907763107. [Google Scholar] [CrossRef]
Huang Z, Shi J, Gao Y, Cui C, Zhang S, Li J, Zhou Y, Cui Q. (2018). HMDD v3.0: A database for experimentally supported human microRNA-disease associations. Nucleic Acids Research 47: D1013–D1017. DOI 10.1093/nar/gky1010. [Google Scholar] [CrossRef]
Jayakumar AR, Tong XY, Shamaladevi N, Barcelona S, Gaidosh G, Agarwal A, Norenberg MD. (2017). Defective synthesis and release of astrocytic thrombospondin-1 mediates the neuronal TDP-43 proteinopathy, resulting in defects in neuronal integrity associated with chronic traumatic encephalopathy: In vitro studies. Journal of Neurochemistry 140: 645–661. DOI 10.1111/jnc.13867. [Google Scholar] [CrossRef]
Jensen L, Jørgensen LH, Bech R, Frandsen U, Schrøder H. (2016). Skeletal muscle remodelling as a function of disease progression in amyotrophic lateral sclerosis. BioMed Research International 2016: 1–12. DOI 10.1155/2016/5930621. [Google Scholar] [CrossRef]
Ji YJ, Ugolino J, Brady NR, Hamacher-Brady A, Wang J. (2017). Systemic deregulation of autophagy upon loss of ALS-and FTD-linked C9orf72. Journal of Autophagy 13: 1254–1255. DOI 10.1080/15548627.2017.1299312. [Google Scholar] [CrossRef]
Khoo SK, Petillo D, Kang UJ, Resau JH, Berryhill B, Linder J, Forsgren L, Neuman LA, Tan AC. (2012). Plasma-based circulating MicroRNA biomarkers for Parkinson’s disease. Journal of Parkinson’s Disease 2: 321–331. DOI 10.3233/JPD-012144. [Google Scholar] [CrossRef]
Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. (2007). A MicroRNA feedback circuit in midbrain dopamine neurons. Science 317: 1220–1224. DOI 10.1126/science.1140481. [Google Scholar] [CrossRef]
Kocerha J, Kouri N, Baker M, Finch NC, DeJesus-Hernandez M, Gonzalez J, Chidamparam K, Josephs KA, Boeve BF, Graff-Radford NR, Crook J, Dickson DW, Rademakers R. (2011). Altered microRNA expression in frontotemporal lobar degeneration with TDP-43 pathology caused by progranulin mutations. BMC Genomics 12: 527. DOI 10.1186/1471-2164-12-527. [Google Scholar] [CrossRef]
Koval ED, Shaner C, Zhang P, du Maine X, Fischer K, Tay J, Chau BN, Wu GF, Miller TM. (2013). Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice. Human Molecular Genetics 22: 4127–4135. DOI 10.1093/hmg/ddt261. [Google Scholar] [CrossRef]
Kovanda A, Leonardis L, Zidar J, Koritnik B, Dolenc-Groselj L, Ristic Kovacic S, Curk T, Rogelj B. (2018). Differential expression of microRNAs and other small RNAs in muscle tissue of patients with ALS and healthy age-matched controls. Scientific Reports 8: 5609. DOI 10.1038/s41598-018-23139-2. [Google Scholar] [CrossRef]
Kozomara A, Griffiths-Jones S. (2013). miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research 42: D68–D73. DOI 10.1093/nar/gkt1181. [Google Scholar] [CrossRef]
Kurland LT, Mulder DW. (1955). Epidemiologic investigations of Amyotrophic Lateral Sclerosis: 2. Familial aggregations indicative of dominant inheritance Part II. Neurology 5: 249. DOI 10.1212/WNL.5.4.249. [Google Scholar] [CrossRef]
Kwiatkowski TJ, Bosco DA, LeClerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH. (2009). Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323: 1205–1208. DOI 10.1126/science.1166066. [Google Scholar] [CrossRef]
Kye MJ, Niederst ED, Wertz MH, Gonçalves IdCG, Akten B, Dover KZ, Peters M, Riessland M, Neveu P, Wirth B, Kosik KS, Sardi SP, Monani UR, Passini MA, Sahin M. (2014). SMN regulates axonal local translation via miR-183/mTOR pathway. Human Molecular Genetics 23: 6318–6331. DOI 10.1093/hmg/ddu350. [Google Scholar] [CrossRef]
Le MN, Chan JLK, Rosenberg SA, Nabatian AS, Merrigan KT, Cohen-Solal KA, Goydos JS. (2010). The glutamate release inhibitor Riluzole decreases migration, invasion, and proliferation of melanoma cells. Journal of Investigative Dermatology 130: 2240–2249. DOI 10.1038/jid.2010.126. [Google Scholar] [CrossRef]
Lee YH, Na HS, Jeong SY, Jeong SH, Park HR, Chung J. (2011). Comparison of inflammatory microRNA expression in healthy and periodontitis tissues. BIOCELL 35: B43–B49. DOI 10.32604/biocell.2011.35.043. [Google Scholar] [CrossRef]
Leggio L, Vivarelli S, L’Episcopo F, Tirolo C, Caniglia S, Testa N, Marchetti B, Iraci N. (2017). microRNAs in Parkinson’s disease: From pathogenesis to novel diagnostic and therapeutic approaches. International Journal of Molecular Sciences 18: 2698. DOI 10.3390/ijms18122698. [Google Scholar] [CrossRef]
Liao S, Ruiz Y, Gulzar H, Yelskaya Z, Ait Taouit L, Houssou M, Jaikaran T, Schvarts Y, Kozlitina K, Basu-Roy U, Mansukhani A, Mahajan SS, Chen S. (2017). Osteosarcoma cell proliferation and survival requires mGluR5 receptor activity and is blocked by Riluzole. PLoS One 12: e0171256. DOI 10.1371/journal.pone.0171256. [Google Scholar] [CrossRef]
Liu Y, Pattamatta A, Zu T, Reid T, Bardhi O, Borchelt DR, Yachnis AT, Ranum LPW. (2016). C9orf72 BAC mouse model with motor deficits and neurodegenerative features of ALS/FTD. Neuron 90: 521–534. DOI 10.1016/j.neuron.2016.04.005. [Google Scholar] [CrossRef]
Lukiw WJ. (2007). Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport 18: 297–300. DOI 10.1097/WNR.0b013e3280148e8b. [Google Scholar] [CrossRef]
Maciotta Rolandin S, Meregalli M, Torrente Y. (2013). The involvement of microRNAs in neurodegenerative diseases. Frontiers in Cellular Neuroscience 7: 265. [Google Scholar]
Mackenzie IR, Neumann M. (2016). Molecular neuropathology of frontotemporal dementia: Insights into disease mechanisms from postmortem studies. Journal of Neurochemistry 138: 54–70. DOI 10.1111/jnc.13588. [Google Scholar] [CrossRef]
Magri F, Vanoli F, Corti S. (2018). miRNA in spinal muscular atrophy pathogenesis and therapy. Journal of Cellular and Molecular Medicine 22: 755–767. [Google Scholar]
Mahishi LH, Hart RP, Lynch DR, Ratan RR. (2012). miR-886-3p levels are elevated in Friedreich ataxia. Journal of Neuroscience 32: 9369–9373. DOI 10.1523/JNEUROSCI.0059-12.2012. [Google Scholar] [CrossRef]
Maimon R, Ionescu A, Bonnie A, Sweetat S, Wald-Altman S, Inbar S, Gradus T, Trotti D, Weil M, Behar O, Perlson E. (2018). miR126-5p downregulation facilitates axon degeneration and NMJ disruption via a non-cell-autonomous mechanism in ALS. Journal of Neuroscience 38: 5478–5494. DOI 10.1523/JNEUROSCI.3037-17.2018. [Google Scholar] [CrossRef]
Majounie E, Renton AE, Mok K, Dopper EGP, Waite A, Rollinson S, Chiò A, Restagno G, Nicolaou N, Simon-Sanchez J, van Swieten JC, Abramzon Y, Johnson JO, Sendtner M, Pamphlett R, Orrell RW, Mead S, Sidle KC, Houlden H, Rohrer JD, Morrison KE, Pall H, Talbot K, Ansorge O, Hernandez DG, Arepalli S, Sabatelli M, Mora G, Corbo M, Giannini F, Calvo A, Englund E, Borghero G, Floris GL, Remes AM, Laaksovirta H, McCluskey L, Trojanowski JQ, Van Deerlin VM, Schellenberg GD, Nalls MA, Drory VE, Lu CS, Yeh TH, Ishiura H, Takahashi Y, Tsuji S, Le Ber I, Brice A, Drepper C, Williams N, Kirby J, Shaw P, Hardy J, Tienari PJ, Heutink P, Morris HR, Pickering-Brown S, Traynor BJ. (2012). Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: A cross-sectional study. Lancet Neurology 11: 323–330. DOI 10.1016/S1474-4422(12)70043-1. [Google Scholar] [CrossRef]
Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, Komure O, Matsuura S, Kobatake K, Morimoto N, Abe K, Suzuki N, Aoki M, Kawata A, Hirai T, Kato T, Ogasawara K, Hirano A, Takumi T, Kusaka H, Hagiwara K, Kaji R, Kawakami H. (2010). Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465: 223–226. DOI 10.1038/nature08971. [Google Scholar] [CrossRef]
Masliah-Planchon J, Pasmant E, Luscan A, Laurendeau I, Ortonne N, Hivelin M, Varin J, Valeyrie-Allanore L, Dumaine V, Lantieri L, Leroy K, Parfait B, Wolkenstein P, Vidaud M, Vidaud D, Bièche I. (2013). MicroRNAome profiling in benign and malignant neurofibromatosis type 1-associated nerve sheath tumors: Evidences of PTEN pathway alterations in early NF1 tumorigenesis. BMC Genomics 14: 473. DOI 10.1186/1471-2164-14-473. [Google Scholar] [CrossRef]
Matamala JM, Arias-Carrasco R, Sanchez C, Uhrig M, Bargsted L, Matus S, Maracaja-Coutinho V, Abarzua S, van Zundert B, Verdugo R, Manque P, Hetz C. (2018). Genome-wide circulating microRNA expression profiling reveals potential biomarkers for amyotrophic lateral sclerosis. Neurobiology of Aging 64: 123–138. DOI 10.1016/j.neurobiolaging.2017.12.020. [Google Scholar] [CrossRef]
Mateen FJ, Carone M, Sorenson EJ. (2010). Patients who survive 5 years or more with ALS in Olmsted County, 1925-2004. Journal of Neurology, Neurosurgery & Psychiatry 81: 1144–1146. DOI 10.1136/jnnp.2009.201251. [Google Scholar] [CrossRef]
Mathworks IJMI, Natick. (2014). MATLAB: R2014a. [Google Scholar]
McKee AC, Gavett BE, Stern RA, Nowinski CJ, Cantu RC, Kowall NW, Perl DP, Hedley-Whyte ET, Price B, Sullivan C, Morin P, Lee HS, Kubilus CA, Daneshvar DH, Wulff M, Budson AE. (2010). TDP-43 proteinopathy and motor neuron disease in chronic traumatic encephalopathy. Journal of Neuropathology & Experimental Neurology 69: 918–929. DOI 10.1097/NEN.0b013e3181ee7d85. [Google Scholar] [CrossRef]
Morel L, Regan M, Higashimori H, Ng SK, Esau C, Vidensky S, Rothstein J, Yang Y. (2013). Neuronal exosomal miRNA-dependent translational regulation of astroglial glutamate transporter GLT1. Journal of Biological Chemistry 288: 7105–7116. DOI 10.1074/jbc.M112.410944. [Google Scholar] [CrossRef]
Mosakhani N, Sarhadi V, Panula P, Partinen M, Knuutila S. (2017). Narcolepsy patients’ blood-based miRNA expression profiling: miRNA expression differences with Pandemrix vaccination. Acta Neurologica Scandinavica 136: 462–469. DOI 10.1111/ane.12749. [Google Scholar] [CrossRef]
Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IRA. (2009). A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 132: 2922–2931. DOI 10.1093/brain/awp214. [Google Scholar] [CrossRef]
Oskarsson B, Horton DK, Mitsumoto H. (2015). Potential environmental factors in Amyotrophic Lateral Sclerosis. Neurologic Clinics 33: 877–888. DOI 10.1016/j.ncl.2015.07.009. [Google Scholar] [CrossRef]
Ozsait B, Komurcu-Bayrak E, Levula M, Erginel-Unaltuna N, Kähönen M, Rai M, Lehtimäki T, Laaksonen R. (2010). Niemann–Pick type C fibroblasts have a distinct microRNA profile related to lipid metabolism and certain cellular components. Biochemical and Biophysical Research Communications 403: 316–321. DOI 10.1016/j.bbrc.2010.11.026. [Google Scholar] [CrossRef]
Pascut D, Tamini S, Bresolin S, Giraudi P, Basso G, Minocci A, Tiribelli C, Grugni G, Sartorio A. (2018). Differences in circulating microRNA signature in Prader-Willi syndrome and non-syndromic obesity. Endocrine Connections 7: 1262–1274. [Google Scholar]
Patel VD, Capra JA. (2017). Ancient human miRNAs are more likely to have broad functions and disease associations than young miRNAs. BMC Genomics 18: 672. DOI 10.1186/s12864-017-4073-z. [Google Scholar] [CrossRef]
Pergande MR, Cougnoux A, Rathnayake RA, Porter FD, Cologna SM. (2019). Differential proteomics reveals miR-155 as a novel indicator of liver and spleen pathology in the symptomatic Niemann-Pick disease, type C1 mouse model. Molecules 24: 994. DOI 10.3390/molecules24050994. [Google Scholar] [CrossRef]
Prajapati P, Sripada L, Singh K, Bhatelia K, Singh R, Singh R. (2015). TNF-α regulates miRNA targeting mitochondrial complex-I and induces cell death in dopaminergic cells. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 1852: 451–461. DOI 10.1016/j.bbadis.2014.11.019. [Google Scholar] [CrossRef]
Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72: 257–268. DOI 10.1016/j.neuron.2011.09.010. [Google Scholar] [CrossRef]
Ricci C, Marzocchi C, Battistini S. (2018). MicroRNAs as biomarkers in amyotrophic lateral sclerosis. Cells 7: 219. DOI 10.3390/cells7110219. [Google Scholar] [CrossRef]
Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX, Rahmani Z, Krizus A, McKenna-Yasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, Van den Bergh R, Hung WY, Bird T, Deng G, Mulder DW, Smyth C, Laing NG, Soriano E, Pericak–Vance MA, Haines J, Rouleau GA, Gusella JS, Horvitz HR, Brown RHJr. (1993). Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362: 59–62. DOI 10.1038/362059a0. [Google Scholar] [CrossRef]
Rowland LP. (2001). How amyotrophic lateral sclerosis got its name: The clinical-pathologic genius of Jean-Martin Charcot. Archives of Neurology 58: 512–515. DOI 10.1001/archneur.58.3.512. [Google Scholar] [CrossRef]
Russell AP, Wada S, Vergani L, Hock MB, Lamon S, Léger B, Ushida T, Cartoni R, Wadley GD, Hespel P, Kralli A, Soraru G, Angelini C, Akimoto T. (2013). Disruption of skeletal muscle mitochondrial network genes and miRNAs in amyotrophic lateral sclerosis. Neurobiology of Disease 49: 107–117. DOI 10.1016/j.nbd.2012.08.015. [Google Scholar] [CrossRef]
Sangwan S, Eisenberg DS. (2016). Perspective on SOD1 mediated toxicity in Amyotrophic Lateral Sclerosis. Postepy Biochemii 62: 362–369. [Google Scholar]
Seco-Cervera M, González-Rodríguez D, Ibáñez-Cabellos JS, Peiró-Chova L, Pallardó FV, García-Giménez JL. (2018). Small RNA-seq analysis of circulating miRNAs to identify phenotypic variability in Friedreich’s ataxia patients. Scientific Data 5: 180021. DOI 10.1038/sdata.2018.21. [Google Scholar] [CrossRef]
Sedani A, Cooper DN, Upadhyaya M. (2012). An emerging role for microRNAs in NF1 tumorigenesis. Human Genomics 6: 23. DOI 10.1186/1479-7364-6-23. [Google Scholar] [CrossRef]
Shang Y, Huang EJ. (2016). Mechanisms of FUS mutations in familial amyotrophic lateral sclerosis. Brain Research 1647: 65–78. DOI 10.1016/j.brainres.2016.03.036. [Google Scholar] [CrossRef]
Sperling S, Aung T, Martin S, Rohde V, Ninkovic M. (2017). Riluzole: A potential therapeutic intervention in human brain tumor stem-like cells. Oncotarget 8: 96697–96709. DOI 10.18632/oncotarget.18043. [Google Scholar] [CrossRef]
Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. (2008). TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319: 1668–1672. DOI 10.1126/science.1154584. [Google Scholar] [CrossRef]
Taguchi Y, Wang H. (2017). Genetic association between amyotrophic lateral sclerosis and cancer. Genes 8: 243. DOI 10.3390/genes8100243. [Google Scholar] [CrossRef]
Taguchi Y, Wang H (2018a). Exploring microRNA biomarkers for Parkinson’s disease from mRNA expression profiles. Cells 7: 245. DOI 10.3390/cells7120245. [Google Scholar] [CrossRef]
Taguchi YH, Wang H (2018b). Exploring microRNA biomarker for amyotrophic lateral sclerosis. International Journal of Molecular Sciences 19: 1318. DOI 10.3390/ijms19051318. [Google Scholar] [CrossRef]
Takahashi I, Hama Y, Matsushima M, Hirotani M, Kano T, Hohzen H, Yabe I, Utsumi J, Sasaki H. (2015). Identification of plasma microRNAs as a biomarker of sporadic Amyotrophic Lateral Sclerosis. Molecular Brain 8: 67. DOI 10.1186/s13041-015-0161-7. [Google Scholar] [CrossRef]
Takuse Y, Watanabe M, Inoue N, Ozaki R, Ohtsu H, Saeki M, Katsumata Y, Hidaka Y, Iwatani Y. (2017). Association of IL-10-regulating microRNAs in peripheral blood mononuclear cells with the pathogenesis of autoimmune thyroid disease. Immunological Investigations 46: 590–602. DOI 10.1080/08820139.2017.1322975. [Google Scholar] [CrossRef]
Tarver JE, Sperling EA, Nailor A, Heimberg AM, Robinson JM, King BL, Pisani D, Donoghue PCJ, Peterson KJ. (2013). miRNAs: Small genes with big potential in metazoan phylogenetics. Molecular Biology and Evolution 30: 2369–2382. DOI 10.1093/molbev/mst133. [Google Scholar] [CrossRef]
Toivonen JM, Manzano R, Oliván S, Zaragoza P, García-Redondo A, Osta R, Cai H. (2014). MicroRNA-206: A potential circulating biomarker candidate for amyotrophic lateral sclerosis. PLoS One 9: e89065. DOI 10.1371/journal.pone.0089065. [Google Scholar] [CrossRef]
Tremblay C, St-Amour I, Schneider J, Bennett DA, Calon F. (2011). Accumulation of transactive response DNA binding protein 43 in mild cognitive impairment and Alzheimer disease. Journal of Neuropathology & Experimental Neurology 70: 788–798. DOI 10.1097/NEN.0b013e31822c62cf. [Google Scholar] [CrossRef]
Turner MR, Hardiman O, Benatar M, Brooks BR, Chio A, de Carvalho M, Ince PG, Lin C, Miller RG, Mitsumoto H, Nicholson G, Ravits J, Shaw PJ, Swash M, Talbot K, Traynor BJ, Van den Berg LH, Veldink JH, Vucic S, Kiernan MC. (2013). Controversies and priorities in amyotrophic lateral sclerosis. Lancet Neurology 12: 310–322. DOI 10.1016/S1474-4422(13)70036-X. [Google Scholar] [CrossRef]
Van Mossevelde S, van der Zee J, Gijselinck I, Sleegers K, De Bleecker J, Sieben A, Vandenberghe R, Van Langenhove T, Baets J, Deryck O, Santens P, Ivanoiu A, Willems C, Bäumer V, Van den Broeck M, Peeters K, Mattheijssens M, De Jonghe P, Cras P, Martin JJ, Cruts M, De Deyn PP, Engelborghs S, Van Broeckhoven C. (2017). Clinical evidence of disease anticipation in families segregating a C9orf72 repeat expansion. JAMA Neurology 74: 445–452. DOI 10.1001/jamaneurol.2016.4847. [Google Scholar] [CrossRef]
Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. (2009). Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323: 1208–1211. DOI 10.1126/science.1165942. [Google Scholar] [CrossRef]
Visser J, de Jong JV, de Visser M. (2008). The history of progressive muscular atrophy: Syndrome or disease? Neurology 70: 723–727. DOI 10.1212/01.wnl.0000302187.20239.93. [Google Scholar] [CrossRef]
Wakabayashi K, Mori F, Kakita A, Takahashi H, Utsumi J, et al. (2014). Analysis of microRNA from archived formalin-fixed paraffin-embedded specimens of amyotrophic lateral sclerosis. Acta Neuropathologica Communications 2: 173. DOI 10.1186/s40478-014-0173-z. [Google Scholar] [CrossRef]
Waller R, Goodall EF, Milo M, Cooper-Knock J, Da Costa M, Hobson E, Kazoka M, Wollff H, Heath PR, Shaw PJ, Kirby J. (2017). Serum miRNAs miR-206, 143-3p and 374b-5p as potential biomarkers for amyotrophic lateral sclerosis (ALS). Neurobiology of Aging 55: 123–131. DOI 10.1016/j.neurobiolaging.2017.03.027. [Google Scholar] [CrossRef]
Wang H (2016a). Efficacies of treatments for anti-NMDA receptor encephalitis. Frontiers in Bioscience 21: 651–663. DOI 10.2741/4412. [Google Scholar] [CrossRef]
Wang H (2016b). Predicting microRNA biomarkers for cancer using phylogenetic tree and microarray analysis. International Journal of Molecular Sciences 17: 773. DOI 10.3390/ijms17050773. [Google Scholar] [CrossRef]
Wang H. (2017). Anti-NMDA receptor encephalitis and vaccination. International Journal of Molecular Sciences 18: 193. DOI 10.3390/ijms18010193. [Google Scholar] [CrossRef]
Wang H. (2019). Phylogenetic analysis to explore the association between anti-NMDA receptor encephalitis and tumors based on microRNA biomarkers. Biomolecules 9: 572. DOI 10.3390/biom9100572. [Google Scholar] [CrossRef]
Wang H. (2020). MicroRNA, diabetes mellitus and colorectal cancer. Biomedicines 8: 530. DOI 10.3390/biomedicines8120530. [Google Scholar] [CrossRef]
Wang H, Hung SL. (2012). Phylogenetic tree selection by the adjusted k-means approach. Journal of Applied Statistics 39: 643–655. DOI 10.1080/02664763.2011.610442. [Google Scholar] [CrossRef]
Wang LT, Chiou SS, Liao YM, Jong YJ, Hsu SH. (2014). Survival of motor neuron protein downregulates miR-9 expression in patients with spinal muscular atrophy. Kaohsiung Journal of Medical Sciences 30: 229–234. DOI 10.1016/j.kjms.2013.12.007. [Google Scholar] [CrossRef]
Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. (2008). The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of β-site amyloid precursor protein-cleaving enzyme 1. Journal of Neuroscience 28: 1213–1223. DOI 10.1523/JNEUROSCI.5065-07.2008. [Google Scholar] [CrossRef]
Wertz MH, Winden K, Neveu P, Ng SY, Ercan E, Sahin M. (2016). Cell-type-specific miR-431 dysregulation in a motor neuron model of spinal muscular atrophy. Human Molecular Genetics 25: 2168–2181. DOI 10.1093/hmg/ddw084. [Google Scholar] [CrossRef]
Wu CW, Dong YJ, Liang QY, He XQ, Ng SSM, Chan FKL, Sung JJY, Yu J, Guan XY. (2013). MicroRNA-18a attenuates DNA damage repair through suppressing the expression of ataxia telangiectasia mutated in colorectal cancer. PLoS One 8: e57036. DOI 10.1371/journal.pone.0057036. [Google Scholar] [CrossRef]
Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y. (2010). DNA methylation mediated by a microRNA pathway. Molecular Cell 38: 465–475. DOI 10.1016/j.molcel.2010.03.008. [Google Scholar] [CrossRef]
Yin H, Fan Z, Li X, Wang J, Liu W, Wu B, Ying Z, Liu L, Liu Z, Li J. (2016). Phylogenetic tree-informed microRNAome analysis uncovers conserved and lineage-specific miRNAs in Camellia during floral organ development. Journal of Experimental Botany 67:2641–2653. [Google Scholar]
Zhang Z, Falaleeva M, Agranat-Tamir L, Pages A, Eyras E, Sperling J, Sperling R, Stamm S. (2013). The 5’ untranslated region of the serotonin receptor 2C pre-mRNA generates miRNAs and is expressed in non-neuronal cells. Experimental Brain Research 230: 387–394. DOI 10.1007/s00221-013-3458-8. [Google Scholar] [CrossRef]
Zhao JP, Diao S, Zhang BY, Niu BQ, Wang QL, Wan XC, Luo YQ, Buratti E. (2012). Phylogenetic analysis and molecular evolution patterns in the MIR482-MIR1448 polycistron of Populus L. PLoS One 7: e47811. DOI 10.1371/journal.pone.0047811. [Google Scholar] [CrossRef]
Zhou Y, Liu S, Öztürk A, Hicks GG. (2014). FUS-regulated RNA metabolism and DNA damage repair: Implications for amyotrophic lateral sclerosis and frontotemporal dementia pathogenesis. Rare Diseases 2: e1003895. DOI 10.4161/rdis.29515. [Google Scholar] [CrossRef]
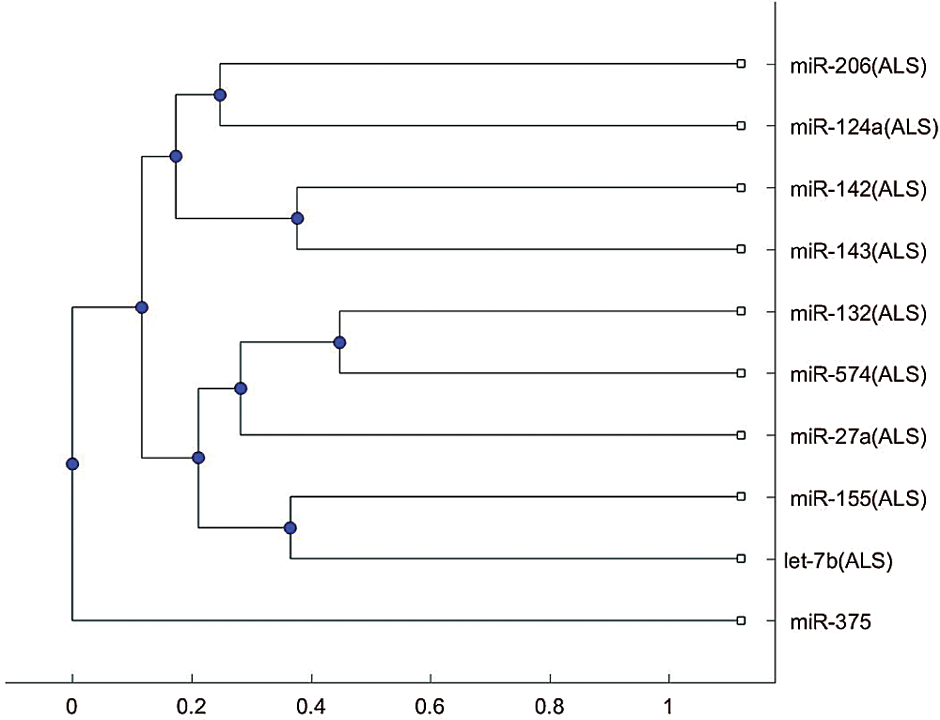
Figure s1: Phylogenetic tree based on Jukes-Cantor distance and the average linkage method.
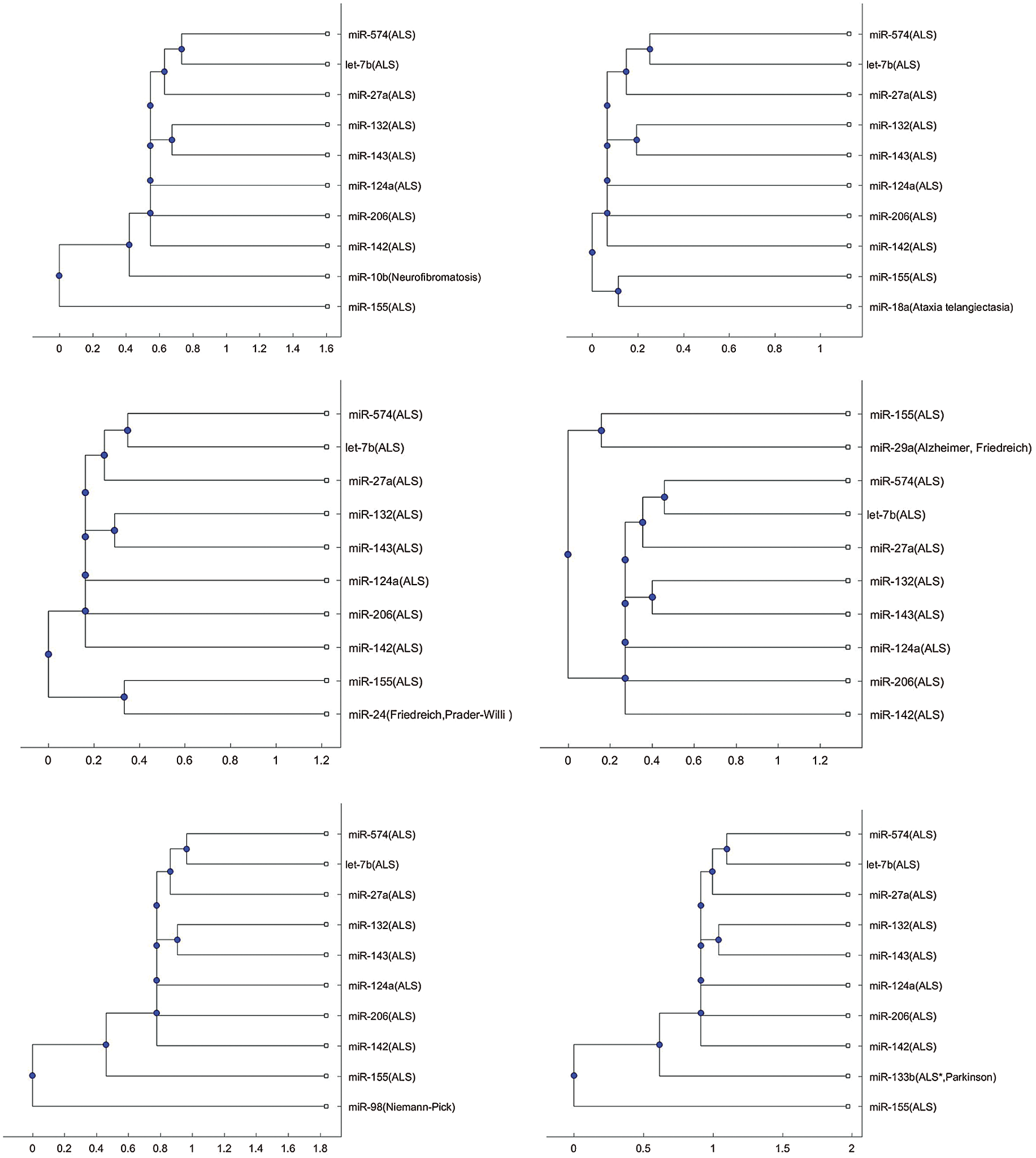
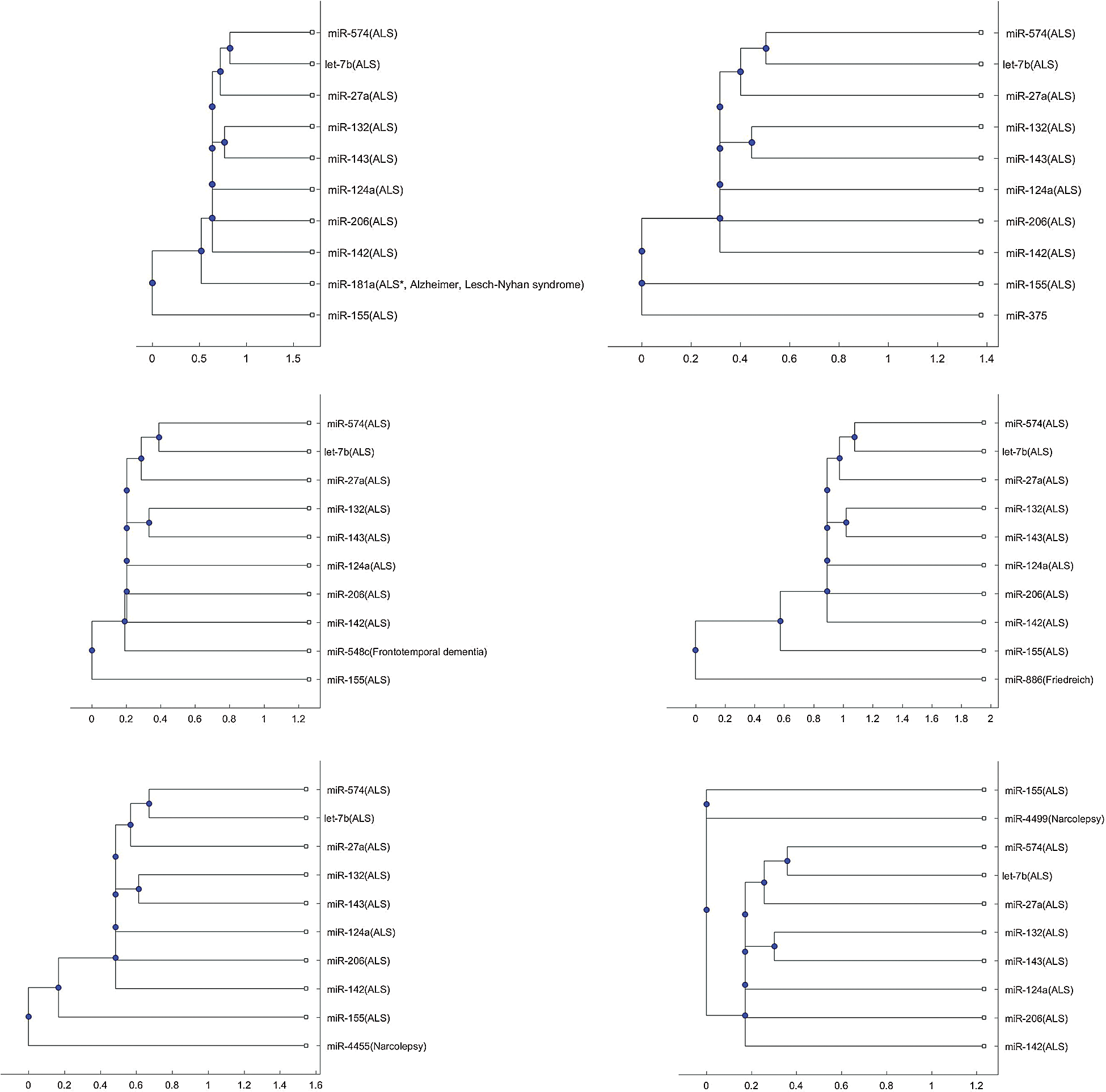
Figure s2: Phylogenetic tree based on alignment-score distance and the median linkage method.
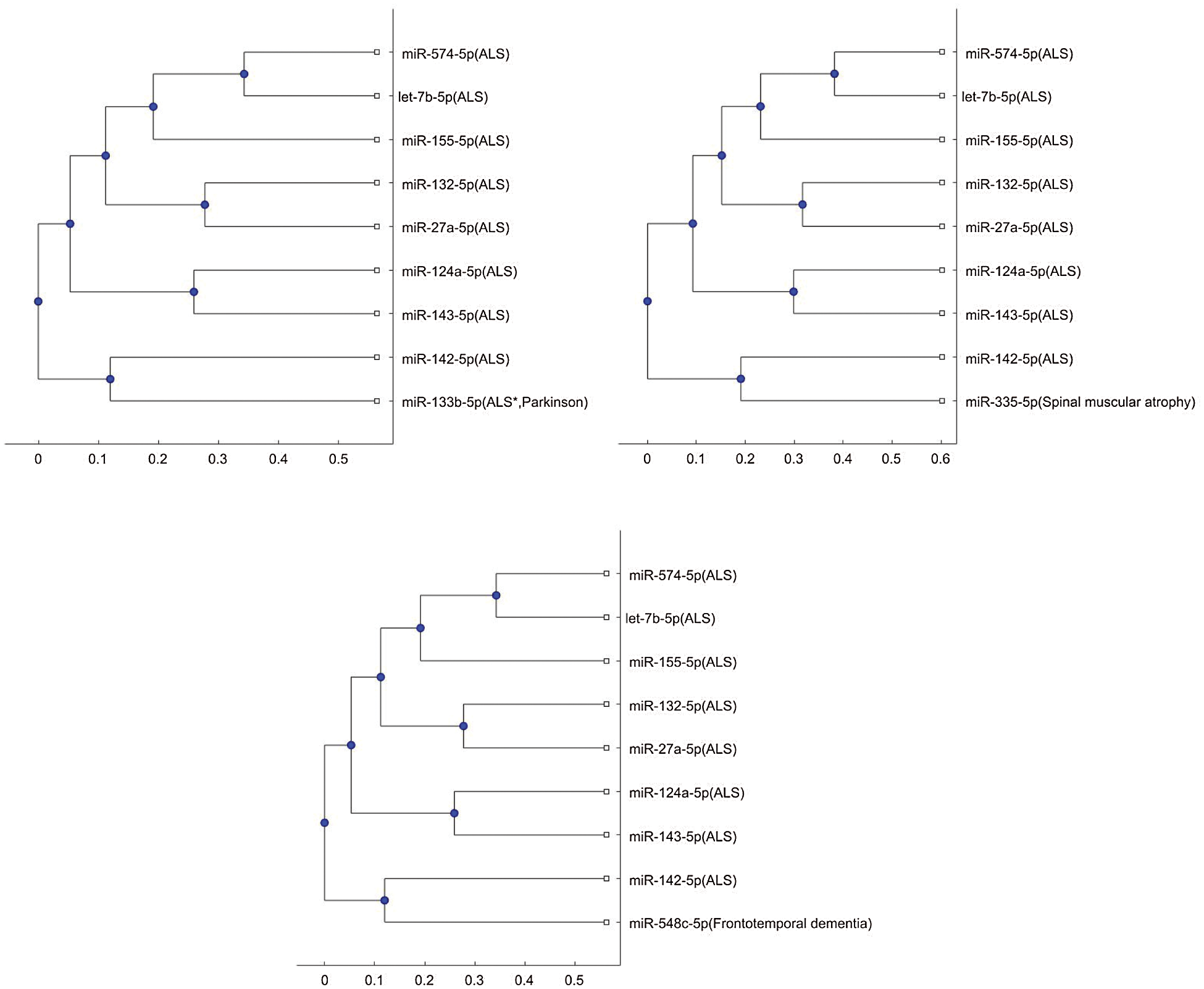
Figure S3: Phylogenetic tree for 3p- or 5p- miRNA sequences based on Jukes-Cantor distance and the average linkage method.
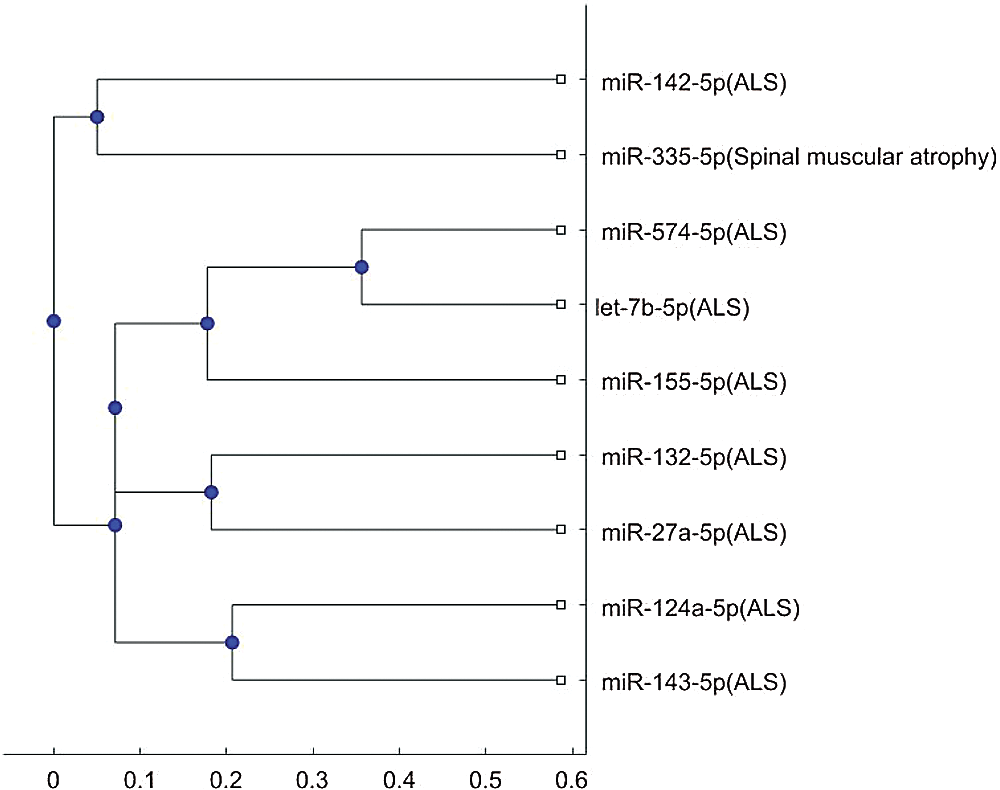
Figure S4: Phylogenetic tree for 3p- or 5p- miRNA sequences based on alignment-score distance and the median linkage method.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |