DOI:10.32604/biocell.2021.014756

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.014756 |  www.techscience.com/journal/biocell |
| Article |
Silencing of Astrocyte Elevated Gene-1 (AEG-1) inhibits the proliferative and invasive potential through interaction with Exostosin-1 (EXT-1) in primary and metastatic colon cancer cells
1Department of Medical Biotechnology, Faculty of Allied Health Sciences, Chettinad Academy of Research and Education (CARE), Chettinad Hospital and Research Institute (CHRI), Chennai, India
2Bioinformatics Centre, Department of RDAP, North-Eastern Hill University, Tura, India
3Department of Food Science and Nutrition, CAMS, Sultan Qaboos University, Muscat, Oman
4Ageing and Dementia Research Group, Sultan Qaboos University, Muscat, Oman
5Department of Pharmacology, JSS College of Pharmacy, JSS Academy of Higher Education & Research, Mysuru, India
6Research & Policy Department, World Innovation Summit for Health (WISH), Qatar Foundation, Doha, Qatar
7Q3CG Research Institute (QRI), Research & Policy Division, Ypsilanti, MI 48917, USA
*Address correspondence to: Surajit Pathak, surajit.pathak@gmail.com
Received: 27 October 2020; Accepted: 23 December 2020
Abstract: Colon cancer is the third major cause of cancer deaths, accounting for about 8% in terms of mortality globally. The present study aims to explore the effect of silencing Astrocyte Elevated Gene-1 (AEG-1), a metastasis mediating factor, and how it interacts with Exostosin-1 (EXT-1) protein to inhibit the proliferative and invasive potential in colon cancer cells. Forward siRNA transfection was performed using AEG-1 siRNA in SW480 and SW620 colon cancer cell lines, and the expression levels of mRNA and protein were analyzed by Real-time PCR and Immunofluorescence. A simple bioinformatics approach was carried out to identify the possible interactions between AEG-1 and EXT-1 using Easy Networks and Pathway Commons Database. Cell survival and clonal efficiency were determined using Cell Counting Kit-8 assay and clonogenic assay, apoptosis using flow cytometry analysis, migration and invasion using scratch and Transwell assays, respectively. Forward siRNA transfection significantly suppressed the expression of AEG-1 in mRNA and protein levels on SW480 and SW620 colon cancer cells. From our results, we found that EXT-1 mRNA and protein level was significantly upregulated in AEG-1 siRNA transfected cells. Moreover, treatment with AEG-1 siRNA inhibited the proliferation, clonogenic ability, migration, and invasion and also induced apoptosis. Through the bioinformatic approach, our data analyses pointed towards the crosstalk between AEG-1 and EXT-1 mediated through Patched-1 (PTCH-1) protein. Our current results demonstrated that silencing AEG-1 can restrain cell proliferation, migration, and invasion, ultimately leading to apoptosis. In AEG-1 siRNA transfected cells, PTCH-1 activity might be modulated by several genes and, in turn, affects the EXT-1 activity. Collectively, these observations not only provide insight into the interplay between AEG-1 and EXT-1 but also suggest that AEG-1 may represent a possible candidate therapeutic target through interaction with EXT-1 in colon cancer.
Keywords: Oncogene; EXT-1; siRNA; Colon cancer; AEG-1
Colon cancer (CC) is considered the third major cause of cancer-death, accounting for about 8% in terms of mortality globally (Budisan et al., 2019). Till now, there are no effective treatments available to reduce the risk of CC, and the rate of metastasis for this cancer seems to be very high (Vogelstein and Kinzler, 2004). Activities of several signaling pathways like Wnt/β-catenin, PI3K/Akt, MAPK and NF-κB are found to be correlated with colon carcinogenesis, which provides potential biomarkers in CC (Lièvre et al., 2010; Grady and Pritchard, 2014). Identifying the potential therapeutic targets is highly needed to efficiently inhibit the proliferation, invasiveness, and metastasis of CC cells (Akagi et al., 2013). Metadherin (MTDH) or Astrocyte Elevated Gene-1 (AEG-1) has been shown to have an upraised expression in fetal astrocytes after subsequent infection by HIV-1 or gp-120 treatment (Zhong et al., 2003). The location of AEG-1 is highly significant in several malignancies such as glioma, breast cancer, and hepatocellular carcinoma (HCC). However, a copious amount of investigation is necessary on AEG-1 to distinguish the entire biochemical as well as molecular properties. Localization of AEG-1 is not only limited in the membrane, but also includes cytoplasm, nucleus, and endoplasmic reticulum (ER). It is reported to be overexpressed in most cancers, including CC, and promotes tumor cell proliferation, invasion, and metastasis (Hu et al., 2009; Li et al., 2017; Yao et al., 2014; Zhu et al., 2015). It is also evident that increased AEG-1 expression is in correlation with the Union for International Cancer Control (UICC) (Song et al., 2010). AEG-1 is considered as a downstream target of H-ras oncogene and plays a significant role in H-ras-mediated carcinogenesis (Lee et al., 2006). Comprehensively, these observations stipulate that AEG-1 could act as a possible fundamental gene in controlling the progression of the tumor (Lee et al., 2008). In some studies, oncogene AEG-1 was found to impede apoptosis and enhance migration and invasion via the PI3K/Akt signaling pathway (Noch and Khalili, 2013). Apoptosis, also known as type-I cell death, is an eminently controlled physiological process that plays a significant role in maintaining normal colonic epithelium (Watson and Pritchard, 2000). Increasing evidence was shown in the disruption between the equilibrium of growth and the rate of cell apoptosis during CC formation. Inhibition of apoptosis could possibly cause a disparity in human intestinal epithelial cell homeostasis, which associates with the genesis of CC and its deprived reaction to therapies (Abraha and Ketema, 2016). Mutations in onco- and tumor-suppressor genes like APC, K-RAS, AEG-1, P53 and PIK3CA were reported to lead to inefficacious apoptosis mechanisms (Armaghany et al., 2012). Continual genomic alteration due to the imbalance in CC may also give rise to variations in genes, which has been found to be involved in controlling apoptosis (Stoian et al., 2014). Li et al. (2018) suggested that AEG-1 may function via the expression of numerous apoptosis-related proteins in CC cells. Through some studies, AEG-1 overexpression enhanced migration and matrix invasive ability in HCC, HeLa, and neuroblastoma cells. In vivo studies have also proven that AEG-1 overexpression resulted in metastatic tumors in nude mice models of human liver cells (Byoung et al., 2009; Gnosa et al., 2016; Emdad et al., 2006). Though various signaling pathways and small molecules have been discovered, additional research is required to elucidate the complete mechanism of invasion and metastasis.
For decades, it is well known that heparan sulfate proteoglycans (HSPGs) are intricated in the development of cancer at various stages. Studies have proven that depletion of HSPGs could be responsible for tumor growth and invasion, which substantially inhibit invasion and metastasis of renal carcinoma cells (Qazi et al., 2016). In particular, various CC critical genes, including Exostosin-1 (EXT-1), exhibit a strong relationship with HSPGs. EXT-1, a well-known tumor suppressor, is a type-II transmembrane glycoprotein with glycosyl-transferase activities required for the biosynthesis of HS, and its localization is limited to ER (Busse et al., 2007). This EXT-1 is a co-polymerase that is accountable for HS formation. Both EXT-1 and EXT-2 protein forms a hetero-oligomeric complex, which led to an assemblage in Golgi. Some mutations of EXT-1 and EXT-2 have been reported to be a significant pathogenic effect of hereditary multiple exostoses, distinguished by several cartilage-capped malignancies (Pacifici, 2017).
The reason behind investigating both AEG-1 and EXT-1 in our study is that AEG-1 is oncogenic because it plays a significant role in the activation of diverse signaling pathways involved in cancer proliferation and invasion. It is also a pivotal intermediate for malignant tumors and a major converging point of a complex network of cancer pathways and EXT-1, being a tumor suppressor could help in the biosynthesis of HSPGs. Although studies have reported that HS backbone synthesis is mediated by its biosynthetic enzyme EXT-1, the involvement of EXT-1 in CC metastasis is not well understood (Busse-Wicher et al., 2014). Thus, the main aim of our current study was to investigate the effect of silencing AEG-1, a metastasis mediating factor and how it interacts with EXT-1 protein to inhibit the proliferative and invasive potential in CC cells. To the best of our knowledge, this is the first study to find the potential relationship between AEG-1 and EXT-1 in CC cells. In addition, in silico analysis was also performed to find the possible biological interactions between AEG-1 and EXT-1.
CC primary and metastatic cell lines SW480 and SW620 were acquired from NCCS, Pune, India. There was no mycoplasma infection or changes in cellular morphology seen during the usage of these cell lines. Culturing was done with Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% Fetal Bovine Serum (FBS), 1% glutamine, and 1% penicillin-streptomycin at 37°C in 5% CO2. Materials including DMEM, FBS, trypsin, antibiotic-antimycotic solution, Opti-MEM and Phosphate-Buffered Saline (PBS) were obtained from GIBCO (Thermo Fisher Scientific).
Transfection of SW480 and SW620 with MTDH/AEG-1 Silencer Pre-designed siRNA (Ambion, Thermo Fisher Scientific) was carried out with the help of Lipofectamine RNAiMAX Transfection Reagent acquired from Invitrogen, Thermo Fisher Scientific using the protocol specified by manufacturer’s instructions. Seeding was done in Opti-MEM without antibiotic, and after it reaches 30%–50% confluence, forward transfection was performed for the adherent cells. Transfected cells were collected after 24 h, and it was utilized for further experiments.
The Cell Counting Kit-8 (CCK-8, Sigma-Aldrich) was used to determine cell proliferation. A total of 2000 transiently transfected cells were seeded into 96-well plates and grown overnight at different time intervals of 24 h, 48 h, and 72 h. CCK-8 (10 µL) reagent was added to each well individually and kept for incubation at 37°C for a period of 4 h. The resulting absorbance was measured at 450 nm with a microplate reader (Robonik Touch Well Reader, India).
A total of 1000 transiently transfected cells were seeded in a 6 well plate followed by incubation. The total time of incubation for this assay was 12 days at 37°C, after which they were stained using 0.5% crystal violet at room temperature (RT) for just 15 min. Colonies with more than 50 cells as a minimum for scoring were counted under an inverted phase-contrast microscope (Leica Microsystems).
Transient transfection was carried out using AEG-1 siRNA, and the cells were resuspended 24 h post-treatment in 1X binding buffer. Cell apoptosis was examined using the kit obtained from BD Pharmingen. The FITC Annexin-V Apoptosis Detection Kit (Cat. No #556547) was used according to the manufacturer’s instructions. BD FACS Canto™ (BD Biosciences) was used to determine the percentage of apoptotic cells. Annexin-V FITC and Propidium Iodide (PI), each of 5 μL were used to stain the cells at RT in dark for 10–15 min, and analysis was carried out.
A total of 75,000 transfected cells were seeded in 24-well plates and grown overnight. Opti-MEM was used to starve the cells for 24 h and subsequently, after confluency, a scratch was made with a sterile 200 μL tip across the cell monolayer. After scratching, unattached cells were washed off and day 0 pictures were taken in Leica Image viewer. 24 h, 48 h, and 72 h post-scratching images were taken in 4X magnification. Using Image J software (Polygon selection mode), the area of the wound was measured.
Transfected cells were starved using Opti-MEM for 24 h prior to seeding in the Transwell chambers. Matrigel (Cat. No. #356234, Corning) was used to pre-coat the Transwell chambers (Corning). In the upper chamber of a Transwell, 2 × 104 cells were added in 100 μL serum-free medium and the lower chamber contained 500 μL of 10% FBS DMEM medium. After incubation for about 48 h at 37°C, the cells in the upper chamber were removed. Those that invaded through the membranes were fixed using methanol and further stained with Giemsa. Images were acquired in Leica Image viewer and invading cells were counted under the microscope in five random fields.
Gene expression analysis using qPCR
Cells were treated with TRIzol reagent (Invitrogen, USA) in order to extract RNA. Quantity and purity check of total RNA for all samples was determined using NanoDrop at 260 nm and 280 nm, respectively. Reverse transcriptase reaction was performed using the Eurogentec Reverse Transcriptase PCR kit (Eurogentec, Belgium). The mRNA levels of human normal colon RNA (Cat. No. #AM7986, Ambion) and colon cancer cells were assessed using qPCR. The qPCR was performed using Eurogentec TakyonTM ROX SYBR Mastermix, dTTP Blue kit (Takyon, Eurogentec, Belgium) for genes with primer sequences mentioned in Tab. 1. The reaction setup (50°C for 2 min, Carryover prevention; 95°C for 3 min, Takyon activation; 95°C for 10 s, Denaturation and 60°C for 1 min, Annealing/Extension) was carried out in ABI 7500 Fast Real-Time PCR System (Applied Biosystems) and threshold cycle value (Ct) was acquired. Normalization of target gene Ct value with β-actin expression was done to obtain the ΔCt values and fold change was calculated.
Table 1: List of primer sequences used to identify the gene expression patterns in qPCR

SW480 and SW620 cells were grown on coverslips and transfected with AEG-1 siRNA. After 24 h, cold PBS was used to wash the cells, and further, cells were fixed using chilled methanol for about 10 min at −20°C. Permeabilization of cells was carried out with 0.1% Triton X-100 for 15 min at RT. After blocking in 2% bovine serum albumin (BSA) for about 1 h at RT, the coverslips were stained with the desired concentration of primary antibodies AEG-1 (1:250) (Catalog #40-6400-Invitrogen, Thermo Fisher Scientific), EXT-1 (1:500) (Catalog No. # PA5-76041), and BAX (1:500) (Catalog No. sc-7480). Incubation for overnight at 4°C and further treated by Alexa FluorTM 633 goat anti-rabbit (IgG) secondary antibody (Catalog #A-21070-Invitrogen, Thermo Fisher Scientific) for about 1 h at RT protected from light (Invitrogen, 1:400). DAPI (Sigma Aldrich) was used for nuclear counterstaining followed by mounting the coverslips on glass slides using a mounting medium, and then images were taken in EVOS FLoid Cell Imaging Station (Invitrogen, Thermo Fisher Scientific).
Bioinformatic approach using Easy Networks and Pathway Commons database
The network interaction data was visualized through esyN (“easy networks”) (Bean et al., 2014). In esyN, the interaction network was predicted by utilizing human interaction data of both kinds, genetic and physical, available in HumanMine (Smith et al., 2012) and BioGRID (Oughtred et al., 2019) with the set of genes of interest. Pathway Commons (Rodchenkov et al., 2020), on the other hand, integrates the expression and interaction data from 22 databases using Systems Biology Graphical Notation (SBGN) (Le et al., 2009) and PATIKAweb (Dogrusoz et al., 2009) tool.
The statistical significance was assessed using Student’s t-test with Graph Pad Prism 5.0 software and P < 0.05 was considered to be statistically significant. Data are expressed as mean ± standard deviation. All experiments were carried out in triplicates.
Silencing of AEG-1 upregulate EXT-1 expression in CC cells
SW480 and SW620 cells were cultured in 10% FBS DMEM and subcultured for further experiments after reaching about 80% confluence. The mRNA and protein expression levels of AEG-1 were assessed in SW480 and SW620 cells. The expression pattern of AEG-1 was analyzed by qPCR and immunofluorescence, respectively. Results from qPCR demonstrated that AEG-1 expression was found to be significantly upregulated in CC cells compared to the normal colon cells confirming it is an oncogene (Fig. 1a) whereas the cells transfected with AEG-1 siRNA showed significant downregulation in both SW480 and SW620 cells (Figs. 1c and 1d). Immunofluorescence analysis displayed that the fluorescence intensity of AEG-1 protein was found to be decreased in AEG-1 siRNA transfected cells indicating significantly lower expression of protein in both SW480 (Figs. 2a and 2b) and SW620 cells (Figs. 2c and 2d). Further examination for the function of AEG-1 in CC cell behavior was by using these cells in subsequent experiments. Alongside, as studies have exhibited that EXT-1 acts by suppressing tumor growth in many kinds of tumor cells including CC and further studies have stipulated that EXT-1 majorly participates in HS biosynthesis contributing to invasiveness and metastasis of cancer cells, we thus identified the EXT-1 expression in CC cells. It was found to be significantly downregulated in CC cells compared to the normal colon cells, confirming it is a tumor suppressor (Fig. 1b). In this study, we surprisingly found that EXT-1 expression was significantly upregulated in AEG-1 siRNA transfected cells (mRNA level) in both SW480 and SW620 cells (Figs. 1e and 1f). Immunofluorescence analysis was performed to confirm further in protein level, where the results displayed that the fluorescence intensity of EXT-1 protein was found to be increased in AEG-1 siRNA transfected cells indicating significantly higher expression of protein in both SW480 (Figs. 2e and 2f) and SW620 cells (Figs. 2g and 2h).
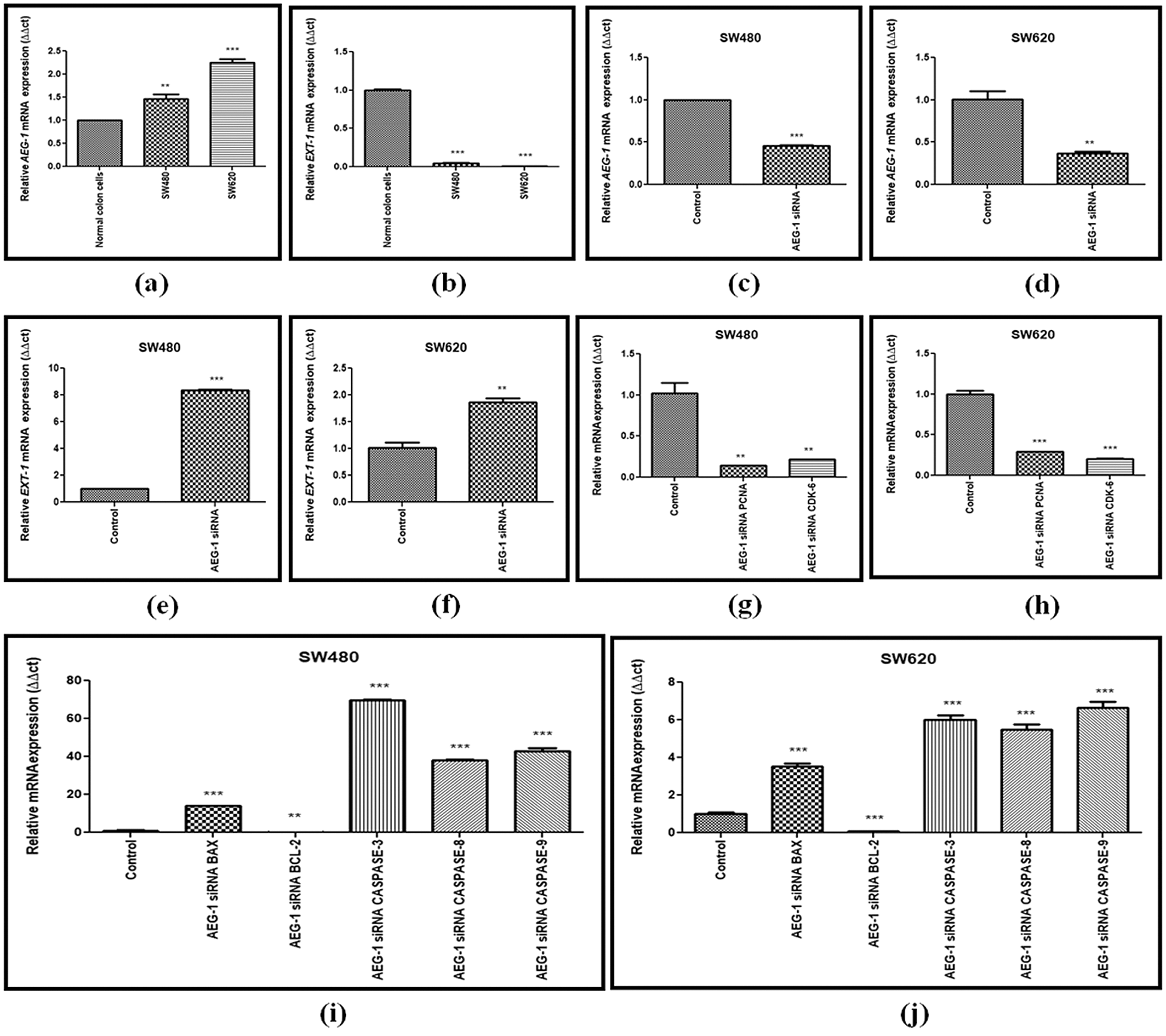
Figure 1: (a) Basal level expression of AEG-1 in normal colon cells and colon cancer cells (SW480 and SW620). (b) Basal level expression of EXT-1 in normal colon cells and colon cancer cells (SW480 and SW620). (c) Downregulation of AEG-1 after silencing with AEG-1 siRNA in SW480 cells and (d) SW620 cells. (e) Upregulation of EXT-1 after silencing with AEG-1 siRNA in SW480 cells and (f) SW620 cells. (g) Expression of PCNA and CDK-6 after treatment with AEG-1 siRNA supporting proliferation studies in SW480 cells and (h) SW620 cells. (i) Expression of apoptotic and anti-apoptotic markers (BAX, BCL-2, CASPASE-3, 8 and 9) after treatment with AEG-1 siRNA in SW480 cells and (j) SW620 cells (P < 0.05)
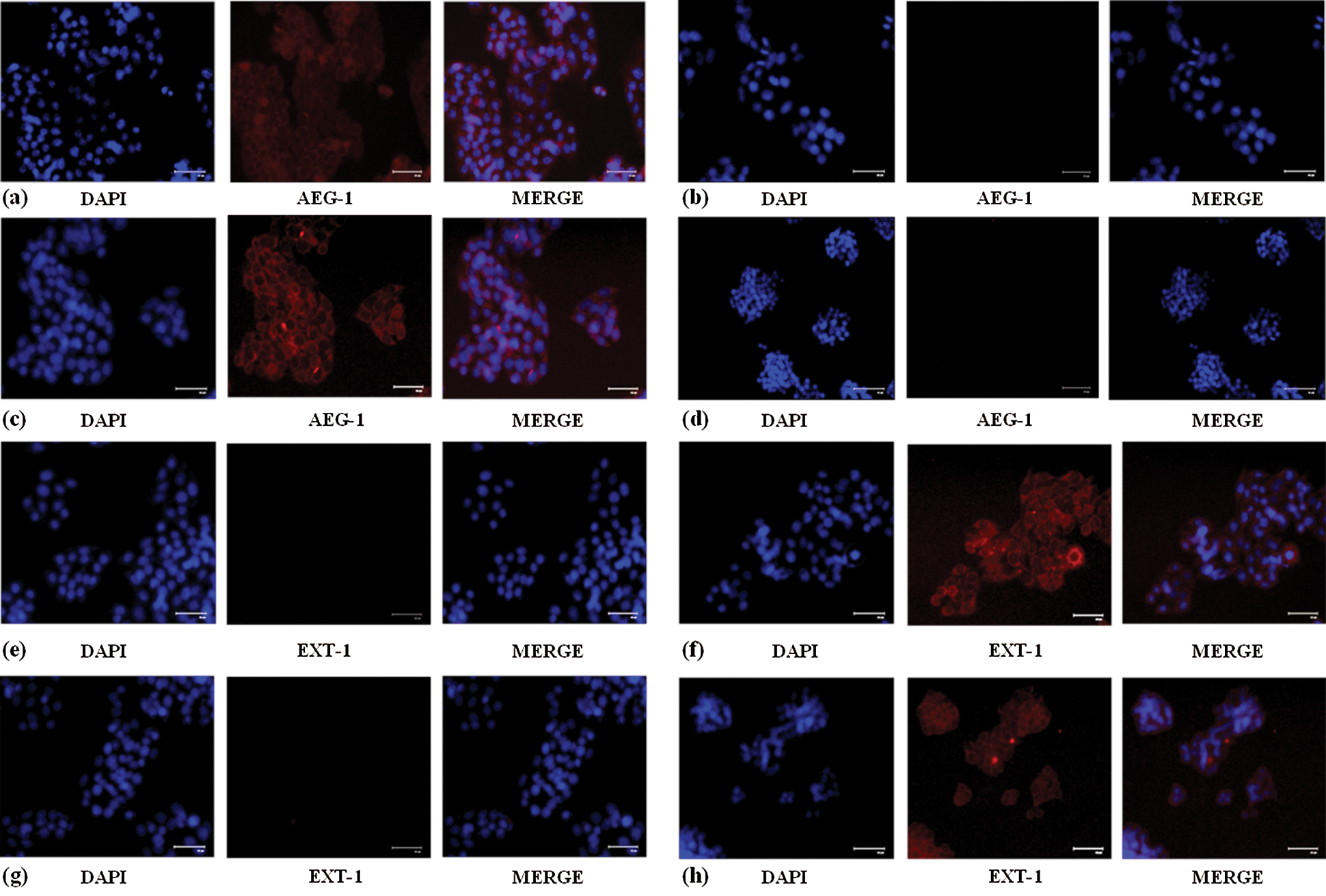
Figure 2: (a) Increased AEG-1 protein expression in control SW480 cells compared to the (b) cells treated with AEG-1 siRNA. (c) Increased AEG-1 protein expression in control SW620 cells compared to the (d) cells treated with AEG-1 siRNA. (e) Decreased EXT-1 protein expression in control SW480 cells compared to the (f) cells treated with AEG-1 siRNA. (g) Decreased EXT-1 protein expression in control SW620 cells compared to the (h) cells treated with AEG-1 siRNA
Molecular interactions of AEG-1 and EXT-1 proteins through Patched-1 protein
From esyN and Pathway Commons data integration and analyses, it was apparent that AEG-1 and EXT-1 proteins were capable to perturb each other’s function in more than one way (Figs. 3a and 3b). Both the protein–protein interaction network data analyses pointed towards the crosstalk between AEG-1 and EXT-1 mediated through Patched-1 (PTCH-1). Further, the PTCH-1 activity and expression might be modulated by a number of genes as evident from the figure and in turn, affects the AEG-1 and/or EXT-1 activity.
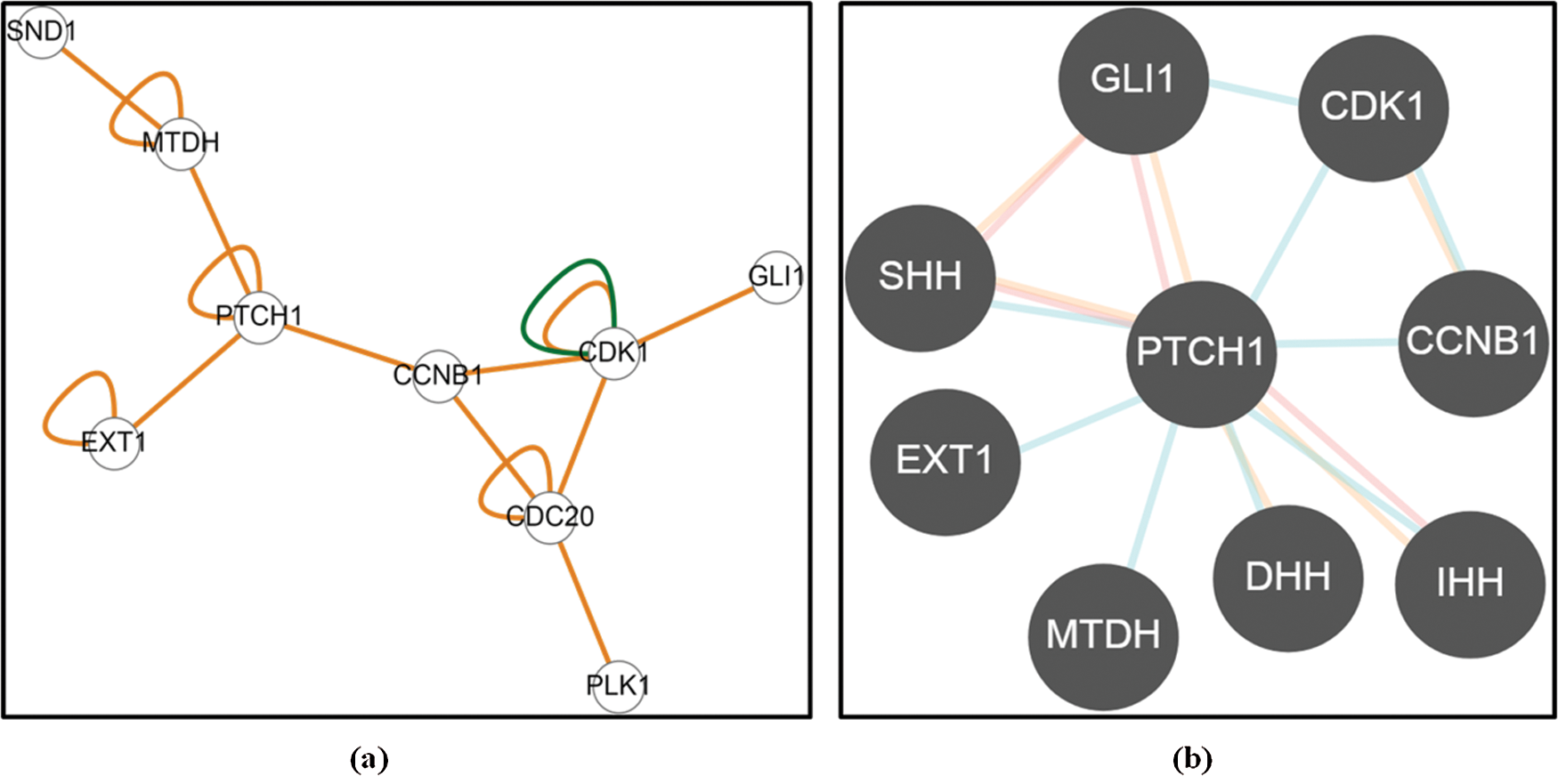
Figure 3: Predictive Molecular interactions between MTDH/AEG-1 and EXT-1 through (a) Easy Networks database and (b) Pathway Commons database
Silencing of AEG-1 inhibits CC cellular proliferation in vitro
AEG-1 silencing effects on the growth of SW480 and SW620 cells in vitro were detected. Detection was done by CCK-8 assay. Our results displayed that the proliferation of SW480 and SW620 transfected cells was significantly suppressed in a time-dependent manner (Figs. 4a and 4b). Average Inhibition Rate (in %) was calculated using a formula: [(OD value of control group–OD value of experimental group)/OD value of control group] x 100%. Inhibition rate of SW480 cells were 16.3% (24 h), 52.2% (48 h), 54.9% (72 h) and SW620 cells were 4.6% (24 h), 41.3% (48 h), and 41.6% (72 h). To support proliferation studies, the expression of proliferation markers like proliferating-cell nuclear antigen (PCNA) and cyclin-dependent kinase 6 (CDK-6) was observed in qPCR and was found to be significantly downregulated after AEG-1 siRNA transfection in SW480 and SW620 cells (Figs. 1g and 1h). Colony formation assay further confirmed that knockdown of AEG-1 displayed a significant reduction in the colony-forming ability of SW480 and SW620 cells (Figs. 4c and 4d). Collectively, these results suggest/strongly supported that in absence of AEG-1, both the number and the size of colonies are suppressed.
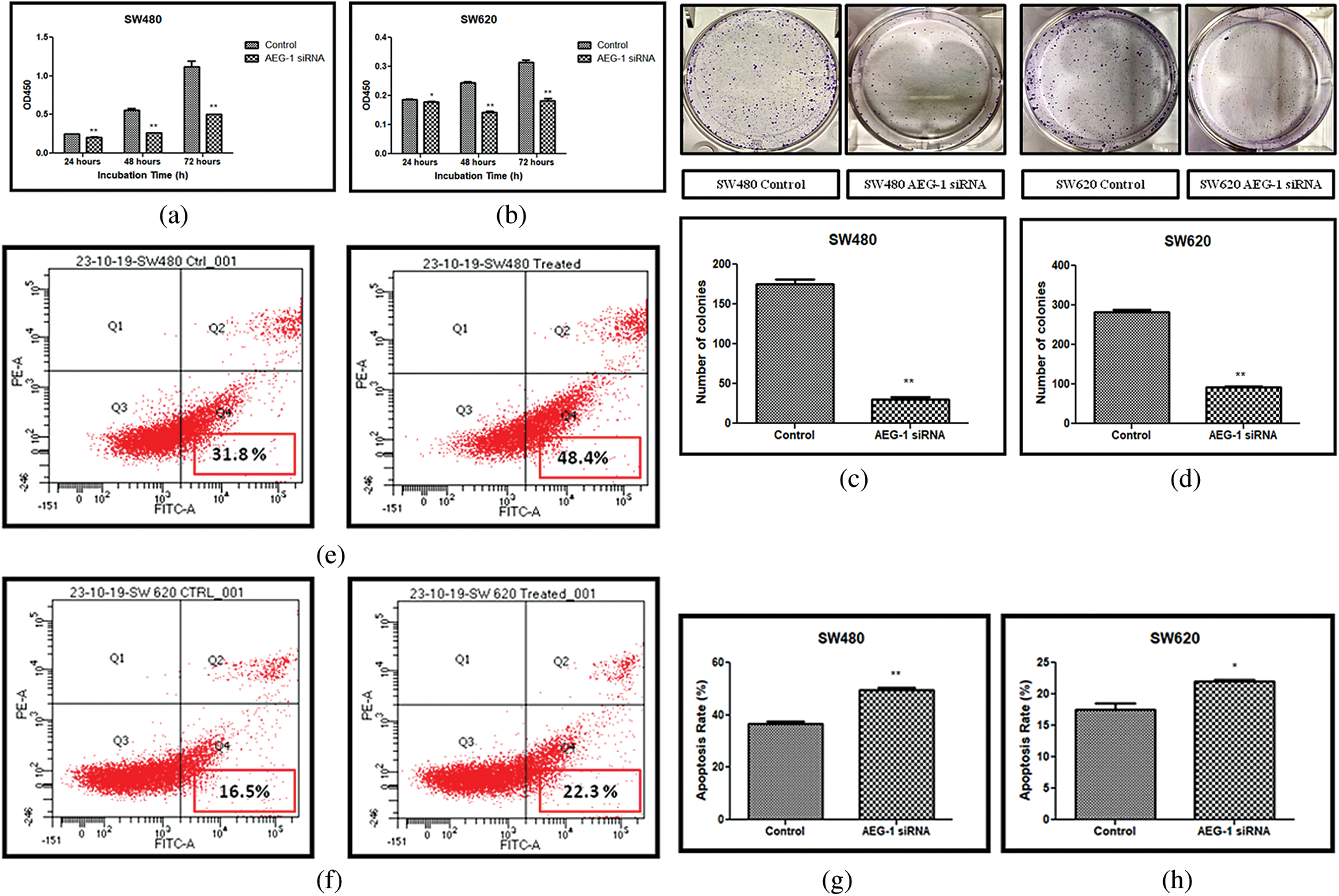
Figure 4: (a) Cell proliferation assay before and after treatment with AEG-1 siRNA in SW480 cells and (b) SW620 cells. (c) Colony-forming assay before and after treatment with AEG-1 siRNA in SW480 cells and (d) SW620 cells. (e) FACS analysis using Annexin-V/FITC in SW480 cells and (f) SW620 cells. (g) Graph plotted for apoptosis detection rate (in %) before and after treatment with AEG-1 siRNA in SW480 cells and (h) SW620 cells (P < 0.05)
Silencing of AEG-1 increases apoptosis-associated proteins in CC cells
Apoptosis which is vitally important in cell growth suppression was detected by Annexin-V FITC/PI staining by flow cytometry. As presented in the Figs. 4e and 4f. the flow cytometry analysis was carried out and the early apoptosis rate of SW480 and SW620 control cells was found to be 31.8% and 16.5% respectively whereas the AEG-1 siRNA transfected cells were 48.4% and 22.3% respectively (Figs. 4e and 4f). Apoptosis detection rate (in %) was plotted in Y-axis and cells before and after treatment with AEG-1 siRNA in X-axis for SW480 and SW620 cells (Figs. 4g and 4h). To support apoptosis analysis, the expression of apoptosis markers like bcl-2-like protein 4 (BAX), Caspase-3, Caspase-8 and Caspase-9 was observed in qPCR and were found to be upregulated whereas anti-apoptotic B-cell lymphoma-2 (BCL-2) marker was found to be downregulated in AEG-1 siRNA transfected cells compared to control cells (Figs. 1i and 1j). Furthermore, the expression of proteins related to apoptosis was determined by immunofluorescence analysis. Immunofluorescence results showed that the fluorescent intensity of BAX was found to be increased in AEG-1 siRNA transfected cells indicating significantly higher expression of protein in both SW480 (Figs. 5a and 5b) and SW620 cells (Figs. 5c and 5d). These results suggest that AEG-1 knockdown may induce apoptosis. This results in inhibition of the cellular proliferation indicating that AEG-1 may function through apoptosis-related protein expression in CC cells.
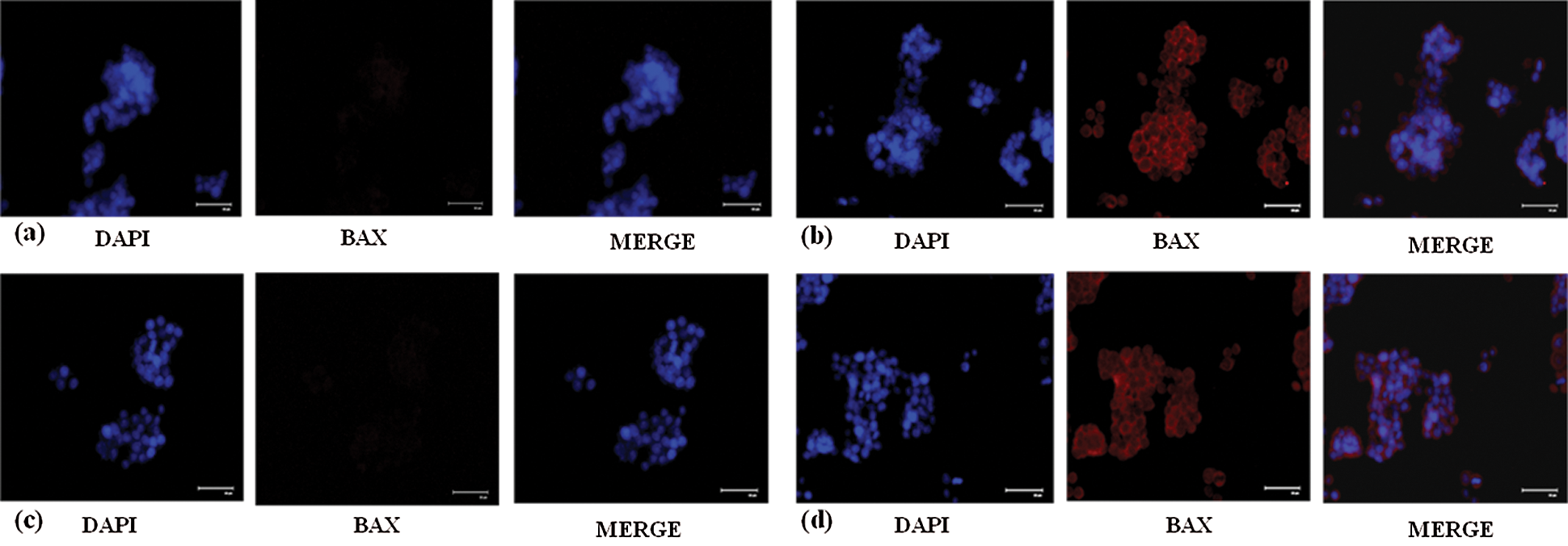
Figure 5: (a) Decreased expression of BAX protein in control SW480 cells compared to the (b) cells treated with AEG-1 siRNA. (c) Decreased expression of BAX protein in control SW620 cells compared to the (d) cells treated with AEG-1 siRNA
AEG-1 knockdown inhibits the migration capability of CC cells
Scratch assay was performed to examine the effect of AEG-1 knockdown on the migration ability of SW480 and SW620 cells (Figs. 6a and 6b). Silencing of AEG-1 in CC cells significantly slowed down the wound closure and also inhibited migration in AEG-1 siRNA transfected cells in a time-dependent manner (Figs. 6c and 6d). As Collagen type I alpha 1 chain (COL1A1) expression appeared to be associated with invasion and metastasis, we observed the role of COL1A1 in migration by qPCR and were found to be significantly downregulated in AEG-1 siRNA transfected cells compared to control cells (Figs. 7a and 7b). The results thus revealed that suppression of COL1A1 in AEG-1 siRNA transfected cells could attenuate the migration capabilities in SW480 and SW620 cells.
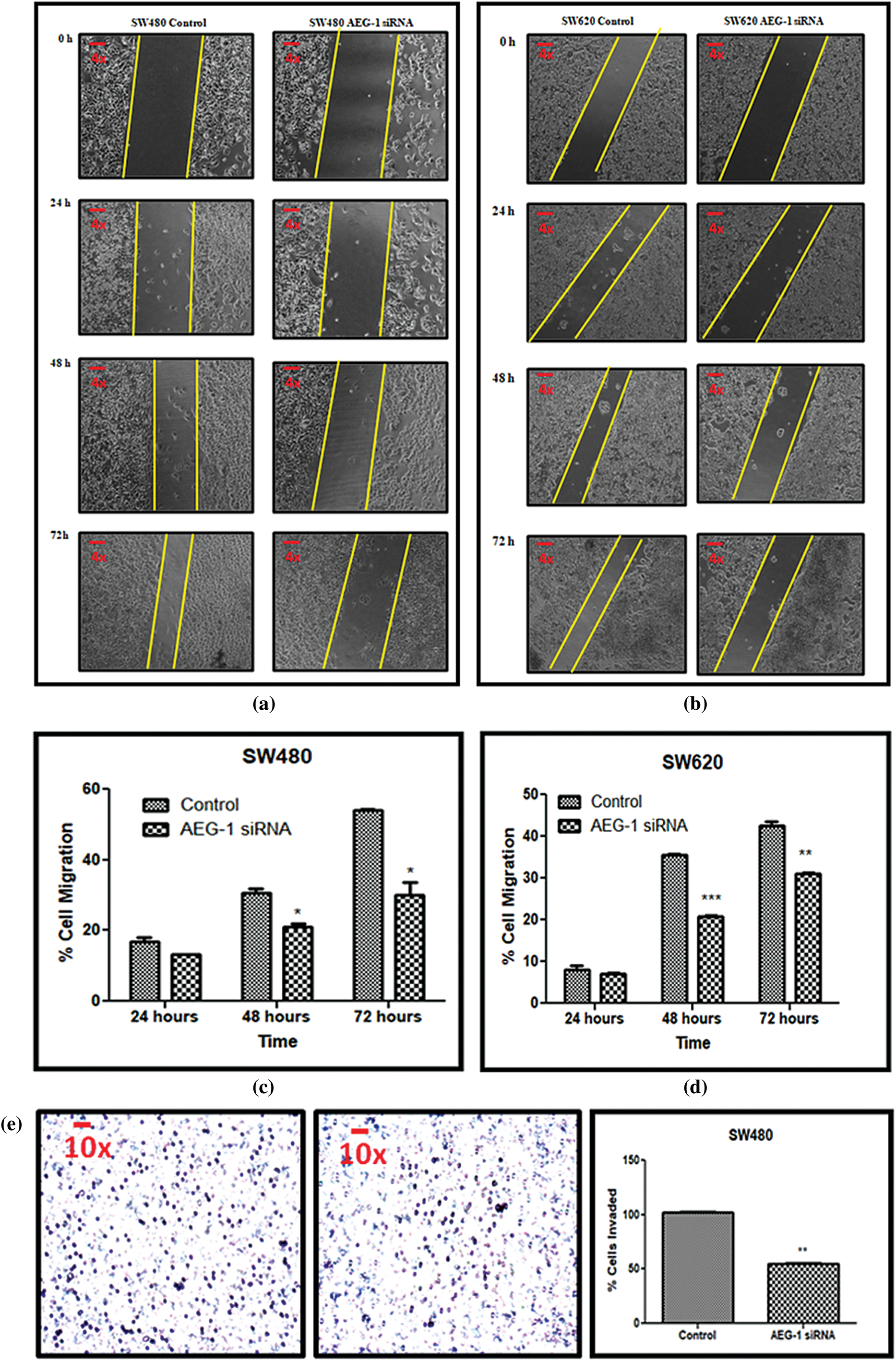
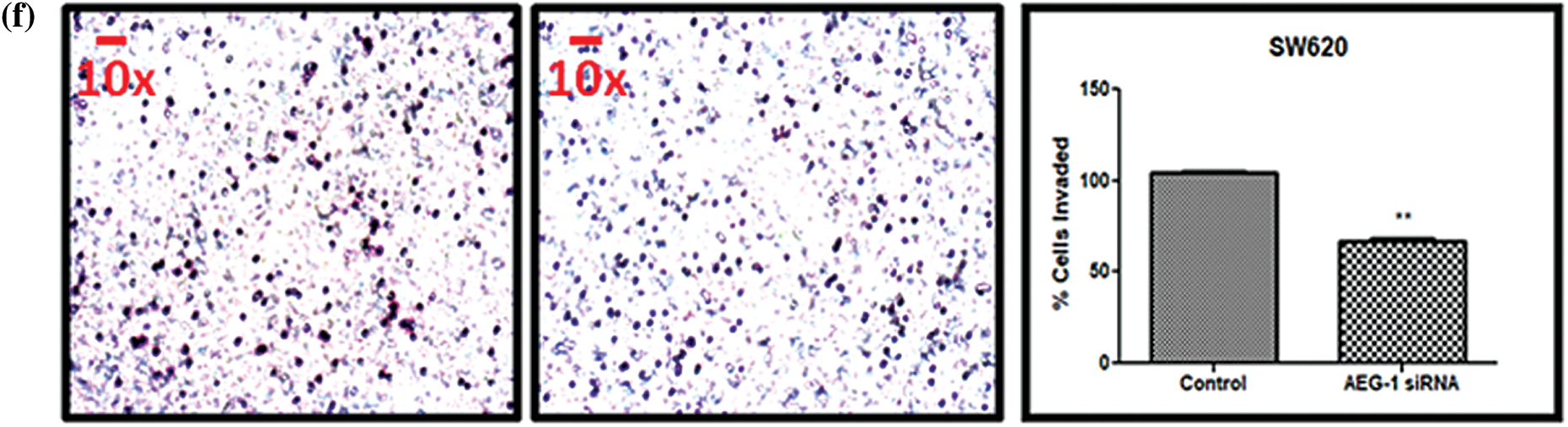
Figure 6: (a) Migration of SW480 cells and (b) SW620 cells before and after treatment with AEG-1 siRNA at 0 h, 24 h, 48 h, and 72 h. (c) Scratch assay before and after treatment with AEG-1 siRNA at different time intervals in SW480 cells and (d) SW620 cells. (e) Transwell assay for invasive ability before and after treatment with AEG-1 siRNA in SW480 cells and (f) SW620 cells (P < 0.05)
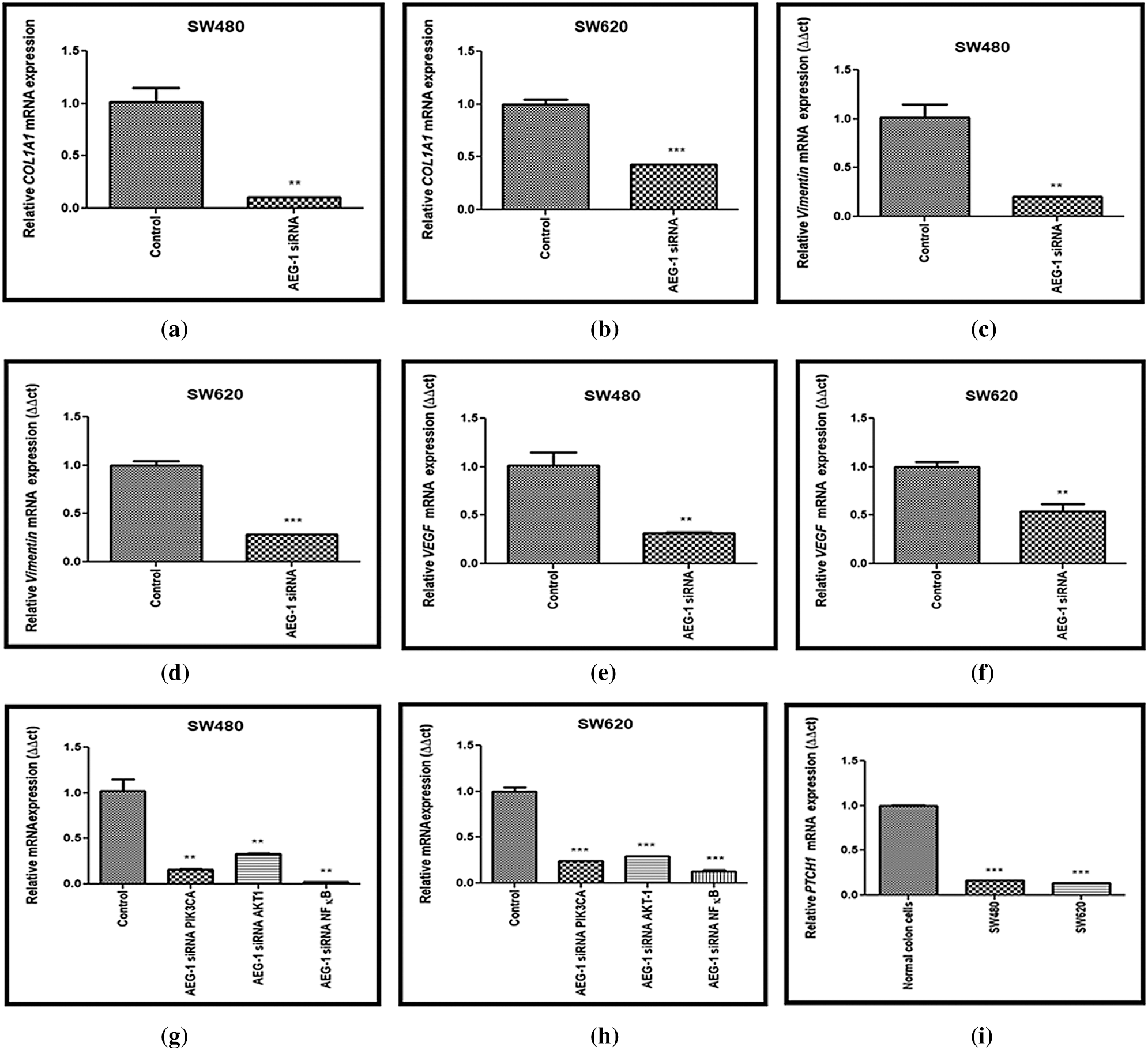
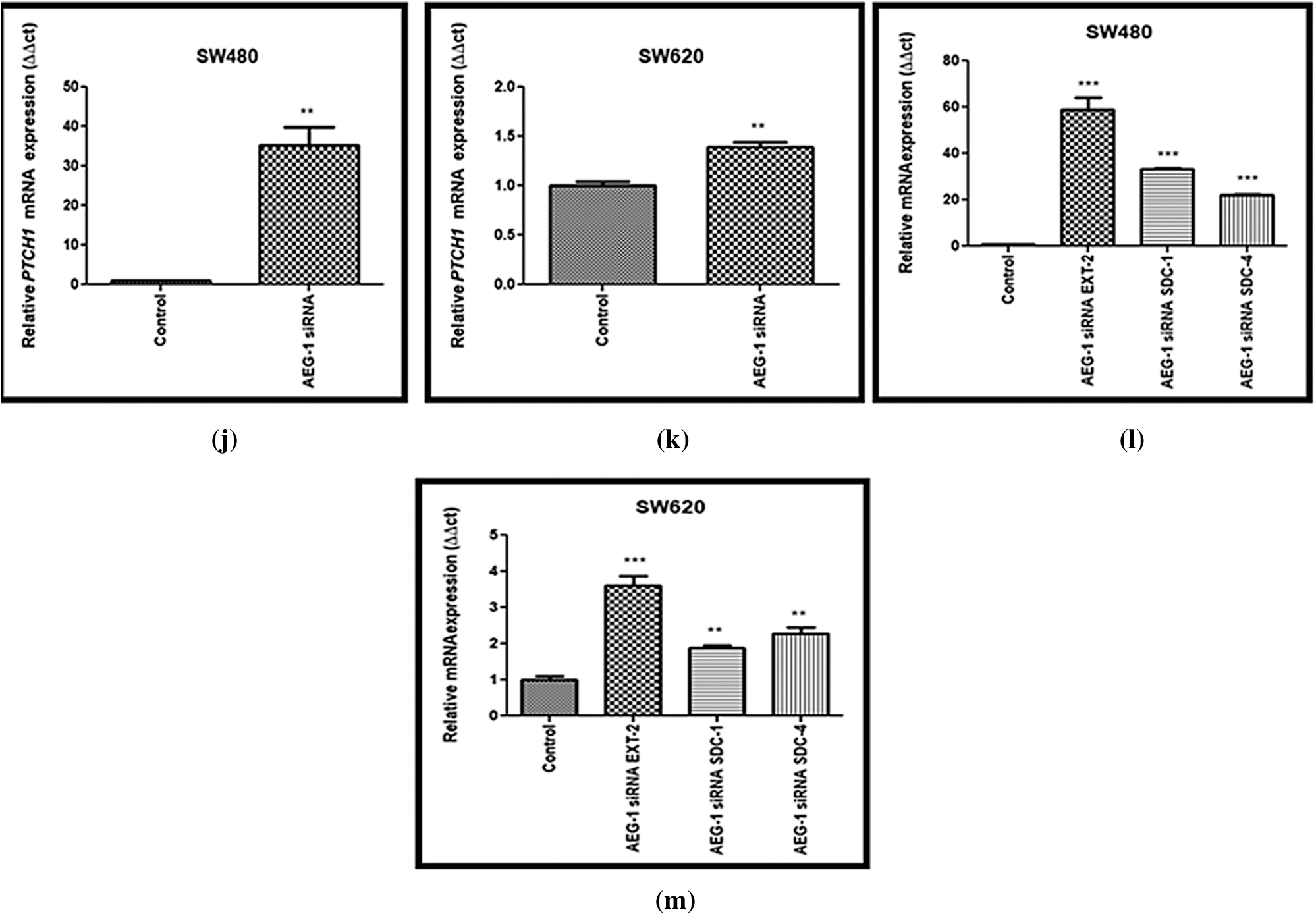
Figure 7: (a) Downregulation of COL1A1 after treatment with AEG-1 siRNA supporting migration studies in SW480 cells and (b) SW620 cells. (c) Downregulation of Vimentin after treatment with AEG-1 siRNA supporting invasion studies in SW480 cells and (d) SW620 cells. (e) Downregulation of VEGF after treatment with AEG-1 siRNA in SW480 cells and (f) SW620 cells. (g) Downregulation of oncogenes (PIK3CA, AKT-1, NF-κB) after treatment with AEG-1 siRNA in SW480 cells and (h) SW620 cells. (i) Basal level expression of PTCH-1 in normal colon cells and colon cancer cells (SW480 and SW620). (j) Upregulation of PTCH-1 after silencing with AEG-1 siRNA in SW480 cells and (k) SW620 cells (l) Upregulation of EXT2, SDC-1 and SDC-4 after silencing with AEG-1 siRNA in SW480 cells and (m) SW620 cells (P < 0.05)
AEG-1 knockdown inhibits invasive potential in CC cells
In addition to migration, we observed the invasive potential of both SW480 and SW620 cells (Figs. 6e and 6f). From the results of the Transwell assay, it was clearly displayed that count of invasive cells was found to be significantly decreased in AEG-1 siRNA transfected cells. To support invasive analysis, we investigated the AEG-1 knockdown effect on the expression of key epithelial-to-mesenchymal transition (EMT)-related marker vimentin. As the expression of vimentin plays a crucial role in the invasion and metastasis of CC, it is considered as one of the finest indicators of EMT in carcinogenesis. Thus, we noted that the expression of vimentin was significantly decreased following AEG-1 siRNA treatment in SW480 and SW620 cells compared to control cells (Figs. 7c and 7d).
Knockdown of AEG-1 downregulates potent oncogenes and suppresses angiogenesis
The molecular mechanism of AEG-1-mediated CC cell growth was investigated. Our study examined the expression of potent oncogenes which includes Phosphatidylinositol-4,5-bisphosphate 3-kinase (PIK3CA), Protein Kinase-B (AKT-1) and Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB). We identified that the expression of PIK3CA, AKT-1 and NF-κB was significantly downregulated in AEG-1 siRNA transfected cells compared to control (Figs. 7g and 7h). This indicated that the abnormal expression of oncogenes which was found to be reduced in AEG-1 siRNA transfected cells led to inhibition of tumorigenesis. In addition, we further identified the vascular endothelial growth factor (VEGF) expression in AEG-1 siRNA transfected cells as its well-known that CC cells secrete a high level of VEGF and promote angiogenesis to maintain the nutrition and oxygen supply in the tumor microenvironment. Thus, our results revealed that AEG-1 knockdown significantly suppressed VEGF expression in AEG-1 siRNA transfected cells compared to control cells (Figs. 7e and 7f).
Altered EXT-1 expression in AEG-1 siRNA transfected cells upregulate potent genes involved in metastasis of CC cells
The expression level of PTCH-1 with tumor suppressor properties involved in relation with EXT-1 in invasion and metastasis process was also identified using qPCR (Fig. 7i). We found that the expression of PTCH-1 was also found to be significantly upregulated in AEG-1 siRNA transfected cells (Figs. 7j and 7k). In addition, we observed the expression of EXT-2 (tumor suppressor), Syndecan-1 (SDC-1) and Syndecan-4 (SDC-4) which are HSPGs belonging to the syndecan proteoglycan family. As these syndecans can interact with the surrounding microenvironment through HS chains, they play a major role in the cancer invasion and metastasis process. The expression level of EXT-2, SDC-1 and SDC-4 was observed in qPCR and it was found to be significantly upregulated in AEG-1 siRNA transfected cells, which confirms that it might inhibit CC invasion and metastasis (Figs. 7l and 7m).
CC is considered one of the common malignant tumors, and the prevalence rate begins to increase globally in recent times. Even though there are substantial improvements in diagnostic and treatment protocols including chemotherapy, radiotherapy, and surgery, still the rational clinical outcome remains dissatisfying for the patients suffering from CC (Chen et al., 2008; Compton, 2003). Thus, it is mandatory to attain a greater understanding of this disease in order to initiate a novel therapeutic approach for CC.
As previous studies demonstrated that AEG-1 expression seems to be much higher in CC cell lines compared to normal colon cells (Gnosa et al., 2016), we performed loss-of-function assay via AEG-1 siRNA for investigating its role in cell behavior (including proliferation, apoptosis, migration, and invasion) in SW480 and SW620 cells in vitro. In this study, we have also transiently transfected AEG-1 siRNA into SW480 and SW620 cells and the expression pattern of AEG-1 were found to be efficiently downregulated after transfection. We observed inhibition in cellular proliferation and reduction in colony numbers in SW480 and SW620 AEG-1 siRNA transfected cells, confirming that the proliferation was controlled by AEG-1 expression. Meanwhile, reduced EXT-1 expression was also found in CC cells when compared to normal colon cells. Further, alteration of EXT-1 expression (mRNA and protein level) in AEG-1 siRNA transfected cells was observed. In addition, we also found that cells with low AEG-1 expression showed decreased proliferation, clonogenic ability, migration and invasion and induced apoptosis in vitro.
We then identified the possible interaction networks between AEG-1 and EXT-1 proteins using esyN and Pathway Commons databases. It was evident from the data analyses that AEG-1 and EXT-1 expression is modulated through PTCH-1. There are previous studies that revealed that CC metastasis and PTCH-1 expression are inversely correlated, and loss of its expression stimulated the metastatic potential (You et al., 2010). Our data are in line with these results to describe that PTCH-1, a receptor of the Hedgehog pathway, was found to be significantly upregulated in AEG-1 siRNA transfected cells confirming the inhibition of metastasis. Thus, PTCH-1 activity might be modulated by several genes and in turn, it affects EXT-1 activity.
Apoptosis is a vigorous cellular self-destruction process that not only aims to restrict viral replication by destroying the infected cells but in addition, it imparts to restrict dissemination (Jorgensen et al., 2017). As siRNA has been reported to promote apoptosis in tumor cells, we have also investigated whether AEG-1 silencing could promote apoptosis or not by analyzing the phosphatidylserine membrane. Cells undergoing early apoptosis were found to be significantly increased in AEG-1 siRNA transfected groups when compared to control groups in both SW480 and SW620 cells. However, we found that siRNA did not influence the cells undergoing late apoptosis. Thus, these results indicate that AEG-1 siRNA transfected cells decreased the proliferation of SW480 and SW620 cells by inducing apoptosis. Pro- and anti-apoptotic BCL-2 family proteins regulate the permeabilization of the external membrane of mitochondria. Here, we found that AEG-1 could induce a significant upregulation in pro-apoptotic BAX and also a significant downregulation in anti-apoptotic BCL-2 directing to increased apoptosis in AEG-1 siRNA transfected cells. Balancing between the pro- and anti-apoptotic proteins does determine whether a cell undergoes apoptosis or not. We are in plan to induce apoptosis in near future by using a targeted AEG-1 inhibitor, in order to attain the goal of treating CC cells.
Further, we investigated whether AEG-1 siRNA-mediated silencing affected migration and invasion of CC cells. From our results, we observed that siRNA-mediated silencing of AEG-1 expression in SW480 and SW620 cells decreased migration and invasive potential. Consistent with previous in vitro results in other cell types (Wang et al., 2017), AEG-1 silencing resulted in decreased cell migration and invasion in SW480 and SW620 cells.
From earlier studies, it is much familiar that there is a strong relationship between HS and cancer. HSPGs play a pivotal role in cell adhesion, invasiveness, and metastasis of cancer cells. There seem to have conflicting reports pertaining to the role of HS in cancer where it could inhibit or promote invasion and metastasis (Tímár et al., 2002; Sanderson, 2001). Interactions of proteins in tumor cells could solely depend on the structural properties of HS. Tumor cells generally modify their cell surface HS profile either by differential expression of specific PG protein cores or by modification of the structure of HS chains (Hull et al., 2017; Blackhall et al., 2003; Theocharis et al., 2016). There are several mRNA levels of HS biosynthesis-related enzymes or genes reported in most of the cancers, including CC (Fernández-Vega et al., 2015; Suhovskih et al., 2015), glioma (Ushakov et al., 2017), breast and prostate cancer (Mao et al., 2016; Fernández-Vega et al., 2013).
EXT-1 is majorly involved in the biosynthesis of HS (Okada et al., 2010) and both EXT-1 and EXT-2 possess in vitro HS polymerase activity (Busse et al., 2007; Senay et al., 2000). It has been reported that EXT-1 expression is able to extricate HS-deficient mutant Chinese Hamster Ovary cells, which harbor missense mutations in EXT-1 (Lin et al., 2000). Evident results pointed out that EXT-1 is essential for HS biosynthesis by performing in vivo studies in the EXT-1 knockout mouse model (Kenneally et al., 2000). Even though there are studies that show modifications in the EXT’s expression level and it further affects tumor progression and metastasis, still no clear research has been performed on how these changes affect chain elongation of HS which eventually control the biological function of EXTs (Sembajwe et al., 2018). Continued investigations on the biosynthetic enzymes related to HS could help us to inhibit the metastasis of tumor cells.
To the best of our knowledge, our present study is the first to identify the potential relationship between AEG-1 and EXT-1 in CC cells. Interestingly, this is the first report to show that AEG-1 silencing could inhibit cell proliferation and invasive potential through EXT-1. Also, we found that EXT-1 mRNA and protein expression was upregulated in AEG-1 siRNA transfected cells. Silencing AEG-1 expression could be able to interfere the expression of EXT-1 through PTCH-1, thereby affecting cell adhesion and cell-ECM interactions which further led to inhibition of invasion and metastasis. These findings suggest that AEG-1 might have the ability to regulate proliferative and invasive potential in AEG-1 siRNA transfected cells indirectly through regulating the expression of EXT-1. Till now the molecular mechanisms underlying these observations have not been fully studied. Our present results could add to the body of evidence that loss of SDC-1 and SDC-4 is a typical feature of cancer progression and incidence of metastasis (Li et al., 2019). In fact, our results showed that along with upregulation of EXT-2 (essential for the HSPG biosynthesis), SDC-1 and SDC-4 which are HSPGs belonging to syndecan proteoglycan family was also found to be upregulated in AEG-1 siRNA transfected cells confirming that cells may lead to inhibition of metastasis. Further studies on protein levels of these HSPG related genes might be required to conclude its firm role in inhibiting metastasis.
Our results demonstrated that AEG-1 siRNA could reduce cell proliferation, migration, and invasion of both SW480 and SW620, finally leading to apoptosis. Our current results indicated that AEG-1 was significantly correlated with CC cell proliferation. Inhibition of AEG-1 could represent a novel therapeutic target for the prevention and treatment of CC. Through this study, we report, the link between the expressions of EXT-1 to AEG-1 in CC cells. Silencing of AEG-1 increased the EXT-1 expression, which in turn may inhibit cell proliferation and invasion. Results from in silico interaction studies pointed towards the crosstalk between AEG-1 and EXT-1 mediated through PTCH-1. In our experimental data, PTCH-1 activity was modulated by AEG-1 silencing, and in turn, it affects the expression of EXT-1. These observations from the investigation not only provide insight into the interplay between AEG-1 and EXT-1 during CC but further open up new perspectives related to the function of EXT-1 under normal and pathological states as well. Further extensive studies are necessary to determine the mechanisms underlying the increased expression of EXT-1 in AEG-1 silenced CC cells and its role in relation to inhibit cell proliferation and invasion. Collectively, from our data, we can conclude that AEG-1 could be a possible therapeutic target candidate for CC through the interaction with EXT-1.
Further extensive in vivo studies are required to uncover the mechanism of action of AEG-1 protein in colon carcinogenesis. However, finding the signaling cascade of how AEG-1 siRNA transfected cells inhibit metastasis through EXT-1 would be of great interest in the future.
Acknowledgement: We would like to thank the Chettinad Academy of Research and Education for providing us the facilities to carry out the research work. The language editing done by The Editing Refinery, MD, USA, is highly acknowledged.
Availability of Data and Materials: Readers can access the data upon request to Corresponding author.
Author Contributions: Conceptualization–SP and SS; Supervision–SP and AB; Methodology–SP, SS, and SM; Writing (Original draft preparation)–SS; (Review and Editing)–SP, AB, SS, MWQ, SBC, and MME; Bioinformatic analysis–SKN; Data analysis and Statistical part–SS. All authors revised and approved the content of the manuscript.
Funding Statement: This study was supported by a grant from the Department of Science and Technology (DST)–Science and Engineering Research Board (SERB) (EMR/2017/001877) and Lady Tata Memorial Trust (LTMT) for providing the fellowship.
Conflicts of Interest: The authors declare no conflicts of interest.
Abraha AM, Ketema EB. (2016). Apoptotic pathways as a therapeutic target for colorectal cancer treatment. World Journal of Gastrointestinal Oncology 8: 583–591. DOI 10.4251/wjgo.v8.i8.583. [Google Scholar] [CrossRef]
Akagi Y, Kinugasa T, Adachi Y, Shirouzu K. (2013). Prognostic significance of isolated tumor cells in patients with colorectal cancer in recent 10-year studies. Molecular and Clinical Oncology 1: 582–592. DOI 10.3892/mco.2013.116. [Google Scholar] [CrossRef]
Armaghany T, Wilson JD, Chu Q, Mills G. (2012). Genetic alterations in colorectal cancer. Gastrointestinal Cancer Research 5: 19–27. [Google Scholar]
Bean DM, Heimbach J, Ficorella L, Micklem G, Oliver SG, Favrin G. (2014). esyN: Network building, sharing and publishing. PLoS One 9: 1–5. [Google Scholar]
Blackhall FH, Merry CLR, Lyon M, Jayson GC, Folkman J, Javaherian K, Gallagher JT. (2003). Binding of endostatin to endothelial heparan sulphate shows a differential requirement for specific sulphates. Biochemical Journal 375: 131–139. DOI 10.1042/bj20030730. [Google Scholar] [CrossRef]
Budisan L, Gulei D, Jurj A, Braicu C, Zanoaga O, Cojocneanu R, Berindan-Neagoe I. (2019). Inhibitory effect of CAPE and kaempferol in colon cancer cell lines—possible implications in new therapeutic strategies. International Journal of Molecular Sciences 20: 1199. [Google Scholar]
Busse M, Feta A, Presto J, Wilén M, Grønning M, Kjellén L, Kusche-Gullberg M. (2007). Contribution of EXT1, EXT2, and EXTL3 to heparan sulfate chain elongation. Journal of Biological Chemistry 282: 32802–32810. DOI 10.1074/jbc.M703560200. [Google Scholar] [CrossRef]
Busse-Wicher M, Wicher KB, Kusche-Gullberg M. (2014). The exostosin family: Proteins with many functions. Matrix Biology 35: 25–33. DOI 10.1016/j.matbio.2013.10.001. [Google Scholar] [CrossRef]
Byoung KY, Emdad L, Su ZZ, Villanueva A, Chiang DY, Mukhopadhyay ND, Sarkar D. (2009). Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. Journal of Clinical Investigation 119: 465–477. DOI 10.1172/JCI36460. [Google Scholar] [CrossRef]
Chen YC, Hsu HS, Chen YW, Tsai TH, How CK, Wang CY, Chiou SH. (2008). Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS One 3: 1–14. [Google Scholar]
Compton CC. (2003). Colorectal carcinoma: Diagnostic, prognostic, and molecular features. Modern Pathology 16: 376–388. DOI 10.1097/01.MP.0000062859.46942.93. [Google Scholar] [CrossRef]
Dogrusoz U, Cetintas A, Demir E, Babur O. (2009). Algorithms for effective querying of compound graph-based pathway databases. BMC Bioinformatics 10: D623. DOI 10.1186/1471-2105-10-376. [Google Scholar] [CrossRef]
Emdad L, Sarkar D, Su ZZ, Randolph A, Boukerche H, Valerie K, Fisher PB. (2006). Activation of the nuclear factor κB pathway by astrocyte elevated gene-1: Implications for tumor progression and metastasis. Cancer Research 66: 1509–1516. DOI 10.1158/0008-5472.CAN-05-3029. [Google Scholar] [CrossRef]
Fernández-Vega I, García O, Crespo A, Castañón S, Menéndez P, Astudillo A, Quirós LM. (2013). Specific genes involved in synthesis and editing of heparan sulfate proteoglycans show altered expression patterns in breast cancer. BMC Cancer 13: 2893. DOI 10.1186/1471-2407-13-24. [Google Scholar] [CrossRef]
Fernández-Vega I, García-Suárez O, García B, Crespo A, Astudillo A, Quirós LM. (2015). Heparan sulfate proteoglycans undergo differential expression alterations in right sided colorectal cancer, depending on their metastatic character. BMC Cancer 15: 1–20. DOI 10.1186/1471-2407-15-1. [Google Scholar] [CrossRef]
Gnosa S, Capodanno A, Murthy RV, Jensen LDE, Sun XF. (2016). AEG-1 knockdown in colon cancer cell lines inhibits radiation-enhanced migration and invasion in vitro and in a novel in vivo zebrafish model. Oncotarget 7: 81634–81644. DOI 10.18632/oncotarget.13155. [Google Scholar] [CrossRef]
Grady WM, Pritchard CC. (2014). Molecular alterations and biomarkers in colorectal cancer. Toxicologic Pathology 42: 124–139. DOI 10.1177/0192623313505155. [Google Scholar] [CrossRef]
Hu G, Wei Y, Kang Y. (2010). The multifaceted role of MTDH/AEG-1 in cancer progression. Clinical Cancer Research 15: 5615–5620. DOI 10.1158/1078-0432.CCR-09-0049. [Google Scholar] [CrossRef]
Hull EE, Montgomery MR, Leyva KJ. (2017). Epigenetic regulation of the biosynthesis & enzymatic modification of heparan sulfate proteoglycans: Implications for tumorigenesis and cancer biomarkers. International Journal of Molecular Sciences 18: 1361. [Google Scholar]
Jorgensen I, Rayamajhi M, Miao EA. (2017). Programmed cell death as a defence against infection. Nature Reviews Immunology 17: 151–164. DOI 10.1038/nri.2016.147. [Google Scholar] [CrossRef]
Kenneally M, Harkin P, Jung M, Maclaughlin DT, Donahoe PK, Maheswaran S. (2000). Copyright 2000 by the American Society for Biochemistry and Molecular Biology, Inc. Biochemistry, 617–638. [Google Scholar]
Le NN, Hucka M, Mi H, Moodie S, Schreiber F, Sorokin A, Schmidt E. (2009). The systems biology graphical notation. Nature Biotechnology 27: 735–741. DOI 10.1038/nbt.1558. [Google Scholar] [CrossRef]
Lee SG, Su ZZ, Emdad L, Sarkar D, Fisher PB. (2006). Astrocyte elevated gene-1 (AEG-1) is a target gene of oncogenic Ha-ras requiring phosphatidylinositol 3-kinase and c-Myc. Proceedings of the National Academy of Sciences of the United States of America 103: 17390–17395. DOI 10.1073/pnas.0608386103. [Google Scholar] [CrossRef]
Lee SG, Su ZZ, Emdad L, Sarkar D, Franke TF, Fisher PB. (2008). Astrocyte elevated gene-1 activates cell survival pathways through PI3K-Akt signaling. Oncogene 27: 1114–1121. DOI 10.1038/sj.onc.1210713. [Google Scholar] [CrossRef]
Li JW, Huang CZ, Li JH, Yuan JH, Chen QH, Zhang WF, Lu LG. (2018). Knockdown of metadherin inhibits cell proliferation and migration in colorectal cancer. Oncology Reports 40: 2215–2223. [Google Scholar]
Li K, Li L, Wu X, Yu J, Ma H, Zhang R, Wang W. (2019). Loss of SDC1 expression is associated with poor prognosis of colorectal cancer patients in northern China. Disease Markers 5: 1–7. [Google Scholar]
Li M, Dai Y, Wang L, Li L. (2017). Astrocyte elevated gene-1 promotes the proliferation and invasion of breast cancer cells by activating the Wnt/β-catenin signaling pathway. Oncology Letters 13: 2385–2390. DOI 10.3892/ol.2017.5695. [Google Scholar] [CrossRef]
Lièvre A, Blons H, Laurent-Puig P. (2010). Oncogenic mutations as predictive factors in colorectal cancer. Oncogene 29: 3033–3043. DOI 10.1038/onc.2010.89. [Google Scholar] [CrossRef]
Lin X, Wei G, Shi Z, Dryer L, Esko JD, Wells DE, Matzuk MM. (2000). Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Developmental Biology 224: 299–311. DOI 10.1006/dbio.2000.9798. [Google Scholar] [CrossRef]
Mao X, Gauche C, Coughtrie MWH, Bui C, Gulberti S, Merhi-Soussi F, Fournel-Gigleux S. (2016). The heparan sulfate sulfotransferase 3-OST3A (HS3ST3A) is a novel tumor regulator and a prognostic marker in breast cancer. Oncogene 35: 5043–5055. DOI 10.1038/onc.2016.44. [Google Scholar] [CrossRef]
Noch EK, Khalili K. (2013). The role of AEG-1/MTDH/LYRIC in the pathogenesis of central nervous system disease. Advances in Cancer Research. 1st ed., pp. 120. Elsevier Inc. [Google Scholar]
Okada M, Nadanaka S, Shoji N, Tamura J, Kitagawa H. (2010). Biosynthesis of heparan sulfate in EXT1-deficient cells. Biochemical Journal 428: 463–471. DOI 10.1042/BJ20100101. [Google Scholar] [CrossRef]
Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, Tyers M. (2019). The BioGRID interaction database: 2019 update. Nucleic Acids Research 47: D529–D541. DOI 10.1093/nar/gky1079. [Google Scholar] [CrossRef]
Pacifici M. (2017). Hereditary multiple exostoses: New insights into pathogenesis, clinical complications, and potential treatments. Current Osteoporosis Reports 15: 142–152. DOI 10.1007/s11914-017-0355-2. [Google Scholar] [CrossRef]
Qazi H, Shi ZD, Song JW, Cancel LM, Huang P, Zeng Y, Roberge S, Munn L, Tarbell JM. (2016). Heparan sulfate proteoglycans mediate renal carcinoma metastasis. International Journal of Cancer 139: 2791–2801. DOI 10.1002/ijc.30397. [Google Scholar] [CrossRef]
Rodchenkov I, Babur O, Luna A, Aksoy BA, Wong JV, Fong D, Sander C. (2020). Pathway Commons 2019 Update: Integration, analysis and exploration of pathway data. Nucleic Acids Research 48: D489–D497. [Google Scholar]
Sanderson RD. (2001). Heparan sulfate proteoglycans in invasion and metastasis. Seminars in Cell & Developmental Biology 12: 89– 98. DOI 10.1006/scdb.2000.0241. [Google Scholar] [CrossRef]
Sembajwe LF, Katta K, Grønning M, Kusche-Gullberg M. (2018). The exostosin family of glycosyltransferases: mRNA expression profiles and heparan sulphate structure in human breast carcinoma cell lines. Bioscience Reports 38: 287. DOI 10.1042/BSR20180770. [Google Scholar] [CrossRef]
Senay C, Lind T, Muguruma K, Tone Y, Kitagawa H, Sugahara K, Kusche-Gullberg M. (2000). The EXT1/EXT2 tumor suppressors: Catalytic activities and role in heparan sulfate biosynthesis. EMBO Reports 1: 282–286. DOI 10.1093/embo-reports/kvd045. [Google Scholar] [CrossRef]
Smith RN, Aleksic J, Butano D, Carr A, Contrino S, Hu F, Micklem G. (2012). InterMine: A flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics 28: 3163–3165. DOI 10.1093/bioinformatics/bts577. [Google Scholar] [CrossRef]
Song H, Li C, Li R, Geng J. (2010). Prognostic significance of AEG-1 expression in colorectal carcinoma. International Journal of Colorectal Disease 25: 1201–1209. DOI 10.1007/s00384-010-1009-3. [Google Scholar] [CrossRef]
Stoian M, State N, Stoica V, Radulian G. (2014). Apoptosis in colorectal cancer. Journal of Medicine and Life 7: 160–164. [Google Scholar]
Suhovskih AV, Domanitskaya NV, Tsidulko AY, Prudnikova TY, Kashuba VI, Grigorieva EV. (2015). Tissue-specificity of heparan sulfate biosynthetic machinery in cancer. Cell Adhesion & Migration 9: 452–459. DOI 10.1080/19336918.2015.1049801. [Google Scholar] [CrossRef]
Theocharis AD, Skandalis SS, Neill T, Multhaupt HAB, Hubo M, Frey H, Karamanos NK. (2015). Insights into the key roles of proteoglycans in breast cancer biology and translational medicine. Biochimica et Biophysica Acta-Reviews on Cancer 1855: 276–300. [Google Scholar]
Tímár J, Lapis K Dudás J, Kopper L et al. (2002). Proteoglycans and tumor progression: Janus-faced molecules with contradictory functions in cancer. Seminars in Cancer Biology 12: 173–186. DOI 10.1016/S1044-579X(02)00021-4. [Google Scholar] [CrossRef]
Ushakov VS, Tsidulko AY, De La Bourdonnaye G, Kazanskaya GM, Volkov AM, Kiselev RS, Grigorieva EV. (2017). Heparan sulfate biosynthetic system is inhibited in human glioma due to EXT1/2 and HS6ST1/2 down-regulation. International Journal of Molecular Sciences 18: 2301. DOI 10.3390/ijms18112301. [Google Scholar] [CrossRef]
Vogelstein B, Kinzler KW. (2004). Cancer genes and the pathways they control. Nature Medicine 10: 789–799. DOI 10.1038/nm1087. [Google Scholar] [CrossRef]
Wang J, Chen X, Tong M. (2017). Knockdown of astrocyte elevated gene-1 inhibited cell growth and induced apoptosis and suppressed invasion in ovarian cancer cells. Gene 616: 8–15. DOI 10.1016/j.gene.2017.03.024. [Google Scholar] [CrossRef]
Watson AJM, Pritchard DM. (2000). Lessons from genetically engineered animal models VII. Apoptosis in intestinal epithelium: Lessons from transgenic and knockout mice. American Journal of Physiology–Gastrointestinal and Liver Physiology 278: 1–5. DOI 10.1152/ajpgi.2000.278.1.G1. [Google Scholar] [CrossRef]
Yao Y, Gu X, Liu H, Wu G, Yuan D, Yang X, Song Y. (2014). Metadherin regulates proliferation and metastasis via actin cytoskeletal remodelling in non-small cell lung cancer. British Journal of Cancer 111: 355–364. DOI 10.1038/bjc.2014.267. [Google Scholar] [CrossRef]
You S, Zhou J, Chen S, Zhou P, Lv J, Han X, Sun Y. (2010). PTCH1, a receptor of Hedgehog signaling pathway, is correlated with metastatic potential of colorectal cancer. Upsala Journal of Medical Sciences 115: 169–175. DOI 10.3109/03009731003668316. [Google Scholar] [CrossRef]
Zhong SZ, Chen Y, Chul KD, Chao W, Simm M, Volsky DJ, Fisher PB. (2003). Customized rapid subtraction hybridization (RaSH) gene microarrays identify overlapping expression changes in human fetal astrocytes resulting from human immunodeficiency virus-1 infection or tumor necrosis factor-α treatment. Gene 306: 67–78. DOI 10.1016/S0378-1119(03)00404-9. [Google Scholar] [CrossRef]
Zhu HD, Liao JZ, He XX, Li PY. (2015). The emerging role of astrocyte-elevated gene-1 in hepatocellular carcinoma (Review). Oncology Reports 34: 539–546. DOI 10.3892/or.2015.4024. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |