DOI:10.32604/biocell.2021.014354

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.014354 |  www.techscience.com/journal/biocell |
| Review |
Development of high yield and tomato yellow leaf curl virus (TYLCV) resistance using conventional and molecular approaches: A review
1Institute of Tropical Agriculture and Food Security, Universiti Putra Malaysia, Serdang, 43400, Malaysia
2Department of Crop Science, Faculty of Agriculture, Universiti Putra Malaysia, Serdang, 43400, Malaysia
3Department of Plant Protection, Faculty of Agriculture, Universiti Putra Malaysia, Serdang, 43400, Malaysia
*Address correspondence to: Mohd Y. Rafii, mrafii@upm.edu.my
Received: 19 September 2020; Accepted: 21 December 2020
Abstract: Tomato (Solanum lycopersicum L.) belonging to the family Solanaceae is the second most consumed and cultivated vegetable globally. Since the ancient time of its domestication, thousands of cultivated tomato varieties have been developed targeting an array of aspects. Among which breeding for yield and yield-related traits are mostly focused. Cultivated tomato is extremely genetically poor and hence it is a victim for several biotic and abiotic stresses. Among the biotic stresses, the impact of viral diseases is critical all over tomato cultivating areas. Improvement of tomato still largely rely on conventional methods worldwide while molecular approaches, particularly Marker Assisted Selection (MAS) has become popular across the globe as a fast, low cost and precise tool which is essential in present day plant breeding. In this review paper, breeding tomato for high yield and viral disease resistance, particularly to tomato yellow leaf curl virus disease (TYLCVD) using conventional and molecular approaches will be discussed. Lining up of this set of information will be useful to those who are interested in tomato variety development with high yielding and TYLCVD resistance.
Keywords: Molecular markers; Tomato yellow leaf curl virus disease; Resistance breeding; Solanum lycopersicum; High yield
Tomato (Solanum lycopersicum L.) is an annual crop that is considered the second most consumed vegetable in the world, with production exceeding 180 million tons cultivated on over 4.8 million hectares (Food, 2018). It belongs to the Solanaceae family, which consists of about 3000–4000 species with approximately 96 genera across three subfamilies of economically important crops such as eggplant, tobacco, petunia, potato, and pepper. The global tomato industry is valued at more than fifty billion dollars (Mattoo and Handa, 2017). Tomato market comprises of fresh market and processing industries. Considering the volume consumed, tomato contributes significantly to the dietary intake of essential vitamins and minerals (Willcox et al., 2003). Tomatoes, both processed and fresh market types, are a rich source of the dietary antioxidant lycopene, having the ability to protect cells from cancer (Giovannucci, 1999). The mountainous regions of the Andes, which comprise of the current Chile, Ecuador, and Peru, were believed to be the origin of tomato (Dhaliwal et al., 2020). The wild relatives and cultivated tomato had a similar karyotype and chromosome number of 2n = 24 (Foolad, 2007). With the domestication process started around 600 years ago (Rick, 1978) genetic variability of cultivated tomato is narrowed. This is because, during the domestication and evolution process, the cultivated tomato went through several genetic tailbacks as a result of extreme inbreeding and imposed selective breeding.
Hence, these events cause a reduction in genetic variation among the cultivated tomato. Therefore, it is estimated that the cultivated tomato contains approximately 5% less genome of the genetic variability of their wild relatives (Dhaliwal et al., 2020).
At the end of the 19th century (late 1800’s) farmers were used to the cultivation of open-pollinated cultivars (Heirlooms), while in the middle of the 20th century breeding of hybrid cultivars were initiated where ‘Single Cross’ being the first-ever tomato hybrid cultivar (Bai and Lindhout, 2007). After the invention of hybrid cultivars in mid of the 20th century, the whole tomato industry was dominated by various tomato hybrids. In 1994—1st transgenic cultivar ‘Flavr-Savr’TM (Acquaah, 2012) was released. However, its popularity decreased within a few years of release due to unfavorable qualities. From the time of development of heirlooms up to transgenics, the tomato crop is highly affected by a number of diseases and environmental factors. Among these, viral diseases are considerably devastative. In particular, tomato yellow leaf curl disease (TYLCD) transmitted by Bemisia tabaci is a major viral disease across the world (Lapidot and Friedmann, 2002). Graphical plant–virus Interaction of TYLCV is reported by Prasad et al. (2020). TYLCD is a global limitation to tomato production and remains one of the most devastating viral diseases of tomato. Most of the cultivated tomato is susceptible to this disease showing symptoms such as flower abortion, severe stunting, leaves cupping, curling, and yellowing, which can result in up to 100% yield losses (Yan et al., 2018). However, many accessions of tomato wild relatives have been identified as TYLCV resistant sources (Yan et al., 2018). Meanwhile, Firdaus et al. (2012) has reported wild relatives of tomato with resistance to TYLCV vector (S. pennellii Correll, S. habrochaites S. Knapp, S. habrochaites f. glabratum, S. pimpinellifolium L., S. chilense (Dunal) Reiche). However, TYLCV has become difficult to control due to several reasons. One major reason is the polyphagous nature of TYLCV, where this virus-host range from many commercial crops viz., lisianthus (Eustoma grandiflorum (Raf.) Shinners), Petunia Juss., common bean (Phaseolus vulgaris L.), tobacco (Nicotiana tabacum L.), chili pepper (C. chinense Jacq.), sweet pepper (Capsicum annuum L.) and tomato (S. lycopersicum), and several common weeds (Diaz-Pendon et al., 2010). Up to now, introgression of TYLCV resistant genes from wild relatives to elite lines of cultivated tomato has paved way for the breeding of high-yielding tomato cultivars with TYLCV resistance (Vidavski et al., 2008).
Varietal improvement program of tomato during the last century is based on several standard breeding techniques such as hybridization followed by pedigree selection and backcrossing of preferred characters from one parent into another that resulted in the generation of improvement tomato hybrids and varieties with high yield and quality. Improvement in tomato occurred as a result of continuous exploitation of germplasm and incorporation of desirable genes into the genetic background of elite cultivars. Traditional breeding has not only developed genotypes with dominant and monogenic resistance for controlling certain plant pathogens or a combination of resistance F1 hybrids but has also aided the acquisition of good agronomic traits such as increase shelf-life and firmness suitable for long distant shipping, well-adapted genotypes, uniform ripening, earliness, fruit setting, and high fertility rate. Hence, the objectives of this review are to describe the recent methods of developing a durable high yielding tomato genotype with resistance to tomato yellow leaf curl virus (TYLCV) through conventional and molecular approaches and to comprehensively review the available information on TYLCV and yield genes, marker-assisted selection, gene transformation, and QTL analysis.
Breeding Goals of Tomato across the Time
The tomato industry has two main categories, viz. fresh market and processing tomatoes (Arah et al., 2015), which makes the breeding objectives to be dynamic and varied across time. For example, tomato breeding in the 1970s mainly focused on yield; however, this scenario changed in the 1980s, where the major goal was focused mainly on shelf life. During the 1990s, the trend of tomato breeding was focused on taste, and in the 2000s, much attention has focused on the nutritional qualities and development of resistant varieties against pests and diseases (Osei et al., 2018). Apart from the variability along the time, there is a considerable variation in tomato breeding goals depend on the location, need of the community, and available resources (Acquaah, 2012; Rick, 1988).
The earliest approach in tomato breeding was achieved along with the domestication process, where farmers selected tomato cultivars with larger fruit size, lesser rates of dormancy, and higher rates of self-pollination. With the characteristic of self-pollination, earlier tomato cultivars were open-pollinated, and the selection was based on the uniform fruit shape, color, and size. Hence, these selected genotypes are referred to as “heirlooms” which were more or less similar to their parents (Watson, 1996; Bai and Lindhout, 2007). Towards the mid of the twentieth century, the new era of tomato breeding was commenced with the development of hybrid tomato cultivars, over leading the popularity of heirlooms. Hybrids are cultivars with combinations of commercially desirable characteristics or traits. The phenomenon called hybrid vigor or heterosis is generally associated with an increased yield (Bradshaw, 2016). Apart from hybrid vigor, hybrids are preferred to be developed by the breeders for uniformity, protection from unauthorized reproduction, and better resistance to pests and diseases (Fentik, 2017; Acquaah, 2012). With the development of the first hybrid tomato cultivar “Single Cross” (Bai and Lindhout, 2007) in 1946; breeders, producers, and growers were plagued by the benefits associated with the hybrid cultivars, and since both the fresh market and processing tomato industries were dominated by hybrid varieties (Fufa et al., 2009).
Conventional Methods of Tomato Breeding
Before the development of molecular markers and genetic engineering tools, tomato breeding mostly depended on conventional methods. With the domestication, which began in South America, breeding methods such as selection (Mass selection) of better cultivars and introduction to the rest of the world was accomplished by the local farmers (Lin et al., 2014). The pedigree method is another method used in tomato breeding where controlled crossing and selection of superior plants from early generations (F2 generation), and continued until genetic purity is reached (Kalloo, 2012). Also, hybridization followed by pedigree selection is one of the most common methods used in tomato breeding, while back cross-breeding is used to transfer traits such as disease resistance to cultivated tomato (Fentik, 2017). Generally, most of the early conventional breeding programs were practiced as combinations of backcross followed by the pedigree selection, which resulted in more successful cultivars. Casali and Tigchelaar (1975) proposed the single seed descendant method specially focused on low heritable traits. However, these findings showed that a single seed descendant method was not efficient as compared with the pedigree selection method.
Among the conventional techniques, hybrid breeding is the most intensively used technique in the development of new tomato varieties, hence, almost all commercial cultivated tomato cultivars are hybrids (van de Wiel et al., 2010). This hybrid development was achieved using complementary inbred lines as two parental lines. Hybrid vigor or heterosis is the phenomenon that adds value to the hybrids. Generally, due to the hybrid vigor, hybrids are more superior in many qualities than their parents. Though the percentage of contribution is not fully defined, over-dominance and additive effects are attributed to the hybrid vigor (Birchler et al., 2010; Chen, 2013). These hybrids are popular and accepted by many growers around the world for the fact that they allow an easy combination of many economically important traits such as disease-resistant genes that are dominant in gene action.
In hybrid production, the removal of anthers from the female plant (emasculation) is done manually. This tedious and laborious procedure has been replaced by the discovery of different techniques. As a solution, a set of nuclear recessive genes responsible for male sterility has been studied by Kaul (1991). However, this technique was not fully practiced or commercialized due to the nonheritable nature of the male-sterile character. In hybrid production, combining ability analysis is important for the selection of the best hybrids derived from crosses between selected inbred lines and the elite lines (Peralta et al., 2006). Kaushik and Dhaliwal (2018) reported the screening of best cross combinations based on multiple trait performance, including fruit yield and resistance to tomato yellow leaf curl virus (TYLCV), using diallele analysis. The selected hybrids can be further evaluated in multi-location replicated trials to short-list the broadly adapted crosses with lower genotype × environment interaction. Peirce (1991) revealed that G × E interaction is strongly significant to the marketable yield. The effect of G × E for different fruit quality and plant characteristics have not given consistent results in all attempts. Even with many advancements of conventional breeding methods, it is obvious the difficulty of combining many desirable characteristics together into a single variety. On the other hand, the gene action of a quantitative trait is difficult to predict because the ultimate output or level of expression of traits are governed by multiple genes and are highly influenced by the environment (Osei et al., 2018).
Embryo rescue is another method where callus is raised from the excised embryos and regenerating plants. This method was reported as the only successful method for the cross between Tomato (Solanum lycopersicum) and Solanum peruvianum L. (Poysa, 1990). Similarly, Acquaah (2012) reported the use of embryo rescue between crosses of the cultivated tomato and wild species S. peruvianum and S. chilense to use in gene transfer from wild relatives to the cultivated tomato. The monosomic line is another method used in tomato breeding where the production of aneuploid plants with extra chromosomes from donor species with desirable genes (Pertuzé et al., 2003). Mutagenesis also plays a considerable role in tomato breeding, and yet the method is being used in many countries. Mutations are induced by exposing plant parts to ionizing radiation viz., X-rays and gamma-rays or mutagenesis by chemicals viz., EMS (Ethyl Methyl Sulfonate) and sodium azide (Oladosu et al., 2016). Rick et al. (1996) found that mutations of some monogenic traits of tomato were induced by mutagens. Most of the fruit mutants originated from the early cultivars/lines of S. lycopersicum out of which major mutations were used in the improvement of tomato fruit quality (Peralta et al., 2006). There is a continuous rise in the number of developed mutant tomatoes (IAEA, 2020).
Although desirable mutants could be transferred to the elite cultivars by successive backcrosses after about 10 generations, however, most mutants were not used due to the association of unfavorable traits linked with the gene of interest (pleiotropic effect) (Young and Tanksley, 1989). TILLING (Targeting Induced Local Lesions) in genomes has-facilitated the way to identify an array of new mutants in tomatoes with still unrevealed gene actions (Comai and Henikoff, 2006).
Grafting is one of the most focused breeding techniques practiced in many countries in recent times. Rivard and Louws (2008) have reported the cultivation of high-quality heirloom varieties by grafting without the burden of soil-borne pathogens viz., bacterial wilt. Apart from the well-known benefit of grafting as a method to control soil-borne diseases, there are many other benefits viz., tolerance to drought, heat, and salinity stresses (Singh et al., 2017). Also, Rouphael et al. (2018) discussed the efficiency of water, nutrient uptake, photosynthesis, and powerful defense mechanisms in grafted tomatoes. Flores et al. (2010) investigated that the rootstock (cv. Radja) was able to induce both tomato fruit yield and fruit quality traits of the scion.
Marker-Assisted Selection (MAS) as a Tool of Tomato Breeding
Marker-assisted selection is a DNA based marker used in plant breeding for three major purposes viz., (i) Accumulation of favorable alleles by through generations tracing of either recessive or dominant desirable alleles (ii) Identification of desirable individuals genotype from segregated breeding population/lines based on either part of allelic composition or entire genome (iii) Introgression of favorable alleles from a donor parent into elite cultivar by breaking the undesirable linkage loci. The general terms used in modern molecular breeding techniques include genome-wide selection (GWS) or genomic selection (GS), marker-assisted pedigree selection (MAPS), marker-assisted selection (MAS), marker-assisted backcrossing (MABC) and marker-assisted recurrent selection (MARS). Among the solanaceous crops, tomato is one of the most studied based on genomic and genetic studies. MAS is an indirect method of selection for the trait based on the genotype of an associated marker instead of the trait of interest (Osei et al., 2018). For the past few decades, MAS is the most popularly used technique by breeders to develop new cultivars (Banga and Banga, 1998). Also, MAS is a constructive way for gene pyramiding or identification of quantitative trait loci (QTL), especially for low heritability traits (Jacob et al., 2016).
Different categories of linked markers have been identified in the tomato breeding program following the simple Mendelian inheritance. These markers were useful to select the plants or lines with traits of interest. There are over 1300 markers that have been identified in tomatoes (Osei et al., 2018). They include morphological (viz., leaf shape, seedling anthocyanin, and fruit shape), physiological (viz., days to flowering, self-incompatibility/male sterility), pest and disease-resistant traits. However, the setback associated with the genetic markers such as dominance, epistasis, pleiotropic effects, penetrance, and expressivity leads to the lowering of selection efficiency of breeding programs (Peralta et al., 2006). After genetic markers in 1970–1980, a set of isozyme markers was developed as second-generation markers. These isozyme markers were low in diversity and hence could not identify closely related genotypes (Foolad et al., 1993).
Molecular MAS has become an essential tool for crop improvement over the phenotypic selection. MAS is useful to select genes of interest from the donor or wild relative (foreground selection) to the recurrent parent and at the same time can recover the recurrent genome (background selection) by breaking the linkage drag (Osei et al., 2018). In this technique, tightly linked molecular markers are used to select the gene of interest (Usman et al., 2018; Usman et al., 2017).
With the invention of DNA markers such as RFLPs (Restricted Fragment Length Polymorphism) and AFLPs (Amplified Fragment Length Polymorphism), a lot of constraints associated with genetic linkage maps were successfully overcome (Foolad, 2007). Then, PCR (Polymerase Chain Reaction) based markers such as CAPS (Cleaved Amplified Polymorphic Sequence) and SCAR (Sequence Characterized Amplified Region) were used as DNA markers in the marker-assisted selection in molecular breeding (Bai et al., 2004). Recently developed high-resolution molecular markers such as SNP (Single Nucleotide Polymorphism) and InDels (Insertion Deletions) (Yang et al., 2004; Foolad, 2007) have allowed breeders to distinguish the differences within and among closely related species. The latest updates of the newest molecular markers of tomato can be obtained from the SOL Genomics Network, 2020 (SGN) website (https://solgenomics.net/community/links/related_sites.pl, retrieved June 15, 2020).
Bernatzky and Tanksley (1986) published the first linkage map of tomato, and since then, several improvements have been affiliated to the map bringing more usefulness to the breeders. The density of the tomato molecular linkage map is being gradually expanded across time with the untiring effort of scientists in several related disciplines all over the world. Most of the genes and QTLs responsible for commercially desirable traits (viz., fruit quality, yield, and disease resistance) are found in wild relatives, and MAS is a supporting tool to transfer those genes and QTLs to cultivated tomato (Tanksley and McCouch, 1997). However, earlier developed molecular markers such as RFLPs, CAPS, or AFLPs have not been able to distinguish between closely related cultivars and close wild relatives. The discovery of SNP markers would be helpful to mitigate the limitations associated with such molecular markers. Major QTL identification of high-resolution fine maps will facilitate the use of MAS in breeding new cultivars in the future (Foolad, 2007).
ILs are interspecific lines with a single ‘exotic’ chromosome segment introgressed from wild relatives and the rest of the genome of the recipient (Eshed and Zamir, 1995). ILs have high potential in QTL identification than those interspecific crosses used in the early days of MAS. So far, several ILs have developed, including S. habrochaites (Monforte and Tanksley, 2000), S. lycopersicoides Dunal–Peruvian wolfpeach, and S. sitiens I.M.Johnst (Canady et al., 2006) and S. pennellii (Eshed and Zamir, 1995; Tomato Genetics Resource Center, 2020). These ILs or pre-breeding lines facilitate plant breeders to pyramid many commercially important traits such as high yield, resistance to pests, diseases, and abiotic stresses together into a single cultivar (Fridman et al., 2004). Among the mapping populations, ILs may be useful for developing NILs (Near Isogenic Lines) for fine mapping and cloning of genes and QTLs controlling traits like fruit weight (Frary et al., 2000). Apart from this, IL populations were found to be used for marker-assisted selection of QTLs responsible for the yield of tomato (Foolad, 2007; Gur and Zamir, 2004).
Use of in molecular markers in advancement of yield components of tomato
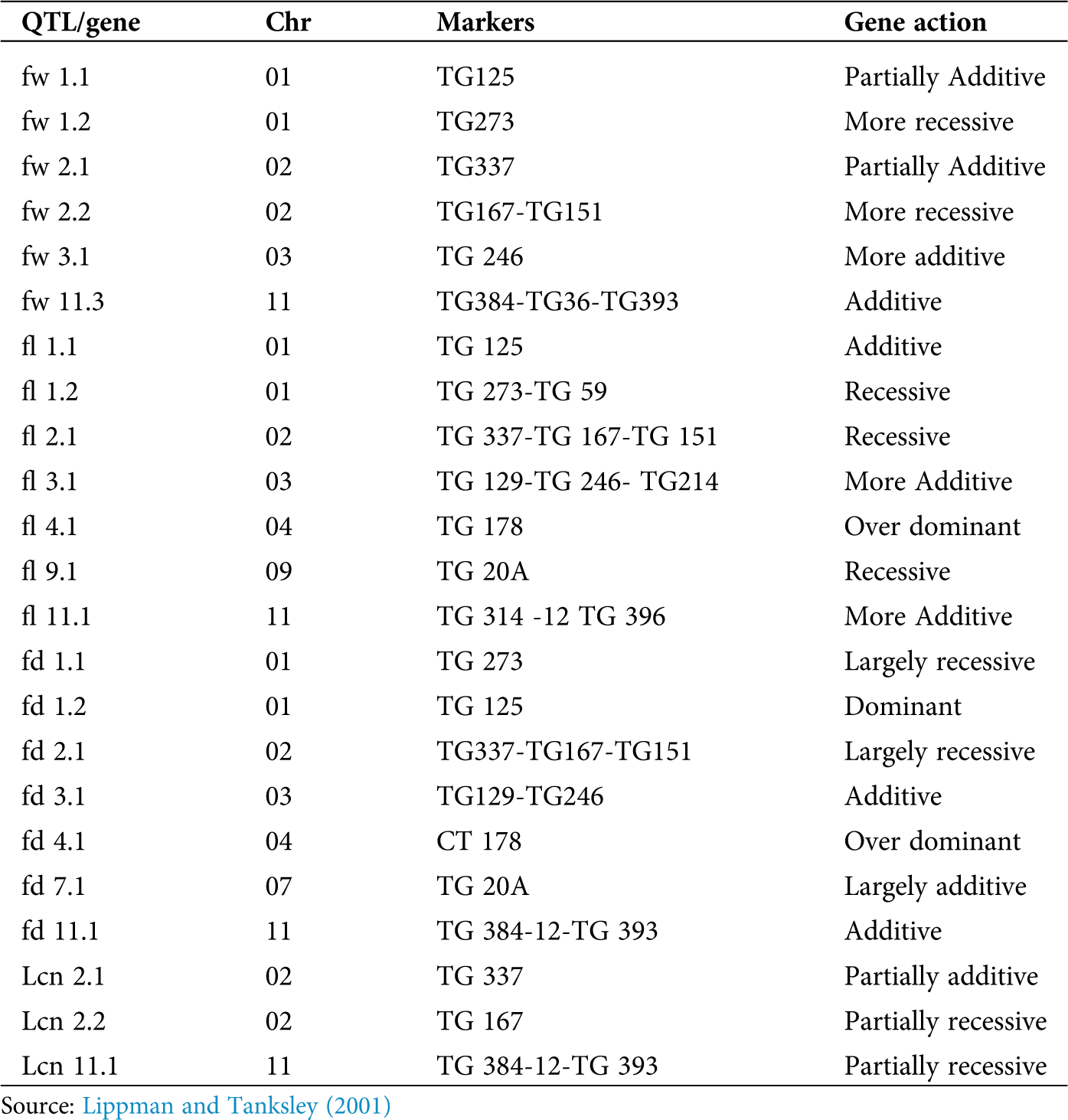
Fruit size is a highly heritable character that can be achieved through conventional breeding or phenotypic selection (Kemble and Gardner, 1992). Besides the high heritability, tomato fruit size is controlled by multiple genes and highly influence environmental conditions to greater proportions (Causse et al., 2004). Molecular markers allow the dissection of such quantitative traits into discrete QTL which can be located on a genetic map. Recombinants of QTLs may have led to big-fruited heirlooms (fruit weight ~1000 g) while QTLs fw1.1, fw2.1, fw 2.2, fw 3.1 and fw 3.2 and fw 11.3 may have led to medium-sized tomatoes which do not affect locule number (Foolad, 2007; Grandillo et al., 1999). Cultivated tomato bears large fruits while the wild relatives bear small berries (Bauchet and Causse, 2012). The first-ever map-based cloning of a QTL for fruit size fw2.2 was conducted on tomato (Frary et al., 2000), and this particular locus is responsible for most of the variation in fruit size (Lippman and Tanksley, 2001) (Tab. 1). Very limited studies have been done on QTL of fruit yield in tomatoes due to low heritability and the complexity of the character. In addition, it is difficult to measure or predict fruit yield which is affected by various complex genetic and environmental factors (Foolad, 2007).
Table 1: Summary of markers associated with important yield-related parameters
Conventional approaches in controlling viral diseases
To date, breeding resistance varieties seem the best approach for controlling viral diseases. At present, there are many commercial varieties available with partial resistance to viruses (Glick et al., 2009). Pico et al. (1999) developed several tomato advanced breeding lines using the backcrossing method with S. chillence as the donor parent. In general, tropics and subtropics are good breeding grounds for viruses to outbreak. To control the viral outbreak from growing beyond the economic threshold level, combinations of management practices and the development of resistant varieties are a prerequisite. Among these, modified cultural practices such as crop rotation, use of virus-free planting materials, use of virus-host resistance and their insect-vectors, cross-protection, and application of insecticides to physical and chemical control of vector are important (Tripathi and Verma, 2017). An efficient environmental and user-friendly approach of resistant breeding for tomato is the combination of vector control (by conventional and chemical methods) and breeding resistant varieties.
Pest resistant traits of tomato as an approach to control virus transmitting vectors
A study conducted by Mutschler et al. (1996) showed that L. pennellii accession LA716 secretes acyl sugars by type-IV glandular trichomes on the leaf surface, which act as oviposition deterrents for Silverleaf whitefly (i.e., TYLCV vector). Similarly, retardation in oviposition and the number of nymphs of whitefly have been observed in tomato genotypes with high acyl sugar content (Neiva et al., 2019). Oliveira et al. (2020) have recently reported a repellence effect and non-preference to oviposition by whiteflies in tomato genotypes with high zingiberene content obtained at F2BC2 from an interspecific cross between S. lycopersicum × S. habrochaites var. hirsutum. In another study, Lawson et al. (1997) included a pest-resistant gene into cultivated tomatoes; however, this attempt failed due to linkage drag showing the complex gene function. At present, farmers control pests by applying pesticides, however, more pest-resistant gene incorporation to tomato cultivars is expected with the advancement of MAS together with the potential and applicable conventional breeding approaches (Foolad, 2007).
Disease resistant traits of tomato as an approach to control virus transmitting vectors
Viral diseases are limiting factors affecting crop productivity, especially in tomatoes, due to the unavailability of antiviral control measures (Hanson et al., 2000). Among the top viral diseases affecting tomato, TBSV (tomato bushy stunt virus), TSWV (tomato spotted wilt virus), and TYLCV (Tomato Yellow Leaf Curl Virus) were considered the most destructive (Scholthof et al., 2011). PCR-based markers are available only for few disease-resistant traits due to the low polymorphism, which is a limiting factor in exploiting MAS for resistant breeding in tomatoes. However, the discovery of SNP markers seems to be helpful to mitigate this constraint. Major QTL identification using fine maps with high resolution will facilitate the use of MAS in breeding new cultivars ahead (Foolad, 2007). Schuch et al. (1991) showed that breeding of virus and insect-resistant cultivars is difficult solely by the traditional approach. Agrama and Scott (2006) have identified resistant genes responsible for TYLCV and ToMV (tomato mosaic virus) and the tightly linked markers with the resistant genes.
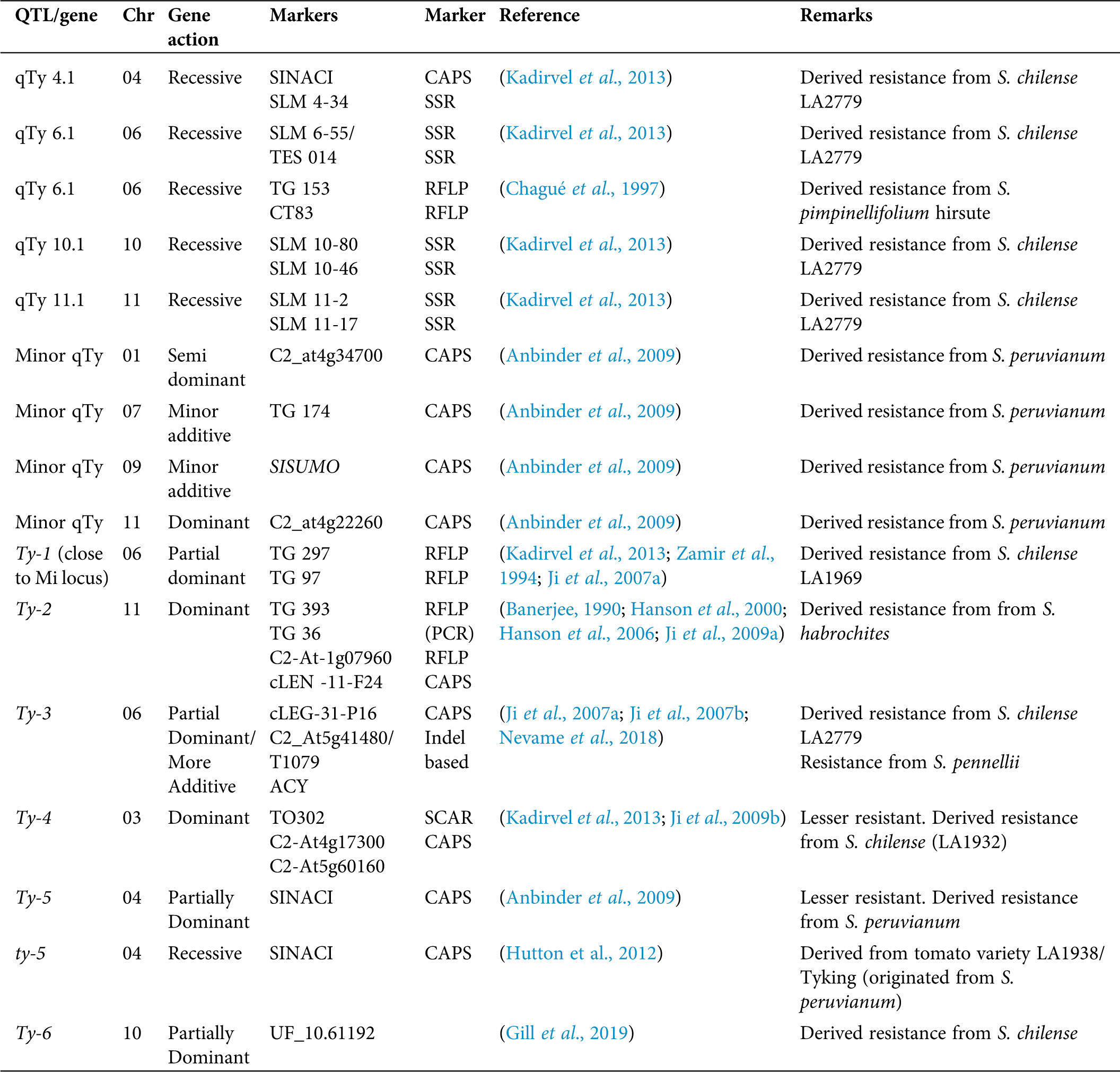
Apart from the conventional breeding approach, MAS, transgenic approach, and pathogen-induced resistance is common attempts of resistant variety development (Saidi and Warade, 2008). Hamilton et al. (1999) reported the identification of resistant genes (Cmr) for cucumber mosaic virus (CMV) in tomato and Patil et al. (2002) identified the resistant gene (Pot 1) for potyviruses, Bruening and Lyons (2000) identified three resistant genes ToMV Tm-1, Tm-2, and Tm-22 while Martin et al. (1993) reported Sw 5 spotted wilt virus (TSWV) resistant gene in tomato. However, the use of these resistant genes and QTLs in the development of virus-resistant tomato cultivars are rarely reported (Foolad, 2007). Apart from the nematode resistance, the Mi gene in the tomato genome has been reported with the resistant action against two biotypes of whitefly (B. tabaci) (Osei et al., 2018). There are many records on R genes and QTLs for the tomato yellow leaf curl virus, such as qTy 4.1, 6.1, 10.1, and 11.1 (Prasanna et al., 2015; Kadirvel et al., 2013).
Lapidot and Friedmann (2002) stated that none of the chemical and physical barriers would help-in controlling whitefly during a severe outbreak, and the best approach would be the development of resistant varieties using classical or genetic engineering to control TYLCV in tomato. Up to now, there are six Ty genes that act independently (Gorovits et al., 2017; Dhaliwal et al., 2020) and have been incorporated into commercial cultivated tomato by introgression from its wild relatives (Singh et al., 2019). Kumar et al. (2014) reported the successful pyramiding of the Ty-2 gene to two TYLCV susceptible cultivars and the production of crosses with TYLCV resistance throughout the lifecycle.
Among the TYLCV resistant loci so far identified, Prasanna et al. (2015) explained that TYLCV disease-resistant gene Ty-3 contributed a vital role for broad-spectrum resistance after gene pyramiding, and this would be utilized in TYLCV prevalence areas as a potential genetic resource for tomato hybrid breeding programs. Research done in Guatemala reported that tomato inbred lines with both Ty-3 and Ty-4 genes had a higher level of resistance to TYLCVD compared with lines with only Ty-3 (Nakhla et al., 2004; Vidavski, 2007). Nevame et al. (2018) reported a new molecular marker for the Ty-3 gene and stated that tomato hybrid carrying Ty-2 and Ty-3 resistance genes can mitigate the effect of the virus as compared to a single gene. Dhaliwal et al. (2020) have reviewed the importance of possessing TYLCV resistant genes in both parents when developing an effective resistance in tomato hybrids since Ty genes contribute partial dominance.
According to Gill et al. (2019), the resistance incurred by Ty 6 major gene located on chromosome 10 thrives with the presence of the TYLCV resistant gene Ty 3 and Ty 5. In a comprehensive study by Yan et al. (2018), 138 out of 708 wild tomato accessions tested using two different inoculation methods were resistant to TYLCD. In addition, they identified allelic polymorphism in Ty1/Ty3 gene using VIGS (virus-induced gene silencing) and allele mining in few S. chilense accessions. Their findings will pave the way for tomato breeders to develop new tomato cultivars with TYLCV resistance. Ammara et al. (2015) developed transgenic tomato plants with RNAi (RNA interference) based resistance against TYLCV Oman strain and the associated beta satellite. Though the developed transgenic plants were not immune to TYLCD, they conferred the reduction of disease symptom severity.
Similar efforts of producing TYLCD resistant transgenic tomato plants expressing TYLCV capsid protein have been reported by Singh et al. (2019). It has been reported that the association of TYLCV with the beta satellites of other plant viruses altering the gene action of already identified resistant (Ty) genes. A recent finding by Gelbart et al. (2020) has revealed such association of cotton leaf curl Gezira Beta satellite with TYLCV, which compromises the gene action of resistance covered by Ty-1 gene in tomato. In addition to the major genes and QTLs presented in Tab. 2, Zamir et al. (1994) mapped two modifier genes in chromosomes 3 and 7, respectively, associated with the action of the Ty-1 gene. Similarly, Kadirvel et al. (2013) reported that the QTLs 4.1 and 10.1 in chromosomes 4 and 10 contain virus-resistant candidate genes such as CTV 22 and eukaryotic translation initiation factor 4E. The majority of findings on the usage of molecular markers are based on TYLCV resistant markers as summarized in Tab. 2.
Table 2: Summary of important markers associated with TYLCV resistant /QTLS genes
Future direction of tomato cultivar development for high yield and viral disease—resistant traits
In addition to unexploited knowledge on gene function, repulsion linkages between clustered resistance loci (Scott, 2004), linkage drag, and low resolution of the linkage maps are major constraints in TYLCV resistance breeding (Foolad, 2007). In the future, a combination of traditional or conventional knowledge together with a re-sequenced genetic map with high-resolution markers like SNPs and InDels will be helpful to minimize the present-day constraints in tomato breeding (Foolad, 2007). More disease-resistant genes in tomato wild relatives will unveil in the future, and breeders will be able to develop new cultivars with multiple disease resistance and higher fruit quality through breeding by design (Bai et al., 2004). The transition of tomato breeding from traditional breeding techniques to the new “omics” era will allow breeders to expand their knowledge on gene expression and metabolism of each gene efficiently and effectively (Peleman and van der Voort, 2003).
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) technology is a milestone in molecular breeding where the outcome is quite similar to that obtained from conventional breeding but with a very short time (Bortesi and Fischer, 2015). This is a cheap, fast, and reliable method. Changes in the regulatory region of genes responsible for tomato yield have been made with SDN-1 (site-directed nuclease-1) mutant using the use of CRISPR technology. This makes positive changes to increase the variation of the regulatory region of the gene of interest. This has boosted the tomato yield in a short time. Hence, CRISPR technology is expected to be a powerful tool to achieve goals in tomato breeding (Gao, 2018). Similar efforts on producing TYLCV resistant tomato cultivars using CRISPR/ Cas 9 technology have been made by Tashkandi et al. (2018). The advanced molecular technologies, tools, strategies, conventional approaches, and agronomic practices should go hand in hand to see the success of future tomato variety development programs irrespective of the breeding goals. Similarly, no exception for the development of high yielding and TYLCD resistant tomato varieties. Especially with the upcoming unpredictable environment conditions such as extreme climates, pest and disease outbreaks and their complex interactions could be addressed only by these integrated approaches.
Acknowledgement: Tharangani Welegama is grateful to Sri Lanka Council for Agricultural Research Policy (SLCARP) for the scholarship rendered for her PhD Degree at Universiti Putra Malaysia.
Author Contribution: All authors equally contributed to manuscript preparation.
Funding Statement: The authors express their acknowledgements of the Long-term Research Grant Scheme (LRGS), Ministry of Higher Education, Malaysia, Project No. LRGS/1/2019/UKM/5, Vote No. 6300242 for the financial support to conduct activities on this research program.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Acquaah G (2012). Breeding tomato. Principles of Plant Genetics and Breeding. pp. 667–678. John Wiley & Sons, Chichester, UK. [Google Scholar]
Agrama HA, Scott JW (2006). Quantitative trait loci for Tomato yellow leaf curl virus and Tomato mottle virus resistance in tomato. Journal of the American Society for Horticultural Science 131: 267–272. DOI 10.21273/JASHS.131.2.267. [Google Scholar] [CrossRef]
Anbinder I, Reuveni M, Azari R, Paran I, Nahon S, Shlomo H, Chen L, Lapidot M, Levin I (2009). Molecular dissection of Tomato leaf curl virus resistance in tomato line TY172 derived from Solanum peruvianum. Theoretical and Applied Genetics 119: 519–530. DOI 10.1007/s00122-009-1060-z. [Google Scholar] [CrossRef]
Arah IK, Amaglo H, Kumah EK, Ofori H (2015). Preharvest and postharvest factors affecting the quality and shelf life of harvested tomatoes: A mini review. International Journal of Agronomy 2015: 1–6. DOI 10.1155/2015/478041. [Google Scholar] [CrossRef]
Bai Y, Lindhout P (2007). Domestication and breeding of tomatoes: What have we gained and what can we gain in the future? Annals of Botany 100: 1085–1094. DOI 10.1093/aob/mcm150. [Google Scholar] [CrossRef]
Bai Y, Feng X, van der Hulst R, Lindhout P (2004). A set of simple PCR markers converted from sequence specific RFLP markers on tomato chromosomes 9 to 12. Molecular Breeding 13: 281–287. DOI 10.1023/B:MOLB.0000022535.82602.79. [Google Scholar] [CrossRef]
Banerjee MK (1990). Transfer of tomato leaf curl virus resistance from Lycopersicon hirsutum f. glabratum to L. esculentum. Plant Breeding 105: 156–159. DOI 10.1111/j.1439-0523.1990.tb00469.x. [Google Scholar] [CrossRef]
Banga SS, Banga SK (1998). Hybrid Cultivar Development. pp. 436. Springer-Verlag, Berlin. [Google Scholar]
Bauchet G, Causse M (2012). Genetic diversity in tomato (Solanum lycopersicum) and its wild relatives. Genetic Diversity in Plants 8: 134–162. [Google Scholar]
Bernatzky R, Tanksley SD (1986). Toward a saturated linkage map in tomato based on isozymes and random cDNA sequences. Genetics 112: 887–898. DOI 10.1093/genetics/112.4.887. [Google Scholar] [CrossRef]
Birchler JA, Yao H, Chudalayandi S, Vaiman D, Veitia RA (2010). Heterosis. Plant Cell 22: 2105–2112. DOI 10.1105/tpc.110.076133. [Google Scholar] [CrossRef]
Bortesi L, Fischer R (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnology Advances 33: 41–52. DOI 10.1016/j.biotechadv.2014.12.006. [Google Scholar] [CrossRef]
Bradshaw JE (2016). Genetic basis of heterosis and inbred line versus hybrid cultivars. Plant Breeding: Past, Present and Future. Cham: Springer. [Google Scholar]
Bruening G, Lyons J (2000). The case of the FLAVR SAVR tomato. California Agriculture 54: 6–7. DOI 10.3733/ca.v054n04p6. [Google Scholar] [CrossRef]
Canady MA, Ji Y, Chetelat RT (2006). Homeologous recombination in Solanum lycopersicoides introgression lines of cultivated tomato. Genetics 174: 1775–1788. DOI 10.1534/genetics.106.065144. [Google Scholar] [CrossRef]
Casali VW, Tigchelaar EC (1975). Breeding progress in tomato with pedigree selection and single descent. Journal of the American Society for Horticultural Science 100: 362–364. [Google Scholar]
Causse M, Duffe P, Gomez MC, Buret M, Damidaux R, Zamir D, Gur A, Chevalier C, Lemaire-Chamley M, Rothan C (2004). A genetic map of candidate genes and QTLs involved in tomato fruit size and composition. Journal of Experimental Botany 55: 1671–1685. DOI 10.1093/jxb/erh207. [Google Scholar] [CrossRef]
Chagué V, Mercier JC, Guenard M, De Courcel A, Vedel F (1997). Identification of RAPD markers linked to a locus involved in quantitative resistance to TYLCV in tomato by bulked segregant analysis. Theoretical and Applied Genetics 95: 671–677. DOI 10.1007/s001220050611. [Google Scholar] [CrossRef]
Chen ZJ (2013). Genomic and epigenetic insights into the molecular bases of heterosis. Nature Reviews Genetics 14: 471–482. DOI 10.1038/nrg3503. [Google Scholar] [CrossRef]
Comai L, Henikoff S (2006). TILLING: Practical single-nucleotide mutation discovery. Plant Journal 45: 684–694. DOI 10.1111/j.1365-313X.2006.02670.x. [Google Scholar] [CrossRef]
Dhaliwal MS, Jindal SK, Sharma A, Prasanna HC (2020). Tomato yellow leaf curl virus disease of tomato and its management through resistance breeding: A review. Journal of Horticultural Science and Biotechnology 95: 425–444. DOI 10.1080/14620316.2019.1691060. [Google Scholar] [CrossRef]
Diaz-Pendon JA, Canizares MC, Moriones E, Bejarano ER, Czosnek H, Navas-Castillo J (2010). Tomato yellow leaf curl viruses: Ménage à trois between the virus complex, the plant and the whitefly vector. Molecular Plant Pathology 11: 441–450. DOI 10.1111/j.1364-3703.2010.00618.x. [Google Scholar] [CrossRef]
Ammara U, Mansoor S, Saeed M, Amin I, Briddon RW, Al-Sadi AM (2015). RNA interference-based resistance in transgenic tomato plants against Tomato yellow leaf curl virus-Oman (TYLCV-OM) and its associated betasatellite. Virology Journal 12: 38. DOI 10.1186/s12985-015-0263-y. [Google Scholar] [CrossRef]
Eshed Y, Zamir D (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162. DOI 10.1093/genetics/141.3.1147. [Google Scholar] [CrossRef]
Fentik DA (2017). Review on genetics and breeding of tomato (Lycopersicon esculentum Mill). Advances in Crop Science and Technology 5: 306. [Google Scholar]
Firdaus S, van Heusden AW, Hidayati N, Supena EDJ, Visser RG, Vosman B (2012). Resistance to Bemisia tabaci in tomato wild relatives. Euphytica 187: 31–45. DOI 10.1007/s10681-012-0704-2. [Google Scholar] [CrossRef]
Flores FB, Sanchez-Bel P, Estan MT, Martinez-Rodriguez MM, Moyano E, Morales B, Campos JF, Garcia-Abellán JO, Egea JI, Fernández-Garcia N, Romojaro F, Bolarín MC (2010). The effectiveness of grafting to improve tomato fruit quality. Scientia Horticulturae 125: 211–217. DOI 10.1016/j.scienta.2010.03.026. [Google Scholar] [CrossRef]
Food FAO (2018). Agriculture Organization of the United Nations. FAOSTAT: Statistics database. http://www.fao.org/faostat/en/#data/QC/visualize. [Google Scholar]
Foolad MR (2007). Genome mapping and molecular breeding of tomato. International Journal of Plant Genomics 2007: 1–52. DOI 10.1155/2007/64358. [Google Scholar] [CrossRef]
Foolad MR, Jones RA, Rodriguez RL (1993). RAPD markers for constructing intraspecific tomato genetic maps. Plant Cell Reports 12: 293–297. DOI 10.1007/BF00237139. [Google Scholar] [CrossRef]
Frary A, Nesbitt TC, Frary A, Grandillo S, Van Der Knaap E, Cong B (2000). fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88. DOI 10.1126/science.289.5476.85. [Google Scholar] [CrossRef]
Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D (2004). Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305: 1786–1789. DOI 10.1126/science.1101666. [Google Scholar] [CrossRef]
Fufa F, Hanson P, Dagnoko S, Dhaliwal M (2009). AVRDC-The World Vegetable Center tomato breeding in sub-Saharan Africa: Lessons from the past, present work, and future prospects. In I All Africa Horticultural Congress 911, 87–98. [Google Scholar]
Gao C (2018). The future of CRISPR technologies in agriculture. Nature Reviews Molecular Cell Biology 19: 275–276. DOI 10.1038/nrm.2018.2. [Google Scholar] [CrossRef]
Gelbart D, Chen L, Alon T, Dobrinin S, Levin I, Lapidot M (2020). The recent association of a DNA betasatellite with Tomato yellow leaf curl virus in Israel—A new threat to tomato production. Crop Protection 128: 104995. DOI 10.1016/j.cropro.2019.104995. [Google Scholar] [CrossRef]
Gill U, Scott JW, Shekasteband R, Ogundiwin E, Schuit C, Francis DM, Sim SC, Smith H, Hutton SF (2019). Ty-6, a major begomovirus resistance gene on chromosome 10, is effective against Tomato yellow leaf curl virus and Tomato mottle virus. Theoretical and Applied Genetics 132: 1543–1554. DOI 10.1007/s00122-019-03298-0. [Google Scholar] [CrossRef]
Giovannucci E (1999). Tomatoes, tomato-based products, lycopene, and cancer: Review of the epidemiologic literature. Journal of the National Cancer Institute 91: 317–331. DOI 10.1093/jnci/91.4.317. [Google Scholar] [CrossRef]
Glick E, Levy Y, Gafni Y (2009). The viral etiology of tomato yellow leaf curl disease—A review. Plant Protection Science 45: 81–97. DOI 10.17221/26/2009-PPS. [Google Scholar] [CrossRef]
Gorovits R, Moshe A, Amrani L, Kleinberger R, Anfoka G, Czosnek H (2017). The six Tomato yellow leaf curl virus genes expressed individually in tomato induce different levels of plant stress response attenuation. Cell Stress and Chaperones 22: 345–355. DOI 10.1007/s12192-017-0766-0. [Google Scholar] [CrossRef]
Grandillo S, Ku HM, Tanksley SD (1999). Identifying the loci responsible for natural variation in fruit size and shape in tomato. Theoretical and Applied Genetics 99: 978–987. DOI 10.1007/s001220051405. [Google Scholar] [CrossRef]
Gur A, Zamir D (2004). Unused natural variation can lift yield barriers in plant breeding. PLoS Biology 2: e245. DOI 10.1371/journal.pbio.0020245. [Google Scholar] [CrossRef]
Hamilton CM, Frary A, Xu Y, Tanksley SD, Zhang HB (1999). Construction of tomato genomic DNA libraries in a binary-BAC (BIBAC) vector. Plant Journal 18: 223–229. DOI 10.1046/j.1365-313X.1999.00433.x. [Google Scholar] [CrossRef]
Hanson PM, Bernacchi D, Green S, Tanksley SD, Muniyappa V, Padmaja AS, Chen HM, Kuo G, Fang D, Chen JT (2000). Mapping a wild tomato introgression associated with tomato yellow leaf curl virus resistance in a cultivated tomato line. Journal of the American Society for Horticultural Science 125: 15–20. DOI 10.21273/JASHS.125.1.15. [Google Scholar] [CrossRef]
Hanson P, Green SK, Kuo G (2006). Ty-2, a gene on chromosome 11 conditioning geminivirus resistance in tomato. Tomato Genetics Cooperative Reports 56: 17–18. [Google Scholar]
Hutton SF, Scott JW, Schuster DJ (2012). Recessive resistance to Tomato yellow leaf curl virus from the tomato cultivar Tyking is located in the same region as Ty-5 on chromosome 4. HortScience 47: 324–327. DOI 10.21273/HORTSCI.47.3.324. [Google Scholar] [CrossRef]
IAEA (2020). IAEA mutant database. Vienna: International Atomic Energy Agency; 2015. http://mvd.iaea.org/. [Google Scholar]
Jacob C, Carrasco B, Schwember AR (2016). Advances in breeding and biotechnology of legume crops. Plant Cell, Tissue and Organ Culture 127: 561–584. DOI 10.1007/s11240-016-1106-2. [Google Scholar] [CrossRef]
Ji Y, Schuster DJ, Scott JW (2007a). Ty-3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Molecular Breeding 20: 271–284. [Google Scholar]
Ji Y, Scott JW, Schuster DJ (2009a). Toward fine mapping of the Tomato yellow leaf curl virus resistance gene Ty-2 on chromosome 11 of tomato. HortScience 44: 614–618. [Google Scholar]
Ji Y, Scott JW, Hanson P, Graham E, Maxwell DP (2007b). Sources of resistance, inheritance, and location of genetic loci conferring resistance to members of the tomato-infecting begomoviruses. Tomato Yellow Leaf Curl Virus Disease. pp. 343–362. Dordrecht: Springer. [Google Scholar]
Ji Y, Scott JW, Schuster DJ, Maxwell DP (2009b). Molecular mapping of Ty-4, a new Tomato yellow leaf curl virus resistance locus on chromosome 3 of tomato. Journal of the American Society for Horticultural Science 134: 281–288. [Google Scholar]
Kadirvel P, De la Peña R, Schafleitner R, Huang S, Geethanjali S, Kenyon L, Tsai W, Hanson P (2013). Mapping of QTLs in tomato line FLA456 associated with resistance to a virus causing tomato yellow leaf curl disease. Euphytica 190: 297–308. [Google Scholar]
Kalloo G (2012). Genetic improvement of tomato. Vol. 14, pp. 457. Springer-Verlag, Berlin. [Google Scholar]
Kaul MLH (1991). Reproductive biology in tomato. Genetic Improvement of Tomato. pp. 39–50. Berlin, Heidelberg: Springer. [Google Scholar]
Kaushik P, Dhaliwal M (2018). Diallel analysis for morphological and biochemical traits in tomato cultivated under the influence of tomato leaf curl virus. Agronomy 8: 153. [Google Scholar]
Kemble JM, Gardner RG (1992). Inheritance of shortened fruit maturation in the cherry tomato Cornell 871213-1 and its relation to fruit size and other components of earliness. Journal of the American Society for Horticultural Science 117: 646–650. [Google Scholar]
Kumar A, Tiwari KL, Datta D, Singh M (2014). Marker assisted gene pyramiding for enhanced Tomato leaf curl virus disease resistance in tomato cultivars. Biologia Plantarum 58: 792–797. [Google Scholar]
Lapidot M, Friedmann M (2002). Breeding for resistance to whitefly-transmitted geminiviruses. Annals of Applied Biology 140: 109–127. [Google Scholar]
Lawson DM, Lunde CF, Mutschler MA (1997). Marker-assisted transfer of acylsugar-mediated pest resistance from the wild tomato, Lycopersicon pennellii, to the cultivated tomato. Lycopersicon esculentum. Molecular Breeding 3: 307–317. [Google Scholar]
Lin T, Zhu G, Zhang J, Xu X, Yu Q, Zheng Z, Zhang Z, Lun Y, Li S, Wang X, Huang Z, Li J, Zhang C, Wang T, Zhang Y, Wang A, Zhang Y, Lin K, Li C, Xiong G, Xue Y, Mazzucato A, Causse M, Fei Z, Giovannoni JJ, Chetelat RT, Zamir D, Städler T, Li J, Ye Z, Du Y, Huang S (2014). Genomic analyses provide insights into the history of tomato breeding. Nature Genetics 46: 1220–1226. [Google Scholar]
Lippman Z, Tanksley SD (2001). Dissecting the genetic pathway to extreme fruit size in tomato using a cross between the small-fruited wild species Lycopersicon pimpinellifolium and L. esculentum var Giant Heirloom. Genetics 158: 413–422. [Google Scholar]
Martin G, Brommonschenkel S, Chunwongse J, Frary A, Ganal M, Spivey R, Wu T, Earle E, Tanksley S (1993). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262: 1432–1436. DOI 10.1126/science.7902614. [Google Scholar] [CrossRef]
Mattoo A, Handa A (2017). Modelling crop growth and yield in tomato cultivation Kenneth J. Boote, University of Florida, USA. Achieving Sustainable Cultivation of Tomatoes. pp. 25–44. Cambridge: Burleigh Dodds Science Publishing. [Google Scholar]
Monforte AJ, Tanksley SD (2000). Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: A tool for gene mapping and gene discovery. Genome 43: 803–813. DOI 10.1139/g00-043. [Google Scholar] [CrossRef]
Mutschler MA, Doerge RW, Liu SC, Kuai JP, Liedl BE, Shapiro JA (1996). QTL analysis of pest resistance in the wild tomato Lycopersicon pennellii: QTLs controlling acylsugar level and composition. Theoretical and Applied Genetics 92: 709–718. DOI 10.1007/BF00226093. [Google Scholar] [CrossRef]
Nakhla MK, Sorensen A, Mejía L, Ramírez P, Karkashian JP, Maxwell DP (2004). Molecular characterization of tomato-infecting begomoviruses in Central America and development of DNA-based detection methods. I International Symposium on Tomato Diseases 695, pp. 277–288. [Google Scholar]
Neiva IP, Silva AAD, Resende JF, Carvalho RDC, Oliveira AMSD, Maluf WR (2019). Tomato genotype resistance to whitefly mediated by allelochemicals and Mi gene. Chilean Journal of Agricultural Research 79: 124–130. DOI 10.4067/S0718-58392019000100124. [Google Scholar] [CrossRef]
Nevame AYM, Xia L, Nchongboh CG, Hasan MM, Alam MA, Yongbo L, Wenting Z, Yafei H, Emon RM, Ismail MR, Efisue A, Gang S, Wenhu L, Longting S (2018). Development of a new molecular marker for the resistance to tomato yellow leaf curl virus. BioMed Research International 2018: 1–10. DOI 10.1155/2018/8120281. [Google Scholar] [CrossRef]
Oladosu Y, Rafii MY, Abdullah N, Hussin G, Ramli A, Rahim HA, Miah G, Usman M (2016). Principle and application of plant mutagenesis in crop improvement: A review. Biotechnology & Biotechnological Equipment 30: 1–16. DOI 10.1080/13102818.2015.1087333. [Google Scholar] [CrossRef]
Oliveira JRFD, Resende JTVD, Roberto SR, Silva PRD, Rech C, Nardi C (2020). Tomato breeding for sustainable crop systems: High levels of zingiberene providing resistance to multiple arthropods. Horticulturae 6: 34. DOI 10.3390/horticulturae6020034. [Google Scholar] [CrossRef]
Osei MK, Prempeh R, Adjebeng-Danquah J, Opoku JA, Danquah A, Danquah E, Blay E, Adu-Dapaah H (2018). Marker-Assisted Selection (MASA fast-track tool in tomato breeding. Recent Advances in Tomato Breeding and Production. IntechOpen. [Google Scholar]
Patil RS, Davey MR, Power JB, Cocking EC (2002). Effective protocol for Agrobacterium-mediated leaf disc transformation in tomato (Lycopersicon esculetum Mill.). NISCAIR Online Periodicals Repository, 339–343. [Google Scholar]
Peirce LC (1991). Selection systems for tomato improvement. Genetic Improvement of Tomato. pp. 59–71. Berlin, Heidelberg: Springer. [Google Scholar]
Peleman JD, Van der Voort JR (2003). Breeding by design. Trends in Plant Science 8: 330–334. DOI 10.1016/S1360-1385(03)00134-1. [Google Scholar] [CrossRef]
Peralta IE, Spooner DM, Razdan MK, Mattoo AK (2006). History, origin and early cultivation of tomato (Solanaceae). Genetic Improvement of Solanaceous Crops 2: 1–27. [Google Scholar]
Pertuzé RA, Ji Y, Chetelat RT (2003). Transmission and recombination of homeologous Solanum sitiens chromosomes in tomato. Theoretical and Applied Genetics 107: 1391–1401. DOI 10.1007/s00122-003-1384-z. [Google Scholar] [CrossRef]
Pico B, Ferriol M, Diez MJ, Nuez F (1999). Developing tomato breeding lines resistant to tomato yellow leaf curl virus. Plant Breeding 118: 537–542. DOI 10.1046/j.1439-0523.1999.00427.x. [Google Scholar] [CrossRef]
Poysa V (1990). The development of bridge lines for interspecific gene transfer between Lycopersicon esculentum and L. peruvianum. Theoretical and Applied Genetics 79: 187–192. DOI 10.1007/BF00225950. [Google Scholar] [CrossRef]
Prasad A, Sharma N, Hari-Gowthem G, Muthamilarasan M, Prasad M (2020). Tomato yellow leaf curl virus: Impact, challenges, and management. Trends in Plant Science 25: 897–911. DOI 10.1016/j.tplants.2020.03.015. [Google Scholar] [CrossRef]
Prasanna HC, Sinha DP, Rai GK, Krishna R, Kashyap SP, Singh NK, Singh M, Malathi VG (2015). Pyramiding T y-2 and T y-3 genes for resistance to monopartite and bipartite tomato leaf curl viruses of India. Plant Pathology 64: 256–264. DOI 10.1111/ppa.12267. [Google Scholar] [CrossRef]
Rick CM (1978). The tomato. Scientific American 239: 76–89. DOI 10.1038/scientificamerican0878-76. [Google Scholar] [CrossRef]
Rick CM (1988). Tomato-like nightshades: Affinities, autoecology, and breeders’ opportunities. Economic Botany 42: 145–154. DOI 10.1007/BF02858915. [Google Scholar] [CrossRef]
Rick CM, Gifford EM, Chetelat RT (1996). Genetics, anatomy and genealogy of obscuravenosa, a monogenic tomato leaf trait. Tomato Molecular Biology Symposium. UC-Berkeley. [Google Scholar]
Rivard CL, Louws FJ (2008). Grafting to manage soilborne diseases in heirloom tomato production. HortScience 43: 2104–2111. DOI 10.21273/HORTSCI.43.7.2104. [Google Scholar] [CrossRef]
Rouphael Y, Kyriacou MC, Colla G (2018). Vegetable grafting: A toolbox for securing yield stability under multiple stress conditions. Frontiers in Plant Science 8: 2255. DOI 10.3389/fpls.2017.02255. [Google Scholar] [CrossRef]
Saidi M, Warade SD (2008). Tomato breeding for resistance to Tomato spotted wilt virus (TSWVAn overview of conventional and molecular approaches. Czech Journal of Genetics and Plant Breeding 44: 83–92. DOI 10.17221/47/2008-CJGPB. [Google Scholar] [CrossRef]
Scholthof KBG, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders T, Candresse T, Ahlquist P, Hemenway C, Foster GD (2011). Top 10 plant viruses in molecular plant pathology. Molecular Plant Pathology 12: 938–954. DOI 10.1111/j.1364-3703.2011.00752.x. [Google Scholar] [CrossRef]
Schuch W, Kanczler J, Robertson D, Hobson G, Tucker G, Grierson D, Bright S, Bird CR (1991). Fruit quality characteristics of transgenic tomato fruit with altered polygalacturonase activity. HortScience 26: 1517–1520. DOI 10.21273/HORTSCI.26.6.751E. [Google Scholar] [CrossRef]
Scott JW (2004). Perspectives on tomato disease resistance breeding: Past, present, and future. I International Symposium on Tomato Diseases695: 217–224. [Google Scholar]
Singh H, Kumar P, Chaudhari S, Edelstein M (2017). Tomato grafting: A global perspective. HortScience 52: 1328–1336. DOI 10.21273/HORTSCI11996-17. [Google Scholar] [CrossRef]
Singh RK, Rai N, Singh AK, Kumar P, Singh B (2019). A critical review on Tomato leaf curl virus resistance in tomato. International Journal of Vegetable Science 25: 373–393. DOI 10.1080/19315260.2018.1520379. [Google Scholar] [CrossRef]
Sol Genomics Network (2020) https://solgenomics.net/community/links/related_sites.pl. [Google Scholar]
Tanksley SD, McCouch SR (1997). Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 277: 1063–1066. DOI 10.1126/science.277.5329.1063. [Google Scholar] [CrossRef]
Tashkandi M, Ali Z, Aljedaani F, Shami A, Mahfouz MM (2018). Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signaling & Behavior 13: e1525996. DOI 10.1080/15592324.2018.1525996. [Google Scholar] [CrossRef]
Tomato Genetics Resource Center (2020) http://tgrc.ucdavis.edu. [Google Scholar]
Tripathi S, Verma R (2017). Management of viral diseases through conventional approaches in India. A Century of Plant Virology in India. pp. 729–755. Singapore: Springer. [Google Scholar]
Usman MG, Rafii MY, Martini MY, Yusuff OA, Ismail MR, Miah G (2018). Introgression of heat shock protein (Hsp70 and sHsp) genes into the Malaysian elite chilli variety Kulai (Capsicum annuum L.) through the application of marker-assisted backcrossing (MAB). Cell Stress and Chaperones 23: 223–234. DOI 10.1007/s12192-017-0836-3. [Google Scholar] [CrossRef]
Usman MG, Rafii MY, Martini MY, Yusuff OA, Ismail MR, Miah G (2017). Molecular analysis of Hsp70 mechanisms in plants and their function in response to stress. Biotechnology and Genetic Engineering Reviews 33: 26–39. DOI 10.1080/02648725.2017.1340546. [Google Scholar] [CrossRef]
van de Wiel CCM, Schaart JG, Niks RE, Visser RGF (2010). Traditional Plant Breeding Methods 338, Wageningen UR Plant Breeding. [Google Scholar]
Vidavski FS (2007). Exploitation of resistance genes found in wild tomato species to produce resistant cultivars; pile up of resistant genes. Tomato Yellow Leaf Curl Virus Disease. pp. 363–372. Dordrecht: Springer. [Google Scholar]
Vidavski F, Czosnek H, Gazit S, Levy D, Lapidot M (2008). Pyramiding of genes conferring resistance to Tomato yellow leaf curl virus from different wild tomato species. Plant Breeding 127: 625–631. DOI 10.1111/j.1439-0523.2008.01556.x. [Google Scholar] [CrossRef]
Watson B (1996). Taylor’s guide to heirloom vegetables. New York, NY. [Google Scholar]
Willcox JK, Catignani GL, Lazarus S (2003). Tomatoes and cardiovascular health. Critical Reviews in Food Science and Nutrition 43: 1–18. DOI 10.1080/10408690390826437. [Google Scholar] [CrossRef]
Yan Z, Pérez-de-Castro A, Díez MJ, Hutton SF, Visser RGF, Wolters AM, Bai A, Li Y (2018). Resistance to tomato yellow leaf curl virus in tomato germplasm. Frontiers in Plant Science 9: 1198. DOI 10.3389/fpls.2018.01198. [Google Scholar] [CrossRef]
Yang W, Bai X, Kabelka E, Eaton C, Kamoun S, van der Knaap E (2004). Discovery of single nucleotide polymorphisms in Lycopersicon esculentum by computer aided analysis of expressed sequence tags. Molecular Breeding 14: 21–34. DOI 10.1023/B:MOLB.0000037992.03731.a5. [Google Scholar] [CrossRef]
Young ND, Tanksley SD (1989). RFLP analysis of the size of chromosomal segments retained around the Tm-2 locus of tomato during backcross breeding. Theoretical and Applied Genetics 77: 353–359. DOI 10.1007/BF00305828. [Google Scholar] [CrossRef]
Zamir D, Ekstein-Michelson I, Zakay Y, Navot N, Zeidan M, Sarfatti M, Eshed Y, Harel E, Pleban T, van-Oss H, Kedar N, Rabinowitch HD, Czosnek H (1994). Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, TY-1. Theoretical and Applied Genetics 88: 141–146. DOI 10.1007/BF00225889. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |