DOI:10.32604/biocell.2021.014499

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.014499 |  www.techscience.com/journal/biocell |
| Article |
Genome-wide identification and expression analysis of Aux/IAA gene family in strawberry (Fragaria vesca)
1School of Biological and Environmental Engineering, Xi’an University, Xi’an, 710065, China
2Rural Science and Technology Development Center, Xi’an, 710054, China
3School of Basic Medical Sciences, Southwest Medical University, Luzhou, 646000, China
4Citrus Research Institute, Southwest University-Chinese Academy of Agricultural Sciences, Chongqing, 400712, China
*Address correspondence to: Qiang Li, liqiang@cric.cn
#These authors contributed equally to this work
Received: 02 October 2020; Accepted: 14 January 2021
Abstract: Auxin signaling and its components (Auxin/Indole-3-Acetic Acid (Aux/IAA)) are critical for plant growth and development. Here, we performed a genome-wide annotation and identified twenty-one Aux/IAA genes in strawberry (Fragaria vesca). Most FveIAAs were located on chromosomes 1, 2, 4, 5, and 6, while no FveIAAs were found in chromosomes 3 and 7. Phylogenetic analysis divided these genes into nine subfamilies. Most FveIAAs contained the DNA-binding and Aux/IAA domains, as well as motifs I–IV. There were 2–6 exons in the FveIAA genes based on the gene structure analysis. Also, we found that four pairs of FveIAA genes underwent segment duplications. Moreover, four pairs of orthologous genes were observed between strawberry and Arabidopsis. Cis-element analysis in the promoter region indicated that FveIAAs may be involved in light, phytohormones, stress responses, and growth processes. Prediction of protein-protein interaction revealed that 17 of 21 FveIAA proteins were involved in the auxin-related signaling pathways. Additionally, FveIAAs showed tissue-specific expression and responded to IAA treatment. Thus, this systematic annotation of the FveIAA family would provide a fundamental basis for further functional and evolutionary analysis and to understanding the role of FveIAAs in strawberry growth and development.
Keywords: FveIAA; Auxin; Indole-3-Acetic Acid; Fragaria vesca
Auxin, a plant hormone, modulates plant growth and development by regulating the expression of Gretchen Hagen 3 (GH3), Auxin Response Factor (ARF), Indole-3-acetic Acid (Aux/IAA), and Small Auxin Up RNA (SAUR) gene families (Abbas et al., 2016; Aloni et al., 2006; Esmon et al., 2006; Mattsson et al., 2003; Mishra et al., 2009; Tiryaki, 2009). In the presence of cycloheximide, a translational inhibitor, auxin induces the expression of Aux/IAA genes. The degradation of Aux/IAA protein through the 26S proteasome pathway is induced by the auxin transport inhibitor response 1 (TIR1), which regulates the expression of auxin-responsive genes by releasing ARFs (Farcot et al., 2015; Hu et al., 2015b).
There are four conserved domains present in Aux/IAA genes with domain I containing a leucine repeat motif (LXLXLX) as a potent transcriptional repressor; domain II inducing Aux/IAA protein degradation; domain III constituting a βαα-DNA recognition motif; domain IV representing an acidic region (Liscum and Reed, 2002). Domains III and IV are also known to induce the homodimerization and heterodimerization between the ARFs and the Aux/IAA proteins (Mano and Nemoto, 2012). ARFs modulate the expression of auxin- responsive genes by specifically binding to the AuxRE (TGTCTC) sequence in their promoter region (Kim et al., 1997; Ulmasov et al., 1997). Aux/IAA proteins suppress the activity of ARF by interacting with the DNA-bound ARF partner protein through domains III and IV. Additionally, Aux/IAA proteins are directed towards the nucleus via two localization signals (Retzer et al., 2014; Wu et al., 2012). Genomic analyses have identified Aux/IAA gene family in the following plants: 29 in Arabidopsis, 31 in rice (Oryza sativa), 26 in tomato (Solanum lycopersicon), 27 in cucumber (Cucumis sativus), and 34 in maize (Zea mays) and other species, including Medicago truncatula, Populus trichocarpa, Vitis vinifera, etc. (Audran-Delalande et al., 2012; Cakir et al., 2013; Dreher et al., 2006; Gan et al., 2013; Jain et al., 2006; Kalluri et al., 2007; Wang et al., 2010; Wu et al., 2012). Studies on the functional analysis of IAAs in plant growth and development revealed the role of iaa3/shy2, iaa7/axr2, iaa14/slr, iaa17/axr3, iaa28 in lateral root formation in Arabidopsis (Fukaki et al., 2002; Knox et al., 2003; Mai et al., 2011; Timpte et al., 1994). In rice, three Aux/IAA members: OsIAA1, OsIAA11, and OsIAA23, were involved in regulating root development (Ni et al., 2011; Thakur et al., 2001; Zhu et al., 2012). In tomato, the under-expression of Sl-IAA9 affected leaf morphogenesis and the fruit set process (Wang et al., 2005). The downregulation of Sl-IAA15 resulted in reduced apical dominance, a lower trichome number, dark green leaves, and increased lateral root formation (Deng et al., 2012). Also, reduced fertilization and altered fruit development were observed due to the silencing of the Sl-IAA27 gene (Bassa et al., 2012). Another study revealed the role of the Sl-IAA17 transcriptional repressor in controlling fruit size by regulating endoreduplication-related cell expansion (Su et al., 2014).
Strawberry, a delicious and healthy food, constitutes an important fruit crop worldwide. Fragaria vesca is diploid (2n = 14), has a small genome size (<240 Mb), a relatively short reproductive cycle (14–15 weeks), and its genome sequence is available; thus, it is considered a model plant for studying transformation (Shulaev et al., 2011). Strawberry fruits are non-climacteric as they do not undergo ethylene-induced ripening. Previous studies have confirmed the role of auxin in the fruit set, development, and ripening in strawberry (Kang et al., 2013; Nitsch, 1950); however, its molecular regulation mechanisms remain unclear.
Past studies on plant genome sequences have enabled the genome-wide analyses of several multigenic protein families. Here, we used the public databases to conduct the genome-wide analyses of the strawberry Aux/IAA family, including genomic organization, the conserved protein domains, comparative phylogenetic analyses, prediction of protein structural motifs, putative cis-regulatory elements within promoters, subcellular location, and protein-protein interactions (PPI). We studied the expression of the Aux/IAA members in different organs at different developmental stages of the fruit and the expression of FveIAAs post-IAA treatment. Thus, this systematic annotation of the FveIAA family would provide a fundamental basis for the functional and evolutionary analysis, and to understand the role of the FveIAAs in strawberry growth and development.
Aux/IAA genes in Fragaria vesca
The strawberry genome and proteome were downloaded from Phytozome V12 (Goodstein et al., 2012) to perform exhaustive data mining of the FveIAA family. The IAA protein sets from A. thaliana were obtained from The Arabidopsis Information Resource (TAIR) (Swarbreck et al., 2008). Default parameters and cutoff value 0.01 was used for the hidden Markov Model (HMM) profiles to identify the Aux/IAA genes from the F. vesca genome (Eddy, 1998).The presence of conserved domains in the candidate Aux/IAA genes was evaluated using PFAM V32 and SMART tools (Finn et al., 2016; Schultz et al., 1998). NCBI CDD was used to examine the uniqueness of the obtained sequences for the Aux/IAA domains. ProtParam software was used to determine the molecular weight (MW) and the isoelectric point (pI) and of the FveIAA proteins. The annotated Aux/IAAs of strawberry were labeled as ‘FveIAA’ followed by a number representing their chromosomal orders.
In silico characterization of FveIAAs
Based on the strawberry genome database, we mapped all FveIAA genes to strawberry chromosomes using Circos (An et al., 2015). The chromosomal location of the FveIAA family was visualized using MapChart V2.1 (Voorrips, 2002). The gene structure of FveIAA genes was extracted from Phytozome and visualized using GSDS V2.0 (Hu et al., 2015a). MEME V5.1 and PFAM were used to detect the conserved motifs and functional domains, respectively (Bailey et al., 2009; Finn et al., 2016). A Multiple Collinearity Scan toolkit (MCScanX) with default parameters was used to study the gene duplication events (Wang et al., 2013). A syntenic analysis map was built using Dual System Plotter to study the relationship between the orthologous Aux/IAA genes obtained from strawberry and other organisms. STRING V11.0 with a threshold of 0.7 was used to construct the PPI network (Szklarczyk et al., 2017). The promotors (1500 to 1 bp before ATG) of the FveIAA genes were obtained from Phytozome, and the cis-elements were predicted by PlantCARE (Lescot et al., 2002), and subcellular localization of FveIAAs was predicted by WoLF PSORT (Horton et al., 2007).
Phylogeny of FveIAAs and AtIAAs
ClustalW and DNAMAN were used for multiple sequence alignment of the complete FveIAA and AtIAA protein sequences. The neighbor-joining (NJ) phylogenetic tree was built using MEGA V7.0 (Kumar et al., 2016), with a poison model and 1000 bootstrap replications. FveIAA protein sequences from Arabidopsis (Dreher et al., 2006) were obtained from Phytozome (Goodstein et al., 2012).
Plant growth condition and hormonal treatments
The strawberry (F. vesca f. semperflorens) plants were procured from Xi’an University, Xi’an, China. The plants were grown in a greenhouse at a temperature of 20°C–25°C with a 14-h light/10-h dark cycle and relative humidity of 70%–85%. During the first week after anthesis, more than 100 small green (SG) fruits on 50 strawberry plants were labeled. Fruits were collected on days 7, 14, 25 days post fertilization, at three different stages: SG, BG (Big green), and R (Red), respectively. At each stage, sampling of uniformly sized fruits (N = 10) was done in triplicates. Small cubes of the receptacle (pulp) (0.5–0.8 cm3) were flash-frozen in liquid nitrogen and stored at −80°C. The other tissues were collected from healthy strawberry plants (N = 10) at the flowering stage and analyzed in triplicates. The newly growing and fully expanded leaves were used for the IAA treatments. The branch containing three-piece leaves were cut from the strawberry plants and dipped in 10 μM IAA (pH 6.6) solution. The branches treated with H2O served as the control. The samples were collected 0, 1, 6, and 12 h after treatment. For each treatment, 24 branches were sampled from 12 different plants. All samples were flash-frozen in liquid nitrogen and stored at −80°C for RNA extraction.
RNA isolation and qRT-PCR analysis
TRIZOL Reagent was used to isolate total RNAs from the frozen samples (100–200 mg), followed by treatment with Turbo DNA-free TM kit to eliminate DNA contamination. After reverse transcription into cDNA, qRT-PCR was performed with the reaction mixture (20 μL) containing 1 μL forward/reverse specific primer (10 μM), 1 μL cDNA (30 ng/μL), and 10 μL SYBR Green Master Mix on an ABI Quant Studio tm 6 Flex Real-Time PCR System. The cycle parameters included 95°C for 3 min; 40 cycles at 95°C for 20 s, 58°C for 30 s, and 72°C for 30 s; 71 cycles increasing from 60°C to 95°C at 0.5°C per cycle for 30 s. FvActin (GenBank accession no. AB116565.1) was used as the internal reference to calculate the relative fold differences, and the data were analyzed using a previously described comparative CT method (2−ΔΔCt) (Su et al., 2015). A P-value < 0.05 was regarded as statistically significant. NCBI primer blast was used to design the primers for qRT-PCR (Suppl. Tab. S1) using F. vesca mRNA as the reference database.
Identification of Aux/IAA family genes in the strawberry genome
We identified and characterized 21 Aux/IAA genes in the F. vesca genome, which were labeled FveIAA1-21 based on their chromosomal localization. In-depth analysis of each predicted FveIAA, including chromosomal localization, gene length, deduced protein length, MW, pI, and exon numbers, was performed (Tab. 1). The FveIAA genes encode 181 (FveIAA16) to 370 amino acids (aa) (FveIAA3) with corresponding MWs of 20.45 to 39.84 kDa. The pI ranged from 5.23 (FveIAA5) to 8.61 (FveIAA10), indicating that different Aux/IAA proteins might function under different pH conditions. Sixteen FveIAAs were located in the nucleus, except FveIAA10, FveIAA11, FveIAA16, and FveIAA18, which were located in the chloroplasts; FveIAA12 was located in the mitochondria (Suppl. Tab. S2).
Table 1: Aux/IAA gene family in strawberry
Chromosomal location and duplications of FveIAA genes
The 21 identified FveIAA genes were distributed across five chromosomes, mainly on chromosomes 1, 2, 4, 5, and 6, but were not found on chromosomes 3 and 7 (Fig. 1). These genes did not show random distribution on the chromosome due to gene clusters and hot regions. The unequal distribution of FveIAA genes suggested genetic variation during the evolutionary process (Li et al., 2020a, Li et al., 2019; Li et al., 2015). Thus, the segmental duplication and tandem duplication events were investigated to explore potential gene duplication within the strawberry genome. Four duplicated gene pairs (FveIAA2 and FveIAA4, FveIAA3 and FveIAA15, FveIAA7 and FveIAA11, FveIAA8 and FveIAA20) of FveIAAs, all occurring on different chromosomes with a possibility of segmental duplication, were observed in F. vesca (Fig. 2A). We hypothesized that apart from expanding the FveIAA gene family size, these segmental gene duplications also increased their functional diversity. We constructed a comparative syntenic map of F. vesca and Arabidopsis to further explore the phylogenetic mechanism of the F. vesca IAA gene family. A syntenic relationship was discovered between four FveIAA genes and Arabidopsis (Fig. 2B), indicating the importance of these genes from the Aux/IAA gene family during the evolutionary process.

Figure 1: Chromosomal localization of FveIAA genes.

Figure 2: Segmentally duplicated gene pairs in F. vesca.
Multiple sequence alignment and conserved domains
Protein motif analysis by PFAM and sequence alignment revealed that 20 FveIAAs had four characteristic conserved domains: I–III and IX (Fig. 3).The domain I of most FveIAA proteins had a highly conserved LXLXLX motif, which served as a protein transcriptional repressor except FveIAA12, which was a non-canonical Aux/IAA protein missing domain I, also found in tomato and Medicago truncatula, indicating that these proteins may play specialized roles while mediating auxin response during plant growth and development. Most FveIAA proteins exhibited two distinct types of nuclear localization signals (NLS): A typical NLS (at the end of domain IV) and a bipartite NLS (between domains I and II).

Figure 3: Multiple sequence alignment of the FveIAA gene family.
The phylogeny of FveIAAs and AtIAAs
A phylogenetic NJ tree was built based on the complete sequence alignment of 21 FveIAAs and 29 AtIAAs to explore the phylogenetic relationship between the Aux/IAA proteins of F. vesca and A. thaliana (Suppl. Tabs. S3 and S4). Based on the phylogenetic distribution, IAA proteins were classified into nine major groups (labeled 1 to 9) with well-supported bootstrap values (Fig. 4). Also, we analyzed the identity of the predicted Aux/IAA protein sequences between Arabidopsis and strawberry predicted based on the orthologs between these two species. We found no organism-specific group from the tree. In the common clades, there was an unequal distribution of the IAAs from the two organisms. For instance, Group 4 contained one FveIAA and four AtIAAs. Also, there was an unequal distribution of the FveIAAs in the nine groups. Groups 1–6, 8, and 9 included twenty FveIAA members (largest), which were located in the nucleus and the chloroplast, while Group 7 contained one gene (FveIAA12), which was located in the mitochondria (Suppl. Tab. S2).

Figure 4: Phylogenetic relationships of IAA proteins between A. thaliana and F. vesca.
Conserved motifs and exon-intronic structures
We performed phylogenic and conserved motif analysis of the FveIAAs to explore the relationship between motifs and evolution and to identify the conserved regions (Fig. 5A). Twenty-one FveIAAs containing 2–10 conserved motifs were detected (Fig. 5B and Suppl. Fig. S1). Motifs 1 and 2 were common in all 21 FveIAAs, indicating that they were essential for basic functions of FveIAAs, while the other eight motifs were more or less missing from the FveIAAs. structure analysis showed that the gene structure of most of the FveIAA genes was identical to the AtIAAs, which included 3–5 exons and 2–4 introns, except for FveIAA15 (six exons and five introns) and FveIAA5 and FveIAA16 (each containing two exons and one intron) (Fig. 5C). Exon-intron structure analysis provides critical insights into the process of evolution of gene families. Motif composition, arrangements, and gene structures were consistent with the phylogenetic tree (Fig. 5).
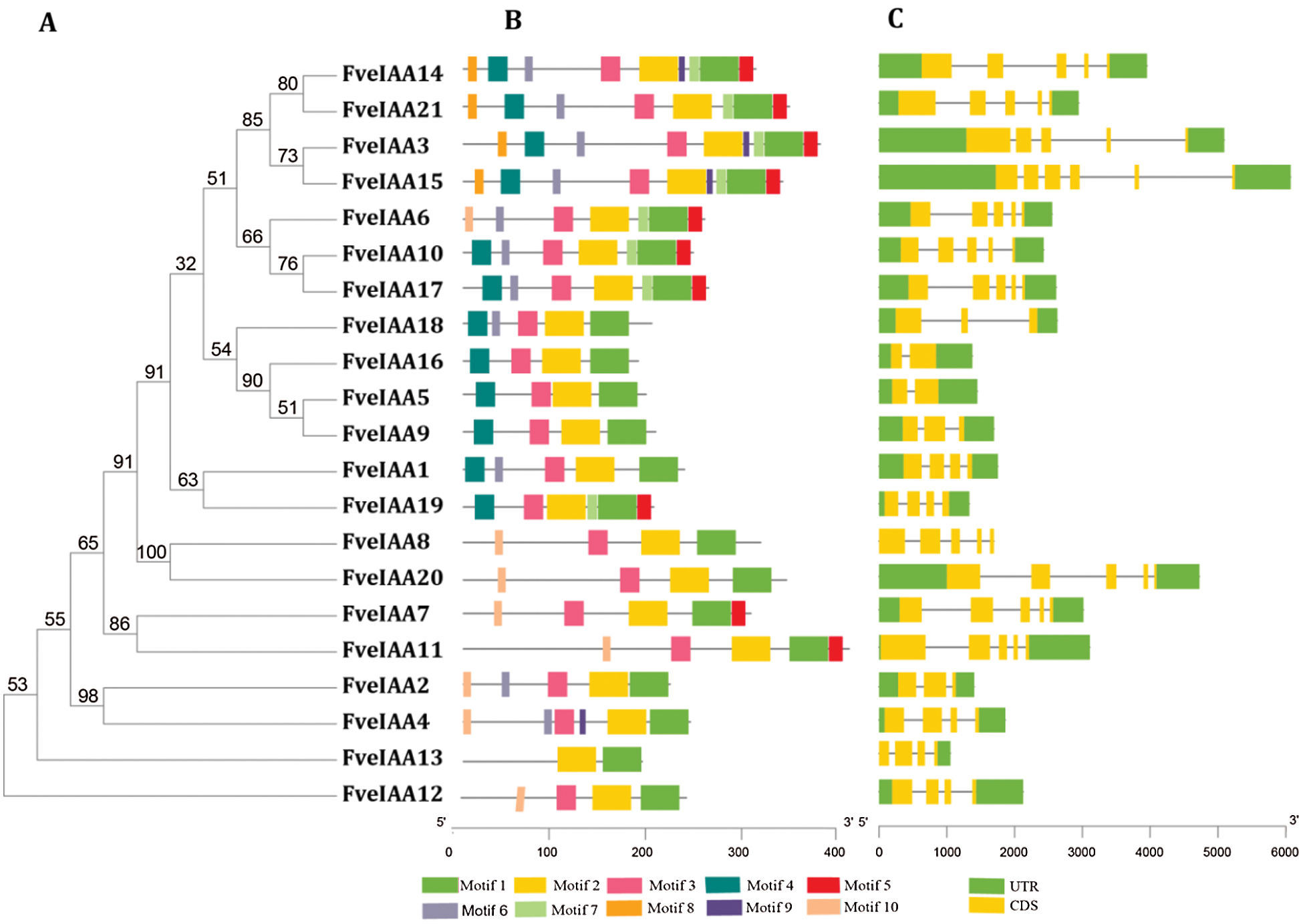
Figure 5: Phylogeny, motifs, and exon-intronic structures of FveIAAs.
Cis-element analysis and subcellular localization prediction
The interaction between cis-elements and the corresponding trans-acting factors is known to promote gene regulation. PlantCARE was used to determine the probable cis-regulatory elements within the promoter region of the FveIAA genes to understand possible regulatory patterns of the FveIAAs. We found that all 21 FveIAAs promoter sequences contained several light-responsive elements, indicating that FveIAAs played a critical role in strawberry morphogenesis. Additionally, we found hormonal response-related cis-regulatory elements, such as auxin, methyl jasmonate (MeJA), salicylic acid (SA), abscisic acid (ABA), and gibberellins (GA), as well as stress-responsive elements, including anaerobic induction, defense, drought, and low temperature in the promoter region of most FveIAA genes (Fig. 6). Some of the FveIAA genes contained tissue-specific elements (endosperm, meristem, and seed-specific activation) and circadian control elements.

Figure 6: Putative cis-regulatory elements in the promoter region of FveIAA genes.
Prediction of PPI networks of FveIAA family
A complete interaction network of the FveIAA family and its interacting proteins were predicted using the STRING tool to investigate the functions of the FveIAAs. Seventeen FveIAAs were identified in the interaction network except FveIAA4, FveIAA5, FveIAA6, and FveIAA20 (Fig. 7). Protein-protein relationship analysis revealed that most FveIAAs interacted with each other indicating collaborative functioning, such as in FveIAA1, FveIAA13, and FveIAA19. Also, the direct interaction between FveIAAs and the nodal proteins suggested the possibility of occasional indirect interaction between the FveIAAs, such as XP_004293901.1, a common core protein, induced indirect interaction between FveIAA19, FveIAA1, FveIAA15, FveIAA17, and FveIAA7 (Fig. 7). Functional annotation revealed that most of the common interacting proteins in the PPI network were involved in auxin-related signaling pathways (Suppl. Tab. S5).

Figure 7: Putative interaction network of FveIAA proteins in F. vesca.
Expression analyses of the Aux/IAA genes in F. vesca organs
We investigated the spatial-specific expression pattern of the 21 FveIAAs in different organs, including stems, roots, flowers, leaves, and fruits, to investigate the physiological function of FveIAAs (Fig. 8). All the FveIAAs were detected in different organs, except for FveIAA21, whose expression was downregulated in all organs. All the FveIAAs showed tissue-specific expression patterns in F. vesca. Most FveIAAs showed high stem-specific mRNA expression compared with other organs, except FveIAA4, FveIAA5, FveIAA13, and FveIAA18 whose expression level were upregulated in the fruit; FveIAA2, FveIAA9, FveIAA10, and FveIAA17 which showed root-specific expressions; FveIAA3, FveIAA11, and FveIAA16 also showed flower-specific expression; FveIAA1, FveIAA19, and FveIAA14, which showed leaf-specific expression. Transcriptional analysis of the FveIAAs showed tissue-specific expression in F. versa, suggesting that FveIAA genes might play distinct roles in different organs during strawberry development.

Figure 8: Heatmap of transcription profiling individual FveIAA genes in different organs.
We detected the expression pattern of FveIAAs in leaves at 1, 6, and 12 h of post-IAA treatment using qRT-PCR to test the responsiveness to exogenous auxin stimuli (Fig. 9). We found that except for FveIAA21, which was downregulated in all organs, all other genes were auxin-responsive. Exogenous auxin upregulated the expression of most FveIAA genes after 1 h and 6 h; however, the expression was restored to near pre-stress levels after 12 h in FveIAA2, FveIAA3, FveIAA5, FveIAA7, FveIAA8, FveIAA14, FveIAA16 and FveIAA18. Also, elevated expression of FveIAA4, FveIAA10, FveIAA13, FveIAA17 and FveIAA20 was observed at all time points, including 12 h after treatment. In contrast, the expression of FveIAA9, FveIAA11, and FveIAA15 were downregulated by auxin at every time point. Thus, the complexity of auxin-regulated gene expression was reflected in the diverse pattern of expression of the FveIAA genes post-treatment with auxin.

Figure 9: Relative expression of FveIAAs under auxin treatment.
Auxin, a plant hormone, is critical for plant growth and development (Liu et al., 2017; Mano and Nemoto, 2012; Tiryaki, 2009). Aux/IAAs regulate the transcription of auxin-responsive genes that are involved in variable aspects of plant growth and development (Golan et al., 2013; Liscum and Reed, 2002; Luo et al., 2018). Therefore, to elucidate the function of strawberry IAAs in stimulating specific auxin responses, we performed a genome-wide comprehensive survey of the Aux/IAA gene family in strawberry. In this study, 21 strawberry IAA genes were identified and labeled based on their chromosomal location. Fewer duplication resulted in fewer FveIAA genes compared with other species, such as Arabidopsis, tomato, rice, and maize (Audran-Delalande et al., 2012; Dreher et al., 2006; Jain et al., 2006; Wang et al., 2010) (Suppl. Tab. S6). Next, we assessed the conserved structural domains of the strawberry Aux/IAA proteins. The amino acid sequence analysis revealed four highly conserved domains between the FveIAA gene family and Arabidopsis, suggesting the possibility of similar functions.
Previous studies have suggested that phylogenetic analysis not only helps elucidate phylogenetic relationships but also predicts putative functions of various genes, which helps in the selection of candidate genes (Horton et al., 2007; Xu et al., 2018; Zhai et al., 2014). Here, the F. vesca Aux/IAA gene family members were divided into nine groups based on their sequence similarity. The phylogenetic tree between strawberry and Arabidopsis showed that all the FveIAAs had orthologs in Arabidopsis (Fig. 2). Comparative genome analysis of the Aux/IAA genes in F. vesca and Arabidopsis confirmed gene duplication (segmental duplication) in at least four pairs of FveIAA genes and the absence of tandem duplications. Segmental and tandem duplication events are critical for the expansion of the gene families (He et al., 2019; Kramer et al., 2004; Li et al., 2020b; Zhu et al., 2014). Typically, angiosperm evolution is associated with the whole-genome duplication events leading to the expansion of gene families (Kawai et al., 2014; Yamada et al., 2019). Also, the cis-element analysis confirmed the presence of cis-regulatory elements associated with hormone response, tissue-specific, and stress response on most of the strawberry Aux/IAA gene promoter sequences.
Plants are frequently exposed to abiotic and biological stresses, such as cold, desiccation, salinity, and hormones during the developmental stages (Kim et al., 2015; Ku et al., 2018; Mishra and Richa, 2016). One study reported that Aux/IAA as transcriptional regulators might promote Auxin signal transposition directly (Mano and Nemoto, 2012). We analyzed the promoter cis-elements of the FveIAA genes family and found that several hormone-responsive stress elements were present in the promoter region. Thus, we analyzed the expression of the FveIAA genes in the strawberry seedlings post-IAA treatment and found that most of the FveIAA genes were responsive to IAA treatment, despite the absence of corresponding cis-elements in some of the genes, indicating the possibility of indirect regulation of these genes. The results also substantiated the involvement of FveIAA in auxin signaling pathways.
Expression patterns of FveIAAs were investigated in different organs using real-time PCR to study their physiological functions, especially fruit development (Fig. 8). Some FveIAA genes showed organ-specific expression patterns, indicating their differential roles during strawberry development. FveIAA3, FveIAA4, FveIAA5, FveIAA11, FveIAA13, FveIAA16, and FveIAA18 showed preferential expression in flower and/or fruit, suggesting their importance in improving fruit-related agronomic traits in strawberry. Interestingly, during the fruit development and ripening stage, we observed a rapid increase in the transcription levels of FveIAA4 and FveIAA5 from the SG to the BG stage and maintained a high expression throughout the fruit ripening. While FveIAA13 and FveIAA18 showed the highest expression levels in the R stage (fruit maturity period). Previous studies have demonstrated the involvement of several Aux/IAA genes in the fruit developmental processes (Pattison et al., 2014); however, there is a scarcity of information on the regulation of fruit ripening regulation by Aux/IAA proteins, which needs to be further studied (Luo et al., 2018; Ori, 2019). Here, fruit-specific expression pattern and response to auxin indicated a novel role of these genes in regulating fruit development and ripening in strawberry. FveIAA21 exhibited a downregulated expression, which indicated it might have a different role during plant growth and development. Most FveIAA family genes showed higher expression in stem compared with other organs, indicating that Aux/IAAs could be vital for stem development.
Thus, we identified twenty-one putative candidate Aux/IAA genes in F. vesca in this study. FveIAAs were localized across five chromosomes of F. vesca and were divided into nine groups. All members had high homology and conserved domains, but they were different in a way. The study of the synteny analysis and phylogenetic relationships between F. vesca and Arabidopsis provided valuable information about the evolutionary characteristics of FveIAA genes. Protein motif architecture and PPI analysis indicated that FveIAA genes played a role in gene regulation and protein interaction net. Thus, FveIAA genes probably played an important role during strawberry development via the auxin signal transduction pathway. Based on tissue-specific expression and IAA treatment response, FveIAA4, FveIAA5, FveIAA13, and FveIAA18 were involved in fruit formation and ripening. These results would help decipher the biological roles of the Aux/IAA family in F. vesca.
Author Contribution: The authors confirm contribution to the paper as follows: Conceptualization: QL and LS; methodology: XZ and XL; software: Formal analysis, LS, HY and JZ; data curation: QL; writing—original draft preparation: LS and QL; writing—review and editing: QL; project administration: QL. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analysed during this study are included in this published article and its supplementary information files.
Ethics Approval: The experiments involved in this manuscript have no ethics related issues.
Supplementary Material: The supplementary material is available online at DOI 10.32604/biocell.2021.014499.
Funding Statement: The work was supported by the Natural Science Foundation of China (31701935), the Agricultural technology R&D Project of Xi’an City (20NYYF0037), the Natural Science Foundation of Chongqing (cstc2020jcyj-msxmX1064). We also gratefully thank funding from the Plant Biotechnology and Germplasm Conservation Project (XAWLKYTD017) and Key Disciplines of Botany of Xi’an City (103060002).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Abbas M, Hernandez-Garcia J, Blazquez MA, Alabadi D (2016). Reduction of IAA methyltransferase activity compensates for high-temperature male sterility in Arabidopsis. New Biotechnology 33: 435. DOI 10.1016/j.nbt.2015.10.058. [Google Scholar] [CrossRef]
Aloni R, Aloni E, Langhans M, Ullrich CI (2006). Role of auxin in regulating Arabidopsis flower development. Planta 223: 315–328. DOI 10.1007/s00425-005-0088-9. [Google Scholar] [CrossRef]
An JY, Lai J, Sajjanhar A, Batra J, Wang CW, Nelson CC (2015). J-Circos: An interactive Circos plotter. Bioinformatics 31: 1463–1465. DOI 10.1093/bioinformatics/btu842. [Google Scholar] [CrossRef]
Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M (2012). Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant and Cell Physiology 53: 659–672. DOI 10.1093/pcp/pcs022. [Google Scholar] [CrossRef]
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren JY, Li WW, Noble WS (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Research 37: W202–W208. DOI 10.1093/nar/gkp335. [Google Scholar] [CrossRef]
Bassa C, Mila I, Bouzayen M, Audran-Delalande C (2012). Phenotypes associated with down-regulation of Sl-IAA27 support functional diversity among Aux/IAA family members in tomato. Plant and Cell Physiology 53: 1583–1595. DOI 10.1093/pcp/pcs101. [Google Scholar] [CrossRef]
Cakir B, Kilickaya O, Olcay AC (2013). Genome-wide analysis of Aux/IAA genes in Vitis vinifera: Cloning and expression profiling of a grape Aux/IAA gene in response to phytohormone and abiotic stresses. Acta Physiologiae Plantarum 35: 365–377. DOI 10.1007/s11738-012-1079-7. [Google Scholar] [CrossRef]
Deng W, Yang YW, Ren ZX, Audran-Delalande C, Mila I, Wang XY, Song HL, Hu YH, Bouzayen M, Li ZG (2012). The tomato SlIAA15 is involved in trichome formation and axillary shoot development. New Phytologist 194: 379–390. DOI 10.1111/j.1469-8137.2012.04053.x. [Google Scholar] [CrossRef]
Dreher KA, Brown J, Saw RE, Callis J (2006). The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714. DOI 10.1105/tpc.105.039172. [Google Scholar] [CrossRef]
Eddy SR (1998). Profile hidden Markov models. Bioinformatics 14: 755–763. DOI 10.1093/bioinformatics/14.9.755. [Google Scholar] [CrossRef]
Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E (2006). A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proceedings of the National Academy of Sciences of the United States of America 103: 236–241. DOI 10.1073/pnas.0507127103. [Google Scholar] [CrossRef]
Farcot E, Lavedrine C, Vernoux T (2015). A modular analysis of the auxin signalling network. PLoS One 10: e0122231. DOI 10.1371/journal.pone.0122231. [Google Scholar] [CrossRef]
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A (2016). The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Research 44: D279–D285. DOI 10.1093/nar/gkv1344. [Google Scholar] [CrossRef]
Fukaki H, Tameda S, Masuda H, Tasaka M (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant Journal 29: 153–168. DOI 10.1046/j.0960-7412.2001.01201.x. [Google Scholar] [CrossRef]
Gan DF, Zhuang D, Ding F, Yu ZZ, Zhao Y (2013). Identification and expression analysis of primary auxin-responsive Aux/IAA gene family in cucumber (Cucumis sativus). Journal of Genetics 92: 513–521. DOI 10.1007/s12041-013-0306-3. [Google Scholar] [CrossRef]
Golan G, Betzer R, Wolf S (2013). Phloem-specific expression of a melon Aux/IAA in tomato plants alters auxin sensitivity and plant development. Frontiers in Plant Science 4: 329. DOI 10.3389/fpls.2013.00329. [Google Scholar] [CrossRef]
Goodstein DM, Shu SQ, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Research 40: D1178–D1186. DOI 10.1093/nar/gkr944. [Google Scholar] [CrossRef]
He YR, Jia RR, Qi JJ, Chen SC, Lei TG, Xu LZ, Peng AH, Yao LX, Long Q, Li ZG, Li Q (2019). Functional analysis of citrus AP2 transcription factors identified CsAP2-09 involved in citrus canker disease response and tolerance. Gene 707: 178–188. DOI 10.1016/j.gene.2019.04.021. [Google Scholar] [CrossRef]
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007). WoLF PSORT: Protein localization predictor. Nucleic Acids Research 35: W585–W587. DOI 10.1093/nar/gkm259. [Google Scholar] [CrossRef]
Hu B, Jin JP, Guo AY, Zhang H, Luo JC, Gao G (2015a). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31: 1296–1297. DOI 10.1093/bioinformatics/btu817. [Google Scholar] [CrossRef]
Hu W, Zuo J, Hou XW, Yan Y, Wei YX, Liu JH, Li MY, Xu BY, Jin ZQ. (2015b). The auxin response factor gene family in banana: Genome-wide identification and expression analyses during development, ripening, and abiotic stress. Frontiers in Plant Science 6: 742. DOI 10.3389/fpls.2015.00742. [Google Scholar] [CrossRef]
Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP (2006). Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Functional & Integrative Genomics 6: 47–59. DOI 10.1007/s10142-005-0005-0. [Google Scholar] [CrossRef]
Kalluri UC, Difazio SP, Brunner AM, Tuskan GA (2007). Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biology 7: 59. DOI 10.1186/1471-2229-7-59. [Google Scholar] [CrossRef]
Kang CY, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu ZC (2013). Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25: 1960–1978. DOI 10.1105/tpc.113.111732. [Google Scholar] [CrossRef]
Kawai Y, Ono E, Mizutani M (2014). Expansion of specialized metabolism-related superfamily genes via whole genome duplications during angiosperm evolution. Plant Biotechnology 31: 579–584. DOI 10.5511/plantbiotechnology.14.0901a. [Google Scholar] [CrossRef]
Kim J, Harter K, Theologis A (1997). Protein-protein interactions among the Aux/IAA proteins. Proceedings of the National Academy of Sciences of the United States of America 94: 11786–11791. DOI 10.1073/pnas.94.22.11786. [Google Scholar] [CrossRef]
Kim JM, Sasaki T, Ueda M, Sako K, Seki M (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Frontiers in Plant Science 6: 114. DOI 10.3389/fpls.2015.00114. [Google Scholar] [CrossRef]
Knox K, Grierson CS, Leyser O (2003). AXR3 and SHY2 interact to regulate root hair development. Development 130: 5769–5777. DOI 10.1242/dev.00659. [Google Scholar] [CrossRef]
Kramer EM, Jaramillo MA, di Stilio VS (2004). Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166: 1011–1023. DOI 10.1534/genetics.166.2.1011. [Google Scholar] [CrossRef]
Ku YS, Sintaha M, Cheung MY, Lam HM (2018). Plant hormone signaling crosstalks between biotic and abiotic stress responses. International Journal of Molecular Sciences 19: 3206. DOI 10.3390/ijms19103206. [Google Scholar] [CrossRef]
Kumar S, Stecher G, Tamura K (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. DOI 10.1093/molbev/msw054. [Google Scholar] [CrossRef]
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30: 325–327. DOI 10.1093/nar/30.1.325. [Google Scholar] [CrossRef]
Li Q, Dou WF, Qi JJ, Qin XJ, Chen SC, He YR (2020a). Genomewide analysis of the CIII peroxidase family in sweet orange (Citrus sinensis) and expression profiles induced by Xanthomonas citri subsp. citri and hormones. Journal of Genetics 99: 10. DOI 10.1007/s12041-019-1163-5. [Google Scholar] [CrossRef]
Li Q, Hu AH, Dou WF, Qi JJ, Long Q, Zou XP, Lei TG, Yao LX, He YR, Chen SC (2019). Systematic analysis and functional validation of Citrus XTH genes reveal the role of Csxth04 in Citrus bacterial canker resistance and tolerance. Frontiers in Plant Science 10: 1109. DOI 10.3389/fpls.2019.01109. [Google Scholar] [CrossRef]
Li Q, San Clemente H, He YR, Fu YY, Dunand C (2020b). Global evolutionary analysis of 11 gene families part of Reactive Oxygen Species (ROS) gene network in four Eucalyptus species. Antioxidants 9: 257. DOI 10.3390/antiox9030257. [Google Scholar] [CrossRef]
Li Q, Yu H, Cao PB, Fawal N, Mathe C, Azar S, Cassan-Wang H, Myburg AA, Grima-Pettenati J, Marque C, Teulieres C, Dunand C (2015). Explosive tandem and segmental duplications of multigenic families in Eucalyptus grandis. Genome Biology and Evolution 7: 1068–1081. DOI 10.1093/gbe/evv048. [Google Scholar] [CrossRef]
Liscum E, Reed JW (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Molecular Biology 49: 387–400. DOI 10.1023/A:1015255030047. [Google Scholar] [CrossRef]
Liu KD, Yuan CC, Feng SX, Zhong ST, Li HL, Zhong JD, Shen CJ, Liu JX (2017). Genome-wide analysis and characterization of Aux/IAA family genes related to fruit ripening in papaya (Carica papaya L.). BMC Genomics 18: 351. DOI 10.1186/s12864-017-3722-6. [Google Scholar] [CrossRef]
Luo J, Zhou JJ, Zhang JZ (2018). Aux/IAA gene family in plants: Molecular structure, regulation, and function. International Journal of Molecular Sciences 19: 259. DOI 10.3390/ijms19010259. [Google Scholar] [CrossRef]
Mai YX, Wang L, Yang HQ (2011). A gain-of-function mutation in IAA7/AXR2 confers late flowering under short-day light in Arabidopsis. Journal of Integrative Plant Biology 53: 480–492. DOI 10.1111/j.1744-7909.2011.01050.x. [Google Scholar] [CrossRef]
Mano Y, Nemoto K (2012). The pathway of auxin biosynthesis in plants. Journal of Experimental Botany 63: 2853–2872. DOI 10.1093/jxb/ers091. [Google Scholar] [CrossRef]
Mattsson J, Ckurshumova W, Berleth T (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiology 131: 1327–1339. DOI 10.1104/pp.013623. [Google Scholar] [CrossRef]
Mishra BS, Singh M, Aggrawal P, Laxmi A (2009). Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. PLoS One 4: e4502. DOI 10.1371/journal.pone.0004502. [Google Scholar] [CrossRef]
Mishra RC, Richa GA (2016). Constitutive over-expression of rice ClpD1 protein enhances tolerance to salt and desiccation stresses in transgenic Arabidopsis plants. Plant Science 250: 69–78. DOI 10.1016/j.plantsci.2016.06.004. [Google Scholar] [CrossRef]
Ni J, Wang GH, Zhu ZX, Zhang HH, Wu YR, Wu P (2011). OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice. Plant Journal 68: 433–442. DOI 10.1111/j.1365-313X.2011.04698.x. [Google Scholar] [CrossRef]
Nitsch JP (1950). Growth and morphogenesis of the strawberry as related to auxin. American Journal of Botany 37: 211–215. DOI 10.1002/j.1537-2197.1950.tb12183.x. [Google Scholar] [CrossRef]
Ori N (2019). Dissecting the biological functions of ARF and Aux/IAA genes. Plant Cell 31: 1210–1211. DOI 10.1105/tpc.19.00330. [Google Scholar] [CrossRef]
Pattison RJ, Csukasi F, Catala C (2014). Mechanisms regulating auxin action during fruit development. Physiologia Plantarum 151: 62–72. DOI 10.1111/ppl.12142. [Google Scholar] [CrossRef]
Retzer K, Butt H, Korbei B, Luschnig C (2014). The far side of auxin signaling: Fundamental cellular activities and their contribution to a defined growth response in plants. Protoplasma 251: 731–746. DOI 10.1007/s00709-013-0572-1. [Google Scholar] [CrossRef]
Schultz J, Milpetz F, Bork P, Ponting CP (1998). SMART, a simple modular architecture research tool: Identification of signaling domains. Proceedings of the National Academy of Sciences of the United States of America 95: 5857–5864. DOI 10.1073/pnas.95.11.5857. [Google Scholar] [CrossRef]
Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, Burns P, Davis TM, Slovin JP, Bassil N, Hellens RP, Evans C, Harkins T, Kodira C, Desany B, Crasta OR, Jensen RV, Allan AC, Michael TP, Setubal JC, Celton JM, Rees DJG, Williams KP, Holt SH, Rojas JJR, Chatterjee M, Liu B, Silva H, Meisel L, Adato A, Filichkin SA, Troggio M, Viola R, Ashman TL, Wang H, Dharmawardhana P, Elser J, Raja R, Priest HD, Bryant DW, Fox SE, Givan SA, Wilhelm LJ, Naithani S, Christoffels A, Salama DY, Carter J, Girona EL, Zdepski A, Wang WQ, Kerstetter RA, Schwab W, Korban SS, Davik J, Monfort A, Denoyes-Rothan B, Arus P, Mittler R, Flinn B, Aharoni A, Bennetzen JL, Salzberg SL, Dickerman AW, Velasco R, Borodovsky M, Veilleux RE, Folta KM (2011). The genome of woodland strawberry (Fragaria vesca). Nature Genetics 43: 109–116. DOI 10.1038/ng.740. [Google Scholar] [CrossRef]
Su L, Diretto G, Purgatto E, Danoun S, Zouine M, Li Z, Roustan JP, Bouzayen M, Giuliano G, Chervin C (2015). Carotenoid accumulation during tomato fruit ripening is modulated by the auxin-ethylene balance. BMC Plant Biology 15: 114. DOI 10.1186/s12870-015-0495-4. [Google Scholar] [CrossRef]
Su LY, Bassa C, Audran C, Mila I, Cheniclet C, Chevalier C, Bouzayen M, Roustan JP, Chervin C (2014). The Auxin Sl-IAA17 transcriptional repressor controls fruit size via the regulation of endoreduplication-related cell expansion. Plant and Cell Physiology 55: 1969–1976. DOI 10.1093/pcp/pcu124. [Google Scholar] [CrossRef]
Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, Radenbaugh A, Singh S, Swing V, Tissier C, Zhang P, Huala E (2008). The Arabidopsis Information Resource (TAIRGene structure and function annotation. Nucleic Acids Research 36: D1009–D1014. DOI 10.1093/nar/gkm965. [Google Scholar] [CrossRef]
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C (2017). The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Research 45: D362–D368. DOI 10.1093/nar/gkw937. [Google Scholar] [CrossRef]
Thakur JK, Tyagi AK, Khurana JP (2001). OsIAA1, an Aux/IAA cDNA from rice, and changes in its expression as influenced by auxin and light. DNA Research 8: 193–203. DOI 10.1093/dnares/8.5.193. [Google Scholar] [CrossRef]
Timpte C, Wilson AK, Estelle M (1994). The Axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 138: 1239–1249. DOI 10.1093/genetics/138.4.1239. [Google Scholar] [CrossRef]
Tiryaki I (2009). Biosynthesis, metabolism and signaling pathway of auxin in plants. Philippine Agricultural Scientist 92: 243–253. [Google Scholar]
Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. DOI 10.1105/tpc.9.11.1963. [Google Scholar] [CrossRef]
Voorrips RE (2002). MapChart: Software for the graphical presentation of linkage maps and QTLs. Journal of Heredity 93: 77–78. DOI 10.1093/jhered/93.1.77. [Google Scholar] [CrossRef]
Wang H, Jones B, Li ZG, Frasse P, Delalande C, Regad F, Chaabouni S, Latche A, Pech JC, Bouzayen M (2005). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17: 2676–2692. DOI 10.1105/tpc.105.033415. [Google Scholar] [CrossRef]
Wang YJ, Deng DX, Bian YL, Lv YP, Xie Q (2010). Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays L.). Molecular Biology Reports 37: 3991–4001. DOI 10.1007/s11033-010-0058-6. [Google Scholar] [CrossRef]
Wang YP, Li JP, Paterson AH (2013). MCScanX-transposed: Detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 29: 1458–1460. DOI 10.1093/bioinformatics/btt150. [Google Scholar] [CrossRef]
Wu J, Peng Z, Liu SY, He YJ, Cheng L, Kong FL, Wang J, Lu G (2012). Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Molecular Genetics and Genomics 287: 295–311. DOI 10.1007/s00438-012-0675-y. [Google Scholar] [CrossRef]
Xu C, Ye H, Qiu W, Lin H, Chen Y, Zhang H, Liao M (2018). Phylogenetic classification of hemagglutinin gene of H9N2 avian influenza viruses isolated in China during 2012–2016 and evaluation of selected candidate vaccine strains. Poultry Science 97: 3023–3030. DOI 10.3382/ps/pey154. [Google Scholar] [CrossRef]
Yamada K, Davydov BII, Salamin GN (2019). Duplication history and molecular evolution of the rbcS multigene family in angiosperms. Journal of Experimental Botany 70: 6127–6139. DOI 10.1093/jxb/erz363. [Google Scholar] [CrossRef]
Zhai W, Zhao Y, Zhang LX, Li XJ (2014). Structural and phylogenetic analysis of Pto-type disease resistance gene candidates in Hevea brasiliensis. Genetics and Molecular Research 13: 4348–4360. DOI 10.4238/2014.June.10.2. [Google Scholar] [CrossRef]
Zhu Y, Wu NN, Song WL, Yin GJ, Qin YJ, Yan YM, Hu YK (2014). Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biology 14: DOI 10.1186/1471–2229-14-93. [Google Scholar]
Zhu ZX, Liu Y, Liu SJ, Mao CZ, Wu YR, Wu P (2012). A gain-of-function mutation in OsIAA11 affects lateral root development in rice. Molecular Plant 5: 154–161. DOI 10.1093/mp/ssr074. [Google Scholar] [CrossRef]
Supplementary materials
Figure S1. The motif weblog visualization of FveIAAs. MEME was used to elucidate and visualize the conserved motifs of FveIAAs. The different colors of amino acids are classified according to the chemical properties of amino acids.
Table S1. The primers used in this study.
Table S2. Subcellular localization analysis of FveIAAs.
Table S3. List and sequences of FveIAAs annotated in this study.
Table S4. List and sequences of AtIAAs for the phylogenic analysis.
Table S5. List of interacting proteins of FveIAAs.
Table S6. Summary of IAA numbers in some species.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |