DOI:10.32604/biocell.2021.015282

www.techscience.com/journal/biocell
| Biocell DOI:10.32604/biocell.2021.015282 |  www.techscience.com/journal/biocell |
| Article |
Genome-wide identification of WRKY gene family and expression analysis under abiotic stresses in Andrographis paniculata
1Zhejiang Province Key Laboratory of Plant Secondary Metabolism and Regulation, College of Life Sciences and Medicine, Zhejiang Sci-Tech University, Hangzhou, 310018, China
2Department of Agronomy, University of Agriculture, Faisalabad, 38040, Pakistan
3Tianjin Tasly Modern TCM Resources Co., Ltd., Tianjin, 300410, China
*Address correspondence to: Ling Xu, lxu@zstu.edu.cn; Zongsuo Liang, liangzs@zstu.edu.cn
Received: 06 December 2020; Accepted: 18 January 2021
Abstract: Andrographis paniculata (A. paniculata) is a Chinese herbal medicine that clears away heat, reduces inflammation, protects the liver, and promotes choleretics. The WRKYs of A. paniculata are still not well characterized, although many WRKYs have been identified in various plant species. In the present study, 59 A. paniculata WRKY (ApWRKY) genes were identified and renamed on the basis of their respective chromosome distribution. These ApWRKYs were divided into three groups via phylogenetic analysis according to their WRKY domains and combined with WRKY of Arabidopsis. The 59 identified ApWRKY transcription factors were non-uniformity distributed on 23 chromosomes of A. paniculata. From the structural analysis of the conserved motifs, different ApWRKYs structures showed different biological functions, and the ApWRKY transcription factor had certain species-specificity in the evolutionary process. The expression patterns of the 41 ApWRKYs were examined through quantitative real-time PCR (qRT-PCR) in various tissues and under abiotic stresses (salt). The results showed that most of the ApWRKY had different reactions to salt treatment. In addition, the content of the four main secondary metabolites in A. paniculata leaves was determined under salt stress. The results show that under a low concentration of salt treatment, the synthesis of andrographolide can be improved.
Keywords: A. paniculata; ApWRKY; Secondary metabolites; Diterpene lactones; Andrographolide; Abiotic stress
Andrographis paniculata (Burm. f.) Nees is an herbaceous plant, which is commonly known as the ‘King of Bitters’ in the Acanthaceae family. A. paniculata is wildly distributed in Southern Asia (Dai et al., 2019) and widely distributed in Southern China. In China, A. paniculata, also known as Chuanxinlian in Chinese, is used for medicine for the dry ground part. Andrographolide, a natural diterpene lactone, is the main active compound distributed in A. paniculata (Zhang et al., 2019). It contains the active phytochemicals from the aerial parts (leaves and stems), such as diterpenoids and 2’-oxygenated flavonoids, including andrographolide (AP), neoandrographolide (NAP), 14-deoxy-11,12didehydroandrographolide (DHAP), 14-deoxyandrographolide (DOAP), isoandrographolide, homoandrographolide, rographolidegraphan, andrographosterin, and stigmasterol (Chao and Lin, 2010).
In Arabidopsis, WRKY is almost as complicated as the well-known transcription factor families such as myeloblastosis (MYB) and NAC. Transcription factors can be involved in regulating various developmental and physiological processes of plants by directly or indirectly binding cis-acting elements of genes involved in signal transduction process of stress.
Abiotic stress responses and gene regulation have been studied in a number of plant species, including Arabidopsis, rice, maize, and tomato. Several families of genes are particularly associated with significant improvements in abiotic stress tolerance, including the WRKY, NAC, and ethylene response factor (ERF) gene families (Shen et al., 2017, Huang et al., 2015, Fan et al., 2016). Previous studies have demonstrated that WRKY genes are expressed strongly and rapidly in response to particular abiotic stresses, including wounding, waterlogging, drought, and salt stress (Madhunita and Ralf, 2014). As the seventh largest transcription factor family in higher plants, WRKY is generally involved in various biological processes, including plant growth and development as well as biological and abiotic stress responses (Zhang and Wang, 2005). Therefore, the analysis of the function of the WRKY transcription factor has always been one of the hotspots in the study of plant functional genes. SPF1 protein isolated from sweet potato was the first WRKY protein discovered (Ishiguro and Nakamura, 1994). Subsequently, researchers detected numerous WRKY family members in crops, including Arabidopsis, rapeseed, and rice, among which 74 and 126 WRKY family members were detected in Arabidopsis and rice, respectively (Berri et al., 2009).
The genome size of A. paniculata is 269 Mb, gene annotation predicted 25,428 protein-coding genes. Full genome sequencing of A. paniculata was completed in 2019 (Sun et al., 2019). However, to date, no genome-wide characterization of the WRKY family has been conducted in A. paniculata. So, it is of the utmost interest to carry out a genome-wide survey of this gene family in A. paniculata. This study focused on the whole genome-wide identification and expression analyses of WRKY in A. paniculata under abiotic stresses. The phylogenetic relationship of the WRKY proteins was investigated according to the WRKY information from Arabidopsis and rice. The genomic structure, chromosome localization, conserved domain, gene structure, and other structural features were also explored. However, 41 WRKY genes were selected for analyzing their expression patterns in response to abiotic stresses using quantitative real-time PCR. The objective of this study was to establish a solid foundation for understanding the regulatory mechanism of WRKY genes and to explore new strategies for the improvement in A. paniculata.
Identification and sequence analysis of WKRY genes in A. paniculata
The A. paniculata genome sequences were downloaded from a database (Sun et al., 2019). The WRKY sequence data of Arabidopsis were obtained from TAIR release 10 http://plants.ensembl.org/Arabidopsis_thaliana/Info/Index. A Hidden Markov Model (HMM) search was performed against the A. paniculata protein database using the WRKY-domain PF03106 http://hmmer.org/ An e-value ≤ 1.1e-25 was used as the criterion, The primary candidate sequences were submitted in the Pfam database http://pfam.xfam.org and NCBI Conserved Domains Database https://www.ncbi.nlm.nih.gov/cdd for verification. Finally, 59 WRKY proteins were screened out in the A. paniculata genome (Tab. 1). The isoelectric point and protein molecular weight of WRKY proteins in A. paniculata were predicted using the pI/Mw tool in ExPASy proteomics server http://web.expasy.org/protparam.
Multiple sequence alignment and phylogenetic analysis
A ClustalW analysis was used with the default option to implement the multiple sequence alignment on the WRKY domain sequences of the WRKY genes from two species, namely, A. paniculata and Arabidopsis. A Maximum Likelihood (ML) phylogenetic tree in MEGA X with the following parameters: Use all sites and bootstrap analysis with 1000 replicates for the reliability of interior branches (Kumar et al., 2018). The sequences for phylogenetic trees are shown in Suppl. Tab. S1.
Chromosomal locations of the ApWRKY genes
The location of ApWRKY was obtained by using Perl script to extract mRNA locations from A. paniculata GFF files, and Mapchart2.32 software was used to map to 24 chromosomes in the A. paniculata genome (Voorrips, 2002).
Protein properties and sequence analysis
Fifty-nine ApWRKYs were analyzed by MEME online software for conversed motif prediction with the following criteria (http://meme-suite.org/tools/meme): Select the site distribution: Zoops, 20 motifs with an optimum motif width between 10 and 50 residues, and any number of repetitions. The location information of CDS and UTR in the mRNA of ApWRKYs was screened from the GFF file of the whole genome of A. paniculata, and the gene structure was identified.
Plant materials and abiotic stress treatments
The medicinal plant A. paniculata was obtained from Zhejiang Province, China (30.26N,120.21E.) The plant material used in this study was formally identified by Dr. Ling Xu (Zhejiang Sci-Tech University, Hangzhou, China) and Prof. Zongsuo Liang (Zhejiang Sci-Tech University, Hangzhou, China). A. paniculata seeds were sown in plastic pots (2:1:1 ratio) of nutritive soil, perlite, and vermiculite, and are grown in normal sunlight at an average temperature of 30°C. Three-month-old plants were employed for the abiotic stress treatments, as previously described (Talei et al., 2013). A total of 9 pots were cultivated, one plant for each pot, including 3 pots in the control group, 3 pots treated with 100 mL 50 mmol/L NaCl, and 3 pots treated with 100 mL 100 mmol/L NaCl. After 9 days of treatment, the roots, stems, and leaves were sampled. Samples were snap-frozen in liquid nitrogen and stored at −80°C until they were employed for total RNA extraction. All experiments were repeated three times.
Total RNA extraction and cDNA synthesis
RNA was extracted using the TIANGEN kit, for tissue-specific expression, RNA was extracted from different tissues, including the roots, stems, and leaves. RNA integrity was examined through agarose gel electrophoresis, and RNA purity was determined based on the OD260/OD280 and OD260/OD230 nm ratios. The RNA extracted was reversely transcribed into cDNA using the TaKaRa (Dalian, China) reverse transcription kit. The cDNA was then diluted 10-fold to be employed as a template for qRT-PCR analysis.
Transcriptional analysis by real-time quantitative PCR (RT-qPCR)
RT-qPCRs were performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme, China) and carried out in triplicate for each tissue sample. Gene-specific primers (Suppl. Tab. S2) were designed using Real-time PCR (TaqMan) Primer and Probes Design Tool https://www.genscript.com/tools/real-time-pcr-taqman-primer-design-tool. The length of amplicons is between 150 bp and 200 bp. ApUBC was selected as a reference gene as described previously (Li et al., 2013). We selected 59 genes for RT-qPCR experiments, of which 18 genes were expressed very low. So, we used the remaining 41 genes for mapping. Three independent biological replicates were performed. The obtained cDNA was used as a template for the RT-qPCR analysis using the QuantStudio TM Flex6 System (ABI, Alexandria, America). Briefly, standardization of gene expression data was performed from three biological replicates as described. The 2−ΔΔCt method was used to achieve results for relative quantification. For statistical analysis, analysis of variance (ANOVA) was done calculated using SPSS (Version 20.0, IBM, USA).
Metabolite extraction and HPLC analysis
The whole plant was treated with NaCl, and the contents of andrographolide (AP, NAP, DHAP and DOAP) under different salt stress concentrations were determined by HPLC (Fig. 7). The extraction and analysis of secondary metabolites were carried out according to the following methods (Bindu et al., 2020), and improvements were made on this basis. Four diterpenoid lactones (AP, NAP, DHAP and DOAP) were investigated. Preparation of test solution: 0.2 g of A. paniculata powder (passed through a 60-mesh sieve) was accurately weighed, placed in a stoppered Erlenmeyer flask, filled with 10 mL of methanol, tightly stoppered, weighed, immersed for 30 min, and then ultrasonically extracted for 30 min, when the solution temperature reaches room temperature, make up the weight with methanol, centrifuge, take the supernatant, pass through a 0.22 μm filter membrane, and continue the filtrate as the test solution. Waters HPLC system (Milford, MA, USA) contained a 1525 binary pump, an automatic sample injector, and a Waters 2998 photodiode array detector (PDA). HPLC separation was performed with a SunFire C18 column (4.6 mm × 250 mm, 5 µm particle size) at 30°C. Empower 3 software (Milford, MA, USA) was used for data acquisition and analysis. The sample injection volume was 20 µL; the PDA detection wavelength for the diterpenoids was 255 nm. Separation was achieved by elution using a linear gradient with solvent-A (acetonitrile) and solvent-B (0.4% phosphoric acid solution) gradient elution: 0–35 min, 5–45% A(v/v), 35–60 min, 45–52% A (v/v), 60–62 min, 52–5% A (v/v) flow rate: 1 mL/min. Standards of secondary metabolite compounds were purchased from Shanghai Bio-Technology Co., Ltd., Shanghai, China.
Identification and classification of WRKY gene family of A. paniculata
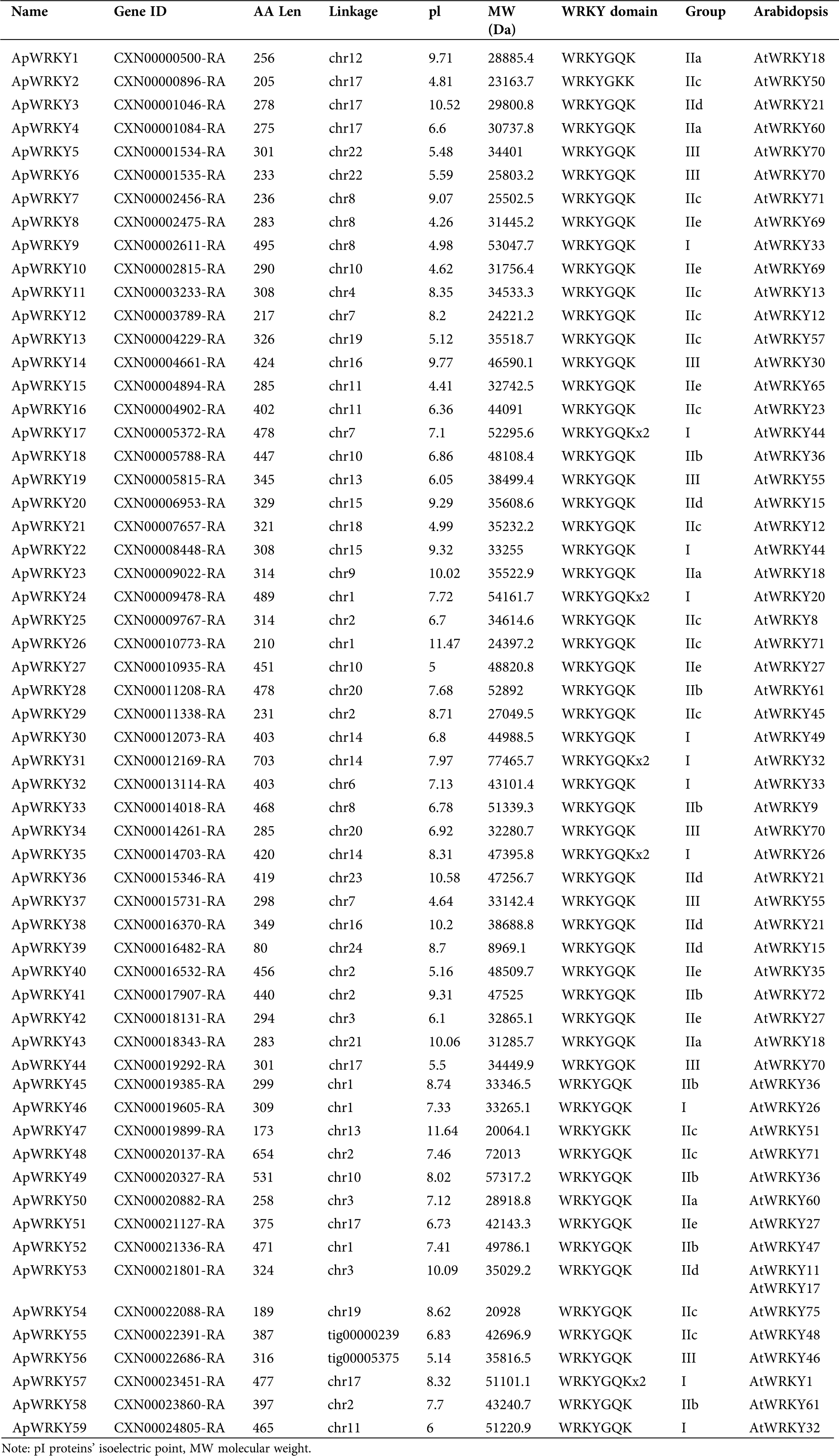
In total, 67 WRKY genes were predicted protein sequences without a WRKY domain were excluded. After comparison to a Hidden Markov Model (HMM) search using the WRKY-domain PF03106, a total of 59 WRKY proteins were identified in the A. paniculata genome. The 59 A. paniculata WRKY proteins ranged from 80 (ApWRKY39) to 703 (ApWRKY31) amino acid (aa) in length, with an average length of approximately 352 aa. The molecular weights (MWs) ranged from 8969.9 Da (ApWRKY39) to 77465.7 Da (ApWRKY31). The isoelectric points (pIs) of the WKRY proteins ranged from 4.26 (ApWRKY8) to 11.64 (ApWRKY47), with 26 pIs <7 and the remaining pIs >7 (Tab. 1). Similar observations were made in sesame (Li et al., 2017), which showed 71 SiWRKYs with pIs ranging from 4.81 to 9.74 and MWs ranging from 14.44 kDa to 125.94 kDa. Total 59 ApWRKY proteins contained one or two identical WRKYGQK domains (Tab. 1). Although the WRKYGQK domain is highly conserved in WRKY protein, ApWRKY2 and ApWRKY47 differ from other ApWRKYs in that glutamine residues are replaced by lysine residues (Meng et al., 2016, Eulgem et al., 2000, He et al., 2012). This change was also observed in Arabidopsis and another plant WRKY family.
Table 1: Sequence features of ApWRKYs in A. paniculata
Phylogenetic analysis of the ApWRKY genes
To further analyze the evolution of these ApWRKYs, we constructed a phylogenetic tree based on a total of 131 WRKYs (72 from A. thaliana and 59 from A. paniculata distributed in different groups) (Fig. 1).
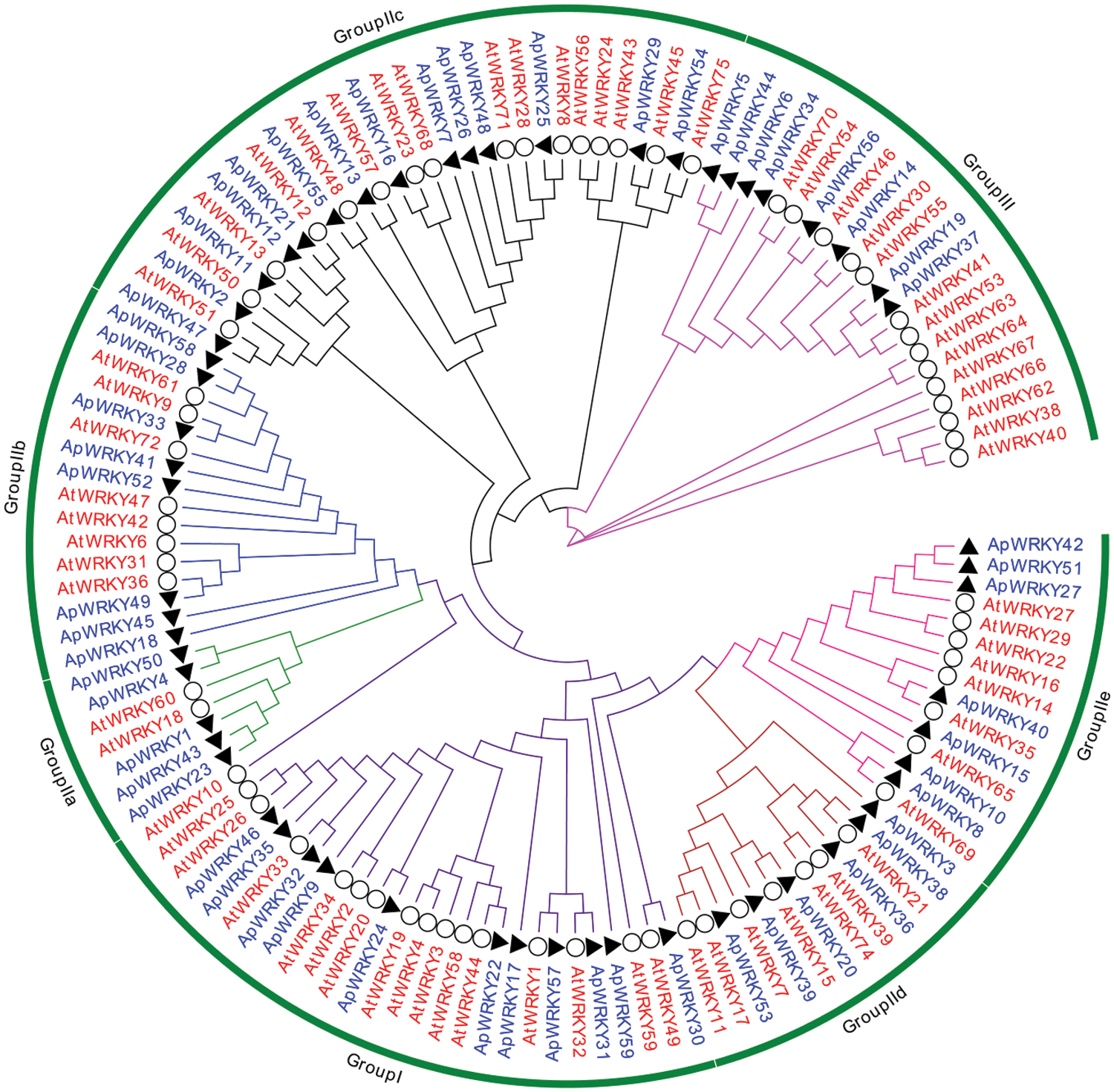
Figure 1: Phylogenetic analysis of WRKYs proteins from A. paniculata and A. thaliana.
Combined with the classification of Arabidopsis WRKY transcription factors, the conserved domains of candidate ApWRKY were identified and analyzed. The results showed that WRKY family protein sequences of A. paniculata can also be divided into three Groups I, II, and III, of which there are 11 ApWRKYs domains in Group I, and five ApWRKYs (ApWRKY17, ApWRKY24, ApWRKY31, ApWRKY35, ApWRKY57) have two WRKYGQK conserved domain structures (Figs. 1 and 2; Tab. 1). Forty ApWRKYs were distributed in Group II, which was further classified into five subgroups: IIa, IIb, IIc, IId and IIe, which contained 5, 8, 14, 6 and 7 ApWRKYs, respectively. The remaining 8 ApWRKYs belonged to Group III. Overall results indicated that different WRKY subgroups contained different evolutionary rates.
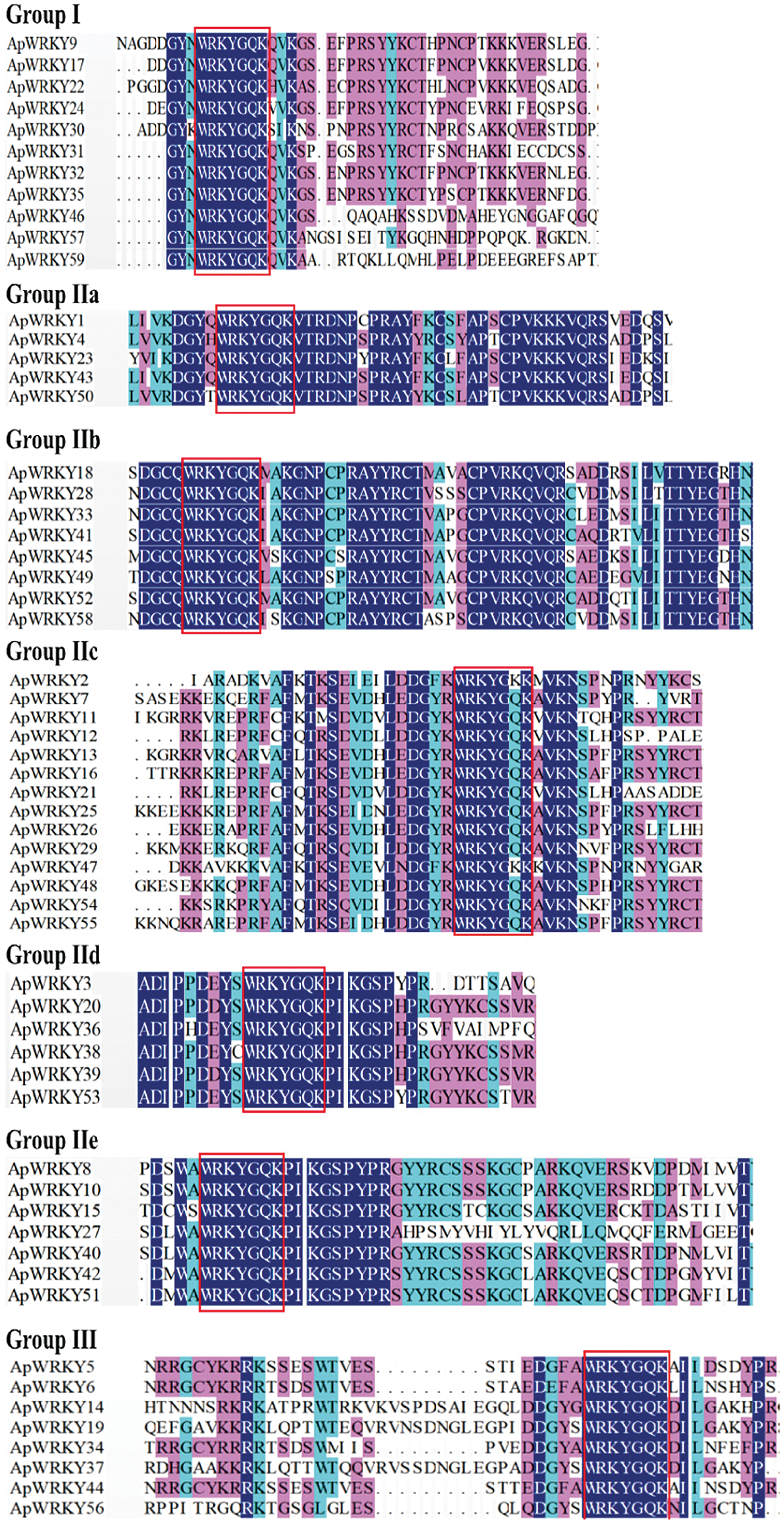
Figure 2: Multiple sequence alignment of the WRKY domain from ApWRKYs.
In this study, ApWRKYs transcription factors could be divided into three groups, among which the number of Group II could be further subdivided into five subgroups at most. By analyzing the structural characteristics of the WRKY domain, it was found that most ApWRKY proteins in Group I had two WRKYGQK conserved domains without variation. However, it was found in Group IIc that the WRKYGQK conserved domain of ApWRKY2 and ApWRKY47 were mutated from ‘Q’ to ‘K’ (WRKYGKK) (Fig. 2) (Chanwala et al., 2020, Qin et al., 2020). There is one WRKYGQK conservative domain for each ApWRKY in Group III.
Distribution of ApWRKY genes from A. paniculata chromosomes
Except that two WRKY genes ApWRKY55 and ApWRKY56 could not be located on any of the chromosomes, namely ApWRKY55 and ApWRKY56, the others distributed on chromosomes 1 to 24 (Fig. 3, Tab. 1). Chr2 and Ch17 contained the greatest number of A. paniculata 6 ApWRKY genes, whereas Chr4, Chr6, Chr9, Chr12, Chr18, Chr21, and Chr24 contained only one gene each. The other chromosomes varied from 2 to 5 genes. The analysis showed that both ApWRKY15 and ApWRKY16 were on Chr11, with a distance difference of 30403 bases. Similarly, both ApWRKY5 and ApWRKY6 were on Chr22, with a distance difference of 6,166 bases. The preliminary judgment was the tandem repeat between genes, which needs further analysis (Fig. 3).
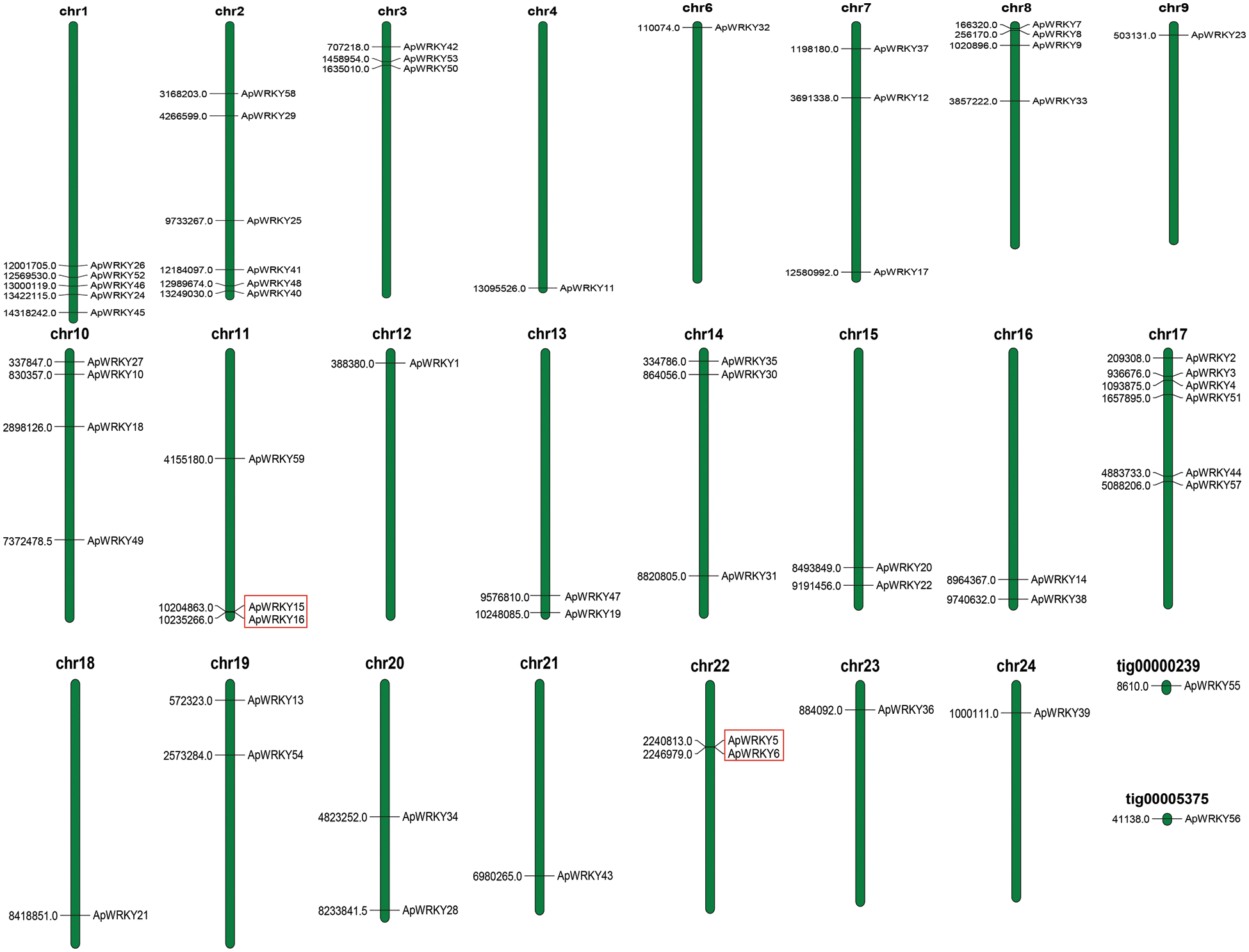
Figure 3: Distribution of ApWRKY genes within the A. paniculata chromosomes (Chrs).
Motif composition and gene structure of A. paniculata WRKY gene families
The structure diagram of ApWRKY protein was constructed based on the results of MEME motif analysis. Among them, motif 1, motif 2, and motif 3 were widely distributed on 46, 49, and 39 ApWRKYs proteins, respectively. It may be related to the important function of the WRKY transcription factor. It was found that the ApWRKY members of each group usually had similar motif composition. For example, motif 18 was unique to Group IIb, and motif 6 was specific to Groups IIa and IIb. Further, each ApWRKY contained its specificity; for example, Motif16 only existed in ApWRKY; in ApWRKY17, ApWRKY24, and ApWRKY35, there were two motifs, 1 and 2 (Fig. 4b). ApWRKY protein showed a similar motif arrangement in the subgroup, indicating that the protein structure is conserved in a specific subfamily. The function of most conservative motifs remained to be clarified. In general, the conservative motif composition and similar gene structure of the same WRKY members, as well as phylogenetic tree analysis results, strongly supported the credibility of this group of taxa.
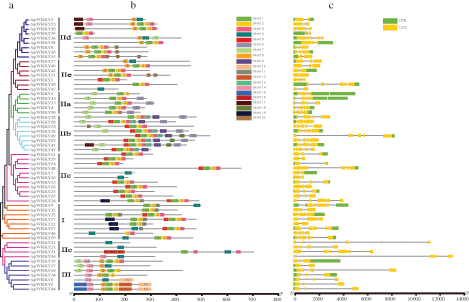
Figure 4: Phylogenetic relationships, the architecture of conserved protein motifs and gene structure in WRKY genes from A. paniculata.
The exons of all the identified ApWRKY genes were examined to gain more insight into the evolution of the WRKY family in A. paniculata (Fig. 4c). The mRNA structure of all the identified ApWRKY were also examined. All ApWRKY genes possessed two to seven CDS (nine with two CDS regions, 20 with three CDS regions, 13 with four CDS regions, 11 with five CDS regions, five with six CDS regions, and one with seven CDS regions). Further observation revealed that ApWRKY mRNA in the three CDS regions accounted for 1/6 of the universal. mRNA within the same group usually had similar structures; for example, all Group III members contained almost three CDS regions.
Differential expression of ApWRKY gene under salt stress
In order to verify whether the expression of ApWRKY gene was affected by abiotic stress, we selected 41 ApWRKY genes (Fig. 5) from 59 A. paniculata WRKY genes, and the transcription level was relatively higher in different tissues. To illustrate the analysis results, we divided 41 ApWRKY genes into four clusters. In Cluster I and Cluster IIa, 21 ApWRKY transcription factors were sensitive to NaCl in the stems of A. paniculata. In Cluster IIb and Cluster IIc, 20 ApWRKY transcription factors were sensitive to NaCl in the roots of A. paniculata (Fig. 5). Most of the WRKY genes were upregulated in the roots and stems of A. paniculata after treatment, while WRKY transcript levels were depressed in the leaves. These findings indicated that WRKY gene transcripts were mainly induced in the roots and stems of A. paniculata after salt treatment. In the roots and stems, the expression level of most WRKY genes increased with the increase of Na+ level, with some exceptions including ApWRKY9, ApWRKY20, ApWRKY53 and ApWRKY59. Suggesting that WRKY genes play a crucial role in the salt stress tolerance of A. paniculata. In addition, the transcript levels of some WRKY genes, such as ApWRKY9, ApWRKY10 and ApWRKY16, were significantly induced in A. paniculata leaves under low salinity conditions.
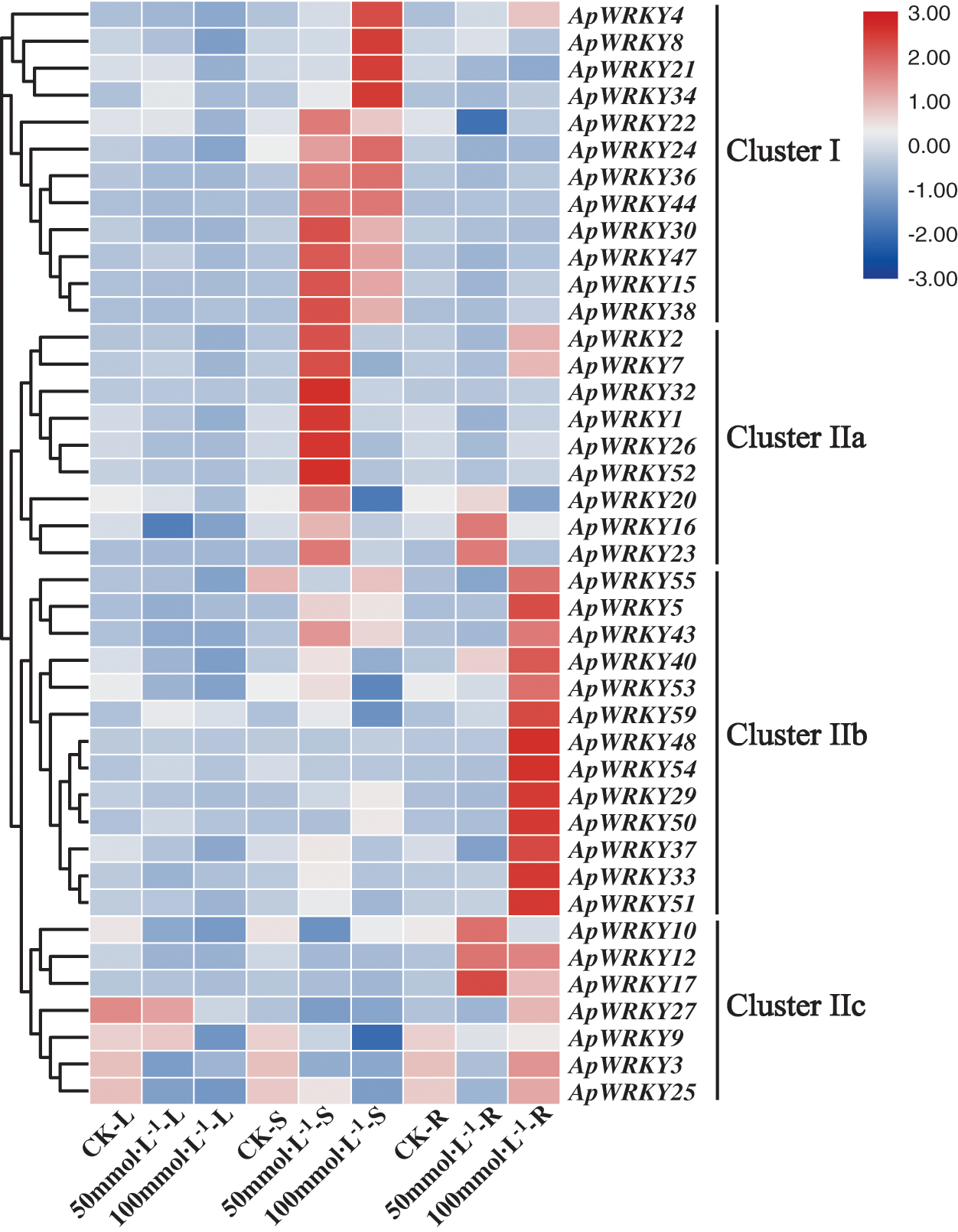
Figure 5: Expression patterns of 41 ApWRKY genes in root (R), stem (S), leaf (L) with A. paniculata.
In this study, the expression of 20 ApWRKY genes was significantly altered under the salinity stress (Fig. 6a). The greatest increase in expression (nearly 45-fold) was detected for under 100 mmol/L NaCl treatment as compared with the control in roots. ApWRKY10, ApWRKY16 and ApWRKY25 transcript levels were downregulated in the leaves of A. paniculata under salinity stress (Fig. 6c). ApWRKY17, ApWRKY29, ApWRKY33, ApWRKY37, ApWRKY48, ApWRKY50, ApWRKY51, and ApWRKY54 were significantly upregulated in the roots in response to different levels of salinity stress. ApWRKY29, ApWRKY48, ApWRKY50, and ApWRKY54 genes clearly reduced transcript levels in the leaves under the 50 mmol/L salinity concentration and even decreased to undetectable levels in roots. In contrast, ApWRKY29, ApWRKY48, ApWRKY50, and ApWRKY54 genes were significantly induced, and their high levels were maintained in the roots under high salinity stress. The transcripts of ApWRKY4 and ApWRKY34 exhibited an obvious increase in the stems with the enhancement of salinity concentration (Fig. 6b). In addition, under 50 mmol/L NaCl stress, the transcription level in the stems of A. paniculata was significantly increased compared with the control group (Fig. 6c). The salt tolerance mechanism of different ApWRKY genes was obviously different.
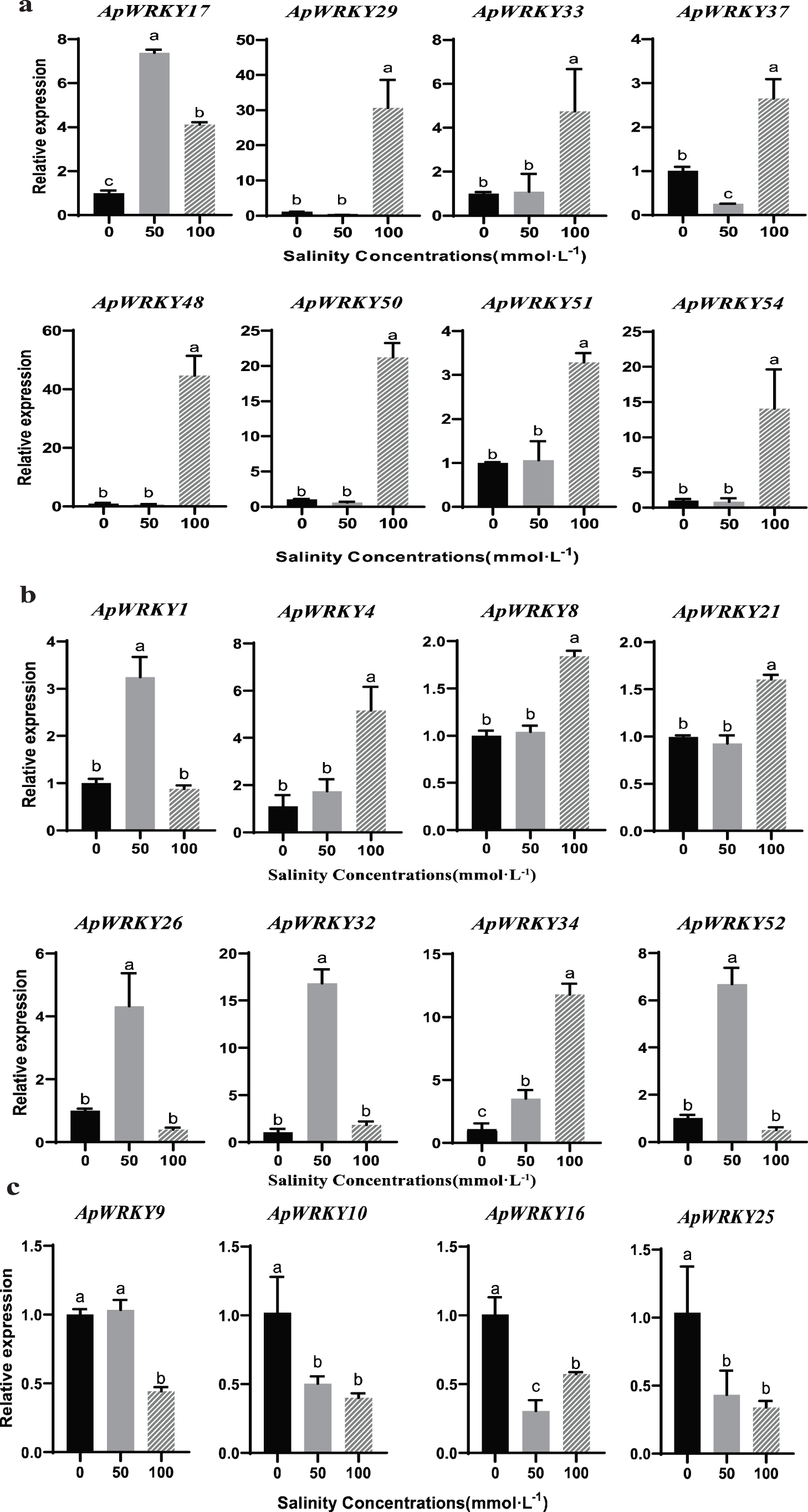
Figure 6: The expression patterns of selected genes in response to NaCl in different tissues of A. paniculata.
The active ingredient changes of A. paniculata under salt stress
Stems and leaves of A. paniculata, which are regarded as a promising system with which to produce AP, NAP, DHAP, and DOAP. Expression levels of biosynthesis genes were different after treatment with different concentrations of salt.
The accumulation of secondary metabolism in these environments was quantified. Interestingly, we found that the content of AP, NAP, DHAP and DOAP were obviously elevated by 50 mmol/L NaCl treatment after, and the contents of all four lactones are the highest. However, after 100 mmol/L NaCl stress, the contents of four andrographolides were lower than those of the control group. The results showed that a certain concentration of salt could promote or inhibit the biosynthesis of diterpenoid lactones in Andrographis paniculata leaves. According to Figs. 6 and 7, there can be seen that in the stem, under the salt stress of 50 mmol/L, ApWRKY1, 26, 32 and 52 are significantly up-regulated in the stem, while ApWRKY9 is significantly up-regulated in the leaf, which binds to under 50 mmol/L salt stress, A. paniculata has the highest content of active ingredients. We speculate that ApWRKY1, 9, 26, 32 and 52 may be involved in the synthesis of four kinds of andrographolides.
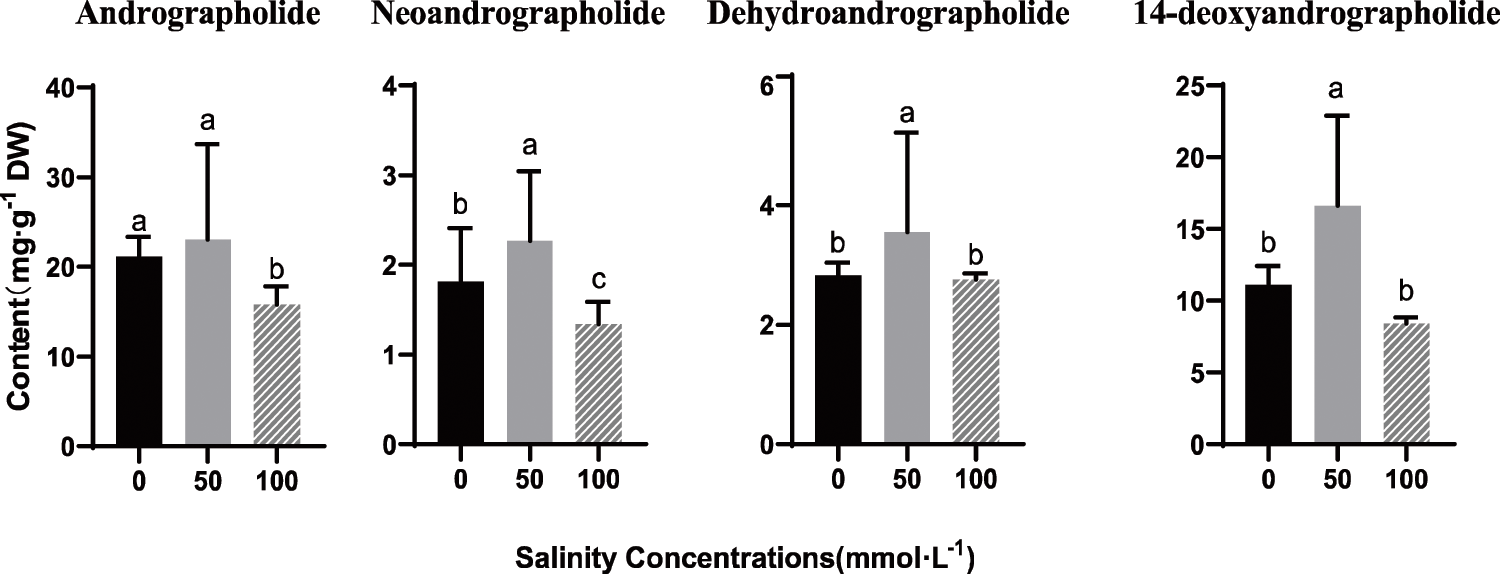
Figure 7: Effects of NaCl on accumulations of andrographolide, neoandrographolide, 14-deoxyandrographolide, and dehydroandrographolide in leaves of A. paniculata.
WRKY transcription factors participate in many physiological and biochemical processes in higher plants, not only in the growth and development processes of plant stem elongation, seed development, leaf senescence, root development, and trichome formation but also in the biological and abiotic stress processes of plants (Eulgem et al., 2000). Since WRKY genes play an important regulatory role in plant defense and disease resistance, understanding these genes is of great significance to plant research, plant breeding, and crop improvement. From the perspective of evolutionary relationship, the clustering results of the WRKY protein phylogenetic tree are highly consistent with the classification. The structure of the ApWRKY transcription factor phylogenetic tree is basically the same as that of the Salvia miltiorrhiza WRKY transcription factor phylogenetic tree of Salvia miltiorrhiza and sunflower (Yu et al., 2018, Li et al., 2020), Group IId, Group IIe, and Group III members clustered on the same branch, indicating that A. paniculata Group IId, Group IIe, and Group III WRKY transcription factors evolved from Group I in succession. In terms of structure, the ApWRKY transcription factor protein is highly conserved, but there are also variations in conserved domains, indicating that the ApWRKY transcription factor has diversity in the course of evolution. ApWRKY transcription factors of Group IIc have two different conserved domain structural differentiations. Proteins with mutations in several domains cluster together, indicating that these ApWRKY transcription factors may have similar functions, which is consistent with the presence of SiWRKY60 in sesame seeds in Group IIc (Li et al., 2017). In Arabidopsis, barley, and tomatoes, a similar phenomenon occurs in Group IIe members (Huang et al., 2012, Mangelsen et al., 2008, Eulgem et al., 2000).
In terms of conserved motifs, there were two conserved motif structures in Group I, and the differentiation of different domains appears, indicating that different structures may have different biological functions. Similar structure and differentiation were also found in the WRKY transcription factor protein of Caragana intermedia, also in Group I, the same phenomenon also appeared in Arabidopsis, cucurbitaceous species (Jiao et al., 2018, Eulgem et al., 2000), indicating that the ApWRKY transcription factor has a certain species specificity in the evolution process.
The expression level of WRKY genes is usually closely related to biological stress. Comparative expression analysis of the WRKY genes family can understand the potential function of WRKY in the process of plant development. In the present study, high ApWRKYs transcript levels were observed in both roots, stems, and leaves under salinity treatment. In this study, ApWRKY has different expression patterns in different tissues, which is consistent with the results of other plants such as poplar (Jiang et al., 2020), sunflower (Liu et al., 2020), and Artemisia annua (De et al., 2020). It can be seen from Fig. 5 that only a small part of WRKY genes is not expressed. among which while the ApWRKY10, ApWRKY16, and ApWRKY25 genes were obviously downregulated in the leaves under salinity stress (Eulgem et al., 2000). These findings indicated that these specific expression level changes might have helped protect the cell membranes system in A. paniculata under salinity stress (Nan et al., 2020). Moreover, most of the ApWRKY genes were induced to express in the roots of A. paniculata under the salinity stress, implying a tissue-specific expression in response to the salinity condition or a possible fact that the root was the tissue directly exposed to the treatment (Goyal et al., 2020, Zhao et al., 2019). According to research by Chen et al. (2010), in Arabidopsis, AtWRKY18, AtWRKY40, AtWRKY60 are expressed under abiotic stress, and ApWRKY1, 4, 23, 43, 50 transcription factors in the same branch may also form a highly interactive regulatory network (Fig. 1). By acting as a transcriptional activator or repressor to regulate gene expression in A. paniculata defense and stress response. Research by Li et al. (2011) and Jiang and Deyholos (2009) showed that WRKY25, WRKY26 and WRKY33 are part of a complex transcription network that regulates responses to specific abiotic stresses. The author predicts that the gene functions of ApWRKY32 and ApWRKY35 in the same branch are similar. Zou et al. (2010) found that AtWRKY34 is specifically expressed in pollen. After low-temperature treatment, the gene expression is upregulated and reversely regulates the low-temperature sensitivity of mature Arabidopsis pollen. The author predicts that ApWRKY9, which is closely related to its relationship (Fig. 1), may also be sensitive to low-temperature stress. While negatively regulating the low-temperature sensitivity of a certain tissue. Although the functions of most WRKY genes in A. paniculata remain unknown, phylogenetic and expression analyses provide a solid foundation for future functional studies.
In addition, the content of the four main secondary metabolites in A. paniculata leaves was determined under salt stress. From the results, The accumulation of diterpenoid lactone was the highest in A. paniculata under 50 mmol/L NaCl stress, indicating that salt stress in this adversity environment improves the effective components of andrographolide (Talei et al., 2013, Talei et al., 2015, Chanwala et al., 2020), which is a further reasonable and efficient breeding of A. paniculata. Studies have shown that in Salvia miltiorrhiza, SmWRKY2 can activate the up-regulated expression of genes related to the tanshinone biosynthesis pathway, thereby increasing the content of tanshinone in the hairy roots of salvia (Deng et al., 2019). OsWRKY76 overexpressed in rice can reduce the production of rice phytoalexins (Yokotani et al., 2013). The main secondary metabolites of A. paniculata come from the leaves. The analysis in Fig. 5 shows that the four genes ApWRKY3, 9, 25 and 27 are upregulated in the leaves. It is speculated that these four genes may be involved in the regulation of diterpenoids in the leaves. These genes, therefore, can be used as candidate genes for the study of andrographolide biosynthetic pathways. A similar phenomenon occurs when the rice transcription factor OsWRKY51 gene can be induced by abscisic acid (ABA). It can inhibit the transduction of gibberellins (GA) signals in rice cells by combining with the OsWRKY transcription factor. It is an important integration factor in the two signal pathways of ABA and GA (Xie et al., 2006). Among the 37 WRKY transcription factor genes expressed in Arabidopsis roots, 12 genes are specifically expressed in the tissue cells of the mature zone of the roots and may be involved in the morphogenesis of Arabidopsis roots (Birnbaum et al., 2003).
In this study, using the latest genome data of A. paniculata, 59 WRKY family members of A. paniculata were systematically identified and screened, and chromosome mapping was carried out. Through the construction of the phylogenetic tree, the locations of the conserved domains and CDS and UTR regions of the genes were used to identify ApWRKY. The specific expression analysis of the ApWRKY gene family in different tissues under salt stress was carried out, and the effect of different concentrations of A. paniculata on the secondary metabolites under salt stress was also studied. It provides a basis for further research on the functions of A. paniculata WRKY family members, as well as a reference for the salt stress mechanism of A. paniculata and the genetic improvement of abiotic stress.
We would like to thank Basharat Ali at the Department of Agronomy, University of Agriculture, Faisalabad, Pakistan, for the help with study suggestion. This study was supported by the pilot project of Zhejiang Province’s major agricultural technology collaborative promotion plan of China (Grant No. 2018XTTGYC03) and Zhejiang Province’s Basic Public Welfare Research Project of China (Grant No. LGN19H280004).
Author Contributions: QW, LX and ZL conceived and designed this study. QW and WZ conducted experiments. BA, LX and XZ contributed the analytical tool. QW wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in the manuscript. Data on preliminary tests are also provided in separate files as supplementary materials (Supplementary Tables and Figures). Thus, the readers can freely access all the published data with proper acknowledgment and citation. Besides, all the raw data sets generated or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary Material: The supplementary material is available online at DOI 10.32604/biocell.2021.015282.
Funding Statement: This study was supported by the pilot project of Zhejiang Province’s major agricultural technology collaborative promotion plan of China (Grant No. 2018XTTGYC03) and Zhejiang Province’s Basic Public Welfare Research Project of China (Grant No. LGN19H280004).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Berri S, Abbruscato P, Faivre RO, Brasileiro ACM, Fumasoni I, Satoh K, Kikuchi S, Mizzi L, Morandini P, Pè ME, Piffanelli P (2009). Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biology 9: 120–142. DOI 10.1186/1471-2229-9-120. [Google Scholar] [CrossRef]
Bindu V, Srinath M, Shailaja A, Giri C (2020). Proteome analysis and differential expression by JA driven elicitation in Andrographis paniculata (Burm. f.) Wall. ex Nees using Q-TOF-LC-MS/MS. Plant Cell, Tissue, and Organ Culture 140: 489–504. DOI 10.1007/s11240-019-01741-0. [Google Scholar] [CrossRef]
Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960. DOI 10.1126/science.1090022. [Google Scholar] [CrossRef]
Chanwala J, Satpati S, Dixit A, Parida A, Giri MK, Dey N (2020). Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress. BMC Genomics 21: 231–247. DOI 10.1186/s12864-020-6622-0. [Google Scholar] [CrossRef]
Chao W, Lin B (2010). Isolation and identification of bioactive compounds in Andrographis paniculata (Chuanxinlian). Chinese Medicine 5: 17–32. DOI 10.1186/1749-8546-5-17. [Google Scholar] [CrossRef]
Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X (2010). Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biology 10: 281–196. DOI 10.1186/1471-2229-10-281. [Google Scholar] [CrossRef]
Dai Y, Chen S, Chai L, Zhao J, Wang Y, Wang Y (2019). Overview of pharmacological activities of Andrographis paniculata and its major compound andrographolide. Critical Reviews in Food Science and Nutrition 591: S17–S29. DOI 10.1080/10408398.2018.1501657. [Google Scholar] [CrossRef]
De PA, Caretto S, Quarta A, Di SG, Sbrocca I, Mita G, Frugis G (2020). Genome-wide identification of WRKY genes in Artemisia annua: Characterization of a putative ortholog of AtWRKY40. Plants 9: 12. DOI 10.3390/plants9121669. [Google Scholar] [CrossRef]
Deng C, Hao X, Shi M, Fu R, Wang Y, Zhang Y, Zhou W, Feng Y, Makunga NP, Kai G (2019). Tanshinone production could be increased by the expression of SmWRKY2 in Salvia miltiorrhiza hairy roots. Plant Science 284: 1–8. DOI 10.1016/j.plantsci.2019.03.007. [Google Scholar] [CrossRef]
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000). The WRKY superfamily of plant transcription factors. Trends in Plant Science 5: 199–206. DOI 10.1016/S1360-1385(00)01600-9. [Google Scholar] [CrossRef]
Fan W, Hai M, Guo Y, Ding Z, Tie W, Ding X, Yan Y, Wei Y, Liu Y, Wu C, Shi H, Li K, Hu W (2016). The ERF transcription factor family in cassava: Genome-wide characterization and expression analyses against drought stress. Scientific Reports 6: 37379–37391. DOI 10.1038/srep37379. [Google Scholar] [CrossRef]
Goyal P, Manzoor MM, Vishwakarma RA, Sharma D, Dhar MK, Gupta S (2020). A comprehensive transcriptome-wide identification and screening of WRKY gene family engaged in abiotic stress in Glycyrrhiza glabra. Scientific Reports 10: 373–391. DOI 10.1038/s41598-019-57232-x. [Google Scholar] [CrossRef]
He H, Dong Q, Shao Y, Jiang H, Zhu S, Cheng B, Xiang Y (2012). Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Report 31: 1199–1217. DOI 10.1007/s00299-012-1241-0. [Google Scholar] [CrossRef]
Huang XS, Li KQ, Xu XX, Yao ZH, Jin C, Zhang SL (2015). Genome-wide analysis of WRKY transcription factors in white pear (Pyrus bretschneideri) reveals evolution and patterns under drought stress. BMC Genomics 16: 1104–1118. DOI 10.1186/s12864-015-2233-6. [Google Scholar] [CrossRef]
Huang S, Gao Y, Liu J, Peng X, Niu X, Fei Z, Cao S, Liu Y (2012). Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Molecular Genetics and Genomics 287: 495–513. DOI 10.1007/s00438-012-0696-6. [Google Scholar] [CrossRef]
Ishiguro S, Nakamura K (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5’ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Molecular & General Genetics: MGG 244: 562–571. DOI 10.1007/BF00282746. [Google Scholar] [CrossRef]
Jiang Y, Tong S, Chen N, Liu B, Bai Q, Chen Y, Bi H, Zhang Z, Lou S, Tang H, Liu J, Ma T, Liu H (2020). The PalWRKY77 transcription factor negatively regulates salt tolerance and ABA signaling in Populus. Plant Journal: For Cell and Molecular Biology 9: 1495. DOI 10.1111/tpj.15109. [Google Scholar] [CrossRef]
Jiang Y, Deyholos MK (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Molecular Biology 69: 91–105. DOI 10.1007/s11103-008-9408-3. [Google Scholar] [CrossRef]
Jiao Z, Sun J, Wang C, Dong Y, Xiao S, Gao X, Cao Q, Li L, Li W, Gao C (2018). Genome-wide characterization, evolutionary analysis of WRKY genes in Cucurbitaceae species and assessment of its roles in resisting to powdery mildew disease. PLoS One 13: e0199851. DOI 10.1371/journal.pone.0199851. [Google Scholar] [CrossRef]
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. DOI 10.1093/molbev/msy096. [Google Scholar] [CrossRef]
Li D, Liu P, Yu J, Wang L, Dossa K, Zhang Y, Zhou R, Wei X, Zhang X (2017). Genome-wide analysis of WRKY gene family in the sesame genome and identification of the WRKY genes involved in responses to abiotic stresses. BMC Plant Biology 17: 1–19. DOI 10.1186/s12870-017-1099-y. [Google Scholar] [CrossRef]
Li SJ, Fu QT, Chen LG, Huang WD (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233: 1237–1252. DOI 10.1007/s00425-011-1375-2. [Google Scholar] [CrossRef]
Li Z, Guo J, Li Y, He R, Wei J, Xu H (2013). Selection of internal reference genes in real-time quantitative PCR analysis of Andrographis paniculata. Journal of Guangzhou University of Traditional Chinese Medicine 30: 240–244. DOI 10.13359/j.cnki.gzxbtcm.2013.02.032. [Google Scholar] [CrossRef]
Li J, Islam F, Huang Q, Wang J, Zhou W, Xu L, Yang C (2020). Genome-wide characterization of WRKY gene family in Helianthus annuus L. and their expression profiles under biotic and abiotic stresses. PLoS One 15: 12. DOI 10.1371/journal.pone.0241965. [Google Scholar] [CrossRef]
Liu AK, Liu CL, Lei HY, Wang ZJ, Zhang M, Yan XR, Yang G, Ren JH (2020). Phylogenetic analysis and transcriptional profiling of WRKY genes in sunflower (Helianthus annuus L.Genetic diversity and their responses to different biotic and abiotic stresses. Industrial Crops and Products 148: 112268. DOI 10.1016/j.indcrop.2020.112268. [Google Scholar] [CrossRef]
Madhunita B, Ralf O (2014). WRKY transcription factors. Plant Signaling & Behavior 9: e27700. DOI 10.4161/psb.27700. [Google Scholar] [CrossRef]
Mangelsen E, Kilian J, Berendzen KW, Kolukisaoglu UH, Harter K, Jansson C, Wanke D (2008). Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 9: 194–211. DOI 10.1186/1471-2164-9-194. [Google Scholar] [CrossRef]
Meng D, Li Y, Bai Y, Li M, Cheng L (2016). Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress. Plant Physiology and Biochemistry 103: 71–83. DOI 10.1016/j.plaphy.2016.02.006. [Google Scholar] [CrossRef]
Nan H, Lin Y, Liu J, Huang H, Li W, Gao L (2020). Genome-wide analysis of the WRKY transcription Factor gene family and their response to salt stress in rubber tree. Tropical Plant Biology. DOI 10.1007/s12042-020-09268-x. [Google Scholar] [CrossRef]
Qin Z, Hou F, Li A, Dong S, Wang Q, Zhang L (2020). Transcriptome-wide identification of WRKY transcription factor and their expression profiles under salt stress in sweetpotato (Ipomoea batatas L.). Plant Biotechnology Reports 14: 599–611. DOI 10.1007/s11816-020-00635-4. [Google Scholar] [CrossRef]
Shen JB, Lv B, Luo LQ, He JM, Mao CJ, Xi DD, Ming F (2017). Corrigendum: The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Scientific Reports 7: 46890–46891. DOI 10.1038/srep46890. [Google Scholar] [CrossRef]
Sun WK, Leng L, Yin Q, Xu M, Huang M, Xu Z, Zhang Y, Yao H, Wang C, Xiong C, Chen S, Jiang C, Xie N, Zheng X, Wang Y, Song C, Peters RJ, Chen S (2019). The genome of the medicinal plant Andrographis paniculata provides insight into the biosynthesis of the bioactive diterpenoid neoandrographolide. Plant Journal 97: 841–857. DOI 10.1111/tpj.14162. [Google Scholar] [CrossRef]
Talei D, Valdiani A, Maziah M, Sagineedu SR, Abiri R (2015). Salt stress-induced protein pattern associated with photosynthetic parameters and andrographolide content in Andrographis paniculata Nees. Bioscience, Biotechnology, and Biochemistry 79: 51–58. DOI 10.1080/09168451.2014.963499. [Google Scholar] [CrossRef]
Talei D, Valdiani A, Maziah M, Sagineedu SR, Saad MS, Surguchov A (2013). Analysis of the anticancer phytochemicals in Andrographis paniculata Nees. under salinity stress. BioMed Research International 2013: 319047. DOI 10.1155/2013/319047. [Google Scholar] [CrossRef]
Voorrips RE (2002). MapChart: Software for the graphical presentation of linkage maps and QTLs. Journal of Heredity 93: 77–78. DOI 10.1093/jhered/93.1.77. [Google Scholar] [CrossRef]
Xie Z, Zhang ZL, Zou XL, Yang GX, Komatsu S, Shen QX (2006). Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant Journal 46: 231–242. DOI 10.1111/j.1365-313X.2006.02694.x. [Google Scholar] [CrossRef]
Yokotani N, Sato Y, Tanabe S (2013). WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. Journal of Experimental Botany 64: 5085–5097. DOI 10.1093/jxb/ert298. [Google Scholar] [CrossRef]
Yu HZ, V WL, Yang DF, Hou ZN, Liang ZS (2018). Transcriptional profiles of SmWRKY family genes and their putative roles in the biosynthesis of tanshinone and phenolic acids in Salvia miltiorrhiza. International Journal of Molecular Sciences 19: 1593–1600. DOI 10.3390/ijms19061593. [Google Scholar] [CrossRef]
Zhang Y, Wang L (2005). The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evolutionary Biology 5: 1–12. DOI 10.1186/1471-2148-5-1. [Google Scholar] [CrossRef]
Zhang JJ, Gao TT, Wang Y, Wang JL, Guan W, Wang YJ, Wang CN, Liu JF, Jiang B (2019). Andrographolide exerts significant antidepressant-like effects involving the hippocampal BDNF system in mice. International Journal of Neuropsychopharmacology 22: 585–600. DOI 10.1093/ijnp/pyz032. [Google Scholar] [CrossRef]
Zhao K, Zhang D, Lv K, Zhang X, Cheng Z, Li R, Zhou B, Jiang T (2019). Functional characterization of poplar WRKY75 in salt and osmotic tolerance. Plant Science 289: 110259. DOI 10.1016/j.plantsci.2019.110259. [Google Scholar] [CrossRef]
Zou C, Jiang W, Yu D (2010). Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. Journal of Experimental Botany 61: 3901–3914. DOI 10.1093/jxb/erq204. [Google Scholar] [CrossRef]
Supplementary Materials
Table S1. List of sequences used for phylogenetic tree construction.
Table S2. List of RT-qPCR specific primers used for ApWRKY expression studies.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |