DOI:10.32604/biocell.2021.015537

| BIOCELL DOI:10.32604/biocell.2021.015537 |  |
| Review |
The bacteriophage mu lysis system–A new mechanism of host lysis?
1Department of Biotechnology, School of Life Science & Biotechnology, Adamas University, Kolkata, 700126, India
2Institute for Skeletal Aging & Orthopedic Surgery, Hallym University-Chuncheon Sacred Heart Hospital, Chuncheon-si, 24252, Korea
3Department of Zoology, Fakir Mohan University, Balasore, 56020, India
*Address correspondence to: Rudra Prasad Saha, rudrasaha@gmail.com; Sang-Soo Lee, 123sslee@gmail.com; Chiranjib Chakraborty, drchiranjib@yahoo.com
#Authors contributed equally
Received: 25 December 2020; Accepted: 28 February 2021
Abstract: Bacteriophages are viruses that infect bacteria and can choose any one of the two alternative pathways for infection, i.e., lysis or lysogeny. Phage lysis is one of the conventional biological processes required to spread infection from one bacterium to another. Our analysis suggests that in the paradigm bacteriophage Mu, six proteins might be involved in host cell lysis. Mu has a broad host range, and Mu-like phages were found in both Gram-negative and Gram-positive bacteria. An analysis of the genomes of Mu and Mu-like phages could be useful in elucidating the lysis mechanism in this group of phages. A detailed review of the various mechanisms of phage lysis and different proteins associated with the process will help researchers understand the phage biology and their life cycle in different bacteria. The recent increase in the number of multidrug-resistant (MDR) strains of bacteria and the usual long-term nature of new drug development has encouraged scientists to look for alternative strategies like phage therapy and the discovery of new lysis mechanisms. Understanding the lysis mechanism in the Mu-like phages could be exploited to develop alternative therapeutics to kill drug-resistant pathogenic bacteria. In this review article, we have analyzed the phage Mu-mediated host lysis system, which is unknown till now, and our analysis indicates a possibility of the existence of a new lysis mechanism operating in Mu.
Keywords: Bacteriophage Mu; Host cell lysis; Endolysin; Holin; Spanin
The lysis of bacterial hosts by bacteriophages is a highly regulated process, controlled by several proteins, which act sequentially to disrupt the multi-layers of the bacterial membrane in a highly coordinated manner (Fig. 1). It has been proposed that two phage proteins play a crucial responsibility in phage lysis, and they are the endolysin and holin (Young, 1992). Endolysins induce the lysis pathway by breaking the peptidoglycan (PG) layer (Cahill and Young, 2019), but they need another group of proteins for access to the PG layer, and this class of proteins is known as holins. Holins are membrane-bound proteins, which form large pores in the inner membrane and allow the passage of endolysin to the PG layer (Young, 1992). Another phage-derived protein spanin is found to disrupt the outer membrane of Gram-negative bacteria (Kongari et al., 2018). In recent years, phage-derived endolysins and other PG hydrolases that disrupt the bacterial cell wall were found highly efficient in the treatment of bacterial infections, especially for Gram-positive bacteria (Fischetti, 2018; Kashani et al., 2018). The use of phage lytic proteins has shown promising results in clearing Methicillin-resistant Staphylococcus aureus (MRSA) infection in animal models (Gutiérrez et al., 2018). The unique ability of endolysins to quickly kill bacteria has enabled researchers to engineer them to be utilized as antibacterial or biocontrol agents in food preservation, bioprocess, fermentation, biotechnology, medicine, and others (Oliveira et al., 2012). The applications of endolysins were perceived to be only useful in Gram-positive bacteria as these bacteria lack the outer membrane of Gram-negative bacteria and, therefore, endolysins cannot directly access the PG layer in Gram-negative bacteria from outside. However, phage-derived novel endolysins were recently discovered that kill Gram-negative bacteria (Larpin et al., 2018; Lood et al., 2015).
The bacteriophage Mu is known to infect and kill a wide range of bacterial hosts (Paolozzi and Ghelardini, 2006). Moreover, several Mu-like phages were discovered in both Gram-negative and Gram-positive bacteria (Braid et al., 2004; Toussaint, 2013). However, the host-lysis mechanism of phage Mu is still unknown despite knowing the phage for more than 50 years by the scientific community (Taylor, 1963; Harshey, 2012; Saha et al., 2013). Our bioinformatics analysis of Mu genes revealed that at least seven gene products spanning early (kil), middle (19 and 20), and late regions (lys, 23/23a, and 25) of the Mu genome might be involved in host lysis. Our analysis also indicates a possibility that a new lysis mechanism may be operating in phage Mu because, in addition to putative holin, endolysin, and spanin proteins, other putative lysis-related proteins are also found in the genome (Faelen and Toussaint, 1973; Mathee and Howe, 1993; Morgan et al., 2002; Summer et al., 2007; Pastagia et al., 2013). Understanding how lysis proteins operate in Mu/Mu-like phages may provide us useful information that can be utilized to genetically engineer new lysis proteins or broad-range lytic phages for controlling multidrug-resistant bacterial infections.
The Bacteriophage Lysis System
Endolysins induce the lysis pathway by breaking the peptidoglycan layer (Cahill and Young, 2019). But these proteins generally do not possess any signal sequence and are unable to pass the cytoplasmic membrane by themselves (Cahill and Young, 2019). They need another group of proteins for access to the peptidoglycan layer, and this class of proteins is known as holins. Holins are membrane-bound proteins, which form large pores or holes in the cytoplasmic or inner membrane and allow passage for endolysin to the peptidoglycan layer (Young, 1992). For more than two decades, researchers perceived this mechanism as the sole pathway for phage lysis (Figs. 1A, 1B, and 1E). However, the holin-endolysin theory lost its universality upon the discovery of a new category of proteins, pinholin-SAR (signal-arrest-release) endolysin, that emerged as alternative candidates for phage lysis (Figs. 1C, 1D and 1E). Despite having similarities in the naming pattern, the molecular mechanisms of action of these two sets of enzymes are radically different (Young, 2013). SAR endolysin does not need holins for their transport to the peptidoglycan layer, and instead, they are reported to carry the N-terminal SAR sequence, which enables them to transport through the membrane by bacterial sec system (São-José et al., 2000). So, SAR endolysins could be transported to the periplasm in a holin-independent manner. However, these membrane-bound SAR endolysins are in inactive form unless they are released from the membrane and refolds in a proper active conformation (Sun et al., 2009; Xu et al., 2005). Their release from the membrane is correlated with their activation, and this is done by rapid membrane depolarization (Cahill and Young, 2019). The rapid membrane depolarization is mainly dependent on another group of proteins, i.e., pinholins. Instead of transporting endolysin, pinholins form small pores in the membrane, which help in rapid depolarization of the membrane and consequently activation of the SAR endolysins (Young, 2013). Another class of proteins named antiholins inactivate the holin assembly until the phage is ready to enter the lytic cycle (Wang et al., 2000; Park et al., 2006).
In addition to the holin-endolysin theory, some other lytic proteins play a pivotal role in the lysis of Gram-negative bacteria, named spanins (Kongari et al., 2018). They are associated with the disruption of the third layer, i.e., the outer membrane of Gram-negative bacteria. There are two classes of spanin proteins (one-component and two-component systems) reported so far. The two-component spanin complex consists of two proteins, i.e., o-spanin and i-spanin. O-spanin is an outer membrane lipoprotein, whereas i-spanin is an essential cytoplasmic membrane protein, and these two proteins are connected by a periplasmic domain (Young, 2013). One-component spanin system is known as u-spanin, which consists of a C-terminal transmembrane domain as well as an N-terminal outer membrane signal. The primary function of the spanins is to disrupt the outer membrane of Gram-negative bacteria and to fuse the inner and outer membranes for facilitating host cell lysis.
Unlike the phages of Gram-negative bacteria, lytic phages of Gram-positive bacteria do not need spanin proteins to promote cell lysis (Fig. 1F). However, it has been observed that overexpression of lytic genes (holin and endolysin) from Lactobacillus fermentum temperate bacteriophage phiPYB5 exhibited a broad range of lytic spectrum and can induce lysis in both Gram-positive and as well as Gram-negative bacteria (Wang et al., 2008). These proteins could potentially be exploited as a therapeutic method against a broad range of pathogenic bacteria. Some endolysins secreted by phages that infect Gram-positive bacteria (e.g., Lactobacillus plantarum phage φg1e, Oenococcus oeni phage fOg44, and Bacillus cereus phage TP21-L, etc.) contain signal sequences (intrinsic) that permit them to go through the bacterial inner membrane (Kakikawa et al., 2002; São-José et al., 2000). Lysins or endolysins released by phages of Gram-positive bacteria usually have modular conformation defined by N-terminal catalytic domains (one or more with different specificities) and C-terminal cell wall-binding domains (Braid et al., 2004; Fenton et al., 2010; Fischetti, 2005; Pastagia et al., 2013).
Also, specific enzymes of mycobacteriophage have been identified that are known to target the complex cell wall of Mycobacterium. The lytic enzymes of mycobacteriophage mainly comprise lipolytic enzymes that target the mycolic acid-containing outer membrane of the bacteria and peptidoglycan hydrolases that are responsible for breaking the mycobacterial peptidoglycan layer (Fig. 1G) (Catalão and Pimentel, 2018). The mycobacteriophage Ms6 that infects Mycobacterium smegmatis has several proteins that control host lysis-Gp1 assists the transport of the endolysin (LysA) utilizing the host sec system, and the lipolytic LysB protein that disrupts the OM of mycobacteria (Catalão and Pimentel, 2018).
Unlike dsDNA lytic phages, ssRNA and ssDNA lytic phages do not possess multiple lysis proteins. Instead, the host lysis process was facilitated by a ‘single-gene lysis’ (Sgl) protein that mainly inhibits bacterial cell wall synthesis (Chamakura and Young, 2019). The sgl genes share little to no sequence similarity to each other and are sometimes found more than one per phage genome (Chamakura et al., 2020; Chamakura and Young, 2020).

Figure 1: Schematic diagram of the typical lysis process in Gram-negative, Gram-positive, and mycobacteria by phage lytic proteins.
To identifying the lysis genes that are present in bacteriophage Mu, the genome sequence of Mu has been downloaded from the Nucleotide database (Benson et al., 2013) (GenBank: AF083977.1) and was analyzed for the genes that may affect the host lysis process (Braid et al., 2004; NCBI Resource Coordinators, 2018; Morgan et al., 2002). All Mu protein sequences were searched for homologs by the BLAST program against the UniProtKB database using the UniProt server (Altschul et al., 1990; Pundir et al., 2017; Uniprot Consortium, 2019). The presence of any conserved domain in protein sequence was searched by the NCBI CDD (conserved domain database) server (Marchler-Bauer et al., 2017). The presence of transmembrane domains and signal sequences in the Mu proteins were searched using TMpred and Protter servers (Hofmann and Stoffel, 1993; Omasits et al., 2014).
The bacteriophage Mu dsDNA genome spans 36,717 base pairs comprising at least 56 genes that are divided into three regions based on their expression timings in life-cycle - early, middle, and late (Morgan et al., 2002) (Fig. 2). The genes present in the early region are involved in controlling lytic-lysogenic choice, transposition, and replication, middle region genes are required for transcriptional regulation, and late region genes are responsible for encoding structural proteins of the phage (Choi et al., 2014; Harshey, 1988; Morgan et al., 2002). The region between muB and muC genes consists of 16 other genes (kil, 6, 7, 8, 9, gam, 11, 12, 13, 14, 15, gemA, mor, 18, 19, and 20) (Fig. 2), spanning about 5.5 kb region, whose functions are not essential for the phage Mu and was named as the “semi-essential” (SE) region (Morgan et al., 2002; Paolozzi, 1987). The host-lysis mechanism by bacteriophage Mu is still a mystery, and we predicted that at least seven gene products spanning early (kil), middle (19 and 20), and late (lys, 23/23a, and 25) regions of Mu’s genome may be involved in host-lysis (Tab. 1, Fig. 2). Interestingly, three (kil, 19, and 20) out of these seven predicted lysis genes fall within the SE region of the Mu genome.
In bacteriophage Mu, the expression of the Mu kil gene causes the death of host cells in the absence of phage replication (van de Putte et al., 1977; Westmaas et al., 1976). Expression of the kil gene induces striking morphological changes in the host cell structure, which becomes enlarged and mostly spherical, resulted in cell death (Waggoner et al., 1989). Although there is no clear idea of how the expression of the kil gene leads to cell death, some data suggested that the lethal effect of kil comes from its interaction with the cell wall, which resulted in abnormal cell wall synthesis (Goosen and van de Putte, 1984; Paolozzi, 1987). The deletion of a region of the Mu genome stretching from 4.4 kb to 7.2 kb (from kil to gam gene) resulted in tiny plaques after an abnormally long latent period of 4–5 h (Pastagia et al., 2013) compared to the normal period of 1 h (Goosen et al., 1982). This extended latent period can be substantially reduced by expressing SE genes from a recombinant plasmid, and even only expression of the kil gene product alone is sufficient to restore the latent period almost to normal (Goosen et al., 1982; Waggoner et al., 1989). Bacteriophage λ and Racprophage each hold a gene, also known as kil (Conter et al., 1996; Haeusser et al., 2014). However, these kil genes of λ or Rac phages do not have any sequence or functional similarity with the Mu kil gene (Waggoner et al., 1989; Haeusser et al., 2014). The expression of Rac kil stops FtsZ ring formation, which inhibits cell division that results in filamentation of the cell (Conter et al., 1996). The product of bacteriophage λ kil inhibits host cell division via the ZipA-dependent inhibition of FtsZ joining (Haeusser et al., 2014).

Figure 2: Schematic diagram of the partial Mu genome. Early (grey), middle (pink), and late (sky blue) genes are shown.
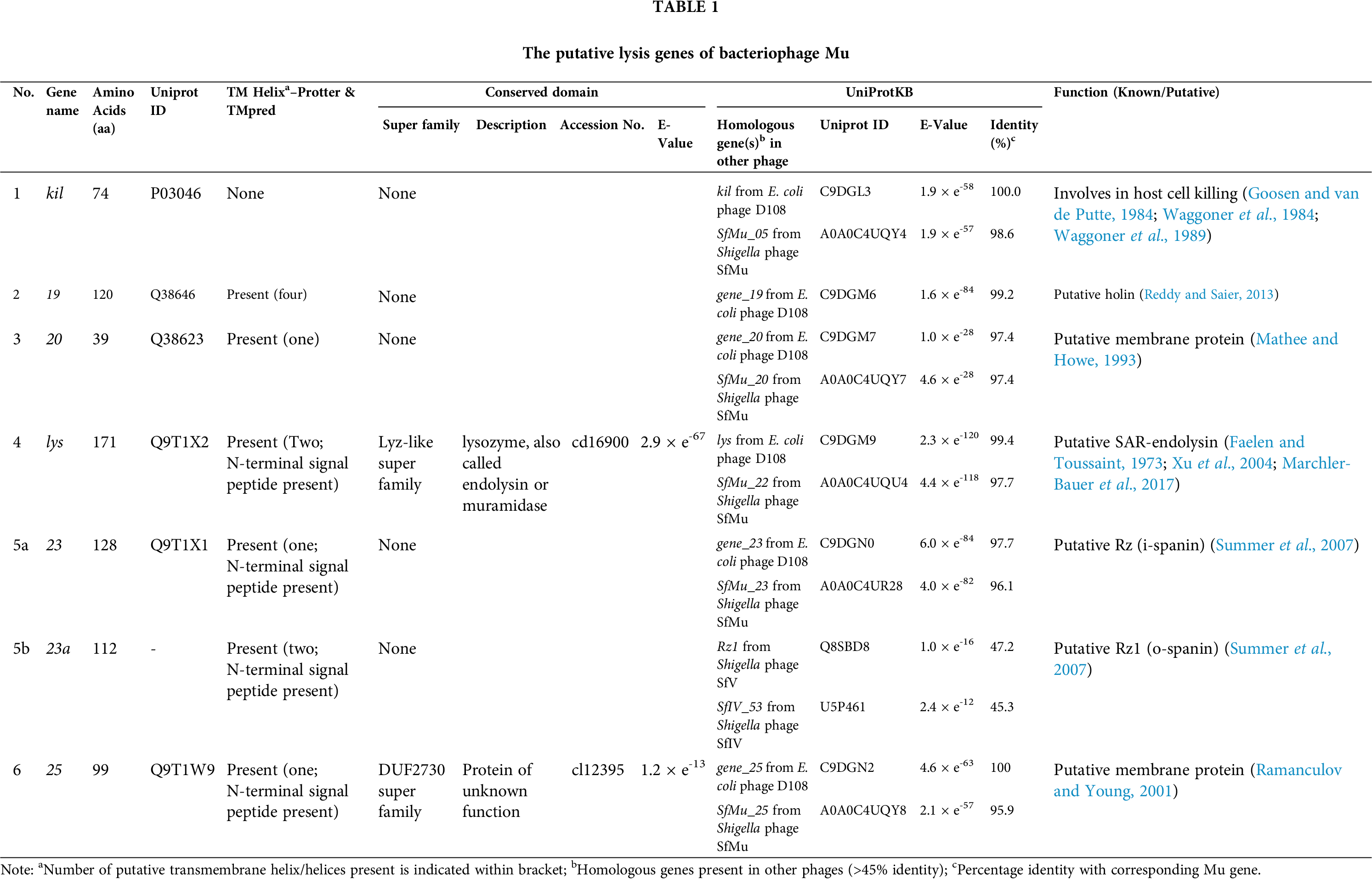
Mor (middle operon regulator) protein that acts as the activator for the Pm promoter transcribes 4 genes–18, 19, 20, and C (Fig. 2). The C protein activates the Plys promoter that transcribes 10 genes, from lys to G genes (Howe, 1998; Margolin and Howe, 1990; Margolin et al., 1989; Swapna et al., 2015; Zha et al., 1994). The lys gene product of Mu was found to be essential for efficient host cell lysis, although the mechanism is not known (Faelen and Toussaint, 1973). The lys gene appears to encode the Mu endolysin protein due to its high similarity with endolysins found in other phages (Tab. 1). The NCBI CDD search of Mu lys gene product identified it as a member of the Lyz-like superfamily that includes proteins with lysozyme-like domains, for example, lysozyme, endolysin, pesticin, etc. (Marchler-Bauer et al., 2017). We also found that the lys gene sequence has an N-terminal signal-arrest-release (SAR) sequence, which is a characteristic of SAR-endolysins (Xu et al., 2004). The UniProt server also annotated the Mu lys gene product as a putative SAR-endolysin (Tab. 1).
Interestingly, three out of four gene products (18, 19, and 20) of Pm promoter induced transcript were found to have some role in host cell lysis, as deletion of these genes resulted in 6–22 min delay in host cell lysis, but that did not affect plating efficiency and burst sizes. Therefore these three genes were identified as ‘non-essential’ genes for Mu (Mathee and Howe, 1993). The 18 gene product does not appear to have any transmembrane domain or signal-sequence, and therefore, unlikely to have any effect on the host lysis system, as no role within the host lysis system has yet been established. The 19 gene product was predicted to be similar to E. coli FhuB protein that scavenges iron and increases pathogenicity of the host, thereby resulted in a fitness advantage (Crosa, 1989). The 19 gene product has four transmembrane helices and a highly charged hydrophilic C-terminal end as predicted by our bioinformatic analysis and appears to be a new type of holin as most of the holins known were usually consist of up to 3 transmembrane helices (Park et al., 2006; To and Young, 2014; White et al., 2011). However, putative holins with four transmembrane helices have been identified in Mycobacterium phage Acadian, TM4, and in others too (Reddy and Saier, 2013). Holins usually have a simple membrane topology with membrane-spanning helices and a highly charged C-terminal end (Young and Bläsi, 1995; Reddy and Saier, 2013). Based on the membrane topology holins were mainly segregated into three classes–I, II, and III (Park et al., 2006; To and Young, 2014). Class I holin has three transmembrane domains (e.g., phage λ S holin, phage P2 Y holin, etc.), class II has two transmembrane domains (e.g., phage 21 S21 holin, etc.), and class III has a single transmembrane domain (e.g., phage T4 T holin, etc.) (Park et al., 2006; To and Young, 2014; White et al., 2011). Interestingly some class I and class II holin genes encode two proteins–one acts like a holin and the other as antiholin (White et al., 2011). Phage λ S holin gene (class I holin) encodes two proteins, S107 and S015, where S107 acts as an antiholin by inhibiting S105 mediated membrane disruption (Park et al., 2006). The additional positive charge in the N-terminal of S107 restricts the movement of its first transmembrane domain through the membrane, and in that conformation, it can inhibit the S105 holin (Park et al., 2006). Similarly, in the phage 21 S21 holin gene (class II holin) encodes two proteins, S2171, and S2168, where S2171 and S2168 are antiholin and holin, respectively (Park et al., 2006). The 20 gene product is a putative membrane protein with a single transmembrane helix and might also be involved in host lysis as deletion of the gene resulted in delayed host lysis (Mathee and Howe, 1993) (Tab. 1). The presence of transmembrane helix and N-terminal charge residues are characteristic features of an antiholin protein, which are also the characteristic features of gene 20 product (Young and Bläsi, 1995).
The 23 gene product (23/23a overlapping genes) was predicted to be the equivalent of phage spanin (i-spanin and o-spanin) proteins (Summer et al., 2007). Our bioinformatic analysis also finds that the product of gene 25 is a transmembrane protein and may have some role in the host lysis. The TMpred software finds the presence of one transmembrane domain in the gene 25 product (Tab. 1). Also, the bioinformatic analysis revealed both 23/23a and 25 gene products have N-terminal signal sequences (Tab. 1). In T4 phage, rI gene encodes an antiholin that possesses an N-terminal signal sequence similar to gene 25 product in Mu (Ramanculov and Young, 2001). Therefore, the gene 25 product may either function as an antiholin or it may have a novel function involving host lysis (Young and Bläsi, 1995).
It is not clear whether phage Mu has a holin-endolysin or SAR-endolysin-pinholin system. The lys gene product has all the features to be a SAR-endolysin protein, and the 19 gene product is predicted to be a holin, but there is no known SAR-endolysin-holin system in bacteria. Therefore, the gene 19 product may also function as a pinholin to complete the SAR-endolysin-pinholin pairing (Fig. 1C). However, in phage P1, the Lyz protein which encodes a SAR-endolysin found to be sufficient to lyse the host cell even though the phage expresses a holin (LydA) and an antiholin (LydB) (Young, 2014). Moreover, in phage P1, the lyz gene is not clustered with holin lydA or antiholin lydB genes (Young, 2014), similar to phage Mu lys gene (late region gene), which is also not clustered with the putative holin 19 gene (middle region gene) (Morgan et al., 2002), indicating that the Mu Lys protein may also function in a holin-independent manner similar to P1 Lyz. At this point, it is not clear whether the gene 20 or gene 25 product functions as an antiholin or they might have a novel lysis function. Further experiments are necessary to ascertain their functions. Unlike any other phage, the Mu early (kil) and middle (19 and 20) region genes, although ‘semi-essential’, have some indirect effects on the phage lytic cycle as deletion of these gene delays host lysis (Mathee and Howe, 1993; Pastagia et al., 2013). Together, the Mu lysis system is appeared to be less similar to other lambdoid and T4 systems (Young, 1992), and the data suggests that there is a possibility of a new lysis mechanism operating in phage Mu which needs to be uncovered for a better understanding of the underlying mechanism.
Several Mu-like transposable phages were discovered not only in Gram-negative bacteria, e.g., Escherichia coli phages D108 and Sp18 (Hayashi et al., 2001; Hull et al., 1978), Pseudomonas aeruginosa phages D3112, B3, PaMx73 and H70 (Braid et al., 2004; Cazares et al., 2014; Roncero et al., 1990), Haemophilus infuenzae phage FluMu (Fleischmann et al., 1995), Neisseria meningitides phages Pnm1, Pnm2 and MuMenB (Klee et al., 2000; Masignani et al., 2001), Salmonella typhi phage SalMu (Parkhill et al., 2001), Photorhabdus luminescens phage PhotoMu (Duchaud et al., 2003), Chromobacterium violaceum phage ChromoMu (Brazilian National Genome Project Consortium, 2003), Burkholderia cenocepacia phage BcepMu (Summer et al., 2004), Burkholderia cepacia phage KS10 (Goudie et al., 2008), Rhodobacter capsulatus phage RcapMu (Fogg et al., 2011), Haemophilus parasuis phage SuMu (Zehr et al., 2012), Shigella flexneri phage SfMu (Jakhetia and Verma, 2015), Mannheimia haemolytica phage 3927AP2 (Niu et al., 2015), etc., but also in Gram-positive bacteria, e.g., Deinococcus radiodurans phage RadMu (White et al., 1999), Syntrophobotulus glycolicus phage SglyMu-1 (Toussaint, 2013), Bacillus alcalophilus phage BalMu (Yang et al., 2015), etc. (Tab. 2). These Mu-like phages all carry proteins that control lysogenic or lytic lifestyle, ‘semi-essential’ genes with unknown functions, the transposition activator B as well as transposase A genes, head and tail genes, lysis genes, etc. Overall, till now, more than 50 Mu-like phages were discovered that reside or infect as prophages in both Gram-negative and Gram-positive bacteria, indicating that they are widespread mobile genetic elements in nature (Braid et al., 2004; Toussaint, 2013). Unfortunately, no work has been done on the lysis-system operating in these Mu-like phages infecting Gram-negative and Gram-positive bacteria. Therefore, analysis of these Mu-like phage genomes is essential and would help us to understand how the lysis machinery working in this unique group of bacteriophages.
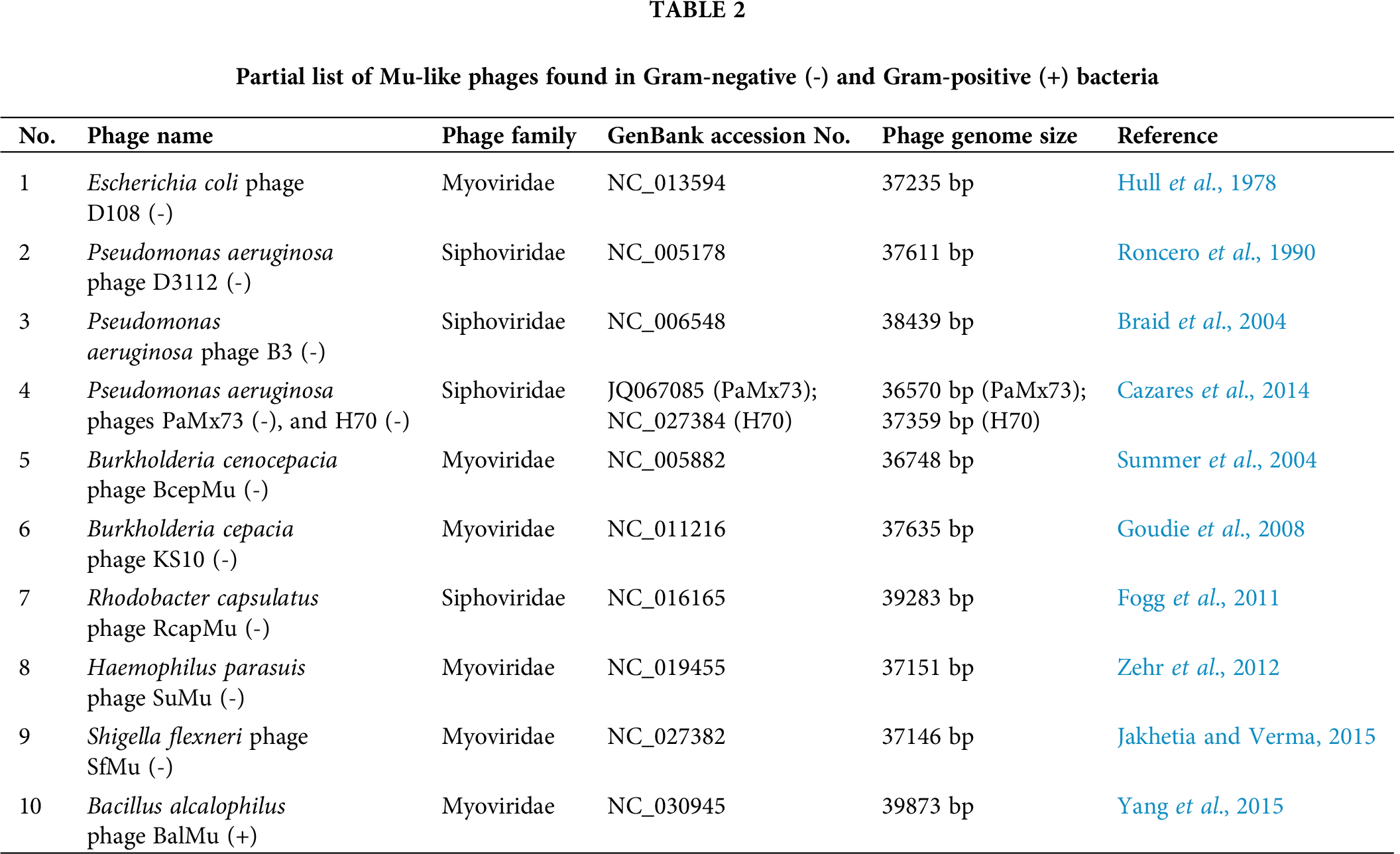
All predicted lysis genes in phage Mu were found to be highly similar (>97%; Tab. 1) to Escherichia phage D108 homologs even though these two phages have dissimilar host ranges and were hetero-immune (Hull et al., 1978). The Shigella phage SfMu is also found to be very close to phage Mu as most of its proteins have >95% sequence similarities with phage Mu (Tab. 1) (Jakhetia and Verma, 2015). In other Mu-like phages, FluMu (Haemophilus), D3112 (Pseudomonas), and PaMx73 (Pseudomonas), most of the genes were found to be conserved, including semi-essential and lysis genes (Cazares et al., 2014; Morgan et al., 2002). Similarly, several other Mu-like phages in Gram-positive and Gram-negative bacteria (e.g., Pseudomonas phage B3, Mannheimia phage 3927AP2, Syntrophobotulus phage SglyMu, Bacillus phage BalMu, etc.) were found to encode proteins, including lysis proteins, that are homologous to phage Mu proteins (Braid et al., 2004; Niu et al., 2015; Toussaint, 2013; Yang et al., 2015), although, in some Mu-like phages variations in the early and middle regions of the genomes were observed (Morgan et al., 2002; Niu et al., 2015). Further analysis of the early, middle and late regions of the Mu-like phages is required to understand how these unique phages evolved with time especially in the context of Gram-negative and Gram-positive bacteria.
Phage Therapy by Lytic Phage, or Endolysin Protein
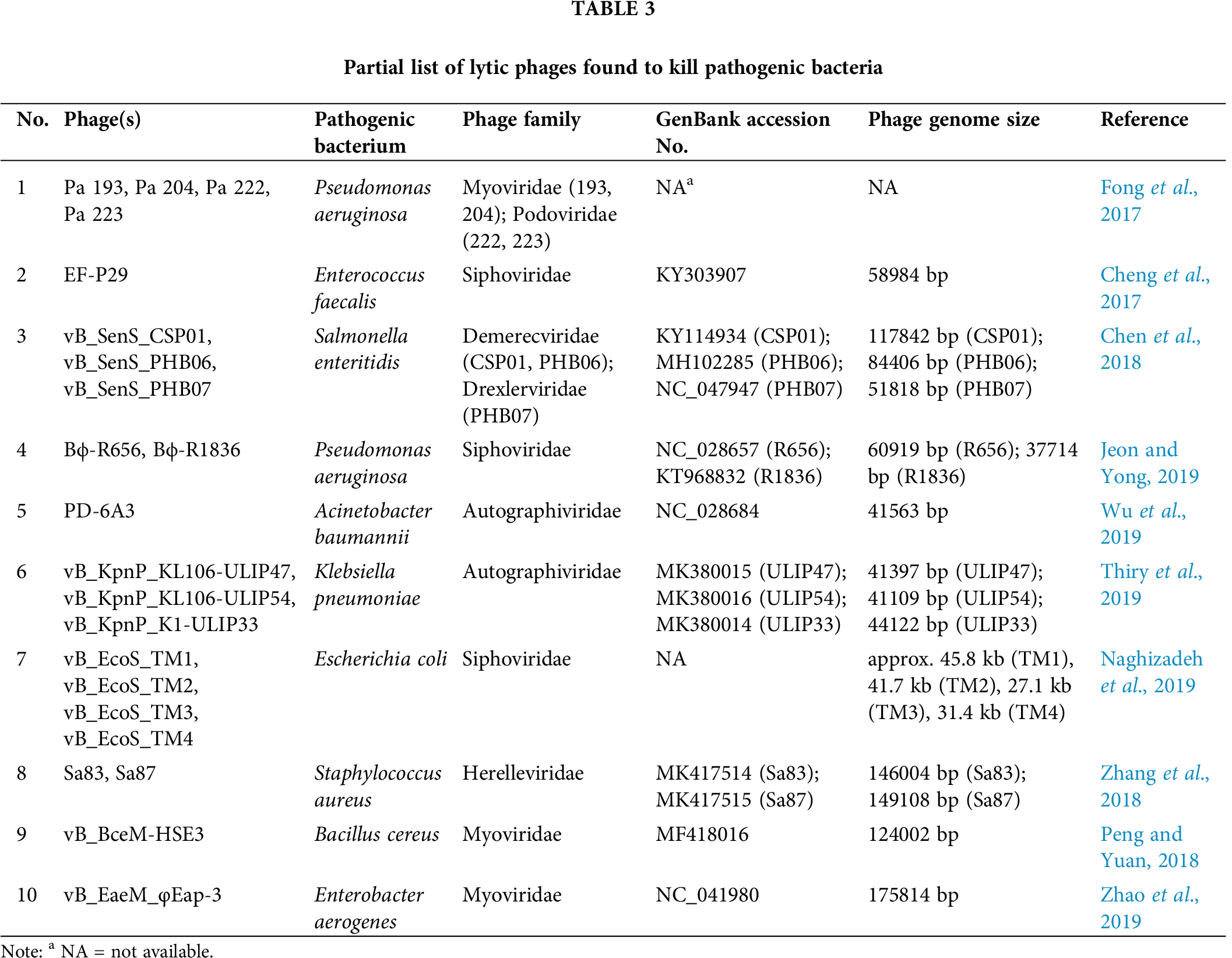
Various lytic phages or phage-derived endolysin proteins were found to successfully reduce or clear the fatal/multidrug-resistant infections by several pathogenic bacteria, e.g., Pseudomonas aeruginosa (Fong et al., 2017), Enterococcus faecalis (Cheng et al., 2017), Salmonella enteritidis (Chen et al., 2018), Pseudomonas aeruginosa (Jeon and Yong, 2019), Acinetobacter baumannii (Wu et al., 2019), Klebsiella pneumoniae (Thiry et al., 2019), Escherichia coli (Naghizadeh et al., 2019), Staphylococcus aureus (Zhang et al., 2018), Salmonella typhimurium (Seo et al., 2018), Bacillus cereus (Peng and Yuan, 2018), Enterobacter aerogenes (Zhao et al., 2019), Mycobacterium abscessus (Dedrick et al., 2019), etc (Tab. 3). Phage-derived endolysins and other peptidoglycan hydrolases that disrupt the bacterial cell wall are found to be highly efficient in the treatment of bacterial infections, especially for Gram-positive bacteria (Fischetti, 2018; Kashani et al., 2018; Nelson et al., 2001). The use of phage lytic proteins has shown promising results in clearing Methicillin-resistant Staphylococcus aureus (MRSA) infection in animal models without any apparent side effects (Gutiérrez et al., 2018). The distinctive capability of endolysins to quickly kill bacteria has enabled researchers to engineer them to be utilized as antibacterial or biocontrol agents in food preservation, bioprocess and fermentation, biotechnology, medicine, and others (Oliveira et al., 2012). Although the activity of endolysin is restricted in Gram-negative bacteria due to the existence of an impermeable outer membrane, the development of genetically engineered endolysins is in full force that kills Gram-negative bacteria (Briers and Lavigne, 2015; Briers et al., 2007; Briers et al., 2011; Vázquez et al., 2018). Also, phage-derived novel endolysins were found recently that kill Gram-negative bacteria (Larpin et al., 2018; Lood et al., 2015). The endolysin proteins are being engineered so that they can penetrate the outer membrane of Gram-negative bacteria and reach the peptidoglycan layer. It has been shown that the addition of a highly positive-charged peptide at any of the end of an exogenous lysin/endolysin can destabilize the outer membrane of Gram-negative bacteria to get access to the peptidoglycan layer which results in the killing of the bacteria (Larpin et al., 2018; Lood et al., 2015). These modified proteins work optimally at low pH when the terminal peptides become highly protonated, which destabilizes the outer membrane by interfering with the stabilizing divalent cations that interact with the negatively charged phosphate groups in the outer membrane (Larpin et al., 2018). Moreover, it has been observed that in recent times a considerable amount of progress has been done, specifically about the mechanism of action and detailed comprehension of bacteriophage lysis proteins, which could be considered as an important lead towards the development of alternative therapeutics specially designed to combat new multi-drug resistant pathogenic bacteria. For example, recently, researchers have developed a new generation of lytic proteins by adding selected peptides to endolysins named Artilysin (Rodríguez-Rubio et al., 2016). Artilysins are engineered endolysins fused with permeabilizing peptides that help the fusion protein to penetrate the outer membrane of Gram-negative bacteria and degrade the peptidoglycan layer resulting in the death of the bacteria (Briers et al., 2014). Artilysins are extremely bactericidal and found to be especially useful in killing a wide range of multidrug-resistant bacteria (Briers et al., 2014; Rodríguez-Rubio et al., 2016). It has been noted that these novel lytic proteins have a broad range of activities for both Gram-positive and Gram-negative bacteria.
Interestingly, Mu is the most efficient transposable element known (Mizuuchi, 1983; Walker and Harshey, 2020) and can infect and lyse several hosts (Escherichia coli, Shigella sonnei, Citrobacter freundii, Erwinia, and Enterobacter) (Paolozzi and Ghelardini, 2006). Moreover, Mu-like phages were discovered in a wide range of Gram-negative and Gram-positive bacteria (Hull et al., 1978; Hayashi et al., 2001; Toussaint, 2013). The lysis-system operating in Mu/Mu-like phages is predicted to be unique, and the presence of several putative lysis-related genes (holin/pinholin, SAR-endolysin, spanin, etc.) bolsters the hypothesis (Tab. 1). Deciphering the lysis mechanism operating in this group of phages may help us to design highly efficient lysis proteins that would kill a broad range of pathogenic bacteria.
Most phages require one or more of some specialized classes of proteins (holin/pinholin, endolysin, and spanin) that are designated to disrupt different layers of the cell envelope of the bacterial host (Young, 2014; Howard-Varona et al., 2017). Whereas, in simple phages (ssDNA and ssRNA phages), another diverse class of enzymes (single-gene lysis or Sgl proteins) is involved in host cell lysis. Phage encoded Sgl proteins are unique and are usually unrelated to each other (Chamakura and Young, 2019). Moreover, it has been shown that the lysis process has more important aspects than just the “rupture” of the cell wall, and a detailed understanding of phage lytic functions would be helpful in the development of advanced alternative drugs to combat present and new bacterial pathogens.
The phage Mu can infect and kill several different bacterial hosts (Paolozzi and Ghelardini, 2006), and Mu-like phages can lyse both Gram-positive and Gram-negative hosts (Hull et al., 1978; Hayashi et al., 2001; Toussaint, 2013). The lytic machinery of bacteriophage Mu is unknown, and our analysis reveals the presence of several putative lysis genes in Mu. The presence of ‘semi-essential’ lysis proteins (kil, 19, and 20 gene products) may help Mu phage to disrupt the integrity of the host cell membranes and, therefore, may assist in the host lysis process by the Mu SAR-endolysin Lys. The presence of these non-essential lysis genes might be the key element for the Mu/Mu-like phages for killing a wide range of hosts, both Gram-negative and Gram-positive bacteria. Therefore, understanding how the lysis system operates in Mu/Mu-like bacteriophages is of utmost importance as these proteins (e.g., endolysin, etc.) can be directly utilized to kill a broad range of bacteria, particularly multidrug-resistant bacteria. The possibility of the existence of a new lysis mechanism in phage Mu would be enticing enough for the researchers to uncover the long-standing mystery in this paradigm phage Mu.
Author Contributions: Conceptualization, SS, ARS, and RPS; Methodology, SS and ARS; Validation, AS and MKS; Formal Analysis, SS, MB, and AD; Investigation, SS, AD, and ARS; Resources, SS, MKS, and RPS; Data Curation, AS, AD, and CC; Writing–Original Draft Preparation, AS, ARS, and RPS; Writing–Review & Editing, ARS, MB, and CC; Visualization, RPS and CC; Supervision, RPS, SSL, and CC; Project Administration, RPS and CC; Funding Acquisition, ARS, SSL.
Ethics Approval: The research has not involved any animal or human.
Funding Statement: This research was supported by Hallym University Research Fund and by Basic Science Research Program through the National Research Foundation of Korea (NRF) Funded by the Ministry of Education (NRF-2020R1C1C1008694 & NRF-2020R1I1A3074575). The authors would like to thank Adamas University for providing computer support for the bioinformatic analysis.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990). Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. [Google Scholar]
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2013). GenBank. Nucleic Acids Research 41: D36–D42. [Google Scholar]
Braid MD, Silhavy JL, Kitts CL, Cano RJ, Howe MM (2004). Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. Journal of Bacteriology 186: 6560–6574. [Google Scholar]
Brazilian National Genome Project Consortium (2003). The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proceedings of the National Academy of Sciences of the United States of America 100: 11660–11665. [Google Scholar]
Briers Y, Lavigne R (2015). Breaking barriers: Expansion of the use of endolysins as novel antibacterials against Gram-negative bacteria. Future Microbiology 10: 377–390. [Google Scholar]
Briers Y, Volckaert G, Cornelissen A, Lagaert S, Michiels CW, Hertveldt K, Lavigne R (2007). Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages φKZ and EL. Molecular Microbiology 65: 1334–1344. [Google Scholar]
Briers Y, Walmagh M, Lavigne R (2011). Use of bacteriophage endolysin EL188 and outer membrane permeabilizers against Pseudomonas aeruginosa. Journal of Applied Microbiology 110: 778–785. [Google Scholar]
Briers Y, Walmagh M, Van Puyenbroeck V, Cornelissen A, Cenens W, Aertsen A, Oliveira H, Azeredo J, Verween G, Pirnay JP, Miller S, Volckaert G, Lavigne R (2014). Engineered endolysin-based “Artilysins” to combat multidrug-resistant gram-negative pathogens. mBio 5: e01379–14. [Google Scholar]
Cahill J, Young R (2019). Phage lysis: Multiple genes for multiple barriers. Advances in Virus Research 103: 33–70. [Google Scholar]
Catalão MJ, Pimentel M (2018). Mycobacteriophage lysis enzymes: Targeting the mycobacterial cell envelope. Viruses 10: 428. [Google Scholar]
Cazares A, Mendoza-Hernandez G, Guarneros G (2014). Core and accessory genome architecture in a group of Pseudomonas aeruginosa Mu-like phages. BMC Genomics 15: 1146. [Google Scholar]
Chamakura K, Young R (2019). Phage single-gene lysis: Finding the weak spot in the bacterial cell wall. Journal of Biological Chemistry 294: 3350–3358. [Google Scholar]
Chamakura KR, Tran JS, O’Leary C, Lisciandro HG, Antillon SF, Garza KD, Tran E, Min L, Young R (2020). Rapid de novo evolution of lysis genes in single-stranded RNA phages. Nature Communications 11: 6009. [Google Scholar]
Chamakura KR, Young R (2020). Single-gene lysis in the metagenomic era. Current Opinion in Microbiology 56: 109–117. [Google Scholar]
Chen Y, Sun E, Song J, Tong Y, Wu B (2018). Three Salmonella enterica serovar Enteritidis bacteriophages from the Siphoviridae family are promising candidates for phage therapy. Canadian Journal of Microbiology 64: 865–875. [Google Scholar]
Cheng M, Liang J, Zhang Y, Hu L, Gong P, Cai R, Zhang L, Zhang H, Ge J, Ji Y, Guo Z, Feng X, Sun C, Yang Y, Lei L, Han W, Gu J (2017). The bacteriophage EF-P29 efficiently protects against lethal vancomycin-resistant Enterococcus faecalis and alleviates gut microbiota imbalance in a murine bacteremia model. Frontiers in Microbiology 8: 837. [Google Scholar]
Choi W, Saha RP, Jang S, Harshey RM (2014). Controlling DNA degradation from a distance: A new role for the Mu transposition enhancer. Molecular Microbiology 94: 595–608. [Google Scholar]
Conter A, Bouche J-P, Dassain M (1996). Identification of a new inhibitor of essential division gene ftsZ as the kil gene of defective prophage Rac. Journal of Bacteriology 178: 5100–5104. [Google Scholar]
Crosa JH (1989). Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiological Reviews 53: 517–530. [Google Scholar]
Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, Spencer H (2019). Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nature Medicine 25: 730–733. [Google Scholar]
Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux-Eude C, Chandler M, Charles JF, Dassa E, Derose R, Derzelle S, Freyssinet G, Gaudriault S, Médigue C, Lanois A, Powell K, Siguier P, Vincent R, Wingate V, Zouine M, Glaser P, Boemare N, Danchin A, Kunst F (2003). The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nature Biotechnology 21: 1307–1313. [Google Scholar]
Faelen M, Toussaint A (1973). Isolation of conditional defective mutants of temperate phage Mu-1 and deletion mapping of the Mu-1 prophage. Virology 54: 117–124. [Google Scholar]
Fenton M, McAuliffe O, O’Mahony J, Coffey A (2010). Recombinant bacteriophage lysins as antibacterials. Bioengineered Bugs 1: 9–16. [Google Scholar]
Fischetti VA (2005). Bacteriophage lytic enzymes: Novel anti-infectives. Trends in Microbiology 13: 491–496. [Google Scholar]
Fischetti VA (2018). Development of phage lysins as novel therapeutics: a historical perspective. Viruses 10: 310. [Google Scholar]
Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM (1995). Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269: 496–512. [Google Scholar]
Fogg PC, Hynes AP, Digby E, Lang AS, Beatty JT (2011). Characterization of a newly discovered Mu-like bacteriophage, RcapMu, in Rhodobacter capsulatus strain SB1003. Virology 421: 211–221. [Google Scholar]
Fong SA, Drilling A, Morales S, Cornet ME, Woodworth BA, Fokkens WJ, Psaltis AJ, Vreugde S, Wormald PJ (2017). Activity of bacteriophages in removing biofilms of Pseudomonas aeruginosa isolates from chronic rhinosinusitis patients. Frontiers in Cellular and Infection Microbiology 7: 418. [Google Scholar]
Goosen N, van de Putte P (1984). Hek: An Escherichia coli function involved in functional expression of the kil gene of bacteriophage Mu. Molecular and General Genetics 196: 170–172. [Google Scholar]
Goosen N, van de Putte P (1984). Hek: An Escherichia coli function involved in functional expression of the kil gene of bacteriophage Mu. Molecular and General Genetics 196: 170–172. [Google Scholar]
Goosen T, Giphart-Gassler M, van de Putte P (1982). Bacteriophage Mu DNA replication is stimulated by non-essential early functions. Molecular and General Genetics 186: 135–139. [Google Scholar]
Goudie AD, Lynch KH, Seed KD, Stothard P, Shrivastava S, Wishart DS, Dennis JJ (2008). Genomic sequence and activity of KS10, a transposable phage of the Burkholderia cepacia complex. BMC Genomics 9: 615. [Google Scholar]
Gutiérrez D, Fernández L, Rodríguez A, García P (2018). Are phage lytic proteins the secret weapon to kill Staphylococcus aureus? mBio 9: e01923–01917. [Google Scholar]
Haeusser DP, Hoashi M, Weaver A, Brown N, Pan J, Sawitzke JA, Thomason LC, Court DL, Margolin W (2014). The Kil peptide of bacteriophage λ blocks Escherichia coli cytokinesis via ZipA-dependent inhibition of FtsZ assembly. PLoS Genetics 10: e1004217. [Google Scholar]
Harshey RM (1988). Phage Mu. The Bacteriophages, 193–234. [Google Scholar]
Harshey RM (2012). The Mu story: How a maverick phage moved the field forward. Mobile DNA 3: 21. [Google Scholar]
Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H (2001). Complete genome sequence of enterohemorrhagic Escherichia coli O157: H7 and genomic comparison with a laboratory strain K-12. DNA Research 8: 11–22. [Google Scholar]
Hofmann K, Stoffel W (1993). TMbase-A database of membrane spanning proteins segments. Biological Chemistry Hoppe-Seyler 374: 166. [Google Scholar]
Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB (2017). Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME Journal 11: 1511–1520. [Google Scholar]
Howe MM (1998). Bacteriophage Mu. In: Stephan JWB, Christopher MT, Nigel LB (eds.pp. 65–80. Molecular Microbiology, Springer, Berlin, Hidelberg. [Google Scholar]
Hull RA, Gill GS, Curtiss R, 3rd (1978). Genetic characterization of Mu-like bacteriophage D108. Journal of Virology 27: 513–518. [Google Scholar]
Jakhetia R, Verma NK (2015). Identification and molecular characterisation of a Novel Mu-like bacteriophage, SfMu, of Shigella flexneri. PLoS One 10: e0124053. [Google Scholar]
Jeon J, Yong D (2019). Two novel bacteriophages improve survival in Galleria mellonella infection and mouse acute pneumonia models infected with extensively drug-resistant Pseudomonas aeruginosa. Applied and Environmental Microbiology 85: e02900–02918. [Google Scholar]
Kakikawa M, Yokoi Kj, Kimoto H, Nakano M, Kawasaki KI, Taketo A, Kodaira KI (2002). Molecular analysis of the lysis protein Lys encoded by Lactobacillus plantarum phage φg1e. Gene 299: 227–234. [Google Scholar]
Kashani HH, Schmelcher M, Sabzalipoor H, Hosseini ES, Moniri R (2018). Recombinant endolysins as potential therapeutics against antibiotic-resistant Staphylococcus aureus: Current status of research and novel delivery strategies. Clinical Microbiology Reviews 31: e00071–00017. [Google Scholar]
Klee SR, Nassif X, Kusecek B, Merker P, Beretti JL, Achtman M, Tinsley CR (2000). Molecular and biological analysis of eight genetic islands that distinguish Neisseria meningitidis from the closely related pathogen Neisseria gonorrhoeae. Infection and Immunity 68: 2082–2095. [Google Scholar]
Kongari R, Rajaure M, Cahill J, Rasche E, Mijalis E, Berry J, Young R (2018). Phage spanins: Diversity, topological dynamics and gene convergence. BMC Bioinformatics 19: 326. [Google Scholar]
Larpin Y, Oechslin F, Moreillon P, Resch G, Entenza JM, Mancini S (2018). In vitro characterization of PlyE146, a novel phage lysin that targets Gram-negative bacteria. PLoS One 13: e0192507. [Google Scholar]
Lood R, Winer BY, Pelzek AJ, Diez-Martinez R, Thandar M, Euler CW, Schuch R, Fischetti VA (2015). Novel phage lysin capable of killing the multidrug-resistant gram-negative bacterium Acinetobacter baumannii in a mouse bacteremia model. Antimicrobial Agents and Chemotherapy 59: 1983–1991. [Google Scholar]
Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH (2017). CDD/SPARCLE:functional classification of proteins via subfamily domain architectures. Nucleic Acids Research 45: D200–D203. [Google Scholar]
Margolin W, Howe MM (1990). Activation of the bacteriophage Mu lys promoter by Mu C protein requires the sigma 70 subunit of Escherichia coli RNA polymerase. Journal of Bacteriology 172: 1424–1429. [Google Scholar]
Margolin W, Rao G, Howe M (1989). Bacteriophage Mu late promoters: four late transcripts initiate near a conserved sequence. Journal of Bacteriology 171: 2003–2018. [Google Scholar]
Masignani V, Giuliani MM, Tettelin H, Comanducci M, Rappuoli R, Scarlato V (2001). Mu-like Prophage in serogroup B Neisseria meningitidis coding for surface-exposed antigens. Infection and Immunity 69: 2580–2588. [Google Scholar]
Mathee K, Howe MM (1993). The bacteriophage Mu middle operon: Essential and nonessential functions. Virology 196: 712–721. [Google Scholar]
Mizuuchi K (1983). In vitro transposition of bacteriophage Mu: A biochemical approach to a novel replication reaction. Cell 35: 785–794. [Google Scholar]
Morgan GJ, Hatfull GF, Casjens S, Hendrix RW (2002). Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. Journal of Molecular Biology 317: 337–359. [Google Scholar]
Naghizadeh M, Karimi Torshizi MA, Rahimi S, Dalgaard TS (2019). Synergistic effect of phage therapy using a cocktail rather than a single phage in the control of severe colibacillosis in quails. Poultry Science 98: 653–663. [Google Scholar]
NCBI Resource Coordinators (2018). Database resources of the national center for biotechnology information. Nucleic Acids Research 46: D8–D13. [Google Scholar]
Nelson D, Loomis L, Fischetti VA (2001). Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proceedings of the National Academy of Sciences of the United States of America 98: 4107–4112. [Google Scholar]
Niu YD, Cook SR, Wang J, Klima CL, Hsu YH, Kropinski AM, Turner D, McAllister TA (2015). Comparative analysis of multiple inducible phages from Mannheimia haemolytica. BMC Microbiology 15: 175. [Google Scholar]
Oliveira H, Azeredo J, Lavigne R, Kluskens LD (2012). Bacteriophage endolysins as a response to emerging foodborne pathogens. Trends in Food Science & Technology 28: 103–115. [Google Scholar]
Omasits U, Ahrens CH, Müller S, Wollscheid B (2014). Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30: 884–886. [Google Scholar]
Paolozzi L (1987). The SE region, Phage Mu, 53–62. [Google Scholar]
Paolozzi L, Ghelardini P (2006). The bacteriophage Mu. In: Calendar R (ed.pp. 469–496. The Bacteriophages. Oxford University Press, New York, USA. [Google Scholar]
Park T, Struck DK, Deaton JF, Young R (2006). Topological dynamics of holins in programmed bacterial lysis. Proceedings of the National Academy of Sciences of the United States of America 103: 19713–19718. [Google Scholar]
Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O’Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG (2001). Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413: 848–852. [Google Scholar]
Pastagia M, Schuch R, Fischetti VA, Huang DB (2013). Lysins: the arrival of pathogen-directed anti-infectives. Journal of Medical Microbiology 62: 1506–1516. [Google Scholar]
Peng Q, Yuan Y (2018). Characterization of a novel phage infecting the pathogenic multidrug-resistant Bacillus cereus and functional analysis of its endolysin. Applied Microbiology and Biotechnology 102: 7901–7912. [Google Scholar]
Pundir S, Martin MJ, O’Donovan C (2017). UniProt protein knowledgebase. In: Cathy HW, Cecilia NA, Karen ER (eds.pp. 41–55. Protein Bioinformatics. Humana Press, New York, USA. [Google Scholar]
Ramanculov E, Young R (2001). An ancient player unmasked: T4 rI encodes a t-specific antiholin. Molecularrr Microbiology 41: 575–583. [Google Scholar]
Reddy BL, Saier MH,Jr (2013). Topological and phylogenetic analyses of bacterial holin families and superfamilies. Biochimica et Biophysica Acta 1828: 2654–2671. [Google Scholar]
Rodríguez-Rubio L, Chang WL, Gutiérrez D, Lavigne R, Martínez B, Rodríguez A, Govers SK, Aertsen A, Hirl C, Biebl M, Briers Y, García P (2016). Artilysation’of endolysin λSa2lys strongly improves its enzymatic and antibacterial activity against streptococci. Scientific Reports 6: 35382. [Google Scholar]
Roncero C, Darzins A, Casadaban MJ (1990). Pseudomonas aeruginosa transposable bacteriophages D3112 and B3 require pili and surface growth for adsorption. Journal of Bacteriology 172: 1899–1904. [Google Scholar]
Saha RP, Lou Z, Meng L, Harshey RM (2013). Transposable prophage Mu is organized as a stable chromosomal domain of E. coli. PLoS Genetics 9: e1003902. [Google Scholar]
São-José C, Parreira R, Vieira G, Santos MA (2000). The N-terminal region of the Oenococcus oeniBacteriophage fOg44 Lysin behaves as a bona fide signal peptide in Escherichia coli and as a cis-inhibitory element, preventing lytic activity on Oenococcal cells. Journal of Bacteriology 182: 5823–5831. [Google Scholar]
Seo BJ, Song ET, Lee K, Kim JW, Jeong CG, Moon SH, Son JS, Kang SH, Cho HS, Jung BY, Kim WI (2018). Evaluation of the broad-spectrum lytic capability of bacteriophage cocktails against various Salmonella serovars and their effects on weaned pigs infected with Salmonella typhimurium. Journal of Veterinary Medical Science 80: 851–860. [Google Scholar]
Summer EJ, Berry J, Tran TA, Niu L, Struck DK, Young R (2007). Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. Journal of Molecular Biology 373: 1098–1112. [Google Scholar]
Summer EJ, Gonzalez CF, Carlisle T, Mebane LM, Cass AM, Savva CG, LiPuma J, Young R (2004). Burkholderia cenocepacia phage BcepMu and a family of Mu-like phages encoding potential pathogenesis factors. Journal of Molecular Biology 340: 49–65. [Google Scholar]
Sun Q, Kuty GF, Arockiasamy A, Xu M, Young R, Sacchettini JC (2009). Regulation of a muralytic enzyme by dynamic membrane topology. Nature Structural & Molecular Biology 16: 1192–1194. [Google Scholar]
Swapna G, Kumari V, Nagaraja V (2015). Different modes of transactivation of bacteriophage Mu late promoters by transcription factor C. PLoS One 10: e0129504. [Google Scholar]
Taylor AL (1963). Bacteriophage-induced mutation in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 50: 1043–1051. [Google Scholar]
Thiry D, Passet V, Danis-Wlodarczyk K, Lood C, Wagemans J, De Sordi L, van Noort V, Dufour N, Debarbieux L, Mainil JG, Brisse S, Lavigne R (2019). New bacteriophages against emerging lineages ST23 and ST258 of Klebsiella pneumoniae and efficacy assessment in Galleria mellonella larvae. Viruses 11: 411. [Google Scholar]
To KH, Young R (2014). Probing the structure of the S105 hole. Journal of Bacteriology 196: 3683–3689. [Google Scholar]
Toussaint A (2013). Transposable Mu-like phages in Firmicutes: New instances of divergence generating retroelements. Research in Microbiology 164: 281–287. [Google Scholar]
UniProt Consortium (2019). UniProt: A worldwide hub of protein knowledge. Nucleic Acids Research 47: D506–D515. [Google Scholar]
van de Putte P, Westmaas GC, Wijffelman C (1977). Transfection with Mu-DNA. Virology 81: 152–159. [Google Scholar]
Vázquez R, García E, García P (2018). Phage lysins for fighting bacterial respiratory infections: A new generation of antimicrobials. Frontiers in Immunology 9: 2252. [Google Scholar]
Waggoner BT, Marrs CF, Howe MM, Pato ML (1984). Multiple factors and processes involved in host cell killing by bacteriophage Mu: Characterization and mapping. Virology 136: 168–185. [Google Scholar]
Waggoner BT, Sultana K, Symonds N, Karlok MA, Pato ML (1989). Identification of the bacteriophage Mu kil gene. Virology 173: 378–389. [Google Scholar]
Walker DM, Harshey RM (2020). Deep sequencing reveals new roles for MuB in transposition immunity and target-capture, and redefines the insular Ter region of E. coli. Mobile DNA 11: 26. [Google Scholar]
Wang IN, Smith DL, Young R (2000). Holins: the protein clocks of bacteriophage infections. Annual Review of Microbiology 54: 799–825. [Google Scholar]
Wang S, Kong J, Zhang X (2008). Identification and characterization of the two-component cell lysis cassette encoded by temperate bacteriophage φPYB5 of Lactobacillus fermentum. Journal of Applied Microbiology 105: 1939–1944. [Google Scholar]
Westmaas GC, van der Maas WL, van de Putte P (1976). Defective prophages of bacteriophage Mu. Molecular and General Genetics 145: 81–87. [Google Scholar]
White O, Eisen JA, Heidelberg JF, Hickey EK, Peterson JD, Dodson RJ, Haft DH, Gwinn ML, Nelson WC, Richardson DL, Moffat KS, Qin H, Jiang L, Pamphile W, Crosby M, Shen M, Vamathevan JJ, Lam P, McDonald L, Utterback T, Zalewski C, Makarova KS, Aravind L, Daly MJ, Minton KW, Fleischmann RD, Ketchum KA, Nelson KE, Salzberg S, Smith HO, Venter JC, Fraser CM (1999). Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286: 1571–1577. [Google Scholar]
White R, Chiba S, Pang T, Dewey JS, Savva CG, Holzenburg A, Pogliano K, Young R (2011). Holin triggering in real time. Proceedings of the National Academy of Sciences of the United States of America 108: 798–803. [Google Scholar]
Wu M, Hu K, Xie Y, Liu Y, Mu D, Guo H, Zhang Z, Zhang Y, Chang D, Shi Y (2019). A novel phage PD-6A3, and its endolysin Ply6A3, with extended lytic activity against Acinetobacter baumannii. Frontiers in Microbiology 9: 3302. [Google Scholar]
Xu M, Arulandu A, Struck DK, Swanson S, Sacchettini JC, Young R (2005). Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science 307: 113–117. [Google Scholar]
Xu M, Struck DK, Deaton J, Wang IN, Young R (2004). A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proceedings of the National Academy of Sciences of the United States of America 101: 6415–6420. [Google Scholar]
Yang J, Kong Y, Li X, Yang S (2015). A novel transposable Mu-like prophage in Bacillus alcalophilus CGMCC 1.3604 (ATCC 27647). Virologica Sinica 30: 63–65. [Google Scholar]
Young R (1992). Bacteriophage lysis: Mechanism and regulation. Microbiology and Molecular Biology Reviews 56: 430–481. [Google Scholar]
Young R (2013). Phage lysis: Do we have the hole story yet? Current Opinion in Microbiology 16: 790–797. [Google Scholar]
Young R (2014). Phage lysis: Three steps, three choices, one outcome. Journal of Microbiology 52: 243–258. [Google Scholar]
Young R, Bläsi U (1995). Holins: Form and function in bacteriophage lysis. FEMS Microbiology Reviews 17: 191–205. [Google Scholar]
Zehr ES, Tabatabai LB, Bayles DO (2012). Genomic and proteomic characterization of SuMu, a Mu-like bacteriophage infecting Haemophilus parasuis. BMC Genomics 13: 331. [Google Scholar]
Zha J, Zhao Z, Howe MM (1994). Identification and characterization of the terminators of the lys and P transcripts of bacteriophage Mu. Journal of Bacteriology 176: 1111–1120. [Google Scholar]
Zhang G, Zhao Y, Paramasivan S, Richter K, Morales S, Wormald PJ, Vreugde S (2018). Bacteriophage effectively kills multidrug resistant Staphylococcus aureus clinical isolates from chronic rhinosinusitis patients. International Forum of Allergy & Rhinology 8: 406–414. [Google Scholar]
Zhao J, Zhang Z, Tian C, Chen X, Hu L, Wei X, Li H, Lin W, Jiang A, Feng R, Yuan J, Yin Z, Zhao X (2019). Characterizing the biology of lytic bacteriophage vb_eaem_φeap-3 infecting multidrug-resistant Enterobacter aerogenes. Frontiers in Microbiology 10: 420. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |