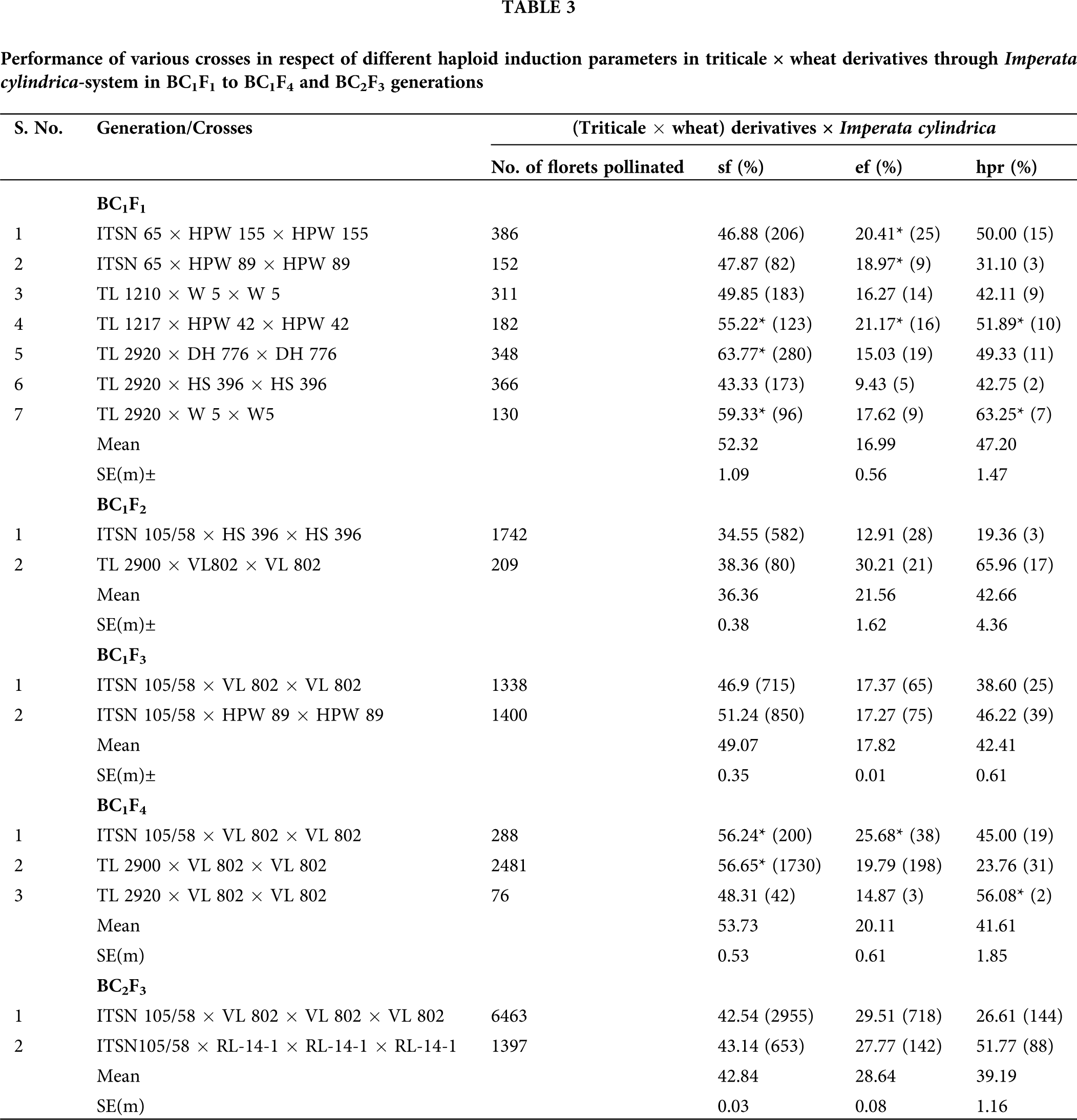
DOI:10.32604/biocell.2021.015735

| BIOCELL DOI:10.32604/biocell.2021.015735 |  |
| Article |
Doubled haploid production in advanced back cross generations and molecular cytogenetic characterization of rye chromatin in triticale × wheat derived doubled haploid lines
1AICRP(MULLaRP), Directorate of Research, CAU, Imphal, India
2Molecular Cytogenetics and Tissue Culture Laboratory, CSKHPKV, Palampur, India
3Department of Agricultural Biotechnology, CSKHPKV, Palampur, India
4Mountain Research Center for Field Crops, Sher-e-Kashmir University of Agricultural Sciences and Technology, Khudwani, India
*Address correspondence to: Muniyandi Samuel Jeberson, samuel8142@gmail.com; Shabir Hussain Wani, shabirhwani@skuastkashmir.ac.in
Received: 09 January 2021; Accepted: 24 May 2021
Abstract: The rye genome has shown potential for improvement of bread wheat, where wheat-rye substitutions and translocations have been and are frequently used in resistance breeding. Crosses belongs to different generations viz., BC1F1, BC1F2, BC1F3, BC1F4 and BC2F3 of triticale × wheat derived were used for different haploid induction parameters using Gogon grass (Imperata cylindrica) as a pollen source. The percentage of pseudo seed formation ranged from 34.55% for BC1F2 to 63.77 for BC1F1 crosses, the haploid embryo formation ranges from 9.43% for BC1F1 to 30.2% for BC1F2, the haploid plant generation ranges from 19.36% for BC1F2 to 63.25% for BC1F1. Four doubled haploids were developed from ITSN 105/58 × VL 802 × VL 802 of BC2F3 underwent molecular cytogenetic analyses using the probes, viz., rye genomic rDNA, pSc 119 and pAs1. FISH and GISH analysis revealed an IBL.1RS translocation and substitution of 5R chromosome instead of the 5D chromosomes in these doubled haploids.
Keywords: Haploid; Triticale × wheat backcross; FISH; GISH; Rye chromatin
Abbreviations
| FISH: | Fluorescence in situ hybridization |
| GISH: | Genotypic in situ hybridization |
| 2,4-D: | 2,4-dinitrophenylhydrazine |
| SSC: | Saline sodium citrate, |
| BSA: | Bovine serum albumin, |
| DAPI: | 4’,6-diamidino-2-phenylindole |
Among the food grain crops of the world, wheat (Triticum aestivum L. em Thell) is pre-eminent regarding its antiquity and importance as a food of humankind (Arjona et al., 2020). There has been a tremendous increase in wheat production in India since the time of the Green Revolution. This has been possible with the introduction of the dwarf wheat genotypes that carrying the dwarfing genes viz. Rht-B1b and Rht-D1b from Norin-10-Brevor-14 background (Borner et al., 1996) and the genes for photo-insensitivity from Mexican spring wheat. Wheat improvement has led the country’s efforts to reach the status of self-sufficiency in food grain production (Rani and Mor, 2020). However, at present, production is affected by climate change and increase in pest and diseases infestation (Dar et al., 2020; Wani et al., 2020). To address this problem, new disease resistant genes need to be introgressed from cultivars/wild resources (Pietrusińska et al., 2018; Klymiuk et al., 2019). In the mountainous regions of India, wheat is generally grown under diverse and rainfed conditions (Dar et al., 2020) Thus, drought becomes the major constraint, followed by frost stress and susceptibility to various diseases, which drastically reduce wheat production in these areas. Therefore, the breeding objectives must be essentially comprised of the development of high yielding varieties resistant to abiotic (drought and frost) and biotic (rusts and powdery mildew) stresses prevalent in this region. Winter wheat and rye are the important sources can be used for the transfer of resistance for biotic and abiotic stresses to spring wheat.
Triticale (× Triticosecale Wittmack) may be used as a bridging species to accomplish the transfer of the rye chromatin into the background of wheat. Because of rye chromosome complement, triticales have many agronomic attributes not found in wheat (Merker, 1984). To isolate promising recombinants from the segregating populations, careful selection of parents is required on the part of breeders for triticale × wheat hybridization programs. To achieve faster desirable results, doubled haploid breeding has a significant advantage over conventional breeding, where it leads to the production of completely homozygous plants in just a single step (1 year). In contrast, the conventional breeding approach takes about 7–8 years to isolate stable lines from the crosses. Bread wheat has been improved using the rye genome as a potential resource, where wheat-rye translocations and substitutions have been and are commonly used in resistance breeding (Rabinovich, 1998), and the 1B/1R wheat-rye translocation is incorporated in most of the wheat currently grown around the world (Heslop-Horrison et al., 1990). Doubled haploid breeding helps to attain homozygous populations from the triticale × wheat hybrids and their backcrosses following chromosome elimination approach through the use of Imperata cylindrica-mediated systems. It is further required to enhance the precision and efficiency of selection amongst the newly developed wheat doubled haploids to identify alien chromatin/genes introgressed with minimal introgression of undesirable genes. The molecular cytogenetic approach gives various powerful and novel tools such as genomic in situ hybridization (GISH) and fluorescence in situ hybridization (FISH) which can help in the physical mapping of introgressed chromatin/genes into wheat genome. The present investigation envisages the development of triticale × wheat-derived doubled haploids involving triticale and elite wheat lines following intergeneric hybridization with I. cylindrica and identification and characterization of rye chromatin introgressed into wheat genome (developed through DH breeding) through FISH and further isolation of wheat like doubled haploids having the desired rye chromatin with minimal undesirable genes.
Triticale × wheat (Tab. 1) derived populations of different backcross generations used for doubled haploid production following chromosome elimination approach by using Imperata cylindrica pollen and further utilized to detect rye chromatin introgression using GISH and FISH techniques. The present work was carried out in the Molecular Cytogenetics Laboratory of Department of Crop Improvement, CSK HP Agricultural University, Palampur, H.P, India.

Emasculation was done three days before anthesis by removing the anthers manually. The next day, fresh pollen from I. cylindrica was collected and applied to the feathery stigma of the emasculated spikes. On third day, the spikes were injected with a 2,4-D solution of 250 mg/L concentration (Pratap and Chaudhary, 2007) at the base of the uppermost internode using a syringe fitted with a fine hypodermic disposable needle. Petroleum jelly (Vaseline-Hindustan Lever, Ltd.) was used for sealing the injection holes. The injections were repeated for two more consecutive days to ensure proper seed and embryo formation. Murashige and Skoog medium (Murashige and Skoog, 1962) was used to rescue haploid embryos. This medium was supplemented with 0.5 mg/L kinetin, 150 mg/L glutamine, 20 mg/L each arginine, cysteine and leucine, and solidified agar. The pollinated spikes were harvested from the tiller base after 18–20 days of pollination. The embryo carrying seeds were identified using the technique of Bains et al. (1998). The embryos were removed under strict aseptic conditions and placed on the culture medium in the test tubes. Cultured immature embryos were given cold treatment at 4°C temperature in dark for first 24 h. After that, they were incubated in the dark in the Plant Growth Chamber at 25 ± 1°C for regeneration for about a week till the roots and shoots initiated. The regenerated plantlets were then transferred to the other section of the Plant Growth Chamber at 25 ± 1°C with 10/14 h light/dark profile for plants’ proper development. The haploid plantlets were transferred to rooting medium for profuse rooting, then potted in soil mixture for hardening and later treated with 0.1% colchicine solution for chromosome doubling. The haploid plantlets were treated with colchicine at three to five tiller stages according to the method given by Inagaki (1985) with slight modifications. Each haploid plant’s crown was submerged in a 0.1% colchicine solution supplemented with 1.5% dimethyl sulphoxide at 20°C for 5 h. The treated plants were kept in the running tap water for 20 min, then potted in soil and maintained in the cage house up to maturity.
Observations were recorded with respect to haploid induction traits on per cent basis as follows:
These data were transformed to definite value using Arcsine transformation. The significant difference for various haploid induction parameters, namely pseudo seed formation, embryo formation and haploid regeneration frequencies was analysed by simple t-test.
Genomic and fluorescence in situ hybridization procedure
Four doubled haploid lines derived from (ITSN 105/58 × VL 802) × VL 802 crosses of BC2F3 generation of the present investigations were utilized in this study for molecular cytogenetic analysis following genomic in situ hybridization (GISH), and Fluorescence in situ hybridization (FISH) approaches (Yamamoto and Mukai, 1989; Mukai et al., 1993) to identify and characterize the introgressed rye chromatin and isolate wheat like recombinants with less undesirable genes. Molecular Probes (Tab. 2) viz., Genomic probe of rye, Ribosomal DNA probe (pTa 71) and repetitive DNA sequences probes (pSc119 and pAs1) were used to detect and characterize the alien introgressions (Yamamoto and Mukai, 1989). All the probes were labelled with the haptens viz., biotin-16-dUTP (Vitamin H) and digoxigenin-11-dUTP (Steroid) following the nick translation protocol given by (Maniatis et al., 1975). Detection of the labelled sites was executed by the fluorophores viz., fluorescein-conjugated streptavidin and rhodamine-conjugated anti-digoxigenin.

Hybridization signals were detected with an Olympus fluorescence microscope equipped with a filter for FITC (fluorescein isothiocyanate), a filter for rhodamine and a triple-band filter set for FITC, DAPI (4’,6-diamidino-2-phenylindole) and rhodamine. Images were captured using an Olympus CCD (charge-coupled device) camera.
The data were transformed to definite value using Arcsine transformation (Warton and Hui, 2011). The significant difference for various haploid induction parameters, namely pseudo seed formation, embryo formation and haploid regeneration frequencies was analysed by simple t-test.
The pseudo seed formation data are provided below (Tab. 3). The data were transferred from percentage to absolute value for easy analysis using arcsine transformation. The pseudo seed formation was lower in the crosses of BC1F2, whereas higher in the crosses of BC1F4 generations. Variation in the pseudo seed formation might be due to genotype specificity in the crosses. The embryo formation was lower in the crosses of BC1F1, whereas higher in the crosses of BC2F3 generations. It may happen because all the pseudo seeds might not be containing the haploid embryo due to the chromosomal disharmony between the triticale × wheat derivatives and Imperata cylindrica. Although successful wheat haploid formation after crossing with I. cylindrica has been reported through cytological evidence of fertilization of parental gametes and further complete elimination of I. cylindrica chromosomes. The haploid plant regeneration was lower in the crosses of BC1F3 and higher in the crosses of BC1F1 generations. The recovery of doubled haploids was less after colchicine treatment due to the mortality of haploid plants. The cross (ITSN 105/58 × VL 802) × VL 802 × VL 802 of BC2F3 generation was yielded four doubled haploids. The pseudo seed formation data are provided below (Tab. 3). The data were transferred from percentage to absolute value for easy analysis using Arcsine transformation. The pseudo seed formation was lower in the crosses of BC1F2, whereas higher in the crosses of BC1F4 generations. Variation in the pseudo seed formation might be due to genotype specificity in the crosses. The embryo formation was lower in the crosses of BC1F1, whereas higher in the crosses of BC2F3 generations. This may happen because all the pseudo seeds might not be containing the haploid embryo due to the chromosomal disharmony between the triticale × wheat derivatives and Imperata cylindrica. Although successful wheat haploid formation after crossing with I. cylindrica has been reported through cytological evidence as the elimination of complete set of I. cylindrica chromosomes. The haploid plant regeneration was lower in the crosses of BC1F3 and higher in the crosses of BC1F1 generations. The recovery of doubled haploids was less after colchicine treatment due to mortality of haploid plants. The cross (ITSN 105/58 × VL 802) × VL 802 × VL 802 of BC2F3 generation was yielded four doubled haploids.
FISH and GISH analysis in triticale × wheat-derived doubled haploids
The four doubled haploid lines TWDH 1, TWDH 2, TWDH 4 and TWDH 5 (Figs. 1–4) derived from the BC2F3s of ITSN 105/58 × VL 802 × VL 802 were also subjected to molecular cytogenetic analysis using probes viz., rye genomic rDNA, pSc119 and pAs1 (Tab. 4). The lines TWDH 1, TWDH 2 (Figs. 5 and 6), and TWDH 4 were possessing 1BL.1RS translocation. The photographic plates of spikes and seeds of TWDHs are given in Figs. 1 to 4. The GISH analysis revealed that 1RS replaced the translocation chromosome’s short arms due to its obvious satellite. The line TWDH 1 (Fig. 2), apart from 1BL.1RS translocation, showed 5D (5R) chromosome substitution and the line TWDH 5 exhibited 5D (5R) chromosome substitution.

Figure 1: Spike and seeds of the doubled haploid, TWDH 1.

Figure 2: Spike and seeds of the doubled haploid, TWDH 2.

Figure 3: Spike and seeds of the doubled haploid, TWDH 4.

Figure 4: Spike and seeds of the doubled haploid, TWDH 5.

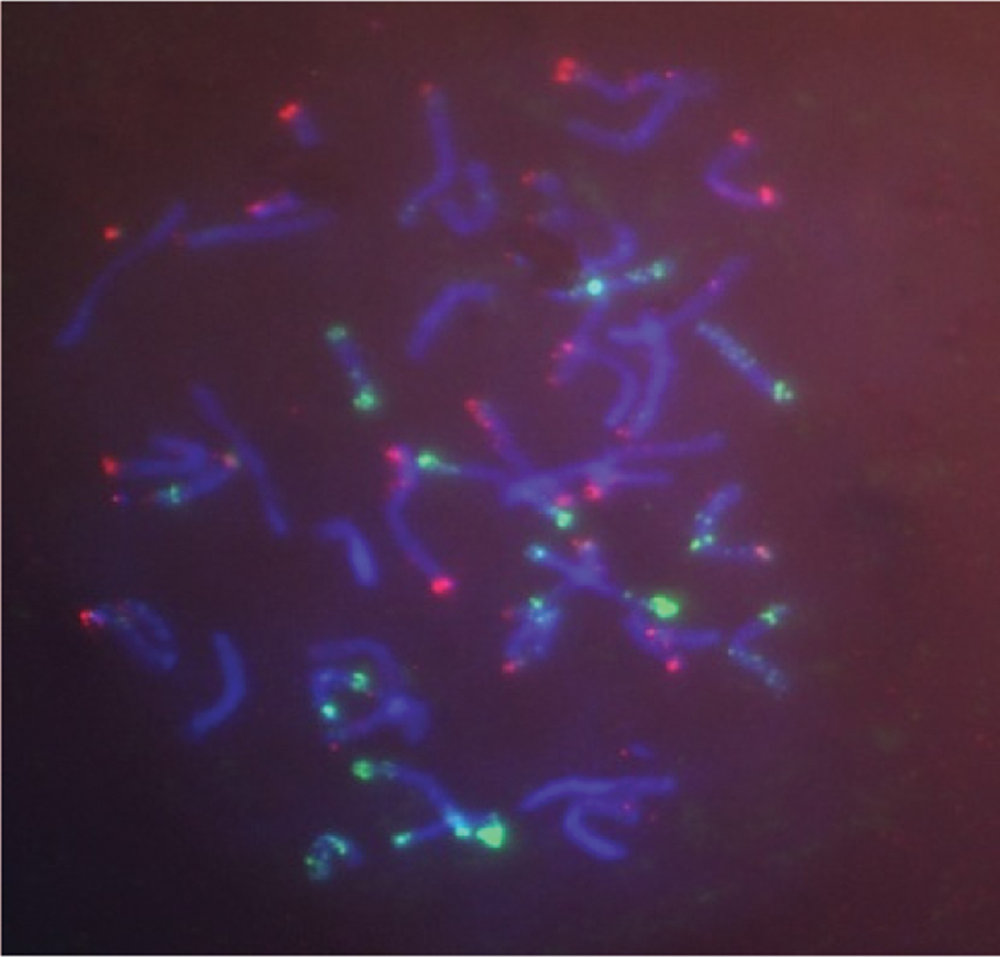
Figure 5: Detection of 1BL. 1RS translocation in triticale × wheat derived doubled haploid bread wheat line, TWDH 2 with the probe Bio: pAs1 (green), Dig: pSc 119 (red).

Figure 6: Detection of 1 BL. 1 RS translocation and 5D (5R) substitution in triticale × wheat derived doubled haploid bread wheat line, TWDH 1 with the probe Bio: rye genomic (green), Dig: rDNA (red).
The good triticale response to Imperata cylindrica induced haploid induction has also been reported in the few studies carried out earlier. Kishore et al. (2011) had reported 21.3% pseudo seed formation in BC1F1 and 46.8% in BC1F2, and Pratap and Chaudhary (2007) had reported similar results. Chaudhary et al. (2015) findings of enhanced production of pseudo seed (30.2 to 56.3%) were also corroborated with the present investigation. The successful seed set obtained in this investigation (62.79%) was close to the value reported by Matzk and Mahn (1994) with 12 wheat genotypes. Percent of seed set represents the efficiency of emasculation and pollination procedures. The lack of embryos in the F1s of triticale × wheat in the current study indicated that either fertilization did not take place or embryo development was stalled at an early stage. The problem seemed to be accentuated due to the chromosomal imbalance in the gametes produced by triticale × wheat F1s. With the idea of improving chromosomal balance in the gametes of triticale × wheat crosses, the F1s were advanced to the subsequent generations by backcrossing to wheat or selfing. Advanced generations (BC1F1, BC1F2, BC1F3, BC1F4 and BC2F3) were used for attempting crosses with maize employing 250 ppm 2, 4-D hormonal treatment. This kind of hormonal modification was used by Pratap et al. (2004); Chaudhary et al. (2002) with 20% pseudo seed production. However, variation was observed even within a group of lines derived from the same triticale × wheat cross where some lines had very low to no response to maize-mediated induction. Again, a probable reason may be the imbalanced chromosome complement of these lines. These results are in correspondence with the previous studies by Gill et al. (2008). They reported low seed set and no embryo development in F1 lines crossed with maize. Overall, the approach has high feasibility as a rapid technique for generating chromosome transfers between triticale and wheat. Successful seed formation was reported by Pratap et al. (2005) in all the 15 triticale × wheat hybrids fertilized with maize, Imperata cylindrica, pearl millet, sorghum pollen which suggests that these species effect fertilization in triticale × wheat hybrids, thereby stimulating seed formation. Similar studies were conducted by Mahato and Chaudhary (2015) involving seven diverse durum wheat genotypes and using two composite varieties of Himalayan maize, viz., Bajaura Makka and Early Composite and a wild grass, I. cylindrica, as pollen sources. Their result showed that I. cylindrica performed better for haploid induction in durum wheat over maize in terms of pseudo seed formation (46.93%), embryo formation (38.06%), haploid regeneration (40.42%) and haploid formation efficiency (7.44%). Authors opined the use of durum wheat × I. cylindrica as a superior technique over the maize-mediated system, and its large-scale use could open a new horizon in the sphere of durum wheat doubled haploid breeding programme.
Very few investigations have been reported to produce Doubled Haploid (DH) on triticale × wheat cross using I. cylindrica. Several technical problems affect the haploid embryo production efficiency. The genotypes pollinated with I. cylindrica pollen coupled with the post-pollination treatments efficiently produced haploids and doubled haploids. However, in cross combinations, the frequency of both haploid embryo development and DH production varied considerably. Imperata cylindrica was crossable with all the triticale × wheat-derived F1 hybrids, as shown by pseudo seed formation and embryo formation in all wheat genotypes. These hybrids’ parental lines did not carry recessive crossability alleles kr1 and kr2 except for variety C 306. Thus, the wheat × maize system, as demonstrated earlier (Singh et al., 2005; Bakos et al., 2005), is independent of crossability alleles kr1 and kr2. Similarly, I. cylindrica also independent of crossability problem observed by Jamwal et al. (2016) in triticale × wheat recombinants used for DH production. Many possible reasons for the effect of the environment on haploid embryo recovery have been reported in other similar investigation (Pienaar et al., 1996). The variation in glasshouse condition could affect the pollen viability and wheat fertilization. Similarly, environment influenced durum wheat embryo survival in a genotypically dependent manner (Donoughue and Bennett, 1994). As reported, the performance of cultivar ‘Rampton Rivet’ for embryo recovery was significantly better in a 20°C growth room than in an unheated glasshouse as compared to cultivars ‘Wakona’ and ‘Chinese Spring’. Campbell et al. (1998) evaluated the effects of temperature and light intensity on wheat genotypes crossed with maize pollen and showed that both could significantly affect haploid embryo numbers. They also reported that the light intensity of 1000 µmol/m2 s produced the greatest number of embryos (38% of florets pollinated) compared to the optimal temperature (22/17°C) embryo recovery. Overall germination rates were still low (~40% of all embryos) irrespective of variety. Low germination rates (43%) of wheat × maize cross-derived embryos were consistently observed (Inagaki and Tahir, 1990). Similarly, the non-development of embryos into plants in this investigation appeared due to vitrification or hyperhydricity. Badiyal et al. (2016) study also corroborated with present investigation for the haploid embryo formation using I. cylindrica as a pollen source. The chromosome elimination study (Komeda et al., 2007) indicated that I. cylindrica chromosomes were eliminated during the first mitotic cell division, whereas maize chromosomes were eliminated after two to three cell divisions. This gives a clear picture of the haploid nature of embryos obtained.
The present investigation of plant regeneration ranged from 19.36% to 65.69% for different triticale × wheat-derived generations. One of the factors limiting the further development of a germinated embryo to a plantlet is the embryos’ deficiency in one of their polarity (poles) to obtain shoot or root induction. In such cases, meristems are not properly formed (Lefebvre and Devaux, 1996). More florets can be pollinated for getting more haploid plants. Earlier investigations have revealed that spikelet positions (lower, middle, and upper) determined the success ratios for embryo initiation (Martins-Lopes et al., 2001). However, Bitsch et al. (1998) previously observed that embryo initiation was found to be distributed evenly all over the wheat spike. Similar results were obtained by Tayeng et al. (2012) in wheat × wheat recombinants using I. cylindrica as a pollen source. In a normal wheat × maize crossing process, 25–30 florets are generally pollinated per spike, and the synchronization of flowering decides the increase in the number of florets to be pollinated. Better space planting of the wheat plants might increase the number of spikes for pollination. Similar results of efficient haploid induction by I. cylindrica were obtained by Rather et al. (2014) for 21 F1 wheat crosses assessed for their haploid induction efficiency when crossed with four Indian and one Japanese accession of I. cylindrica. Authors concluded that both wheat and I. cylindrica genotypes influenced haploid induction and the accession Ic-Aru, performed better as a pollen source.
FISH and GISH analysis in triticale × wheat-derived doubled haploids
In the present investigation, doubled haploid line TWDH2 possessed IBL.IRS translocation, amber colour seed and good spike type and the line TWDH1 showed 1BL. 1 RS translocation and 5R (5D) substitution. A similar result was obtained by Carvalho et al. (2009) and Efremova et al. (2014) in wheat × rye crosses using fluorescent in situ hybridization (FISH) performed with genomic DNA probes for genomic in situ hybridization (GISH) from rye. Among the 55 plants, a wheat-rye translocation was detected in one plant after GISH. Recombinant chromosomes were identified using probes pTa71 and pSc119.2. Badaeva et al. (2002) reported that the repetitive DNA probe pAs1 was not only hybridized well with D-genome chromosomes of wheat but was also successfully used in the identification of specific chromosomes having different colour bands. In our investigation, the identification of D chromosomes present in the line TWDH2 was made using the probe pAs1. Tan et al. (2009) used FISH and suggested that line 15-3-2 possessed all 14 D-genome chromosomes and chromosome 5U, concluding that line 15-3-2 was a new synthetic wheat – Aegilops biuncialis partial amphiploid, and could be used to transfer the disease resistance genes to wheat. Georgiev (2008) reported that after GISH, it was obtained that the mutant forms K1 and K2 of Triticum aestivum carried the 1B/1R chromosome translocation. Altuntepe and Jauhar (2001) study of GISH in haploid plants derived from durum substitution lines also supports the current investigation results. An et al. (2015) developed WR49-1 using sequential GISH (genomic in situ hybridization), mc-FISH (multicolour fluorescence in situ hybridization), mc-GISH (multicolour GISH) and EST (expressed sequence tag)-based marker analysis, WR49-1 proved to be a new wheat-rye 6R disomic addition line. Yang et al. (2016) observed that the newly developed wheat-rye addition line N9436B possessed two rye chromosomes. A similar result of wheat-rye translocation T1RS.1BL was obtained by Ren et al. (2017) from the progeny of the crossing of the wheat cultivar Mianyang11-1 and a Chinese local rye variety, Weining. Two novel translocation lines were identified by molecular cytogenetic analysis and the results also revealed that the pSc119.2 signals of 5AL were absent in both lines along with the pSc119.2 signals of 4AL of RT828-11. Liu et al. (2017) was also observed one 1B(1R) substitution line and five 1BL.1RS whole-arm translocation lines. Most of the recombinant lines were associated with important alien chromatin translocation like 1BL/1RS, substitutions 1R (1D), 5R (5D) and combination of both, i.e., 1BL/1RS + 5R (5D) and in some cases presence of more than 4 rye chromosomes (Jeberson et al., 2021).
Very few investigations have been conducted using Imperata cylindrica as a pollen sources for crossing different generations of triticale × wheat derivatives. Therefore, from the above study, it can be inferred that I. cylindrica is a useful pollen source for efficient doubled haploid production in triticale × wheat advanced generations. Doubled haploids possessing an alien introgression of rye chromatin were identified through the molecular cytogenetic analysis viz., FISH and GISH technique, which are efficient techniques for detecting alien addition, translocation, and substitution lines. The developed doubled haploid lines can be released as varieties for use in future wheat breeding programmes.
Acknowledgement: The authors are thankful to the CSKHPKV, Palampur, Himachal Pradesh for providing necessary facilities to conduct this research.
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Author Contribution: The authors confirm contribution to the paper as follows: study conception and design: Muniyandi Samuel Jeberson, Harinder Kumar Chaudhary; data collection: Muniyandi Samuel Jeberson; analysis and interpretation of results: Muniyandi Samuel Jeberson, Harinder Kumar Chaudhary, Rakesh Kumar Chahota; draft manuscript preparation: Muniyandi Samuel Jeberson, Harinder Kumar Chaudhary, Rakesh Kumar Chahota, Shabir Hussain Wani. All authors reviewed the results and approved the final version of the manuscript.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
An D, Zheng Q, Luo Q, Ma P, Zhang H et al. (2015). Molecular cytogenetic identification of a new wheat-rye 6R chromosome disomic addition line with powdery mildew resistance. PLoS One 10: e0134534. DOI 10.1371/journal.pone. [Google Scholar] [CrossRef]
Altuntepe MD, Jauhar PP, (2001). Production of durum wheat substitution haploids from durum × maize crosses and their cytological characterization. Genome 44: 137–142. DOI 10.1139/gen-44-1-137. [Google Scholar] [CrossRef]
Arjona JM, Royo C, Dreisigacker S, Ammar K, Subirà J, Villegas D (2020). Effect of allele combinations at Ppd-1 loci on durum wheat grain filling at contrasting latitudes. Journal of Agronomy and Crop Science 206: 64–75. [Google Scholar]
Badaeva ED, Amosova AV, Muravenko OV, Samatadze TE, Chikida NN, Zelenin AV, Friebe B, Gill BS (2002). Genome differentiation in Aegilops. 3. Evolution of the D-genome cluster. Plant System and Evolution 231: 163–190. DOI 10.1007/s006060200018. [Google Scholar] [CrossRef]
Badiyal A, Chaudhary HK, Jamwal NS, Bhatt AK, Hussain W (2016). Comparative assessment of different auxin analogues on haploid induction in triticale × wheat derived backcross generations. Agricultural Research Journal 53: 157–168. [Google Scholar]
Bains NS, Mangat GS, Singh K, Nanda GS (1998). A simple technique for the identification of embryo carrying seeds from wheat × maize crosses prior to dissection. Plant Breeding 117: 191–192. DOI 10.1007/s10681-005-5812-9. [Google Scholar] [CrossRef]
Bakos F, Jager K, Barnabas B (2005). Regeneration of haploid plants after distant pollination of wheat via zygote rescue. Acta Biologica Cracoviensia 47: 167–171. [Google Scholar]
Bitsch C, Groger S, Lelley T (1998). Effect of parental genotypes on haploid embryo and plantlet formation in wheat × maize crosses. Euphytica 103: 319–323. DOI 10.1023/a:1018654000521. [Google Scholar] [CrossRef]
Borner A, Plaschke J, Korzun V, Worland AJ (1996). The relationship between the dwarfing gene of wheat and rye. Euphytica 89: 69–75. DOI 10.1007/BF00015721. [Google Scholar] [CrossRef]
Campbell AW, Griffin WB, Conner AJ, Rowarth JS, Burritt DJ (1998). The effects of temperature and light intensity on embryo numbers in wheat doubled haploid production through wheat × maize crosses. Annals of Botany 82: 29–33. DOI 10.1006/anbo.1998.0641. [Google Scholar] [CrossRef]
Carvalho A, Martin A, Heslop-Harrison JS, Guedes-Pinto H, Lima-Brito J (2009). Identification of the spontaneous 7BS/7RL intergenomic translocation in one F1 multigeneric hybrid from the Triticeae tribe. Plant Breeding 128: 105–108. DOI 10.1111/j.1439-0523.2008.01519.x. [Google Scholar] [CrossRef]
Chaudhary HK, Mahato A, Kaila V, Rather SA, Tayeng T (2015). Dihaploid induction in tetraploid durum wheat (Triticum durum L.) using pollen of Imperata cylindrica. Czech Journal of Genetics and Plant Breeding 51: 142–147. [Google Scholar]
Chaudhary HK, Sethi GS, Singh S, Pratap A, Sharma S (2005). Efficient haploid induction in wheat by using pollen of Imperata cylindrica. Plant Breeding 124: 96–98. DOI 10.1111/j.1439-0523.2004.01034. [Google Scholar] [CrossRef]
Chaudhary HK, Singh S, Sethi GS (2002). Interactive influence of wheat and maize genotypes on haploid induction in winter x spring wheat hybrids. Journal of Genetics and Breeding 56: 259–266. DOI 10.1007/s10681-005-5812-9. [Google Scholar] [CrossRef]
Dar SB, Kanth RH, Raja W, Bangroo SA, Rashid Z, Dar ZA, Wani SH (2020). Performance of wheat variety Shalimar Wheat-2 under rainfed conditions of temperate Kashmir as influenced by sowing dates and nitrogen levels. Journal of Cereal Research 12: 129–136. [Google Scholar]
Donoughue OLS, Bennett MD (1994). Comparative responses of tetraploid wheats pollinated with Zea mays L. and Hordeum bulbosum L. Theoretical and Applied Genetics 87: 673–680. DOI 10.1007/BF00222892. [Google Scholar] [CrossRef]
Efremova T, Trubacheeva N, Chumanova E, Badaeva E, Rosseeva I et al. (2014). Development and characterization of wheat-rye lines combining T1RS·1BL translocation and 5R (5D) chromosome substitution or T1RS·1BL and T5AS·5RL translocations. Cereal Research Communications 42: 547–557. DOI 10.1556/CRC.2014.0013. [Google Scholar] [CrossRef]
Georgiev S (2008). Activity of rRNA genes from rye chromosome in translocation mutant forms of Triticum aestivum L. Biotechnology and Biotechnology Equipment 22: 679–683. DOI 10.1080/13102818.2008.10817535. [Google Scholar] [CrossRef]
Gill RS, Puja, Dhindsa GS, Bains NS (2008). Maize-induced haploid production from triticale × wheat crosses. Indian Journal of Genetics 68: 387–390. [Google Scholar]
Heslop-Harrison JS, Leitch AR, Schwanzacher T, Anamthawat-Jonsson K, (1990). Detection and characterization of 1BL/1R translocation in hexaploid wheat. Heredity 65: 385–392. DOI 10.1038/hdy.1990.108. [Google Scholar] [CrossRef]
Inagaki MN (1985). Chromosome doubling of the wheat haploids obtained with crosses with Hordeum bulbosum L. Japanese Journal of Breeding 35: 193–195. DOI 10.1270/jsbbs1951.35.193. [Google Scholar] [CrossRef]
Inagaki M, Tahir M (1990). Comparison of haploid production frequencies in wheat varieties crossed with Hordeum bulbosum L. and maize. Japanese Journal of Breeding 40: 209–216. DOI 10.1270/jsbbs1951.45.157. [Google Scholar] [CrossRef]
Jamwal NS, Chaudhary HK, Badiyal A, Hussain W (2016). Factors influencing crossability among triticale and wheat and its subsequent effect along with hybrid necrosis on haploid induction, Acta Agriculturae Scandinavica, Section B. Soil & Plant Science 66: 282–289. [Google Scholar]
Jeberson MS, Chaudhary HK, Chahota RK, Ashokkumar K (2021). Identification of alien chromosome/chromatin introgressions in triticale × wheat derived stable lines through molecular cytogenetic analysis. Journal of Current Opinion in Crop Science 2: 1–10. [Google Scholar]
Kishore N, Chaudhary HK, Chahota RK, Kumar V, Sood SP, Jeberson S, Tayeng T (2011). Relative efficiency of the maize and Imperata cylindrica mediated chromosome elimination approaches for induction of haploids of wheat-rye derivatives. Plant Breeding 130: 192–194. [Google Scholar]
Klymiuk V, Fatiukha A, Huang L, Wei ZZ, Kis-Papo T et al. (2019). Durum wheat as a bridge between wild emmer wheat genetic resources and bread wheat. In: Applications of Genetic and Genomic Research in Cereals, pp. 201–230. Woodhead Publishing. [Google Scholar]
Komeda N, Chaudhary HK, Mukai Y (2007). Cytological evidence for chromosome elimination in wheat × Imperata cylindrica hybrids. Genes and Genetics Systems 82: 241–248. DOI 10.1266/ggs.82.241. [Google Scholar] [CrossRef]
Lefebvre D, Devaux P (1996). Doubled haploids of wheat from wheat × maize crosses: Genotypic influence, fertility and inheritance of the IBL.IRS chromosome. Theoretical and Applied Genetics 93: 1267–1273. DOI 10.1007/BF00223459. [Google Scholar] [CrossRef]
Liu C, Song Q, Zhang H, Yang Z, Hu Y (2017). Molecular cytogenetic characterization and phenotypic evaluation of new wheat-rye lines derived from hexaploid triticale ‘Certa’ × common wheat hybrids. Plant Breeding 136: 809–819. DOI 10.1111/pbr.12523. [Google Scholar] [CrossRef]
Mahato A, Chaudhary HK (2015). Relative efficiency of maize and Imperata cylindrica for haploid induction in Triticum durum following chromosome elimination-mediated approach of doubled haploid breeding. Plant Breeding 134: 379–383. DOI 10.1111/pbr.12288. [Google Scholar] [CrossRef]
Maniatis TA, Jeffrey A, Kleid DG (1975). Nucleotide sequence of the rightward operator of phage λ. Proceedings of the National Academy of Sciences of the United States of America 72: 1184–1188. DOI 10.1073/pnas.72.3.1184. [Google Scholar] [CrossRef]
Martins-Lopes PF, Guedes-Pinto H, Pinto-Carnide O, Snape J (2001). The effect of spikelet position on the success frequencies of wheat haploid production using the maize cross system. Euphytica 121: 265–271. DOI 10.1023/A:1012091610195. [Google Scholar] [CrossRef]
Matzk F, Mahn A (1994). Improved techniques for haploid production in wheat using chromosome elimination. Plant Breeding 113: 125–129. DOI 10.1111/j.1439-0523.1994.tb00714.x. [Google Scholar] [CrossRef]
Merker A (1984). The rye genome in wheat breeding. Hereditas 100: 183–191. DOI 10.1111/j.1601-5223. [Google Scholar] [CrossRef]
Mukai Y, Friebe B, Hatchett JH, Yamamoto M, Gill BS (1993). Molecular cytogenetic analysis of radiation-induced wheat- rye terminal and intercalary chromosomal translocation and the detection of rye chromatin specifying resistance to Hessian fly. Chromosoma 102: 88–95. DOI 10.1007/BF00356025. [Google Scholar] [CrossRef]
Murashige T, Skoog F (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiology of Plants 15: 473–497. DOI 10.1111/j.1399-3054.1962.tb08052.x. [Google Scholar] [CrossRef]
Pienaar R, Horn M, Lesch AJG (1996). Doubled haploids. In: Raupp WJ (eds.Annual Wheat Newsletter, vol. 42, pp. 199. [Google Scholar]
Pietrusińska AL, Żurek MO, Piechota UR, Słowacki PI, Smolińska KI (2018). Searching for diseases resistance sources in old cultivars, landraces and wild relatives of cereals. A review. Agronomy Science 73: 45–60. [Google Scholar]
Pratap A, Chaudhary HK (2007). Comparative studies on crossability among triticale and wheat gene pools under different climatic regimes. Journal of Genetics and Breeding 2: 141–143. DOI 10.17221/12991-CJGPB. [Google Scholar] [CrossRef]
Pratap A, Sethi GS, Chaudhary HK (2004). Comparative performance of androgenesis and maize-mediated systems of polyploidy induction in wheat and triticale. Journal of Genetics and Breeding 58: 311–318. [Google Scholar]
Pratap A, Sethi GS, Chaudhry HK (2005). Relative efficiency of different Gramineae genera for haploid induction in triticale and triticale × wheat hybrids through chromosome elimination technique. Plant Breeding 124: 147–153. DOI 10.1111/j.1439-0523.2004.01059.x. [Google Scholar] [CrossRef]
Rabinovich SV (1998). Importance of wheat-rye translocations for breeding modern cultivars of Triticum aestivum L. Euphytica 100: 323–340. DOI 10.1023/A:1018361819215. [Google Scholar] [CrossRef]
Rani A, Mor S (2020). Trends in food grains production in India: Analysis of India vis-à-vis North India. Plant Archives 20: 3592–3599. [Google Scholar]
Rather SA, Chaudhary HK, Kaila V (2014). Influence of different wheat and Imperata cylindrica genetic backgrounds on haploid induction efficiency in wheat doubled haploid breeding. Czech Journal of Genetics and Plant Breeding 50: 195–200. DOI 10.17221/105/2013-cjgpb. [Google Scholar] [CrossRef]
Ren T, Tang Z, Fu S, Yan B, Tan F, Ren Z, Li Z (2017). Molecular cytogenetic characterization of novel wheat-rye T1RS.1BL translocation lines with high resistance to diseases and great agronomic traits. Frontiers in Plant Science 8: 799. DOI 10.3389/fpls.2017.00799. [Google Scholar] [CrossRef]
Singh N, Behl RK, Punia MS (2005). Effect of genotypic background on haploid production through embryo rescue in wheat × maize crosses. Plant Soil Environment 51: 193–196. [Google Scholar]
Tan F, Zhou J, Yang Z, Zhang Y, Pan L, Ren Z (2009). Characterization of new synthetic wheat-Aegilops biuncialis partial amphiploid. African Journal of Biotechnology 8: 3215–3218. DOI 10.5897/AJB09.359. [Google Scholar] [CrossRef]
Tayeng T, Chaudhary HK, Kishore N (2012). Enhancing doubled haploid production efficiency in wheat (Triticum aestivum L. em. Thell) by in vivo colchicine manipulations in Imperata cylindrica-mediated chromosome elimination approach. Plant Breeding 131: 574–578. DOI 10.1111/j.1439-0523.2012.01986.x. [Google Scholar] [CrossRef]
Wani SH, Choudhary JR, Choudhary M, Rana M, Gosal SS (2020). Recent advances in genomics assisted breeding for drought stress tolerance in major cereals. Journal of Cereal Research 12: 1–12. [Google Scholar]
Warton DI, Hui FKC (2011). The arcsine is asinine: The analysis of proportions in ecology. Ecology 92: 3–10. DOI 10.2307/29779568. [Google Scholar] [CrossRef]
Yamamoto M, Mukai Y (1989). Application of fluorescence in situ hybridization to molecular cytogenetics of wheat. Wheat Information Service 69: 30–32. [Google Scholar]
Yang W, Wang C, Chen C, Wang Y, Zhang H, Liu X, Ji W (2016). Molecular cytogenetic identification of a wheat-rye 1R addition line with multiple spikelets and resistance to powdery mildew. Genome 59: 277–288. DOI 10.1139/gen-2015-0129. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |