DOI:10.32604/biocell.2021.09220

| Biocell DOI:10.32604/biocell.2021.09220 |  |
| Article |
High level of circPTN promotes proliferation and stemness in gastric cancer
1Department of Gastroenterology, Shanghai Changzheng Hospital, The Second Military Medical University, Shanghai, 200001, China
2Department of Gastroenterology, Shanghai First People’s Hospital, Shanghai, 200003, China
3Department of Gastroenterology, Shanghai Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai, 200003, China
*Address correspondence to: Liang Zhu, zhu_liang486@126.com
#Contributing equally to this work
Received: 26 November 2019; Accepted: 14 April 2020
Abstract: Increasing evidence proves that circular RNAs (circRNAs) play an important role in regulating the biological behaviors of tumors. The central purpose of this research was to investigate the functions of circRNA in gastric cancer. The utilization of real-time PCR was to test circPTN expression in gastric cancer cells. Cell counting colony formation assays, CCK-8 assay, and EdU assay were used to investigate proliferation. Transwell assay was applied to investigate migration. We discovered that circPTN was highly expressed in gastric cancer cells. Low expression of circPTN inhibits gastric cancer cell proliferation and migration. Elevated expression of circPTN promotes gastric cancer cell proliferation and migration. Moreover, we discovered that circPTN could accelerate self-renewal and increase the expression of stemness markers. The results of our study suggested that a high level of circPTN expression promotes the proliferation and stemness of gastric cancer cells.
Keywords: circPTN; Gastric cancer; Proliferation and stemness
Gastric cancer is considered to be one of the most common malignant tumors in the human digestive system, which ranks fourth in all malignant tumors worldwide and ranks third in mortality (Cancer Genome Atlas Research Network, 2014; Miller et al., 2016). Early symptoms of gastric cancer manifest lack clear clinical signs and sensitive biomarkers. In a large number of patients, the diagnosis of gastric cancer is often delayed (Leung et al., 2008; Shi et al., 2018). Although clinical treatment has made great progress, the five-year survival rate of patients with advanced gastric cancer is still low (Bornschein et al., 2011). Thus, a deeper understanding of the latent pathological mechanisms is imperative for the development of therapies of gastric cancer.
Circular RNAs (circRNAs) are considered to be a ubiquitous endogenous non-coding RNA in eukaryotic cells (Du et al., 2017b). CircRNAs have been reported to be useful as prognostic markers and targets for developing new treatments (Du et al., 2017b). Nevertheless, studies have indicated that some circRNAs could translate proteins (Pamudurti et al., 2017; Yang et al., 2017). Existing research showed that circRNA plays the crucial functions in the part of regulating expressions of genes in cells (Ashwal-Fluss et al., 2014; Zhang et al., 2009; Zheng et al., 2016). Recently, research demonstrated the discrepant expressions of circRNAs in GC tissues, which included 107 up-regulated and 201 down-regulated circRNAs (Kearney et al., 2017). In accordance with data, FNDC3B circular RNA accelerates the migrated and invaded capabilities in gastric cancer cells (Cai et al., 2012; Hong et al., 2019; Zhang et al., 2009). In addition, novel computational methods for the prediction of circRNA-disease associations were performed (Kearney et al., 2017). The IBNPKATZ looks like a useful biomedical research tool for predicting potential circRNA-disease associations (Kearney et al., 2017). However, we know extremely little about the biological functions of circPTN in the progression of gastric cancer. This study is aimed to investigate the functions of circPTN on cell proliferation and stemness in GC.
China Center for Type Culture Collection served as the channel for getting GC cell lines, including AGS, BGC-823, MGC-803, and SGC-7901. Cells were cultivated in 10% FBS in RPMI-1640 medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). And they were cultured with 5% CO2 at 37°C.
A circPTN template with an artificial flanking sequence was synthesized and inserted into pcDNA 3.1. For transient transfection, siRNA or plasmid was added to Optifectamine 2000 (Invitrogen, Carlsbad, CA, USA) in OptiMEM (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). It is transfected into cells in the form of a complex. The medium was changed after 6 h. And the whole RNA and protein were extracted within two days. In order to stabilize transfection, GenePharma prepared a circPTN lentiviral package (Bornschein et al., 2011). Cells were cultivated in a 24-well plate with 1 × 105 before lentivirus transfection. 24 h later, replaced the original medium with 2 mL fresh RPMI-1640 medium containing 6 g/mL polybrene, and add an appropriate amount of virus suspension at 37°C. After 4 h, 2 mL fresh medium was added to dilute polybrene. Continue to cultivate until the lentivirus contained fluorescent protein was detected in 72 h. After that, the transfected cells were picked out with 5 μg/mL puromycin for a fortnight.
Quantitative RT-PCR (reverse transcription–polymerase chain reaction)
Total RNA in cells was abstracted with Trizol regent (Invitrogen, Carlsbad, CA, USA). The quality and consistence of RNA were tested by utilizing a microplate reader. 2000 ng of RNA was reversed through a reverse transcription kit (Takara, Shiga Prefecture, Japan). RNA amplification was then carried out through SYBR Premix Ex TaqTM II (Takara, Shiga Prefecture, Japan) under the defined procedures. U6 is an internal reference gene. In the end, the expressions were analyzed by utilizing the 2−ΔΔCt method (Bornschein et al., 2011).
The adoption of cell lysate containing protease inhibitor was to abstract the whole proteins form cells. Protein consistence was tested by BCA (bicinchoninic acid) Protein Assay kit. Western buffer was added to the protein. The protein was denatured by boiling at 100°C for 10 min. 20 μL sample was added to each well of 12% SDS-PAGE (Sodium dodecyl sulfate-polyacrylamide gel electrophoresis) gel at 100 V for 120 min. The gel was applied to a PVDF (polyvinylidene fluoride) membrane with electrophoresed at 120 V for 90 min. Block with skim milk for 1 h at RT (Reverse Transcription). The primary antibody was cultivated on the membrane at 4°C overnight. After rinsed with PBST (Phosphate Buffered Saline) three times, the secondary antibody was cultivated on the membrane for two hours at RT (Reverse Transcription). After rinsed by PBST (Phosphate Buffered Saline), developing solution was added to the membrane. The outcomes of the experiment were observed through a chemiluminometer.
The cells were seeded into 98-well plates. In the 0–5 days, 10 μL of CKK-8 (Cell Counting Kit-8) solution (Dojindo, Kyushu, Japan) was added per well per day. On the fifth day, the cells were cultivated for two hours at 37°C. In the end, the absorbance of each well was measured at 450 nm by a microplate reader.
Six-well plates were adopted to cultivated cells at 37°C for 10 days. Cells were fixed with 1% paraformaldehyde. The colonies were stained with 0.1% crystal violet dye (RIBOBIO, Guangzhou, Guangdong, China).
Cell migration was analyzed by a Transwell chamber (8.0-μm pore size; 6.5-mm diameter, Corning, NYC, USA). Transfected cells were cultured on the upper chamber with Matrigel-coated of 24-well BioCoat Matrigel Invasion Chambers (Becton Dickinson and Company, Franklin Lakes, NJ, Jiangsu, China) and in serum-free RPMI 1640. After 24 h, cells in the upper chambers were not crystal violet stained until the medium was removed, and the upper chamber cells were scraped.
The adoption of 96-well plates was to cultivate cells. Then 100 μL of medium containing 20 μM EdU (5-Ethynyl-2’-deoxyuridine) was added to each well. The cells were cultivated for two hours at 37°C, 5% CO2. Following, they were fixed with 4% paraformaldehyde. Wash on a shaker with 0.5% Triton-X-100 PBS for 20 min. Cell staining was conducted utilizing the EdU (5-ethynyl-2’-deoxyuridine) Apollo DNA in vitro kit (RIBOBIO, Nanjing, Jiangsu, China) (Bornschein et al., 2011). In the end, cell proliferation was tested under an immunofluorescence microscope.
All data are expressed as mean ± standard deviation. All data were statistically analyzed by Student’s t-test or one-way analysis of variance. The most significant value is the probability of p < 0.05.
circPTN is upregulated in gastric cell lines
For the sake of disclosing the functions of circPTN in gastric cancer, we devised a divergent primer based on circRNA junction sequences and specifically targeted circPTN to precisely test circPTN expressions through RT-qPCR analysis in line with the circBase database (Glazar et al., 2014). For testing whether circPTN exerted crucial functions in GC, we tested circPTN expression in cells utilizing qRT-PCR. Then, we discovered that circPTN was highly expressed in gastric cancer cells in contrast with normal cells (Fig. 1A). Outcomes indicated that increased circPTN expression was observed in gastric cancer. Thus, we investigated whether circPTN controls the biological behavior of gastric cancer.
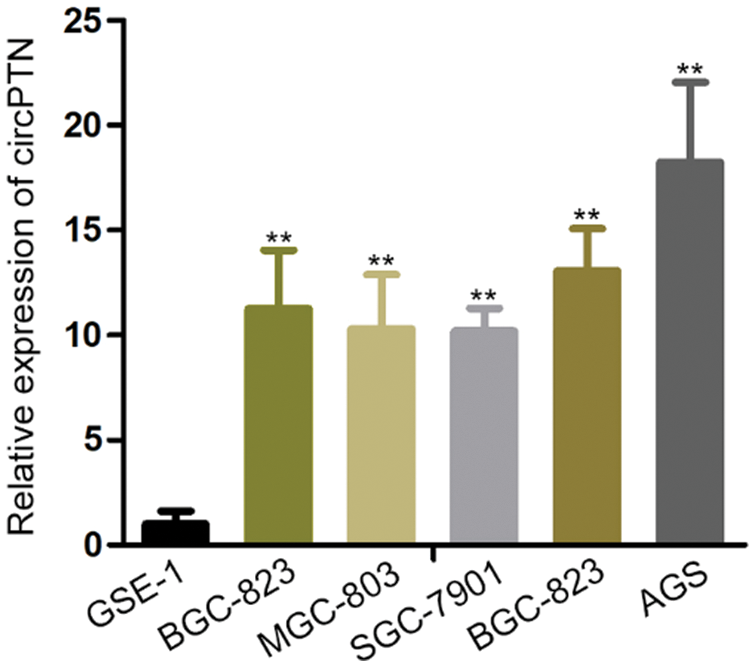
Figure 1: circPTN is upregulated in gastric cancer tissues and gastric cell lines.
(A) The expression of circPTN was observed in gastric cell lines. **p < 0.01.
circPTN promotes proliferation of GC
The circPTN was proved that is highly expressed in gastric cells. We tried to discover the functions of circPTN in the GC. Though previous studies have reported that circPTN is an oncogene that accelerates cell proliferation, we aimed to prove this effect in gastric cancer cells. Following, we set up the stable circPTN knockdown and overexpression systems by transfecting BGC-823 and AGS cells with lentivirus (Figs. 2A and 2B). By performing a colony formation experiment, we discovered that cell proliferation was restrained after silencing circPTN, while cell proliferation was increased after overexpressed circPTN (Figs. 2C and 2D). It demonstrated that silenced circPTN could suppress cell growth in gastric cancer.
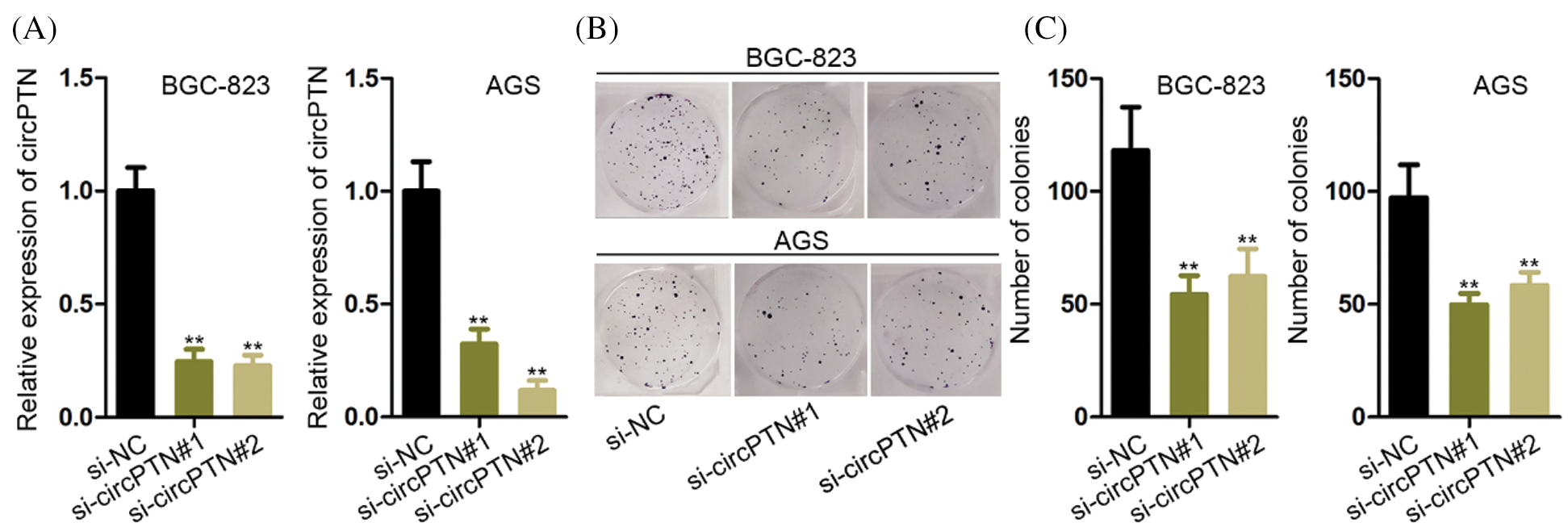
Figure 2: circPTN promotes the proliferation of GC.
(A) qRT-PCR detection of circPTN knockdown and overexpression efficiency. (B) Colony formation experiments were utilized to test cell proliferation after knockdown of circPTN. (C) Colony formation experiments were utilized to test cell proliferation after overexpression of circPTN. **p < 0.01.
circPTN promotes migration of GC
The circPTN was certified that it promoted migration in gastric cells. Similarly, we used stable circPTN knockdown and overexpression systems by transfecting BGC-823 and AGS cells with lentivirus. By performing a Transwell experiment, we discovered that cell migration was reduced after silencing circPTN, while cell migration was increased after overexpressed circPTN (Figs. 3A and 3B). It demonstrated that silenced circPTN could suppress cell migration in gastric cancer.
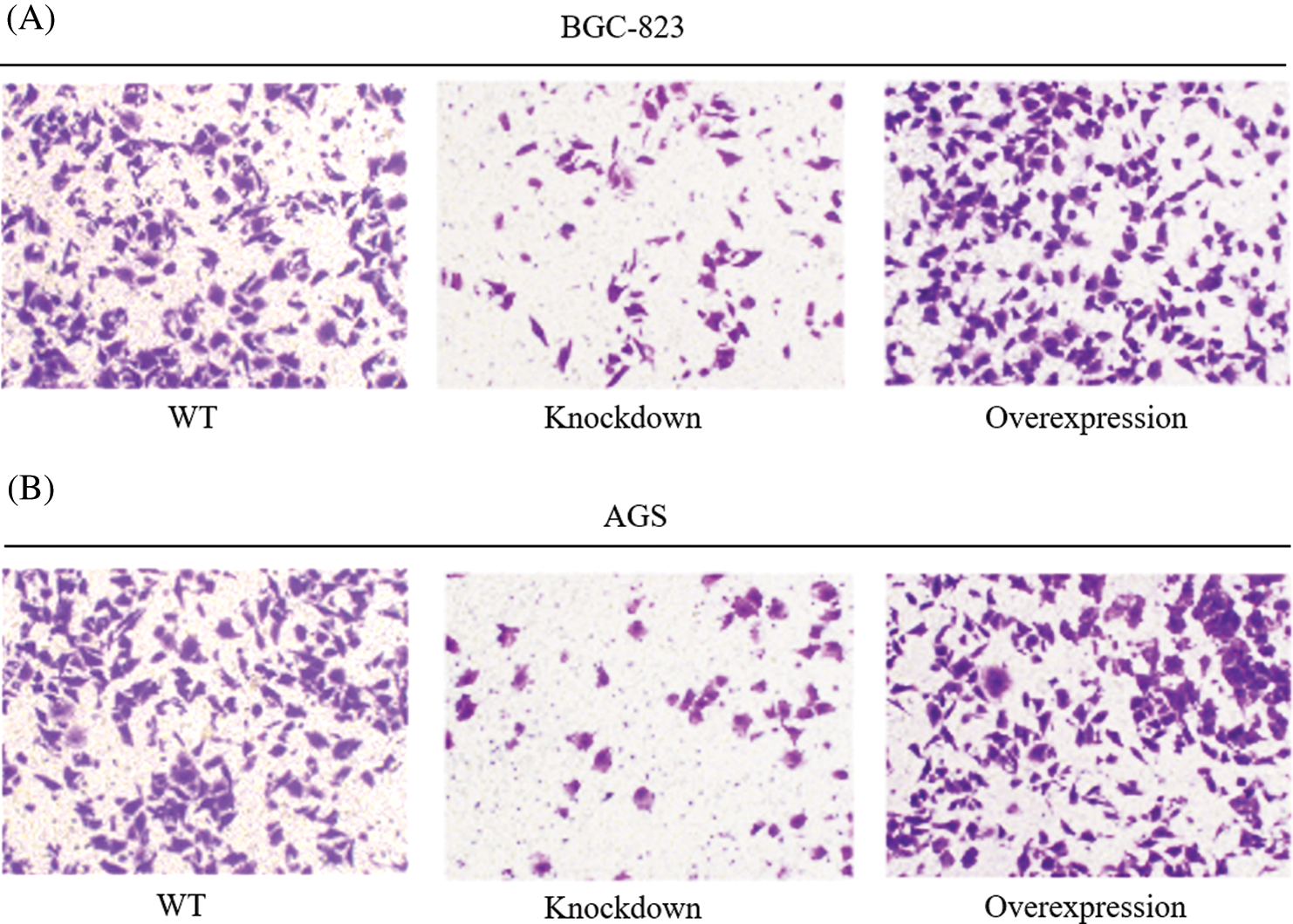
Figure 3: circPTN promotes the migration of GC.
Transwell experiments were utilized to test cell migration after knockdown and overexpression of circPTN in BGC-823 (A) and AGS cells (B). **p < 0.01.
The knockdown of circPTN inhibits the progression of gastric cancer
In order to ulteriorly test the functions of circPTN in gastric cancer cells, we conducted CCK-8 and EdU staining experiments in BGC-823 and AGS cells. We could evidently see that cell viability and EdU positive cells were declined (Figs. 4A and 4B). It indicated that cell proliferation capability was inhibited when circPTN was knocked down. By the large, we concluded that circPTN could induce gastric cancer cell proliferation.
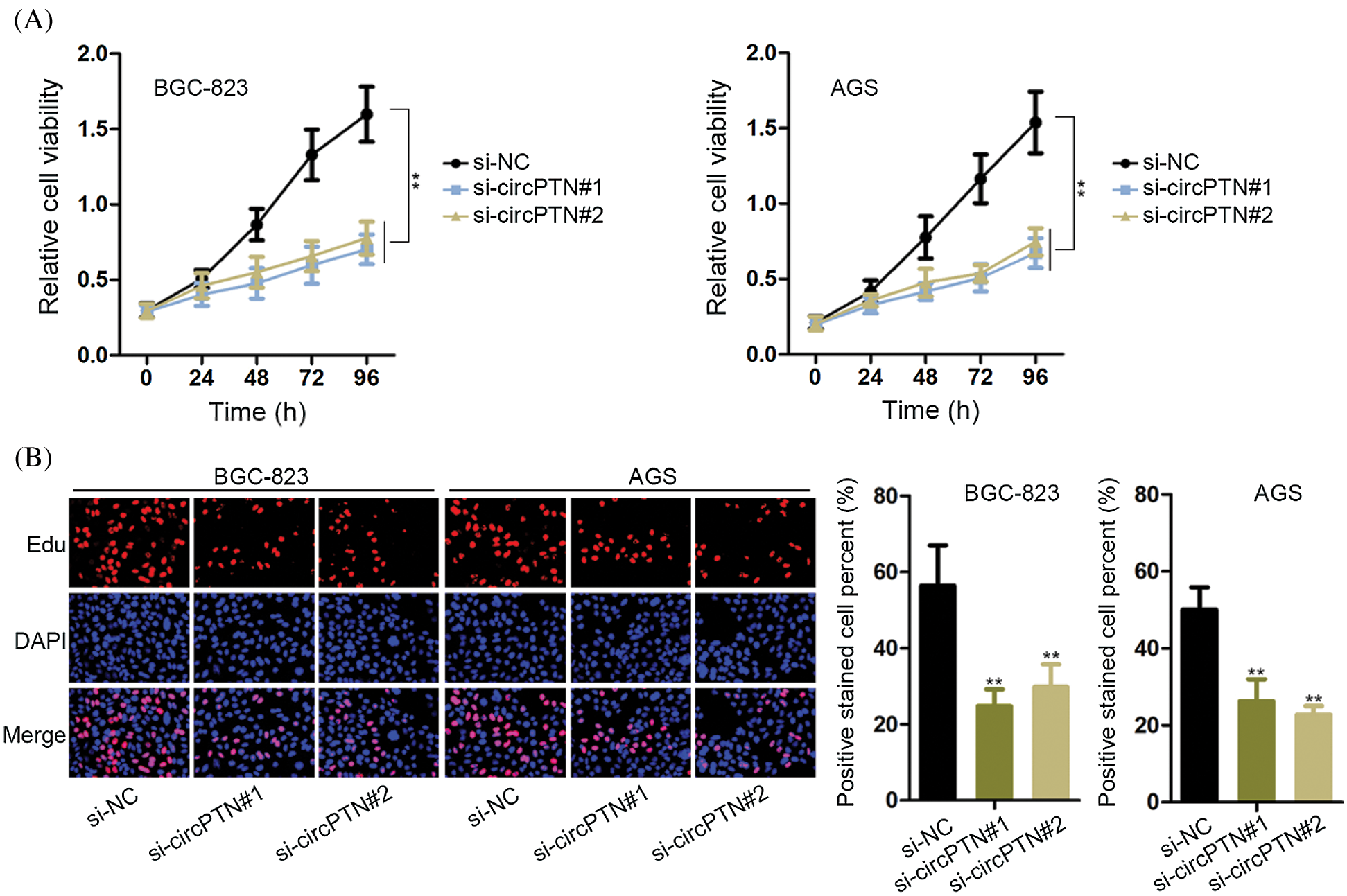
Figure 4: The knocking down of circPTN decreased gastric cancer cell growth.
(A) CCK-8 assay was utilized to detect cell proliferation after knockdown of circPTN. **p < 0.01. (B) EdU experiments were utilized to test cell proliferation after knockdown of circPTN. **p < 0.01.
circPTN promotes stemness of gastric cancer
We discovered that previous research had described that circPTN was related to the regulation of stemness or self-renewal (Aponte and Caicedo, 2017). Moreover, countless evidence demonstrated that gastric-stem cell (GSCs), clusters of cancer cells, possess the capacity for self-renewal (Bartfeld and Koo, 2017). Thus, we hypothesized that circPTN might influence self-renewal. We tested stemness markers, which included Nestin, CD133, CD133, SOX2, and SOX9 in BGC-823 and AGS cells. As we thought, the expression of stemness markers determined that these markers were positively associated with the circPTN level (Fig. 5A). Moreover, the outcomes demonstrated that circPTN could exert functions in regulating self-renewal. We examined the protein levels of stemness markers (CD133 and SOX2) in BGC-823 and AGS cells. Consequently, we discovered that the protein levels of these markers were significantly declined in cells, demonstrating that circPTN might exert the crucial roles in regulating stemness (Fig. 5B). Taken together, these outcomes indicated that circPTN may regulate self-renewal.
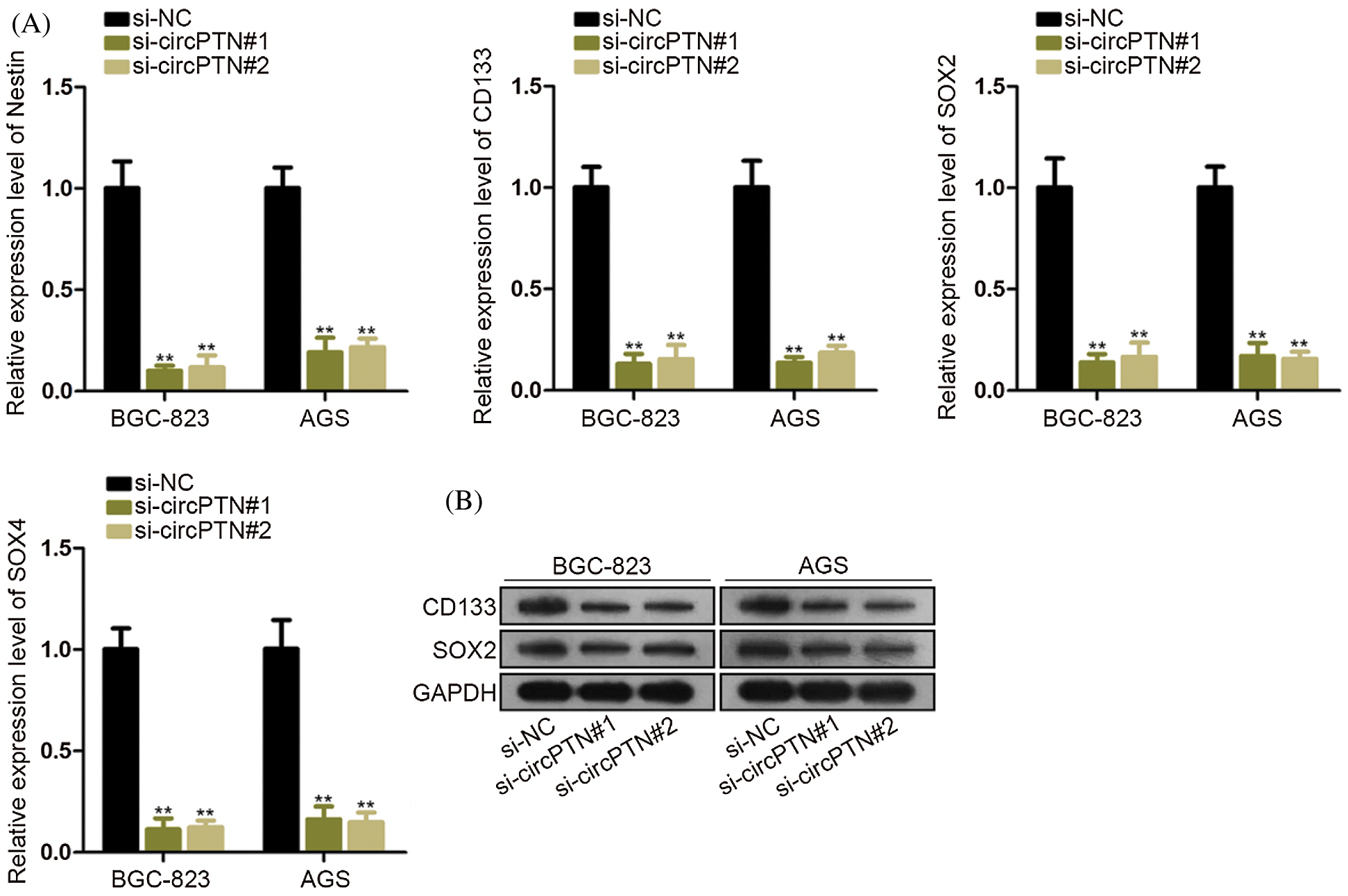
Figure 5: Decreasing circPTN inhibited the stemness of gastric cancer.
(A) qRT-PCR detection of mRNA levels of stem stemness markers. (B) Western bolt detection of protein levels of stem stemness markers. **p < 0.01.
GC is a malignancy with high morbidity and mortality (Choi and Kim, 2016). In the early stages, on account of the symptoms of GC are not distinct, the prognosis is poor. Recently, a considerable number of carcinogenic genes or tumor suppressors were proved to exert vital regulators in GC development (Song et al., 2017). Nevertheless, the functions of these entities in the control mechanisms involved in GC are little known. Thus, exploring new modulators and therapeutic targets is critical to comprehend the specific molecular mechanisms behind GC development. CircRNAs were discovered more than 30 years ago. But since Memczak et al. (2013) and Hansen et al. (2013) reported, circRNA have been recently attracted general interests (Hansen et al., 2013; Memczak et al., 2013). Lots of researches have reported that, in addition to its action as a miRNA sponge, circRNA may interact with RNA binding proteins (Du et al., 2017a; Du et al., 2016), regulate transcription (Li et al., 2015), and convert to proteins (Yang et al., 2018). The landscape of circRNAs in the human is characterized by tissue specificity and structure stability (Yang et al., 2018). The generation of circRNA is different from linear transcripts. The transcriptome is cleaved into a covalently closed loop by reverse splicing. Our research discovered that circPTN was highly expressed in gastric cancer cells. After that, CCK-8, colony formation, and EdU experiments proved that the high expression of circPTN could accelerate cell proliferation and the cell cycle in gastric cancer. Low expression of circPTN could inhibit proliferation and the cell cycle in gastric cancer. Furthermore, a Transwell assay was used to investigate migration. We discovered that the high expression of circPTN could accelerate cell migration in gastric cancer. Low expression of circPTN could inhibit migration in gastric cancer. Moreover, we discovered that circPTN could expedite stem cells of gastric cancer through the marker of stem cells. In short, this research discovered circPTN might make a significant impact on tumorigenesis and offered a new idea for curing gastric cancer.
Availability of Data and Materials: Data available on request from the authors.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Aponte PM, Caicedo A (2017). Stemness in cancer: Stem cells, cancer stem cells, and their microenvironment. Stem Cells International 2017: 5619472. DOI 10.1155/2017/5619472. [Google Scholar] [CrossRef]
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014). circRNA biogenesis competes with pre-mRNA splicing. Molecular Cell 56: 55–66. DOI 10.1016/j.molcel.2014.08.019. [Google Scholar] [CrossRef]
Bartfeld S, Koo BK (2017). Adult gastric stem cells and their niches. Wiley Interdisciplinary Reviews: Developmental Biology 6: e261. DOI 10.1002/wdev.261. [Google Scholar] [CrossRef]
Bornschein J, Rokkas T, Selgrad M, Malfertheiner P (2011). Gastric cancer: Clinical aspects, epidemiology and molecular background. Helicobacter 16: 45–52. DOI 10.1111/j.1523-5378.2011.00880.x. [Google Scholar] [CrossRef]
Cai C, Rajaram M, Zhou X, Liu Q, Marchica J, Li J, Powers RS (2012). Activation of multiple cancer pathways and tumor maintenance function of the 3q amplified oncogene FNDC3B. Cell Cycle 11: 1773–1781. DOI 10.4161/cc.20121. [Google Scholar] [CrossRef]
Cancer Genome Atlas Research Network (2014). Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513: 202–209. [Google Scholar]
Choi YJ, Kim N (2016). Gastric cancer and family history. Korean Journal of Internal Medicine 31: 1042–1053. DOI 10.3904/kjim.2016.147. [Google Scholar] [CrossRef]
Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB (2017a). Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. European Heart Journal 38: 1402–1412. DOI 10.1093/eurheartj/ehx501.P440. [Google Scholar] [CrossRef]
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Research 44: 2846–2858. DOI 10.1093/nar/gkw027. [Google Scholar] [CrossRef]
Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB (2017b). Identifying and Characterizing circRNA-Protein Interaction. Theranostics 7: 4183–4191. DOI 10.7150/thno.21299. [Google Scholar] [CrossRef]
Glazar P, Papavasileiou P, Rajewsky N (2014). circBase: A database for circular RNAs. RNA 20: 1666–1670. DOI 10.1261/rna.043687.113. [Google Scholar] [CrossRef]
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388. DOI 10.1038/nature11993. [Google Scholar] [CrossRef]
Hong Y, Qin H, Li Y, Zhang Y, Zhuang X, Liu L, Lu K, Li L, Deng X, Liu F, Shi S, Liu G (2019). FNDC3B circular RNA promotes the migration and invasion of gastric cancer cells via the regulation of E-cadherin and CD44 expression. Journal of Cellular Physiology 234: 19895–19910. DOI 10.1002/jcp.28588. [Google Scholar] [CrossRef]
Kearney MF, Wiegand A, Shao W, Mcmanus WR, Bale MJ, Luke B, Maldarelli F, Mellors JW, Coffin JM (2017). Ongoing HIV replication during ART reconsidered. Open Forum Infectious Diseases 4: ofx173. DOI 10.1093/ofid/ofx173. [Google Scholar] [CrossRef]
Leung WK, Wu MS, Kakugawa Y, Kim JJ, Yeoh KG, Goh KL, Wu KC, Wu DC, Sollano J, Kachintorn U, Gotoda T, Lin JT, You WC, Ng EK, Sung JJ (2008). Screening for gastric cancer in Asia: Current evidence and practice. Lancet Oncology 9: 279–287. DOI 10.1016/S1470-2045(08)70072-X. [Google Scholar] [CrossRef]
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nature Structural & Molecular Biology 22: 256–264. DOI 10.1038/nsmb.2959. [Google Scholar] [CrossRef]
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, Le Noble F, Rajewsky N (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338. DOI 10.1038/nature11928. [Google Scholar] [CrossRef]
Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A (2016). Cancer treatment and survivorship statistics, 2016. CA: A Cancer Journal for Clinicians 66: 271–289. DOI 10.3322/caac.21349. [Google Scholar] [CrossRef]
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S (2017). Translation of CircRNAs. Molecular Cell 66: 9–21. DOI 10.1016/j.molcel.2017.02.021. [Google Scholar] [CrossRef]
Shi X, Wang X, Hua Y (2018). LncRNA GACAT1 promotes gastric cancer cell growth, invasion and migration by regulating MiR-149-mediated of ZBTB2 and SP1. Journal of Cancer 9: 3715–3722. DOI 10.7150/jca.27546. [Google Scholar] [CrossRef]
Song Z, Wu Y, Yang J, Yang D, Fang X (2017). Progress in the treatment of advanced gastric cancer. Tumour Biology 39: 1010428317714626. [Google Scholar]
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z (2017). Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Research 27: 626–641. DOI 10.1038/cr.2017.31. [Google Scholar] [CrossRef]
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, Huang S, Xie B, Zhang N (2018). Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. JNCI: Journal of the National Cancer Institute 110: 304–315. [Google Scholar]
Zhang X, Liu S, Hu T, Liu S, He Y, Sun S (2009). Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 50: 490–499. DOI 10.1002/hep.23008. [Google Scholar] [CrossRef]
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S (2016). Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature Communications 7: 11215. DOI 10.1038/ncomms11215. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |