DOI:10.32604/biocell.2021.016316

| BIOCELL DOI:10.32604/biocell.2021.016316 |  |
| Article |
Genome-wide identification and in silico gene expression analysis of the related to ABI3/VP1 (RAV) transcription factor family in barley (Hordeum vulgare L.)
Department of Molecular Biology and Genetics, Istanbul University, Istanbul, 34134, Turkey
*Address correspondence to: Cüneyt Uçarlı, ucarlicu@istanbul.edu.tr
Received: 25 February 2021; Accepted: 13 April 2021
Abstract: RAV (Related to ABI3/VP1) transcription factors are unique members of the AP2-ERF superfamily with AP2 and B3 domains and play important roles in the regulation of seed germination, plant growth, and stress response. In the study, 7 RAV genes, named HvRAVs, were identified in barley based on the available genome sequences. While five of the seven HvRAVs were located on chromosome 3, HvRAV5 and HvRAV7 were located on chromosome 1 and 4, respectively. Six of the predicted HvRAVs were intron-less, except HvRAV2, which had one intron. HvRAV proteins have shown basic, instable, and hydrophilic properties. The AP2 domain specific RAYD and WLG motifs were detected in all HvRAV proteins. Besides, B3 repression domain, R/KLFGV, is also found in the C-terminal of HvRAVs. HvRAVs were found to have stress-related cis-acting elements, including MYB, MYC, and W-BOX. HvRAV2 was predicted to have no GARE motifs, TATCCCA or TAACAA(G/A), and LTREs. Under drought conditions, the expression level of HvRAVs did not significantly change in a drought-sensitive barley genotype, whereas HvRAV5 and HvRAV7 were dramatically down-regulated in a drought-tolerant genotype. Expression of HvRAV5 was also inhibited by salinity. HvRAV7 was strongly induced by plant pathogen attack. Only HvRAV6 was induced by exogenous gibberellin application and during the germination process. Interestingly, HvRAV6 transcript was detected higher than other HvRAVs in all stress and control conditions as well as during germination. In silico analyses have shown that HvRAVs play a role in response to different abiotic and biotic stresses as well as in plant development. However, the extensive biological roles of HvRAV genes in plant development and in response to abiotic and biotic stresses need further investigation.
Keywords: Barley; RAV; AP2 domain; B3 domain; AP2/ERF family; Cis-acting elements
Transcription factors (TFs), sequence-specific DNA binding proteins, play central roles in plant growth, development, hormone response, and in response to biotic and abiotic stresses by regulating target genes (Liu et al., 2013). The transcription factor RAVs (Related to ABI3/VP1) are plant-specific and belong to the APETALA2/Ethylene Responsive Factor (AP2/ERF) family, which plays a crucial role in the regulation of biotic and abiotic stress-responsive gene expression (Du et al., 2016). The AP2/ERF superfamily is characterized by the AP2/ERF DNA binding domain, which consists of about 60 to 70 highly conserved amino acids (aa) (Sakuma et al., 2002). The AP2/ERF superfamily is divided into 5 subfamilies, including APETALA2 (AP2), Related to ABI3/VP1 (RAV), Dehydration-Responsive Element Binding (DREB), Ethylene-Responsive Factors (ERF), and soloist (Guo et al., 2016). RAV TFs were first identified in the ABA insensitive-3 (abi3) mutant from Arabidopsis and the viviparous1 (vp1) mutant from Zea mays (Giraudat et al., 1992; Suzuki et al., 1997). RAV proteins contain two different DNA-binding domains, the AP2 and B3 domains, differently from other AP2/ERF TFs, which have only AP2 domain (Feng et al., 2005). The AP2 and B3 DNA-binding domains recognize consensus CAACA and CACCTG sequences, respectively, in Arabidopsis, so plant RAVs with both AP2 and B3 domains show much higher DNA binding affinity (Kagaya et al., 1999; Wei et al., 2018, Xie et al., 2019).
RAV proteins serve as negative regulators in plant development by repressing the transcriptional activity and forming complexes with other co-repressors. Besides, RAVs have important functions in abiotic and biotic stress responses in plants (Matías-Hernández et al., 2014). It has been found that the 15-aa peptide sequence (GNSKTLRLFGVNMEC), named B3 repression domain (BRD), had a repressive activity among the B3 super-family, including auxin response factor (ARF), abscisic acid-insensitive3 (ABI3), high-level expression of sugar inducible (HSI), related to ABI3/VP1 (RAV) and reproductive meristem (REM). Deletions in BRD of some B3 proteins revealed that only a short peptide composed of five amino acids, R/KLFGV, is essential to maintain the repressive activity of the BRD domain (Ikeda and Ohme-Takagi, 2009). A quite similar sequence, MLFGV, has been found in Arabidopsis RAV3 and RAV3-like TFs (Causier et al., 2012).
The number of TFs in the RAV family is relatively less in comparison to other subfamilies of the AP2/ERF superfamily, i.e., in cauliflower, 146 of 171 AP2/ERF TFs belong to the ERF family, whereas 15 and 9 TFs were classified into the AP2 and RAV subfamily, respectively (Li et al., 2017). RAV family members play important roles in many different physiological and developmental pathways, including hormone signaling (ethylene and brassinosteroids), flowering, leaf senescence, bud outgrowth, and response to stress in several plant species (Matías-Hernández et al., 2014). There is little information in the literature about the characterization, expression patterns, and functional roles of barley RAV genes.
Barley (Hordeum vulgare L.) ranks fourth after rice, wheat, and maize in terms of cultivated area and production among cereals (FAO, 2018). Barley is majorly consumed as animal feed (seeds or as forage crop) and used as malt in the production of alcohol and non-alcoholic beverages, and a minor portion of production is used in the human diet (Uçarlı and Gürel, 2020). Barley is grown in different environmental conditions with a higher tolerance against biotic and abiotic stresses than other crops (Dawson et al., 2015). Besides its agronomic importance, barley is a well-studied plant in terms of genetics, functional genomics, and breeding for many researchers in the field of cereal crops.
In the present study, genome-wide bioinformatics analyses were conducted to predict cDNA and amino acid sequences of RAV TF family members in barley. Gene structure, cis-acting elements related to abiotic and biotic stress, and highly conserved motifs were determined in HvRAVs. Physicochemical properties of HvRAV proteins were also predicted as well as secondary structures. The phylogenetic trees were generated, and the evolutionary distances were computed between HvRAV proteins and also compared with other monocots. The expression pattern of HvRAV genes at the germination stage and their expression profiles under different stresses, including drought, salinity, and plant-pathogen attacks, were determined using data obtained from distinct microarray experiments. The results will be useful in further study of the RAV family in cereals, especially in the Triticeae tribe.
In addition to the barley genome database, Transcription Factor Databases, including plantTFDB (http://planttfdb.cbi.pku.edu.cn/) and iTAK (http://itak.feilab.net/cgi-bin/itak/db_browse.cgi), were used to identify the sequences of barley RAV TFs as well as some monocot RAVs including Oryza sativa, Triticum aestivum, Zea mays and Brachypodium distachyon (Jin et al., 2014; Zheng et al., 2016). BLAST programs (BLASTP, TBLASTN, and BLASTN) which are available on the IPK barley genome database sourced from the assembly Morex V2.0 2019 (http:// webblast.ipk-gatersleben.de/barley/), Gramene (http: //ensembl.gramene.org/Hordeumvulgare/Info/Index) and Phytozome (https://phytozome.Jgi.doe.gov/pz/ portal.html) were used to increase the extent of the transcription factor database search results and to determine the location of the RAV genes on barley chromosomes.
Gene structure, cis-acting element analysis, and motif recognition
Gene structure was analyzed using the Gene Structure Display Server websites (http://gsds.cbi.pku.edu.cn/) (Hu et al., 2015). AP2 and B3 domains were predicted using the domain analysis programs Simple Modular Architecture Research Tool (SMART (http://smart.embl-heidelberg.de/)) based on the STRING database (Letunic and Bork, 2018). Conserved motifs of HvRAV TFs were investigated with web-based sequence analysis tool MEME Suite 5.1.1 (http://meme-suite.org/tools/meme) with the following parameters: optimum width 6–50 amino acids, any number of repetitions of a motif, the maximum number of motifs set at 10, and all other parameters set at default (Bailey et al., 2009). Cis-acting elements, including abiotic and biotic stress-related, and ABA- and GA-responsive elements were identified using plant cis-acting element databases PlantCARE (Lescot et al., 2002), and PLACE (Higo et al., 1999) in putative promoter regions (2-kb upstream sequences of the transcriptional start site) of HvRAV genes. Putative nuclear localization signal (NLS) sequences were predicted using a cNLS mapper with a cut-off score higher than 3 (Kosugi et al., 2009).
Physiochemical characterization, secondary and three-dimensional structure analysis
The physical and chemical properties of HvRAVs, including molecular weight (MW), theoretical isoelectric point (pI), instability index (II), aromaticity, and grand average of hydropathicity (GRAVY), were determined by ProtParam tools (http://web.expasy.org/ protparam/). Putative Kinase-specific prediction of protein phosphorylation sites was conducted using NetPhos 3.1 (Blom et al., 2004). Prediction of secondary structure elements and generation of 3-D models of HvRAV proteins were performed with Phyre2 server (Kelley et al., 2015).
Multiple sequence alignment and phylogenetic analysis
The identified protein sequences of HvRAVs and monocot RAVs were aligned using the Clustal T-Coffee program with default parameters (Notredame et al., 2000). The phylogenetic trees were generated by the neighbor-joining (NJ) statistical method with 1000 bootstrap replications using MEGA X software (Kumar et al., 2018). The evolutionary distances were computed using the Poisson correction method with pairwise deletion.
Gene expression profile analysis
Gene Expression profile barley under stress conditions and during germination was downloaded from Gene Expression Omnibus using data obtained from different Affymetrix Barley1 GeneChip microarray experiments (dataset IDs: Drought stress, GSE15970; salt stress, GSE3097; cold stress, GSE10329; heat stress, GSE23896; fungal stress GSE33401 and GSE33407; germination, GSE9365; gibberellin (GA) and abscisic acid (ABA), GSE18785). The probe sequences of the microarray chip were blasted with cDNA sequences of barley RAV genes. The four RAV genes, namely HvRAV1, HvRAV5, HvRAV6, and HvRAV7, hit the four-probe sequences contig7483_at, contig19699_at, contig7483_at, and Hs16k21u_at, respectively. Expression intensities under the control, drought, salinity, cold, and heat conditions were used for expression analysis of HvRAV genes under abiotic stresses. In addition, expression intensities obtained from the treatment of barley plants with plant pathogens, including Fusarium graminearum, Cochliobolus sativus, and Puccinia hordei, were used to determine the expression profiles of HvRAV genes under biotic stresses. Heat maps were generated using TIGR Multiple Experiment Viewers (Saeed et al., 2003).
Identification of the RAV genes in barley
In the barley genome, seven putative RAV genes were identified using BLAST searches of the plant transcription factor and barley databases (Tab. 1). The coding sequence (CDS) lengths of the predicted barley RAV genes varied, ranging from 927 to 1245 bp. All HvRAV genes have both AP2 and B3 domains. While AP2 domains of seven HvRAV proteins are 66 aa, the length of B3 domains varied from 99 to 113 residues. It has been found that five HvRAV genes, namely HvRAV1, HvRAV2, HvRAV3, HvRAV4, and HvRAV6 located on chromosome 3 and are very close to each other, while HvRAV5 and HvRAV7 are located on chromosomes 1 and 4, respectively. All barley RAV genes have one exon except HvRAV2, which is disrupted by a 676-bp intron.
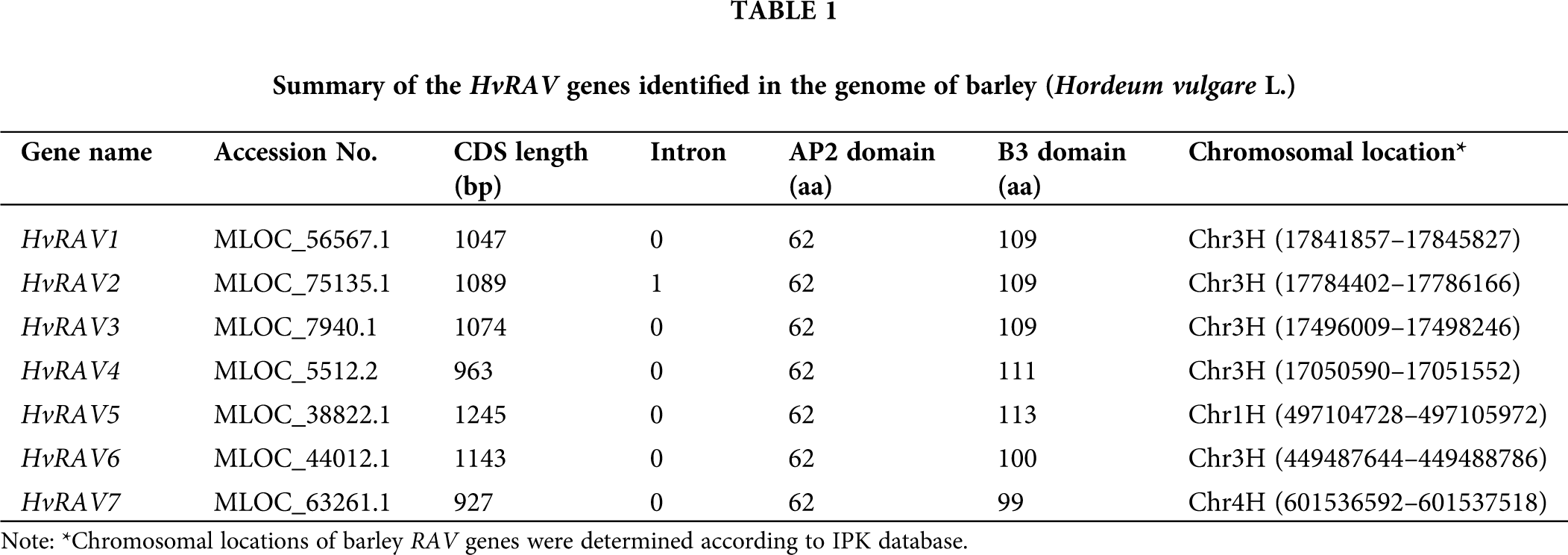
Physiochemical characterization, secondary and three-dimensional structure analysis
Detailed information on the physicochemical characteristics of HvRAV proteins is shown in Tab. 2. Amino acid lengths of the predicted barley RAV proteins ranged from 308 to 362 aa, and the molecular weight (MW) from 33.53 (HvRAV7) to 44.70 (HvRAV5) kDa. The theoretical isoelectric point (pI) of the five HvRAV proteins was calculated around 9.5, indicating basic character, while the theoretical pI value of HvRAV4 and HvRAV7 was calculated as 5.5 considering as acidic protein. Only HvRAV2 was classified as a stable protein according to the instability index (II) value by 36.95 among the barley RAV proteins (II higher than 40 in other HvRAVS). Grand average of hydropathicity (GRAVY) of HvRAV proteins was measured between –0.35 and –0.68, indicating hydrophilic proteins. The aliphatic index (AI) values of barley RAV proteins ranged from 59.88 (HvRAV4) to 69.53 (HvRAV6), thus suggesting protein thermostability. While the net charge of two HvRAVs, namely HvRAV4 and HvRA7, predicted negatively by –4.36 and –3.90, respectively, the rest of HvRAV proteins varied from 9.08 and 18.57. The highest Kinase-specific phosphorylation sites were predicted on HvRAV5 with 57 sites.

Three-dimensional structure of whole HvRAV protein, as well as binding domains AP2 and B3, were generated using Phyre2 server (Fig. 1A). Secondary structure analysis showed that all HvRAV sequences have 3–4 stranded β-sheet that are antiparallel with one α-helix, relating to the AP2 domain, with a confidence value of 100% (Fig. 1B). It was predicted that the B3 domain contains 5–6 β-sheets that form an open barrel structure with 2 α-helixes in barley RAV proteins. The total number of helixes and strands ranged 4–9 and 9–12, respectively, in seven HvRAV proteins. The percentage of disordered amino acid residues among members of the HvRAVs varied from 47% to 52%, meaning that almost half of the HvRAV proteins were unfolded (Fig. 1B). The linker region between AP2 and B3 domain was found to have 1 or 2 helixes in all barley RAVs except HvRAV3. On the other hand, only HvRAV7 was found to have one-stranded beta sheet on the linker region among the barley RAVs.
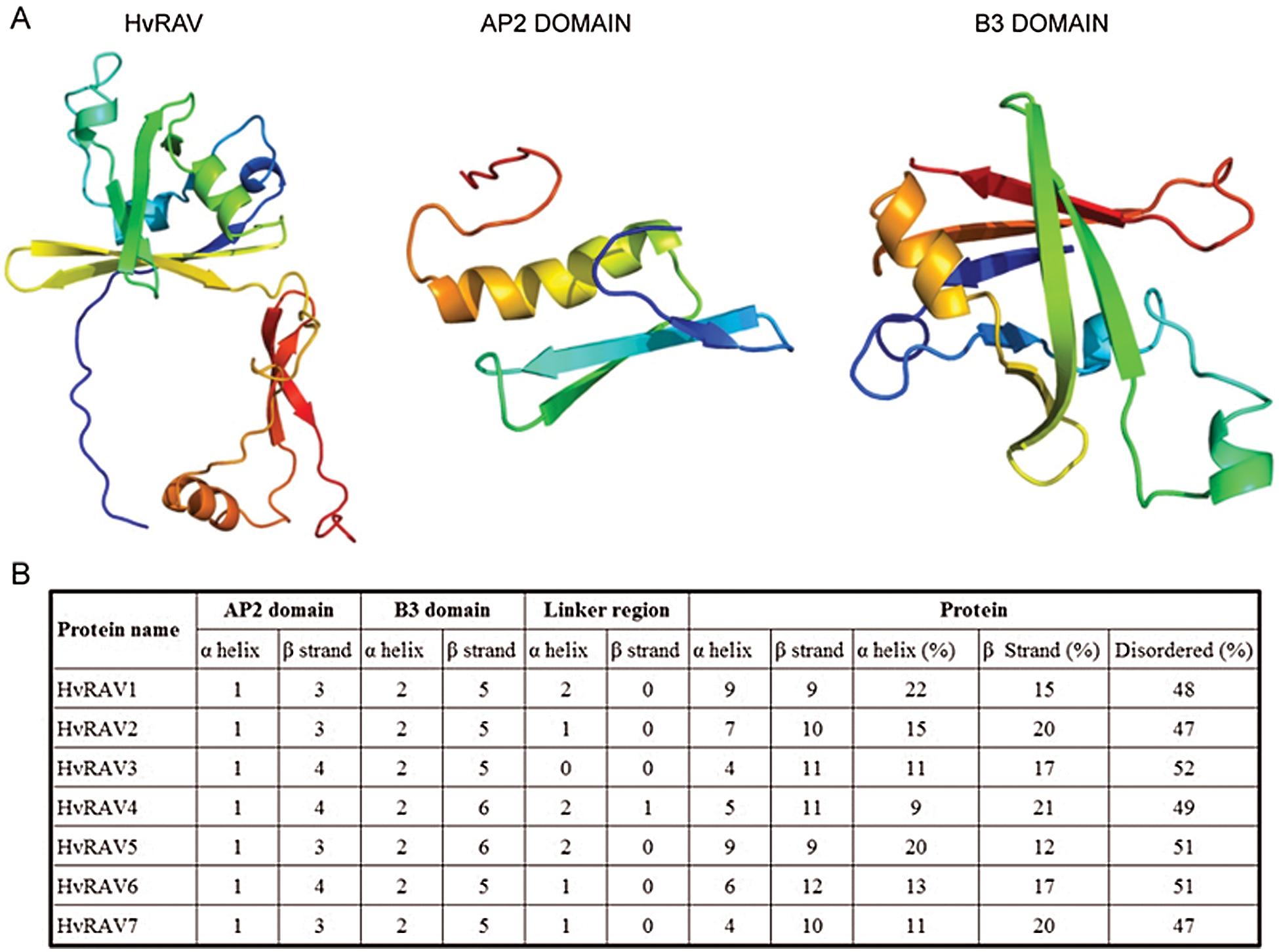
Figure 1: Three–dimensional and secondary structures of HvRAVs.
(A) Predicted 3D structures of HvRAV1 protein and highly conserved AP2 and B3 domains modeled at 100% confidence level by using Phyre2 server. (B) Secondary structures of HvRAV proteins and AP2 and B3 domains according to the Phyre 2 server.
Analysis of cis-acting elements in the putative promoter regions of the HvRAV genes
Analysis of the promoter regions of HvRAV genes showed the presence of different cis-acting elements in different numbers, which may play a role in abiotic and biotic stress, as well as hormone responsiveness, including ABA and GA (Tab. 3). All HvRAV genes contained one or more ABA-responsive elements (ABREs) except HvRAV5. Seven HvRAV genes contain at least one Dehydration-response element (DRE) site, which has a role in response to drought, salinity, and cold stress. Ethylene-responsive-element-binding factors (ERFs) participate in transcriptional regulation of abiotic and biotic stresses and specifically bind to the GCC-BOX with the core sequence of AGCCGCC. The GCC-BOX motif was only predicted on the promoter region of HvRAV5 and HvRAV6 among seven HvRAVs. Cis-acting elements MYB, MYC, and W-BOX, involved in the abiotic stress responsiveness, with high numbers have been found on the promoter of HvRAVs. It has been predicted that promoter regions of HvRAVs contained two or five low-temperature response elements (LTREs) except HvRAV2, which had no LTRE. TC-rich repeats with the core ATTTTC sequence have a crucial role in defense and biotic stress response, were determined on the promoter of HvRAVs. TATCCCA motif, one of the Gibberellin-responsive elements (GARE), has been found only on the promoter regions of HvRAV2 with 2 copies and HvRAV6 with1 copy, while another GARE motif TAACAA(G/A) found on promoter regions of HvRAV1, HvRAV3, HvRAV4, and HvRAV5. No GAREs were detected on the promoter region of HvRAV7.
Multiple sequence alignment and phylogenetic analysis
Multiple sequence alignment (MSA) of seven barley RAV proteins was displayed in Fig. 2. Multiple Sequence alignment showed that all barley RAV transcription factors contained highly conserved AP2 and B3 domains. In the AP2 domain of barley RAV proteins, highly conserved RAYD and YRG elements were predicted. A highly conserved 18 amino acid core region of RAYD was predicted to form an α-helix in the C terminal region of the AP2 domains. A highly conserved YRG element, containing mostly hydrophilic and basic 19–20 amino acids, was predicted to form 2 β sheets in the N terminal region of the AP2 domain. It was also detected that the YRG motif has been replaced with the R(K)F(Y)K motif in AP2 domains of barley RAVs. MSA analysis demonstrated that seven barley RAV proteins contain a WLG motif in the AP2 domain. B3 repression domain (BRD) containing R/KLFGV residues was detected in the C-terminal of all HvRAVs. All barley RAV proteins have been found to have at least one bipartite nuclear localization signal (NLS) sequence with scores ranging from 3.0 to 6.1; thus, HvRAVs are predicted to be localized in both the nucleus and the cytoplasm. Besides, HvRAV1, HvRAV5, and HVRAV6 have monopartite NLS sequences GWFKRQCMA, HAVWKKRCVDF, and VAFKKQCIE, respectively, in the C-terminal region of the proteins (Fig. 2).

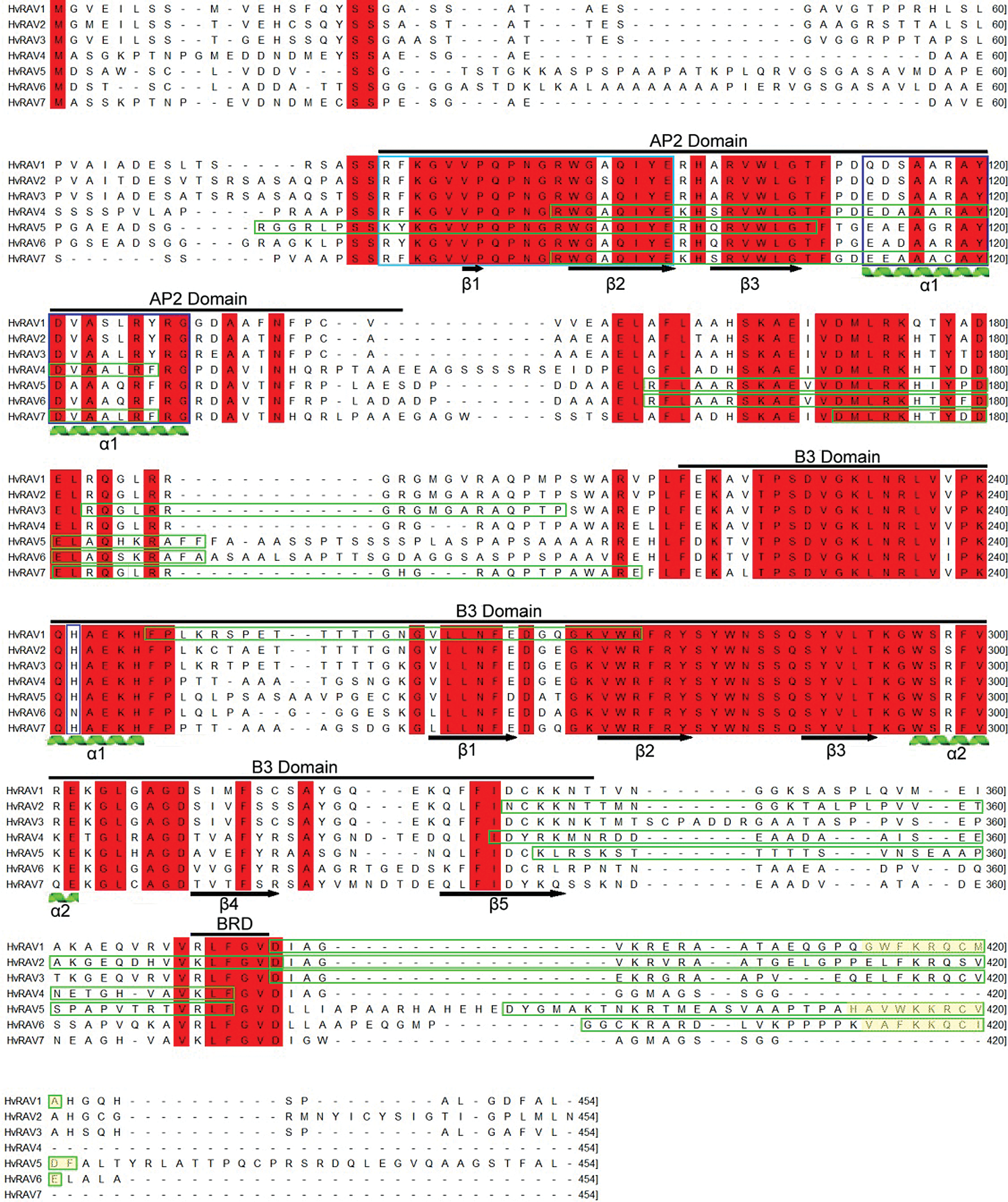
Figure 2: Multiple sequence alignment of seven barley RAV proteins according to the Clustal T-Coffee program.
Domain AP2, domain B3, and B3 repression domain (BRD) are marked above the corresponding sequences with a black line. Fully conserved regions are indicated in red highlight. In AP2 and B3 domains, common beta-sheet (β) and alpha-helix (α) are underlined with arrow and helix, respectively. Bipartite nuclear localization signals (NLS) are shown in green rectangles, and monopartite ones are in yellow highlight. Highly conserved YRG motif (light blue rectangle) and RAYD element (dark blue rectangle) are predicted in the AP2 domain.
The MEME motif search tool was conducted to find motifs in barley RAV proteins. In total, 6 conserved motifs were identified, lengths ranging from 20–50 amino acids in all RAV proteins (Fig. 3). While 50-aa-length motif 1 including F/YKG and WLG sequences was observed on the AP2 domain, the motifs 3, 4, and 5 were predicted on the B3 domain of barleys RAVs. Interestingly, a F/YRG sequence was found at the terminal of motif 1. The highly conserved RAYD (motif 1) element in AP2/ERF superfamily was converted to CAYD in HvRAV7. Motif 2 specifying a linker region between AP2 and B3 domains was highly conserved. Motif 6 was located in the C terminal region of RAV protein and containing the BRD element R/KLFGV.
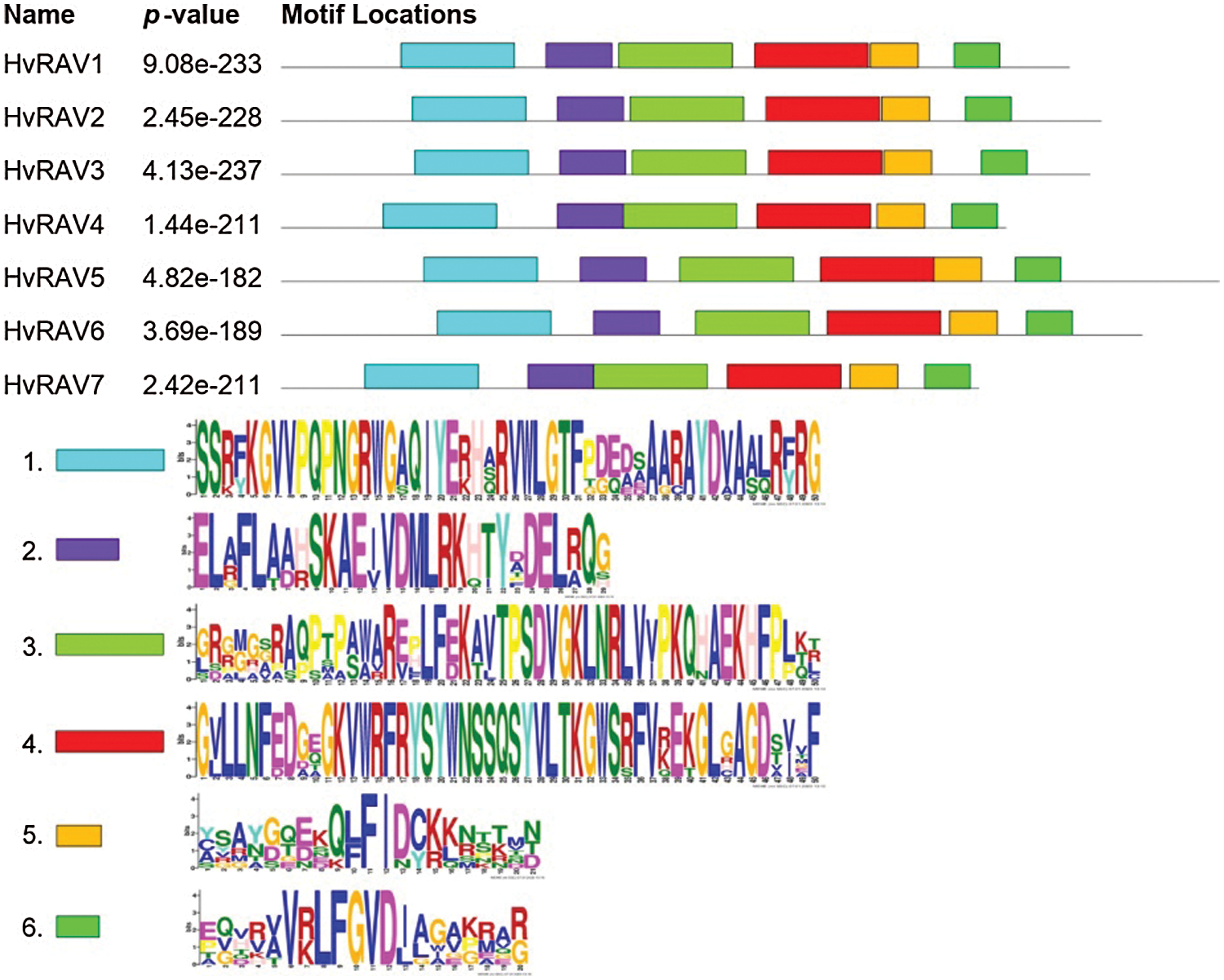
Figure 3: Distribution of conserved motifs in barley RAV proteins.
Sites and number of motif distribution in genes; E-value and statistical significance of motif.
A phylogenetic tree of seven barley RAV transcription factors was generated based on the full-length amino acid sequences with the neighbor-joining method. According to the phylogenetic analysis, barley RAV proteins were distributed into three groups: Group I, including HvRA1, HvRAV2, and HvRAV3; Group II, including HvRAV4 and HvRAV7; and Group III, with HvRAV5 and HvRAV6 (Fig. 4A). To evaluate evolutionary relationships between RAV proteins in monocots, a phylogenetic analysis of the 7 barley RAVs, 4 Rice RAVs, 7 wheat RAVs, 3 maize RAVs, and 6 Brachypodium RAVs were undertaken based on a phylogenetic tree generated with a statistical method using full-length amino acid sequences of RAV proteins. Based on the phylogenetic analysis of RAV proteins, some barley RAVs show closer evolutionary status with RAVs of different species. Barley RAV3 (HvRAV3) was more closely related to wheat Ta390BE2090B.1 than other barley RAVs (Fig. 4B). Amino acid composition of AP2 and B3 domains in monocots was aligned in five monocots. It is predicted that AP2 domains were highly conserved with YRG, WLG, and RAYD elements. AP2 domains contain 2–4 beta-sheets with one helix. B3 domains of monocots also demonstrated highly conserved regions. In general, monocots have 2 helixes in the B3 domain, but rice RAV (Os01g04750.1) has an extra alpha-helix in the N-terminal region of the domain.
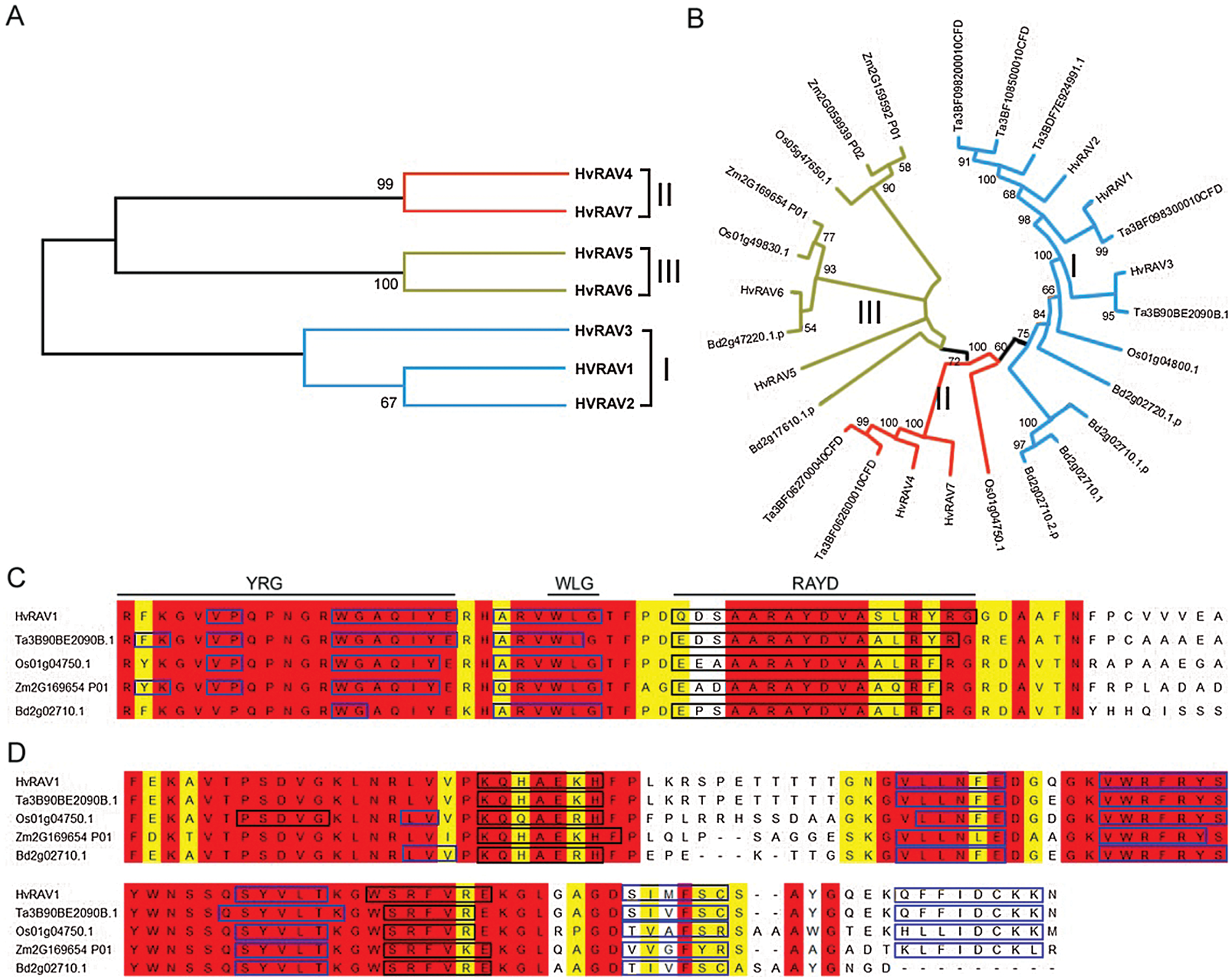
Figure 4: Phylogenetic tree and multiple sequence alignment of RAV gene.
(A) The phylogenetic analysis of HvRAV proteins in barley. (B) Phylogenetic relationship of the RAV families of monocots including Hordeum vulgare (HvRAV1), Triticum aestevium (Ta3B90BE2090B.1), Oryza sativa (Os01g04750.1), Zea mays (Zm2G169654 P01), and Brachypodium distachyon (Bd2g02710.1). Multiple sequence alignment of AP2 (C) and B3 (D) domains. YRG and RAYD elements and WLG motif are marked above the corresponding sequences in black lines in AP2 domains. Fully conserved regions and only single amino acid changes are indicated in red and yellow highlight, respectively. Black and blue rectangles represent alpha-helix and beta-sheet, respectively.
Gene expression profile analysis of barley RAVs
The data obtained from distinct microarray experiments (GEO numbers are mentioned in Materials and Methods section) were used to monitor the expression profiles of the HvRAV transcription factors, namely HvRAV1, HvRAV5, HvRAV6, and HVRAV7, in response to abiotic and biotic stresses as well as gibberellic acid (GA) and abscisic acid (ABA) treatments (Fig. 5).
Drought-sensitive and tolerant barley genotypes were treated with drought stress in 10% AWC (available water capacity) soil for 0, 1, 3, and 5 days. While the expression of the four HvRAV genes was not significantly changed in response to drought stress in drought-sensitive genotype, the drought stress significantly down-regulated the expression of HvRAV5 and HVRAV7 with 14.3- and 25-fold, respectively, in drought-tolerant genotype (Fig. 5A). In another study, 17-day-old barley plants were exposed to 100 mM NaCl for 0, 3, 8, and 27 h. The expression of HvRAV5 was changed with a significant decrease at 27 h of NaCl treatment (Fig. 5B). The expression of the other HvRAV genes did not change or slightly decreased depending on exposure time. The changes in expression of HvRAV genes under low temperatures were investigated by subjecting the 7-old-day barley seedlings of cv. Dicktoo to chilling (4°C for 5 days), subzero (–4°C for 8 days), and deacclimation (20°C for 3 days) conditions. The mRNA level of HvRAV1 did not change in all cold conditions. On the other hand, the chilling and subzero treatments resulted in the upregulation of the HvRAV7 gene with 2.9- and 6.3-fold, respectively, while the expression of HvRAV5 was upregulated under subzero and deacclimation conditions compared to the control conditions (Fig. 5C). Developing barley caryopses were harvested at 0, 0.5, 3, and 6 h during heat stress (42°C). Heat stress has been found to decrease the expression of HvRAV5 and HvRAv7 at 0.5 h of treatment, but by time expression level was upregulated. On the other hand, the expression of HvRAV1 and HvRAV6 almost did not change during heat stress (Fig. 5C). The expression profile of HvRAVs in response to biotic stress was also monitored using gene chip arrays. Hordeum vulgare ssp. spontaneum was inoculated with Cochliobolus sativus, causing spot blotch, and Puccinia hordei, causing leaf rust, for 0, 12, 24, 36, and 48 h. The expression of HvRAV7 was up-regulated at 12 h of both pathogen attacks, but by the time mRNA level has been decreased. Besides, it has been found that the pathogen attacks have affected the expression patterns of HvRAV6 and HvRAV7 antagonistically. In another study, the spikes of cv. Morex inoculated with Fusarium graminearum, causing head blight in cereals for 0, 24, 48, 72, 96, and 144 h. HvRAV7 has been found to express differentially at different time points after F. gramineraum inoculation, while the expression of HvRAV1 and HvRAV6 were not significantly changed (Fig. 5D).
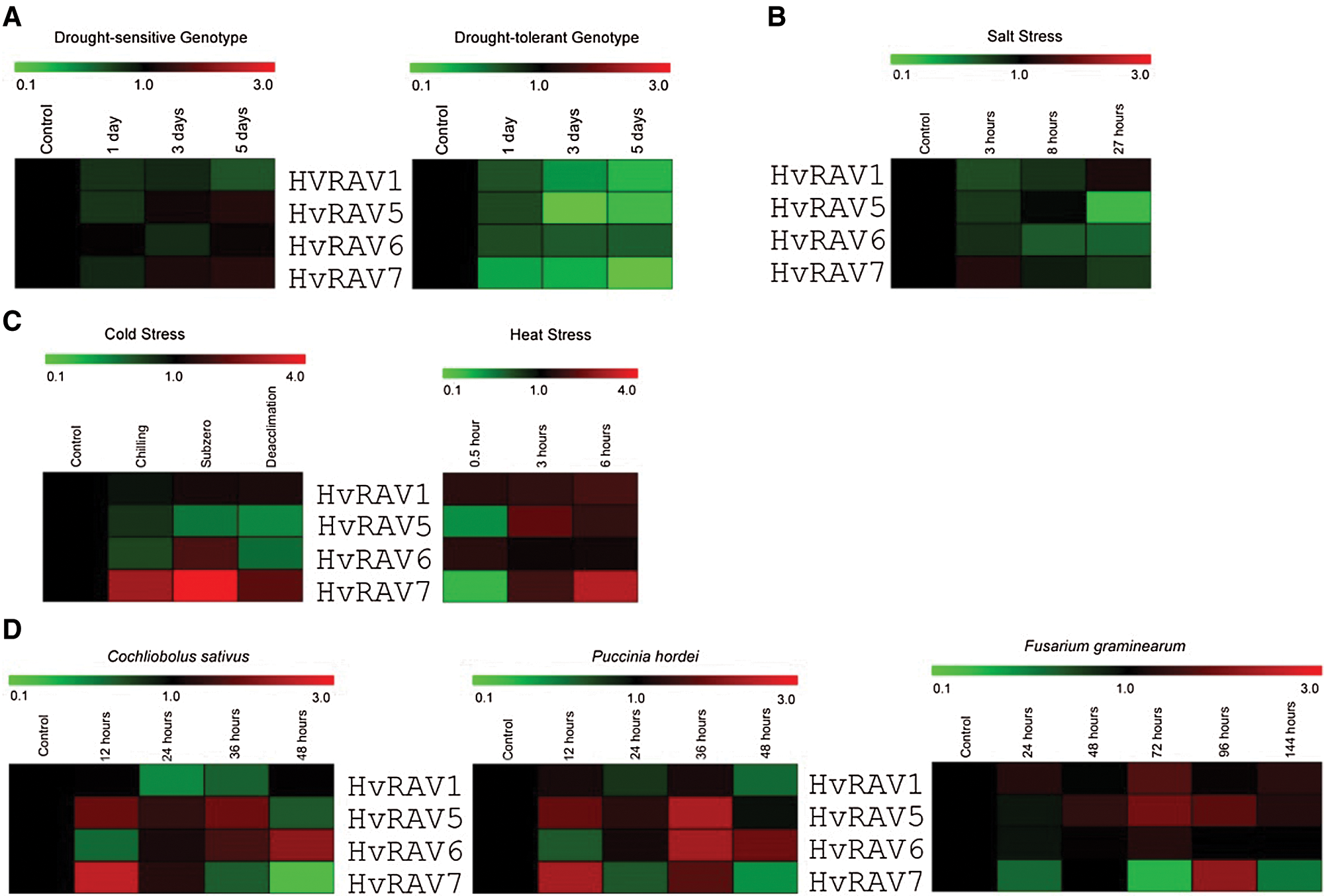
Figure 5: Expression profiles of the four HvRAV genes, namely HvRAV1, HvRAV5, HvRAV6, and HvRAV7, in response to abiotic and biotic stresses.
Heat map represents the differential gene expression of HvRAV genes in response to (A) drought stress, (B) salt stress, (C) cold and heat stress, (D) pathogen attacks, including C. sativus, P. hordei, and F. graminearum. While the red color indicates the upregulation of the HvRAVs, the green color shows the downregulation of the HvRAVs.
The expression profile of HvRAVs was investigated during germination at 0, 24, 48, and 72 h after imbibition (HAI) in endosperm and embryo tissues. Only HvRAV6 was observed to differentially expressed in endosperm and embryo tissues during the germination of barley seeds among the HvRAVs. HvRAV6 was dramatically upregulated and reached the highest peak at 24 HAI but then decreased in endosperm tissues. In embryo tissues, the mRNA level of HvRAV6 rapidly increased at 24 HAI and maintained a high transcript level during the germination process (Fig. 6A). It has been observed that HvRAV6 was upregulated in response to exogenous GA (1 µM) treatment, whereas its expression did not significantly change after exogenous ABA (50 µM) treatment in barley aleurone cells. SLN1 is a DELLA protein, which is a repressor in the G signaling pathway. In sln1 mutants, the transcript level of HvRAV6 was lower than GA-treated samples. In addition, the transcript level of HvVAV6 has been found very high in all conditions when compared to the other three barley RAVs.
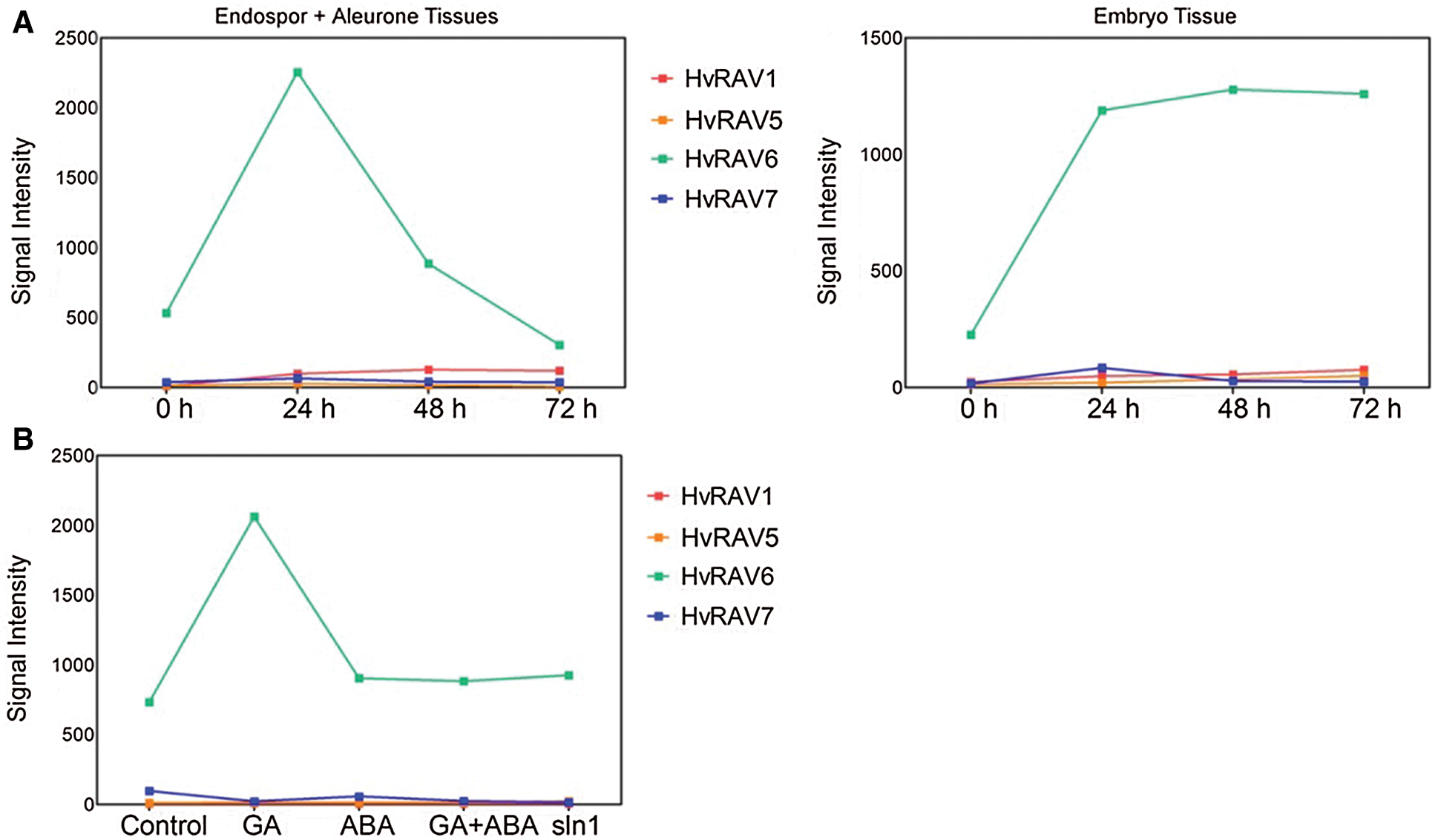
Figure 6: Signal intensities of HvRAV genes obtained from microarray experiments.
(A) Signal intensity of HvRAV genes in endosperm+aleurone and embryo tissues during germination at different hours. (B) Signal intensities of HvRAV genes after GA and/or ABA treatment and in sln1 mutants.
Characterization of the barley RAVs
RAV (Related to ABI3/VP1) proteins are members of the APETALA2/Ethylene Responsive Factor (AP2/ERF) family and play a crucial role in response to biotic and abiotic stresses by regulating gene expression as well as seed germination (Zhao et al., 2017; Wei et al., 2018). In barley, seven RAV genes were identified based on barley genome and transcription factor databases. In plants, the number of RAVs is less than AP2/ERF superfamily members, including AP2, B3, and DREB. Barley has predicted to have 19 HvAP2, 41 DREB, and 54 HvERF. Besides, in Arabidopsis, 146 AP2/ERF genes are distributed into 6 RAV, 18 AP2, 57 DREB, and 65 ERF (Guo et al., 2016). The RAVs are unique transcription factors with two DNA binding domains, including AP2 and B3 in AP2/ERF superfamily. All seven HvRAVs were predicted to have both AP2 (62 aa length) and B3 domains ranging from 99 to 113 aa. Generally, RAVs have no introns, i.e., sesame and soybean have 4 and 5 RAV genes, respectively, with only single exons (Dossa et al., 2016; Zhao et al., 2017). However, there are some exceptions in some members of RAVs. Analysis of gene structures in Medicago RAV revealed that one of three RAV genes contains a single intron, whereas the others have no intron (Agarwal et al., 2016). Similarly, the analysis showed that HvRAV genes also have only one exon, except HvRAV2 has two exons in barley. It has been reported that introns play role in the regulation of gene expression as well as gene evolution (Rose, 2008). A low number of intron or intronless characteristics of RAV genes may result in a lesser number of RAVs in plants. Interestingly, five of the seven barley RAVs have been found to locate on chromosome 3. In addition, HvRAV1, HvRAV2, HvRAV3, and HvRAV4 were predicted at the telomeric end of chromosome 3. Two barley RAVs, HvRAV5 and HvRAV7, were located on the chromosome 1 and 4, respectively. Similarly, in cotton, some chromosomes contain more than two RAV genes, i.e., 4 RAVs on chromosome 13. Besides, six of eleven RAV genes (three pairs) are located as a tandem at the telomere ends of chromosomes. Analysis revealed that these RAV genes originated from tandem duplication events (Liu and Zhang, 2017).
Seven HvRAV proteins ranged from 33.53 to 44.70 kDa in molecular weight (MW) with hydrophilic and thermostable properties. Major of the HvRAVs were predicted to have basic characteristics with around 9.5 pI and positive charge, except HvRAV4 and HvRAV7 with 5.5 pI. All the soybean GmRAVs were predicted basic characteristics with higher than 7 pI (Zhao et al., 2017). Only HvRAV2 was predicted as a stable protein according to instability index value (36.95) among HvRAV proteins. The HvRAV proteins were calculated to have a high number of kinase-specific phosphorylation (KSP) sites, i.e., HvRAV5 had 57 KSP sites. Protein phosphorylation is a post-translational modification that regulates the function of a protein and plays a crucial role in cellular control of biological processes (Blom et al., 2004). A high number of phosphorylation sites in HvRAVs may promote the interaction with the co-repressor proteins and bind to cis-regulatory elements of stress-responsive genes.
Multiple sequence alignment revealed that all predicted barley RAV transcription factors contained highly conserved AP2 and B3 DNA binding domains. Highly conserved YRG and RAYD elements were predicted on the N-terminal and C-terminal of the AP2 domain of barley RAV proteins, respectively. The YRG element contains basic and hydrophilic 20 amino acids in the N-terminal region of the AP2 domain and plays an important role in DNA binding activity (Okamuro et al., 1997). In the study, the YRG motif has been found to replace with the R(K)F(Y)K motif in AP2 domains of barley RAVs (Fig. 2). Similar results have been also observed in the RAVs of other plants including wheat, rice, maize, Brachypodium distachyon (Fig. 3C). It has been reported that the YRG motif was highly conserved in the AP2 domain of other AP2/ERF superfamily members including ERF, DREB, and AP2 in Triticum aestivum and Arabidopsis thaliana (Zhuang et al., 2011). Another highly conserved region in the AP2 domain is RAYD element which is about 40 aa lengths and plays a crucial role in protein-protein interactions and DNA binding through α-helix (Okamuro et al., 1997). The 18-amino-acid core region of the RAYD element was predicted to form an α-helix in the C- terminal region of the AP2 domain in barley RAVs. But the arginine (R) residue of RAYD motif was replaced with cysteine (C), thus forming CAYD motif in HvRAV7. The RAYD motif was also detected in the AP2 domains of other monocots RAVs including wheat, rice, and maize. In AP2 domains of barley AP2/ERF superfamily, RAYD motif was highly conserved, but also replaced with CAYD, MAYD, LAYD, RAHD, and RAND in DREB subfamily, as well as EAYD, KAYD, GAYD, and RAYI in AP2 subfamily (Guo et al., 2016) WLG is another important motif in AP2 domain and part of the β3 sheet. All barley RAVs contained highly conserved WLG residues, same as the ERF and DREB subfamilies. However, the W (tryptophan) residue of WLG was replaced with another aromatic and hydrophilic residue tyrosine (Y) in the AP2 subfamily. Similar results have been reported in maize, rice, and Arabidopsis (Nakano et al., 2006).
RAV TFs have an additional B3 DNA binding domain, which consists of around 110 residues, as well as a single AP2 domain (Nakano et al., 2006). B3 domain is widespread in plant genomes and contains seven-stranded β-sheet that constitutes an open β-barrel with two short α-helices at the ends of the barrel (Waltner et al., 2005). Similarly, two short α-helices and 5–6 β-sheets were predicted in the B3 domains of barley. The β1, β2, and β3 are on highly conserved sequences and located between two short α-helices (Fig. 2). AP2/ERF super family TFs can function as transcriptional activator and repressor proteins. Two major repressor motifs are the ERF-associated amphiphilic repression (EAR) motif (DLNxxP or LxLxLx) and the B3 repression domain (BRD) motif (GNSKTLRLFGVNMEC) in the C-terminal (Ohta et al., 2001; Ikeda and Ohme-Takagi, 2009; Matías-Hernández et al., 2014). It has been reported that only a short peptide of five amino acids, R/KLFGV, is important as a transcriptional repression activity in B3 proteins. In this study, BRD, containing R/KLFGV residues, was found in the C-terminal of all HvRAVs as well as in some monocot RAVs including wheat, rice, maize. Repression domains (RD) function as transcriptional repression activity by interacting with the co-repressor proteins as well as by binding DNA directly. Causier et al. (2012) have found that variations of the R/KLFGV including MLFGV, RLFGI, and RLFGF showed interaction with co-repressor TPL (TOPLESS), while mutations in leucine (L) and phenylalanine (F) of the RLFGV sequence, new RSSGV motif, and prevented interaction with TPL in Arabidopsis.
Gene expression in response to abiotic and biotic stresses
To evaluate the potential functions of the HvRAV genes in response to biotic and abiotic stresses, the expression patterns of HvRAV1, HvRAV5, HvRAV6, and HvRAV7 were investigated using available microarray gene expression data. The RAV genes play a crucial role in response to environmental stresses, including salinity, drought, and pathogen attacks, by regulating the expression of target genes (Liu and Zhang, 2017). Chen et al. (2016) have reported RAV orthologs Bradi2g1760 and Bradi2g47220 were significantly induced by salinity stress, while another ortholog Bradi2g02720 was repressed by salt stress at 3 time points (1, 2, and 3 h). On the contrary, in barley, HvRAVs were not induced by salinity stress, even expression of HvRAV5 significantly decreased at 27 h after 100 mM NaCl (Fig. 5B). The HvRAV genes exhibited different expression patterns under long-term drought stress in drought-sensitive and drought-tolerant barley genotypes. While the expression level of the four HvRAV genes did not show considerable changes in drought-sensitive genotype, the drought stress significantly down-regulated the expression of HvRAV5 and HVRAV7 with 14.3- and 25-fold, respectively, in drought-tolerant genotype. The RAV’s negative regulation in response to salinity and drought has been reported using both rav mutants and RAV–overexpressing transgenic plants (Zhao et al., 2017). Under salt conditions, RAV-overexpressing transgenic plants showed more inhibited seed germination than the wild type, while rav mutants exhibited higher seed germination as well as better growth after germination compared to wild types in the Arabidopsis. In addition, transpirational water loss was found higher in RAV-overexpressing transgenic plants than in wild types under drought conditions (Fu et al., 2014). On the contrary, positive effects of heterologous expression of some RAV genes in Arabidopsis also have been reported, i.e., overexpression of soybean GmRAV3 and pepper CaRAV1 improved salinity and drought tolerance in transgenic Arabidopsis (Sohn et al., 2006; Zhao et al., 2017). The contradictory results in expression patterns also have suggested that homologous RAV genes may exhibit various responses against the specific stresses in different levels. There is no clear explanation about the various responses of RAV genes against salinity and drought. Being involved in different signaling and regulation pathways may result in various responses in similar stress conditions.
Barley HvRAV7 was expressed highly after long-term chilling and subzero treatment (Fig. 5C). In another microarray experiment (GSE41513), HvRAV6 and HvRAV7 were immediately induced by cold treatment within hours in both roots and shoot tissues of barley. Similarly, RAV genes have been reported to be induced by cold in Brassica napus and Chinese cabbage (Zhuang et al., 2011; Li et al., 2013). On the other hand, Fu et al. (2014) have reported that RAV1 and RAV2 genes were downregulated by cold in Arabidopsis. High temperature also changed the expression profile of HvRAV genes irregularly. The expression of HvRAV5 and HvRAv7 was repressed at 0.5 h after heat treatment but then beginning to increase in barley caryopses (Fig. 5C). BraRAv14, a member of the Chinese cabbage RAV subfamily, has been shown to be induced by heat stress (38°C) and reached peak level at 4 h of treatment (Li et al., 2013). In another study, it has been reported that the expression level of tea CsRAV (Cs088-RAV) was increased in response to heat stress in 4 tea cultivars compared to the controls. However, expression profiles of the Cs088-RAV were varied in 4 tea cultivars in low- and high-temperature stress (Wu et al., 2015). The same RAV gene may show different responses to adverse temperature changes in cultivars of the plants.
Plants defend themselves against a wide variety of pathogens including fungi, bacteria, and viruses by regulating multiple signal pathways and inducing defense-related genes (Sohn et al., 2006). Transcription factors have significant roles in controlling the expression of these target genes. Sohn et al. (2006) showed that over-expression of pepper CaRAV1 strongly induced some pathogenesis-related genes including PR1, PR2 and PR5. This result conferred resistance to bacterial pathogen Pseudomonas syringae in transgenic Arabidopsis. RAV transcription factors play role in disease resistance against various plant pathogens by inducing the genes involved in melatonin, a signal molecule in plant defense mechanism, biosynthesis, thus improved the endogenous melatonin level (Lee et al., 2014). The cassava MeRAV1 and MeRAV2 have been shown to improve the disease resistance against cassava bacterial blight by activating melatonin biosynthesis genes and increasing the melatonin level (Wei et al., 2018). In barley, HvRAV7 was induced by fungal pathogens C. sativus and P. hordei at early hours after inoculation but then decreased. HvRAV6 showed an antagonistic expression pattern compared to HvRAv7 after both pathogen attacks (Fig. 5D). The transcript level of the HvRAv7 gene demonstrated to oscillate after each 24 h (for 144 h): decreasing mRNA level at 24 h and increasing at 48 h after F. graminearum treatment in barley cv. Morex. In long-term conditions, it has been reported that only BnRAV12 was significantly over-expressed among 15 BrRAVs in response to fungal plant pathogen L. biglobosa treatment at 7 d after inoculation in Brasscica napus (Owji et al., 2017). The activation of defense responses in plants is initiated by host recognition of primary pathogen-derived elicitors. Different RAV genes are involved in response to different pathogen attacks. Distinct pathogen elicitor activates different signal transduction pathways and defense genes and transcription factors. Besides, the type and number of pathogen-responsive cis-acting elements in the promoter region may lead to differential expression of some RAV genes. In barley, RAV genes have been predicted to have a different number of pathogen-responsive cis-acting elements including W-BOX, GT1, TC-rich repeats and GCC-BOX (Tab. 3). HvRAV6 has been predicted to contain 14 W-BOX, 5 GT1 and 2 GCC-BOX binding elements, whereas HvRAV7 has a lower number of W-BOX and GT1, and no GCC-BOX.
Gene expression during germination and in response to ABA and GA treatments
Arabidopsis RAV1 (AtRAV1) has been reported to regulate the ABA signaling negatively during seed germination and establishment of the seedling. AtRAV1 repressed the expression of ABI3, ABI4, and ABI5, which are ABA-responsive genes during seed germination. On the other hand, exogenous application of ABA also suppressed the expression of AtRAV1 (Feng et al., 2014). In barley aleurone cells, it has been observed that ABA treatment did not significantly change the expression of barley RAVs (Fig. 6B). Opposite results also have been reported in the literature. Exogenous ABA application induced the soybean RAVs and overexpression of GmRAV3 improved the plant insensitivity in response to exogenous ABA during seed germination and early seedling development in Arabidopsis (Zhao et al., 2017). These different results emphasize the functional diversity of RAVs in plants. On the other hand, exogenous application of GA, which promotes seed germination, significantly induced the expression of HvRAV6 compared to the controls. SLN1 (Slender1) is a DELLA protein, which negatively regulates the GA signaling pathway in barley (Chandler et al., 2002). Chen et al. (2010) have reported that the genes induced by GA and highly expressed genes in sln1 mutant are highly similar genes. GA mostly regulates the expression of TFs via SLN1 in the barley aleurone cells. The mRNA level of HvRAV6 was dramatically increased after GA treatment (Fig. 6B). Interestingly, however, in sln1 mutant barley, the transcript level of HvRAV6 as high as ABA treated samples and lower than GA treated samples. The results may suggest that HvRAV6 was also regulated by GA but not dependent on SLN1.
In barley, the transcript level of the HvRAV6 gene was dramatically increased and reached peak point at 24 h after imbibition in endosperm tissues, but then decreased until the end of the germination process (Fig. 6A). In embryo tissues, mRNA level of HvRAV6 began to increase by imbibition, reached the highest level at 48 h after imbibition, and remained stable and high level for the next 24 h during post-germination. However, the transcripts of three RAV genes (HvRAV1, HvRAV5, and HvRAV7) were only detectable at a low level and their expression did not change during germination in both tissues. On the other hand, the transcript level of HvRAV6 was detected very high when compared to HvRAV1, HvRAV5, and HvRAV7 in both tissues and plant hormone (ABA and GA) treatments.
RAVs are unique members of the AP2/ERF superfamily with two DNA binding domains, AP2 and B3. They have important roles in the regulation of different plant processes including germination and stress responses. There is little information about barley RAV genes. In this study, 7 RAV genes were identified and characterized using bioinformatics tools and databases in barley. Conserved motifs and protein characteristics are determined using bioinformatics tools. Cis-acting elements that have a role in stress were predicted in the promoter region of HvRAV genes. Based on the microarray analysis, the expression profiles of 4 HvRAVs (HvRAV1, HvRAV5, HvRAV6, and HvRAV7) were determined under adverse environmental conditions, including drought, salinity, low-and high temperature, and pathogen attack. Besides, the transcript level of HvRAVs was also determined in germination and in response to GA and ABA treatments. In silico analyses have demonstrated that HvRAVs play roles in response to different abiotic and biotic stress conditions as well as plant growth. However, the extensive biological roles of HvRAV genes in plant development and in response to abiotic and biotic stresses need further investigation.
Availability of Data and Materials: All data was collected from the public database.
Authors’ Contribution: The author confirms sole responsibility for the following: study conception and design, data collection, analysis and interpretation of results, and manuscript preparation.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Agarwal G, Garg V, Kudapa H, Doddamani D, Pazhamal LT, Khan AW, Thudi M, Lee SH, Varshney RK (2016). Genome-wide dissection of AP2/ERF and HSP90 gene families in five legumes and expression profiles in chickpea and pigeonpea. Plant Biotechnology Journal 14: 1563–1577. [Google Scholar]
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Research 37: W202–208. [Google Scholar]
Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S (2004). Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4: 1633–1649. [Google Scholar]
Causier B, Ashworth M, Guo W, Davies B (2012). The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiology 158: 423–438. [Google Scholar]
Chandler PM, Marion-Poll A, Ellis M, Gubler F (2002). Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiology 129: 181–190. [Google Scholar]
Chen K, Tian S, Yandell BS, Kaeppler SM, An YC (2010). Loss-of-function of DELLA protein SLN1 activates GA signaling in barley aleurone. Acta Physiologiae Plantarum 32: 789–800. [Google Scholar]
Chen L, Han J, Deng X, Tan S, Li L, Li L, Zhou J, Peng H, Yang G, He G, Zhang W (2016). Expansion and stress responses of AP2/EREBP superfamily in Brachypodium distachyon. Scientific Reports 6: 21623. [Google Scholar]
Dawson IK, Russell J, Powell W, Steffenson B, Thomas WT, Waugh R (2015). Barley: A translational model for adaptation to climate change. New Phytologist 206: 913–931. [Google Scholar]
Dossa K, Wei X, Li D, Fonceka D, Zhang Y, Wang L, Yu J, Boshou L, Diouf D, Ciss N, Zhang X (2016). Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress. BMC Plant Biology 16: 171. [Google Scholar]
Du C, Hu K, Xian S, Liu C, Fan J Tu J, Fu T (2016). Dynamic transcriptome analysis reveals AP2/ERF transcription factors responsible for cold stress in rapeseed (Brassica napus L.). Molecular Genetics and Genomics 291: 1053–1067. [Google Scholar]
FAO (2018). FAOSTAT statistical database. http://www.fao.org/faostat/en/#data/QC. [Google Scholar]
Feng CZ, Chen Y, Wang C, Kong YH, Wu WH, Chen YF (2014). Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant Journal 80: 654–668. [Google Scholar]
Feng JX, Liu D, Pan Y, Gong W, Ma L, Luo JC, Deng XW, Zhu YX (2005). An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Molecular Biology 59: 853–868. [Google Scholar]
Fu MJ, Kang HK, Son SH, Kim SK, Nam KH (2014). A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiology 55: 1892–1904. [Google Scholar]
Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261. [Google Scholar]
Guo B, Wei Y, Xu R, Lin S, Luan H, Lv C, Zhang X, Song X, Xu R (2016). Genome-wide analysis of APETALA2/ethylene-responsive factor (AP2/ERF) gene family in barley (Hordeum vulgare L.). PLoS One 11: 1–17. [Google Scholar]
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999). Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Research 27: 297–300. [Google Scholar]
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31: 1296–1297. [Google Scholar]
Ikeda M, Ohme-Takagi M (2009). A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiolology 50: 970–975. [Google Scholar]
Jin JP, Zhang H, Kong L, Gao G, Luo JC (2014). PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Research 42: D1182–D1187. [Google Scholar]
Kagaya Y, Ohmiya K, Hattori T (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Research 27: 470–478. [Google Scholar]
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10: 845–858. [Google Scholar]
Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H (2009). Six classes of nuclear localization signals specific to different binding grooves of importin α. Journal of Biological Chemistry 284: 478–485. [Google Scholar]
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. [Google Scholar]
Lee HY, Byeon Y, Back K (2014). Melatonin as a signal molecule triggering defense responses against pathogen attack in Arabidopsis and tobacco. Journal of Pineal Research 67: 262–268. [Google Scholar]
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouz P, Rombauts S (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30: 325–327. [Google Scholar]
Letunic I, Bork P (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Research 46: D493–D496. [Google Scholar]
Li H, Wang Y, Wu M, Li L, Li C, Han Z, Yuan J, Chen C, Song W, Wang C (2017). Genome-wide identification of AP2/ERF transcription factors in cauliflower and expression profiling of the ERF family under salt and drought stresses. Frontiers in Plant Science 8: 946. [Google Scholar]
Li MY, Wang F, Jiang Q, Li R, Ma J, Xiong AS (2013). Genome-wide analysis of the distribution of AP2/ERF transcription factors reveals duplication and elucidates their potential function in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Molecular Biology Reporter 31: 1002–1011. [Google Scholar]
Liu C, Zhang T (2017). Expansion and stress responses of the AP2/EREBP superfamily in cotton. BMC Genomics 18: 118. [Google Scholar]
Liu S, Wang X, Wang H, Xin H, Yang X, Yan J, Li J, Tran LS, Shinozaki K, Yamaguchi-Shinozaki K, Qin F (2013). Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genetics 9: 119–129. [Google Scholar]
Matías-Hernández L, Aguilar-Jaramillo AE, Marín-González E, Suárez-López P, Pelaz S (2014). RAV genes: Regulation of floral induction and beyond. Annual Botany 114: 1459–1470. [Google Scholar]
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology 140: 411–432. [Google Scholar]
Notredame C, Higgins DG, Heringa J (2000). T-Coffee: A novel method for fast and accurate multiple sequence alignment. Journal of Molecular Biology 302: 205–217. [Google Scholar]
Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968. [Google Scholar]
Okamuro JK, Caster B, Villarroel R, van Montagu M, Jofuku KD (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 94: 7076–7081. [Google Scholar]
Owji H, Hajiebrahimi A, Seradj H, Hemmati S (2017). Identification and functional prediction of stress responsive AP2/ERF transcription factors in Brassica napus by genome-wide analysis. Computational Biology and Chemistry 71: 32–56. [Google Scholar]
Rose AB (2008). Intron-mediated regulation of gene expression. Current Topics in Microbiology and Immunology 326: 277–290. [Google Scholar]
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J (2003). TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378. [Google Scholar]
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochemical and Biophysical Research Communications 290: 998–1009. [Google Scholar]
Sohn KH, Lee SC, Jung HW, Hong JK, Hwang BK (2006). Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Molecular Biology 61: 897–915. [Google Scholar]
Suzuki M, Kao CY, McCarty DR (1997). The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9: 799–807. [Google Scholar]
Uçarlı C, Gürel F (2020). Differential physiological and molecular responses of three-leaf stage barley (Hordeum vulgare L.) under salt stress within hours. Plant Biotechnology Reports 14: 89–97. [Google Scholar]
Waltner JK, Peterson FC, Lytle BL, Volkman BF (2005). Structure of the B3 domain from Arabidopsis thaliana protein At1g16640. Protein Science 14: 2478–2483. [Google Scholar]
Wei Y, Chang Y, Zeng H, Liu G, He C, Shi H (2018). RAV transcription factors are essential for disease resistance against cassava bacterial blight via activation of melatonin biosynthesis genes. Journal of Pineal Research 64: e12454. [Google Scholar]
Wu ZJ, Li XH, Liu ZW, Li H, Wang YX, Zhuang J (2015). Transcriptome-based discovery of AP2/ERF transcription factors related to temperature stress in tea plant (Camellia sinensis). Functional & Integrative Genomics 15: 741–752. [Google Scholar]
Xie Z, Nolan TM, Jiang H, Yin Y (2019). AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Frontiers in Plant Science 10: 228. [Google Scholar]
Zhao SP, Xu ZS, Zheng WJ, Zhao W, Wang YX, Yu TF, Chen M, Zhou YB, Min DH, Ma YZ, Chai SC, Zhang XH (2017). Genome-wide analysis of the RAV family in soybean and functional identification of GmRAV-03 involvement in salt and drought stresses and exogenous ABA treatment. Frontiers in Plant Science 8: 905. [Google Scholar]
Zheng Y, Jiao C, Sun H, Rosli HG, Pombo MA, Zhang P, Banf M, Dai X, Martin GB, Giovannoni JJ, Zhao PX, Rhee SY, Fei Z (2016). iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Molecular Plant 9: 1667–1670. [Google Scholar]
Zhuang J, Chen JM, Yao QH, Xiong F, Sun CC, Zhou XR, Zhang J, Xiong AS (2011). Discovery and expression profile analysis of AP2/ERF family genes from Triticum aestivum. Molecular Biology Reports 38: 745–753. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |