DOI:10.32604/biocell.2022.016346

| BIOCELL DOI:10.32604/biocell.2022.016346 |  |
| Article |
An active RUNX1-ID1/ID3 axis governs differentiation and chemoresistance of cancer stem cell population in epithelial ovarian cancer cells
1Imaging Cell Signaling & Therapeutics Laboratory, Advanced Centre for Treatment, Research and Education in Cancer, Tata Memorial Centre, Navi Mumbai, 410210, India
2Homi Bhabha National Institute, Mumbai, 400094, India
*Address correspondence to: Pritha Ray, pray@actrec.gov.in
Received: 27 February 2021; Accepted: 17 May 2021
Abstract: Progression, relapse, and therapy resistance are the most challenging features of cancer therapy that have been postulated to be driven by Cancer Stem Cell (CSC) population. This enigmatic subpopulation of cancer cells has therefore emerged as promising therapeutic candidate. We earlier reported enrichment of CSC-like side population (SP) with increasing resistance towards Cisplatin and Paclitaxel either alone or in combination in epithelial ovarian cancer (EOC) cells. This SP population is a small proportion of the total population of cancer cells characterised with high expression of drug transporters, a unique feature of stem cells and thereby can be isolated through their efflux properties of DNA binding dyes. While the bulk non-SP (NSP) population of the cancer cells lack overexpression of the drug transporters and thus can be identified as the dye containing population. In this study, we show that increased expression of Runt related transcription factor 1 (RUNX1) maintains undifferentiated state of CSC-like SP cells through upregulation of inhibitors of DNA binding/differentiation genes (ID1 and ID3) in late cisplatin-paclitaxel resistant cells. Higher RUNX1 expression was found to correlate with decreased median overall survival and disease-free survival in The Cancer Genome Atlas (TCGA) data set of high grade serous ovarian cancer (HGSOC) patients. The protein-protein interaction network analysis of 397 upregulated genes in RUNX1-high samples of TCGA data show significant enrichment of pathways known to negatively regulate CSC differentiation. Intriguingly RUNX1 inhibition not only induces CSC differentiation but also downregulates anti-apoptotic protein BCL2 in both SP and NSP cells and potentiates cytotoxic effects of Cisplatin-Paclitaxel in chemoresistant EOC cells. Inhibition of BCL2 through Venetoclax treatment, a small molecule BH3 mimic, sensitized these cells to platinum taxol treatment. Altogether, our data reveal new regulatory roles by RUNX1 to modulate CSC differentiation via ID1 and ID3 and to promote chemoresistance through BCL2 upregulation.
Keywords: Cancer stem cells; Differentiation; RUNX1; ID1; ID2; BCL2; Chemoresistance; Ovarian cancer
The cancer stem cell (CSC) model of tumor progression recognizes CSC population at top of hierarchically organized tumor bulk, accounting for the phenotypic and functional heterogeneity in majority of human cancers (Batlle and Clevers, 2017; Kreso and Dick, 2014; Vermeulen et al., 2012). These elusive CSCs not only exhibit the stemness properties of self-renewal and differentiation, but are also shown to be quiescent, highly tumorigenic and are intrinsically resistant to both chemotherapy and radiotherapy (Abdullah and Chow, 2013; Arnold et al., 2020; Rycaj and Tang, 2014; Zhao, 2016). Intriguingly, both pre-clinical and clinical evidence suggest that CSCs are not only crucial for tumor initiation and progression, but also play important role in therapy resistance and disease recurrence due to their ability to endure the therapeutic interventions (Jin et al., 2017; Yang et al., 2020).
Therapeutic strategies to eradicate CSCs either focus on direct targeting of CSCs through cell surface CSC biomarkers or by modulating stemness signaling pathways and CSC niche. While transcriptional modulators such as MYC, NOTCH3, β-catenin, NFκB and RUNX1 are crucial for CSC self-renewal (Yang et al., 2020), the inhibitor of DNA binding/inhibitor of differentiation proteins (ID1/ID2/ID3/ID4) are dominant negative regulators of CSC differentiation, which act by sequestering basic-helix loop helix proteins and forming transcriptionally inactive heterodimers (Roschger and Cabrele, 2017). However, most of these pathways, which are crucial for maintenance of CSC phenotype, are known to play important role in survival and therapy resistance of non-CSCs. Though several potential therapeutics against CSCs have entered clinical trials, it is unclear whether these therapies exclusively eliminate the CSCs or both CSC and non-CSC populations. Their effect on CSC plasticity also remains poorly understood. Thus, elucidating the underlying mechanisms and identifying the critical regulators and pathways are of great importance.
The residual tumor cells post chemo/radio therapy are found to be enriched of CSCs in several human malignancies such as, acute myeloid leukemia (AML) (Boyd et al., 2018; Gerber et al., 2012; Van Rhenen et al., 2005), breast cancer (Lagadec et al., 2012; Lagadec et al., 2010; Lu et al., 2017), lung cancer (Hamilton and Olszewski, 2013), colorectal cancer (Dylla et al., 2008), hepatocellular carcinoma (Tan et al., 2009), prostate cancer (Chen et al., 2019), glioblastoma (Ramakrishnan et al., 2020), and ovarian cancer (Thakur and Ray, 2017; Wang et al., 2018; Zhao et al., 2020). We recently reported enrichment of CSC population (CSC-like side population or spheroids) with increasing resistance in Cisplatin-Paclitaxel resistant cellular models of Epithelial Ovarian cancer (EOC) cells. Intriguingly, these SP cells possessing augmented Insulin-like growth factor receptor 1 (IGF1R) at the onset of resistance in these models showed faster tumorigenicity compared to SP cells of highly or late resistant cells (Singh et al., 2016). Further we identified that RUNT related transcription factor 1 (RUNX1), showed increased expression with increasing resistance and inhibition of RUNX1 sensitized both early and late resistant cells to Cisplatin-Paclitaxel (Dhadve et al., 2020). Till date various studies have uncovered the role of RUNX1 in disease progression and therapy resistance of EOC but none has shed light on its role in CSC phenotype in EOC (Ge et al., 2014; Keita et al., 2013; Xiao et al., 2020).
In this study, we investigated the role of RUNX1 and ID proteins upon differentiation and chemoresistance properties of CSCs isolated from platinum-taxol resistant EOC cells. Increased RUNX1 expression in high grade serous (HGS) Ovarian Serous Cystadenocarcinoma Firehose Legacy data set of The Cancer Genome Atlas (TCGA) negatively correlated with median overall survival and median disease-free survival. In our cellular models, enhanced RUNX1 levels was found to retain the CSC-like SP cells in undifferentiated state by regulating two members of the inhibitor of differentiation gene family, ID1 and ID3. In parallel, RUNX1 inhibition downregulates anti-apoptotic protein BCL2 in both total population and SP cells. Abrogating RUNX1 or BCL2 activities by respective small molecular inhibitors potentiated cytotoxic effects of Cisplatin-Paclitaxel in these chemoresistant EOC cells. To the best of our knowledge, this is the first report on new regulatory roles by RUNX1 in modulating CSC differentiation and chemoresistance, the critical factors for development and maintenance of platinum-taxol resistance in epithelial ovarian cancer cells.
Paclitaxel, Cisplatin, α-tubulin, anti-mouse, and anti-rabbit tagged with horseradish peroxidase were purchased from Sigma-Aldrich (USA). BCL2 and RUNX1 and were purchased from Abcam (UK). ID1, ID2, ID3 and ID4 were purchased from Santacruz Biotech (USA). Ro5-3335 was purchased from Merck Millipore (USA). BCL2 inhibitor ABT-119 (Venetoclax) was purchased from Selleck Chemicals Llc (USA).
Core Binding Factor Beta (CBFβ) knock down A2780-DualLR cells (Dhadve et al., 2020), A2780 and OAW42 platinum-taxol resistance models and SKOV3 were cultured as described earlier. The models were categorized into sensitive cells (Parental), and Cisplatin-Paclitaxel late resistant cells (DualLR) stages based on their resistant indices (Singh et al., 2014).
Immunofluorescence and Western blotting
Immunofluorescence for RUNX1 was performed and images were acquired using Carl Zeiss, LSM 780 microscope (Singh et al., 2014). Whole cell lysates were prepared as described earlier and western blotting was performed for RUNX1, ID1, ID2, ID3, ID4, BCL2 and α-tubulin (Gaikwad et al., 2015).
Bioinformatics analysis of TCGA data
The online tools and resources The Cancer Genome Atlas (TCGA; http://www.cbioportal.org) and GEPIA (http://gepia.cancer-pku.cn/) were used to study the relationship between RUNX1 expression levels and OC patient overall survival and disease-free survival rate in HGS Ovarian Serous Cystadenocarcinoma, Firehose Legacy data set of TCGA (Cerami et al., 2012; Gao et al., 2013). The RUNX1 mRNA expression-based stratification of OC patients were done in two groups using quartile segregation, namely RUNX1 low (Q1) and RUNX1 high (Q3) using cBioPortal tool. The differential gene expression analysis between RUNX1 low (Q1) and RUNX1 high (Q3) was done using cBioPortal tool. The ClueGO version 2.5.5, Cytoscape plug-in software (http://apps.cytoscape.org/apps/cluego), was used to obtain the protein-protein interaction network of the pathways significantly enriched (GOBiologicalProcess UniProt, KEGG and the Reactome pathway databases) in the 253 upregulated genes (log ratio change 1.5-fold up, q < 0.05, false discovery rate) in RUNX1 high group. The Two-sided hypergeometric test with Bonferroni step down correction method and p-value cut-off of <0.05 was used in Cytoscape analysis (Bindea et al., 2009).
Real-time PCR was performed using SYBR Green (Invitrogen) using primers for BCL2 (Forward: 5’-TCGCCCTGTGGATGACTGA-3’ and Reverse: 5’-CAGAGACAGCCAGGAGAAATCA-3’) and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Forward: 5’-TGCACCACCAACTGCTTAGC-3’ and Reverse: 5’-GGCATGGACTGTGGTCATGAG-3’) and GAPDH was used for normalization (Singh et al., 2014).
CBFβ silencing by lentiviral mediated sh-RNA constructs
CBFβ-knock down lentiviral cassette was developed using a target sequence against CBFβ (5’-CCGCGAGTGTGAGATTAAGTA-3’). A2780-DualLR cells were transduced with lentiviruses and stable cells were FACS sorted using eGFP as a marker (Dhadve et al., 2020).
DyeCycle violet side population assay
Dye exclusion DyeCycle Violet (DCV) side population assay was used to isolate the CSC like cells called the side population (SP) and non-CSCs called non-side population (NSP) form EOC cells as described previously (Singh et al., 2016). The cells were treated with Ro5-3335 for 12 h prior to the DCV assay.
Cytotoxicity 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay for cisplatin-paclitaxel (72 h) or Ro5-3335 or ABT-199 alone (12 h) or the combinations were performed following published protocols (Gaikwad et al., 2015).
All the data represent the mean ± SEM of at least three independent experiments and were analyzed for significance using unpaired Student’s t-test. P-value ≤ 0.05 was considered as significant.
Increased RUNX1 expression show strong association with decreased overall survival and enrichment of oncogenic pathways in HGS EOC Firehose Legacy TCGA data
Our previous results showed positive association between CSC phenotype and RUNX1 expression with acquirement of chemoresistance in A2780 and OAW42 Cisplatin-Paclitaxel resistant cellular models of EOC cells (Dhadve et al., 2020; Singh et al., 2016). Since CSC phenotype shows high degree of cellular plasticity and is governed by a network of aberrant transcriptional modulators including RUNX1, we first investigated the association between RUNX1 mRNA expression and clinical outcome in Ovarian Serous Cystadenocarcinoma Firehose Legacy TCGA dataset. The EOC tumors showed high levels of RUNX1 transcripts as compared to the normal ovarian tissue (Fig. 1A). Next, we categorized the RUNX1 mRNA expression into two groups, RUNX1 low (Q1) and RUNX1 high (Q3) by quartile segregation using cBioPortal tool (Fig. 1B) and found strong association of both poor overall survival and poor disease-free survival with high RUNX1 expression (Figs. 1C and 1D). The median overall survival for RUNX1 high group was 43.89 months as compared to the 52.4 months in RUNX1 low group (Log-rank-Mantel-Cox test, P = 0.0155) (Fig. 1C). Similarly, median disease-free survival was low in RUNX1 high group, 15.64 months as compared to 20.83 months in RUNX1 low group (Log-rank-Mantel-Cox test, P = 0.0284) (Fig. 1D). The Hazard Ratio (Mantel-Haenszel RUNX1 high/RUNX1 low) was found to be 1.469 and 1.412 for overall survival and disease-free survival, respectively (Figs. 1C and 1D). This data prompted us to investigate for differentially expressed genes among RUNX1-low and RUNX1-high groups in the TCGA Ovarian Serous Cystadenocarcinoma Firehose Legacy data set. Around 7997 genes were found to be differentially expressed between RUNX1 high vs. RUNX1 low groups with q-value < 0.05 (Fig. 1E and Suppl. File 1), of which 481 genes were differentially expressed with log ratio change of either 1.5-fold up or 1.5-fold low (q < 0.05) between RUNX1 high vs. RUNX1 low groups (Suppl. File 2). Among these 481 genes, 84 were found to be upregulated in RUNX1 low group and 397 were upregulated in RUNX1 high group (Suppl. File 2). Next, protein-protein interaction network analysis of the 397 upregulated genes (log ratio change 1.5-fold up, q < 0.05) in RUNX1 high group was carried out using the ClueGo, (a Cytoscape plug-in) software (http://apps.cytoscape.org/apps/cluego). Further similar analysis of this interaction using GOBiologicalProcess UniProt, KEGG and the Reactome databases revealed significant enrichment (P < 0.05) of pathways inclusive of embryonic morphogenesis, skeletal system development, integrin, and non-integrin cell-extracellular matrix interactions (ECM), ECM degradation, cytokine secretion, vascular development, regulation of epithelial cell proliferation, tumor cellular response to necrosis factor alpha (TNFα) and apoptosis (Fig. 2 and Suppl. File 3). Intriguingly, the embryonic morphogenesis protein-protein interaction network showed enrichment of pathways which are involved in negative regulation of both the Bone Morphogenetic Protein (BMP) signaling and transforming growth factor beta (TGFβ)/SMAD signaling known promoter of CSC differentiation (Jin et al., 2017; Yang et al., 2020). These analysis brought up an unexplored functional consequence of RUNX1 overexpression in addition to its known role in disease progression and therapy resistance of EOC (Ge et al., 2014; Keita et al., 2013; Xiao et al., 2020).
Increased RUNX1 expression in chemoresistant EOC cells maintains the CSC-like SP phenotype
We observed significantly enhanced RUNX1 expression in A2780-DualLR (A2780 Cisplatin-Paclitaxel late resistant cells), OAW42-DualLR (OAW42 Cisplatin-Paclitaxel late resistant cells) compared to their sensitive counterparts (A2780 and OAW42) and in inherently Cisplatin resistant SKOV3 cells (Figs. 3A and 3B). Strong DNA binding activity of RUNX1 requires hetero-dimerization with CBFβ which can be inhibited by a small molecule, Ro5-3335 (Cunningham et al., 2012). To gain further insights of RUNX1 role in EOC CSC phenotype, we used a flowcytometry based Dye Cycle Violet (DCV) dye efflux assay to identify and isolate the CSC-like SP cells which are characterized by high expression of membrane transporters such as ATP-binding cassette (ABC) transporter proteins, a distinctive feature of CSCs (Telford et al., 2007). Briefly all the cells, however only the SP cells are able to efflux DCV out due to overexpressed membrane transporters and thus appear as low intensity side population in the scatter plot take up the DCV dye passively. The SP cells are gated using Verapamil (an inhibitor of ABC transporter family proteins) since these cells get remitted from the scatter plot in verapamil pre-treated cells (Garcia-Escarp et al., 2004). These SP cells are known to be enriched with CSC population (Challen and Little, 2006).
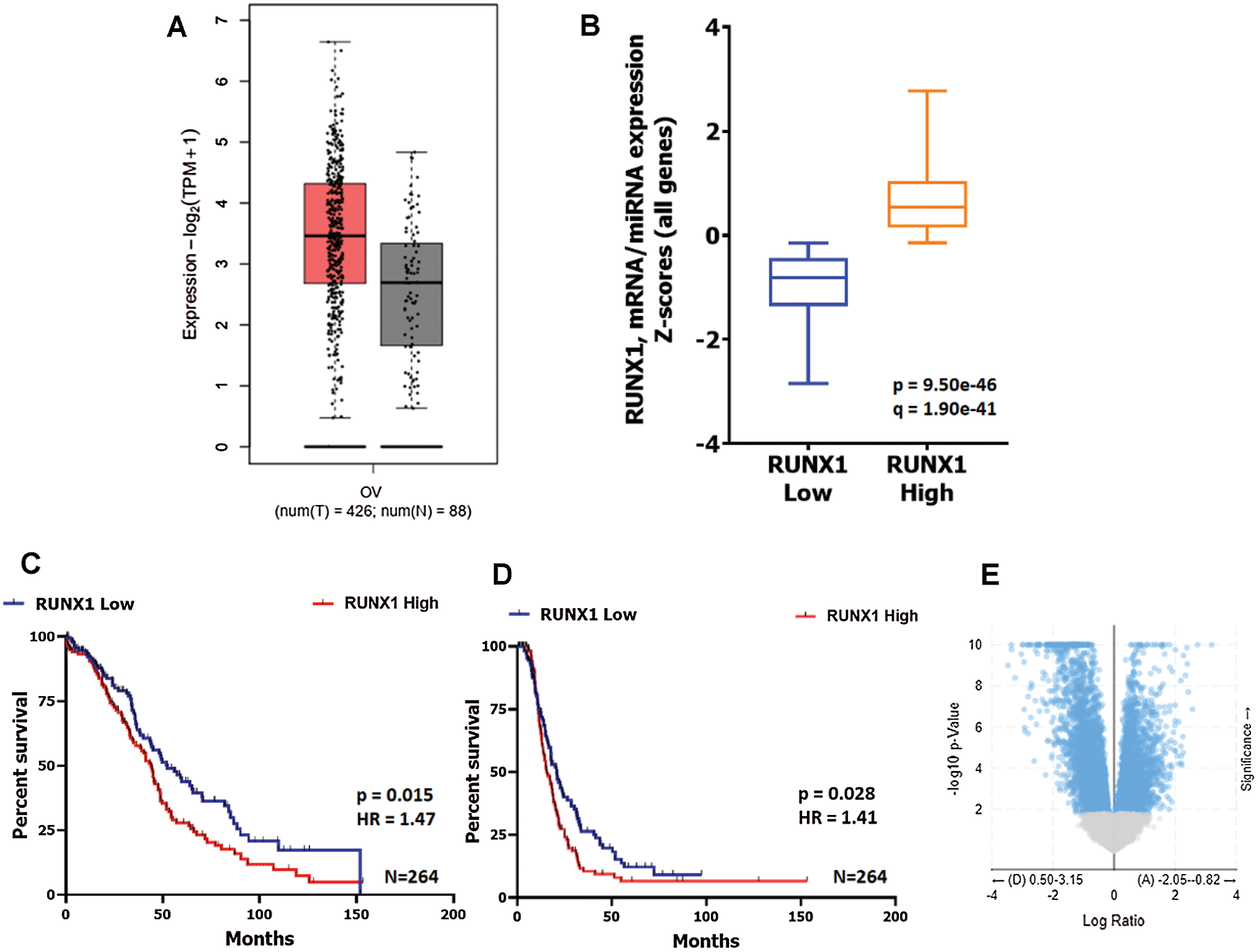
Figure 1: Increased RUNX1 expression show strong association of both poor overall survival and poor disease-free survival in HGS EOC TCGA data.
A. RUNX1 mRNA expression in HGS EOC is higher than in normal tissue. Data source: TCGA Ovarian Serous Cystadenocarcinoma, Firehose Legacy (n = 426) and GTEx (n = 88). B. Graphical representation of RUNX1 mRNA expression-based segregation of TCGA HGS EOC tumors into two groups, RUNX1 high (Q1) and RUNX1 low (Q2) using cBioPortal tool. C–D. Kaplan-meier survival plot showing strong association of both poor overall survival and poor disease-free survival with high RUNX1 expression respectively. The median overall survival for RUNX1 high group was 43.89 months as compared to the 52.4 months in RUNX1 low group (Log-rank-Mantel-Cox test, P = 0.0115). The median disease-free survival was low in RUNX1 high group, 15.64 months as compared to 20.83 months in RUNX1 low group (Log-rank-Mantel-Cox test, P = 0.0284). E. Volcano plot showing differential gene expression (mRNA) between RUNX1 high and RUNX1 low groups plotted using cBioPortal tool.
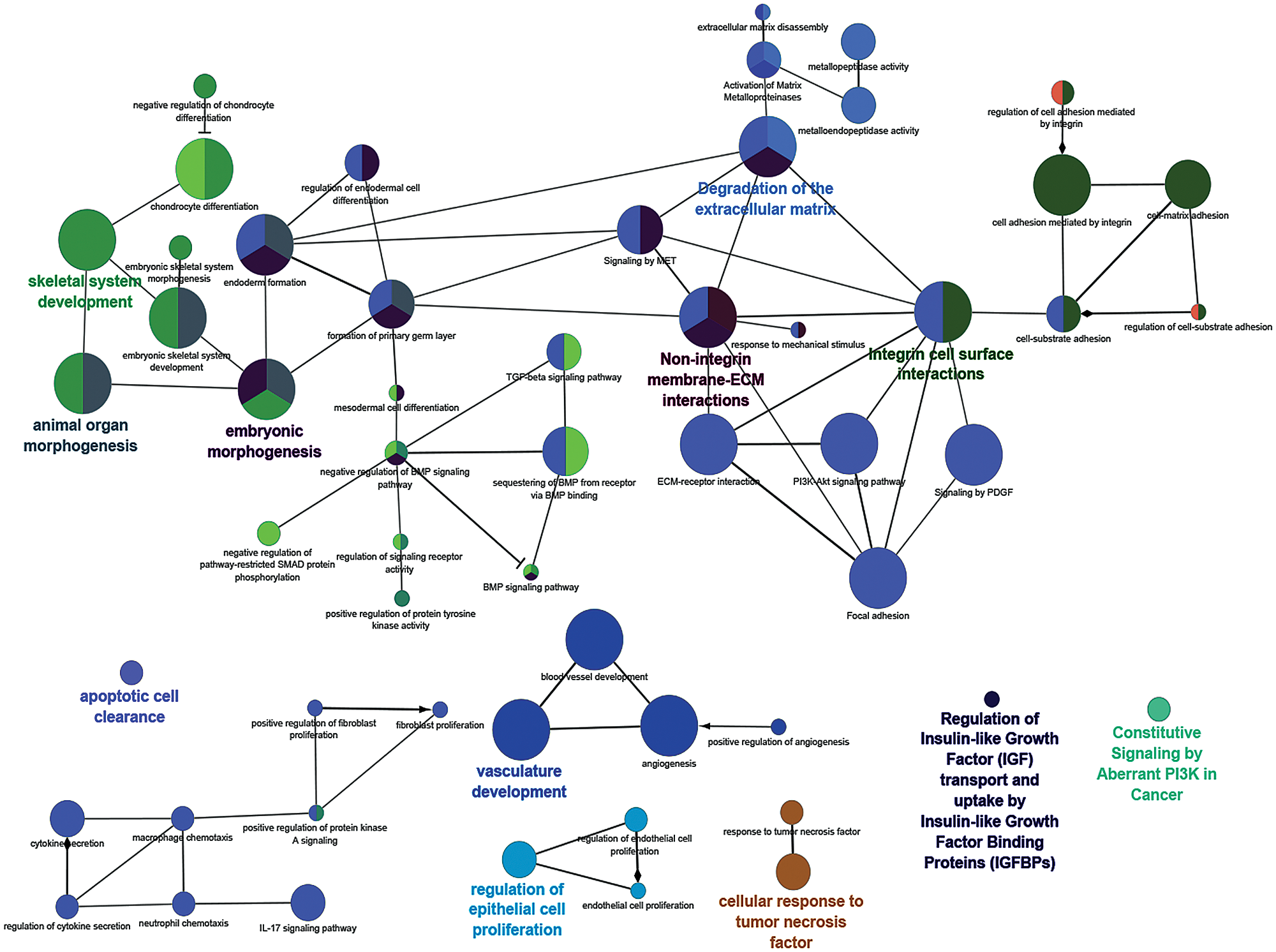
Figure 2: Interaction network analysis of the pathways significantly enriched in the genes upregulated in RUNX1 high expression group.The significantly upregulated 397 genes (Fold log ratio <1.5 and q > 0.05) in RUNX1 high group were analysed for their interaction and functional enrichment (P > 0.05) in the pathways from the GOBiologicalProcess UniProt, KEGG and Reactome database using the ClueGO application in the Cytoscape software. Different colours of one single node denote interaction with multiple pathways depending upon the number of colours present in the node. The larger size of a node indicates higher number of interacting partners.
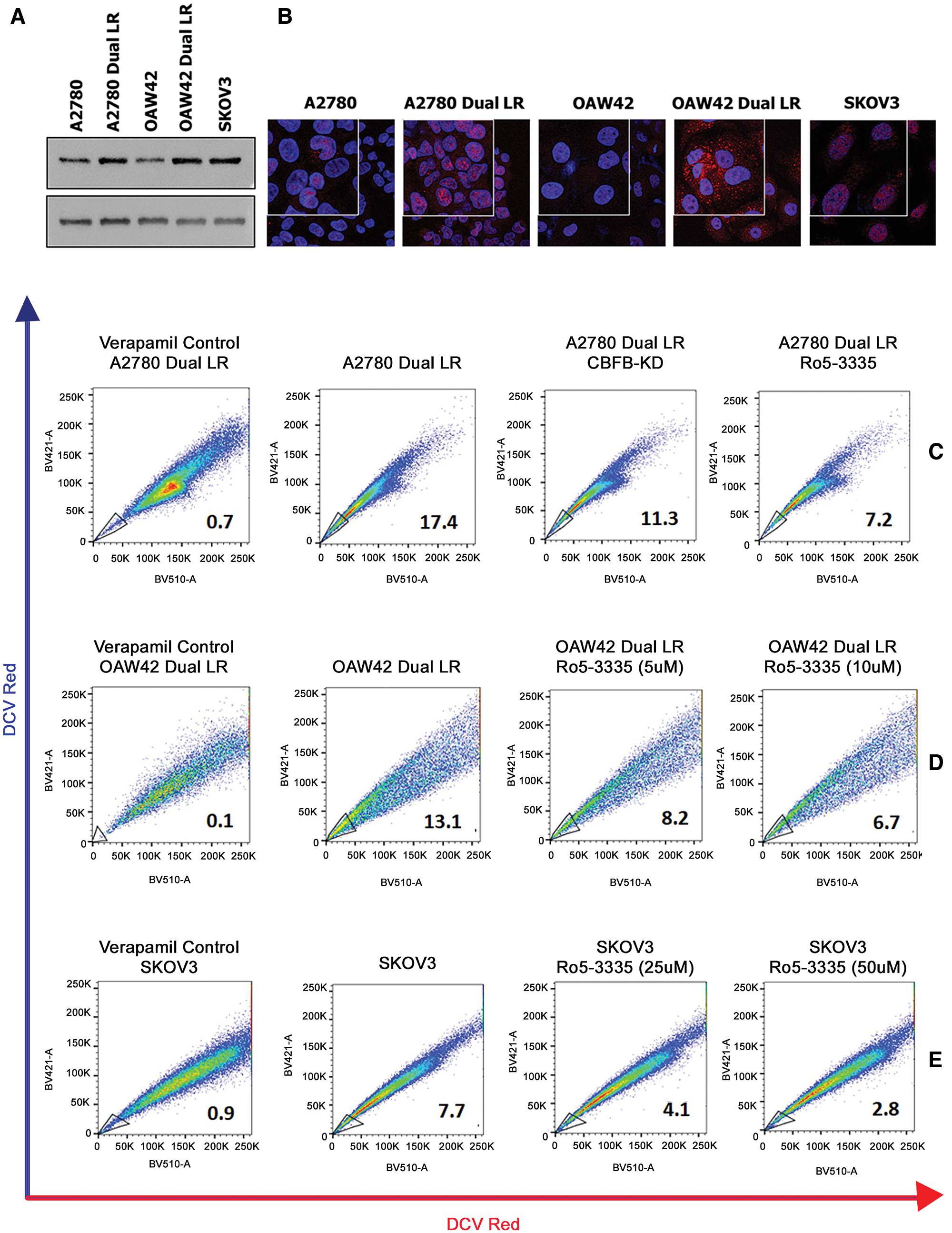
Figure 3: Transcriptional inhibition of RUNX1 abrogates CSC-like SP cells in EOC cells.
(A–B) Western blot and Immunofluorescence showing increased expression of RUNX1 in Cisplatin-Paclitaxel resistant (A2780-DualLR and OAW42-DualLR) cells and inherently Cisplatin resistant SKOV3 OC cells, respectively. (C) Perturbation of RUNX1 transcriptional activity either by CBF knockdown or Ro5-3335 treatment reduced SP population from 17.0% to 11.3% and 7.2% respectively in A2780-DualLR cells. (D–E) Dose dependent decrease in SP population in OAW42-DualLR cells from 13.1% to 8.2% (5 µM) and 6.7 (10 µM) and SKOV3 cells from 7.7% to 4.1% (25 µM) and 2.8% (50 µM) post Ro5-3335 treatment, respectively.
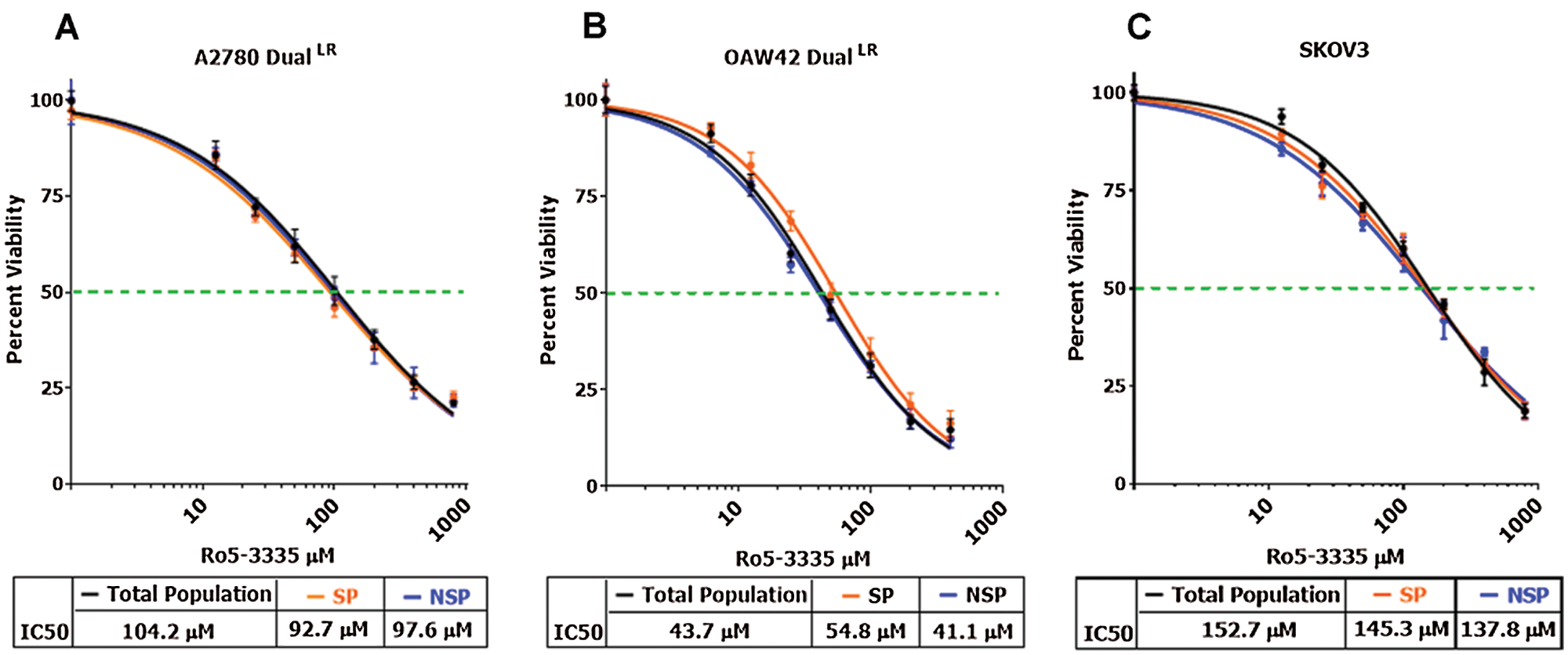
Figure 4: RUNX1 inhibition did not affect survival of TP, NSP and SP cells differentially.
(A–C) MTT cell cytotoxicity assay showing no significant difference in cell viability of TP, NSP and SP cells post Ro5-3335 treatment in A2780-DualLR, OAW42-DualLR and SKOV3 cells, respectively.
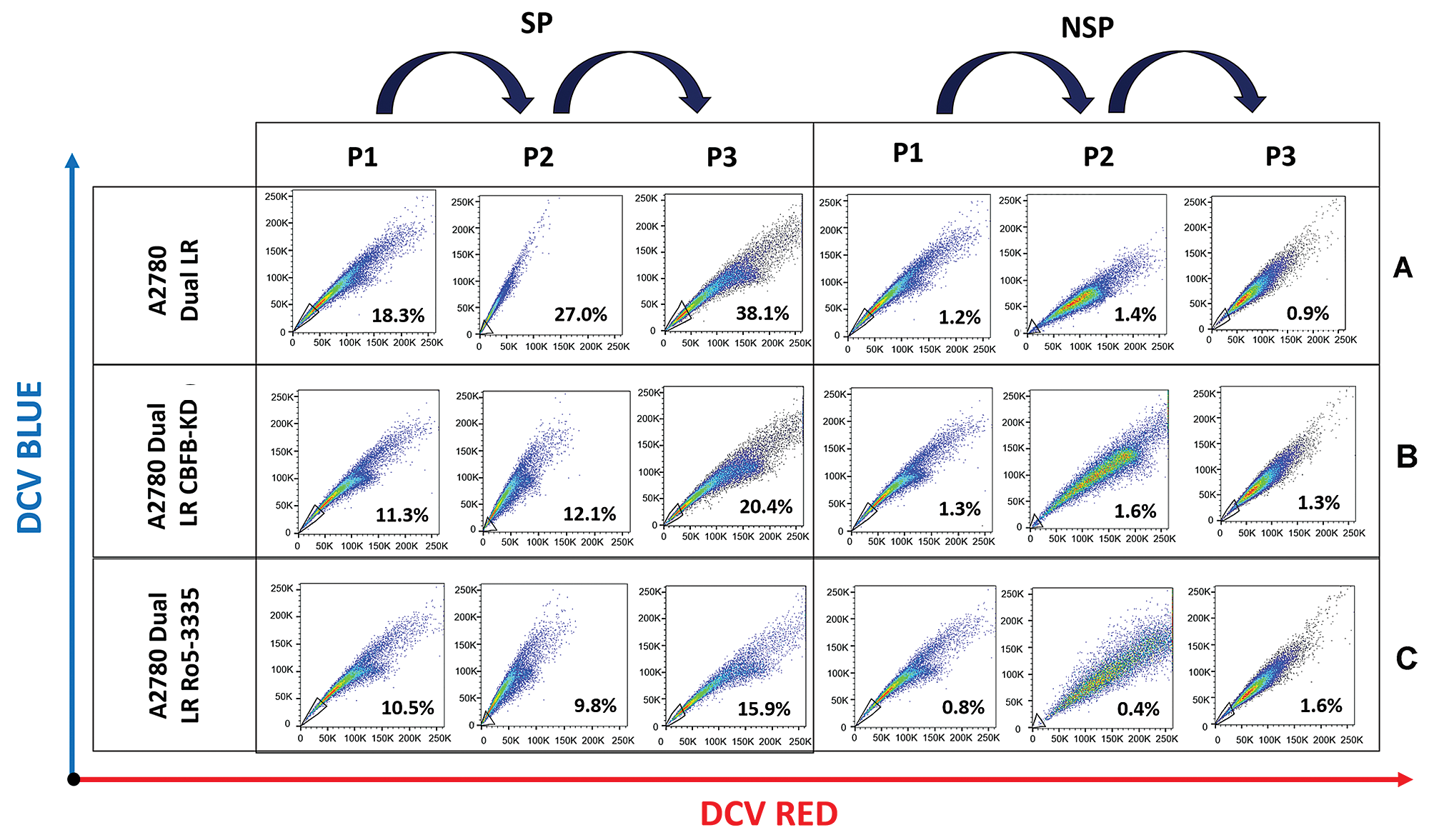
Figure 5: RUNX1 inhibition induces differentiation of CSC-like SP population into NSP population in A2780-dualLR cells.
(A) FACS analysis of differentiating ability of SP and NSP cells monitored over three subsequent passages by DCV staining, showing enrichment of SP population from 17.0% to 38.1% in control A2780-DualLR cells. (B–C) RUNX1 inhibition (CBFβ-KD and Ro5-3335) showed impaired enrichment of SP cells respectively. CBFβ-KD A2780-DualLR cells showed inherently lower SP population (11.3%), which increased to only 20.4% post third sort. Ro5-3335 treated A2780-DualLR cells, a mere increase from 10.5% to 15.9% in SP population post third sort. (A–C) No enrichment of SP population was observed in NSP cells in subsequent passages of Control A2780-DualLR, CBFβ-KD A2780-DualLR and Ro5-3335 treated A2780-DualLR cells.
In our study, RUNX1 inhibition either using shRNA against CBFβ or using Ro5-3335, a pharmacological inhibitor of RUNX1-CBFβ heterodimerization, showed significant inhibition of CSC-like SP cells in A2780-DualLR, OAW42-DualLR and SKOV3 cells (Figs. 3C–3E). CBFβ knockdown (CBFβ-KD) or Ro5-3335 (50 µM) treated A2780-DualLR cells possessed only 11.3% and 7.2% SP population respectively compared to the 17.4% SP in untreated A2780-DualLR cells (Fig. 3C). Further, OAW42-DualLR and SKOV3 cells showed dose dependent decrease in SP population, from 13.1% to 8.2% (5 µM) and 6.7 (10 µM) (Fig. 2D) and from 7.7% to 4.1% (25 µM) and 2.8% (50 µM), respectively (Fig. 3E). Since RUNX1 is known to regulate survival of CSCs of AML and triple negative breast cancer cells, we sought to determine the similar property of RUNX1 for the growth of SP cells in chemoresistant EOC cells. To our surprise RUNX1 inhibition (Ro5-3335 treatment) did not affect survival of the SP cells compared to the total and NSP of A2780-DualLR, OAW42-DualLR and SKOV3 cells (Figs. 4A–4C).
RUNX1 promotes undifferentiated state of CSC-like SP cells in chemoresistant EOC cells
Since RUNX1 inhibition reduced CSC-like SP cells without affecting viability and our protein-protein interaction network analysis showed significant enrichment of negative regulators of CSC differentiation, we investigated differentiation ability of SP cells, an important property of CSC population.
To assess differentiation ability, we repeatedly sorted SP from Ro5-3335 treated A2780-DualLR or CBFβ-KD A2780-DualLR cells and cultured for three generations as reported earlier (Thakur and Ray, 2017). Intriguingly, we did not observe an absolute persistence of (100%) SP population from first to third sorting, rather a gradual enrichment of CSC-like SP cells was observed (from 18.3% to 38.1%), with simultaneous decrease in NSP cells (from 83.0% to 61.9%) (Fig. 5A). However, CBFβ-KD A2780-DualLR cells showed inherently lower SP population (11.3%), which increased to only 20.4% post third sort (Fig. 5B). Similarly, in Ro5-3335 treated A2780-DualLR cells, a mere increase of 4.6% (from 11.3% to 15.9%) CSC population was observed post third sort (Fig. 5C). These data clearly indicated a possible positive role of RUNX1 in differentiation of CSCs. To identify the underlying molecular regulators, we checked the levels of four well known basic helix loop helix (bHLH) ID proteins (ID1, ID2, ID3 and ID4), which are reported to maintain undifferentiated state of CSCs in many cancers (Roschger and Cabrele, 2017). Among the four ID genes, ID1 and ID3 showed increased expression in SP cells compared to that of NSP and total population (TP) of A2780-DualLR cells, whereas ID2 expression was significantly low and ID4 was strongly expressed across TP, NSP and SP of A2780-DualLR cells (Fig. 6A). Significant upregulation of ID1, ID2 and ID3 gene expression was found in RUNX1 high group of TCGA data set, whereas ID4 showed down regulation (Fig. 6B). Genetic (CBFβ-KD) or pharmacological (Ro5-3335) inhibition of RUNX1 in A2780-DualLR cells showed significant decrease in ID1 and ID3 expression in SP cells compared to control SP cells (Fig. 6C) thus indicating that RUNX1 maintains the undifferentiated state of CSC-like SP cells in chemoresistant EOC cells through upregulation of ID1 and ID3.
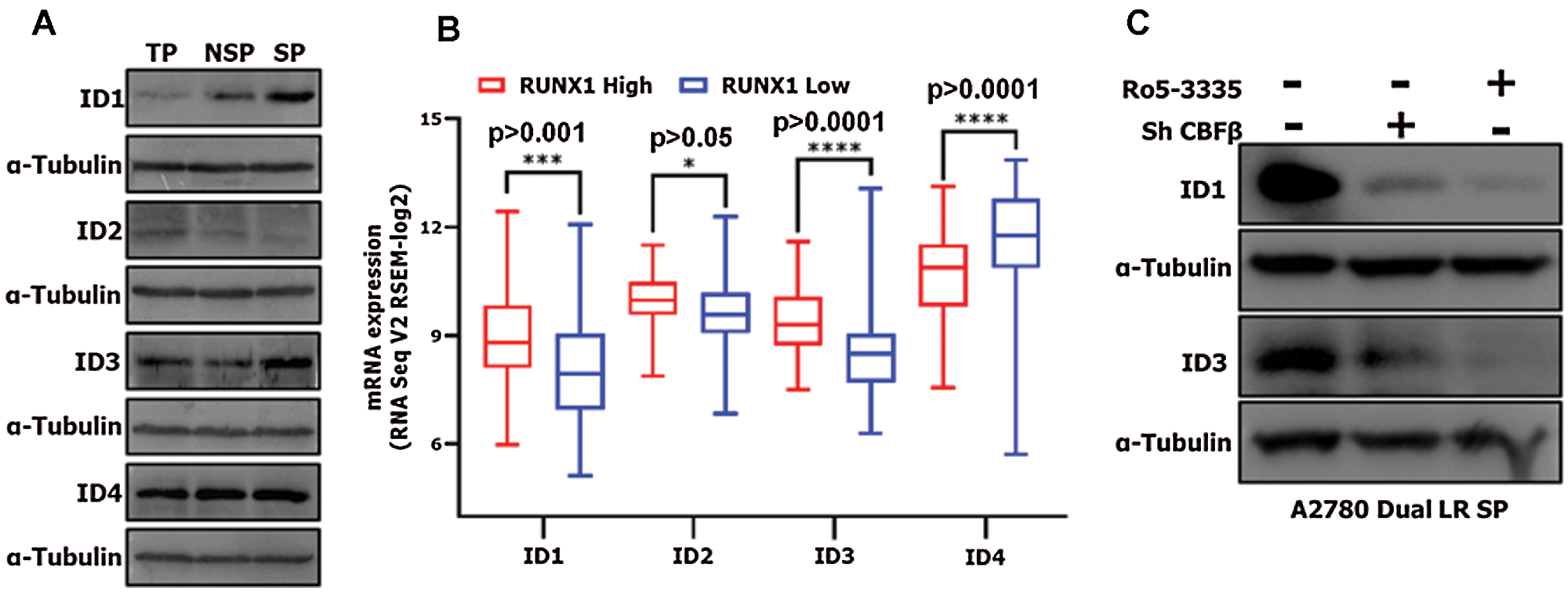
Figure 6: RUNX1 promotes CSC-like SP phenotype through regulation of ID1 and ID3 in A2780-DualLR cells.
(A) Western blot of ID1, ID2, ID3 and ID4 expression in SP, NSP and TP population of A2780-DualLR cells. ID1 and ID3 showed significantly increased expression in SP cells as compared to both TP and NSP. (B) mRNA expression of ID1, ID2, ID3 and ID4 genes in RUNX1 high and RUNX1 low groups for HGS EOC TCGA data. (C) CBFβ-KD and Ro5-3335 treatment showed significant downregulation of ID1 and ID3 in SP cells isolated from A2780-DualLR cells.
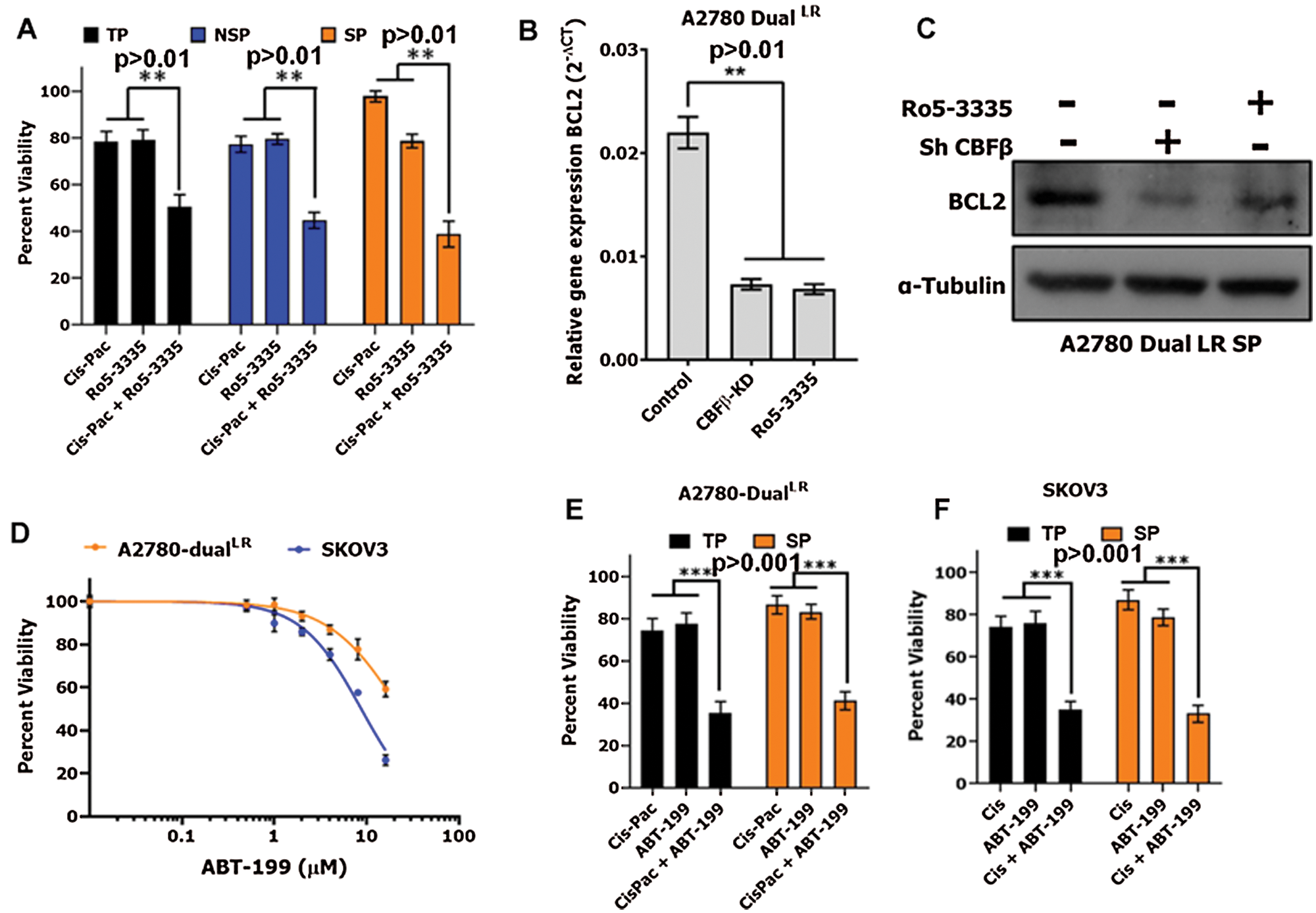
Figure 7: RUNX1 inhibition potentiates cytotoxic effects of Cisplatin-Paclitaxel through downregulation of BCL2.
(A) MTT cell cytotoxicity assay showing increased sensitively of TP, NSP and SP cells towards Cisplatin-Paclitaxel post Ro5-3335 treatment in A2780-DualLR cells. (B–C) RUNX1 inhibition either by CBFβ-KD or Ro5-3335 treatment reduced BCL2 mRNA expression and protein levels in TP and SP population of A2780-DualLR cells, respectively. (D) Dose response inhibition curve for ABT-199 (Venetoclax) for A2780-DualLR and SKOV3 cells. (E–F) MTT cell cytotoxicity assay showing increased sensitivity of TP and SP cells towards Cisplatin-Paclitaxel post ABT-199 treatment in A2780-DualLR cells (E) and Cisplatin post ABT-199 treatment in SKOV3 cells (F).
Loss of BCL2 expression upon RUNX1 inhibition potentiates cytotoxic effects of Cisplatin-Paclitaxel
Cancer stem cells that are intrinsically drug resistant in nature are hypothesized to be a prime cause of disease relapse. Since, we observed that RUNX1 inhibition did not induce cell death in CSC-like SP cells as well as in NSPs, and our previous data along with others showed that RUNX1 inhibition sensitizes the tumor cells including EOC cells to Cisplatin (Dhadve et al., 2020; Xiao et al., 2020), we were curious to assess the similar effect on SP cells. The SP, NSP and total population (TP) of A2780-DualLR cells were treated with either Ro5-3335 (at IC20 concentration) or Cisplatin-Paclitaxel (at IC20 concentration) or in combination (Ro5-3335 and Cisplatin-Paclitaxel). The SP cells isolated from untreated A2780-DualLR cells showed marked increased in chemoresistance as compared to their respective NSP and TP cells (Fig. 7A). RUNX1 perturbation by Ro5-3335 treatment showed enhanced chemosensitization to Cisplatin-Paclitaxel in all TP, SP and NSP (Fig. 7A). Next, we checked the status of anti-apoptotic protein BCL2, a known downstream target of RUNX1 that promotes cell survival (Ge et al., 2014; Xiao et al., 2020). Interestingly RUNX1 inhibition down regulated BCL2 levels in both TP and SP cells of A2780-DualLR cells (Figs. 7B and 7C). Next, we investigated the effect of BCL2 inhibition on Cisplatin-Paclitaxel resistance of SP and TP cells. We used a BCL2 specific BH3-mimetic small molecular inhibitor, ABT-199 (Venetoclax), which binds BH3-domain of BCL2 and displaces the BH3-doamin containing pro-apoptotic proteins, which otherwise are sequestered by BCL2. ABT-199 does not affect the BCL2 protein levels (Juarez-Salcedo et al., 2019). IC20 dose for ABT-199 was determined for both A2780-DualLR cells and Cisplatin resistant SKOV3 cells (Fig. 7D). Combination treatment of IC20 dosage of Cisplatin-Paclitaxel and ABT-199 showed significant chemosensitization of both TP and SP cells as compared to the single agent treatment in A2780-DualLR cells (Fig. 7E). Similar chemosensitization of both TP and SP cells was also observed in SKOV3 cells (Fig. 7F). Altogether, these results suggest impairing RUNX1 activity abrogates expression of anti-apoptotic protein BCL2 and promotes chemosensitization both CSC-like SP cells and total population in chemoresistant EOC cells.
The CSC phenotype has long been studied as a cellular state that replenish the bulk tumor cells by self-renewing and possess high tumor initiating ability along with therapy resistance characteristics. The hope of targeting this small pool of cells for eradicating the tumor thereby preventing relapse of the disease is becoming stronger in recent times with many groups devising new strategies (Yang et al., 2020). Several master transcriptional regulators, such as Myc, β-Catenin, NFκB, STAT, RUNX and NOTCH/CSL have emerged as new therapeutic candidates to target CSCs (Jin et al., 2017). Recent evidences suggest that CSCs are much more dynamic and show extreme cellular plasticity bestowed by aberrant activation of transcriptional modulators of CSC phenotype (Mal et al., 2020). Therefore, it is crucial to identify the underlying molecular regulators that control specific functional properties like self-renewal, differentiation, chemoresistance, etc., in CSCs. Using indigenously developed isogenic cellular models of Cisplatin-Paclitaxel resistance and SKOV3 Cisplatin resistant cells, we herein report that RUNX1, a master transcriptional regulator, control the differentiation of CSCs through ID1 and ID3 proteins. We have recently showed that RUNX1 expression positively increased with increasing resistance in these cellular models (Dhadve et al., 2020). Using a dye exclusion functional assay for serial enrichment of SP cells we showed that, impaired RUNX1 expression promoted differentiation of SP cells into NSP cells and decreased expression of anti-apoptotic protein BCL2 in A2780-DualLR, OAW42-DualLR and SKOV3 cells. Genetic or pharmacological inhibition of RUNX1 down regulated ID1 and ID3 expression in the SP cells collected from A2780-DualLR cells, which have the highest expression of ID1 and ID3 among SP, NSP and total populations and among all the ID proteins. Intriguingly, analysis of TCGA datasets of ovarian cancer indicated a positive relation between ID1 and ID3 proteins with the RUNX1 high cohort of patients. Finally, we showed that ABT-199, a BH3- mimetic BCL2 specific small molecular inhibitor sensitized both SP and TP cells to Cisplatin-Paclitaxel. Our study therefore brings out a novel aspect of RUNX1 regulating the cancer stem cell differentiation and maintenance of inherent resistant characteristics in epithelial ovarian cancer cells which could potentially be translatable in clinic. Such RUNX1 driven regulatory aspects of various CSC phenotypes may exist for other malignancies as well.
Runt related transcription factor 1 (RUNX1), an important molecule in etiology of leukemogenesis has been shown to play important role in tumor initiation and progression of solid tumors such as pancreatic cancer (Cheng et al., 2017; Liu et al., 2020), gastric cancer (Mitsuda et al., 2018), uterine cancer (De Sousa et al., 2014; Planaguma et al., 2011), ovarian cancer (Ge et al., 2014; Keita et al., 2013), triple-negative breast cancer (Ferrari et al., 2014), oral and skin squamous cell carcinoma (Sarper et al., 2018; Scheitz et al., 2012). RUNX1 is known to promote tumor metastasis by activating Wnt/β-catenin pathway in colorectal cancer (Li et al., 2019). DMBA/TPA induced skin tumors in mice showed overexpression of RUNX1 in skin epithelium, papilomas, and squamous cell carcinomas. RUNX1 mediated suppression of negative regulators of STAT3 signaling SOCS3 and SOCS4 was shown to be essential for tumor initiation and maintenance of CSCs (Scheitz et al., 2012). RUNX1 is known to suppress the CSC phenotype in estrogen/progesterone-positive breast carcinoma by direct inhibition of ZEB1 expression (Hong et al., 2018). RUNX1 is also known to be overexpressed in ovarian tumor samples, moreover hypomethylation of RUNX1 gene post chemotherapy has been observed in OC (Keita et al., 2013). Using publicly available TCGA data on HGSOC patients we have shown here that high RUNX1 expression is associated with both decreased median overall survival and disease-free survival. We identified a total 481 genes that were differentially expressed in RUNX1 low vs. RUNX1 high tumor samples. The interaction network analysis of 397 genes in RUNX1 high group using GOBiologicalProcess UniProt, KEGG and the Reactome databases, revealed significant enrichment of pathways associated with metastasis and negative regulation of CSC differentiation (BMP7 and β-catenin signaling), both of which are major hurdles in cancer treatment (Sneddon et al., 2006; Tate et al., 2012). While role of RUNX1 in regulation of metastatic properties of EOC cells is well known, its implication in CSC phenotype in EOC remains elusive. Our analysis thus indicated that RUNX1 might play definitive role in CSC differentiation and other properties.
Using indigenously developed isogenic cellular models of Cisplatin-Paclitaxel, we earlier demonstrated presence of functionally distinctive CSC phenotype during acquirement chemoresistance in EOC cells. The CSCs at the onset of resistance are regulated by increased insulin like growth factor receptor and showed faster tumorigenicity than CSCs isolated from late resistant cells with hyper-active AKT (Singh et al., 2016). We have also reported concomitant increase in expression of transcription factor RUNX1 with increasing resistance in these cellular models (Dhadve et al., 2020). Herein we show that both genetic and pharmacological inhibition of RUNX1 significantly attenuated the CSC-like SP population in chemoresistant EOC cells without differentially affecting the cell survival of SP cells. CSCs are characterized by their ability of self-renewal and differentiation that enable them to recapitulate the tumor bulk. RUNX1 is known to maintain the hair follicle stem cell and skin squamous cell carcinoma CSC pool by activating STAT3 signaling and elevated p21 levels (Hoi et al., 2010). Interestingly, serial enrichment of SP cells over three passages showed no absolute persistence of SP cells in A2780-DualLR cells, rather enriched SP cells showed ability to differentiate into NSP cells. Genetic or pharmacological inhibition of RUNX1 impaired such enrichment of the SP cells, indicating RUNX1 is critical to maintain the undifferentiated state of SP cells. The transcriptional modulators that maintain self-renewal of CSCs, whereas ID proteins, the dominant negative regulators of bHLH transcription factors, inhibit the differentiation pathways. In human colon cancer cells ID1 and ID3 were shown maintain self-renewal of CSCs via p21 increasing resistance against Oxaliplatin (O’brien et al., 2012). Inhibition of ID1 in Glioblastoma multiform was shown to reduced self-renewal of CSCs but promoted enrichment of cells with progenitor-associated markers like Olig2 with high proliferative capacity (Barrett et al., 2012). In TNBC ID1 and ID3 were shown to promote CSC phenotype by negative regulation of Roundabout Axon Guidance Receptor Homolog 1 (Robo1) leading to activation of a Myc transcriptional program (Teo et al., 2020). In this study SP cells isolated from A2780-DualLR cells showed significantly higher levels of ID1 and ID3 as compared to the NSP and TP cells. This observation is supported by the TCGA data analysis where we observed positive correlation between increased transcript levels of ID1 and ID3 with increased RUNX1 expression. Inhibition of RUNX1 significantly downregulated ID1 and ID3 levels in SP cells thus, suggesting that elevated RUNX1 levels promote undifferenced SP phenotype in chemoresistant EOC cells through members of inhibitors of differentiation gene family. While ID2 has also been shown to maintain CSC phenotype in Glioblastoma and Acute myeloid leukemia, ID4 is mainly downregulated in many cancer types through promoter methylation (Roschger and Cabrele, 2017). We have not observed any significant change in ID2 and ID4 in SP cells in our study.
CSCs show intrinsic chemoresistance properties these enable them to evade therapeutic interventions and ultimately leading to relapse of the disease. The spheroid culture derived CSCs from A2780, SKOV3 and PA1 EOC cells showed increased levels of drug transporters ABCB1 and ABCG2 that imparted resistance against Paclitaxel and Doxorubicin in tumor xenograft (Kim et al., 2016). Similarly, CSC-like spheroid cells derived from human EOC cells and primary cultures were shown to be highly tumorigenic, invasive, and resistant against Cisplatin and Paclitaxel (Eyre et al., 2014; Liao et al., 2014). As expected, SP cells isolated from A2780-DualLR cells showed marked increased in platinum-taxol resistance compared to TP and SP. Interestingly, RUNX1 inhibition significantly sensitized the TP, SP and NSP of A2780-DualLR cells to Cisplatin-Paclitaxel. The sensitization of SP cells to Cisplatin-Paclitaxel was 2-fold higher than NSP and TP cells. Mechanistically RUNX1 inhibition downregulated the anti-apoptotic protein BCL2 in both total population and SP cells. Moreover, ABT-199, an BH3-mimetic BCL2 specific small molecular inhibitor potentiated the cytotoxic effects of Cisplatin-Paclitaxel in these chemoresistant EOC cells.
Though RUNX1 has been recognized as one of the key players in tumor initiation and progression of epithelial origin cancers including EOC, its widespread role in regulating hallmarks of cancer and specifically CSC differentiation and chemoresistance has not been elucidated till date. Here we identify RUNX1/ID1/ID3 as a key molecular pathway maintaining the CSC differentiation in platinum-taxol resistant EOC cells, whereas RUNX1 dependent BCL2 imparted Cisplatin-Paclitaxel chemoresistance. Therefore, modulating CSC differentiation or chemoresistance through RUNX1 inhibition might emerge as an attractive strategy to target CSCs in cancer, however the effect of RUNX1 inhibition on CSC self-renewal and quiescence still remains unexplored and thus warrants the further investigation. RUNX1, therefore can arise as a potential therapeutic target for platinum-resistant EOC tumors by reducing CSC population and re-sensitizing the bulk of the tumor to platinum agents.
Acknowledgement: We acknowledge funding support from Advanced Centre for Treatment, Research and Education in Cancer-Tata Memorial Centre and Advanced Centre for Treatment, Research and Education in Cancer-Tata Memorial Centre for fellowship to ACD. Dr. Syed K. Hasan, Advanced Centre for Treatment, Research and Education in Cancer-Tata Memorial Centre, gifted ABT-199.
Availability of Data and Materials: The datasets analysed during in this study are available in the HGS Ovarian Serous Cystadenocarcinoma, Firehose Legacy data set of TCGA (https://www.cbioportal.org/study/summary?id = ov_tcga).
Authors’ Contribution: The authors confirm contribution to the paper as follows: PR conceptualized, designed the experiments, and supervised the project. ACD designed and performed the experiments. ACD and PR analyzed, interpreted the data, wrote, reviewed, and/or revised the manuscript.
Supplementary Material: The supplementary material is available online at DOI https://10.32604/biocell.2022.016346.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Abdullah LN, Chow EK (2013). Mechanisms of chemoresistance in cancer stem cells. Clinical and Translational Medicine 2: 3. [Google Scholar]
Arnold CR, Mangesius J, Skvortsova I, Ganswindt U (2020). The role of cancer stem cells in radiation resistance. Frontiers in Oncology 10: 164. [Google Scholar]
Barrett LE, Granot Z, Coker C, Iavarone A, Hambardzumyan D et al. (2012). Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. Cancer Cell 21: 11–24. [Google Scholar]
Batlle E, Clevers H (2017). Cancer stem cells revisited. Nature Medicine 23: 1124–1134. [Google Scholar]
Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M et al. (2009). ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25: 1091–1093. [Google Scholar]
Boyd AL, Aslostovar L, Reid J, Ye W, Tanasijevic B et al. (2018). Identification of chemotherapy-induced leukemic-regenerating cells reveals a transient vulnerability of human AML recurrence. Cancer Cell 34: 483–498. [Google Scholar]
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO et al. (2012). The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discovery 2: 401–404. [Google Scholar]
Cunningham L, Finckbeiner S, Hyde RK, Southall N, Marugan J et al. (2012). Identification of benzodiazepine Ro5-3335 as an inhibitor of CBF leukemia through quantitative high throughput screen against RUNX1-CBFbeta interaction. Proceedings of the National Academy of Sciences of the United States of America 109: 14592–14597. [Google Scholar]
Challen GA, Little MH (2006). A side order of stem cells: The SP phenotype. Stem Cells 24: 3–12. [Google Scholar]
Chen F, Chen X, Ren Y, Weng G, Keng PC et al. (2019). Radiation-induced glucocorticoid receptor promotes CD44+ prostate cancer stem cell growth through activation of SGK1-Wnt/beta-catenin signaling. Journal of Molecular Medicine 97: 1169–1182. [Google Scholar]
Cheng Y, Yang H, Sun Y, Zhang H, Yu S et al. (2017). RUNX1 promote invasiveness in pancreatic ductal adenocarcinoma through regulating miR-93. Oncotarget 8: 99567–99579. [Google Scholar]
De Sousa VP, Chaves CB, Huguenin JF, Moreira FC, De Reis BS et al. (2014). ERM/ETV5 and RUNX1/AML1 expression in endometrioid adenocarcinomas of endometrium and association with neoplastic progression. Cancer Biology & Therapy 15: 888–894. [Google Scholar]
Dhadve AC, Hari K, Rekhi B, Jolly MK, De A, Ray P (2020). Decoding molecular interplay between RUNX1 and FOXO3a underlying the pulsatile IGF1R expression during acquirement of chemoresistance. Biochimica et Biophysica Acta (BBA)—Molecular Basis of Disease 1866: 165754. [Google Scholar]
Dylla SJ, Beviglia L, Park IK, Chartier C, Raval J et al. (2008). Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS One 3: e2428. [Google Scholar]
Eyre R, Harvey I, Stemke-Hale K, Lennard TW, Tyson-Capper A, Meeson AP (2014). Reversing paclitaxel resistance in ovarian cancer cells via inhibition of the ABCB1 expressing side population. Tumour Biology 35: 9879–9892. [Google Scholar]
Ferrari N, Mohammed ZM, Nixon C, Mason SM, Mallon E et al. (2014). Expression of RUNX1 correlates with poor patient prognosis in triple negative breast cancer. PLoS One 9: e100759. [Google Scholar]
Gaikwad SM, Thakur B, Sakpal A, Singh RK, Ray P (2015). Differential activation of NF-kappaB signaling is associated with platinum and taxane resistance in MyD88 deficient epithelial ovarian cancer cells. International Journal of Biochemistry and Cell Biology 61: 90–102. [Google Scholar]
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling 6: pl1. [Google Scholar]
Garcia-Escarp M, Martinez-Munoz V, Sales-Pardo I, Barquinero J, Domingo JC et al. (2004). Flow cytometry-based approach to ABCG2 function suggests that the transporter differentially handles the influx and efflux of drugs. Cytometry A 62: 129–138. [Google Scholar]
Ge T, Yin M, Yang M, Liu T, Lou G (2014). MicroRNA-302b suppresses human epithelial ovarian cancer cell growth by targeting RUNX1. Cellular Physiology and Biochemistry 34: 2209–2220. [Google Scholar]
Gerber JM, Smith BD, Ngwang B, Zhang H, Vala MS et al. (2012). A clinically relevant population of leukemic CD34(+)CD38(-) cells in acute myeloid leukemia. Blood 119: 3571–3577. [Google Scholar]
Hamilton G, Olszewski U (2013). Chemotherapy-induced enrichment of cancer stem cells in lung cancer. Journal of Bioanalysis & Biomedicine 213: S9. [Google Scholar]
Hoi CS, Lee SE, Lu SY, Mcdermitt DJ, Osorio KM et al. (2010). Runx1 directly promotes proliferation of hair follicle stem cells and epithelial tumor formation in mouse skin. Molecular and Cellular Biology 30: 2518–2536. [Google Scholar]
Hong D, Fritz AJ, Finstad KH, Fitzgerald MP, Viens AL et al. (2018). RUNX1 suppresses breast cancer stemness and tumor growth, bioRxiv 315093. [Google Scholar]
Jin X, Jin X, Kim H (2017). Cancer stem cells and differentiation therapy. Tumour Biology 39: 1010428317729933. [Google Scholar]
Juarez-Salcedo LM, Desai V, Dalia S (2019). Venetoclax: Evidence to date and clinical potential. Drugs in Context 8: 212574. [Google Scholar]
Keita M, Bachvarova M, Morin C, Plante M, Gregoire J et al. (2013). The RUNX1 transcription factor is expressed in serous epithelial ovarian carcinoma and contributes to cell proliferation, migration and invasion. Cell Cycle 12: 972–986. [Google Scholar]
Kim DK, Seo EJ, Choi EJ, Lee SI, Kwon YW et al. (2016). Crucial role of HMGA1 in the self-renewal and drug resistance of ovarian cancer stem cells. Experimental & Molecular Medicine 48: e255. [Google Scholar]
Kreso A, Dick JE (2014). Evolution of the cancer stem cell model. Cell Stem Cell 14: 275–291. [Google Scholar]
Lagadec C, Vlashi E, Della Donna L, Dekmezian C, Pajonk F (2012). Radiation-induced reprogramming of breast cancer cells. Stem Cells 30: 833–844. [Google Scholar]
Lagadec C, Vlashi E, Della Donna L, Meng Y, Dekmezian C et al. (2010). Survival and self-renewing capacity of breast cancer initiating cells during fractionated radiation treatment. Breast Cancer Research 12: R13. [Google Scholar]
Li Q, Lai Q, He C, Fang Y, Yan Q et al. (2019). RUNX1 promotes tumour metastasis by activating the Wnt/beta-catenin signalling pathway and EMT in colorectal cancer. Journal of Experimental and Clinical Cancer Research 38: 334. [Google Scholar]
Liao J, Qian F, Tchabo N, Mhawech-Fauceglia P, Beck A et al. (2014). Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism. PLoS One 9: e84941. [Google Scholar]
Liu S, Xie F, Gan L, Peng T, Xu X et al. (2020). Integration of transcriptome and cistrome analysis identifies RUNX1-target genes involved in pancreatic cancer proliferation. Genomics 112: 5343–5355. [Google Scholar]
Lu H, Chen I, Shimoda LA, Park Y, Zhang C et al. (2017). Chemotherapy-induced Ca2+ release stimulates breast cancer stem cell enrichment. Cell Reports 18: 1946–1957. [Google Scholar]
Mal A, Bukhari AB, Singh RK, Kapoor A, Barai A et al. (2020). EpCAM-mediated cellular plasticity promotes radiation resistance and metastasis in breast cancer. Frontiers in Cell and Developmental Biology 8: 597673. [Google Scholar]
Mitsuda Y, Morita K, Kashiwazaki G, Taniguchi J, Bando T et al. (2018). RUNX1 positively regulates the ErbB2/HER2 signaling pathway through modulating SOS1 expression in gastric cancer cells. Scientific Reports 8: 6423. [Google Scholar]
O’brien CA, Kreso A, Ryan P, Hermans KG, Gibson L et al. (2012). ID1 and ID3 regulate the self-renewal capacity of human colon cancer-initiating cells through p21. Cancer Cell 21: 777–792. [Google Scholar]
Planaguma J, Liljestrom M, Alameda F, Butzow R, Virtanen I et al. (2011). Matrix metalloproteinase-2 and matrix metalloproteinase-9 codistribute with transcription factors RUNX1/AML1 and ETV5/ERM at the invasive front of endometrial and ovarian carcinoma. Human Pathology 42: 57–67. [Google Scholar]
Ramakrishnan V, Xu B, Akers J, Nguyen T, Ma J et al. (2020). Radiation-induced extracellular vesicle (EV) release of miR-603 promotes IGF1-mediated stem cell state in glioblastomas. EBioMedicine 55: 102736. [Google Scholar]
Roschger C, Cabrele C (2017). The Id-protein family in developmental and cancer-associated pathways. Cell Communication and Signaling 15: 7. [Google Scholar]
Rycaj K, Tang DG (2014). Cancer stem cells and radioresistance. International Journal of Radiation Biology 90: 615–621. [Google Scholar]
Sarper SE, Inubushi T, Kurosaka H, Ono Minagi H, Kuremoto KI et al. (2018). Runx1-Stat3 signaling regulates the epithelial stem cells in continuously growing incisors. Scientific Reports 8: 10906. [Google Scholar]
Scheitz CJ, Lee TS, Mcdermitt DJ, Tumbar T (2012). Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO Journal 31: 4124–4139. [Google Scholar]
Singh RK, Dhadve A, Sakpal A, De A, Ray P (2016). An active IGF-1R-AKT signaling imparts functional heterogeneity in ovarian CSC population. Scientific Reports 6: 36612. [Google Scholar]
Singh RK, Gaikwad SM, Jinager A, Chaudhury S, Maheshwari A, Ray P (2014). IGF-1R inhibition potentiates cytotoxic effects of chemotherapeutic agents in early stages of chemoresistant ovarian cancer cells. Cancer Letters 354: 254–262. [Google Scholar]
Sneddon JB, Zhen HH, Montgomery K, Van De Rijn M, Tward AD et al. (2006). Bone morphogenetic protein antagonist gremlin 1 is widely expressed by cancer-associated stromal cells and can promote tumor cell proliferation. Proceedings of the National Academy of Sciences of the United States of America 103: 14842–14847. [Google Scholar]
Tan S, Chen JS, Sun LJ, Yao HR (2009). Selective enrichment of hepatocellular cancer stem cells by chemotherapy. Journal of International Medical Research 37: 1046–1056. [Google Scholar]
Tate CM, Pallini R, Ricci-Vitiani L, Dowless M, Shiyanova T et al. (2012). A BMP7 variant inhibits the tumorigenic potential of glioblastoma stem-like cells. Cell Death & Differentiation 19: 1644–1654. [Google Scholar]
Telford WG, Bradford J, Godfrey W, Robey RW, Bates SE (2007). Side population analysis using a violet-excited cell-permeable DNA binding dye. Stem Cells 25: 1029–1036. [Google Scholar]
Teo WS, Holliday H, Karthikeyan N, Cazet AS, Roden DL et al. (2020). ID proteins promote a cancer stem cell phenotype in mouse models of triple negative breast cancer via negative regulation of Robo1. Frontiers in Cell and Developmental Biology 8: 552. [Google Scholar]
Thakur B, Ray P (2017). Cisplatin triggers cancer stem cell enrichment in platinum-resistant cells through NF-kappaB-TNFalpha-PIK3CA loop. Journal of Experimental and Clinical Cancer Research 36: 164. [Google Scholar]
Van Rhenen A, Feller N, Kelder A, Westra AH, Rombouts E et al. (2005). High stem cell frequency in acute myeloid leukemia at diagnosis predicts high minimal residual disease and poor survival. Clinical Cancer Research 11: 6520–6527. [Google Scholar]
Vermeulen L, de Sousa E, Melo F, Richel DJ, Medema JP (2012). The developing cancer stem-cell model: Clinical challenges and opportunities. Lancet Oncology 13: e83–e89. [Google Scholar]
Wang Y, Zong X, Mitra S, Mitra AK, Matei D, Nephew KP (2018). IL-6 mediates platinum-induced enrichment of ovarian cancer stem cells. JCI Insight 3: e122360. [Google Scholar]
Xiao L, Peng Z, Zhu A, Xue R, Lu R et al. (2020). Inhibition of RUNX1 promotes cisplatin-induced apoptosis in ovarian cancer cells. Biochemical Pharmacology 180: 114116. [Google Scholar]
Yang L, Shi P, Zhao G, Xu J, Peng W et al. (2020). Targeting cancer stem cell pathways for cancer therapy. Signal Transduction and Target Therapy 5: 8. [Google Scholar]
Zhao J (2016). Cancer stem cells and chemoresistance: The smartest survives the raid. Pharmacology & Therapeutics 160: 145–158. [Google Scholar]
Zhao Y, He M, Cui L, Gao M, Zhang M et al. (2020). Chemotherapy exacerbates ovarian cancer cell migration and cancer stem cell-like characteristics through GLI1. British Journal of Cancer 122: 1638–1648. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |