DOI:10.32604/biocell.2022.016953

| BIOCELL DOI:10.32604/biocell.2022.016953 |  |
| Review |
miR-153 as biomarker for cancer—functional role as tumor suppressor
1Faculty of Applied Sciences and Biotechnology, Shoolini University, Solan, 173229, India
2Department of Biotechnology, MMEC, Maharishi Markandeshwar (Deemed to be University), Mullana, 133207, India
3Maharishi Markandeshwar University, Kumarhatti, Solan, 173229, India
4Department of Chemistry, Physical Sciences, Mizoram University, Aizawl, 796004, India
5Department of Kinesiology, California State University, Sacramento, CA 95819, USA
6Laboratory on the Mechanism and Regulation of Protein Synthesis, Eunice K. Shriver, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892, USA
*Address correspondence to: Reena V. Saini, reenavohra10@mmumullana.org; Jagpreet S. Nanda, jagpreet.nanda@nih.gov
Received: 13 April 2021; Accepted: 09 June 2021
Abstract: MicroRNA-153 (miR-153), belongs to a class of small non-coding RNA. It is a critical regulator of gene expression at the post-transcriptional level which interacts with the functional mRNA at 3’UTR region and suppresses the expression of the mRNA. More recently, it has become apparent that changes in the miR-153 expression lead to invasion, metastasis, angiogenesis and various types of tumor progression. This review summarizes the connection between dysregulation of miR-153 and various types of cancer progression. miR-153 regulates various signaling pathways to inhibit the proliferation and induce apoptosis in the cancer cell and also show synergistic activity with anticancer drugs. In addition to this, the oncogenic behavior of miR-153 and their use as a potential biomarker in cancer was also reviewed.
Keywords: Biomarker; Cancer; miR-153; Oncogenic; Tumor suppressor
| Abbreviations | |
| ABCE1: | ATP-binding cassette E1 |
| AKT: | protein kinase B |
| ALK-5: | activin receptor-like kinase 5 |
| ANG1: | angiopoietin 1 |
| As2O3: | arsenic trioxide |
| ATF3: | activating transcription factor 3 |
| BC: | breast cancer |
| Bcl-2: | B cell lymphoma 2 |
| CAM: | chorioallantoic membrane |
| CAR T: | chimeric antigen receptor T cells |
| CC: | colon cancer |
| CML: | chronic myeloid leukemia |
| CXCR4: | C-X-C chemokine receptor type 4 |
| DCVs: | dense-core vesicles |
| EGFR: | epidermal growth factor receptor |
| EMT: | epithelial-mesenchymal transition |
| EOC: | epithelial ovarian cancer |
| ERG: | ETS-related gene |
| ESCC: | esophageal squamous cell carcinoma |
| FOXO1: | forkhead box protein O1 |
| FOXO3: | forkhead box protein O3 |
| GBM: | glioblastoma multiform |
| GBMSc: | glioblastoma multiforme stem cells |
| GFAP: | glial fibrillary acidic protein |
| hCMECs: | human cerebral microvascular endothelial cells |
| HIF1: | hypoxia-inducible factor 1 |
| HUVECs: | human umbilical vein endothelial cells |
| IA: | islet antigen |
| IDO1: | indoleamine 2,3-dioxygenase 1 |
| IFNγ-: | interferon γ |
| IGF: | insulin like growth factor |
| IGFBP-3: | insulin-like growth factor-binding protein 3 |
| IRE1: | inositol-requiring enzyme 1 |
| KLF5: | Kruppel-like factor 5 |
| LC: | lung cancer |
| lncRNA: | long non-coding RNA |
| LSCC: | laryngeal squamous cell carcinoma |
| MALT1: | mucosa-associated lymphoid tissue lymphoma translocation translocation protein 1 |
| MAP2: | mitogen-activated protein 2 |
| Mcl-1: | myeloid cell leukemia 1 |
| MDR-1: | multiple drug resistant |
| miR-153: | microRNA 153 |
| miRNAs: | microRNA |
| mLST8: | mTOR associated protein, LST8 homolog |
| MMP-9: | metalloprotease-9 |
| mSIN1: | mammalian stress-activated protein kinase-interacting protein |
| mToRC2: | mechanistic target of rapamycin complex 2 |
| NHOst: | normal osteoblast cells |
| NPC: | nesopharyngeal cancer |
| NSCLC: | non-small cell lung cancinoma |
| nt: | nucleotides |
| p-AKT: | phosphorylated AKT |
| PC: | pancreatic cancer |
| PDAC: | pancreatic ductal adenocarcinoma |
| PTPRN: | protein tyrosine phosphatase receptor |
| Rictor: | rapamycin-insensitive companion of mammalian target of ramamycin |
| RUNX2: | runt-related transcription factor 2 |
| SNHG15: | small nucleolar RNA host gene 15 |
| TGFBR2: | TGF-β receptor 2 |
| TGF-β: | transforming growth factor-β |
| TGFβRI: | TGF-β type I receptor |
| TGFβRII: | TGF-β type II receptor |
| TNBC: | triple negative breast cancer |
| TNM: | tumor node metastasis |
| TRBS: | transactivating response RNA-binding protein |
| VEGFA: | vascular endothelial growth factor |
| Wnt: | wingless-related Integration sites |
| XBP1: | X-box binding protein 1 |
| XIST: | X inactive-specific transcript |
| ZEB2: | zinc finger E-box binding homeobox 2 |
MicroRNAs (miRNAs) represent highly conserved small non-coding RNAs of ~18–25 nucleotides (nt) in length. RNA polymerase II transcribes a hairpin structure known as pri-micro RNAs which is further processed by Drosha (RNAse III) into pre-micro RNA (70–100 nt). The pre-micro RNA is exported by Exportin-5 into the cytoplasm where it is further processed by RNAse III Dicer. RNAse III Dicer acts along with TRBS (transactivating response RNA-Binding Protein), and generates a double-stranded RNA (dsRNA) of 18–25 nt in length known as miRNA (Fig. 1) (Iorio and Croce, 2012). This miRNA binds to the 3’ untranslated regions (3’ UTR) of messenger RNA (mRNA). Its binding to the 3’ UTR either degrade the mRNA or exhibit translational repression and, thus, regulates the gene expression at post-transcriptional stage (Bartel, 2004; Engels and Hutvagner, 2006).
Many of these miRNAs are integral part of diverse cellular physiologies, like metabolism, apoptosis, cellular proliferation and differentiation, invasion, migration, and drug resistance (Kloosterman and Plasterk, 2006). It is predicted that nearly 60% gene expression in human is directly controlled by miRNAs (Kloosterman and Plasterk, 2006). It is known that abnormal expression (down-regulation or over-expression) of miRNAs can lead to cell progression and angiogenesis which further leads to tumor formation or cancer (Lu et al., 2005). It was observed that various miRNAs can act either as potent oncogenes or tumor suppressor genes, and thus their dysregulation could trigger cancer (Lu et al., 2005; Esquela-Kerscher and Slack, 2006).
miRNA can be defined by two factors; (1) share similar promoter with encoded target genes, (2) spliced out of the transcript and processed into mature miRNA. Some miRNA encoded in a palindromic region of the genomic intron and known as non-intronic or intergenic miRNA because the promoter is situated in the antisense direction to the gene, such as let-7c. However, intronic miRNA share similar promoter with gene transcript such as miR-208 (Gao et al., 2012).
In this review, we are discussing about miR-153 and its functional role in tumor apoptosis. miR-153, a conserved intragenic miRNA, is located on chromosome 2 in the intron 19 of Islet antigen (IA)-2 and on chromosome 7 of IA-2β in the intronic region between exon 19 and 20 (Mandemakers et al., 2013). Both IA (2 and 2β) are the integral transmembrane proteins of synaptic vesicles and dense-core vesicles (DCVs) (Xu et al., 2015), which are expressed in neuroendocrine cells throughout the body (adrenals, pancreatic islets, gastrointestinal and in the brain) (Solimena et al., 1996; Nishimura et al., 2009; Gomi et al., 2013). In these cells, miR-153 functions as both tumor suppressor and tumor mediator and, this function varies from cell to cell.
miR-153 and Its Dysregulation in Cancer
Dysregulation of various miRNAs has been found in different categories of cancer. Such dysregulations can be caused by transcriptional dysregulation, mutation, deletion, amplification, and epigenetic changes (Chen et al., 2012; Croce, 2009). Several reports have suggested that miR-153 is dysregulated in several human cancers and mostly these dysregulations are the major factors responsible for tumorigenesis.
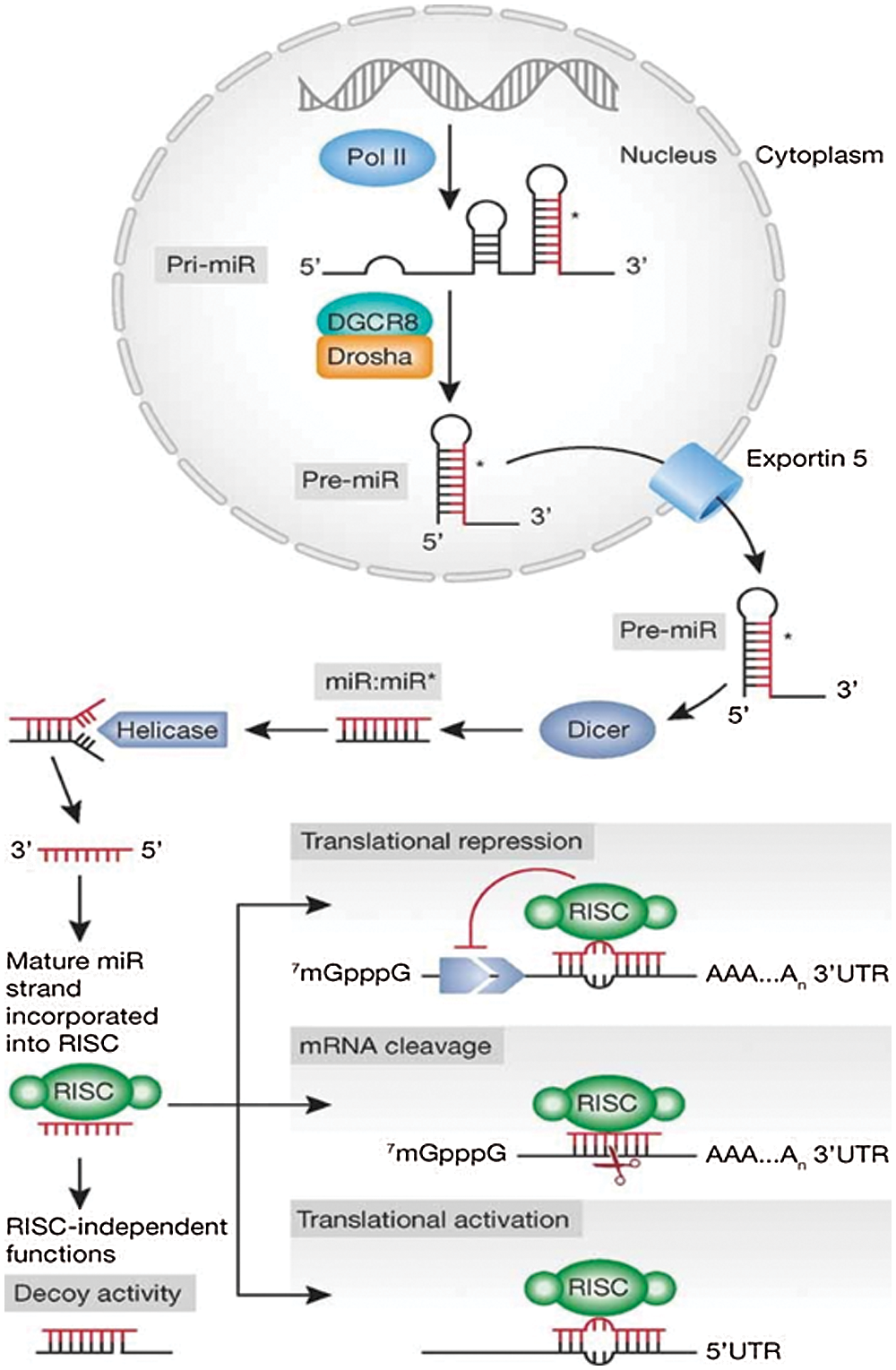
Figure 1: Schematic representation of miRNA synthesis and mechanism of action. Reprinted with permission from (EMBO Molecular Medicine 2012, 4, 143-159) Copyright (2012) EMBO Press.
Using in silico and methylation-sensitive PCR amplification, Bao et al. (2012) showed three methylation sites in cytosine in two genes transcribing miR-153. One cytosine is located at 434 bp upstream in the miR-153-1 gene and other two are located at 2721 bp and 4028 bp upstream in miR-153-2 gene of mature miR-153 gene promoter, and can dysregulate miR-153 by repressing its expression in HEK-293cells. The levels of methylation were found to be inversely proportional to the expression of miR-153, and thus regulate the expression of the target 3’ UTR of holocarboxylase synthetase and histone biotinylation in HEK-293 cells. Methylation generally depends upon folate and on methyl donors. So, it might be possible that epigenetic synergies between biotin and folate mediated the gene silencing, and it can be speculated that epigenetic regulation is involved in the dysregulation of miR-153 in various tissue (Bao et al., 2012).
miR-153 is found to be dysregulated in many types of cancer. In breast cancer (BC) MDA-MB-231 cell line, its expression was found to be decreased by ~30 fold as compared to the non-malignant mammary epithelial cell (Zuo et al., 2019; Wang et al., 2019a). Overexpression and downregulation of miR-153 can lead to tumor formation depending upon the presence of miR-153 in various tissues and their action on various mRNAs. On self-regulation, miR-153 can control the tumor formation, angiogenesis and can leads to apoptosis of cancer cells, like in MDA-MB-231 cells. The upregulation of miR-153 expression can decrease the invasion behavior of BC cells and can also inhibit the tumor growth (Zuo et al., 2019; Wang et al., 2019a). Overexpression of miR-153 can inhibit epithelial to mesenchymal transition (EMT) in BC cells via the downregulation of the EMT-related transcription factors, like zinc finger E-box binding homeobox 2 (ZEB2), Notch1, TWIST, ETS-related gene (ERG), and SNA1 (Li et al., 2015).
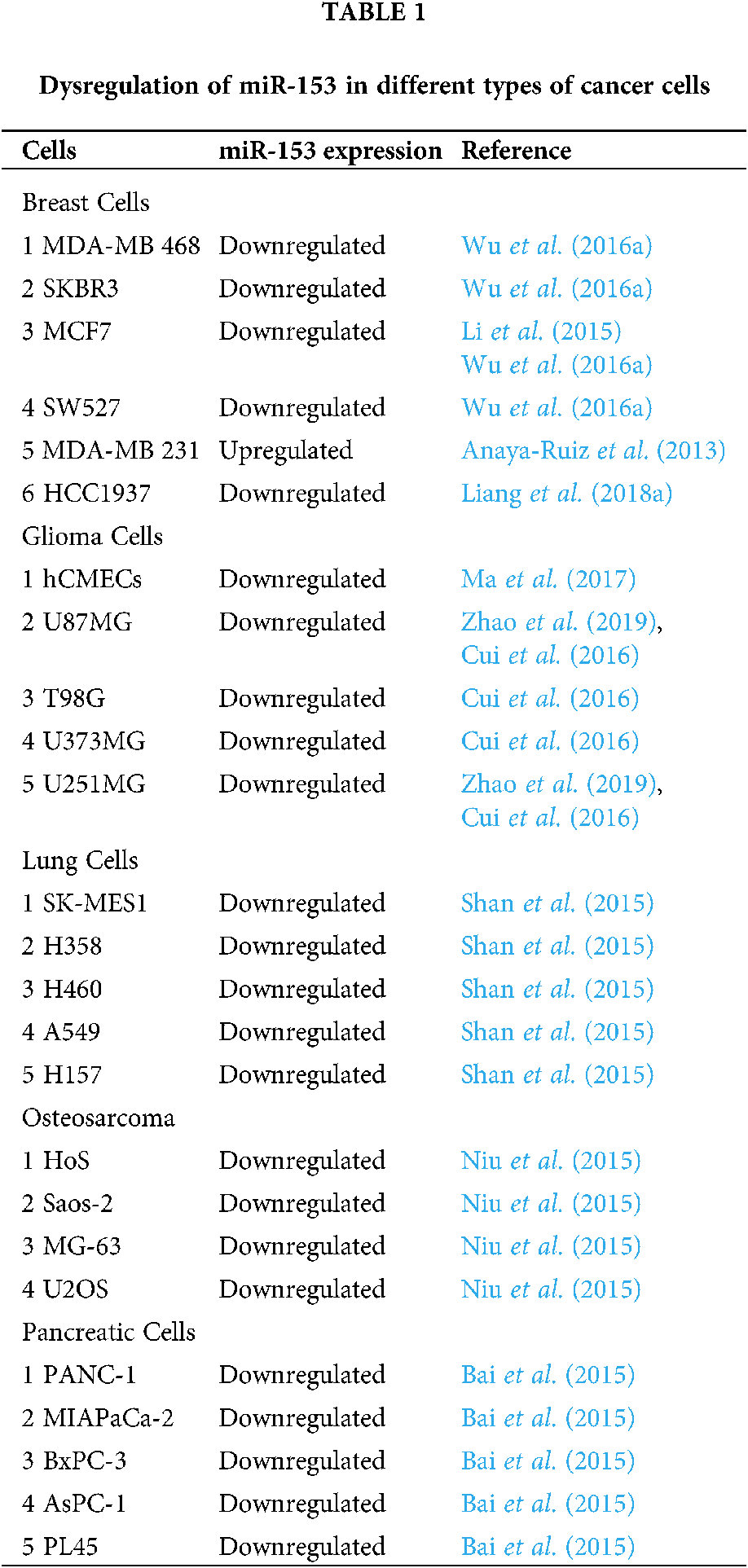
Similarly, downregulation of miR-153 is observed in endometrial cancer (Myatt et al., 2010), in epithelial cancer cells that underwent EMT (Xu et al., 2013), in glioblastoma multiform (GBM) relative to normal brain tissues (Zhao et al., 2013), and in glioma tissues and cell lines (U87MG, T98G, U373MG, and U251MG cell lines) (Cui et al., 2016; Zhao et al., 2019). As compared to the normal human cerebral microvascular endothelial cells (hCMECs) in primary astrocyte cells, miR-153 was found to be dysregulated in glioma induced hCMECs (Ma et al., 2017). The miR-153 is also dysregulated in K562 drug-tolerant cells as well as in CD+34 cells (Liu et al., 2012), in lung cancer (LC) tissue and its cell lines (A549, H460, and H157) (Yuan et al., 2015; Shan et al., 2015), in nasopharyngeal cancer (13-9B cell line) (Guo et al., 2019), in laryngeal squamous cell carcinoma (Liu et al., 2016a) and, in osteosarcoma tissue and other cell lines (U2OS, Saos-2, HOS, and MG-63) in comparison to normal osteoblast cells (NHOst) (Niu et al., 2015). Similarly, its downregulation is observed in ovarian tumors (Kim et al., 2010; Zhou et al., 2015; Bai et al., 2015) and can be used as a diagnostic biomarker for distinguishing grade 1–3 ovarian tumors by International Federation of Gynecology and Obstetrics- (Kim et al., 2010). The expression of miR-153 is also seen to be altered in the pancreatic ductal adenocarcinoma (PADC) cell line, whereas, the normal expression was found in the pancreatic duct epithelial cell line. Dysregulated miR-153 was also found in patients with advanced TNM tumor stage (Tumor Node Metastasis is a staging system), tumor recurrence, lymph node metastasis and tumor differentiation (Bai et al., 2015). The altered expression of miR-153 in various cells is shown in Tab. 1.
Role of miR-153 in Tumor Suppression
BC patients with low miR-153 expression have poorer prognosis than the patients with higher expression of miR-153. Overexpressed miR-153 can reduce the invasion, migration, proliferation, tube formation ability, and EMT in BC cells (BT-549, SK-BR-3 cells, MDA-MB-231 cells (Zuo et al., 2019; Wang et al., 2019a). It was shown that miR-153 decreased the invasive behavior of MCF-7 cells and suppressed the EMT by downregulating Vimentin and upregulating cadherin, and thus inhibited the tumor growth. To determine the function of miR-153 as a tumor suppressor, Li et al. (2015) have increased the expression of miR-153 in MCF-7 and MDA-MB-231 cells, and found suppression of their invasive behavior and lower proliferation rate. High levels of miR-153 downmodulated the EMT-associated transcription factors, like ZEB2, Notch1, TWIST, ERG, and SNA1 in BC cells (Li et al., 2015). In one of the studies, it was shown that miR-153 targeted the Runt-related transcription factor 2 (RUNX2) gene which is responsible for BC, bone metastasis, and osteogenesis (Vishal et al., 2017). Upregulation of RUNX2 led to lymph node metastasis and reduced the suppression mechanism of miR-153, thereby further led to increased invasion, migration, proliferation, and EMT in BC cells viz BT-549 and SK-BR-3 cells (Zuo et al., 2019).
The transforming growth factor-β (TGF-β) signalling pathway induces EMT via TGF-β receptor 2 (TGFBR2), and thus increases the malignancy of BC cells. The increased expression of TGFBR2 is found to be associated with the overall low survival rate of BC patients (Wei et al., 2015). It has been shown through luciferase reporter assay that 3’UTR of TGFBR2 mRNA has two target sites for miR-153 which knockdown the expression of TGFBR2 (Fig. 2) (Wang et al., 2019a).
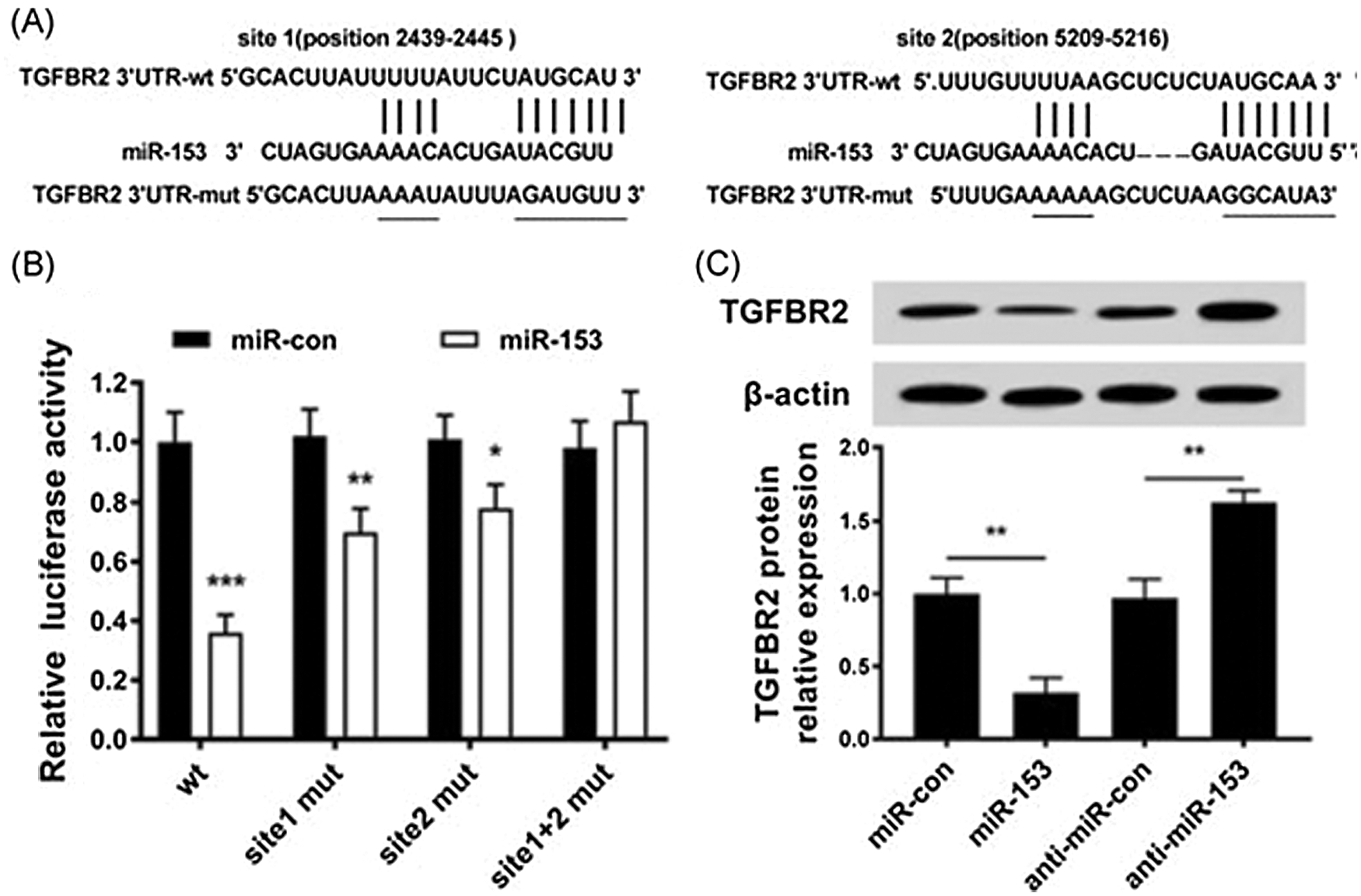
Figure 2: Expression of TGFBR2 regulated by miR-153. (A). miR-153 and TGFBR2 3’-UTR targeting binding sites; (B). Luciferase activity in cells. (C). Level of TGFBR2 protein in transfected cells with miR-153 mimics or miR-153 inhibitors. Reprinted (Journal of Cellular Biochemistry) with permission from John Wiley and Sons (License number 5020561012043).
miR-153 also suppresses the angiogenesis by targeting the vascular endothelial growth factor/hypoxia-inducible factor 1α (VEGFA/HIF1α) axis. Liang et al. (2018b) showed that hypoxia triggered the ER stress which further followed the inositol-requiring enzyme 1[a]/X-box binding protein spliced (IRE1α/XBP1) pathway, and increased the expression of miR-153 and downregulated VEGFA/HIF1α at the post-transcriptional level (Liang et al., 2018b). miR-153 gene is located in the gene encoding for PTPRN (Protein Tyrosine Phosphatase Receptor Type N or Islet Antigen 2) and PTPRN2 (or Islet Antigen 2β) and its promoter region has XBP1-binding site. They demonstrated using chromatin immunoprecipitation assay that under hypoxia, XBP1 and PTPRN interacted with each other and activated PTPRN promoter which induced the miR-153 expression (Fig. 3) (Liang et al., 2018b).
miR-153 has been shown to have a direct impact on angiogenesis by regulating the levels of Angiopoietin 1 (ANG1) which is a glycoprotein involved in the development of new blood vessels and sprouting of endothelial cells. Clinical data shows around four-fold high expression level of ANG1 in BC tissue versus the matched adjacent normal tissue. High levels of ANG1 in tumor is due to the elimination of the repressor complex BRCA1/CtIP/ZBRK1 from the promoter of ANG1 (Furuta et al., 2006). ANG1 mRNA expression triggers migration of HUVECs (human umbilical vein endothelial cells) and the tube formation ability as shown in BC cell lines (MCF7, MDA-MB-231 and HCC1937). The bioinformatic analysis showed that miR-153 can bind at three sites in the 3’UTR of ANG1 mRNA and can downregulate its expression, thereby suppressing the HUVECs migration and tube formation ability (Liang et al., 2018a).
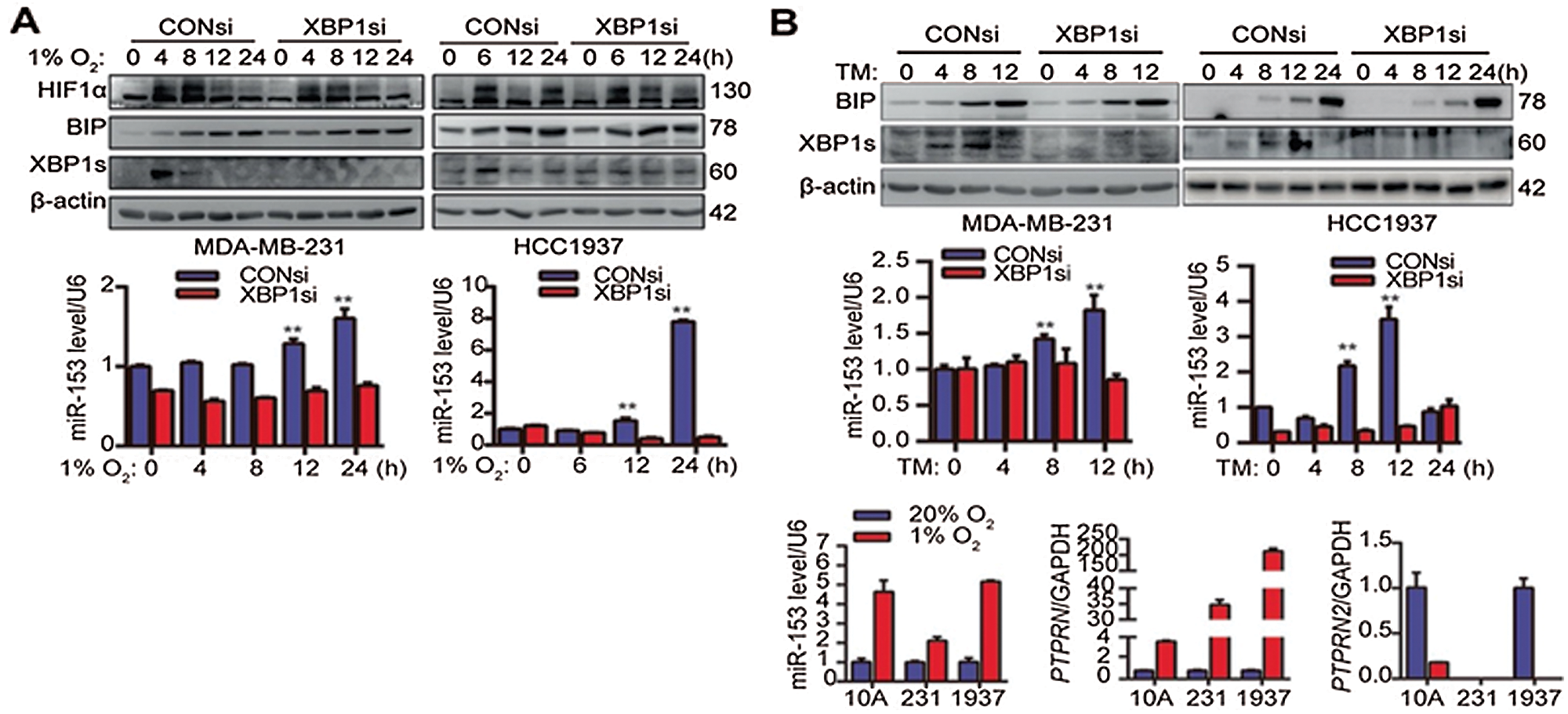
Figure 3: X-box binding protein 1 induces miR-153 expression by binding to the PTPRN promoter. (A) XBP1 knockdown suppresses the miR-153 upregulation induced by hypoxia. (B) XBP1 knockdown suppress the miR-153 upregulation induced by TM. (C) Hypoxia induces expression of miR-153 and PTPRN, but not PTPRN2, at the mRNA level. Reprinted (Oncogene 2018, 37, 1961-1975) with permission from Nature Research, Copyright (2018) (License number CC BY-NC-SA 4.0).
miR-153 could also bind to the mRNA of transcription factor kruppel-like factor 5 (KLF5). KLF5 serves as an oncogene in different types of human cancer (Ghaleb et al., 2005; Dong and Chen, 2009) by inhibiting apoptosis and promoting cell survival via multiple mechanisms (Li et al., 2014a; Li et al., 2014b; Nakaya et al., 2014). miR-153 can target the 3’UTR region of KLF5; therefore, it was speculated that miR-153 could suppress the expression of KLF5 (Liu et al., 2016b). Very importantly, it was shown that in triple-negative BC (TNBC) which accounts for ~15% of all BC, mifepristone (a synthetic progesterone receptor) inhibited the cell proliferation and survival through miR-153 via suppressing KLF5 expression (Liu et al., 2016b). Derivatives of mifepristone, FZU-00,003 also induced the miR-153 expression at a lower dosage than the mifepristone and inhibited the KLF5 expression in TNBC (Liu et al., 2020). Similarly, the relationship between miR-153 and KLF5 was seen in laryngeal squamous cell carcinoma (LSCC). The upregulation of miR-153 in LSCC promotes the inhibition of cell proliferation and progression. Therefore, it was speculated that inhibition of cell proliferation is due to the suppression of KLF5 (Liu et al., 2016b).
miR-153 also inhibits the TNBC invasion, migration and proliferation by regulating ZEB2/EMT axis. ZEB2 is a protein which functions to transform tumor cells into a mesenchymal phenotype that results in EMT. Shi et al. (2019) confirmed by Luciferase reporter assay that miR-153 could bind at 3’UTR region of ZEB2 (Fig. 4). miR-153 targets SNA1 and ZEB2 at 3’UTR sites and downregulate their expression in epithelial cancer (HNH and HN12 cells). SNA1 and ZEB2 are transcriptional repressors of E-cadherin and its downregulation leads to EMT (Xu et al., 2013). miR-153 ectopic expression also prevents the EMT induced by TGF-β and reverts epithelial-like phenotype from mesenchymal-like cells (Xu et al., 2013). In this way, miR-153 could be used to regulate the TNBC.
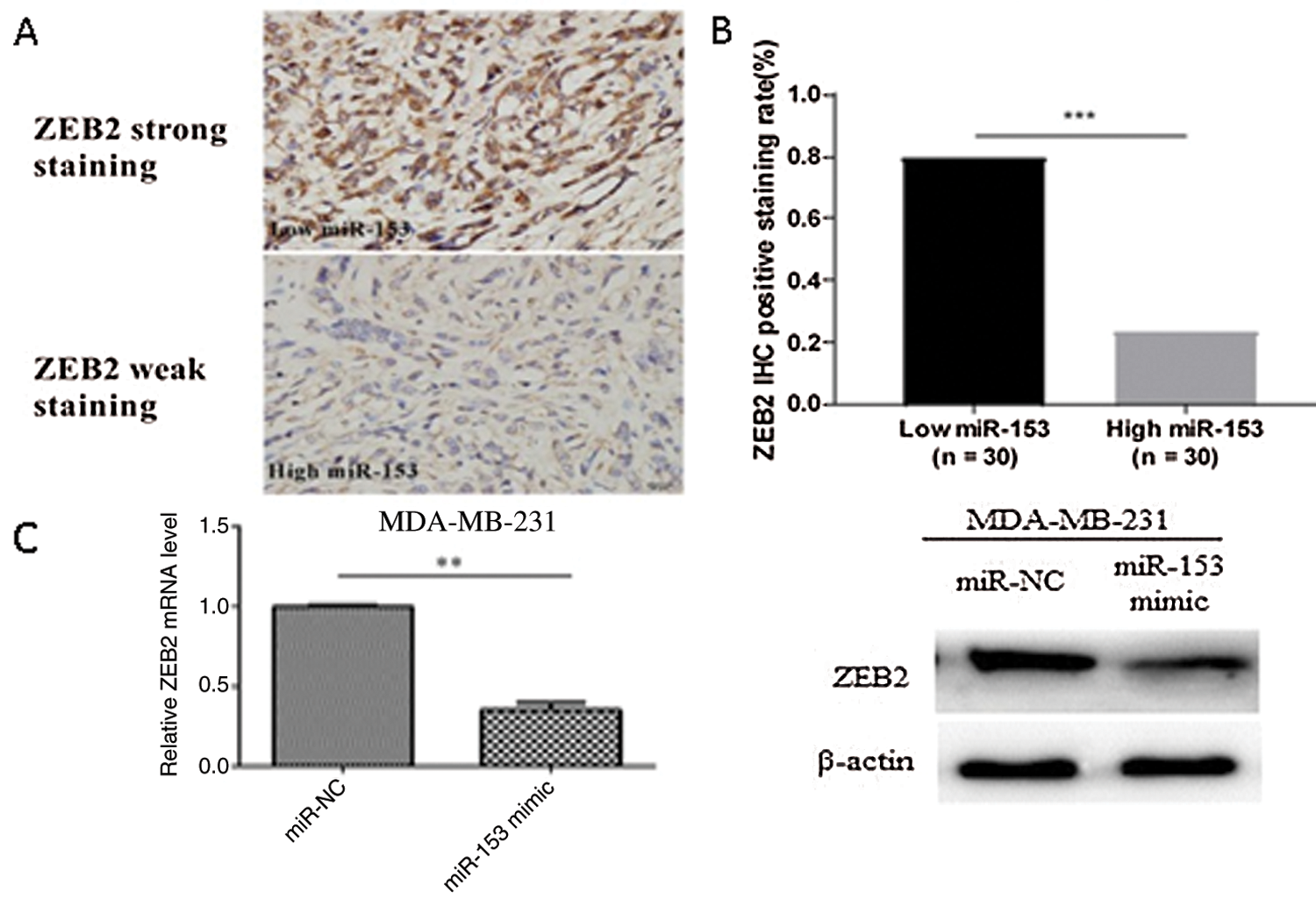
Figure 4: ZEB as a direct target of miR-153. (A) ZEB2 expression level with low and high miR-153 expression. (B) Correlation between ZEB2 and miR-153 in TNBC. (E) Expression of ZEB2 mRNA and protein in MDA-MB-231 cells. Reprinted (OncoTargets and Therapy 2019, 12, p. 9611) with permission from Dovepress, Copyright (2019) (License number CC BY-NC 3.0).
miR-153 has been shown to reduce cell proliferation and induce the cell death via reducing the expression of myeloid cell leukemia 1 (Mcl-1) and B cell lymphoma 2 (Bcl-2) in GBM by targeting their mRNAs (Xu et al., 2010). Glioblastoma Multiforme stem cells (GBMSc) transfected with miR-153 showed the lower expression of CD133 and nestin but a higher expression of glial fibrillary acidic protein (GFAP) and mitogen-activated protein 2 (MAP2), indicating that miR-153 can enhance GBMSc differentiation and tends to direct the cells towards neuron fates and astrocyte. It was speculated by Zhao et al. (2013) that miR-153 suppresses the GBMSCs activity by suppressing Dvl-3 in canonical Wnt (Wingless-related Integration sites) signaling which affects the cell proliferation and stemness properties of GBM-SCs (Zhao et al., 2013).
In glioma cells, miR-153 downregulates the rapamycin-insensitive companion of mammalian target of rapamycin (Rictor) protein expression which is responsible for mToRC2 (mechanistic Target of Rapamycin Complex 2) activation (p-Akt ser 473) that promotes glioma cell proliferation and migration (Sarbassov et al., 2004; Jacinto et al., 2006). Generally, in glioma tissues the level of miR-153 is dramatically downregulated than the normal brain tissues and, the mTOR activity and expression of Rictor protein are upregulated. Therefore, Rictor is the direct target of miR-153, and overexpression of this miRNA exerts a pro-apoptotic activity in glioma cells (U87MG and U373MG) (Cui et al., 2016). As already discussed, SNAI1 promotes EMT, represses the E-cadherin expression and endows normal cells with the ability to metastasize (Zhao et al., 2019). SNA1 is also overexpressed in glioma cells and promotes cell invasion ability. The number of invasive cells in the glioma cell line (U87MG and U251) is decreased with the increase in the level of miR-153 (Zhao et al., 2019).
miR-153 is shown to be negatively regulated by the small nucleolar RNA host gene 15 (SNHG15) which is a long non-coding RNA (Ma et al., 2017). The expression of SNHG15 is high in endothelial cells which are induced by glioma. SNHG15 increases the levels of VEGFA and Cdc42, whereas down-regulates the expression of miR-153 and promotes the cancer cell proliferation, tube formation, and migration. miR-153 binds with SNHG15 at its 3’UTR site and hinder the Cdc42 and VEGFA expressions. The knockdown of SNG15 hinders the proliferation, tube formation and migration of endothelial cell. All these suggest that for glioma anti-angiogenesis therapy, miR-153 could be a potential target (Ma et al., 2017).
In colon cancer (CC), high level of indoleamine 2,3-dioxygenase 1 (IDO1) is inversely correlated with CC patient survival and can be upregulated by interferon-γ (IFN-γ). IDO1 degrades the tryptophan in the kynurenine pathway leading to the formation of tumor microenvironment. Huang et al. (2018) has downregulated the expression of IDO1 by targeting its 3’UTR site with miR-153 that further enhances the chimeric antigen receptor T cells (CAR T) based immunotherapy against CC. Therefore, both miR-153 and CAR T cell shows a synergistic effect against CC (Fig. 5) (Huang et al., 2018).
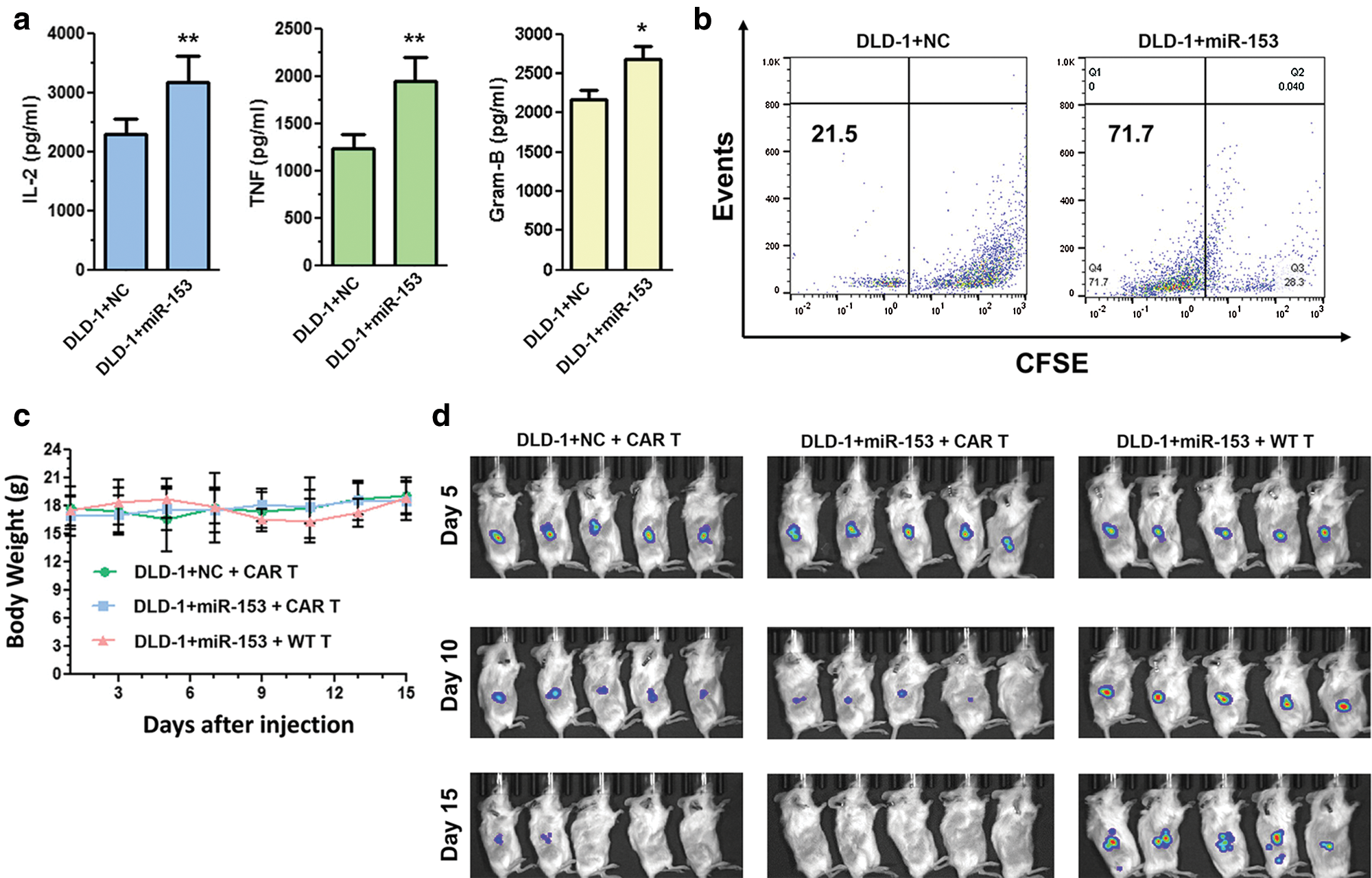
Figure 5: Overexpressed miR-153 enhances the CAR T cells growth and inhibit tumor growth in colon cancer cells. (a) Cytokine production of CAR T cells co-cultured with DLD-1 cells with or without miR-153. (b) CFSE based proliferation of CAR T cells cultured with DLD-1 + NC or DLD-1 + miR-153. (c) Body weight of NSG mice injected with tumor cells and T cells. (d) Xenograft tumor growth in NSG mice inoculated with DLD-1 + NC or DLD-1 + miR-153 cells and treated with CAR T or WT T cells. Reprinted (Journal of Hematology and Oncology 2018, 11, 1-12) with permission from Springer link, Copyright (2018) (License number CC BY 4.0).
In LC it was shown that the forced expression of miR-153 inhibited the cell proliferation and migration by targeting AKT (Protein Kinase B, PKB), leading to the apoptosis of LC cells. AKT is a cytosolic signal transduction protein, and its activation leads to cell proliferation, tissue invasion, survival and angiogenesis by regulating various pathways and promote carcinogenesis (Hutchinson et al., 2001; Altomare and Testa, 2005). Overexpression of phosphorylated AKT (p-AKT) in clinical non-small cell LC (NSCLC) specimens relates with poor prognosis of NSCLS patients (Tang et al., 2006). The expression level of miR-153 is significantly lower in the LC tissues with high AKT expression is high. So miR-153 based therapy can be applied to treat the LC patients with decreased endogenous miR-153 and increased AKT level (Yuan et al., 2015).
The expression of miR-153 is also inversely correlated with the level of ADAM19, and the overexpression of ADAM19 is correlated with NSCLC lymph node status and advanced tumor stage. High level of ADAM19 inhibits the function of miR-153 and it is also a target for miR-153 in NSCLC. miR-153 downregulates the ADAM19 expression, and inhibits migration and invasion in A549 and H157 cell lines (Shan et al., 2015). Jagged1 is a ligand for Notch signaling pathway, shows higher expression in NSCLC and responsible for the progression of cancer, poor prognosis and stemness properties of LC cells. By targeting Jagged1, miR-153 successfully inhibits the stem-like properties and suppresses LC growth (Fig. 6) (Zhao et al., 2020).
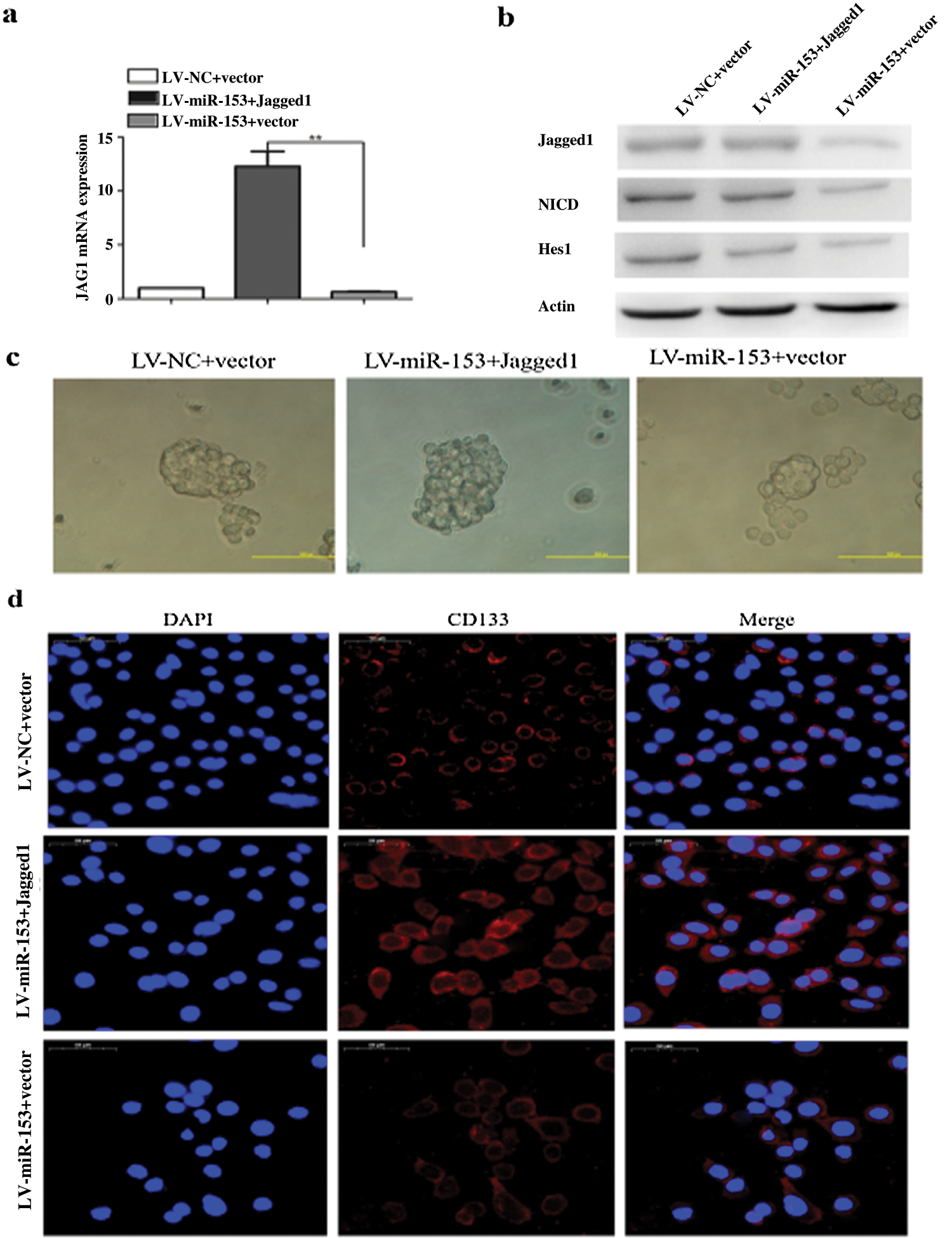
Figure 6: miR-153 suppressed Jagged1/Notch pathway and reduced lung carcinoma cell stemness. (a) Jagged1 mRNA expression. (b) Protein expression of Jagged1, NCID, and Notch target gene Hes1. (c) Tumor sphere formation capacity of SPC-A-1 cells was analyzed after Jagged1 overexpression. (d) Stem cell marker CD133 expression. Reprinted (Stem Cell Research & Therapy 2020, 11, 1-12) with permission from BMC (License number CC BY 4.0).
One of the key mediators required for the epithelial to mesenchymal transition is the SNAI1 protein. The levels of miR-153 and SNAI1 protein expressions are also inversely correlated in esophageal squamous cell carcinoma (ESCC). In ESCC, miR-153 targets the 3’UTR of SNAI1, and downregulates its expression that further inhibits the ESCC cell’s migration and invasion ability (Zuo et al., 2016). Zuo et al. (2016) have performed xenograft assay on 6-week-old female nude mice and injected 106 OE-21 cells overexpressing miR-153 mimic and C-X-C chemokine receptor type 4 (CXCR4). Mice were observed for lung metastasis by bioluminescence imaging using an IVIS Imaging System for 56 days. They observed metastasis in OE21/CXCR4 treated animals; whereas, animals treated with OE21/miR-153 shows significant attenuation of lung colonization and greatly reduce lung metastasis (Zuo et al., 2016).
miR-153 could also modulate the invasion and cell proliferation via regulating TGF-β2 expression. The 3’UTR region of TGF-β2 has sites for miR-153 binding, and it has been shown that overexpression of miR-153 is associated with a reduction in mRNA and protein levels of TGF-β2 (Guo et al., 2019). TGF-β2 induces EMT in nasopharyngeal cancer (NPC) through the smad2 pathway. Upregulation of miR-153 significantly increases the caspase-3 and 9 activities, which promotes the Bax/Bcl2 protein expression and suppresses the TGF-β2 expression by targeting the TGF-β2/smad2 pathway, thus induces apoptosis in NPC (13-9B cells) (Guo et al., 2019). As an example, in osteosarcoma tissue and its cell lines, miR-153 suppressed the TGF-β2 expression, and inhibited the cell proliferation and invasion in MG-63 cells (Niu et al., 2015). Bone releases the TGF-β growth factor, which is known to induce cell proliferation in various cancers (Perera et al., 2010; Schedlich et al., 2013). It promotes cancer metastasis by regulating the composition of proteolysis, inflammatory responses, and extracellular matrix (Kato et al., 2009; Taylor et al., 2013; Lyu et al., 2014). TGF-β was also shown to stimulate MG-63 cell growth via ALK-5, the type-1 receptor of TGF-β, with a concomitant induction of growth factor mainly the platelet-derived growth factor AA (Matsuyama et al., 2003). The induction in growth of ALK-5 (Activin receptor-like kinase 5) expressing MG-63 by TGF-β also involved the unexpected induction of c-Myc. In another study, TGF-β also led to the activation of the Raf/MAPK pathway that is closely linked with cell invasion and cell growth (Janda et al., 2002). Insulin-like growth factor-binding protein 3 (IGFBP-3) expression was also observed to be induced by TGF-β2, which further induced the action of insulin like growth factor (IGF) via high-affinity binding (Schedlich et al., 2013). Due to the involvement of TGF-β signaling in EMT and cell proliferation, it is considered as a therapeutic target in osteosarcoma as well (Hou et al., 2014; Sung et al., 2014). TGF can also induce IGFBP-3, EGFR (Epidermal growth factor receptor) and P-SMAD3 expressions, but the overexpression of miR-153 represses the expression of all of them (Niu et al., 2015). According to some previous studies, overexpression of long non-coding RNA (lncRNA) X inactive-specific transcript (XIST) is linked with metastasis, tumorigenesis and shows resistance towards chemotherapy and radiotherapy (Yang et al., 2018; Li et al., 2019; Gao et al., 2019). It was also found by Wen et al. (2020) that under oxidative stress expression of XIST was increased whereas expression of miR-153 was decreased in osteosarcoma tumor tissues. They also speculated that miR-153 is a potential target of XIST and recommended to target XIST-miR-153 axis for osteosarcoma therapy.
Many oncogenes (HMGA2, UNC45A, ACTN4, and PIK3R1) involved in ovarian cancer are the predicted targets for miR-153, thereby suggesting its tumor suppressor role. miR-153 is differentially expressed in various histological subtypes of ovarian cancer that include endometrioid, serous clear cell, and mucinous carcinoma. Overexpressed miR-153 in mucinous carcinoma also represents a marker to discriminate mucinous carcinoma from endometroid, serous, and clear cell carcinoma (Kim et al., 2010). In ovarian cancer, upregulated miR-153 inhibits the cell proliferation and invasion of the OVCAR3 cell line. For this, miR-153 binds to the 3’UTR region of SET7/ZEB2, and promotes its mRNA degradation. By downregulating SET7 and ZEB2, miR-153 inhibits the proliferation in EOC (Epithelial ovarian cancer) cells and suppresses EMT (Zaman et al., 2012). miR-153 also targets the AKT1 in EOC to inhibit proliferation, migration, and colony formation since PI3K/AKT pathway inhibition and EOC tumor growth is negatively correlated (Li et al., 2017).
Human pancreatic ductal adenocarcinoma (PDAC) is usually diagnosed at a late stage and have early metastasis. Generally, the expression of miR-153 is remarkably lower in PDAC cell lines in comparison to normal pancreatic duct epithelial cell lines. The transcription of miR-153 is low in the patients with tumor differentiation, tumor recurrence, lymph node metastasis, and advanced TNM tumor stage. On upregulation, the level of miR-153 prevents the PDAC cell invasion and migration by binding 3’UTR of SNA1 (Bai et al., 2015), which is considered as a crucial pro-metastatic agent in PDAC (Chen et al., 2014).
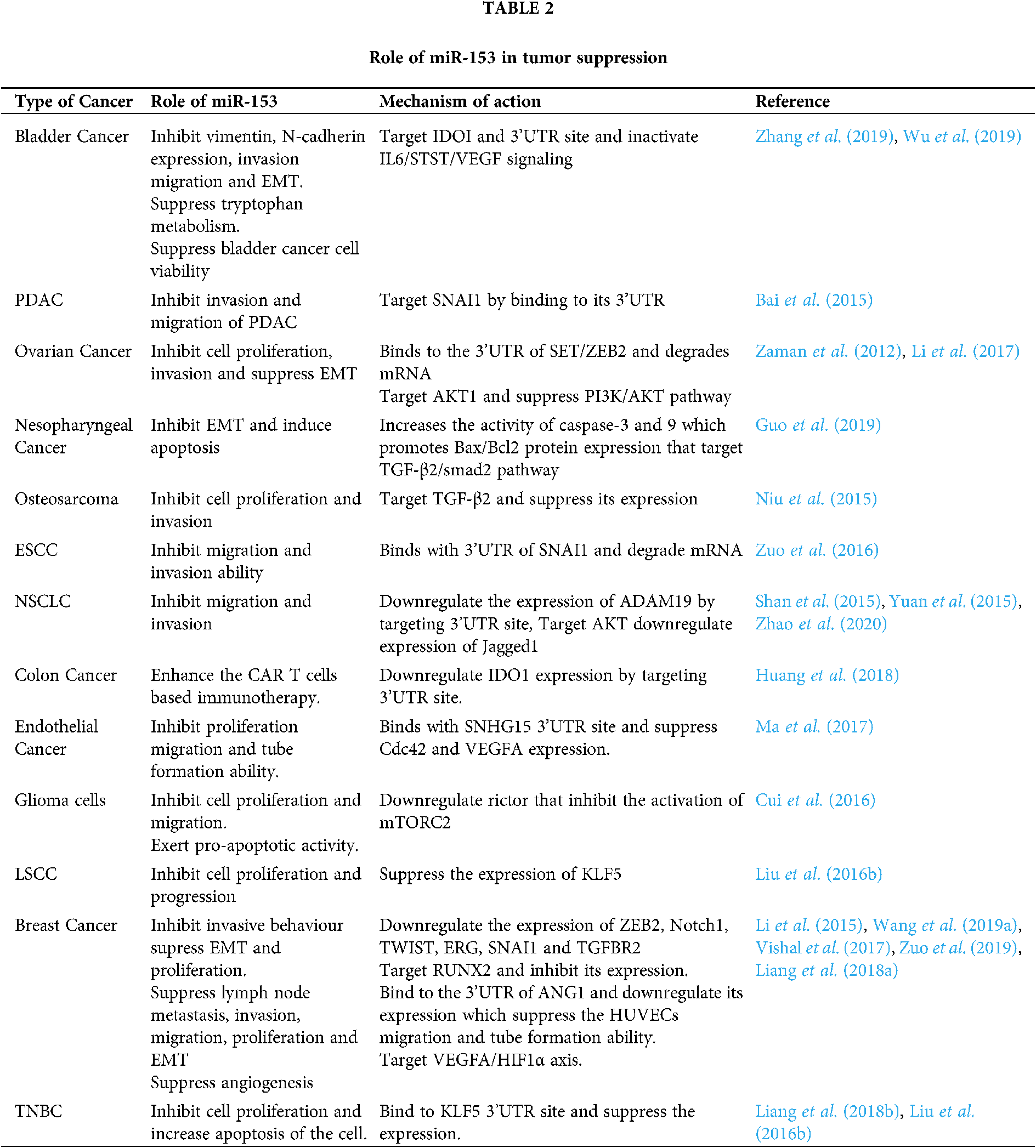
In the case of bladder cancer, miR-153 overexpression inhibits the vimentin and N-cadherin expression. Overexpressed miR-153 also suppresses the tryptophan metabolism by targeting IDOI to inactivate IL6/STAT3/VEGF signaling. IDOI plays an important role in tryptophan metabolism and has the complementary target sequence for miR-153 at its 3’UTR. Thus miR-153 inhibits the tumor cell growth, invasion, migration, and EMT in bladder cancer. It also suppresses the angiogenesis of chorioallantoic membrane (CAM) and human umbilical vein endothelial cells (HUVEC) (Zhang et al., 2019). miR-153 elevates the expression of Mcl-1, Bcl-2, and forkhead box protein O1 (FOXO1) protein, and suppresses the bladder cancer cell viability (Wu et al., 2019). Different roles of miR-153 are summarized in Tab. 2.
Role of miR-153 in Cancer Drug Resistance
In chronic myeloid leukemia (CML) therapy, arsenic trioxide (As2O3) has been considered very effective in combination with other drugs, but resistance towards the As2O3 was also developed by CML cells. It is known that CML CD34+ cells are insensitive towards the various chemotherapeutic drugs, and drug therapy might enforce cells with some molecular characters similar to CD34+ cells (Huang et al., 2018). Generally, the expression of miR-153 is low in K562 drug-tolerant cells as well as in CD34+ cells. So, forced expression of miR-153 with the treatment of As2O3 synergistically induced apoptosis in K562 drug-resistant cells, suggesting that miR-153 inhibition plays an important role in drug resistance. So, rescuing the miR-153 expression in drug-resistant cells may give benefit to eradicate CML (Liu et al., 2012).
miR-153 also inhibits the cell proliferation and invasion in NSCLC by knocking down the ATP- binding cassette E1 (ABCE1), and suppresses the multiple drug resistant genes (MDR-1) expression. In LC, gefitinib-resistant cancer is one of the major problems and the cells with gefitinib resistance have a high expression of ABCE1. So, by knocking down ABCE1 expression, miR-153 can regulate gefitinib resistance (Wang et al., 2019b).
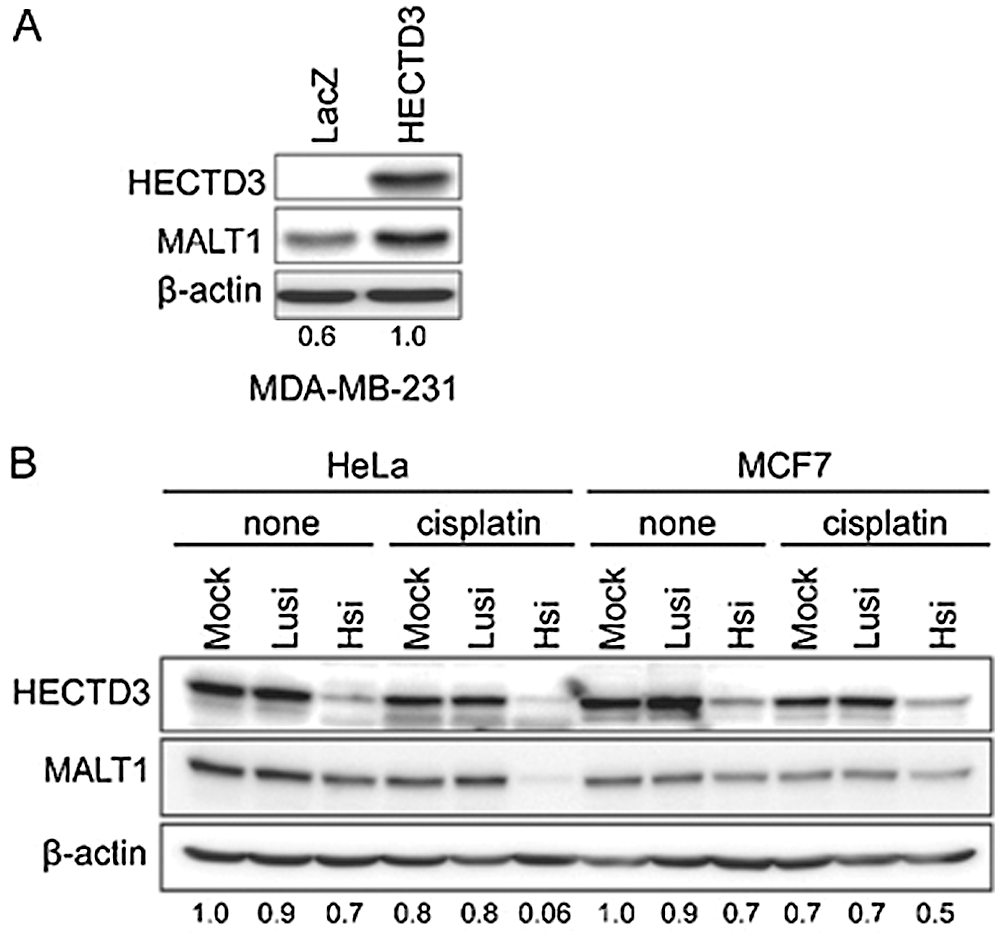
Figure 7: HECTD3 increases the MALT1 protein stability. (A) Overexpression of HECTD3 increased the endogenous MALT1 protein level in MDA-MB-231. (B) Knockdown of HECTD3 in both HeLa and MCF7 decreased the endogenous protein levels of MALT1, especially when the cells were treated with cisplatin. Reprinted (Neoplasia 2013, 15, pp.39-IN15) with permission from Elsevier (License number CC BY-NC-ND 3.0).
In Pancreatic cancer (PC) chemoresistance (gemcitabine-resistance) is developed with the overexpression of SNAIl. SNAIl is a pro-metastatic factor that induces EMT and thus participates in chemoresistance. miR-153 reverses gemcitabine resistance in PC cells by targeting SNAIl and increased the sensitivity for gemcitabine (Liu et al., 2017). PC also shows resistance towards radiotherapy however the mechanism of resistance is still unknown. A recent study reveals that JAG1 is responsible to enhance radiotherapy resistance in PC. Overexpressed JAG1 was directly targeted by miR-153 and further mediates the resistance towards the radiotherapy (Zhao et al., 2021).
HECTD3, a E3 ubiquitin ligase promotes BC cell survival by hampering apoptosis through caspase 8 (Li et al., 2013a), and shields the tumor cells from cisplatin mediated apoptosis through MALT1 (Mucosa-associated lymphoid tissue lymphoma translocation protein 1) (Fig. 7) (Li et al., 2013b). Generally, in the absence of HECTD3, the stability of MALT1 is significantly reduced by cisplatin. However, the presence of HECTD3 promotes MALT1 non-degradative polyubiquitination, prevents the proteasomal degradation of MALT1, and escalates the protein stability, thereby conferring cisplatin resistance (Li et al., 2013b). On the other hand, miR-153 promotes the cisplatin mediated apoptosis by suppressing HECTD3. The combined therapy of chemotherapeutic drug and miR-153 induces cytotoxicity in BC cells (Wu et al., 2016a).
Although miR-153 has been identified as a cancer inhibitor in various types of cancer, it can also serve the role in tumor progression. Zhang et al. (2013) showed that upregulation of miR-153 in colorectal cancer cells (SW480 and SW620) led to the enhanced invasiveness and resistance towards the chemotherapy. Upregulation of miR-153 enhances the matrix metalloprotease-9 (MMP-9) enzyme activity that further enhances the invasiveness of colorectal cancer cells. Zhang et al. (2013) have also speculated that miR-153 enhances the resistance to oxaliplatin by altering the effect of forkhead box protein O3 (FOXO3a). FOXO3a is responsible for upregulating pro-apoptotic genes, like Bin and PUMA, and initiation of apoptotic programs via downregulation of the antiapoptotic genes as well (You and Mak, 2005). The increased expression of miR-153 downregulates the FOXO3a transcription and further reduces their protein level as miR-153 directly interacts with the 3’UTR of FOXO3a (Zhang et al., 2013; Moradi Marjaneh et al., 2019).
Regulation of different pathways by miR-153
Various signaling pathways play an important role in tumorigenesis. miRNA-153 has emerged as an EMT controller in cancer development through transcription network regulation. It downregulates the expression of transcription factors associated with EMT, like TWIST, Nitch1, ERG, SNAI1, and ZEB2 (Li et al., 2015). The phenomenon of EMT is associated with loss of cell-cell contact and interference with an intracellular tight junction that facilitates the change in the phenotype from epithelial to mesenchymal (Wu et al., 2016b). EMT can be stimulated by various growth factor signaling, tumor-stromal cell interaction, and cytokines. Various transcription factors, such as ZEB1, ZEB2, Twist, Snail, and Slug induces EMT through various cell signaling pathways.
TGF-β, Notch, and Wnt/β-catenin pathways are generally known to induce EMT in cancer. In TGF-β pathway, TGFR2 gets activated and phosphorylates TGFR1, thereby promoting the activation of the Smad pathway. Further, TGF-β turns on type II receptor (TGFβRII) which phosphorylates type I receptor (TGFβRI), and activates Smad3. Upon phosphorylation, Smad3 engages Smad4 and translocates into the nucleus where they activate EMT-inducers and suppress ID2 by interacting with activating transcription factor 3 (ATF3) and leads to high Twist expression and EMT (Wu et al., 2016b).
The binding of the Wnt ligand with a transmembrane receptor of Frizzled family initiates the Wnt signaling pathway, and promotes the destruction of E-cadherin, cell-cell adhesion, and APC complex which lead to β-catenin stabilization. In the nucleus, β-catenin form the β-catenin/TCF/LEF transcriptional complex, and direct the Wnt signaling-targeted genes for transactivation and subsequently promote EMT (Wu et al., 2016b). Notch signaling also initiates EMTvia interaction with Jagged1. Following activation, Notch represses E-cadherin by Slug, and induces EMT (Zhao et al., 2020). miR-153 can suppress EMT by modifying these pathways via targeting key mediators.
VEGFA and ANG1, both are proangiogenic factors. ANG1 interacts with tyrosine-protein kinase receptor Tie2 and initiate phosphorylation of Tie2 that leads to the sprouting of endothelial cells, blood vessels maturation and angiogenesis (Hawighorst et al., 2002). miR-153 inhibits the expression and secretion of ANG1, and regulates the ANG1/Tie2 signaling pathway (Liang et al., 2018a).
PI3K pathway is a key regulator for cell survival and proliferation. PTEN is known as a tumor suppressor gene. It negatively regulates the PI3K signaling pathway and inhibits its activation. But mutation or inactivation of PTEN drives the activation of PI3K that further activates the PKB/Akt and initiates tumorigenesis (Cully et al., 2006). Akt is also phosphorylated by mTORC2 at Ser473 that further initiates cell proliferation, metabolism, survival, and cytoskeletal organization (Li et al., 2016). The main components of mTORC2 are mTOR, mSIN1 (mammalian stress-activated protein kinase-interacting protein), Rictor, mLST8 (mTOR associated protein, LST8 homolog), and Protor1. Rictor plays a vital role in mTORC2 activation and is associated with mLST8 and Protor1 to construct the mTORC2 complex. Depletion of Rictor by miR-153 regulates this pathway, and results in apoptosis of cancer cells (Masri et al., 2007; Cui et al., 2016). Further, it also suppresses the AKT/p-AKT and ceases the PI3K/Akt pathway (Yuan et al., 2015). miR-153 regulates PI3K/Akt signaling pathway by altering the expression of the RUNX2 transcription factor, Rictor and Akt.
miR-153 as a Biomarker in Cancer
Over the past few years, biomarkers based targeted therapeutic technique has gained great attention and focuses on the understanding that how an individual’s genetic environment helps to determine the best treatment method for cancer. Biomarkers are biological molecules such as peptides, proteins, hormones, lipids, nucleic acid (DNA, RNA, siRNA, miRNA, etc.) (Gutschner et al., 2018).
miR-153 is involved in various biological processes related to cancer. As we have already discussed that miR-153 can act as a tumor suppressor in various types of cancer, but some studies also show that miR-153 also acts as an oncogene and helps in tumor progression. Anaya-Ruiz et al. (2013) have reported high expression of miR-153 in MDA-MB-231 cells which results in high proliferation of these cells and development of BC. However, silencing of miR-153 affects cell viability and leads to apoptosis of MDA-MB-231 cells (Anaya-Ruiz et al., 2013). Along with this, miR-153 also show high expression in TNBC and can be used as a biomarker (M’hamed et al., 2017). Similarly, Wu et al. (2013) also reported high expression of miR-153 in prostate cancer where it inactivates the function of the PTEN tumor suppressor gene and promotes cell proliferation by activating PI3K/Akt pathway (Wu et al., 2013; Bi et al., 2019). Additionally, miR-153 is also highly expressed in mucinous carcinoma (a subtype of ovarian carcinoma), whereas the other subtypes of ovarian carcinoma, such as endometrioid, serous, and clear cell carcinoma show less expression of miR-153. Therefore, it can be used as a marker to differentiate the histological subtypes of ovarian cancer (Kim et al., 2010).
Bai et al. (2015) has reported lower expression of miR-153 in PDAC patients with advance TNM tumor stage lymph node metastasis, poor tumor differentiation and tumor recurrence, thereby suggesting that lower expression of miR-153 plays a crucial role in PDAC development and progression. They have also suggested that miR-153 is an independent prognostic biomarker to predict the 3-year survival of PDAC patients (Bai et al., 2015). A recent study by Baber et al. (2021) shows that miR-153 expression was low in the peripheral blood of oral squamous cell carcinoma patients than normal individuals and speculated that expression of miR-153 in the serum can function as a diagnostic biomarker (Baber et al., 2021). The meta-analysis data also shows the correlation between low mir-153 expression and poor overall survival of cancer patient and suggest miR-153 as an effective biomarker for various type of cancer (Huang et al., 2020).
The function of miR-153 in various cancer cell lines is still unexplored, but the present studies give evidence that miR-153 serve as both tumor suppressor and oncogene; therefore, it could serve as a potential predictive biomarker in cancer.
The importance of miR-153 in the regulation of various physiological processes has become evident in the past few years. Our current knowledge provides pieces of evidence about the oncogenic and tumor suppressor role of miR-153 gene and the platform to further develop diagnosis tools, and therapeutic strategies for curing cancer. In the future, we expect some novel techniques to introduce miR-153 or its inhibitors into cancer cells to inhibit cancer progression by regulating the cellular pathways. Further, we need to develop integrated approaches to incorporate miR-153 into clinical intervention so that cancer patients can be benefited.
Acknowledgement: Authors acknowledge the support from Maharishi Markandeshwar (Deemed to be University) Mullana and Shoolini University, Solan.
Authors’ Contribution: The authors confirm contribution to the paper as follows: study conception and design: JSN, RVS; data collection: ST, AKS, JD; draft manuscript preparation: ST, VS, PB. All authors compared different studies and approved the final version of the manuscript.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Altomare DA, Testa JR (2005). Perturbations of the AKT signaling pathway in human cancer. Oncogene 24: 7455–7464. [Google Scholar]
Anaya-Ruiz M, Cebada J, Delgado-López G, Sanchez-Vazquez ML, Perez-Santos JLM (2013). miR-153 silencing induces apoptosis in the MDA-MB-231 breast cancer cell line. Asian Pacific Journal of Cancer Prevention 14: 2983–2986. [Google Scholar]
Baber S, Bayat M, Mohamadnia A, Shamshiri A, Amini Shakib P, Bahrami N (2021). Role of miR153 and miR455-5p expression in oral squamous cell carcinoma isolated from plasma. Asian Pacific Journal of Cancer Prevention 22: 157–161. [Google Scholar]
Bai Z, Sun J, Wang X, Wang H, Pei H, Zhang Z (2015). MicroRNA-153 is a prognostic marker and inhibits cell migration and invasion by targeting SNAI1 in human pancreatic ductal adenocarcinoma. Oncology Reports 34: 595–602. [Google Scholar]
Bao B, Rodriguez-Melendez R, Zempleni J (2012). Cytosine methylation in miR-153 gene promoters increases the expression of holocarboxylase synthetase, thereby increasing the abundance of histone H4 biotinylation marks in HEK-293 human kidney cells. Journal of Nutritional Biochemistry 23: 635–639. [Google Scholar]
Bartel DP (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [Google Scholar]
Bi CW, Zhang GY, Bai Y, Zhao B, Yang H (2019). Increased expression of miR-153 predicts poor prognosis for patients with prostate cancer. Medicine 98: e16705–e16708. [Google Scholar]
Chen PS, Su JL, Hung MC (2012). Dysregulation of microRNAs in cancer. Journal of Biomedical Science 19: 1–8. [Google Scholar]
Chen J, Xu H, Zou X, Wang J, Zhu Y et al. (2014). Snail recruits Ring1B to mediate transcriptional repression and cell migration in pancreatic cancer cells. Cancer Research 74: 4353–4363. [Google Scholar]
Croce CM (2009). Causes and consequences of microRNA dysregulation in cancer. Nature Reviews Genetics 10: 704–714. [Google Scholar]
Cui Y, Zhao J, Yi L, Jiang Y (2016). microRNA-153 targets mTORC2 component rictor to inhibit glioma cells. PLoS One 11: e0156915. [Google Scholar]
Cully M, You H, Levine AJ, Mak TW (2006). Beyond PTEN mutations: The PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nature Reviews Cancer 6: 184–192. [Google Scholar]
Dong JT, Chen C (2009). Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cellular and Molecular Life Sciences 66: 2691–2706. [Google Scholar]
Engels BM, Hutvagner G (2006). Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene 25: 6163–6169. [Google Scholar]
Esquela-Kerscher A, Slack FJ (2006). Oncomirs—microRNAs with a role in cancer. Nature Reviews Cancer 6: 259–269. [Google Scholar]
Furuta S, Wang JM, Wei S, Jeng YM, Jiang X, Gu B, Chen PL, Eva YHL, Lee WH (2006). Removal of BRCA1/CtIP/ZBRK1 repressor complex on ANG1 promoter leads to accelerated mammary tumor growth contributed by prominent vasculature. Cancer Cell 10: 13–24. [Google Scholar]
Gao X, Qiao Y, Han D, Zhang Y, Ma N (2012). Enemy or partner: Relationship between intronic microRNAs and their host genes. IUBMB Life 64: 835–840. [Google Scholar]
Gao W, Gao J, Chen L, Ren Y, Ma J (2019). Targeting XIST induced apoptosis of human osteosarcoma cells by activation of NF-kB/PUMA signal. Bioengineered 10: 261–270. [Google Scholar]
Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Vincent WY (2005). Krüppel-like factors 4 and 5: The Yin and Yang regulators of cellular proliferation. Cell Research 15: 92–96. [Google Scholar]
Gomi H, Kubota-Murata C, Yasui T, Tsukise A, Torii S (2013). Immunohistochemical analysis of IA-2 family of protein tyrosine phosphatases in rat gastrointestinal endocrine cells. Journal of Histochemistry & Cytochemistry 61: 156–168. [Google Scholar]
Guo G, Zhang Y, Hu L, Bian X (2019). MicroRNA 153 affects nasopharyngeal cancer cell viability by targeting TGF β2. Oncology Letters 17: 646–651. [Google Scholar]
Gutschner T, Richtig G, Haemmerle M, Pichler M (2018). From biomarkers to therapeutic targets—The promises and perils of long non-coding RNAs in cancer. Cancer and Metastasis Reviews 37: 83–105. [Google Scholar]
Hawighorst T, Skobe M, Streit M, Hong YK, Velasco P, Brown LF, Riccardi L, Lange-Asschenfeldt B, Detmar M (2002). Activation of the tie2 receptor by angiopoietin-1 enhances tumor vessel maturation and impairs squamous cell carcinoma growth. American Journal of Pathology 160: 1381–1392. [Google Scholar]
Hou CH, Lin FL, Tong KB, Hou SM, Liu JF (2014). Transforming growth factor alpha promotes osteosarcoma metastasis by ICAM-1 and PI3K/Akt signaling pathway. Biochemical Pharmacology 89: 453–463. [Google Scholar]
Huang Q, Xia J, Wang L, Wang X, Ma X et al. (2018). miR-153 suppresses IDO1 expression and enhances CAR T cell immunotherapy. Journal of Hematology & Oncology 11: 1–12. [Google Scholar]
Huang M, Li C, Kong F, Wu Y, Yuan Q, Hu L (2020). Prognostic and clinicopathological significance of MicroRNA-153 in human cancers: A meta-analysis. Medicine 99: e22833. [Google Scholar]
Hutchinson J, Jin J, Cardiff RD, Woodgett JR, Muller WJ (2001). Activation of Akt (protein kinase B) in mammary epithelium provides a critical cell survival signal required for tumor progression. Molecular and Cellular Biology 21: 2203–2212. [Google Scholar]
Iorio MV, Croce CM (2012). MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Molecular Medicine 4: 143–159. [Google Scholar]
Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B (2006). SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127: 125–137. [Google Scholar]
Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, Beug H, Grünert S (2002). Ras and TGFβ cooperatively regulate epithelial cell plasticity and metastasis: Dissection of Ras signaling pathways. Journal of Cell Biology 156: 299–314. [Google Scholar]
Kato M, Putta S, Wang M, Yuan H, Lanting L et al. (2009). TGF-β activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nature Cell Biology 11: 881–889. [Google Scholar]
Kim TH, Kim YK, Kwon Y, Heo JH, Kang H, Kim G, An HJ (2010). Deregulation of miR-519a, 153, and 485-5p and its clinicopathological relevance in ovarian epithelial tumours. Histopathology 57: 734–743. [Google Scholar]
Kloosterman WP, Plasterk RH (2006). The diverse functions of microRNAs in animal development and disease. Developmental Cell 11: 441–450. [Google Scholar]
Li Y, Kong Y, Zhou Z, Chen H, Wang Z et al. (2013a). The HECTD3 E3 ubiquitin ligase facilitates cancer cell survival by promoting K63-linked polyubiquitination of caspase-8. Cell Death & Disease 4: e935. [Google Scholar]
Li Y, Chen X, Wang Z, Zhao D, Chen H et al. (2013b). The HECTD3 E3 ubiquitin ligase suppresses cisplatin-induced apoptosis via stabilizing MALT1. Neoplasia 15: 39–IN15. [Google Scholar]
Li X, Liu X, Xu Y, Liu J, Xie M, Ni W, Chen S (2014a). KLF5 promotes hypoxia-induced survival and inhibits apoptosis in non-small cell lung cancer cells via HIF-1α. International Journal of Oncology 45: 1507–1514. [Google Scholar]
Li Q, Dong Z, Zhou F, Cai X, Gao Y, Wang LW (2014b). Krüppel-like factor 5 promotes lung tumorigenesis through upregulation of Sox4. Cellular Physiology and Biochemistry 33: 1–10. [Google Scholar]
Li W, Zhai L, Zhao C, Lv S (2015). miR-153 inhibits epithelial-mesenchymal transition by targeting metadherin in human breast cancer. Breast Cancer Research and Treatment 150: 501–509. [Google Scholar]
Li X, Wu C, Chen N, Gu H, Yen A, Cao L, Wang E, Wang L (2016). PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 7: 33440. [Google Scholar]
Li W, Wang M, Meng B, Yu J, Chen Q, Li H, Liu Y (2017). MicroRNA-153 regulated AKT1 expression and suppressed cell proliferation of epithelial ovarian cancer cells. International Journal of Clinical and Experimental Pathology 10: 7417. [Google Scholar]
Li H, Cui J, Xu B, He S, Yang H, Liu L (2019). Long non coding RNA XIST serves an oncogenic role in osteosarcoma by sponging miR 137. Experimental and Therapeutic Medicine 17: 730–738. [Google Scholar]
Liang H, Ge F, Xu Y, Xiao J, Zhou Z, Liu R, Chen C (2018a). miR-153 inhibits the migration and the tube formation of endothelial cells by blocking the paracrine of angiopoietin 1 in breast cancer cells. Angiogenesis 21: 849–860. [Google Scholar]
Liang H, Xiao J, Zhou Z, Wu J, Ge F et al. (2018b). Hypoxia induces miR-153 through the IRE1α-XBP1 pathway to fine tune the HIF1α/VEGFA axis in breast cancer angiogenesis. Oncogene 37: 1961–1975. [Google Scholar]
Liu L, Chen R, Huang S, Wu Y, Li G, Zhang B, Liu Q, Yin D, Liang Y (2012). miR-153 sensitized the K562 cells to As2O3-induced apoptosis. Medical Oncology 29: 243–247. [Google Scholar]
Liu JY, Lu JB, Xu Y (2016a). MicroRNA 153 inhibits the proliferation and invasion of human laryngeal squamous cell carcinoma by targeting KLF5. Experimental and Therapeutic Medicine 11: 2503–2508. [Google Scholar]
Liu R, Shi P, Nie Z, Liang H, Zhou Z et al. (2016b). Mifepristone suppresses basal triple-negative breast cancer stem cells by down-regulating KLF5 expression. Theranostics 6: 533. [Google Scholar]
Liu F, Liu B, Qian J, Wu G, Li J, Ma Z (2017). miR-153 enhances the therapeutic effect of gemcitabine by targeting Snail in pancreatic cancer. Acta biochimica et biophysica Sinica 49: 520–529. [Google Scholar]
Liu R, Chen H, Zhao P, Chen CH, Liang H, Yang C, Zhou Z, Zhi X, Liu S, Chen C (2020). Mifepristone Derivative FZU-00,003 Suppresses Triple-negative Breast Cancer Cell Growth partially via miR-153-KLF5 axis. International Journal of Biological Sciences 16: 611. [Google Scholar]
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J et al. (2005). MicroRNA expression profiles classify human cancers. Nature 435: 834–838. [Google Scholar]
Lyu X, Fang W, Cai L, Zheng H, Ye Y et al. (2014). TGFβR2 is a major target of miR-93 in nasopharyngeal carcinoma aggressiveness. Molecular Cancer 13: 1–14. [Google Scholar]
Ma Y, Xue Y, Liu X, Qu C, Cai H, Wang P, Li Z, Li Z, Liu Y (2017). SNHG15 affects the growth of glioma microvascular endothelial cells by negatively regulating miR-153. Oncology Reports 38: 3265–3277. [Google Scholar]
Mandemakers W, Abuhatzira L, Xu H, Caromile LA, Hébert SS et al. (2013). Co-regulation of intragenic microRNA miR-153 and its host gene Ia-2 β: Identification of miR-153 target genes with functions related to IA-2β in pancreas and brain. Diabetologia 56: 1547–1556. [Google Scholar]
Masri J, Bernath A, Martin J, Jo OD, Vartanian R, Funk A, Gera J (2007). mTORC2 activity is elevated in gliomas and promotes growth and cell motility via overexpression of rictor. Cancer Research 67: 11712–11720. [Google Scholar]
Matsuyama S, Iwadate M, Kondo M, Saitoh M, Hanyu A et al. (2003). SB-431542 and Gleevec inhibit transforming growth factor-β-induced proliferation of human osteosarcoma cells. Cancer Research 63: 7791–7798. [Google Scholar]
M’hamed IF, Privat M, Trimeche M, Penault-Llorca F, Bignon YJ, Kenani A (2017). miR-10b, miR-26a, miR-146a And miR-153 expression in triple negative vs. non triple negative breast cancer: Potential biomarkers. Pathology & Oncology Research 23: 815–827. [Google Scholar]
Moradi Marjaneh R, Khazaei M, Ferns GA, Avan A, Aghaee-Bakhtiari SH (2019). MicroRNAs as potential therapeutic targets to predict responses to oxaliplatin in colorectal cancer: From basic evidence to therapeutic implication. IUBMB Life 71: 1428–1441. [Google Scholar]
Myatt SS, Wang J, Monteiro LJ, Christian M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S, Lam EW (2010). Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Research 70: 367–377. [Google Scholar]
Nakaya T, Ogawa S, Manabe I, Tanaka M, Sanada M et al. (2014). KLF5 regulates the integrity and oncogenicity of intestinal stem cells. Cancer Research 74: 2882–2891. [Google Scholar]
Nishimura T, Kubosaki A, Ito Y, Notkins AL (2009). Disturbances in the secretion of neurotransmitters in IA-2/IA-2β null mice: Changes in behavior, learning and lifespan. Neuroscience 159: 427–437. [Google Scholar]
Niu G, Li B, Sun L, An C (2015). MicroRNA-153 inhibits osteosarcoma cells proliferation and invasion by targeting TGF-β2. PLoS One 10: e0119225. [Google Scholar]
Perera M, Tsang CS, Distel RJ, Lacy JN, Ohno-Machado L et al. (2010). TGF-β1 interactome: Metastasis and beyond. Cancer Genomics-Proteomics 7: 217–229. [Google Scholar]
Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2004). Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Current Biology 14: 1296–1302. [Google Scholar]
Shan N, Shen L, Wang J, He D, Duan C (2015). MiR-153 inhibits migration and invasion of human non-small-cell lung cancer by targeting ADAM19. Biochemical and Biophysical Research Communications 456: 385–391. [Google Scholar]
Schedlich LJ, Yenson VM, Baxter RC (2013). TGF-β-induced expression of IGFBP-3 regulates IGF1R signaling in human osteosarcoma cells. Molecular and Cellular Endocrinology 377: 56–64. [Google Scholar]
Shi D, Li Y, Fan L, Zhao Q, Tan B, Cui G (2019). Upregulation Of miR-153 inhibits triple-negative breast cancer progression by targeting ZEB2-mediated EMT and contributes to better prognosis. OncoTargets and Therapy 12: 9611. [Google Scholar]
Solimena M, Dirkx RJr, Hermel JM, Pleasic-Williams S, Shapiro JA, Caron L, Rabin DU (1996). ICA 512, an autoantigen of type I diabetes, is an intrinsic membrane protein of neurosecretory granules. EMBO Journal 15: 2102–2114. [Google Scholar]
Sung JY, Park SY, Kim JH, Kang HG, Yoon JH, Na YS, Kim YN, Park BK (2014). Interferon consensus sequence-binding protein (ICSBP) promotes epithelial-to-mesenchymal transition (EMT)-like phenomena, cell-motility, and invasion via TGF-β signaling in U2OS cells. Cell Death & Disease 5: e1224. [Google Scholar]
Tang JM, He QY, Guo RX, Chang XJ (2006). Phosphorylated Akt overexpression and loss of PTEN expression in non-small cell lung cancer confers poor prognosis. Lung Cancer 51: 181–191. [Google Scholar]
Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, Schiemann WP (2013). TGF-β upregulates miR-181a expression to promote breast cancer metastasis. Journal of Clinical Investigation 123: 150–163. [Google Scholar]
Vishal M, Swetha R, Thejaswini G, Arumugam B, Selvamurugan N (2017). Role of Runx2 in breast cancer-mediated bone metastasis. International Journal of Biological Macromolecules 99: 608–614. [Google Scholar]
Wang J, Liang S, Duan X (2019a). Molecular mechanism of miR-153 inhibiting migration, invasion and epithelial-mesenchymal transition of breast cancer by regulating transforming growth factor beta (TGF-β) signaling pathway. Journal of Cellular Biochemistry 120: 9539–9546. [Google Scholar]
Wang L, Lv X, Fu X, Su L, Yang T, Xu P (2019b). MiR-153 inhibits the resistance of lung cancer to gefitinib via modulating expression of ABCE1. Cancer Biomarkers 25: 361–369. [Google Scholar]
Wei CY, Tan QX, Zhu X, Qin QH, Zhu FB, Mo QG, Yang WP (2015). Expression of CDKN1A/p21 and TGFBR2 in breast cancer and their prognostic significance. International Journal of Clinical and Experimental Pathology 8: 14619. [Google Scholar]
Wen JF, Jiang YQ, Li C, Dai XK, Wu T, Yin WZ (2020). LncRNA-XIST promotes the oxidative stress-induced migration, invasion, and epithelial-to-mesenchymal transition of osteosarcoma cancer cells through miR-153-SNAI1 axis. Cell Biology International 44: 1991–2001. [Google Scholar]
Wu Z, He B, He J, Mao X (2013). Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. Prostate 73: 596–604. [Google Scholar]
Wu X, Li L, Li Y, Liu Z (2016a). MiR-153 promotes breast cancer cell apoptosis by targeting HECTD3. American Journal of Cancer Research 6: 1563. [Google Scholar]
Wu Y, Sarkissyan M, Vadgama JV (2016b). Epithelial-mesenchymal transition and breast cancer. Journal of Clinical Medicine 5: 13. [Google Scholar]
Wu J, Zhang P, Wang X, Liu X (2019). MiRNA-153 regulates cell viability and the cell cycle in bladder cancer. International Journal of Clinical and Experimental Medicine 12: 5933–5937. [Google Scholar]
Xu J, Liao X, Wong C (2010). Downregulations of B-cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. International Journal of Cancer 126: 1029–1035. [Google Scholar]
Xu Q, Sun Q, Zhang J, Yu J, Chen W, Zhang Z (2013). Downregulation of miR-153 contributes to epithelial-mesenchymal transition and tumor metastasis in human epithelial cancer. Carcinogenesis 34: 539–549. [Google Scholar]
Xu H, Abuhatzira L, Carmona GN, Vadrevu S, Satin LS, Notkins AL (2015). The Ia-2β intronic miRNA, miR-153, is a negative regulator of insulin and dopamine secretion through its effect on the Cacna1c gene in mice. Diabetologia 58: 2298–2306. [Google Scholar]
Yang C, Wu K, Wang S, Wei G (2018). Long non-coding RNA XIST promotes osteosarcoma progression by targeting YAP via miR-195-5p. Journal of Cellular Biochemistry 119: 5646–5656. [Google Scholar]
You H, Mak TW (2005). Crosstalk between p53 and FOXO transcription factors. Cell Cycle 4: 37–38. [Google Scholar]
Yuan Y, Du W, Wang Y, Xu C, Wang J et al. (2015). Suppression of AKT expression by mi R-153 produced anti-tumor activity in lung cancer. International Journal of Cancer 136: 1333–1340. [Google Scholar]
Zaman MS, Maher DM, Khan S, Jaggi M, Chauhan SC (2012). Current status and implications of microRNAs in ovarian cancer diagnosis and therapy. Journal of Ovarian Research 5: 1–11. [Google Scholar]
Zhang L, Pickard K, Jenei V, Bullock MD, Bruce A et al. (2013). miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Research 73: 6435–6447. [Google Scholar]
Zhang W, Mao S, Shi D, Zhang J, Zhang Z et al. (2019). MicroRNA-153 decreases tryptophan catabolism and inhibits angiogenesis in bladder cancer by targeting indoleamine 2, 3-dioxygenase 1. Frontiers in Oncology 9: 619. [Google Scholar]
Zhao S, Deng Y, Liu Y, Chen X, Yang G, Mu Y, Zhang D, Kang J, Wu Z (2013). MicroRNA-153 is tumor suppressive in glioblastoma stem cells. Molecular Biology Reports 40: 2789–2798. [Google Scholar]
Zhao W, Yin CY, Jiang J, Kong W, Xu H, Zhang H (2019). MicroRNA-153 suppresses cell invasion by targeting SNAI1 and predicts patient prognosis in glioma. Oncology Letters 17: 1189–1195. [Google Scholar]
Zhao G, Zhang Y, Zhao Z, Cai H, Zhao X et al. (2020). MiR-153 reduces stem cell-like phenotype and tumor growth of lung adenocarcinoma by targeting Jagged1. Stem Cell Research & Therapy 11: 1–12. [Google Scholar]
Zhao Z, Shen X, Zhang D, Xiao H, Kong H, Yang B, Yang L (2021). miR-153 enhances the therapeutic effect of radiotherapy by targeting JAG1 in pancreatic cancer cells. Oncology Letters 21: 1–8. [Google Scholar]
Zhou J, Xie M, Shi Y, Luo B, Gong G et al. (2015). MicroRNA-153 functions as a tumor suppressor by targeting SET7 and ZEB2 in ovarian cancer cells. Oncology Reports 34: 111–120. [Google Scholar]
Zuo J, Wang D, Shen H, Liu F, Han J, Zhang X (2016). MicroRNA-153 inhibits tumor progression in esophageal squamous cell carcinoma by targeting SNAI1. Tumor Biology 37: 16135–16140. [Google Scholar]
Zuo Z, Ye F, Liu Z, Huang J, Gong Y (2019). MicroRNA 153 inhibits cell proliferation, migration, invasion and epithelial mesenchymal transition in breast cancer via direct targeting of RUNX2. Experimental and Therapeutic Medicine 17: 4693–4702. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |