DOI:10.32604/biocell.2021.014728

| Biocell DOI:10.32604/biocell.2021.014728 |  |
| Article |
STMN1 promotes the proliferation and inhibits the apoptosis of acute myeloid leukemiacells by activating the PI3K/Akt pathway
1Department of Hematology, The Fourth People’s Hospital of Shenyang, Geriatrics Center, Shenyang, 110031, China
2The Fourth People’s Hospital of Shenyang, Geriatrics Center, Shenyang, 110031, China
*Address correspondence to: Peng Yang, yppp1981_1@163.com
Received: 24 October 2020; Accepted: 22 December 2020
Abstract: Recent studies have shown that the microtubule disrupting protein Stathmin 1 (STMN1) is differentially expressed in AML patients and healthy control. The aim of this study was to explore the effects and molecular mechanism of STMN1 in AML. Here, the expression of STMN1 in peripheral blood cells (PBMCs) and bone marrow of AML patients and healthy volunteers was detected by RT-PCR and Western blot. STMN1 expression was regulated by transfected with STMN1 overexpressed plasmid or shRNA in two human leukemia cell lines K562 and HL60. Cell proliferation was examined by CCK8 and Edu staining. Annexin V and TUNEL assays were applied to test cell apoptosis. Flow cytometry was used to test the cell cycle distribution. The activation of the PI3K signaling pathway and the expression levels of cell cycle and cell apoptosis-related protein were determined by Western blot. In this study, we found that STMN1 was overexpressed in PBMCs and bone marrow of AML patients. STMN1 expression was closely related to FAB subtypes, risk stratification, disease-free survival, and overall survival of AML. Functional assays showed that overexpression of STMN1 in HL60 and K562 cells enhanced cell proliferation, decreased cell apoptosis, and caused G1 phase arrest. In contrast, suppression of STMN1reduced cell proliferation and enhanced cell apoptosis in both HL60 and K562 cells. Moreover, the PI3K/Akt pathway was activated by STMN1, while suppression of STMN1 dysregulated the PI3K/Akt pathway and upregulating the levels of caspases3 and Bax expression. In conclusion, STMN1 was confirmed to promote the proliferation and inhibit the apoptosis of HL60 and K562 cells by modulating the PI3K/Akt pathway. STMN1 might be a novel molecular target for treating AML.
Keywords: STMN1; AML; PI3K/Akt
As the most common type of acute leukemia, the pathophysiology of acute myeloid leukemia (AML) involves the maturational arrest of bone marrow cells and the activation or inactivation of genes that contribute to genomic instability (Ghiaur et al., 2015). Currently, cytogenetic markers are considered as the most important factors for risk classification and selecting a treatment for AML patients (De Kouchkovsky and Abdul-Hay, 2016). In addition, the identification of new biomarkers has increased our understanding of the molecular mechanism of AML, and those markers can be used to predict the results of therapy (De Kouchkovsky and Abdul-Hay, 2016). Therefore, it remains important to identify new therapeutic markers and targets for AML.
Stathmin 1 (STMN1), also known as oncoprotein 18 (Op18), is a cytosolic protein that mediates microtubule destabilization (Gavet et al., 1998). STMN1 regulates microtubule dynamics and assists in assembling the mitotic spindle (Horwitz et al., 1997; Honnappa et al., 2003; Steinmetz, 2007). Recent findings show that STMN1 is highly expressed in various types of human cancers and plays critical roles in regulating tumor progression (Rana et al., 2008; Belletti and Baldassarre, 2011). In addition, STMN1 induces cell proliferation, migration, and differentiation in several types of solid tumors, andSTMN1 expression is related to the prognosis of tumor patients (Hsieh et al., 2010; Zheng et al., 2010; Zhang et al., 2019). A high level of STMN1 expression is significantly associated with vascular invasion in lungsquamous cell carcinoma (Bao et al., 2017) and chemoresistance in gastric cancer (Bai et al., 2017). A meta-analysis revealed that STMN1 is more highly expressed in cancer patients than non-cancer individuals and that overexpression of STMN1 is associated with the degree of a tumor’s malignancy (Rong et al., 2016). Recently, an AML-array revealed that the levels of STMN1 expression were different in AML patients with the M1 or M2 subtypes when compared with healthy control subjects (Handschuh et al., 2018). Furthermore, another study showed that the levels of STMN1 mRNA expression in relapsed patients with acute leukemia (AL) were increased when compared with the levels in healthy volunteers, and no significant difference was found between STMN1 mRNA expression in AML patients and acute lymphoblastic leukemia (ALL) patients (Xu et al., 2013). Although STMN1 has been proposed as a potential prognostic marker for different types of tumors, the mechanism by whichSTMN1 participates in the initiation and development of tumors and affects the overall survival time and prognosis of AML has not been identified.
Phosphoinositide 3-kinase (PI3K), Akt (protein kinase B/PKB), and mammalian target of rapamycin (mTOR) all participate in a signaling pathway that mediates many key functions in cells (Sapon-Cousineau et al., 2020). The G1/S cell cycle phase is mediated by PI3K/Akt, which causes an increase in cyclin D1 expression (Liang and Slingerland, 2003). Akt phosphorylates and inactivates Bad, leading to the promotion of cell survival and activation of the anti-apoptotic proteins Bcl-2 and Bcl-xL (Datta et al., 1999). Caspases are effectors of cell apoptosis. Once activated by an initiating signal, Caspase-9 activates Caspase-3, and causes cell apoptosis (Porter and Jänicke, 1999). More importantly, inactivation of the PI3K/Akt signaling
pathway is highly related to an up-regulation of Bax expression and activation of Caspase-3 (Liu et al., 2010), and PI3K/Akt pathway are aberrantly activated in human cancers, including AML, via the phosphorylation of PI3K and Akt proteins (Nepstad et al., 2020). Upon activation, PI3K/Akt pathway contributes to metabolic reprogramming (Elstrom et al., 2004) and plays crucial roles in cell proliferation, differentiation, and survival in AML (Nepstad et al., 2020). Moreover, activation of the PI3K/Akt pathway is related to a decreased overall disease survival rate(Min et al., 2003; Nepstad et al., 2018; Nepstad et al., 2019) and is used as a prognostic factor in AML (Tamburini et al., 2007). A recent study showed that STMN1 is can serve as a substitute marker of PI3K pathway activity and is related to tumor recurrence (Jiang et al., 2018).
Here, we found that STMN1 expression was increased in PBMCs and bone marrow of AML patients and was closely related to FAB subtypes, risk stratification, disease-free survival, and overall survival of AML patients. Functionally, overexpression of STMN1enhanced cell proliferation and reduced cell apoptosis rates of both HL60 and K562 cells. We also found that the PI3K/Akt pathway could be activated by STMN1. When taken together, our data suggest STMN1 as a potential prognostic marker for AML patients and also a promising therapeutic target for AML.
Clinical samples and cell culture
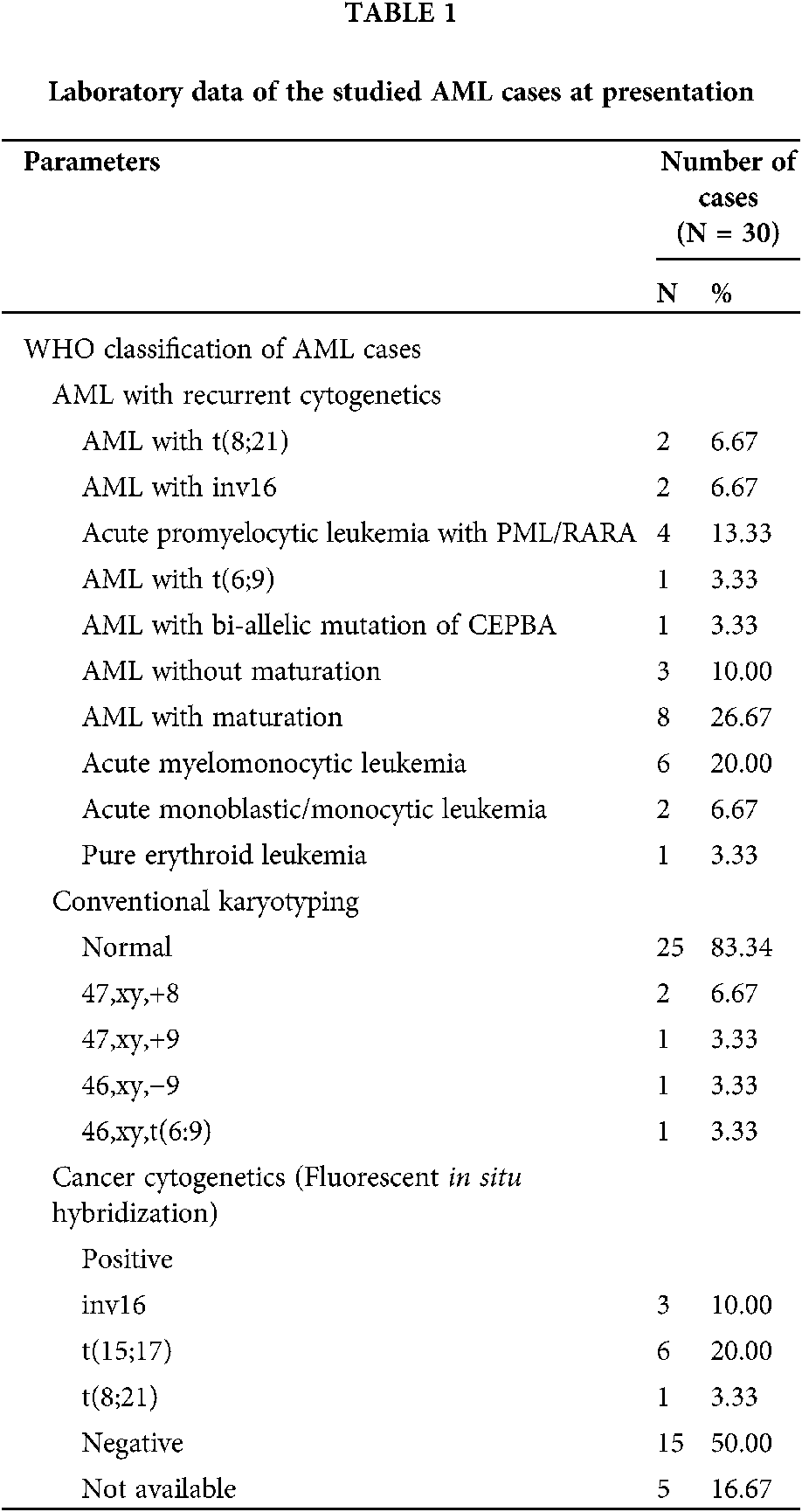
A total of 30 AML patients and 10 volunteers with external spinal injury, and who were diagnosed without AML were enrolled in this study. All the AML patients were diagnosed according to ELN criteria (Creutzig et al., 2012), and their AML subtypes were classified according to the French-American-British (FAB) classification system. Twenty-two patients were classified as M0–M3 AML and 8 patients were classified as M3–M7 AML. The patients were diagnosed according to the WHO classification (2016), and percentages are shown in Tab. 1. All clinical samples were collected from patients at the Fourth People’s Hospital of Shenyang, and each AML patient and healthy volunteer provided their written informed consent for study participation. The study protocol was approved by the Institute Research Ethics Committee at the Fourth People’s Hospital of Shenyang. Detailed clinical characteristics of the AML patients are shown in Tab. 1. Samples of Bone marrow tissues were collected from de novo AML patients (30 cases) by Ficoll-Hypaque (GE, Cat. No. 71-7167-00 AG) density gradient centrifugation. Samples of normal bone marrow were collected from 10healthy donors. PBMCs were isolated from the venous blood of AML patients and healthy control subjects by Ficoll density gradient centrifugation.
Human promyelocytic leukemia HL60 cells and human chronic myelogenic leukemic cells K562 were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in RPMI-1640 medium (Thermo Fisher Scientific, Cat. No. A1049101) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific, Cat. No. 10100147C) at 37°C in an incubator containing 95% air and 5% CO2.
The human STMN1 gene was subcloned into a pcDNA3.1 vector (Thermo Fisher Scientific, Cat. No. V79520) by double restriction enzyme digestion. Control or STMN1 shRNA was subcloned into a pRS shRNA vector (Origene, Cat. No. TR20003) by double restriction enzyme digestion. All the plasmids were confirmed by DNA sequencing. For plasmid transfection, 5 × 105 cells suspended in 1 mL of growth medium without antibiotics were added to each well of a 6-well plate. Lipofectamine 2000 (Thermo Fisher Scientific, Cat. No. 11668-500) was used for transient transfection. Briefly, the plasmid (4 μg for each well of a 6-well plate) or Lipofectamine 2000 (10 μL for each well of a 6-well plate) was diluted with 250 μL of Opti-MEM (Thermo Fisher Scientific, Cat. No. 31985088). After 5 min of incubation, the diluted plasmid and diluted Lipofectamine 2000 were combined and incubated for another 15–20 min. The mixture was then added to cells, and the cells were cultured for the indicated time.
Total RNA was extracted from PBMCs or bone marrow using Trizol reagent (Takara, Cat. No. 9109) according to the manufacturer’s instructions. HL60 and K562 cells (5 × 105) were transfected for 48 h with pcDNA3.1 (vector control), pcDNA-STMN1, pRS Control shRNA (shCTRL) or pRS STMN1 shRNA (shSTMN1) by Lipofectamine 2000, cells were collected and extracted by Trizol reagent. First-strand cDNA was synthesized using Bestar qPCR RT mix (DBI, Cat. No. 220), and the mRNA was amplified by using Bestar qPCR Master Mix kit (DBI, Cat. No. 2043) on an ABI real-time PCR system (7500). The PCR procedure was performed as follows: 95°C for 2 min, followed by 40 cycles of 95°C for 20 s, 60°C for 20 s, and 72°C for 20 s. The specific primers used were GAPDH: (forward) 5’ TGTTCGTCATGGGTGTGAAC3’, (reverse) 5’ ATGGCATGGACTGTGGTCAT 3’; STMN1: (forward) 5’ TCAGCCCTCGGTCAAAAGAAT 3’, (reverse) 5’ TTCTCGTGCTCTCGTTTCTCA 3’. The data were analyzed according to the 2−∆∆Ct method.
HL60 or K562 cells (1 × 104 in 100 μL of culture medium) were seeded into the wells of a 96-well plate and transfected with pcDNA3.1 (vector control), pcDNA-STMN1, pRS Control shRNA (shCTRL) or pRS STMN1 shRNA (shSTMN1) using Lipofectamine 2000. After 24, 48 or 72 h of transfection, 10 µL of CCK-8 solution (Sigma, Cat. No. 96992, 1:10 dilution) was added to each well, and the cells were incubated for another 2 h at 37°C. The absorbance of each well at 450 nm was determined with a microplate reader.
Edu staining for detection of cell proliferation
HL60 or K562 cells (2 × 105) were seeded into the wells of a 12-well plate and transiently transfected with pcDNA3.1 (vector control), pcDNA-STMN1, pRS Control shRNA (shCTRL), or pRS STMN1 shRNA (shSTMN1) using Lipofectamine 2000. After 48 h of transfection, the cells were cytocentrifuged onto SuperFrost Plus glass slides at 450 × g for 10 min using a Shandon Cytospin 4 cytocentrifuge. A Click-iTEdU Imaging Kit (Thermo Fisher Scientific, Cat. No. C10350) was used to detect Edu positive cells. Images were obtained with a fluorescence microscope.
HL60 or K562 cells (5 × 105) were seeded into the wells of a 6-well plate and transiently transfected with pcDNA3.1 (vector control), pcDNA-STMN1, pRS Control shRNA (shCTRL), or pRS STMN1 shRNA (shSTMN1) using Lipofectamine 2000. After 48 h of transfection, the cells were centrifuged at 1000 rpm for 5 min and washed once with PBS. The cells were then fixed in pre-cooled 70% ethanol and stored at −20°C overnight.The nextday, the cells were pelleted by centrifugation, washed once with PBS, and stained with 400 μL of PI/RNase solution (PI: 50 μg/mL, RNase: 100 μg/mL, Cell Signaling Technology, Cat. No. 4087) for 20 min at 37°C in the dark. The cell cycle distribution was determined by flow cytometry.
HL60 or K562 cells (2 × 105) were seeded into the wells of a 12-well plate and transfected with pcDNA3.1 (vector control), pcDNA-STMN1, pRS Control shRNA (shCTRL), or pRS STMN1 shRNA (shSTMN1) using Lipofectamine 2000. After 48 h of transfection, the cells were centrifuged at 1500 × g for 10 min and collected, after which they were fixed and double-stained with Annexin V and PI (BD Biosciences, Cat. No. 556547). The results were acquired by a flow cytometer and analyzed by FlowJo software.
HL60 or K562 cells (2 × 105) were plated into the wells of a 12-well plate and transiently transfected with pcDNA3.1 (vector control), pcDNA-STMN1, pRS Control shRNA (shCTRL), or pRS STMN1 shRNA (shSTMN1) using Lipofectamine 2000. After 48 h of transfection, the cells were fixed by 4% paraformaldehyde (PFA) and permeabilized with 0.25% Triton X-100. After being washed twice with deionized water, the cells were incubated with DNase I solution for 30 min and then treated with TdT reaction cocktail for 60 min at 37°C. After being washed 3 times with 3% BSA in PBS, the cells were incubated with Click-iT reaction cocktail (Thermo Fisher Scientific, Cat. No. C10338) for 30 min and then washed one tome in 3% BSA dissolved in PBS.
Bone marrow samples collected from patients or healthy control subjects were lysed in RIPA buffer (Sigma, Cat. No.R0278). HL60 or K562 cells were added into the wells of a 6-well plate and transiently transfected with pcDNA3.1 (vector control), pcDNA-STMN1, pRS Control shRNA (shCTRL), or pRS STMN1 shRNA (shSTMN1) using Lipofectamine 2000. After 48 h of transfection, the cells were lysed by RIPA lysis buffer, and the total proteins were extracted. The protein concentration in each extract with determined using a BCA protein assay kit (Sigma, Cat. No. B9643). Next, an equal amount of total protein (20 μg) from each extract was separated by 10% SDS-PAGE, and the protein bands were transferred onto PVDF membrane (Millipore, Cat. No. IPVH00010), which were subsequently blocked with 5% non-fat milk for 1 h. The membranes were then washed with 1 × TBST and incubated with the following antibodies: STMN1 (Novus, Cat. No. NBP1-76798), cyclin B1 (Santa Cruz, Cat. No. sc-245), cyclin D1 (Cell Signaling Technology, Cat. No. 2922), Caspase3 (Cell Signaling Technology, Cat. No. 9662), Bcl-2 (Cell Signaling Technology, Cat. No. 423), Bax (Cell Signaling Technology, Cat. No. 2772), PTEN (Cell Signaling Technology, Cat. No. 9552), AKT (Cell Signaling Technology, Cat. No. 9272), p-AKT (Cell Signaling Technology, Cat. No. 4060), PI3K (Cell Signaling Technology, Cat. No. 4255), p-PI3K (Cell Signaling Technology, Cat. No. 17366), and GAPDH (Cell Signaling Technology, Cat. No. 97166) at 4°C overnight. Next, the membranes were washed three times with 1 × TBST and incubated with an HRP conjugated anti-rabbit (Abcam, Cat. No. ab6721) or anti-mouse (Abcam, Cat. No. ab6728) secondary antibodies. Immunostaining of the target proteins was visualized by enhanced chemiluminescence (Thermo Fisher Scientific, Cat. No. 32106).
All assays were repeated at least three times, and results were shown as a mean value ± standard error of the mean (S.E.M.). All data were analyzed using GraphPad Prism Software (Prism 7.0). Comparisons of normally distributed data between two groups were performed using the Student’s t-test. One-way analysis of variance (ANOVA) followed by Bonferroni’s post hoc test was used to determine the statistical significance of differences between groups. A p-value < 0.05 was considered to be a significant difference.
STMN1 expression is induced in AML patients
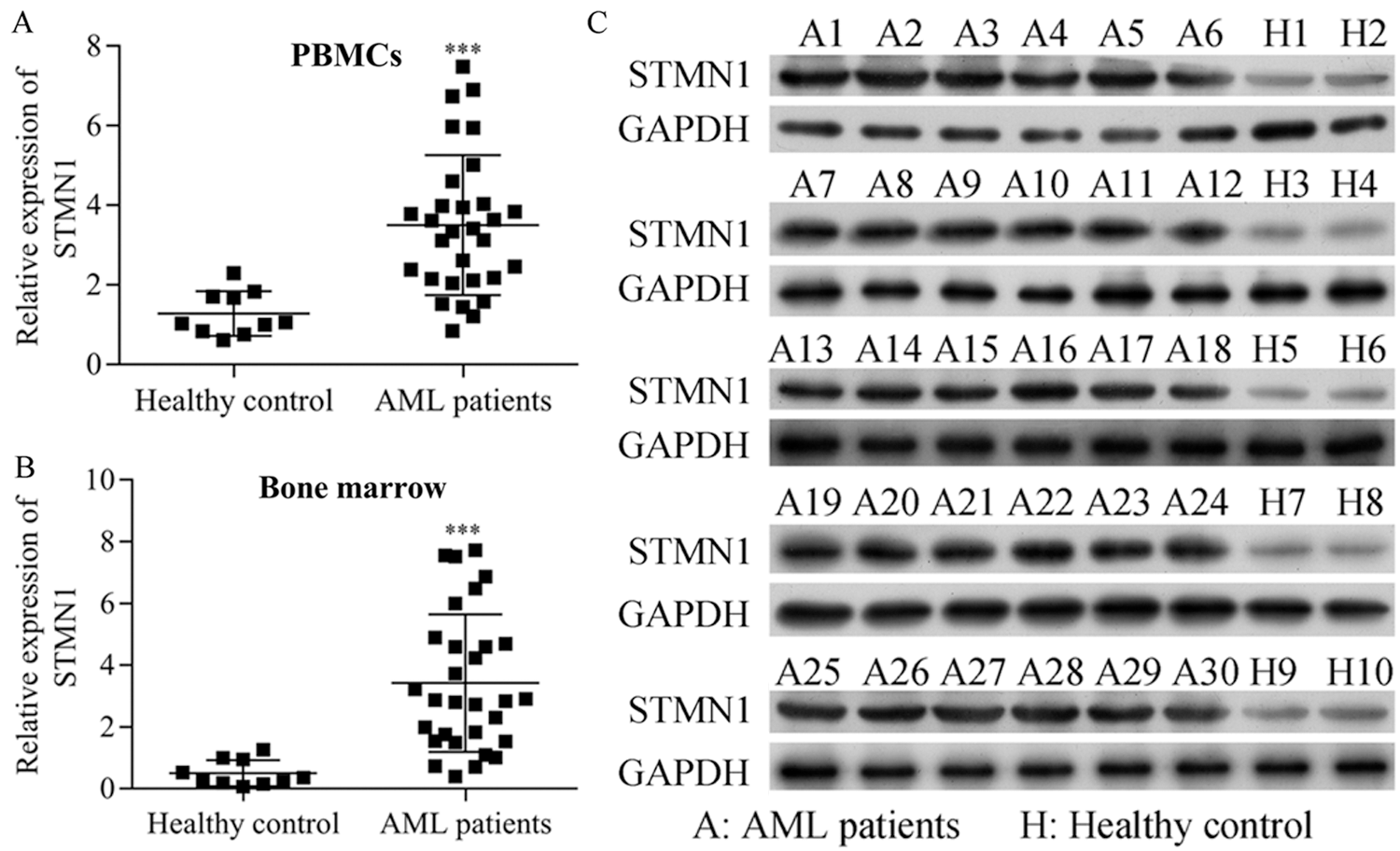
Figure 1: STMN1 levels were increases in AML patients.
(A and B) PBMCs (A) or bone marrow samples (B) were collected from 30 patients with AML and 10 healthy control subjects, and the total RNA was extracted for RT-PCR assay. (C) The total proteins were extracted from bone marrow from 30 patients with AML and 10 healthy control subjects, and STMN1 expression was detected by Western blot. GAPDH served as a loading control. ***p < 0.001, compared with healthy control.
The expression of STMN1 in the PBMCs and bone marrow from 30 patients with AML and 10 healthy control was detected, and our analysis showed that the levels of STMN1 mRNA expression were significantly elevated in the PBMCs and bone marrow of AML patients when compared with their levels in healthy control subjects (Figs. 1A and 1B). Additionally, the levels of STMN1 protein expression were also elevated in AML patients (Fig. 1C).
STMN1 expression was closely related to the disease-free survival time and overall survival time of AML patients
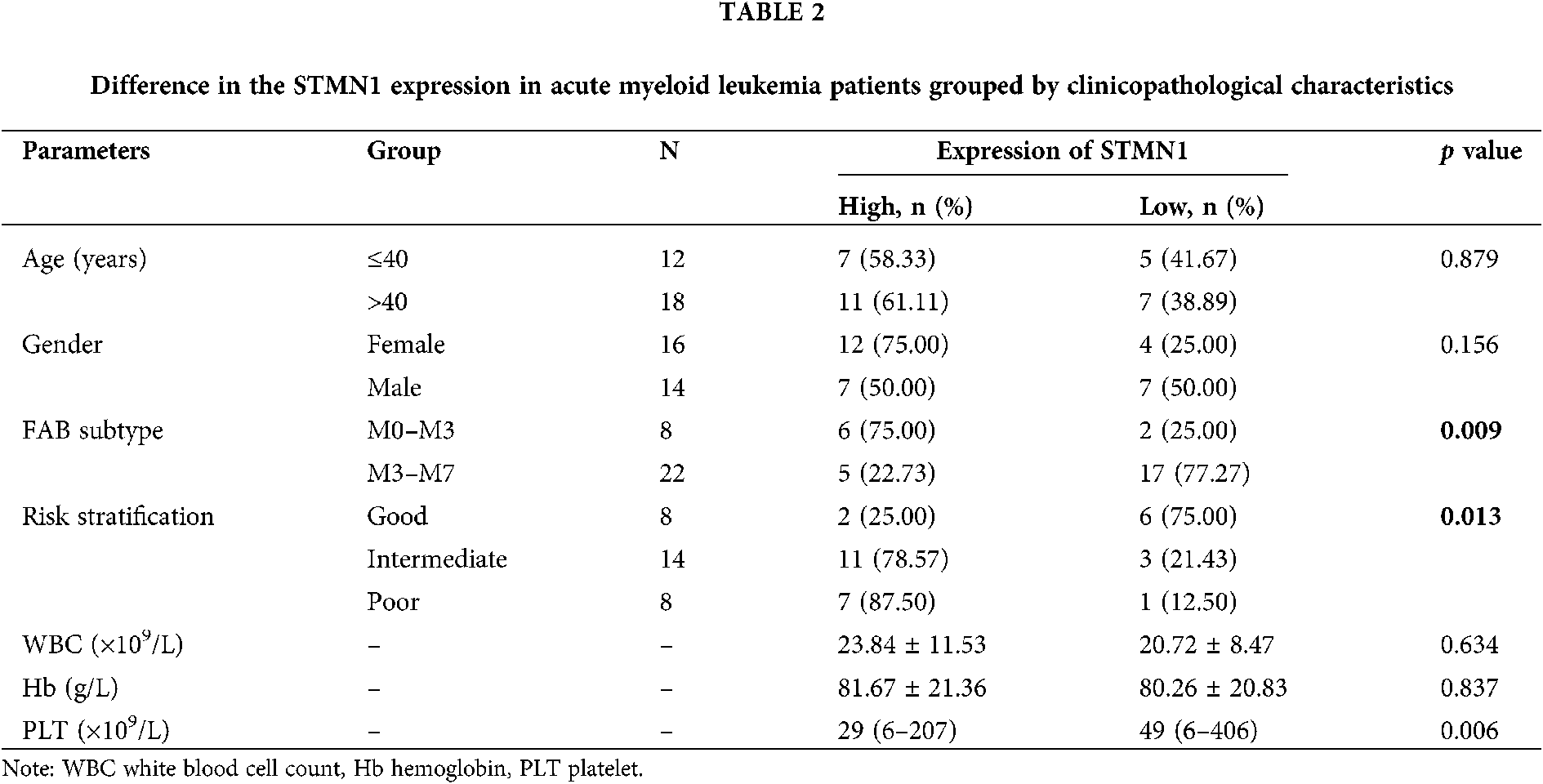
We found that STMN1 expression was not related to patient age, sex, or white blood cell (WBC) (p > 0.05). We next examined whether STMN1 was correlated with the patient’s prognosis and found that STMN1 levels were positively correlated with a patient’s rench-American-British (FAB) classification and risk stratification (Tab. 2). Moreover, a high level of SMTN1 expression was found to correlate with shorter disease-free survival and overall survival time (Figs. 2A and 2B).These results indicated that STMN1, as an independent prognostic factor, was adversely associated with the clinical prognosis of AML patients.
Overexpression of STMN1 induces cell proliferation and regulates cell cycle progression
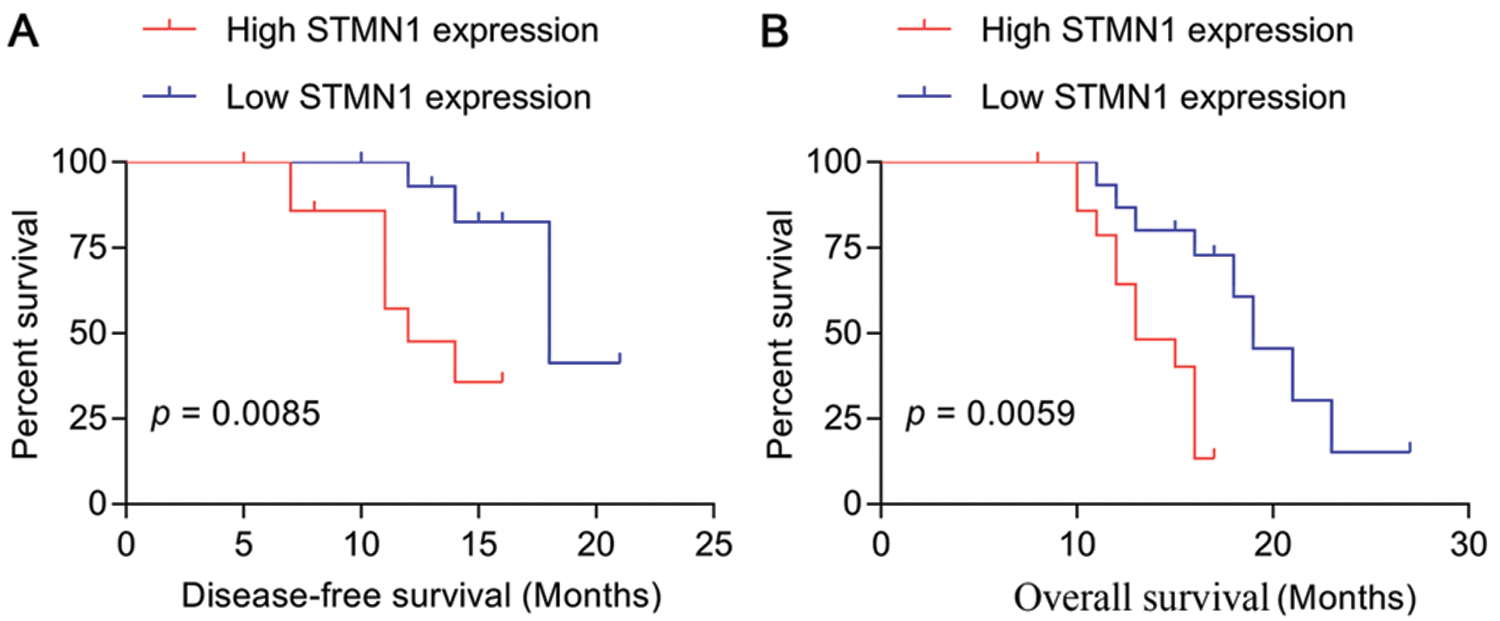
Figure 2: STMN1 expression was related to disease-free survival and overall survival times.
(A and B) The disease-free survival and overall survival times were summarized by survival curves.
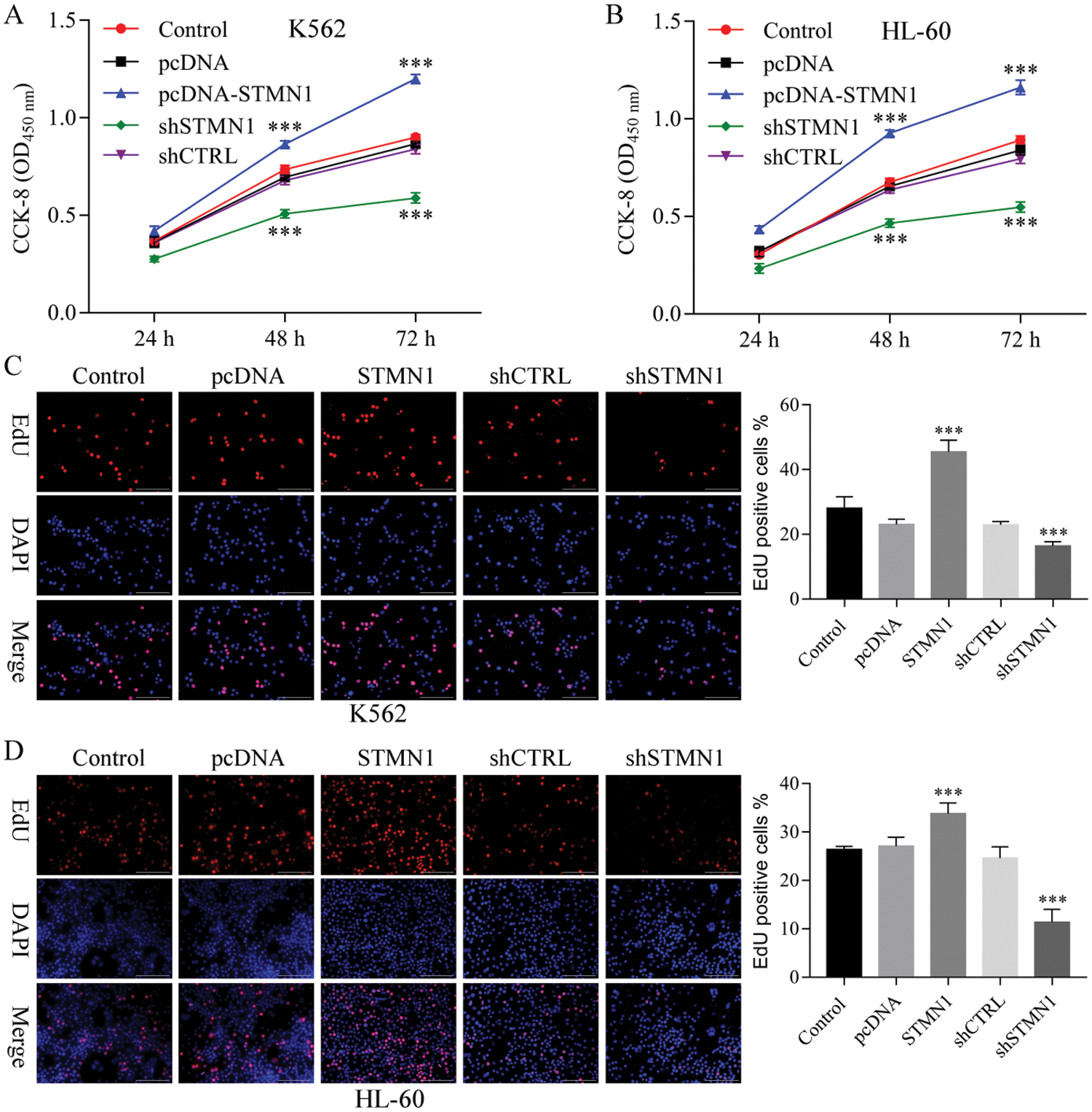
Figure 3: STMN1 induced cell proliferation.
(A and B) K562 cells (A) or HL60 cells (B) were seeded into the wells of a 96-well plate and transfected with plasmid. OD450 values were determined at 24, 48, and 72 h posttransfection. (C and D) K562 cells (C) or HL60 cells (D) were transfected with the indicated plasmid were fixed and stained with EdU. The percentage of EdUpositive cells was calculated after examining 20 fields for each group (number of EdU-positive cells/total cells × 100%). Results are based on data obtained from three independent experiments. ***p < 0.001, compared with control.
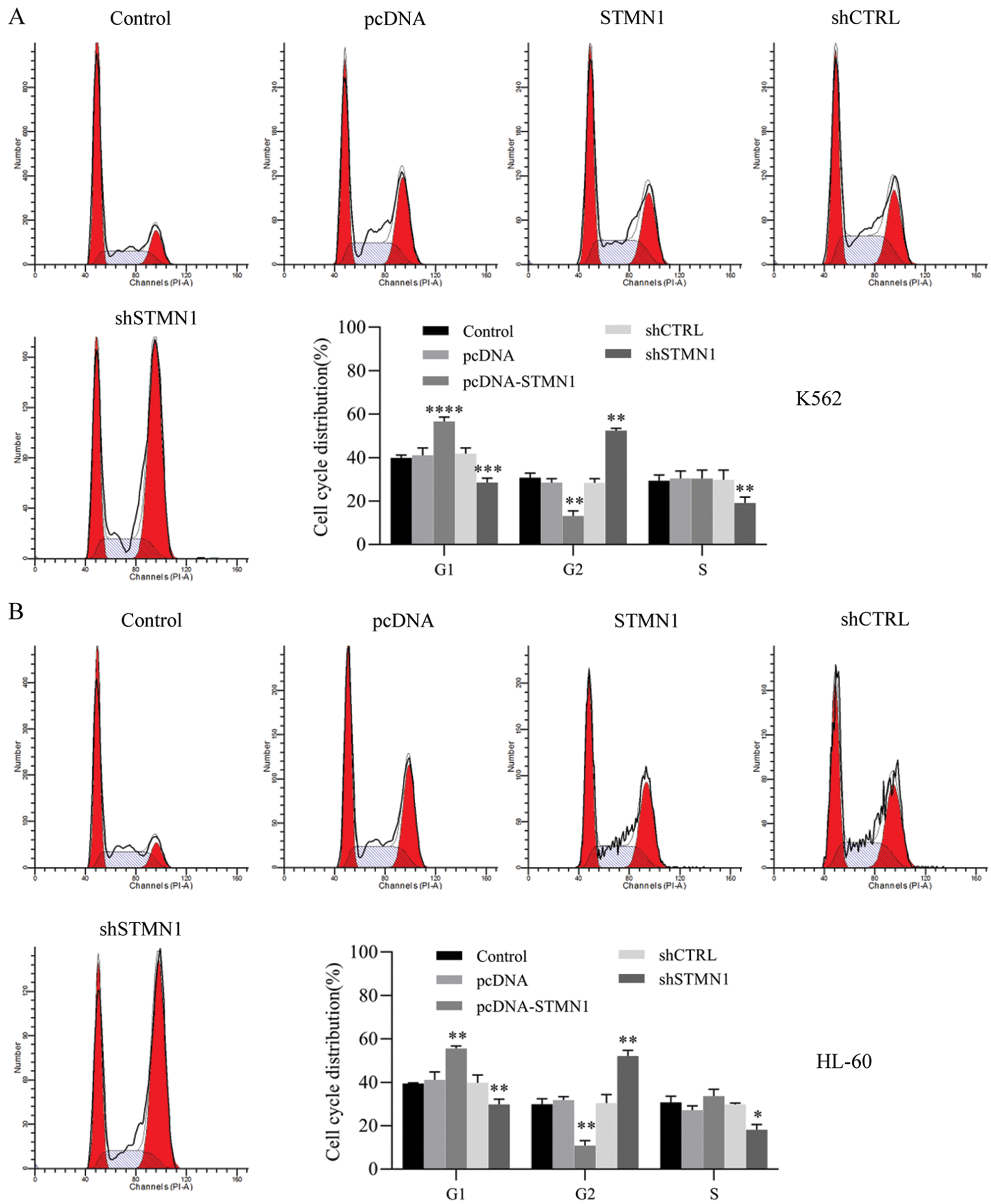
Figure 4: Inhibition of STMN1 induced G2/M phase arrest.
(A and B) K562 cells (A) or HL60 cells (B) were transfected with the indicated, fixed for PI staining, and analyzed by flow cytometry. Results are based on data obtained from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, compared with control.
To further examine the effects of STMN1 on AML progression, we used two human leukemia cell lines (K562 and HL60) to clarify the effect of STMN1 on cell proliferation. We found that overexpression of STMN1 induced the proliferation of both K562 and HL60 cells as detected by CCK8 (Figs. 3A and 3B). In addition, suppression of STMN1 by shRNA significantly reduced cell proliferation (Figs. 3A and 3B). Moreover, Edu staining results showed that overexpression of STMN1 increased the numbers of Edu-positive cells, while inhibition of STMN1 decreased the numbers of Edu-positive K562 and HL60 cells (Figs. 3C and 3D).
Next, we examined the cell cycle distribution induced by STMN1. Overexpression of STMN1 induced G1 phase arrest and suppression of STMN1 accumulations ofthe G2 phase in both K562 and HL60 cells (Figs. 4A and 4B).
These data indicated that overexpression of STMN1 enhanced cell proliferation and suppression of STMN1 reduced cell proliferation. In addition, cells that overexpressed STMN1 were arrested in the G1 phase of cell division, while cells with suppressed levels of STMN1 expression were arrested in the G2/M phase.
Inhibition of STMN1 enhances cell apoptosis
Results of Annexin V assays revealed that when compared with control (shCTRL), suppression of STMN1 significantly increased the rate of apoptosis in HL60 and K562 cells from 21.2% to 43.6% and from 15.9% to 33.2%, respectively (early and late apoptosis, the sum of the second and fourth quadrants). We also found that overexpression of STMN1 reduced the apoptosis rate to 50% in both K562 and HL60 cells (Figs. 5A and 5B).
Moreover, we performed TUNEL assays to further confirm the effects of STMN1 on cell apoptosis. As shown in (Figs. 6A and 6B), inhibition of STMN1 increased the numbers of TUNEL-positive cells among populations of both HL60 and K562 cells, and overexpression of STMN1 reduced the numbers of TUNEL-positive cells in both cell populations (Figs. 6A and 6B). These data indicated that suppression of STMN1 induced a significant increase in apoptosis.
STMN1 activates thePI3K/Akt pathway
To help identify the signal pathway involved in STMN1-induced cell proliferation and apoptosis reduction, we detected the expression of phosphorylated PI3K and Akt. Western blot analysis revealed that STMN1 activated the PI3K/Akt pathway, while inhibition of STMN1 reduced the phosphorylation of PI3K/Akt (Figs. 7A–7D). We next examined the levels of Caspase3, Bcl-2, Bax, and PTEN expression, which are related to cell apoptosis. Our results showed that overexpression of STMN1 enhanced Bcl-2 expression and reduced the levels of Bax, PTEN, and caspase-3 expression (Figs. 7A–7D). Additionally, suppression of STMN1 induced the levels of the proapoptotic factor Bax and reduced levels of anti-apoptotic factor Bcl-2 (Figs. 7A–7D). Furthermore, we found that the cellular levels of cell cycle-related protein cyclin B1 accumulated, suggesting a delay in cell cycle progression in STMN1 suppressed cells. As a regulator of the cell cycle through the G1 to S phase, the levels of cyclin D1 increased in STMN1 overexpressing cells and decreased in STMN1 repressed cells (Figs. 7A–7D).
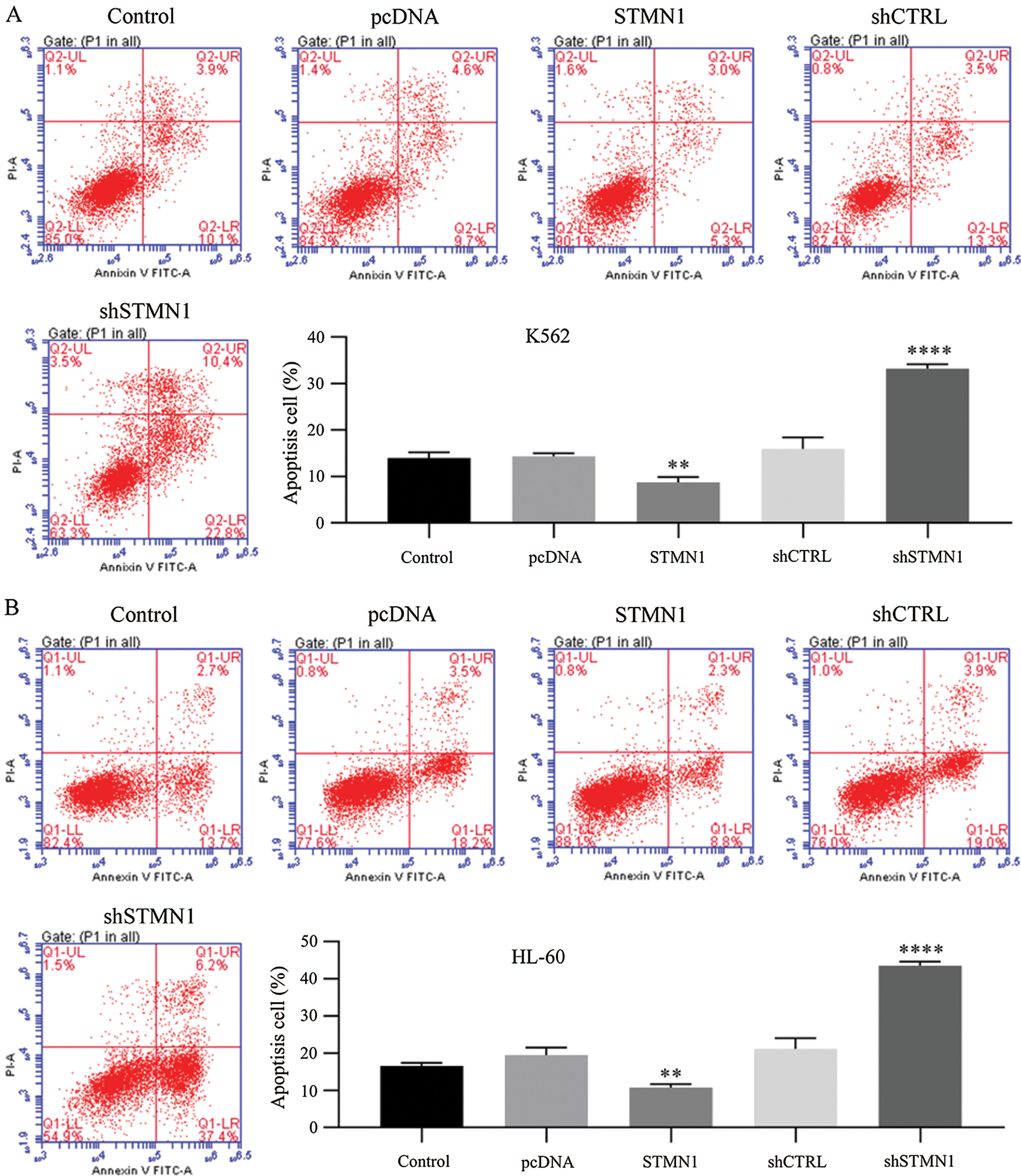
Figure 5: Inhibition of STMN1 enhanced cell apoptosis.
(A and B) K562 cells (A) or HL60 cells (B) were transfected with the indicated plasmid and collected for Annexin V staining. The percentage of apoptotic cells was calculated based on the second and fourth quadrants in the Annexin V assay. Results are based on data obtained from three independent experiments. **p < 0.01, ****p < 0.001, compared with control.
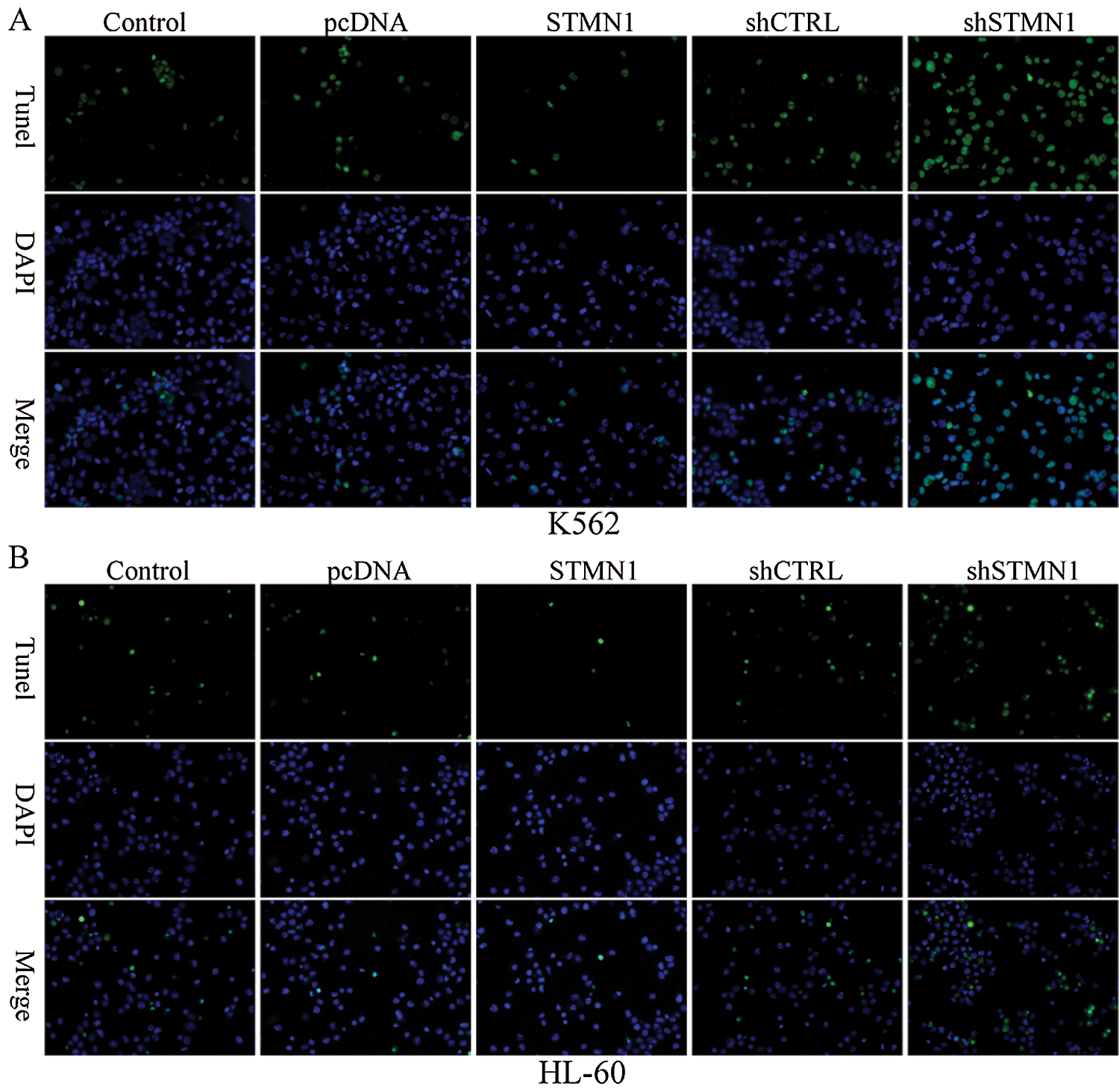
Figure 6: Inhibition of STMN1 enhances cell apoptosis.
K562 cells (A) or HL60 cells (B) were transfected with the indicated plasmid and fixed for TUENL staining. The respective images are shown.
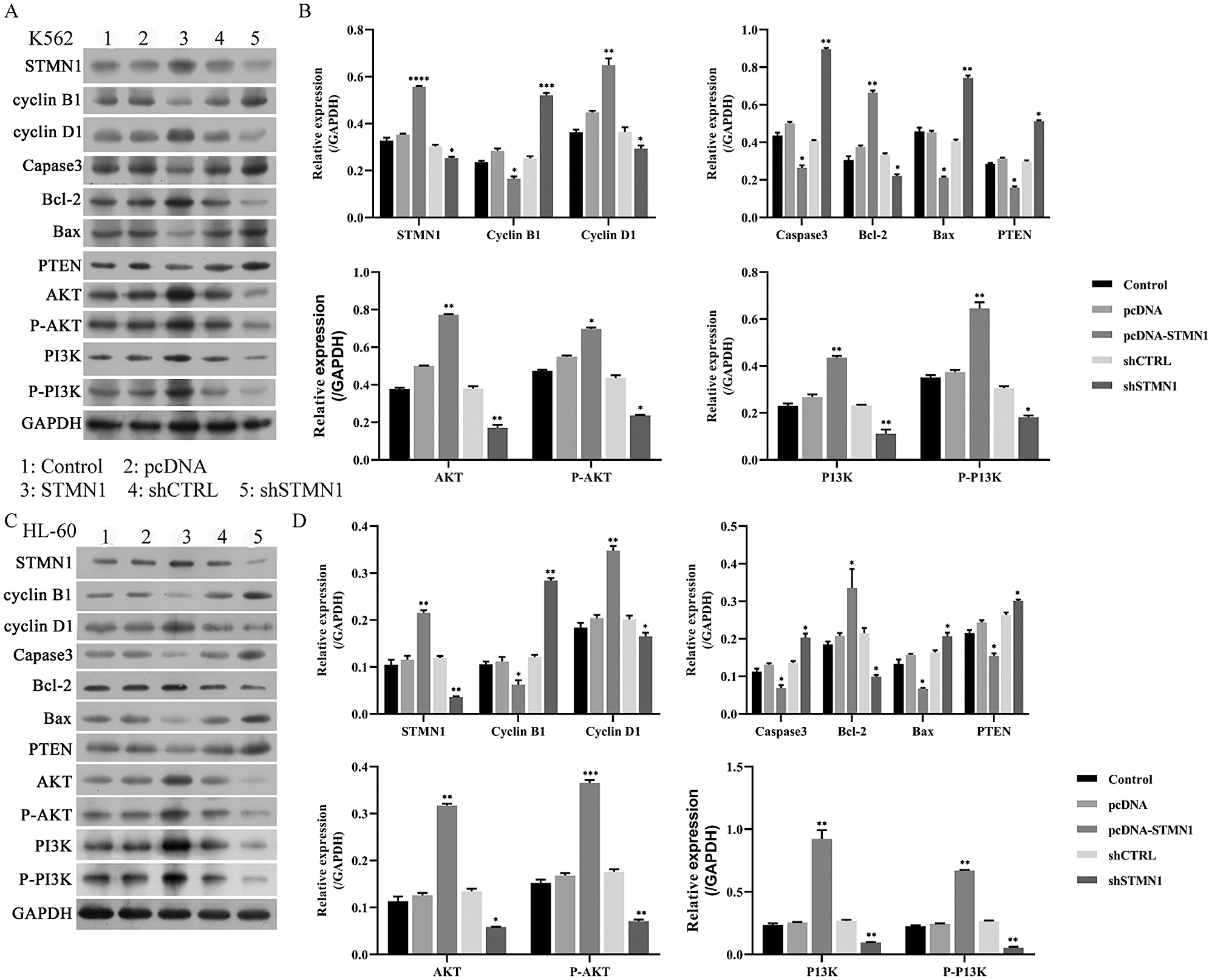
Figure 7: STMN1 activated the PI3K/Akt pathway.
(A–D) K562 cells or HL60 cells were transfected with the plasmid, and their cellular levels of STMN1, cyclin B1, cyclin D1, Caspase 3, Bcl-2, Bax, PTEN, AKT, p-AKT, p-PI3K, PI3K were determined by Western blot (A and C). GAPDH served as a loading control. The relative levels of expression are based on data obtained from three independent experiments (B and D). A: Control, B: pCDNA, C: pCDNA-STMN1, D: shCTRL, E: shSTMN1. *p < 0.05, **p < 0.01, ***p < 0.001, compared with control.
We found that STMN1 was more highly expressed in the PBMCs and bone marrow of AML patients than in healthy control subjects. Overexpression of STMN1 induced cell proliferation and reduced cell apoptosis, while suppression of STMN1 inhibited cell proliferation and enhanced cell apoptosis. Furthermore, suppression of STMN1 dysregulated the PI3K/Akt pathway and promoted the cellular expression of Caspases3 and the pro-apoptotic factor Bax, but reduced the levels of the anti-apoptotic factor, Bcl-2. Thus, we showed that STMN1 playedpromotive roles in AML.
STMN1, a phosphoprotein, belongs to the stathmin family of proteins and helps to mediate microtubule dynamics by destabilizing microtubules (Segerman et al., 2003). STMN1 is highly expressed in many neoplasms and closely correlates with the malignancy of tumors (Sherbet and Cajone, 2005). STMN1 was recently found to be highly expressed in AML patients (Rana et al., 2008; Handschuh et al., 2018). Here, we showed that the levels of STMN1 expression were significantly elevated in the bone marrow and PBMCs of AML patients when compared with healthy control subjects. Moreover, STMN1 levels were found to be associated with an advanced tumor stage (FAB subtypes) and a patient’s risk stratification. More importantly, an increased level of SMTN1 was associated with a shorter disease-free survival time and overall survival time. We thus showed that STMN1 levels were adversely associated with the clinical prognosis of AML patients. STMN1 has been found to be a potential prognostic marker for patients with different types of tumors, including colon cancer (Zhang et al., 2016), gastric cancer (Bai et al., 2017), pancreatic ductal adenocarcinoma (Li et al., 2015), meningioma (Wang et al., 2017a), gallbladder carcinoma (Bo et al., 2017), osteosarcoma (Zhao et al., 2018), esophageal squamous cell carcinoma (Ni et al., 2017), cholangiocarcinoma (Watanabe et al., 2014), hepatoma (Hsieh et al., 2010) and non-small cell lung cancer (Nie et al., 2015; Wang et al., 2017b). Our data and previous reports indicated that STMN1 may not be only a prognostic marker for solid cancer but could also serve as a prognostic marker for patients with leukemia.
Uncontrolled proliferation is a key hall mark of tumorigenesis. STMN1 plays an essential role in the dynamics of the mitotic spindle, which allows chromosome segregation and cell division (Cajone and Sherbet, 1999). STMN1 also regulates cell proliferation by controlling cell cycle progression (Lu et al., 2014). In many types of tumors, STMN1 enhances cell proliferation and accelerates the cell cycle (Biaoxue et al., 2016). Previous findings showed that inhibition of STMN1 led to an accumulation of G2/M phase cells, and a decrease in the proliferation of K562 leukemic cells (Luo et al., 1994; Marklund et al., 1996). Here, we found that STMN1 enhanced cell proliferation in two human leukemia cell lines (K562 and HL60), and suppression of STMN1 induced G2/M arrest. Moreover, the expression of cyclin D1, a regulator of the cell cycle through G1 to S (Stacey, 2003), was increased in STMN1 overexpressing cells and decreased in STMN1 repressed cells. Cyclin B1 is one of the main protein kinases that becomes activated and serves as a master regulator of the M phase transition during the cell cycle (Schnittger and De Veylder, 2018). We found that inhibition of STMN1 arrested cells at the G2/M phase resulted in an accumulation of cyclin B1 in AML cells. Our results support previous roles proposed for STMN1 in leukemia and suggest that STMN1 might be a pivotal factor that contributes to leukemia progression.
Escape from apoptosis is an essential step for malignant tumor progression (Hanahan and Weinberg, 2011). The Bcl-2 protein family, which includes anti-apoptotic proteins and pro-apoptotic proteins, controls activation of the intrinsic apoptosis pathway (Del Poeta et al., 2003; Ashkenazi et al., 2017). In the present study, we showed that inhibition of STMN1 caused a significant amount of apoptosis, while overexpression of STMN1 repressed cell apoptosis in AML cells. We also found that inhibition of STMN1 induced the expression of pro-apoptotic factor Bax and reduced the levels of anti-apoptotic factor Bcl-2, suggesting that inhibition of STMN1 activated the intrinsic apoptosis pathway in AML. Moreover, Caspase-3 activation caused the final destruction of cells. We showed that the level of Caspase-3 in HL60 and K562 cells were enhanced by suppressing STMN1 expression, suggesting that inhibition of STMN1 induced by Caspase-3 activation may cause apoptosis.
The PI3K/Akt pathway is abnormally upregulated in cancers, including AML, and plays an essential role in AML progression (Park et al., 2010; Nepstad et al., 2020). PI3K signaling regulates various cellular processes, such as the signaling required for cell proliferation, apoptosis, migration, glucose metabolism, and angiogenesis, which all participated in cancer progression (Vanhaesebroeck et al., 2010). The PI3K/Akt/mTOR has been found to be activated in >60% of AML patients and is associated with decreased overall survival time (Xu et al., 2003; Tamburini et al., 2007). After activated by the PI3K pathway, Akt phosphorylates a variety of downstream substrates that play roles in regulating tumorigenesis (Manning and Cantley, 2007). A recent study showed that the PI3K pathway mediates STMN1 expression, and STMN1 levels are related to tumor recurrence (Jiang et al., 2018). Additionally, STMN1 expression is a confirmed marker of activation of the oncogenic PI3K-AKT-mTOR pathway (Saal et al., 2007; Andersen et al., 2010; Trovik et al., 2010; Karst et al., 2011; Howitt et al., 2013). In this study, we revealed that STMN1 activates PI3K/Akt pathway, while inhibition of STMN1 reduces the phosphorylation of PI3K/Akt. Therefore, we provided evidence that STMN1 induces activation of the PI3K pathway in AML.
In conclusion, we found that the levels of STMN1 expression in PBMCs and bone marrow from AML patients were higher than those levels in healthy controls subjects. We also found that a high level of SMTN1 expression was related to shorter disease-free survival and overall survival times. Mechanistically, STMN1 increased the proliferation rate and decreased the apoptosis rate of AML cells by activating the PI3K/Akt pathway. STMN1 might serve as a new prognostic marker or therapeutic target for AML.
Availability of Data and Materials: The datasets used during the current study are available from the corresponding author on reasonable request.
Author Contribution: The authors confirm contribution to the paper as follows: Study conception and design: Peng Yang; data collection: Peng Yang and Zhiying Zou; analysis and interpretation of results: Zhiying Zou and Xuling Gao; draft manuscript preparation: Peng YANG and Xu-ling Gao. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: The study protocol was approved by the Institute Research Ethics Committee at the Fourth People’s Hospital of Shenyang at 25 August 2019, Approval No. 201908251.
Funding Statement: The author(s) received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Andersen JN, Sathyanarayanan S, Di Bacco A, Chi A, Zhang T, Chen AH, Dolinski B, Kraus M, Roberts B, Arthur W, Klinghoffer RA, Gargano D, Li L, Feldman I, Lynch B, Rush J, Hendrickson RC, Blume-Jensen P, Paweletz CP (2010). Pathway-based identification of biomarkers for targeted therapeutics: Personalized oncology with PI3K pathway inhibitors. Science Translational Medicine 2: 43ra55. [Google Scholar]
Ashkenazi A, Fairbrother WJ, Leverson JD, Souers AJ (2017). From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors. Nature Reviews Drug Discovery 16: 273–284. DOI 10.1038/nrd.2016.253. [Google Scholar] [CrossRef]
Bai T, Yokobori T, Altan B, Ide M, Mochiki E, Yanai M, Kimura A, Kogure N, Yanoma T, Suzuki M, Bao P, Kaira K, Asao T, Katayama A, Handa T, Gombodorj N, Nishiyama M, Oyama T, Ogata K, Kuwano H (2017). High STMN1 level is associated with chemo-resistance and poor prognosis in gastric cancer patients. British Journal of Cancer 116: 1177–1185. DOI 10.1038/bjc.2017.76. [Google Scholar] [CrossRef]
Bao P, Yokobori T, Altan B, Iijima M, Azuma Y, Onozato R, Yajima T, Watanabe A, Mogi A, Shimizu K, Nagashima T, Ohtaki Y, Obayashi K, Nakazawa S, Bai T, Kawabata-Iwakawa R, Asao T, Kaira K, Nishiyama M, Kuwano H (2017). High STMN1 expression is associated with cancer progression and chemo-resistance in lung squamous cell carcinoma. Annals of Surgical Oncology 24: 4017–4024. DOI 10.1245/s10434-017-6083-0. [Google Scholar] [CrossRef]
Belletti B, Baldassarre G (2011). Stathmin: A protein with many tasks. New biomarker and potential target in cancer. Expert Opinion on Therapeutic Targets 15: 1249–1266. DOI 10.1517/14728222.2011.620951. [Google Scholar] [CrossRef]
Rong BX, Liu H, Gao WL, Yang SY (2016). Overexpression of stathmin promotes metastasis and growth of malignant solid tumors: A systemic review and meta-analysis. Oncotarget 7: 78994–79007. DOI 10.18632/oncotarget.12982. [Google Scholar] [CrossRef]
Bo X, Wang J, Fu Q, Wang Y, Liu H, Xu J (2017). Stathmin 1 expression predicts prognosis and benefits from adjuvant chemotherapy in patients with gallbladder carcinoma. Oncotarget 8: 108548–108555. DOI 10.18632/oncotarget.19625. [Google Scholar] [CrossRef]
Cajone F, Sherbet GV (1999). Stathmin is involved in S100A4-mediated regulation of cell cycle progression. Clinical & Experimental Metastasis 17: 865–871. DOI 10.1023/A:1006778804532. [Google Scholar] [CrossRef]
Creutzig U, van den Heuvel-Eibrink MM, Gibson B, Dworzak MN, Adachi S, de Bont E, Harbott J, Hasle H, Johnston D, Kinoshita A, Lehrnbecher T, Leverger G, Mejstrikova E, Meshinchi S, Pession A, Raimondi SC, Sung L, Stary J, Zwaan CM, Kaspers GJL, Reinhardt D (2012). Diagnosis and management of acute myeloid leukemia in children and adolescents: Recommendations from an international expert panel. Blood 120: 3187–3205. DOI 10.1182/blood-2012-03-362608. [Google Scholar] [CrossRef]
Datta SR, Brunet A, Greenberg ME (1999). Cellular survival: A play in three Akts. Genes and Development 13: 2905–2927. DOI 10.1101/gad.13.22.2905. [Google Scholar] [CrossRef]
De Kouchkovsky I, Abdul-Hay M (2016). Acute myeloid leukemia: A comprehensive review and 2016 update. Blood Cancer Journal 6: e441–e441. DOI 10.1038/bcj.2016.50. [Google Scholar] [CrossRef]
Del Poeta G, Venditti A, Del Principe MI, Maurillo L, Buccisano F, Tamburini A, Cox MC, Franchi A, Bruno A, Mazzone C, Panetta P, Suppo G, Masi M, Amadori S (2003). Amount of spontaneous apoptosis detected by Bax/Bcl-2 ratio predicts outcome in acute myeloid leukemia (AML). Blood 101: 2125–2131. DOI 10.1182/blood-2002-06-1714. [Google Scholar] [CrossRef]
Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, Thompson CB (2004). Akt stimulates aerobic glycolysis in cancer cells. Cancer Research 64: 3892–3899. DOI 10.1158/0008-5472.CAN-03-2904. [Google Scholar] [CrossRef]
Gavet O, Ozon S, Manceau V, Lawler S, Curmi P, Sobel A (1998). The stathmin phosphoprotein family: Intracellular localization and effects on the microtubule network. Journal of Cell Science 111: 3333–3346. [Google Scholar]
Ghiaur G, Wroblewski M, Loges S (2015). Acute myelogenous leukemia and its microenvironment: A molecular conversation. Seminars in Hematology 52: 200–206. DOI 10.1053/j.seminhematol.2015.03.003. [Google Scholar] [CrossRef]
Hanahan D, Weinberg RA (2011). Hallmarks of cancer: The next generation. Cell 144: 646–674. DOI 10.1016/j.cell.2011.02.013. [Google Scholar] [CrossRef]
Handschuh L, Kaźmierczak M, Milewski MC, Góralski M, Łuczak M, Wojtaszewska M, Uszczyńska-Ratajczak B, Lewandowski K, Komarnicki M, Figlerowicz M (2018). Gene expression profiling of acute myeloid leukemia samples from adult patients with AML-M1 and -M2 through boutique microarrays, real-time PCR and droplet digital PCR. International Journal of Oncology 52: 656–678. [Google Scholar]
Honnappa S, Cutting B, Jahnke W, Seelig J, Steinmetz MO (2003). Thermodynamics of the Op18/stathmin-tubulin interaction. Journal of Biological Chemistry 278: 38926–38934. DOI 10.1074/jbc.M305546200. [Google Scholar] [CrossRef]
Horwitz SB, Shen HJ, He L, Dittmar P, Neef R, Chen J, Schubart UK (1997). The microtubule-destabilizing activity of metablastin (p19) is controlled by phosphorylation. Journal of Biological Chemistry 272: 8129–8132. DOI 10.1074/jbc.272.13.8129. [Google Scholar] [CrossRef]
Howitt BE, Nucci MR, Drapkin R, Crum CP, Hirsch MS (2013). Stathmin-1 expression as a complement to p16 helps identify high-grade cervical intraepithelial neoplasia with increased specificity. American Journal of Surgical Pathology 37: 89–97. DOI 10.1097/PAS.0b013e3182753f5a. [Google Scholar] [CrossRef]
Hsieh SY, Huang SF, Yu MC, Yeh TS, Chen TC, Lin YJ, Chang CJ, Sung CM, Lee YL, Hsu CY (2010). Stathmin1 overexpression associated with polyploidy, tumor-cell invasion, early recurrence, and poor prognosis in human hepatoma. Molecular Carcinogenesis 49: 476–487. DOI 10.1002/mc.20627. [Google Scholar] [CrossRef]
Jiang W, Huang S, Song L, Wang Z (2018). STMN1, a prognostic predictor of esophageal squamous cell carcinoma, is a marker of the activation of the PI3K pathway. Oncology Reports 39: 834–842. [Google Scholar]
Karst AM, Levanon K, Duraisamy S, Liu JF, Hirsch MS, Hecht JL, Drapkin R (2011). Stathmin 1, a marker of PI3K pathway activation and regulator of microtubule dynamics, is expressed in early pelvic serous carcinomas. Gynecologic Oncology 123: 5–12. DOI 10.1016/j.ygyno.2011.05.021. [Google Scholar] [CrossRef]
Li J, Hu G, Kong F, Wu K, Song K, He J, Sun W (2015). Elevated STMN1 expression correlates with poor prognosis in patients with pancreatic ductal adenocarcinoma. Pathology & Oncology Research 21: 1013–1020. DOI 10.1007/s12253-015-9930-y. [Google Scholar] [CrossRef]
Liang J, Slingerland JM (2003). Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle 2: 339–345. DOI 10.4161/cc.2.4.433. [Google Scholar] [CrossRef]
Liu JJ, Liu WD, Yang HZ, Zhang Y, Fang ZG, Liu PQ, Lin DJ, Xiao RZ, Hu Y, Wang CZ, Li XD, He Y, Huang RW (2010). Inactivation of PI3k/Akt signaling pathway and activation of caspase-3 are involved in tanshinone I-induced apoptosis in myeloid leukemia cells in vitro. Annals of Hematology 89: 1089–1097. DOI 10.1007/s00277-010-0996-z. [Google Scholar] [CrossRef]
Lu Y, Liu C, Xu YF, Cheng H, Shi S, Wu CT, Yu XJ (2014). Stathmin destabilizing microtubule dynamics promotes malignant potential in cancer cells by epithelial-mesenchymal transition. Hepatobiliary & Pancreatic Diseases International 13: 386–394. DOI 10.1016/S1499-3872(14)60038-2. [Google Scholar] [CrossRef]
Luo XN, Mookerjee B, Ferrari A, Mistry S, Atweh GF (1994). Regulation of phosphoprotein p18 in leukemic cells. Cell cycle regulated phosphorylation by p34cdc2 kinase. Journal of Biological Chemistry 269: 10312–10318. DOI 10.1016/S0021-9258(17)34062-0. [Google Scholar] [CrossRef]
Manning BD, Cantley LC (2007). AKT/PKB signaling: Navigating downstream. Cell 129: 1261–1274. DOI 10.1016/j.cell.2007.06.009. [Google Scholar] [CrossRef]
Marklund U, Larsson N, Gradin HM, Brattsand G, Gullberg M (1996). Oncoprotein 18 is a phosphorylation-responsive regulator of microtubule dynamics. EMBO Journal 15: 5290–5298. [Google Scholar]
Min YH, Eom JI, Cheong JW, Maeng HO, Kim JY, Jeung HK, Lee ST, Lee MH, Hahn JS, Ko YW (2003). Constitutive phosphorylation of Akt/PKB protein in acute myeloid leukemia: Its significance as a prognostic variable. Leukemia 17: 995–997. DOI 10.1038/sj.leu.2402874. [Google Scholar] [CrossRef]
Nepstad I, Hatfield KJ, Grønningsæter IS, Aasebø E, Hernandez-Valladares M, Hagen KM, Rye KP, Berven FS, Selheim F, Reikvam H, Bruserud Ø (2019). Effects of insulin and pathway inhibitors on the PI3K-Akt-mTOR phosphorylation profile in acute myeloid leukemia cells. Signal Transduction and Targeted Therapy 4: 20. DOI 10.1038/s41392-019-0050-0. [Google Scholar] [CrossRef]
Nepstad I, Hatfield KJ, Grønningsæter IS, Reikvam H (2020). The PI3K-Akt-mTOR signaling pathway in human acute myeloid leukemia (AML) cells. International Journal of Molecular Sciences 21: 2907. DOI 10.3390/ijms21082907. [Google Scholar] [CrossRef]
Nepstad I, Hatfield KJ, Tvedt THA, Reikvam H, Bruserud Ø (2018). Clonal heterogeneity reflected by PI3K-AKT-mTOR signaling in human acute myeloid leukemia cells and its association with adverse prognosis. Cancers 10: 332. DOI 10.3390/cancers10090332. [Google Scholar] [CrossRef]
Ni PZ, He JZ, Wu ZY, Ji X, Chen LQ, Xu XE, Liao LD, Wu JY, Li EM, Xu LY (2017). Overexpression of Stathmin 1 correlates with poor prognosis and promotes cell migration and proliferation in oesophageal squamous cell carcinoma. Oncology Reports 38: 3608–3618. [Google Scholar]
Nie W, Xu MD, Gan L, Huang H, Xiu Q, Li B (2015). Overexpression of Stathmin 1 is a poor prognostic biomarker in non-small cell lung cancer. Laboratory Investigation 95: 56–64. DOI 10.1038/labinvest.2014.124. [Google Scholar] [CrossRef]
Park S, Chapuis N, Tamburini J, Bardet V, Cornillet-Lefebvre P, Willems L, Green A, Mayeux P, Lacombe C, Bouscary D (2010). Role of the PI3K/AKT and mTOR signaling pathways in acute myeloid leukemia. Haematologica 95: 819–828. DOI 10.3324/haematol.2009.013797. [Google Scholar] [CrossRef]
Porter AG, Jänicke RU (1999). Emerging roles of caspase-3 in apoptosis. Cell Death & Differentiation 6: 99–104. DOI 10.1038/sj.cdd.4400476. [Google Scholar] [CrossRef]
Rana S, Maples PB, Senzer N, Nemunaitis J (2008). Stathmin 1: A novel therapeutic target for anticancer activity. Expert Review of Anticancer Therapy 8: 1461–1470. DOI 10.1586/14737140.8.9.1461. [Google Scholar] [CrossRef]
Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, Maurer M, Koujak S, Ferrando AA, Malmström P, Memeo L, Isola J, Bendahl PO, Rosen N, Hibshoosh H, Ringnér M, Borg Å, Parsons R (2007). Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proceedings of the National Academy of Sciences of the United States of America 104: 7564–7569. DOI 10.1073/pnas.0702507104. [Google Scholar] [CrossRef]
Sapon-Cousineau V, Sapon-Cousineau S, Assouline S (2020). PI3K inhibitors and their role as novel agents for targeted therapy in lymphoma. Current Treatment Options in Oncology 21: 51. DOI 10.1007/s11864-020-00746-8. [Google Scholar] [CrossRef]
Schnittger A, De Veylder L (2018). The dual face of cyclin B1. Trends in Plant Science 23: 475–478. DOI 10.1016/j.tplants.2018.03.015. [Google Scholar] [CrossRef]
Segerman B, Holmfeldt P, Morabito J, Cassimeris L, Gullberg M (2003). Autonomous and phosphorylation-responsive microtubule-regulating activities of the N-terminus of Op18/stathmin. Journal of Cell Science 116: 197–205. DOI 10.1242/jcs.00205. [Google Scholar] [CrossRef]
Sherbet GV, Cajone F (2005). Stathmin in cell proliferation and cancer progression. Cancer Genomics Proteomics 2: 227–237. [Google Scholar]
Stacey DW (2003). Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Current Opinion in Cell Biology 15: 158–163. DOI 10.1016/S0955-0674(03)00008-5. [Google Scholar] [CrossRef]
Steinmetz MO (2007). Structure and thermodynamics of the tubulin–stathmin interaction. Journal of Structural Biology 158: 137–147. DOI 10.1016/j.jsb.2006.07.018. [Google Scholar] [CrossRef]
Tamburini J, Elie C, Bardet V, Chapuis N, Park S, Broët P, Cornillet-Lefebvre P, Lioure B, Ugo V, Blanchet O, Ifrah N, Witz F, Dreyfus F, Mayeux P, Lacombe C, Bouscary D (2007). Constitutive phosphoinositide 3-kinase/Akt activation represents a favorable prognostic factor in de novo acute myelogenous leukemia patients. Blood 110: 1025–1028. DOI 10.1182/blood-2006-12-061283. [Google Scholar] [CrossRef]
Trovik J, Wik E, Stefansson I, Carter SL, Beroukhim R, Øyan AM, Kalland KH, Akslen LA, Salvesen HB (2010). Stathmin is superior to AKT and phospho-AKT staining for the detection of phosphoinositide 3-kinase activation and aggressive endometrial cancer. Histopathology 57: 641–646. DOI 10.1111/j.1365-2559.2010.03661.x. [Google Scholar] [CrossRef]
Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B (2010). The emerging mechanisms of isoform-specific PI3K signalling. Nature Reviews Molecular Cell Biology 11: 329–341. DOI 10.1038/nrm2882. [Google Scholar] [CrossRef]
Wang H, Li W, Wang G, Zhang S, Bie L (2017a). Overexpression of STMN1 is associated with the prognosis of meningioma patients. Neuroscience Letters 654: 1–5. DOI 10.1016/j.neulet.2017.06.020. [Google Scholar] [CrossRef]
Wang M, Li W, Xing X, Zhang D, Lei J, Li G (2017b). BRCA1 and STMN1 as prognostic markers in NSCLCs who received cisplatin-based adjuvant chemotherapy. Oncotarget 8: 80869–80877. DOI 10.18632/oncotarget.20715. [Google Scholar] [CrossRef]
Watanabe A, Suzuki H, Yokobori T, Tsukagoshi M, Altan B, Kubo N, Suzuki S, Araki K, Wada S, Kashiwabara K, Hosouchi Y, Kuwano H (2014). Stathmin1 regulates p27 expression, proliferation and drug resistance, resulting in poor clinical prognosis in cholangiocarcinoma. Cancer Science 105: 690–696. DOI 10.1111/cas.12417. [Google Scholar] [CrossRef]
Xu JP, Hu JD, Li J, Liu TB, Lin MH (2013). Expression and significance of stathmin1 in acute leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi 21: 1105–1110. [Google Scholar]
Xu Q, Simpson SE, Scialla TJ, Bagg A, Carroll M (2003). Survival of acute myeloid leukemia cells requires PI3 kinase activation. Blood 102: 972–980. DOI 10.1182/blood-2002-11-3429. [Google Scholar] [CrossRef]
Zhang D, Dai L, Yang Z, Wang X, LanNing Y (2019). Association of STMN1 with survival in solid tumors: A systematic review and meta-analysis. International Journal of Biological Markers 34: 108–116. DOI 10.1177/1724600819837210. [Google Scholar] [CrossRef]
Zhang HQ, Guo X, Guo SQ, Wang Q, Chen XQ, Ni XN, Guo LS (2016). STMN1 in colon cancer: Expression and prognosis in Chinese patients. European Review for Medical and Pharmacological Sciences 20: 2038–2044. [Google Scholar]
Zhao C, Li H, Wang L, Sun W (2018). An immunohistochemical study of Stathmin 1 expression in osteosarcoma shows an association with metastases and poor patient prognosis. Medical Science Monitor 24: 6070–6078. DOI 10.12659/MSM.910953. [Google Scholar] [CrossRef]
Zheng P, Liu YX, Chen L, Liu XH, Xiao ZQ, Zhao L, Li GQ, Zhou J, Ding YQ, Li JM (2010). Stathmin, a new target of PRL-3 identified by proteomic methods, plays a key role in progression and metastasis of colorectal cancer. Journal of Proteome Research 9: 4897–4905. DOI 10.1021/pr100712t. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |