DOI:10.32604/biocell.2022.021542

| BIOCELL DOI:10.32604/biocell.2022.021542 |  |
| Article |
Response of MaHMA2 gene expression and stress tolerance to zinc stress in mulberry (Morus alba L.)
1School of Biology and Technology, Jiangsu University of Science and Technology, Zhenjiang, 212018, China
2Department of Materials Science and Engineering, Suzhou University of Science and Technology, Suzhou, 215004, China
3Nanjing University of Finance & Economics, Nanjing, 210023, China
4Sericultural Research Institute, Anhui Academy of Agricultural Sciences, Hefei, 230061, China
*Address correspondence to: Mengdi Zhao, mdzhao@usts.edu.cn; Weiguo Zhao, wgzsri@126.com
#These authors contributed equally to this work
Received: 20 January 2022; Accepted: 03 March 2022
Abstract: HMA2 (heavy metal ATPase 2) plays a crucial role in extracellular and intracellular Zn2+ transport across biomembranes, maintaining ion homeostasis, and playing an important role in the normal physiological metabolism, growth, and development of plants. In our study, a novel HMA2 gene, named MaHMA2, was isolated and cloned from white mulberry (Morus alba L.). The gene sequence obtained was 1,342 bp long, with an open reading frame of 1,194 bp, encoding a protein of 397 amino acids, with a predicted molecular mass of 42.852 kD and an isoelectric point of 7.53. This protein belonged to the PIB-type ATPase transport protein family. We analyzed the expression of the MaHMA2 gene by quantitative real-time PCR. The results showed that the level of MaHMA2 gene expression decreased to a Zn concentration of 800 mg/kg. Malondialdehyde and proline levels increased and responded to increasing Zn when the MaHMA2 gene was silenced, whereas the activities of peroxidase and superoxide dismutase tended to increase in response to increasing Zn2+ ion stress concentrations but were lower in the gene-silenced plants. These findings suggested that the MaHMA2 gene played an active role in the tolerance response of mulberry to Zn stress.
Keywords: Mulberry; Zn stress; MaHMA2 gene; VGIS; Physiological; Biochemical
Zinc (Zn) plays an important role in many biological processes, including respiratory electron transport chain, photosynthesis, oxidative stress protection, hormone signaling, cell wall metabolism, and pollen formation (Rizwan et al., 2019). Zn Deficiencies or excess can have adverse effects on plants. Soil organic matter has a strong adsorption capacity for Zn, which significantly reduces Zn effectiveness. Zn bioavailability is reduced at a high soil pH and under other soil conditions, leading to Zn deficiency in the plant (Wu et al., 2020). Plants exhibit a range of injurious effects, such as inhibition of photosynthesis, reduced lignification, the inhibition of protein synthesis, reduced pollen viability, and decreased disease resistance in response to Zn-deficiency. Due to this host of injuries, plant leaves suffer chlorosis, susceptibility to deformation, slow growth and development, reduced yield, and poor tolerance to abiotic stresses. As a consequence of the rapid development of industrial, agricultural, and mining activities, heavy metal elements such as Zn are deposited in increasing quantities in soils, leading to heavy metal pollution; thus, endangering the growth and development of plants and animals and posing a threat to human health, and planting Zn-tolerant mulberry species is a very effective bioremediation measure (Qin et al., 2018). The Zn content of Chinese soils ranges from 3 to 790 mg/kg, with an average of 100 mg/kg, which is twice the world average, and the Zn content of soils near zinc mining areas is as high as 4025 mg/kg (Liu et al., 2020). Excessive Zn concentrations cause cycling between the reduced and oxidized Zn+ states of Zn ions to generate highly toxic hydroxyl radicals, which, in turn, damage macromolecules (Wu et al., 2016). Therefore, it is imperative that plants maintain Zn homeostasis for normal growth and development.
To maintain Zn homeostasis, plants have evolved a tightly regulated network to control the uptake and transport of Zn. Although excess Zn is generally highly toxic to plants, some plants can still grow normally in environments with considerable Zn2+ contamination through hyper-accumulation of Zn2+ in their tissues to levels toxic to most other plant spp. This suggests the existence/deployment of Zn-tolerance mechanisms in Zn-hyperaccumulating plants that allows them to survive in Zn2+-contaminated soils (Guzmán-Rangel et al., 2017). Studying Zn2+-tolerance mechanisms in plants is important for agricultural production and the phytoremediation of Zn2+-contaminated soils (Versieren et al., 2016).
The top priority in phytoremediation is to improve plant tolerance to heavy metals (Wu et al., 2021). This can be achieved through various routes, such as increasing the activity of stress-related antioxidant enzymes, promoting compartmentalization, and increasing the quantities of metal-chelating molecules synthesized. The heavy metal-transporting ATPases (HMAs) are transport proteins that actively transport heavy metal cations across membranes in a process driven by ATP hydrolysis. The HMA gene is widely distributed in plants, and HMA1, HMA3, and HMA4 have been identified and isolated from plants and shown to play important roles in plant responses to environmental stresses (Takahashi et al., 2012). The different HMA proteins are selective for specific heavy metal ion transport. For instance, HMA2-4 are Zn/Cd transporters, whereas HMA5-8 are Cu+/Ag+ transporters. The metal specificity of the HMA1 transporter is still not completely understood. The HMAs are predicted to be localized at the plasma membrane, chloroplast, Golgi membrane (HMA7), or tonoplast (HMA3) (Blindauer and Leszczyszyn, 2010; Romero-Isart and Vasák, 2002; Leszczyszyn et al., 2013). The gene sequences, protein structural domains, and conserved motif patterns among members of the different subgroups are closely related to the phylogenetic diversity, reflecting the fact that they all have different metal ion transport-related functions and differences in the specific heavy metal ions transported (Lee et al., 2007; Cun et al., 2014).
Virus-induced gene silencing (VIGS) is an RNA interference-based technique widely used to down-regulate various plant-specific transcripts and knocks-down the expression of target genes using modified plant viral genomes (Zhang et al., 2015). VIGS has been developed as a new reverse genetics technique for the rapid functional characterization of plant genes. So far, VIGS systems have been established with RNA viruses, DNA viruses, satellite viruses, and DNA satellite molecules as vectors (Geuten et al., 2013). Compared with transgenic plant techniques such as knockout and antisense, VIGS does not require the construction of transgenic plants. It has a short cycle time, is simple to perform, and obtains novel phenotypes quickly and at a low cost.
The mulberry plant (Morus alba L.) is native to central and northern China, and is now cultivated from the northeast to southwest provinces and regions of China (Zhao et al., 2007). The soil environment conditions in these areas are harsh and usually heavily overloaded with heavy metals, thus, providing a poor soil environment for the large-scale cultivation of mulberry. Although research on HMA gene expression in response to heavy metal stress has been reported in other plant spp., little is known about how mulberry plant HMA genes help in response to metal stress such as Zn (Imran et al., 2016). In this study, the response of MaHMA2 gene expression and stress tolerance to Zn stress in mulberry was investigated through a bioinformatics analysis and molecular approaches. This work serves as a foundation for understanding the MaHMA2 gene regulatory effect in mulberry under Zn stress and help breed mulberry variety to withstand heavy metal contaminated soils (Fan et al., 2018).
Plant materials and growth conditions
Morus alba seedlings were obtained from the National Mulberry Germplasm Nursery at Jiangsu University of Science and Technology, Zhenjiang, China. The seedlings were transplanted in pots containing vermiculite and loamy soil (pH 7.0). When the seedlings reached about 20 cm above soil, they were randomly divided into five groups. The groups were treated with ZnSO4·5H2O at concentrations of 0 (control), 100, 200, 400 and 800 mg/kg to represent different Zn stress treatments. Three biological replicates of each treatment group were set up and sampled simultaneously after five days of Zn treatment. For the VIGS suppression of the MaHMA2 gene, M. alba seeds were sown in seedling trays, and when the two cotyledons had emerged, they were injected with either recombinant virus pTRV2-MaHMA2, empty pTRV2 vector, or buffer. Thirty days after the inoculation, seedlings with uniform growth and condition were selected and transplanted into pots containing vermiculite and loamy soil (pH 7.0) with the ZnSO4·5H2O concentrations described above. The seedlings injected with buffer were set as the blank control, pTRV2 empty vector as the negative control and the recombinant virus pTRV2-MaHMA2 as the experimental group. A volume (100 mL) of Murashige & Skoog (MS) nutrient solution was added to each pot at a fixed time every day for five days and sampled at the same time point, with three biological replicates (three seedlings per replicate) of each group.
Primers for PCR, quantitative real-time PCR (qPCR), and vector construction were designed using OLIGO Primer Analysis Software v.7 (Molecular Biology Insights, Inc., USA) and based on the coding sequence (CDS) of the MaHMA2 gene (GenBank: MT444766.1) obtained from the transcriptome library. Nucleotide sequences were assembled using DNAStar software (DNAStar, USA). The BLAST online tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to compare and analyze sequence correlation data. Sequence comparisons were performed using the Clustal X 2.0 alignment program and DNAMAN software (version 8.0) (Thompson et al., 1997). Expasy online tool (http://web.expasy.org/compute_pi/) was used to calculate the molecular masses and predict the isoelectric points of the predicted proteins. The online SWISS-MODEL prediction software (http://swissmodel.expasy.org/) was used for the 3D structure studies of MaHMA2 (Zhao et al., 2011). MEGA6 software was used to construct phylogenetic trees using the maximum likelihood (ML) method (Tamura et al., 2013).
RNA isolation and cloning of the cDNA
Total RNA was extracted from fresh leaf samples using the RNAiso Plus reagent (Takara, China), according to the manufacturer’s protocol. The concentration and purity of total RNA were checked on a Nanodrop spectrophotometer, and its quality was examined by electrophoresis on 1.0% agarose gel. The extracted total RNA was reverse transcribed using PrimeScript™ RT reagent Kit (Takara, China), following the manufacturer’s instructions to obtain the cDNAs.
The target MaHMA2 gene amplification was performed according to the manufacturer’s instructions, using the PCR Amplification Kit (Monad Biotech Co., China). The primer sequences were: MaHMA2-F: 5’-ATGCCTCTCGATGGAGTCAAAGAGG-3’, MaHMA2-R:5’-ATCCACCAGTAAACC-3’. The reaction mixture consisted of 4 μL cDNA, 2 μL MaHMA2-F (10 μM), 2 μL MaHMA2-R (10 μM), 25 μL 2 × Taq mixture, with ddH2O added to a final volume of 50 μL. Amplification was carried out using the following PCR cycling conditions: denaturation at 95°C, 3 min; 35 cycles of denaturation at 95°C, 15 s; annealing at 60°C, 20 s; extension at 72°C, 1 min, then extension at 72°C for 5 min. The PCR-amplified target fragments were cloned into pMD 18-T vector using Vector Cloning Kit (Takara BioInc., Japan) following the manufacturer’s instructions, and transformed into E. coli TOP10 receptor cells by thermal excitation. Positive colonies were selected and sequenced for verification.
Three-step qPCR was performed using the FastStart Universal SYBR Green Master Mix kit (Roche, USA) in a LightCycler® 96 Real-Time PCR System (Roche, USA), according to the kit instructions. β-Actin was used as the internal reference gene (Jiang et al., 2015), and primer sequences were as follows: qMaHMA2-F: 5’-TCACAAAGGCTGCAACTTCC-3’, qMaHMA2-R: 5’-CATCATCAAGAAGAGACCGAA-3’. The qPCR reaction mixture consisted of 10 μL FastStart Universal® SYBR Green Master, 1 μL qMaHMA2-F (10 μM), 1 μL qMaHMA2-R (10 μM), 2 μL cDNA, and the addition of ddH2O to make a final volume of 20 μL. Amplification was achieved using the following PCR cycling conditions: 95°C denaturation, 1 min, 35 cycles: denaturation at 95°C, 20 s; annealing at 60°C, 20 s; extension at 72°C, 30 s.
Physiological and biochemical index detection
Concentrations of soluble protein (determined by the bicinchoninic acid (BCA) assay), proline (PRO) and malondialdehyde (MDA), as well as superoxide dismutase (SOD) and peroxidase (POD) activities, were determined according to the appropriate assay kits (Comin, China). Proline, MDA and protein content, SOD and POD activities were determined as described in Liang et al. (2017).
Each study was replicated three times independently, and each data point shown represents the mean ± standard deviation (SD). Data were analyzed by analysis of variance (ANOVA), with multiple comparisons between samples conducted using Duncan’s multiple range test. Statistical analyses were performed using Excel 2013 software (Microsoft, USA) and SPSS Statistics 19.0 software (SPSS Inc., USA).
Cloning and verification of the MaHMA2 gene in mulberry
The HMA2 gene amplified from white mulberry had a total length of 1,342 bp and an ORF of 1,194 bp, encoding a protein of 397 amino acid residues (Fig. S1), with a predicted molecular mass of 42.852 kD and an isoelectric point of 7.53, which was named MaHMA2. There are eight members of the mulberry HMA gene family, and we compared the nucleic acid sequences and amino acid sequences of each of the eight genes, as shown in Figs. S2 and S3. NCBI online Blast sequence comparison showed that the amino acid family encoded by the MaHMA2 gene belonged to several families, such as the HAD-like superfamily, the zntA superfamily, and the ATPase-IB2_Cd superfamily (Fig. S4), which have certain functions in Zn ion uptake and transport, achieve the coordination of metal homeostasis in plants, and promotion of growth and development. The tertiary protein structure of MaHMA2 was predicted by the online software SWISS-MODEL (Fig. S5). The predicted results indicated that MaHMA2 had a high degree of structural homology with the P1B-type, ATPase family of heavy metal transport proteins.
Homology-based alignment and phylogenetic analysis of the MaHMA2
The amino acid sequence encoded by the MaHMA2 gene of M. alba was compared with nine other species. The results showed up to 90% identity between this clade and Morus atropurpurea and Morus notabilis, and more than 70% with other species (Fig. S6). The HMA2 amino acid sequences of different species were selected to construct a phylogenetic tree. Among the 21 species compared, the Guangdong mulberry species ‘Da Shi’ mulberry line was found to be the most similar to Sichuan mulberry (EXC27875.1 M. notabilis) and least similar to grape (CBI40117.3 Vitis vinifera), walnut (XP_018856913.1 Juglans regia) and wild tea tree (XP_028114667.1 Camellia sinensis) (Fig. 1). Based on the bioinformatics analysis, homology, and phylogeny with other plant species, the closest genetic relationship with MaHMA2 was the HMA2 gene of M. notabilis (at 97% homology) with a more distant relationship (at 97% homology) to V. vinifera. Other species, such as J. regia and C. sinensis, showed lower homologies, the lowest of which was with V. vinifera. These data suggest that the HMA2 gene is highly conserved across species and may have evolved in response to different growing conditions and environmental factors yet had retained considerable homology with MaHMA2.
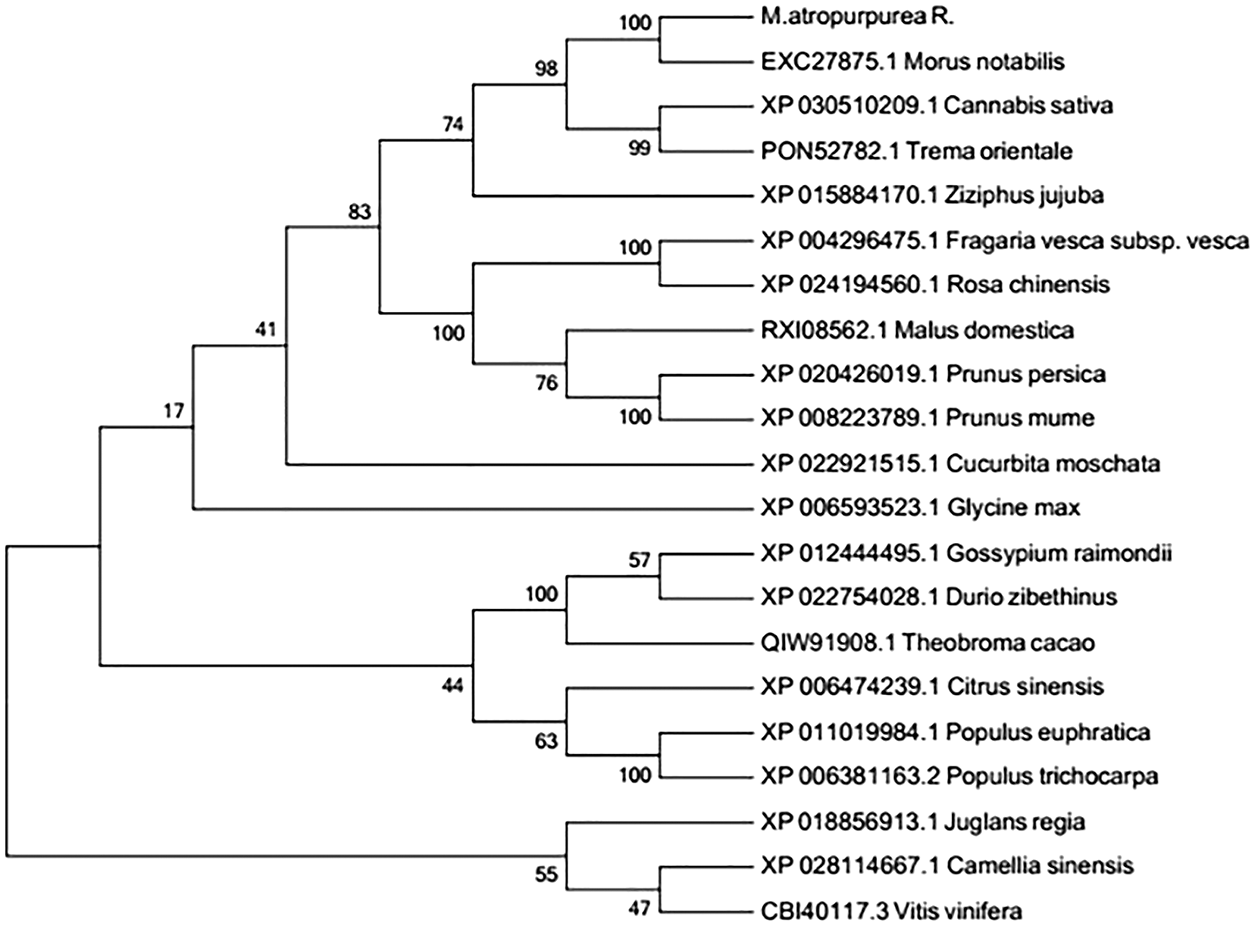
Figure 1: The MaHMA2 homologous sequence phylogenetic tree.
Expression levels of MaHMA2 under different Zn concentrations
The expression of the MaHMA2 gene first increased and then decreased in response to increasing Zn concentration. Relative to the control (0 mg/kg), at Zn 200 mg/kg, the gene expression increased to 5.67-fold, from which point the gene expression gradually decreased as Zn concentration were further increased (Fig. 2). These findings suggest that MaHMA2 may function in the uptake and translocation of Zn ions and that the expression of the gene was upregulated in the presence of low to moderate Zn concentrations but was then downregulated once Zn ion concentrations exceeded a threshold value.
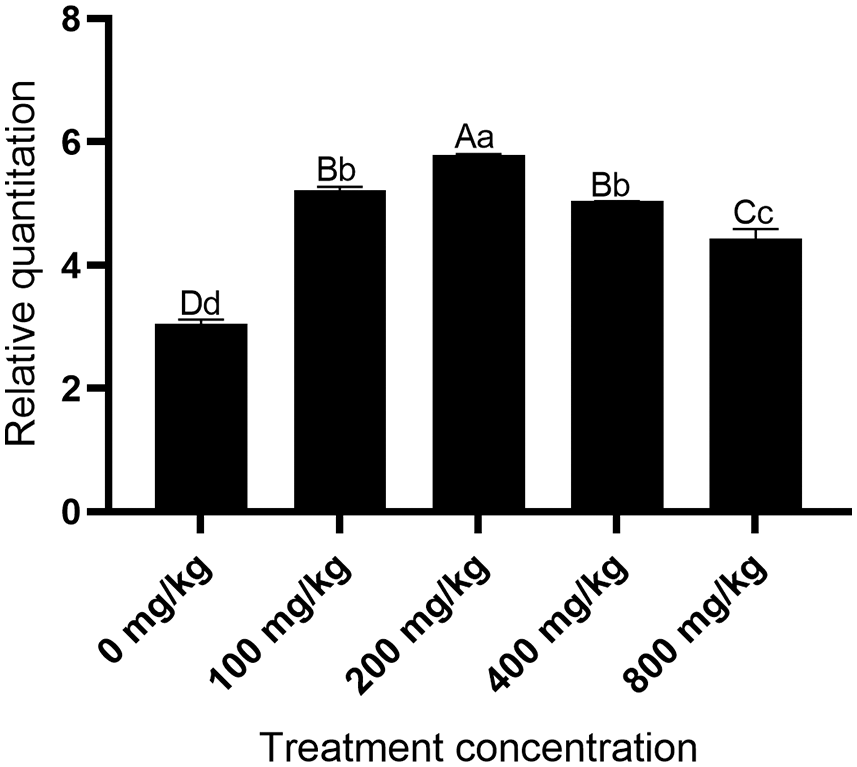
Figure 2: Relative levels of MaHMA2 gene expression in response to increasing Zn concentration stress. The individual data points are mean ± standard deviation (SD) of the three biological replicates. Bars with different letters indicate a significant difference between expression levels at different concentrations (p ≤ 0.05) on the basis of Duncan’s multiple range test.
Expression level under different Zn concentrations after MaHMA2 gene silencing
The expression levels of the MaHMA2 gene were then measured in gene-silenced VIGS transgenic plants grown under different concentrations of Zn. Empty pTRV2 vector was used as a negative control (CK) (Fig. 3). Under different concentrations of Zn stress, gene-silenced pTRV2-MaHMA2-VIGS plants showed a slight increase in expression relative to the 0 mg/kg VIGS plants followed by a gradual decrease with increasing Zn concentration; at 100 mg/kg Zn, expression of the gene-silenced plants was about 52% that of the empty pTRV2 plants. The expression of the MaHMA2 gene reached its highest value at a Zn stress concentration of 200 mg/kg, indicating that Zn stress affected the expression of the MaHMA2 gene, promoting the adaptation of mulberry to Zn ion stress.
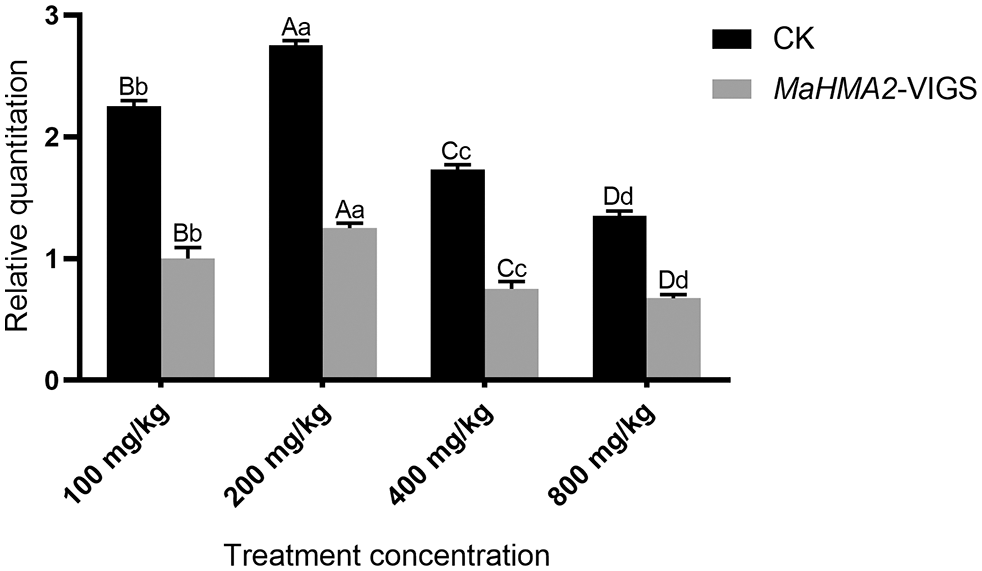
Figure 3: The expression level of MaHMA2 gene under Zn stress with and without MaHMA2 silencing. The individual data points are the mean ± standard deviation (SD) of the three biological replicates. CK means the negative control using empty pTRV2. MaHMA2-VIGS refers to the silencing of MaHMA2 gene in the experimental group. Bars with different letters indicate a significant difference between expression levels at different concentrations (p ≤ 0.05) on the basis of Duncan’s multiple range test.
Physiological and biochemical indexes under different Zn stress
The extent of lipid peroxidation, achieved due to the stress-induced accumulation of reactive oxygen species (ROS), was detected by assaying the concentration of MDA (Fig. 4A). As the concentration of Zn increased, plants injected with buffer as a blank control (wild type, WT) or those injected with empty pTRV2 vector as a negative CK showed no obvious increase in MDA concentration in response to increasing Zn concentration, but the gene-silenced pTRV2-MaHMA2-VIGS plants showed significantly higher levels of MDA which responded to increasing Zn concentration, indicating that MaHMA2 gene silencing increased the MDA content (reflecting the stress suffered) of the transgenic plants. Proline is widely distributed in plant tissues, and it is known that increased proline concentration reflects stress tolerance to some extent, with stress-tolerant varieties tending to accumulate more proline. Proline concentrations in the blank control, pTRV2 null-negative control, and gene-silenced pTRV2-MaHMA2-VIGS plants all showed an increasing trend with increasing Zn concentration. As with MDA, however, the proline concentration in the gene silenced pTRV2-MaHMA2-VIGS plants was significantly higher than that in the two non-silenced treatments (Fig. 4B).
Under abiotic stress, the activity of antioxidant enzymes such as POD and SOD, in plants facilitates stress tolerance by decreasing the stress-induced accumulation of ROS. As the concentration of Zn increased, the POD and (especially) SOD activity of the wild-type and blank control plants increased, but there was a significant decrease in the activity of both enzymes in the gene-silenced pTRV2-MaHMA2-VIGS plants (Figs. 4C and 4D). This finding indicated that Zn stress increased oxidative stress and hence increased the activity of POD and SOD to help plants reduce the ROS levels adapt to the stress. As the Zn concentration increased, the protein concentration was significantly lower in gene-silenced pTRV2-MaHMA2-VIGS plants than in the blank control (Fig. 4E). The soluble protein content of control and gene-silenced pTRV2-MaHMA2-VIGS plants gradually increased with increasing Zn concentrations and reached a maximum at 800 mg/kg. The soluble protein concentration of the samples is commonly used to express enzyme activity.
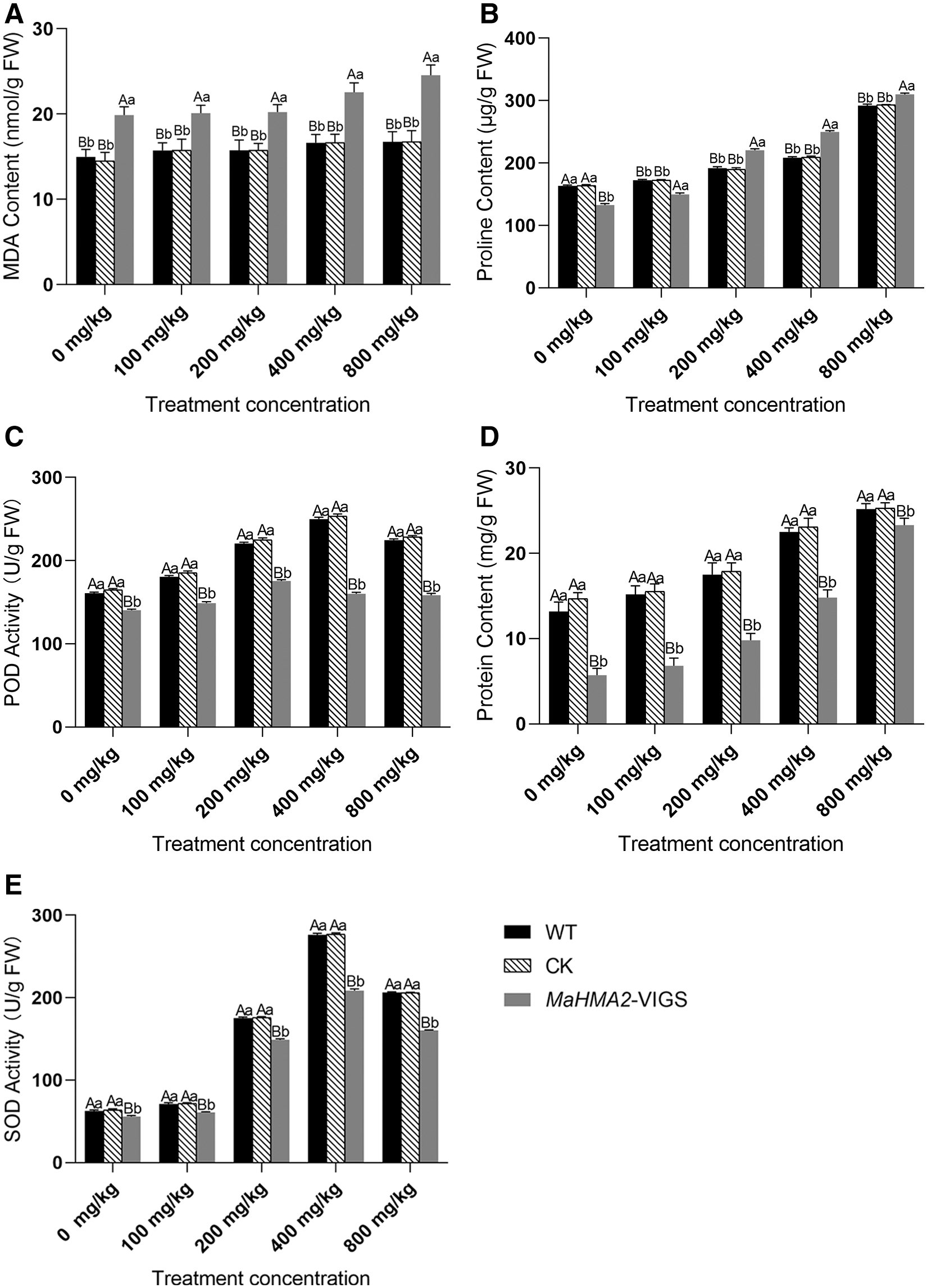
Figure 4: Physiological and biochemical indexes of mulberry in response to different Zn concentrations. A: MDA content; B: Proline content; C: POD activity; D: SOD activity; E: Protein content. The individual data points are mean ± standard deviation (SD) of the three biological replicates. WT (wild type) represents plants injected with buffer as the blank control, CK means negative control plants injected with empty pTRV2, and MaHMA2-VIGS means gene-silenced plants. Bars with different letters indicate a significant difference between expression levels at different concentrations (p ≤ 0.05) on the basis of Duncan’s multiple range test.
Plants have evolved complex molecular, cellular, and whole-plant mechanisms to cope with various abiotic stresses (Matsui et al., 2019). Previous studies have shown that HMAs play an important role in regulating plant responses to metal ion stress. Mulberry is an ecologically and economically important perennial woody plant affected by the adverse effects of various stresses on growth and productivity (Kim et al., 2021). Higher plants usually adapt to biotic and abiotic stresses by activating a cascade of molecular networks involving stress perception, signal transduction, and the expression of specific stress-related genes. In this study, a cDNA sequence encoding MaHMA2 was cloned from mulberry leaves with a full length of 1342 bp and an ORF of 1194 bp, encoding a protein with 397 amino acids. The predicted protein’s estimated molecular weight and isoelectric point (pI) were 42.852 kDa and 7.53, respectively. The predicted protein tertiary structure of the amino acid sequence of the MaHMA2 gene was similar to the tertiary structure of the P-type ATPase family of Zn transporter proteins. Phylogenetic analysis showed that MaHMA2 was closely related to M. notabilis and more distantly related to grape, walnut, and wild tea tree.
In the model organism Arabidopsis thaliana, HMA2 is annotated as a Zn/Cd transport protein (Wong et al., 2009). qRT-PCR revealed that the expression of MaHMA2 was sensitive to changes in Zn concentrations. This is in agreement with transcriptome sequencing results of Zn stress in our laboratory, which indicated a large difference in expression of the MaHMA2 gene in the experimental group that was stressed by Zn compared with the untreated group (data not shown). The HMA2 in arabidopsis is a Zn2+-dependent ATPase that is also activated by Cd2+ and, to a lesser extent, by other divalent heavy metals (Pb2+, Ni2+, Cu2+, and Co2+) (Eren and Argüello, 2004). The observed increase in MaHMA2 expression was aimed at increasing the transport of Zn to above-ground parts and provide for the study of the regulation mechanism of mulberry resistance to heavy metal stress and the cultivation of the mulberry. qRT-PCR analysis of MaHMA2 expression in response to Zn applications showed that compared to the control, the expression of the MaHMA2 gene was first increased, reaching the maximum value at 200 mg/kg, and then gradually decreased. However, even at 800 mg/kg, the expression was maintained higher than control levels. Partial suppression of MaHMA2 expression was achieved after VIGS treatment of M. alba with pTRV2-MaHMA2 (ca. 2-fold reduction). Interestingly, the expression pattern of MaHMA2 in the modified seedlings (MaHMA2-VIGS) in response to Zn treatments was very similar to that observed in WT, although proportionately reduced at all Zn concentrations.
In our research, the mobilization of plant defense in response to Zn treatments was studied indirectly from physiological (MDA, proline, protein synthesis) and enzymatic (POD, SOD) markers of stress. The results indicated that while Zn treatments did not induce significant differences in WT MDA (indicator of oxidative stress) levels, changes in the levels of the other markers (proline, protein synthesis, POD and SOD) confirmed that Zn treatments caused a significant mobilization of plant defense responses, especially at 200–800 mg/kg. The partial repression of MaHMA2 in MaHMA2-VIGS seedlings led to significant alterations in the plant response to Zn. Relative to the WT response, MDA production was elevated while protein content, POD and SOD activities were reduced at all Zn concentrations. The sensitivity of MaHMA2 gene expression to external Zn concentrations and its effects on the mobilization of plant defense responses, suggests that MaHMA2 is involved in the maintenance of plant Zn homeostasis to attenuate Zn stress.
In conclusion, we treated mulberry species with varied concentrations of Zn to induce Zn stress in mulberry plants. We isolated and characterized a novel MaHMA2 gene RNA sequence technology and confirmed that Zn stress induced the gene. We found that silencing MaHMA2 with VIGS could improve Zn tolerance by regulating physiological changes, phenotypic traits and Zn stress-related genes in mulberry. Our study revealed the function of MaHMA2 in mulberry and laid the foundation for further understanding the mechanisms of plant responses to Zn stress.
Acknowledgement: We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Author Contributions: W. G. Zhao, R. X. Li, and M. D. Zhao conceived this study. L. Wang, Q. X. Du, D. Y. Zheng, P. L. Li, P. Guo and M. M. Wu performed the experiments. L. Wang and Y. S. Shi wrote the manuscript. W. G. Zhao and M. Ackah, and Z. Yin revised the original manuscript. All authors read and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Ethical Approval: This article does not contain any studies involving animals or human participants as research objects.
Funding Statement: This work was supported by the China Agriculture Research System of MOF and MARA, National Key R&D Program of China, the key projects of International Scientific and Technological Innovation Cooperation (Grant No. 2021YFE0111100), the Guangxi Innovation-Driven Development Project (Grant No. AA19182012-2), and the Zhenjiang Science and Technology Support Project (Grant No. GJ2021015).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Blindauer CA, Leszczyszyn OI (2010). Metallothioneins: Unparalleled diversity in structures and functions for metal ion homeostasis and more. Natural Product Reports 27: 720–741. DOI 10.1039/b906685n. [Google Scholar] [CrossRef]
Cun P, Sarrobert C, Richaud P, Chevalier A, Soreau P, Auroy P, Gravot A, Baltz A, Leonhardt N, Vavasseur A (2014). Modulation of Zn/Cd P1B2-ATPase activities in Arabidopsis impacts differently on Zn and Cd contents in shoots and seeds. Metallomics 6: 2109–2116. DOI 10.1039/c4mt00182f. [Google Scholar] [CrossRef]
Eren E, Argüello JM (2004). Arabidopsis HMA2, a divalent heavy metal-transporting PIB-type ATPase, is involved in cytoplasmic Zn2+ homeostasis. Plant Physiology 136: 3712–3723. DOI 10.1104/pp.104.046292. [Google Scholar] [CrossRef]
Fan W, Guo Q, Liu C, Liu X, Zhang M, Long D, Xiang Z, Zhao A (2018). Two mulberry phytochelatin synthase genes confer zinc/cadmium tolerance and accumulation in transgenic Arabidopsis and tobacco. Gene 645: 95–104. DOI 10.1016/j.gene.2017.12.042. [Google Scholar] [CrossRef]
Geuten K, Viaene T, Vekemans D, Kourmpetli S, Drea S (2013). Analysis of developmental control genes using virus-induced gene silencing. Methods in Molecular Biology 975: 61–69. DOI 10.1007/978-1-62703-278-0_5. [Google Scholar] [CrossRef]
Guzmán-Rangel G, Versieren L, Qiu H, Smolders E (2017). Additive toxicity of zinc and arsenate on barley (Hordeum vulgare) root elongation. Environmental Toxicology and Chemistry 36: 1556–1562. DOI 10.1002/etc.3674. [Google Scholar] [CrossRef]
Imran QM, Falak N, Hussain A, Mun B-G, Sharma A et al. (2016). Nitric oxide responsive heavy metal-associated gene AtHMAD1 contributes to development and disease resistance in Arabidopsis thaliana. Frontiers in Plant Science 7: 1712. DOI 10.3389/fpls.2016.01712. [Google Scholar] [CrossRef]
Jiang X, Yao F, Li X, Jia B, Zhong G, Zhang J, Zou X, Hou L (2015). Molecular cloning, characterization and expression analysis of the protein arginine N-methyltransferase 1 gene (As-PRMT1) from Artemia sinica. Gene 565: 122–129. DOI 10.1016/j.gene.2015.04.004. [Google Scholar] [CrossRef]
Kim JS, Jeon BW, Kim J (2021). Signaling peptides regulating abiotic stress responses in plants. Frontiers in Plant Science 12: 704490. DOI 10.3389/fpls.2021.704490. [Google Scholar] [CrossRef]
Lee S, Kim YY, Lee Y, An G (2007). Rice P1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiology 145: 831–842. DOI 10.1104/pp.107.102236. [Google Scholar] [CrossRef]
Leszczyszyn OI, Imam HT, Blindauer CA (2013). Diversity and distribution of plant metallothioneins: A review of structure, properties and functions. Metallomics: Integrated Biometal Science 5: 1146–1169. DOI 10.1039/c3mt00072a. [Google Scholar] [CrossRef]
Liang QY, Wu YH, Wang K, Bai ZY, Liu QL, Pan YZ, Zhang L, Jiang BB (2017). Chrysanthemum WRKY gene DgWRKY5 enhances tolerance to salt stress in transgenic chrysanthemum. Scientific Reports 7: 4799. DOI 10.1038/s41598-017-05170-x. [Google Scholar] [CrossRef]
Liu YM, Liu DY, Zhao QY, Zhang W, Chen XX, Xu SJ, Zou CQ (2020). Zinc fractions in soils and uptake in winter wheat as affected by repeated applications of zinc fertilizer. Soil and Tillage Research 200: 104612. DOI 10.1016/j.still.2020.104612. [Google Scholar] [CrossRef]
Matsui A, Nakaminami K, Seki M (2019). Biological function of changes in RNA metabolism in plant adaptation to abiotic stress. Plant & Cell Physiology 60: 1897–1905. DOI 10.1093/pcp/pcz068. [Google Scholar] [CrossRef]
Qin F, Liu G, Huang G, Dong T, Liao Y, Xu X (2018). Zinc application alleviates the adverse effects of lead stress more in female Morus alba than in males. Environmental and Experimental Botany 146: 68–76. DOI 10.1016/j.envexpbot.2017.10.003. [Google Scholar] [CrossRef]
Rizwan M, Ali S, Rehman MZU, Maqbool A (2019). A critical review on the effects of zinc at toxic levels of cadmium in plants. Environmental Science and Pollution Research International 26: 6279–6289. DOI 10.1007/s11356-019-04174-6. [Google Scholar] [CrossRef]
Romero-Isart N, Vasák M (2002). Advances in the structure and chemistry of metallothioneins. Journal of Inorganic Biochemistry 88: 388–396. DOI 10.1016/s0162-0134(01)00347-6. [Google Scholar] [CrossRef]
Takahashi R, Bashir K, Ishimaru Y, Nishizawa NK, Nakanishi H (2012). The role of heavy-metal ATPases, HMAs, in zinc and cadmium transport in rice. Plant Signaling & Behavior 7: 1605–1607. DOI 10.4161/psb.22454. [Google Scholar] [CrossRef]
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. DOI 10.1093/molbev/mst197. [Google Scholar] [CrossRef]
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–4882. DOI 10.1093/nar/25.24.4876. [Google Scholar] [CrossRef]
Versieren L, Evers S, de Schamphelaere K, Blust R, Smolders E (2016). Mixture toxicity and interactions of copper, nickel, cadmium, and zinc to barley at low effect levels: Something from nothing? Environmental Toxicology and Chemistry 35: 2483–2492. DOI 10.1002/etc.3380. [Google Scholar] [CrossRef]
Wong CKE, Jarvis RS, Sherson SM, Cobbett CS (2009). Functional analysis of the heavy metal binding domains of the Zn/Cd-transporting ATPase, HMA2, in Arabidopsis thaliana. The New Phytologist 181: 79–88. DOI 10.1111/j.1469-8137.2008.02637.x. [Google Scholar] [CrossRef]
Wu B, Luo S, Luo H, Huang H, Xu F, Feng S, Xu H (2021). Improved phytoremediation of heavy metal contaminated soils by Miscanthus floridulus under a varied rhizosphere ecological characteristic. Science of the Total Environment 808: 151995. DOI 10.1016/j.scitotenv.2021.151995. [Google Scholar] [CrossRef]
Wu C, Dun Y, Zhang Z, Li M, Wu G (2020). Foliar application of selenium and zinc to alleviate wheat (Triticum aestivum L.) cadmium toxicity and uptake from cadmium-contaminated soil. Ecotoxicology and Environmental Safety 190: 110091. DOI 10.1016/j.ecoenv.2019.110091. [Google Scholar] [CrossRef]
Wu X X, Chen J L, Xu S, Zhu ZW, Zha DS (2016). Exogenous 24-epibrassinolide alleviates zinc-induced toxicity in eggplant (Solanum melongena L.) seedlings by regulating the glutathione-ascorbate-dependent detoxification pathway. Journal of Horticultural Science and Biotechnology 91: 412–420. DOI 10.1080/14620316.2016.1162030. [Google Scholar] [CrossRef]
Zhang C, Wu Z, Li Y, Wu J (2015). Biogenesis, function, and applications of virus-derived small RNAs in plants. Frontiers in Microbiology 6: 1237. DOI 10.3389/fmicb.2015.01237. [Google Scholar] [CrossRef]
Zhao D, Zhou C, Kong F, Tao J (2011). Cloning of phytoene desaturase and expression analysis of carotenogenic genes in persimmon (Diospyros kaki L.) fruits. Molecular Biology Reports 38: 3935–3943. DOI 10.1007/s11033-010-0510-7. [Google Scholar] [CrossRef]
Zhao WG, Zhou ZH, Miao XX, Zhang Y, Wang SB, Huang JH, Xiang H, Pan YL, Huang YP (2007). A comparison of genetic variation among wild and cultivated Morus Species (Moraceae: Morus) as revealed by ISSR and SSR markers. Biodiversity and Conservation 16: 275–290. DOI 10.1007/s10531-005-6973-5. [Google Scholar] [CrossRef]
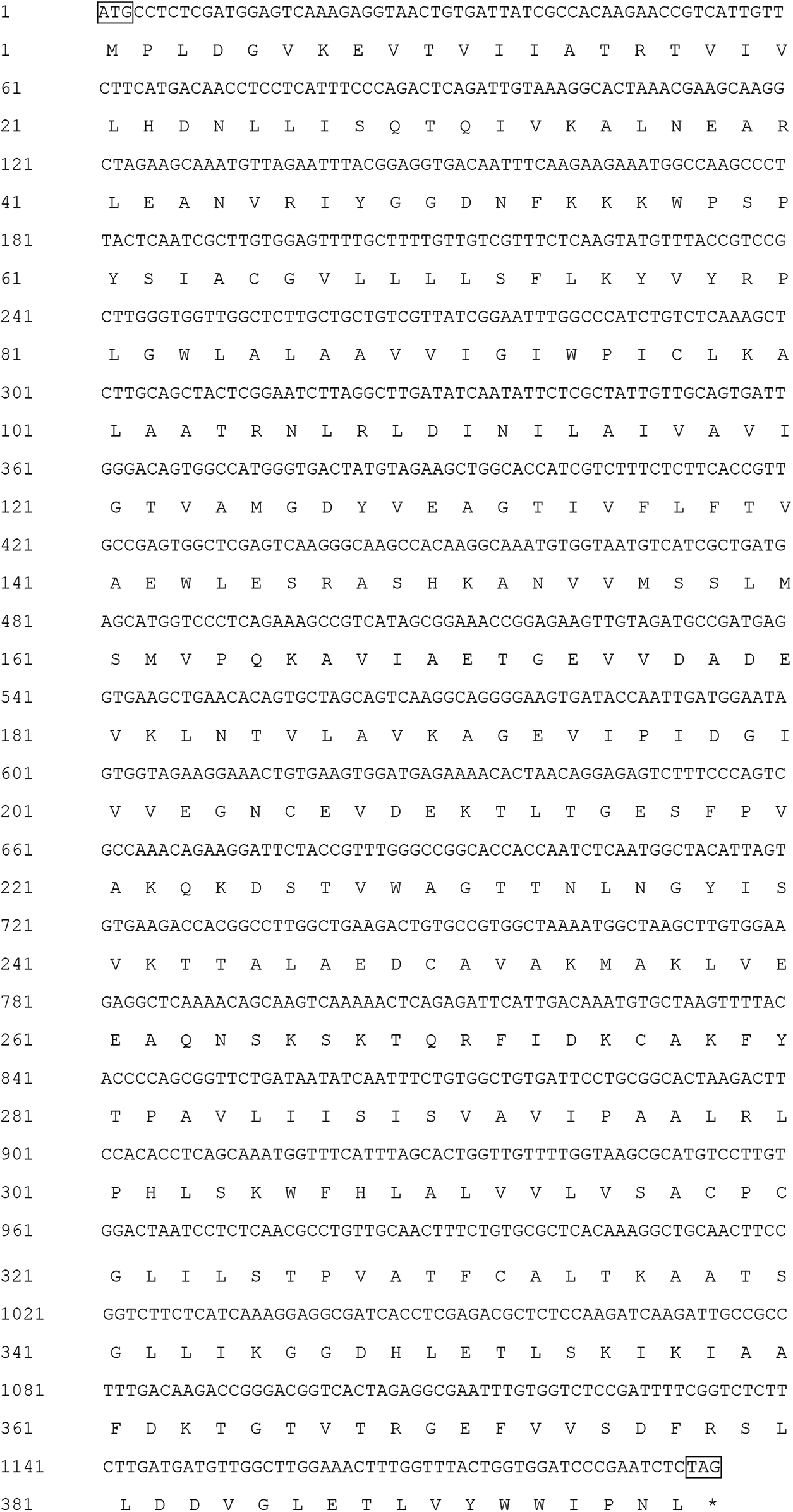
Supplementary Figure S1: The MaHMA2 gene cDNA sequence and the predicted amino acid sequence. ATG indicates the start codon; TAG indicates the stop codon.
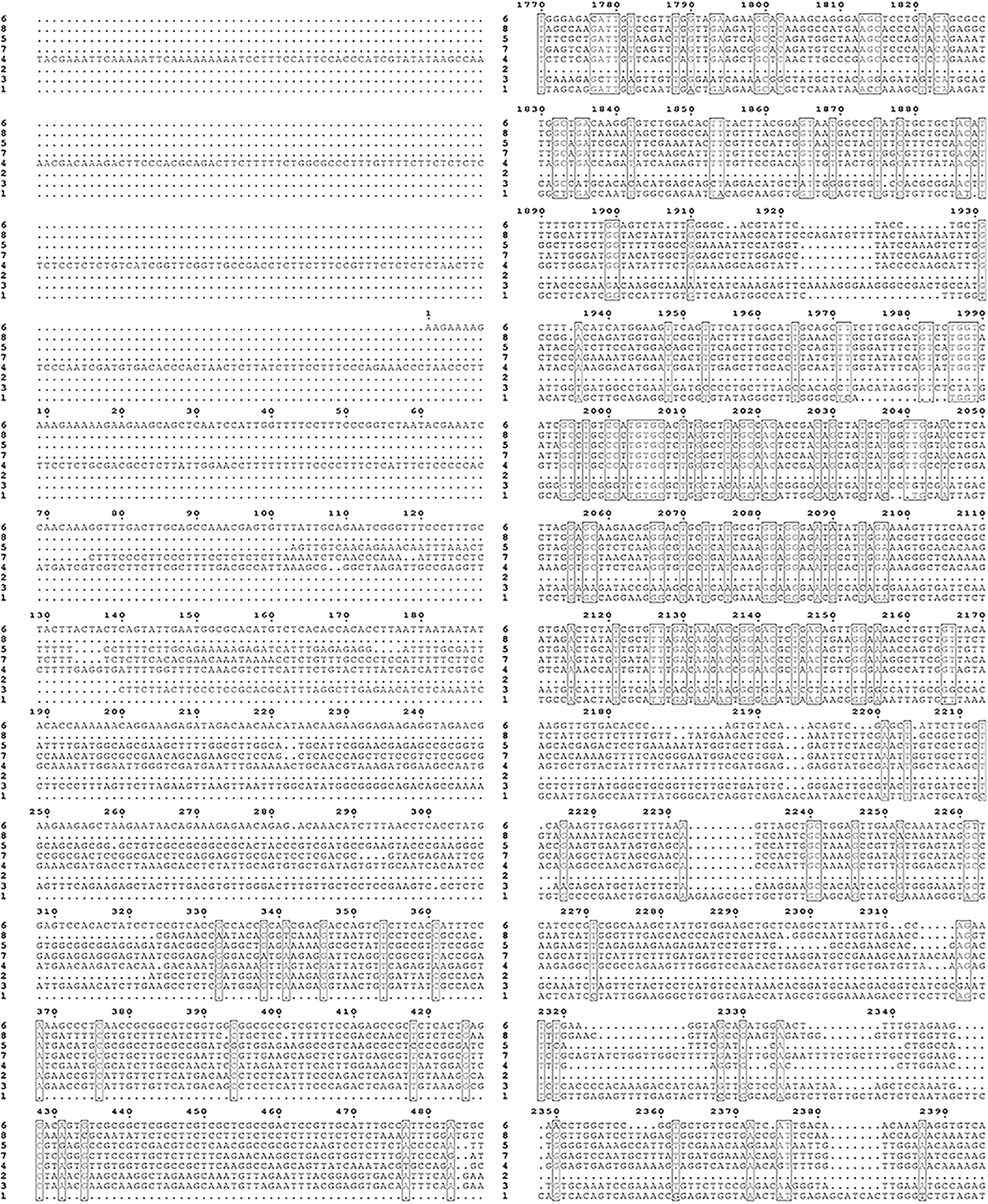
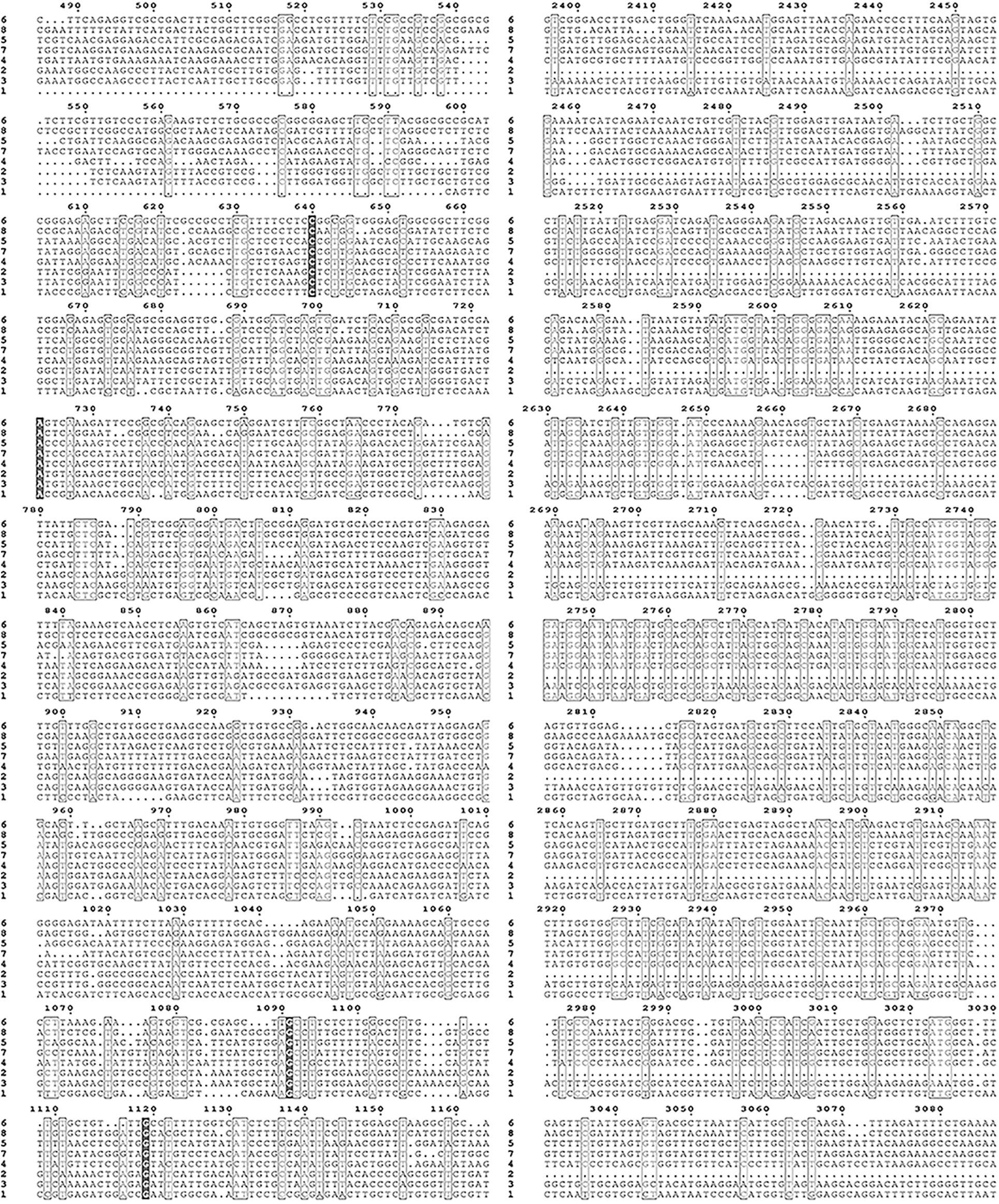
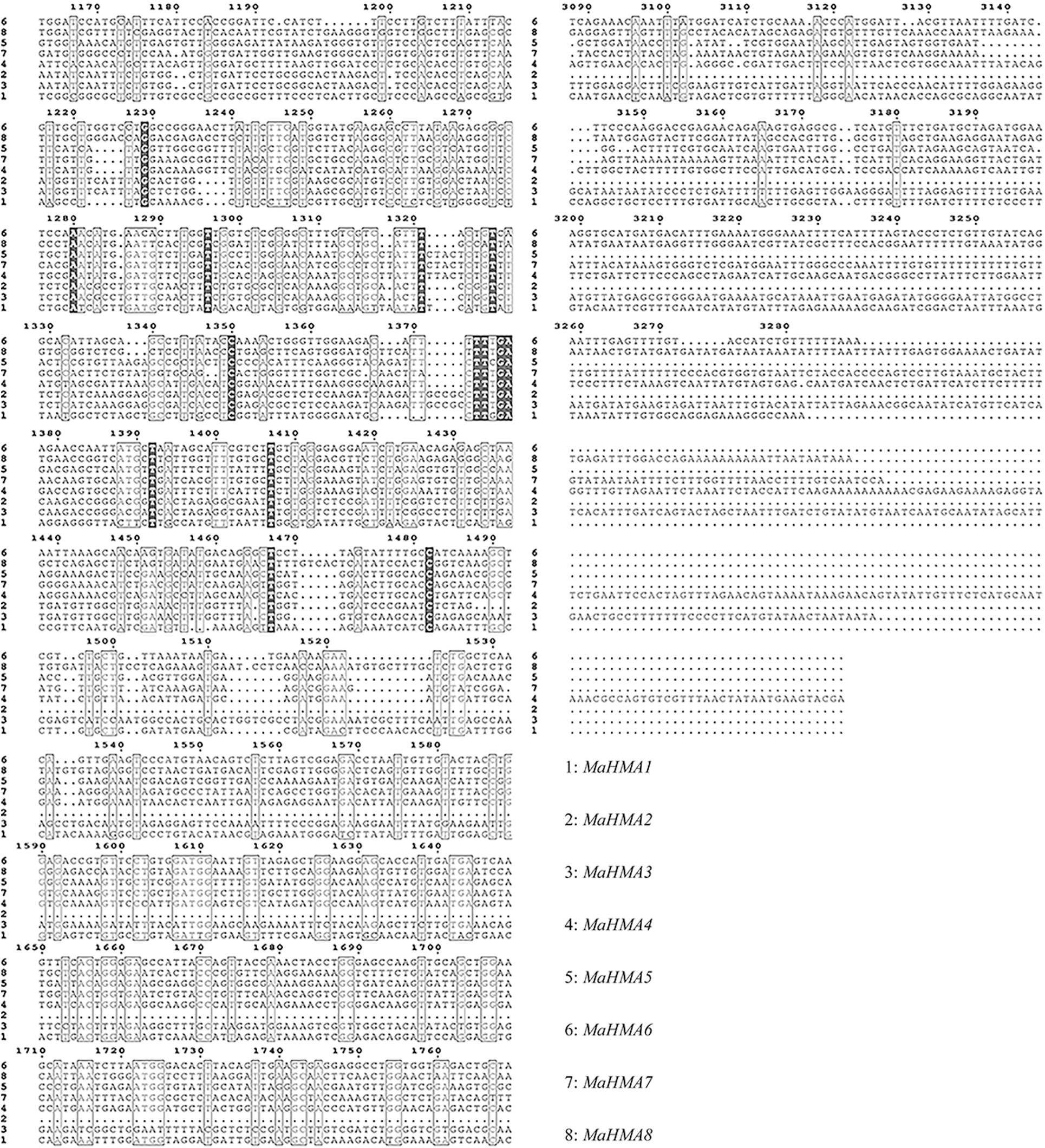
Supplementary Figure S2: Nucleic acid sequence alignment of the HMAs gene family of mulberry.
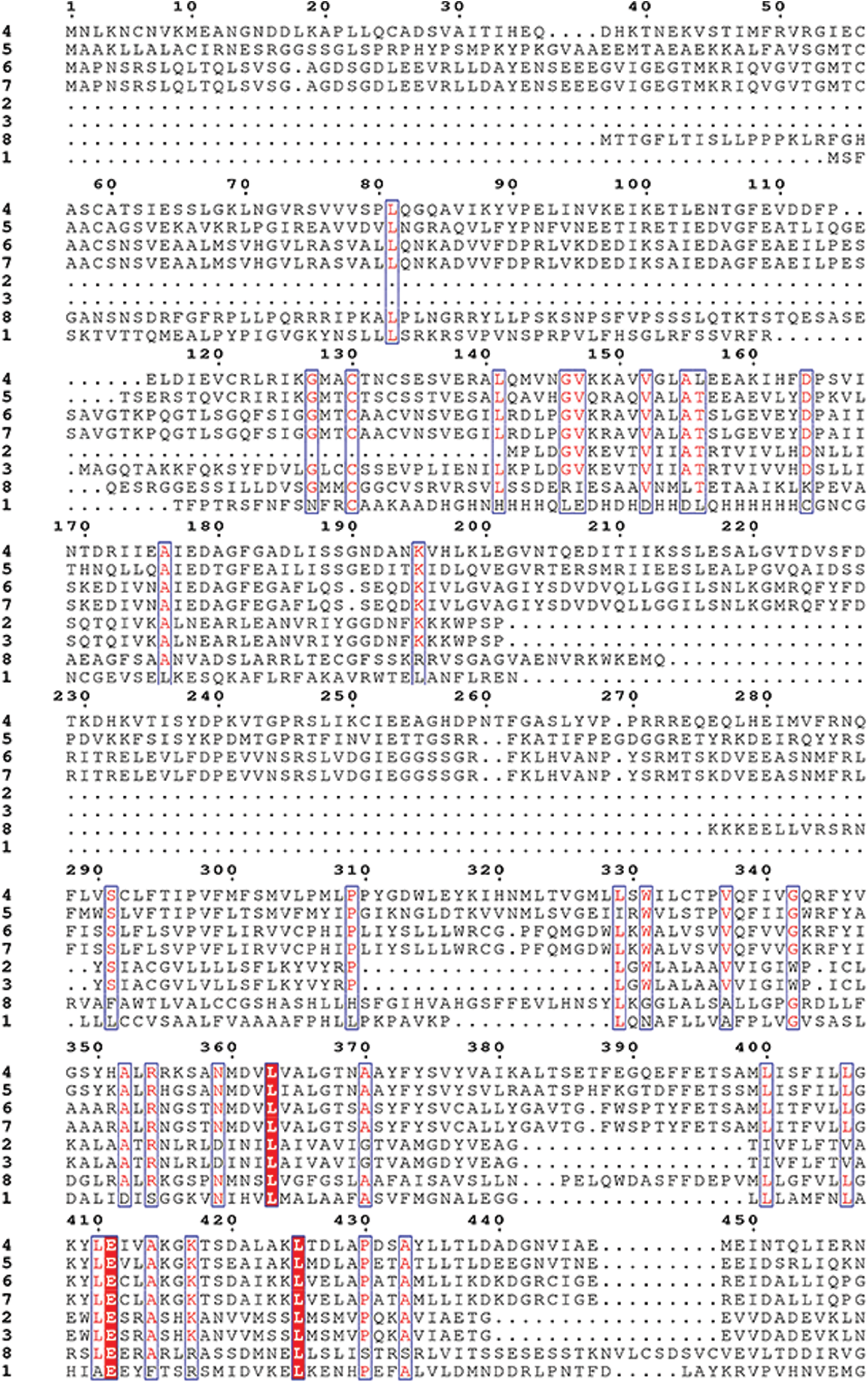
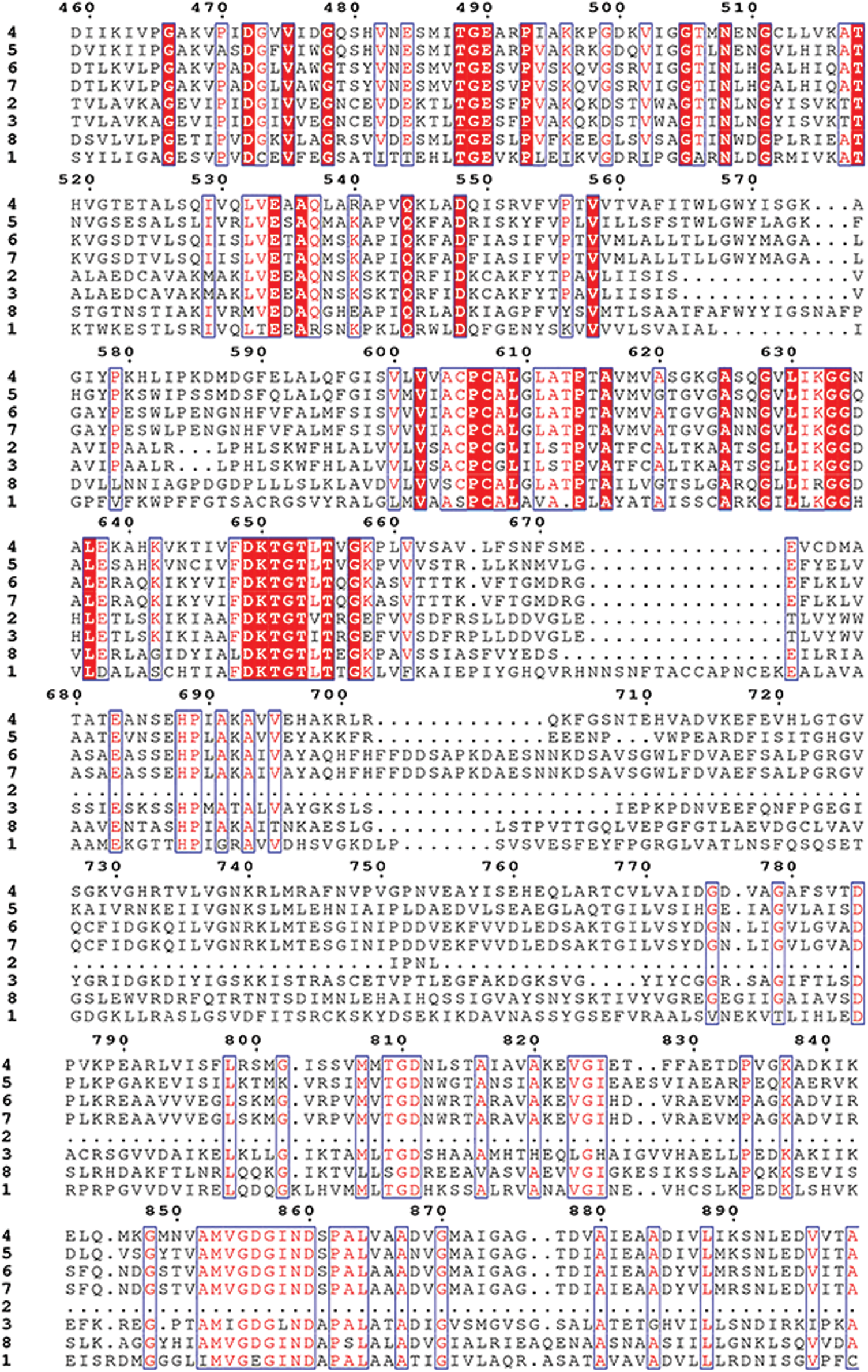

Supplementary Figure S3: Amino acid sequence alignment of the HMAs gene family of mulberry.
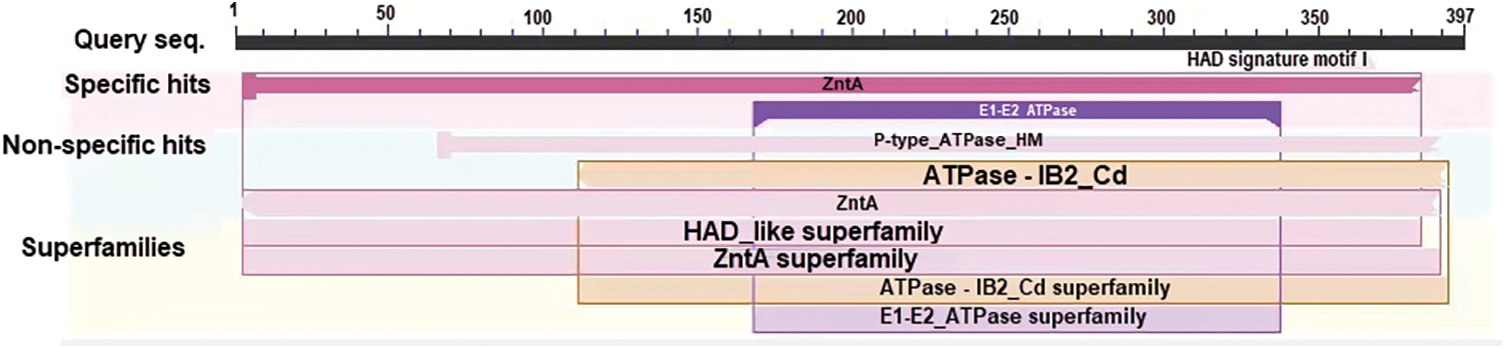
Supplementary Figure S4: The detection of conserved domains of superfamilies related with metal ion binding and transport.
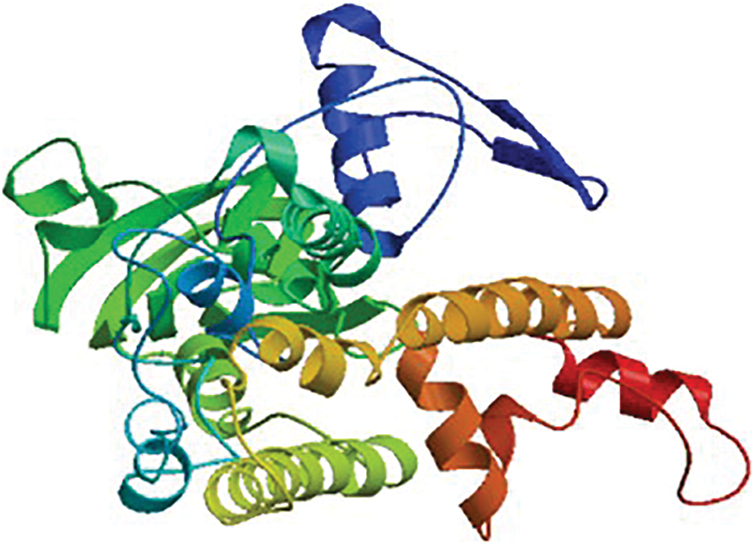
Supplementary Figure S5: The MaHMA2 protein tertiary structure.
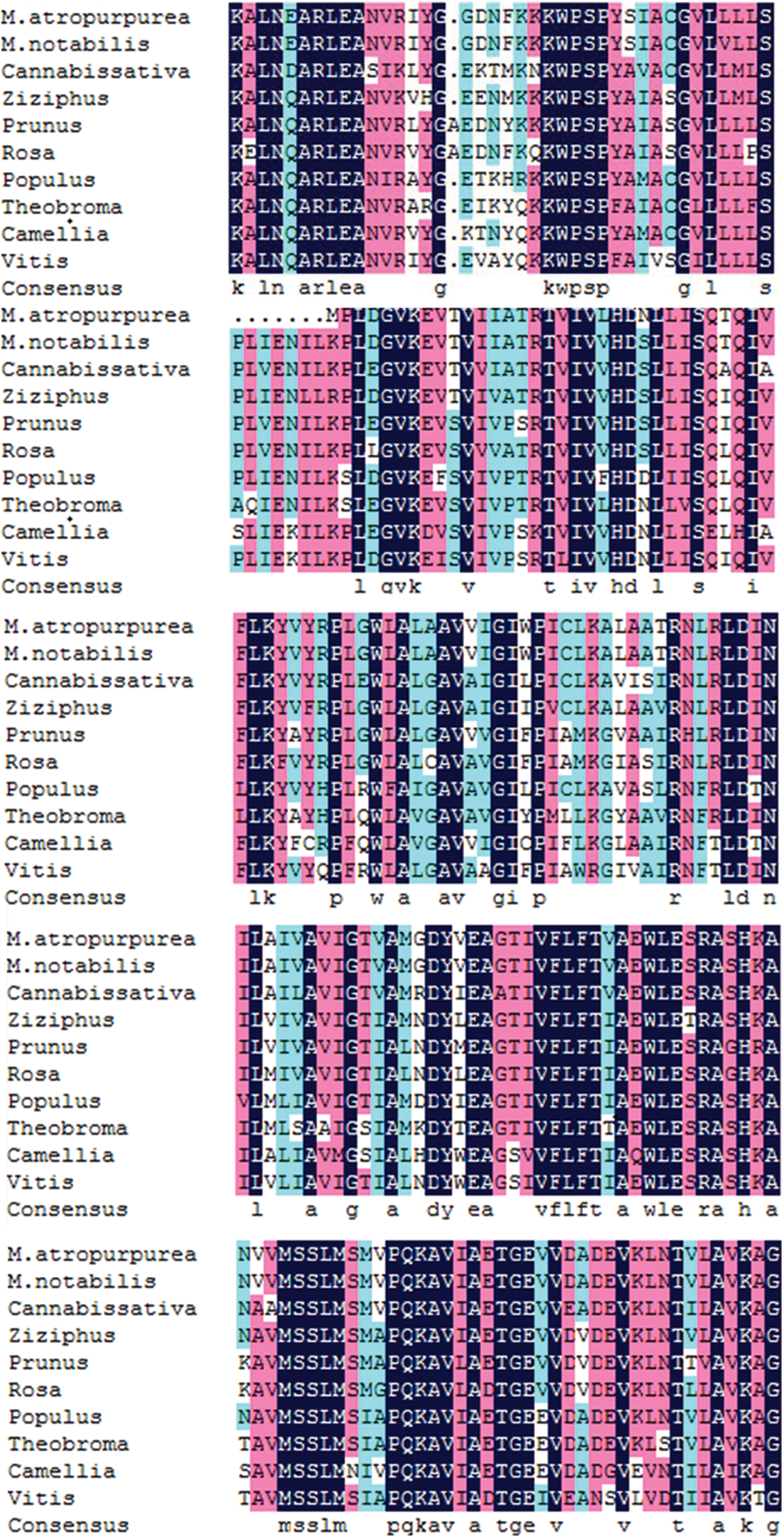
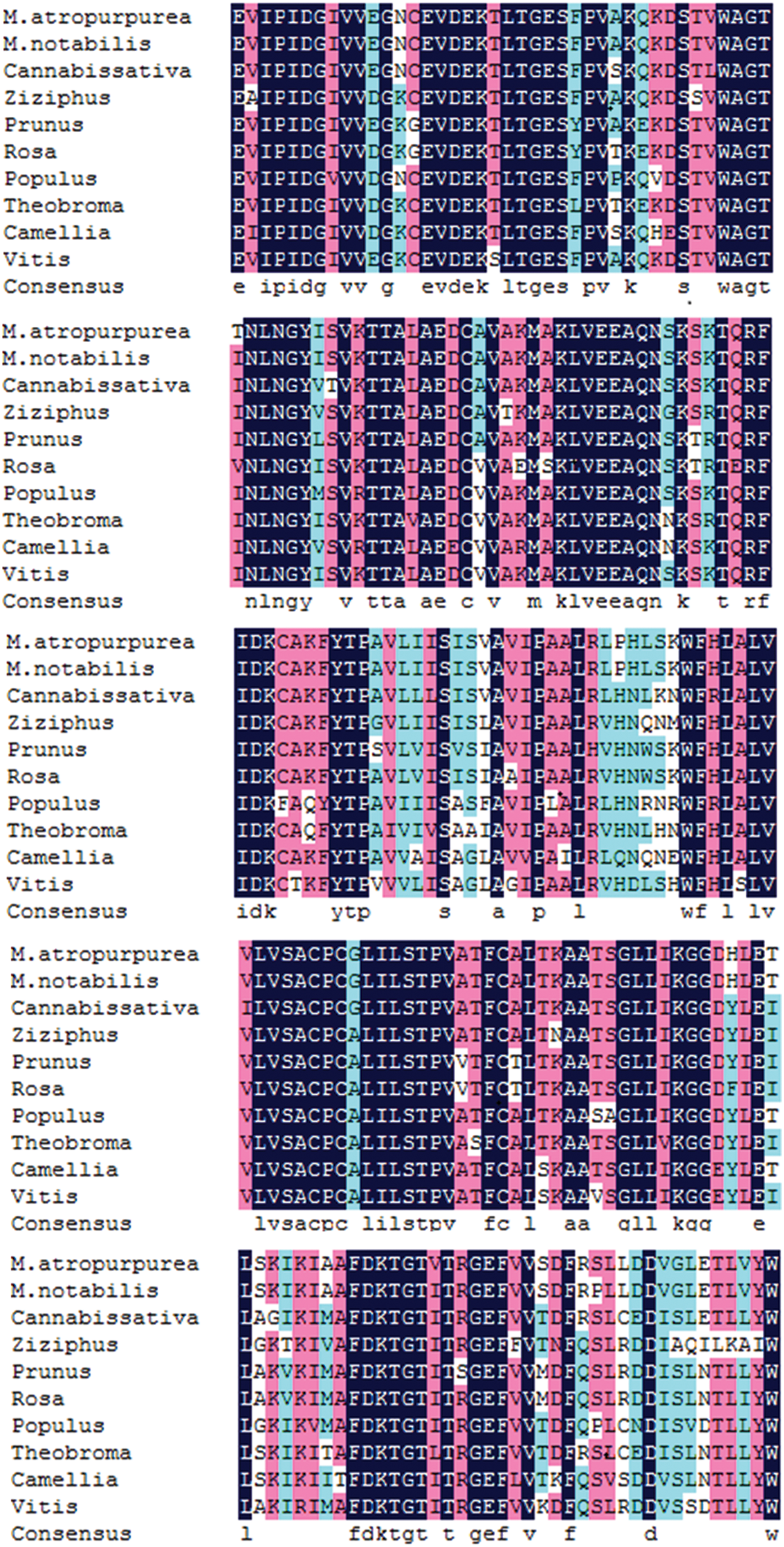
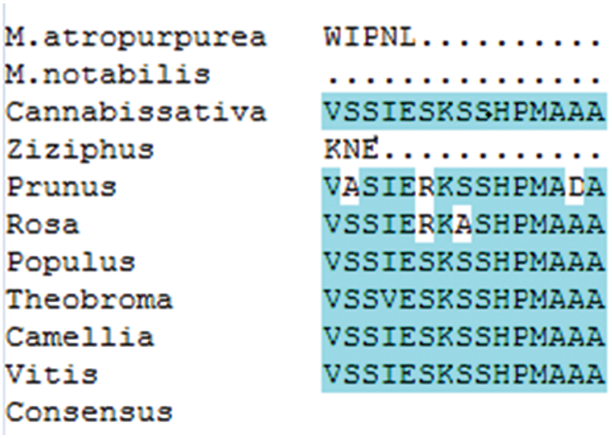
Supplementary Figure S6: The MaHMA2 gene encodes a multiple sequence alignment of amino acids.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |