DOI:10.32604/biocell.2022.020154

| BIOCELL DOI:10.32604/biocell.2022.020154 |  |
| Review |
Exosomes: Key tools for cancer liquid biopsy
1Dipartimento di Farmacia-DIFAR, Università di Genova, Genoa, 16132, Italy
2Laboratory of Molecular Nephrology, IRCCS Istituto Giannina Gaslini, Genoa, 16147, Italy
*Address correspondence to: Isabella Panfoli, panfoli@difar.unige.it
Received: 07 November 2021; Accepted: 11 February 2022
Abstract: Precision medicine is based on the identification of biomarkers of tumor development and progression. Liquid biopsy is at the forefront of the ability to gather diagnostic and prognostic information on tumors, as it can be noninvasively performed prior or during treatment. Liquid biopsy mostly utilizes circulating tumor cells, or free DNA, but also exosomes. The latter are nanovesicles secreted by most cell types, found in any body fluid that deliver proteins, nucleic acids and lipids to nearby and distant cells with a unique homing ability. Exosomes function in signalling between the tumor microenvironment and the rest of the body, promoting metastasis, immune remodelling and drug resistance. Exosomes are emerging as a key tool in precision medicine for cancer liquid biopsy, as they efficiently preserve their biomarker cargo. Moreover, exosomes strongly resemble the parental cell, which can help in assessing the oxidative and metabolic state of the donor cell. In this respect, exosomes represent one of the most promising new tools to fight cancer. This review will discuss the clinical applications of profiling exosomal proteins and lipids by high-throughput proteomics and metabolomics, and nucleic acids by next generation sequencing, as well as how this may allow cancer diagnosis, therapy response monitoring and recurrence detection.
Keywords: Cancer; Exosome; Liquid biopsy; Precision medicine
Cancer is the second leading cause of death globally, but early diagnosis, prognosis and monitoring of therapy response in primary stages remain challenging (The global challenge of cancer, 2020). Late diagnosis and metastasis are a major cause of cancer deaths: the former is responsible for about 67% of solid tumor deaths (Dillekas et al., 2019). Although cancer screening and detection technologies, as well as good treatment protocols are currently available, their limitations are many. There is the need for novel molecular biomarkers of the pre-invasive stage and for non-invasive bioptic procedures.
Traditional biopsies and surgical procedures are invasive and potentially harmful for complications. Their limitations include small sample size, unrepeatability and tumor inaccessibility such as for example, after surgery (Wang et al., 2017; Palmirotta et al., 2018). Also, samples may not reflect intrinsic tumor heterogeneity, which occurs at both genetic and phenotypic level throughout the different clinical stages (Siravegna et al., 2017). The identification of metastatic biomarkers appears promising in shedding light into the mechanisms that control metastasis, to reduce cancer mortality. Notably, no metastasis-exclusive mutations have been found; instead, evidence suggests that metastasis is promoted by oncogenic driver mutations occurring via epigenetic mechanisms elicited in response to selective pressure of therapies, that interact with various physiological pathways (Patel et al., 2021).
‘Liquid biopsy’ is a non-invasive test for genomic or proteomic assay of cancer-derived components in peripheral blood or body fluids (Poulet et al., 2019). In the field of clinical oncology, the analysis of tumours using biomarker sources circulating in biofluids is drawing great interest as a safe alternative to solid biopsies (Quandt et al., 2017) and an invaluable tool in precision medicine (Jiménez-Zenteno and Cerf, 2020). Conventional biopsies involve analysis of material obtained from biopsied samples. Advantages of tissue biopsies are the possibility to comprehensively reflect the tumor genetic profile than tissue biopsy, and analyse both the tumor and its microenvironment, facilitating the clinical decision-making. Disadvantages of tissue biopsies are invasiveness, difficulty in accessing the tumor cells, and lack of seriality when multiple analyses are necessary such as for example to monitor tumor progression. Liquid biopsies are fast, little costly minimally invasive, allow serial sampling, and monitoring of tumor heterogeneity. With respect to tissue biopsies, the relatively easy-to-obtain nature of bioliquids makes them an attractive alternative source for clinical application (Poulet et al., 2019). Data from liquid biopsy are reproducible and highly specific, allowing to detect premalignant and early-stage cancers, to monitor response to treatment and therapy resistance (Quandt et al., 2017). Cancer death rates have dropped by 26% between 1999 and 2018, according to the last update on cancer deaths (An Update on Cancer Deaths in the United States. Atlanta, GA: US Department of Health and Human Services, Centers for Disease Control and Prevention, Division of Cancer Prevention and Control; 2020), however, such improvement in cancer survival rates due the use of preventative measurements and precision medicine, mainly occurred in high-income countries (Girardi et al., 2019). By contrast, access to tumor screening tests and cancer treatments in low-income countries are inadequate, with worse cancer outcomes and higher incidence rates (Kamaraju et al., 2020). Early care and diagnosis need expensive bench top equipment and extensive training. Provided liquid biopsy option is implemented as portable user-friendly integrated microfluidic apparatuses this would render affordable and accessible the benefit of liquid biopsies even for low-income countries (Contreras-Naranjo et al., 2017). Of course, before these devices can be used in clinical settings, validation and standardization of procedures and sample collection, storage and characterization must be achieved. Although liquid biopsy is not yet a routine test in clinical oncology practice, its application appears promising (Cescon et al., 2020). Liquid biopsies are informative in early diagnostic screening and hopefully could predict recurrence or metastasis of malignancies. In fact, the organizations and foundations recognizing the impact of the global use of liquid biopsy have gathered in the International Liquid Biopsy Standardization Alliance (ILSA). ILSA has the scope to promote standardization-based programs and support resources and regulatory aspects in the field (Connors et al., 2020).
The cancer-derived analytes can help in the early cancer diagnosis, assessment of tumor stage, identification of the micrometastases and of targets for tailored therapy, evaluation of recurrence risk, monitoring of treatment response. In patients with advanced stage disease, liquid biopsy can enable timely management of recurrences and choice of the appropriate treatment when resistance to targeted therapy is detected. In fact, liquid biopsy allows the dynamic follow-up of changes in tumour biology. Liquid biopsies may include detection of circulating tumor cells (CTCs), circulating cell-free DNA (cfDNA) and circulating tumour DNA (ctDNA), metabolites, tumor-educated platelets (TEPs), and exosomes (Exo) released by tumor cells.
Circulating tumor cells (CTCs) are cancer cells that shed from tumors and enter the circulatory system (Jimenez-Zenteno and Cerf, 2020). CTCs do not have defined morphological characteristics and are extremely rare, their frequency being lower than 10 cells/mL of blood, moreover their survival in the bloodstream is limited (Palmirotta et al., 2018; Poulet et al., 2019). CTC are found in the blood of patients with common tumors, but not in healthy subjects or patients with non-malignant diseases (Allard et al., 2004). CTCs spread through circulation and may reside in specific permissive tissues, including bone marrow, in which case they are termed disseminated tumour cells (DTCs), always related to molecular residual disease (Sai and Xiang, 2018). CTCs are believed to be the metastatic precursors, having detached from the primary tumor site during the metastatic process that can establish metastatic loci in permissive niches (Masuda et al., 2016). CTCs can be different from the primary tumor cells, having evolved under environmental and drug selection pressure (Ma and Jeffrey, 2020). CTCs exist in both single and clustered forms, which play different roles in metastasis: they form aggregates with other tumor cells or blood cells such as platelets, which protect them from the immune system (Heeke et al., 2019). The CTC clustering in the circulation was shown to be also promoted by plakoglobin, a cell adhesion protein whose elevated expression is associated with higher breast cancer metastatic potential, and low survival rates (Lu et al., 2015). On the other hand, diminished plakoglobin expression promoted epithelial to mesenchymal transition (EMT) (Lu et al., 2015), in turn playing a pivotal role in all stages of cancer progression, and metastasis (Brabletz et al., 2018). CTC can be detected in the peripheral blood of at least 10% of early-stage breast cancer and in up to 80% of metastatic breast cancer patients (Xu et al., 2018). The presence of less than 5 cells/7.5 mL of blood before treatment was shown to be an independent predictor of progression-free survival in patients with metastatic breast cancer (Cristofanilli et al., 2004). This result was confirmed by a meta-analysis on 50 studies with 6712 breast cancer patients, showing that therapy significantly reduced CTC-positive rate, which is in turn associated with longer progression-free survival (Yan et al., 2017).
CTCs isolation is technically challenging. Current CTC capture methods are either “label-free”, i.e., based on their physicalproperties (e.g., size, density, mechanical and electrical properties) or affinity-based (e.g., utilizing antibodies against protein markers) (Ferreira et al., 2016). Among the latter, an efficient biomarker for CTC capture in breast cancer patients’ blood is the epithelial membrane protein 2 (EMP2) cancer-promoting protein, highly expressed in the epithelial malignancies (Chen et al., 2019). An automated enrichment and immunocytochemical detection system for CTCs CellSearch Circulating Tumor Cell Kits, as EpCAM+/CK+/CD45-cells, in 7.5 mL of blood. The most used marker is in fact theepithelial cell adhesion molecule (EpCAM) (Ferreira et al., 2016; Andree et al., 2016). To date, the CellSearch Circulating Tumor Cell Kit (Menarini Silicon Biosystems, Firenze, Italy) assay, using immunomagnetic technique and cytometry imaging is the only US Food and Drug Administration (FDA) approved CTC diagnostic technology (Palmirotta et al., 2018). A blood test, called CancerSEEK, can detect cancer types using combined assays for genetic alterations and protein biomarkers. CancerSEEK tests were positive in a median of 70% of eight common cancer types, among which ovary, liver, pancreas (Cohen et al., 2018).
Platelets were found to be ‘educated’ (Tumor-Educated Platelets, TEPs) by local and systemic conditions in a number of diseases altering their transcriptome, including cancer. In fact, platelets can sequester RNAs released by cancer cells. The RNA profiles of TEPs from cancer patients are altered, as compared with healthy donors (In’t Veld and Wurdinger, 2019). Hence, TEPRNA repertoire appears a promising biomarker source (Best et al., 2017).
Cell-free nucleic acids include DNA (cfDNA), and RNAs. cfDNA is short, fragmented DNA released from apoptotic or necrotic cancer cells able to provide information on cancer cells genetic and epigenetic mutations (Chen et al., 2020). However, due to its short half-life in the circulation, cfDNA applicability is limited, moreover, its sequencing needs highly sensitive assays such as next-generation sequencing (NGS) (Jimenez-Zenteno and Cerf, 2020). The development of novel techniques for the analysis of low-abundance DNA has allowed to exploit the potential of circulating tumor DNA (ctDNA) as a marker of cancer disease burden, predictor of therapy response or resistance (Cescon et al., 2020). TruSight™ Oncology 500 circulating tumor DNA (Illumina, San Diego, CA) assay enables comprehensive genomic profiling of ctDNA for liquid biopsy profiling assay designed to identify known and emerging tumor biomarkers, including small variants, splice variants, and fusions. Importantly, the TruSightTM Oncology 500 measures TMB and microsatellite instability (MSI), by sequencing cancer-related genes (Wang et al., 2017). However, as its abundance is low and quality often poor, ctDNA extraction needs improvement and ctDNA-based diagnostic tests still need validation.
A major component of the cell secretome is that of extracellular vesicles (EVs). According to the updated guidelines of the International Society for Extracellular Vesicles of 2018 (MISEV2018), EV is a collective term for the lipid-bound nanoparticle, not able to replicate, released by cells and present in all bodily fluids (Thery et al., 2018). The EVs molecular cargo (i.e., proteins, lipids, nucleic acids, and metabolites) plays a signalling role between the cell of origin and the recipient cells (Raposo and Stoorvogel, 2013; van Niel et al., 2018) in a diverse array of functions, thereby comprising an inter-kingdom communication (Chen et al., 2021). EV secretion is an evolutionarily conserved constitutive and regulated process. After release into the extracellular space, EVs reach their target cells where their information is conveyed. Uptake of EVs seems dependent on the recipient cell and can require specific receptors, or involve direct fusion to the plasma membrane, phagocytosis, or endocytosis either clathrin- or caveolin-dependent (Stahl et al., 2019). Therefore, differently from the previous MISEV guidelines, that distinguished exosomes (20–150 nm, deriving from multivesicular bodies), microvesicles (100–1,000 nm, directly budding form the plasmamembrane), and apoptotic bodies (1,000–5,000 nm) (Raposo and Stoorvogel, 2013; Lotvall et al., 2014) the collective term “EVs” is now recommended. EVs can be classified based on their size and biogenesis, as apoptotic bodies (800–5000 nm), microvesicles (MV, 100–1000 nm), and exo (Exo, 30–120 nm) (Maas et al., 2017). Although some markers, such as principally tetraspannins (CD9, CD81, and CD63), and annexins are enriched on Exo (Vlassov et al., 2012; Dear et al., 2013; Abramowicz et al., 2016), there can be some overlap in EV size and markers, therefore classification should primarily based on the size and biogenesis (Thery et al., 2018). It has been suggested to name EVs that do not precipitate at 10,000×g small EVs (sEVs, i.e., exosomes), and large EVs (lEVs, i.e., MV, apoptosomes and oncosomes) those that do (Kowal et al., 2016). MV, also referred to as ectosomes, derive from direct budding from the plasma membrane. Apoptotic bodies are released from apoptotic or tumor cells. Exo, in particular, originate by inward budding of late endosomes/multivesicular bodies (MVB) that release their contents in the extracellular environment fusing with the plasma membrane (Kowal et al., 2014). Exo are found in a vast range of biological fluids as well as in cell culture media (Rashed et al., 2017; van Niel et al., 2018). Exo are released by a variety of mammalian cells, thereby including cancer cells, that were shown to release more Exo than healthy cells (Sun et al., 2018). Recently, Exo have gained interest as biomarkers source for cancer diagnosis and treatment, due to their valuable cargo of proteins, RNAs and microRNAs (miRNAs), DNA and lipids, along with their role as mediators of intercellular communication, immune response and disease development (Chaput and Thery, 2011; Keerthikumar et al., 2016; Rashed et al., 2017). EVs have become a widely studied novel source of disease biomarkers and possible therapeutic agents (Cocucci and Meldolesi, 2015; Maas et al., 2017; Panfoli and Bruschi, 2020). Numerous studies have explored the immense therapeutic potential of Exo that can be collected from bodily fluids by minimally invasive procedures and can be precisely controlled by nanoscale particle assays (Kalluri and LeBleu, 2020). By proteomic and biochemical analyses, we have reported a novel characteristic of human mesenchymal stem cell (MSC) and urinary Exo, i.e., they can consume oxygen to aerobically synthesize ATP (Bruschi et al., 2015; Panfoli et al., 2016; Bruschi et al., 2016). Such metabolic capacity appears consistent with the Exo prolonged permanence in the circulation (Bruschi et al., 2018).
The choice of the proper method for isolation and analysis of clinical grade Exo is challenging (Li et al., 2017; Zhang et al., 2020). There is still lack of standard procedures for single biological fluids for specific downstream clinical diagnostics applications. The current isolation techniques rely either on size differences between EVs (ultracentrifugation, precipitation, filtration, chromatography), or on specific surface markers (immunoaffinity-based methods) (Li et al., 2020). The gold-standard is ultracentrifugation, whoseonly drawback is low Exo recovery. Precipitation is simple and fast, allowing one-step EV isolation, but lacks selectivity setting purity low. Filtration through membranes with appropriate pore size has been used, often combined with ultracentrifugation. However, clogging effects can lower Exo yields. Density gradient centrifugation (on either sucrose or iodixanol) isolates EVs based on in size, mass, and density: it is efficient in separating single EV subpopulations and EVs from contaminant aggregates. Size-exclusion chromatography (SEC) has also been used to separate Exo from protein aggregates, generally after a centrifugation or filtration step (Nordin et al., 2015). Immunoaffinity-based approaches utilize antibody coated magnetic beads to capture Exo expressing specific proteins (Kowal et al., 2016). Exo isolation protocols often combine different methods. A study examined the comparative efficiencies the efficacy of six protocols among which some combinations, for the isolation of urinary Exo: the purest Exo proteins for subsequent proteomic analysis came from ultracentrifugation, or ultracentrifugation on a 30% sucrose cushion, while nano-membrane ultrafiltration centrifugal concentrator (Vivaspin 20) performed less efficiently (Alvarez et al., 2012). The commercial Exo isolation kits (ExoQuick™ Exosome precipitation), that use polymeric additives to precipitate Exo with a standard centrifuge, according to the Authors, need some modifications in order toyield high quality Exo RNAs (Alvarez, 2014). Consistently, a study comparing the efficiency of six commercial kits (exoEasy, ExoQuick, Exo-spin, ME kit, ExoQuick Plus and Exo-Flow) in obtaining pure Exo from healthy sera, found that the highest purity was achieved with ExoQuick Plus and exoEasy, while the lowest with ME kit and ExoQuick (Macias et al., 2019). The isolation of Exo from serum by differential ultracentrifugation and by commercial Exo-spin™ columns was compared recently, and it was found that Exo isolated from blood with either Exo-spin™ or ultracentrifugation were similar, but yields were lower with Exo-spin™ (Małys et al., 2021). Also, residual matrices may influence downstream analyses (Paolini et al., 2016). Optimization is needed to maximize yield and minimize impurities, in function of the biological sample. Microfluidics offers incredible potential, enabling rapid isolation and analysis of clinical grade Exo for diagnostic applications (He and Zeng, 2016). A significant step toward diagnostic accuracy for optimal clinical applications are the microfluidic-based systems, able to perform integrated on-chip exosome isolation, detection, and quantification within a single device. These on-chip multiplexed assays platforms offer several advantages in terms of timing, and sensitivity when processing of small sample volumes, as in the clinical settings (Contreras-Naranjo et al., 2017). These versatile platforms for Exo separation in lab-on-a-chip format can be integrated with multiple processes in a single instrument, reducing the risk of cross-contamination (Liu et al., 2017). The most used of these approaches is immunoaffinity captureof Exo by antibodies immobilized on solid surfaces. A microfluidic platform for multi-scale filtration has been developed (Chen et al., 2020). Acoustic trapping of microparticles on seed microbeads against the flow direction inside a capillary been performed (Wu et al., 2017). Downstream analysis to assess Exo quantity and purity typically involves size characterization by dynamic light scattering (DLS), TEM microscopy to observe morphology, surface marker analysis by immunoassays, and characterization of nucleic acid content (Oosthuyzen et al., 2013; Veziroglu and Mias, 2020).
Exo are drawing attention in the field of liquid biopsy, as they bear an enormous potential to enable personalized medicine (He and Zeng, 2016; Zhou et al., 2020; Li et al., 2021). Already in 2010, the American Society of Clinical Oncology suggested that blood Exo would be a better choice than CTCs for monitoring cancer progression (Pawlowski et al., 2010). In fact, a higher level of blood Exo is found in tumor patients, respect to healthy individuals, independently of tumor histology (Zhong et al., 2021). Exo can be efficiently isolated from a small amount of human bodily fluids and easily classified by homogeneous size, cup-shaped appearance and specific protein markers which can be exploited to separate them from other subcellular vesicles (Li et al., 2020). Moreover, Exo are remarkably stable in the circulation and their cargo is protected from enzymatic degradation by the lipid membrane. Moreover, the release of EVs, among which Exo, is enhanced in cancer (Inal et al., 2013).
The Exo cargo can be investigated by high throughput technologies, such as proteomics and NGS (Abramowicz et al., 2016; Veziroglu and Mias, 2020) and provide a lot of potential information on the parent cancer cell in a simple manner. Moreover, Exo conceivably come from heterogeneous cancer cells, therefore their cargo would not only represent the primary, but also the metastatic tumor cells, as well as the crosstalk among the cancer cells and their environment. Therefore, Exo could unveil the molecular tools enabling of the original cancer to disseminate and metastasize (Fig. 1). In fact, tumor-derived Exo are believed to promote cancer progression and metastasis, by setting the premetastatic niche priming organs for cancer metastasis (Adem et al., 2020). It has been long known that melanoma-derived Exopromote the process of metastasis by modulating the invasive capacity of malignant cells (Wu et al., 2019; Gowda et al., 2020). Exosomal miR-23a plays an important role in promoting tumor progression (Cui et al., 2018).
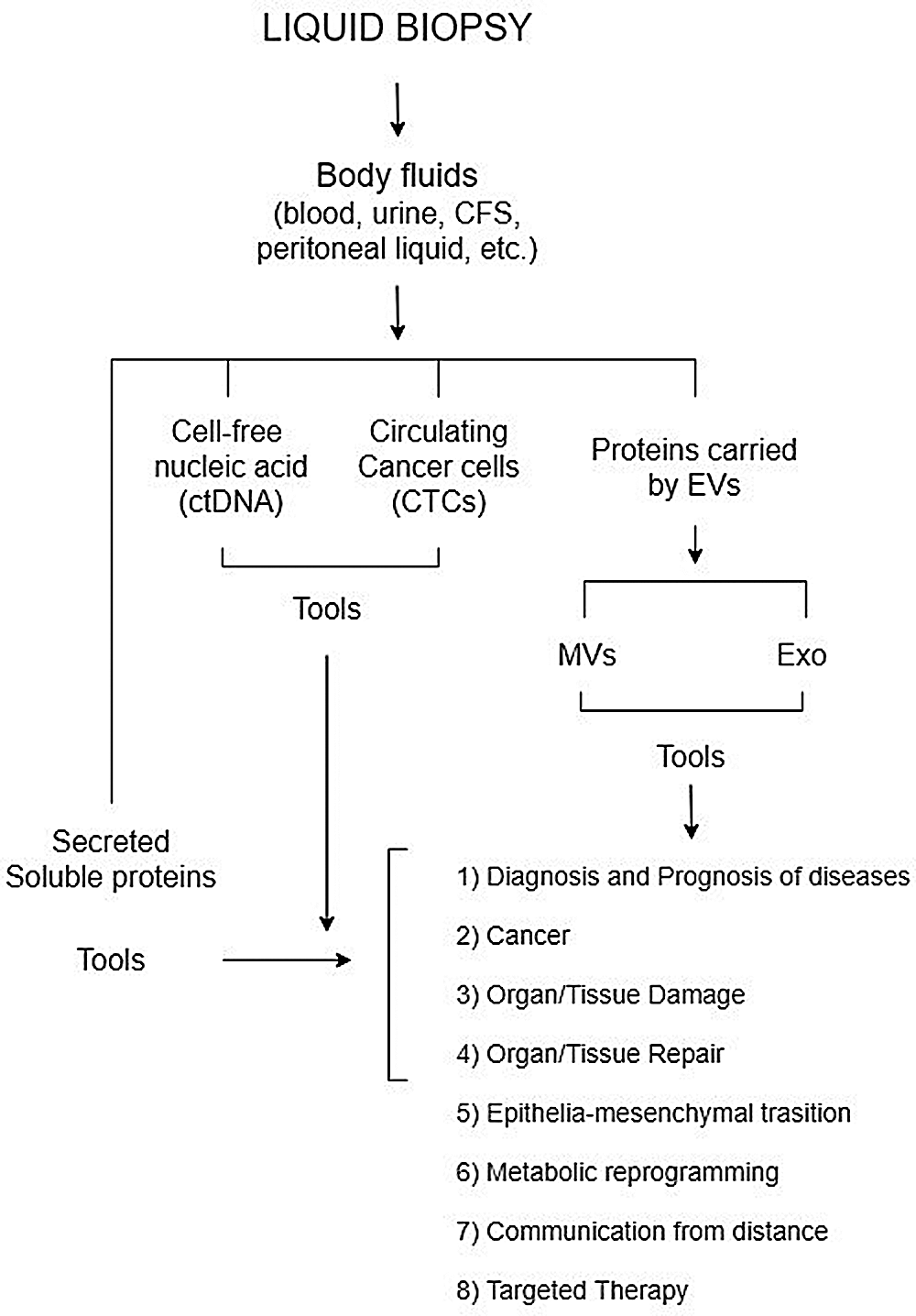
Figure 1: Figure illustrates the use of extracellular vesicle proteins for liquid biopsy.
There has been a tremendous increase in publications on the use of Exo for cancer management (Zhou et al., 2020). Exo are emerging as a novel tool in the field of precision medicine (Kim et al., 2015) and for cancer liquid biopsy (Li et al., 2021). In fact, Exo contain a wealth of information both on their membranes and as a cargo for diagnosis, response to therapy, metastasis, recurrence assessment of tumors.
The use of Exo for liquid biopsy has been demonstrated to hold a great potential for a variety of cancers. Exo are emerging as a valuable source of biomarkers in the effort to improve the prognosis of breast cancer, presently the second cause of cancer mortality in women worldwide (Meng et al., 2019; Wang et al., 2021). One of the recently identified Exo protein biomarkers for the diagnosis of breast cancer was CD82, a tetraspanin that is specifically expressed in Exo microdomains (TEMs) (Wang et al., 2019). The first knowledge base for exosome-based biomarker discovery for breast cancer ExoBCD has recently been developed, that identified 36 promising biomarkers, among which the most promising were IGF1R and FRS2 (Wang et al., 2021). Exo were also shown to participate in a non-targeting effect of breast radiotherapy, i.e., promotion of secondary tumorigenesis through their action on the endothelium: Exo isolated form conditioned media from X-ray irradiated human breast cancer cells (MCF-7) promoted angiogenesis in human umbilical vein endothelial cells (HUVECs), in vitro (Jabbari et al., 2019).
Exo appear also promising in liquid biopsy for ovarian cancer, the most lethal gynaecologic malignancy, for early diagnosis, and even in general population screening (Giannopoulou et al., 2018). In fact, Exo released by the primary ovarian tumor play a pivotal role in metastatic invasion, preparing the pre-metastatic niche (Feng et al., 2019). In particular, the lipidomic profile of exosomes from human ovarian cancer cells (SKOV-3) and ovarian epithelial cancer cell line (HOSEPiC), had a different and individual lipid profile (Cheng et al., 2020) Moreover, in ovarian cancer Exo can be isolated from both ascites and blood.
Exo are valuable diagnostic/prognostic biomarkers for lung cancer, the leading cause of cancer-related death worldwide, due to failure to timely detect the disease the leading cause of cancer-related death worldwide, due to failure to timely detect the disease (Cui et al., 2018). The abundance of miR-3182 in blood Exo was shown to be able of distinguishing non-small cell lung cancer (NSCLC) from benign lung tumours with high sensitivity and specificity, suggesting the potential use of miR-3182 as a biomarker for early NSCLC diagnosis (Visan et al., 2022). A panel of six up-regulated miRNAs (miR-19b-3p, miR-21-5p, miR-221-3p, miR-409-3p, miR-425-5p and miR-584-5p) was developed for the diagnosis of lung adenocarcinoma in peripheral plasma. In particular, the first three cited miRNAs were significantly upregulated in Exo from samples from lung adenocarcinoma (Zhou et al., 2017).
The potential of urinary Exo for urologic tumour lipid biopsy has also been exploited. Prostate Cancer (PCa) has a great prevalence, and there is the lack of specific prognostic tests to differentiate aggressive from indolent forms. Exo mRNA profiling has been proposed as diagnostic procedure for PCa as an alternative to PSA-based screening, which appears inadequate and responsible of over diagnosis. A quantitative lipidomic analysis of urinary Exo was performed in 15 prostate cancer patients and 13 healthy controls: nine lipid species were found to be significantly different among the two groups, besides, an alteration in sphingolipids was observed (Skotland et al., 2017). A high-throughput, profiling platform for spherical nucleic acid-based miRNA (Scano-miR bioassay) was developed, able to identify the molecular signature specific for very high-risk aggressive PCa (Alhasan et al., 2016).
Pancreatic cancer (PC) is a latent lethal malignancy, whose late diagnosis contributes to its poor prognosis. Several studies recently reported the promising diagnostic value of serum Exo in PC early detection and diagnosis (Lu and Risch, 2016). The potential of blood Exo RNAs in PC early detection was confirmed by a recent study showing that PC patients have a distinct blood Exo RNA signature, respect to individuals without PC. In particular, the Exo levels of HIST2H2AA3, LUZP6 and HLA-DRA were considered a signature able to distinguish with high sensitivity and specificity PC patients from healthy controls (Wu et al., 2021). Serum Exo from most PC patients were shown to express selected protein markers (CD44v6, Tspan8, EpCAM, MET and CD104, expressed in Exo of PC cell lines), and to contain miRNAs (miR-1246, miR-4644, miR-3976 and miR-4306), which significantly improve PC diagnosis (Madhavan et al., 2015). A case-control study profiled eight long RNAs (FGA, KRT19, HIST1H2BK, ITIH2, MARCH2, CLDN1, MAL2 and TIMP1) from blood extracellular vesicles from subjects with pancreatic ductal adenocarcinoma (PDAC), respect to controls without PDAC: by a support vector machine algorithm, the study identified a d-signature able to identify PDAC at a resectable stage, which could sensibly improve the PDAC prognosis (Yu et al., 2020). Interestingly, urine Exo also were shown to contain several markers associated with PC (Madhavan et al., 2015). The possibility to find diagnostic biomarkers that are not exclusive of the urinary tract and the kidney in the urinary Exo, holds an immense potential for liquid biopsy (Franzen et al., 2016; Panfoli, 2017). Long non-coding RNA (lncRNA) CCHE1 plays a role in ovarian cancer metastasis, as demonstrated by its silencing, which suggests its potential role as a biomarker for its diagnosis (Chen et al., 2020).
The potential of Exo as biomarker source for therapeutic applications has been highlighted also for liver diseases. Exo are released by hepatocytes as part of a crosstalk among them and non parenchymal cells (hepatic stellate cells, liver sinusoidal endothelial cells, and cholangiocytes). In liver, Exo function in regulating regeneration upon liver injury but can also play a role in priming the tumour microenvironment promoting cancer progression (Sung et al., 2018).
CRC-derived Exo were shown to promote metabolic reprogramming and immuno-suppressive signals facilitating the formation of the pre-metastatic niche (Mannavola et al., 2019). Therefore, as CRC cells secrete Exo that convey tumorigenic signals to stromal cells, Exo represent a promising target of liquid biopsy also for colorectal cancer (CRC) since its early phases.
Melanoma is one of the most aggressive cancers, with growing incidence rates. Lack of markers for the early detection of the disease still prevents proper treatment. Melanoma-derived Exo are known to promote the process of metastasis by setting the pre-metastatic niche (Gowda et al., 2020). A study isolated simultaneously CTCs and Exo from blood samples using the dual-utilization OncoBean (DUO) device and found that both express melanoma-associated genes (Kang et al., 2020). Moreover, blood of melanoma patients possessed three times more exosomal protein mL−1 than healthy donors (Kang et al., 2020).
Exo-based diagnosis appears promising also for brain cancer, as it could allow bypassing tissue biopsy, especially in patients with high surgical risk. Exo isolated from serum of 96 high-grade glioma patients could reliably detect the tumorigenic epidermal growth factor receptor variant III (EGFRvIII), typical of high-grade gliomas (Manda et al., 2018).
Exo from dendritic cells (DCs), named dexosomes were shown to transfer the antigen-MHC complexes to naïve DCs, as they express the major histocompatibility complex class I/II (MHC I/II). This ability has been exploited to ex vivo induce DCs to present tumorantigens, and use them as cancer vaccines. These were shown to be safe for administration into patients to induce a tumor-specific immune response, in two phase I and one phase II clinical trials. In particular, in malignant melanoma, dexosomes elicited NK-related immune responses (Bol et al., 2019).
Advantages of Exosomes for Liquid Biopsy
The use of Exo for cancer liquid biopsy has several advantages, respect to TEPs, CTCs and ctDNA (Li et al., 2021). In fact, TEP RNAs can be affected by drugs or immunological status. CTCs are phenotypically heterogeneous and require rapid processing after isolation (Abramowicz et al., 2016). By contrast, is the availability of integrated microfluidic platforms for cheap and rapid separation of Exo, which require small sample volumes and allowing automation (Li et al., 2020). Limitations of ctDNA regard its chemical characteristics and its variable concentration in blood, in fact it accounts for a fraction of cfDNA, which mostly derives from non-malignant cells. ctDNA can also be influenced by multiple tumor, and anatomical factors, as well as unrelated somatic mutations (Li et al., 2021). Notably, it is possible to detect cancer cell DNA aberrations also using Exo. A disadvantage in the use of Exo is the technical difficulty isolation and detection techniques, and the impossibility to image them acquiring morphological information as can be done on CTCs (Masuda et al., 2016).
The most important advantage of using Exo for liquid biopsies is the possibility to identify all at the same time proteins, nucleic acids and lipids (Cheng et al., 2020) specifically packed so to convey an integrated information for the homing at distance from the primary tumor site, meantto build the premetastatic niche (Masuda et al., 2016), also reflecting the stromal along with the malignant cells. Furthermore, in this respect Exo offer the possibility to identify surface vs. cargo proteins. Notably, the proteins the Exo carry mediate a non-conventional form of protein secretion, as they lack a signal peptide (Inal et al., 2013) and interestingly among them the five complexes of the redox chain were found functionally expressed (Bruschi et al., 2015; Panfoli et al., 2016; Bruschi et al., 2016). Consistently, ExoCarta (http://www.exocarta.org), reports the expression of several of the subunits of the F1Fo-ATP synthase and of the respiratory chain complexes. Human urinary and umbilical cord mesenchymal stem cell Exo produce ATP and display a respiratory ability independent of whole mitochondria (Bruschi et al., 2015; Panfoli et al., 2016; Bruschi et al., 2018). Such ecto-ATP can be metabolizedto adenosine by ectonucleotidases such as CD73, expressed on extracellular vesicles, presumably as a part of inflammatory signalling mechanism (Schneider et al., 2021). Therefore, besides bearing a potential for biomarker for cancer liquid biopsy, Exo can convey information on the bioenergetic and oxidative state of the individual: this was shown to be the case for Mesenchymal Stem Cell exosomes from preterm and term newborns (Panfoli et al., 2016). In fact, stem cells produce Exo that promote angiogenesis and appear to be a promising tool for the treatment of ischemic diseases (Babaei and Rezaie, 2021).
The identification and selection of valuable biomarkers from biofluids remains a challenge. Liquid biopsy is a novel, minimally invasive emerging technique alternative to tissue biopsy for malignancies (Martins et al., 2021), for the advantage of allowing serial sampling. In particular, the potential of Exo for liquid biopsy is undebatable, due to their unique potential in revealing the role of a specific combination of lipids, nucleic acids and surface/cargo proteins in cancer dissemination and metastasis, the most important cause of cancer death (Dillekas et al., 2019). There is the need for standardization of Exo isolation and storage protocols from the different body fluids, to reach analytical consistency, and for validation from clinical trials to support applicability in monitoring cancer. Nonetheless, it can be foreseen that new/standardized methods for Exo capture and analysis of Exo from body fluids will be implemented, and, after demonstration of feasibility, their use for liquid biopsy may hopefully be inserted into guidelines.
Author Contribution: Conceptualization, Isabella Panfoli; literature search and data analysis, Maurizio Bruschi; writing -original draft, review and editing, Isabella Panfoli; critical revision of the work, Giovanni Candiano. All authors have read and agreed to the published version of the manuscript.
Funding Statement: This study was supported by the Italian Ministry of Health-Cinque per mille and Ricerca Corrente to Istituto Giannina Gaslini and Fondazione Malattie Renali del Bambino OLNUS.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Abramowicz A, Widlak P, Pietrowska M (2016). Proteomic analysis of exosomal cargo: The challenge of high purity vesicle isolation. Molecular Biosystem 12: 1407–1419. DOI 10.1039/C6MB00082G. [Google Scholar] [CrossRef]
Adem B, Vieira PF, Melo SA (2020). Decoding the biology of exosomes in metastasis. Trends in Cancer 6: 20–30. DOI 10.1016/j.trecan.2019.11.007. [Google Scholar] [CrossRef]
Alhasan AH, Scott AW, Wu JJ, Feng G, Meeks JJ, Thaxton CS, Mirkin CA (2016). Circulating microRNA signature for the diagnosis of very high-risk prostate cancer. PNAS 113: 10655–10660. DOI 10.1073/pnas.1611596113. [Google Scholar] [CrossRef]
Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW (2004). Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clinical Cancer Research 10: 6897–6904. DOI 10.1158/1078-0432.CCR-04-0378. [Google Scholar] [CrossRef]
Alvarez ML (2014). Isolation of urinary exosomes for RNA biomarker discovery using a simple, fast, and highly scalable method. Methods in Molecular Biology 1182: 145–170. DOI 10.1007/978-1-4939-1062-5. [Google Scholar] [CrossRef]
Alvarez ML, Khosroheidari M, Kanchi Ravi R, DiStefano JK (2012). Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney International 82: 1024–1032. DOI 10.1038/ki.2012.256. [Google Scholar] [CrossRef]
Andree KC, van Dalum G, Terstappen LW (2016). Challenges in circulating tumor cell detection by the CellSearch system. Molecular Oncology 10: 395–407. DOI 10.1016/j.molonc.2015.12.002. [Google Scholar] [CrossRef]
Babaei M, Rezaie J (2021). Application of stem cell-derived exosomes in ischemic diseases: Opportunity and limitations. Journal of Translational Medicine 19: 196. DOI 10.1186/s12967-021-02863-w. [Google Scholar] [CrossRef]
Best MG, Vancura A, Wurdinger T (2017). Platelet RNA as a circulating biomarker trove for cancer diagnostics. Journal of Thrombosis and Haemostasis 15: 1295–1306. DOI 10.1111/jth.13720. [Google Scholar] [CrossRef]
Bol KF, Schreibelt G, Rabold K, Wculek SK, Schwarze JK et al. (2019). The clinical application of cancer immunotherapy based on naturally circulating dendritic cells. Journal for Immunotherapy of Cancer 7: 109. DOI 10.1186/s40425-019-0580-6. [Google Scholar] [CrossRef]
Brabletz T, Kalluri R, Nieto MA, Weinberg RA (2018). EMT in cancer. Nature Reviews Cancer 18: 128–134. DOI 10.1038/nrc.2017.118. [Google Scholar] [CrossRef]
Bruschi M, Ravera S, Santucci L, Candiano G, Bartolucci M et al. (2015). The human urinary exosome as a potential metabolic effector cargo. Expert Review of Proteomics 12: 425–432. DOI 10.1586/14789450.2015.1055324. [Google Scholar] [CrossRef]
Bruschi M, Santucci L, Ravera S, Bartolucci M, Petretto A, Calzia D, Ghiggeri GM, Ramenghi LA, Candiano G, Panfoli I (2018). Metabolic signature of microvesicles from umbilical cord mesenchymal stem cells of preterm and term infants. PROTEOMICS-Clinical Applications 12: e1700082. DOI 10.1002/prca.201700082. [Google Scholar] [CrossRef]
Bruschi M, Santucci L, Ravera S, Candiano G, Bartolucci M et al. (2016). Human urinary exosome proteome unveils its aerobic respiratory ability. Journal of Proteomics 136: 25–34. DOI 10.1016/j.jprot.2016.02.001. [Google Scholar] [CrossRef]
Cescon DW, Bratman SV, Chan SM, Siu LL (2020). Circulating tumor DNA and liquid biopsy in oncology. Nature Cancer 1: 276–290. DOI 10.1038/s43018-020-0043-5. [Google Scholar] [CrossRef]
Chaput N, Thery C (2011). Exosomes: Immune properties and potential clinical implementations. Seminars in Immunopathology 33: 419–440. DOI 10.1007/s00281-010-0233-9. [Google Scholar] [CrossRef]
Chen Q, Yao L, Burner D, Minev B, Lu L, Wang M, Ma W (2019). Epithelial membrane protein 2: A novel biomarker for circulating tumor cell recovery in breast cancer. Clinical and Translational Oncology 21: 433–442. DOI 10.1007/s12094-018-1941-1. [Google Scholar] [CrossRef]
Chen X, Wang L, Lou J (2020). Nanotechnology strategies for the analysis of circulating tumor DNA: A review. Medical Science Monitor 26: e921040. DOI 10.12659/MSM.921040. [Google Scholar] [CrossRef]
Chen Y, Xu Y, Zhong H, Yuan H, Liang F, Liu J, Tang W (2021). Extracellular vesicles in Inter-Kingdom communication in gastrointestinal cancer. American Journal of Cancer Research 11: 1087–1103. [Google Scholar]
Cheng L, Zhang K, Qing Y, Li D, Cui M, Jin P, Xu T (2020). Proteomic and lipidomic analysis of exosomes derived from ovarian cancer cells and ovarian surface epithelial cells. Journal of Ovarian Research 13: 9. DOI 10.1186/s13048-020-0609-y. [Google Scholar] [CrossRef]
Cocucci E, Meldolesi J (2015). Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends in Cell Biology 25: 364–372. DOI 10.1016/j.tcb.2015.01.004. [Google Scholar] [CrossRef]
Cohen JD, Li L, Wang Y, Thoburn C, Afsari B et al. (2018). Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359: 926–930. DOI 10.1126/science.aar3247. [Google Scholar] [CrossRef]
Connors D, Allen J, Alvarez JD, Boyle J, Cristofanilli M et al. (2020). International liquid biopsy standardization alliance white paper. Critical Reviews in Oncology/Hematology 156: 103112. DOI 10.1016/j.critrevonc.2020.103112. [Google Scholar] [CrossRef]
Contreras-Naranjo JC, Wu HJ, Ugaz VM (2017). Microfluidics for exosome isolation and analysis: Enabling liquid biopsy for personalized medicine. Lab on a Chip 17: 3558–3577. DOI 10.1039/C7LC00592J. [Google Scholar] [CrossRef]
Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J et al. (2004). Circulating tumor cells, disease progression, and survival in metastatic breast cancer. New England Journal of Medicine 351: 781–791. DOI 10.1056/NEJMoa040766. [Google Scholar] [CrossRef]
Cui S, Cheng Z, Qin W, Jiang L (2018). Exosomes as a liquid biopsy for lung cancer. Lung Cancer 116: 46–54. [Google Scholar]
Dear JW, Street JM, Bailey MA (2013). Urinary exosomes: A reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics 13: 1572–1580. [Google Scholar]
Dillekas H, Rogers MS, Straume O (2019). Are 90% of deaths from cancer caused by metastases? Cancer Medicine 8: 5574–5576. [Google Scholar]
Feng W, Dean DC, Hornicek FJ, Shi H, Duan Z (2019). Exosomes promote pre-metastatic niche formation in ovarian cancer. Molecular Cancer 18: 124. [Google Scholar]
Ferreira MM, Ramani VC, Jeffrey SS (2016). Circulating tumor cell technologies. Molecular Oncology 10: 374–394. [Google Scholar]
Franzen CA, Blackwell RH, Foreman KE, Kuo PC, Flanigan RC, Gupta GN (2016). Urinary exosomes: The potential for biomarker utility, intercellular signaling and therapeutics in urological malignancy. Journal of Urology 195: 1331–1339. DOI 10.1016/j.juro.2015.08.115. [Google Scholar] [CrossRef]
Giannopoulou L, Zavridou M, Kasimir-Bauer S, Lianidou ES (2018). Liquid biopsy in ovarian cancer: The potential of circulating miRNAs and exosomes. Traslational Research 205: 77–91. DOI 10.1016/j.trsl.2018.10.003. [Google Scholar] [CrossRef]
Girardi F, Allemani C, Coleman MP (2019). Worldwide trends in survival from common childhood brain tumors: A systematic review. Journal of Global Oncology 5: 1–25. DOI 10.1200/JGO.19.00140. [Google Scholar] [CrossRef]
Gowda R, Robertson BM, Iyer S, Barry J, Dinavahi SS, Robertson GP (2020). The role of exosomes in metastasis and progression of melanoma. Cancer Treatment Reviews 85: 101975. DOI 10.1016/j.ctrv.2020.101975. [Google Scholar] [CrossRef]
He M, Zeng Y (2016). Microfluidic exosome analysis toward liquid biopsy for cancer. Journal of Laboratory Automation 21: 599–608. DOI 10.1177/2211068216651035. [Google Scholar] [CrossRef]
Heeke S, Mograbi B, Alix-Panabieres C, Hofman P (2019). Never travel alone: The crosstalk of circulating tumor cells and the blood microenvironment. Cells 8: 714. DOI 10.3390/cells8070714. [Google Scholar] [CrossRef]
In’t Veld S, Wurdinger T (2019). Tumor-educated platelets. Blood 133: 2359–2364. DOI 10.1182/blood-2018-12-852830. [Google Scholar] [CrossRef]
Inal JM, Kosgodage U, Azam S, Stratton D, Antwi-Baffour S, Lange S (2013). Blood/plasma secretome and microvesicles. Biochimica Biophysica Acta 1834: 2317–2325. DOI 10.1016/j.bbapap.2013.04.005. [Google Scholar] [CrossRef]
Jabbari N, Nawaz M, Rezaie J (2019). Bystander effects of ionizing radiation: Conditioned media from X-ray irradiated MCF-7 cells increases the angiogenic ability of endothelial cells. Cell Communication and Signaling 17: 165. DOI 10.1186/s12964-019-0474-8. [Google Scholar] [CrossRef]
Jimenez-Zenteno AK, Cerf A (2020). Liquid biopsy based on circulating cancer-associated cells: Bridging the gap from an emerging concept to a mainstream tool in precision medicine. Advanced Biosystems 4: e1900164. DOI 10.1002/adbi.201900164. [Google Scholar] [CrossRef]
Kalluri R, LeBleu VS (2020). The biology, function, and biomedical applications of exosomes. Science 367: eaau6977. [Google Scholar]
Kamaraju S, Drope J, Sankaranarayanan R, Shastri S (2020). Cancer prevention in low-resource countries: An overview of the opportunity. American Society of Clinical Oncology Educational Book 40: 1–12. DOI 10.1200/EDBK_280625. [Google Scholar] [CrossRef]
Kang YT, Hadlock T, Lo TW, Purcell E, Mutukuri A et al. (2020). Dual-isolation and profiling of circulating tumor cells and cancer exosomes from blood samples with melanoma using immunoaffinity-based microfluidic interfaces. Advanced Science 7: 2001581. DOI 10.1002/advs.202001581. [Google Scholar] [CrossRef]
Keerthikumar S, Chisanga D, Ariyaratne D, Al Saffar H, Anand S et al. (2016). ExoCarta: A web-based compendium of exosomal cargo. Journal of Molecular Biology 428: 688–692. DOI 10.1016/j.jmb.2015.09.019. [Google Scholar] [CrossRef]
Kim DK, Lee J, Simpson RJ, Lotvall J, Gho YS (2015). EVpedia: A community web resource for prokaryotic and eukaryotic extracellular vesicles research. Seminars in Cell & Developmental Biology 40: 4–7. DOI 10.1016/j.semcdb.2015.02.005. [Google Scholar] [CrossRef]
Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Thery C (2016). Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. PNAS 113: E968–77. DOI 10.1073/pnas.1521230113. [Google Scholar] [CrossRef]
Kowal J, Tkach M, Thery C (2014). Biogenesis and secretion of exosomes. Current Opinion in Cell Biology 29: 116–125. DOI 10.1016/j.ceb.2014.05.004. [Google Scholar] [CrossRef]
Li G, Tang W, Yang F (2020). Cancer liquid biopsy using integrated microfluidic exosome analysis platforms. Biotechnology Journal 15: e1900225. DOI 10.1002/biot.201900225. [Google Scholar] [CrossRef]
Li P, Kaslan M, Lee SH, Yao J, Gao Z (2017). Progress in exosome isolation techniques. Theranostics 7: 789–804. DOI 10.7150/thno.18133. [Google Scholar] [CrossRef]
Li S, Yi M, Dong B, Tan X, Luo S, Wu K (2021). The role of exosomes in liquid biopsy for cancer diagnosis and prognosis prediction. International Journal of Cancer 148: 2640–2651. DOI 10.1002/ijc.33386. [Google Scholar] [CrossRef]
Liu C, Guo J, Tian F, Yang N, Yan F, Ding Y, Wei J, Hu G, Nie G, Sun J (2017). Field-free isolation of exosomes from extracellular vesicles by microfluidic viscoelastic flows. ACS Nano 11: 6968–6976. DOI 10.1021/acsnano.7b02277. [Google Scholar] [CrossRef]
Lotvall J, Hill AF, Hochberg F, Buzas EI, di Vizio D et al. (2014). Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the international society for extracellular vesicles. Journal of Extracellular Vesicles 3: 26913. DOI 10.3402/jev.v3.26913. [Google Scholar] [CrossRef]
Lu L, Risch HA (2016). Exosomes: Potential for early detection in pancreatic cancer. Future Oncology 12: 1081–1090. DOI 10.2217/fon-2015-0005. [Google Scholar] [CrossRef]
Lu L, Zeng H, Gu X, Ma W (2015). Circulating tumor cell clusters-associated gene plakoglobin and breast cancer survival. Breast Cancer Research and Treatment 151: 491–500. DOI 10.1007/s10549-015-3416-1. [Google Scholar] [CrossRef]
Ma N, Jeffrey SS (2020). Deciphering cancer clues from blood. Science 367: 1424–1425. DOI 10.1126/science.abb0736. [Google Scholar] [CrossRef]
Maas SLN, Breakefield XO, Weaver AM (2017). Extracellular vesicles: Unique intercellular delivery vehicles. Trends in Cell Biology 27: 172–188. DOI 10.1016/j.tcb.2016.11.003. [Google Scholar] [CrossRef]
Macias M, Rebmann V, Mateos B, Varo N, Perez-Gracia JL, Alegre E, Gonzalez A (2019). Comparison of six commercial serum exosome isolation methods suitable for clinical laboratories. Effect in cytokine analysis. Clinical Chemistry and Laboratory Medicine 57: 1539–1545. DOI 10.1515/cclm-2018-1297. [Google Scholar] [CrossRef]
Madhavan B, Yue S, Galli U, Rana S, Gross W et al. (2015). Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. International Journal of Cancer 136: 2616–2627. DOI 10.1002/ijc.29324. [Google Scholar] [CrossRef]
Małys MS, Aigner C, Schulz SM, Schachner H, Rees AJ, Kain R, (2021). Isolation of Small Extracellular Vesicles from Human Sera. International Journal of Molecular Sciences 22, DOI 10.3390/IJMS22094653. [Google Scholar] [CrossRef]
Manda SV, Kataria Y, Tatireddy BR, Ramakrishnan B, Ratnam BG, Lath R, Ranjan A, Ray A (2018). Exosomes as a biomarker platform for detecting epidermal growth factor receptor-positive high-grade gliomas. Journal of Neurosurgery 128: 1091–1101. DOI 10.3171/2016.11.JNS161187. [Google Scholar] [CrossRef]
Mannavola F, Salerno T, Passarelli A, Tucci M, Interno V, Silvestris F (2019). Revisiting the role of exosomes in colorectal cancer: Where are we now? Frontiers in Oncology 9: 521. DOI 10.3389/fonc.2019.00521. [Google Scholar] [CrossRef]
Martins I, Ribeiro IP, Jorge J, Goncalves AC, Sarmento-Ribeiro AB, Melo JB, Carreira IM (2021). Liquid biopsies: Applications for cancer diagnosis and monitoring. Genes 12: 349. DOI 10.3390/genes12030349. [Google Scholar] [CrossRef]
Masuda T, Hayashi N, Iguchi T, Ito S, Eguchi H, Mimori K (2016). Clinical and biological significance of circulating tumor cells in cancer. Molecular Oncology 10: 408–417. DOI 10.1016/j.molonc.2016.01.010. [Google Scholar] [CrossRef]
Meng Y, Sun J, Wang X, Hu T, Ma Y, Kong C, Piao H, Yu T, Zhang G (2019). Exosomes: A promising avenue for the diagnosis of breast cancer. Technology in Cancer Research and Treatment 18: 1533033818821421. DOI 10.1177/1533033818821421. [Google Scholar] [CrossRef]
The global challenge of cancer (2020). Nature Cancer 1: 1–2. DOI 10.1038/s43018-019-0023-9. [Google Scholar] [CrossRef]
Nordin JZ, Lee Y, Vader P, Mager I, Johansson HJ et al. (2015). Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine: Nanotechnology, Biology, and Medicine 11: 879–883. DOI 10.1016/j.nano.2015.01.003. [Google Scholar] [CrossRef]
Oosthuyzen W, Sime NE, Ivy JR, Turtle EJ, Street JM et al. (2013). Quantification of human urinary exosomes by nanoparticle tracking analysis. Journal of Physiology 591: 5833–5842. DOI 10.1113/jphysiol.2013.264069. [Google Scholar] [CrossRef]
Palmirotta R, Lovero D, Cafforio P, Felici C, Mannavola F, Pelle E, Quaresmini D, Tucci M, Silvestris F (2018). Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Therapeutic Advances in Medical Oncology 10: 1758835918794630. DOI 10.1177/1758835918794630. [Google Scholar] [CrossRef]
Panfoli I (2017). Cancer exosomes in urine: A promising biomarker source. Translational Cancer Research 6: S1389–S1393. DOI 10.21037/tcr.2017.10.17. [Google Scholar] [CrossRef]
Panfoli I, Bruschi M (2020). The good and bad sides of exosomes: Pre-metastatic niche formation, cancer biomarker and therapy carriers. Journal of Cancer Metastasis and Treatment 6: 35. DOI 10.20517/2394-4722.2020.50. [Google Scholar] [CrossRef]
Panfoli I, Ravera S, Podesta M, Cossu C, Santucci L et al. (2016). Exosomes from human mesenchymal stem cells conduct aerobic metabolism in term and preterm newborn infants. FASEB Journal 30: 1416–1424. DOI 10.1096/fj.15-279679. [Google Scholar] [CrossRef]
Paolini L, Zendrini A, di Noto G, Busatto S, Lottini E, Radeghieri A, Dossi A, Caneschi A, Ricotta D, Bergese P (2016). Residual matrix from different separation techniques impacts exosome biological activity. Scientific Reports 6: 23550. DOI 10.1038/srep23550. [Google Scholar] [CrossRef]
Patel SA, Rodrigues P, Wesolowski L, Vanharanta S (2021). Genomic control of metastasis. British Journal of Cancer 124: 3–12. DOI 10.1038/s41416-020-01127-6. [Google Scholar] [CrossRef]
Pawlowski TL, Spetzler D, Tinder T, Kimbrough J, Deng T, Kim J, Ellis P, Tyrell A, Kennedy P, Kuslich C (2010). Circulating exosomes may provide a more sensitive platform to monitor disease progression compared to circulating tumor cells. Journal of Clinical Oncology 28: 10580. DOI 10.1200/jco.2010.28.15_suppl.10580. [Google Scholar] [CrossRef]
Poulet G, Massias J, Taly V (2019). Liquid biopsy: General concepts. Acta Cytological 63: 449–455. DOI 10.1159/000499337. [Google Scholar] [CrossRef]
Quandt D, Dieter Zucht H, Amann A, Wulf-Goldenberg A, Borrebaeck C et al. (2017). Implementing liquid biopsies into clinical decision making for cancer immunotherapy. Oncotarget 8: 48507–48520. DOI 10.18632/oncotarget.17397. [Google Scholar] [CrossRef]
Raposo G, Stoorvogel W (2013). Extracellular vesicles: Exosomes, microvesicles, and friends. Journal of Cell Biology 200: 373–383. DOI 10.1083/jcb.201211138. [Google Scholar] [CrossRef]
Rashed MH, Bayraktar E, Helal GK, Abd-Ellah MF, Amero P, Chavez-Reyes A, Rodriguez-Aguayo C (2017). Exosomes: From garbage bins to promising therapeutic targets. International Journal of Molecular Science 18: 538. DOI 10.3390/ijms18030538. [Google Scholar] [CrossRef]
Sai B, and Xiang J, (2018). Disseminated tumour cells in bone marrow are the source of cancer relapse after therapy. Journal of Cellular and Molecular Medicine 22: 5776. DOI 10.1111/JCMM.13867. [Google Scholar] [CrossRef]
Schneider E, Winzer R, Rissiek A, Ricklefs I, Meyer-Schwesinger C et al. (2021). CD73-mediated adenosine production by CD8 T cell-derived extracellular vesicles constitutes an intrinsic mechanism of immune suppression. Nature Communications 12: 5911. DOI 10.1038/s41467-021-26134-w. [Google Scholar] [CrossRef]
Siravegna G, Marsoni S, Siena S, Bardelli A (2017). Integrating liquid biopsies into the management of cancer. Nature Review in Clinical Oncology 14: 531–548. DOI 10.1038/nrclinonc.2017.14. [Google Scholar] [CrossRef]
Skotland T, Ekroos K, Kauhanen D, Simolin H, Seierstad T, Berge V, Sandvig K, Llorente A (2017). Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. European Journal of Cancer 70: 122–132. DOI 10.1016/j.ejca.2016.10.011. [Google Scholar] [CrossRef]
Stahl AL, Johansson K, Mossberg M, Kahn R, Karpman D (2019). Exosomes and microvesicles in normal physiology, pathophysiology, and renal diseases. Pediatric Nephrology 34: 11–30. DOI 10.1007/s00467-017-3816-z. [Google Scholar] [CrossRef]
Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Yuan W (2018). Effect of exosomal miRNA on cancer biology and clinical applications. Molecular Cancer 17: 147. DOI 10.1186/s12943-018-0897-7. [Google Scholar] [CrossRef]
Sung S, Kim J, Jung Y (2018). Liver-derived exosomes and their implications in liver pathobiology. International Journal of Molecular Sciences 19: 3715. DOI 10.3390/ijms19123715. [Google Scholar] [CrossRef]
Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018A position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. Journal of Extracell Vesicles 7: 1535750. DOI 10.1080/20013078.2018.1535750. [Google Scholar] [CrossRef]
van Niel G, D'Angelo G, Raposo G (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology 19: 213–228. DOI 10.1038/nrm.2017.125. [Google Scholar] [CrossRef]
Veziroglu EM, Mias GI (2020). Characterizing extracellular vesicles and their diverse RNA contents. Frontiers in Genetics 11: 700. DOI 10.3389/fgene.2020.00700. [Google Scholar] [CrossRef]
Visan KS, Lobb RJ, Wen SW, Bedo J, Lima LG et al. (2022). Blood-derived extracellular vesicle-associated miR-3182 detects non-small cell lung cancer patients. Cancers (Basel) 14: 257. [Google Scholar]
Vlassov AV, Magdaleno S, Setterquist R, Conrad R (2012). Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochimica et Biophysica Acta 1820: 940–948. DOI 10.1016/j.bbagen.2012.03.017. [Google Scholar] [CrossRef]
Wang J, Chang S, Li G, Sun Y (2017). Application of liquid biopsy in precision medicine: Opportunities and challenges. Frontiers of Medicine 11: 522–527. DOI 10.1007/s11684-017-0526-7. [Google Scholar] [CrossRef]
Wang X, Chai Z, Pan G, Hao Y, Li B et al. (2021). ExoBCD: A comprehensive database for exosomal biomarker discovery in breast cancer. Briefings in Bioinformatics 22: bbaa088. [Google Scholar]
Wang X, Zhong W, Bu J, Li Y, Li R, Nie R, Xiao C, Ma K, Huang X (2019). Exosomal protein CD82 as a diagnostic biomarker for precision medicine for breast cancer. Molecular Carcinogenesis 58: 674–685. DOI 10.1002/mc.22960. [Google Scholar] [CrossRef]
Wu M, Ouyang Y, Wang Z, Zhang R, Huang PH et al. (2017). Isolation of exosomes from whole blood by integrating acoustics and microfluidics. PNAS 114: 10584–10589. DOI 10.1073/pnas.1709210114. [Google Scholar] [CrossRef]
Wu M, Wang G, Hu W, Yao Y, Yu XF (2019). Emerging roles and therapeutic value of exosomes in cancer metastasis. Molecular Cancer 18: 53. DOI 10.1186/s12943-019-0964-8. [Google Scholar] [CrossRef]
Wu Y, Zeng H, Yu Q, Huang H, Fervers B, Chen ZS, Lu L (2021). A circulating exosome RNA signature is a potential diagnostic marker for pancreatic cancer, a systematic study. Cancers 13: 2565. DOI 10.3390/cancers13112565. [Google Scholar] [CrossRef]
Xu L, Jia S, Li H, Yu Y, Liu G et al. (2018). Characterization of circulating tumor cells in newly diagnosed breast cancer. Oncology Letters 15: 2522–2528. [Google Scholar]
Yan WT, Cui X, Chen Q, Li YF, Cui YH, Wang Y, Jiang J (2017). Circulating tumor cell status monitors the treatment responses in breast cancer patients: A meta-analysis. Scientific Reports 7: 43464. DOI 10.1038/srep43464. [Google Scholar] [CrossRef]
Yu S, Li Y, Liao Z, Wang Z, Qian L et al. (2020). Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut 69: 540–550. DOI 10.1136/gutjnl-2019-318860. [Google Scholar] [CrossRef]
Zhang Y, Bi J, Huang J, Tang Y, Du S et al. (2020). Exosome: A review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. International Journal of Nanomedicine 15: 6917–6934. [Google Scholar]
Zhong Y, Li H, Li P, Chen Y, Zhang M et al. (2021). Exosomes: A new pathway for cancer drug resistance. Frontiers in Oncology 11: 1–9. DOI 10.3389/fonc.2021.743556. [Google Scholar] [CrossRef]
Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y, Shao Y, Zheng S (2020). Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduction and Targeted Therapy 5: 144. DOI 10.1038/s41392-020-00258-9. [Google Scholar] [CrossRef]
Zhou X, Wen W, Shan X, Zhu W, Xu J et al. (2017). A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget 8: 6513–6525. DOI 10.18632/oncotarget.14311. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |