DOI:10.32604/biocell.2022.021107

| BIOCELL DOI:10.32604/biocell.2022.021107 |  |
| Review |
CRISPR accelerates the cancer drug discovery
1School of Life Science, Anhui Medical University, Hefei, 230000, China
2Institute for Molecular Medicine Finland, Helsinki Institute of Life Science, University of Helsinki, Helsinki, 00014, Finland
3Department of Anatomy, Faculty of Medicine, University of Helsinki, Helsinki, 00014, Finland
*Address correspondence to: Kecheng Zhou, zhoukecheng@ahmu.edu.cn
#Equal contribution
Received: 28 December 2021; Accepted: 10 March 2022
Abstract: Emerging cohorts and basic studies have associated certain genetic modifications in cancer patients, such as gene mutation, amplification, or deletion, with the overall survival prognosis, underscoring patients’ genetic background may directly regulate drug sensitivity/resistance during chemotherapies. Understanding the molecular mechanism underpinning drug sensitivity/resistance and further uncovering the effective drugs have been the major ambition in the cancer drug discovery. The emergence and popularity of CRISPR/Cas9 technology have reformed the entire life science research, providing a precise and simplified genome editing tool with unlimited editing possibilities. Furthermore, it presents a powerful tool in cancer drug discovery, which hopefully facilitates us with a rapid and reliable manner in developing novel therapies and understanding the molecular mechanisms of drug sensitivity/resistance. Herein, we summarized the application of CRISPR/Cas9 in drug screening, with the focus on CRISPR/Cas9 mediated gene knock-out, gene knock-in, as well as transcriptional modification. Additionally, this review provides the concerns, cautions, and ethnic considerations that need to be taken when applying CRISPR in the drug discovery.
Keywords: CRISPR/Cas9; Cancer drug discovery; Drug sensitivity/Resistance; Novel cancer therapies
Around the world, tremendous efforts and resources are being invested in discovering and developing anticancer drugs/therapies. Conventional anticancer drug discovery focused on cytotoxic compounds (Gavande et al., 2016), selecting agents that exhibit significant cytotoxic effects on tumor cell lines in vitro and further induced tumor regression in animal models (Liu et al., 2017b). Although this strategy has achieved significant success, the recent developments in molecular biology and an understanding of the pharmacology of cancer at a molecular level have challenged researchers to come up with target-based drugs (Baudino, 2015; Connors, 1996).
During the historical events in cancer drug discovery, identifying key genetic signatures to increase the drug effects has always been one major task and goal (Hyter et al., 2018; Roti and Stegmaier, 2012). Loss-of-Function (LOF) and Gain-of-Function (GOF) are among the most commonly employed strategies to deconvolve the key gene roles in the drug response (Plenge et al., 2013). LOF mainly includes inhibition of gene expression (such as siRNA/shRNA-based transcript degradation/blocking) and total knockout of certain genes from the genome (e.g., via CRISPR/Cas9, zinc-finger nucleases, or transcription activator-like effector nucleases (TALENs).
Conversely, GOF is usually achieved by cDNA expression. However, the overexpression system by different plasmids may bring artificial readout since the expression level is much higher than the endogenous one (Plenge et al., 2013). Recent advances in CRISPR/Cas9 medicated gene-activation/gene knock-in can manipulate gene expression at the endogenous level, which is promising in overcoming these obstacles (Konermann et al., 2015). In this review, we summarize the applications of CRISPR/Cas9 in drug discovery, which functions in a more feasible and precise manner, including several potentials in both LOF and GOF research strategies. Additionally, we provide the concerns and cautions that must be taken practically.
LOF and the Principle of CRISPR/Cas9 System
The most adaptable and popular method for LOF study is siRNA-mediated gene knockdown guided by sequence complementarity because siRNAs and transfection reagents are commercially available at a reasonable cost and the practical procedures are relatively easy (Jackson and Linsley, 2010), it additionally displays potential in therapeutics development as it blocks the synthesis of disease-causing proteins (Wittrup and Lieberman, 2015). Nevertheless, the siRNA-mediated gene silencing is transient, may come together with poor reproducibility and serious off-target effect (Jackson and Linsley, 2010), occasionally it even lead to entirely wrong results (Neumeier and Meister, 2021).
Thus, siRNAs are commonly used to conduct pilot experiments. On the contrary, specific stable gene silencing techniques are usually employed in massive numbers of protein functional studies, especially considering the increasing usage of CRISPR/Cas9 techniques which have become widespread in the past few years (Zhan et al., 2019).
The Clustered Regular Interspaced Short Palindromic Repeats (CRISPR) is an adaptive immune system in bacteria and related organisms (Jinek et al., 2012; Sharma et al., 2021). CRISPR/Cas9 consists of programmed single-stranded guide RNA (sgRNA) and a Cas9 endonuclease, which produces double-stranded DNA breakage (DSB) at the sequence-specific site and disrupts one particular area of a gene, making the cell permanently incapable of producing the protein of interest (Fig. 1A) (Doudna and Charpentier, 2014; Sharma et al., 2021). It has shown great potential in biological research, even in treating specific human diseases (Sharma et al., 2021). However, it is worthy to note that even after Cas9 mediated gene modification, some proteins that are truncated at the N-terminus may still be encoded and can retain partial functions of the original protein (Sharpe and Cooper, 2017). CRISPR/Cas9 overcomes the limitations of conventional genome engineering predecessors, such as Zinc-finger nucleases and transcription activator-like effector nucleases (TALENs), by far simplifying the whole experimental steps (Fig. 1B) (Gaj et al., 2013). Besides, Cas9 makes a double-strand break in the DNA, which is repaired by the cell’s error-prone DNA repair machinery, improving the gene inhibition efficiency compared with siRNA or shRNA (Gaj et al., 2013; Makarova et al., 2011).
The CRISPR/Cas9 system is localized by sgRNA, particularly the first 20 bases of sgRNA, which significantly reduces the experimental cost and period for mammalian gene editing by simply designing different sgRNAs (Jiang and Doudna, 2017; Zhan et al., 2019). Moreover, the Cas9 (mediating nucleic acid endocytosis function) and sgRNA (mediating localization function) of the CRISPR/Cas9 system are independent. Therefore, Cas9 protein and multiple sgRNAs could be expressed simultaneously in the cell (Ceasar et al., 2016; Doudna and Charpentier, 2014), increasing the research application (Jinek et al., 2013). CRISPR/Cas9 library built by this strategy has been used to identify new biological mechanisms of drug resistance and explore single or combined drug effects in the cancer viability (Doudna, 2020; Kurata et al., 2018).
CRISPR/Cas9 Mediated Gene Knock-Out
CRISPR/Cas9 generates accurate DNA damage on the genome, usually double-stranded breaks (DSBs) (Jinek et al., 2012; Yang et al., 2020). Cells have several DNA repair mechanisms for DSBs, the most common one is linking the two broken DNA terminals together without any modification, namely nonhomologous DNA end joining (NHEJ) (Fig. 1C) (Ceccaldi et al., 2016; Makarova et al., 2020). Since CRISPR/Cas9 machinery commonly deletes a couple of nucleotides from the genome, the directly linking repairment by NHEJ usually results in the frameshift change for the original DNA sequence, causing the permanent loss of functions for a particular gene/genomic region, namely “gene knockout (KO)” (Ran et al., 2013). For this reason, gene knockout is the primary and also well-developed application of the CRISPR/Cas9 system in the genome/gene editing (Ceasar et al., 2016; Doudna and Charpentier, 2014; Jinek et al., 2012). We have recently employed CRISPR/Cas9 to generate LAPTM4B KO cell lines; our studies further revealed that the lysosomal protein LAPTM4B plays a critical role in lysosomal leucine uptake and the mTORC1 signaling activation (Zhou et al., 2018).

Figure 1: Introduction of CRISPR/Cas9 and the principle. (A) The gene-editing capability of CRISPR relies on Cas9 protein, guide RNA, and PAM sequence. PAM: Protospacer adjacent motif. (B) An overview of CRISPR/Cas9, TALEN, and ZFN. TALEN: Transcription activator-like effector nucleases; ZFN: Zinc-finger nucleases; RVD: Repeat variable di-residues. (C) The functional diagram of NHEJ and HDR. Repairment by NHEJ directly links two DNA terminals or with a random insertion, commonly causing gene knockout. On the contrary, repairment by HDR relies on template DNA, resulting in “error-free” gene knock-in. NHEJ: Nonhomologous DNA end joining; HDR: Homology-directed repair.
CRISPR/Cas9 Medicated Knock-In
Another mechanism of double-strand break repair (DSBR) is the homology-directed repair (HDR) (Ceccaldi et al., 2016; Doudna and Charpentier, 2014). HDR can achieve precise insertion, deletion, or mutation of base pairs using homologous sequences as donor templates (Fig. 1C) (Yang et al., 2020). In this process named “gene knock-in”, cells were transfected with plasmids containing Cas9 and sgRNAs, together with a plasmid that accommodates homology arms flanking the targeted genomic locus where the knocked-in sequence is to be inserted, such as a point mutation, protein tag, or fluorescence (Fig. 2A) (Banan, 2020). Therefore, CRISPR/Cas9 mediated gene knock-in can produce accurate modification or label specific genes with fluorescence at the endogenous level (Banan, 2020; Doudna and Charpentier, 2014). Previous studies have generated cells with start codon mutation on LAPTM4B isoforms by using HDR to deconvolve the functional dissimilar between LAPTM4B isoforms (Zhou et al., 2020). By utilizing CRISPR/Cas9 mediated gene knock-in, we labeled LAPTM4B with GFP to visualize the endogenous protein localization and found LAPTM4B distributes on multivesicular bodies (Dichlberger et al., 2021). We believe endogenously labeling oncogene with fluorescence, together with a high-throughput drug screen platform and automatic/semi-automatic image/data analysis, will significantly speed up the drug discovery process and emerge promising usage both academically and industrially (Fig. 2A). However, it is noteworthy that the high-throughput imaging-based drug screen method is time-consuming and costly, and more efforts are warranted to improve the flexibility and reduce the experimental cost.
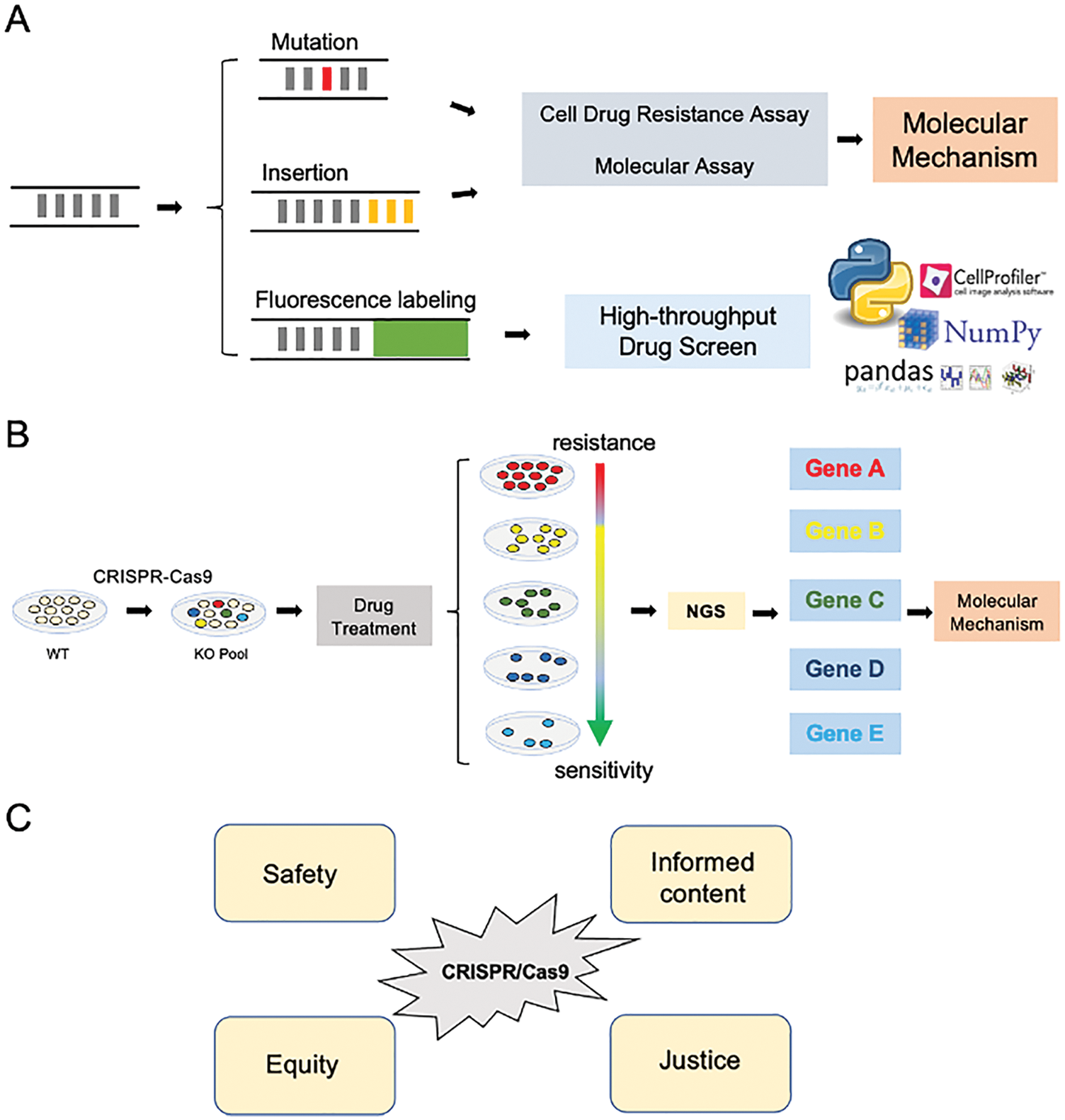
Figure 2: The application of CRISPR/Cas9 in cancer drug discovery. (A) CRISPR/Cas9 mediated gene knock-in can induce gene mutation, insertion, and fluorescence labeling for a particular gene of interest. Cells with mutant/inserted genes could be utilized in drug resistance assay, together with molecular experiments, to deconvolute the biological mechanism of drug resistance/sensitivity. Cells labeled fluorescence can be used in the high-throughput drug screen, the fluorescent images can be processed by “Cell Profiler” to convert image data to digital data. The digital data further be calculated by “Python” using Pandas/Numpy modules; the image and digital data facilitate discovering the specific drugs for the gene of interest. (B) Gene knockout (KO) cell pool was generated by CRISPR/Cas9 method, which undergoes drug treatment. Drug resistance/sensitivity was assessed by cell viability. Selected cells are sequenced by NGS, which gives insights into drug resistance/sensitivity-related genes and the related molecular mechanism. NGS: Next-generation sequencing. (C) The ethnic consideration of CRISPR/Cas9 when conducting the cancer drug discovery projects.
Unlike NHEJ, which could occur at any stage in the cell cycle, HDR occurs only in the S and G2 phases of the cell cycle because HDR relies on sister chromatids as a template for the repair (Yang et al., 2020). Furthermore, the knock-in strategy shows potential in cells and organoids. Artegiani et al. recently developed a CRISPR/Cas9-mediated homology-independent organoids transgenesis, making it possible to conduct a fast and efficient generation of knock-in human organoids (Artegiani et al., 2020).
CRISPR/Cas9 Mediated Transcriptional Repression
As an RNA-guided DNA endonuclease, Cas9 can be easily programmed to target new sites by altering the guide RNA sequence (Ceasar et al., 2016). Recently, Larson et al. (2013) developed a novel methodology to design and construct sgRNAs for transcriptional interference, which could be utilized for modifying any gene of interest in a flexible, efficient, and accurate manner. Excitingly, this strategy can be adapted for high-throughput studies on gene functions, even in various organisms, displaying several advantages compared with the RNA interference (Larson et al., 2013).
CRISPR/Cas9 Mediated Transcriptional Activation
Alternatively, CRISPR-Cas9 has been employed in mediating efficient transcriptional activation at endogenous loci (Joung et al., 2017). Recently, in a screen to study BRAF inhibitors resistance, Konermann et al. (2015) have synthesized a library with more than 70 thousand guides to active different RefSeq coding isoforms, intriguingly, the gene signature generated from this screen are consistent with previous reports, indicating the potential usage and the robustness of CRISPR/Cas9 mediated transcriptional activation in drug discovery (Konermann et al., 2015). Meanwhile, CRISPR shows promise in enhancing gene expression by modulating endogenous regulatory genomic elements in vivo, developed as a proof-of-concept study in mice models expressing Cas9 and transcriptional transactivation domains (Schoger et al., 2020).
The New Era of CRISPR/Cas9 in Drug Discovery
Human Genome Project provides us the reference DNA sequence at the whole genome level (Collins et al., 2003; Green et al., 2015); however, the function of encoding genes or non-coding regions, and how are they related to health and diseases are yet to be uncovered (Gonzaga-Jauregui et al., 2012). CRISPR/Cas9 emerged as an essential tool in studying these biological processes (Fig. 2B) (Zhan et al., 2019). Moreover, the current emergence of CRISPR/Cas9 technology enables the possibility in conducting large-scale genetic manipulation to investgate drug resistance, thus revealing physiological gene function and exploring the protentional drugs (Fig. 2B) (Larson et al., 2013).
CRISPR/Cas9 facilitates constructing transgenic animal models, laying a good foundation and severing as an in vivo platform for drug target discovery and validation (Ma et al., 2020; Ryu et al., 2018), as the early “knockout” animal models by zinc-finger nucleases or TALEN are time-consuming and costly (Whitelaw et al., 2016). In summary, CRISPR/Cas9 technology enables drug developers and researchers to screen drug targets in a high-efficient and orientated manner, accelerating the drug discovery process by various gene manipulation, such as gene knockout or knock-in, site-specific mutation, and homologous recombination.
Cautions Need to Be Taken on CRISPR/Cas9 in Drug Discovery
CRISPR/Cas9-mediated gene modifications have been widely used recently. However, the downside of CRISPR/Cas9 is that establishing a genetically modified cell line may be time-consuming, especially for the low-proliferative cells, because the whole procedure includes plasmids construction and transfection, pre-selection, single-cell clone expansion, and multiple validation experiments at DNA level, transcript level or protein level (Larson et al., 2013).
Due to the relatively long period of single-clone expansion (usually 1–2 months), genome-modified cells may undergo certain reprogram to compensate for the gene KO effects (Liu et al., 2017a; Rodríguez-Rodríguez et al., 2019; Rossi et al., 2015). As a result, phenotypes observed in the eventual KO clone may result from reprogramming mechanisms rather than directly induced by the gene of interest (Rossi et al., 2015). Therefore, during the cancer drug discovery, it is plausible that GOF approaches are additionally employed to rescue the phenotypes of gene KO to confirm that the phenotypes observed are dependent on the protein of interest.
Another primary concern is the off-target effect of the CRISPR/Cas9 system. However, Cas9 is supposed to be directed by 20 nt guide sequence of sgRNA, and the protospacer adjacent motif (PAM) (Anders et al., 2014), 3–5 base pair mismatched in the PAM-distal part of the sgRNA guiding sequence may induce off-target cleavage activity (Pattanayak et al., 2013). Extensive efforts and strategies have been proposed to minimize the off-target cleavage, e.g. designing more precise sgRNAs to improve target specificity, selecting sgRNAs in promoters, enhancers, and genes as far as possible to improve the target efficiency (Zhang et al., 2015). Moreover, in silico methods are developed to predict/screen the potential off-target sites, such as “OffScan” which serves as a standard and rapid CRISPR off-target sites detection tool (Cui et al., 2020).
Due to the promise of CRISPR/Cas9 in developing as novel therapeutics, the concern regarding off-target effect drives researchers’ interest in uncovering specific inhibitors. Several inhibitors have been found to date, e.g., AcrIIC1 is capable of disabling Cas9’s nucleases, and ArcIIC3 prevents DNA targeting and thus induces dimerization of Cas9 (Harrington et al., 2017).
The off-target effect promotes us to consider the ethical concern on CRISPR in animal or human germline/embryo projects although it gives great promise in cancer drug discovery. International Summit on Human Gene Editing acknowledges that safety, informed content, justice, and equity are the primary concern when conducting genome-editing research (Fig. 2C) (Araki and Ishii, 2014; Chan et al., 2015).
Discussion and Future Directions
Cancer treatment with the stratifying prognosis remains challenging despite enormous research in improving the treatment outcome. Identifying essential genes regulating drug resistance is the critical question, imperative for understanding the biological progress of drug resistance and developing novel cancer therapeutics, whether by utilizing FDA-approved drugs or synthesizing new molecular inhibitors.
CRISPR/Cas9 technology has altered and revolutionized biological research by presenting a precise, simple, and versatile tool with multiple potentials in gene knockout, gene knock-in, active/inactive transcriptional modification. Recent studies report that several CRISPR-Cas proteins display impressive usage in genome/transcription level manipulations and resolving critical scientific and social problems, e.g., Cas1-Cas2-mediated spacer integration facilitates CRISPR-Cas establishing immunity (Xiao et al., 2017), Cas12 based lateral flow assay helps detect SARS-CoV-2, providing an efficient and robust alternative to the current RT-PCR test during COVID-19 pandemic (Broughton et al., 2020).
Recently, many novel techniques underwent rapid development, such as high-content confocal microscopy, high-throughput drug screen system, and automated robotic systems. Moreover, novel bioinformatic resources are vital in data analysis and prediction, e.g., calculation and prediction algorithms, imaging analysis software, data analysis, and assembling tools. We believe that with the advances mentioned above and newly developed multi-omics assay (genomic, transcriptomic, proteomics, metabolomics) to study the biological mechanism, the CRISPR/Cas9 system is revolutionizing drug discovery with new cancer therapeutics in the visible venue.
Author Contributions: YR, JW, ML, and KZ jointly wrote this review. YR and JW drafted the manuscript. ML contributed to the in-depth discussion and conception. KZ developed the structure, designed the review’s scope, and helped draft the manuscript. All authors read and approved the final manuscript.
Funding Statement: This work was supported by the Start-Up funding from Anhui Medical University (KZ 0801033201), and the Doctoral Research Funding from the Cancer Society of Finland (KZ).
Conflicts of Interest: The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Anders C, Niewoehner O, Duerst A, Jinek M (2014). Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513: 569–573. DOI 10.1038/nature13579. [Google Scholar] [CrossRef]
Araki M, Ishii T (2014). International regulatory landscape and integration of corrective genome editing into in vitro fertilization. Reproductive Biology and Endocrinology 12: 1–12. DOI 10.1186/1477-7827-12-108. [Google Scholar] [CrossRef]
Artegiani B, Hendriks D, Beumer J, Kok R, Zheng X et al. (2020). Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nature Cell Biology 22: 321–331. DOI 10.1038/s41556-020-0472-5. [Google Scholar] [CrossRef]
Banan M (2020). Recent advances in CRISPR/Cas9-mediated knock-ins in mammalian cells. Journal of Biotechnology 308: 1–9. DOI 10.1016/j.jbiotec.2019.11.010. [Google Scholar] [CrossRef]
Baudino T (2015). Targeted cancer therapy: The next generation of cancer treatment. Current Drug Discovery Technologies 12: 3–20. DOI 10.2174/1570163812666150602144310. [Google Scholar] [CrossRef]
Broughton J, Deng X, Yu G, Fasching C, Servellita V et al. (2020). CRISPR-Cas12-based detection of SARS-CoV-2. Nature Biotechnology 38: 870–874. DOI 10.1038/s41587-020-0513-4. [Google Scholar] [CrossRef]
Ceasar SA, Rajan V, Prykhozhij SV, Berman JN, Ignacimuthu S (2016). Insert, remove or replace: A highly advanced genome editing system using CRISPR/Cas9. Biochimica et Biophysica Acta-Molecular Cell Research 1863: 2333–2344. DOI 10.1016/j.bbamcr.2016.06.009. [Google Scholar] [CrossRef]
Ceccaldi R, Rondinelli B, D’Andrea AD (2016). Repair pathway choices and consequences at the double-strand break. Trends in Cell Biology 26: 52–64. DOI 10.1016/j.tcb.2015.07.009. [Google Scholar] [CrossRef]
Chan S, Donovan PJ, Douglas T, Gyngell C, Harris J, Lovell-Badge R, Mathews DJ, Regenberg A, Hinxton Group (2015). Genome editing technologies and human germline genetic modification: The hinxton group consensus statement. The American Journal of Bioethics: AJOB 15: 42–47. DOI 10.1080/15265161.2015.1103814. [Google Scholar] [CrossRef]
Collins FS, Morgan M, Patrinos A (2003). The human genome project: Lessons from large-scale biology. Science 300: 286–290. DOI 10.1126/science.1084564. [Google Scholar] [CrossRef]
Connors T (1996). Anticancer drug development: The way forward. Oncologist 1: 180–181. DOI 10.1634/theoncologist.1-3-180. [Google Scholar] [CrossRef]
Cui Y, Liao X, Peng S, Tang T, Huang C, Yang C (2020). OffScan: A universal and fast CRISPR off-target sites detection tool. BMC Genomics 21: 8–11. DOI 10.1186/s12864-019-6241-9. [Google Scholar] [CrossRef]
Dichlberger A, Zhou K, Bäck N, Nyholm T, Backman A, Mattjus P, Ikonen E, Blom T (2021). LAPTM4B controls the sphingolipid and ether lipid signature of small extracellular vesicles. Biochimica et Biophysica Acta-Molecular and Cell Biology of Lipids 1866: 158855. DOI 10.1016/j.bbalip.2020.158855. [Google Scholar] [CrossRef]
Doudna JA (2020). The promise and challenge of therapeutic genome editing. Nature 578: 229–236. DOI 10.1038/s41586-020-1978-5. [Google Scholar] [CrossRef]
Doudna JA, Charpentier E (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. DOI 10.1126/science.1258096. [Google Scholar] [CrossRef]
Gaj T, Gersbach CA, Barbas CF (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in Biotechnology 31: 397–405. DOI 10.1016/j.tibtech.2013.04.004. [Google Scholar] [CrossRef]
Gavande NS, Vandervere-Carozza PS, Hinshaw HD, Jalal SI, Sears CR et al. (2016). DNA repair targeted therapy: The past or future of cancer treatment? Pharmacology & Therapeutics 160: 65–83. DOI 10.1016/j.pharmthera.2016.02.003. [Google Scholar] [CrossRef]
Gonzaga-Jauregui C, Lupski JR, Gibbs RA (2012). Human genome sequencing in health and disease. Annual Review of Medicine 63: 35–61. DOI 10.1146/annurev-med-051010-162644. [Google Scholar] [CrossRef]
Green ED, Watson JD, Collins FS (2015). Human genome project: Twenty-five years of big biology. Nature 526: 29–31. DOI 10.1038/526029a. [Google Scholar] [CrossRef]
Harrington LB, Doxzen KW, Ma E, Liu JJ, Knott GJ et al. (2017). A broad-spectrum inhibitor of CRISPR-Cas9. Cell 170: 1224–1233.e15. DOI 10.1016/j.cell.2017.07.037. [Google Scholar] [CrossRef]
Hyter S, Hirst J, Pathak H, Pessetto ZY, Koestler DC et al. (2018). Developing a genetic signature to predict drug response in ovarian cancer. Oncotarget 9: 14828–14848. DOI 10.18632/oncotarget.23663. [Google Scholar] [CrossRef]
Jackson AL, Linsley PS (2010). Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nature Reviews Drug Discovery 9: 57–67. DOI 10.1038/nrd3010. [Google Scholar] [CrossRef]
Jiang F, Doudna JA (2017). CRISPR-Cas9 structures and mechanisms. Annual Review of Biophysics 46: 505–529. DOI 10.1146/annurev-biophys-062215-010822. [Google Scholar] [CrossRef]
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA et al. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. DOI 10.1126/science.1225829. [Google Scholar] [CrossRef]
Jinek M, East A, Cheng A, Lin S, Ma E et al. (2013). RNA-programmed genome editing in human cells. Elife 2: e00471. DOI 10.7554/eLife.00471.009. [Google Scholar] [CrossRef]
Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ et al. (2017). Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nature Protocols 12: 828–863. DOI 10.1038/nprot.2017.016. [Google Scholar] [CrossRef]
Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588. DOI 10.1038/nature14136. [Google Scholar] [CrossRef]
Kurata M, Yamamoto K, Moriarity BS, Kitagawa M, Largaespada DA (2018). CRISPR/Cas9 library screening for drug target discovery. Journal of Human Genetics 63: 179–186. DOI 10.1038/s10038-017-0376-9. [Google Scholar] [CrossRef]
Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS et al. (2013). CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nature Protocols 8: 2180–2196. DOI 10.1038/nprot.2013.132. [Google Scholar] [CrossRef]
Liu J, Zhou Y, Qi X, Chen J, Chen W et al. (2017a). CRISPR/Cas9 in zebrafish: An efficient combination for human genetic diseases modeling. Human Genetics 136: 1–12. DOI 10.1007/s00439-016-1739-6. [Google Scholar] [CrossRef]
Liu Z, Delavan B, Roberts R, Tong W (2017b). Lessons learned from two decades of anticancer drugs. Trends in Pharmacological Sciences 38: 852–872. DOI 10.1016/j.tips.2017.06.005. [Google Scholar] [CrossRef]
Ma X, Shang X, Qin X, Lu J, Liu M et al. (2020). Characterization of organic anion transporting polypeptide 1b2 knockout rats generated by CRISPR/Cas9: A novel model for drug transport and hyperbilirubinemia disease. Acta Pharmaceutica Sinica B 10: 850–860. DOI 10.1016/j.apsb.2019.11.007. [Google Scholar] [CrossRef]
Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nature Reviews Microbiology 9: 467–477. DOI 10.1038/nrmicro2577. [Google Scholar] [CrossRef]
Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS et al. (2020). Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nature Reviews Microbiology 18: 67–83. DOI 10.1038/s41579-019-0299-x. [Google Scholar] [CrossRef]
Neumeier J, Meister G (2021). siRNA Specificity: RNAi mechanisms and strategies to reduce off-target effects. Frontiers in Plant Science 11: 526455. DOI 10.3389/fpls.2020.526455. [Google Scholar] [CrossRef]
Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA et al. (2013). High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nature Biotechnology 31: 839–843. DOI 10.1038/nbt.2673. [Google Scholar] [CrossRef]
Plenge RM, Scolnick EM, Altshuler D (2013). Validating therapeutic targets through human genetics. Nature Reviews Drug Discovery 12: 581–594. DOI 10.1038/nrd4051. [Google Scholar] [CrossRef]
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA et al. (2013). Genome engineering using the CRISPR-Cas9 system. Nature Protocols 8: 2281–2308. DOI 10.1038/nprot.2013.143. [Google Scholar] [CrossRef]
Rodríguez-Rodríguez DR, Ramírez-Solís R, Garza-Elizondo MA, Garza-Rodríguez MDL, Barrera-Saldaña HA (2019). Genome editing: A perspective on the application of CRISPR/Cas9 to study human diseases. International Journal of Molecular Medicine 43: 1559–1574. DOI 10.3892/ijmm.2019.4112. [Google Scholar] [CrossRef]
Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S et al. (2015). Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524: 230–233. DOI 10.1038/nature14580. [Google Scholar] [CrossRef]
Roti G, Stegmaier K (2012). Genetic and proteomic approaches to identify cancer drug targets. British Journal of Cancer 106: 254–261. DOI 10.1038/bjc.2011.543. [Google Scholar] [CrossRef]
Ryu J, Prather RS, Lee K (2018). Use of gene-editing technology to introduce targeted modifications in pigs. Journal of Animal Science and Biotechnology 9: 5. DOI 10.1186/s40104-017-0228-7. [Google Scholar] [CrossRef]
Schoger E, Carroll KJ, Iyer LM, McAnally JR, Tan W et al. (2020). CRISPR-mediated activation of endogenous gene expression in the postnatal heart. Circulation Research 126: 6–24. DOI 10.1161/CIRCRESAHA.118.314522. [Google Scholar] [CrossRef]
Sharma G, Sharma AR, Bhattacharya M, Lee SS, Chakraborty C (2021). CRISPR-Cas9: A preclinical and clinical perspective for the treatment of human diseases. Molecular Therapy 29: 571–586. DOI 10.1016/j.ymthe.2020.09.028. [Google Scholar] [CrossRef]
Sharpe JJ, Cooper TA (2017). Unexpected consequences: Exon skipping caused by CRISPR-generated mutations. Genome Biology 18: 1–4. DOI 10.1186/s13059-017-1240-0. [Google Scholar] [CrossRef]
Whitelaw CBA, Sheets TP, Lillico SG, Telugu BP (2016). Engineering large animal models of human disease. The Journal of Pathology 238: 247–256. DOI 10.1002/path.4648. [Google Scholar] [CrossRef]
Wittrup A, Lieberman J (2015). Knocking down disease: A progress report on siRNA therapeutics. Nature Reviews Genetics 16: 543–552. DOI 10.1038/nrg3978. [Google Scholar] [CrossRef]
Xiao Y, Ng S, Nam HK, Ke A (2017). How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration. Nature 550: 137–141. DOI 10.1038/nature24020. [Google Scholar] [CrossRef]
Yang H, Ren S, Yu S, Pan H, Li T et al. (2020). Methods favoring homology-directed repair choice in response to CRISPR/Cas9 induced-double strand breaks. International Journal of Molecular Sciences 21: 1–20. DOI 10.3390/ijms21186461. [Google Scholar] [CrossRef]
Zhan T, Rindtorff N, Betge J, Ebert MP, Boutros M (2019). CRISPR/Cas9 for cancer research and therapy. Seminars in Cancer Biology 55: 106–119. DOI 10.1016/j.semcancer.2018.04.001. [Google Scholar] [CrossRef]
Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH (2015). Off-target effects in CRISPR/Cas9-mediated genome engineering. Molecular Therapy Nucleic Acids 4: e264. DOI 10.1038/mtna.2015.37. [Google Scholar] [CrossRef]
Zhou K, Dichlberger A, Ikonen E, Blom T (2020). Lysosome associated protein transmembrane 4B-24 is the predominant protein isoform in human tissues and undergoes rapid, nutrient-regulated turnover. American Journal of Pathology 190: 2018–2028. DOI 10.1016/j.ajpath.2020.07.003. [Google Scholar] [CrossRef]
Zhou K, Dichlberger A, Martinez-Seara H, Nyholm TKM, Li S et al. (2018). A ceramide-regulated element in the late endosomal protein LAPTM4B controls amino acid transporter interaction. ACS Central Science 4: 548–558. DOI 10.1021/acscentsci.7b00582. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |