DOI:10.32604/biocell.2022.021290

| BIOCELL DOI:10.32604/biocell.2022.021290 |  |
| Article |
Uridine dynamic administration affects the circadian variation of bile acid metabolism in high-fat-diet-fed mice
1CAS Key Laboratory of Agri-Ecological Processes in Subtropical Region, Institute of Subtropical Agriculture, the Chinese Academy of Sciences, Hunan Provincial Engineering Research Center for Healthy Livestock and Poultry Production, Changsha, 410125, China
2Institute of Biological Resources, Jiangxi Academy of Sciences, Nanchang, 330096, China
3University of Chinese Academy of Sciences, Beijing, 100049, China
*Address correspondence to: Xin Wu, wuxin@isa.ac.cn
#These authors contributed equally to this work.
Received: 06 January 2022; Accepted: 11 April 2022
Abstract: High-fat diet (HFD) is demonstrated to disturb the bile acid metabolism. The rhythm of bile acid metabolism can also be affected by uridine, whose metabolism exhibits a daily rhythm. However, the mechanism of dynamic uridine administration affecting bile acid during HFD remains unclear. In this study, C57BL/6J mice were fed HFD (the control group; CON) or HFD with oral administration of uridine in the daytime (DUR) and nighttime (NUR) to investigate the mechanism of the effect of uridine on the bile acid. This study showed that the mRNA expression of uridine transporters and circadian clock genes in the jejunum was affected by zeitgeber time (ZT) (P < 0.001). Genes related to the metabolism of pyrimidines in the liver showed a high dependence on daily rhythm (P < 0.01), and DUR remarkably up-regulated the expression of ribonucleotide reductase regulatory subunit M2 (RRM2) (P < 0.05) compared to the CON group. Importantly, the mRNA expression of bile acids nuclear receptors, bile acid synthesis, and transporters in the liver showed significantly rhythmically changed (P < 0.05), and the expression of cholesterol 7-alpha-hydroxylase (CYP7A1), fibroblast growth factor receptor 4 (FGFR4), Na+/taurocholate co transporting polypeptide (NTCP), and bile salt export pump (BSEP) mRNAs of mice with uridine administration increased significantly (P < 0.05). The mRNA expression of the transporters of cholesterol and bile acids in the ileum was also affected by ZT (P < 0.01) and significantly dependent on uridine administration (P < 0.05). The expression of FXR and SHP was significantly affected by ZT and uridine, respectively. In conclusion, dynamic administration of uridine could regulate the rhythm of gene expression of pyrimidine and bile acid metabolism in the liver and ileum of HFD-fed mice, which contributed to the further study of circadian rhythmic physiological and pathological changes of bile acids.
Keywords: Diurnal rhythm; Uridine; Dynamic; Bile acids metabolism; High-fat diet
Circadian rhythm is a rhythmic pattern of approximately 24 h exhibited in most organisms (Chowdhury et al., 2019). The circadian clock system resides in the suprachiasmatic nucleus and peripheral tissues, which controls physiological functions and behaviors in the body through a complex program of circadian clock genes and proteins, which present rhythmically in nearly all cell types and expressions (Bass, 2012; Chen et al., 2019; Green et al., 2008). Daily rhythms are typically measured as activity vs. rest in animals and have environmental cues, such as light and temperature, and are called zeitgebers (Merrow et al., 2005). Importantly, the circadian machinery is frequently due to the variation in nutrition mode, including feeding time or energy level of diet (high-fat diet, HFD) (Asher and Sassone, 2015; Hatori et al., 2012; Johnston et al., 2016; Kohsaka et al., 2007), which leads to the disruption of rhythmic metabolism of glucose and lipids (Appiakannan et al., 2019; Hatsumi et al., 2018; Honma et al., 2016; Sun et al., 2015). Several studies have demonstrated that in the HFD-induced obesity mice model, the expression of core circadian clock genes such as circadian locomotor output cycles kaput (CLOCK) and brain and muscle Arnt-like protein-1 is altered and is associated with lipid metabolic changes (Turek et al., 2005).
The expression of genes responsible for nucleotide metabolism is under the control of the circadian clock (Fustin et al., 2012). Uridine, as one of the nucleosides, can stimulate nucleotide transport (Xie et al., 2019), maintain intestinal development (Li et al., 2016), improve the growth performance of suckling piglets and weaned piglets (Li et al., 2019; Wu et al., 2020), and suppress the function of intestinal stem cells (Liu et al., 2020). Importantly, uridine can regulate lipid metabolism (Le et al., 2014a, Le et al., 2014b); short-term uridine administration could prevent drug-induced liver lipid accumulation (Le et al., 2014a), while long-term uridine administration induced severe liver lipid accumulation in mice (Urasaki et al., 2016). Most interestingly, dynamic uridine administration affected the diurnal variations in liver nucleotide, cholesterol, and lipid metabolism (Liu et al., 2019; Zhang et al., 2018b), indicating that dynamic uridine administration may provide a reference for lipid metabolism disorders.
As a lipid emulsifier, bile acids are important for lipid metabolism (Ye et al., 2018), and bile acid metabolism also shows circadian rhythm (Eggink et al., 2017); the profile of bile acid metabolism is regulated by a high-fat diet (Muhammad et al., 2016; Yoshitsugu et al., 2019). Of note, there is a close relationship between bile acid metabolism and uridine, and bile might be involved in plasma uridine clearance (Deng et al., 2017). Besides, bile acid and uridine may be rhythmically linked, and the changes in uridine concentration due to meal times could affect bile acid levels (Deng et al., 2017). However, the mechanism of uridine supplementation affecting the bile acid metabolism in HFD-fed mice is not very well-studied. Therefore, this study was carried out to investigate the influence of the dynamic administration of uridine on the metabolism of bile acids in HFD-fed mice.
Animal and experimental design
This study was approved by the animal welfare committee of the Institute of Subtropical Agriculture, Chinese Academy of Sciences (China, Changsha).
A total of 72 male C57BL/6J mice (from SLAC Laboratory Animal Central, Changsha, China) aged 7-weeks with similar weight were randomly assigned into three groups (n = 24): (1) the CON group included mice fed with HFD containing 60% kcal fat (D12492, Research Diet); (2) the DUR group included mice fed with HFD and oral administration with 400 mg/kg uridine (Meiya Co., Ltd., Hangzhou, China) during ZT0-ZT12; (3) the NUR group included mice fed HFD and oral supplemented with 400 mg/kg uridine during ZT12-ZT24. The dosage of uridine was as described in a previous study (Le et al., 2014b). Mice were housed with ad libitum access to food and water (temperature, 20°C ± 2°C; relative humidity, 45% ± 5%); the “lights on” condition was designated as ZT 0 (ZT0; 8:00 am of local time), and “lights off” as ZT 12 (ZT12, 8:00 pm of local time). The duration of the experiment was 14 days. Mice were sacrificed by cervical dislocation, starting at ZT4 at 6-h intervals during the day (ZT 4, 10, 16, and 22, six mice for each time point), and all samples of liver, jejunum, and ileum were snap-frozen in liquid nitrogen and stored at −80°C until analysis.
RNA extraction and real-time quantitative PCR (RT-qPCR) assay
The RT-qPCR processing was consistent with our previous study (Liu et al., 2019); all primers were designed using Primer-BLAST on the National Center for Biotechnology Information website and are presented in Table 1.

All statistical tests were performed using SPSS 22 software (IBM SPSS Statistics 22, USA). Each ZT was analyzed by one-way ANOVA, and time-course changes in gene expression were analyzed by two-way ANOVA (Uridine × ZT), followed by Tukey’s post-hoc tests. The difference was considered to be significant when P < 0.05.
Dynamic administration of uridine alters the expression of mRNA related to pyrimidine metabolism in the liver of high fat diet-fed mice
The liver is an essential organ for the de novo synthesis and homeostasis of pyrimidine, and uridine significantly impacts hepatic cellular function (Le et al., 2013). As presented in Fig. 1, the expression of pyrimidine metabolism-related genes was extremely significantly affected by ZT (P < 0.01). The expression of uridine phosphorylase 1 (UPP1) mRNA decreased significantly at ZT4 both in DUR and NUR groups (P < 0.05). Besides, DUR and NUR groups had increased expression of ribonucleotide reductase regulatory subunit M2 (RRM2) at ZT22 and ZT4 (P < 0.05), respectively. Of note, uridine administration had a significant influence on the expression of UPP1 and RRM2 (P < 0.05) and on the expression of CMPK2 (0.05 < P < 0.1). In addition, mRNA expression of cytidine 5'-triphosphate synthetase (CTPS) and cytidine 5'-triphosphate synthetase2 (CTPS2) also showed rhythmic changes.
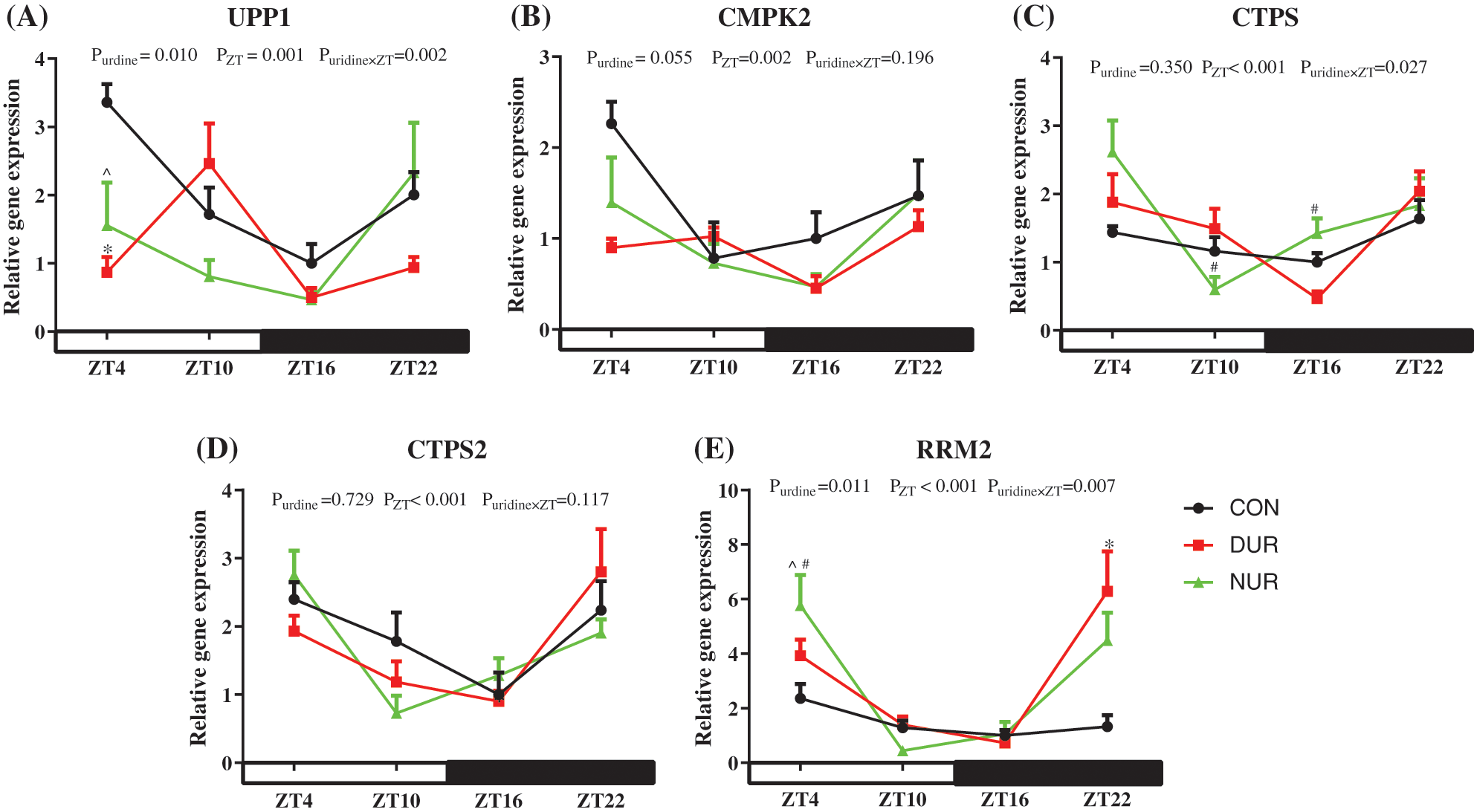
Figure 1: Dynamic administration of uridine alters the mRNA expression of proteins associated with pyrimidines metabolism in the liver of high-fat-diet (HFD)-fed mice. (A–E) Changes in expression of genes, which were normalized to β-actin gene expression at ZT4 in the HFD group. UPP1 encodes the enzyme that converts uridine and uracil, CMPK2 encodes a protein that converts UMP to UTP, CTPS, and CTPS2 encode enzymes to convert UTP to CTP, and the protein encoded by RRM2 converts UDP to dUDP. Values are presented as mean ± SEM; *P < 0.05 indicates a significant difference between CON and DUR groups; ^P < 0.05 indicates a significant difference between CON and NUR groups; #P < 0.05 indicates a significant difference between DUR and NUR groups.
Dynamic administration of uridine alters the mRNA expression of uridine transporters and the circadian clock in the jejunum of high fat diet-fed mice
As shown in Fig. 2, the relative expression of sodium-coupled nucleoside transporter 1 (SLC28A1), sodium-coupled nucleoside transporter 3 (SLC28A3), and equilibrative nucleoside transporter 1 (SLC29A1) showed time dependence (P < 0.001). Relative to the CON group, the DUR group exhibited significantly down-regulated expression of SLC28A1 at ZT4, ZT10, and ZT22, while the NUR group exhibited decreased expression of SLC28A1 at ZT22; the DUR group, compared to the CON group, showed lower expression of SLC28A3 at ZT4. Meanwhile, the expression of SLC29A1 at ZT22 was higher in the DUR group tin han the CON group. Concurrently, the expression of SLC28A1 mRNA in the jejunum was highly significantly affected by uridine administration (P < 0.01). ZT significantly affected the expression of CLOCK and period circadian regulator 2 (PER2) (P < 0.001) mRNAs, and DUR treatment decreased the expression of CLOCK mRNA at ZT16 while the NUR group had a down-regulated expression of CLOCK at ZT10 (P < 0.05). The mRNA expression of CLOCK mRNA in the jejunum was significantly affected by uridine administration (P = 0.0027).
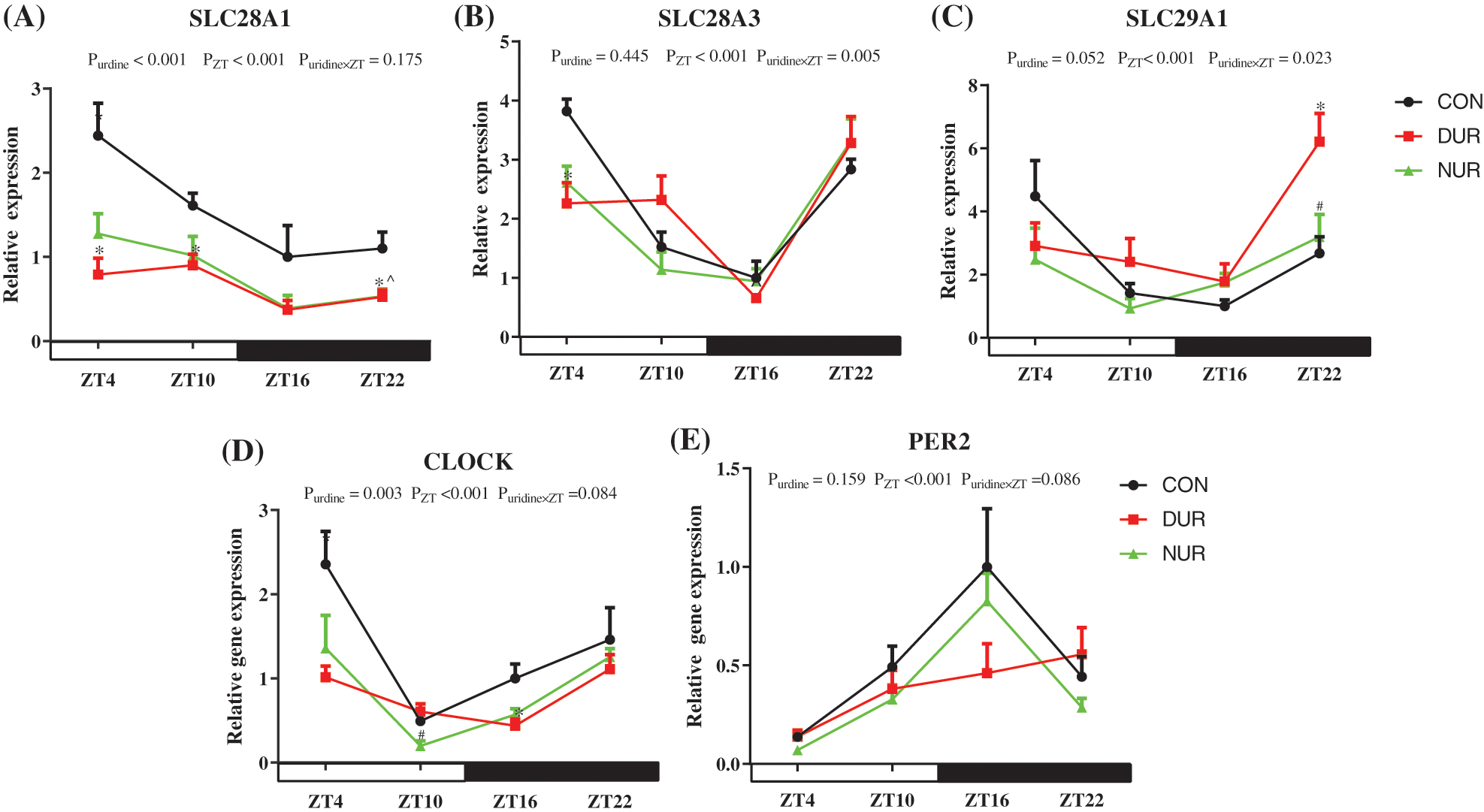
Figure 2: Dynamic administration of uridine alters the mRNA expression of uridine transporters, including circadian locomotor output cycles kaput (CLOCK) and period circadian regulator 2 (PER2) in the jejunum in HFD-fed mice. (A–E) Changes in expression of genes, which were normalized to β-actin gene expression at ZT4 in the high fat diet-fed (HFD) group. (Values are presented as mean ± SEM; *P < 0.05 between CON and DUR groups; ^P < 0.05 between CON and NUR groups; #P < 0.05 between DUR and NUR groups). (SLC28A1: sodium-coupled nucleoside transporter 1, SLC28A3: sodium-coupled nucleoside transporter 3, SLC29A1: equilibrative nucleoside transporter 1).
Dynamic administration of uridine alters the mRNA expression of proteins associated with bile acid metabolism in the liver of HFD mice
As shown in Fig. 3, ZT significantly affected mRNA expressions of farnesoid X receptor (FXR), small heterodimer partner (SHP), cholesterol 7-alpha-hydroxylase (CYP7A1), fibroblast growth factor receptor 4 (FGFR4), Na+/taurocholate co transporting polypeptide (NTCP), and bile salt export pump (BSEP) in the liver (P < 0.05). Importantly, the expression of CYP7A1, FGFR4, NTCP, and BSEP mRNAs showed a strong dependence on uridine administration (P < 0.05). The expression of FXR was significantly up-regulated at ZT4 in NUR vs. the DUR group (P < 0.05). The expression of SHP was significantly up-regulated at ZT4 and ZT10 in NUR vs. the CON group (P < 0.05); this mRNA was down-regulated at ZT4 while up-regulated at ZT10 in DUR vs. the CON group (P < 0.05). Compared to the CON group, the expression of CYP7A1 was significantly up-regulated at ZT22 in NUR and down-regulated at ZT16 in DUR groups (P < 0.05). The expression of FGFR4 was up-regulated at ZT4 and ZT10 in NUR compared to DUR and CON groups, and significantly up-regulated at ZT22 in both DUR and NUR groups compared to the CON group (P < 0.05). The expression of NTCP was significantly up-regulated at ZT4 and ZT22 in the NUR group when compared with DUR and CON groups and the CON group, respectively (P < 0.05). The expression of BSEP was significantly up-regulated at ZT4 and ZT16 in the NUR groups compared to DUR and CON groups and significantly up-regulated at ZT22 both in NUR and DUR groups vs. the CON group (P < 0.05).
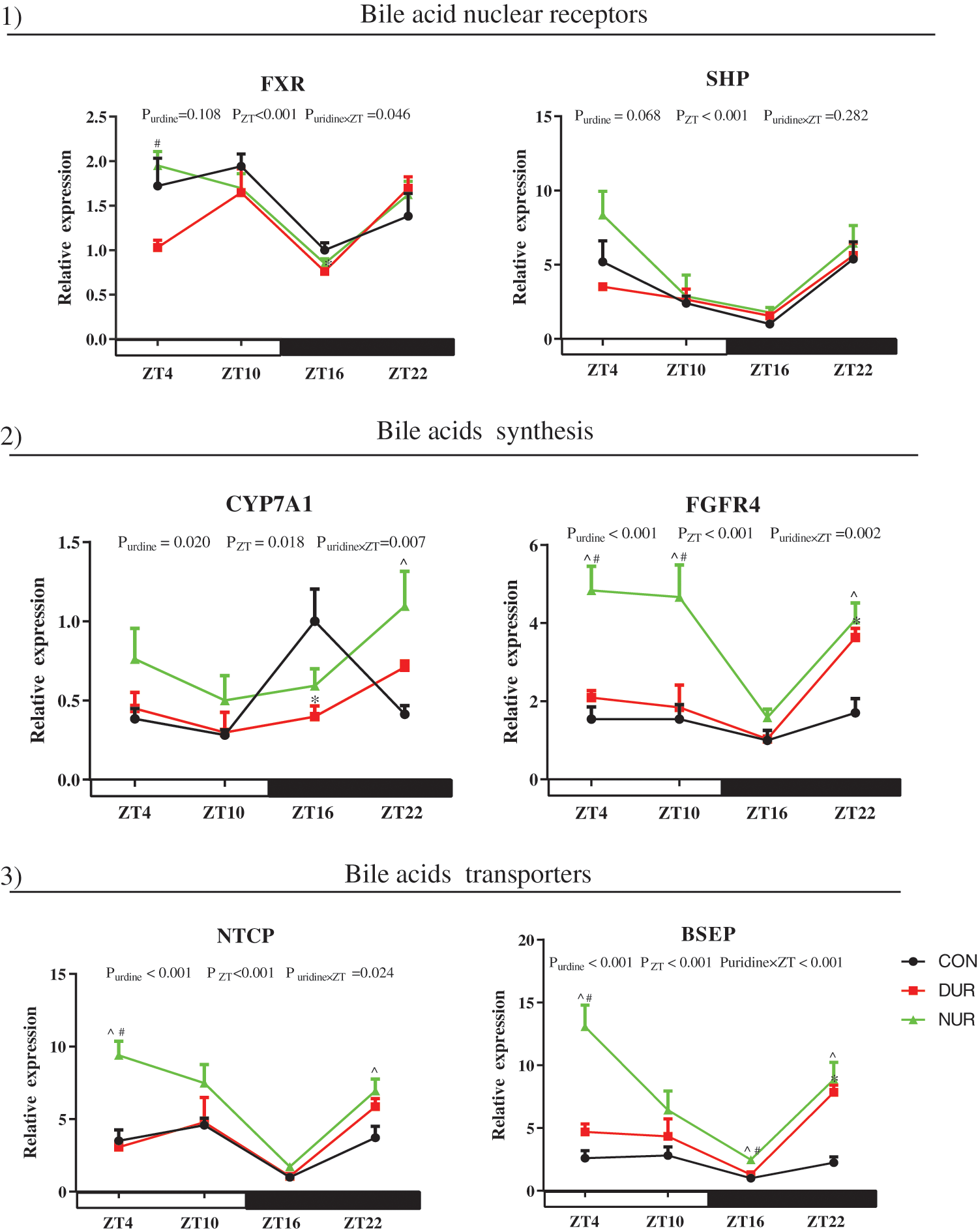
Figure 3: Dynamic administration of uridine alters the mRNA expression of bile acid metabolism in liver. mRNA expression of 1) bile acids nuclear receptors, 2) proteins related to bile acid synthesis, 3) bile acid transporters. Values are presented as mean ± SEM; *P < 0.05 between CON and DUR groups; ^P < 0.05 between CON and NUR groups; #P < 0.05 between DUR and NUR groups. (BSEP: bile salt export pump; CYP7A1: cholesterol 7-alpha-hydroxylase; FXR: farnesoid X receptor; FGFR4: fibroblast growth factor receptor 4; NTCP: Na+/taurocholate co transporting polypeptide; SHP: small heterodimer partner).
Dynamic administration of uridine alters the mRNA expression of cholesterol and bile acid metabolism in the ileum of HFD mice
As presented in Fig. 4, the mRNA expression of microsomal triglyceride transfer protein (MTTP), NPC1 like intracellular cholesterol transporter 1 (NPC1L1), ATP binding cassette subfamily G member 5 (ABCG5), apical sodium-dependent bile acid transporter (ASBT), ileum bile acid-binding protein (IBABP), and FXR were significantly affected by ZT (P < 0.05). At the same time, the expression of NPC1L1, ABCG5, ATP binding cassette subfamily A member 1 (ABCA1), ASBT, and SHP were also affected by uridine administration (P < 0.05).
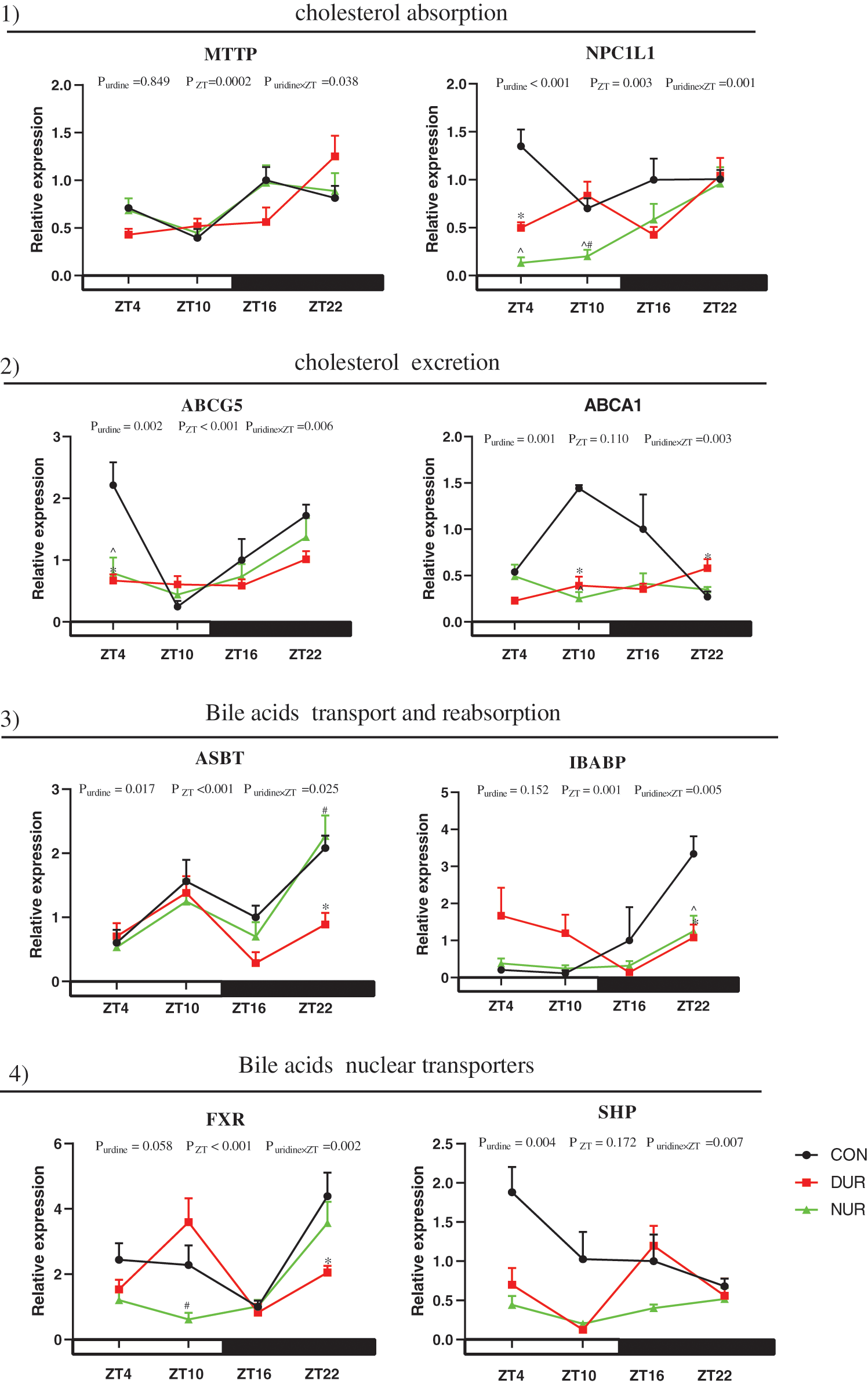
Figure 4: Dynamic administration of uridine alters the expression of proteins associated with cholesterol and bile acid metabolism in the ileum of HFD mice. mRNA expression of proteins related to 1) cholesterol absorption, 2) cholesterol excretion, 3) bile acid transport and reabsorption, 4) bile acid nuclear receptors. Values are presented as mean ± SEM; *P < 0.05 between CON and DUR groups; ^P < 0.05 between CON and NUR groups; #P < 0.05 between DUR and NUR groups (ABCG5: ATP binding cassette subfamily G member 5; ABCA1: ATP binding cassette subfamily A member 1; ASBT: apical sodium-dependent bile acid transporter; FXR: farnesoid X receptor; IBABP: ileum bile acid-binding protein; MTTP: microsomal triglyceride transfer protein; NPC1L1: NPC1 like intracellular cholesterol transporter 1; SHP: small heterodimer partner).
Uridine contributes to systemic metabolism, which could influence the bile acid metabolism, although how dynamic administration of uridine affects bile acid during HFD remains to be unraveled. The present study investigated the influence of uridine dynamic administration on bile acid metabolism in HFD-fed mice by analyzing the expression level of genes related to pyrimidines metabolism, uridine transporters, circadian clock, and bile acid metabolism.
The circadian expression of genes involved in pyrimidine nucleotide metabolism in mice has been reported earlier (Fustin et al., 2012). Solute carrier families 28 and 29 (SLC28 and SLC29) are essential participants in the pyrimidine nucleotides transport (Gray et al., 2004; Young, 2016). The relative expression of SLC28A1, SLC28A3, and SLC29A1 was time-dependent, and the results of our study suggest that pyrimidine transporters may be regulated by a biological clock, concurrent with a previous study of the possible role of uridine in rhythmically regulating the expression of gene related to the nucleotide metabolism (Zhang et al., 2018b). Furthermore, the expression of SLC28A1 mRNA in the jejunum was significantly affected by uridine supplementation, different from that reported in a previous study that uridine supplementation increased the expression of SLC28A1 in the duodenum mucosa of weaned piglets (Xie et al., 2019), which might be due to differences in physiology between weaned piglets and adult mice or between duodenum mucosa and jejunum.
Per2 and CLOCK are key circadian clock genes (Wallace et al., 2018; Xiang et al., 2018); results of this study showed that ZT has a significant effect on the expression of these genes, consistent with that reported in a previous study (Wallace et al., 2018). However, the interaction between uridine administration and circadian rhythm still needs further investigation (Liu et al., 2019).
Pyrimidine nucleotide metabolism shows rhythmicity at the mRNA level (Ferrell and Chiang, 2015; Osborne et al., 1983). In the current study, the expression of UPP1 mRNA was significantly decreased at ZT4 while that of RRM2 at ZT22 and ZT4 increased significantly both in DUR and NUR groups. Our results also showed that the mRNA expression of pyrimidines (in this study, uridine) metabolism-related genes was significantly affected by ZT. These results suggest that the gene regulation related to pyrimidine metabolism possibly arises from an intertwined relationship between feeding rhythms and the circadian clock. However, the trend of mRNA expression between uridine transporter in the jejunum and pyrimidine metabolism-related genes in the liver were different, which may be due to a time lag between intestinal transport and metabolism in the liver (Frazer et al., 2004).
Bile acid and uridine metabolism have circadian rhythmicity, and HFD is thought to impair bile acid synthesis and excretion (Duane et al., 1984; Fustin et al., 2012). Our previous studies revealed that uridine administration affects lipid metabolism in high fat diet-fed mice and early-weaned piglets (Liu et al., 2019; Zhang et al., 2019; Liu et al., 2021). Bile acids play an important role in lipid metabolism and promote the digestion and absorption of lipid substances (Joyce et al., 2014; Qi et al., 2015; Ye et al., 2018). FXR, having a potential significance in the regulation of the diurnal rhythm of bile acid, is involved in cholesterol biosynthesis and homeostasis of bile acids by regulating the expression of CYP7A1 (Cariello et al., 2017; Denson et al., 2001; Zhong et al., 2019) and the participation of FXR and FGF19. FGFR4 is also important in bile acids homeostasis, and mice lacking FGFR4 exhibited an elevated expression of liver CYP7A1 (Yu et al., 2005; Yu et al., 2000). The current study results showed that ZT has a significant effect on the expression of FXR, SHP, CYP7A1, and FGFR4 in the liver, indicating that the regulation of bile acid synthesis has a circadian rhythm similar to that reported in the previous study (GaLman et al., 2005; Gilberstadt et al., 1991). NTCP, as the primary uptake transporter of conjugated bile acids in the liver, plays a pivotal role in bile acid metabolism (Donkers et al., 2017), and BSEP is an efflux transporter and also of importance in the secretion of bile salts (Fukuda et al., 2014). We also observed that the expression of NTCP and BSEP was affected by ZT, indicating the regulation of bile acid transporters has a circadian rhythm, in agreement with previous reports (Janecke et al., 2011; Ma et al., 2009; Zhang et al., 2018a). Importantly, the expression of CYP7A1, FGFR4, NTCP, and BSEP showed a strong dependence on uridine administration, similar to that observed in our former study, which showed that uridine supplementation affects the synthesis and transport of bile acids (Zhang et al., 2018b).
Bile acids are recycled through the enterohepatic circulation (Meng et al., 2017; Roberts et al., 2002); cholesterol is required for bile acid synthesis, and the ileum is an important site for bile acid transport and absorption (Sklan et al., 1976). In this study, the expression of MTTP, NPC1L1, and ABCG5 mRNAs was significantly affected by ZT, and the results showed the rhythmicity of cholesterol absorption and excretion, consistent with that reported in previous studies (Akashi et al., 2017; Pan et al., 2013). Furthermore, ASBT participates in the active reabsorption of bile acids jointly with IBABP (Dawson, 2017). A bile acid-binding protein in the cytoplasm of ileal epithelial cells, IBABP is important to the reabsorption of bile salts (Shneider et al., 1995). The relative expression of ASBT and IBABP was also affected by ZT. The analysis of mRNA expression in the gut further validated the rhythmicity of bile acid enterohepatic circulation, consistent with that reported previously (Ho, 1976). Excess bile acids can be highly toxic to mammalian cells; thus, the pool size of bile acid is tightly regulated (Zhou and Hylemon, 2014). Therefore, bile acid biosynthesis in the intestine is controlled through changes in the expression of FXR and SHP, which are important for bile acid regulation. Importantly, the expression of NPC1L1, ABCG5, ABCA1, ASBT, and SHP was affected by uridine administration. Our results indicate that uridine administration affects the absorption and excretion of cholesterol and bile acids, and uridine may be indirectly involved in the regulation of bile acid synthesis, and the mechanism needs further examination.
To conclude, dynamic uridine administration affected circadian variation in the mRNA expression of bile acid metabolism in HFD-fed mice, showing that dynamic uridine administration may provide a reference to improve lipid metabolism disorders.
Availability of Data and Materials: The datasets generated during and analysed during the current study are available from the corresponding author on reasonable request.
Author Contribution: The authors confirm contribution to the paper as follows: study conception and design: Xin Wu, Chunyan Xie; data collection: Tiantian Zhou, Yumei Zhang; analysis and interpretation of results: Tiantian Zhou, Yumei Zhang, Juan Zhang; draft manuscript preparation: Tiantian Zhou; draft manuscript modification: Zhenya Zhai. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: The experimental protocol and procedures for this study were reviewed and approved by the Guidelines for Laboratory Animal Ethics Committee of the Chinese Academy of Sciences. The animal experiments were approved by the animal care and use committee of the Institute of Subtropical Agriculture, Chinese Academy of Science (No. ISA-2020-18).
Funding Statement: This paper was jointly supported by grants from the Science and Technology Projects of Hunan Province (2019RS3020, 2019RS3021), Jiangxi Provincial Innovation and Entrepreneurship Projects.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Akashi M, Matsumura R, Matsuo T, Kubo Y, Node K (2017). Hypercholesterolemia causes circadian dysfunction: A potential risk factor for cardiovascular disease. EBioMedicine 20: 127–136. DOI 10.1016/j.ebiom.2017.04.034. [Google Scholar] [CrossRef]
Appiakannan HS, Kestyus DR, Weber ET (2019). Effects of high fat diet and chronic circadian challenge on glucocorticoid regulation in C57BL/6J mice. Physiology & Behavior 204: 100–105. DOI 10.1016/j.physbeh.2019.01.014. [Google Scholar] [CrossRef]
Asher G, Sassone PC (2015). Time for food: The intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161: 84–92. DOI 10.1016/j.cell.2015.03.015. [Google Scholar] [CrossRef]
Bass J (2012). Circadian topology of metabolism. Nature 491: 348–356. DOI 10.1038/nature11704. [Google Scholar] [CrossRef]
Cariello M, Piccinin E, Garcia-Irigoyen O, Sabbà C, Moschetta A (2017). Nuclear receptor FXR, bile acids and liver damage: Introducing the progressive familial intrahepatic cholestasis with FXR mutations. Biochimica et Biophys Acta 1864: 1308–1318. DOI 10.1016/j.bbadis.2017.09.019. [Google Scholar] [CrossRef]
Chen H, Gao L, Yang D, Xiao Y, Zhang M et al. (2019). Coordination between the circadian clock and androgen signaling is required to sustain rhythmic expression of Elovl3 in mouse liver. Journal of Biological Chemistry 294: 7046–7056. DOI 10.1074/jbc.RA118.005950. [Google Scholar] [CrossRef]
Chowdhury D, Wang C, Lu AP, Zhu HL (2019). Understanding quantitative circadian regulations are crucial towards advancing chronotherapy. Cells 8: 883. DOI 10.3390/cells8080883. [Google Scholar] [CrossRef]
Dawson AP (2017). Roles of ileal ASBT and OSTα-OSTβ in regulating bile acid signaling. Digestive Diseases 35: 261–266. DOI 10.1159/000450987. [Google Scholar] [CrossRef]
Deng Y, Wang ZV, Gordillo R, An Y, Zhang C et al. (2017). An adipo-biliary-uridine axis that regulates energy homeostasis. Science 355: eaaf5375. DOI 10.1126/science.aaf5375. [Google Scholar] [CrossRef]
Denson LA, Sturm E, Echevarria W, Zimmerman TL, Makishima M et al. (2001). The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology 121: 140–147. DOI 10.1053/gast.2001.25503. [Google Scholar] [CrossRef]
Donkers JM, Zehnder B, van Westen GJP, Kwakkenbos MJ, IJzerman AP et al. (2017). Reduced hepatitis B and D viral entry using clinically applied drugs as novel inhibitors of the bile acid transporter NTCP. Scientific Reports 7: 15307. DOI 10.1038/s41598-017-15338-0. [Google Scholar] [CrossRef]
Duane WC, Levitt DG, Mueller SM, Behrens JC (1984). Regulation of bile acid synthesis in man. Presence of a diurnal rhythm. Journal of Clinical Investigation 72: 1930–1936. DOI 10.1172/JCI111157. [Google Scholar] [CrossRef]
Eggink HM, Oosterman JE, de Goede P, de Vries EM, Foppen E et al. (2017). Complex interaction between circadian rhythm and diet on bile acid homeostasis in male rats. Chronobiology International 34: 1–15. DOI 10.1080/07420528.2017.1363226. [Google Scholar] [CrossRef]
Ferrell JM, Chiang JYL (2015). Circadian rhythms in liver metabolism and disease. Acta Pharmaceutica Sinica B 5: 113–122. DOI 10.1016/j.apsb.2015.01.003. [Google Scholar] [CrossRef]
Frazer DM, Inglis HR, Wilkins SJ, Millard KN, Steele TM et al. (2004). Delayed hepcidin response explains the lag period in iron absorption following a stimulus to increase erythropoiesis. Gut 53: 1509–1515. DOI 10.1136/gut.2003.037416. [Google Scholar] [CrossRef]
Fukuda H, Nakanishi T, Tamai T (2014). More relevant prediction for in vivo drug interaction of candesartan cilexetil on hepatic bile acid transporter BSEP using sandwich-cultured hepatocytes. Drug Metabolism and Pharmacokinetics 29: 94–96. DOI 10.2133/dmpk.DMPK-13-NT-049. [Google Scholar] [CrossRef]
Fustin JM, Doi M, Yamada H, Komatsu R, Okamura H (2012). Rhythmic nucleotide synthesis in the liver: Temporal segregation of metabolites. Cell Reports 1: 341–349. [Google Scholar]
GaLman C, Angelin B, Rudling M (2005). Bile acid synthesis in humans has a rapid diurnal variation that is asynchronous with cholesterol synthesis. Gastroenterology 129: 1445–1453. DOI 10.1053/j.gastro.2005.09.009. [Google Scholar] [CrossRef]
Gilberstadt ML, Bellinger LL, Lindblad S, Duane WC (1991). Liver denervation does not alter the circadian rhythm of bile acid synthesis in rats. American Journal of Physiology 261: 799–802. DOI 10.1152/ajpgi.1991.261.5.G799. [Google Scholar] [CrossRef]
Gray JH, Owen RP, Giacomini KM (2004). The concentrative nucleoside transporter family, SLC28. Pflugers Arch: European Journal of Physiology 447: 728–734. DOI 10.1007/s00424-003-1107-y. [Google Scholar] [CrossRef]
Green CB, Takahashi JS, Bass J (2008). The meter of metabolism. Cell 134: 728–742. DOI 10.1016/j.cell.2008.08.022. [Google Scholar] [CrossRef]
Hatori M, Vollmers C, Zarrinpar A, Ditacchio L, Bushong EA et al. (2012). Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metabolism 15: 848–860. DOI 10.1016/j.cmet.2012.04.019. [Google Scholar] [CrossRef]
Hatsumi S, Hanzawa F, Kim D, Sun S, Laurent T et al. (2018). Delayed first active-phase meal, a breakfast-skipping model, led to increased body weight and shifted the circadian oscillation of the hepatic clock and lipid metabolism-related genes in rats fed a high-fat diet. PLoS One 13: e0206669. DOI 10.1371/journal.pone.0206669. [Google Scholar] [CrossRef]
Ho KJ (1976). Circadian distribution of bile acid in the enterohepatic circulatory system in hamsters. Journal of Lipid Research 17: 600–604. DOI 10.1016/S0022-2275(20)41731-6. [Google Scholar] [CrossRef]
Honma K, Hikosaka M, Mochizuki K, Goda T (2016). Loss of circadian rhythm of circulating insulin concentration induced by high-fat diet intake is associated with disrupted rhythmic expression of circadian clock genes in the liver. Metabolism Clinical & Experimental 65: 482–491. DOI 10.1016/j.metabol.2015.12.003. [Google Scholar] [CrossRef]
Janecke A, Zhang YKJ, Guo GL, Klaassen CD (2011). Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS One 6: e16683. DOI 10.1371/journal.pone.0016683. [Google Scholar] [CrossRef]
Johnston JD, Ordovas JM, Scheer FA, Turek FW (2016). Circadian rhythms, metabolism, and chrononutrition in rodents and humans. Advances in Nutrition 7: 399–406. DOI 10.3945/an.115.010777. [Google Scholar] [CrossRef]
Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF et al. (2014). Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proceedings of the National Academy of Sciences 111: 7421–7426. DOI 10.1073/pnas.1323599111. [Google Scholar] [CrossRef]
Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C et al. (2007). High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metabolism 6: 414–421. DOI 10.1016/j.cmet.2007.09.006. [Google Scholar] [CrossRef]
Le TT, Urasaki Y, Pizzorno G (2014a). Uridine prevents fenofibrate-induced fatty liver. PLoS One 9: e87179. DOI 10.1371/journal.pone.0087179. [Google Scholar] [CrossRef]
Le TT, Urasaki Y, Pizzorno G (2014b). Uridine prevents tamoxifen-induced liver lipid droplet accumulation. BMC Pharmacology and Toxicology 15: 27. DOI 10.1186/2050-6511-15-27. [Google Scholar] [CrossRef]
Le TT, Ziemba A, Urasaki Y, Hayes E, Brotman S et al. (2013). Disruption of uridine homeostasis links liver pyrimidine metabolism to lipid accumulation. Journal of Lipid Research 54: 1044–1057. DOI 10.1194/jlr.M034249. [Google Scholar] [CrossRef]
Li B, Chen Z, Zhou H, Yin Y, Wu X (2016). Effects of dietary supplementation with uridine monophosphate on performance and intestinal morphology of weanling piglets. Journal of Animal Science 94: 62–65. [Google Scholar]
Li G, Xie C, Wang Q, Wan D, Zhang Y et al. (2019). Uridine/UMP metabolism and their function on the gut in segregated early weaned piglets. Food & Function 10: 4081–4089. DOI 10.1039/C9FO00360F. [Google Scholar] [CrossRef]
Liu Y, Guo S, Xie C, Niu K, de Jonge H et al. (2020). Uridine inhibits the stemness of intestinal stem cells in 3D intestinal organoids and mice. RSC Advances 10: 6377–6387. DOI 10.1039/C9RA07742A. [Google Scholar] [CrossRef]
Liu Y, Xie C, Zhai Z, Deng ZY, de Jonge HR et al. (2021). Uridine attenuates obesity, ameliorates hepatic lipid accumulation and modifies the gut microbiota composition in mice fed with a high-fat diet. Food & Function 12: 1829–1840. DOI 10.1039/D0FO02533J. [Google Scholar] [CrossRef]
Liu Y, Zhang Y, Yin J, Ruan Z, Wu X et al. (2019). Uridine dynamic administration affects circadian variations in lipid metabolisms in the liver of high-fat-diet-fed mice. Chronobiology International 36: 1258–1267. DOI 10.1080/07420528.2019.1637347. [Google Scholar] [CrossRef]
Ma K, Xiao R, Tseng HT, Shan L, Fu L et al. (2009). Circadian dysregulation disrupts bile acid homeostasis. PLoS One 4: e6843. DOI 10.1371/journal.pone.0006843. [Google Scholar] [CrossRef]
Meng W, Wei M, Shao Y, Liu QR, Wu QZ et al. (2017). Bile acid profiles within the enterohepatic circulation in a diabetic rat model after bariatric surgeries. AJP Gastrointestinal and Liver Physiology 314: G537–G546. [Google Scholar]
Merrow M, Spoelstra K, Roenneberg T (2005). The circadian cycle: Daily rhythms from behaviour to genes. EMBO Reports 6: 930–935. DOI 10.1038/sj.embor.7400541. [Google Scholar] [CrossRef]
Muhammad NA, Bassis CM, Zhang L, Zaidi S, Varani J et al. (2016). Calcium reduces liver injury in mice on a high-fat diet: Alterations in microbial and bile acid profiles. PLoS One 11: e0166178. DOI 10.1371/journal.pone.0166178. [Google Scholar] [CrossRef]
Osborne WR, Hammond WP, Dale DC (1983). Canine cyclic hematopoiesis is associated with abnormal purine and pyrimidine metabolism. Journal of Clinical Investigation 71: 1348–1355. DOI 10.1172/JCI110887. [Google Scholar] [CrossRef]
Pan X, Jiang XC, Hussain MM (2013). Impaired cholesterol metabolism and enhanced atherosclerosis in clock mutant mice. Circulation 128: 1758–1769. DOI 10.1161/CIRCULATIONAHA.113.002885. [Google Scholar] [CrossRef]
Qi Y, Jiang C, Cheng J, Krausz KW, Li T et al. (2015). Bile acid signaling in lipid metabolism: Metabolomic and lipidomic analysis of lipid and bile acid markers linked to anti-obesity and anti-diabetes in mice. Biochimica et Biophysica Acta-Molecular and Cell Biology of Lipids 1851: 19–29. DOI 10.1016/j.bbalip.2014.04.008. [Google Scholar] [CrossRef]
Roberts MS, Magnusson BM, Burczynski FJ, Weiss M (2002). Enterohepatic circulation–Physiological, pharmacokinetic and clinical implications. Clinical Pharmacokinetics 41: 751–790. DOI 10.2165/00003088-200241100-00005. [Google Scholar] [CrossRef]
Shneider BL, Dawson PA, Christie DM, Hardikar W, Wong MH, Suchy FJ (1995). Cloning and molecular characterization of the ontogeny of a rat ileal sodium-dependent bile acid transporter. Journal of Clinical Investigation 95: 745–754. DOI 10.1172/JCI117722. [Google Scholar] [CrossRef]
Sklan D, Budowski P, Hurwitz S (1976). Site of bile acid absorption in the rat. Lipids 11: 467–471. DOI 10.1007/BF02532837. [Google Scholar] [CrossRef]
Sun L, Wang Y, Song Y, Cheng XR, Xia S et al. (2015). Resveratrol restores the circadian rhythmic disorder of lipid metabolism induced by high-fat diet in mice. Biochemical & Biophysical Research Communications 458: 86–91. DOI 10.1016/j.bbrc.2015.01.072. [Google Scholar] [CrossRef]
Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G et al. (2005). Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308: 1043–1045. DOI 10.1126/science.1108750. [Google Scholar] [CrossRef]
Urasaki Y, Pizzorno G, Le TT (2016). Chronic uridine administration induces fatty liver and pre-diabetic conditions in Mice. PLos One, 11: e0146994. DOI 10.1371/journal.pone.0146994. [Google Scholar] [CrossRef]
Wallace E, Wright S, Schoenike B, Roopra A, Rho JM et al. (2018). Altered circadian rhythms and oscillation of clock genes and sirtuin 1 in a model of sudden unexpected death in epilepsy. Epilepsia Journal of the International League Against Epilepsy 59: 1527–1539. DOI 10.1111/epi.14513. [Google Scholar] [CrossRef]
Wu X, Gao LM, Liu YL, Xie C, Cai L et al. (2020). Maternal dietary uridine supplementation reduces diarrhea incidence in piglets by regulating the intestinal mucosal barrier and cytokine profiles. Journal of the Science of Food and Agriculture 100: 3709–3718. DOI 10.1002/jsfa.10410. [Google Scholar] [CrossRef]
Xiang R, Cui Y, Wang Y, Xie T, Yang X et al. (2018). Circadian clock gene Per2 downregulation in non‐small cell lung cancer is associated with tumour progression and metastasis. Oncology Reports 40: 3040–3048. DOI 10.3892/or.2018.6704. [Google Scholar] [CrossRef]
Xie C, Wang Q, Li G, Fan Z, Wu X (2019). Dietary supplement with nucleotides in the form of uridine monophosphate or uridine stimulate intestinal development and promote nucleotide transport in weaned piglets. Journal of the Science of Food and Agriculture 99: 6018–6113. DOI 10.1002/jsfa.9850. [Google Scholar] [CrossRef]
Ye Z, Cao C, Liu Y, Cao P, Li Q (2018). Digestion fates of different edible oils vary with their composition specificities and interactions with bile salts. Food Research International 111: 281–290. DOI 10.1016/j.foodres.2018.05.040. [Google Scholar] [CrossRef]
Yoshitsugu R, Kikuchi K, Iwaya H, Fujii N, Hori S et al. (2019). Alteration of bile acid metabolism by a high-fat diet is associated with plasma transaminase activities and glucose intolerance in rats. Journal of Nutritional Science and Vitaminology 65: 45–51. DOI 10.3177/jnsv.65.45. [Google Scholar] [CrossRef]
Young DJ (2016). The SLC28 (CNT) and SLC29 (ENT) nucleoside transporter families: A 30-year collaborative odyssey. Biochemical Society Transactions 44: 869–876. DOI 10.1042/BST20160038. [Google Scholar] [CrossRef]
Yu C, Wang F, Jin C, Huang X, McKeehan WL (2005). Independent repression of bile acid synthesis and activation of c-Jun N-terminal kinase (JNK) by activated hepatocyte fibroblast growth factor receptor 4 (FGFR4) and bile acids. Journal of Biological Chemistry 280: 17707–17714. DOI 10.1074/jbc.M411771200. [Google Scholar] [CrossRef]
Yu C, Wang F, Kan M, Jin C, Jones RB et al. (2000). Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. Journal of Biological Chemistry 275: 15482–15489. DOI 10.1074/jbc.275.20.15482. [Google Scholar] [CrossRef]
Zhang F, Duan Y, Xi L, Wei M, Shi A et al. (2018a). The influences of cholecystectomy on the circadian rhythms of bile acids as well as the enterohepatic transporters and enzymes systems in mice. Chronobiology International 35: 673–690. DOI 10.1080/07420528.2018.1426596. [Google Scholar] [CrossRef]
Zhang K, Liu Y, Zhang Y, Zhang J, Deng Z et al. (2018b). Dynamic oral administration of uridine affects the diurnal rhythm of bile acid and cholesterol metabolism-related genes in mice. Biological Rhythm Research 50: 543–552. DOI 10.1080/09291016.2018.1474844. [Google Scholar] [CrossRef]
Zhang Y, Guo S, Xie C, Wang R, Zhang Y et al. (2019). Short-term oral UMP/UR administration regulates lipid metabolism in early-weaned piglets. Animals 9: 610. DOI 10.3390/ani9090610. [Google Scholar] [CrossRef]
Zhong D, Xie Z, Huang B, Zhu S, Wang G et al. (2019). Ganoderma lucidum polysaccharide peptide alleviates hepatoteatosis via modulating bile acid metabolism dependent on FXR-SHP/FGF. Cell Physiology and Biochemistry 49: 1163–1179. [Google Scholar]
Zhou H, Hylemon PB (2014). Bile acids are nutrient signaling hormones. Steroids 86: 62–68. DOI 10.1016/j.steroids.2014.04.016. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |