DOI:10.32604/biocell.2022.021368

| BIOCELL DOI:10.32604/biocell.2022.021368 |  |
| Review |
Biomechanics of transendothelial migration by cancer cells
Université Grenoble Alpes, CNRS, LIPhy, Grenoble, 38000, France
*Address correspondence to: Claude Verdier, claude.verdier@univ-grenoble-alpes.fr
Received: 15 January 2022; Accepted: 27 April 2022
Abstract: Cancer metastasis is still a major social issue with limited knowledge of the formation of tumors and their growth. In addition the formation of metastases is very difficult to understand, since it involves very complex physical mechanisms such as cellular interactions and cell rheology, which are flow-dependent. Previous studies investigated transendothelial migration using sophisticated techniques such as microfluidics, traction force microscopy (TFM) or Atomic Force Microscopy (AFM), combined with physical modeling. Here we summarize recent results and suggest new ways to investigate the precise mechanisms used by cancer cells to undergo transendothelial migration.
Keywords: Rheology; Deformation; Forces; Adhesion; Biophysics
Cancer arises as tumors are formed within the body and grow in size because cells behave abnormally and divide rapidly. Tumors can be localized due to the pressure exerted on the surrounding medium (Deptuła et al., 2020), and possibly can be destroyed using chemo- or radiotherapy. Unfortunately, before and after the operation, cancer cells manage to escape from the initial tumor and penetrate into the blood stream where they can be transported for large distances, until they reach a distant organ (colon, breast, skin, bladder, bone), i.e., a soil (Fidler, 2003; Yang et al., 2020). Once in this location, cancer cells (CCs) interact with the vessel walls covered by endothelial cells (ECs) as shown in Fig. 1. It is known from other works on leukocytes that rolling motion (Alon et al., 1997) can first occur due to weak interactions between ligands on ECs and receptors on leukocytes or CCs (Dabagh et al., 2020). After rolling has taken place, the next step is secondary adhesion when strong forces are produced to balance the flow forces. Then new bonds are formed involving integrins, immunoglobulins (Laurent et al., 2014; Jin et al., 2021) leading to larger forces through catch bonds (Kong et al., 2009; Yeoman et al., 2021). The activation of these adhesion proteins can be long, up to hours (Haddad et al., 2010). One of the important questions is to determine which molecules are involved and whether they are common to all cancers. The final two steps are CC migration towards the endothelial junctions, and extravasation through the gap. This process involves both chemical signaling and mechanical effects (Mierke, 2021; Arefi et al., 2020), but is not so well understood. Due to the interest of biophysicists, new tools are now available to quantify the interactions involved in these dynamic processes (Michor et al., 2011), as well as the measurement of cell mechanical properties (Cross et al., 2008; Lekka et al., 2012; Rianna and Radmacher, 2017; Zbiral et al., 2022). The viewpoint is organized as follows. Recent results concerning new techniques developed for the investigation of transendothelial migration are presented in the next part, while future promising researches are proposed, in relation with essential biological needs. Finally, conclusions are drawn.
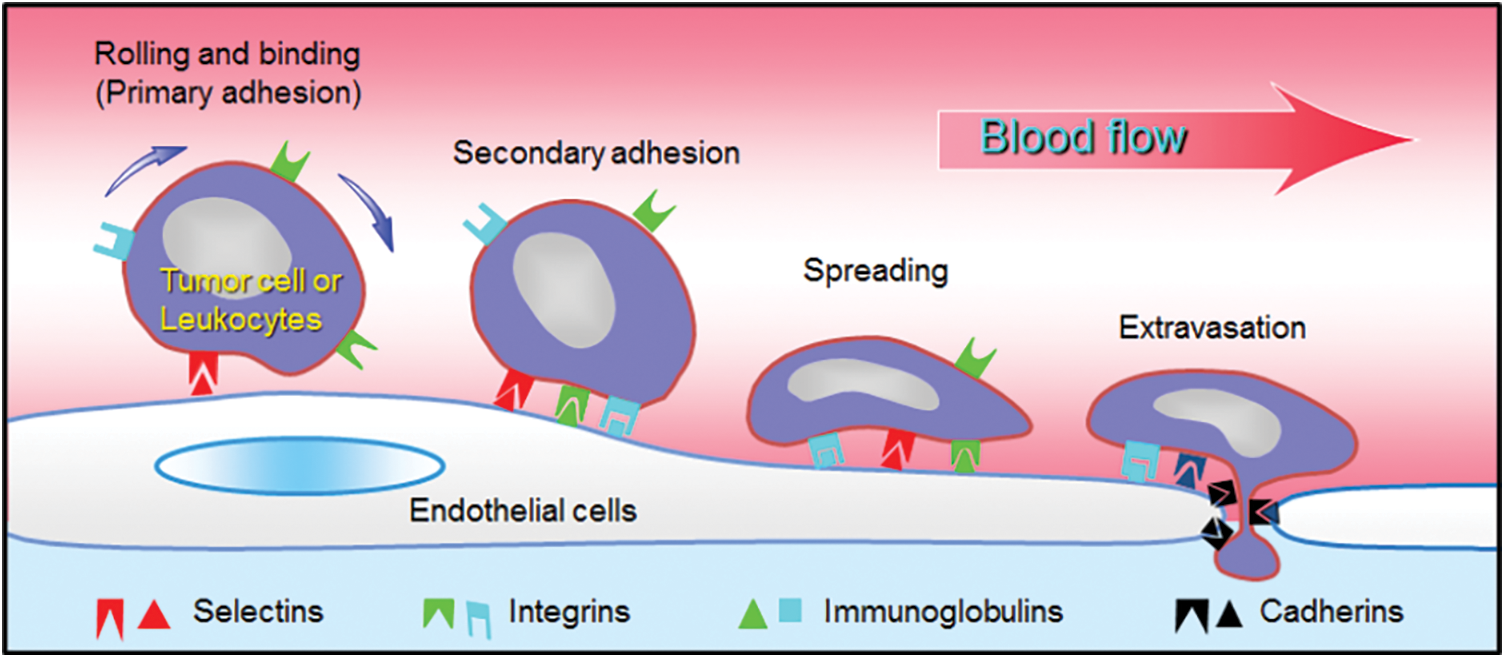
Figure 1: Extravasation process. Different steps used by leukocytes or tumor cells to interact with the endothelium. Sketch of the possible molecules involved.
As discussed above, it seems essential to understand what mechanisms are used by cancer cells to a) resist the flow in order to adhere to the endothelium; b) form strong bonds, i.e., receptor-ligand ones; c) migrate along the soft endothelium; d) be able to deform in order to pass through tight junctions, in other words to modify their rheological properties rapidly.
Flow chambers and microfluidics
Flow chambers or microfluidics devices have been designed since the 80’s. Usually microchannels are made of PDMS (typical dimensions between 5 to 200 µm) where fluid is driven at constant flow rate or imposed pressure. Different geometries are used. Surfaces can be coated with proteins or cells under such confinement so that it is possible to study cell-surface or cell-cell interactions. In particular interactions between the endothelium and circulating cells (leukocytes or cancer cells) can be studied.
The role of flow was initially found important for the binding of cells at low shear rates, but for higher rates, the lift force detaches cells and they are unable to adhere to the endothelium (Lawrence et al., 1987; Couzon et al., 2009). Another important aspect is the alignment of endothelial cells under flow. Usually, after 12 to 24 hours, ECs align in the direction of flow, depending on the shear stress (typically 0.2 to 2 Pa) and the actin cytoskeleton follows this trend (Chien, 2006). However, signaling pathways involving CCM proteins and β1-integrins can actually produce an opposite effect with ECs aligned perpendicular to the flow direction (Jilkova et al., 2014). Regarding cancer cells, the role of higher flow rate is decisive to enhance axial spreading of cancer cells within the endothelium, as compared to radial spreading (Chotard-Ghodsnia et al., 2007). Finally, flow affects the overexpression of cellular adhesion molecules (CAMs) like E-selectins, ICAM-1 and VCAM-1, through the NFκB pathway, but this effect is ruled out at higher shear stresses (Haddad et al., 2010).
Finally, new platforms enable to control and measure forces, while visualising cancer cell extravasation (Coughlin and Kamm, 2020) and promise to become outstanding tools for therapy.
Cell-cell interactions using AFM
AFM (Atomic Force Microscopy) in liquid environment can be used in Single Cell Force Spectroscopy (SCFS) mode to measure adhesion of living cells in near-physiological conditions. Cell-substrate or cell-cell interactions can be measured directly The cell is attached to the cantilever and comes in contact with another cell, then is pulled away after a given contact time. This is a good method to probe the presence of receptor-ligand interactions and it enables to characterize how force rates can affect the dissociation of bonds (i.e., detachment forces).
For example, cell-cell interactions involving receptor-ligand bonds have shown the role of LFA-1 and ICAMs (Wojcikiewicz et al., 2006). In the case of adhesion of tumor cells to the endothelium, the expression of ICAM-1 on CCs has been confirmed (Laurent et al., 2014) and CD43 and MUC1 were shown to be the relevant ligands (Rajan et al., 2017). It appears that more invasive bladder cancer cells use the latter CAMs simultaneously in order to bind more efficiently and a reduction of around 70% of cancer cell adhesion has been obtained when blocking these two molecules with antibodies. Moreover, CD43 and MUC1 are associated with ICAM-1 with a stronger connection with the cytoskeleton in the case of CD43, whereas MUC1 is more likely to form tethers when detaching. However other molecules are involved in CC adhesion to the endothelium, so no general trend can be proposed. Ultimately, as CCs transmigrate through the endothelium, they express β1 integrins or P-selectins that bind with Extra-Cellular Matrix (ECM) proteins to migrate further (Mierke et al., 2011; Le Cigne et al., 2016; Dao et al., 2021).
Altogether, AFM-based measurements have possible clinical implications, since they allow to characterize adhesion molecules relevant during the transmigration process.
Traction Force Microscopy and cell migration
Another possible way to explore the physics of cancer is to find how invasive cells can exert forces on the surrounding medium. Such methods–called Traction Force Microscopy (TFM)–have been developed in the years 2000 on two-dimensional substrates using the displacement of beads embedded in elastic gels onto which cells adhere, then an inverse problem is solved to determine traction stresses (Butler et al., 2002; Schwarz et al., 2002; Ambrosi et al., 2009).
This is important here, since invasive cancer cells migrate differently as compared to normal cells and exert less stress in order to move faster (Peschetola et al., 2013). This technique also proved to be quite efficient to determine the forces exerted by cancer cells as they transmigrate through an endothelium layer (grown as a circular patch on a 10 kPa gel, see Figs. 2A and 2B). In such a case, the horizontal (shear) forces exerted by CCs are small compared to other ones at the edges of the patch (Figs. 2C and 2D). This reveals that forces crucial for transmigration are vertical ones, necessary to pull the cell through the junction. They can be related to the strength of bonds between CAMs located at the cell invadopodium (intense green levels in Fig. 2A, Rajan, 2016) and ECM proteins on the gel surface below (fibronectin in this case).
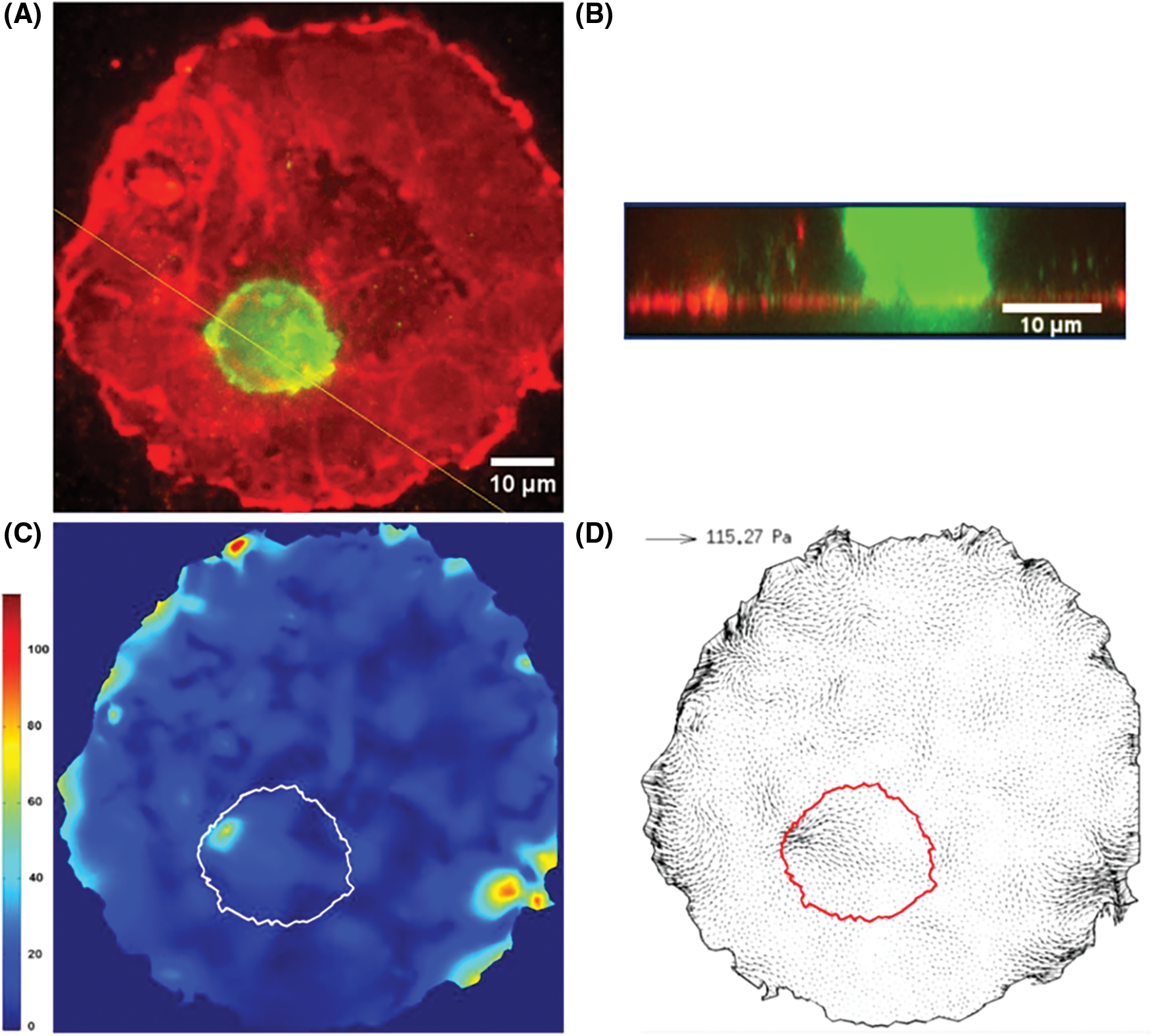
Figure 2: 2D Traction Force Microscopy performed when a CC interacts with the EC monolayer. A) Fluorescence image of ECs (red) and CC (green). B) Confocal side view, following the yellow line in A. C) Traction stresses (Pa), the white line represents the cell contour. D) Stress vectors with maximum value indicated. The cell contour is in red (Rajan, 2016).
Therefore, it is important to continue in this direction and explore this process using 3D TFM developed in recent studies, using elastic gels (Legant et al., 2013; Fertin et al., 2019) or nonlinear matrices (Jorge-Peñas et al., 2017; Song et al., 2020). Clinical applications could also benefit from these in vitro studies, allowing to test various drugs.
AFM has another added value, which is its ability to indent soft substrates and obtain force curves. AFM cantilevers built with a specific tip (pyramid, sphere, etc.) probe cells in a controlled manner. The analysis of such force curves allows to calculate the elastic (or viscoelastic) moduli. This is particularly relevant in this study as cancer cells have been known to be less rigid as compared to usual cells (Cross et al., 2008; Lekka et al., 2012). On the other hand, cells need to be rigid enough to push through the EC junctions. Therefore precise viscoelastic measurements (G’, elastic modulus, G’’, loss modulus) of CCs in contact with various substrates are to be carried out.
Earlier results have shown the adaptation of CC stiffness when plated on different elastic gels: cells usually spread more, with a higher elasticity, on stiffer substrates (Solon et al., 2007). Similarly, viscoelastic effects are enhanced (Abidine et al., 2021) and fT, the typical crossover frequency (such that G’(fT) = G’’(fT)) is reduced for low elasticity substrates or when in contact with the endothelium (Abidine et al., 2018). This demonstrates how the microenvironment (i.e., the endothelium) leads to a glassy-like cell response. Thus cancer cells use this key mechanism to modify their rheology quite rapidly (see Fig. 2A) and relocalize their rigid actin domains to push through the endothelial junction. Nuclear stiffness is also an important determinant of the ability of cancer cells to undergo transmigration. By combining Brillouin confocal microscopy (BCM) and confocal reflectance quantitative phase microscopy (QPM), Roberts et al., 2021 found that the cells and their nuclei soften upon extravasation while the nuclear membranes remain soft for at least 24 hours.
Finally, the ability of cancer cells to extravasate through the tight endothelial junctions depends on crosstalk between CCs and ECs during contact (Haddad et al., 2010; Stojak et al., 2020), implies β-catenins, E-cadherins, tight junction proteins and is mediated by reactive oxygen species (Haidari et al., 2013). There has been attempts to block TEM of breast cancer cells using cadherins or tight junction protein inhibitors (Bednarek et al., 2020).
To conclude, AFM is a versatile tool enabling to carry out precise cell rheological measurements in close-to physiological environments. It can be adapted to study physiological/pathological processes and therefore promises to answer questions relevant for clinical studies.
Modeling cell rheology processes
Modeling cell mechanical processes has been a source of interest within the biophysics community for a very long time so only a few features will be addressed here. There is a large number of cellular models, going from vesicles, composite or deformable objects (Jadhav et al., 2005), tensegrity models (Ingber, 1993), active drops (Joanny et al., 2013) that can be used to model cells depending on the problem. Flow effects are also included (Verdier et al., 2009) and cell interactions are usually treated using the stochastic behavior of cell bonds that can form or break based on previous theories (Kramers, 1940; Evans and Ritchie, 1997). This results in a force vs. loading rate relationship, being able to explain AFM data as well as flow effects. Such models are therefore particularly relevant for the study of transmigration of cancer cells through the endothelial wall.
Attempts considering cell-cell interactions (i.e., the contact of cells) and deformations using chemo-mechanical models, have been proposed (Arefi et al., 2020) but are too few, probably because they involve numerous mechanical aspects, such as the dynamics of invadopodia protrusions (Kim et al., 2022). These simulations lead to a vast number of parameters to be determined experimentally or tuned, and this remains a major challenge.
Therefore, future models and simulations could use deep learning to try and identify the model parameters roles in order to build a smaller parameter landscape and get a deeper understanding of the transmigration process.
New physical tools (microfluidics, AFM, force-based methods, enriched modeling) have been developed or improved in the past twenty years and promise to give a better understanding of the mechanisms at play during cancer cell transmigration. New platforms are now available, capable of measuring forces under flow, with simultaneous microscopic observations of the mechanisms involved in such processes. These new tools bring a higher added value for clinical applications, because they allow to test various drugs in vitro, using organ-on-a chip devices. Recently, the quantification of 3D forces (TFM) developed during cell interactions in complex media has made significant progresses. Still more in vitro experimental data is necessary, and needs to be collected in view of models better adapted to a 3D cell environment. Such models have reached a state of sophistication that should help select the relevant parameters sometimes hidden within the vast biological data pool.
Acknowledgement: The author is thankful to A Duperray and VM Laurent for fruitful discussions, and to VS Rajan for help with the TFM analysis.
Availability of Data and Materials: Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Author Contribution: The author confirms sole responsibility for the following: study conception and design, data collection, analysis and interpretation of results, and manuscript preparation.
Funding Statement: The author is greatful to the Grenoble Nanoscience Foundation, the ANR “TRANSMIG” (Grant No. 12-BS09-020-01), and the LabeX Tec21 (Grant No. ANR-11-LABX-0030).
Conflicts of Interest: The author declares that he has no conflicts of interest to report regarding the present study.
Abidine Y, Constantinescu A, Laurent VM, Rajan VS, Michel R et al. (2018). Mechanosensitivity of cancer cells in contact with soft substrates using AFM. Biophysical Journal 114: 1165–1175. DOI 10.1016/j.bpj.2018.01.005. [Google Scholar] [CrossRef]
Abidine Y, Giannetti A, Revilloud J, Laurent VM, Verdier C (2021). Viscoelastic properties in cancer: From cells to spheroids. Cells 10: 1704. DOI 10.3390/cells10071704. [Google Scholar] [CrossRef]
Alon R, Chen S, Puri KD, Finger EB, Springer TA (1997). The kinetics of L-selectin tethers and the mechanics of selectin-mediated rolling. Journal of Cell Biology 138: 1169–1180. DOI 10.1083/jcb.138.5.1169. [Google Scholar] [CrossRef]
Ambrosi D, Duperray A, Peschetola V, Verdier C (2009). Traction patterns of tumor cells. Journal of Mathematical Biology 58: 163–181. DOI 10.1007/s00285-008-0167-1. [Google Scholar] [CrossRef]
Arefi SMA, Tsvirkun D, Verdier C, Feng JJ (2020). A biomechanical model for the transendothelial migration of cancer cells. Physical Biology 17: 036004. DOI 10.1088/1478-3975/ab725c. [Google Scholar] [CrossRef]
Bednarek R, Selmi A, Wojkowska D, Karolczak K, Popielarski M et al. (2020). Functional inhibition of F11 receptor (F11R/junctional adhesion molecule-A/JAM-A) activity by a F11R-derived peptide in breast cancer and its micro- environment. Breast Cancer Research and Treatment 179: 325–335. DOI 10.1007/s10549-019-05471-x. [Google Scholar] [CrossRef]
Butler JP, Tolic-Nørrelykke IM, Fabry B, Fredberg JJ (2002). Traction fields, moments, and strain energy that cells exert on their surroundings. American Journal of Physiolology–Cell Physiology 282: C595–605. DOI 10.1152/ajpcell.00270.2001. [Google Scholar] [CrossRef]
Chien S (2006). Molecular basis of rheological modulation of endothelial functions: Importance of stress direction. Biorheology 43: 95–116. [Google Scholar]
Chotard-Ghodsnia R, Haddad O, Leyrat A, Drochon A, Verdier C et al. (2007). Morphological analysis of tumor cell/endothelial cell interactions under shear flow. Journal of Biomechanics 40: 335–344. DOI 10.1016/j.jbiomech.2006.01.001. [Google Scholar] [CrossRef]
Coughlin MF, Kamm RD (2020). The use of microfluidic platforms to probe the mechanism of cancer cell exravasation. Advanced Healthcare Materials 9: e1901410. DOI 10.1002/adhm.201901410. [Google Scholar] [CrossRef]
Couzon C, Duperray A, Verdier C (2009). A critical stress to detach cancer cells in microchannels. European Biophysical Journal 38: 1035–1047. DOI 10.1007/s00249-009-0506-1. [Google Scholar] [CrossRef]
Cross SE, Jin YS, Tondre J, Wong R, Rao J, Gimzewski JK (2008). AFM-based analysis of human metastatic cancer cells. Nanotechnology 19: 384003. DOI 10.1088/0957-4484/19/38/384003. [Google Scholar] [CrossRef]
Dabagh M, Gounley J, Randles A (2020). Localization of rolling and firm-adhesive interactions between circulating tumor cells and the microvasculature wall. Cellular Molecular Bioengineering 13: 141–154. DOI 10.1007/s12195-020-00610-7. [Google Scholar] [CrossRef]
Dao L, Blaue C, Franz CM (2021). Integrin α2β1 as a negative regulator of the laminin receptors α6β1 and α6β4. Micron 148: 103106. DOI 10.1016/j.micron.2021.103106. [Google Scholar] [CrossRef]
Deptuła P, Łysik D, Pogoda K, Cieśluk M, Namiot A et al. (2020). Tissue Rheology as a possible complementary procedure to advance histological diagnosis of colon cancer. ACS Biomaterials Science & Engineering 6: 5620–5631. DOI 10.1021/acsbiomaterials.0c00975. [Google Scholar] [CrossRef]
Evans E, Ritchie K (1997). Dynamic strength of molecular adhesion bonds. Biophysical Journal 72: 1541–1555. DOI 10.1016/S0006-3495(97)78802-7. [Google Scholar] [CrossRef]
Fertin A, Laforgue L, Laurent VM, Usson Y, Duperray A et al. (2019). Displacement fields using correlation methods as a tool to investigate cell migration in 3D collagen gels. Journal of Microscopy 275: 172–182. DOI 10.1111/jmi.12825. [Google Scholar] [CrossRef]
Fidler IJ (2003). The pathogenesis of cancer metastasis: The ‘seed and soil’ hypothesis revisited. Nature Reviews Cancer 3: 453–458. DOI 10.1038/nrc1098. [Google Scholar] [CrossRef]
Haddad O, Chotard-Ghodsnia R, Verdier C, Duperray A (2010). Tumor cell/endothelial cell tight contact upregulates endothelial adhesion molecule expression mediated by NFκB: Differential role of the shear stress. Experimental Cell Research 316: 615–626. DOI 10.1016/j.yexcr.2009.11.015. [Google Scholar] [CrossRef]
Haidari M, Zhang W, Wakame K (2013). Disruption of endothelial adherens junction by invasive breast cancer cells is mediated by reactive oxygen species and is attenuated by AHCC. Life Sciences 93: 994–1003. DOI 10.1016/j.lfs.2013.10.027. [Google Scholar] [CrossRef]
Ingber DE (1993). Cellular tensegrity: Defining new rules of biological design that govern the cytoskeleton. Journal of Cell Science 104: 613–627. DOI 10.1242/jcs.104.3.613. [Google Scholar] [CrossRef]
Jadhav S, Eggleton CD, Konstantopoulos K (2005). A 3-D computational model predicts that cell deformation affects selectin-mediated leukocyte rolling. Biophysical Journal 88: 96–104. DOI 10.1529/biophysj.104.051029. [Google Scholar] [CrossRef]
Jilkova ZM, Lisowska J, Manet S, Verdier C, Deplano V et al. (2014). CCM proteins control endothelial β1 integrin dependent response to shear stress. Biology Open 3: 1228–1235. DOI 10.1242/bio.201410132. [Google Scholar] [CrossRef]
Jin Y, Liu W, Wang F, Wang M, Xu K et al. (2021). Tissue factor potentiates adherence of breast cancer cells to human umbilical vein endothelial cells under static and flow conditions. Cell Adhesion and Migration 15: 74–83. DOI 10.1080/19336918.2021.1898709. [Google Scholar] [CrossRef]
Joanny JF, Kruse K, Prost J, Ramaswamy S (2013). The actin cortex as an active wetting layer. European Physical Journal E 36: 52. DOI 10.1140/epje/i2013-13052-9. [Google Scholar] [CrossRef]
Jorge-Peñas A, Bové H, Sanen K, Vaeyens MM, Steuwe C et al. (2017). 3D full-field quantification of cell-induced large deformations in fibrillar biomaterials by combining non-rigid image registration with label-free second harmonic generation. Biomaterials 136: 86–97. DOI 10.1016/j.biomaterials.2017.05.015. [Google Scholar] [CrossRef]
Kim MC, Li R, Abeyaratne R, Kamm RD, Asada HH (2022). A computational modeling of invadopodia protrusion into an extracellular matrix fiber network. Scientific Reports 12: 1231. DOI 10.1038/s41598-022-05224-9. [Google Scholar] [CrossRef]
Kong F, García AJ, Mould AP, Humphries MJ, Zhu C (2009). Demonstration of catch bonds between an integrin and its ligand. Journal of Cell Biology 185: 1275–1284. DOI 10.1083/jcb.200810002. [Google Scholar] [CrossRef]
Kramers HA (1940). Brownian motion in a field of force and the diffusion model of chemical reactions. Physica VII: 284–304. DOI 10.1016/S0031-8914(40)90098-2. [Google Scholar] [CrossRef]
Laurent VM, Duperray A, Sundar VR, Verdier C (2014). Atomic force microscopy reveals a role for endothelial cell ICAM-1 expression in bladder cancer cell adherence. PLoS One 9: e98034. DOI 10.1371/journal.pone.0098034. [Google Scholar] [CrossRef]
Lawrence MB, McIntire LV, Eskin LV (1987). Effect of flow on polymorphonuclear leukocyte-endothelial cell adhesion. Blood 70: 1284–1290. DOI 10.1182/blood.V70.5.1284.1284. [Google Scholar] [CrossRef]
Le Cigne A, Chièze L, Beaussart A, El-Kirat-Chatel S, Dufrêne YF et al. (2016). Analysis of the effect of LRP-1 silencing on the invasive potential of cancer cells by nanomechanical probing and adhesion force measurements using atomic force microscopy. Nanoscale 8: 7144–7154. DOI 10.1039/C5NR08649C. [Google Scholar] [CrossRef]
Legant WR, Choi CK, Miller JS, Shao L, Gao L et al. (2013). Multidimensional traction force microscopy reveals out-of-plane rotational moments about focal adhesions. Proceedings of the National Academy of Science 110: 881–886. DOI 10.1073/pnas.1207997110. [Google Scholar] [CrossRef]
Lekka M, Gil D, Pogoda K, Dulińska-Litewka J, Jach R et al. (2012). Cancer cell detection in tissue sections using AFM. Archives of Biochemistry and Biophysics 518: 151–156. DOI 10.1016/j.abb.2011.12.013. [Google Scholar] [CrossRef]
Michor F, Liphardt J, Ferrari M, Widom J (2011). What does physics have to do with cancer. Nature Reviews Cancer 11: 657–670. DOI 10.1038/nrc3092. [Google Scholar] [CrossRef]
Mierke CT (2021). Viscoelasticity acts as a marker for tumor extracellular matrix characteristics. Frontiers in Cell and Developmental Biology 9: 785138. DOI 10.3389/fcell.2021.785138. [Google Scholar] [CrossRef]
Mierke CT, Frey B, Fellner M, Herrmann M, Fabry B (2011). Integrin α5β1 facilitates cancer cell invasion through enhanced contractile forces. Journal of Cell Science 124: 369–383. DOI 10.1242/jcs.071985. [Google Scholar] [CrossRef]
Peschetola V, Laurent VM, Duperray A, Michel R, Ambrosi D et al. (2013). Time-dependent traction force microscopy for cancer cells as a measure of invasiveness. Cytoskeleton 70: 201–214. DOI 10.1002/cm.21100. [Google Scholar] [CrossRef]
Rajan VS (2016). Adhesion and Transendothelial Migration of Cancer Cells (PhD Thesis). Université Grenoble-Alpes. [Google Scholar]
Rajan VS, Laurent VM, Verdier C, Duperray A (2017). Unraveling the receptor-ligand interactions between bladder cancer cells and the endothelium using AFM. Biophysical Journal 112: 1246–1257. DOI 10.1016/j.bpj.2017.01.033. [Google Scholar] [CrossRef]
Rianna C, Radmacher M (2017). Comparison of viscoelastic properties of cancer and normal thyroid cells on different stiffness substrates. European Biophysical Journal 46: 309–324. DOI 10.1007/s00249-016-1168-4. [Google Scholar] [CrossRef]
Roberts AB, Zhang J, Singh VR, Nikolić M, Moeendarbary E et al. (2021). Tumor cell nuclei soften during transendothelial migration. Journal of Biomechanics 121: 110400. DOI 10.1016/j.jbiomech.2021.110400. [Google Scholar] [CrossRef]
Schwarz US, Balaban NQ, Riveline D, Bershadsky A, Geiger B et al. (2002). Calculation of forces at focal adhesions from elastic substrate data: The effect of localized force and the need for regularization. Biophysical Journal 83: 1380–1394. DOI 10.1016/S0006-3495(02)73909-X. [Google Scholar] [CrossRef]
Solon J, Levental I, Sengupta K, Georges PC, Janmey PA (2007). Fibroblast adaptation and stiffness matching to soft elastic substrates. Biophysical Journal 93: 4453–4461. DOI 10.1529/biophysj.106.101386. [Google Scholar] [CrossRef]
Song D, Dong L, Gupta M, Li L, Klaas O et al. (2020). Recovery of tractions exerted by single cells in three-dimensional nonlinear matrices. Journal of Biomechanical Engineering 142: 081012. DOI 10.1115/1.4046974. [Google Scholar] [CrossRef]
Stojak M, Milczarek M, Kurpinska A, Suraj-Prazmowska J, Kaczara P et al. (2020). Protein disulphide isomerase A1 is involved in the regulation of breast cancer cell adhesion and transmigration via lung microvascular endothelial cells. Cancers 12: 2850. DOI 10.3390/cancers12102850. [Google Scholar] [CrossRef]
Verdier C, Couzon C, Duperray A, Singh P (2009). Modeling cell interactions under flow. Journal of Mathematical Biology 58: 235–259. DOI 10.1007/s00285-008-0164-4. [Google Scholar] [CrossRef]
Wojcikiewicz EP, Abdulreda MH, Zhang X, Moy VT (2006). Force spectroscopy of LFA-1 and its ligands, ICAM-1 & ICAM-2. Biomacromolecules 7: 3188–3195. DOI 10.1021/bm060559c. [Google Scholar] [CrossRef]
Yang C, Tian Y, Zhao F, Chen Z, Su P et al. (2020). Bone microenvironment and osteosarcoma metastasis. International Journal of Molecular Science 21: 6985. DOI 10.3390/ijms21196985. [Google Scholar] [CrossRef]
Yeoman B, Shatkin G, Beri P, Banisadr A, Katira P et al. (2021). Adhesion strength and contractility enable metastatic cells to become adurotactic. Cell Reports 34: 108816. DOI 10.1016/j.celrep.2021.108816. [Google Scholar] [CrossRef]
Zbiral B, Weber A, Toca-Herrera J (2022). Measuring mechanical properties of breast cancer cells with atmic force microscopy. Methods in Molecular Biology 2471: 323–343. DOI 10.1007/978-1-0716-2193-6. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |