DOI:10.32604/biocell.2021.014834

| Biocell DOI:10.32604/biocell.2021.014834 |  |
| Article |
Elp3 modulates neural crest and colorectal cancer migration requiring functional integrity of HAT and SAM domains
1Department of R&D, Biotech & Science Company of UP, Guangzhou Branch, Guangzhou, 510000, China
2State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, 650223, China
3Laboratory of Developmental Disease and Natural Medicine, Medical Faculty, Kunming University of Science and Technology, Kunming, 650000, China
*Address correspondence to: Xiangcai Yang, shuiyuejuehshi@126.com; Jiejing Li, jjli@kmust.edu.cn
Received: 03 November 2020; Accepted: 23 December 2020
Abstract: Cell migration is a finely tuned biological process that often involves epithelial-mesenchymal transition (EMT). EMT is typically characterized by the upregulation of mesenchymal markers such as Snail1. This process has been shown to be of critical importance to normal developmental processes, including neural crest migration and invasion. Interestingly, similar mechanisms are utilized in disease processes, such as tumor metastasis and migration. Notably, EMT and EMT-like processes confer tumor cells with the ability to migrate, invade, and adopt stem cell-like properties that largely account for immunosuppression and tumor recurrence. Therefore, identifying sensitive EMT markers may contribute to cancer prognosis and diagnosis in many facets. Previously, we showed that Elp3 plays an essential role during neural crest migration by stabilizing Snail1. In the current study, we further elucidate the molecular mechanism underlying colorectal cancer migration. We found that mElp3 exerted an identical function to xElp3 in modulating neural crest migration, and both HAT and SAM domains are imperative during this migratory process. Interestingly, overexpression of mElp3 in SW480 cells promoted cell migration and invasion, and we further showed that Elp3 stabilized Snail1 requiring integrity of both SAM and HAT domains. Our findings warrant further exploration of the specific target of Elp3 in cancer cells.
Keywords: Elp3; Neural crest; Colorectal cancer cells; Migration; HAT domain; SAM domain
Colorectal cancer (CRC) is a major public health issue in China and a leading cause of death worldwide (Ferlay et al., 2015). Although surgery is the mainstay of curative treatment for CRC, metastatic spread of tumor cells often occurs in refractory circumstances. Metastasis eventually causes cancer-related death in CRC cases (Ling et al., 2015). Therefore, it is necessary to identify molecular targets favoring prognostics and diagnostics.
Elongator protein 3 (Elp3) is the catalytic core of the Elongator complex. Together with hyperphosphorylated RNA polymerase II, it plays an essential role in transcriptional elongation (Otero et al., 1999; Wittschieben et al., 1999b; Kim et al., 2002). Elp3 contains an N-terminal radical S-adenosylmethionine (SAM) binding domain and a C-terminal histone acetyltransferase (HAT) domain. The HAT domain is well characterized and required for catalyzing acetylation of histones H3 and H4 (Wittschieben et al., 1999a; Kim et al., 2002). However, the SAM domain is otherwise. SAM contains an iron-sulfur (FeS) cluster motif, which is similar to the catalytic domain of SAM radical enzymes (Chinenov, 2002; Paraskevopoulou et al., 2006). The radical SAM domain is engaged in various catalytic reactions, including methylations, isomerization, sulfur insertion, ring formation, anaerobic oxidation, and protein radical formation (Sofia et al., 2001). It has been reported that Elp3 is important for paternal DNA demethylation since knockdown of Elp3 impairs paternal DNA demethylation, which depends on its radical SAM domain (Okada et al., 2010; Boal et al., 2011; Grove et al., 2011). SAM is more often a structural domain as it functions by recognizing and binding FeS clusters (Greenwood et al., 2009).
By employing the Xenopus neural crest developmental model, a classical in vivo cell migratory and invasive system (Le Douarin et al., 2004; Tucker, 2004), we found that Elp3 regulated cell migration through inhibition β-transducin repeat-containing protein (β-Trcp)-mediated ubiquitination of Snail1, which is a key inducer and regulator of neural crest migration as well as malignant epithelial-mesenchymal transition (EMT) property (Peinado et al., 2007; Theveneau and Mayor, 2012). As a result, Elp3 stabilized Snail1 and consequently transactivated N-cadherin to promote cell migration (Yang et al., 2016). It is noteworthy that Elp3 was also found to regulate neuron motility via acetylation of α-tubulin in mouse and Caenorhabditis elegans neurons (Creppe et al., 2009; Nguyen et al., 2010; Solinger et al., 2010). Moreover, depletion of Elp3 significantly impaired the migration of cancer cells and neural crest cells (Close et al., 2006; Close et al., 2012; Yang et al., 2016). Thus, in the present study, we explored Elp3 function in CRC cells since it has been detected in multiple clinical stages of CRC (Ladang et al., 2015; Atlas, 2020).
Herein, we showed that SAM and HAT domains play an imperative role in Elp3 interacting with and stabilizing Snail and, in turn, regulating the EMT procedure. Furthermore, we unraveled that the SAM and HAT domains play identical but different modulations in Elp3-driven EMT procedure in vivo and in vitro.
Embryo microinjection and whole-mount in situ hybridization
In vitro fertilization, embryo culture, whole-mount in situ hybridization, preparation of mRNA, and microinjection were carried out as described previously (Sive et al., 2000; Desimone et al., 2005). After fertilization of Xenopus embryos in vitro, we removed their jelly-coats with 2% cysteine (pH 8.0) in 0.3 X Modified Barth’s Saline (MBS): 88 mM NaCl, 1 mM KCl, 2.5 mM NaHCO3, 0.3 mM CaNO3, 0.41 mM CaCl2, 0.82 mM MgSO4, and 15 mM HEPES (pH 7.6) with antibiotic. Whole embryos were maintained in 0.3X MBS (in an incubator at 21°C–23°C) until the required stage for microinjection and/or in situ hybridization and subsequently photographed with a Leica MZ16 FA (Wetzlar, Germany). The antisense morpholino oligo (MO) sequence for xElp3 5’-TTCATGTTGCCCGATGTTCCGCTAG-3’ was obtained from Gene Tools (Philomath, OR, USA). Capped mRNAs for microinjection were synthesized using the SP6 mMessage mMachine Kit (Ambion, Austin, TX, USA). MO and mRNA were injected into the dorsal region of 2- to 4-cell stage embryos. MO was injected at 25 ng per embryo, and Xenopus and mouse Elp3 mRNA (0.2 ng/cell) were injected into one cell of 4-cell stage embryos with Elp3 MO. For in situ hybridization, the Slug, FoxD3, and Twist probes were used as described previously (Li et al., 2011).
The full-length Xenopus laevis and mouse Elp3 coding regions were obtained using PCR according to sequences in NCBI (accession NM_001095505 and NM_001253812.1) and then cloned into pCS2+ (the expression vector was a gift from Dr. Ralph A.W. Rupp (Howard Hughes Medical Institute, Chevy Chase, MD, USA)) and pCS2+-C-Flag/pCS2+-N-Myc vectors (our lab constructed those two vectors based on pCS2+), respectively. For the Co-immunoprecipitation (co-IP) assays and overexpression, truncated mouse Elp3 constructs (radical SAM and HAT domain) were subcloned into the pCMV-3Tag-8vector (Agilent Technologies, Santa Clara) (Okada et al., 2010). Full-length mouse Snail1 constructs were obtained by PCR according to sequences in the NCBI database (accession NM_011427) and cloned into a pCS2+-N-Myc vector.
For cell culture, 293 and SW480 CRC were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (both from Gibco, Waltham, MA, USA) in an atmosphere with 5% CO2 at 37°C. The cells were seeded in 6-well plates and allowed to reach 60%–70% confluence prior to transfection. The 293 and SW480 cells were transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. At 48 h post-transfection, the samples were subjected to Western Blot (WB) analysis and/or invasion assay.
Co-IP assay and immunoblotting
For Snail1/mElp3 co-IP, 293 cells were transfected into 6-well plates with the indicated plasmids. At 48 h post-transfection, the cells were lysed in 700 μL of lysis buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 5 mM EDTA at pH 8.0, and 1% Triton X-100) containing a protease inhibitor mixture (EDTA-free protease inhibitor cocktail; Roche, Basel, Switzerland) for 30 min on ice. Following centrifugation at 14000 rpm and 4°C for 15 min, the supernatant was incubated with Flag-M2 beads (ANTI-FLAG M2 Affinity Gel; A2220; Sigma-Aldrich, St. Louis, MO, USA) at 4°C for 4 h and then washed three times with the lysis buffer at 4°C for 5 min. The bound proteins were eluted with SDS-loading buffer at 95°C for 10 min and then subjected to 12% SDS-PAGE and WB analysis. The antibodies used were as follows: Anti-FLAG (F7425; 1:5000, Sigma-Aldrich), anti-Myc (C3956; 1:5000; Sigma-Aldrich), and anti-Elp3 (HPA025812; 1:5000; Sigma-Aldrich). Horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG (Pierce Biotechnology, Waltham, MA, USA) was used as the secondary antibody.
For other cell extraction immunoblots, cells were prepared using modified RIPA buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 0.5% SDS) in the presence of a proteinase inhibitor cocktail (cOmplete™ Mini; Roche), then centrifuged at 4°C and 12000 rpm for 10 min. The supernatant containing protein fraction was collected. Protein concentration was measured using a bicinchoninic acid protein assay kit (Applygen, Beijing, China). Fifty micrograms of proteins were separated on a 12% SDS-PAGE and transferred to a 0.20 μm PVDF membrane (Millipore, Burlington, MA, USA). The membrane was blocked at room temperature with 5% BSA for 1–2 h and incubated with antibodies to anti-FLAG (F7425; 1:5000; Sigma-Aldrich), anti-Myc (C3956; 1:5000; Sigma-Aldrich), anti-Elp3 (HPA025812; 1:5000; Sigma-Aldrich), and anti-α-tubulin (2144; 1:5000; Cell Signaling Technology, Danvers, MA, USA) overnight at 4°C. After washing with TBST six times, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1:10,000; Gt anti-Ms IgG (H+L) secondary antibody and Gt anti-Rb IgG (H+L) secondary antibody; Invitrogen) for 2 h at room temperature. The proteins were detected using a SuperSignal TM West Pico PLUS stable peroxide solution (Thermo Fisher Scientific, Waltham, MA, USA) and exposed to X-ray film. The integrated densities of bands were quantified using Image J software (National Institutes of Health, Bethesda, MD, USA).
SW480 cells transfected with the indicated plasmids were seeded in Millicell Hanging Cell Culture inserts (Millipore) with or without Matrigel after transfection for 48 h and subsequently incubated for 36 h. The cells were fixed with 4% PFA for 15 min, stained with hematoxylin for 5 min, and stained with eosin for 40 s (H&E staining kit; Boster, China). Cells on the seeded side were removed with a cotton swab, and cells adhering to the lower surface were counted under a microscope (EVOS XL Core Imaging System AMEX1000 at 20X magnification; Thermo Fisher Scientific) in five randomly selected fields. Analyses were performed using Microsoft Excel and GraphPad Prism 6 software (GraphPad Software, San Diego, CA, USA).
GraphPad Prism 6 software was used for statistical analysis. Data are expressed as means ± SEMs, and comparisons were performed using the two-tailed Student’s t-test. P-values < 0.05 or 0.01 were considered statistically significant or very significant, respectively. All experiments were carried out at least three times, and the samples were analyzed in triplicate.
mElp3 exerted an identical function to xElp3 in modulating neural crest migration
It is well known that the Elp3 gene product is conserved range from Saccharomyces cerevisiae and Drosophila melanogaster to Caenorhabditis elegans and Arabidopsis thaliana (Wittschieben et al., 1999a). We found that Xenopus and mouse Elp3 are highly conserved (protein identities: 89%). Previously, we showed that cranial neural crest (CNC) cells of Elp3 morphants failed to emigrate from the medial region of the injected side (Figs. 1a and 1d) (Yang et al., 2016). Co-injection of Xenopus Elp3 mRNA with Elp3, targeting MO, restored CNC cell migration (Figs. 1b and 1d). Meanwhile, co-injection of mouse Elp3 mRNA with Elp3-specific MO rescued the migration of CNC cells resembling xElp3 mRNA (Figs. 1c and 1d).

Figure 1: mElp3 is functionally identical to its counterpart in Xenopus laevis. Elp3 MO (25 ng) with or without Elp3 mRNA was injected into one cell of four-cell-stage embryos, and whole-mount in situ hybridization with probes for neural crest markers (Slug) was processed at St.19-21 (a–c). LacZ mRNA was co-injected to trace the injected sides (stained red on the corresponding sides). The statistical data are shown in (d). Mouse Elp3 mRNA (0.2 ng/cell) was injected into one cell of four-cell-stage embryos with Elp3 MO. LacZ mRNA was co-injected to trace the injected sides (stained red on the corresponding sides). Abbreviations: Elp3, Elongator protein 3; MO, morpholino oligo.
As mentioned above, the Xenopus neural crest developmental model is a classical system of in vivo cell migration (Le Douarin et al., 2004). Elp3 is a potential prognostic marker in CRC. It has been wildly detected in multiple clinical stages of CRC (Ladang et al., 2015; Atlas, 2020). To further our understanding of Elp3’s function during cancerous migration, we subsequently utilized CRC cell line SW480 to impel our investigation.
Overexpression of mElp3 promoted the migration and invasion of SW480 cells
To assess whether Elp3 has any potential effects on malignant CRC, we performed Boyden chamber assays to study cell migration and invasion (Fig. 2). The migration and invasive abilities of SW480 cells were considerably promoted by forced expression of mElp3, as larger numbers of cells migrated through the chamber with or without Matrigel (Figs. 2a–2c). At the same time, overexpression of either the SAM or HAT domain promoted SW480 migration and invasion to a less extent in comparison to wild-type mElp3 (Figs. 2a–2c). Thus, wild type mElp3, the SAM domain, and the HAT domain were all able to impact migration and invasion of SW480 cells to different degrees.
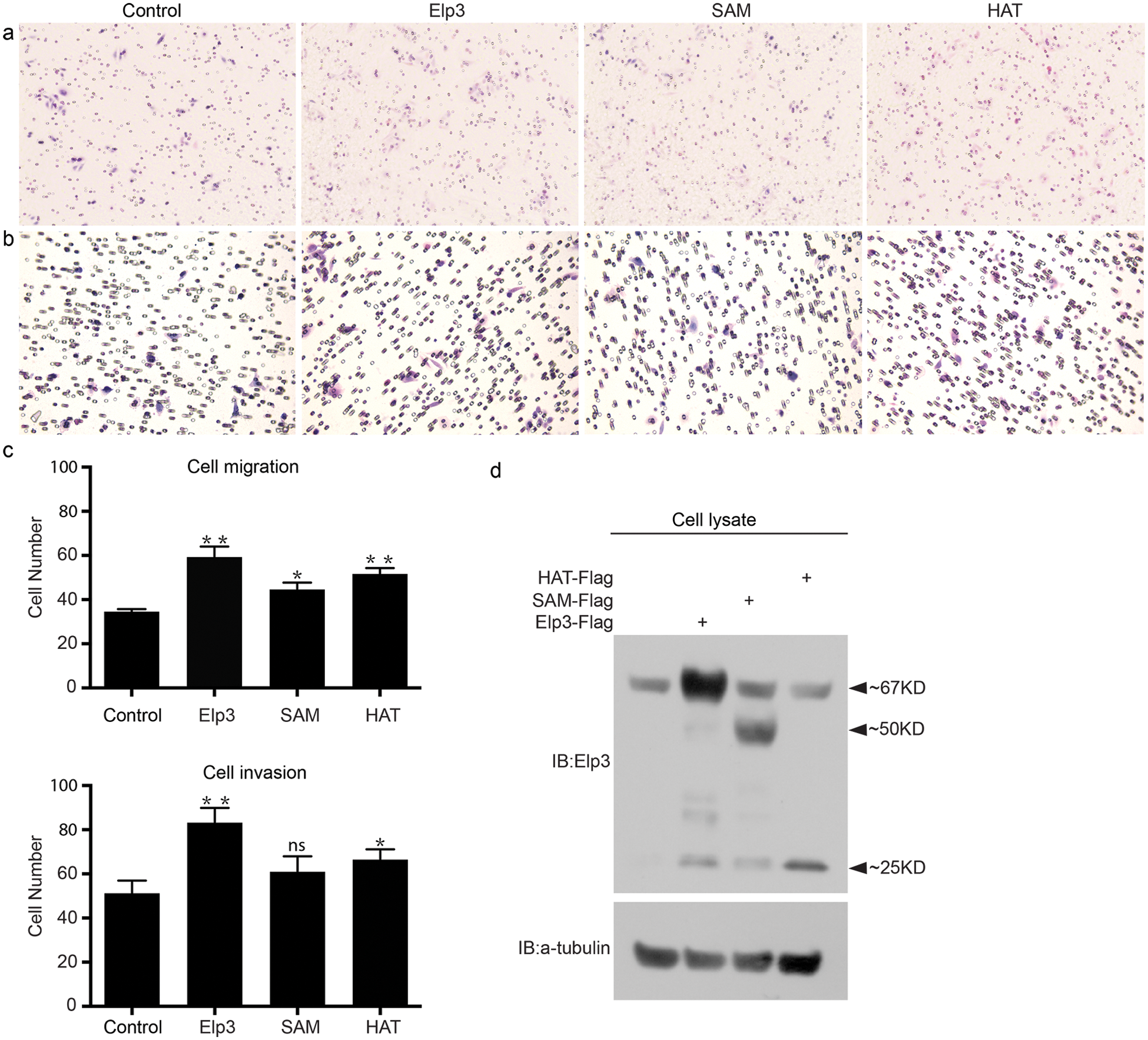
Figure 2: Elp3 promotes SW480 cell invasion. Transwell assays were carried out for SW480 cells transfected with the control vector or indicated plasmids, as described in Materials and methods. Images of cells (stained with hematoxylin-eosin and photographed at 20X magnification using the EVOS XL Core Imaging System AMEX1000) that migrated (a) or invaded through the Matrigel (b) in a representative experiment are shown, and results of 5 different regions per experiment are summarized in 3 independent experiments (c), *P < 0.05 and **P < 0.01. The transfection efficiency results are presented (d).
mElp3 stabilized mSnail1 requiring the whole structure of Elp3 protein
Snail1, a key inducer of EMT and regulator of neural crest migration, is a labile protein because it is dynamically modulated via the ubiquitin-proteasome pathway (Li et al., 2017). We previously found that Elp3 interacts with Snail1, and forced expression of Elp3 and Snail1 in 293 cells dramatically increased the protein stability of Snail1 by inhibiting its ubiquitination (Yang et al., 2016). As mentioned previously, mElp3 contains an N-terminal radical SAM binding domain and a C-terminal HAT domain. Therefore, to further investigate the affinity between the SAM/HAT domains and mSnail1, we constructed two corresponding truncated forms without either the SAM or HAT domains (Fig. 3a). In co-IP assays carried out in 293 cells, the SAM and HAT domains exhibited relatively feebler interactions with mSnail1 compared to wild-type mElp3 (Fig. 3b). We also used a series of cycloheximide-based protein chases to test the protein stability of Snail1 with the indicated Elp3 plasmids in 293 cells. Snail1 was clearly stabilized in the presence of Elp3 (Figs. 3c and 3d). While SAM or HAT truncation could not stabilize the Snail protein as wild-type mElp3 did. In addition, co-expression of HAT with Snail1 appeared to accelerate the turnover of Snail. These findings indicated that the whole structure of the Elp3 protein is necessary in order to interact with and stabilize Snail1.

Figure 3: Truncated forms and mutants of mElp3 modulate stabilization of the Snail1 protein. (a) Schematic representation of the mElp3 constructs. (b) Co-IP assays showing the interaction between mSnail1 and the mElp3 truncated forms. (c and d) Biochemical comparison of wild-type mElp3 and truncated forms influence of the stability of mSnail1 using 293 cells that were transiently transfected with the indicated plasmids. At 48 h after transfection, CHX was added to all the samples, and the cells were then harvested at specific time periods (0, 2, 4, and 6 h). The relative quantity of mSnail1 was determined by western blotting of the anti-Myc antibody. The loading control was indicated by α-tubulin. The relative expression of Snail1 was quantified densitometrically and subsequently normalized against α-tubulin. Abbreviations: WCL, whole-cell lysate; IB, immunoblot; IP, immunoprecipitation; CHX, cycloheximide.
Both the HAT and SAM domain are imperative during CNC migration
To recapitulate the above findings in the Xenopus neural crest model, we introduced the aforementioned mutants of Elp3 to advance our research on the SAM and HAT domains (Fig. 3a). Consistent with the biochemical phenotype, we observed that overexpression of mElp3 promoted the migration of neural crest cells in comparison to the contralateral un-injected side (Figs. 4a and 4d). In no contrast, overexpression of either the SAM (Figs. 4b and 4e) or HAT (Figs. 4c and 4e) domains significantly inhibited neural crest migration. This was identical to the downregulation of Elp3 per se (Figs. 1a and 1d). These findings suggest that both the SAM and HAT functional domains are required for the regulation of cell migration.

Figure 4: Overexpression of mElp3 truncated forms and mutants inhibit cranial neural crest migration in Xenopus. Mouse Elp3 mRNA (0.2 ng/cell), mElp3 truncated mRNA (1 ng/cell) were injected into one cell of four-cell-stage embryos (a–c). Whole-mount in situ hybridization with probes for neural crest markers (Slug, FoxD3, and Twist) was processed at St.19-21. LacZ mRNA was co-injected to trace the injected sides (stained red on the corresponding sides). The statistics are shown in d and e.
Elp3 is a catalytic subunit of the Elongator complex. It has been shown that Elp3 plays versatile functions in a number of biological processes, including transcriptional regulation, tRNA modification, and protein acetylation. We previously reported that Elp3 is required for neural crest migration in Xenopus embryos. Accordingly, we identified that Elp3 interacts with and stabilizes Snail1 through inhibition of β-Trcp-induced ubiquitination. Our research provided insight into the potential mechanism by which Elp3 promotes cell migration (Yang et al., 2016). Given that migration plays an essential role in CRC invasion and metastasis, which are the main causes of mortality in patients, it is necessary to explore the role of Elp3 in CRC cell migration. Consequently, we found that Elp3 enhanced SW480 cell migration and invasion. Aside from biochemical findings, endogenous research showed that truncated SAM or HAT domain forms inhibit neural crest migration in the Xenopus embryos (Fig. 4) but promote cell migration to a lesser extent than full-length Elp3 (Fig. 2).
We found that Snail1 preferentially bands with the SAM domain relative to the HAT domain. However, both the SAM and HAT domains exhibited less baiting ability than full-length Elp3 (Figs. 3a and 3b). In line with biochemical observations, endogenous overexpression of either the SAM or HAT domain inhibited migration of neural crest cells (Figs. 4b, 4c and 4e). This finding indicated that Elp3 interacts with and stabilizes Snail1 depending on its structural integrity.
Elp3 plays a pivotal role in neural crest cell migration during normal developmental procedures as well as in cancerous migration (Close et al., 2006; Close et al., 2012). Moreover, our data implied that Elp3 regulated cell migration through mechanisms other than integration with stabilized Snail1, and for instance, could be responsible for acetylating regulation. Previously, it was demonstrated that Elp3 is capable of driving cell migration during neurogenesis through the acetylation of α-tubulin (Creppe et al., 2009; Solinger et al., 2010). In turn, acetylation of α-tubulin facilitates motor-based trafficking in neurons. This eventually facilitates cell migration by inducing cell polarization (Creppe et al., 2009; Li et al., 2012, 2015). At the same time, there is a large body of proving that acetylation of relevant migration regulators ardently takes place during cell migration. PCAF-mediated acetylation of ACAP4 is essential for CCL18-induced breast cancer cell migration and invasion (Rehman et al., 2018). Simultaneously, acetylation of GATA3 by CBP inhibited the migration and invasion of lung adenocarcinoma cells (Li et al., 2018). Moreover, in line with acetylation, blocking the deacetylation of sirtuin2 promoted migration by restoring the acetylation of HSP90 (Min et al., 2018). Along with HSP90, Elp3 was shown to cis-transcriptionally regulate HSP70 through the acetylation of histone H3 and is capable of promoting cell migration in breast cancer (Han et al., 2008; Sims et al., 2011). Altogether, our research warrants further exploration of the specific targets of Elp3 in terms of acetylating regulation, such as HSP70 and HSP90, as well as the diagnostic and prognostic function of Elp3 in CRC.
Author Contribution: X. Y. and J. L. designed the experiments. X. Y., Y. X., S. M., J. L. performed the experiments. X. Y., Y. X. and J. L. analyzed the data and wrote the manuscript.
Availability of Data and Materials: The analyzed datasets generated during the present study are available from the corresponding author upon reasonable request.
Ethics Approval: The care of Xenopus laevis (Nasco), in vitro fertilization procedure and manipulation of embryos were performed accordingly to standard protocols. All of the animal protocols were approved by the Ethics Committee of Kunming Institute of Zoology, Chinese Academy of Sciences (Permit No. SYDW-2006–006).
Funding Statement: This project was supported by the National Natural Science Foundation of China (Grant Nos. 81760503 and 81760660), the Applied Basic Research Key Project of Yunnan (Grant Nos. 2018FE001(-318) and 2018FE001(-123)), and the Yunnan Health Science and Technology Plan Projects (Grant No. 2016NS207).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Atlas THP (2020). ELP3 Tissue Atlas. https://www.proteinatlas.org/ENSG00000134014-ELP3/tissue. [Google Scholar]
Boal AK, Grove TL, McLaughlin MI, Yennawar NH, Booker SJ, Rosenzweig AC (2011). Structural basis for methyl transfer by a radical SAM enzyme. Science 332: 1089–1092. DOI 10.1126/science.1205358. [Google Scholar] [CrossRef]
Close P, Gillard M, Aélie L, Jiang Z, Papuga J, Hawkes N, Nguyen L, Chapelle JP, Bouillenne F, Svejstrup J, Fillet M, Chariot A (2012). DERP6 (ELP5) and C3ORF75 (ELP6) regulate tumorigenicity and migration of melanoma cells as subunits of Elongator. Journal of Biological Chemistry 287: 32535–32545. DOI 10.1074/jbc.M112.402727. [Google Scholar] [CrossRef]
Close P, Hawkes N, Cornez I, Creppe C, Lambert CA, Rogister B, Siebenlist U, Merville MP, Slaugenhaupt SA, Bours V, Svejstrup JQ, Chariot A (2006). Transcription impairment and cell migration defects in elongator-depleted cells: Implication for familial dysautonomia. Molecular Cell 22: 521–531. DOI 10.1016/j.molcel.2006.04.017. [Google Scholar] [CrossRef]
Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O, Laguesse S, Cornez I, Rahmouni S, Ormenese S, Belachew S, Malgrange B, Chapelle JP, Siebenlist U, Moonen G, Chariot A, Nguyen L (2009). Elongator controls the migration and differentiation of cortical neurons through acetylation of α-tubulin. Cell 136: 551–564. DOI 10.1016/j.cell.2008.11.043. [Google Scholar] [CrossRef]
Chinenov Y (2002). A second catalytic domain in the Elp3 histone acetyltransferases: A candidate for histone demethylase activity? Trends in Biochemical Sciences 27: 115–117. DOI 10.1016/S0968-0004(02)02058-3. [Google Scholar] [CrossRef]
Desimone DW, Davidson LA, Marsden M, Alfandari D (2005). The Xenopus embryo as a model system for studies of cell migration. Methods of Molecular Biology 294: 235–245. [Google Scholar]
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2015). Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer 136: E359–E386. DOI 10.1002/ijc.29210. [Google Scholar] [CrossRef]
Greenwood C, Selth LA, Dirac-Svejstrup AB, Svejstrup JQ (2009). An iron-sulfur cluster domain in Elp3 important for the structural integrity of elongator. Journal of Biological Chemistry 284: 141–149. DOI 10.1074/jbc.M805312200. [Google Scholar] [CrossRef]
Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ (2011). A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science 332: 604–607. DOI 10.1126/science.1200877. [Google Scholar] [CrossRef]
Han Q, Lu J, Duan J, Su D, Hou X, Li F, Wang X, Huang B (2008). Gcn5- and Elp3-induced histone H3 acetylation regulates hsp70 gene transcription in yeast. Biochemical Journal 409: 779–788. DOI 10.1042/BJ20070578. [Google Scholar] [CrossRef]
Kim JH, Lane WS, Reinberg D (2002). Human Elongator facilitates RNA polymerase II transcription through chromatin. Proceedings of the National Academy of Sciences of the United States of America 99: 1241–1246. DOI 10.1073/pnas.251672198. [Google Scholar] [CrossRef]
Ladang A, Rapino F, Heukamp LC, Tharun L, Shostak K, Hermand D, Delaunay S, Klevernic I, Jiang Z, Jacques N, Jamart D, Migeot V, Florin A, Göktuna S, Malgrange B, Sansom OJ, Nguyen L, Büttner R, Close P, Chariot A (2015). Elp3 drives Wnt-dependent tumor initiation and regeneration in the intestine. Journal of Experimental Medicine 212: 2057–2075. DOI 10.1084/jem.20142288. [Google Scholar] [CrossRef]
Le Douarin NM, Creuzet S, Couly G, Dupin E (2004). Neural crest cell plasticity and its limits. Development 131: 4637–4650. DOI 10.1242/dev.01350. [Google Scholar] [CrossRef]
Li J, Qu J, Shi Y, Perfetto M, Ping Z, Christian L, Niu H, Mei S, Zhang Q, Yang X, Wei S (2017). Nicotinic acid inhibits glioma invasion by facilitating Snail1 degradation. Scientific Reports 7: 43173. DOI 10.1038/srep43173. [Google Scholar] [CrossRef]
Li J, Shi Y, Sun J, Zhang Y, Mao B (2011). Xenopus reduced folate carrier regulates neural crest development epigenetically. PLoS One 6: e27198. DOI 10.1371/journal.pone.0027198. [Google Scholar] [CrossRef]
Li L, Liu B, Zhang X, Ye L (2015). The oncoprotein HBXIP promotes migration of breast cancer cells via GCN5-mediated microtubule acetylation. Biochemical and Biophysical Research Communications 458: 720–725. DOI 10.1016/j.bbrc.2015.02.036. [Google Scholar] [CrossRef]
Li L, Wei D, Wang Q, Pan J, Liu R, Zhang X, Bao L (2012). MEC-17 deficiency leads to reduced α-tubulin acetylation and impaired migration of cortical neurons. Journal of Neuroscience 32: 12673–12683. DOI 10.1523/JNEUROSCI.0016-12.2012. [Google Scholar] [CrossRef]
Li X, Jin J, Yang S, Xu W, Meng X, Deng H, Zhan J, Gao S, Zhang H (2018). GATA3 acetylation at K119 by CBP inhibits cell migration and invasion in lung adenocarcinoma. Biochemical and Biophysical Research Communications 497: 633–638. DOI 10.1016/j.bbrc.2018.02.120. [Google Scholar] [CrossRef]
Ling H, Pickard K, Ivan C, Isella C, Ikuo M, Mitter R, Spizzo R, Bullock MD, Braicu C, Pileczki V, Vincent K, Pichler M, Stiegelbauer V, Hoefler G, Almeida MI, Hsiao A, Zhang X, Primrose JN, Packham GK, Liu K, Bojja K, Gafà R, Xiao L, Rossi S, Song JH, Vannini I, Fanini F, Kopetz S, Zweidler-McKay P, Wang X, Ionescu C, Irimie A, Fabbri M, Lanza G, Hamilton SR, Berindan-Neagoe I, Medico E, Mirnezami AH, Calin GA, Nicoloso MS (2015). The clinical and biological significance of MIR-224 expression in colorectal cancer metastasis. Gut 65: 977–989. DOI 10.1136/gutjnl-2015-309372. [Google Scholar] [CrossRef]
Min JS, Kim JC, Kim JA, Kang I, Ahn JK (2018). SIRT2 reduces actin polymerization and cell migration through deacetylation and degradation of HSP90. Biochimica et Biophysica Acta (BBA)—Molecular Cell Research 1865: 1230–1238. DOI 10.1016/j.bbamcr.2018.06.005. [Google Scholar] [CrossRef]
Nguyen L, Humbert S, Saudou F, Chariot A (2010). Elongator—An emerging role in neurological disorders. Trends in Molecular Medicine 16: 1–6. DOI 10.1016/j.molmed.2009.11.002. [Google Scholar] [CrossRef]
Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y (2010). A role for the elongator complex in zygotic paternal genome demethylation. Nature 463: 554–558. DOI 10.1038/nature08732. [Google Scholar] [CrossRef]
Otero G, Fellows J, Li Y, de Bizemont T, Dirac AMG, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ (1999). Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Molecular Cell 3: 109–118. DOI 10.1016/S1097-2765(00)80179-3. [Google Scholar] [CrossRef]
Paraskevopoulou C, Fairhurst SA, Lowe DJ, Brick P, Onesti S (2006). The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Molecular Microbiology 59: 795–806. DOI 10.1111/j.1365-2958.2005.04989.x. [Google Scholar] [CrossRef]
Peinado H, Olmeda D, Cano A (2007). Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nature Reviews Cancer 7: 415–428. DOI 10.1038/nrc2131. [Google Scholar] [CrossRef]
Song X, Liu W, Yuan X, Jiang J, Wang W, Mullen MK, Zhao X, Zhang Y, Liu F, Du S, Rehman A, Tian R, Li J, Frost A, Song Z, Green HN, Henry C, Liu X, Ding X, Wang D, Yao X, Wu J (2018). Acetylation of ACAP4 regulates CCL18-elicited breast cancer cell migration and invasion. Journal of Molecular Cell Biology 10: 559–572. DOI 10.1093/jmcb/mjy058. [Google Scholar] [CrossRef]
Sims JD, McCready J, Jay DG (2011). Extracellular heat shock protein (Hsp)70 and Hsp90α assist in matrix metalloproteinase-2 activation and breast cancer cell migration and invasion. PLoS One 6: e18848. DOI 10.1371/journal.pone.0018848. [Google Scholar] [CrossRef]
Sive HL, Grainger RM, Harland RM (2000). Early Development of Xenopus Laevis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE (2001). Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: Functional characterization using new analysis and information visualization methods. Nucleic Acids Research 29: 1097–1106. DOI 10.1093/nar/29.5.1097. [Google Scholar] [CrossRef]
Solinger JA, Paolinelli R, Klöß H, Scorza FB, Marchesi S, Sauder U, Mitsushima D, Capuani F, Stürzenbaum SR, Cassata G, Shen K (2010). The Caenorhabditis elegans elongator complex regulates neuronal α-tubulin acetylation. PLoS Genetics 6: e1000820. DOI 10.1371/journal.pgen.1000820. [Google Scholar] [CrossRef]
Theveneau E, Mayor R (2012). Neural crest delamination and migration: From epithelium-to-mesenchyme transition to collective cell migration. Developmental Biology 366: 34–54. DOI 10.1016/j.ydbio.2011.12.041. [Google Scholar] [CrossRef]
Tucker RP (2004). Neural crest cells: A model for invasive behavior. International Journal of Biochemistry & Cell Biology 36: 173–177. DOI 10.1016/S1357-2725(03)00243-7. [Google Scholar] [CrossRef]
Wittschieben BØ, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ (1999a). A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Molecular Cell 4: 123–128. DOI 10.1016/S1097-2765(00)80194-X. [Google Scholar] [CrossRef]
Wittschieben BØ, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ (1999b). A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Molecular Cell 4: 123–128. DOI 10.1016/S1097-2765(00)80194-X. [Google Scholar] [CrossRef]
Yang X, Li J, Zeng W, Li C, Mao B (2016). Elongator Protein 3 (Elp3) stabilizes Snail1 and regulates neural crest migration in Xenopus. Scientific Reports 6: 26238. DOI 10.1038/srep26238. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |