DOI:10.32604/biocell.2021.015340

| BIOCELL DOI:10.32604/biocell.2021.015340 |  |
| Article |
Lysophosphatidylcholine acyltransferase 1 is involved in the regulation of exosome secretion and uptake in colorectal cancer cells
1Anorectal Surgery, Yuncheng City Central Hospital, Yuncheng, 044000, China
2Department of Gastrointestinal and Pancreatic Surgery, Shanxi Provincial People’s Hospital, Taiyuan, 030012, China
*Address correspondence to: Shaofei Chang, sy4960343@163.com
Received: 10 December 2020; Accepted: 01 February 2021
Abstract: Lysophosphatidylcholine acyltransferase 1 (LPCAT1) is a phospholipid acyltransferase that promotes phospholipid synthesis and plasma membrane reconstruction. Exosomes play an important role in tumor metastasis. The release and uptake of exosomes are key steps of their functions and depend on plasma membrane fusion and plasma membrane receptors, respectively. The purpose of this study was to explore whether LPCAT1-induced plasma membrane remodeling would change the secretion and uptake behavior of exosomes in tumor cells. We first confirmed the abnormally high expression of LPCAT1 in colorectal cancer cells by quantitative real-time PCR (qPCR) and Western blot analysis. Then, SW620 cells were used as exosome source cells, and SW480 cells were used as exosome receiver cells. Exosomes from SW620 cells could effectively promote the migration of SW480 cells. When LPCAT1 expression was reduced via siRNA knockdown in the source cells, the secretion of exosomes was downregulated, thus weakening the pro-migratory effects of exosomes on target cells. Conversely, when LPCAT1 was knocked down in target cells, the uptake of exosomes in target cells also decreased sharply. These results undoubtedly revealed that LPCAT1 is functionally associated with the release and internalization of exosomes in colorectal cancer cells and could affect the paracrine effects of exosomes, preliminarily extending the classical metabolic function of LPCAT1 to exosome-related pathways.
Keywords: LPCAT1; Exosomes; Colorectal cancer
| Abbreviations | |
| HSP70: | heat shock 70 KDa protein |
| LPCAT1: | lysophosphatidylcholine acyltransferase 1 |
| MVBs: | multivesicular bodies |
| NTA: | nanoparticle tracking analysis |
| PCR: | polymerase chain reaction |
| qRT-PCR: | quantitative reverse transcription-PCR |
| RIPA: | radio-immunoprecipitation assay |
| TEM: | transmission electron microscopy |
| TSG101: | tumor susceptibility gene 101 protein |
Lysophosphatidylcholine acyltransferase 1 (LPCAT1) is an important enzyme involved in complex lipid metabolism. It is widely found in plants and animals and has acyltransferase and acetyltransferase activities (Wang and Tontonoz, 2019). It is encoded by the Lpcat1 gene, and the protein is located in the endoplasmic reticulum, the Golgi body, and other organelles (Bouchoux et al., 2011; Moessinger et al., 2011). In recent years, with the deepening of the research on its structure, function, and expression regulation, it has been found that it is involved in the synthesis of alveolar surfactant (Harayama et al., 2009; Lin et al., 2015; Nakanishi et al., 2006), and it is related to a variety of tumors, retinal degeneration, neurological diseases, and many other diseases. LPCAT1 expression is increased in various tumor tissues and is positively correlated with a malignant degree, cancer stage, and tumor grade (Du et al., 2017; Grupp et al., 2013; Mansilla et al., 2009; Wei et al., 2019). Dependent on its biological characteristics, LPCAT1 has the ability to remodel the plasma membrane. When the expression of LPCAT1 is inhibited, the levels of the oncogenic receptors located on the plasma membrane are reduced accordingly (Bi et al., 2019).
Exosomes are membranous vesicles with a diameter of about 30–100 nm. After the fusion of multivesicular bodies (MVBs) and the cell membrane, exosomes can be released into the extracellular matrix. They are widely distributed and originated and can be secreted by a variety of cells (Vlassov et al., 2012). Exosomes consist of proteins, lipids, and nucleic acids, and are involved in many physiological processes, for instance, the immune response, antigen presentation, intercellular communication, and protein and RNA transport (Shokrollahi et al., 2019; Simons and Raposo, 2009). Besides, it is a crucial tool for intercellular material and information exchange. Tumor cell exosomes usually have autocrine and paracrine effects, which are involved in the release and uptake of exosomes (Joo et al., 2020; Mills et al., 2019; Oliveira et al., 2020). The release of exosomes depends on plasma membrane fusion, while the uptake of exosomes by most cells depends on specific receptors in the plasma membrane (McKelvey et al., 2015; Salunkhe et al., 2020).
Therefore, the abnormal expression of LPCAT1 may be associated with exosomal signal transduction. We explored the relationship between LPCAT1 and exosome release and uptake, as well as the autocrine or paracrine effects of exosomes on colorectal cancer cells.
The human colorectal cancer cell lines, including HCT116, SW620, and SW480, were obtained from the Cell Resource Center, Peking Union Medical College. Human colon fibroblast CCD-18Co cells were purchased from The American Type Culture Collection (CRL-1459). All the cells were cultured in high-glucose DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% (v/v) fetal bovine serum (FBS; Invitrogen) at 37°C with 5% CO2.
Cells were plated in 24-well or 6-well plates and transfected with 10 nM siRNAs (Ruibo Bio, Guangzhou, China) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations.
Cells were cultured in DMEM supplemented with 10% exosome-depleted FBS for 48 h. Exosomes from the cultured supernatant were prepared using the ExoQuick-TC kit described by the manufacturer (System Biosciences, Mountain View, CA, USA). In general, the culture medium was centrifuged at 3000 × g for 15 min to remove apoptotic cells and cell debris. 3.3 mL exosome precipitation solution was added to every 10 mL culture supernatant, then frozen overnight, centrifuged at 10000 × g for 30 min, and the supernatant was discarded. The exosomes were suspended in 50 μL PBS and stored at −80°C or used directly.
Quantitative analysis of exosomes
Nanoparticle tracking analysis (NTA) was used to detect the prepared exosome samples. In few words, exosomes resuspended in 50 μL PBS, which were detected by NanoSight NS-300 (Malvern Panalytical, Malvern, UK), were further diluted to 300-fold to obtain between 20 and 100 objects per frame. Each sample was measured in triplicate via the camera, whose acquisition time is 30 s. Meanwhile, the detection threshold was set to 3. There are at least 200 complete analyses in each video. NTA 2.3 software was applied to collect and analyze the result.
Transmission electron microscopy (TEM)
Exosomes were isolated and resuspended in 100 μL PBS, and 50 μL of the suspension was placed onto formvar carbon-coated copper grids at room temperature for 20 min. The excess suspension was removed using filter paper. Exosomes were stained with 2% phosphotungstic acid (PTA) at room temperature for 1 min. The grids were fixed with 2% glutaraldehyde at room temperature for 5 min. Images were obtained with a transmission electron microscope (JEM-1230, Jeol, Ltd., Japan).
Uptake of PKH67-labeled exosomes by receptor cells
With lipophilic tracer PKH67 solution (Sigma-Aldrich, St. Louis, MO, USA), the 40 μg of protein equivalents of exosomes were incubated at 37°C for 10 min. The PKH67-labeled exosomes were purified by using Amicon® Ultra-15 filter (#UFC901024,10 kDa MWCO) according to the manufacturer’s suggestion (Merck Millipore, Danvers, MA, USA). The cells were seeded on 6-well plates (1 × 104 cells/well) and incubated for 24 h. The PKH67 labeled exosomes of 20 μg were added to the receptor cell culture medium and incubated at 37°C for 4 h. Receptor cells were washed with PBS three times, fixed with 4% paraformaldehyde at room temperature for 10 min, and permeated with 0.1% Triton X-100 for 5 min. Then the cells were stained with DAPI and visualized through a fluorescence microscope (IX71, Olympus, Japan).
The preparation of protein and Western blot
The RIPA lysis buffer (Beyotime Institute of Biotechnology, Beijing, China) was used to extract total exosomal and cellular protein. The BCA protein assay kit (Beyotime Institute of Biotechnology, Beijing, China) was applied to determine the protein concentrations of cell and exosome lysates. As for the analyses of Western blot, the cellular and 10 µg/well exosomal proteins were separated by electrophoresis in 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and finally transferred to polyvinylidene difluoride membrane (PVDF) (Amersham, Buckinghamshire, UK). In TBS-T, 5% skimmed milk (10 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.5% Tween 20) was used to block the membrane at 4°C overnight and to rinse it three times (10 min/time). After doing this, using the specific primary antibodies (1:1000 dilution) to incubate the membrane for 3 h at room temperature, and using the second antibody (1:5000 dilution) to incubate it for 1 h. ECL developer (Thermo Scientific) was added, and exposure imaging was performed through Odyssey gel imaging system. The specific antibodies to LPCAT1 (ab214034), CD81 (ab109201), HSP70 (ab181606), Calnexin (ab22595) and TSG101 (ab125011), were purchased from Abcam (Cambridge, UK). β-actin antibody (#4970) was acquired from Cell Signaling Technology (Danvers, MA, USA).
By using TRIzol reagent (Invitrogen, CA, USA), total RNA can be extracted. And by using a Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific, CA, USA), reverse transcription of cDNA was carried out. On ABI 7500 Real-Time PCR System (Applied Biosystems, Warrington, UK), Gene-specific PCR amplification was performed with Power SYBR Green Master Mix. After the expression of GAPDH was normalized, relative gene expression can be calculated via the 2−ΔΔCt method.
An All-in-One miRNA qRT-PCR Detection Kit (GeneCopoeia, MD, USA) was applied to detect the expression of miRNA. To put it shortly, in order to add poly-A tails to the 3’-end of miRNAs, total RNA was treated with poly-A polymerase.
To reverse transcribe miRNA tailed poly-A, M-MLV RTase was used with a universal adaptor PCR primer. Then, the expression of microRNA was measured by performing real-time qPCR using SYBR Green. RNU6B was adopted as an internal control for miRNAs in cells. miRNAs in exosomes were normalized to synthetic spike-in control Caenorhabditis elegans cel-miR-39-3p, which was added to all exosome isolates prior to RNA isolation.
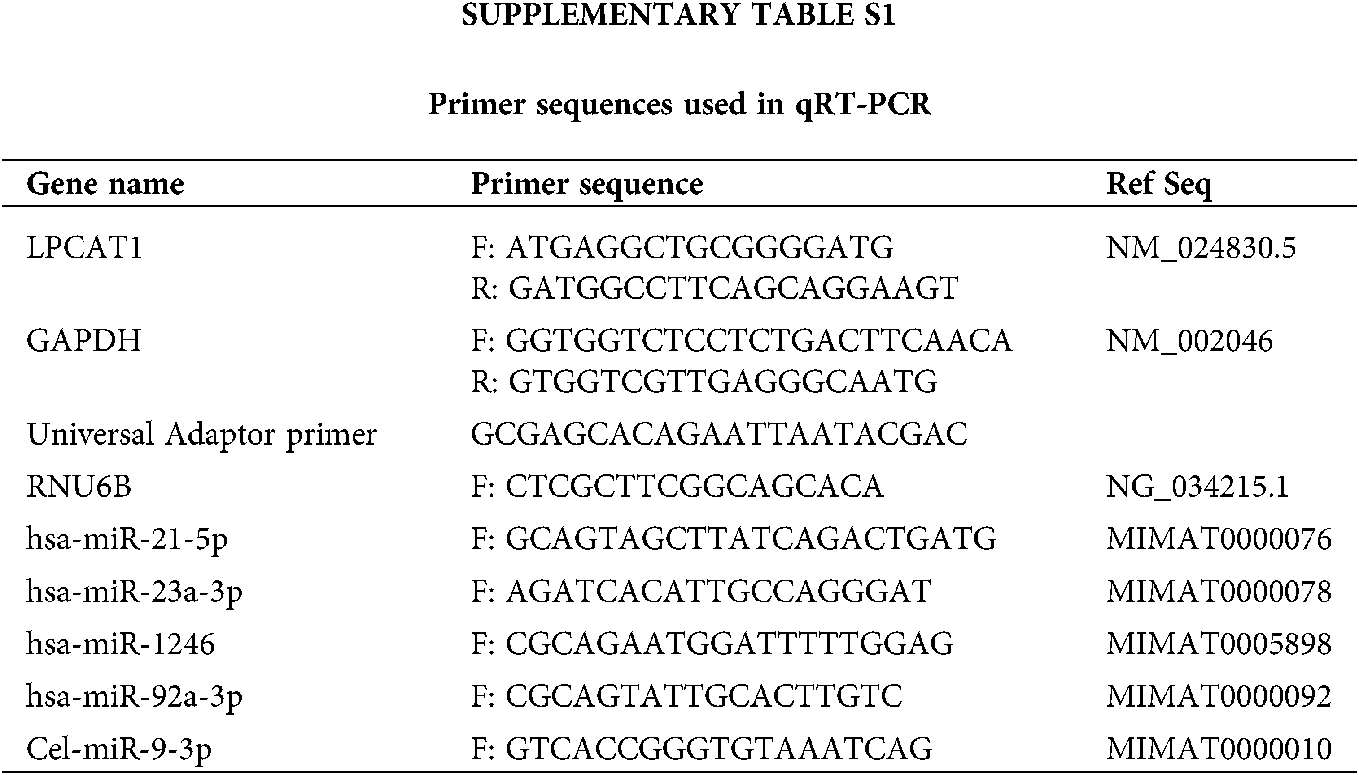
In Suppl. Tab. S1, primers for RT-qPCR sequences are listed.
Cell proliferation capacity was measured using the CCK8 assay. The digested cells, 3 × 103 cells/well, were seeded into the 96-well plate. Incubate in a 5% CO2 incubator at 37°C for various time points, discard the original culture medium, add 100 µL of the mixture of CCK8 reagent (DoJinDo Laboratories, Japan) and medium at the ratio of 1:9, and incubate in the incubator for additional 1 h. The absorbance values of 450 nm wavelength were determined by an enzyme labeling instrument (Multimode Reader, PerkinElmer, USA).
Transwell cell migration assay was performed with 24-well transwell chamber plates with 8 μm-pore size polycarbonate membranes (BD Biosciences), which were used for analyzing the ability of cellular migration. Cells in serum-free DMEM can be put into the upper chamber, and DMEM, which contains 20% FBS, was applied to the lower chamber. After 18 h, residual cells could be removed from the upper chamber. Then, cells were fixed with methanol, as well as stained with 4', 6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Science). After washing with PBS, under an inverted microscope, the number of cells passing through the membrane was counted (IX71, Olympus, Japan). Three random microscopic fields were selected for carrying out statistical analysis.
miRNA loading into exosomes by electroporation
Briefly, the exosome pellet was resuspended in 0.1 × PBS and gently mixed with the appropriated µL of miRNA (10 nM) in a final volume of 400 μL into 0.4 cm electroporation cuvettes. Exosomes were then electroporated using a Gene Pulser II Electroporator (Bio-Rad, Hercules, CA, USA) at 300 V and 25 μF of capacitance. Lastly, exosomes were incubated in ice for 30 min to allow the exosome membrane to be fully restored. To get rid of free nucleic acids, exosomes were diluted with cold PBS and isolated again by ultracentrifugation at 120,000 × g for 90 min at 15°C.
All data were processed via SPSS21.0 software and expressed as mean ± SD. In the meantime, the normality test of Shapiro-Wilk (W) was performed. In terms of non-normally distributed data, the nonparametric Wilcoxon signed-rank test was applied to assess the statistical differences among groups. When the data were close to a normal distribution, multiple sets of data were compared by using one-way ANOVA. In addition, in order to analyze the multiple comparisons of means, the LSD method was adopted. P < 0.05 has a significant meaning of statistics.
Abnormal expression of LPCAT1 in colon cancer tissues and cells
First, we found abnormally high expression of LPCAT1 (Fig. 1A) in colorectal tissues in the TGCA-based UALCAN online database (http://ualcan.path.uab.edu/index.html). Then, LPCAT1 expression was compared between the normal intestinal cell line CCD18-Co and several intestinal cancer cell lines (SW620, SW480, and HCT116). Both mRNA and protein levels of LPCAT1 were significantly higher in colorectal cells than in normal cells (Figs. 1B–1D).

Figure 1: Abnormal expression of LPCAT1 in colon cancer tissues and cells.
(A) LPCAT1 expressed in normal and colorectal tumor tissues. Data were acquired from UALCAN by analyzing the TCGA samples. (B) qRT-PCR analysis of LPCAT1 mRNA expression in colorectal cancer cells and normal colon cells. (C–D) Western blot analysis of LPCAT1 mRNA and protein expression in colorectal cancer cells and normal colon cells. One-way analysis of variance with Tukey post-hoc analysis. *P < 0.05 and **P < 0.01.
LPCAT1 level affects the exosome secretion
We compared the amounts of exosomes secreted by CCD18-Co and colon cancer cell lines (SW620, SW480, and HCT116) under the same conditions by collecting exosomes from identical volumes of culture supernatant at the corresponding time points. The number of exosomes secreted by the three colorectal cancer cell lines was apparently higher than that secreted by the CCD18-Co cell line (Fig. 2A). LPCAT1 remodels the plasma membrane, but it is not known whether LPCAT1 affects exosome secretion. Therefore, we used siRNA to knock down the expression of LPCAT1 in three colorectal cancer cell lines (Figs. 2B and 2C) and found that with the downregulation of LPCAT1, the cell viability did not change significantly at 24 h but decreased significantly at 48 h (Figs. 2D–2F). In order to exclude the influence of cell number on exosomes, we chose to extract exosomes within 24 h after siRNA treatment. The results showed that when LPCAT1 was knocked down, the number of exosomes in the three cells was significantly reduced (Fig. 2G). These results suggested that the expression of LPCAT1 in colorectal cancer cells is positively correlated with exosome secretion.

Figure 2: Correlation between the secretion of exosomes and the expression of LPCAT1.
(A) Exosome concentrations derived from different cells were quantified by NTA. (B, C) The effect of siRNAs on mRNA and protein expression of PFDN1 was examined by qPCR and Western blot. (D–F) Three kinds of colorectal cancer cells (HCT116, SW480, and SW620) were transfected with LPCAT1 siRNA (10 nM). CCK-8 was used to detect cell viability at 24 h and 48 h, respectively. (G) Bar graphs show the effect of LPCAT1 siRNAs on the concentration of exosomes. The relative exosome concentration was normalized to the cell number. Comparisons among groups were determined by one-way ANOVA with Tukey post-hoc analysis. *P < 0.05 and **P < 0.01. si, siRNA; si#1 and si#2, two different siRNA duplexes against LPCAT1.
LPCAT1 affects the ability of exosomes from source cells to promote migration
The exosomes of tumor cells can promote the migration of target cells through autocrine and paracrine effects, and the strength of this ability usually depends on the characteristics of the source cells (Jabbari et al., 2020; Whiteside, 2016; Zhang and Grizzle, 2014). We compared the migration ability of three kinds of colorectal cancer cells, and the results showed that the migration ability of SW480 cells was the weakest, while SW620 showed the strongest migration ability (Figs. 3A and 3B). SW480 cells were selected as the target cells, and the exosomes derived from SW480, HCT116, and SW620 cells were incubated and co-cultured. The results showed that the exosomes from SW480 cells could not effectively promote migration of SW480 cells themselves, while the exosomes of HCT116 and SW620 cells could promote the migration of SW480 cells (Figs. 3C and 3D), and the ability to promote migration was consistent with that of their own cells. In addition, three types of cell-derived exosomes did not effectively promote the proliferation of SW480 cells (Fig. 3E). Therefore, SW620 cells, which produce exosomes with the strongest pro-migratory effects, were selected as exosome source cells, and SW480, which exhibit weak migration capacity, were selected as target cells for our research model.
When the expression of LPCAT1 was knocked down by siRNA in SW620 source cells, the migration promoting capacity of exosomes was significantly lower than that of wild-type exosomes (Figs. 3F and 3G). One of the reasons for this result may be that LPCAT1 affects the amount of exocrine secretion. In addition, is it possible for LPCAT1 to enhance the pro-migratory effects of exosomes by changing their composition? We first detected the morphology and the relative content of marker proteins (CD81, HSP70, and TSG101) in the exosomes of SW620 cells after LPCAT1 knockdown (Figs. 4A and 4B). We selected miRNA markers of colorectal cancer exosomes (containing mir-1246, mir-23a, mir-21, and mir-92a), which are well recognized (Desmond et al., 2019). These miRNAs are upregulated in colorectal cancer cells and the serum of colorectal cancer patients and are positively correlated with the ability of tumor metastasis. The quantitative PCR results showed that the relative contents of these miRNAs in source cells were significantly higher than those in CCD18-Co and SW480 cells, both in the cells and in exosomes (Figs. 4C and 4D). However, knockdown of LPCAT1 in SW620 cells did not significantly change the relative content of these four miRNAs in cells and exosomes (Figs. 4E and 4F). These results suggested that LPCAT1 may have more influence on exosome secretion.

Figure 3: LPCAT1 affects the ability of exosomes from source cells to promote migration.
(A, B) Cell migration ability was tested by Transwell cell migration assay (Magnification, 200×; Scale bar, 50 µm). The quantifications of cell migration are presented in the histogram. (C, D) The indicated exosomes from different cells were collected under the same conditions (the same number of initial cells, the same culture time, and the same volume of culture medium), resuspended in 50 μL PBS, and co-cultured with SW480 cells for 24 h. The migration ability of SW480 cells in each group was detected by Transwell cell migration assay (Magnification, 200×; Scale bar, 50 µm). The control group was treated with PBS. (E) CCK-8 detection of SW620 cell activity at different time points via co-incubation with exosomes from each of the three types of colorectal cancer cells. (F, G) The exosomes from NC siRNA- or LPCAT1 siRNA-treated SW620 cells were collected and co-cultured with SW480 cells for 24 h. The migration ability of SW480 cells in each group was detected by Transwell cell migration assay (Magnification, 200×; Scale bar, 50 µm). One-way ANOVA with Tukey post-hoc analysis. *P < 0.05 and **P < 0.01.

Figure 4: Correlation between LPCAT1 expression and exosome characteristics.
(A) Representative scanning electron micrographs of exosomes purified from SW620 cells treated with NC siRNA or LPCAT1 siRNA. (B) Expression of LPCAT1 and the related exosome markers confirmed by Western blot in cells and exosomes (with a 50-μg protein loading volume). (C) Quantitative PCR was used to detect the relative content of four miRNAs in the three kinds of cells. U6 was used as an internal control. (D) Quantitative PCR was used to detect four miRNAs’ relative content in the same amount of exosomes derived from three kinds of cells. Cel-miR-39 was used as the spike-in control. (E, F) Quantitative PCR verification of the differential expression of miRNAs in cells and exosomes derived from SW620 cells treated with NC siRNA or LPCAT1 siRNA. The relative miRNA expression levels in cells were normalized to U6; the relative amount of the miRNAs in exosomes were normalized to spiked-in cel-miR-39. Comparisons among groups were determined by one-way ANOVA with Tukey post-hoc analysis. *P < 0.05 and **P < 0.01.
Effects of LPCAT1 on exosome uptake by target cells
If LPCAT1 is knocked down in SW480 target cells, the migration ability of the cells itself decreases (Figs. 5A and 5B). Interestingly, however, the pro-migratory effects of exosomes derived from SW620 cells on SW480 cells were weakened due to the downregulation of LPCAT1 in target cells (Figs. 5C and 5D). If LPCAT1 knockdown in the source cells reduces exosome secretion and weakens the pro-migratory effects of exosomes, why does LPCAT1 knockdown in target cells also weaken the pro-migratory effects of exosomes? To answer this question, exosomes from SW620 cells were fluorescently labeled with PKH-67, and it was found that the uptake of exosomes was significantly decreased after LPCAT1 was knocked down in SW480 target cells (Fig. 5E). In other words, LPCAT1 expression is positively correlated with the ability to take up exosomes. To further prove this hypothesis, we transferred C. elegans miRNA cel-miR-39 into exosomes derived from SW620 cells by electroporation (Fig. 5F). After the artificial cel-miR-39-rich exocrine was co-cultured with SW480 cells, the qRT-PCR results showed that the cel-miR-39 uptake in the LPCAT1 knock-down group was significantly lower than that in the control group (Fig. 5G). These results suggest that LPCAT1 is involved in the regulation of exosome uptake in colorectal cancer cells.

Figure 5: LPCAT1 is involved in the regulation of exosome uptake by target cells.
(A, B) The migration ability of SW480 cells treated with NC siRNA or LPCAT1 siRNA was detected by Transwell migration assay. (C, D) SW480 cells were pretreated with NC siRNA or LPCAT1 siRNA for 24 h. The migration ability of SW480 cells in each group after the treatment with exosomes was detected by Transwell migration assay (Magnification, 200×; Scale bar, 50 µm). The control group was treated with PBS. (E) Exosomes from SW620 cells were labeled with PKH-67 and incubated with SW480 target cells for 4 h. The uptake of exosomes by SW480 cells in each group was observed by fluorescence microscopy (Magnification, 400×; Scale bar, 50 µm). (F) Transfection of cel-miR-39 into the exosomes derived from SW620 cells by electroporation to prepare exosomes rich in cel-miR-39. Quantitative PCR was used to detect the relative content of cel-miR-39 in exosomes. ET, electroporation. (G) Detection of the relative contents of cel-miR-39 after co-incubation of exosomes rich in cel-miR-39 with SW480 cells treated with NC siRNA or LPCAT1 siRNA. U6 was used as an internal control. Comparisons among groups were determined by one-way ANOVA with Tukey post-hoc analysis. *P < 0.05 and **P < 0.01.
The expression of LPCAT1 is increased in a variety of tumor tissues and is positively correlated with the malignant degree, cancer stage, and tumor grade. LPCAT1 is involved in the regulation of the enzymatic synthesis of lysophosphatidylcholine and unsaturated fatty acid from phosphatidylcholine, while diacylglycerol acyltransferase (DGAT) could synthesize triacylglycerol (TAG) from unsaturated fatty acids and phosphatidylcholine (Pan et al., 2015). TAG is an important molecule in lipid metabolism and energy metabolism, which can provide a large amount of ATP for the growth and survival of tumor cells (Boroughs and DeBerardinis, 2015). Therefore, it has been speculated that LPCAT1 is a very important protease in the lipid metabolism processes needed for the survival of tumor cells and is involved in tumor development. A recent study by Bi et al. (2019) showed that in glioblastoma multiforme (GBM) cells when the expression of LPCAT1 is inhibited by shRNA, the levels of oncogenic receptors on the plasma membrane are reduced, growth factor signaling is blocked, and the growth of cancer cell lines is inhibited, revealing the functional association between oncogenic growth factor receptor signaling and LPCAT1 in dynamic lipid remodeling and metabolic abnormalities.
Tumor metastasis refers to the process in which malignant tumor cells leave the primary tumor, reach distant tissue or organs through various channels, and continue to proliferate and grow, forming metastatic foci. It is a sign of poor prognosis and a leading cause of death in cancer patients. Studies have shown that tumor metastasis is a multi-factor, multi-step, and dynamic process. Many scholars believe that exosomes from tumors and the metastatic microenvironment play an important role in tumor metastasis, and a large number of studies have been carried out on the function of exosomes in tumor metastasis (Milane et al., 2015; Wortzel et al., 2019). Studies have shown that exosomes can be used as biomarkers to predict the potential for metastasis. Exosomes play an important role in determining the organotropism of tumor metastasis (Hoshino et al., 2015), participating in the formation of the microenvironment before tumor metastasis (Costa-Silva et al., 2015), promoting the transformation and migration of the epithelial–mesenchymal tumor cells, affecting the proliferation capacity after tumor metastasis, promoting tumor angiogenesis, and participating in the immune escape of tumors.
Exosomes originate from the membrane cavity or early endosomes in the plasma membrane circulation pathway, which will sink inward to form intraluminal vesicles (ILVs) and further invaginate through intracellular lysosomal particles to form MVBs. The fusion of MVBs and cell membrane leads to the release of tiny vesicles with lipid bilayer membranes and with a diameter of 30–100 nm to the extracellular matrix. The docking to and subsequent fusion with the plasma membrane of MVBs are regulated by Rab, SNAP, and SNARE proteins (Hurley, 2008). The Rab27b and Rab27 effector molecules synaptotagmin-like4 and exophilin5 regulate the docking of MVBs on the plasma membrane (Ostrowski et al., 2010). At the same time, Rab11, Rab31, the R-SNARE protein YKT6, and the V-SNARE protein VAMP7/TIVAMP participate in the fusion of MVBs and plasma membrane (Bobrie et al., 2011; Fader et al., 2009). These results suggest that if plasma membrane remodeling occurs, the occurrence and secretion of exocrine bodies are very likely to be affected. Similarly, theoretically, the following mechanisms affect the absorption and uptake of exosomes: fusion, macropinocytosis, phagocytosis, and receptor- and raft-mediated endocytosis (McKelvey et al., 2015). As the names suggest, receptor-mediated endocytosis and raft-mediated endocytosis require ligands on the exosomes to bind to specific receptors on the cell membrane and microdomains rich in cholesterol and sphingomyelin, respectively. Then plasma membrane remodeling is also likely to affect exosome uptake. In this study, SW620 cells, which have a relatively high migration capacity, were used as exosome source cells, while SW480 cells, which have a relatively low migration capacity, were used as target cells. The decrease of LPCAT1 expression in source cells led to the downregulation of exosome secretion, and in this way, the ability of exosomes to promote the migration of target cells was weakened. Conversely, when LPCAT1 was knocked down in the target cells, the target cells’ uptake of exosomes also decreased sharply. These results undoubtedly suggest that LPCAT1 is involved in both the release and the uptake of exosomes in colorectal cancer cells.
Unfortunately, we have not been able to further elucidate which specific receptors are affected during LPCAT1-induced plasma membrane remodeling and whether these receptors determine the release and internalization of exosomes in colorectal cells. This requires large-scale follow-up screening and more accurate in-depth research. In addition, knockdown of LPCAT1 in SW620 cells did not significantly change the relative content of some characteristic miRNAs (Figs. 4E and 4F), but this does not mean that the expression of other functional miRNAs is not affected. Besides, lncRNAs, proteins, and lipids in exosomes may also be downstream effector molecules of the LPCAT1 regulatory pathway, which is also worth further study.
In summary, we demonstrated that LPCAT1 is associated with exosomal release and internalization in colorectal cancer cells and that it can affect the paracrine effects of exosomes, extending the classical metabolic function of LPCAT1 to the exosome-related pathway.
Authors’ Contribution: HL and SFC conceived and designed the experiments. HL performed the experiments. HL and SFC analyzed and interpreted the results of the experiments. HL and SFC drafted the manuscript. All authors read and approved the final manuscript for publication.
Availability of Data and Material: All data generated or analyzed during this study are included in this published article.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Bi J, Ichu TA, Zanca C, Yang H, Zhang W, Gu Y, Chowdhry S, Reed A, Ikegami S, Turner KM, Zhang W, Villa GR, Wu S, Quehenberger O, Yong WH, Kornblum HI, Rich JN, Cloughesy TF, Cavenee WK, Furnari FB, Cravatt BF, Mischel PS (2019). Oncogene amplification in growth factor signaling pathways renders cancers dependent on membrane lipid remodeling. Cell Metabolism 30: 525–538. [Google Scholar]
Bobrie A, Colombo M, Raposo G, Théry C (2011). Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 12: 1659–1668. [Google Scholar]
Boroughs LK, DeBerardinis RJ (2015). Metabolic pathways promoting cancer cell survival and growth. Nature Cell Biology 17: 351–359. [Google Scholar]
Bouchoux J, Beilstein F, Pauquai T, Guerrera IC, Chateau D, Ly N, Alqub M, Klein C, Chambaz J, Rousset M, Lacorte JM, Morel E, Demignot S (2011). The proteome of cytosolic lipid droplets isolated from differentiated Caco-2/TC7 enterocytes reveals cell-specific characteristics. Biology of the Cell 103: 499–517. [Google Scholar]
Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, Xiang J, Zhang T, Theilen TM, García-Santos G, Williams C, Ararso Y, Huang Y, Rodrigues G, Shen TL, Labori KJ, Lothe IM, Kure EH, Hernandez J, Doussot A, Ebbesen SH, Grandgenett PM, Hollingsworth MA, Jain M, Mallya K, Batra SK, Jarnagin WR, Schwartz RE, Matei I, Peinado H, Stanger BZ, Bromberg J, Lyden D (2015). Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nature Cell Biology 17: 816–826. [Google Scholar]
Desmond BJ, Dennett ER, Danielson KM (2019). Circulating extracellular vesicle microRNA as diagnostic biomarkers in early colorectal cancer—A review. Cancers 12: 52. [Google Scholar]
Du Y, Wang Q, Zhang X, Wang X, Qin C, Sheng Z, Yin H, Jiang C, Li J, Xu T (2017). Lysophosphatidylcholine acyltransferase 1 upregulation and concomitant phospholipid alterations in clear cell renal cell carcinoma. Journal of Experimental & Clinical Cancer Research 36: 66. [Google Scholar]
Fader CM, Sánchez DG, Mestre MB, Colombo MI (2009). TI-VAMP/VAMP7 and VAMP3/cellubrevin: Two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochimica et Biophysica Acta 1793: 1901–1916. [Google Scholar]
Grupp K, Sanader S, Sirma H, Simon R, Koop C, Prien K, Hube-Magg C, Salomon G, Graefen M, Heinzer H, Minner S, Izbicki JR, Sauter G, Schlomm T, Tsourlakis MC (2013). High lysophosphatidylcholine acyltransferase 1 expression independently predicts high risk for biochemical recurrence in prostate cancers. Molecular Oncology 7: 1001–1011. [Google Scholar]
Harayama T, Shindou H, Shimizu T (2009). Biosynthesis of phosphatidylcholine by human lysophosphatidylcholine acyltransferase 1. Journal of Lipid Research 50: 1824–1831. [Google Scholar]
Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D (2015). Tumour exosome integrins determine organotropic metastasis. Nature 527: 329–335. [Google Scholar]
Hurley JH (2008). ESCRT complexes and the biogenesis of multivesicular bodies. Current Opinion in Cell Biology 20: 4–11. [Google Scholar]
Jabbari N, Karimipour M, Khaksar M, Akbariazar E, Heidarzadeh M, Mojarad B, Aftab H, Rahbarghazi R, Rezaie J (2020). Tumor-derived extracellular vesicles: Insights into bystander effects of exosomes after irradiation. Lasers in Medical Science 35: 531–545. [Google Scholar]
Joo HS, Suh JH, Lee HJ, Bang ES, Lee JM (2020). Current knowledge and future perspectives on mesenchymal stem cell-derived exosomes as a new therapeutic agent. International Journal of Molecular Sciences 21: 727. [Google Scholar]
Lin S, Ikegami M, Moon C, Naren AP, Shannon JM (2015). Lysophosphatidylcholine acyltransferase 1 (LPCAT1) specifically interacts with phospholipid transfer protein StarD10 to facilitate surfactant phospholipid trafficking in alveolar type II cells. Journal of Biological Chemistry 290: 18559–18574. [Google Scholar]
Mansilla F, da Costa KA, Wang S, Kruhøffer M, Lewin TM, Orntoft TF, Coleman RA, Birkenkamp-Demtröder K (2009). Lysophosphatidylcholine acyltransferase 1 (LPCAT1) overexpression in human colorectal cancer. Journal of Molecular Medicine: Official Organ of the “Gesellschaft Deutscher Naturforscher und Ärzte” 87: 85–97. [Google Scholar]
McKelvey KJ, Powell KL, Ashton AW, Morris JM, McCracken SA (2015). Exosomes: Mechanisms of uptake. Journal of Circulating Biomarkers 4: 7. [Google Scholar]
Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM (2015). Exosome mediated communication within the tumor microenvironment. Journal of Controlled Release: Official Journal of the Controlled Release Society 219: 278–294. [Google Scholar]
Mills J, Capece M, Cocucci E, Tessari A, Palmieri D (2019). Cancer-derived extracellular vesicle-associated microRNAs in intercellular communication: One cell’s trash is another cell’s treasure. International Journal of Molecular Sciences 20: 6109. [Google Scholar]
Moessinger C, Kuerschner L, Spandl J, Shevchenko A, Thiele C (2011). Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. Journal of Biological Chemistry 286: 21330–21339. [Google Scholar]
Nakanishi H, Shindou H, Hishikawa D, Harayama T, Ogasawara R, Suwabe A, Taguchi R, Shimizu T (2006). Cloning and characterization of mouse lung-type acyl-CoA: lysophosphatidylcholine acyltransferase 1 (LPCAT1). Expression in alveolar type II cells and possible involvement in surfactant production. Journal of Biological Chemistry 281: 20140–20147. [Google Scholar]
Oliveira MC, Caires HR, Oliveira MJ, Fraga A, Vasconcelos MH, Ribeiro R (2020). Urinary biomarkers in bladder cancer: Where do we stand and potential role of extracellular vesicles. Cancers 12: 1400. [Google Scholar]
Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C (2010). Rab27a and Rab27b control different steps of the exosome secretion pathway. Nature Cell Biology 12: 19–30. [Google Scholar]
Pan X, Chen G, Kazachkov M, Greer MS, Caldo KM, Zou J, Weselake RJ (2015). In vivo and in vitro evidence for biochemical coupling of reactions catalyzed by lysophosphatidylcholine acyltransferase and diacylglycerol acyltransferase. Journal of Biological Chemistry 290: 18068–18078. [Google Scholar]
Salunkhe S, Dheeraj B, Chitkara M, Mittal DA (2020). Surface functionalization of exosomes for target-specific delivery and in vivo imaging & tracking: Strategies and significance. Journal of Controlled Release: Official Journal of the Controlled Release Society 326: 599–614. [Google Scholar]
Shokrollahi E, Nourazarian A, Rahbarghazi R, Salimi L, Karbasforush S, Khaksar M, Salarinasab S, Abhari A, Heidarzadeh M (2019). Treatment of human neuroblastoma cell line SH-SY5Y with HSP27 siRNA tagged-exosomes decreased differentiation rate into mature neurons. Journal of Cellular Physiology 234: 21005–21013. [Google Scholar]
Simons M, Raposo G (2009). Exosomes-vesicular carriers for intercellular communication. Current Opinion in Cell Biology 21: 575–581. [Google Scholar]
Vlassov AV, Magdaleno S, Setterquist R, Conrad R (2012). Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochimica et Biophysica Acta 1820: 940–948. [Google Scholar]
Wang B, Tontonoz P (2019). Phospholipid remodeling in physiology and disease. Annual Review of Physiology 81: 165–188. [Google Scholar]
Wei C, Dong X, Lu H, Tong F, Chen L, Zhang R, Dong J, Hu Y, Wu G, Dong X (2019). LPCAT1 promotes brain metastasis of lung adenocarcinoma by up-regulating PI3K/AKT/MYC pathway. Journal of Experimental & Clinical Cancer Research: CR 38: 95. [Google Scholar]
Whiteside TL (2016). Tumor-derived exosomes and their role in cancer progression. Advances in Clinical Chemistry 74: 103–141. [Google Scholar]
Wortzel I, Dror S, Kenific CM, Lyden D (2019). Exosome-mediated metastasis: Communication from a distance. Developmental Cell 49: 347–360. [Google Scholar]
Zhang HG, Grizzle WE (2014). Exosomes: A novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. American Journal of Pathology 184: 28–41. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |