DOI:10.32604/biocell.2022.016916

| BIOCELL DOI:10.32604/biocell.2022.016916 |  |
| Review |
Alteration in the expression of microRNA-21 regulated target genes: Role in breast cancer
1Faculty of Sciences, Shoolini University, Solan, 173229, India
2Department of Biotechnology, MMEC, Maharishi Markandeshwar (Deemed to be University), Ambala, 133207, India
3Maharishi Markandeshwar University, Solan, 173229, India
4Centre of Biotechnology, Maharshi Dayanand University, Rohtak, 124001, India
5School of Biotechnology, Jawaharlal Nehu University, New Delhi, 110067, India
6Biorefining and Advanced Materials Research Center, Scotland’s Rural College (SRUC), Edinburgh, EH9 3JG, UK
7Department of Chemistry, MMEC, Maharishi Markandeshwar (Deemed to be University), Ambala, 133207, India
*Address correspondence to: Adesh K. Saini, sainiade@mmumullana.org; Reena V. Saini, reenavohra10@mmumullana.org
Received: 09 April 2021; Accepted: 07 June 2021
Abstract: Breast cancer, also recognized as the principal cause of cancer-related deaths among women, is the second most familiar and prevalent form of cancer. New diagnostic and prognostic biomarkers that are highly specific are urgently needed for its early prognosis. MicroRNAs (miRNAs), a class of non-coding RNAs, are known to control the biological processes involving transcription, post-transcriptional and covalent modifications, splicing, translation, cell differentiation, proliferation, apoptosis, cancer progression, and invasion. Any dysregulation in miRNA expression, demonstrating their oncogenic and tumor-suppressive functions, contributes to cancer progression. MicroRNA-21 (miR-21), an ‘onco-miR’ in breast cancer, is involved in tumor progression and metastasis by suppressing the activity of the target gene via its interaction with the 3’UTR of the target gene. The upregulation of miR-21 is observed in many instances of breast cancer. Our review aims to summarize the current understanding of miR-21 in the regulation of important cellular functions via regulation of its target genes. We discuss its biosynthesis, oncogenic function in breast cancer, and different methods used for its detection. This will increase the current understanding of the role of miR-21 in breast cancer tumorigenesis, which will offer a perception of using miR-21 as an early detection molecular prognostic and diagnostic biomarker and as a therapeutic target in breast cancer care.
Keywords: Non-coding RNA; microRNAs; Breast cancer; Tumor suppression; Biomarker
Abbreviations
| Her2: | Human epidermal growth factor receptor 2 |
| ncRNAs: | Noncoding RNAs |
| miRNAs: | MicroRNAs |
| piRNA: | PIWI-interacting RNAs |
| circRNAs: | Circular RNAs |
| lncRNAs: | Long noncoding RNAs |
| miR-21: | MicroRNA-21 |
| pre-miR-21: | Precursor miR-21 |
| pri-miRNA: | Primary-miRNA |
| SNP: | Single nucleotide polymorphism |
| m6A: | N(6)-methyladenosine |
| HNRNPA2/B1: | Heterogeneous nuclear ribonucleoprotein A2/B1 |
| RISC: | RNA-induced silencing complex |
| AGO2: | Argonaute 2 |
| miRISC: | miRNA-induced silencing complex |
| Dcp 1: | Decapping protein 1 |
| Dcp 2: | Decapping protein 2 |
| XRN1: | Exoribonuclease 1 |
| qRT-PCR: | Quantitative reverse transcription polymerase chain reaction |
| NGS: | Next-generation sequencing |
| PDCD4: | Programmed cell death 4 |
| TPM1: | Tropomyosin-1 |
| PTEN: | Phosphatase and tensin homolog |
| MASPIN: | Mammary serine protease inhibitor |
| Bcl-2: | B-cell lymphoma 2 |
| LZTFL1: | Leucine zipper transcription factor-like 1 |
| PRMT5: | Protein arginine methyltransferase 5 |
| OSCC: | Oral squamous cell carcinoma |
| 2-DIGE: | Two-dimensional differentiation in-gel electrophoresis |
| anti-miR-21: | Antisense miR-21 |
| PIP: | Phosphatidylinositol phosphate |
| FAP: | Fibroblast activation protein |
| EMT: | Epithelial-mesenchymal transition |
| MAPK: | Mitogen-activated protein kinase |
The second most complex and heterogeneous cancer which brings mortality in 45–55 years older women across the world is breast cancer (Hemmatzadeh et al., 2016; Wang et al., 2017; Becker, 2015; Quan et al., 2020; Ataollahi et al., 2015). Aging is one of the most significant recognized risk factors associated with breast cancer. However, other risk factors such as deficiency of iodine, high estrogen level, obesity, intake of alcohol, menopause, family history, physically inactive and chest radiation exposure are also involved in breast cancer (Yager and Davidson, 2006; Steiner et al., 2008; Stoddard et al., 2008; Ataollahi et al., 2015). Breast cancer occurs in tissues that include mammary glands and ducts, resulting in lumps, swelling, redness, irritation in breast skin and nipples discharge (Ataollahi et al., 2015). The breast cancer cells are categorized into six major subtypes based on their phenotype and gene expression profile, including i) luminal A, ii) luminal B, iii) tumor enriched with human epidermal growth factor receptor 2 (also known as Her2), iv) basal-like (triple-negative), v) normal-like, and vi) claudin-low subtype (Singh and Mo, 2013; Sørlie et al., 2003; Eroles et al., 2012).
It is believed that the mortality rate due to breast cancer can be reduced through the early diagnosis and detection of these six subtypes (Imani et al., 2017). The mammographic screening tool is the most operative tool used for the early diagnosis of tissue-based tumors in the breast (Taplin et al., 2008; Imani et al., 2017). But this tool has certain limitations such as high false-positive results, low reactivity, and low preciseness and, not adequate to diagnose the subtypes, so there is a need for new diagnostic and prognostic biomarkers that are highly specific (Adhami et al., 2018; Imani et al., 2017). Various non-coding RNAs (ncRNAs) that involve microRNAs (miRNAs), PIWI-interacting RNAs (piRNA), circular RNAs (circRNAs), and long non-coding RNAs (lncRNAs) are found to regulate biological processes like transcription, post-transcriptional modifications, covalent modifications, splicing, and translation in the metastasis phase of breast cancer cells (Klinge, 2018). Of all the ncRNAs studied in breast cancer, miRNAs are the most thoroughly studied and have diagnostic potential (Lo et al., 2016).
miRNAs perform a crucial function in cancer research as predictive, diagnostic and, prognostic biomarkers because of their regulatory roles in vital processes of life like cell differentiation, proliferation, apoptosis, cancer progression and invasion (Kim, 2005; O’Day and Lal, 2010; Yu and Cheah, 2017). miRNAs are 15-27 nucleotides, short endogenous ncRNA molecules that are well known for post-transcriptionally regulating gene expression in eukaryotes by base pairing with protein-coding mRNA genes 3’-UTR (Feng and Tsao, 2016; Han et al., 2016; Wilczynska and Bushell, 2015; Adhami et al., 2018). Around half of the human miRNAs are placed on human cancer-related chromosomal regions (fragile sites), enabling them to alter the tumor suppressor or oncogenic pathways (Calin et al., 2004; Hemmatzadeh et al., 2016; Tang et al., 2012; Casalini and Iorio, 2009; Croce, 2009). Various studies have shown the miRNAs involvement as oncogenic/tumor suppressor in the pathogenesis of breast cancer as defined in Tab. 1 (Zhu et al., 2008; Si et al., 2007; Yan et al., 2008; Kong et al., 2012; Hassan et al., 2012; Fabian and Sonenberg, 2012).
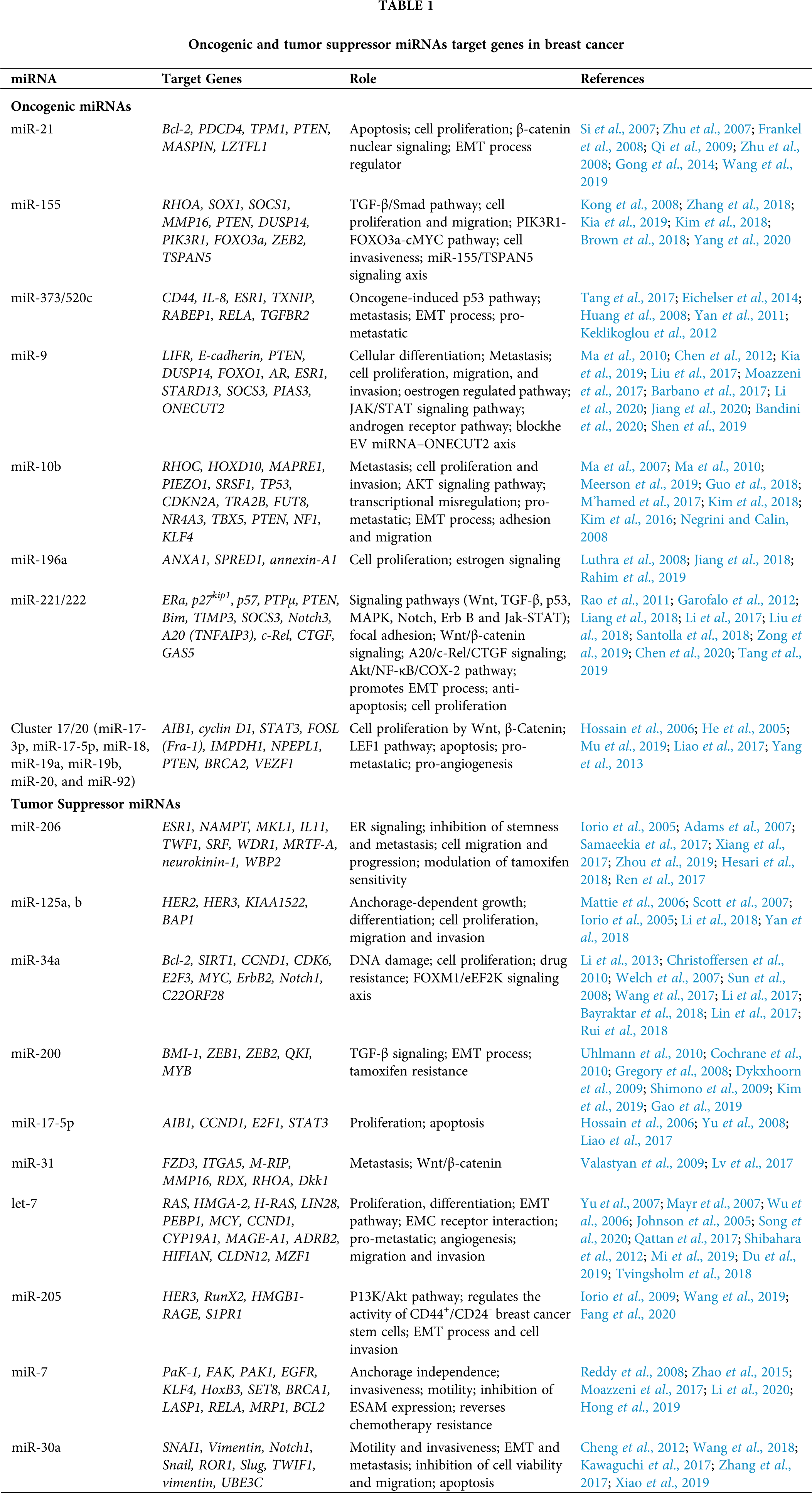
In breast cancer, microRNA-21 (miR-21) is the key miRNA used in the invasion process and facilitates tumor progression and metastasis (Han et al., 2012a; Han et al., 2012b). miR-21, also identified as MIR21, hsa-mir-21, miRNA21, MIRN21, is located on chromosome 17q21.3 encoding 72-nt long precursor miR-21 (pre-miR-21) (Abdel-hamid et al., 2015; Hemmatzadeh et al., 2016). By targeting multiple tumor/metastasis suppressor genes, miR-21 functions as an oncogene (Abdel-hamid et al., 2015; Hemmatzadeh et al., 2016; Selcuklu et al., 2009). The upregulated expression of miRNA-21 has been correlated with lymph node metastasis, advanced tumor levels and poor prognosis; therefore, it can act as a prognostic marker in breast cancer (Yan et al., 2008; Hemmatzadeh et al., 2016; Lee et al., 2011; Li et al., 2016; Shen et al., 2015).
In the nucleus, the RNA polymerase II transcribes non-coding miRNA gene into 100-120 nt long hair pin structured primary-miRNA (pri-miRNA) (Petri and Klinge, 2020; Lee et al., 2004; Yu and Cheah, 2017) (as shown in Fig. 1). The enzymes DROSHA (a class 2 ribonuclease III) and DGCR8 (dsRNA-binding protein) cleave the hair pin structure of pri-miRNA to produce 70 nt pre-miRNA (Filippov et al., 2000; Gregory et al., 2004; Denli et al., 2004). DROSHA (encoded by miRNA machinery gene) is responsible for processing miRNA in the initial phase (Khan et al., 2014). The pri-miRNA is further modified by the N(6)-methyladenosine (m6A) post-transcriptionally, stimulating the beginning of the miRNA biosynthesis process (Alarcón et al., 2015a). The heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2/B1) attaches to m6A in pri-miRNA, assisting DROSHA’s transformation of pri-miRNA into pre-miRNA (Alarcón et al., 2015b). After trimming of pri-miRNA, Exportin 5 (encoded by XPO5) or Exportin 1 (encoded by XPO1) exports pre-miRNA to the cytoplasm for further processing by the Dicer enzyme (Sheng et al., 2018). DROSHA plays a very significant role. The single nucleotide polymorphism (SNP) located in the 3’UTR of DROSHA rs10719 has been linked to breast cancer risk because it abrogates the hsa-miR-1298 binding site in DROSHA, thus affecting its expression (Khan et al., 2014).
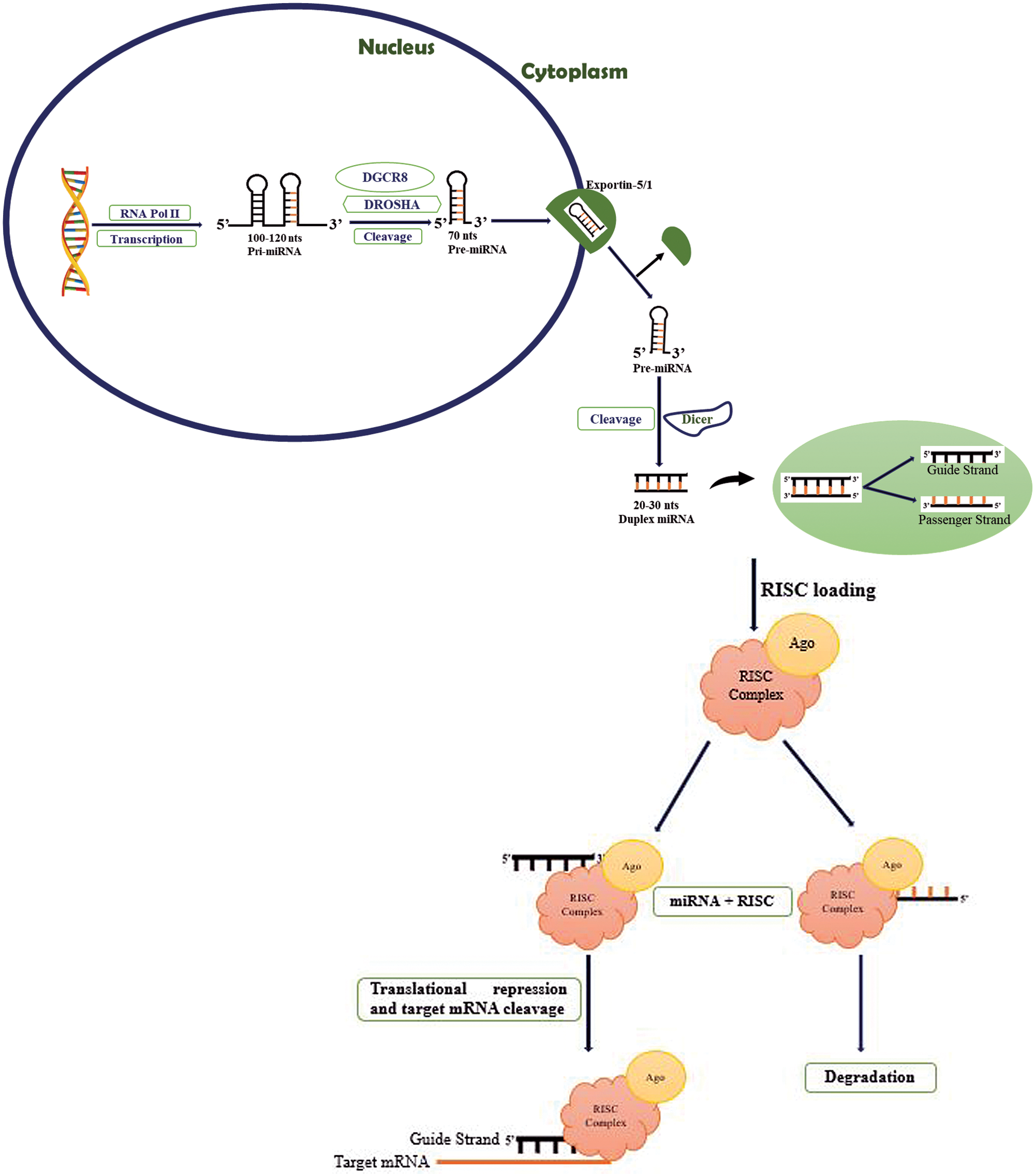
Figure 1: Biosynthesis of miRNA. RNA polymerase II is responsible for the transcription of 100–120 nts pri-miRNA in the nucleus. The pri-miRNA is trimmed into 70 nts pre-miRNA by DROSHA and DGCR8, which is then exported by exportin 5/1 into the cytoplasm, where it is further processed by Dicer enzyme forming 20–30 nts duplex miRNA with the guide (5’ to 3’) and passenger (3’ to 5’) strands. The guide strand interacts with the PAZ domain of the RISC, causing Argonaute 2 (AGO2) to cause post-transcriptional gene silencing and cleavage of the target mRNA, while the RISC’s PIWI domain degrades the passenger strand.
Dicer is a RNase III class of enzyme that cleaves the hair pin region of pre-miRNA to produce 20–30 nts of duplex miRNA composed of two single guide (5’ to 3’) and passenger (3’ to 5’) strands (Kobayashi and Tomari, 2016; Höck and Meister, 2008; Petri and Klinge, 2020). This guide strand joins the PAZ domain of RNA-induced silencing complex (RISC) to mediate post-transcriptional gene silencing of the target mRNA through Argonaute 2 (AGO2, RNase, catalytic part of RISC having “slicer” activity) which cleaves the target mRNA and the PIWI domain of RISC is responsible for the degradation of passenger strand (Höck and Meister, 2008; MacFarlane and Murphy, 2010; Yu and Cheah, 2017).
miRNA-induced silencing complex (miRISC, composed of AGO2 and guide strand) mediate the degradation of target mRNA. miRISC follows Ago-catalyzed, decapping, deadenylation, and exonucleolytic mechanisms (Eulalio et al., 2008; Behm-Ansmant et al., 2006; Wahid et al., 2010). Once miRNA and target mRNA complement each other, AGO2’s endonuclease activity is activated, cleaving the target mRNA (MacFarlane and Murphy, 2010). The miRISC inhibits the translation by obstructing the eIF4F complex, and mRNA circularization process of the target mRNA (Wahid et al., 2010). The GW182 family proteins bind to AGO2 are recruited by miRISC and act as a scaffold for the complex poly(A)-deadenylase PAN2-PAN-3 and CCR4-NOT proteins (MacFarlane and Murphy, 2010; O’Brien et al., 2018). The poly(A)-deadenylation process is initiated by the PAN2-PAN3 complex and completed by the CCR4-NOT protein complex. This process is further enhanced through the interaction of poly(A)-binding protein (PABPC) and GW182 tryptophan (W)-repeats (MacFarlane and Murphy, 2010; O’Brien et al., 2018). The exosome (with 3’-5’ exonuclease activity) mediates the degradation process and the enzymes, decapping protein 1 (Dcp 1) and decapping protein 2 (Dcp 2) facilitate the process by inducing exoribonuclease 1 (XRN1), which target 5’-3’ mRNA degradation (MacFarlane and Murphy, 2010; O’Brien et al., 2018).
Identifying miRNA in breast cancer cells
miRNAs have crucial role in gene regulation, so it is becoming imperative to develop and improve methods that can detect miRNAs because their detection in breast cancer is difficult due to their small size, low abundance, high level of sequence similarities (Chandrasekaran et al., 2019; Ye et al., 2019). The most well-known traditional methods for the detection of miRNA are microarray, in situ hybridization, bead-based flow cytometry, next-generation sequencing (NGS), northern blotting, and quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Ye et al., 2019; van Schooneveld et al., 2015). Out of these methods, northern blotting and in situ hybridization are low throughput methods, whereas microarray, bead-based flow cytometry, qRT-PCR and NGS are high throughput methods (van Schooneveld et al., 2015).
Northern blotting is the most standardized and highly specific method that can detect mature miRNA and their precursors (Ye et al., 2019; Várallyay et al., 2008). Besides these, the northern blotting method has a poor sensitivity and cannot detect the RNAs present in low amounts in few cells (Ye et al., 2019). qRT-PCR method is a highly sensitive and specific method used for evaluating miRNA and for the authentication of data prevailed from other detection platforms (Balcells et al., 2011; Yu and Cheah, 2017). The bead-based flow cytometry method is moderate in sensitivity and strong in specificity, whereas the in situ hybridization method is low in sensitivity and specificity (van Schooneveld et al., 2015). Microarray is a rapid method that can examine plenty of miRNAs from vast numbers of samples, with low sensitivity and specificity (Li and Ruan, 2009; Cissell and Deo, 2009). All these methods recognize only known miRNA structures, but NGS is the latest high sensitivity and specificity approach that enables new miRNAs to be recognized (van Schooneveld et al., 2015; Creighton et al., 2009). In the case of breast cancer, the upregulated levels of miR-21 can also be detected from tissues, blood, and serum (Yu and Cheah, 2017; Savad et al., 2012).
Oncogenic role of miR-21 in breast cancer cell proliferation and metastasis
The “oncomiR” miR-21 is believed to be involved in tumor proliferation and metastasis in breast tissues by controlling the expression of tumor suppressor genes such as programmed cell death 4 (PDCD4), tropomyosin-1 (TPM1), phosphatase and tensin homolog (PTEN), mammary serine protease inhibitor (MASPIN); an apoptosis suppressor gene, B-cell lymphoma 2 (Bcl-2), and leucine zipper transcription factor-like 1 gene [(LZTFL1), a tumor suppressor] (Si et al., 2007; Zhu et al., 2007; Frankel et al., 2008; Qi et al., 2009; Zhu et al., 2008; Wei et al., 2010; Wang et al., 2019) (illustrated in Fig. 2). These genes play a critical role in breast cancer and are targeted by miR-21.
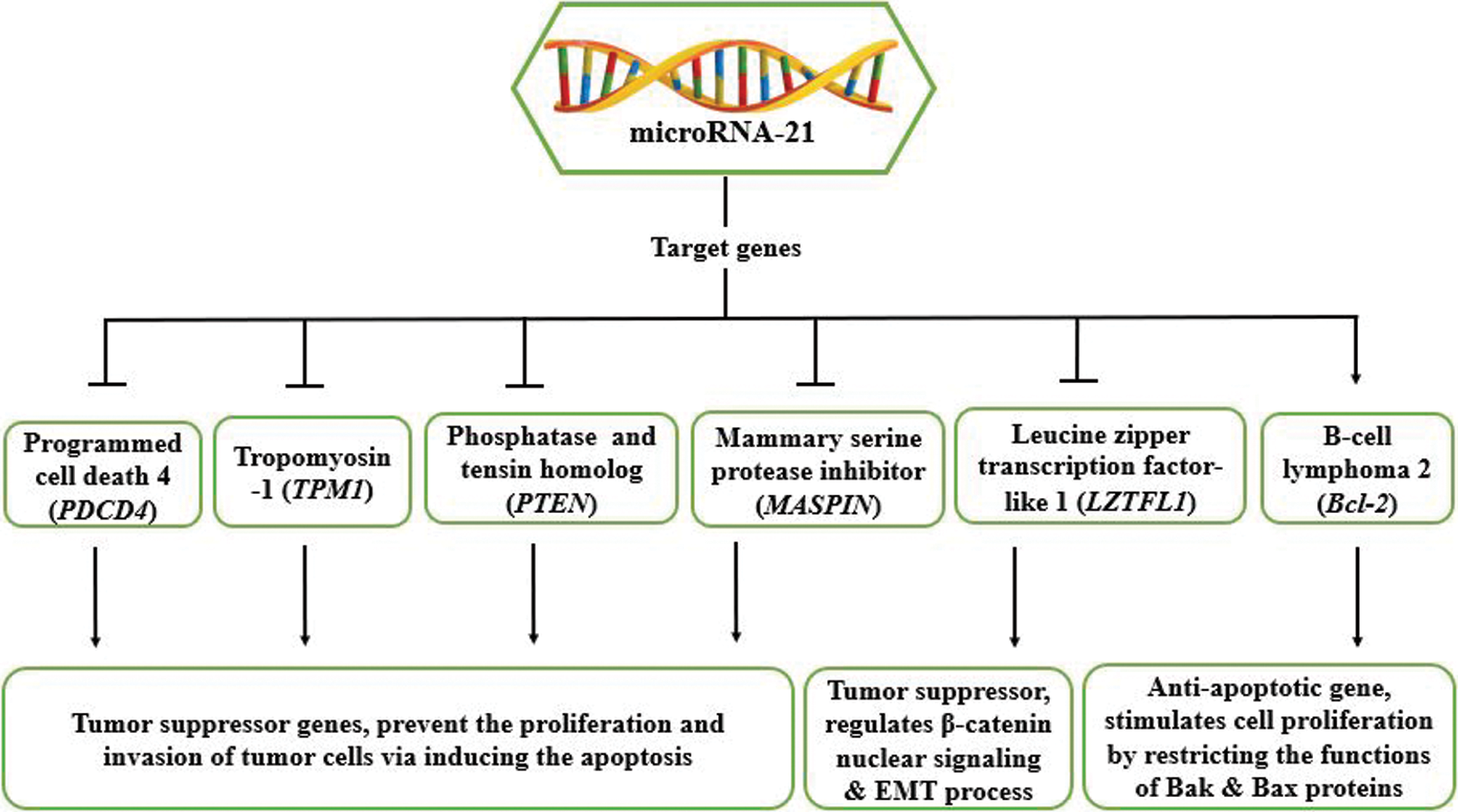
Figure 2: Depicts miR-21’s target genes and their functions in tumorigenesis.
Programmed Cell Death 4 (PDCD4)
In both immune/non-immune cells, the programmed cell death 4 gene is a tumour suppressor (Jiang et al., 2017). The PDCD4 protein contains three domains (two MA-3 domains and the N-terminal domain) that play a role in transcription and translation inhibition (Jiang et al., 2017). Owing to the binding of its MA-3 domains to eIF4A, PDCD4 prevents the translation of mRNAs having structured 5’UTRs, hindering eIF4A binding to eIF4G, even blocking eIF4A’s helicase activity, thus repressing the translation initiation mechanism of many genes, such as p53, Atg5, pro-caspase 3, and LXR-α (Kroczynska et al., 2012; Yang et al., 2004). By interacting with its N-terminal domain with A-myb & c-myb (RNA secondary structures), PDCD4 represses the elongation mechanism of translation (Singh et al., 2011). PDCD4 prevents tumor cell proliferation and invasion via inducing their apoptosis, leading to its upregulation in various tumors like breast, ovarian, gastric, esophageal, lung, hepatocellular, colon and glioma tumors (Powers et al., 2011; Wei et al., 2012; Jiang et al., 2017). The protein arginine methyltransferase 5 (PRMT5) deregulates the activity of the PDCD4 gene, resulting in breast cancer cell proliferation and invasiveness (Powers et al., 2011). miR-21 has been shown to target PDCD4 in tumour cells, thereby downregulating its expression (Sheedy et al., 2010).
PDCD4 is a target of miR-21 for breast cancer (Frankel et al., 2008). The miR-21 binding sites in the PDCD4 3’UTR are shown in Fig. 3A, which shows how miR-21 interacts with the PDCD4 3’UTR. To assess the fact that miR-21 specifically targets PDCD4, the firefly luciferase reporter assay was carried out. Frankel and his colleagues first cloned PDCD4’s 400–500 bp of 3’UTR into the pGL3 vector and then developed single (pGL3-PDCD4MUT1) and double (pGL3-PDCD4MUT2) mutations in the pGL3-PDCD4 seed region. PGL3-PDCD4, pGL3-PDCD4MUT1 & pGL3-PDCD4MUT2 were transfected into HEK293 cells and luciferase activity was tested after 24 h.
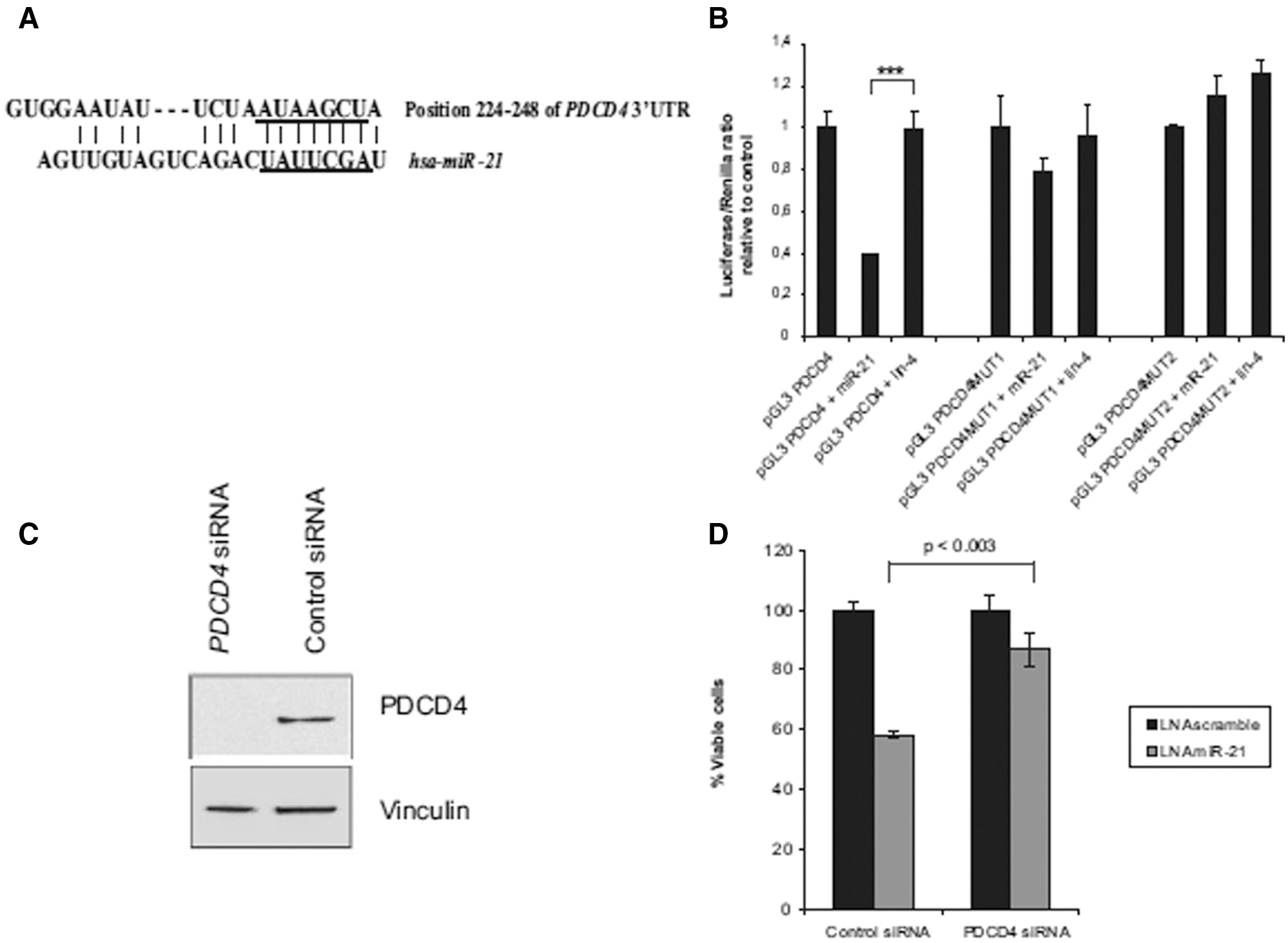
Figure 3: Represents that PDCD4 is a putative target of miR-21. A) shows the interaction of miR-21 with the 3’UTR of PDCD4. B) shows firefly luciferase reporter assay results, where co-transfection of HEK cells was done with pGL3-PDCD4, pGL3-PDCD4MUT1, pGL3-PDCD4MUT2 along with a Renilla luciferase transfection control plasmid, either alone or together with miR-21/lin-4 precursor miRNAs. Shown are relative luciferase values normalized to transfections without miRNA. Data are shown as the mean S.D. of four replicates and are representative of two independent experiments. ***, p_0.001 using a two-tailed t test. C) shows western blotting results where siRNA mediated knockdown of PDCD4 was shown in MCF-7 cells. D) shows the growth assay of PDCD4/control siRNA treated 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide in MCF-7 cells and transfection with inhibitors against miR-21/a scrambled control. Quantification of cell number was done after 5 days of transfection. Data are shown as the mean _ S.D. of three replicates and are representative of three independent experiments. The p value was calculated using a two-tailed t test (Frankel et al., 2008).
Results showed that miR-21 had no effect on pGL3-PDCD4MUT2 regulation but had little effect on pGL3-PDCD4MUT1 regulation (as shown in Fig. 3B), leading to the inference that there is a clear association between miR-21 and PDCD4 seed region 3’UTR. To verify the effects of miR-21 inhibition, they knocked down PDCD4 in MCF-7 cells via siRNA and found that PDCD4 siRNA mediated the antiproliferative effect on MCF-7 cells (see Figs. 3C, D). Thus, revealing the significance of PDCD4 by miR-21 as a putative target for breast cancer (Frankel et al., 2008).
Another research by Zhu et al. (2008) showed that miR-21 interacted with PDCD4’s 3’UTR, resulting in its reduced expression in MDA-MB-231 cells. Anti-miR-21 also led to increased PDCD4 expression, indicating the role of miR-21 in breast cancer cell invasion by directly interacting with the PDCD4 target gene (Zhu et al., 2008).
Tropomyosin-1, also known as TM1 and the actin-binding cytoskeletal protein, is a TPM family isoform that functions as a tumour suppressor gene by preventing cancer cell invasion by persuading apoptosis (Wang et al., 2015; Pan et al., 2017; Qi et al., 2009; Wang et al., 2019). Downregulation of TPM1 has been observed in breast cancer, glioma cancer, renal cell carcinoma, human oral squamous cell carcinoma (OSCC), and cholangiocarcinoma (Wang et al., 2015; Dube et al., 2015; Pan et al., 2017; Bharadwaj and Prasad, 2002; Yang et al., 2013; Ku et al., 2010). The expression of TPM1 in the OSCC is regulated by miR-21 (Pan et al., 2017). In addition, Zhu et al. (2007) in their report, showed that TPM1 is targeted by miR-21 in breast cancer to inhibit the TPM1 mediated apoptosis of cancer cells. They conducted two-dimensional differentiation in-gel electrophoresis, where they found the upregulated TPM1 expression in tumors procured from MCF-7 cells by antisense miR-21 oligonucleotides (anti-miR-21). miR-21 controls TPM1 expression by base-pairing with the V1 variant 3’UTR of TPM1, including miR-21’s putative binding sites. For further evidence, the V1-3’UTR was cloned into a luciferase reporter. Luciferase assay results suggested that anti-miR-21 increased Luc-TPM1-V1-UTR activity, while miR-21 decreased its activity. Moreover, the effect of miR-21 inhibition on TPM1 was also tested at the level of translation. MTT assay revealed that TPM1-V1 repressed the growth of MCF-7 cells in vitro, and soft agar assay results also showed that TPM1-V1 reduced colony formation leading to anchorage-independent growth repression. Altogether, we can conclude that miR-21 uses TPM1 to control its expression in MCF-7 cells as a novel target.
Phosphatase and tensin homolog (PTEN)
PTEN on chromosome 10 is recognized as a tumor suppressor gene coding 403 amino acids protein, which functions as a metabolic regulator of a number of cellular processes like glycogen synthesis, lipid and mitochondrial metabolism, glycolysis, and gluconeogenesis (Chen et al., 2018). It has dephosphorylation activities for both proteins and lipids due to which it is known as dual phosphatase enzyme (Chu and Tarnawski, 2004). PTEN has five domains: 1) N-terminal domain, responsible for the binding of phosphatidylinositol phosphate (PIP) substrates; 2) Phosphatase domain, carries acetylation sites and CX5R signature motif for the enzymatic actions; 3) C2 domain, a regulatory domain which contains a phospholipid-binding site and known for PTEN cell localization; 4) C-tail, which preserves phospho-sites and is responsible for the stability of protein; 5) PDZ domain, which is not well known (as shown in Fig. 4) (Chu and Tarnawski, 2004; Jerde, 2015; Chen et al., 2018). PTEN has regulatory functions in several processes, including cell migration, cell cycle arrest, MAP kinase signaling and angiogenesis (Chu and Tarnawski, 2004). Any mutations in PTEN gene lead to the progression of sporadic breast cancer, renal cell carcinoma, ovarian cancer, lung cancer, thyroid cancer, lymphoma cancer, hepatocellular carcinoma, lymphoma, head & neck cancer, prostate cancer, and glioblastoma (Chu and Tarnawski, 2004; Meng et al., 2007; Halachmi et al., 1998; Shao et al., 1998; Forgacs et al., 1998; Saito et al., 2000). In human hepatocellular cancer, miR-21 is known to regulate the PTEN expression and pathways which are mediated by PTEN (Meng et al., 2007).
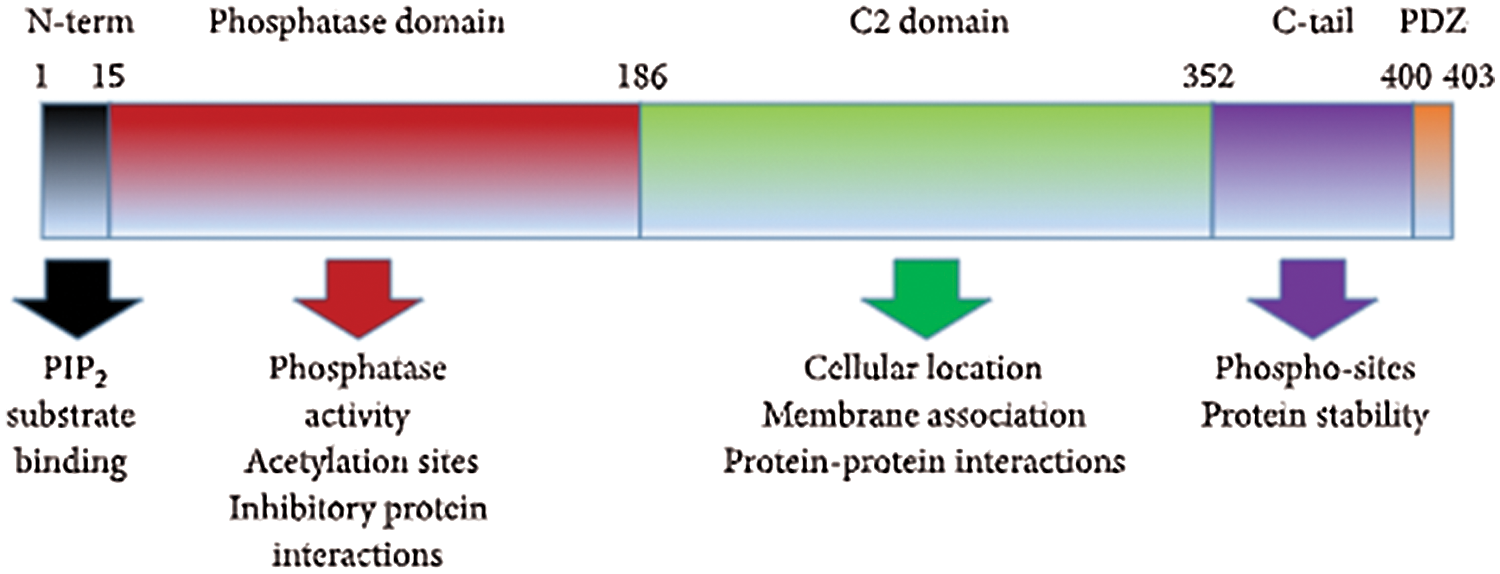
Figure 4: Protein domains of PTEN (Jerde, 2015).
A research conducted by Gong et al. (2014) showed that miR-21 decreases PTEN expression in tumors of breast phyllodes. Like PDCD4 and TPM1, PTEN also has miR-21 binding sequences at 3’UTR (Zhu et al., 2007; Frankel et al., 2008; Gong et al., 2014). PTEN protein levels were increased via miR-21 antisense oligonucleotides while, decreased via miR-21 mimics in stromal fibroblasts, demonstrating the role of miR-21 in targeting PTEN to increase the proliferation of phyllodes tumors cells (Gong et al., 2014). PTEN is known for the regulation of fibroblast activation protein (FAP) function (FAP is familiar as a serine protease and myofibroblast marker to increase the proliferation of phyllodes tumors) (Liu et al., 2012; Gong et al., 2014). The PTEN knockdown raised the protein expression levels of FAP during the myofibroblast differentiation process, suggesting the role of PTEN in proliferation. Moreover, reintroduction of PTEN resulted in a decrease in both PTEN mRNA and protein levels in benign tumors, thus inhibiting the proliferation of tumor cells by miR-21. Together, these findings concluded that miR-21 downregulates the PTEN expression to induce cell proliferation by increasing FAP expression in breast phyllodes tumor (Gong et al., 2014).
Mammary Serine Protease Inhibitor (MASPIN)
Mammary serine protease inhibitor is related to the serpin family (serine protease inhibitor family) (Zhang, 2002). Due to its ability to prevent tumor invasion by inducing apoptosis in tumor cells found on the 18q21.3-q23 chromosome, it is also recognized as a tumor suppressor gene (Berardi et al., 2013; Zhang, 2002). It is also known to prevent the process of angiogenesis (Zhang, 2002). Owing to its nuclear/cytoplasmic location in cancer cells, its expression is downregulated in various cancers like breast, gastric, prostate, melanoma and upregulated in gallbladder, thyroid, colorectal, pancreatic cancers (Berardi et al., 2013).
Maspin decreased MDA-MB-231 tumour cell invasion in the case of breast cancer (Zhu et al., 2008). With Flag-tagged maspin, they transfected MDA-MB-231 cells and confirmed the invasiveness of tumour cells through western blotting. However, by targeting it, the oncomir miR-21 downregulated maspin expression in MDA-MB-231 cells. Maspin also has 3’UTR binding sites for miR-21, so they cloned 3’UTR of maspin into pGL3 vector (luciferase gene) to check the Luc-Maspin-3’UTR luciferase activity. They found that miR-21 repression of luciferase activity was greater than 40%. Furthermore, anti-miR-21 increased the maspin expression, leading to the conclusion that by targeting its 3’UTR, miR-21 inhibits the maspin expression in MDA-MB-231 cells (Zhu et al., 2008).
B-cell Lymphoma 2 gene (Bcl-2)
B-cell lymphoma 2 is a 26 kDa protein present on human chromosome 18q21 with a hydrophobic carboxyl terminus positioned on the outer mitochondrial membrane and an anti-apoptotic gene as well (Lu et al., 1996). It suppresses the caspase-mediated cell death and stimulates cell proliferation by restricting the function of pro-apoptotic proteins, Bak and Bax (Lu et al., 1996; Wickramasinghe et al., 2009). Upregulated levels of bcl-2 gene are found in many cancers like lymphoma, breast, colorectal, thyroid, and cervical (Flangea et al., 2008; Manne et al., 2000; Zhou and Wang, 2015). Research on human glioblastoma U87MG cells revealed that miR-21 overexpression resulted in decreased Bax expression and increased bcl-2 gene expression along with reduced caspase-3 activity (Shi et al., 2010). Similarly, miR-21 controls tumorigenesis by upregulating the expression of the bcl-2 gene in breast cancer as miR-21 was highly overexpressed in breast tumors compared to the matched normal breast tissues 157 human miRNAs analysed. (Si et al., 2007). To investigate the function of miR-21, MCF-7 cells were transfected with anti-miR-21 oligonucleotide.
They showed that anti-miR-21 inhibits in vitro cell growth and tumor growth in the xenograft mouse model. Furthermore, to assess anti-miR-21 induced apoptosis, transfected cells were given Z-VAD-fmk, a caspase inhibitor, to counteract growth inhibition mediated by anti-miR-21. Apoptosis was caused by reduced Bcl-2 protein expression in anti-miR-21 transfected MCF-7 cells. These findings concluded that miR-21 should be used as a therapeutic target for breast cancer because of its essential function in controlling the Bcl-2 gene (Si et al., 2007). In another study, breast cancer cell lines were developed, which are paclitaxel-resistant, and it was shown that down regulation of miR-21 via its mimic enhance the sensitivity against paclitaxel in the developed cell lines and increase the expression of Bcl-2 (Zhao et al., 2015).
Leucine Zipper Transcription Factor-like 1 (LZTFL1)
The 3p21.3 region of the chromosome contains the leucine zipper transcription factor-like 1 (LZTFL-1) gene expressed in epithelial cells of a diversity of normal cells (Wei et al., 2010). LZTFL-1, a cytoplasmic protein, regulates β-catenin nuclear signaling, ciliary protein trafficking, and the epithelial-mesenchymal transition (EMT) process (Wei et al., 2016). It represses EMT by inhibiting the mitogen-activated protein kinase (MAPK) signaling pathway, which is especially important in lung cancer (Wei et al., 2010; Wei et al., 2019). It functions as a tumor suppressor by stabilizing E-cadherin-mediated adherens junctions and inhibiting β-catenin relocation into the nucleus, preventing cell invasion and EMT-mediated breast cancer metastasis (Wang et al., 2014; Wang et al., 2019). Wang et al. (2019) discovered that LZTFL1 could be used as a new target in breast cancer cells via miR-21, facilitating cell invasion and metastasis. miR-21 was found to be upregulated in the plasma of three patient groups and various cell lines.
The level of plasma miR-21 was significantly higher in the breast cancer patient group than the benign breast cancer patient group and healthy control group (see Fig. 5A), suggesting that plasma miR-21 level may be used as a potential diagnostic biomarker for breast cancer. The histopathological examination showed an upregulated level of miR-21 in breast cancer types Her-2+ and luminal B (as shown in Fig. 5B), and in vitro analysis results showed miR-21 upregulate in all cell lines of breast cancer (MDA-MB-231, Hs578T, MCF-7, and SK-BR3) compared to immortalized mammary epithelial (HBL-100) cell line (see Fig. 5C). As demonstrated by luciferase reporter assay, miR-21 directly targets LZTFL1, where miR-21 overexpression resulted in decreased luciferase activity in HEK293T cells transfected with luciferase gene containing either wild-type or mutant 3’UTR of LZTFL1 (see Fig. 6).
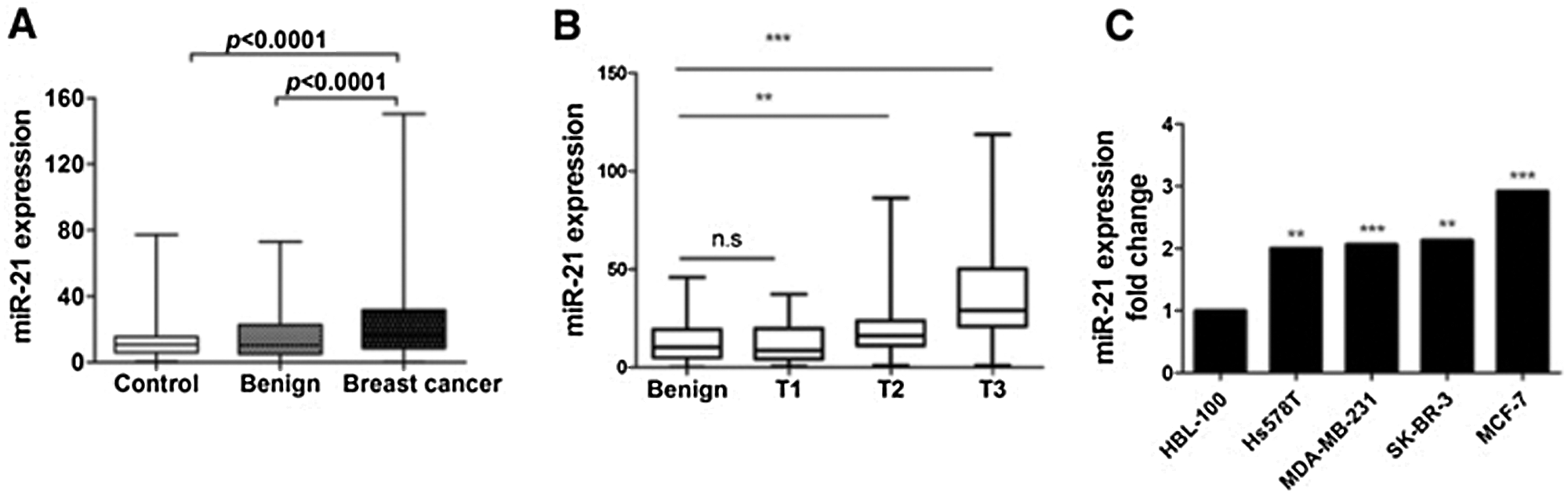
Figure 5: The levels of miR-21 in breast cancer patient plasma and cell lines. (A) Plasma miR-21 levels in 252 breast cancer patients, 127 healthy controls, and 82 benign breast cancer patients (p < 0.0001). (B) Plasma miR-21 levels in luminal A, luminal B, Her-2+ and basal-like types of breast cancer patients (p < 0.05). (C) The mRNA levels of miR-21 in HBL-100, Hs578T, MDA-MB-231, SK-BR3, and MCF-7 cell lines (*p < 0.05, **p < 0.01, ***p < 0.001) (Wang et al., 2019).
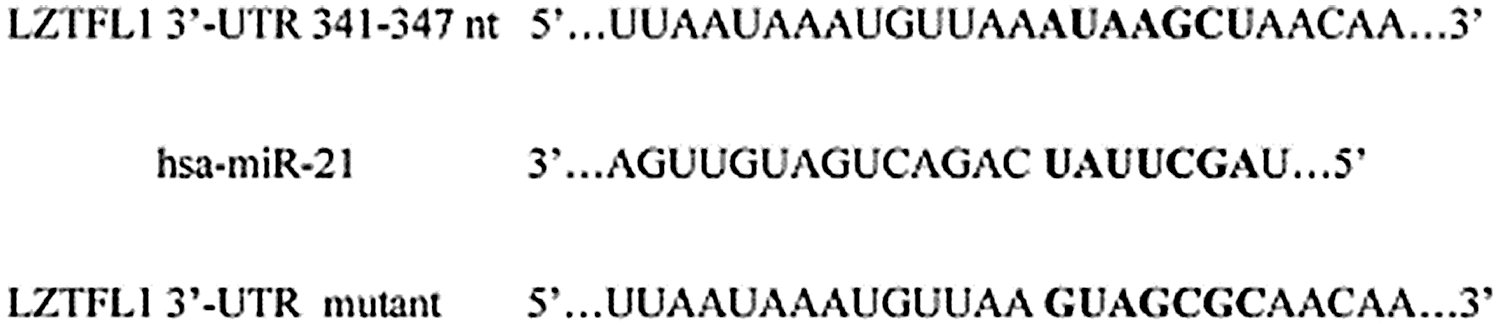
Figure 6: The predicted binding site of miR-21 in the 3’-UTR of wild type and mutant LZTFL1 (Wang et al., 2019).
The miR-21/LZTFL1/-catenin axis induces the EMT process. The effect of miR-21 inhibition and LZTFL1 knockdown on various EMT markers such as N-cadherin, vimentin, E-cadherin and claudin-1. miR-21 inhibition decreased N-cadherin and vimentin expression while increasing E-cadherin and claudin-1 expression, whereas LZTFL1 knockdown reversed the expression of these markers (as shown in Fig. 7). Immunofluorescence assay revealed that translocation of β-catenin in the nucleus was increased by miR-21 which contributed to the transcription of snail and slug transcription factors (EMT markers) (as shown in Fig. 7), while LZTFL1 repressed it. Therefore, miR-21 controls EMT by suppressing the LZTFL1, which increases β-catenin nuclear translocation, resulting in breast cancer proliferation and metastasis. This was also proved in BALB/c nude mice inoculated with Hs578T (human breast cancer) cells. miR-21 overexpression increased tumor size, weight, and volume (see Figs. 8A–8C) as well as lymph node invasion (see Figs. 8D and 8E). Furthermore, overexpression of miR-21 increased metastasis in liver and lung tissues (as shown in Figs. 8F and 8G) (Wang et al., 2019).
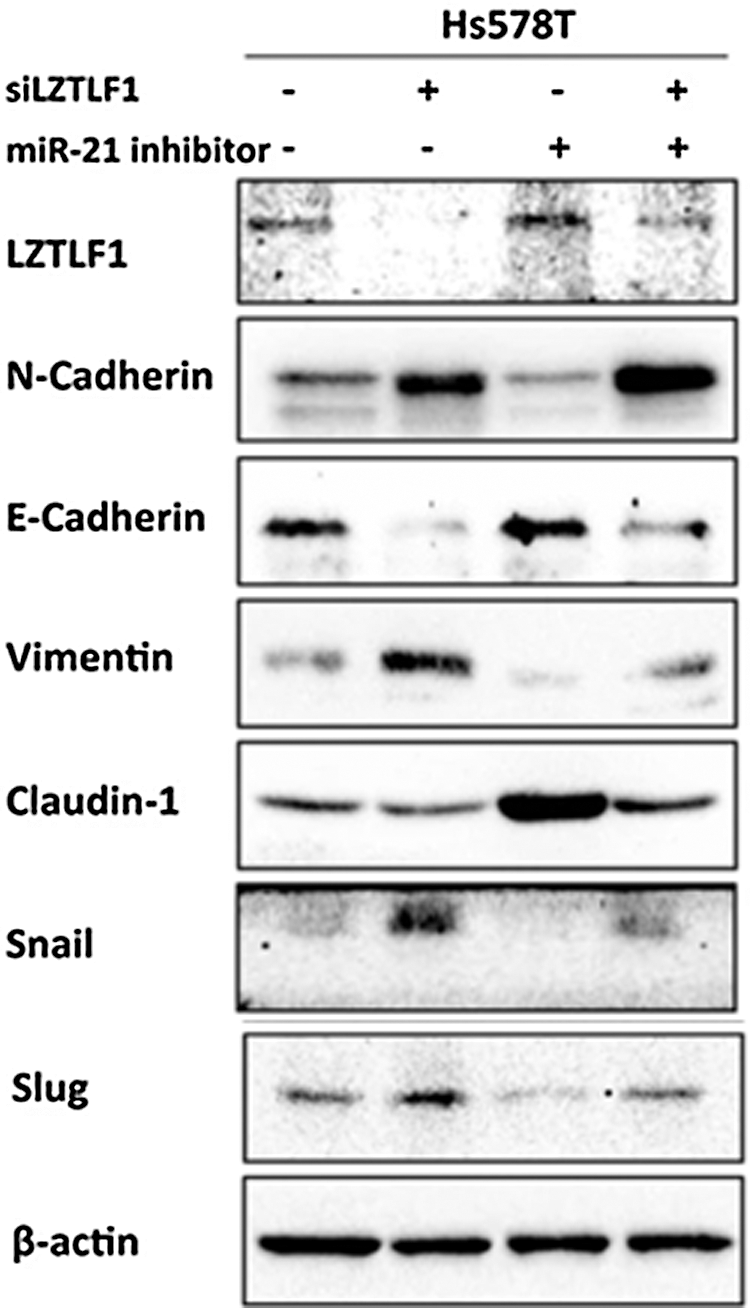
Figure 7: miR-21/LZTFL1 regulates β-catenin nuclear translocation and EMT process. The protein levels of EMT markers in Hs578T cells treated with miR-21 inhibitor, LZTFL1 siRNA alone, or combined for 48 h (Wang et al., 2019).
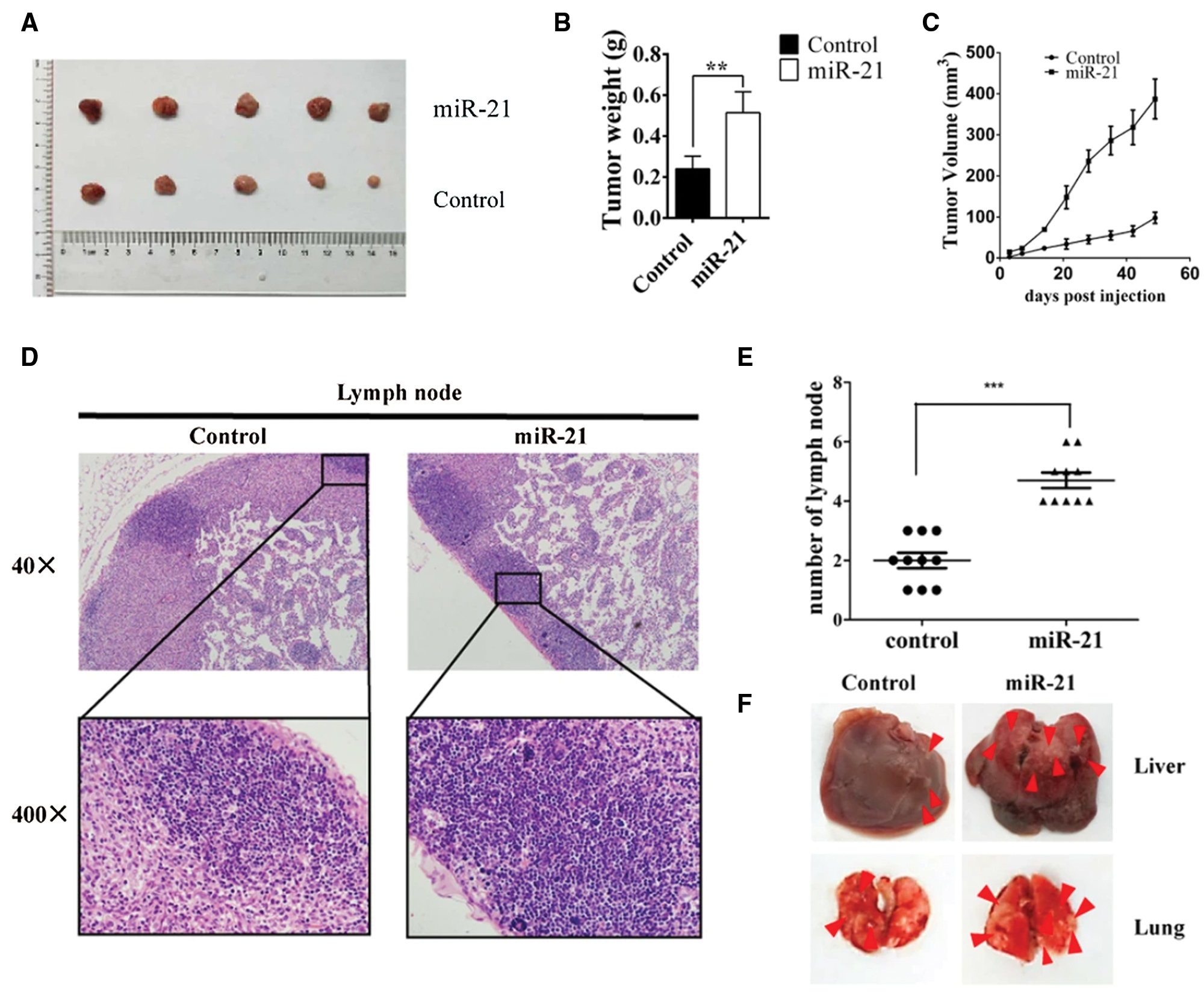
Figure 8: miR-21 promotes breast cancer proliferation and metastasis in vivo. (A) Xenografted tumors were obtained from miR-21-treated Hs578T and control Hs578T cells in situ. (B and C) Tumor weight and volume were observed and recorded in the groups indicated above. (D and E) The number of lymph nodes invaded was determined. (F and G) Liver and lung tissues were obtained, and the metastatic cells were visualized (Wang et al., 2019).
There is an extensive role of ‘oncomiR’ miR-21 in various stages breast cancer viz invasion, development, and metastasis. Different studies have demonstrated miR-21 upregulation in breast cancer cells by targeting multiple genes involved in breast cancer tumorigenesis. As mentioned above, miR-21 overexpression, through the downregulation of tumour suppressor genes, increases cell proliferation (Frankel et al., 2008; Zhu et al., 2007; Gong et al., 2014). miR-21 specifically interacts with PDCD4, TPM1, PTEN, and maspin 3’UTR, contributing to the suppression of apoptotic activities (Frankel et al., 2008; Zhu et al., 2007; Zhu et al., 2008; Gong et al., 2014). It also promotes breast cancer cell proliferation by increasing the bcl-2 gene expression. These studies indicate that alteration in the expression of miR-21 can be a beneficial intervention in the treatment of breast cancer. Various molecular biology-based techniques have been developed to detect the alteration in the expression of miR-21. Besides this, biosensor-based detection of miR-21 is a new area of research that could be more accessible and can help in point-of-care testing (Wang et al., 2020; Meng et al., 2020; Sun et al., 2018). Another sensitive tool to detect miR-21 in breast cancer is DNA–Peptide dendrimer and mass spectrometric method. It would also be essential to understand the role of miRNA in connection with the changing microbiome, which will help in early diagnosis and be used as a therapeutic biomarker (Allegra et al., 2020; Rastogi et al., 2020; Kashyap et al., 2021).
Acknowledgement: Authors acknowledge the support from Maharishi Markandeshwar (Deemed to be University) Mullana and Shoolini University, Solan.
Author Contribution: The authors confirm contribution to the paper as follows: study conception and design: AKS, RVS; data collection: PT, NKS, AC; draft manuscript preparation: PT, VKT, SSS. All authors compared different studies and approved the final version of the manuscript.
Funding Statement: Authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Abdel-hamid NR, Mohammed EA, Abbas AH, Badr FM (2015). MicroRNA-21 expression in primary breast cancer tissue among Egyptian female patients and its correlation with chromosome 17 aneusomy. Molecular Diagnosis & Therapy 19: 365–373. [Google Scholar]
Adams BD, Furneaux H, White BA (2007). The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-α (ERα) and represses ERα messenger RNA and protein expression in breast cancer cell lines. Molecular Endocrinology 21: 1132–1147. [Google Scholar]
Adhami M, Haghdoost AA, Sadeghi B, Afshar RM (2018). Candidate miRNAs in human breast cancer biomarkers: A systematic review. Breast Cancer 25: 198–205. [Google Scholar]
Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF (2015a). HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 162: 1299–1308. [Google Scholar]
Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF (2015b). N 6-methyladenosine marks primary microRNAs for processing. Nature 519: 482–485. [Google Scholar]
Allegra A, Musolino C, Tonacci A, Pioggia G, Gangemi S (2020). Interactions between the MicroRNAs and microbiota in cancer development: Roles and therapeutic opportunities. Cancers (Basel) 4: 805. [Google Scholar]
Ataollahi MR, Sharifi J, Paknahad MR, Paknahad A (2015). Breast cancer and associated factors: A review. Journal of Medicine and Life 8: 6. [Google Scholar]
Balcells I, Cirera S, Busk PK (2011). Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers. BMC Biotechnology 11: 1. [Google Scholar]
Bandini E, Fanini F, Vannini I, Rossi T, Plousiou M et al. (2020). miR-9-5p as a regulator of the androgen receptor pathway in breast cancer cell lines. Frontiers in Cell and Developmental Biology 8: 579160. [Google Scholar]
Barbano R, Pasculli B, Rendina M, Fontana A, Fusilli C et al. (2017). Stepwise analysis of MIR9 loci identifies miR-9-5p to be involved in Oestrogen regulated pathways in breast cancer patients. Scientific Reports 7: 1–2. [Google Scholar]
Bayraktar R, Ivan C, Bayraktar E, Kanlikilicer P, Kabil NN et al. (2018). Dual suppressive effect of miR-34a on the FOXM1/eEF2-kinase axis regulates triple-negative breast cancer growth and invasion. Clinical Cancer Research 24: 4225–4241. [Google Scholar]
Becker S (2015). A historic and scientific review of breast cancer: The next global healthcare challenge. International Journal of Gynecology & Obstetrics 131: S36–S39. [Google Scholar]
Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E (2006). mRNA degradation by miRNAs and GW182 requires both CCR4: NOT deadenylase and DCP1: DCP2 decapping complexes. Genes & Development 20: 1885–1898. [Google Scholar]
Berardi R, Morgese F, Onofri A, Mazzanti P, Pistelli M et al. (2013). Role of maspin in cancer. Clinical and Translational Medicine 2: 1–9. [Google Scholar]
Bharadwaj S, Prasad GL (2002). Tropomyosin-1, a novel suppressor of cellular transformation is downregulated by promoter methylation in cancer cells. Cancer Letters 183: 205–213. [Google Scholar]
Brown CY, Dayan S, Wong SW, Kaczmarek A, Hope CM et al. (2018). FOXP3 and miR-155 cooperate to control the invasive potential of human breast cancer cells by down regulating ZEB2 independently of ZEB1. Oncotarget 9: 27708–27727. [Google Scholar]
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E et al. (2004). Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America 101: 2999–3004. [Google Scholar]
Casalini P, Iorio MV (2009). MicroRNAs and future therapeutic applications in cancer. Journal of Buon 14: S17–S22. [Google Scholar]
Chandrasekaran AR, MacIsaac M, Dey P, Levchenko O, Zhou L, Andres M, Dey BK, Halvorsen K (2019). Cellular microRNA detection with miRacles: microRNA-activated conditional looping of engineered switches. Science Advances 5: eaau9443. [Google Scholar]
Chen CY, Chen J, He L, Stiles BL (2018). PTEN: Tumor suppressor and metabolic regulator. Frontiers in Endocrinology 9: 338. [Google Scholar]
Chen D, Sun Y, Wei Y, Zhang P, Rezaeian AH et al. (2012). LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nature Medicine 18: 1511–1517. [Google Scholar]
Chen Z, Pan T, Jiang D, Jin L, Geng Y, Feng X, Shen A, Zhang L (2020). The lncRNA-GAS5/miR-221-3p/DKK2 axis modulates ABCB1-mediated adriamycin resistance of breast cancer via the Wnt/β-catenin signaling pathway. Molecular Therapy-Nucleic Acids 19: 1434–1448. [Google Scholar]
Cheng CW, Wang HW, Chang CW, Chu HW, Chen CY et al. (2012). MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Research and Treatment 134: 1081–1093. [Google Scholar]
Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M et al. (2010). p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death & Differentiation 17: 236–245. [Google Scholar]
Chu EC, Tarnawski AS (2004). PTEN regulatory functions in tumor suppression and cell biology. Medical Science Monitor 10: RA235–41. [Google Scholar]
Cissell KA, Deo SK (2009). Trends in microRNA detection. Analytical and Bioanalytical Chemistry 394: 1109–1116. [Google Scholar]
Cochrane DR, Howe EN, Spoelstra NS, Richer JK (2010). Loss of miR-200c: A marker of aggressiveness and chemoresistance in female reproductive cancers. Journal of Oncology 2010: 821717. [Google Scholar]
Creighton CJ, Reid JG, Gunaratne PH (2009). Expression profiling of microRNAs by deep sequencing. Briefings in Bioinformatics 10: 490–497. [Google Scholar]
Croce CM (2009). Causes and consequences of microRNA dysregulation in cancer. Nature Reviews Genetics 10: 704–714. [Google Scholar]
Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ (2004). Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235. [Google Scholar]
Du Y, Lin Y, Yin K, Zhou L, Jiang Y, Yin W, Lu J (2019). Single nucleotide polymorphisms of let-7-related genes increase susceptibility to breast cancer. American Journal of Translational Research 11: 1748. [Google Scholar]
Dube S, Yalamanchili S, Lachant J, Abbott L, Benz P et al. (2015). Expression of tropomyosin 1 gene isoforms in human breast cancer cell lines. International Journal of Breast Cancer 2015: 859427. [Google Scholar]
Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A et al. (2009). miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One 4: e7181. [Google Scholar]
Eichelser C, Stückrath I, Müller V, Milde-Langosch K, Wikman H, Pantel K, Schwarzenbach H (2014). Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 5: 9650. [Google Scholar]
Eroles P, Bosch A, Pérez-Fidalgo JA, Lluch A (2012). Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways. Cancer Treatment Reviews 38: 698–707. [Google Scholar]
Eulalio A, Huntzinger E, Izaurralde E (2008). GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nature Structural & Molecular Biology 15: 346. [Google Scholar]
Fabian MR, Sonenberg N (2012). The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nature Structural & Molecular Biology 19: 586. [Google Scholar]
Fang T, Jiang YX, Chen L, Huang L, Tian XH, Zhou YD, Nagle DG, Zhang DD (2020). Coix seed oil exerts an anti-triple-negative breast cancer effect by disrupting miR-205/S1PR1 axis. Frontiers in Pharmacology 11: 1518. [Google Scholar]
Feng YH, Tsao CJ (2016). Emerging role of microRNA-21 in cancer. Biomedical Reports 5: 395–402. [Google Scholar]
Filippov V, Solovyev V, Filippova M, Gill SS (2000). A novel type of RNase III family proteins in eukaryotes. Gene 245: 213–221. [Google Scholar]
Flangea C, Potencz E, Mihăescu R, Gîju S, Anghel A (2008). Bcl-2 expression in Hodgkin’s lymphoma progression. Romanian Journal of Morphology Embryology 49: 357–363. [Google Scholar]
Forgacs E, Biesterveld EJ, Sekido Y, Fong K, Muneer S et al. (1998). Mutation analysis of the PTEN/MMAC1 gene in lung cancer. Oncogene 17: 1557–1565. [Google Scholar]
Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH (2008). Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. Journal of Biological Chemistry 283: 1026–1033. [Google Scholar]
Gao Y, Zhang W, Liu C, Li G (2019). miR-200 affects tamoxifen resistance in breast cancer cells through regulation of MYB. Scientific Reports 9: 1–6. [Google Scholar]
Garofalo M, Quintavalle C, Romano G, croce MC, Condorelli G (2012). miR221/222 in cancer: Their role in tumor progression and response to therapy. Current Molecular Medicine 12: 27–33. [Google Scholar]
Gong C, Nie Y, Qu S, Liao JY, Cui X, Yao H, Zeng Y, Su F, Song E, Liu Q (2014). miR-21 induces myofibroblast differentiation and promotes the malignant progression of breast phyllodes tumors. Cancer Research 74: 4341–4352. [Google Scholar]
Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A et al. (2008). The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biology 10: 593–601. [Google Scholar]
Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R (2004). The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240. [Google Scholar]
Guo D, Guo J, Li X, Guan F (2018). Enhanced motility and proliferation by miR-10b/FUT8/p-AKT axis in breast cancer cells. Oncology Letters 16: 2097–2104. [Google Scholar]
Halachmi N, Halachmi S, Evron E, Cairns P, Okami K et al. (1998). Somatic mutations of the PTEN tumor suppressor gene in sporadic follicular thyroid tumors. Genes, Chromosomes and Cancer 23: 239–243. [Google Scholar]
Han M, Liu M, Wang Y, Chen X, Xu J et al. (2012a). Antagonism of miR-21 reverses epithelial-mesenchymal transition and cancer stem cell phenotype through AKT/ERK1/2 inactivation by targeting PTEN. PLoS One 7: e39520. [Google Scholar]
Han M, Liu M, Wang Y, Mo Z, Bi X, Liu Z, Fan Y, Chen X, Wu C (2012b). Re-expression of miR-21 contributes to migration and invasion by inducing epithelial-mesenchymal transition consistent with cancer stem cell characteristics in MCF-7 cells. Molecular and Cellular Biochemistry 363: 427–436. [Google Scholar]
Han M, Wang F, Gu Y, Pei X, Guo G et al. (2016). MicroRNA-21 induces breast cancer cell invasion and migration by suppressing smad7 via EGF and TGF-β pathways. Oncology Reports 35: 73–80. [Google Scholar]
Hassan O, Ahmad A, Sethi S, Sarkar FH (2012). Recent updates on the role of microRNAs in prostate cancer. Journal of Hematology & Oncology 5. [Google Scholar]
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D et al. (2005). A microRNA polycistron as a potential human oncogene. Nature 435: 828–833. [Google Scholar]
Hemmatzadeh M, Mohammadi H, Jadidi-Niaragh F, Asghari F, Yousefi M (2016). The role of oncomirs in the pathogenesis and treatment of breast cancer. Biomedicine & Pharmacotherapy 78: 129–139. [Google Scholar]
Hesari Z, Nourbakhsh M, Hosseinkhani S, Abdolvahabi Z, Alipour M et al. (2018). Down-regulation of NAMPT expression by mir-206 reduces cell survival of breast cancer cells. Gene 673: 149–158. [Google Scholar]
Höck J, Meister G (2008). The Argonaute protein family. Genome Biology 9: 1–8. [Google Scholar]
Hong T, Ding J, Li W (2019). Mir-7 reverses breast cancer resistance to chemotherapy by targeting MRP1 and BCL2. Onco Targets and Therapy 12: 11097. [Google Scholar]
Hossain A, Kuo MT, Saunders GF (2006). Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Molecular and Cellular Biology 26: 8191–8201. [Google Scholar]
Huang Q, Gumireddy K, Schrier M, Le Sage C, Nagel R et al. (2008). The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nature Cell Biology 10: 202–210. [Google Scholar]
Imani S, Zhang X, Hosseinifard H, Fu S, Fu J (2017). The diagnostic role of microRNA-34a in breast cancer: A systematic review and meta-analysis. Oncotarget 8: 23177. [Google Scholar]
Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A et al. (2009). microRNA-205 regulates HER3 in human breast cancer. Cancer Research 69: 2195–2200. [Google Scholar]
Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R et al. (2005). MicroRNA gene expression deregulation in human breast cancer. Cancer Research 65: 7065–7070. [Google Scholar]
Jerde TJ (2015). Phosphatase and tensin homologue: Novel regulation by developmental signaling. Journal of Signal Transduction 2015: 282567. [Google Scholar]
Jiang CF, Shi ZM, Li DM, Qian YC, Ren Y et al. (2018). Estrogen-induced miR-196a elevation promotes tumor growth and metastasis via targeting SPRED1 in breast cancer. Molecular Cancer 17: 1–8. [Google Scholar]
Jiang M, Zhang W, Zhang R, Liu P, Ye Y et al. (2020). Cancer exosome-derived miR-9 and miR-181a promote the development of early-stage MDSCs via interfering with SOCS3 and PIAS3 respectively in breast cancer. Oncogene 39: 4681–4694. [Google Scholar]
Jiang Y, Jia Y, Zhang L (2017). Role of programmed cell death 4 in diseases: A double-edged sword. Cellular & Molecular Immunology 14: 884–886. [Google Scholar]
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R et al. (2005). RAS is regulated by the let-7 microRNA family. Cell 120: 635–647. [Google Scholar]
Kashyap S, Pal S, Chandan G, Saini V, Chakrabarti S et al. (2021). Understanding the cross-talk between human microbiota and gastrointestinal cancer for developing potential diagnostic and prognostic biomarkers. Seminars in Cancer Biology, S1044–S579X(21)00121-8. DOI 10.1016/j.semcancer.2021.04.020.Epub ahead of print. [Google Scholar]
Kawaguchi T, Yan L, Qi Q, Peng X, Gabriel EM et al. (2017). Overexpression of suppressive microRNAs, miR-30a and miR-200c are associated with improved survival of breast cancer patients. Scientific Reports 7: 1–2. [Google Scholar]
Keklikoglou I, Koerner C, Schmidt C, Zhang JD, Heckmann D et al. (2012). MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-κB and TGF-β signaling pathways. Oncogene 31: 4150–4163. [Google Scholar]
Khan S, Greco D, Michailidou K, Milne RL, Muranen TA et al. (2014). MicroRNA related polymorphisms and breast cancer risk. PLoS One 9: e109973. [Google Scholar]
Kia V, Paryan M, Mortazavi Y, Biglari A, Mohammadi-Yeganeh S (2019). Evaluation of exosomal miR-9 and miR-155 targeting PTEN and DUSP14 in highly metastatic breast cancer and their effect on low metastatic cells. Journal of Cellular Biochemistry 120: 5666–5676. [Google Scholar]
Kim EJ, Kim JS, Lee S, Lee H, Yoon JS et al. (2019). QKI, a miR-200 target gene, suppresses epithelial-to-mesenchymal transition and tumor growth. International Journal of Cancer 145: 1585–1595. [Google Scholar]
Kim J, Siverly AN, Chen D, Wang M, Yuan Y et al. (2016). Ablation of miR-10b suppresses oncogene-induced mammary tumorigenesis and metastasis and reactivates tumor-suppressive pathways. Cancer Research 76: 6424–6435. [Google Scholar]
Kim J, Yao F, Xiao Z, Sun Y, Ma L (2018). MicroRNAs and metastasis: Small RNAs play big roles. Cancer and Metastasis Reviews 37: 5–15. [Google Scholar]
Kim S, Lee E, Jung J, Lee JW, Kim HJ et al. (2018). microRNA-155 positively regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast cancer. Oncogene 37: 2982–2991. [Google Scholar]
Kim VN (2005). MicroRNA biogenesis: Coordinated cropping and dicing. Nature Reviews Molecular Cell Biology 6: 376–385. [Google Scholar]
Klinge CM (2018). Non-coding RNAs in breast cancer: Intracellular and intercellular communication. Non-Coding RNA 4: 40. [Google Scholar]
Kobayashi H, Tomari Y (2016). RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochimica et Biophysica Acta (BBA)–Gene Regulatory Mechanisms 1859: 71–81. [Google Scholar]
Kong W, Yang H, He L, Zhao JJ, Coppola D et al. (2008). MicroRNA-155 is regulated by the transforming growth factor β/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Molecular and Cellular Biology 28: 6773–6784. [Google Scholar]
Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M (2012). microRNAs in cancer management. Lancet Oncology 13: e249–58. [Google Scholar]
Kroczynska B, Sharma B, Eklund EA, Fish EN, Platanias LC (2012). Regulatory effects of programmed cell death 4 (PDCD4) protein in interferon (IFN)-stimulated gene expression and generation of type I IFN responses. Molecular and Cellular Biology 32: 2809–2822. [Google Scholar]
Ku BM, Ryu HW, Lee YK, Ryu J, Jeong JY et al. (2010). 4’-Acetoamido-4-hydroxychalcone, a chalcone derivative, inhibits glioma growth and invasion through regulation of the tropomyosin 1 gene. Biochemical and Biophysical Research Communications 402: 525–530. [Google Scholar]
Lee JA, Lee HY, Lee ES, Kim I, Bae JW (2011). Prognostic implications of microRNA-21 overexpression in invasive ductal carcinomas of the breast. Journal of Breast Cancer 14: 269–275. [Google Scholar]
Lee Y, Kim M, Han J, Yeom KH, Lee S et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO Journal 23: 4051–4060. [Google Scholar]
Li B, Lu Y, Yu L, Han X, Wang H et al. (2017). miR-221/222 promote cancer stem-like cell properties and tumor growth of breast cancer via targeting PTEN and sustained Akt/NF-κB/COX-2 activation. Chemico-Biological Interactions 277: 33–42. [Google Scholar]
Li L, Yuan L, Luo J, Gao J, Guo J, Xie X (2013). MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clinical and Experimental Medicine 13: 109–117. [Google Scholar]
Li M, Pan M, Wang J, You C, Zhao F et al. (2020). MiR-7 reduces breast cancer stem cell metastasis via inhibiting RELA to decrease ESAM expression. Molecular Therapy-Oncolytics 18: 70–82. [Google Scholar]
Li S, Yang X, Yang J, Zhen J, Zhang D (2016). Serum microRNA-21 as a potential diagnostic biomarker for breast cancer: a systematic review and meta-analysis. Clinical and Experimental Medicine 16: 29–35. [Google Scholar]
Li W, Ruan K (2009). MicroRNA detection by microarray. Analytical and Bioanalytical Chemistry 394: 1117–1124. [Google Scholar]
Li X, Zeng Z, Wang J, Wu Y, Chen W et al. (2020). MicroRNA-9 and breast cancer. Biomedicine & Pharmacotherapy 122: 109687. [Google Scholar]
Li Y, Wang Y, Fan H, Zhang Z, Li N (2018). miR-125b-5p inhibits breast cancer cell proliferation, migration and invasion by targeting KIAA1522. Biochemical and Biophysical Research Communications 504: 277–282. [Google Scholar]
Li ZH, Weng X, Xiong QY, Tu JH, Xiao A et al. (2017). miR-34a expression in human breast cancer is associated with drug resistance. Oncotarget 8: 106270. [Google Scholar]
Liang YK, Lin HY, Dou XW, Chen M, Wei XL et al. (2018). MiR-221/222 promote epithelial-mesenchymal transition by targeting Notch3 in breast cancer cell lines. NPJ Breast Cancer 4: 1–9. [Google Scholar]
Liao XH, Xiang Y, Yu CX, Li JP, Li H et al. (2017). STAT3 is required for MiR-17-5p-mediated sensitization to chemotherapy-induced apoptosis in breast cancer cells. Oncotarget 8: 15763. [Google Scholar]
Lin X, Chen W, Wei F, Zhou BP, Hung MC et al. (2017). Nanoparticle delivery of miR-34a eradicates long-term-cultured breast cancer stem cells via targeting C22ORF28 directly. Theranostics 7: 4805. [Google Scholar]
Liu DZ, Chang B, Li XD, Zhang QH, Zou YH (2017). MicroRNA-9 promotes the proliferation, migration, and invasion of breast cancer cells via down-regulating FOXO1. Clinical and Translational Oncology 19: 1133–1140. [Google Scholar]
Liu R, Li H, Liu L, Yu J, Ren X (2012). Fibroblast activation protein: A potential therapeutic target in cancer. Cancer Biology & Therapy 13: 123–129. [Google Scholar]
Liu S, Wang Z, Liu Z, Shi S, Zhang Z et al. (2018). miR-221/222 activate the Wnt/β-catenin signaling to promote triple-negative breast cancer. Journal of Molecular Cell Biology 10: 302–315. [Google Scholar]
Lo PK, Wolfson B, Zhou X, Duru N, Gernapudi R et al. (2016). Non-coding RNAs in breast cancer. Briefings in Functional Genomics 15: 200–221. [Google Scholar]
Lu QL, Abel P, Foster CS, Lalani EN (1996). bcl-2: Role in epithelial differentiation and oncogenesis. Human Pathology 27: 102–110. [Google Scholar]
Luthra R, Singh RR, Luthra MG, Li YX, Hannah C et al. (2008). MicroRNA-196a targets annexin A1: A microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene 27: 6667–6678. [Google Scholar]
Lv C, Li F, Li X, Tian Y, Zhang Y et al. (2017). MiR-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing Wnt signaling antagonists. Nature Communications 8: 1–8. [Google Scholar]
M’hamed IF, Privat M, Trimeche M, Penault-Llorca F, Bignon YJ et al. (2017). miR-10b, miR-26a, miR-146a And miR-153 expression in triple negative vs. non triple negative breast cancer: Potential biomarkers. Pathology & Oncology Research 23: 815–827. [Google Scholar]
Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B et al. (2010). Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nature Biotechnology 28: 341–347. [Google Scholar]
Ma L, Teruya-Feldstein J, Weinberg RA (2007). Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449: 682–688. [Google Scholar]
Ma L, Young J, Prabhala H, Pan E, Mestdagh P et al. (2010). miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nature Cell Biology 12: 247–256. [Google Scholar]
MacFarlane LA, R Murphy P (2010). MicroRNA: Biogenesis, function and role in cancer. Current Genomics 11: 537–561. [Google Scholar]
Manne U, Weiss HL, Grizzle WE (2000). Bcl-2 expression is associated with improved prognosis in patients with distal colorectal adenocarcinomas. International Journal of Cancer 89: 423–430. [Google Scholar]
Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L et al. (2006). Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Molecular Cancer 5: 24. [Google Scholar]
Mayr C, Hemann MT, Bartel DP (2007). Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315: 1576–1579. [Google Scholar]
Meerson A, Eliraz Y, Yehuda H, Knight B, Crundwell M et al. (2019). Obesity impacts the regulation of miR-10b and its targets in primary breast tumors. BMC Cancer 19: 1–0. [Google Scholar]
Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST et al. (2007). MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133: 647–658. [Google Scholar]
Meng T, Shang N, Nsabimana A, Ye H, Wang H, Wang C, Zhang Y (2020). An enzyme-free electrochemical biosensor based on target-catalytic hairpin assembly and Pd@UiO-66 for the ultrasensitive detection of microRNA-21. Analytica Chimica Acta 1138: 59–68. [Google Scholar]
Mi Y, Liu F, Liang X, Liu S, Huang X et al. (2019). Tumor suppressor let-7a inhibits breast cancer cell proliferation, migration and invasion by targeting MAGE-A1. Neoplasma 66: 54–62. [Google Scholar]
Moazzeni H, Najafi A, Khani M (2017). Identification of direct target genes of miR-7, miR-9, miR-96, and miR-182 in the human breast cancer cell lines MCF-7 and MDA-MB-231. Molecular and Cellular Probes 34: 45–52. [Google Scholar]
Mu F, Huang J, Xing T, Jing Y, Cui T et al. (2019). The Wnt/β-catenin/Lef1 pathway promotes cell proliferation at least in part through direct upregulation of miR-17-92 cluster. Frontiers in Genetics 10: 525. [Google Scholar]
Negrini M, Calin GA (2008). Breast cancer metastasis: A microRNA story. Breast Cancer Research 10: 1–4. [Google Scholar]
O’Brien J, Hayder H, Zayed Y, Peng C (2018). Overview of microRNA biogenesis, mechanisms of actions, and circulation. Frontiers in Endocrinology 9: 402. [Google Scholar]
O’Day E, Lal A (2010). MicroRNAs and their target gene networks in breast cancer. Breast Cancer Research 12: 1–0. [Google Scholar]
Pan H, Gu L, Liu B, Li Y, Wang Y et al. (2017). Tropomyosin-1 acts as a potential tumor suppressor in human oral squamous cell carcinoma. PLoS One 12: e0168900. [Google Scholar]
Petri BJ, Klinge CM (2020). Regulation of breast cancer metastasis signaling by miRNAs. Cancer and Metastasis Reviews 39: 837–886. [Google Scholar]
Powers MA, Fay MM, Factor RE, Welm AL, Ullman KS (2011). Protein arginine methyltransferase 5 accelerates tumor growth by arginine methylation of the tumor suppressor programmed cell death 4. Cancer Research 71: 5579–5587. [Google Scholar]
Qattan A, Intabli H, Alkhayal W, Eltabache C, Tweigieri T, Amer SB (2017). Robust expression of tumor suppressor miRNA’s let-7 and miR-195 detected in plasma of Saudi female breast cancer patients. BMC Cancer 17. [Google Scholar]
Qi L, Bart J, Tan LP, Platteel I, van der Sluis T et al. (2009). Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer 9: 1–8. [Google Scholar]
Quan D, Chen K, Zhang J, Guan Y, Yang D, Wu H, Wu S, Lv L (2020). Identification of lncRNA NEAT1/miR-21/RRM2 axis as a novel biomarker in breast cancer. Journal of Cellular Physiology 235: 3372–3381. [Google Scholar]
Rahim A, Afzal M, Naveed AK (2019). Genetic polymorphism of miRNA-196a and its target gene annexin-A1 expression based on ethnicity in Pakistani female breast cancer patients. Pakistan Journal of Medical Sciences 35: 1598. [Google Scholar]
Rastogi YR, Saini AK, Thakur VK, Saini RV (2020). New insights into molecular links between microbiota and gastrointestinal cancers: A literature review. International Journal of Molecular Sciences 9: 3212. [Google Scholar]
Rao X, Di Leva G, Li M, Fang F, Devlin C et al. (2011). MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene 30: 1082–1097. [Google Scholar]
Reddy SD, Ohshiro K, Rayala SK, Kumar R (2008). MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Research 68: 8195–8200. [Google Scholar]
Ren YQ, Wang HJ, Zhang YQ, Liu YB (2017). WBP2 modulates G1/S transition in ER+ breast cancer cells and is a direct target of miR-206. Cancer Chemotherapy and Pharmacology 79: 1003–1011. [Google Scholar]
Rui X, Zhao H, Xiao X, Wang L, Mo L et al. (2018). MicroRNA-34a suppresses breast cancer cell proliferation and invasion by targeting Notch1. Experimental and Therapeutic Medicine 16: 4387–4392. [Google Scholar]
Saito M, Okamoto A, Kohno T, Takakura S, Shinozaki H et al. (2000). Allelic imbalance and mutations of the PTEN gene in ovarian cancer. International Journal of Cancer 85: 160–165. [Google Scholar]
Samaeekia R, Adorno-Cruz V, Bockhorn J, Chang YF, Huang S et al. (2017). miR-206 inhibits stemness and metastasis of breast cancer by targeting MKL1/IL11 pathway. Clinical Cancer Research 23: 1091–1103. [Google Scholar]
Santolla MF, Lappano R, Cirillo F, Rigiracciolo DC, Sebastiani A et al. (2018). miR-221 stimulates breast cancer cells and cancer-associated fibroblasts (CAFs) through selective interference with the A20/c-Rel/CTGF signaling. Journal of Experimental & Clinical Cancer Research 37: 1–2. [Google Scholar]
Savad S, Mehdipour P, Miryounesi M, Shirkoohi R, Fereidooni F, Mansouri F, Modarressi MH (2012). Expression analysis of MiR-21, MiR-205, and MiR-342 in breast cancer in Iran. Asian Pacific Journal of Cancer Prevention 13: 873–877. [Google Scholar]
Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC (2007). Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. Journal of Biological Chemistry 282: 1479–1486. [Google Scholar]
Selcuklu SD, Donoghue MT, Spillane C (2009). miR-21 as a key regulator of oncogenic processes. Biochemical Society Transactions 37: 918–925. [Google Scholar]
Shao X, Tandon R, Samara G, Kanki H, Yano H et al. (1998). Mutational analysis of the PTEN gene in head and neck squamous cell carcinoma. International Journal of Cancer 77: 684–688. [Google Scholar]
Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O’leary JJ et al. (2010). Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nature Immunology 11: 141–147. [Google Scholar]
Shen L, Wan Z, Ma Y, Wu L, Liu F et al. (2015). The clinical utility of microRNA-21 as novel biomarker for diagnosing human cancers. Tumor Biology 36: 1993–2005. [Google Scholar]
Shen M, Dong C, Ruan X, Yan W, Cao M et al. (2019). Chemotherapy-induced extracellular vesicle miRNAs promote breast cancer stemness by targeting ONECUT2. Cancer Research 79: 3608–3621. [Google Scholar]
Sheng P, Fields C, Aadland K, Wei T, Kolaczkowski O et al. (2018). Dicer cleaves 5’-extended microRNA precursors originating from RNA polymerase II transcription start sites. Nucleic Acids Research 46: 5737–5752. [Google Scholar]
Shi L, Chen J, Yang J, Pan T, Zhang S, Wang Z (2010). MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Research 1352: 255–264. [Google Scholar]
Shibahara Y, Miki Y, Onodera Y, Hata S, Chan MS et al. (2012). Aromatase inhibitor treatment of breast cancer cells increases the expression of let-7f, a microRNA targeting CYP19A1. Journal of Pathology 227: 357–366. [Google Scholar]
Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P et al. (2009). Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138: 592–603. [Google Scholar]
Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY (2007). miR-21-mediated tumor growth. Oncogene 26: 2799–2803. [Google Scholar]
Singh P, Wedeken L, Waters LC, Carr MD, Klempnauer KH (2011). Pdcd4 directly binds the coding region of c-myb mRNA and suppresses its translation. Oncogene 30: 4864–4873. [Google Scholar]
Singh R, Mo YY (2013). Role of microRNAs in breast cancer. Cancer Biology & Therapy 14: 201–212. [Google Scholar]
Song X, Liang Y, Sang Y, Li Y, Zhang H et al. (2020). CircHMCU promotes proliferation and metastasis of breast cancer by sponging the let-7 family. Molecular Therapy–Nucleic Acids 20: 518–533. [Google Scholar]
Sørlie T, Tibshirani R, Parker J, Hastie T, Marron JS et al. (2003). Repeated observation of breast tumor subtypes in independent gene expression data sets. Proceedings of The National Academy of Sciences of The United States of America 100: 8418–8423. [Google Scholar]
Steiner E, Klubert D, Knutson D (2008). Assessing breast cancer risk in women. American Family Physician 78: 1361–1366. [Google Scholar]
Stoddard II FR, Brooks AD, Eskin BA, Johannes GJ (2008). Iodine alters gene expression in the MCF7 breast cancer cell line: evidence for an anti-estrogen effect of iodine. International Journal of Medical Sciences 5: 189. [Google Scholar]
Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X (2008). Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. Federation of European Biochemical Societies Letters 582: 1564–1568. [Google Scholar]
Sun X, Wang H, Jian Y, Lan F, Zhang L et al. (2018). Ultrasensitive microfluidic paper-based electrochemical/visual biosensor based on spherical-like cerium dioxide catalyst for miR-21 detection. Biosensors and Bioelectronics 105: 218–225. [Google Scholar]
Tang CP, Zhou HJ, Qin J, Luo Y, Zhang T (2017). MicroRNA-520c-3p negatively regulates EMT by targeting IL-8 to suppress the invasion and migration of breast cancer. Oncology Reports 38: 3144–3152. [Google Scholar]
Tang J, Ahmad A, Sarkar FH (2012). The role of microRNAs in breast cancer migration, invasion and metastasis. International Journal of Molecular Sciences 13: 13414–13437. [Google Scholar]
Tang X, Tu G, Yang G, Wang X, Kang L et al. (2019). Autocrine TGF-β1/miR-200s/miR-221/DNMT3B regulatory loop maintains CAF status to fuel breast cancer cell proliferation. Cancer Letters 452: 79–89. [Google Scholar]
Taplin S, Abraham L, Barlow WE, Fenton JJ, Berns EA et al. (2008). Mammography facility characteristics associated with interpretive accuracy of screening mammography. Journal of the National Cancer Institute 100: 876–887. [Google Scholar]
Tvingsholm SA, Hansen MB, Clemmensen KK, Brix DM, Rafn B et al. (2018). Let-7 microRNA controls invasion-promoting lysosomal changes via the oncogenic transcription factor myeloid zinc finger-1. Oncogenesis 7: 1–5. [Google Scholar]
Uhlmann S, Zhang JD, Schwäger A, Mannsperger H, Riazalhosseini Y et al. (2010). miR-200bc/429 cluster targets PLCγ1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene 29: 4297–4306. [Google Scholar]
Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szász AM et al. (2009). RETRACTED: A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 137: 1032–1046. [Google Scholar]
van Schooneveld E, Wildiers H, Vergote I, Vermeulen PB, Dirix LY et al. (2015). Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Research 17: 1–5. [Google Scholar]
Várallyay E, Burgyán J, Havelda Z (2008). MicroRNA detection by northern blotting using locked nucleic acid probes. Nature Protocols 3: 190. [Google Scholar]
Wahid F, Shehzad A, Khan T, Kim YY (2010). MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochimica et Biophysica Acta (BBA)–Molecular Cell Research 1803: 1231–1243. [Google Scholar]
Wang H, He D, Wan K, Sheng X, Cheng H et al. (2020). In situ multiplex detection of serum exosomal microRNAs using an all-in-one biosensor for breast cancer diagnosis. Analyst 145: 3289–3296. [Google Scholar]
Wang H, Tan Z, Hu H, Liu H, Wu T et al. (2019). microRNA-21 promotes breast cancer proliferation and metastasis by targeting LZTFL1. BMC Cancer 19: 1–3. [Google Scholar]
Wang J, Guan J, Lu Z, Jin J, Cai Y, Wang C, Wang F (2015). Clinical and tumor significance of tropomyosin-1 expression levels in renal cell carcinoma. Oncology Reports 33: 1326–1334. [Google Scholar]
Wang J, Tang C, Yang C, Zheng Q, Hou Y (2019). Tropomyosin-1 functions as a tumor suppressor with respect to cell proliferation, angiogenesis and metastasis in renal cell carcinoma. Journal of Cancer 10: 2220. [Google Scholar]
Wang L, Guo J, Wang Q, Zhou J, Xu C et al. (2014). LZTFL1 suppresses gastric cancer cell migration and invasion through regulating nuclear translocation of β-catenin. Journal of Cancer Research and Clinical Oncology 140: 1997–2008. [Google Scholar]
Wang L, Kang FB, Wang J, Yang C, He DW (2019). Downregulation of miR-205 contributes to epithelial-mesenchymal transition and invasion in triple-negative breast cancer by targeting HMGB1-RAGE signaling pathway. Anti-Cancer Drugs 30: 225. [Google Scholar]
Wang JL, Sun SJ, Zhang J, Wang GN (2017). Prognostic value of circulating microRNA-21 for breast cancer: A systematic review and meta-analysis. Artificial Cells, Nanomedicine, and Biotechnology 45: 1216–1221. [Google Scholar]
Wang X, Qiu H, Tang R, Song H, Pan H et al. (2018). miR-30a inhibits epithelial-mesenchymal transition and metastasis in triple-negative breast cancer by targeting ROR1. Oncology Reports 39: 2635–2643. [Google Scholar]
Wang Y, Zhang X, Chao Z, Kung HF, Lin MC et al. (2017). MiR-34a modulates ErbB2 in breast cancer. Cell Biology International 41: 93–101. [Google Scholar]
Wei N, Liu SS, Chan KK, Ngan HY (2012). Tumour suppressive function and modulation of programmed cell death 4 (PDCD4) in ovarian cancer. PLoS One 7: e30311. [Google Scholar]
Wei Q, Chen Y, Gu YF, Zhao W (2019). Molecular characterization and functional analysis of leucine zipper transcription factor like 1 in zebrafish (Danio rerio). Frontiers in Physiology 10: 801. [Google Scholar]
Wei Q, Chen ZH, Wang L, Zhang T, Duan L et al. (2016). LZTFL1 suppresses lung tumorigenesis by maintaining differentiation of lung epithelial cells. Oncogene 35: 2655–2663. [Google Scholar]
Wei Q, Zhou W, Wang W, Gao B, Wang L, Cao J, Liu ZP (2010). Tumor-suppressive functions of leucine zipper transcription factor-like 1. Cancer Research 70: 2942–2950. [Google Scholar]
Welch C, Chen Y, Stallings RL (2007). MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene 26: 5017–5022. [Google Scholar]
Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y et al. (2009). Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Research 37: 2584–2595. [Google Scholar]
Wilczynska A, Bushell Martin (2015). The complexity of miRNA-mediated repression. Cell Death & Differentiation 22: 22–33. [Google Scholar]
Wu L, Fan J, Belasco JG (2006). MicroRNAs direct rapid deadenylation of mRNA. Proceedings of the National Academy of Sciences of the United States of America 103: 4034–4039. [Google Scholar]
Xiang Y, Liao XH, Yao A, Qin H, Fan LJ et al. (2017). MRTF-A-miR-206-WDR1 form feedback loop to regulate breast cancer cell migration. Experimental Cell Research 359: 394–404. [Google Scholar]
Xiao B, Shi X, Bai J (2019). miR-30a regulates the proliferation and invasion of breast cancer cells by targeting Snail. Oncology Letters 17: 406–413. [Google Scholar]
Yager JD, Davidson NE (2006). Estrogen carcinogenesis in breast cancer. New England Journal of Medicine 354: 270–282. [Google Scholar]
Yan GR, Xu SH, Tan ZL, Liu L, He QY (2011). Global identification of miR-373-regulated genes in breast cancer by quantitative proteomics. Proteomics 11: 912–920. [Google Scholar]
Yan L, Yu MC, Gao GL, Liang HW, Zhou XY et al. (2018). MiR-125a-5p functions as a tumour suppressor in breast cancer by downregulating BAP1. Journal of Cellular Biochemistry 119: 8773–8783. [Google Scholar]
Yan LX, Huang XF, Shao Q, Huang MY, Deng L et al. (2008). MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 14: 2348–2360. [Google Scholar]
Yang HS, Cho MH, Zakowicz H, Hegamyer G, Sonenberg N, Colburn NH (2004). A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Molecular and Cellular Biology 24: 3894–3906. [Google Scholar]
Yang LW, Wu XJ, Liang Y, Ye GQ, Che Y et al. (2020). miR-155 increases stemness and decitabine resistance in triple-negative breast cancer cells by inhibiting TSPAN5. Molecular Carcinogenesis 59: 447–461. [Google Scholar]
Yang W, Wang X, Zheng W, Li K, Liu H, Sun Y (2013). Genetic and epigenetic alterations are involved in the regulation of TPM1 in cholangiocarcinoma. International Journal of Oncology 42: 690–698. [Google Scholar]
Ye J, Xu M, Tian X, Cai S, Zeng S (2019). Research advances in the detection of miRNA. Journal of Pharmaceutical Analysis 9: 217–226. [Google Scholar]
Yu F, Yao H, Zhu P, Zhang X, Pan Q et al. (2007). let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131: 1109–1123. [Google Scholar]
Yu RM, Cheah YK (2017). The roles of miRNAs in human breast cancer and canine mammary tumor. Applied Cancer Research 37: 37. [Google Scholar]
Yu Z, Wang C, Wang M, Li Z, Casimiro MC et al. (2008). A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. Journal of Cell Biology 182: 509–517. [Google Scholar]
Zhang HD, Jiang LH, Sun DW, Li J, Tang JH (2017). miR-30a inhibits the biological function of breast cancer cells by targeting Notch1. International Journal of Molecular Medicine 40: 1235–1242. [Google Scholar]
Zhang M (2002). The role of maspin in tumor progression and normal development, pp. 96–114. Maspin. Landes Bioscience. [Google Scholar]
Zhang W, Chen CJ, Guo GL (2018). MiR-155 promotes the proliferation and migration of breast cancer cells via targeting SOCS1 and MMP16. European Review for Medical and Pharmacological Sciences 22: 7323–7332. [Google Scholar]
Zhao J, Tao Y, Zhou Y, Qin N, Chen C et al. (2015). MicroRNA-7: A promising new target in cancer therapy. Cancer Cell International 15: 1–8. [Google Scholar]
Zhao ZL, Cai Y, Wang YY, Xia CL, Li CX et al. (2015). Effects of miRNA-21 on paclitaxel-resistance in human breast cancer cells. Zhejiang Da Xue Xue Bao. Yi Xue Ban 44: 400–409. [Google Scholar]
Zhou XL, Wang M (2015). Expression levels of survivin, Bcl-2, and KAI1 proteins in cervical cancer and their correlation with metastasis. Genetics and Molecular Research 14: 17059–17067. [Google Scholar]
Zhou Y, Wang M, Tong Y, Liu X, Zhang L et al. (2019). miR-206 promotes cancer progression by targeting full-length neurokinin-1 receptor in breast cancer. Technology in Cancer Research & Treatment 18: 1533033819875168. [Google Scholar]
Zhu S, Si ML, Wu H, Mo YY (2007). MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). Journal of Biological Chemistry 282: 14328–14336. [Google Scholar]
Zhu S, Wu H, Wu F, Nie D, Sheng S et al. (2008). MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Research 18: 350–359. [Google Scholar]
Zong Y, Zhang Y, Sun X, Xu T, Cheng X, Qin Y (2019). miR-221/222 promote tumor growth and suppress apoptosis by targeting lncRNA GAS5 in breast cancer. Bioscience Reports 39: BSR20181859. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |