DOI:10.32604/biocell.2022.018271

| BIOCELL DOI:10.32604/biocell.2022.018271 |  |
| Article |
Potential genomic biomarkers of obesity and its comorbidities for phthalates and bisphenol A mixture: In silico toxicogenomic approach
1Department of Toxicology “Akademik Danilo Soldatović”, University of Belgrade—Faculty of Pharmacy, Belgrade, 11000, Serbia
2Department of Toxicogenomics, GROW School for Oncology and Developmental Biology, Maastricht University, Maastricht, 6221, The Netherlands
*Address correspondence to: Katarina Baralić, katarinab@pharmacy.bg.ac.rs
Received: 12 July 2021; Accepted: 01 September 2021
Abstract: This in silico toxicogenomic study aims to explore the relationship between phthalates and bisphenol A (BPA) co-exposure and obesity, as well as its comorbid conditions, in order to construct a possible set of genomic biomarkers. The Comparative Toxicogenomics Database (CTD; http://ctd.mdibl.org) was used as the main data mining tool, along with GeneMania (https://genemania.org), ToppGene Suite (https://toppgene.cchmc.org) and DisGeNET (http://www.disgenet.org). Among the phthalates, bis(2-ethylhexyl) phthalate (DEHP) and dibutyl phthalate (DBP) were chosen as the most frequently curated phthalates in CTD, which also share similar mechanisms of toxicity. DEHP, DBP and BPA interacted with 84, 90 and 194 obesity-related genes/proteins, involved in 67, 65 and 116 pathways, respectively. Among these, 53 genes/proteins and 42 pathways were common to all three substances. 31 genes/proteins had matching interactions for all three investigated substances, while more than half of these genes/proteins (56.49%) were in co-expression. 7 of the common genes/proteins (6 relevant to humans: CCL2, IL6, LPL, PPARG, SERPINE1, and TNF) were identified in all the investigated obesity comorbidities, while PPARG and LPL were most closely linked to obesity. These genes/proteins could serve as a target for further in vitro and in vivo studies of molecular mechanisms of DEHP, DBP and BPA mixture obesogenic properties. Analysis reported here should be applicable to any mixture of environmental chemicals and any disease present in CTD.
Keywords: Endocrine disruptors; Data mining; Bioinformatics; Toxicology
It has been acknowledged that people are not exposed to a single, but to a great number of chemicals constantly. This exposure usually occurs at low doses, by various routes, from a variety of sources (Kortenkamp, 2008). Many toxicology studies have demonstrated the usefulness of the dose-addition concept in exploring the combined effects of chemicals. Joint effects occur even when all mixture components are present at levels below doses that cause observable effects (Hayes et al., 2019; Kortenkamp, 2007). Considering that most mixtures contain multiple components, quantifying these interactions in terms of risk assessment is not an easy task. It not only includes mining published data and characterizing the mixture in the laboratory, but also in silico hazard analysis and modeling (Hayes et al., 2019).
Phthalates and bisphenol A (BPA) have been used as additives in plastic and are present in many consumer products (Berghuis et al., 2015; Kim and Park, 2014; Rochester, 2013). These substances are not chemically bound to plastic, which enables them to migrate or evaporate into the surrounding environment (Heudorf et al., 2007; Singh and Li, 2012; Kabir et al., 2015). Human exposure to phthalates mainly occurs through foods, because of their uses in wrapping materials and food processing. Similarly, BPA can migrate from polycarbonate plastic, such as epoxy resins that line metal cans for food and beverages (Berghuis et al., 2015; Singh and Li, 2013). People are additionally exposed to phthalates and BPA via ingestion of drinking water, inhalation of contaminated air, as well as dermal absorption (Zarean et al., 2017). Having in mind that phthalates and BPA coexist in natural environments, their combined effects have been investigated in different studies on experimental animals. An acute toxicity study on mice explored the influence of a single dose of DEHP injected subcutaneously on deposition of BPA received as a food supplement. The results of this study indicated that DEHP might increase the deposition of BPA in uterus, ovaries and serum of female and serum and epididymis of male mice compared to the control (Borman et al., 2017). Results of a subchronic toxicity study on rats have demonstrated that, under combined DBP and BPA treatment, the expression levels of the androgen receptor (AR), gonadotropin-releasing hormone receptor (GNRHR), and progesterone hormone receptor (PR) genes were higher compared to the control group, suggesting an additive or a synergistic effect (Zhang et al., 2013). Molecular mechanisms of phthalates and BPA long-lasting effects continue to be investigated and likely involve disruption of epigenetic programming of gene expression during development (Singh and Li, 2012). These chemicals are known to alter cell signaling and metabolic pathways involved in lipid homeostasis. This can result in development of metabolic disorders considered a major global public health problem nowadays, including obesity and its comorbidities (Hatch et al., 2010; Muscogiuri et al., 2017; Stojanoska et al., 2017). Exposure to phthalates and BPA may lead to adipogenesis, affecting serum levels of metabolic hormones, steroid hormone receptors and nuclear receptor signaling pathways in preadipocytes (Muscogiuri et al., 2017; Zarean et al., 2017). Having in mind that toxicogenomics merges measuring families of cellular molecules with bioinformatics and conventional toxicology to investigate the interactions between genes and environmental stress in disease causation (Boverhof and Zacharewski, 2006; Tung et al., 2020; Waters and Fostel, 2004), it can be useful for predicting gene functions in specific molecular pathways and finding genomic biomarkers for further in vitro and in vivo studies (van Breda et al., 2014; Dong et al., 2018). Additionally, it delivers strategies for mixture assessment, considering that all possible chemical, gene, protein, metabolite, and network interactions that may be important in eliciting mixture-specific toxicities can be studied (Boverhof and Zacharewski, 2006).
The aim of the current research was to: (i) explore the relationship between phthalates and BPA mixture and obesity, as well as its comorbidities, by using the toxicogenomics data mining approach; (ii) predict the possible set of genomic biomarkers; and (iii) demonstrate how in silico data mining could be used as preliminary investigation for gene prioritization and setting up a methodology that would be both time and economically viable for further in vitro and in vivo toxicity testing.
The Comparative Toxicogenomics Database (CTD; http://CTD.mdibl.org) was selected as the main data mining tool. Inferences in CTD are mostly based on the information from animal studies. However, since CTD focuses on environmental chemicals and outcomes relevant to human health, genes/proteins of interest are included in the database only if they are also present in humans (Meng et al., 2013). Curators insert the chemical-gene interactions and disease relationships into the CTD following the manual provided by a lead curator. All the captured data include: date of curation, ID of the curator, PubMed ID, interaction, species, chemical, gene/protein, associated diseases and author contact information (Wiegers et al., 2009). Prior to public release on the CTD website, all the curated data are loaded into a database for quality control review. Additionally, quality control in the CTD is feasible because each curated interaction is captured using controlled vocabularies/ontologies with aim to ensure consistency (Davis et al., 2009; Wiegers et al., 2009). Thus, CTD uses official gene symbols and names from the National Center for Biotechnology Information’s (NCBI) Entrez-Gene database, while CTD disease vocabulary uses terms from both MeSH (Medical Subject Headings) and OMIM (Online Mendelian Inheritance of Man) (Davis et al., 2008; Davis et al., 2019). For our analysis, we used: MyVenn CTD tool (http://ctdbase.org/tools/myVenn.go), which is used to explore the relationships among the lists of CTD chemicals, diseases, genes, GO terms or pathways, or any other data; VennViewer CTD tool (http://ctdbase.org/tools/vennViewer.go), which compares associated data sets for up to three chemicals, diseases, or genes, and Set Analyzer CTD tool (http://ctdbase.org/tools/analyzer.go), which performs the analyses such as set-based enrichment for collections of chemicals or genes, and pathway generation for collections of genes.
All curated interactions of phthalates/BPA genes/proteins, phthalates/BPA-obesity/comorbidities in CTD are publicly available, while data are uploaded to the database monthly (Davis et al., 2009; Davis et al., 2011).
The flow chart for the different steps of our analyses is shown in Fig. 1. Data mining the CTD generated a list of phthalates and BPA interacting genes/proteins. These genes/proteins were further analyzed for pathway annotations, gene ontology and protein/disease relationships. SetAnalyser tool was used to retrieve the obesity/gene interactions for phthalates and BPA-induced genes/proteins and molecular pathways these genes/proteins were involved in. Default CTD settings were applied (corrected p-value for this analysis was 0.01). MyVenn and VenViewer tools were used to find the subset of genes/proteins and enriched pathways common to all three substances.
Network of genes/proteins related to the obesity development affected by phthalates and BPA was explored by the GeneMANIA prediction server (https://genemania.org). Among the limited gene network databases, GeneMANIA is one of the most reliable network analyzers. This prediction server is a flexible, user-friendly web interface used for generating hypotheses about gene functions, analyzing gene lists and prioritizing genes for functional assays (Franz et al., 2018).
Gene Ontology (GO) and molecular pathway analysis
ToppGene Suite’s Topp Fun tool (https://toppgene.cchmc.org/enrichment.jsp) generated the list of top 10 obesity-related biological processes, molecular functions and molecular pathways affected by the investigated substances. Default settings were used for the analysis (probability density function method, 0.05 as p-value cutoff value, FDR correction). ToppGene Suite is a one-stop portal for gene list enrichment analysis and candidate gene prioritization based on functional annotations and protein interactions network. Its ToppFun tool detects functional enrichment of the gene list based on Transcriptome, Proteome, Regulome (TFBS and miRNA), Ontologies (GO, Pathway), Phenotype (human disease and mouse phenotype), Pharmacome (Drug-Gene associations), literature co-citation, and other features (Chen et al., 2009).
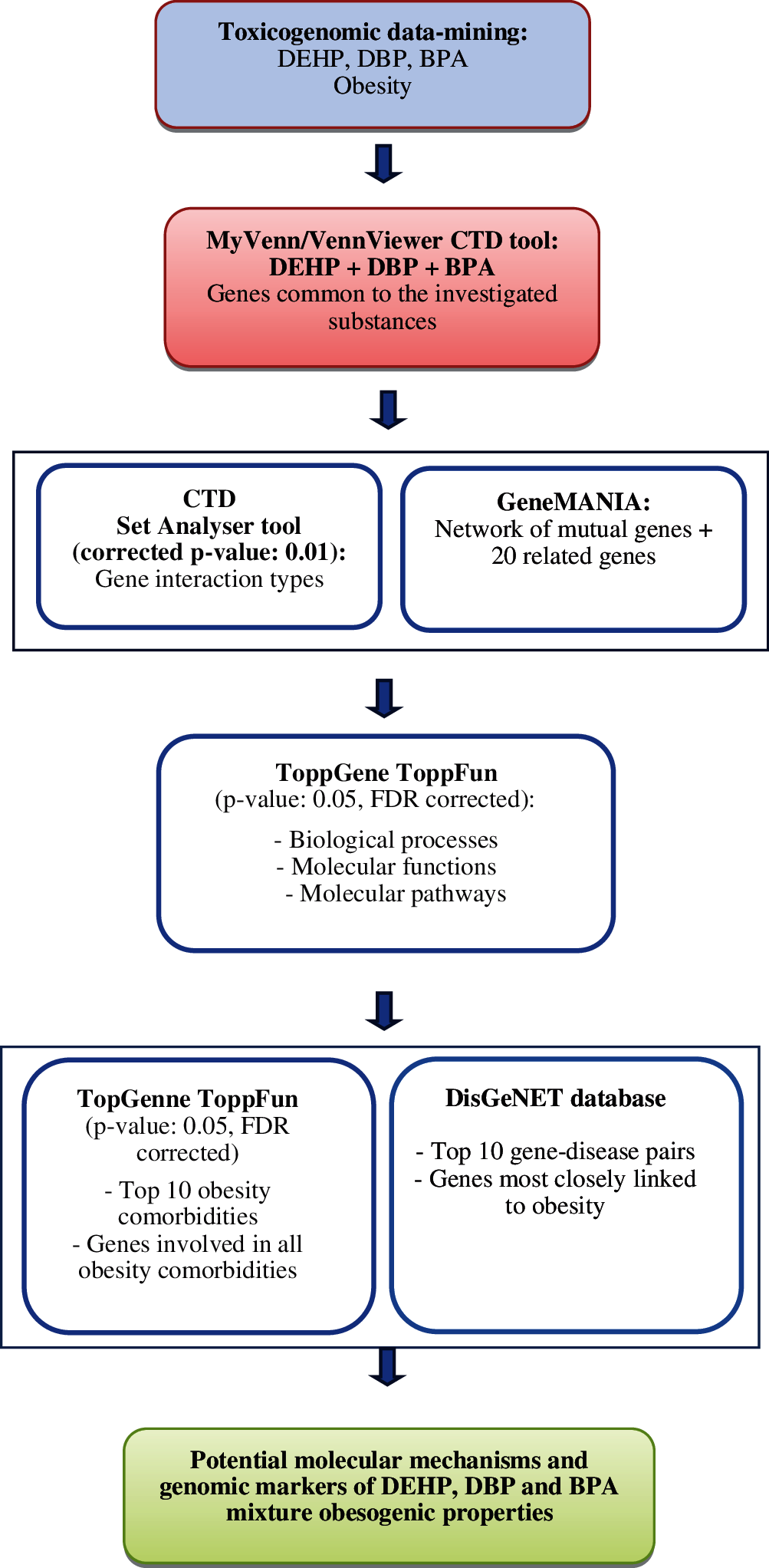
Figure 1: Flow chart explaining the steps of our analysis.
ToppGene Suite’s Topp Fun function was applied to obtain the list of obesity comorbidities in which genes/proteins common to all three substances were involved (default settings). DisGeNET database (http://www.disgenet.org) was used to find the top gene-disease pairs out for the selected genes/proteins present in all the obesity comorbidities obtained from the ToppGene Suite. DisGeNET is one of the largest available collections of genes involved in human diseases. DisGeNET integrates data from expert curated repositories, GWAS catalogues, animal models and the scientific literature. This platform can be helpful for investigating molecular underpinnings of specific human diseases and their comorbidities, analysing the properties of disease genes, generating hypothesis on drug therapeutic action and drug adverse effects, validating computationally predicted disease genes and evaluating text-mining methods performance (Piñero et al., 2016).
Gene interactions—phthalates and BPA
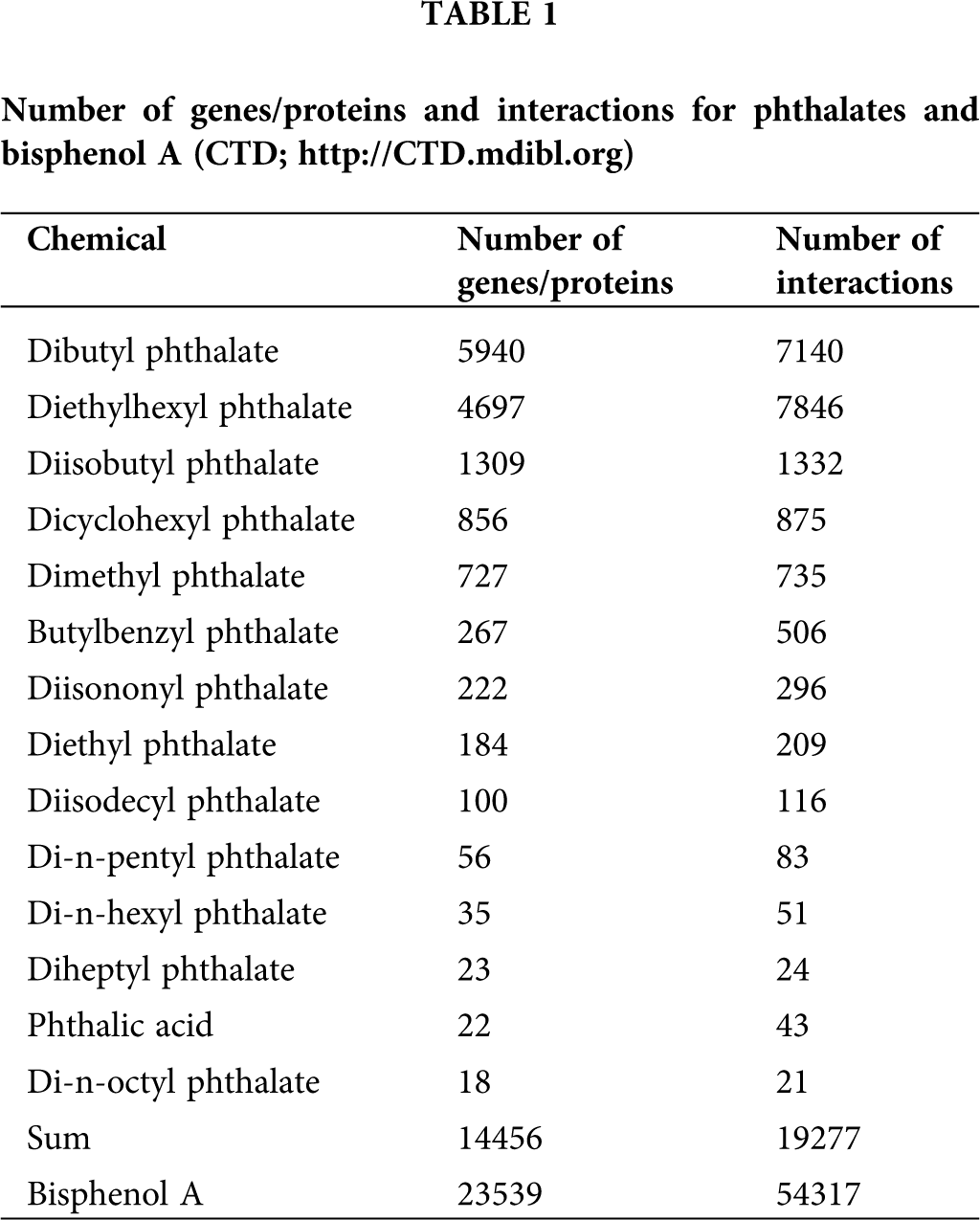
The first step of our analysis included identifying the genes/proteins and gene interactions associated with BPA and 14 phthalates present in the CTD (Tab. 1). The information presented in Tab. 1 was obtained from the Gene tabs on the CTD website for each of the investigated substances across all the species included in the CTD database, since all the genes/proteins are listed in the CTD only if they are also present in human genome (Meng et al., 2013). The obtained data indicated that two phthalates, bis(2-ethylhexyl) phthalate (DEHP) and dibutyl phthalate (DBP), interacted with 4697 and 5940 genes, exhibiting 7846 and 7140 interactions, respectively. Sum of their 14986 interactions accounted for 77.74% of all 19277 phthalate interactions, while sum of the genes/proteins they interacted with was 73.58% of all 14456 genes/proteins phthalates interacted with according to CTD. Furthermore, according to the CTD “Comps” data-tab, DBP was found to be the top comparable phthalate to DEHP, with 1384 common interacting genes/proteins. Thus, as the most frequently curated and comparable phthalates, DEHP (CTD_D004051) and DBP (CTD_D003993) were chosen for toxicogenomic analysis, together with BPA (CTD_C006780), which, with 23539 genes and 54317 interactions, can be considered one of the most curated chemical in the CTD. VenViewer CTD tool revealed that there were 1361 common genes/proteins for DEHP, DBP and BPA, suggesting a possibility that these three substances exhibit similar toxicogenomics and adverse effects on human health. A curated data set for the three investigated substances was further analyzed to illustrate the scope of CTD and highlight its potential applications for toxicity assessment of mixtures.
Genes/proteins connected to the development of obesity
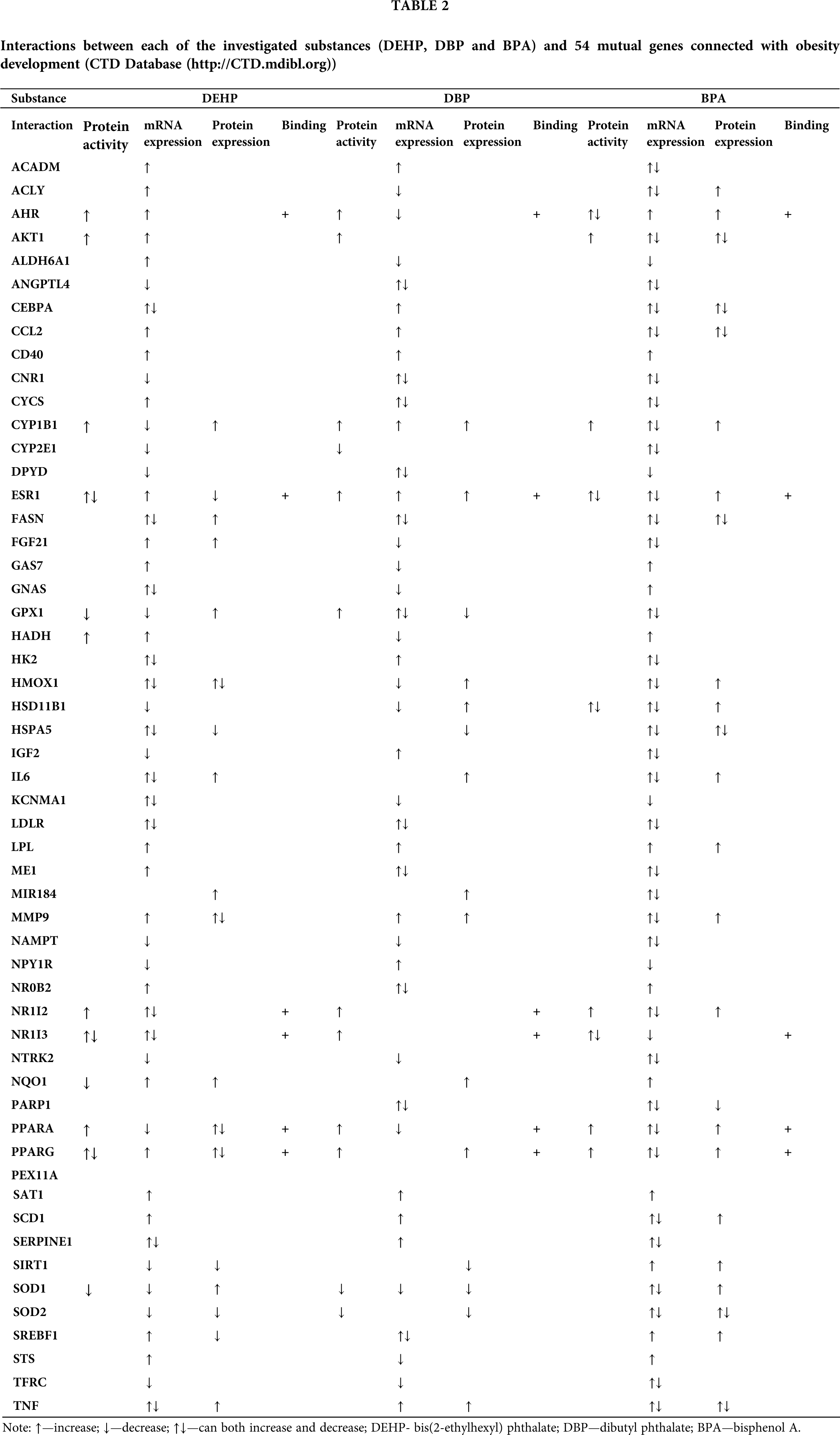
The “Diseases” data-tab on CTD’s webpage lists human pathologies connected to the investigated chemicals and provides candidate genes/proteins that may help explaining the mechanisms underlying the relationships between human pathologies and chemicals. A further analysis of our gene/protein sets was aimed at identifying genes/proteins involved in the development of obesity. According to the CTD, DEHP, DBP and BPA were found to interact with 89, 91 and 186 obesity related-genes/proteins, respectively. Among these, 54 were common to all three investigated substances. The exact manner of the interactions between the investigated substances and the extracted set of 54 mutual genes, including protein activity, mRNA expression, protein expression and binding to the proteins, are presented in Tab. 2.
These 54 common genes/proteins were further analyzed by the CTD, revealing the 31 genes/proteins with matching interactions for all three substances (Tab. 3). This indicates that DEHP, DBP and BPA share a number of common molecular activities and suggests that they might have the capacity to act together in an additive manner.

Obesity—gene interaction network
CTD SetAnalyser tool provided a list of 97 interactions for the obtained 31 genes/proteins. In 72 of these interactions Homo Sapiens was marked as both source and target organism, suggesting the human relevance of these interactions. A Pathway View map of these 31 genes/proteins (Fig. 2) was retrieved using the CTD SetAnalyzer tool to display the gene-gene interactions derived from the BioGRID (database dedicated to the annotation and archival of protein, genetic and chemical interactions) (Chatr-Aryamontri et al., 2017; Davis et al., 2015). The default map was manually configured (merged edges and tree view layout). According to the CTD, all of these interactions were physical, apart from one (SOD1 with SOD2), which was genetic. To provide a more detailed view of molecular networks potentially affected by the investigated substances, GeneMANIA prediction server was used to analyze the interactions between the obtained set of genes/proteins. The tight interaction network between the 31 common genes/proteins was identified, together with 20 related genes/proteins. The total number of links between these 51 genes/proteins was 471 (Fig. 3). These data provide gene to gene (co-expression) and gene to protein (physical interactions) links, pathways the gene is a part of and co-localization of the gene with a high accuracy, revealing that target genes/proteins and their interacting proteins may have the same or similar functions (Osama et al., 2019). These results have demonstrated that 59.49% of our set of genes were in co-expression, meaning that their expression levels are similar across conditions in a gene expression study. Furthermore, 20.78% of the interactions were predicted by the server, 9.48% belonged to the same pathway, 6.29% were in physical interactions, 3.49% were with shared protein domains, 3.28% were in co-localization, while 0.19% were genetic interactions.
Obesity molecular pathways and biological functions
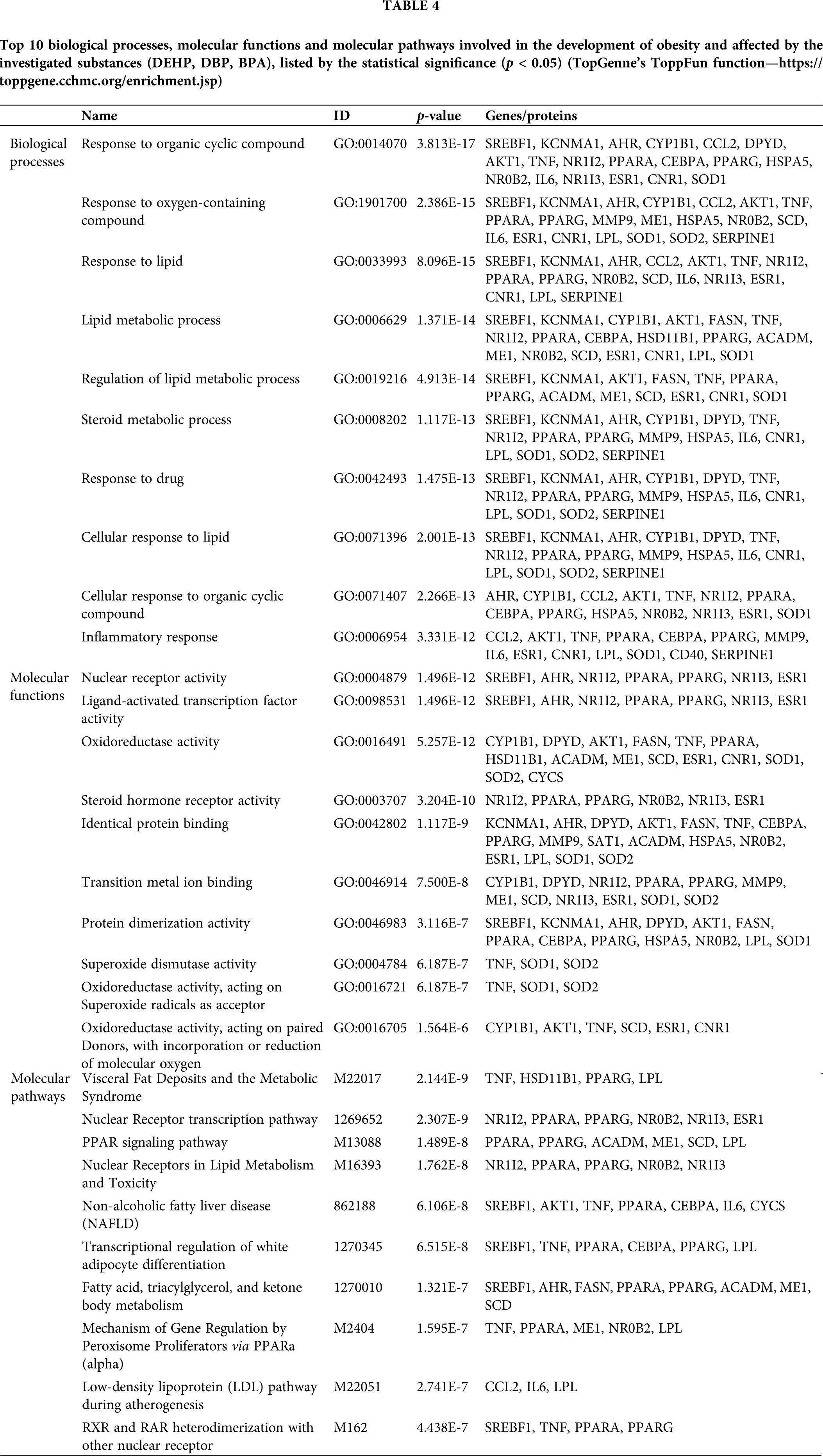
In order to elucidate the biological importance of our set of genes, enriched pathway and gene ontology (GO) analysis was performed. TopGenne’s ToppFun function was used to extract the top 10 biological processes, molecular functions and molecular pathways involved in the development of obesity and affected by the investigated substances (p < 0.05) (Tab. 4).
Investigated genes/proteins and obesity comorbidities
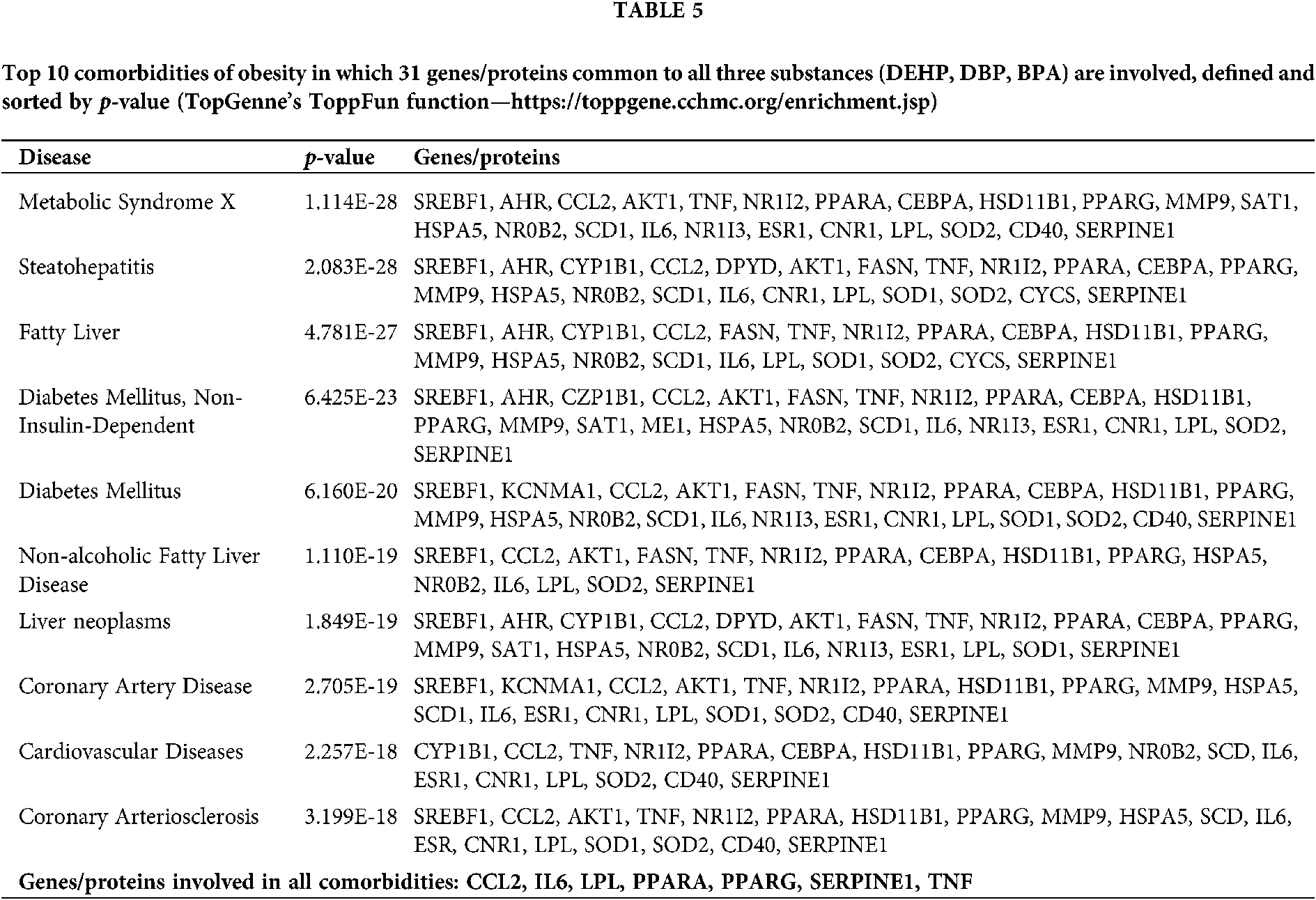
Our set of 31 genes/proteins was further analyzed using the ToppGene’s ToppFun function revealing the top 10 comorbidities of obesity in which these genes/proteins are involved (Tab. 5). Seven genes/proteins (CCL2, IL6, LPL, PPARA, PPARG, SERPINE1 and TNF) were found to be connected with all of the top 10 comorbidities ToppFun had listed.
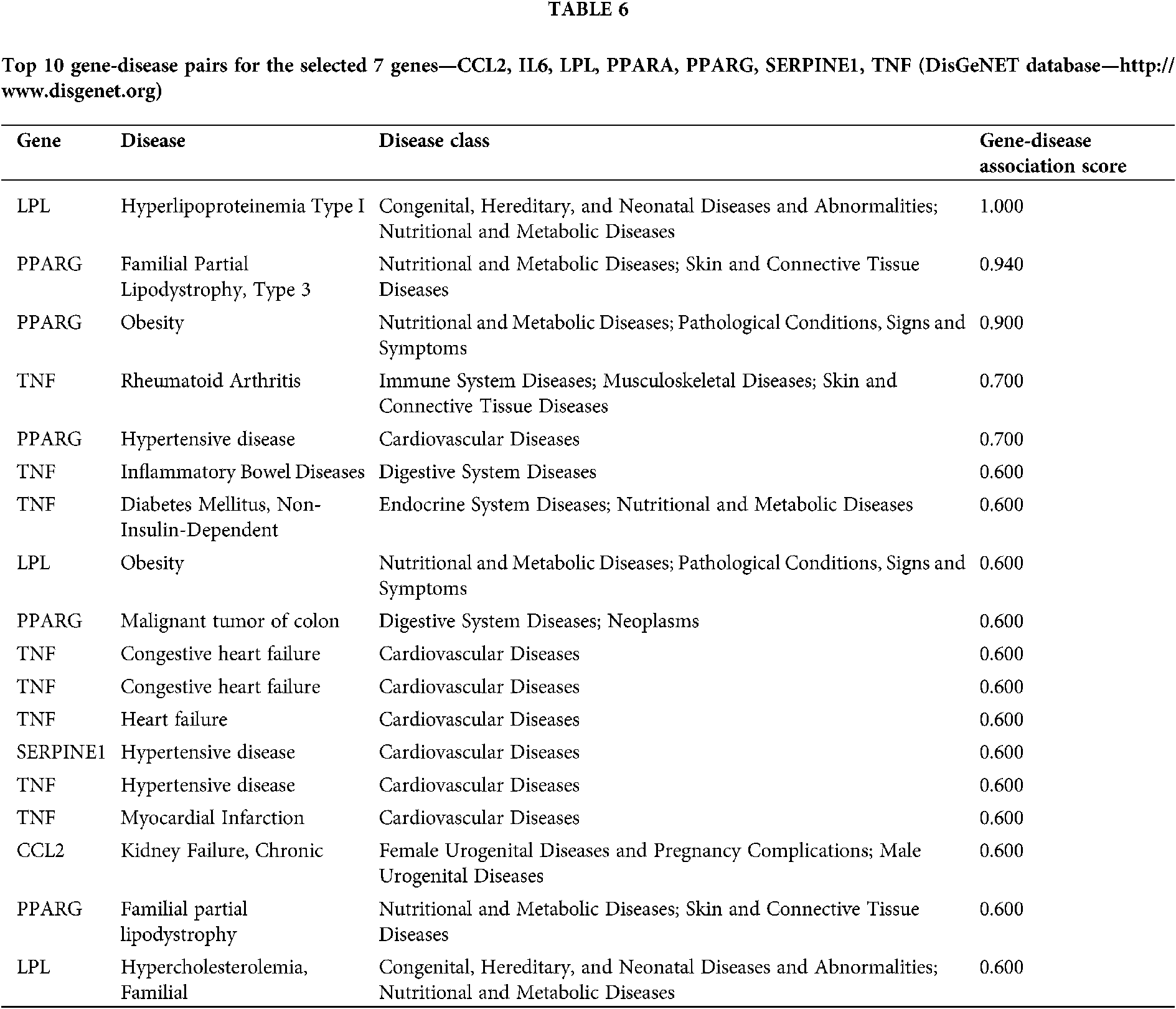
After this, DisGeNET database was used to find the top gene-disease pairs for the selected 7 genes/proteins. A gene-disease association score provided by this database can be used for integration and ranking of gene-disease associations. The DisGeNET score can range from 0 to 1, taking into account the number and type of sources (level of curation, organisms), as well as the number of publications supporting the association. The higher score represents higher association (Piñero et al., 2016). Among the filtered gene-disease pairs, the highest score was found between LPL and Hyperlipoproteinemia Type I (1.000) and PPARG and Familial Partial Lipodystrophy, Type 3 (0.940). For Obesity, PPARG and LPL were found to have the highest association score, 0.900 and 0.600, respectively (Tab. 6).
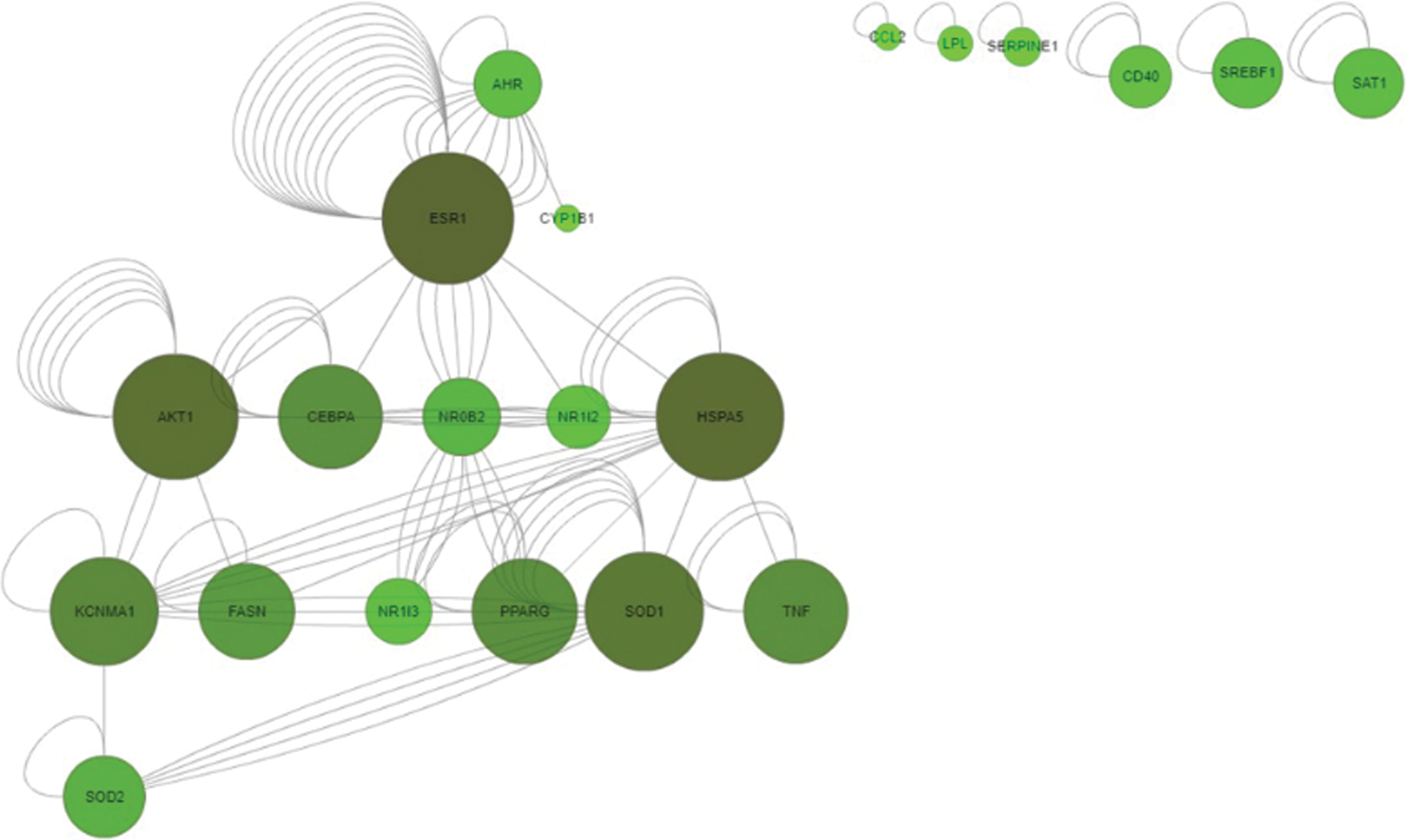
Figure 2: Common gene-gene interactions of 31 investigated genes/proteins obtained by SetAnalyser tool. CTD integrates gene-gene and protein-protein interactions from BioGRID (SetAnalyzer CTD tool—http://ctdbase.org/tools/analyzer.go).
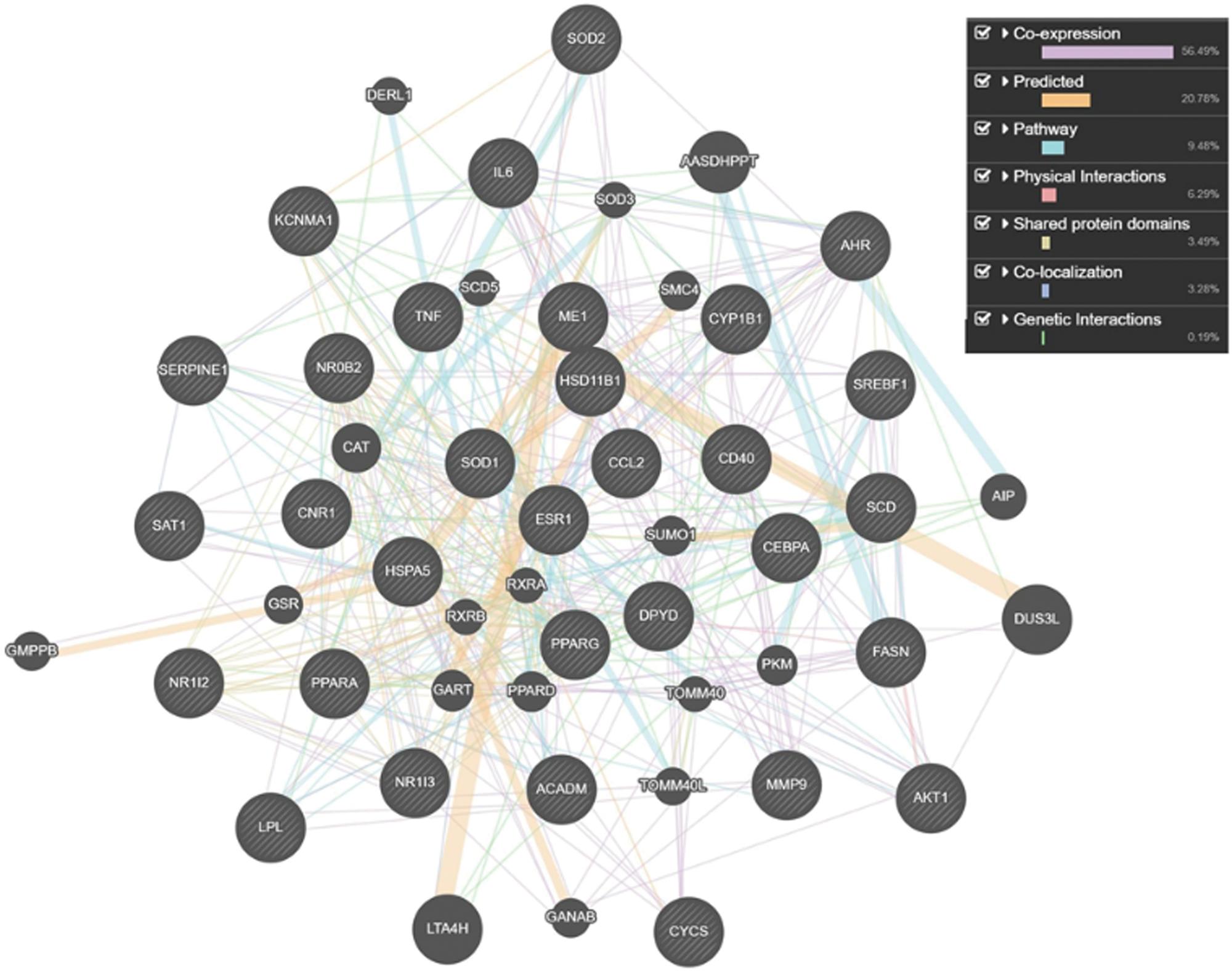
Figure 3: Tight network of 31 genes/proteins affected by the investigated substances (DEHP, DBP and BPA), along with 20 related genes/proteins, and connected to the development of obesity (GeneMania predictive server—https://genemania.org); Co-expression (56.49% of interactions)—purple lines: two genes are linked if their expression levels are similar across conditions in a gene expression study; Predicted by the server (20.78% of interactions)—orange lines: predicted functional relationships between genes, often protein interactions. A major source of predicted data is mapping known functional relationships from another organism via orthology. Two proteins are predicted to interact if their orthologs are known to interact in another organism. Pathway (9.48% of interactions)—light blue lines: two gene products are linked if they participate in the same reaction within a pathway; Physical interactions (6.29% of interactions)—pink lines: two gene products are linked if they were found to interact in a protein-protein interaction study. Shared protein domains (3.49% of interactions)—gray-yellow lines: protein domain data. Two gene products are linked if they have the same protein domain. Co-localization (3.28% of interactions)—blue lines: genes expressed in the same tissue, or proteins found in the same location. Genetic Interactions (0.19% of interactions)—green lines: two genes are functionally associated if the effects of perturbing one gene were found to be modified by perturbations to a second gene.
Linkage between DEHP, DBP and BPA mixture and obesity
In this investigation, in silico approach was applied to elucidate the linkage between the phthalates and BPA mixture and obesity, as well as its comorbidities, and highlight the potential genomic biomarkers. The present study further illustrates the usefulness of computational systems, easily applied as a tool, to help researchers in choosing the most promising biomarkers and exploring the mechanisms of toxicity which could further be investigated by high through-put toxicological screening tests. In this way, targeted research is more easily achieved, which is accompanied by complying with the 3R rule and animal welfare.
Numerous literature data have suggested that phthalates and BPA have significant effects on the obesity development, especially after the prenatal exposure at low doses. For example, Hao et al. (2012) have demonstrated in a mouse animal model that in utero exposure to a low dose (0.05 mg/kg of b.w.) of DEHP metabolite, mono(2-ethylhexyl) phthalate (MEHP), significantly increased body weight and fat pad weight in male offspring at postnatal day 60 and elevated serum cholesterol, glucose and triacylglyceride level (Hao et al., 2012). Likewise, Somm et al. (2009) have confirmed the body weight increase of both male and female offspring after the exposure of pregnant females to 1 mg/L BPA in water from the sixth day of gestation, until the end of lactation (postnatal day 21). Our previous in vivo 28-days subacute toxicity study in Wistar rats has demonstrated the ability of the investigated mixture (50 mg/kg b.w./day DEHP + 50 mg/kg b.w/day DBP + 25 mg/kg b.w./day BPA) to increase the serum glucose level and decrease serum testosterone level even after the shorter exposure time (Baralić et al., 2020a; Baralić et al., 2020b). Also, connection between the three investigated substances (DEHP, DBP and BPA) and obesity has been shown in human biomonitoring studies. In these studies, positive association was found between the body mass index and DEHP and DBP metabolites, as well as BPA (Carwile and Michels, 2011; Harley et al., 2017; Meeker and Ferguson, 2011), along with insulin resistance (Stahlhut et al., 2007). Nonetheless, the possibility that the mixture of the two phthalates and BPA might play a role in the obesity development is still not studied enough. Moreover, it is necessary to specify the particular molecular pathways and explore the mechanisms by which these substances could express their obesogenic properties.
In silico toxicogenomic data mining
After the comparison of our data sets for the three selected substances (DEHP, DBP and BPA), it was concluded that 31 obesity-related genes/proteins had matching interactions for all of them. More than half of these 31 genes/proteins (56.49%) were in co-expression, which indicates a similarity in their expression levels. Any of the aforementioned genes/proteins could potentially be involved in the mechanism of obesity development linked to the investigated mixture and, therefore, could be used in further in vitro and in vivo studies as genomic biomarkers.
In their data mining studies, Singh and Li (2012) have demonstrated that phthalates and BPA exhibit similar toxicogenomic, epigenetic and adverse effects on human health, including obesity. They concluded that five of the top ten toxicity networks of phthalates and BPA were involved in inflammation, while the top four phthalate molecular pathways were included in the regulation of lipid metabolism and PPAR pathway (Singh and Li, 2013, 2012, 2011). They also marked 89 common interacting genes/proteins for DEHP, DBP and BPA (Singh and Li, 2012). However, as these data were collected in 2012, the number of interacting genes for these three substances has significantly increased. Similar observation can be made for the results of Dong et al. (2018), who performed CTD analyses of BPA and DBP as an additional investigation to the in vivo experiment on zebrafish model. In their CTD investigation, they found 4826 and 14737 interactions with different genes/proteins for DBP and BPA, respectively, which is, compared to the number of interactions recorded in our study (7140 for DBP and 54317 for BPA), considerably less.
Molecular pathways and biological processes
Our further analysis has shown that response to lipid, lipid metabolic process and its regulation, as well as steroid metabolic process were among the top 10 listed biological processes connected to the investigated mixture and obesity development. This is in accordance with the recent in silico study by Suvorov et al. (2021), who have also suggested that the sensitivity of lipid metabolism pathways to chemical exposures may be relevant to the current epidemic of metabolic diseases, including obesity, Type 2 diabetes, metabolic syndrome, and non-alcoholic fatty liver disease (Suvorov et al., 2021). Another metabolic process highlighted in the present study was inflammatory response, which is thought to be the link between insulin resistance, obesity and diabetes. It has been shown that increased concentrations of proinflammatory cytokines, TNF-α and IL-6, associated with obesity and type 2 diabetes, might lead to the decrement of insulin signal transduction and interfere with the anti-inflammatory effect of insulin, promoting further inflammation (Dandona et al., 2004). IL-6 signaling furthermore induces IL-10 expression and T cell differentiation to IL-17-expressing T cells, disturbing the definition of cell type specific contributions of inflammatory mediators in not only obesity, but also its comorbidities (Kern et al., 2019). Additionally, TNF-α is involved not only in the inflammatory response, but also apoptosis of adipose tissues and lipid metabolism, by intensifying lipogenesis, insulin signalling and reactive oxygen species (ROS) synthesis. Newly synthesized ROS further induce the production of other inflammatory cytokines and pro-inflammatory transcription factors (e.g., activator protein-1 (AP-1) and nuclear factor kapa B (NF-kB)) which intensify further ROS production and might be regarded as circulus vitiosus (Wang and Trayhurn, 2006; Čolak and Pap, 2021). Fittingly, oxidative stress was also highlighted as one of the important obesity-linked molecular mechanisms extracted in our study, while superoxide dismutase activity oxidoreductase activity (acting on superoxide radicals as acceptor, as well as acting on paired donors, with incorporation or reduction of molecular oxygen) was present among the top molecular functions. Oxidative stress is known to stimulate adipose tissue deposition, including preadipocyte proliferation, adipocyte differentiation and growth (Čolak and Pap, 2021). Furukawa et al. (2004) have stated that oxidative stress in accumulated fat can be regarded as a significant mechanism of metabolic syndrome linked to obesity, and even concluded that redox status in adipose tissue could be used as therapeutic target (Furukawa et al., 2004). Furthermore, our analysis has singled out nuclear and steroid hormone receptor activity among the top molecular functions. This is expected, since all three substances were shown to bind to ESR1 receptor and increase the expression of ESR1 gene mRNA, which was also demonstrated in our study. For example, prenatal BPA exposure was found to provoke obesity development via ERs, while changes in maternal estrogen levels during gestation elevated the number of adipocytes number and weakened their function (Stojanoska et al., 2017). However, it is important to bear in mind that the role of sex steroids in white adipose tissue function is complex, while both concentrations of androgens and estrogens appear to affect the modulation of white adipose tissue function (Newell-Fugate, 2017). On the other hand, molecular pathway analysis has shown that visceral fat deposits and metabolic syndrome, nuclear receptor transcription pathway and PPAR signaling pathway could be highlighted as the top 3 molecular pathways connected to both obesity development and exposure to the investigated mixture.
Choosing the obesity/comorbidities-related genomic biomarkers
Considering that the most metabolic diseases are intertwined, our further investigation listed 10 comorbidities of obesity in which the common 31 genes/proteins were involved, showing that 7 genes/proteins (CCL2, IL6, LPL, PPARA, PPARG, SERPINE1, TNF) were present in the etiology of all these comorbidities.
Our data mining approach has revealed that all three investigated substances interacted with PPARA and PPARG (genes which encode PPARα and PPARγ) in the same manner, while PPAR signaling pathway was among the top 3 molecular pathways connected to the obesity development and exposure to the investigated substances. In the DisGeNET database, PPARG was among the highest gene-disease associations, with a score 0.900 for obesity. While PPARα regulates fatty acid oxidation in several tissues known for their high rates of fatty acid oxidation such as liver, heart, kidney, and brown adipose tissue, PPARγ is responsible for fatty acid uptake and storage by stimulating triglyceride accumulation in adipocytes, NADPH synthesis for lipogenesis, glyceroneogenesis, as well as fatty acid esterification (Kunej et al., 2013; Plutzky, 2011; Singh and Li, 2011). However, PPARα activators exert a variety of metabolic actions, depending on to the species, gender, dose, and timing of exposure. For example, high doses DEHP (100 or 1,000 mg/kg/day) applied in the duration of 13 weeks protected adult mice from diet-induced obesity by promoting fatty acid oxidation and catabolic metabolism by activating PPARα (Feige et al., 2010). On the contrary, in mice expressing human PPARα, perinatal exposure to MEHP in low doses (0.5, 0.25 or 0.05 mg/kg of b.w.) promoted fat accumulation and exacerbated obesity (Hao et al., 2012). Furthermore, other important factors should be considered, such as difference between animals and humans. PPARγ, crucial for white adipose tissue development and adipogenesis, is much more highly expressed in human tissues than PPARα. Also, because of the amino acid sequence within the ligand binding domain, sensitivity of human PPARα activation is lower compared to the rodents’ PPARα (Hurst and Waxman, 2003). Hertz and Bar-Tana (1998) have demonstrated that, unlike in rodents, hypolipidemic effect exerted by peroxisome proliferators in humans is not accompanied by peroxisome proliferation, nor by induction of peroxisomal β-oxidation. They concluded that biological effects exerted by peroxisome proliferators in the human liver are likely to be mediated by a transduction pathway independent of PPARα (Hertz and Bar-Tana, 1998). Thus, taking into account its lower relevance in humans compared to rodents, we have decided to exclude PPARA from our further search for potential biomarkers.
The second gene with the matching interactions for DEHP, DBP and BPA in our study was SERPINE1. This gene encodes plasminogen activator inhibitor-1 (PAI-1), a member of the serine protease inhibitor superfamily. SERPINE1 can be associated with the increased risk for type 2 diabetes and its complications, such as diabetic retinopathy and diabetic coronary artery disease, etc. (Fan et al., 2018). Additionally, Kaur et al. (2010) have suggested that this gene presents a link between obesity and diabetes.
Another gene among the identified potential biomarkers in our study was CCL2. This chemokines gene plays a role in chronic inflammation and, similarly to SERPINE1, compounds the problem of insulin resistance and obesity. CCL2 mRNA was found to be higher expressed in adipocytes of obese individuals (Rakotoarivelo et al., 2020). Both CCL2 and SERPINE1 were among the top 20 genes with the highest numbers of activating and suppressive interactions in the CTD, extracted by the data mining study by Suvorov et al. (2021).
Investigated phthalates and BPA have been found to interact with additional two proinflammatory markers, IL6 and TNF, in the same manner. As mentioned before, these genes are closely linked with chronic low-grade inflammation, so-called meta-inflammation, which occurs in obesity (Sindhu et al., 2015).
Limitations of in silico data mining
Our research has proven the usefulness of freely available web-based analysis tools for toxicogenomic data mining research. However, CTD, ToppGene Suite, DisGeNET or any other functional annotation-based prioritization method has some limitations which confirm that in silico toxicogenomic investigations could serve only as a support to other toxicity testing methods in the overall toxicity testing process. The analysis is heavily dependent on the underlying online sources from which the annotations are retrieved. It also relies upon the quality of interaction data, having in mind all the missing interactions and possible false positive results (Chen et al., 2009). Moreover, the obtained results demonstrate statistical associations between genes impacted by chemicals and those involved in environmental diseases, without the ability to address the dose-response relationship (Harris et al., 2020). It should also be acknowledged that exposure to chemicals is complex and, not only dose, but many other things should be considered, such as route, duration, metabolism, and developmental stage of exposure, as well as all the environmental factors (Davis et al., 2008). As a result of this, many interactions obtained from the CTD could be opposite depending on the type and conditions of the study it was curated from. As shown in Tab. 3, although there were 31 genes/proteins these substances affected in the same manner, some of the interactions were double-natured, showing both activation and inhibition of the interacting gene/protein. Dual-natured interactions could be explained by the unconventional dose-response relationships, the so-called non-monotonic dose-response (NMDR), frequently linked to endocrine disruptive chemicals such as phthalates and BPA. It is characterized by a dose-response curve which slope changes direction within the range of tested doses (Lagarde et al., 2015). In the context of our in silico investigation, this implies that the investigated substances could both increase and decrease the activity of certain genes, depending on the experimental design and applied doses.
All things considered, since various environmental factors might play an important role in the manifestation of environmental diseases, results obtained by in silico data mining represent an insight into further laboratory investigations by which chemical influence on identified gene sets could further be explored by changing external conditions (e.g., high-fat or regular diet, route of exposure, etc.) in order to test all the other impacts on the selected gene set.
The results of in silico study have indicated the presence of 31 obesity-related genes/proteins with matching interactions for DEHP, DBP and BPA, implying that these three substances might act in an additive manner on the development of this disorder. Seven obesity/comorbidities-related genes/proteins (6 of them relevant to humans—CCL2, IL6, LPL, PPARG, SERPINE1, TNF) were selected as potential set of genomic biomarkers for mixture (phthalates and BPA) assessment. Among these, PPARG and LPL were marked as the two genes/proteins most closely linked to obesity. Identification of genomic biomarkers is important for exploring various factors which might influence the development and progression of obesity, among which is exposure to various chemicals. Moreover, identified genes provide the possibility to be used as targets for further translational therapies. Our results have demonstrated that publicly available databases (such as the CTD, ToppGene Suite, DisGeNET and GeneMania) can be used to screen a large number of chemicals and genes/proteins for various effects on molecular responses related to environmental diseases. Furthermore, the analysis reported here should be applicable to almost any mixture of environmental chemicals and any disease present in the CTD in order to explore molecular mechanisms which could further be confirmed by additional toxicity assessment.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article. Additionally, all of the databases and in silico tools used in this study are freely available at: The Comparative Toxicogenomics Database (CTD; http://CTD.mdibl.org); ToppGene Suite (https://toppgene.cchmc.org); GeneMANIA prediction server (https://genemania.org); DisGeNET database (http://www.disgenet.org).
Authors’ Contribution: The authors confirm contribution to the paper as follows: Katarina Baralić: Conceptualization, Methodology, Formal analysis, Investigation, Data Curation, Writing—Original Draft Katarina Živančević: Formal analysis, Investigation, Data Curation, Writing—Original Draft Dragica BoŽIĆ: Methodology, Formal analysis, Investigation, Writing—Review & Editing Danyel Jennen: Writing—Review & Editing, Methodology, Supervision Aleksandra Buha Djordjevic: Writing—Review & Editing, Supervision, Evica Antonijević Miljaković: Formal analysis, Investigation Danijela Đukić-Ćosić: Conceptualization, Visualization, Writing—Review & Editing, Supervision, Project administration.
Ethical Approval: Complete analysis in the present study was conducted in silico. The experiments did not involve human subjects, animals, or plants.
Funding Statement: This work was supported by The Ministry of Education, Science and Technological Development of the Republic of Serbia [Contact Number 451-03-9/2021-14/200161].
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Baralić K, Buha Djordjevic A, Živančević K, Antonijević E, Anđelković M, Javorac D, Ćurčić M, Bulat Z, Antonijević B, Đukić-Ćosić D (2020a). Toxic effects of the mixture of phthalates and bisphenol A—Subacute oral toxicity study in wistar rats. International Journal of Environmental Research and Public Health 17: 746. DOI 10.3390/ijerph17030746. [Google Scholar] [CrossRef]
Baralić K, Jorgovanović A, Živančević K, Antonijević E, Anđelković B, Buha Djordjevic A, Ćurčić M, Đukić-Ćosić D (2020b). Safety assessment of drug combinations used in COVID-19 treatment: in silico toxicogenomic data-mining approach. Toxicology and Applied Pharmacology 406: 115237. DOI 10.1016/j.taap.2020.115237. [Google Scholar] [CrossRef]
Baralić K, Živančević K, Javorac D, Buha Djordjevic A, Anđelković M et al. (2020c). Multi-strain probiotic ameliorated toxic effects of phthalates and bisphenol A mixture in Wistar rats. Food and Chemical Toxicology 143: 111540. [Google Scholar]
Berghuis SA, Bos AF, Sauer PJJ (2015). Developmental neurotoxicity of persistent organic pollutants: An update on childhood outcome Behavior Assessment System for Children Bayley Scales of Infant Development Wechsler Intelligence Scale for Children. Archives of Toxicology 89: 687–709. DOI 10.1007/s00204-015-1463-3. [Google Scholar] [CrossRef]
Borman ED, Vecchi N, Pollock T, deCatanzaro D (2017). Diethylhexyl phthalate magnifies deposition of 14C-bisphenol A in reproductive tissues of mice. Journal of Applied Toxicology 37: 1225–1231. DOI 10.1002/jat.3484. [Google Scholar] [CrossRef]
Boverhof DR, Zacharewski TR (2006). Toxicogenomics in risk assessment: Applications and needs. Toxicological Sciences 89: 352–360. DOI 10.1093/toxsci/kfj018. [Google Scholar] [CrossRef]
Carwile JL, Michels KB (2011). Urinary bisphenol A and obesity: NHANES 2003-2006. Environmental Research 111: 825–830. DOI 10.1016/j.envres.2011.05.014. [Google Scholar] [CrossRef]
Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C et al. (2017). The BioGRID interaction database: 2017 update. Nucleic Acids Research 45: D369–D379. DOI 10.1093/nar/gkw1102. [Google Scholar] [CrossRef]
Chen J, Bardes EE, Aronow BJ, Jegga AG (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Research 37: 305–311. DOI 10.1093/nar/gkp427. [Google Scholar] [CrossRef]
Čolak E, Pap D (2021). The role of oxidative stress in the development of obesity and obesity-related metabolic disorders. Journal of Medical Biochemistry 40: 1–9. DOI 10.5937/jomb0-24652. [Google Scholar] [CrossRef]
Dandona P, Aljada A, Bandyopadhyay A (2004). Inflammation: The link between insulin resistance, obesity and diabetes. Trends in Immunology 25: 4–7. DOI 10.1016/j.it.2003.10.013. [Google Scholar] [CrossRef]
Davis AP, Grondin CJ, Johnson RJ, Sciaky D, McMorran R, Wiegers J, Wiegers TC, Mattingly CJ (2019). The comparative toxicogenomics database: Update 2019. Nucleic Acids Research 47: D948–D954. DOI 10.1093/nar/gky868. [Google Scholar] [CrossRef]
Davis AP, Grondin CJ, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL, Wiegers TC, Mattingly CJ (2015). The comparative toxicogenomics database’s 10th year anniversary: Update 2015. Nucleic Acids Research 43: D914–D920. DOI 10.1093/nar/gku935. [Google Scholar] [CrossRef]
Davis AP, King BL, Mockus S, Murphy CG, Saraceni-Richards C, Rosenstein M, Wiegers T, Mattingly CJ (2011). The comparative toxicogenomics database: Update 2011. Nucleic Acids Research 39: 1067–1072. DOI 10.1093/nar/gkq813. [Google Scholar] [CrossRef]
Davis AP, Murphy CG, Rosenstein MC, Wiegers TC, Mattingly CJ (2008). The comparative toxicogenomics database facilitates identification and understanding of chemical-gene-disease associations: Arsenic as a case study. BMC Medical Genomics 1: 1–12. DOI 10.1186/1755-8794-1-48. [Google Scholar] [CrossRef]
Davis AP, Murphy CG, Saraceni-Richards CA, Rosenstein MC, Wiegers TC, Mattingly CJ (2009). Comparative toxicogenomics database: A knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic Acids Research 37: 786–792. DOI 10.1093/nar/gkn580. [Google Scholar] [CrossRef]
Dong X, Qiu X, Meng S, Xu H, Wu X, Yang M (2018). Proteomic profile and toxicity pathway analysis in zebra fish embryos exposed to bisphenol A and di-n-butyl phthalate at environmentally relevant levels. Chemosphere 193: 313–320. DOI 10.1016/j.chemosphere.2017.11.042. [Google Scholar] [CrossRef]
Fan Q, Li H, Qin Y, Li L, Chen L et al. (2018). Association of SERPINE1 rs6092 with type 2 diabetes and related metabolic traits in a Chinese population. Gene 661: 176–181. DOI 10.1016/j.gene.2018.04.011. [Google Scholar] [CrossRef]
Feige JN, Gerber A, Casals-Casas C, Yang Q, Winkler C et al. (2010). The pollutant diethylhexyl phthalate regulates hepatic energy metabolism via species-specific PPARα-dependent mechanisms. Environmental Health Perspectives 118: 234–241. DOI 10.1289/ehp.0901217. [Google Scholar] [CrossRef]
Franz M, Rodriguez H, Lopes C, Zuberi K, Montojo J, Bader GD, Morris Q (2018). GeneMANIA update 2018. Nucleic Acids Research 46: W60–W64. DOI 10.1093/nar/gky311. [Google Scholar] [CrossRef]
Furukawa S, Fujita T, Shimabukuro M, Iwaki M, Yamada Y, Nakajima Y, Nakayama O, Makishima M, Matsuda M, Shimomura I (2004). Increased oxidative stress in obesity and its impact on metabolic syndrome. Journal of Clinical Investigation 114: 1752–1761. DOI 10.1172/JCI21625. [Google Scholar] [CrossRef]
Hao C, Cheng X, Xia H, Ma X (2012). The endocrine disruptor mono-(2-ethylhexyl) phthalate promotes adipocyte differentiation and induces obesity in mice. Bioscience Reports 32: 619–629. DOI 10.1042/BSR20120042. [Google Scholar] [CrossRef]
Harley KG, Berger K, Rauch S, Kogut K, Henn BC, Calafat AM, Huen K, Eskenazi B, Holland N (2017). Association of prenatal urinary phthalate metabolite concentrations and childhood BMI and obesity. Pediatric Research 82: 405–415. DOI 10.1038/pr.2017.112. [Google Scholar] [CrossRef]
Harris SM, Jin Y, Loch-Caruso R, Padilla IY, Meeker JD, Bakulski KM (2020). Identification of environmental chemicals targeting miscarriage genes and pathways using the comparative toxicogenomics database. Environmental Research 184: 109259. DOI 10.1016/j.envres.2020.109259. [Google Scholar] [CrossRef]
Hatch EE, Nelson JW, Stahlhut RW, Webster TF (2010). Association of endocrine disruptors and obesity: Perspectives from epidemiological studies. International Journal of Andrology 33: 324–331. DOI 10.1111/(ISSN)1365-2605. [Google Scholar] [CrossRef]
Hayes AW, Li R, Hoeng J, Iskandar A, Peistch MC, Dourson ML (2019). New approaches to risk assessment of chemical mixtures. Toxicology Research and Application 3: 1–10. DOI 10.1177/2397847318820768. [Google Scholar] [CrossRef]
Hertz R, Bar-Tana J (1998). Peroxisome proliferator-activated receptor _PPAR. alpha activation and its consequences in humans. Toxicology Letters 102: 85–90. [Google Scholar]
Heudorf U, Mersch-Sundermann V, Angerer J (2007). Phthalates: Toxicology and exposure. The International Journal of Hygiene and Environmental Health 210: 623–634. DOI 10.1016/j.ijheh.2007.07.011. [Google Scholar] [CrossRef]
Hurst CH, Waxman DJ (2003). Activation of PPARα and PPARγ by environmental phthalate monoesters. Toxicological Sciences 74: 297–308. DOI 10.1093/toxsci/kfg145. [Google Scholar] [CrossRef]
Kabir ER, Rahman MS, Rahman I (2015). A review on endocrine disruptors and their possible impacts on human health. Environmental Toxicology and Pharmacology 40: 241–258. DOI 10.1016/j.etap.2015.06.009. [Google Scholar] [CrossRef]
Kaur P, Reis MD, Couchman GR, Forjuoh SN, Greene JFJr, Ase A (2010). SERPINE 1 links obesity and diabetes: A pilot study. The Journal of Proteomics & Bioinformatics 3: 191–199. DOI 10.4172/jpb.1000139. [Google Scholar] [CrossRef]
Kern L, Mittenbühler MJ, Vesting AJ, Ostermann AL, Wunderlich CM, Wunderlich FT (2019). Obesity-induced TNFα and IL-6 signaling: The missing link between obesity and inflammation-driven liver and colorectal cancers. Cancers 11: 1–21. [Google Scholar]
Kim SH, Park MJ (2014). Phthalate exposure and childhood obesity. Annals of Pediatric Endocrinology & Metabolism 19: 69. DOI 10.6065/apem.2014.19.2.69. [Google Scholar] [CrossRef]
Kortenkamp A (2007). Ten years of mixing cocktails: A review of combination effects of endocrine-disrupting chemicals. Environmental Health Perspectives 115: 98–105. DOI 10.1289/ehp.9357. [Google Scholar] [CrossRef]
Kortenkamp A (2008). Low dose mixture effects of endocrine disrupters: Implications for risk assessment and epidemiology. International Journal of Andrology 31: 233–237. DOI 10.1111/j.1365-2605.2007.00862.x. [Google Scholar] [CrossRef]
Kunej T, Skok DJ, Zorc M, Ogrinc A, Michal JJ, Kovac M, Jiang Z (2013). Obesity gene atlas in mammals. Journal of Genomics 1: 45–55. DOI 10.7150/jgen.3996. [Google Scholar] [CrossRef]
Lagarde F, Beausoleil C, Belcher SM, Belzunces LP, Emond C et al. (2015). Non-monotonic dose-response relationships and endocrine disruptors: A qualitative method of assessment. Environmental Health 14: 1–5. DOI 10.1186/1476-069X-14-13. [Google Scholar] [CrossRef]
Meeker JD, Ferguson KK (2011). Relationship between urinary phthalate and bisphenol a concentrations and serum thyroid measures in U.S. adults and adolescents from the national health and nutrition examination survey (NHANES) 2007–2008. Environmental Health Perspectives 119: 1396–1402. DOI 10.1289/ehp.1103582. [Google Scholar] [CrossRef]
Meng Q, Richmond-Bryant J, Lu SE, Buckley B, Welsh WJ et al. (2013). Cardiovascular outcomes and the physical and chemical properties of metal ions found in particulate matter air pollution: A QICAR study. Environmental Health Perspectives 121: 558–564. DOI 10.1289/ehp.1205793. [Google Scholar] [CrossRef]
Muscogiuri G, Barrea L, Laudisio D, Savastano S, Colao A (2017). Obesogenic endocrine disruptors and obesity: Myths and truths. Archives of Toxicology 91: 3469–3475. DOI 10.1007/s00204-017-2071-1. [Google Scholar] [CrossRef]
Newell-Fugate AE (2017). The role of sex steroids in white adipose tissue adipocyte function. Reproduction 153: R133–R149. DOI 10.1530/REP-16-0417. [Google Scholar] [CrossRef]
Osama A, Sati M, Ali S, Ali A, Babikir R et al. (2019). Prediction of four novel SNPs V17M, R11H, A66T, and F57S in the SBDS gene possibly associated with formation of Shwachman-Diamond syndrome, using an insilico approach. BioRxiv 542654. [Google Scholar]
Piñero J, Deu-pons J, Centeno E, Garc J, Sanz F, Furlong LI (2016). DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Research 45: gkw943. [Google Scholar]
Plutzky J (2011). The PPAR-RXR transcriptional complex in the vasculature: Energy in the balance. Circulation Research 108: 1002–1016. DOI 10.1161/CIRCRESAHA.110.226860. [Google Scholar] [CrossRef]
Rakotoarivelo V, Variya B, Langlois M, Ramanathan S (2020). Chemokines in human obesity. Cytokine 127: 154953. DOI 10.1016/j.cyto.2019.154953. [Google Scholar] [CrossRef]
Rochester JR (2013). Bisphenol A and human health: A review of the literature. Reproductive Toxicology 42: 132–155. DOI 10.1016/j.reprotox.2013.08.008. [Google Scholar] [CrossRef]
Sindhu S, Thomas R, Shihab P, Sriraman D, Behbehani K, Ahmad R (2015). Obesity is a positive modulator of IL-6R and IL-6 expression in the subcutaneous adipose tissue: Significance for metabolic inflammation. PLoS One 10: 1–17. DOI 10.1371/journal.pone.0133494. [Google Scholar] [CrossRef]
Singh S, Li SSL (2011). Phthalates: Toxicogenomics and inferred human diseases. Genomics 97: 148–157. DOI 10.1016/j.ygeno.2010.11.008. [Google Scholar] [CrossRef]
Singh S, Li SSL (2012). Bisphenol A and phthalates exhibit similar toxicogenomics and health effects. Gene 494: 85–91. DOI 10.1016/j.gene.2011.11.035. [Google Scholar] [CrossRef]
Singh S, Li SSL (2013). Epigenetic effects of environmental chemicals bisphenol a and phthalates. International Journal of Molecular Sciences 13: 267–278. [Google Scholar]
Somm E, Toulotte A, Cederroth CR, Nef S (2009). Perinatal exposure to Bisphenol A alters early adipogenesis in the rat. Environmental Health Perspectives 117: 1549–1555. DOI 10.1289/ehp.11342. [Google Scholar] [CrossRef]
Stahlhut RW, van Wijngaarden E, Dye TD, Cook S, Swan SH (2007). Concentrations of urinary phthalate metabolites are associated with increased waist circumference and insulin resistance in adult U.S. males. Environmental Health Perspectives 115: 876–882. DOI 10.1289/ehp.9882. [Google Scholar] [CrossRef]
Stojanoska MM, Milosevic N, Milic N, Abenavoli L (2017). The influence of phthalates and bisphenol A on the obesity development and glucose metabolism disorders. Endocrine 55: 666–681. DOI 10.1007/s12020-016-1158-4. [Google Scholar] [CrossRef]
Suvorov A, Salemme V, McGaunn J, Poluyanoff A, Teffera M, Amir S (2021). Unbiased approach for the identification of molecular mechanisms sensitive to chemical exposures. Chemosphere 262: 128362. DOI 10.1016/j.chemosphere.2020.128362. [Google Scholar] [CrossRef]
Tung CW, Jen H, Chia C, Wang C, Shan S, Pinpin W (2020). Leveraging complementary computational models for prioritizing chemicals of developmental and reproductive toxicity concern: An example of food contact materials. Archives of Toxicology 94: 1–10. DOI 10.1007/s00204-019-02641-0. [Google Scholar] [CrossRef]
van Breda SG, Claessen SM, Lo K, van Herwijnen M, Brauers KJ et al. (2014). Epigenetic mechanisms underlying arsenic–Associated lung carcinogenesis. Archives of Toxicology 89: 1959–1969. DOI 10.1007/s00204-014-1351-2. [Google Scholar] [CrossRef]
Wang B, Trayhurn P (2006). Acute and prolonged effects of TNF-α on the expression and secretion of inflammation-related adipokines by human adipocytes differentiated in culture. Pflügers Archiv–European Journal of Physiology 452: 418–427. DOI 10.1007/s00424-006-0055-8. [Google Scholar] [CrossRef]
Waters MD, Fostel JM (2004). Toxicogenomics and systems toxicology: Aims and prospects. Nature Reviews Genetics 5: 936–948. DOI 10.1038/nrg1493. [Google Scholar] [CrossRef]
Wiegers TC, Davis AP, Cohen KB, Hirschman L, Mattingly CJ (2009). Text mining and manual curation of chemical-gene-disease networks for the Comparative Toxicogenomics Database (CTD). BMC Bioinformatics 10: 326. DOI 10.1186/1471-2105-10-326. [Google Scholar] [CrossRef]
Zarean M, Poursafa P, Amin MM, Kelishadi R (2017). Association of endocrine disrupting chemicals, bisphenol A and phthalates, with childhood obesity: A systematic review. Journal of Pediatrics Review 6: 1–10. DOI 10.5812/jpr.11894. [Google Scholar] [CrossRef]
Zhang WZ, Yong L, Jia XD, Li N, Fan YX (2013). Combined subchronic toxicity of bisphenol A and dibutyl phthalate on male rats. Biomedical and Environmental Sciences 26: 63–69. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |