DOI:10.32604/biocell.2022.015726

| BIOCELL DOI:10.32604/biocell.2022.015726 |  |
| Article |
Hsa_circ_0002137 stabled by LIN28B promotes osteosarcoma cell growth through the hsa-miR-1246/BCL2 axis
1Department of Joint and Trauma Surgery, The Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, 510630, China
2Department of Othopaedic Surgery, The Eighth Affiliated Hospital of Sun Yat-sen University, Shenzhen, 518033, China
3Department of Othopaedic Surgery, Dongguan Dongcheng Hospital, Dongguan, 523007, China
4Department of Orthopedic, The First Affiliated Hospital of Anhui Medical University, Hefei, 230022, China
5Key Laboratory for Regenerative Medicine, Ministry of Education, College of Life Science and Technology, Jinan University, Guangzhou, 510632, China
6Department of Plastic Surgery, The Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, 510630, China
*Address correspondence to: Song Jin, jingso@163.com; Kun Wang, wangk@mail.sysu.edu.cn
#These authors contributed equally to this work
Received: 08 January 2021; Accepted: 09 March 2021
Abstract: Circular RNAs (circRNAs) are a novel class of non-coding RNA that have recently shown to have huge capabilities in the regulation of gene expression at the posttranscriptional level. Growing evidence has indicated that circRNAs could serve as competing endogenous RNAs (ceRNAs) to sponge microRNAs (miRNAs) and suppress functions of targeted miRNAs. Osteosarcoma (OS) is the most common malignant primary bone cancer. Hsa_circ_0002137 is upregulated in OS. However, the role of hsa_circ_0002137 in OS remains unclear. Using miRNA pull-down assay, we showed that cir_0002137 sponged hsa-miR-1246, and BCL2 apoptosis regulator (BCL2) mRNA was a potential target of hsa-miR-1246 in human osteosarcoma (HOS) cells. Further, we found that hsa_cir_0002137 could enhance the expression of BCL2 hsa-miR-1246 and promote HOS cell growth through sponging hsa-miR-1246. Moreover, RNA binding protein immunoprecip itation (RIP) assay revealed that lin-28 homolog B (LIN28B) protein associated with hsa_circ_0002137, and LIN28B could increase hsa_circ_0002137 stability and thus accelerate OS cell growth. Our work was the first to study the functions of hsa_circ_0002137, has-miR-1246 and LIN28B in OS, and these results may provide novel therapeutic targets for OS treatment.
Keywords: Osteosarcoma; hsa_circ_0002137; hsa-miR-1246; Proliferation; Apoptosis
Osteosarcoma (OS), which is characterized by high metastatic potential and poor clinical prognosis as well as high recurrence rate (Li et al., 2019; Song and Li, 2018), is widely regarded as the most common malignant primary bone cancer in children and adolescents (Li et al., 2019). Despite the rapid development of therapeutic strategies, the five-year survival rate of OS patients is still unsatisfactory (Li et al., 2019). In order to improve the curative ratio and prognosis of OS, it is urgent to find novel and more effective therapeutic targets with a better understanding of the molecular mechanisms that underlie the occurrence and development of OS (Li and Song, 2017).
CircRNAs are novel endogenous noncoding-RNAs (Wang et al., 2019b) characterized by a covalent closed-loop structure with neither 5’ to 3’ polarity nor polyadenylated tail (Qu et al., 2015). Functional circular RNAs have been proved to be sequestrant agents of RNA binding protein, cytoplasmic microRNA sponges, and nuclear transcription regulators, indicating circular RNA as a participant in the regulatory network of gene expression (Hansen et al., 2013b; He et al., 2017a). Through the ceRNA network, circular RNA can play an important role in the development of various diseases (An et al., 2017; He et al., 2017a; Huang et al., 2016; Wang et al., 2017), including OS (Soghli et al., 2020).
As the target of circRNAs, miRNAs are regulatory molecules that are frequently found to be aberrantly expressed in cancers and may contribute to tumor heterogeneity (Suzuki et al., 2015). A large variety of miRNAs have been demonstrated to be closely associated with OS (Wang et al., 2019a; Wang et al., 2020). Recent efforts have focused on identifying miRNAs as diagnostic biomarkers or therapeutic targets (Kreth et al., 2018; Zhang et al., 2016).
LIN28B (UniProtKB Q6ZN17) contributes to tumorigenesis by regulating the mature of let-7 miRNAs as an RNA-binding protein (RBP) (Balzeau et al., 2017; Wang et al., 2015). Although LIN28B expression is upregulated in OS (Mizushima et al., 2020), the role of LIN28B in OS is unclear. In addition, the effect of LIN28B on circRNA expression remains to be elucidated.
In this study, we investigated the role of hsa_circ_ 0002137 in OS. Our study revealed that hsa_circ_0002137 regulated BCL2 expression by targeting has-miR-1246 and therefore enhanced OS cell growth. LIN28B protein can increase hsa_circ_0002137 stability by interacting with hsa_circ_0002137 and subsequently promote OS cell growth. These results may provide novel therapeutic targets for OS treatment.
HOS cells, MG63 cells, U2OS cells, and hFOB1.19 cells were obtained from the Cell Bank at the Chinese Academy of Sciences (Shanghai, China). All cells are maintained in DMEM (Dulbecco’s modified eagle’s medium) (Gibco BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco BRL) and cultured at 37°C in a 5% CO2 humidified incubator. The 3rd passage HOS cells were used in this study, and HOS cells exhibited a doubling time of 36 h.
According to the protocol of the manufacturer, biotin-labeled probes, small interfering RNAs (siRNAs), vectors, 50 nmol/μL miRNA mimics or inhibitors were transfected into HOS cells by Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA). After incubation with siRNAs, vectors, miRNA mimics or inhibitors, cells were collected for subsequent experiments.
Biotin-labeled hsa-miR-1246 pulldown probe (AAUGGAUU UUUGGAGCAGG) and NC (negative control) pulldown probe were synthesized by HeChuang Biotech Inc., Guangzhou, China. After transfection, miRNA pulldown was carried out to detect whether hsa-miR-1246 could bind to hsa_circ_0002137 and BCL2 mRNA, and the complementary binding between hsa_circ_0002137, hsa-miR-1246 and BCL2 mRNA had been primarily identified by bioinformatics tools. Firstly, hsa-miR-1246 probe (AAUGGAUUUUUGGAGCAGG) was synthesized, at the same time, cells were separated into two groups: for the first group, we applied biotinylated hsa-miR-1246 as probes; for the other one, we applied biotinylated non-related sequence probes as NC group. Then, cell centrifugation was carried out, and we collected the supernatant as input group. For the precipitate, magnetic beads (Thermo Fisher Scientific) were incubated with biotinylated hsa-miR-1246 and NC probes to generate probe-coated beads, they were then incubated with cell lysates at 4°C overnight. After that, the RNAs bound to the beads were isolated. Hsa_circ_0002137, BCL2 mRNA and GAPDH as three different kinds of primers were given respectively to the equal volume of the isolated RNAs from both pulldown and NC groups. The abundance of hsa_circ_0002137, BCL2 mRNA and GAPDH in bound fractions was evaluated by qRT-PCR analysis.
Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, Calif, USA). Subsequently, 1 μg RNA was used for cDNA synthesis performed by PrimeScript II 1st Strand cDNA Synthesis Kit (Takara, Dalian, Liaoning, China). Next, SYBR Premix Ex Taq II (Takara) was used for RT-qPCR analysis performed by a 7500 Fast Real-Time PCR system (Applied Biosystems). Besides, outward-facing primers or divergent primers utilized in RT-qPCR were synthesized by Sangon Biotech (Shanghai, China). Primers used in this study as follows: hsa_circ_0002137 (GeneBank ID: NM_025076, length of the amplicon: 190 bp), 5’-CTTCTGGAGGTATTTCCACTGTC-3’ (forward), 5’- AGAAGAGGCTATGGATGGAGTA-3’ (reverse); LIN28B (loci: 4095-4202, GeneBank ID: NM_001004317.4, length of the amplicon: 108 bp), 5’-GGCAGCAGGTCTGTGAATTA-3’ (forward), 5’-GATCAGCTTACAGGGCTAAGAG-3’ (reverse); BCL2 (loci: 4686-4783, GeneBank ID: NM_000633.3, length of the amplicon: 98 bp), 5’- GGCCAGGGTCAGAGTTAAATAG-3’ (forward), 5’- GGAGGTTCTCAGATGTTCTTCTC-3’ (reverse); GAPDH (loci: 350-486, GeneBank ID: NM_001289745.3, length of the amplicon: 137 bp), 5’-GCACCGTCAAGGCTGAGAAC-3’ (forward), 5’-TG GTGAAGACGCCAGTGGA-3’ (reverse). GAPDH was used as an internal control for RNA expression. The relative expression of candidate genes was normalized to internal control using the 2–ΔΔCt equation. At least three technical repeats were set for RT-PCR verification each time.
Dual-luciferase reporter gene assay
First, three siRNAs for silencing hsa_circ_0002137 expression were synthesized by Sangon (Shanghai, China), screens were carried out to select the effective siRNA. Wild-type (WT) BCL-2 mRNA 3’UTR or a mutant (MUT) sequence of the binding site for hsa-miR-1246 in BCL-2 mRNA 3’UTR was inserted into luciferase reporter gene vectors. After co-transfected with luciferase vectors and siRNA NC, hsa_circ_0002137 siRNA, mimic NC and hsa-miR-1246 mimic respectively, cells seeded into 24-well plates were subjected to luciferase activity determination using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA) at 48 h after transfection.
Proteins were extracted by RIPA buffer (Beyotime Biotechnology, Shanghai, China) with protease and phosphatase inhibitors (Beyotime Biotechnology), quantified, loaded on SDS-PAGE with 30 μg per lane, and transferred onto PVDF membranes (Millipore, Billerica, MA, USA). After the membranes being blocked with 5% defatted milk, the primary antibodies were added to shake the bed at 4°C overnight. Subsequently, the secondary antibody goat anti-rabbit IgG H&L (Southern Biotech, Birmingham, AL, USA, 4050-05, 1:5000) was added to incubate at room temperature for 1 h. After two times incubation, TBST was used to wash the membrane three times. Ultimately, color rendering was performed using hypersensitive ECL (Beyotime Biotechnology). The primary antibodies include BCL2 (UniProtKB Q92934) antibody (Bioss, Beijing, China, #bs-20351R, 1:500), LIN28B antibody (Bioss, #bs-20351R, 1:1000) and internal reference GAPDH antibody (Aksomics, Shanghai, China, #KC-5G5, 1:10000).
Cells were seeded into 96-well plates at 5 × 103 cells per well and cultured for indicated time points. Cell viability was evaluated using Cell Counting Kit-8 (Beyotime Biotechnology, #C0037) according to the manufacturer’s instructions. OD values were read using a microplate reader (BioTek Company, Winooski, VT, USA) at the 450 nm-wavelength. Each time point was repeated in three wells, and the experiment was independently performed three times.
Detection of cell apoptosis by flow cytometry
Cell apoptosis was analyzed by an Annexin V-FITC apoptosis detection kit (BD Biosciences, San Jose, CA, USA). Transfected OS cells were collected and resuspended in 1.25 μL Annexin V-FITC binding buffer. Subsequently, 10 μL PI was added to stain the cells in the dark for about 15 min. At last, the double staining cells were subjected to flow cytometry (FACScan, BD Biosciences) to detect the apoptosis rate.
RIP assay was performed by RNA Binding Protein Immunoprecipitation Kit (Hechuang). Briefly, 1.5 × 107 cells were collected and split. Then cell lysates were incubated with protein A/G beads and LIN28B antibody (Proteintech, # 24017-1-AP, Chicago, IL, USA) or IgG (negative control) at 4°C for 24 h. Next, beads were collected by a magnetic separation device, and RIP products were eluted from beads. Subsequently, RNA was extracted from RIP products and detected by qPCR.
All experiments of this study were repeated in triplicate, and at least three technical repeats were set in each time. Data were expressed as the mean ± standard deviation (SD) and analyzed using unpaired 2-sided Student’s t-test. Statistical analysis was performed using SPSS 22.0 (SPSS Inc., Chicago, IL, USA). p < 0.05 indicated significant difference.
Hsa_circ_0002137 silencing suppresses OS cell growth
The level of hsa_circ_0002137 was detected in HOS cells, MG63 cells, U2OS cells, and hFOB1.19 osteoblast cells. Results showed that the level of hsa_circ_0002137 increased highest in HOS cells compared to that in hFOB1.19 cells, so HOS cells were utilized for the subsequent experiments (Fig. 1A). The sequencing result identified that hsa_circ_0002137 was amplified (Fig. 1B). Then HOS cells were utilized for performing the loss-of-function experiments. Hsa_circ-0002137 siRNAs, which could stably knock-down the expression of hsa_circ_0002137, were used to explore the function of hsa_circ_0002137 in HOS cells. As shown in Fig. 1C, hsa_circ_0002137 expression was significantly reduced by siRNAs. Among the three siRNAs, si 1-hsa_circ_0002137 reduced hsa_circ_0002137 expression most efficiently (Fig. 1C). Thus, si 1-hsa_circ_0002137 was chosen for further study. To assess the role of hsa_circ_0002137 on OS cell proliferation, we carried out a CCK8 assay. Results showed that silencing of hsa_circ_0002137 suppressed the HOS cell proliferation (Fig. 1D). Moreover, apoptosis assay detected by flow cytometry showed that, in HOS cells, hsa_circ_0002137 silencing increased the apoptosis cell rate compared to the negative control group (Fig. 1E). Overall, the results suggested that hsa_circ_0002137 silencing could suppress the OS cell proliferation.
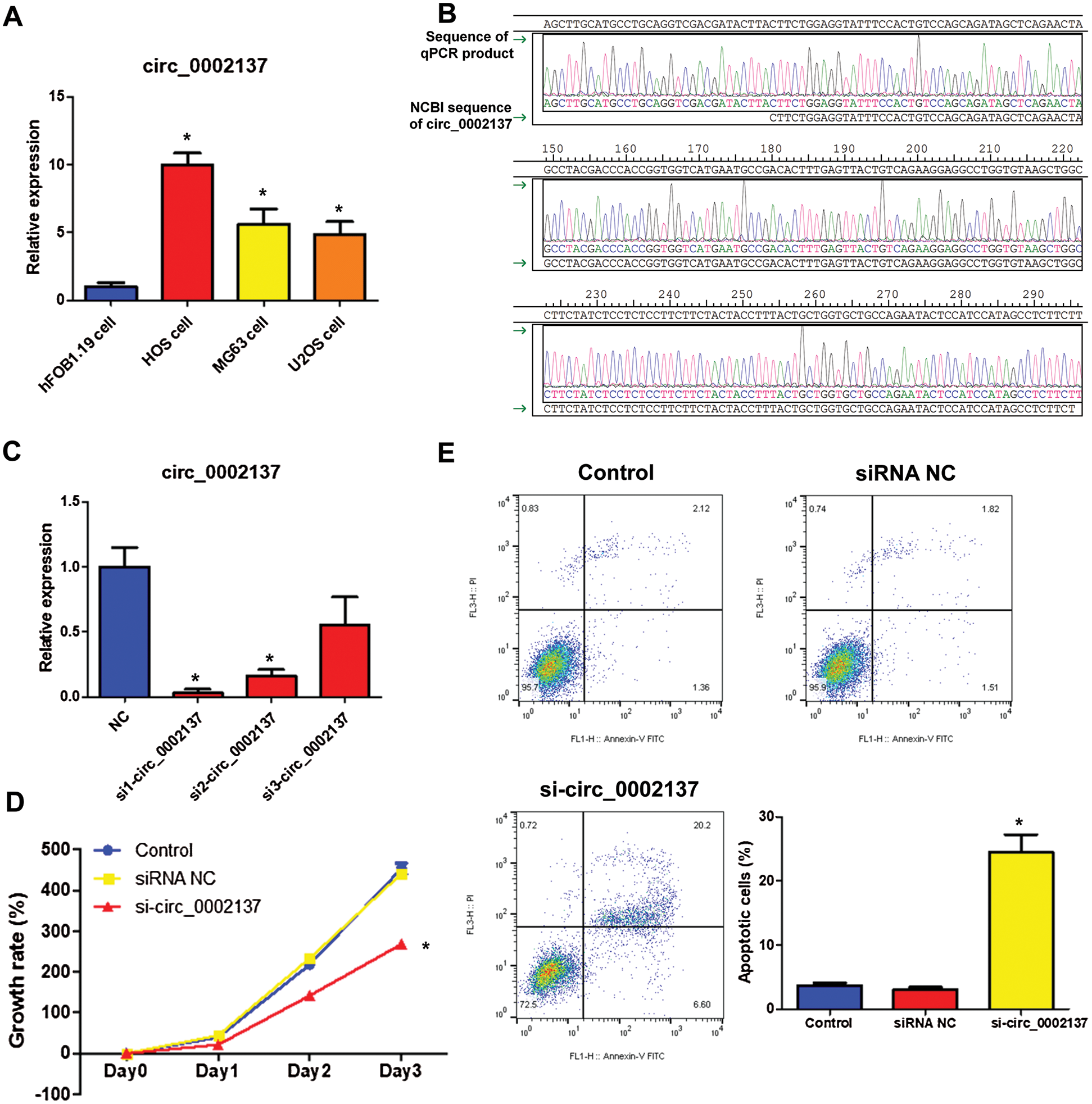
Figure 1: Silencing of hsa_circ_0002137 suppresses HOS cell growth. (A) The level of hsa_circ_0002137 in HOS cells, MG63 cells, U2OS cells and hFOB1.19 cells. (B) Sequencing of qPCR products obtained from A. (C) The level of hsa_circ_0002137 in HOS cells treated with or without siRNAs. (D) The growth rate of HOS cells treated with or without hsa_circ_0002137 siRNA. (E) Flow cytometry analysis of HOS cells for apoptosis and quantification of apoptotic cells per group. Cells were treated with or without hsa_circ_0002137 siRNA. Annexin V labeled with FITC and PI were used to stain cells. NC: negative control; si: siRNA. *indicates a significant difference compared with the NC or Control group (*P < 0.05).
Hsa_circ_0002137 enhances OS cell growth as a sponge for hsa-miR-1246
Functional experiments had revealed the oncogenic role of hsa_circ_0002137 on OS. Further bioinformatics analysis indicated that hsa-miR-1246 can bind with hsa_circ_0002137 (Fig. 2A). In addition, results of miRNA pulldown indicated that hsa-miR-1246 could bind with hsa_circ_0002137 (Fig. 2B). Furthermore, the overexpression of hsa-miR-1246 suppressed HOS proliferation detected by CCK-8 assay, and hsa-miR-1246 inhibitor could reverse the effect of si-hsa_circ_0002137 on HOS proliferation (Fig. 3A). Moreover, overexpression of hsa-miR-1246 induced the apoptosis of HOS cells (Fig. 3B), while hsa-miR-1246 inhibitor could reverse the effect of si-hsa_circ_0002137 on HOS cell apoptosis (Fig. 3B). Taken together, these results suggested that hsa_circ_0002137 sponged hsa-miR-2146 to promote HOS cell growth and suppress HOS cell apoptosis.
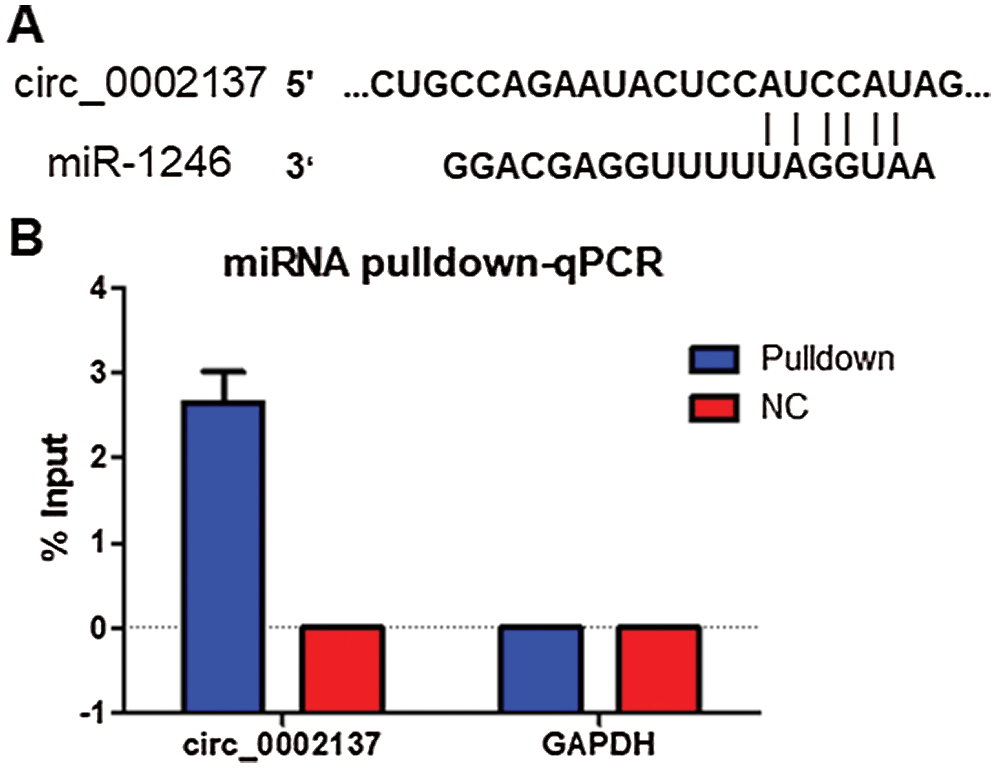
Figure 2: Hsa_circ_0002137 sponges hsa-miR-1246 in HOS cells. (A) Prediction of the hsa-miR-1246 binding site in hsa_circ_0002137 by Starbase. (B) miRNA pulldown and qPCR analysis of the association between hsa_circ_0002137 and hsa-miR-1246 in HOS cells. NC: negative control.
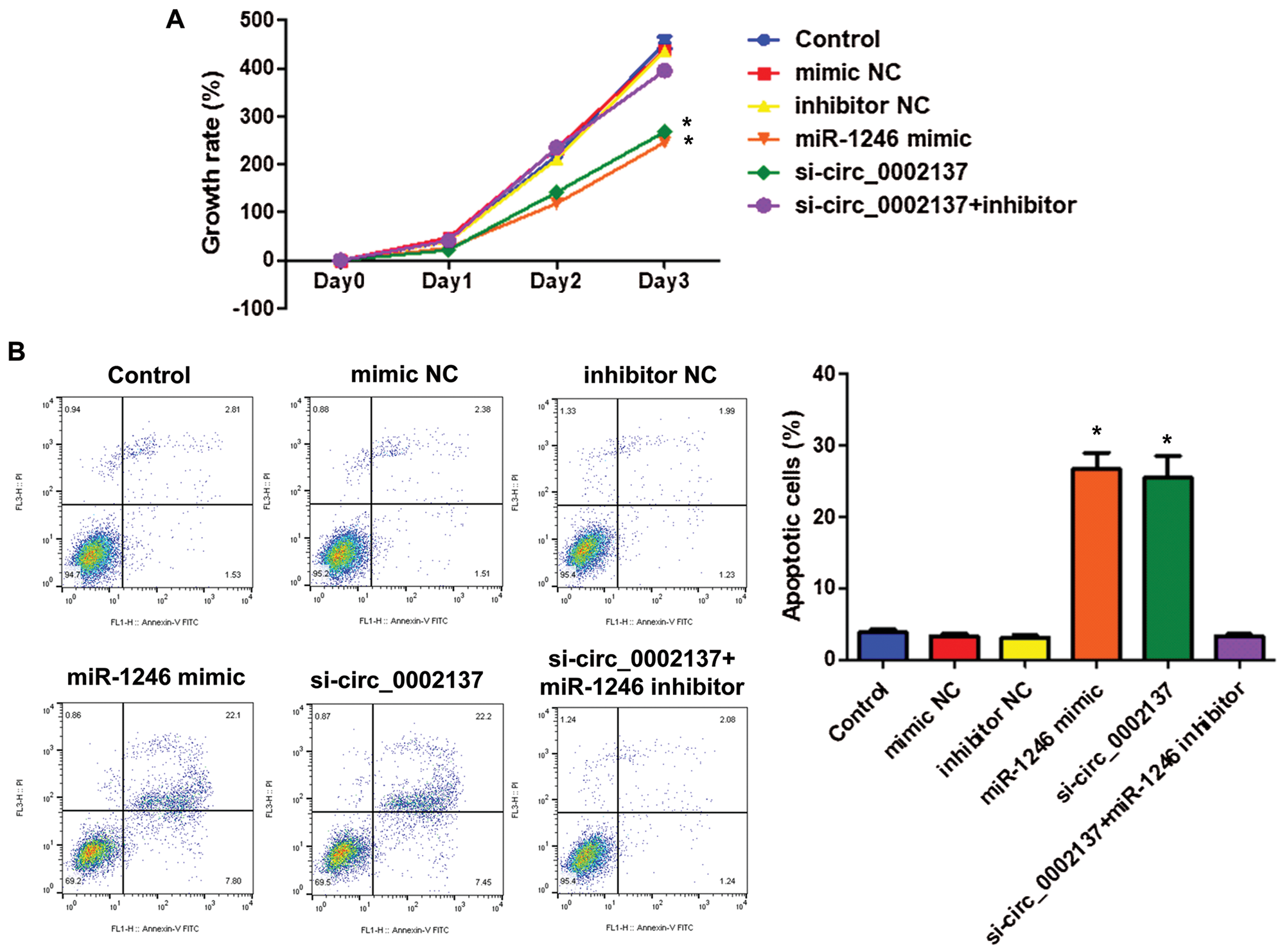
Figure 3: Hsa_circ_0002137 enhances HOS cell growth and suppresses apoptosis by sponging hsa-miR-2146. (A) The growth rate of HOS cells treated with hsa-miR-2146 mimic, inhibitor or hsa_circ_0002137 siRNA. (B) Flow cytometry analysis of HOS cells for apoptosis and quantification of apoptotic cells per group. Cells were treated with hsa-miR-2146 mimic, inhibitor or hsa_circ_0002137 siRNA. Annexin V labeled with FITC and PI were used to stain cells. NC: negative control; si: siRNA. *indicates significant a difference compared with the Control group (*p < 0.05).
Hsa-miR-1246 inhibits OS cell growth by targeting BCL2
To further investigate the downstream target of hsa-miR-1246 in HOS cells, TargetScan (https://www.targetscan.org/) was utilized. The analysis found that BCL2 was the most potential predicted target of hsa-miR-1246. The potential binding site of hsa-miR-1246 in the 3’ UTR of BCL2 mRNA was shown in Fig. 4A. BCL2 suppresses apoptosis and is highly expressed in a variety of cancer cells (Radha and Raghavan, 2017). Using miRNA pulldown, we found that BCL2 mRNA bound with hsa-miR-1246 (Fig. 4B). Subsequently, luciferase reporter assay demonstrated that hsa-miR-1246 negatively regulated BCL2 transcription while results of Western blotting indicated that hsa-miR-1246 decreased BCL2 protein expression in HOS cells (Figs. 4C, 4D). Consistently, the knockdown of hsa_circ_0002137 reduced the transcription activity and protein expression of BCL2 (Figs. 4C and 4D).
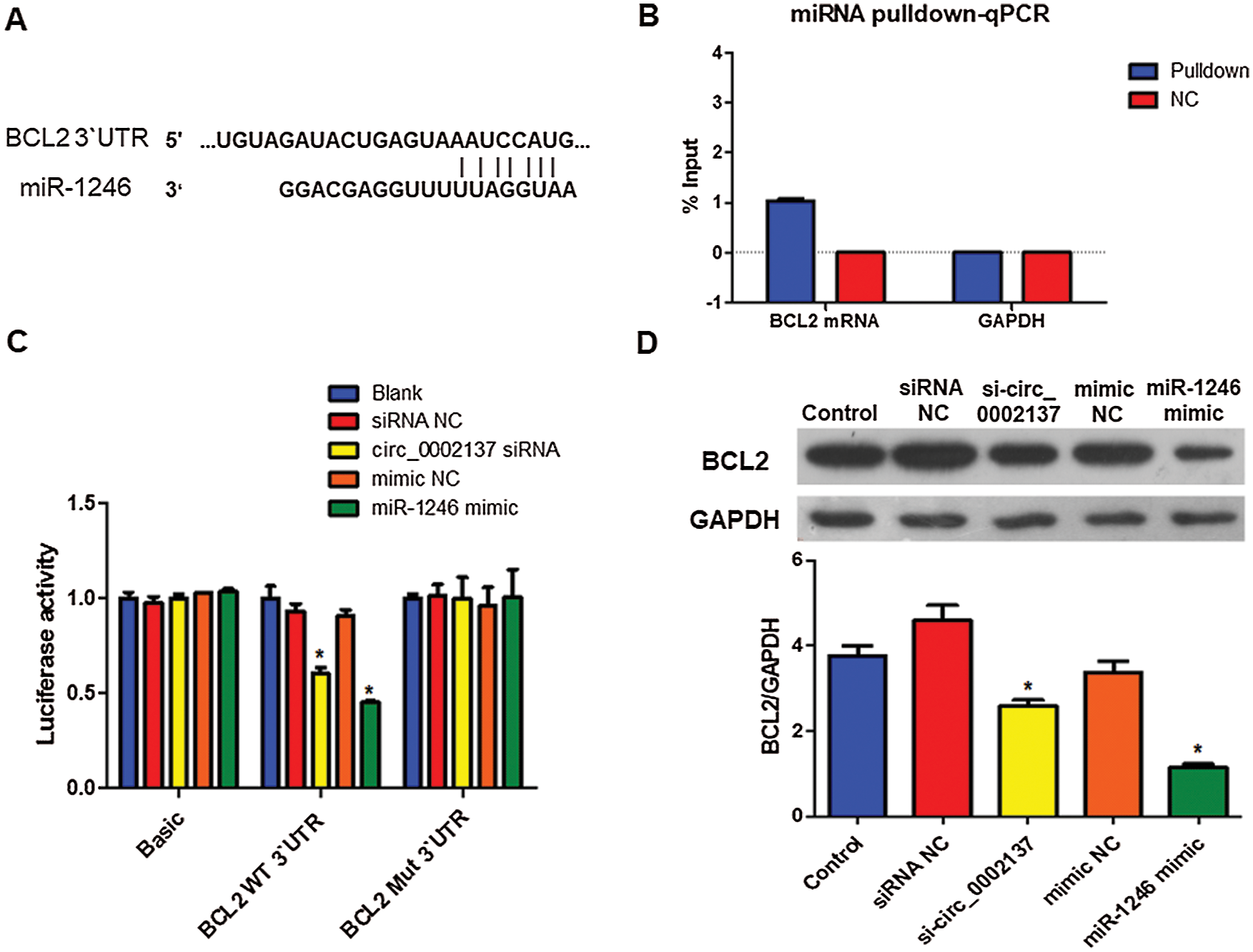
Figure 4: Hsa-miR-2146 targets BCL2 in HOS cells. (A) Prediction of the hsa-miR-2146 binding site in BCL2 miRNA 3’ UTR using Targetscan. (B) miRNA pulldown and qPCR analysis of the association between hsa-miR-1246 and BCL2 mRNA in HOS cells. (C) Luciferase activity of BCL2 in HOS cells treated with hsa-miR-2146 mimic or hsa_circ_0002137 siRNA. Protein abundance of BCL2 in HOS cells treated with hsa-miR-2146 mimic or hsa_circ_0002137 siRNA. NC: negative control; si: siRNA; WT: wild type; Mut: Mutant. *indicates a significant difference compared with the Blank or Control group (*p < 0.05).
Next, the results of the CCK-8 assay found that overexpression of hsa-miR-1246 inhibited HOS cell proliferation. By contrast, BCL2 overexpression attenuated the hsa-miR-1246-mediated inhibition of HOS cell proliferation (Fig. 5A). Apoptosis assay also demonstrated that overexpression of hsa-miR-1246 induced HOS cell apoptosis, whereas BCL2 overexpression abolished the effect of hsa-miR-1246 on HOS cell apoptosis (Fig. 5B). Together, these data suggested that hsa-miR-1246 could suppress HOS cell proliferation by targeting BCL2.
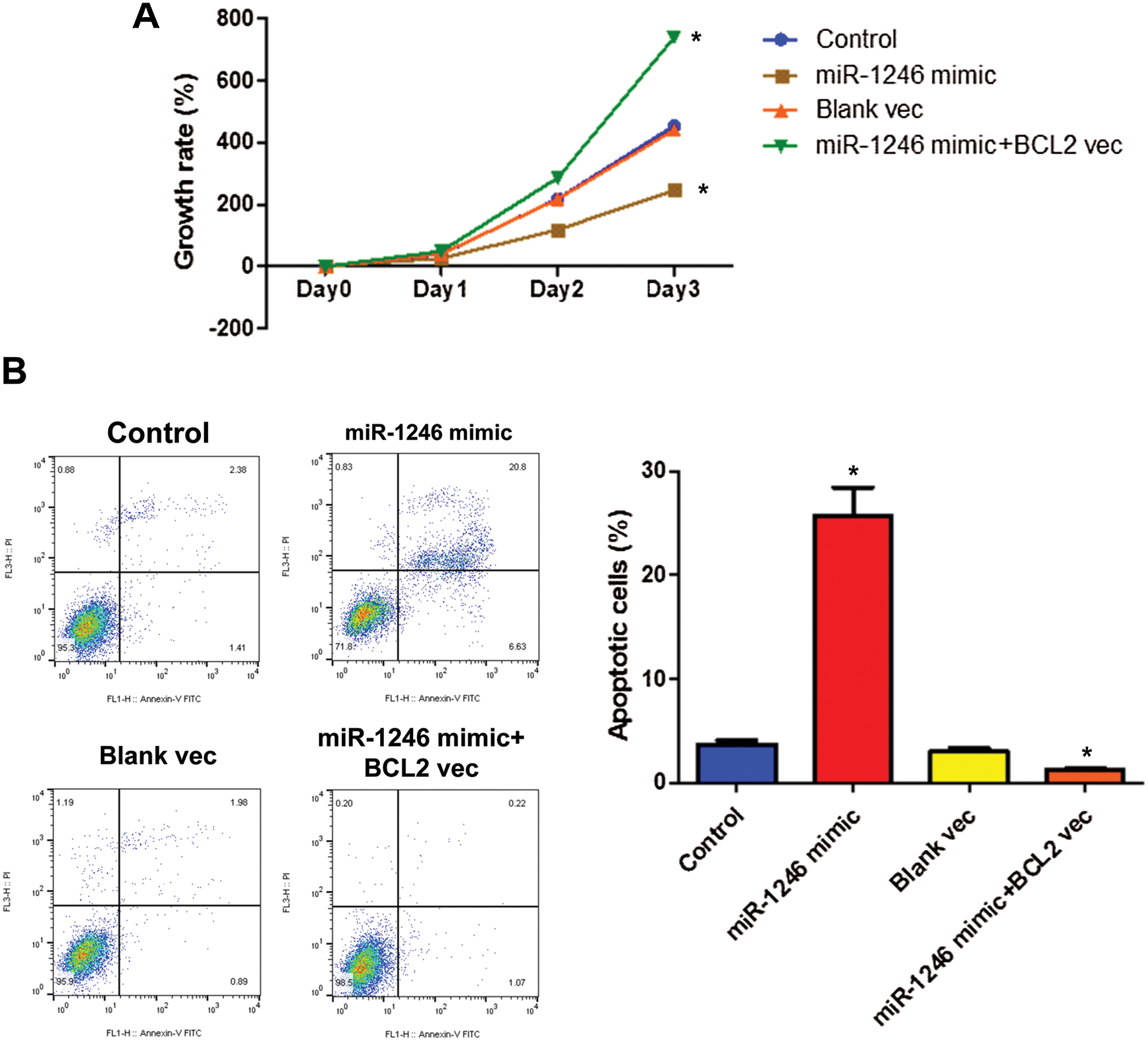
Figure 5: Hsa-miR-2146 inhibits HOS cell growth by targeting BCL2. (A) The growth rate of HOS cells treated with hsa-miR-2146 mimic or BCL2 expression vector. (B) Flow cytometry analysis of HOS cells for apoptosis and quantification of apoptotic cells per group. Cells were treated with hsa-miR-2146 mimic or BCL2 expression vector. Annexin V labeled with FITC and PI were used to stain cells. NC: negative control; vec: expression vector; si: siRNA. *indicates a significant difference compared with the Control group (*p < 0.05).
Hsa_circ_0002137 interacts with LIN28B protein in OS cells
To further interrogate/delineate/investigate the mechanism that governs hsa_circ_0002137 expression, RNA-binding proteins associated with hsa_circ_0002137 were predicted computationally using POSTAR2 (http://lulab.life.tsinghua.edu.cn/postar2/index.php). The analysis suggested that LIN28B can interact with hsa_circ_0002137 (Fig. 6A). Thus, we performed RIP to identify the physical interaction of LIN28B protein with hsa_circ_0002137. As shown in Fig. 6B, LIN28B protein interacted with hsa_circ_0002137 in HOS cells. Similar to hsa_circ_0002137, LIN28B protein level was increased in HOS cells compared to that in hFOB1.19 cells (Fig. 6C). Furthermore, in IHCs we detected enriched LIN28B protein localized in the cytoplasm of HOS cells, whereas it is less abundant in the nuclei (Fig. 6D). Thus, we guessed LIN28B may regulate hsa_circ_0002137 expression at the post-transcriptional level.
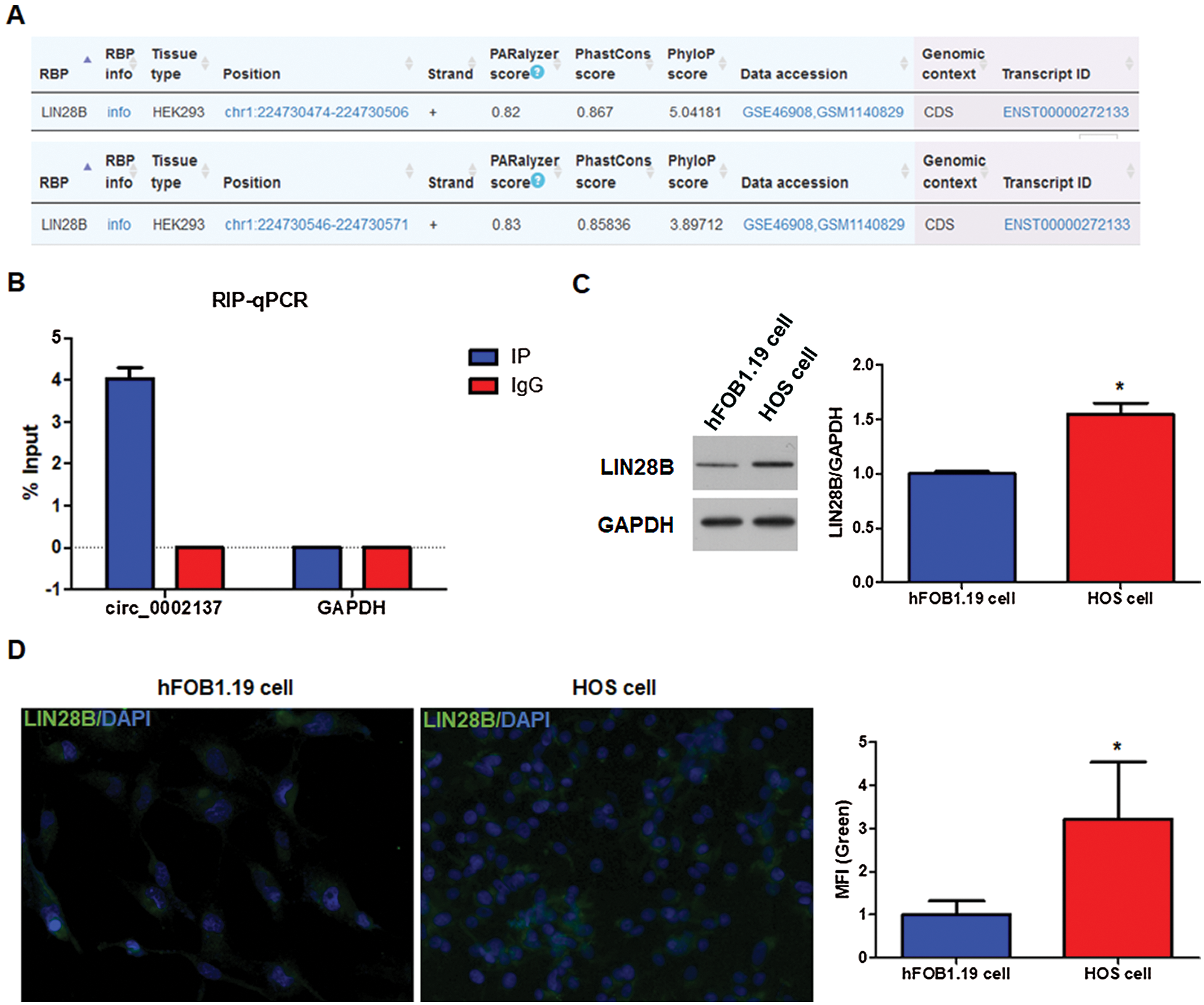
Figure 6: LIN28B interacts with hsa_circ_0002137 and is upregulated in HOS cells. (A) The prediction of interaction between LIN28B and hsa_circ_0002137. (B) RIP and qPCR analysis of the association between LIN28B and hsa_circ_0002137 in HOS cells. (C) The level of LIN28B protein detected by WB in hFOB1.19 cells and HOS cells. (D) The level and location of LIN28B protein detected by IHC in HOS cells. Green: LIN28B; Bule: DAPI. *indicates a significant difference compared with other groups (*p < 0.05).
LIN28B protein increases hsa_circ_0002137 stability in OS cells
To investigate the mechanism of how LIN28B regulates hsa_circ_0002137 expression, LIN28B expression vectors or LIN28B siRNAs were transfected into HOS cells to overexpress or silence LIN28B. Among the three siRNAs, si-LIN28B 1 was found to be the most efficient (Suppl. Figs. S1A and S1C). Therefore, si-LIN28B 1 was used for subsequent experiments. In addition, LIN28B expression vectors could drive LIN28B overexpression effectively in HOS cells (Suppl. Figs. S1B and S1C). Indeed, the overexpression of LIN28B upregulated hsa_circ_0002137 expression, whereas silencing LIN28B reduced the level of hsa_circ_0002137 (Fig. 7A). A previous study has revealed that long non-coding RNA (lncRNA) NEAT1 is stabilized by LIN28B in ovarian cancer (Wu et al., 2018). Considering that LIN28B interacted with hsa_circ_0002137, we hypothesized that LIN28B protein may affect hsa_circ_0002137 stability. Dactinomycin was used to measure hsa_circ_0002137 stability. Results showed that hsa_circ_0002137 level was decreased robustly following the one-hour treatment of dactinomycin at 1 h, and almost diminished at 4 h in the control group (Fig. 7B). In comparison, the hsa_circ_0002137 level in LIN28B-overexpressed cells declined more slowly (Fig. 7B). Consistently, the decay of hsa_circ_0002137 in control cells corresponded to a half-life of 3.1 h, while LIN28B overexpression significantly prolonged the half-life of hsa_circ_0002137 to 4.3 h (Fig. 7B). These data suggested that LIN28B regulated hsa_circ_0002137 abundance by increasing its stability in HOS cells.
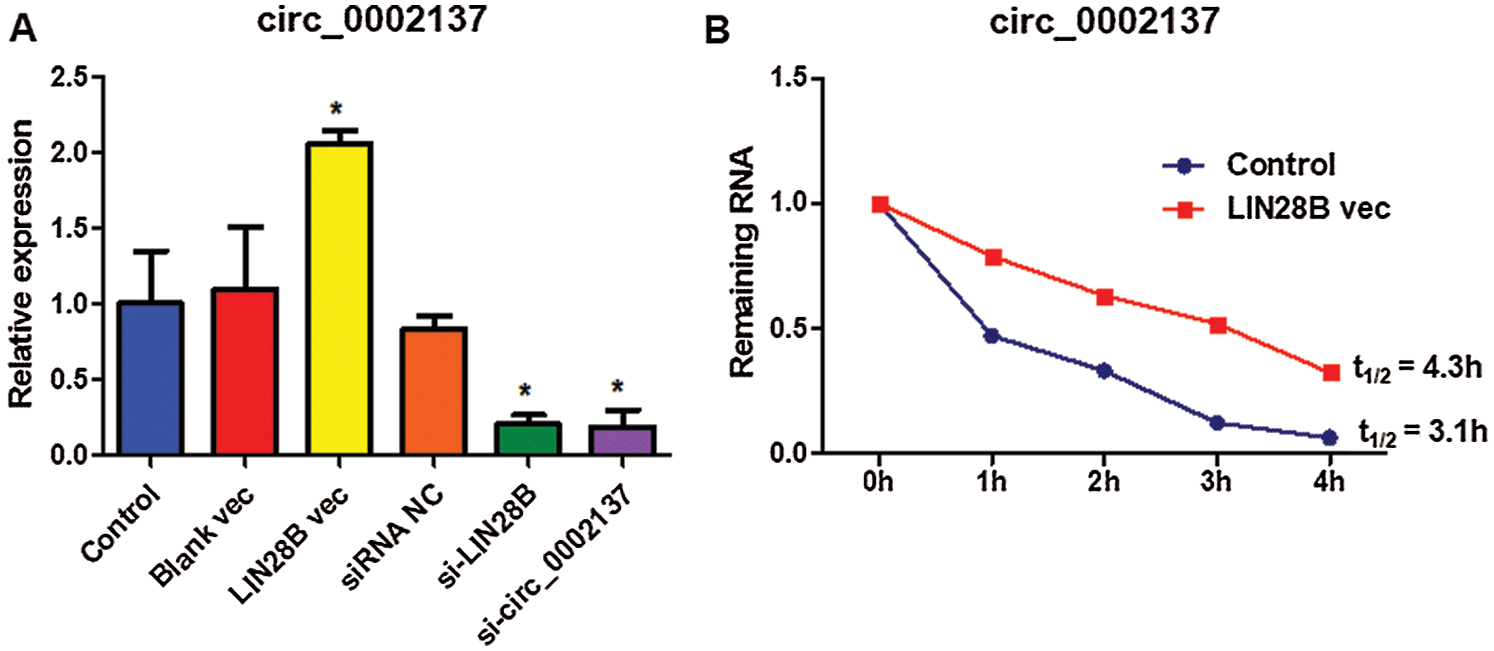
Figure 7: LIN28B protein increases hsa_circ_0002137 stability in HOS cells. (A) The level of hsa_circ_ 0002137 in HOS cells treated with LIN28B vec or LIN28B siRNA. (B) The remaining level of hsa_circ_ 0002137 in HOS cells treated with dactinomycin at 0, 1, 2, 3, and 4 h. NC: negative control; vec: expression vector; si: siRNA. *indicates a significant difference compared with other groups (*p < 0.05).
LIN28B promotes OS cell growth through hsa_circ_0002137
Given our previous finding that hsa_circ_0002137 promotes cell growth (Fig. 3), we next asked whether LIN28B contributes to OS cell growth through hsa_circ_0002137. Results of the CCK8 assay indicated that the overexpression of LIN28B enhanced HOS cell proliferation, whereas the knockdown of LIN28B suppressed HOS cell growth (Fig. 8A). Moreover, silencing hsa_circ_0002137 abolished the effect of overexpressed LIN28B on HOS cell proliferation (Fig. 8A). By contrast, the overexpression of LIN28B reduced HOS cell apoptosis, whereas silence of LIN28B induced HOS cell death (Fig. 8B). Furthermore, hsa_circ_0002137 knockdown attenuated the anti-apoptotic effect of LIN28B in HOS cells (Fig. 8B). Therefore, these results demonstrated that LIN28B enhanced OS cell growth through hsa_circ_0002137.
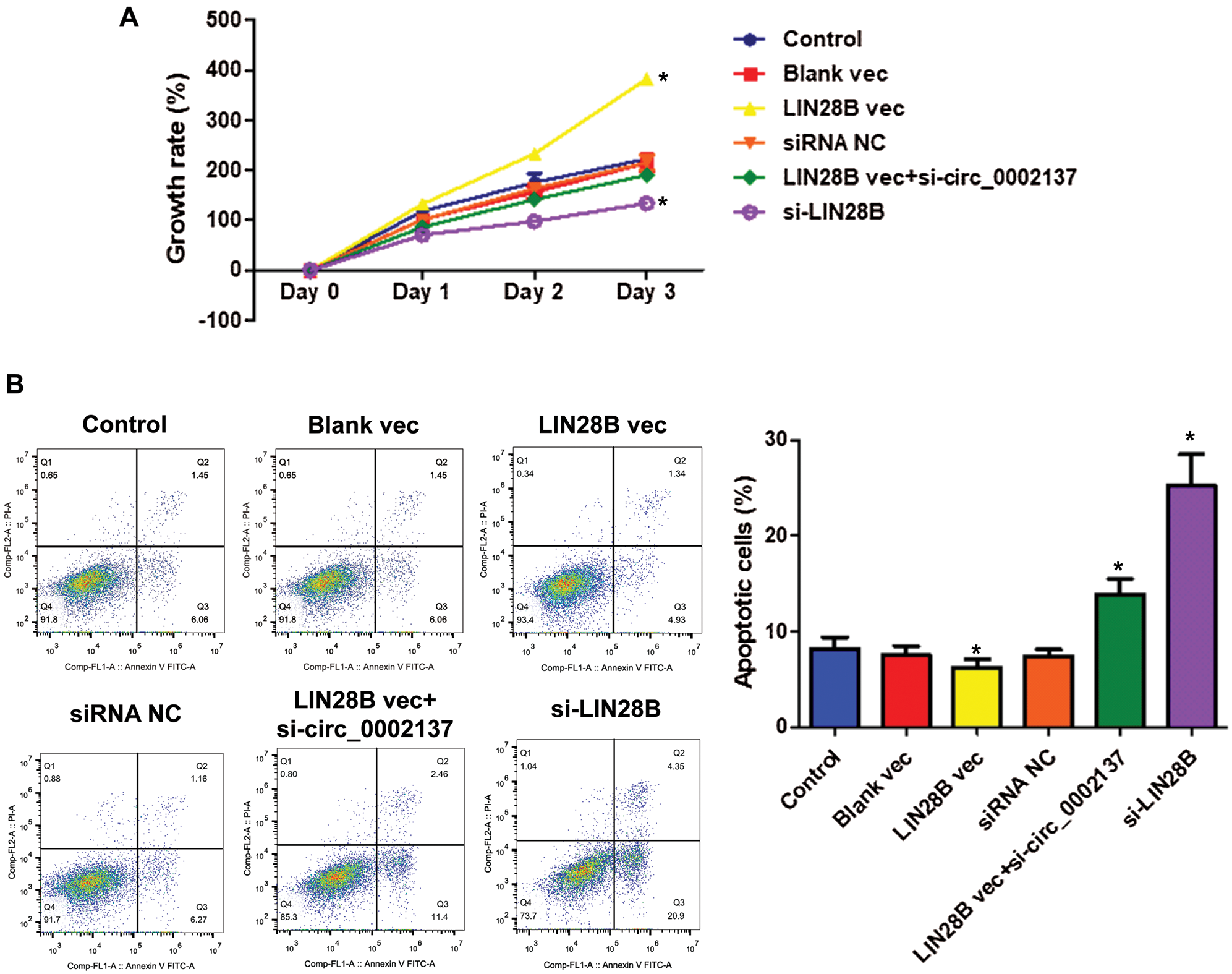
Figure 8: LIN28B suppresses HOS cell growth through hsa_circ_0002137. (A) The growth rate of HOS cells treated with LIN28B vec, LIN28B siRNA or hsa_circ_0002137 siRNA. (B) Flow cytometry analysis of HOS cells for apoptosis and quantification of apoptotic cells per group. Cells were treated with LIN28B vec, LIN28B siRNA or hsa_circ_0002137 siRNA. Annexin V labeled with FITC and PI were used to stain cells. NC: negative control; vec: expression vector; si: siRNA. *indicates a significant difference compared with the control group (*p < 0.05).
Our study revealed that hsa_circ_0002137 enhanced OS cell growth by regulating BCL2 expression by targeting has-miR-1246 and LIN28B protein associated with hsa_circ_0002137 to increase the hsa_circ_0002137 stability and subsequently accelerated OS cell growth. These results may provide novel therapeutic targets for OS treatment.
With the advances of high-throughput sequencing or next-generation sequencing (NGS) and bioinformatics analysis, an increasing number of functional circular RNAs have been discovered and identified in multiple diseases, especially tumorigenesis (Kristensen et al., 2018). CircRNAs may serve as potential diagnostic biomarkers or therapeutic targets as they play important roles in biological processes (Shan et al., 2020; Sheng et al., 2018; Wang et al., 2018). In addition, circRNAs have been widely reported to act as an “miRNA sponge” to regulate its downstream target genes (Hansen et al., 2013a). Several studies have demonstrated the fundamental roles of circRNAs in OS progression by sponging miRNAs. For example, the knockdown of hsa_circ_0001564 suppresses OS proliferation and induces cell cycle arrest and apoptosis by acting miRNA sponge (Song and Li, 2018). As another example, circ-NT5C2 facilitates OS proliferation and metastasis by targeting miR-448 (Liu et al., 2017). Moreover, circ-UBAP2 accelerates OS progression by sponging miR-143 (Zhang et al., 2017). Our present data validated that hsa_circ_0002137 could promote OS cell growth by sponging hsa-miR-1246.
To date, the functions of hsa-miR-1246 remain controversial. In numerous cancer types, hsa-miR-1246 is considered as an onco-miRNA. For example, hsa-miR-1246 enhances metastasis of colorectal cancer via regulating the SPRED2/MAPK pathway (Peng et al., 2019). In breast cancer, exosomal hsa-miR-1246 accelerates tumorigenesis progression and drug resistance by targeting Cyclin-G2 (Li et al., 2017). Moreover, hsa-miR-1246 from the tumor-initiating cell facilitates non-small cell lung cancer progression (Zhang et al., 2016). On the contrary, hsa-miR-1246 induced by p53 suppresses hepatocellular carcinoma cell growth via targeting nuclear factor I/B (Zhang et al., 2015). Here, our study discovered that hsa-miR-1246 inhibited OS cell growth and revealed its contribution to OS for the first time.
During tumor formation, apoptosis, also known as programmed cell death, is usually inhibited, and cancer cells undergo uncontrolled proliferation (Pfeffer and Singh, 2018). BCL2 is an anti-apoptotic protein that may protect cells from a variety of apoptotic stimuli, including cytotoxic drugs, irradiation, heat, or growth factor withdrawal (Piro and Leonardi, 2015). The upregulation of BCL2 has been described in various tumor types, including breast, colon, ovary, and prostate cancers (Delbridge and Strasser, 2015). Moreover, BCL2 could protect cancer cells from apoptosis, such as OS cells (Nedelcu et al., 2008). In addition, numerous miRNAs regulate the biological function of BCL2 in cancer cells. For instance, miR-136 induces gastric cell apoptosis by targeting BCL2 (Yu et al., 2018). Similarly, miR-204-5p triggers apoptosis by targeting BCL2 in prostate cancer cells (He et al., 2017b). In chronic lymphocytic leukemia, miR-15 and miR-16 promote apoptosis by targeting BCL2 (Cimmino et al., 2005). However, the effect of has-miR-1246 on BCL2 is unknown. This study demonstrated that BCL2 was the target of hsa-miR-1246 in OS cells and that has-miR-1246 suppressed tumorigenesis by targeting BCL2 in OS. This mechanism indicated that miRNA-regulated apoptosis plays important roles in the pathogenesis of OS and implies the therapeutic potential of targeting BCL2/hsa-miR-1246 to intervene in tumor cell growth in OS.
LIN28B is a well-recognized oncogene that suppresses the biogenesis of let7 miRNAs (Balzeau et al., 2017). Although LIN28B expression is upregulated in OS, the biological role of LIN28B in OS is unclear (Mizushima et al., 2020). Some studies have revealed the role of LIN28B in RNA stability. For example, lncRNA NEAT1 is stabilized by LIN28B protein in ovarian cancer (Wu et al., 2018). Similarly, LIN28B increases EWS-FLI1 mRNA stability in Ewing sarcoma (Keskin et al., 2020). However, whether LIN28B regulates circRNA stability remains an open question. Here, we found that LIN28B protein increased hsa_circ_0002137 stability by their physical interaction and subsequently accelerated OS cell growth. These findings suggest LIN28B regulates cirRNA stability in a similar way as it regulates RNA and promotes the model that LIN28B participates in accelerating the progression of various cancers by stabilizing RNAs. Disrupting this regulatory network possibly slows down tumor growth and improves the prognosis of cancer.
Osteosarcoma is the most frequent primary malignant solid tumor of bone. At present, the etiology and pathogenesis of OS are not clear. An increasing number of studies have shown that the occurrence, development, and prognosis of OS are closely related to the abnormal expression of oncogenes and anti-oncogenes (Kim et al., 2018; Martin et al., 2012; Xu et al., 2020). Therefore, the molecular mechanisms involved in the pathogenesis of OS will provide a theoretical basis for developing new therapies of OS.
Our study revealed that hsa_circ_0002137 regulated BCL2 expression by targeting has-miR-1246 and therefore enhanced OS cell growth. LIN28B protein can increase hsa_circ_0002137 stability by interacting with hsa_circ_0002137 and subsequently promote OS cell growth. These results may provide novel therapeutic targets for OS treatment.
Availability of Data and Materials: The data of this study are available from the corresponding author upon reasonable request.
Author Contribution:The authors confirm contribution to the paper as follows: study conception and design: FZ, LZ, SJ, KW; data collection: FZ, JZL, RHH, YS; analysis and interpretation of results: FZ, JZL, RHH, YS, KX, SHJ, TZL, WYZ, WDR; draft manuscript preparation: FZ, LZ, SJ, KW. All authors reviewed the results and approved the final version of the manuscript.
Funding Statement:This work was supported by grants from the National Natural Science Foundation of China (81802184), the Natural Science Foundation of Guangdong Province (2017A030310226), and the Fundamental Research Funds for the Central Universities (19ykpy22).
Conflicts of Interest: The authors declare no conflicts of interest.
An Y, Furber KL, Ji S (2017). Pseudogenes regulate parental gene expression via ceRNA network. Journal of Cellular and Molecular Medicine 21: 185–192. DOI 10.1111/jcmm.12952. [Google Scholar] [CrossRef]
Balzeau J, Menezes MR, Cao S, Hagan JP (2017). The LIN28/let-7 pathway in cancer. Frontiers in Genetics 8: 1641. DOI 10.3389/fgene.2017.00031. [Google Scholar] [CrossRef]
Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M (2005). miR and miR-16 induce apoptosis by targeting BCL2. Proceedings of the National Academy of Sciences of the United States of America 102: 13944–13949. [Google Scholar]
Delbridge AR, Strasser A (2015). The BCL-2 protein family, BH3-mimetics and cancer therapy. Cell Death & Differentiation 22: 1071–1080. DOI 10.1038/cdd.2015.50. [Google Scholar] [CrossRef]
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B (2013a). Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388. DOI 10.1038/nature11993. [Google Scholar] [CrossRef]
Hansen TB, Kjems J, Damgaard CK (2013b). Circular RNA and miR-7 in cancer. Cancer Research 73: 5609–5612. DOI 10.1158/0008-5472.CAN-13-1568. [Google Scholar] [CrossRef]
He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W, Li X, Li G, Zeng Z, Tang H (2017a). circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. Journal of Experimental & Clinical Cancer Research 36: 453. DOI 10.1186/s13046-017-0614-1. [Google Scholar] [CrossRef]
He S, Sun H, Lin L, Zhang Y, Chen J, Liang L, Li Y, Zhang M, Yang X, Wang X, Wang F, Zhu F, Chen J, Pei D, Zheng H (2017b). Passive DNA demethylation preferentially up-regulates pluripotency-related genes and facilitates the generation of induced pluripotent stem cells. Journal of Biological Chemistry 292: 18542–18555. DOI 10.1074/jbc.M117.810457. [Google Scholar] [CrossRef]
Huang M, Zhong Z, Lv M, Shu J, Tian Q, Chen J (2016). Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget 7: 47186–47200. DOI 10.18632/oncotarget.9706. [Google Scholar] [CrossRef]
Keskin T, Bakaric A, Waszyk P, Boulay G, Torsello M, Cornaz-Buros S, Chevalier N, Geiser T, Martin P, Volorio A, Iyer S, Kulkarni A, Letovanec I, Cherix S, Cote GM, Choy E, Digklia A, Montemurro M, Chebib I, Nielsen PG, Carcaboso AM, Mora J, Renella R, Suvà ML, Fusco C, Provero P, Rivera MN, Riggi N, Stamenkovic I (2020). LIN28B underlies the pathogenesis of a subclass of Ewing sarcoma LIN28B control of EWS-FLI1 stability. Cell Reports 30: 4567–4583.e5. DOI 10.1016/j.celrep.2019.12.053. [Google Scholar] [CrossRef]
Kim H, Yoo S, Zhou R, Xu A, Bernitz JM, Yuan Y, Gomes AM, Daniel MG, Su J, Demicco EG, Zhu J, Moore KA, Lee DF, Lemischka IR, Schaniel C (2018). Oncogenic role of SFRP2 in p53-mutant osteosarcoma development via autocrine and paracrine mechanism. Proceedings of the National Academy of Sciences of the United States of America 115: E11128–E11137. DOI 10.1073/pnas.1814044115. [Google Scholar] [CrossRef]
Kreth S, Hubner M, Hinske LC (2018). MicroRNAs as clinical biomarkers and therapeutic tools in perioperative medicine. Anesthesia & Analgesia 126: 670–681. DOI 10.1213/ANE.0000000000002444. [Google Scholar] [CrossRef]
Kristensen LS, Hansen TB, Veno MT, Kjems J (2018). Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 37: 555–565. DOI 10.1038/onc.2017.361. [Google Scholar] [CrossRef]
Li JF, Song YZ (2017). Circular RNA GLI2 promotes osteosarcoma cell proliferation, migration, and invasion by targeting miR-125b-5p. Tumour Biology 39: 1010428317709991. [Google Scholar]
Li XJ, Ren ZJ, Tang JH, Yu Q (2018). Exosomal microRNA MiR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cellular Physiology and Biochemistry 44: 1741–1748. DOI 10.1159/000485780. [Google Scholar] [CrossRef]
Li Y, Tang J, Hu Y, Peng Y, Wang J (2019). Expression of miR-664-3p in osteosarcoma and its effects on the proliferation and apoptosis of osteosarcoma cells. Iranian Journal of Public Health 48: 1817–1826. [Google Scholar]
Liu X, Zhong Y, Li J, Shan A (2017). Circular RNA circ-NT5C2 acts as an oncogene in osteosarcoma proliferation and metastasis through targeting miR-448. Oncotarget 8: 114829–114838. DOI 10.18632/oncotarget.22162. [Google Scholar] [CrossRef]
Martin JW, Squire JA, Zielenska M (2012). The genetics of osteosarcoma. Sarcoma 2012: 1–11. DOI 10.1155/2012/627254. [Google Scholar] [CrossRef]
Mizushima E, Tsukahara T, Emori M, Murata K, Akamatsu A, Shibayama Y, Hamada S, Watanabe Y, Kaya M, Hirohashi Y, Kanaseki T, Nakatsugawa M, Kubo T, Yamashita T, Sato N, Torigoe T (2020). Osteosarcoma-initiating cells show high aerobic glycolysis and attenuation of oxidative phosphorylation mediated by LIN28B. Cancer Science 111: 36–46. DOI 10.1111/cas.14229. [Google Scholar] [CrossRef]
Nedelcu T, Kubista B, Koller A, Sulzbacher I, Mosberger I (2008). Livin and Bcl-2 expression in high-grade osteosarcoma. Journal of Cancer Research and Clinical Oncology 134: 237–244. DOI 10.1007/s00432-007-0276-z. [Google Scholar] [CrossRef]
Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, Ji D, Wang Q, Zhang Z, Tang J, Sun Y (2019). Upregulated METTL3 promotes metastasis of colorectal cancer via miR-1246/SPRED2/MAPK signaling pathway. Journal of Experimental & Clinical Cancer Research 38: 394. DOI 10.1186/s13046-019-1408-4. [Google Scholar] [CrossRef]
Pfeffer CM, Singh ATK (2018). Apoptosis: A target for anticancer therapy. International Journal of Molecular Sciences 19: 448. DOI 10.3390/ijms19020448. [Google Scholar] [CrossRef]
Piro F, Leonardi L (2015). Expression of Bcl-2 in canine osteosarcoma. Open Veterinary Journal 5: 27–29. [Google Scholar]
Qu S, Yang X, Li X, Wang J, Gao Y (2015). Circular RNA: A new star of noncoding RNAs. Cancer Letters 365: 141–148. DOI 10.1016/j.canlet.2015.06.003. [Google Scholar] [CrossRef]
Radha G, Raghavan SC (2017). BCL2: A promising cancer therapeutic target. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer 1868: 309–314. DOI 10.1016/j.bbcan.2017.06.004. [Google Scholar] [CrossRef]
Shan G, Shao B, Liu Q, Zeng Y, Fu C (2020). circFMN2 sponges miR-1238 to promote the expression of LIM-homeobox gene 2 in prostate cancer cells. Molecular Therapy-Nucleic Acids 21: 133–146. DOI 10.1016/j.omtn.2020.05.008. [Google Scholar] [CrossRef]
Sheng JQ, Liu L, Wang MR, Li PY (2018). Circular RNAs in digestive system cancer: Potential biomarkers and therapeutic targets. American Journal of Cancer Research 8: 1142–1156. [Google Scholar]
Soghli N, Qujeq D, Yousefi T, Soghli N (2020). The regulatory functions of circular RNAs in osteosarcoma. Genomics 112: 2845–2856. DOI 10.1016/j.ygeno.2020.03.024. [Google Scholar] [CrossRef]
Song YZ, Li JF (2018). Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochemical and Biophysical Research Communications 495: 2369–2375. DOI 10.1016/j.bbrc.2017.12.050. [Google Scholar] [CrossRef]
Suzuki HI, Katsura A, Matsuyama H, Miyazono K (2015). MicroRNA regulons in tumor microenvironment. Oncogene 34: 3085–3094. DOI 10.1038/onc.2014.254. [Google Scholar] [CrossRef]
Wang J, Liu S, Shi J, Li J, Wang S, Liu H, Zhao S, Duan K, Pan X, Yi Z (2019a). The role of miRNA in the diagnosis, prognosis, and treatment of osteosarcoma. Cancer Biotherapy and Radiopharmaceuticals 34: 605–613. DOI 10.1089/cbr.2019.2939. [Google Scholar] [CrossRef]
Wang JY, Yang Y, Ma Y, Wang F, Xue A, Zhu J, Yang H, Chen Q, Chen M, Ye L, Wu H, Zhang Q (2020). Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomedicine & Pharmacotherapy 121: 109627. DOI 10.1016/j.biopha.2019.109627. [Google Scholar] [CrossRef]
Wang L, Wang P, Su X, Zhao B (2019b). Circ_0001658 promotes the proliferation and metastasis of osteosarcoma cells via regulating miR-382-5p/YB-1 axis. Cell Biochemistry and Function 38: 77–86. DOI 10.1002/cbf.3452. [Google Scholar] [CrossRef]
Wang M, Yang Y, Xu J, Bai W, Ren X, Wu H (2018). CircRNAs as biomarkers of cancer: A meta-analysis. BMC Cancer 18: 7. DOI 10.1186/s12885-018-4213-0. [Google Scholar] [CrossRef]
Wang T, Wang G, Hao D, Liu X, Wang D et al. (2015). Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer. Molecular Cancer 14: 3011. DOI 10.1186/s12943-015-0402-5. [Google Scholar] [CrossRef]
Wang Y, Liu J, Jiang Q, Deng J, Xu F, Chen X, Cheng F, Zhang Y, Yao Y, Xia Z, Xu X, Su X, Huang M, Dai L, Yang Y, Zhang S, Yu D, Zhao RC, Wei Y, Deng H (2017). Human adipose-derived mesenchymal stem cell-secreted CXCL1 and CXCL8 facilitate breast tumor growth by promoting angiogenesis. Stem Cells 35: 2060–2070. DOI 10.1002/stem.2643. [Google Scholar] [CrossRef]
Wu Y, Deng Y, Zhu J, Duan YC, Weng WW, Xu MD, Ju XZ, Wu XH (2018). Long noncoding RNA NEAT1, regulated by LIN28B, promotes cell proliferation and migration through sponging miR-506 in high-grade serous ovarian cancer. Cell Death & Disease 9: 1265. DOI 10.1038/s41419-018-0908-z. [Google Scholar] [CrossRef]
Xu N, Xu J, Zuo Z, Liu Y, Yan F, Han C (2020). Downregulation of lncRNA SNHG12 reversed IGF1R-induced osteosarcoma metastasis and proliferation by targeting miR-195-5p. Gene 726: 144145. DOI 10.1016/j.gene.2019.144145. [Google Scholar] [CrossRef]
Yu L, Zhou GQ, Li DC (2018). MiR-136 triggers apoptosis in human gastric cancer cells by targeting AEG-1 and BCL2. European Review for Medical and Pharmacological Sciences 22: 7251–7256. [Google Scholar]
Zhang H, Wang G, Ding C, Liu P, Wang R, Ding W, Tong D, Wu D, Li C, Wei Q, Zhang X, Li D, Liu P, Cui H, Tang H, Ji F (2017). Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget 8: 61687–61697. DOI 10.18632/oncotarget.18671. [Google Scholar] [CrossRef]
Zhang Q, Cao LY, Cheng SJ, Zhang AM, Jin XS, Li Y (2015). microRNA-1246 inhibits the cell growth of human hepatocellular carcinoma cells by targeting NFIB. Oncology Reports 33: 1335–1341. [Google Scholar]
Zhang WC, Chin TM, Yang H, Nga ME, Lunny DP, Lim EKH, Sun LL, Pang YH, Leow YN, Malusay SRY, Lim PXH, Lee JZ, Tan BJW, Shyh-Chang N, Lim EH, Lim WT, Tan DSW, Tan EH, Tai BC, Soo RA, Tam WL, Lim B (2016). Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nature Communications 7: 1367. DOI 10.1038/ncomms11702. [Google Scholar] [CrossRef]
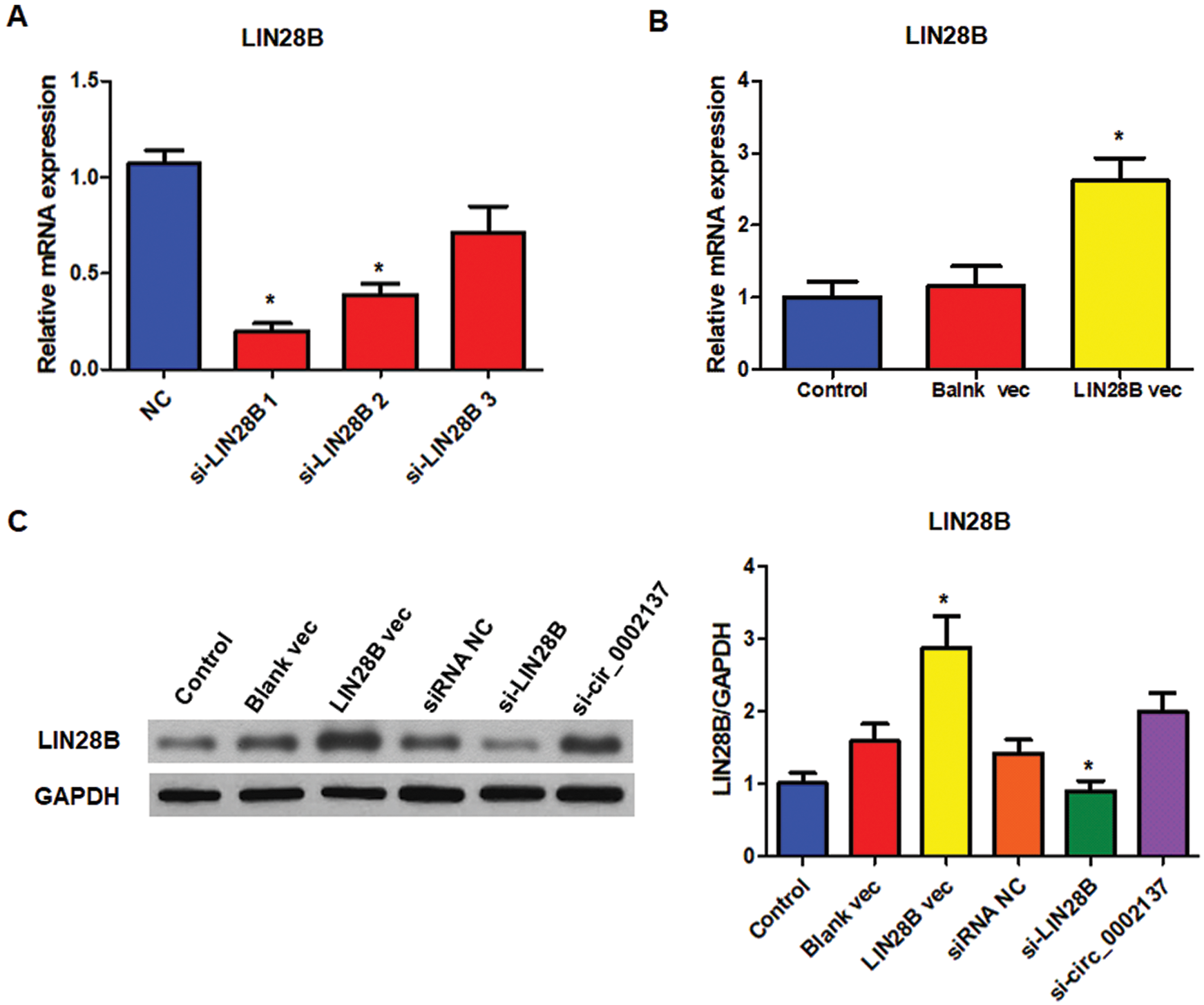
Supplementary Figure S1: Efficiency of LIN28B siRNA and LIN28B expression vector. A The level of LIN28B mRNA in HOS cells treated with or without siRNAs. B The level of LIN28B mRNA in HOS cells treated with or without LIN28B expression vector. C The level of LIN28B protein in HOS cells treated with LIN28B siRNA or LIN28B expression vector. NC: negative control; vec: expression vector; si: siRNA. * indicates significant difference compared with the NC or Control group (*P < 0.05).
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |