DOI:10.32604/biocell.2022.016744

| BIOCELL DOI:10.32604/biocell.2022.016744 |  |
| Article |
Small RNA sequencing revealed aberrant piRNA expression profiles in deciduas of recurrent spontaneous abortion patients
1NHC Key Laboratory of Male Reproduction and Genetics, Guangzhou, 510600, China
2Department of Center Laboratory, Provincial Reproductive Science Institute (Guangdong Provincial Fertility Hospital), Guangzhou, 510600, China
3Human Sperm Bank of Guangdong Province, Guangzhou, 510600, China
4Department of Reproduction Immunology and Genetics, Provincial Reproductive Science Institute (Guangdong Provincial Fertility Hospital), Guangzhou, 510600, China
5Department of Reproduction Medicine, Provincial Reproductive Science Institute (Guangdong Provincial Fertility Hospital), Guangzhou, 510600, China
*Address correspondence to: Weibing Qin, guardqin@163.com; Lixin Zheng, longde882003@163.com
#These authors contributed equally to this work
Received: 23 March 2021; Accepted: 07 May 2021
Abstract: Piwi-interacting RNAs (piRNAs) is a novel class of non-coding RNAs. However, changes in piRNA expression profiles in recurrent spontaneous abortion (RSA) have not yet been investigated. The aim of this study was to identify differentially expressed piRNAs in deciduas of RSA patients. Decidua tissues were collected by curettage from recruited RSA patients and normal early pregnant (NEP) women with their informed consent. Small RNA sequencing was used to evaluate the differences in piRNA expression profiles between RSA and NEP. The present results demonstrated that the counts of total piRNA reads in RSA samples were increased compared with those in NEP samples (0.21% vs. 0.11%). Differential expression analysis identified 29 upregulated piRNAs and 18 downregulated piRNAs in RSA samples. RT-qPCR further confirmed that the expression levels of uniq-109625, uniq-89328, uniq-50651 and uniq-4569 were decreased in 8 RSA tissues, compared with 13 NEP tissues. Otherwise, pi-22628 and uniq-173406 were increased in 8 RSA tissues. Based on GO term and KEGG pathway analysis, we speculate that these piRNAs regulate RSA by targeting extracellular matrix component pathway, cell adhesion pathway and focal adhesion pathway. PiRNAs may be involved in RSA pathogenesis by target genes function on adhesion and extracellular matrix component.
Keywords: Recurrent spontaneous abortion; Normal early pregnancy; piRNA; RNA sequencing; Expression profile
Pregnancy is a complex process, comprising fertilization, implantation, organ and tissue differentiation, and fetal growth, which is effectively controlled by a number of both maternal and fetal factors (Qu et al., 2019; Verma et al., 2019a; Verma et al., 2019b). Recurrent spontaneous abortion (RSA) refers to three or more consecutive spontaneous abortions before 20 weeks of pregnancy and affects approximately 1%–2% of couples who attempt to have a child (Coomarasamy et al., 2015; Raj and Lesley, 2006). There are many factors leading to RSA, such as genetic factors, immune and environmental factors, endocrine disorders, infection and trauma, abnormal anatomy of the reproductive organs, cervical dysfunction, age, lifestyle problems and thrombosis tendency (Pei et al., 2019; Pereza et al., 2017; van den Berg et al., 2012). The causes that are already found cannot explain all the cases. The unsatisfactory clinical efficacy of some RSA cases has become a difficult problem to be solved in the field of gynecology and obstetrics. Therefore, more factors of progression of RSA need to be revealed.
Non-coding RNAs (ncRNAs) are generated from non-coding regions that were previously considered to be junk DNA, without the potential of translation into proteins (Goodrich and Kugel, 2009). NcRNAs, such as microRNAs (miRNAs) and long ncRNAs (lncRNAs), have been identified as crucial regulators of gene expression in RSA, endometriosis, pre-eclampsia, infertility and other reproductive system diseases (Dong et al., 2014; Dong et al., 2017; Qin et al., 2016; Sober et al., 2016; Zhang et al., 2019; Zhao et al., 2018). Piwi-interacting RNAs (piRNAs) are small regulatory RNAs expressed in mammalian germ line cells and have been identified as key players in germ line development. These piRNAs molecules, typically of length 26–32 nt, associate with Piwi proteins of the Argonaute family to form the Piwi-interacting RNA complex. Studies had shown that piRNAs generally exist in clusters on chromosomes, ranging in length from 20 kb to 100 kb, and the density of piRNAs ranges from 40 to 4000. PiRNA cluster was transcribed to form a long single-stranded precursor, and then processed to form mature piRNA of about 26–32 nt (Hirakata and Siomi, 2016; Huang et al., 2017). Although piRNAs were initially considered to be only expressed in germ cells, a growing number of studies identified piRNAs are also abnormally expressed in early embryos (Cui et al., 2018; Russell et al., 2016), indicating that piRNAs may be involved in early embryos development. PiRNA pathway plays a role in animal germ cells and controls TE (Transposable Element) activity. The core of piRNA pathway is a ribonucleo-protein complex composed of a small RNA called piRNA and a protein from the Argonaute nuclease PIWI subfamily. The piRNA pathway relies on the specificity provided by piRNAs to identify TES (Transposable Elements) targets, while the effector function is provided by piwi protein. The piwi-piRNA complex silences TEs at both the transcriptional level (by attracting inhibitory chromatin to modify genomic targets) and the post-transcriptional level (by shearing the transcripts in the cytoplasm) (Lee et al., 2011; Ozata et al., 2019). Damage to piRNA pathway leads to overexpression of TEs, significantly impairs genomic structure, and always leads to germ cell death and infertility (Toth et al., 2016).
In the present study, to improve the understanding of the biological function of piRNAs in pregnancy, the differences in piRNA expression profiles between normal control tissues and RSA decidua tissues were compared using second-generation deep sequencing for small RNAs. Subsequently, the results were validated by reverse transcription quantitative polymerase chain reaction (RT-qPCR). The potential clinical utility of pregnancy associated piRNAs was also assessed by analyzing the association of piRNAs with clinic opathological features of patients with RSA. To the best of our knowledge, the present results provided, for the first time, an overall change of piRNA expression profiles in RSA and an outlook into clinical applications of piRNAs as a clinic opathological target.
In the current study, a total of 8 RSA patients and 13 normal early pregnancy (NEP) patients were recruited at the Family Planning Special Hospital of Guangdong Province between June 2016 and July 2018. Written informed consent was obtained from each RSA patient and control subject before participation in the study. None of the patients had a history of autoimmune conditions, metabolic disorders, hypertension, arterial or venous thrombosis, embryo chromosomal abnormalities, liver or kidney dysfunction, or uterine anomalies. Chromosomal abnormalities were excluded from all couples in the recurrent miscarriage group. 8 RSA patients (age 25.63 ± 4.87 years and gestational age), who had experienced at least three consecutive embryonic losses before the 12th gestational week, were recruited in the RSA group. Meanwhile, 13 clinically normal early pregnant women (age 26.46 ± 4.03 years and gestational age), which were terminated for non-medical reasons and undergoing legal abortions around the 6–12th gestational week were recruited as the control. This study was approved by the Ethics Committee of Family Planning Special Hospital of Guangdong Province [No. 2017(19)]. Chromosome analysis was performed following standard methods for preparing G-banded metaphases from cultured peripheral blood-derived lymphocytes. The chromosomal abnormalities results showed that there were 6 kinds of chromosome aneuploidy in villi of early pregnancy abortion, including trisomy 13, trisomy 16, trisomy 18, trisomy 21, trisomy 22 and X monomer. Other types of chromosome aneuploidy and chromosome terminal monomer or trisomy were also suggested. Tissues were collected immediately after operations. Tissues were divided into 50 mg in each EP tubes and store in −80°C freezer.
Small RNA library construction, sequencing and data analysis
Total RNA used for deep sequencing was extracted from decidua tissues using TRIzol reagent according to the manufacturer’s protocol (Invitrogen), respectively. The concentration of the total RNA was measured by NanoDrop (Thermo Scientific, Wilmington, DE, USA), and the RNA integrity was checked on Bioanalyzer2100 (Agilent, Santa Clara, CA). The library was constructed by using Small RNA Sample Pre Kit (Illumina) after the samples were qualified. The library was constructed by using the special structure of 3’ and 5’ ends of small RNA (complete phosphate group at 5’ end and hydroxyl group at 3’ end). The two ends of small RNA were directly attached to the adaptor with total RNA as the starting sample, and then reverse transcribed to cDNA. Subsequently, the target DNA fragments were separated by PAGE gel electrophoresis after PCR amplification, and the DNA library was recovered by gel cutting. After the construction of the library, the initial quantification was carried out by using Qubit 2.0 to dilute the library to 1 ng/μL, and then the insert size of the library was detected by Agilent 2100. After the insert size met the expectation, the effective concentration of the library was quantified accurately by RT-qPCR (the effective concentration of the library >2 nM) to ensure the quality of the library. After passing the library inspection, the different libraries were pooling according to the effective concentration and the requirement of the target offline data volume of HiSeq/MiSeq sequencing. PiRNA sequences of six species of humans, mouse, rats, drosophilas, zebra fish and platypuses were included in the piRNA Bank (http://pirnabank.ibab.ac.in). As the species to be analyzed belongs to one of these six species, we could first conduct known piRNA analysis. The analysis method was to map the known piRNA with the sequenced reads, and then it used unmap reads of AP to predict, otherwise piRNA could be predicted directly from beginning. The method was referent to the literature (Zhang et al., 2011). The new known piRNAs that could not be matched in the bank are named beginning with ‘uniq’. If the change was ≥2.0 or ≤0.5, the difference was recognized to be significant; the significance was maintained after correction for multiple testing.
Enrichment analysis of piRNA-generation gene
GO term analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of piRNA-generation genes were carried out. Gene Ontology (GO, http://www.geneontology.org/) was the international standard classification system of gene function. GO term analysis method was GO seq. This method was based on Wallenius non-central hyper-geometric distribution, which was different from ordinary HP-geometric distribution in that the probability of extracting an individual from a category was different from that of extracting an individual from another category. This difference in probability was obtained by estimating the preference to gene length, which could more accurately calculate the probability of GO term being enriched by candidate genes.
KEGG enrichment was measured by Rich factor, Q value and the number of genes enriched in this pathway. Rich factor referred to the ratio of the number of differentially expressed genes in the pathway entry to the total number of genes in all annotated genes. Q value was P value after multiple hypothesis test correction. Pathway significant enrichment could determine the main biochemical metabolic pathways and signal transduction pathways in which candidate genes participated (Kanehisa et al., 2008).
Total RNAs were exacted using QiagenReasy mini kit (Qiagen) and reverse transcribed using a Super Script II kit (Invitrogen) according to the manufacturer’s description, respectively. Real-time quantitative PCR was carried out using TB Green Premix EX Taq II (Takara) and analyzed using AB Step One Plus System (Applied Biosystems, AB, USA). The primer pairs used for piRNAs and U6 are shown in Suppl. Table 2. PCR parameters were as follows: 95°C for 20 s, followed by 40 cycles of 95°C for 5 s and 57°C for 30 s. Relative expression level was calculated as 2-(CTU6-CTgene) (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008). The piRNA level was normalized by the housekeeping gene, U6. All experiments were carried out in triplicate. Screen the differential expression of piRNAs following the criteria: P-value ≤ 0.05; fold change ≥ 2.
All values are presented as mean ± SD. Statistical comparisons between groups were analyzed by F-test followed by Student’s t-test. A P-value < 0.05 was considered to indicate a statistically significant difference. Statistical analysis was performed using SPSS software version 17.0 (SPSS, Inc., Chicago, IL, USA).
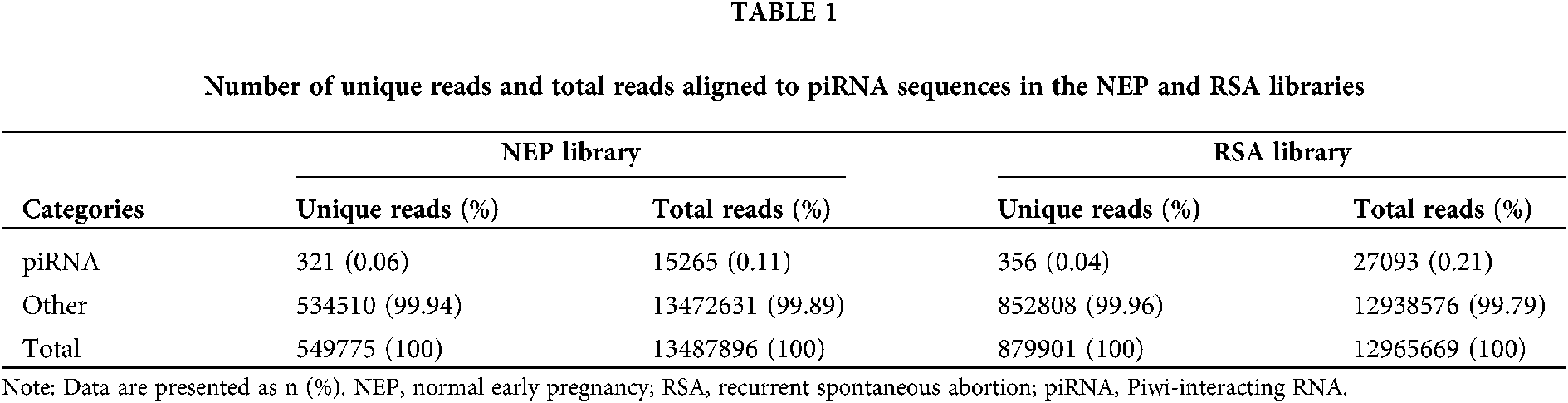
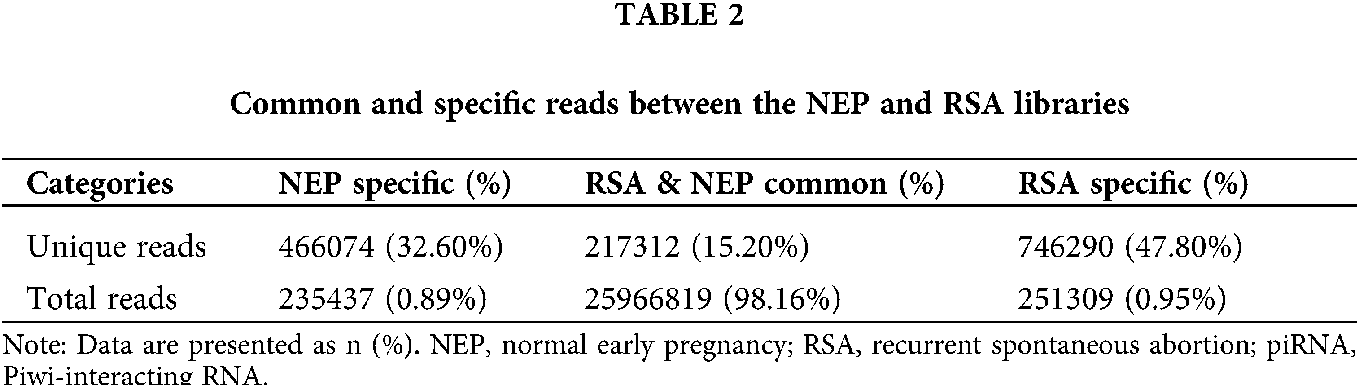
Unique and total reads analysis
To evaluate the overall differences of small RNAs in RSA and control group (NEP), the common and specific reads were analyzed between the RSA library and NEP library, including the number of unique reads (types of reads) and total reads (Table 1). Small RNA sequencing yielded 1429676 unique reads with 26453565 total reads from these two libraries. Among these unique reads, only 67,866 (15.20%) unique reads were shared by the two libraries, whereas 746290 (47.80%) and 466074 (32.60%) unique reads were specific in the RSA library and NEP library, respectively (Table 2). These specific reads were determined to not be abundant, with only 0.95% of total reads in the RSA library and 0.89% of total reads in the NEP library (Table 2). These data indicate that RSA decidua tissues and NEP decidua tissues have diverse small RNA profiles and that specific small RNAs exhibit low expression levels.
Length distribution of small RNAs
In general, the length of small RNAs ranged from 18–35 nt, and the peak of length distribution was beneficial to identify the classes of small RNAs. MiRNAs were concentrated in 21 or 22 nt, whereas piRNAs were concentrated in 26–32 nt (Williams et al., 2015). As depicted in Fig. 1, in the RSA and NEP libraries, the lengths of small RNAs were clustered in two ranges: 18–24 and 25–35 nt, and the most abundant cluster was 18–24 nt with a 22 nt peak point, the canonical length of miRNAs. The percentage of reads within the 18–24 nt cluster demonstrated no notable difference between these two libraries (57.77% vs. 58.55%), indicating that the abundance of miRNAs was not significantly changed in RSA. However, an increased 30–33 nt peak, primarily comprised of piRNAs, was determined in the RSA library compared with the NEP library (33.08% vs. 28.8%), indicating that the piRNA pathway is activated in RSA.
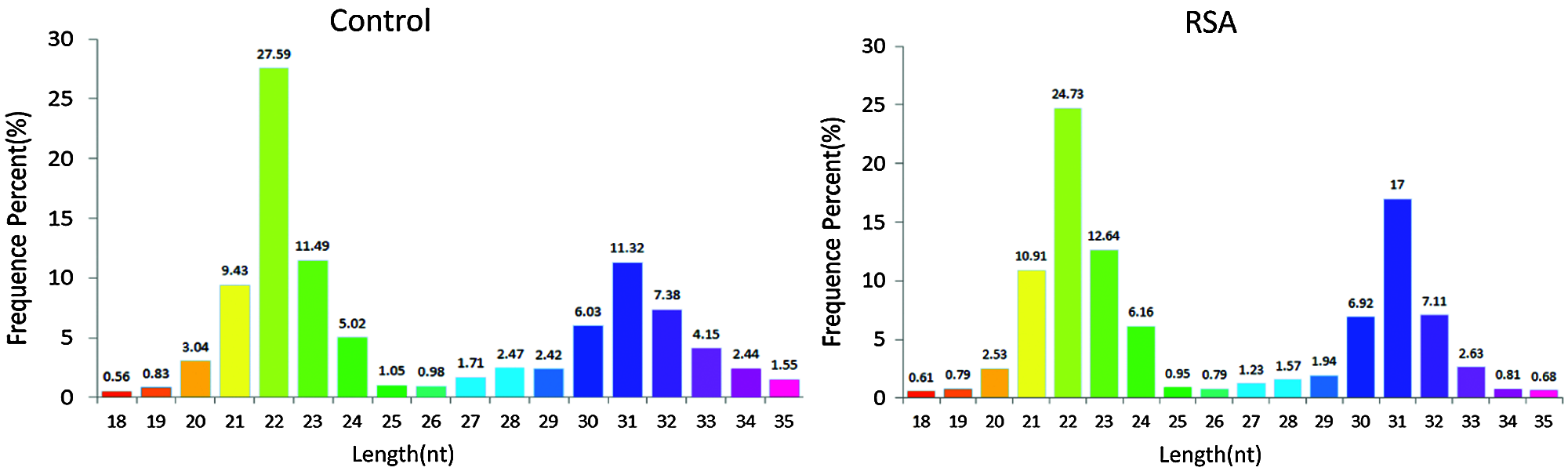
Figure 1: Length distribution of small RNAs. Bar graphs represent the percentage of small RNAs of different lengths from the RSA and NEP libraries. NEP, normal early pregnancy; RSA, recurrent spontaneous abortion.
To analyze the differentially expressed piRNAs between RSA and NEP decidua tissues, piRNAs were first annotated and screened by mapping clean reads to the piRNA database. The results demonstrated that 321 unique reads in the NEP library and 356 unique reads in the RSA library, accounting for 0.06% and 0.04%, respectively, were aligned with piRNA sequences. Although the proportion of unique piRNA reads indicated no notable changes between these two libraries, the counts of total piRNA reads increased from 15265 (0.11%) in the NEP library to 27093 (0.21%) in the RSA library (Table 1), indicating that the overall expression of piRNAs was increased in RSA tissues, compared with NEP tissues, and further implying that piRNAs may be implicated in RSA pathogenesis.
Differentially expressed piRNAs in RSA decidua tissues
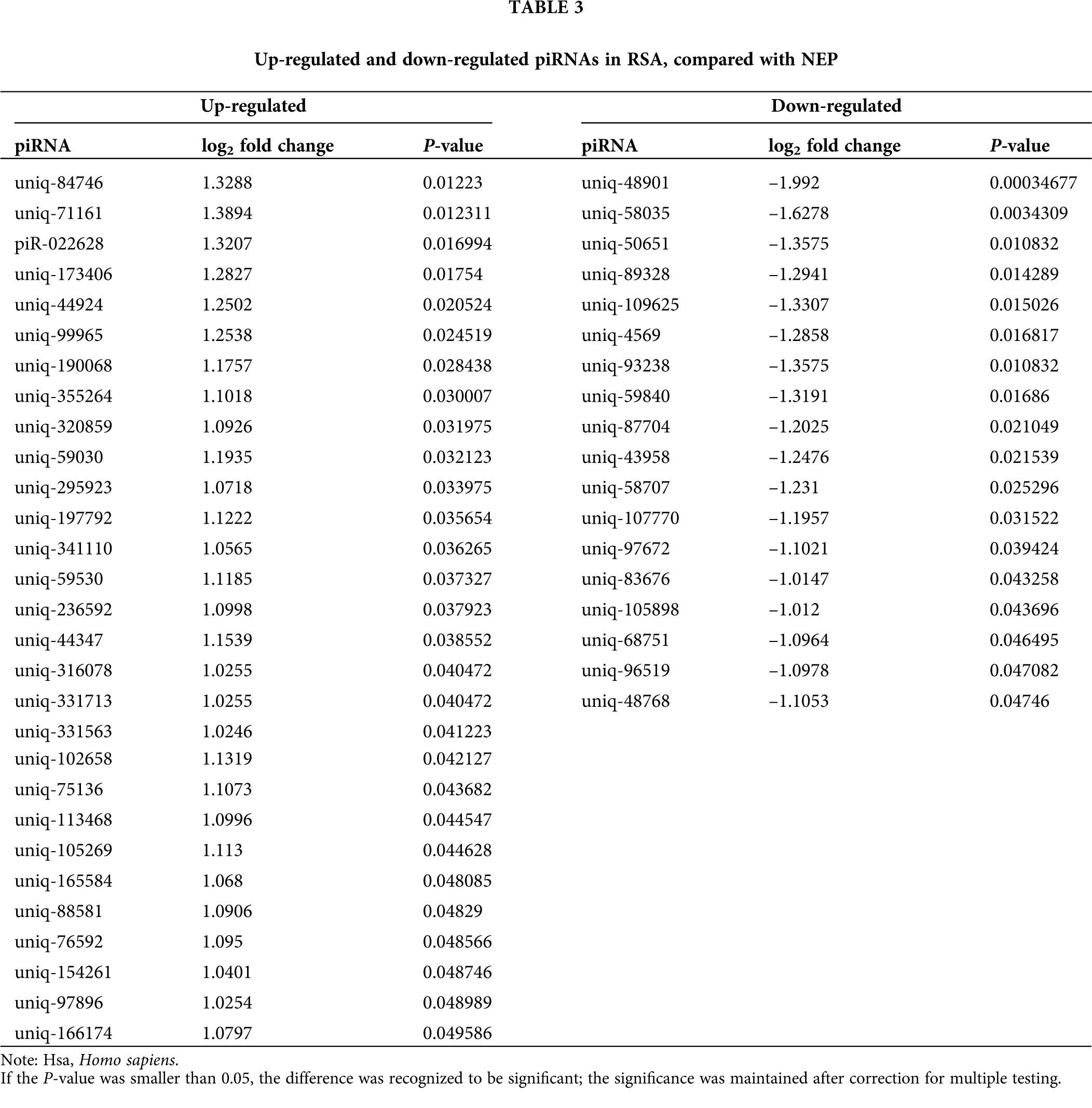
Comparison of the expression levels of piRNAs in RSA and NEP decidua tissues is beneficial for understanding the roles of piRNAs in the pathogenesis of RSA. As depicted in Fig. 2A total of 47 differentially-expressed piRNAs were identified, of which 29 piRNAs were upregulated in RSA decidua tissues, whereas 18 piRNAs were downregulated in RSA decidua tissues. These differentially expressed piRNAs are detailed in Table 3.

Figure 2: Differential expression analysis of piRNAs. (A) Hierarchical clustering represnts the relative expression levels of piRNAs in the RSA and NEP libraries by deep sequencing. The color scale indicates the relative expression levels of piRNAs in the RSA library vs. the NEP library: Red, higher expression; blue, lower expression. (B) Volcano plot of differentially expressed piRNAs. The vertical lines correspond to 2.0-fold up and 0.5-fold down, respectively, and the horizontal line represents a P-value of 0.05. The blue dots represent the piRNAs with no significant difference, and the red dots represent the significantly increased piRNAs. Green dots represent significantly decreased piRNAs. RSA, recurrent spontaneous abortion; NEP, normal early pregnancy; piRNA, Piwi-interacting RNA.
Top two up-regulated piRNAs (pi-22628 and uniq-173406) and top four down-regulated piRNAs (uniq-109625, uniq-89328, uniq-50651 and uniq-4569) were selected for further validation in 8 RSA and 13 NEP decidua tissues by RT-qPCR. The relative expression levels of these six piRNAs, calculated using the 2 –∆Cq method, are depicted in Suppl. Tables 1 and 2. Consistent with the results from small RNA sequencing, the RT-qPCR results demonstrated that the expression levels of six piRNAs were consistent in RSA decidua tissues, compared with NEP decidua tissues (Fig. 3). Notably, the fold changes from the RT-qPCR results were more than those from small RNA sequencing, which may be due to the increased sensitivity of RT-qPCR.
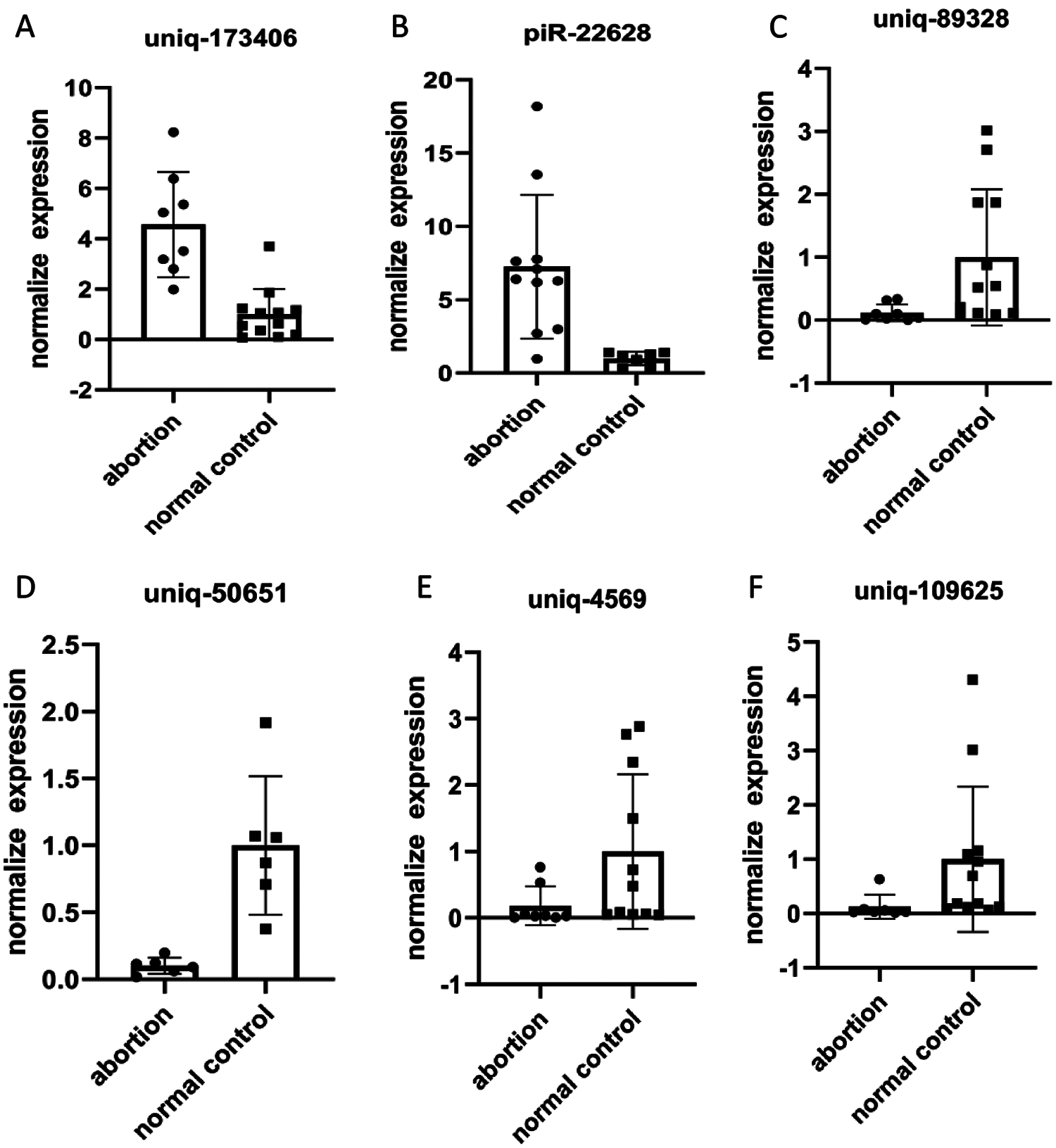
Figure 3: qRT-PCR validation of piRNAs. Differential expression results of piRNAs received by qRT-PCR (A, B, C, D, E and F). Relative piRNA expression levels are normalized against the amount of U6; the results are shown in the graphs. Uniq-109625, uniq-89328, uniq-50651 and uniq-4569 were significantly overexpression in RSA vs. NEP. Otherwise, pi-22628 and uniq-173406 were significantly under expression in RSA vs. NEP.
Enrichment analysis of piRNA-generation genes
GO term results showed that five categories had statistical significance. They were protein binding, positive regulation of biological process, anchoring junction, extracellular matrix component, and cell adhesion molecule binding (Fig. 4). Scatter map of candidate target genes was a graphical display of KEGG pathway enrichment analysis results (Fig. 5). We selected 20 pathway entries with the most significant enrichment to display in this figure (Fig. 5). In KEGG pathway enrichment analysis results, 25 genes of our sequencing results were involved in focal adhesion pathway with big rich factor and small Q-value (Q < 0.05). 15 genes were involved in extracellular matrix receptor interaction (ECM-receptor interaction) with the biggest rich factor and small Q-value (Q < 0.05). GO term results had similarity with KEGG pathway enrichment analysis results. Extracellular matrix component and cell adhesion molecule binding in GO term results were similar to focal adhesion and ECM-receptor interaction in KEGG pathway enrichment analysis results. The above results reveal that piRNAs may link to RSA through adhesion and extracellular matrix.
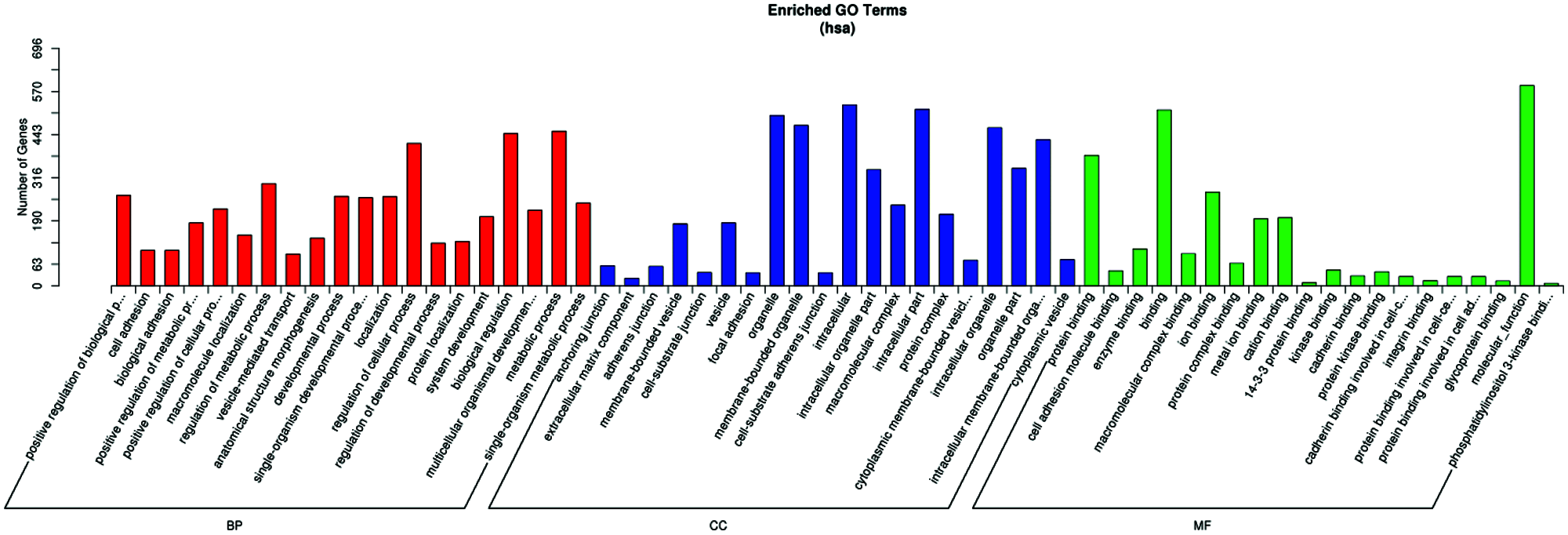
Figure 4: Result of the GO and pathway analysis of the differentially expressed piRNAs. The ontology covers three domains: biological process, cellular component, and molecular function.
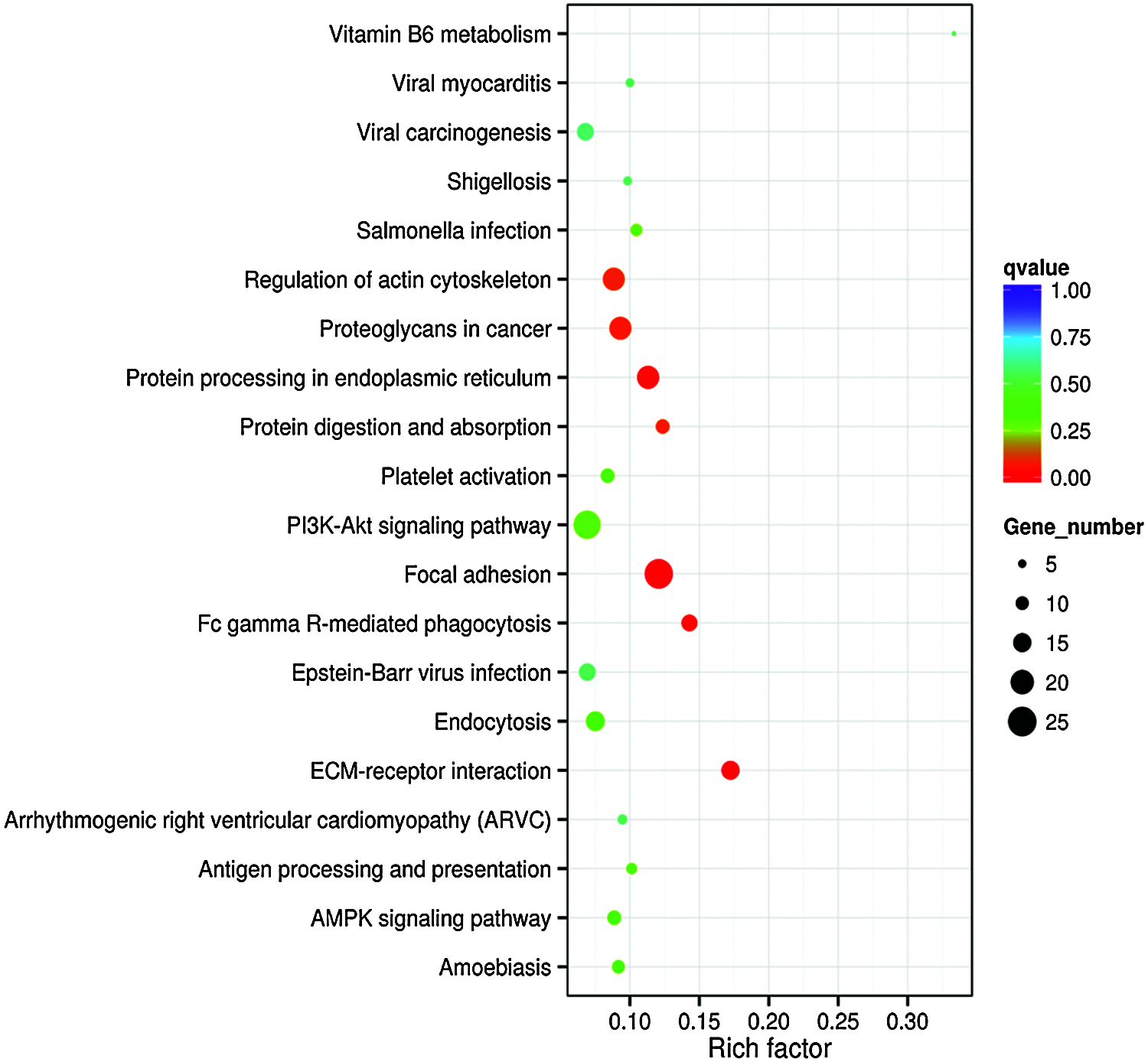
Figure 5: Enrichment scattering map of KEGG candidate target gene. The vertical axis represents the pathway name. The horizontal axis represents the Rich factor. The size of the dot represents the number of candidate target genes in the pathway. The color of the dot corresponds to different Q-value ranges.
It is well known that ncRNAs, as key molecules regulating gene expression, are widely distributed in various tissues (Kaikkonen et al., 2011; Sasaki et al., 2007; Sun et al., 2004). Aberrant expression of ncRNAs is associated with numerous human disorders (de Almeida et al., 2016; Esteller, 2011; Mestdagh et al., 2015). The rapid development of the second-generation deep sequencing technology has provided an unprecedented platform to comprehensively analyze non-coding transcriptomes as well as reveal a number of novel non-coding transcripts in various tissues, organs and disease models. Furthermore, second-generation sequencing has high sensitivity, which can avoid some minor differentially-expressed RNAs being missed. In view of the advantages of second-generation sequencing and our shortage of funds, only 3 pairs of RSA tissues and NEP decidua tissues were used for deep sequencing of small RNAs (≤40 nt). The present data demonstrated that only 15.20% of small RNAs were shared by RSA and NEP decidua tissues, indicating that the expression patterns of small RNAs in RSA and NEP decidua tissues were notably different. These observations further implied that small RNAs may be involved in RSA pathogenesis.
To date, the expression patterns of miRNAs and lncRNAs in RSA, endometriosis, pre-eclampsia, infertility and other reproductive system diseases have been extensively investigated (Dong et al., 2014; Dong et al., 2017; Qin et al., 2016; Sober et al., 2016; Zhang et al., 2019; Zhao et al., 2018). However, piRNAs, as a class of newly identified small ncRNAs, and the expression patterns of piRNAs in RSA remain largely unknown, and the field remains in its infancy. RSA is defined as three or more abortions with the same partner in the first 20 weeks of pregnancy (Cui et al., 2019). Previous reports indicated that cell adhesion has a critical role in RSA (Agostinis et al., 2012; Quenby et al., 2007; Yurdakan et al., 2008). In addition, different types of recurrent miscarriage are associated with varying patterns of adhesion molecule expression in endometrium (Quenby et al., 2007). In women with RSA, the adhesion of trophoblasts to endothelial cells is inhibited. However, the pathogenetic mechanism which inhibits the adhesion of trophoblasts to endothelial cells remains unclear in women with RSA. In our study, 29 piRNAs were up-regulated and 18 piRNAs were down-regulated in RSA, compared with normal controls. We carried out GO and KEGG pathway analysis for the target genes of piRNAs detected. The most relevant two KEGG pathways were focal adhesion and ECM-receptor interaction (extracellular matrix component, ECM), and GO analysis also indicated that piRNAs regulated cell adhesion and extracellular matrix component. Coincidentally, GO and pathway analysis demonstrated that they also regulated protein binding via target genes. Therefore, we speculate that piRNA could affect cell adhesion and extracellular matrix component, and then regulate the occurrence of RSA.
RNA sequencing of the samples of RSA patients and normal controls provided us some piRNAs that may be differently expression by data analysis. Volcanic map and heat map can be used to infer the overall distribution of differential piRNA clusters, which shows 18 down-regulated cluster and 29 up-regulated clusters with statistical significance. As we used RT-qPCR to confirm the results, a total of six piRNAs were found to be up or down-regulated in the samples of RSA patients compared with those in the samples of NP controls. These results suggested that piRNA may involve in the progression of RSA, but the evidence needs to be further confirmed. PIWI Interacting RNA (piRNA) pathway is a conservative defense system that protects the genomic integrity of animal strains from harmful transposable element (Dennis et al., 2019; Ozata et al., 2019). PiRNA biogenesis and their potential roles as part of an epigenetic network are possibly involved in cancer progression. Moreover, potential strategies based on the use of piRNAs and PIWI proteins as diagnostic and prognostic biomarkers as well as for cancer therapeutics are revealed (Assumpcao et al., 2015). Transposable Elements (TEs) have the ability to replicate and insert new genome locations. In addition, aryl-hydrocarbon receptor deficiency may have different effects on testicular and ovarian development through a process involving piRNA-related proteins, piRNAs and transposon elements (Rico-Leo et al., 2016). Moreover, a growing number of studies have shown that piRNAs and PIWI proteins, which are abnormally expressed in various cancers and other diseases, may serve as novel biomarkers and therapeutic targets for disease diagnostics and treatment (Liu et al., 2019).
Collectively, to the best of our knowledge, the present study presented, for the first time, global piRNA expression profiles in RSA and NEP decidua tissues by deep sequencing for small RNAs. Based on the small RNA sequencing data, it was determined that the overall expression levels of piRNAs in RSA decidua tissues were increased or decreased, compared with NEP decidua tissues, implying that piRNAs may be involved in RSA pathogenesis.
In conclusion, the present study is the first to explore the piRNA expression profiles in decidua tissues of RSA. Based on bioinformatics analysis, we speculate that piRNAs may be involved in RSA pathogenesis by target genes function on adhesion and extracellular matrix component. These observations will provide a theoretical basis for piRNA-targeted therapeutic strategies for RSA.
Availability of Data and Materials: The datasets used or analysed during the current study are available from the corresponding author on reasonable request.
Author Contribution: WQ designed the study, drafted the manuscript, and coordinated and participated in every part of the study. JW participated in the design of the study and helped in the study and drafting of the manuscript. LH participated in the design and statistical analyses. XL, HN, YT participated in the design of the study and performed the PCR analyses. LZ, GS and YT helped to write and revise the manuscript and supplied important comments. All of the authors read and approved the final manuscript.
Ethics Approval: This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by Ethics Committee of Family Planning Research Institute of Guangdong Province (No. 2017-19).
Funding Statement: Supported by the National Natural Science Foundation of China Grants (No. 81801523), the Natural Science Foundation of Guangdong Province (Nos. 2017A030313789, 2018A030313528, 2019A1515011984), the Science and Technology Planning Foundation of Guangzhou City (201904010017, 202102080102), Guangdong Province Medical Research Funding (No. A2021269), the Family Planning Research Institute Innovation Team of Guangdong Province grants (C-03), and the Family Planning Research Institute of Guangdong Province Grants (S2018010).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Agostinis C, Bulla R, Tisato V, De Seta F, Alberico S, Secchiero P, Zauli G (2012). Soluble TRAIL is elevated in recurrent miscarriage and inhibits the in vitro adhesion and migration of HTR8 trophoblastic cells. Human Reproduction 27: 2941–2947. DOI 10.1093/humrep/des289. [Google Scholar] [CrossRef]
Coomarasamy A, Williams H, Truchanowicz E, Seed PT, Small R et al. (2015). A randomized trial of progesterone in women with recurrent miscarriages. New England Journal of Medicine 373: 2141–2148. DOI 10.1056/NEJMoa1504927. [Google Scholar] [CrossRef]
Assumpcao CB, Calcagno DQ, Araujo TM, Santos SE, Santos AK, Riggins GJ, Burbano RR, Assumpcao PP (2015). The role of piRNA and its potential clinical implications in cancer. Epigenomics 7: 975–984. DOI 10.2217/epi.15.37. [Google Scholar] [CrossRef]
Cui L, Fang L, Shi B, Qiu S, Ye Y (2018). Spermatozoa expression of piR-31704, piR-39888, and piR-40349 and their correlation to sperm concentration and fertilization rate after ICSI. Reproductive Sciences 25: 733–739. DOI 10.1177/1933719117725822. [Google Scholar] [CrossRef]
Cui Y, He L, Yang CY, Ye Q (2019). iTRAQ and PRM-based quantitative proteomics in early recurrent spontaneous abortion: Biomarkers discovery. Clinical Proteomics 16: 36. DOI 10.1186/s12014-019-9256-y. [Google Scholar] [CrossRef]
de Almeida RA, Fraczek MG, Parker S, Delneri D, O’Keefe RT (2016). Non-coding RNAs and disease: The classical ncRNAs make a comeback. Biochemical Society Transactions 44: 1073–1078. DOI 10.1042/bst20160089. [Google Scholar] [CrossRef]
Dennis C, Brasset E, Vaury C (2019). flam piRNA precursors channel from the nucleus to the cytoplasm in a temporally regulated manner along Drosophila oogenesis. Mobile DNA 10: 28. DOI 10.1186/s13100-019-0170-7. [Google Scholar] [CrossRef]
Dong F, Zhang Y, Xia F, Yang Y, Xiong S, Jin L, Zhang J (2014). Genome-wide miRNA profiling of villus and decidua of recurrent spontaneous abortion patients. Reproduction 148: 33–41. DOI 10.1530/rep-14-0095. [Google Scholar] [CrossRef]
Dong X, Yang L, Wang H (2017). miR-520 promotes DNA-damage-induced trophoblast cell apoptosis by targeting PARP1 in recurrent spontaneous abortion (RSA). Gynecological Endocrinology: The Official Journal of the International Society of Gynecological Endocrinology 33: 274–278. DOI 10.1080/09513590.2016.1266476. [Google Scholar] [CrossRef]
Esteller M (2011). Non-coding RNAs in human disease. Nature Reviews Genetics 12: 861–874. DOI 10.1038/nrg3074. [Google Scholar] [CrossRef]
Goodrich JA, Kugel JF (2009). From bacteria to humans, chromatin to elongation, and activation to repression: The expanding roles of noncoding RNAs in regulating transcription. Critical Reviews in Biochemistry and Molecular Biology 44: 3–15. DOI 10.1080/10409230802593995. [Google Scholar] [CrossRef]
Hirakata S, Siomi MC (2016). piRNA biogenesis in the germline: From transcription of piRNA genomic sources to piRNA maturation. Biochimica et Biophysica Acta 1859: 82–92. DOI 10.1016/j.bbagrm.2015.09.002. [Google Scholar] [CrossRef]
Huang X, Fejes Toth K, Aravin AA (2017). piRNA Biogenesis in Drosophila melanogaster. Trends in Genetics 33: 882–894. DOI 10.1016/j.tig.2017.09.002. [Google Scholar] [CrossRef]
Kaikkonen MU, Lam MT, Glass CK (2011). Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovascular Research 90: 430–440. DOI 10.1093/cvr/cvr097. [Google Scholar] [CrossRef]
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Research 36: D480–484. DOI 10.1093/nar/gkm882. [Google Scholar] [CrossRef]
Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS, Kosik KS (2011). Identification of piRNAs in the central nervous system. RNA 17: 1090–1099. DOI 10.1261/rna.2565011. [Google Scholar] [CrossRef]
Liu Y, Dou M, Song X, Dong Y, Liu S, Liu H, Tao J, Li W, Yin X, Xu W (2019). The emerging role of the piRNA/piwi complex in cancer. Molecular Cancer 18: 123. DOI 10.1186/s12943-019-1052-9. [Google Scholar] [CrossRef]
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. DOI 10.1006/meth.2001.1262. [Google Scholar] [CrossRef]
Mestdagh P, Vandesompele J, Brusselle G, Vermaelen K (2015). Non-coding RNAs and respiratory disease. Thorax 70: 388–390. DOI 10.1136/thoraxjnl-2014-206404. [Google Scholar] [CrossRef]
Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, Zamore PD (2019). PIWI-interacting RNAs: Small RNAs with big functions. Nature Reviews Genetics 20: 89–108. DOI 10.1038/s41576-018-0073-3. [Google Scholar] [CrossRef]
Pei CZ, Kim YJ, Baek KH (2019). Pathogenetic factors involved in recurrent pregnancy loss from multiple aspects. Obstetrics & Gynecology Science 62: 212–223. DOI 10.5468/ogs.2019.62.4.212. [Google Scholar] [CrossRef]
Pereza N, Ostojic S, Kapovic M, Peterlin B (2017). Systematic review and meta-analysis of genetic association studies in idiopathic recurrent spontaneous abortion. Fertility and Sterility 107: 150–159. DOI 10.1016/j.fertnstert.2016.10.007. [Google Scholar] [CrossRef]
Qin W, Tang Y, Yang N, Wei X, Wu J (2016). Potential role of circulating microRNAs as a biomarker for unexplained recurrent spontaneous abortion. Fertility and Sterility 105: 1247–1254. DOI 10.1016/j.fertnstert.2016.01.028. [Google Scholar] [CrossRef]
Qu S, Wang L, Cai A, Cui S, Bai N, Liu N, Kong X (2019). Exploring the cause of early miscarriage with SNP-array analysis and karyotyping. Journal of Maternal-Fetal & Neonatal Medicine 32: 1–10. DOI 10.1080/14767058.2017.1367379. [Google Scholar] [CrossRef]
Quenby S, Anim-Somuah M, Kalumbi C, Farquharson R, Aplin JD (2007). Different types of recurrent miscarriage are associated with varying patterns of adhesion molecule expression in endometrium. Reproductive Biomedicine Online 14: 224–234. [Google Scholar]
Raj R, Lesley R (2006). Recurrent miscarriage. Lancet 368: 601–611. DOI 10.1016/s0140-6736(06)69204-0. [Google Scholar] [CrossRef]
Rico-Leo EM, Moreno-Marín N, González-Rico FJ, Barrasa E, Ortega-Ferrusola C et al. (2016). piRNA-associated proteins and retrotransposons are differentially expressed in murine testis and ovary of aryl hydrocarbon receptor deficient mice. Open Biology 6: 160186. DOI 10.1098/rsob.160186. [Google Scholar] [CrossRef]
Russell SJ, Stalker L, Gilchrist G, Backx A, Molledo G, Foster RA, LaMarre J (2016). Identification of PIWIL1 isoforms and their expression in bovine testes, oocytes, and early embryos. Biology of Reproduction 94: 75. DOI 10.1095/biolreprod.115.136721. [Google Scholar] [CrossRef]
Sasaki YT, Sano M, Ideue T, Kin T, Asai K, Hirose T (2007). Identification and characterization of human non-coding RNAs with tissue-specific expression. Biochem Biophys Res Commun 357: 991–996. DOI 10.1016/j.bbrc.2007.04.034. [Google Scholar] [CrossRef]
Schmittgen TD, Livak KJ (2008). Analyzing real-time PCR data by the comparative C(T) method. Nature Protocols 3: 1101–1108. DOI 10.1038/nprot.2008.73. [Google Scholar] [CrossRef]
Sober S, Rull K, Reiman M, Ilisson P, Mattila P, Laan M (2016). RNA sequencing of chorionic villi from recurrent pregnancy loss patients reveals impaired function of basic nuclear and cellular machinery, 6: 38439. DOI 10.1038/srep38439. [Google Scholar] [CrossRef]
Sun Y, Koo S, White N, Peralta E, Esau C, Dean NM, Perera RJ (2004). Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Research 32: e188. DOI 10.1093/nar/gnh186. [Google Scholar] [CrossRef]
Toth KF, Pezic D, Stuwe E, Webster A (2016). The piRNA pathway guards the germline genome against transposable elements. Non-coding RNA and the Reproductive System 886: 51–77. DOI 10.1007/978-94-017-7417-8_4. [Google Scholar] [CrossRef]
van den Berg MM, van Maarle MC, van Wely M, Goddijn M (2012). Genetics of early miscarriage. Biochimica Et Biophysica Acta 1822: 1951–1959. DOI 10.1016/j.bbadis.2012.07.001. [Google Scholar] [CrossRef]
Verma P, Nair RR, Singh S, Rajender S, Khanna A, Jha RK, Singh K (2019a). High level of apoa1 in blood and maternal fetal interface is associated with early miscarriage. Reproductive Sciences 26: 649–656. DOI 10.1177/1933719118783266. [Google Scholar] [CrossRef]
Verma P, Verma R, Nair RR, Budhwar S, Khanna A, Agrawal NR, Sinha R, Birendra R, Rajender S, Singh K (2019b). Altered crosstalk of estradiol and progesterone with Myeloid-derived suppressor cells and Th1/Th2 cytokines in early miscarriage is associated with early breakdown of maternal-fetal tolerance. American Journal of Reproductive Immunology 81: e13081. DOI 10.1111/aji.13081. [Google Scholar] [CrossRef]
Williams Z, Morozov P, Mihailovic A, Lin C, Puvvula PK, Juranek S, Rosenwaks Z, Tuschl T (2015). Discovery and characterization of piRNAs in the human fetal ovary. Cell Reports 13: 854–863. DOI 10.1016/j.celrep.2015.09.030. [Google Scholar] [CrossRef]
Yurdakan G, Ekem TE, Bahadir B, Gun BD, Kuzey GM, Ozdamar SO (2008). Expression of adhesion molecules in first trimester spontaneous abortions and their role in abortion pathogenesis. Acta Obstet Gynecol Scand 87: 775–782. DOI 10.1080/00016340802177412. [Google Scholar] [CrossRef]
Zhang Y, Liu H, Lv J, Xiao X, Zhu J et al. (2011). QDMR: A quantitative method for identification of differentially methylated regions by entropy. Nucleic Acids Research 39: e58. DOI 10.1093/nar/gkr053. [Google Scholar] [CrossRef]
Zhang Y, Zhou J, Li MQ, Xu J, Zhang JP, Jin LP (2019). MicroRNA-184 promotes apoptosis of trophoblast cells via targeting WIG1 and induces early spontaneous abortion. Cell Death & Disease 10: 223–223. DOI 10.1038/s41419-019-1443-2. [Google Scholar] [CrossRef]
Zhao L, Li J, Huang S (2018). Patients with unexplained recurrent spontaneous abortion show decreased levels of microrna-146a-5p in the deciduae. Annals of clinical and laboratory science 48: 177–182. [Google Scholar]

 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |