DOI:10.32604/biocell.2022.016726

| BIOCELL DOI:10.32604/biocell.2022.016726 |  |
| Article |
Downregulation of hsa_circ_0002198 inhibits keloid fibroblast activities in vitro by reducing NLRP3 inflammasome activity
1Department of Dermatology, Hainan Hospital of Traditional Chinese Medicine, Haikou, 570203, China
2Department of Oncology, Hainan Hospital of Traditional Chinese Medicine, Haikou, 570203, China
*Address correspondence to: Hui Zhang, layihui@foxmail.com
Received: 20 March 2021; Accepted: 07 July 2021
Abstract: The levels of hsa circular RNA_0002198 (hsa_circ_0002198) have been found to be significantly upregulated in keloid dermal fibroblasts. However, the functional role of hsa_circ_0002198 in keloid fibroblasts and the underlying molecular mechanism for its effects have not been reported. In this study, the levels of hsa_circ_0002198 and nucleotide-binding and oligomerization domain, leucine rich repeat and pyrin domain containing 3 (NLRP3) expression in keloid scar tissues and adjacent normal skin tissues were determined by quantitative real-time PCR and western blotting, respectively. In vitro models of keloid tissue were created by culturing primary keloid fibroblasts obtained from patients. A series of functional experiments, including CCK-8 assays, Transwell assays, and ELISA assays were performed to analyze the functional role of hsa_circ_0002198/NLRP3. Our data showed that hsa_circ_0002198 and NLRP3 were upregulated in keloid scar tissues when compared with adjacent normal tissues. Knockdown of hsa_circ_0002198 expression significantly suppressed cell proliferation, migration, and invasion, and those effects could be partially reversed by forced NLRP3 overexpression in keloid fibroblasts. At the molecular level, knockdown of hsa_circ_0002198 downregulated the levels of Col I, α-SMA, and NLRP3 proteins, as well as the levels of TGF-β, IL-1ß, and IL-33, but upregulated caspase 3 expression in keloid fibroblasts. All those effects were partially reversed after NLRP3 overexpression. In conclusion, our results suggest hsa_circ_0002198 as a potential target for treating keloid lesions.
Keywords: Keloid; Fibroblast; hsa_circ_0002198; NLRP3
A keloid scar is regarded as a chronic inflammatory process characterized by an accumulation of fibroblasts and excessive deposition of extracellular matrix (ECM) components, and especially collagen that is deposited due to an abnormal wound healing process (Abergel et al., 1985; Sidgwick and Bayat, 2012; Vincent et al., 2008). The current treatment options for keloids include steroid injections, steroid tape, and surgery with postoperative irradiation; however, there is no consensus concerning a generally accepted treatment regimen, and the cure rate is <30% (Arons, 2008; Ogawa et al., 2007; van Leeuwen et al., 2015). Thus, it is important to better understand the molecular mechanisms underlying keloid pathogenesis in order to improve treatment outcomes.
Several gene regulators, including cytokines and chemokines, have been previously reported to be involved in the initiation and progression of keloid development (Lim et al., 2019). Multifunctional cytokine transforming growth factor-β (TGF-β) is considered as one of the most important cytokines associated with keloid formation, as it helps to regulate cellular growth and differentiation, angiogenesis, adhesion, chemotaxis, and ECM production (Lee et al., 1999; Yang et al., 2014). Related studies have indicated that the wound healing process is initiated by inflammasome activation induced by noxious stimuli in human keratinocytes and immune cells, of which the nucleotide-binding and oligomerization domain, leucine rich repeat and pyrin domain containing 3 (NLRP3) inflammasome is the master regulator of inflammatory responses (Shao et al., 2015; Vinaik et al., 2020a; Zambetti et al., 2012). As a danger-sensing platform, NLRP3 has been shown to facilitate cleaved caspase-1 processing and promote the release of cytokines (TGF-β, IL-1β, and IL-18) in several models of tissue repair (Mack, 2018; Vinaik et al., 2020b). Nevertheless, the role played by NLRP3 inflammasome-mediated inflammation in the pathogenesis of keloids remains largely unclear.
Circular RNAs (circRNAs) are non-coding RNAs characterized by a covalently closed loop chemical structure, which is more stable than the linear structure of coding RNAs (Memczak et al., 2013). CircRNAs are produced by precursor mRNA back-splicing, are widely expressed in mammals, and involved in various diseases (Han et al., 2017; Szabo and Salzman, 2016; Wang et al., 2016; Xu et al., 2015). Recently, Wang et al. (2019a) and Shi et al. (2020) performed bioinformatics analyses which identified differentially expressed circRNAs associated with keloid formation. In agreement with a data analysis performed by Zhang et al. (2020), hsa_circ_0002198 has been identified by bioinformatics tools and verified to be significantly upregulated in keloid dermal fibroblasts. However, the mechanism by which hsa_circ_0002198 regulates the formation of keloid scars, and the association between hsa_circ_0002198 and NLRP3, have not been reported.
In the present study, we first determined the levels of hsa_circ_0002198 and NLRP3 expression in keloid scars and adjacent normal skin tissues. We subsequently performed loss-of-function and rescue studies to explore the roles of hsa_circ_0002198 and NLRP3 in the proliferation, migration, and invasion of human keloid fibroblasts. Our data suggest the existence of a novel signaling mechanism that might be associated with keloid pathogenesis.
Clinical samples and cell culture
Specimens of keloid scar tissue (N = 20) and adjacent normal skin tissue (N = 20) were collected from the chest or face of patients who visited the Department of Dermatology at the Hainan Hospital of Traditional Chinese Medicine. The patients were 22 to 35 years old and did not have any systemic diseases. All patients provided their informed consent for study participation, and the study protocol was approved by the Ethics Committee of Hainan Hospital of Traditional Chinese Medicine.
Primary human keloid fibroblasts were isolated from keloid tissues as previously described (Lei et al., 2019). In brief, we preserved the dermal layer of fibrous tissue by cutting away the epidermis and fat from excised human keloid scar tissues. After cutting the preserved tissue specimens into smaller pieces, the dermal layer of fibrous tissue in each specimen was digested for 6 h in DMEM medium containing 0.25% collagenase Type I (Sigma-Aldrich, C0130, St. Louis, MO, USA). Next, the digested tissue was centrifuged, and the precipitates were cultured in DMEM contained 15% fetal bovine serum (Gibco, 10099141C, Grand Island, NY, USA) at 37°C in a humidified atmosphere containing 5% CO2.
Two small interfering RNAs targeting hsa_circ_0002198 via different sequences (siRNA#1 and siRNA#2), an NLRP3 overexpression plasmid, and the corresponding negative control (NC), were synthesized by Gene Pharma (Shanghai, China). Knockdown of hsa_circ_0002198 in keloid fibroblasts was achieved by transfecting the fibroblasts with siRNA#1 and siRNA#2. In rescue experiments, the keloid fibroblasts were co-transfected with a pcDNA3.1 empty vector (NC and siRNA#1 group) or siRNA#1 with/without NLRP3 (siRNA#1+NLRP3 and siRNA#1 group). All transfections were performed using by using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA), a 50 nM concentration of each siRNA, 2.5 μg of plasmid, and a transfection duration of 48 h.
Total RNA was isolated from tissues or cells using TRIzol reagent (Invitrogen, USA) and reverse transcription was performed using a Prime Script™ RT kit (Takara, Dalian, China) according to the manufacturer’s instructions. Cytoplasmic RNA and nuclear RNA were isolated using a Cytoplasmic & Nuclear RNA Purification Kit (BioBeck Corporation, Canada). Briefly, cells were lysed in lysis buffer J solution; after which, the lysis buffer was centrifuged to produce a cell supernatant (cytoplasmic fraction) and pellet (nuclear fraction). Next, SK buffer was added to the RNA fractions, followed by addition of ethanol reagent. The mixture was then transferred onto a filtration column and the filtered liquor was discarded after centrifugation. The RNA remaining in the filtration column was washed using washing solution followed by centrifugation. Finally, RNA was collected by adding elution buffer and performing a subsequent centrifugation. The filtered liquor was retained and stored at −20°C for further analysis. A 1 µg sample of total RNA was analyzed for its levels of hsa_circ_0002198, PDE7B, and NLRP3 RNA by using SYBR green PCR Master Mix (Toyobo, Japan) on an ABI 7500 fast PCR System (Foster City, CA, USA) using the following conditions: pre-incubation at 95°C for 5 min, followed by 40 cycles of amplification at 95°C for 10 s, 60°C for 20 s, and a final elongation at 72°C for 15 s. Actinomycin D (AAT Bioquest, AAT-17505, Sunnyvale, CA, USA) was used to block transcription when detecting the half-life of RNA. The PCR primer sequences used were as follows: 5’-GTATCACAGGCTGCTGTAGCTCC-3’ and 5’-GCTCTTCGGTGCAGCTACTGG-3’ for hsa_circ_0002198; 5’-CTCGCTTCGGCAGCACA-3’ and 5’-AACGCTTCACGAATTTGCGT-3’ for GAPDH. Relative gene expression was calculated by 2−ΔΔCt method. 5’-TCTCATGCTGCCTGTTCTCA-3’ and 5’-CAAGGAGATGTCGAAGCAGC-3’ for NLRP3; 5’-GGTGTGGCGAAATCTTGTTT-3’ and 5’-TTTTCTTGGTGCCAATCTCC-3’ for PDE7B.
Fluorescence in situ hybridization (FISH)
The FISH assay was performed to detect the subcellular localization of hsa_circ_0002198 with a RiboTM Fluor 488-labeled probe (RiboBio, Guangzhou, China) as described in the manufacturer’s instructions. In brief, cells were inoculated in a 24-well plate at a density of 6 × 104 cells/well, and then fixed with 4% paraformaldehyde for 20 min at room temperature. Next, the cells were incubated overnight with pre-hybridization solution and then washed 3 times with PBST; after which, the nuclei were stained with 4’, 6-diamidino-2-phenylindole (DAPI, Invitrogen) for 5 min. Finally, the cells were observed and photographed under a fluorescence microscope (Olympus, Japan).
The proliferative ability of cells was examined using a Cell Counting kit-8 (CCK-8, Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Briefly, cells from different groups were seeded into 96-well plates (3 × 103 cells per well) and cultured in complete medium. At 0, 24, 48, and 72 h, respectively, 10 μL of sterile CCK-8 solution was added to each well. After another 2 h incubation at 37°C, the colorimetric absorbance of each well at 450 nm was recorded using a microplate reader (Thermo Labsystems, Helsinki, Finland).
The migration and invasion abilities of keloid fibroblasts were determined using 24-well Transwell chambers (Corning, Corning, New York, USA) that had either been pre-coated or not pre-coated with 50 μL (120 μg/mL) of Matrigel, respectively. Briefly, cells were resuspended in FBS-free medium and then seeded in the upper chamber, and 600 µL of culture medium with 20% FBS (Gibco, 10099141C) was added to the lower chamber. After a 24 h incubation, the cells that had penetrated through the membranes were fixed with 4% paraformaldehyde for 20 min and then stained with 0.5% crystal violet. The numbers of migrated or invasive cells were counted under a microscope at × 200 magnification.
Total cellular protein was extracted from tissue samples or cells using RIPA lysis buffer (Beyotime, P0013E, Shanghai, China) containing a protease inhibitor cocktail (Sigma-Aldrich, P9599). The protein concentration in each extract was determined using the bicinchoninic acid (BCA) method. An equal amount of protein from each extract was subjected to 10% SDS-PAGE, and the separated protein bands were transferred onto PVDF membranes, which were subsequently blocked with 5% fat-free milk for 2 h at room temperature. Next, the membranes were incubated with primary antibodies against NLRP3 (Boster, Pleasanton, CA, USA, BA3677), Collagen I (Abcam, Cambridge, UK, ab260043), α-SMA (Abcam, ab124964), Caspase-3 (Abcam, ab32042), and GAPDH (Servicebio, China, GB11002) overnight at 4°C. Next, the membranes were incubated with a horseradish peroxidase-labeled secondary antibody (Abcam, ab150077) for 2 h at room temperature. The immunostained protein bands were visualized with enhanced chemiluminescence reagent (ECL, Bio-Rad, Hercules, CA, USA).
For IF staining, keloid fibroblasts were plated onto glass coverslips and cultured to 80% confluence; after which, they were incubated in complete medium overnight. Next, the fibroblasts were rinsed twice with PBC and fixed with 4% formaldehyde solution for 30 min. The cells were then blocked with 5% bovine serum albumin (BSA, Sigma-Aldrich) for 30 min at room temperature and subsequently incubated with a primary antibody against NLRP3 (Boster, BA3677, Cambridge, UK) overnight at 4°C, followed by incubation with an Alexa-conjugated secondary antibody (Abcam, ab150077) for 2 h. The nucleus was stained with DAPI (Invitrogen), and images were obtained with an LSM510 confocal microscope (Molecular Devices, Sunnyvale, CA, USA).
Enzyme-linked immunosorbent assay
The levels of cytokines (TGF-β, IL-1ß, and IL-33) in the supernatants of keloid fibroblasts from different groups were quantified using commercially available enzyme ELISA kits (R&D Systems, DB100B, DLB50, D3300B, Minneapolis, Minnesota, USA).
All statistical analyses were performed using Graphpad Prism version 6.0 software (GraphPad Software Inc., San Diego, CA, USA), and results are expressed as a mean value ± standard deviation. Differences among multiple groups were assessed by one-way analysis of variance (ANOVA), followed by the Dunnett’s post-test or Turkey’s post-hoc test. P-values < 0.05 were considered statistically significant.
Hsa_circ_0002198 and NLRP3 were highly expressed in keloid scar tissues
The expression levels of hsa_circ_0002198 were significantly higher in samples of keloid scar tissue than in samples of adjacent normal tissue (Fig. 1A). Furthermore, the levels of NLRP3 RNA and protein expression were obviously elevated in the scar tissues when compared with the paired control tissues (Figs. 1B and 1C). These preliminary data suggested that elevated hsa_circ_0002198 and NLRP3 levels might be involved in the regulation of scar hyperplasia.
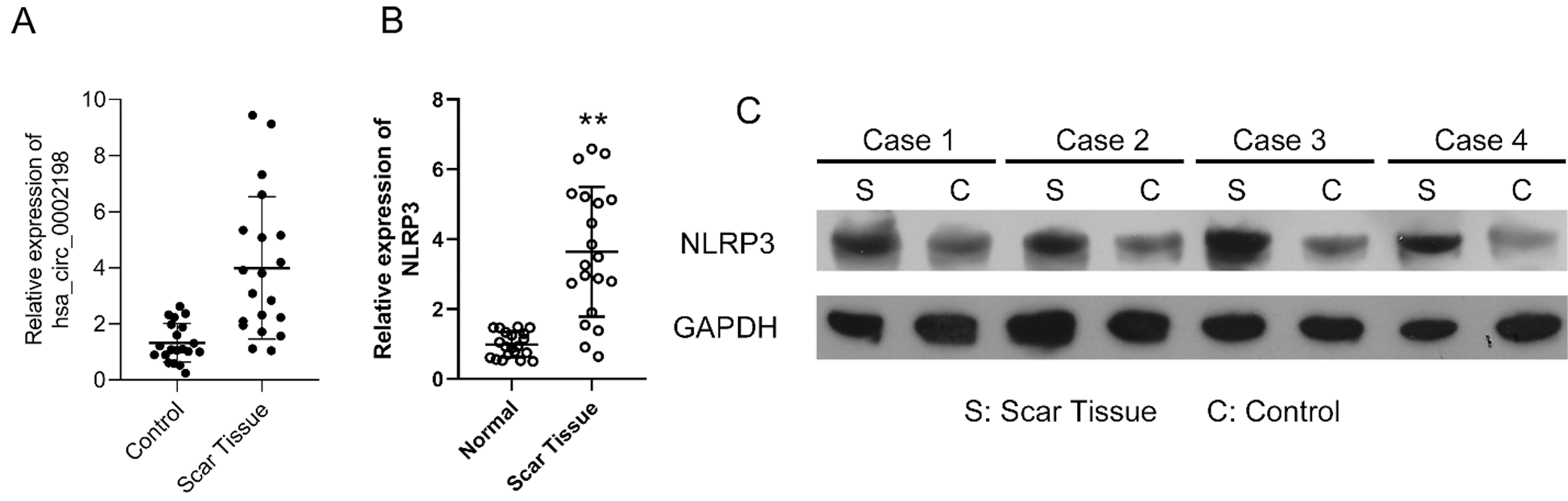
Figure 1: Expression levels of hsa_circ_0002198 and NLRP3 in keloid scar tissues. (A) Hsa_circ_0002198 expression was determined by quantitative real-time PCR analysis. (B and C) A representative analysis of NLRP3 mRNA levels (B) and NLRP3 protein expression (C) in scar tissues and control tissues from four keloid scar patients.
Identification of hsa_circ_0002198
Hsa_circ_0002198 was identified by a half-life comparison and use of a second-generation sequencing method. As shown in Fig. 2A, when measured under the influence of actinomycetes, the half-life of hsa_circ_0002198 was longer than that of its host gene (linear mRNA PDE7B), reflecting the stability of hsa_circ_0002198. Results from second-generation sequencing identified the splice sites (TCAG/GAAA) for hsa_circ_0002198 (Fig. 2B). Further detection showed that hsa_circ_0002198 mainly distributed in cytoplasm (Cy) (Fig. 2C). Moreover, the subcellular localization of hsa_circ_0002198 in cells was further explored using the FISH assay. As depicted in Fig. 2D, hsa_circ_0002198 was mainly expressed in the cytoplasm of keloid fibroblasts rather than in the nucleus. While, we also found that the signal in normal skin fibroblasts was weaker than in keloid fibroblasts (Fig. 2D).
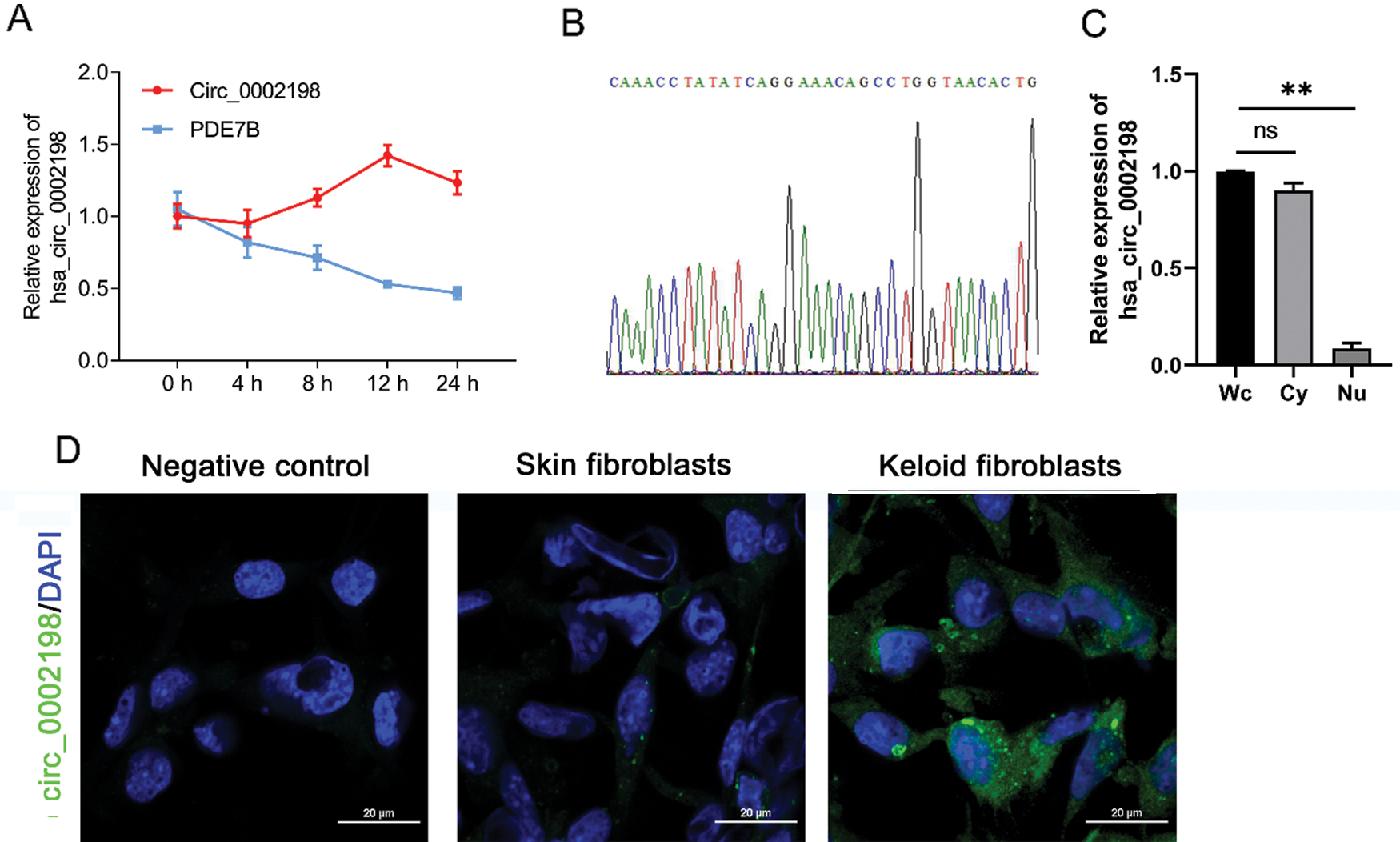
Figure 2: Identification of hsa_circ_0002198. (A) PDE7B mRNA and hsa_circ_0002198 were detected by a half-life comparison. (B) Second-generation sequencing identified the splice sites (TCAG/GAAA) for hsa_circ_0002198. (C) Hsa_circ_0002198 expression in the cytoplasm and nucleus. (D) The localization of hsa_circ_0002198 was detected by an RNA FISH assay. Scale bars: 20 μm. Abbreviations: Wc, whole cell; Cy, cytoplasm; Nu, Nucleus; NS, non-significant; **P < 0.01.
Hsa_circ_0002198 knockdown suppressed cell proliferation, motility, and inflammation in human keloid fibroblasts
To investigate the biological function of hsa_circ_0002198 in scar hyperplasia in vitro, we first established a hsa_circ_0002198 knockdown system by using si-circRNA in keloid fibroblasts. CCK-8 assays showed that knockdown of hsa_circ_0002198 by siRNA#1 or siRNA#2 transfection significantly suppressed the proliferation of keloid fibroblasts (Fig. 3A). Subsequently, results from Transwell assays revealed that the numbers of migratory (Fig. 3B) and invasive (Fig. 3C) keloid fibroblast cells were notably decreased in the siRNA#1 and siRNA#2 transfection group when compared with the NC or blank group. At the molecular level, TGF-β expression was significantly downregulated after knockdown of hsa_circ_0002198 by siRNA#1 or siRNA#2 (Fig. 3D). Moreover, the levels of NLRP3 mRNA expression were downregulated after hsa_circ_0002198 knockdown (Fig. 3E). A western blot analysis showed that knockdown of hsa_circ_0002198 downregulated the levels of Col I, α-SMA, and NLRP3 protein expression, and upregulated caspase 3 expression in keloid fibroblasts (Fig. 3F). The changes in NLRP3 protein expression in keloid fibroblasts were further confirmed by IF assays (Fig. 3G). In addition, ELISA assays showed that the levels of pro-inflammatory cytokines (IL-1ß and IL-33) were decreased following hsa_circ_0002198 knockdown (Figs. 3H and 3I). Finally, quantitative real-time PCR confirmed the downregulation of hsa_circ_0002198 after siRNA#1 or siRNA#2 transfection (Fig. 3J).
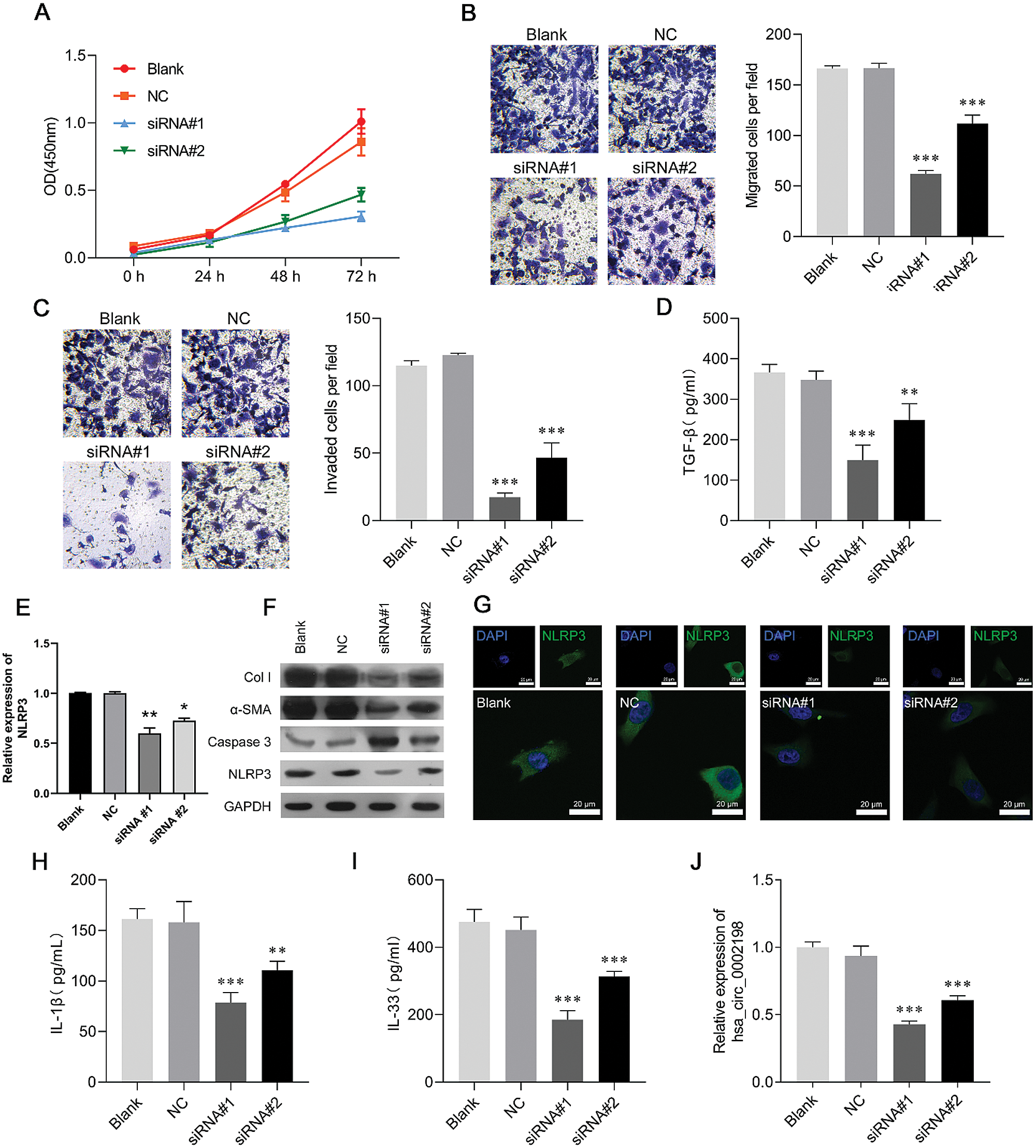
Figure 3: Hsa_circ_0002198 knockdown suppressed cell proliferation, motility, and inflammation in human keloid fibroblasts. Keloid fibroblasts were transfected with si-RNA#1 or siRNA#2, respectively. (A) CCK-8 assays were performed to analyze proliferation of the transfected cells. (B) The migration and (C) invasion capabilities of the above transfected cells were determined by Transwell assays. (D) The levels of cytokines TGF-β were measured by ELISA. (E) NLRP3 expression was detected by qRT-PCR. (F) The levels of collagen 1, α-SMA, caspase 3, and NLRP3 protein expression were detected by western blotting. (G) Immunofluorescence images of NLRP3 in the cytoplasm and nucleus of keloid fibroblasts. (H–I) The levels of pro-inflammatory cytokines (IL-1ß and IL-33) were analyzed by ELISA. (J) Hsa_circ_0002198 expression in the above transfected cells was determined by real-time PCR analysis. Data are expressed as mean ± SD. **P < 0.01, ***P < 0.001, compared with NC.
Restoration of NLRP3 partially reversed the effects of hsa_circ_0002198 on human keloid fibroblast cell proliferation, motility, and inflammation
As NLRP3 was upregulated in scar tissues and could be suppressed by knockdown of hsa_circ_0002198, it seemed reasonable that NLRP3 might be a downstream regulator in hsa_circ_0002198 knockdown cells that attenuates cell functions in keloid fibroblasts. To help validate this expectation, keloid fibroblasts were co-transfected with siRNA#1 and the NLRP3 overexpression plasmid. A subsequent series of functional experiments showed that overexpression of NLRP3 partially reversed the suppressive effects of hsa_circ_0002198 on keloid fibroblast cell proliferation (Fig. 4A), migration, (Fig. 4B), and invasion (Fig. 4C). The decreases in TGF-β levels, induced by hsa_circ_0002198 knockdown, were recovered by NLRP3 overexpression (Fig. 4D). Expression of NLRP3 mRNA was suppressed by hsa_circ_0002198 siRNA, while that loss of expression was recovered after NLRP3 was overexpressed (Fig. 4E). As expected, NLRP3 overexpression partially reversed the effects of hsa_circ_0002198 knockdown on Col I, α-SMA, caspase-3, and NLRP3 expression in keloid fibroblasts, as reflected by western blot analyses (Fig. 4F) and IF staining of NLRP3 (Fig. 4G). In addition, restoration of NLRP3 expression attenuated the effects of hsa_circ_0002198 knockdown on the levels of IL-1ß (Fig. 4H) and IL-33 (Fig. 4I). However, NLRP3 overexpression did not affect hsa_circ_0002198 expression in the siRNA#1 transfection group (Fig. 4J).
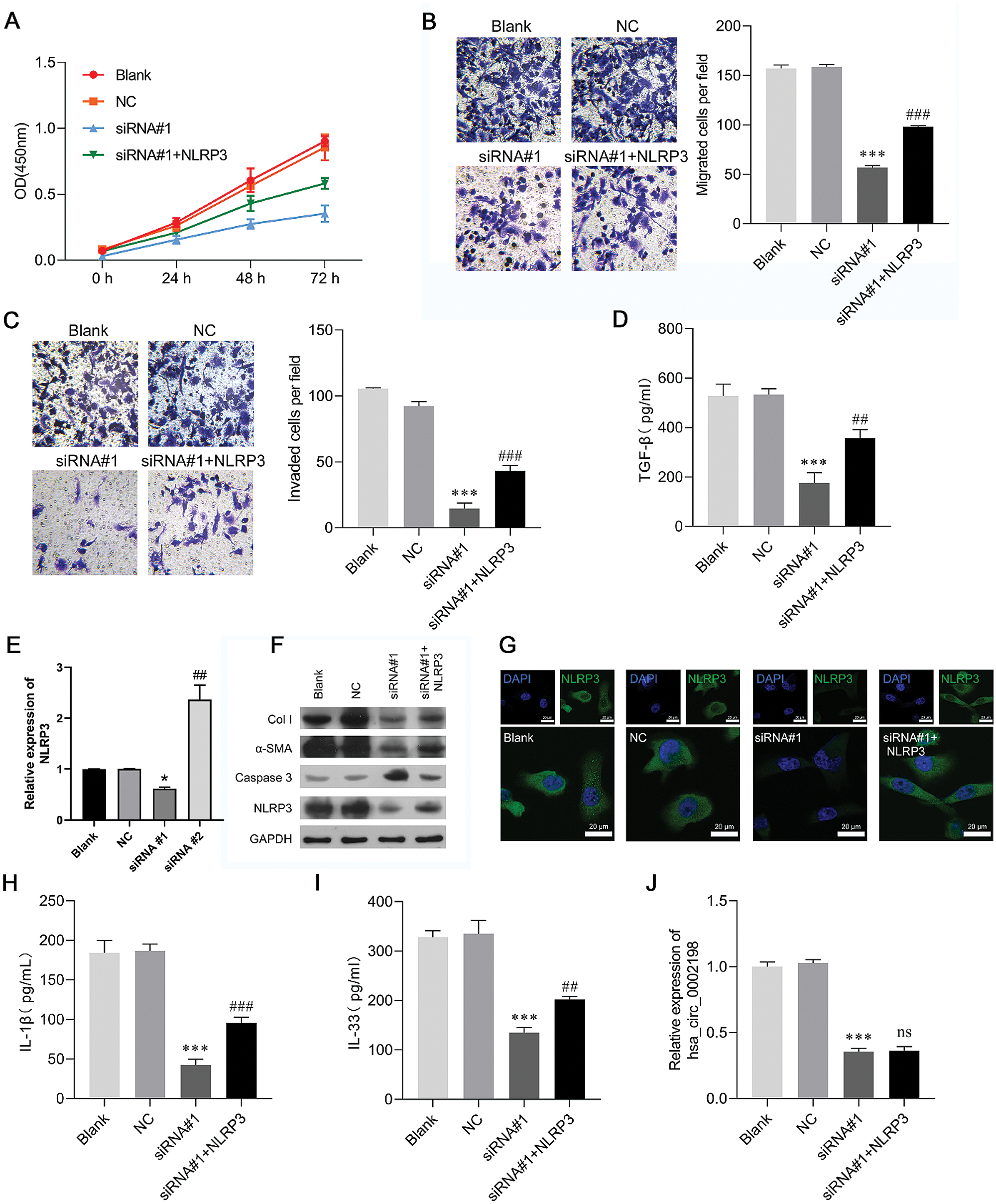
Figure 4: Restoration of NLRP3 expression partially reversed the effects of hsa_circ_0002198 on human keloid fibroblast cell proliferation, motility, and inflammation. Keloid fibroblasts were co-transfected with si-RNA#1 and the NLRP3 overexpression plasmid. (A) CCK-8 assays were performed to analyze the proliferation of transfected cells. (B) The migration and (C) invasion capabilities of the transfected cells were determined by Transwell assays. (D) The levels of cytokines TGF-β were measured by ELISA. (E) NLRP3 expression was detected by qRT-PCR. (F) The levels of Collagen 1, α-SMA, Caspase 3, and NLRP3 protein expression were detected by western blotting. (G) Immunofluorescence images of NLRP3 in the cytoplasm and nucleus of keloid fibroblasts. (H–I) The levels of pro-inflammatory cytokines (IL-1ß and IL-33) were analyzed by ELISA. (J) Hsa_circ_0002198 expression in transfected cells was determined by quantitative real-time PCR analysis. Data are expressed as mean ± SD. **P < 0.01, ***P < 0.001, compared with NC or siRNA#1.
Here, we observed that hsa_circ_0002198 expression was upregulated in keloid scar tissues when compared with adjacent normal tissues. This finding was in agreement with data reported by Zhang et al. (2020), who performed an RNA-Seq data analysis showing that hsa_circ_0002198 expression was significantly elevated in human keloid dermal fibroblasts when compared with normal dermal fibroblasts. However, our current study is the first explore the cellular functions of has_circ_0002198 in keloid fibroblasts. In this study, results from loss-of-function assays indicated that knockdown of hsa_circ_0002198 significantly suppressed the proliferation, migration, and invasion of keloid fibroblasts. Similarly, a previous study reported that circ_101238 knockdown inhibited growth, and promoted the apoptosis of keloid fibroblasts (Yang et al., 2020).
Non-coding RNAs widely participate in mediating the process of keloid formation. LncRNA HOXA11-AS was shown to regulate the progression of keloids via miR-124-3p/Smad5 signaling (Jin et al., 2019). Other non-coding RNAs, including miR-152-3p (Wang et al., 2019b), miR-21 (Wu et al., 2019), and miR-203 (Shi et al., 2018) are differently expressed in keloid tissues and contribute to the formation of keloids. In our study, we found that knockdown of hsa_circ_0002198 downregulated the levels of TGF-β, Col I, α-SMA, and NLRP3 proteins, upregulated caspase 3 expression, and reduced the levels of pro-inflammatory cytokines (IL-1ß and IL-33) in keloid fibroblasts. Fibroblasts are the primary effector cells of keloid disease tissue, where they display some typical pathological features (excessive proliferation, disordered apoptosis, and secretion of large amounts of ECM) (Mofikoya et al., 2007). Thus, it is not hard to understand that elevated pro-apoptotic caspase-3 levels induced by knockdown of hsa_circ_0002198 were associated with suppressed cell proliferation. An abnormally high level of TGF-β signaling has been shown to be required for the initiation and progression of keloids (Zhu et al., 2016). By up-regulating α-smooth muscle actin (α-SMA) and collagen 1 as the major components of ECM (He et al., 2015), TGF-β1 stimulates normal dermal fibroblasts to differentiate into myofibroblasts (Varga and Abraham, 2007). Thus, we confirmed that knockdown of hsa_circ_0002198 could inhibit the secretion of large amounts of ECM in keloid fibroblasts. In addition, a keloid scar is currently considered to be a chronic inflammatory process rather than a benign skin tumor, which is also defined as a highly inflamed, hyperproliferative pathological scar (Da Cunha Colombo Tiveron et al., 2018). In this study, knockdown of hsa_circ_0002198 decreased the levels of pro-inflammatory cytokines (IL-1ß and IL-33), suggesting that hsa_circ_0002198 silencing attenuates the chronic inflammatory process associated with keloid inflammatory etiopathogenesis.
To further confirm the regulatory effect of hsa_circ_0002198 on inflammasome activation, we determined the expression of NLRP3 and explored its relationship with hsa_circ_0002198. As expected, our data showed that NLRP3 protein levels were upregulated in keloid scar tissues when compared with adjacent normal tissues. Moreover, in vitro experiments demonstrated that overexpression of NLRP3 could partially reverse the effects of hsa_circ_0002198 on keloid fibroblast cell proliferation, migration, and invasion, as well as the levels of TGF-β, Col I, α-SMA, and caspase 3 proteins in human keloid fibroblasts. A recent study by Lee et al. (2020) not only showed that NLRP3 inflammasomes are activated in keloid fibroblasts and involved in chronic inflammatory processes, but also that NLRP3-dependent IL-1β release may aggravate tissue damage, prolong inflammatory responses, and adversely affect remodeling by inducing continuous myofibroblast differentiation in keloids (Lee et al., 2020). Accumulated evidence has elucidated how circRNAs help to regulate NLRP3 inflammasomes in various diseases, such as depression (Zhang et al., 2019), preeclampsia (Li et al., 2020b), diabetes mellitus (Cheng et al., 2019), and traumatic brain injury (Li et al., 2020a). Here, we demonstrated that hsa_circ_0002198 regulates NLRP3 inflammasomes that are involved in the pathogenesis of keloids.
In conclusion, our data showed that the levels of hsa_circ_0002198 and NLRP3 expression were both significantly upregulated in keloid scar tissues when compared with adjacent normal tissues. Furthermore, we demonstrated that knockdown of hsa_circ_0002198 suppressed the proliferation, migration, invasion, inflammation, and ECM components of keloid fibroblasts by downregulating NLRP3 expression. Taken together, our data suggest hsa_circ_0002198 as a novel target for treating patients with keloids. However, the details mechanism of hsa_circ_0002198 in regulation NLRP3 was not investigated in this study, and it need further exploration.
Availability of Data and Materials: The datasets used or analyzed during the current study are available from the corresponding authors on reasonable request.
Authors’ Contribution: The authors confirm contribution to the paper as follows: study conception and design: H.Z., L.F.; data collection: J.J., X.C., F.Z.; analysis and interpretation of results: H.Z., J.J.; draft manuscript preparation: H.Z., L.F. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: All patients provided their informed consent for study participation, and the study protocol was approved by the Ethics Committee of Hainan Hospital of Traditional Chinese Medicine (HNSZYY-2020-LL-016).
Funding Statement: This study was supported by The Natural Science Foundation of Hainan Province (Project No. 817338); Hainan Province in 2019–2021 Provincial Key Subject Construction Project of Traditional Chinese Medicine (The Oncology Department) (No. S100111.401).
Conflicts of Interest: The authors declare that they have no competing interests.
Abergel RP, Pizzurro D, Meeker CA, Lask G, Matsuoka LY et al. (1985). Biochemical composition of the connective tissue in keloids and analysis of collagen metabolism in keloid fibroblast cultures. Journal of Investigative Dermatology 84: 384–390. [Google Scholar]
Arons JA (2008). The results of surgical excision and adjuvant irradiation for therapy-resistant keloids: A prospective clinical outcome study. Plastic and Reconstructive Surgery 121: 685–686. [Google Scholar]
Cheng J, Liu Q, Hu N, Zheng F, Zhang X et al. (2019). Downregulation of hsa_circ_0068087 ameliorates TLR4/NF-κB/NLRP3 inflammasome-mediated inflammation and endothelial cell dysfunction in high glucose conditioned by sponging miR-197. Gene 709: 1–7. [Google Scholar]
da Cunha Colombo Tiveron LR, da Silva IR, da Silva MV, Peixoto AB, Rodrigues DBR, Rodrigues VJr (2018). High in situ mRNA levels of IL-22, TFG-beta, and ARG-1 in keloid scars. Immunobiology 223: 812–817. [Google Scholar]
Han D, Li J, Wang H, Su X, Hou J et al. (2017). Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 66: 1151. [Google Scholar]
He T, Bai X, Yang L, Fan L, Li Y et al. (2015). Loureirin B inhibits hypertrophic scar formation via inhibition of the TGF-u03b21-ERK/JNK pathway. Cellular Physiology & Biochemistry 37: 666–676. [Google Scholar]
Jin J, Zhai HF, Jia ZH, Luo XH (2019). Long non-coding RNA HOXA11-AS induces type I collagen synthesis to stimulate keloid formation via sponging miR-124-3p and activation of Smad5 signaling. American Journal of Physiology—Cell Physiology 317: C1001–C1010. [Google Scholar]
Lee S, Kim SK, Park H, Lee YJ, Park SH et al. (2020). Contribution of autophagy-Notch1-mediated NLRP3 inflammasome activation to chronic inflammation and fibrosis in keloid fibroblasts. International Journal of Molecular Sciences 21: 8050. [Google Scholar]
Lee TY, Chin GS, Kim WJ, Chau D, Gittes GK, Longaker MT (1999). Expression of transforming growth factor beta 1, 2, and 3 proteins in keloids. Annals of Plastic Surgery 43: 179–184. [Google Scholar]
Lei R, Shen J, Zhang S, Liu A, Chen X et al. (2019). Inactivating the ubiquitin ligase Parkin suppresses cell proliferation and induces apoptosis in human keloids. Journal of Cellular Physiology 234: 16601–16608. [Google Scholar]
Li H, Lu C, Yao W, Xu L, Zhou J, Zheng B (2020a). Dexmedetomidine inhibits inflammatory response and autophagy through the circLrp1b/miR-27a-3p/Dram2 pathway in a rat model of traumatic brain injury. Sedentary Life and Nutrition 12: 21687–21705. [Google Scholar]
Li X, Yang R, Xu Y, Zhang Y (2020b). Circ_0001438 participates in the pathogenesis of preeclampsia via the circ_0001438/miR-942/NLRP3 regulatory network. Placenta 104: 40–50. [Google Scholar]
Lim KH, Itinteang T, Davis PF, Tan ST (2019). Stem cells in keloid lesions: A review. Plastic and Reconstructive Surgery Global Open 7: e2228. [Google Scholar]
Mack M (2018). Inflammation and fibrosis. Matrix Biology 68-69: 106–121. [Google Scholar]
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338. [Google Scholar]
Mofikoya BO, Adeyemo WL, Abdus-Salam AA (2007). Keloid and hypertrophic scars: A review of recent developments in pathogenesis and management. Nigerian Quarterly Journal of Hospital Medicine 17: 134–139. [Google Scholar]
Ogawa R, Miyashita T, Hyakusoku H, Akaishi S, Kuribayashi S, Tateno A (2007). Postoperative radiation protocol for keloids and hypertrophic scars: Statistical analysis of 370 sites followed for over 18 months. Annals of Plastic Surgery 59: 688–691. [Google Scholar]
Shao BZ, Xu ZQ, Han BZ, Su DF, Liu C (2015). NLRP3 inflammasome and its inhibitors: A review. Frontiers in Pharmacology 6: 262. [Google Scholar]
Shi J, Yao S, Chen P, Yang Y, Qian M et al. (2020). The integrative regulatory network of circRNA and microRNA in keloid scarring. Molecular Biology Reports 47: 201–209. [Google Scholar]
Shi K, Qiu X, Zheng W, Yan D, Peng W (2018). MiR-203 regulates keloid fibroblast proliferation, invasion, and extracellular matrix expression by targeting EGR1 and FGF2. Biomedicine & Pharmacotherapy 108: 1282–1288. [Google Scholar]
Sidgwick GP, Bayat A (2012). Extracellular matrix molecules implicated in hypertrophic and keloid scarring. Journal of the European Academy of Dermatology and Venereology 26: 141–152. [Google Scholar]
Szabo L, Salzman J (2016). Detecting circular RNAs: Bioinformatic and experimental challenges. Nature Reviews Genetics 17: 679–692. [Google Scholar]
van Leeuwen MC, Stokmans SC, Bulstra AE, Meijer OW, Heymans MW et al. (2015). Surgical excision with adjuvant irradiation for treatment of keloid scars: A systematic review. Plastic and Reconstructive Surgery Global Open 3: e440. [Google Scholar]
Varga J, Abraham D (2007). Systemic sclerosis: A prototypic multisystem fibrotic disorder. Journal of Clinical Investigation 117: 557–567. [Google Scholar]
Vinaik R, Abdullahi A, Barayan D, Jeschke MG (2020a). NLRP3 inflammasome activity is required for wound healing after burns. Translational Research 217: 47–60. [Google Scholar]
Vinaik R, Barayan D, Auger C, Abdullahi A, Jeschke MG (2020b). Regulation of glycolysis and the Warburg effect in wound healing. JCI Insight 5: e138949. [Google Scholar]
Vincent AS, Phan TT, Mukhopadhyay A, Lim HY, Halliwell B, Wong KP (2008). Human skin keloid fibroblasts display bioenergetics of cancer cells. Journal of Investigative Dermatology 128: 702–709. [Google Scholar]
Wang J, Wu H, Xiao Z, Dong X (2019a). Expression profiles of lncRNAs and circRNAs in keloid. Plastic and Reconstructive Surgery Global Open 7: e2265. [Google Scholar]
Wang K, Long B, Liu F, Wang JX, Liu CY et al. (2016). A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. European Heart Journal 37: 2602–2611. [Google Scholar]
Wang R, Bai Z, Wen X, Du H, Zhou L et al. (2019b). MiR-152-3p regulates cell proliferation, invasion and extracellular matrix expression through by targeting FOXF1 in keloid fibroblasts. Life Sciences 234: 116779. [Google Scholar]
Wu J, Fang L, Cen Y, Qing Y, Chen J, Li Z (2019). MiR-21 regulates keloid formation by downregulating Smad7 via the TGF-β/Smad signaling pathway. Journal of Burn Care & Research 40: 809–817. [Google Scholar]
Xu H, Guo S, Li W, Yu P (2015). The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Scientific Reports 5: 12453. [Google Scholar]
Yang D, Li M, Du N (2020). Effects of the circ_101238/miR-138-5p/CDK6 axis on proliferation and apoptosis keloid fibroblasts. Experimental and Therapeutic Medicine 20: 1995–2002. [Google Scholar]
Yang F, Chen Y, Shen T, Guo D, Dakhova O (2014). Stromal TGF-β signaling induces AR activation in prostate cancer. Oncotarget 5: 10854. [Google Scholar]
Zambetti LP, Laudisi F, Licandro G, Ricciardi-Castagnoli P, Mortellaro A (2012). The rhapsody of NLRPs: Master players of inflammation… and a lot more. Immunologic Research 53: 78–90. [Google Scholar]
Zhang Y, Huang R, Cheng M, Wang L, Chao J et al. (2019). Gut microbiota from NLRP3-deficient mice ameliorates depressive-like behaviors by regulating astrocyte dysfunction via circHIPK2. Microbiome 7: 116. [Google Scholar]
Zhang Z, Yu K, Liu O, Xiong Y, Yang X et al. (2020). Expression profile and bioinformatics analyses of circular RNAs in keloid and normal dermal fibroblasts. Experimental Cell Research 388: 111799. [Google Scholar]
Zhu HY, Bai WD, Li C, Zheng Z, Guan H et al. (2016). Knockdown of lncRNA-ATB suppresses autocrine secretion of TGF-beta2 by targeting ZNF217 via miR-200c in keloid fibroblasts. Scientific Reports 6: 24728. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |