DOI:10.32604/biocell.2022.018167

| BIOCELL DOI:10.32604/biocell.2022.018167 |  |
| Article |
miR-181b promotes the oncogenesis of renal cell carcinoma by targeting TIMP3
1Department of Urology, First Affiliated Hospital of Gannan Medical University, Ganzhou, 341000, China
2Department of Urology, The First People’s Hospital of Xiangtan City, Xiangtan, 411101, China
3Department of General Surgery, Shenzhen SAMII Medical Center, Shenzhen, 518118, China
*Address correspondence to: Xin Zhong, ystbj301@163.com
#These authors contributed equally to this work
Received: 04 June 2021; Accepted: 22 September 2021
Abstract: Renal cell carcinoma (RCC) has a poor prognosis due to limited diagnosis and treatment. Thus, it is necessary to find novel prognostic biomarkers and therapeutic targets. The aberrant expression of microRNAs plays an important role in RCC oncogenesis. Tissue inhibitors of metalloproteinase 3 (TIMP3) acts as a downstream target of miR-181b. The aim of this study was to understand the role and molecular mechanism of miR-181b in RCC oncogenesis. The results showed that miR-181b expression was significantly higher in RCC tumour tissues, especially in those with significant invasion or metastasis. miR-181b overexpression promoted proliferation and migration of the RCC cell line 786-O, while miR-181b knockdown had the opposite effect. In addition, miR-181b was inversely correlated with TIMP3 expression in RCC tumour tissues. miR-181b overexpression reduced TIMP3 expression in RCC cell line 786-O or OS-RC-2, while miR-181b knockdown had the inverse effect. Mechanistically, a luciferase reporter assay confirmed the binding sites of miR-181b on the 3’-UTR of TIMP3, confirming the targeting effect of miR-181b on TIMP3. Overall, miR-181b promotes the development and progression of RCC by targeting TIMP3 expression, indicating the potential use of miR-181b in the diagnosis and treatment of RCC.
Keywords: microRNA; microRNA-181b; Tissue inhibitors of metalloproteinase 3; Renal cell carcinoma; 3’-UTR region
The incidence of renal cell carcinoma (RCC) is highest in patients with renal malignant tumours, with approximately 338,000 new cases and 143,000 deaths in 2012 worldwide (Bhatt and Finelli, 2014; Torre et al., 2015). Most early RCC could be cured by surgery (Ljungberg et al., 2015). However, approximately 30% of RCCs are diagnosed at advanced stages with metastasis (Zhai et al., 2017). For these RCC patients in later stages, there is no effective therapeutic strategies and the prognosis is still very poor (Baker, 2016). Therefore, it is urgent to find more novel methods for early diagnosis and treatment of RCC. It is necessary to reveal the molecular mechanisms regarding the occurrence and development of RCC, thereby promoting the identification of effective prognostic biomarkers and therapeutic targets for RCC.
An increasing number of studies have shown that abnormal expression of microRNAs (referred to as miRNAs) plays a key role in the growth and metastasis of RCC (Chen et al., 2017b; Liu et al., 2017; Xiao et al., 2017; Zhang et al., 2019a). Therefore, by elucidating the biological function and relevant mechanism of miRNAs in RCC, we can provide new viewpoints for the diagnosis and treatment of RCC.
miRNAs are nonprotein-coding RNA molecules that have a length of 19–25 nt and regulate physiological and pathological processes in organisms by binding to the 3’-untranslated regions (3’-UTR) of targeted mRNAs downstream. miRNAs exert a significant effect on oncogenesis (Piotrowski et al., 2021; Tan et al., 2021). At present, miRNA expression has been observed in various tumours and is closely related to tumour growth (Jiang et al., 2016), metastasis (Wang et al., 2016), angiogenesis (Lu et al., 2015), and other malignant biological characteristics; thus, it is considered an indispensable regulator of tumorigenesis. Therefore, miRNAs detection is beneficial for early clinical diagnosis; moreover, modulating their expression in tumours may have potential clinical values.
In the human genome, the miR-181 family consists of four highly conserved mature miRNA molecules, miR-181a, b, c, and d. miR-181b is aberrantly expressed in a variety of tumours, with functions mainly involved in pathophysiological processes including cell cycle control, cell invasion and metastasis. Previous research has shown that miR-181b promotes the tumorigenesis of colorectal cancer by targeting PDCD4 (Liu et al., 2016). In addition, the epigenetic silencing of miR-181b promotes the tumorigenicity of colorectal cancer by targeting RASSF1A (Zhao et al., 2016). miR-181b promotes the chemoresistance of breast cancer by regulating the expression of BIM (Zheng et al., 2016). Similar results were reported in other investigations (Tian et al., 2016; Zhang et al., 2019b). However, the role of miR-181b in RCC remains unknown.
Tissue inhibitors of metalloproteinase-3 (TIMP3) is a member of the TIMP family and maintains cell stability in cancer by preventing extracellular matrix (ECM) degradation, thereby inhibiting tumour growth and metastasis. Hence, TIMP3 functions as a tumour suppressor and has poor expression in cancerous tissues (Kong et al., 2016; Qi and Anand-Apte, 2015; Su et al., 2017). A previous study found that TIMP3 expression is low in RCC tissues (Masson et al., 2010); furthermore, TIMP3 is closely related to the proliferation and metastasis of RCC (Chen et al., 2017a). Previous studies revealed that the 3’-UTR of TIMP3 mRNA has a specific binding sequence for miR-181b (Heath et al., 2018; Wang et al., 2010; Zhou et al., 2016). In summary, we hypothesized that miR-181b promoted tumour development by targeting TIMP3 in RCC cells.
Our study showed that miR-181b expression was significantly increased in RCC tissues. Based on human histology and in vitro cytology studies, this work suggested that miR-181b is not only directly responsible for the growth and migration of RCC, but also negatively correlated with the expression of TIMP3. The above data provide novel clues for the diagnosis and treatment of RCC.
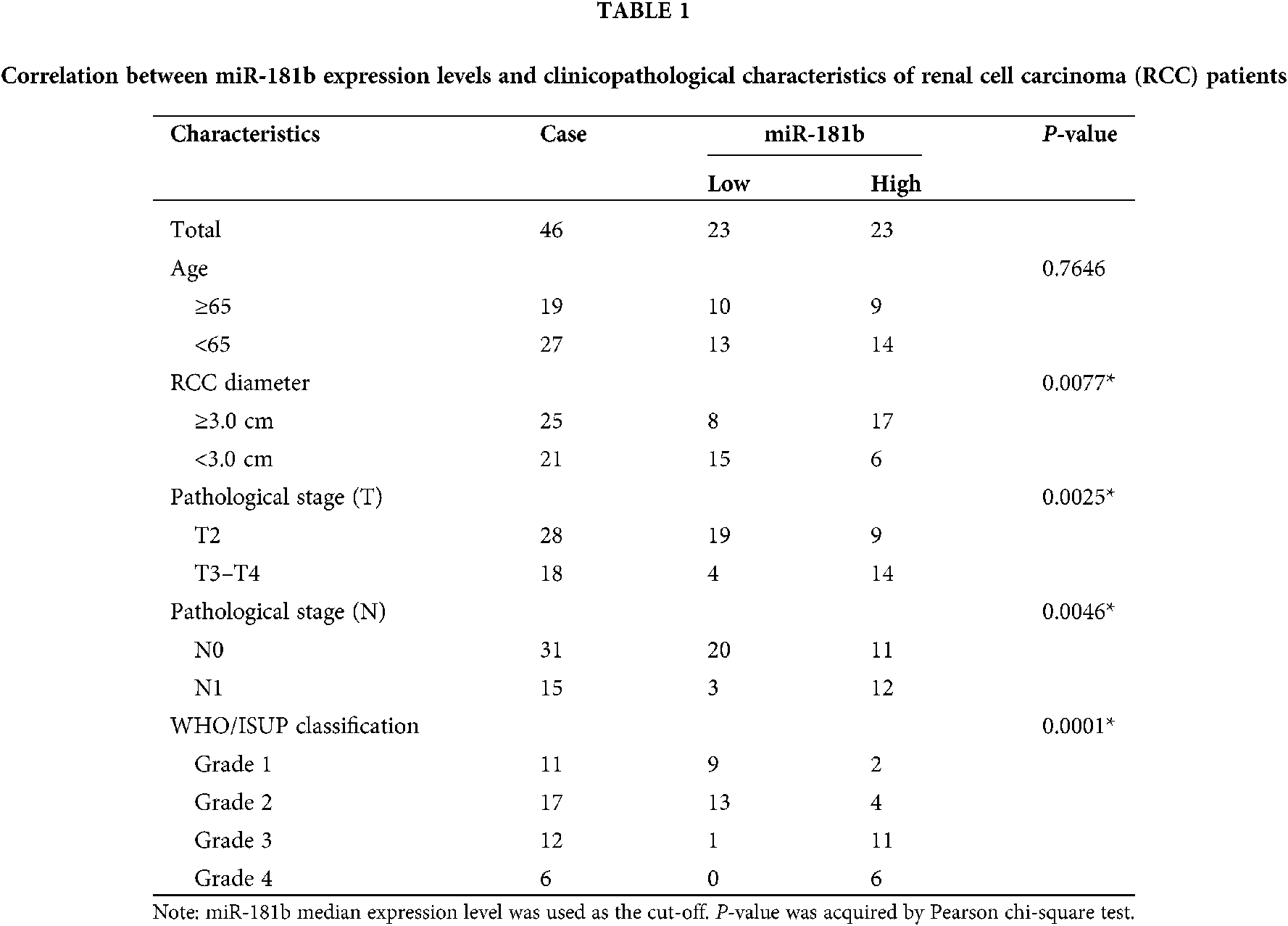
A total of 46 pairs of RCC tissues and adjacent noncancerous tissues were obtained with the informed consent via surgical resection at First Affiliated Hospital of Gannan Medical University, Gannan, Jiangxi, China, from January 2012 to May 2017. All the 46 samples were clear cell renal cell carcinoma. The WHO/ISUP grade distribution was following: Grade 1, 11 (23.9%); Grade 2, 17 (36.9%); Grade 3, 12 (26.1%); and Grade 4, 6 (13%). The age range of the patients is 31–77 years, and the median age is 52 years. There were 29 male patients and 17 female patients. All tissue specimens were histopathologically examined by three independent pathologists. Freshly tissue specimens were frozen in liquid nitrogen and stored at −80°C until use. All the clinical samples were acquired with informed consent from the participants. The Institutional Review Board of First Affiliated Hospital of Gannan Medical University reviewed and approved this study. The clinical studies were conducted according to the principles expressed in the Helsinki Declaration.
Human normal renal tubular epithelial cell line HK-2 and RCC cell lines OS-RC-2, 786-O and Caki-1 were obtained from the American Type Culture Collection (ATCC). The cell lines have been authenticated using STR profiling. HK-2 was cultured in Dulbecco’s Modified Eagle’s Medium/F12 (DMEM/F12) (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% heat-inactivated foetal bovine serum (FBS; Thermo Fisher Scientific). OS-RC-2, 786-O and Caki-1 were cultured in RPMI-1640 Medium (Thermo Fisher Scientific) along with 10% FBS. The whole cells were maintained at 37°C in an atmosphere containing 5% CO2.
Quantitative real-time PCR (qRT-PCR) assays
Total RNA was extracted and purified by the TRIzol method. SYBR real-time fluorescent quantitative kit was used for real-time quantitative PCR (qRT-PCR) detection. The reaction was performed in 25 µL system, and reaction condition was as following: 50°C for 30 min, 94°C for 1 min, 57°C for 1 min, and 72°C for 7 min. β-actin were used as an internal reference gene. The designed primer sequences for qRT-PCR were as follows: miR-181b, 5’-CCAGCTGGGCTCACTGAACAATGA-3’ (sense) and 5’- CAACTGGTGTCGTGGAGTCGGC-3’ (anti-sense); TIMP3, 5’-GATCAAAGTGGAAAACCAGCTT -3’ (sense) and 5’-AGATGGAGGTTCTCATGTAAGC-3’ (anti-sense); β-actin, 5’-GGGAAATCGTGCGTGACATTAAG-3’ (sense) and 5’-TGTGTTGGCGTACAGGTCTTTG-3’ (anti-sense). Relative expression was calculated using the 2−ΔΔCt method.
qRT-PCR was performed using SYBR Premix Ex TaqTM kit (TakaRa, Tokyo, Japan) and ABI7500 PCR system (Applied Biosystems, Thermo, MA, USA).
Plasmid construction and transfection
miR-181b full-length sequences were PCR amplified using Phusion HSII Flash High-Fidelity PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer’s protocol. miR-181b mimics (RiboBio, Guangzhou, Guangdong, China) and miR-181b inhibitors (anti-miR-181b-5p, RiboBio) were obtained from GenePharma (Shanghai, China). In different groups, miR-181b mimics, miR-mimic control, miR-181b inhibitors and miR-inhibitor control in serum-free ACCELL medium were transfected into 786-O cells with a final concentration of 200 nM. Transfection was performed by using Lipofectamine 3000 (Invitrogen) according to manufacturer’s protocol. The transfection efficiency of miR-181b mimics or miR-181b inhibitors was verified by qPCR.
To assess the cell proliferation, Cell Counting Kit-8 (CCK-8) assays was performed. For CCK-8 assays, the cells in each group were plated into 96-well plates with 2,000 cells/well. At the indicated time, 10 μL of CCK-8 reagent (Dojindo, Kumamoto, Japan) was added into the treated cells. Following incubation for 2 h, the optical density at 450 nm (OD450) was measured using Varioskan Flash reader (Thermo).
The migration of HCC cells was measured using the Transwell assay. For Transwell assay, the treated cells suspended in serum-free DMEM supplemented with 1 mg/mL mitomycin C (inhibitor of cell proliferation) were seeded onto the upper chamber of the Transwell. DMEM containing 20% FBS was added to lower chamber. After 36 h incubation, the cells migrating into lower surface of the inserts were fixed, stained with 1% crystal violet, and photographed. Migration was measured by counting the number of stained cells.
Whole-cell lysates of cells with the indicated interventions were prepared from 6-well plates. Lysates were located onto and separated on 10% SDS-PAGE gels, and transferred to polyvinylidene fluoride membranes, which were incubated with the antibodies against TIMP3 and β-actin (Cell Signaling Technology, Danvers, MA, USA). HRP-linked secondary antibodies were used to visualize immunoreactivity using a chemiluminescence system (Amersham Image 600, General Electric, Boston, MA, USA).
The sequence 3’-UTR of TIMP3 was cloned into pMIR expression vectors (Promega, Madison, USA) including luciferase genes to construct pMIR-TIMP3-WT, pMIR-TIMP3-MUT or pMIR-3’-UTR control plasmid as described previously (Heath et al., 2018). The luciferase reporter plasmid together with TK-Renilla luciferase plasmid (pRL-TK-Renilla, Promega) was co-transfected into the indicated 786-O cells. After 2 days’ transfection, luciferase activity in cell lysates was measured in a luminometer (Promega) according to manufacturer’s protocol.
A total of 20 male BALB/c nude mice (age: six weeks; weight: 20–22.5 g) were obtained from the Slaccas Experimental Animal Centre (Shanghai, China). The mice were housed in a specific pathogen-free facility with barrier in which the room temperature was 20–30°C and the humidity 60%–80% and fed a SPF mouse chow and sterile water. miR-181b inhibitors-transfected and control 786-O cells were inoculated subcutaneously on the ventral side of the right rib at the density of 2 × 106 cells per mouse. The health and tumour volume growth of all mice were monitored daily. Tumour volumes were calculated using the following formula: volume (cm3) = length (L; cm) × width (W; cm)2/2. The largest tumour volume was ~1 cm3. 30 days later, all tumour-bearing mice were anesthetized with isoflurane (2%, inhalation anaesthesia), and then sacrificed via cervical dislocation. All mice were considered dead when their hearts and breathings stopped. Their tumours were removed and weighted. These experimental protocols were approved by the Institutional Animal Care and Use Committee of First Affiliated Hospital of Gannan Medical University.
Immunohistochemistry (IHC) assessment
The tissue sections were prepared, incubated overnight at 4°C with human TIMP3 antibody (1:200) (Abcam; Cambridge Science Park, UK) and then visualized using a biotin–streptavidin detection system (Dako, Glostrup, Denmark) according to manufacturer’s protocol. TIMP3 staining was graded semi-quantitatively. The intensity of TIMP3 reactivity of tumour cells was graded as 0 (no stain), 1 (weak stain), 2 (clear stain), or 3 (strong stain). The intensity and abundance (fraction) were multiplied to obtain total immunoreactivity score.
All statistical analyses were performed using the GraphPad Prism Software 6. For comparisons, Wilcoxon signed-rank test, Wilcoxon rank sum test, Pearson, one-way ANOVA test or Student’s t-test were performed as indicated. Bonferroni test was used for post-hoc multiple comparisons of one-way ANOVA. Pearson correlation analysis and Pearson chi-square test were performed for correlations. P < 0.05 was considered as statistically significant.
miR-181b was overexpressed in RCC specimens
miR-181b expression levels in 46 pairs of RCC tissues and adjacent noncancerous tissues were detected by qRT-PCR. As shown in Fig. 1A, miR-181b was significantly overexpressed in RCC tissue specimens compared with adjacent noncancerous tissues. The analyses of the correlation between miR-181b levels and clinicopathological characteristics of RCC patients showed that high miR-181b levels were positively correlated with larger RCC tumours, advanced pathological T/N stage and higher WHO/ISUP classification (Table 1). Moreover, 18 RCC specimens with metastasis had higher miR-181b expression than 28 nonmetastatic RCC specimens (Fig. 1B). Similarly, compared with 21 RCC specimens with diameters less than 3 cm, 25 RCC specimens with diameters greater than 3 cm showed higher miR-181b expression (Fig. 1C). In addition, miR-181b expression levels in the normal renal tubular epithelial cell line HK-2 and the RCC cell lines OS-RC-2, 786-O and Caki-1 were detected by qRT-PCR. As shown in Fig. 1D, miR-181b was also obviously overexpressed in RCC cell lines.
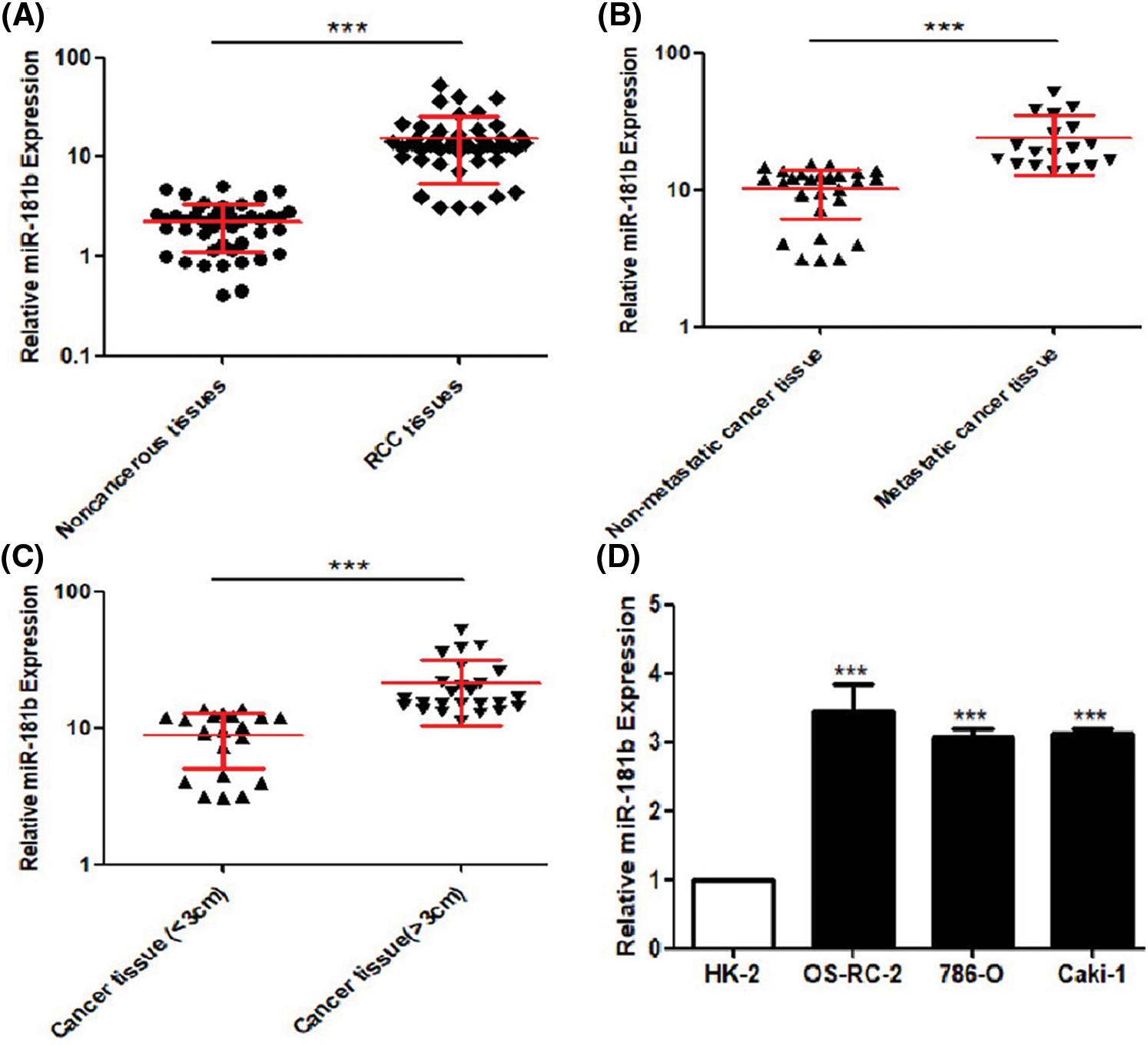
Figure 1: miR-181b is overexpressed in RCC specimens. (A) miR-181b expression levels in 46 pairs of RCC tissues and adjacent noncancerous tissues. Results are presented as median with interquartile range. ***P < 0.001 by Wilcoxon signed-rank test. (B) miR-181b expression levels in 18 RCC specimens with metastasis and 28 nonmetastatic RCC specimens. Results are presented as median with interquartile range. ***P < 0.001 by Wilcoxon rank sum test. (C) miR-181b expression levels in 25 RCC specimens with diameter more than 3 cm and 21 RCC specimens with diameter less than 3 cm. Results are presented as median with interquartile range. ***P < 0.001 by Wilcoxon rank sum test. (D) miR-181b expression levels in normal renal tubular epithelial cell line HK-2 and RCC cell lines OS-RC-2, 786-O and Caki-1. Results are presented as mean ± SEM from three independent experiments. **P < 0.01, ***P < 0.001 by one-way ANOVA test.
miR-181b overexpression enhanced the proliferation, invasion, and migration of RCC cells, whereas miR-181b silencing was contrary
To explore the roles of miR-181b in RCC, we overexpressed or silenced miR-181b by transducing miR-181b mimics or miR-181b inhibitors into 786-O cells. The transfection efficiency of miR-181b mimics and miR-181b inhibitors was measured by detecting the mRNA levels (Fig. 2A). CCK-8 assays showed that miR-181b overexpression enhanced the proliferation of 786-O cells (Fig. 2B). In addition, Transwell assays demonstrated that miR-181b overexpression increased the number of migratory cells (Figs. 2C and 2D). In contrast, miR-181b silencing showed the opposite results in cell proliferation and migration (Figs. 2A–2D). The experimental data in OS-RC-2 cells also showed similar results (Figs. 2E–2H).
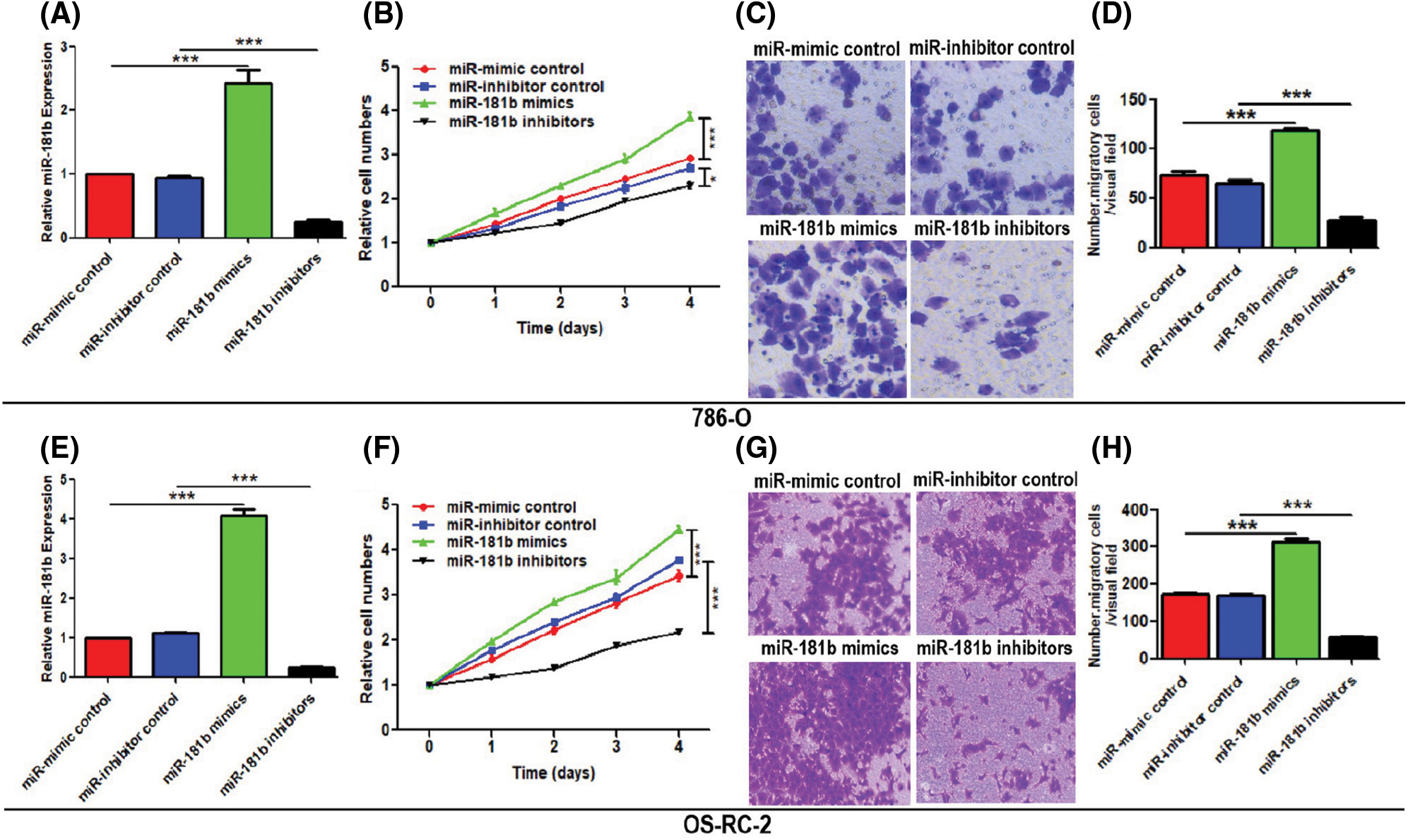
Figure 2: miR-181b promotes the proliferation and migration of RCC cells. (A) miR-181b expression in 786-O cells transfected with miR-mimic control, miR-inhibitor control, miR-181b mimics and miR-181b inhibitors. (B) Cell proliferation of treated 786-O cells was evaluated by CCK-8 assays. (C–D) The migration of treated 786-O cells was evaluated by Transwell assays. Scale bar: 100 μm. (E–H) Repetition of operations as described for (A–D) on OS-RC-2. Results are presented as mean ± SEM from three independent experiments. *P < 0.05, ***P < 0.001 by Student’s t-test. Scr, scrambled control; miR-Cont, miRNA mimics control; cont, control group.
miR-181b inhibited the expression of TIMP3
Next, we investigated whether miR-181b could negatively regulate TIMP3 expression in vivo and in vitro. TIMP3 expression levels in RCC tissues and adjacent noncancerous tissues were observed. Representative images of surgically resected RCC tumour and corresponding HE-staining were presented in Fig. 3A. As shown in Figs. 3B, 3C, and 3E, protein, and mRNA levels of TIMP3 were downregulated in RCC tissue specimens compared with adjacent noncancerous tissues. The correlation analyses between TIMP3 protein levels and miR-181b levels in RCC tissues showed that TIMP3 levels were inversely associated with miR-181b levels in RCC tissues (r = −0.8747, P < 0.0001) (Fig. 3D), demonstrating the negative regulation of miR-181b on TIMP3 in vivo. Next, we continued to observe the regulatory function of miR-181b in TIMP3 in vitro. The TIMP3 protein levels in miR-181b-overexpressing and -silencing 786-O or OS-RC-2 cells were investigated by Western Blotting. As shown in Figs. 3F and 3G, TIMP3 protein levels were decreased in miR-181b-overexpressing cells and increaseed in miR-181b-silencing cells. Similarly, TIMP3 mRNA levels were decreased in miR-181b-overexpressing 786-O cells and increaseed in miR-181b-silencing 786-O cells (Fig. 3H). Next, we further clarified the significance of miR-181b in TIMP3 luciferase activity. Luciferase reporter assays showed that miR-181b overexpression reduced the luciferase activity of 786-O cells, which did not occur in the pMIR-3’-UTR control-transfected cells or pMIR-TIMP3-MUT plasmid-transfected cells that lacks the effective binding region of TIMP3 (Fig. 3I).
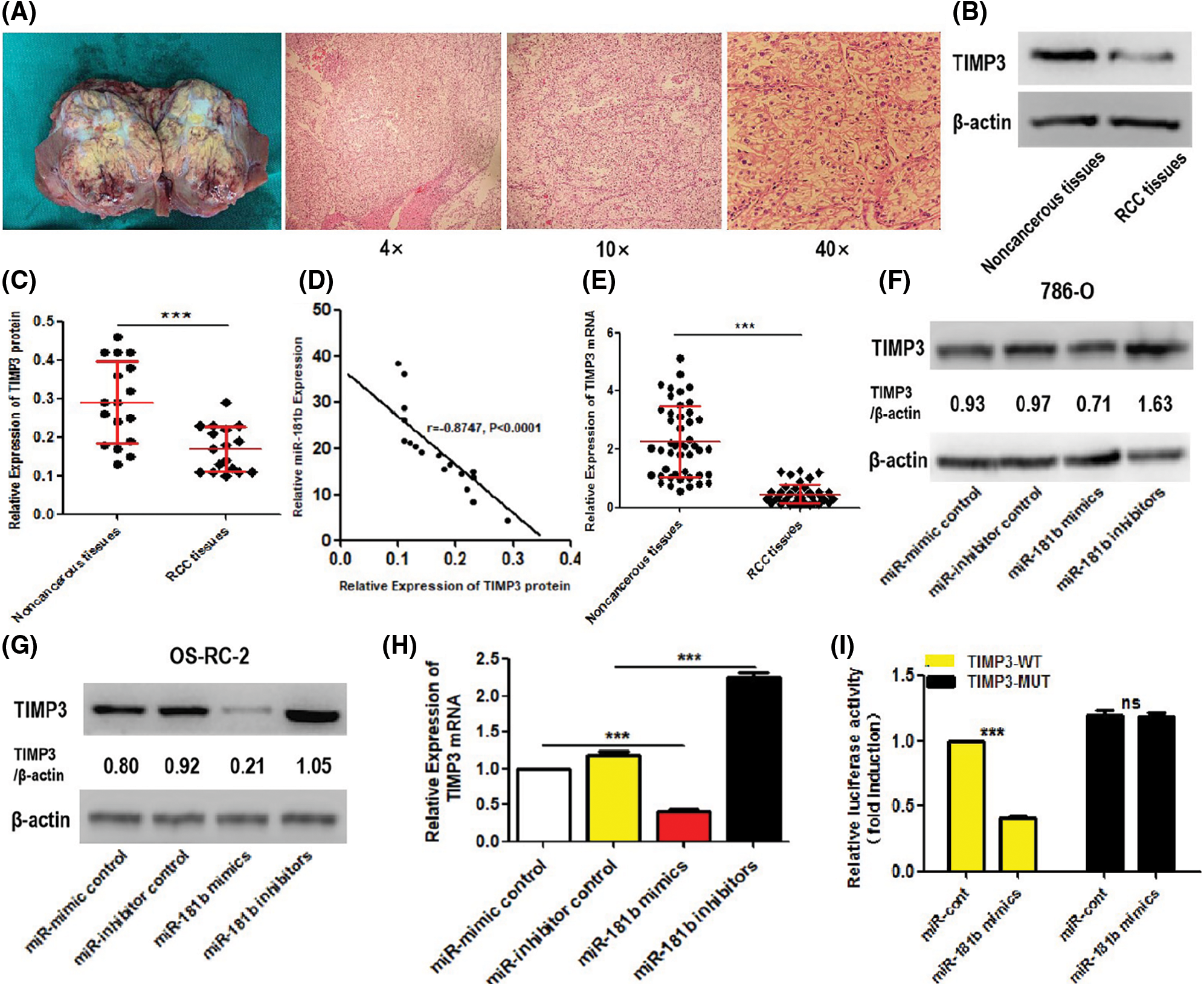
Figure 3: miR-181b suppresses the expression of TIMP3. (A) Representative images of surgically resected RCC tumour and corresponding HE-staining. (B–C) protein expression levels of TIMP3 in 17 pairs of RCC tissues and adjacent noncancerous tissues. Results are presented as median with interquartile range. ***P < 0.001 by Wilcoxon signed-rank test. (D) The association between TIMP3 protein levels and miR-181b levels in RCC tissues was analysed. N = 17, r = −0.8747, P < 0.0001 by Pearson correlation analysis. (E) mRNA expression levels of TIMP3 in 46 pairs of RCC tissues and adjacent noncancerous tissues. Results are presented as median with interquartile range. ***P < 0.001 by Wilcoxon signed-rank test. (F–G) After transfection with corresponding plasmids for 72 h, protein levels of TIMP3 in miR-181b-overexpressing, miR-181b-silencing and control 786-O or OS-RC-2 cells was measured by Western Blotting. Data are representative results from 3 independent assays with unanimous trends. (H) After transfection with corresponding plasmids for 72 h, mRNA levels of TIMP3 in miR-181b-overexpressing, miR-181b-silencing and control 786-O cells was evaluated by qRT-PCR assays. Results are presented as mean ± SEM from three independent experiments. ***P < 0.001 by one way ANOVA test. (I) After transfection with pMIR-TIMP3-WT or pMIR-TIMP3-MUT plasmid (including miR-181b mimics and miR-mimic control) in 786-O cells for 48 h, the luciferase activity of each group was analysed by Dual-Luciferase Reporter Assays. Results are presented as mean ± SEM from three independent experiments. ***P < 0.001 by one way ANOVA test. ns, no significance.
miR-181b silencing suppressed the xenograft tumours of RCC and enhanced TIMP3 expression in vivo
To explore the role of targeting miR-181b in RCC, we injected miR-181b-silencing and control 786-O cells subcutaneously into nude mice. Xenograft volumes were measured daily after injection. As shown in Figs. 4A–4C, miR-181b silencing significantly decreased the growth curve and weights of RCC xenograft tumours, supporting the inhibitory effect of miR-181b silencing on RCC growth. Of note, miR-181b silencing significantly enhanced TIMP3 protein expression in xenograft tumours (Figs. 4D–4F).
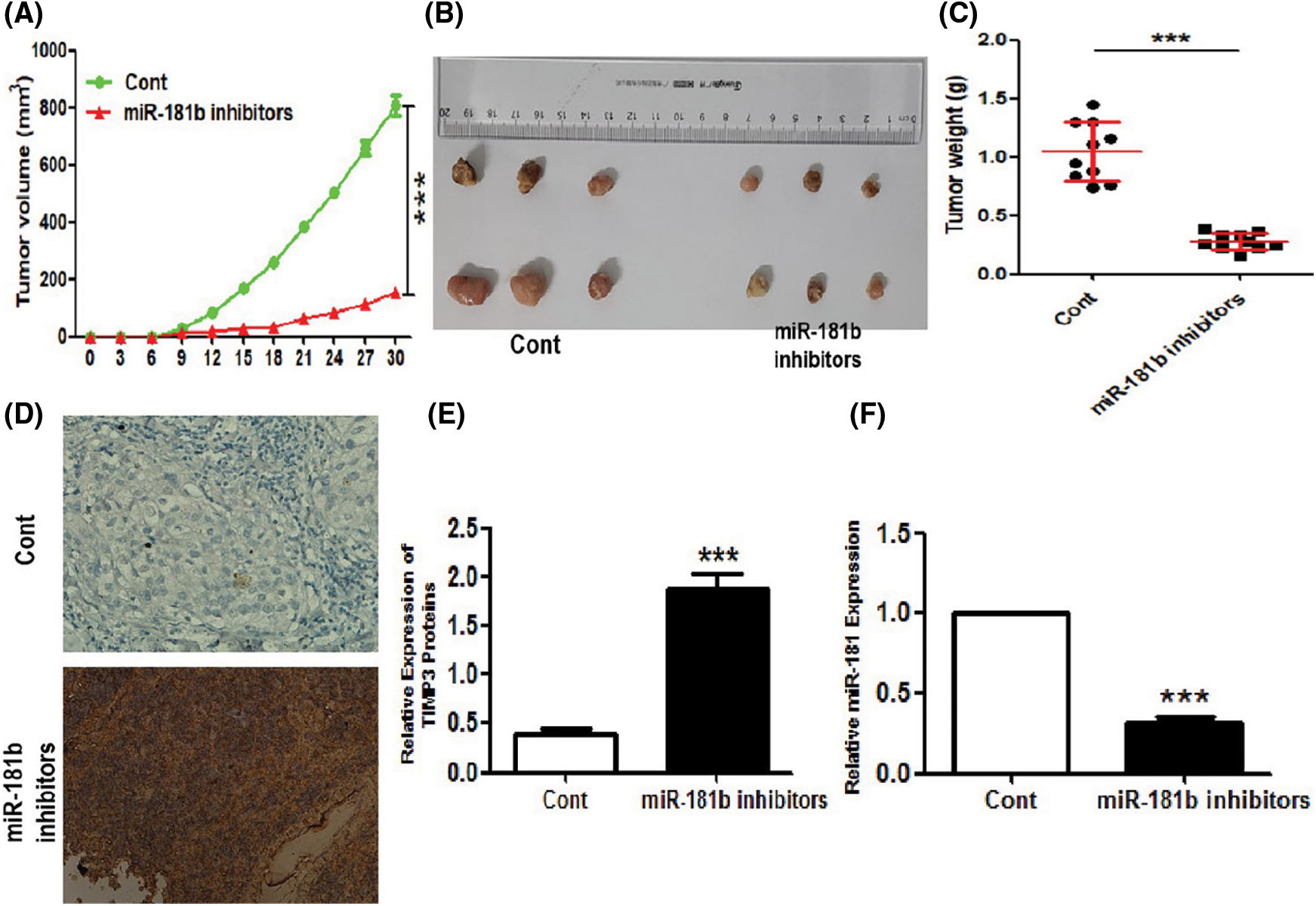
Figure 4: miR-181b knockdown represses RCC xenograft tumours and enhances TIMP3 expression in vivo. miR-181b-silencing or control 786-O cells was inoculated into nude mice. 1, 2, 3 and 4 weeks later, tumour-bearing mice were killed, and tumours were removed and weighted. (A) The scatter plot indicates the growth curves of tumour volumes in two groups (N = 8). (B) Representative images of the removed tumours. (C) The statistical graph indicates the quantitative results of the tumour weights at day 30 (N = 10). Results are presented as mean ± SEM. ***P < 0.001 by Student’s t-test. (D–E) Protein levels of TIMP3 in tumour tissues from subcutaneous xenograft tumours were evaluated by immunohistochemistry (IHC) staining (N = 10). (F) miR-181b expression levels of xenograft tumours in two groups (N = 10). Results are presented as mean ± SEM. ***P < 0.001 by Student’s t-test. Cont, control 786-O cells.
The increasing development in genomic and transcriptomic have authenticated many genetic and epigenetic aberrations in the carcinogenesis and progression of RCC (Miao et al., 2018; Thoma, 2016). The significant role of miRNA in the carcinogenesis has attracted a lot of attentions. Of note, an increasing number of miRNAs have been proved to be related to RCC. The expression of miR-30a-5p decreases in Clear cell renal cell carcinoma (ccRCC) cells and tissues (Chen et al., 2017b). In addition, low miR-30a-5p levels are significantly associated with poor prognosis of ccRCC patients, and miR-30a-5p can prevent ccRCC proliferation and invasion (Chen et al., 2017b). miR-218 suppresses RCC migration and invasion through targeting caveolin-2 involved in focal adhesion pathway (Zhang et al., 2019a). Pseudohypoxia induced by inactivation of miR-126 enhances RCC migration and therapeutic resistance (Liu et al., 2017). miR-144-3p promotes ccRCC proliferation, metastasis and sunitinib resistance by downregulating ARID1A (Xiao et al., 2017). In addition, the effects of miR-372, miR-200b and miR-106a-5p on the growth and function of RCC were also reported in relevant studies (Huang et al., 2015; Li et al., 2019; Pan et al., 2017). These previous studies support the key roles of miRNAs in the pathogenesis of RCC. In addition to the reported miRNAs, there may exist other miRNAs involved in the occurrence and development of RCC.
In this study, we focused on miR-181b, which is widely reported to be related to the growth and function of malignant tumours (Liu et al., 2016; Tian et al., 2016; Zhang et al., 2019b; Zhao et al., 2016; Zheng et al., 2016) and specifically binds to the 3’-UTR of RCC inhibitors. However, reports regarding the role of miR-181b in RCC are still absent. As expected, miR-181b was upregulated in RCC tissues and cell lines compared with noncancerous tissues and a normal renal tubular epithelial cell line, respectively. Moreover, with the growth and metastasis of RCC, miR-181b levels in cancer tissues were further increased. Correlation analyses between miR-181b expression and clinicopathological features revealed that an increase in miR-181b levels indicated poorer overall survival. Previous studies have shown several miRNAs that can predict the prognosis of RCC (Chen et al., 2017b; Liu et al., 2017; Wu et al., 2021; Xie et al., 2018). Therefore, the above data support the notion that miR-181b may be a promising prognostic biomarker for RCC.
Functional assays in vitro showed that the upregulation of miR-181b enhanced the proliferation and migration of RCC cells. In contrast, miR-181b downregulation inhibited the proliferation and migration of RCC cells. Nude mouse xenografts experiments, in vivo, showed that miR-181b knockdown suppressed RCC growth. Several miRNAs have been shown to modulate the carcinogenesis of PTC (Chen et al., 2017b; Liu et al., 2017; Xiao et al., 2017; Zhang et al., 2019a). These results suggest that miR-181b may be a potential therapeutic target for RCC.
A bioinformatics study revealed that the 3’-UTR of TIMP3 mRNA has a specific binding sequence for miR-181b, suggesting that TIMP3 may be a downstream target of miR-181b. miR-181b expression was inversely associated with TIMP3 expression in RCC tissues, which indicates the negative regulatory effect of miR-181b on TIMP3 expression. In addition, cytology assays revealed that the upregulation of miR-181b inhibited TIMP3 expression while the downregulation of miR-181b promoted TIMP3 expression. More importantly, a luciferase reporter assay illustrated that miR-181b can negatively regulate TIMP3 luciferase activity in RCC cells by binding to the 3’-UTR, which explains the inhibitory effect of miR-181b on TIMP3 expression in RCC cells. Similarly, in vivo experiments showed that miR-181b downregulation promoted TIMP3 expression levels in nude mouse xenografts. The above data indicate that the negative regulatory effect on TIMP3 at least partially accounts for the roles of miR-181b in the pathogenesis of RCC. The detailed mechanisms underlying the regulation of TIMP3 by miR-181b need further research. Our current working model is illustrated in Fig. 5. Here, it is difficult to co-transfect miR-181b mimics and TIMP3 overexpression vectors. In addition, there is a lack of specific inhibitors targeting TIMP3. Thus, the related rescue experiments cannot be effectively displayed in this study. The above limitations are expected to be addressed in future studies.
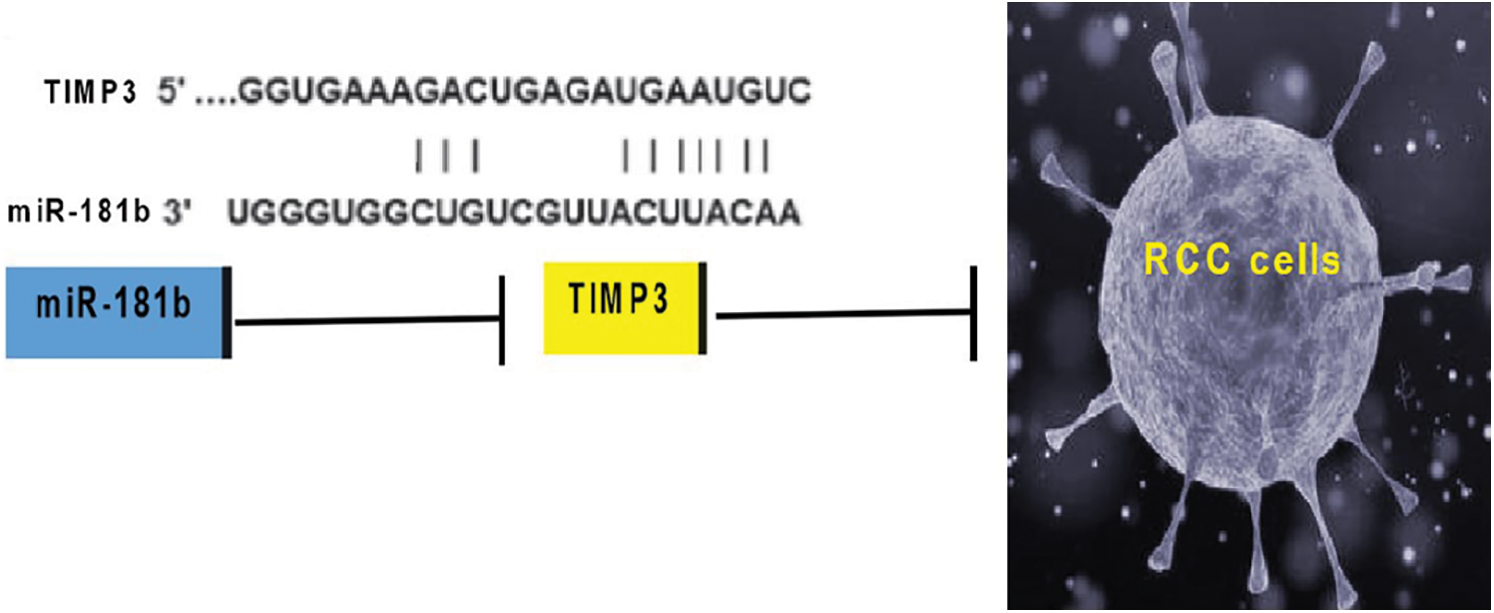
Figure 5: The current working model regarding miR-181b-regulated RCC oncogenesis.
The detection of multiple miRNAs is helpful for the early clinical diagnosis of RCC. Moreover, targeting or mimicking their expression in RCC tissues may have potential clinical values. In this study, our study identified miR-181b as a carcinogenic miRNA in RCC, which is a promising prognostic biomarker for RCC. More importantly, targeting miR-181b may be a potential effective strategy in repressing RCC. Based on these results, the diagnosis and treatment of RCC may be further improved in future.
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Author Contribution: Y. Z. and X. Z. conceived and designed experiments; Y. Z. and L. Z. performed experiments, analyzed data, prepared figures, and helped with writing of the manuscript; Y. Z. and X. Z. wrote the manuscript; X. Z. edited the article.
Ethics Approval: The human study protocol was approved by the Institutional Review Board of First Affiliated Hospital of Gannan Medical University (Gannan, China; Approval No. 2012-R269; 8/1/2012) and written informed consent was obtained from all the participants for their tissues to be used for the purposes of this research. Animal experimental protocols were approved by the Institutional Animal Care and Use Committee of First Affiliated Hospital of Gannan Medical University (Approval No. 2012-R269; 8/1/2012).
Funding Statement: This work was supported by grants Basic Scientific Research Projects of Fujian Provincial Public Welfare Scientific Research Institutes (2016R1029-2).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Baker H (2016). Sunitinib as adjuvant therapy for renal cell carcinoma. Lancet Oncology 17: e485. DOI 10.1016/S1470-2045(16)30511-3. [Google Scholar] [CrossRef]
Bhatt JR, Finelli A (2014). Landmarks in the diagnosis and treatment of renal cell carcinoma. Nature Reviews Urology 11: 517–525. DOI 10.1038/nrurol.2014.194. [Google Scholar] [CrossRef]
Chen J, Gu Y, Shen W (2017a). MicroRNA-21 functions as an oncogene and promotes cell proliferation and invasion via TIMP3 in renal cancer. European Review for Medical and Pharmacological Sciences 21: 4566–4576. [Google Scholar]
Chen Z, Zhang J, Zhang Z, Feng Z, Wei J et al. (2017b). The putative tumor suppressor microRNA-30a-5p modulates clear cell renal cell carcinoma aggressiveness through repression of ZEB2. Cell Death & Disease 8: e2859. DOI 10.1038/cddis.2017.252. [Google Scholar] [CrossRef]
Heath JM, Esmerats JF, Khambouneheuang L, Kumar S, Simmons R et al. (2018). Mechanosensitive microRNA-181b regulates aortic valve endothelial matrix degradation by targeting TIMP3. Cardiovascular Engineering and Technology 9: 141–150. DOI 10.1007/s13239-017-0296-z. [Google Scholar] [CrossRef]
Huang X, Huang M, Kong L, Li Y (2015). miR-372 suppresses tumour proliferation and invasion by targeting IGF 2 BP 1 in renal cell carcinoma. Cell Proliferation 48: 593–599. DOI 10.1111/cpr.12207. [Google Scholar] [CrossRef]
Jiang Q, Zhang Y, Zhao M, Li Q, Chen R et al. (2016). miR-16 induction after CDK4 knockdown is mediated by c-Myc suppression and inhibits cell growth as well as sensitizes nasopharyngeal carcinoma cells to chemotherapy. Tumor Biology 37: 2425–2433. DOI 10.1007/s13277-015-3966-1. [Google Scholar] [CrossRef]
Kong L, Zhang P, Li W, Yang Y, Tian Y et al. (2016). KDM1A promotes tumor cell invasion by silencing TIMP3 in non-small cell lung cancer cells. Oncotarget 7: 27959–27974. DOI 10.18632/oncotarget.8563. [Google Scholar] [CrossRef]
Li Y, Guan B, Liu J, Zhang Z, He S et al. (2019). MicroRNA-200b is downregulated and suppresses metastasis by targeting LAMA4 in renal cell carcinoma. EBioMedicine 44: 439–451. DOI 10.1016/j.ebiom.2019.05.041. [Google Scholar] [CrossRef]
Liu W, Chen H, Wong N, Haynes W, Baker CM et al. (2017). Pseudohypoxia induced by miR-126 deactivation promotes migration and therapeutic resistance in renal cell carcinoma. Cancer Letters 394: 65–75. DOI 10.1016/j.canlet.2017.02.025. [Google Scholar] [CrossRef]
Liu Y, Uzair-Ur-Rehman, Guo Y, Liang H, Cheng R et al. (2016). miR-181b functions as an oncomiR in colorectal cancer by targeting PDCD4. Protein & Cell 7: 722–734. DOI 10.1007/s13238-016-0313-2. [Google Scholar] [CrossRef]
Ljungberg B, Bensalah K, Canfield S, Dabestani S, Hofmann F et al. (2015). EAU guidelines on renal cell carcinoma: 2014 update. European Urology 67: 913–924. DOI 10.1016/j.eururo.2015.01.005. [Google Scholar] [CrossRef]
Lu Z, Zhang W, Gao S, Jiang Q, Xiao Z et al. (2015). MiR-506 suppresses liver cancer angiogenesis through targeting sphingosine kinase 1 (SPHK1) mRNA. Biochemical and Biophysical Research Communications 468: 8–13. DOI 10.1016/j.bbrc.2015.11.008. [Google Scholar] [CrossRef]
Masson D, Rioux-Leclercq N, Fergelot P, Jouan F, Mottier S et al. (2010). Loss of expression of TIMP3 in clear cell renal cell carcinoma. European Journal of Cancer 46: 1430–1437. DOI 10.1016/j.ejca.2010.01.009. [Google Scholar] [CrossRef]
Miao D, Margolis CA, Gao W, Voss MH, Li W et al. (2018). Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 359: 801–806. DOI 10.1126/science.aan5951. [Google Scholar] [CrossRef]
Pan YJ, Wei LL, Wu XJ, Huo FC, Mou J et al. (2017). MiR-106a-5p inhibits the cell migration and invasion of renal cell carcinoma through targeting PAK5. Cell Death & Disease 8: e3155. DOI 10.1038/cddis.2017.561. [Google Scholar] [CrossRef]
Piotrowski I, Zhu X, Saccon TD, Ashiqueali S, Schneider A et al. (2021). miRNAs as biomarkers for diagnosing and predicting survival of head and neck squamous cell carcinoma patients. Cancers 13: 3980. DOI 10.3390/cancers13163980. [Google Scholar] [CrossRef]
Qi JH, Anand-Apte B (2015). Tissue inhibitor of metalloproteinase-3 (TIMP3) promotes endothelial apoptosis via a caspase-independent mechanism. Apoptosis 20: 523–534. DOI 10.1007/s10495-014-1076-y. [Google Scholar] [CrossRef]
Su CW, Su BF, Chiang WL, Yang SF, Chen MK et al. (2017). Plasma levels of the tissue inhibitor matrix metalloproteinase-3 as a potential biomarker in oral cancer progression. International Journal of Medical Sciences 14: 37–44. DOI 10.7150/ijms.17024. [Google Scholar] [CrossRef]
Tan X, Ren S, Fu MZ, Ren S, Yang C et al. (2021). microRNA-196b promotes esophageal squamous cell carcinogenesis and chemoradioresistance by inhibiting EPHA7, thereby restoring EPHA2 activity. American Journal of Cancer Research 11: 3594. [Google Scholar]
Thoma C (2016). When worlds collide—integrating metabolic and transcriptomic data in ccRCC. Nature Reviews Urology 13: 126. DOI 10.1038/nrurol.2016.16. [Google Scholar] [CrossRef]
Tian F, Shen Y, Chen Z, Li R, Lu J et al. (2016). Aberrant miR-181b-5p and miR-486-5p expression in serum and tissue of non-small cell lung cancer. Gene 591: 338–343. DOI 10.1016/j.gene.2016.06.014. [Google Scholar] [CrossRef]
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J et al. (2015). Global cancer statistics, CA: A. Cancer Journal for Clinicians 65: 87–108. DOI 10.3322/caac.21262. [Google Scholar] [CrossRef]
Wang B, Hsu SH, Majumder S, Kutay H, Huang W et al. (2010). TGFβ-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene 29: 1787–1797. DOI 10.1038/onc.2009.468. [Google Scholar] [CrossRef]
Wang XH, Li LJ, Sun GX, Wu ZP, Li JF et al. (2016). Expressions of miR-132, miR-134, and miR-485 in rat primary motor cortex during transhemispheric functional reorganization after contralateral seventh cervical spinal nerve root transfer following brachial plexus avulsion injuries. NeuroReport 27: 12–17. DOI 10.1097/WNR.0000000000000485. [Google Scholar] [CrossRef]
Wu J, Zhang F, Zhang J, Sun Z, Hao C et al. (2021). A novel miRNA-based model can predict the prognosis of clear cell renal cell carcinoma. Technology in Cancer Research & Treatment 20: 15330338211027923. [Google Scholar]
Xiao W, Lou N, Ruan H, Bao L, Xiong Z et al. (2017). MiR-144-3p promotes cell proliferation, metastasis, sunitinib resistance in clear cell renal cell carcinoma by downregulating ARID1A. Cellular Physiology and Biochemistry 43: 2420–2433. DOI 10.1159/000484395. [Google Scholar] [CrossRef]
Xie M, Lv Y, Liu Z, Zhang J, Liang C et al. (2018). Identification and validation of a four-miRNA (miRNA-21-5p, miRNA-9-5p, miR-149-5p, and miRNA-30b-5p) prognosis signature in clear cell renal cell carcinoma. Cancer Management and Research 10: 5759. DOI 10.2147/CMAR. [Google Scholar] [CrossRef]
Zhai W, Sun Y, Guo C, Hu G, Wang M et al. (2017). LncRNA-SARCC suppresses renal cell carcinoma (RCC) progression via altering the androgen receptor (AR)/miRNA-143-3p signals. Cell Death & Differentiation 24: 1502–1517. DOI 10.1038/cdd.2017.74. [Google Scholar] [CrossRef]
Zhang H, Li W, Gu W, Yan Y, Yao X et al. (2019a). MALAT1 accelerates the development and progression of renal cell carcinoma by decreasing the expression of miR-203 and promoting the expression of BIRC5. Cell Proliferation 52: E154. DOI 10.1111/cpr.12640. [Google Scholar] [CrossRef]
Zhang X, Yu J, Zhao C, Ren H, Yuan Z et al. (2019b). MiR-181b-5p modulates chemosensitivity of glioma cells to temozolomide by targeting Bcl-2. Biomedicine & Pharmacotherapy 109: 2192–2202. DOI 10.1016/j.biopha.2018.11.074. [Google Scholar] [CrossRef]
Zhao LD, Zheng WW, Wang GX, Kang XC, Qin L et al. (2016). Epigenetic silencing of miR-181b contributes to tumorigenicity in colorectal cancer by targeting RASSF1A. International Journal of Oncology 48: 1977–1984. DOI 10.3892/ijo.2016.3414. [Google Scholar] [CrossRef]
Zheng Y, Lv X, Wang X, Wang B, Shao X et al. (2016). miR-181b promotes chemoresistance in breast cancer by regulating Bim expression. Oncology Reports 35: 683–690. DOI 10.3892/or.2015.4417. [Google Scholar] [CrossRef]
Zhou Q, Zheng X, Chen L, Xu B, Yang X et al. (2016). Smad2/3/4 pathway contributes to TGF-β-induced miRNA-181b expression to promote gastric cancer metastasis by targeting Timp3. Cellular Physiology and Biochemistry 39: 453–466. DOI 10.1159/000445638. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |