DOI:10.32604/biocell.2022.016962

| BIOCELL DOI:10.32604/biocell.2022.016962 |  |
| Article |
LncRNA-POIR knockdown promotes hepatocellular carcinoma sensitivity to sorafenib through upregulating miR-182-5p and inhibiting autophagy
1Jintan Affiliated Hospital of Jiangsu University, Changzhou, 213200, China
2Tongde Hospital of Zhejiang Province, Hangzhou, 310012, China
*Address correspondence to: Weixin Yu, adam700803@163.com; Qun Xu, 15851900716@163.com
#Jian Xu and Hailong Ge contributed equally to this work
Received: 14 April 2021; Accepted: 24 August 2021
Abstract: Although sorafenib has been found to prolong the survival time of patients with hepatocellular carcinoma (HCC), sorafenib resistance remains an important challenge. Increasing studies have demonstrated that long noncoding RNAs (lncRNAs) contribute to drug resistance in a wide number of cancers. Human periodontal ligament stem cell (PDLSC) osteogenesis impairment-related lncRNA (POIR) is a recently defined lncRNA for which little is known regarding its function. Our study aimed to reveal the role of POIR in the development of HCC cell sorafenib resistance. The level of POIR expression in patients and tumor cells was examined by Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay. CCK-8, EdU, and flow cytometry assay were adopted to examine cell viability, proliferation, and apoptosis, respectively. The autophagy-associated protein expressions were determined by western blotting and autophagic flux analysis. The results of this study exhibited increased POIR in HCC tissues and cells and may be correlated with sorafenib resistance. Knockdown of POIR elevated sorafenib sensitivity by suppressing autophagy in HCC cells. Mechanically, POIR knockdown upregulated miR-182-5p, implying that miR-182-5p mediates POIR regulation. MiR-182-5p overexpression significantly enhanced chemosensitivity to sorafenib, whereas miR-182-5p inhibition had the opposite effect. The sensitization of POIR siRNA to sorafenib was abolished by co-transfection with miR-182-5p inhibitor. Our findings provide a potential target for further clinical treatment of sorafenib-resistant HCC patients.
Keywords: Hepatocellular carcinoma; lncRNA-POIR; miR-182-5p; Resistance; Autophagy
Hepatocellular carcinoma (HCC) is a common malignant liver cancer with an incidence rate from 1.6 per 100,000 individuals to 4.6 per 100,000 individuals, that is rising more quickly compared to other cancers in the world (Cronin et al., 2018; Rawla et al., 2018; Siegel et al., 2019). It has been reported that approximately 71% of cases are potentially preventable due to avoidable risk factors (e.g., smoking, hepatitis B, and C viruses) (Islami et al., 2018). Despite the developments in medical technology which resulted in substantial achievements for the treatment of HCC, the five-year relative survival rate is the lowest of all the liver cancers (18%), second only to pancreatic cancer (9%) (Siegel et al., 2019; Yang et al., 2019). Sorafenib represents a Food and Drug Administration-(FDA) approved first-line drug for treating advanced HCC (Mousa, 2008). Although sorafenib therapy extends the survival time to 10.7 months, compared with the placebo group of 7.9 months, patients with advanced HCC develop a resistance to sorafenib treatment within only a few weeks (Llovet, 2007). Therefore, an understanding of the molecular mechanism associated with sorafenib resistance will be useful for HCC therapy.
Noncoding RNAs (ncRNAs), including short noncoding RNAs and long noncoding RNAs (lncRNAs), have been revealed to be involved in the tumorigenesis and development of HCC (He et al., 2019; Huang et al., 2018; Su et al., 2019). In addition, increasing evidence has shown that ncRNAs also participate in HCC drug resistance (Chen and Xia, 2019; Li et al., 2019). LncRNAs are a type of ncRNA with lengths larger than 200 nt (Wong et al., 2018). In cancer, lncRNAs play a role through a variety of mechanisms, including chromatin remodeling, chromatin interaction, and CeRNAs (Fang and Fullwood, 2016). It has been reported that many lncRNAs are abnormally expressed in HCC and are associated with tumor progression and resistance (Wei et al., 2019). For example, lncRNA HOXA11‑AS has been illustrated to promote HCC progression via sponging miR‑506‑3p as a ceRNA (Liu et al., 2020). In addition, lncRNA CRNDE silencing effectively reduced HCC cell chemotherapy resistance by mediating the epigenetic suppression of CELF2 and LATS2 on multiple tumor suppressor genes (Xie et al., 2020). Long noncoding RNA ZFPM2-AS1 facilitates cell invasion through regulating miR-139/GDF10 axis in HCC (He et al., 2020). In exploring the lncRNAs-related mechanism involved in sorafenib resistance, we found that ENST00000446358 expression was obviously overexpressed in HCC tissues.
Human periodontal ligament stem cell (PDLSC) osteogenesis impairment-related lncRNA (POIR, ENST00000446358) was recently found by RNA-sequencing technology (Wang et al., 2016). However, the specific impact of POIR on tumors remains limited. Recently, only Chen et al. (2021) demonstrated that knockdown of POIR sensitizes HCC cells to sorafenib by suppressing the epithelial-mesenchymal transition. Therefore, we need to further explore the effect of POIR in HCC sorafenib resistance.
Increasing evidence shows that there are many factors identified contributing to sorafenib resistance, including epigenetics, transport processes, regulated cell death, and the tumor microenvironment (Tang et al., 2020). Among them, autophagy is also thought to be an important mechanism of drug resistance. However, the relation between POIR and autophagy has not been reported. Therefore, we want to know whether POIR regulates autophagy in sorafenib resistance.
The previous study has shown that miR-182-5p is the downstream target of POIR (Wang et al., 2016). MiR-182-5p has also been shown to regulate autophagy in other diseases (Mo et al., 2021; Xie et al., 2019). However, it is not clear whether POIR regulates autophagy in HCC by regulating miR-182-5p. In this study, we found that POIR expression was obviously higher in HCC tissues and cell lines by reverse transcription‑quantitative polymerase chain reaction (RT‑qPCR) analysis. The present study also demonstrated that POIR siRNA decreased cell growth and facilitated cell apoptosis in the presence of sorafenib. Additionally, POIR siRNA markedly reversed sorafenib‑induced cell autophagy. Furthermore, POIR silencing sensitized HCC cells to sorafenib by regulating autophagy through miR-182-5p. Therefore, POIR may be employed as a candidate target for the treatment of HCC sorafenib resistance in the future.
Fifty-two pairs of HCC tumor tissues and para-tumor tissues were collected from the Second Affiliated Hospital, School of Medicine, Zhejiang University. This study was approved by the Ethics Committee of the Second Affiliated Hospital, School of Medicine, Zhejiang University. Written informed consent was obtained from HCC patients who underwent surgery. Accession numbers of RNA, DNA and protein sequences used in the manuscript should be provided.
Huh-7, Hep3B, and SUN449 cells were provided and authenticated by ATCC (Manassas, VA, USA). Huh-7 cells were cultivated in DMEM, Hep3B cells were maintained in MEM, and SNU-449 cells were cultivated in RPMI media 1640. These media were purchased from Gibco (Gibco, Carlsbad, CA, USA). All of the cells were maintained in a medium containing 10% fetal bovine serum (FBS, Gibco), 1% penicillin and 1% streptomycin.
RNA isolation and RT‑qPCR were performed as described earlier (Chen et al., 2020). The total RNA was isolated from HCC tissue and cells using TRIzol agent. Next, the cDNA was generated using PrimeScript™ RT reagent Kit with gDNA Eraser (RR047A, Takara, Dalian, China) for lncRNA and MiR-X miRNA First-Strand Synthesis Kit (638315, Takara) for miRNA. RT‑qPCR was conducted using TB Green® Premix Ex Taq™ (RR420A, Takara) according to the specification. The relative gene expression was evaluated by the 2−ΔΔCt method (Livak and Schmittgen, 2001). GAPDH and U6 acted as the internal control, respectively. The sequences of POIR and miR-182 primers utilized in this study are listed in Table 1. Rest primers sequences were listed in Table S1.
Small interfering (si)RNA lnc POIR or scrambled siRNA was synthesized by GenePharma Company (Shanghai, China). miR-182 mimic/inhibitor or a negative mimic/inhibitor control were synthesized by Ribobio, respectively (Ribo, China). The transfection was transfected using Lipofectamine® 2000 (Invitrogen; Carlsbad, CA, USA) according to the instructions. The sequences used in this study are listed in Table 2.
Cytotoxicity assay was assayed using Cell Counting Kit-8 (CCK-8) (Dojindo, Japan). The CCK-8 assay was performed after the seeding of HCC cells (5,000 cells/well) in 96-well plates. Afterward, the cells were treated with 0, 1.25, 2.5, 5, 10, or 20 μM sorafenib for 48 h. The supernatant was discarded and replaced with a fresh 100 µL serum-free medium (containing 10 µL CCK-8 reagents). Absorbance was measured at 450 nm using a microplate reader (BioTek).
Western blotting was performed as previously described. Proteins were lysed in a RIPA buffer (Beyotime, China). Next, the protein concentrations were determined using a bicinchoninic acid protein assay (Beyotime). The lysed protein (40 µg) was electrophoresed with 10% SDS-PAGE and transferred to PVDF membranes (Millipore, USA), followed by incubation with 5% non-fat milk for 2 h. Subsequently, the membranes were incubated with primary antibodies specifically for LC3B (#3868S, Cell Signaling Technology, 1:1000 dilution), P62 (#88588S, CST, 1:1000 dilution), GAPDH (#2118S, CST, 1:2000) at 4°C overnight, followed by an incubation with HRP secondary antibodies (anti-mouse, #7076S; anti-Rabbit, #7074S; 1:2,000, CST) for 2 h at room temperature. The chemiluminescence intensity was evaluated using ECL reagents (Applygen, Beijing, China).
5-Ethynyl-2’-deoxyuridine (EdU) assay
Cell proliferation was analyzed by employing a Click-iTEdU Imaging kit (Invitrogen; Thermo Fisher Scientific, Inc.) as previously described. Briefly, the fixed cells were incubated with 100 µL EdU to detect the positive cells. The cells were counterstained with 100 µL Hoechst 33342 to label the nuclei. Immunofluorescence was observed with a fluorescence microscope at 200× magnification.
mRFP-GFP-LC3 adenovirus was bought from Hanbio (Hanbio, China). The cells that were incubated with an mRFP-GFP-LC3 adenovirus for 24 h were exposed to sorafenib for another 24 h. Autophagic flux was observed with a fluorescent microscope (Olympus, Tokyo, Japan).
The apoptosis assay was performed using an Annexin V-FITC/propidium iodide (PI) apoptosis detection kit according to the user’s guide (Beyotime, Shanghai, China). The harvested cells were suspended and incubated with Annexin V and PI (1:1) in the dark. The analysis of the apoptotic cells was performed using flow cytometry (BD).
Data are presented as the mean ± SD. Statistical analyses were carried out using SPSS software (19.0 revision, IBM, Chicago, IL, USA) and GraphPad Prism (version 7; GraphPad Software, Inc.). Student’s t-test was used to calculate the two group differences, while one-way ANOVA was applied to compare the multiple group differences. P < 0.05 was considered as a statistically significant difference.
POIR was significantly upregulated in HCC tissues and cells, and POIR may be associated with HCC resistance to sorafenib
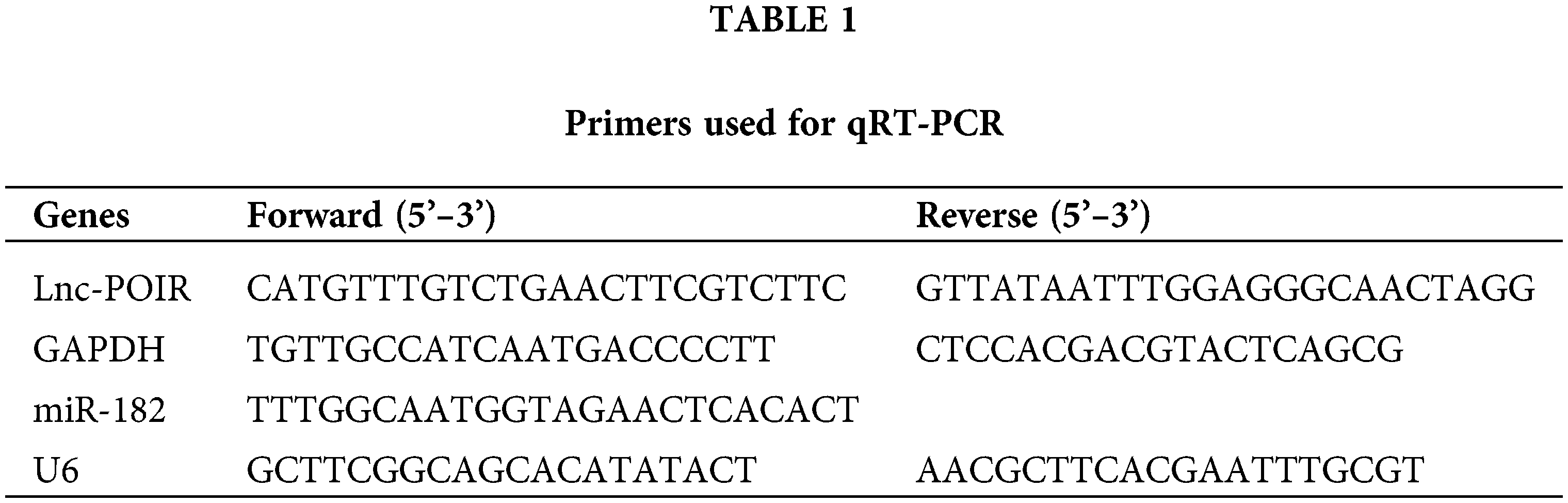
To identify the effect of lncRNAs on the response of HCC cells to sorafenib, we explored the expression profiles of lncRNAs in three pairs of tumor tissues and adjacent tissues from patients with HCC using RT-qPCR (Fig. 1A). Among the 25 lncRNAs, we were interested in POIR. Furthermore, we used RT-qPCR to detect the level of POIR expression in 52 liver cancer tissues. The results showed that POIR expression in the HCC tissues was significantly higher than that in the adjacent tissues (Fig. 1B). The level of POIR expression in each of the 52 cases was shown in Fig. 1C, of which 41 cases exhibited increased POIR expression. These data suggest that POIR may be an oncogene. In addition, we detected the cytotoxicity of sorafenib in HCC cells. The IC50 of SNU449 cells (14.82 ± 0.66 μM), which exhibit mesenchymal phenotype (Xue et al., 2016), was higher than that of Huh7 (6.81 ± 0.25 μM) and Hep3B cells (9.89 ± 0.69 μM), which have epithelial phenotype (Figs. S1A–B). At the same time, we also found that POIR expression was significantly higher in HCC cells than that in the LO2 cells, with POIR expression of SNU449 being the highest, followed by Hep3B and Huh-7 (Fig. S1C). This suggests that POIR may be associated with HCC resistance to sorafenib.
POIR depletion increased sorafenib sensitivity in HCC cells
To assess the effects of POIR on HCC resistance to sorafenib, we constructed POIR siRNA to achieve POIR knockdown. The efficiency of these three HCC cells with POIR siRNA was measured by RT-qPCR (Fig. S1D). When the cells were transfected with POIR siRNA, the cell viability was dramatically inhibited in all three HCC cell lines compared with NC siRNA (NC-si) (Fig. S1E and Table S2). Compared with control, sorafenib could inhibit cell proliferation, as shown by the decrease of the EDU-positive cell ratio (Fig. 1D). POIR siRNA could enhance the inhibitory effect of sorafenib on proliferation (Fig. 1D). Moreover, POIR siRNA could further potentiate the level of cell apoptosis induced by sorafenib (Fig. 1E). Overall, these data suggest that silencing POIR promoted the sensitization of HCC cells to sorafenib.
Figure 1: POIR was significantly upregulated in HCC tissues and may be correlated with sorafenib resistance. (A) Heat map of lncRNAs in three pairs of HCC tissues. (B and C) Relative expression of POIR in 52 pairs of HCC tissues. Among the 52 samples, 41 samples exhibited upregulated POIR. **P < 0.01. (D) The cell proliferation of HCC cells was examined by an EdU assay after treatment with sorafenib (IC50 concentrations) or sorafenib with POIR siRNA. (×200) ***P < 0.001 vs. NC, &&&P < 0.001 vs. sorafenib. (E) The apoptosis of HCC cells was determined by a flow cytometry assay following treatment with sorafenib or sorafenib with POIR siRNA. ***P < 0.001 vs. NC, &&&P < 0.001 vs. sorafenib.
Knockdown of POIR enhanced sorafenib sensitivity by inhibiting autophagy
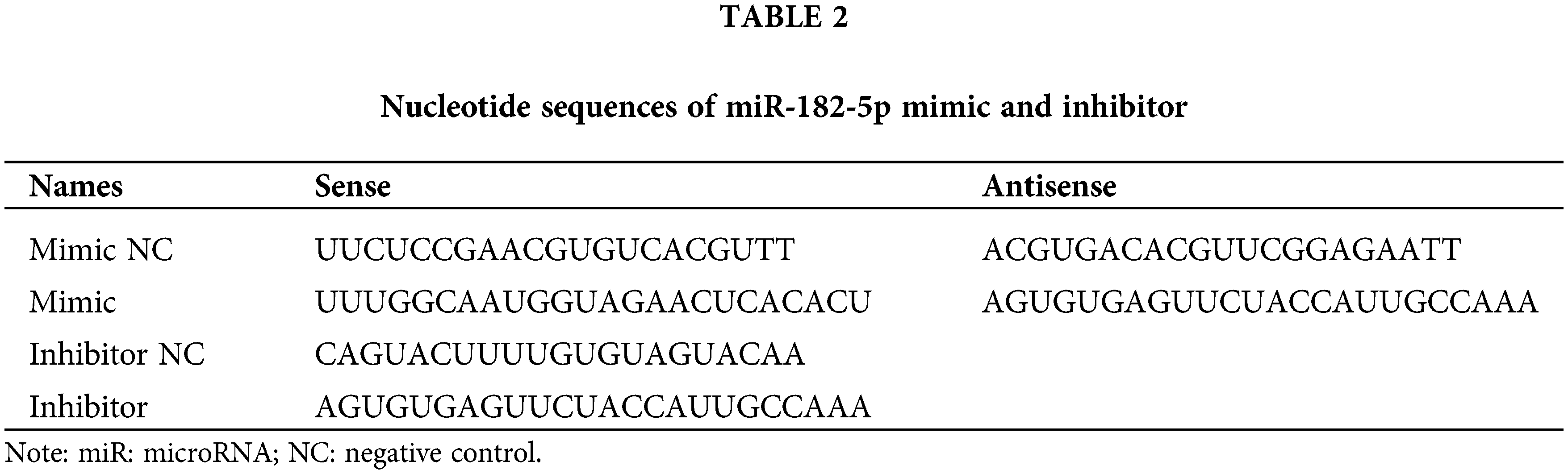
Previous studies have linked autophagy to drug resistance in human cancers (Niu et al., 2017). Therefore, we hypothesized that autophagy contributes to POIR silencing-mediated sorafenib sensitivity. To test this hypothesis, we used 3-methyladenine (3-MA), an autophagy inhibitor. Compared with control, 3-MA treatment enhanced the sensitivity of HCC cells to sorafenib (Fig. 2A). The result indicated that autophagy is involved in sorafenib resistance. To determine whether POIR knockdown mediates sorafenib sensitivity by regulating autophagy in HCC, HCC cells were transfected with or without POIR siRNA. We performed confocal microscopy to observe the change of autophagy flux in transfected HCC cells exposed to sorafenib. The results showed that POIR silencing decreased the autophagy flux induced by sorafenib (Figs. 2B and 2C). The results of the western blot analyses were consistent with the confocal microscopy assay (Fig. 2D), indicating POIR might mediate sorafenib resistance via autophagy.
Figure 2: POIR knockdown attenuated sorafenib-induced autophagy in HCC cells. (A) CCK-8 assay was performed to measure the relative viability of HCC cells in the absence or presence of 3-MA. (B) Representative immunostaining images of LC3 in HCC cells treated with sorafenib and POIR-knockdown. (×1000) (C) The number of GFP-LC3 puncta/cells was quantified in Huh7, Hep3B and SNU449 cells, respectively. (D) The level of P62 and LC3 I/II protein expression in POIR-knockdown HCC cells treated with sorafenib. **P < 0.01, ***P < 0.001 vs. NC, &P < 0.05, &&P < 0.01, &&&P < 0.001 vs. sorafenib.
To further investigate that POIR siRNA increased cell sensitivity to sorafenib by regulating autophagy, we used 3-MA to interfere the autophagic process. Interestingly, in the presence of 3-MA, POIR inhibition no longer influence the cell viability (Fig. 3). Therefore, our findings confirmed that inhibiting POIR increases cell sensitivity to sorafenib by regulating autophagy.
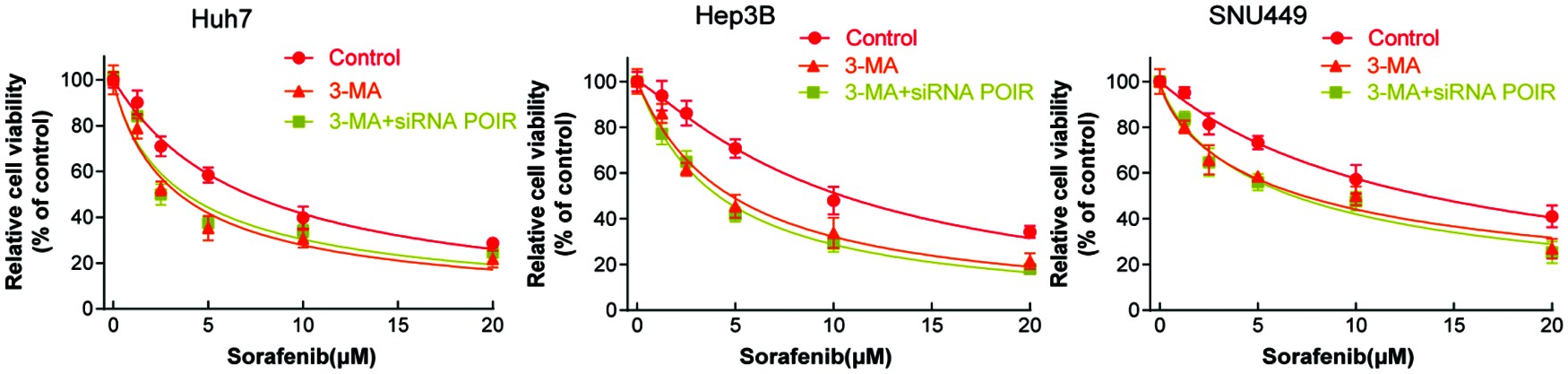
Figure 3: POIR inhibition sensitizes HCC cells to sorafenib via autophagy. HCC cells were treated with 3-MA or 3-MA + POIR siRNA and then exposed to different concentration of sorafenib, then the cell viability was measured by the CCK-8 assay.
MiR-182-5p, as a target of POIR, can regulate sorafenib resistance
It has been reported that POIR acted as a sponge of miR-182-5p and promoted the osteogenesis of PDLSCs (Wang et al., 2016). However, whether POIR mediates sorafenib resistance by regulating miR-182-5p is unknown. Therefore, we hypothesized that POIR siRNA regulates sorafenib sensitivity in HCC by inhibiting autophagy through interacting with miR-182-5p. To further verify the above hypothesis, we first performed miRNA target site prediction using a DIANA software analysis (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php). We found that POIR contains miR-182-5p binding sites (Fig. 4A). We also found that inhibiting POIR by POIR siRNA upregulated miR-182-5p expression in HCC cells (Fig. 4B). Next, we synthesized miR-182-5p mimic and miR-182-5p inhibitor to observe the effect of miR-182-5p on sorafenib resistance. We used CCK-8 and EDU assays to assess their effect. The results demonstrated that miR-182-5p overexpression significantly enhanced sorafenib sensitivity and suppressed cell proliferation compared with NC. Conversely, the downregulation of miR-182-5p inhibited sorafenib sensitivity and promoted the EDU-positive cell ratio (Figs. 4C–4E). The efficiency was detected by RT-qPCR (Fig. 4F).
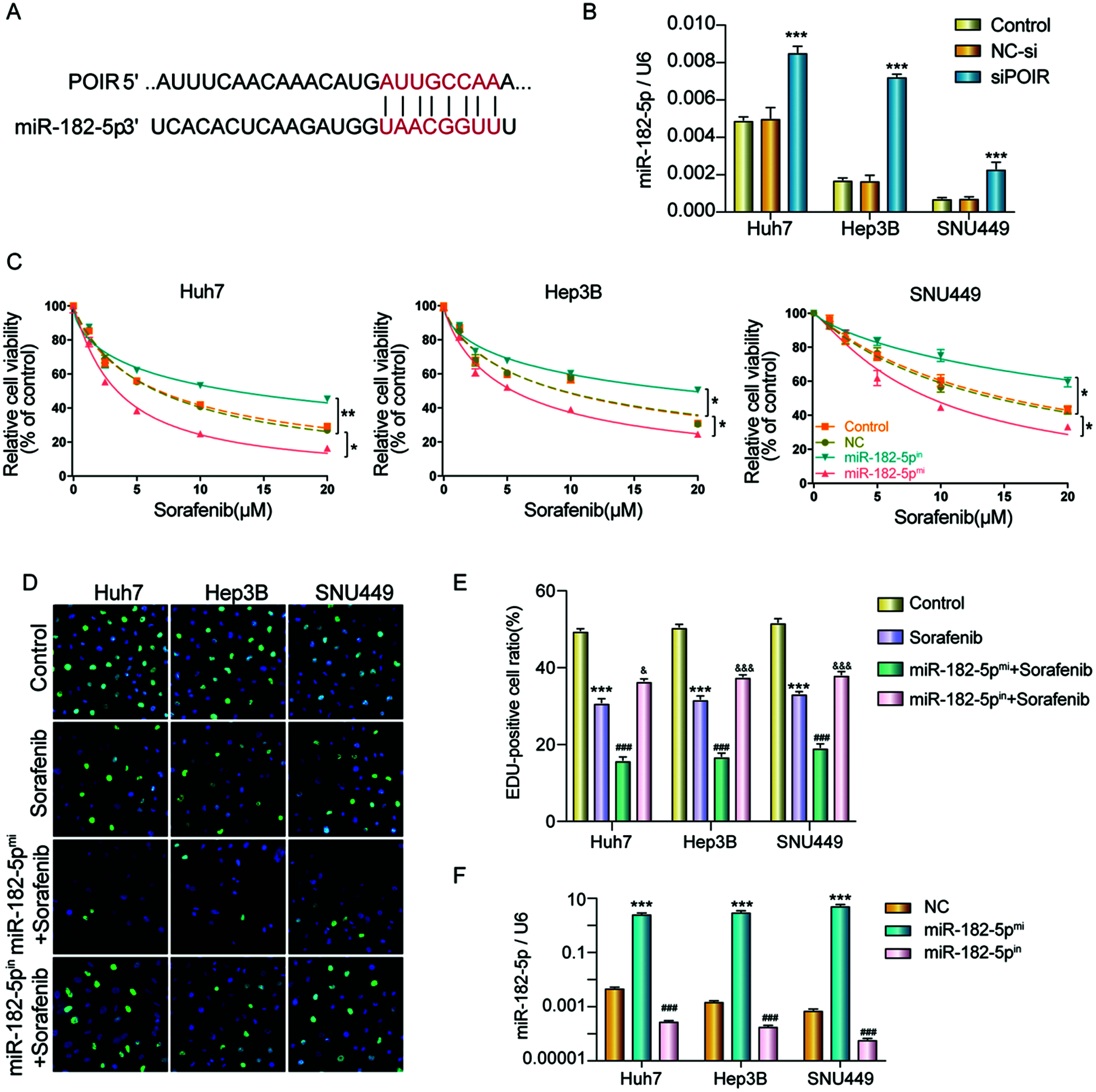
Figure 4: MiR-182-5p functioned as a target of POIR, and miR-182-5p could regulate the sorafenib resistance. (A) Schematic of the miR-182-5p putative target site in the POIR. (B) The effect of POIR on miR-182-5p expression was determined by qPCR. ***P < 0.001 vs. NC. (C) When the cells interfered with miR-182-5p mimic or miR-182-5p inhibitor, respectively, cell viability was measured using a CCK-8 assay in HCC cells treated with sorafenib. *P < 0.05, **P < 0.01. (D and E) The proliferation of HCC cells was examined by EdU assay following treatment with sorafenib or sorafenib combined with mir-182-5p mimic or inhibitor. (×200)***P < 0.001 vs. NC. (F) The efficiency of mir-182-5p mimic or mir-182-5p inhibitor was determined by qPCR. ***P < 0.001 vs. NC.
POIR knockdown regulated sorafenib sensitivity by miR-182-5p and autophagy in SNU449 cells
To further explore the relationship between POIR, miR-182-5p, and autophagy, SNU449 cells were co-transfected with NC siRNA, miR-182-5p inhibitor, POIR siRNA, and POIR siRNA and miR-182 inhibitor, respectively. The CCK-8 assay showed that POIR downregulation increased the sorafenib sensitivity in SNU449 cells, whereas the miR-182-5p inhibitor abolished these effects (Fig. 5A). Furthermore, flow cytometry assay revealed that miR-182-5p inhibitor reversed the effects of POIR knockdown on SNU449 cell apoptosis (Figs. 5B and 5C). Confocal microscopy and Western blot assay showed that the role of POIR siRNA on the autophagy of SNU449 cells could be reversed by a miR-182-5p inhibitor (Figs. 5D–5F). The efficiency of 449 cells with POIR siRNA, miR-182-5p mimic or miR-182-5p inhibitor was measured by RT-qPCR (Fig. 5G). These data indicate that silencing POIR enhanced sorafenib sensitivity via regulating miR-182-5p and autophagy.
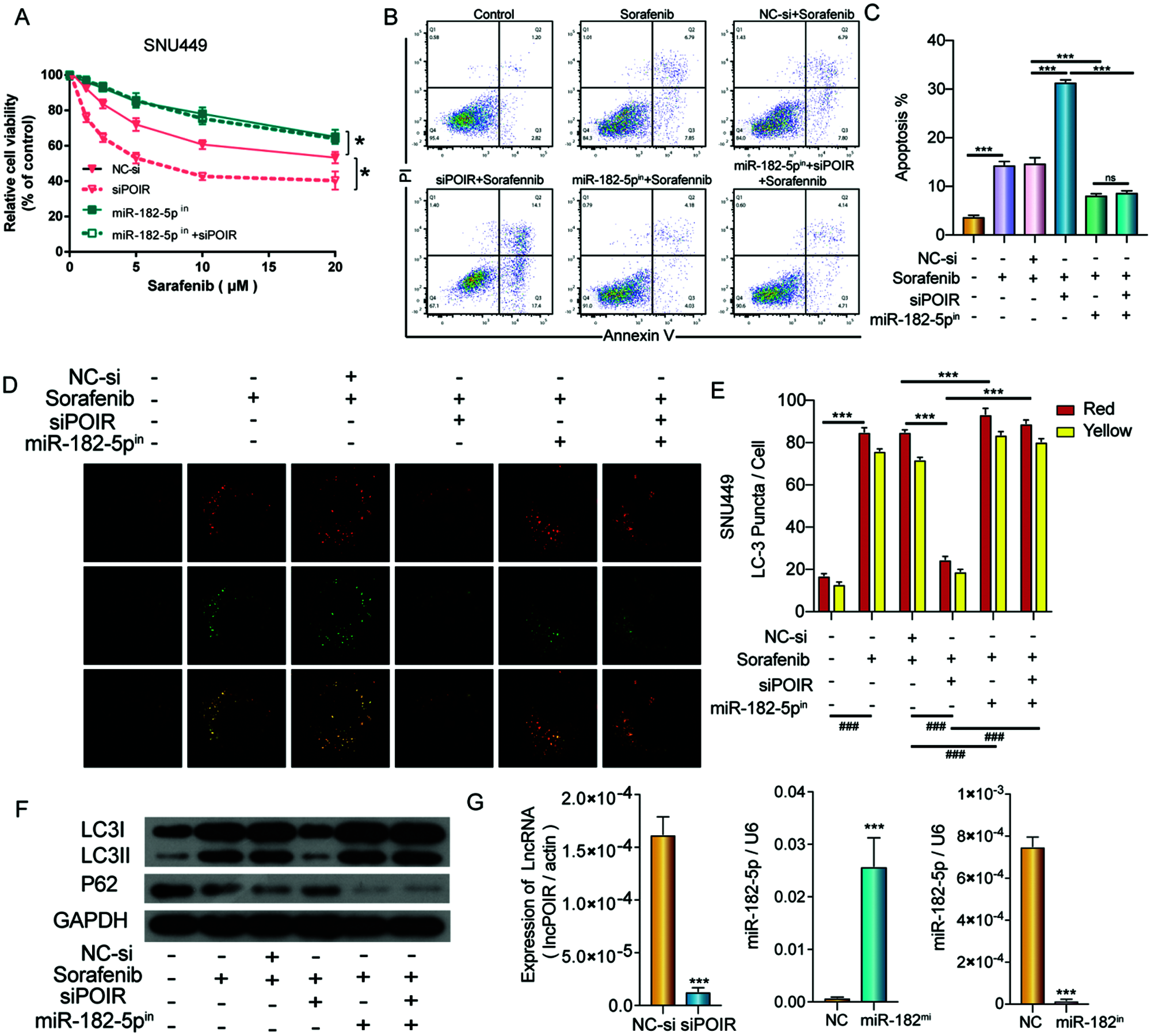
Figure 5: POIR knockdown regulated sorafenib sensitivity by miR-182-5p and autophagy in SNU449 cells. (A) Treatment with miR-182-5p inhibitor reversed the effects of POIR siRNA on SNU449 cells treated with sorafenib. *P < 0.05. (B and C) Treatment with miR-182-5p inhibitor reversed the effects of POIR siRNA on SNU449 apoptosis following sorafenib treatment. ***P < 0.001. (D–F) Treatment with miR-182-5p inhibitor rescued the effects of POIR siRNA on SNU449 autophagy under sorafenib treatment. (×1000) ***P < 0.001. (G) The expression of POIR and miR-182-5p after transfection with POIR siRNA, miR-182-5p mimic, or miR-182-5p inhibitor were determined by qPCR. ***P < 0.001.
Sorafenib, a multi-target kinase inhibitor, can block tumor cell proliferation and angiogenesis through multiple signal pathways. Although sorafenib prolongs survival time and limits its side effects in liver cancer patients, it may also cause resistance which has become an obstacle in extending overall survival time. Several mechanisms were involved in sorafenib resistance, including EMT, autophagy, hypoxia, and epigenetic regulation. Hence, further research is essential to clarify the sorafenib resistance mechanisms involved and identify predictive biomarkers.
Recently, lncRNAs have been demonstrated to play a key role in cancer drug resistance, including HCC. For example, Gao et al. (2021) found that Lnc LEF1-AS1/miR-10a-5p enhances MSI1 expression and promotes chemoresistance in HCC by activating the AKT signaling pathway. Huang et al. (2018) revealed that LncRNA NR2F1-AS1 regulates HCC oxaliplatin resistance by targeting ABCC1 via miR-363. The study by Li et al. (2020a) revealed that downregulation of LINC00467 promoted axitinib sensitivity in HCC through the miR-509-3p/PDGFRA axis. In the current study, we found that POIR was observably upregulated in HCC patients and HCC cell lines. Subsequent loss-of-function assays showed that POIR knockdown enhanced sorafenib sensitivity, implying it is a promising strategy to treat drug resistance by targeting lncRNA. However, how can this strategy be applied to the clinic? Recently, developing effective therapies for silencing (oncogene) or overexpressing (tumor suppressor gene) ncRNA has become an attractive research field (Slack and Chinnaiyan, 2019). Antisense oligonucleotides (ASOs) with chemical modifications therapy is proposed to use to target for oncogene (Arun et al., 2018). However, its validation requires further clinical research.
Autophagy is an important cellular degradation process in which damaged proteins or organelles are encapsulated and sent to lysosomes for degradation and circulation. In cancer, autophagy plays a double-edged role. In some cases, autophagy is a protective mechanism for cancer cells exposed to multiple anticancer drugs, and inhibiting autophagy augments the effect of anti-cancer drugs on cancer cells (Choi, 2012; Doherty and Baehrecke, 2018; Yang et al., 2011). In other cases, autophagy induced by a chemotherapeutic drug is considered to be an antitumor mechanism (Sui et al., 2013). Thus, we want to know the effect of autophagy in HCC sorafenib resistance and whether POIR was involved in the regulation of tumor resistance through autophagy mechanisms. To test conjecture, we first treated the cells with 3-MA and found that 3-MA enhanced the effect of sorafenib. Secondly, sorafenib promoted autophagy, which is consistent with previous studies (Park et al., 2010). The effect of sorafenib in autophagy was reversed by POIR silencing by detecting autophagy flux and autophagy-related markers. Importantly, in the presence of 3-MA, POIR inhibition no longer influence the cell viability. These results suggest that POIR regulates tumor resistance through autophagy.
Recent evidence suggests that long non-coding RNAs participate in tumorigenesis and drug resistance by regulating microRNAs (Jiang et al., 2020; Wei et al., 2019). Similar to our results, previous studies have confirmed that POIR acts as a sponge of miR-182-5p (Wang et al., 2016). MiR-182-5p is reported to function as a putative oncogenic or tumor-suppressive factor and plays a role in various cancers (Gu et al., 2020; Li et al., 2020b; Yan et al., 2020). In this study, we found that miR-182-5p overexpression resulted in the sensitization of HCC cells to sorafenib, whereas miR-182 inhibition led to elevated sorafenib resistance of HCC cells. More importantly, we found that miR-182-5p inhibitor could reverse the decreased autophagy and increased apoptosis effect induced by POIR deficiency. Therefore, it is expected to be a valuable strategy for combination therapy of POIR with sorafenib for HCC. However, further vivo experiments and mechanism studies, such as the reasons for the high expression of POIR and how to regulate autophagy, are still needed for verification.
In conclusion, the findings from this study demonstrate that POIR functioned as an oncogene in HCC, and knockdown of POIR sensitized HCC cells to sorafenib by regulating miR-182-5p and autophagy. Thus, POIR may serve as a potential therapeutic target for HCC treatment.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Authors’ Contribution: The authors confirm contribution to the paper as follows: study conception and design: Weixin Yu, Qun Xu; data collection: Chen Chao, Feng Mo, Yu Wang, Dengkui Zhang; analysis and interpretation of results: Xiaoxiao Zheng, Li Zheng. Xuemeilu, Wei Chen; draft manuscript preparation: Jian Xu, Hailong Ge. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: The present study was approved by the Research Ethics Committee of Second Affiliated Hospital, School of Medicine, Zhejiang University (Approval Nos. 2018-238, 20180424).
Funding Statement: The study was supported by Zhejiang Provincial Nature Science Foundation of China (LR20H160001), Key R&D projects of Zhejiang Province (2020C03G5263593), Zhejiang Provincial Ten Thousand Plan for Young Top Talents (2018), Training objects of health innovative talents of Zhejiang Health (2018), Key Project Co-constructed by Zhejiang Province and Ministry (WKJ-ZJ-1916), Natural Science Foundation of China (81972693, 81802383, 81972674, 81673809 and 31900543), Zhejiang Provincial Traditional Chinese Medicine Science and Technology Project (2020ZZ004).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Arun G, Diermeier SD, Spector DL (2018). Therapeutic targeting of long non-coding RNAs in cancer. Trends in Molecular Medicine 24: 257–277. DOI 10.1016/j.molmed.2018.01.001. [Google Scholar] [CrossRef]
Chen BW, Zhou Y, Wei T, Wen L, Zhang YB et al. (2021). lncRNA-POIR promotes epithelial-mesenchymal transition and suppresses sorafenib sensitivity simultaneously in hepatocellular carcinoma by sponging miR-182-5p. Journal of Cellular Biochemistry 122: 130–142. DOI 10.1002/jcb.29844. [Google Scholar] [CrossRef]
Chen SW, Xia XH (2019). Long noncoding RNA NEAT1 suppresses sorafenib sensitivity of hepatocellular carcinoma cells via regulating miR-335-c-Met. Journal of Cellular Physiology 234: 14999–15009. DOI 10.1002/jcp.27567. [Google Scholar] [CrossRef]
Chen YJ, Qiu FH, Huang LC, Liu WP, Li LQ, Ji CH, Zeng XQ, Qiao LL, Liu MQ, Gong XQ (2020). Long non-coding RNA LINC00312 regulates breast cancer progression through the miR-9/CDH1 axis. Molecular Medicine Reports 21: 1296–1303. [Google Scholar]
Choi KS (2012). Autophagy and cancer. Experimental & Molecular Medicine 44: 109–120. DOI 10.3858/emm.2012.44.2.033. [Google Scholar] [CrossRef]
Cronin KA, Lake AJ, Scott S, Sherman RL, Noone AM et al. (2018). Annual Report to the Nation on the Status of Cancer, Part I: National cancer statistics. Cancer 124: 2785–2800. DOI 10.1002/cncr.31551. [Google Scholar] [CrossRef]
Doherty J, Baehrecke EH (2018). Life, death and autophagy. Nature Cell Biology 20: 1110–1117. DOI 10.1038/s41556-018-0201-5. [Google Scholar] [CrossRef]
Fang YW, Fullwood MJ (2016). Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics, Proteomics & Bioinformatics 14: 42–54. DOI 10.1016/j.gpb.2015.09.006. [Google Scholar] [CrossRef]
Gao J, Dai C, Yu X, Yin XB, Zhou F (2021). Long noncoding RNA LEF1-AS1 acts as a microRNA-10a-5p regulator to enhance MSI1 expression and promote chemoresistance in hepatocellular carcinoma cells through activating AKT signaling pathway. Journal of Cellular Biochemistry 122: 86–99. DOI 10.1002/jcb.29833. [Google Scholar] [CrossRef]
Gu CH, Zhao KY, Zhou NC, Liu F, Xie F et al. (2020). UBAC2 promotes bladder cancer proliferation through BCRC-3/miRNA-182-5p/p27 axis. Cell Death & Disease 11: 733. DOI 10.1038/s41419-020-02935-7. [Google Scholar] [CrossRef]
He H, Wang YW, Ye P, Yi DH, Cheng Y, Tang HB, Zhu Z, Wang X, Jin S (2020). Long noncoding RNA ZFPM2-AS1 acts as a miRNA sponge and promotes cell invasion through regulation of miR-139/GDF10 in hepatocellular carcinoma. Journal of Experimental & Clinical Cancer Research 39: 159. DOI 10.1186/s13046-020-01664-1. [Google Scholar] [CrossRef]
He J, Zuo QZ, Hu B, Jin HJ, Wang C et al. (2019). A novel, liver-specific long noncoding RNA LINC01093 suppresses HCC progression by interaction with IGF2BP1 to facilitate decay of GLI1 mRNA. Cancer Letters 450: 98–109. DOI 10.1016/j.canlet.2019.02.033. [Google Scholar] [CrossRef]
Huang H, Chen J, Ding CM, Jin X, Jia ZM, Peng J (2018). Lnc RNA NR 2F1-AS 1 regulates hepatocellular carcinoma oxaliplatin resistance by targeting ABCC 1 via miR-363. Journal of Cellular and Molecular Medicine 22: 3238–3245. DOI 10.1111/jcmm.13605. [Google Scholar] [CrossRef]
Islami F, Goding Sauer A, Miller KD, Siegel RL, Fedewa SA et al. (2018). Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA: A Cancer Journal for Clinicians 68: 31–54. DOI 10.3322/caac.21440. [Google Scholar] [CrossRef]
Jiang WX, Xia J, Xie SD, Zou RM, Pan SY, Wang ZW, Assaraf YG, Zhu XQ (2020). Long non-coding RNAs as a determinant of cancer drug resistance: Towards the overcoming of chemoresistance via modulation of lncRNAs. Drug Resistance Updates 50: 100683. DOI 10.1016/j.drup.2020.100683. [Google Scholar] [CrossRef]
Li R, Xu T, Wang HT, Wu N, Liu F, Jia XJ, Mi J, Lv JZ, Gao HQ (2019). Dysregulation of the miR-325-3p/DPAGT1 axis supports HBV-positive HCC chemoresistance. Biochemical and Biophysical Research Communications 519: 358–365. DOI 10.1016/j.bbrc.2019.08.116. [Google Scholar] [CrossRef]
Li W, He YF, Chen W, Man WL, Fu Q, Tan HT, Guo HQ, Zhou JN, Yang P (2020a). Knockdown of LINC00467 contributed to Axitinib sensitivity in hepatocellular carcinoma through miR-509-3p/PDGFRA axis. Gene Therapy 132: 2557. DOI 10.1038/s41434-020-0137-9. [Google Scholar] [CrossRef]
Li ZX, Zhang LB, Liu ZQ, Huang TX, Wang Y et al. (2020b). miRNA-182 regulated MTSS1 inhibits proliferation and invasion in Glioma cells. Journal of Cancer 11: 5840–5851. DOI 10.7150/jca.47588. [Google Scholar] [CrossRef]
Liu YH, Yan WZ, Zhou DF, Jin GH, Cheng X (2020). Long non-coding RNA HOXA11-AS accelerates cell proliferation and epithelial-mesenchymal transition in hepatocellular carcinoma by modulating the miR-506-3p/Slug axis. International Journal of Molecular Medicine 46: 1805–1815. [Google Scholar]
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. DOI 10.1006/meth.2001.1262. [Google Scholar] [CrossRef]
Llovet J (2007). For the SHARP investigators study group; sorafenib improves survival in advanced hepatocellular carcinoma (HCC); results of a phase III randomized placebo-controlled trial (SHARP) trial. American Society of Clinical Oncology, Vol. 25, No. 1, pp. 1s. [Google Scholar]
Mo YP, Wu HR, Zheng XJ, Xu L, Liu LL, Liu Z (2021). LncRNA CHRF aggravates myocardial ischemia/reperfusion injury by enhancing autophagy via modulation of the miR-182-5p/ATG7 pathway. Journal of Biochemical and Molecular Toxicology 35: e22709. [Google Scholar]
Mousa AB (2008). Sorafenib in the treatment of advanced hepatocellular carcinoma. Saudi Journal of Gastroenterology: Official Journal of the Saudi Gastroenterology Association 14: 40. DOI 10.4103/1319-3767.37808. [Google Scholar] [CrossRef]
Niu LL, Liu LP, Yang SL, Ren JW, Lai PB, Chen GG (2017). New insights into sorafenib resistance in hepatocellular carcinoma: Responsible mechanisms and promising strategies. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer 1868: 564–570. DOI 10.1016/j.bbcan.2017.10.002. [Google Scholar] [CrossRef]
Park MA, Reinehr R, Häussinger D, Voelkel-Johnson C, Ogretmen B, Yacoub A, Grant S, Dent P (2010). Sorafenib activates CD95 and promotes autophagy and cell death via Src family kinases in gastrointestinal tumor cells. Molecular Cancer Therapeutics 9: 2220–2231. DOI 10.1158/1535-7163.MCT-10-0274. [Google Scholar] [CrossRef]
Rawla P, Sunkara T, Muralidharan P, Raj JP (2018). Update in global trends and aetiology of hepatocellular carcinoma. Contemporary Oncology 22: 141–150. DOI 10.5114/wo.2018.78941. [Google Scholar] [CrossRef]
Siegel RL, Miller KD, Jemal A (2019). Cancer statistics. CA: A Cancer Journal for Clinicians 69: 7–34. [Google Scholar]
Slack FJ, Chinnaiyan AM (2019). The role of non-coding RNAs in oncology. Cell 179: 1033–1055. DOI 10.1016/j.cell.2019.10.017. [Google Scholar] [CrossRef]
Su Y, Lv XR, Yin W, Zhou LL, Hu YL, Zhou A, Qi Z (2019). CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Sedentary Life and Nutrition 11: 8182–8203. DOI 10.18632/aging.102312. [Google Scholar] [CrossRef]
Sui X, Chen R, Wang Z, Huang Z, Kong N et al. (2013). Autophagy and chemotherapy resistance: A promising therapeutic target for cancer treatment. Cell Death & Disease 4: e838. DOI 10.1038/cddis.2013.350. [Google Scholar] [CrossRef]
Tang WW, Chen ZY, Zhang WL, Cheng Y, Zhang B et al. (2020). The mechanisms of sorafenib resistance in hepatocellular carcinoma: Theoretical basis and therapeutic aspects. Signal Transduction and Targeted Therapy 5: 87. DOI 10.1038/s41392-020-0187-x. [Google Scholar] [CrossRef]
Wang L, Wu F, Song Y, Li X, Wu Q, Duan Y, Jin Z (2016). Long noncoding RNA related to periodontitis interacts with miR-182 to upregulate osteogenic differentiation in periodontal mesenchymal stem cells of periodontitis patients. Cell Death & Disease 7: e2327. DOI 10.1038/cddis.2016.125. [Google Scholar] [CrossRef]
Wei L, Wang XW, Lv LY, Liu JB, Xing HX, Song YM, Xie MY, Lei TS, Zhang NS, Yang M (2019). The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Molecular Cancer 18: 147. DOI 10.1186/s12943-019-1086-z. [Google Scholar] [CrossRef]
Wong CM, Tsang FHC, Ng IOL (2018). Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications. Nature Reviews Gastroenterology & hepatology 15: 137–151. DOI 10.1038/nrgastro.2017.169. [Google Scholar] [CrossRef]
Xie L, Huang WB, Fang ZH, Ding F, Zou F et al. (2019). Circ ERCC2 ameliorated intervertebral disc degeneration by regulating mitophagy and apoptosis through miR-182-5p/SIRT1 axis. Cell Death & Disease 10: 751. DOI 10.1038/s41419-019-1978-2. [Google Scholar] [CrossRef]
Xie SC, Zhang JQ, Jiang XL, Hua YY, Xie SW, Qin YA, Yang YJ (2020). LncRNA CRNDE facilitates epigenetic suppression of CELF2 and LATS2 to promote proliferation, migration and chemoresistance in hepatocellular carcinoma. Cell Death & Disease 11: 676. DOI 10.1038/s41419-020-02853-8. [Google Scholar] [CrossRef]
Xue F, Liu YH, Zhang HW, Wen Y, Yan L, Tang Q, Xiao EH, Zhang DY (2016). Let-7a enhances the sensitivity of hepatocellular carcinoma cells to cetuximab by regulating STAT3 expression. OncoTargets and Therapy 9: 7253. DOI 10.2147/OTT. [Google Scholar] [CrossRef]
Yan SS, Wang H, Chen XH, Liang CH, Shang WW, Wang L, Li J, Xu DH (2020). MiR-182-5p inhibits colon cancer tumorigenesis, angiogenesis, and lymphangiogenesis by directly downregulating VEGF-C. Cancer Letters 488: 18–26. DOI 10.1016/j.canlet.2020.04.021. [Google Scholar] [CrossRef]
Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR (2019). A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nature Reviews Gastroenterology & Hepatology 16: 589–604. DOI 10.1038/s41575-019-0186-y. [Google Scholar] [CrossRef]
Yang ZJ, Chee CE, Huang S, Sinicrope FA (2011). The role of autophagy in cancer: Therapeutic implications. Molecular Cancer Therapeutics 10: 1533–1541. DOI 10.1158/1535-7163.MCT-11-0047. [Google Scholar] [CrossRef]

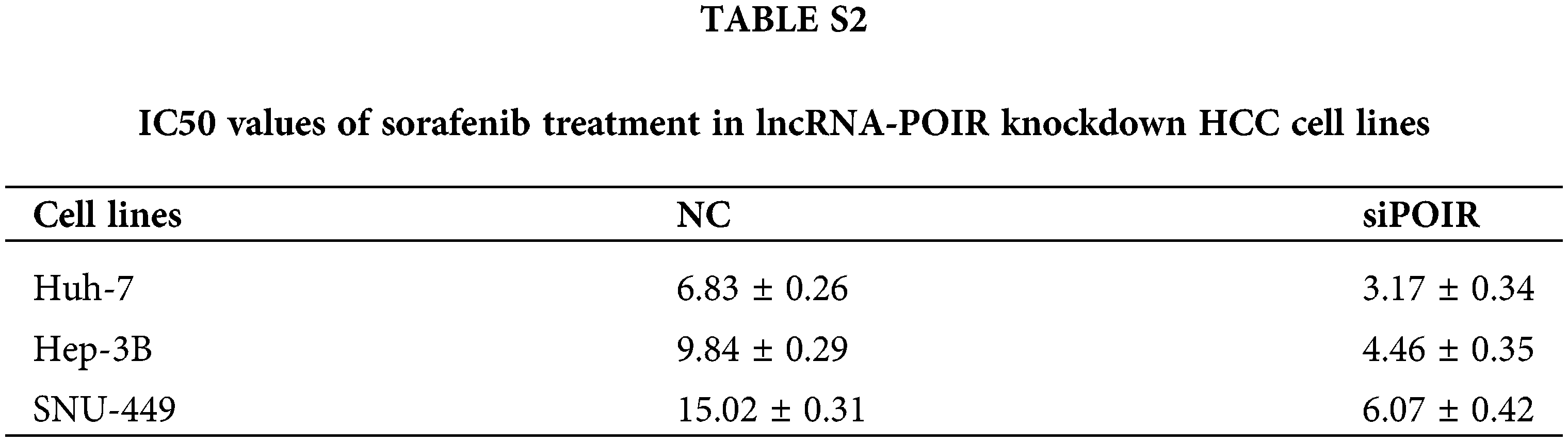
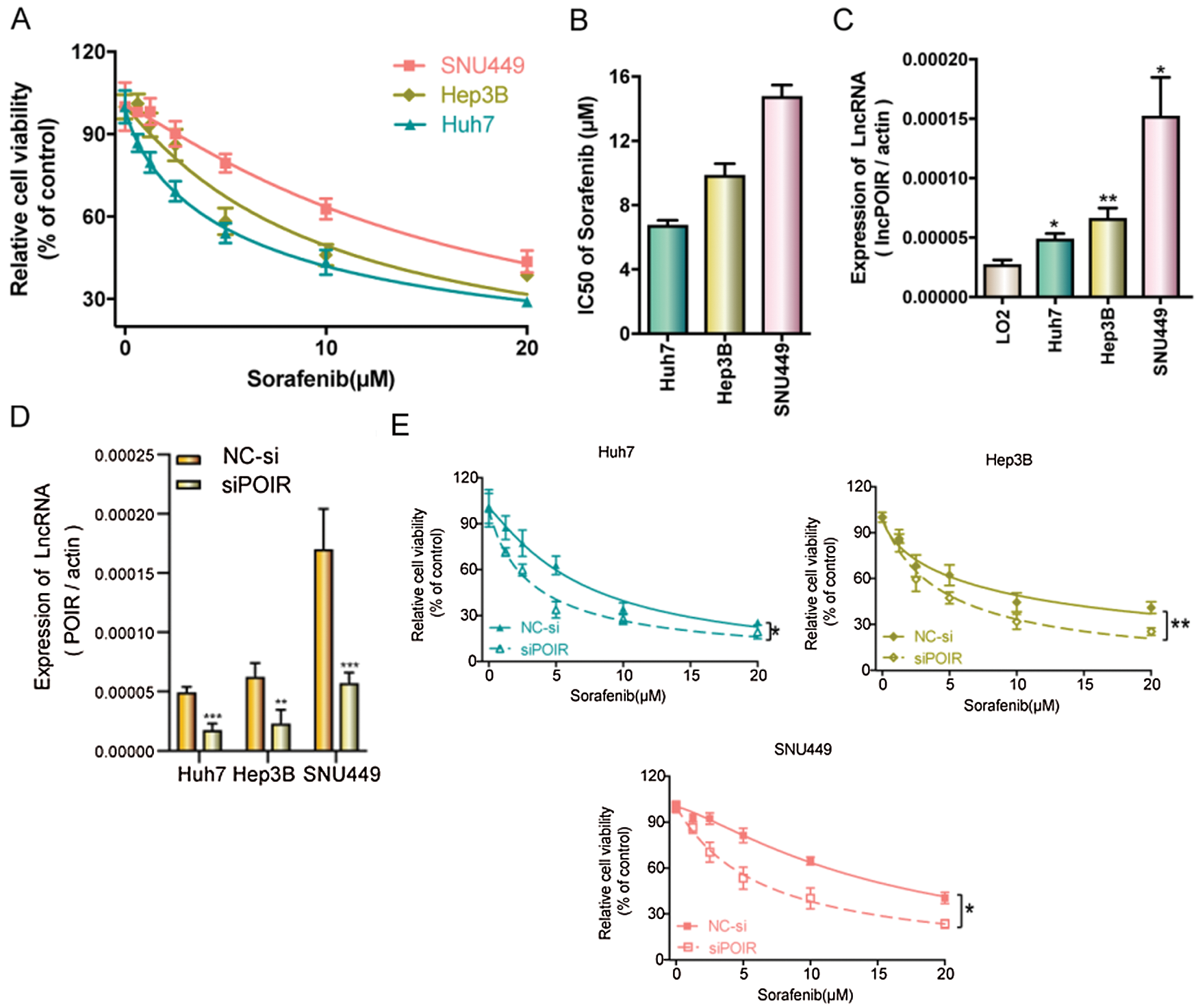
Figure S1: POIR knockdown regulated sorafenib sensitivity by miR-182-5p and autophagy in SNU449 cells.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |