DOI:10.32604/biocell.2022.019111

| BIOCELL DOI:10.32604/biocell.2022.019111 |  |
| Viewpoint |
Expression analysis of OsSERK, OsLEC1 and OsWOX4 genes in rice (Oryza sativa L.) callus during somatic embryo development
1Graduate School of Biotechnology, University of Jember, Jember, 68121, Indonesia
2Faculty of Agriculture, University of Jember, Jember, 68121, Indonesia
3Center Development of Advanced Science and Technology (CDAST), University of Jember, Jember, 68121, Indonesia
4Division of Plant Biosciences, School of Applied BioSciences, College of Agriculture and Life Science, Kyungpook National University, Daegu, 41566, South Korea
*Address correspondence to: Mohammad Ubaidillah, moh.ubaidillah.pasca@unej.ac.id
Received: 03 September 2021; Accepted: 28 October 2021
Abstract: Somatic embryogenesis is an asexual reproduction process that occurs in many plant species, including rice. This process contains several totipotency markers such as Somatic Embryogenesis Receptor-like Kinase (SERK), Leafy Cotyledon1 (LEC1) and WUSCHEL-Related Homeobox4 (WOX4) and also a helpful model for embryo development and clones and transformations. Here, we report the gene expression during somatic embryo development correlates with regeneration frequency in 14 Javanica rice (pigmented and non-pigmented) using modified N6 media supplemented with Kinetin (2.0 mg/L) and NAA (1.0 mg/L). Although there have been advances in understanding the genetic basis of somatic embryogenesis in other varieties, rice is still unexplored, especially during somatic embryo development. Moreover, for the formation of callus induction from immature embryos, 2,4-D (2.0 mg/L, 3.0 mg/L) was used. This study analysed the gene expression of OsSERK, OsWOX4 and OsLEC1 genes through RT-PCR analysis. Higher expression of the OsLEC1 gene indicates that their function may correlate in the in vitro with the high response of rice after transfer to regeneration media. This study found that rice varieties of pigmented rice (MS Pendek and Gogoniti II) and non-pigmented rice (Pandan Ungu) showed high regeneration frequency, showing higher OsLEC1 expression than other varieties because OsLEC1 promotes the maturation of somatic embryos in plant regeneration on day 14. However, the contrast with Genjah nganjuk may be effective because of other regulatory genes. RT-PCR analysis showed OsSERK had less expression level than OsLEC1 and OsWOX4 in the varieties, which correlate with the percentage of plant regeneration, but not for Gogoniti II. In conclusion, the higher percentage of plant regeneration correlates with the higher expression level of OsLEC1 at day 14 of media regeneration of rice.
Keywords: Javanica rice; OsLEC1; OsSERK; OsWOX4; Plant regeneration; Somatic embryos
Somatic embryogenesis is an asexual reproduction process found in many plant species. This is a great model for embryo development and clonal propagation and transformation (Salaün et al., 2021). There are some morphological and biochemical changes in somatic embryogenesis in response to modifications in gene expression patterns (de Oliveira Santos et al., 2005). Somatic embryogenesis involves hormone activity, transcription factors, and epigenetic regulation as a complex mechanism. Despite this, differences in developmental stage and regeneration capacity may be attributable to active genes expressed during somatic embryogenesis (Mahdavi-Darvari et al., 2015). In this work, we have focused on the role of Somatic Embryogenesis Receptor-like Kinase (SERK), Leafy Cotyledon1 (LEC1) and WUSCHEL-Related Homeobox4 (WOX4) genes in rice (Oryza sativa L.) as an integral part of this process.
In Arabidopsis thaliana, WUSCHEL (WUS) is responsible for activating LEC genes. In Gossypium hirsutum, WUS activates GhLEC1, GhLEC2, and GhFUS3 genes. Each stimulation takes place to develop somatic embryogenesis and induce cell differentiation. In addition, high expression of the WUS gene indicates that it is helpful as a valuable gene marker for the initiation of embryogenesis (Kumar and van Staden, 2017). In addition, both zygotic and somatic embryogenesis requires LEC genes, LEC1, LEC2 and FUS3. Functional loss in LEC altered embryonic development substantially (Gulzar et al., 2020). SERK was expressed during the regeneration process in the rye (Secale cereale L.) cultured immature embryos. Nevertheless, its expression is suppressed in later stages (Gruszczyńska and Rakoczy-Trojanowska, 2011). The current study uses this information to understand further somatic embryogenesis regulation in rice and gene expression in SERK, LEC1, and WOX4 to develop somatic embryos.
To improve the quality and quantity of local Indonesian rice in the future, we need to do plant transformation in biotechnology. It is often challenging to get the transformants because not all the local Indonesian rice can regenerate well into plantlets. Some are easy, and some are difficult. Therefore, the existing genetic diversity of local Indonesian rice needs to be studied to obtain clear response information from embryos on culture media through callus induction and plant regeneration through gene expression analysis. Furthermore, each type of rice differs in callus formation and regeneration (Suraiya and Alina, 2018). Thus, in the future, the benefit of plant gene transformation as a selection factor for callus cells will be determined by the efficacy of embryogenic callus formation and plant regeneration in several local rice varieties. However, the response of growth plant regeneration between 14 Javanica rice (pigmented and non-pigmented) related to gene expression in the in vitro culture is still unexplored. Therefore, this study aims to ascertain the gene expression in different efficiency responses of growth plant regeneration using pigmented and non-pigmented Javanica rice. Therefore, this study aims to ascertain the gene expression in different efficiency responses of growth plant regeneration using pigmented and non-pigmented Javanica rice.
Mature seeds of pigmented rice (Super Manggis, MS Pendek, Gogoniti II, Merah SP, Ketan Putri, and Merah Wangi) and non-pigmented rice (Banyuasin, Pandan Ungu, Genjah Nganjuk, Siak Raya, Mendawak, Indragiri, Sereh, and Susu Putih) of rice varieties was provided from farmers listed in Table S1. Healthy mature rice seeds were dehusked and immersed in 50% Clorox bleach (v/v) for 30 min. Then the seeds were thoroughly rinsed at least five times with sterile distilled water to remove residue from the bleach solution. The pH of culture media was adjusted to 5.8 by adding a few drops of 1N NaOH or 1N HCl. The prepared medium was then sterilised in an autoclave at 121°C and 17.5 psi in 30 minutes. Then the 15 seeds per Petri dish with 3 replication were cultured directly on a callus induction media that contained basal N6 media supplemented with 2,4-D (0, 2 or 3 mg/L), casein hydrolysate (0.3 g), sucrose (30 g), and gelrite (4 g) and incubated at 27°C in dark conditions. The callus induction frequency was calculated after one week of culturing using the Eq. (1) below:
Then, after two weeks, high-quality embryogenic calluses were selected and placed on regeneration media (N6 media) supplemented with Kinetin (2 mg/L), NAA (1 mg/L), casamino acids (2 g), myo-Inositol (1 mg/L), sucrose (30 g) and gelrite (4 g), for regeneration and subculture media. Every variety contained three replications with four callus clumps per replication. All cultures were then incubated at 27°C with a photoperiod of 16/8 h (light/dark). The rice embryos were further grown for 14 days on this media regeneration. Then, the percentage of plant regeneration was calculated using the Eq. (2) below:
Total RNA was isolated from 100 mg of callus culture of 14 Javanica rice and utilised for each RNA extraction sample. After fourteen days of growth on the regeneration media, the callus samples were collected and quickly frozen in liquid nitrogen. In this case, we isolated total RNA with some modifications according to the manufacturer’s protocol for the NEXprep™ Plant RNA Mini Kit (Genes Laboratories, Korea). The concentration of total RNA samples was quantified using a Nanodrop (TECAN® Infinite M200 Multi-Detection Microplate Reader Part) with high purity 260 nm/280 nm measurement ranges from 1.8 to 2.2, which can be used for cDNA synthesis and RT-PCR analysis.
cDNA synthesis and PCR analysis
After each sample’s total RNA concentration was confirmed, nuclease-free water was added to the RNA solution until it reached a volume of 6 μL. Then, RNA solutions from each sample were incubated at 65°C for 5 min with PCR. The process of cDNA synthesis must be kept on ice until the total volume becomes 10 μL. After that, 2 μL of 4× DN Master Mix was added to became 8 μL of DNase I reaction solution, and that solution was incubated at 37°C for 5 min by using PCR. Then, the reverse transcription solution was prepared by adding 2 μL of 5× RT Master Mix II to a total volume of 10 μL. This solution was incubated for 15 min at 37°C, followed by 5 min at 50°C, and the 5 min at 98°C by using PCR.
Next, the target cDNA was amplified using the GoTaq® Green Master Mix kit from Promega. PCR analysis was performed on a total volume 10 μL that contained 5 μL of 2 × GoTaq® Green Master Mix, 1 μL cDNA templates, 2 μL Nuclease-free water and 1 μL Forward (F) and Reverse (R) primer to detect the presence of a specific nucleic acid sequences based on Table S2. The PCR was performed by initial denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 53°C for 30 s, extension at 72°C for 1 min, and a final extension at 72°C for 5 min. The PCR product was determined using a 2% agarose gel stained with a green star. The banding patterns were visualized using a UV Transilluminator.
The following reagents (1 μl of 5 μM of each primer, 10 μl of 2× iQ™ SYBR Green Supermix, and 7 μl of nuclease-free water) were mixed on ice in a sterile, nuclease-free microcentrifuge tube. Gently flick the tube while spinning the plate briefly to mix. 19 μl of the mixture was added to the bottom of the tube in each well of the 96-well PCR plate. In addition, 1 μl of each cDNA sample was added to each well. Microseal® “B” Film was used to cover the plate. Gently flick the tube while spinning the plate briefly to mix. The PCR plate was run through an iQ5 multicolour real-time PCR detection system. Parameters for real-time PCR were set up and run based on the primers and the length of the PCR product. The relative expression level of each gene is calculated as fold dilution by using a standard curve. Real-time PCR is used to obtain standard curves using 3 μl of 10-fold dilution, respectively, of cDNA, obtained from the sample. The expression level of each gene is then normalized to the relative expression level of OsActin, a housekeeping gene that serves as a reference control for gene expression in the same sample.
The frequency of callus induction
The mature seeds of fourteen Javanica rice varieties in vitro embryogenic callus induction was studied using an N6 medium as shown in Fig. 1. This basal medium was supplemented with a different concentration of 2,4-D (2 mg/L and 3 mg/L). In addition, N6 medium without 2,4-D was served as a control. In this experiment, the embryogenic callus from 2 mg/L of 2,4-D was used in the regeneration media.
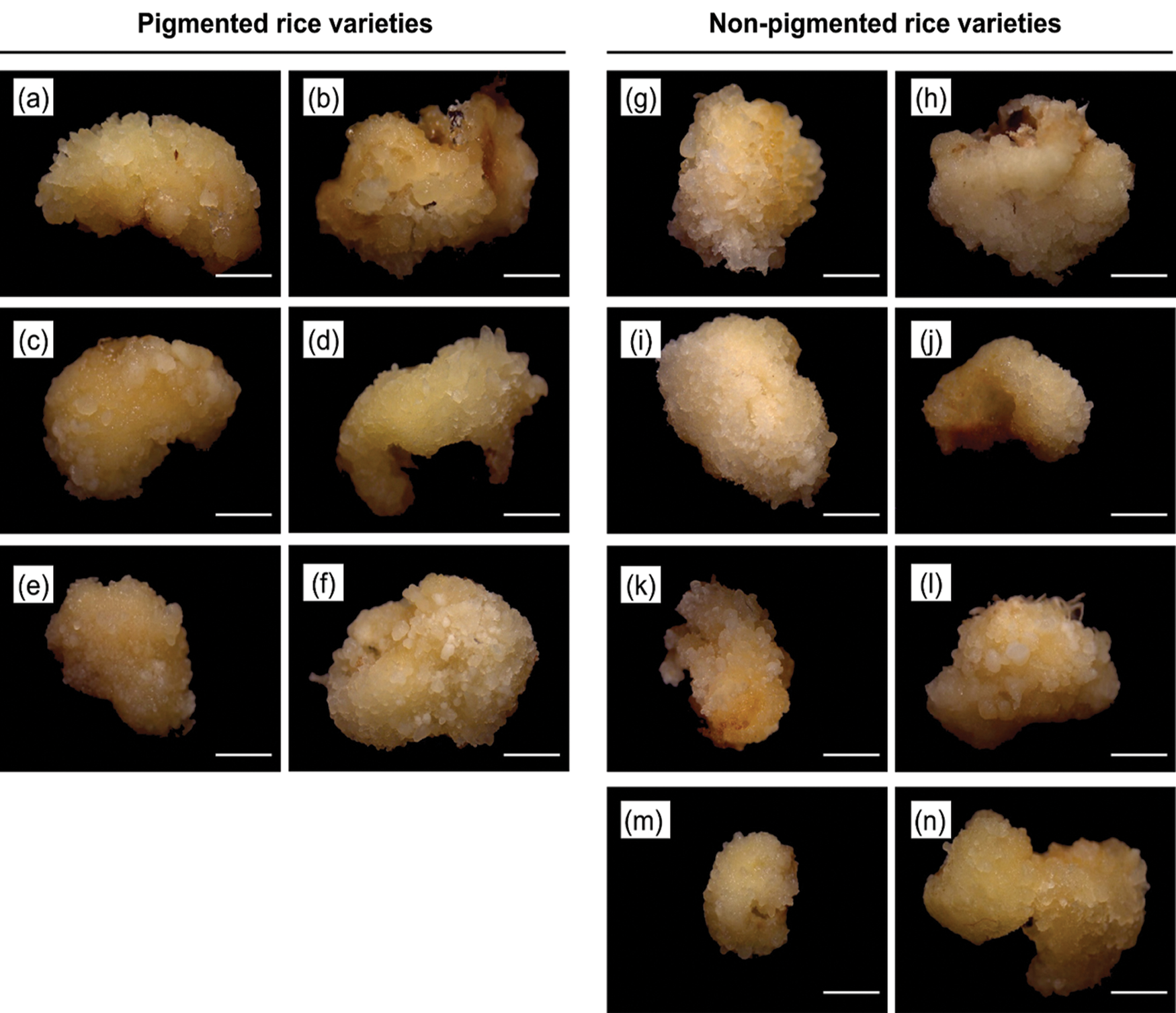
Figure 1: The morphology of embryogenic callus after 7 days on N6 medium supplemented with 2.0 mg/L of 2,4-D. Rice varieties of pigmented rice (a) Super Manggis, (b) MS Pendek, (c) Gogoniti II, (d) Merah SP, (e) Ketan Putri, and (f) Merah Wangi and non-pigmented rice (g) Banyuasin, (h) Pandan Ungu, (i) Genjah Nganjuk, (j) Siak Raya, (k) Mendawak, (l) Indragiri, (m) Sereh, and (n) Susu Putih. Bar: 1 mm.
Once the callus was formed, it could be seen in the scutellum region of all varieties. As shown in Fig. 2, Overall, at 2 mg/L 2,4-D, there are calli ranging from 70% to 100% (Fig. 2a); at 3 mg/L 2,4-D, there are calli ranging from 55% to 80% (Fig. 2b). The increasing concentration of 2,4-D shows a decline in callus induction frequency. In a callus induction concentration of 2 mg/L, MS Pendek, Merah SP, Mendawak, and Susu Putih induced callus at a rate of 100% compared to the media containing 3 mg/L. No callus formation was found on the N6 medium without 2,4-D.
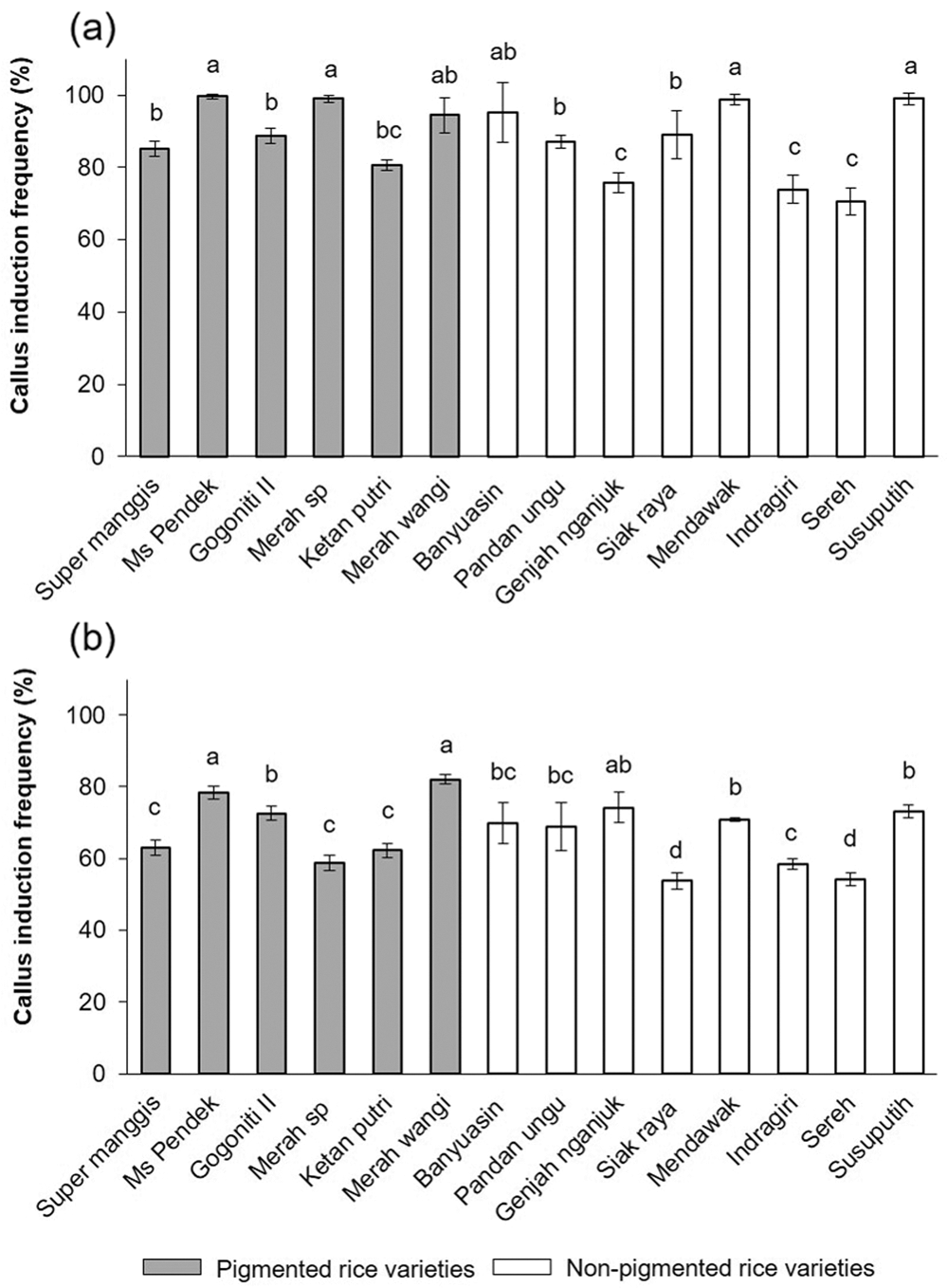
Figure 2: The callus induction frequency was determined in 14 Javanica varieties after 7 days on N6 medium supplemented with different concentrations of 2,4-D (a) 2 mg/L and (b) 3 mg/L. Mean values were taken from average of three replication (n = 3). Mean values ± SD with different letters are statistically different (p ≤ 0.05) according to the Duncan’s multiple range test.
The fresh weight of callus formation
After one week of incubation, the graph in Fig. 3 indicates a significant (p ≤ 0.05) effect of 2,4-D concentrations (white: 2 mg/L; black: 3 mg/L) on the fresh weight of callus formation. The pigmented rice and non-pigmented rice varieties significantly affected (p ≤ 0.05) by the different concentrations of 2,4-D. On the other hand, Ketan Putri and Susu Putih demonstrated no significant difference (p ≥ 0.05) between the concentration of 2,4-D of 2 mg/L and 3 mg/L. Even though these varieties have a lower response to the fresh weight of callus formation, 2 mg/L had a higher response to the fresh weight of callus formation, whereas 3 mg/L had a lower response. Different cultivars responded differently to the increment concentration of 2,4-D. The Duncan’s multiple range test indicates that the mean values of calli formation with the same latter do not differ significantly (p ≤ 0.05).
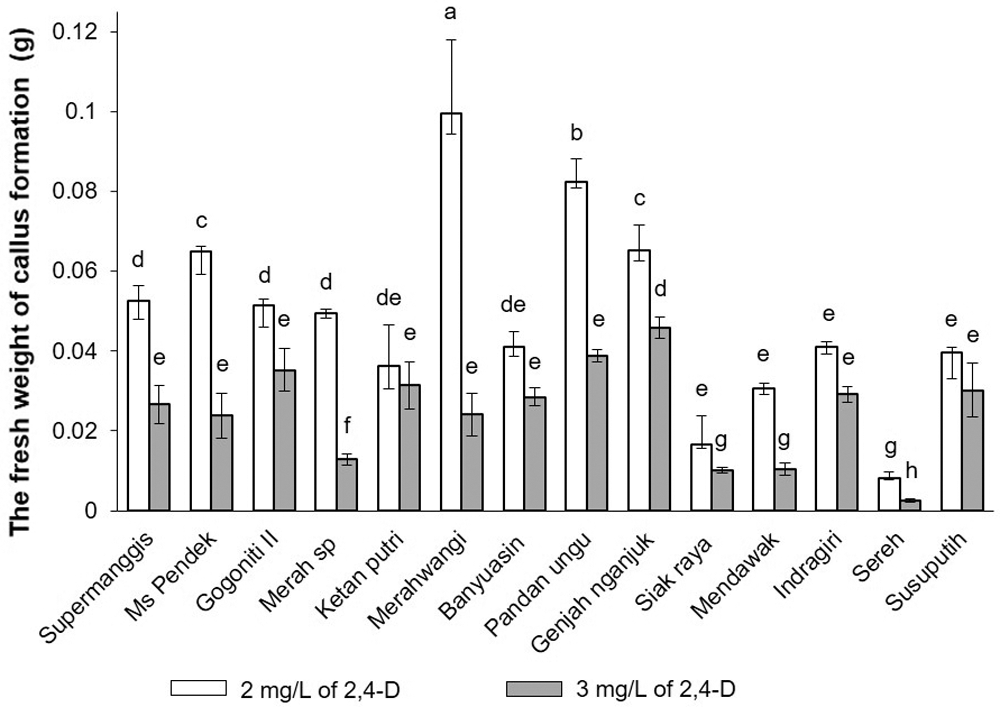
Figure 3: The fresh weight of callus formation was determined in 14 Javanica varieties after 7 days on N6 medium supplemented with different concentrations of and (white) 2 mg/L and (black) 3 mg/L. Mean values were taken from the average of three replications (n = 3). Mean values ± SD with different letters are statistically different (p ≤ 0.05) according to the Duncan’s multiple range test.
Effect of gene expression on regeneration frequency
The gene expression effects the plant regeneration frequency. The graph displayed that GogoNiti II, MS Pendek, Pandan Ungu, and Genjah Nganjuk displayed the highest regeneration frequency in the range of 100% and 90% among 14 Javanica rice significantly different according to Duncan’s multiple range test. Sereh and Siak Raya, on the other hand, have a low regeneration frequency (Fig. 4).

Figure 4: Plant regeneration frequency was determined in 14 Javanica varieties after 14 days on N6 medium supplemented with 2.0 mg/L of Kinetin and 1.0 mg/L of NAA. Mean values were taken from the average of three replications (n = 3). Mean values ± SD with different letters are statistically different (p ≤ 0.05) according to the Duncan’s multiple range test.
MS Pendek, GogoNiti II, Merah Wangi, Genjah Nganjuk, Susu Putih, and Pandan Ungu showed compact, light yellowish colour and globular embryogenic callus with green shoot buds (Fig. 5).

Figure 5: Green spot initiation after 21 days on N6 medium supplemented with 2.0 mg/L of Kinetin and 1.0 mg/L of NAA. Rice varieties of pigmented rice (a) Super Manggis, (b) MS Pendek, (c) Gogoniti II, (d) Merah SP, (e) Ketan Putri, and (f) Merah Wangi and non-pigmented rice (g) Banyuasin, (h) Pandan Ungu, (i) Genjah Nganjuk, (j) Siak Raya, (k) Mendawak, (l) Indragiri, (m) Sereh, and (n) Susu Putih. Bar: 1 mm.
During subculture in media regeneration, the shoot buds further elongated and multiplied vigorously after 45 days due to the availability of an efficient regeneration system (Fig. 6). MS Pendek, GogoNiti II, Pandan Ungu, and Genjah Nganjuk showed the highest response in regeneration frequency. That response can correlate with OsLEC1 expression, which showed higher expression than other varieties, with an inadequate response with plant regeneration frequency.

Figure 6: Plantlet regeneration after 45 days on N6 medium supplemented with 2.0 mg/L of Kinetin and 1.0 mg/L of NAA. Rice varieties of pigmented rice (a) Super Manggis, (b) MS Pendek, (c) Gogoniti II, (d) Merah SP, (e) Ketan Putri, and (f) Merah Wangi and non-pigmented rice (g) Banyuasin, (h) Pandan Ungu, (i) Genjah Nganjuk, (j) Siak Raya, (k) Mendawak, (l) Indragiri, (m) Sereh, and (n) Susu Putih. Bar: 5 mm.
In this study, the higher percentage to plant regeneration frequency correlates to higher expression of OsLEC1 at 14 days in media regeneration of rice excludes Genjah Nganjuk, and this variety does not correlate with the higher expression OsLEC1 (Fig. 7). The analysis correlation may have also been the slight correlation between plant regeneration frequency and gene expression (Fig. 8). This study showed that, for Genjah Nganjuk, this variety does not express the OsSERK gene, but responds well and shows high regeneration growth. The same pattern was observed in MS Pendek and Pandan Ungu variety. Even though the OsSERK gene was expressed lower than OsLEC1 and OsWOX4, it still showed a high plant regeneration frequency.

Figure 7: Gene expression profiles after 14 days of somatic embryo development in 14 Javanica rice. a) Expression of OsLEC1, OsSERK and OSWOX4 was performed from total RNA samples for RT-PCR analysis that were isolated from callus under N6 medium supplemented with 2.0 mg/L of Kinetin and 1.0 mg/L of NAA. OsActin was used as a reference gene. b) Relative expression of OsLEC1, OsSERK and OSWOX4 were normalized to the constitutive gene OsActin. Different letters in columns represent the statistical significance of mean differences at a given time according to the Duncan’s multiple range test (p ≤ 0.05).
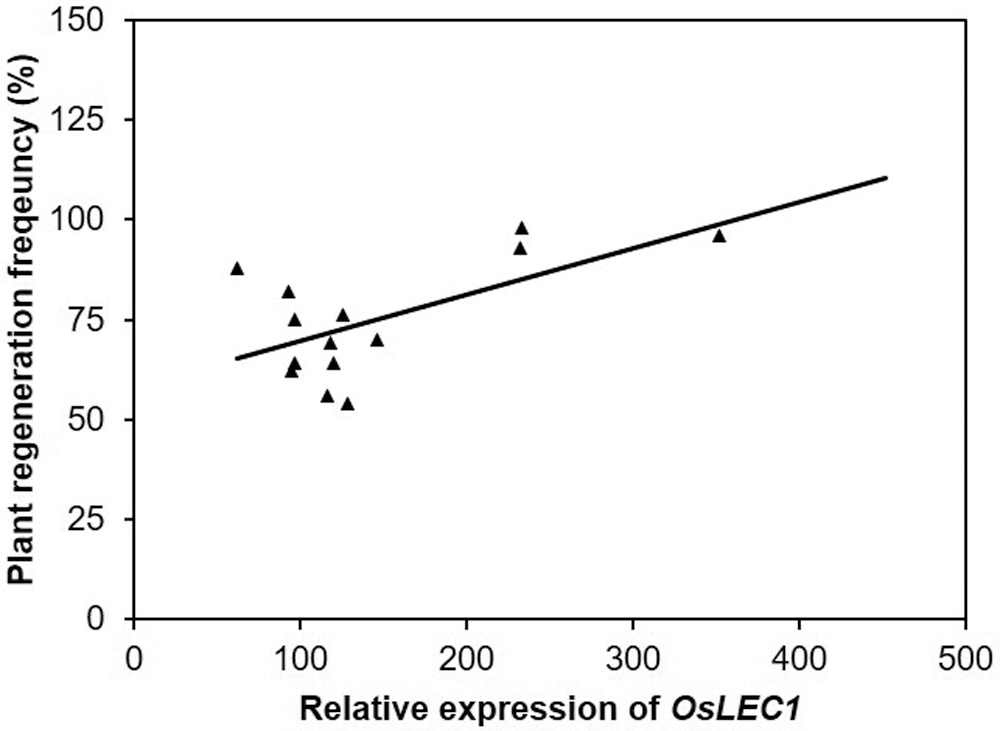
Figure 8: The correlation of plant regeneration frequency versus relative expression of OsLEC1 after 14 days of somatic embryo development. The coefficient of determination, R2 = 0.3728 (P = 58.168).
The callus is an early indicator of a cell’s capacity to regenerate. This is a crucial determinant of the ability of plant regeneration be performed. Moreover, the selection of callus cells is required to evaluate the effectiveness of embryogenic callus production and plant regeneration in many rice varieties. In this study, we discovered how vital callus formation is and how it relates to regeneration. Furthermore, there is a correlation between the frequency of plant regeneration and the fresh weight of callus formation (Figs. 3 and 4). For example, the fresh weight of callus formation increased in rice varieties of Gogoniti II, MS Pendek, Pandan Ungu, and Genjah Nganjuk. Therefore, the regeneration frequency in those varieties also increased. As a result, plant regeneration increases as callus ability increases. However, such as Sereh and Siak Raya, the regeneration frequency lowers with a low fresh weight of callus formation.
As shown in Figs. 1–3, interestingly, despite using the same concentration, 14 varieties of Javanica rice respond differently. From callus formation to plant regeneration, different varieties responded differently. These results are similar to those reported by Suraiya and Alina (2018). The reason for this is the influence of genetic background. During the callus induction to plant regeneration, the genetic background of rice had a significant influence on how well the plantlet performs in tissue culture.
As a result, we examined gene expressions during somatic embryo development. SERK, LEC, and WUS are all expressed during somatic embryogenesis in various plant species (Kumar and van Staden, 2017). Gogoniti II, MS Pendek, Gogoniti II, and Pandan Ungu were easy to regenerate and had a high accumulation of OsLEC1. This means that expressing the level of OsLEC1 in callus cultures affects plant regeneration because each genetic background has its unique characteristics were found to have the highest frequency. Furthermore, in this variety, the higher the level of OsLEC1 expression, the higher the expression level of OsLEC1. Because of this, OsLEC1 has been discovered to be an essential gene in rice regeneration.
This study demonstrated that OsLEC1 expression was found to be increased in varieties with a high rate of plant regeneration during the 14-day media regeneration. Thus, the expression corresponds to plant regeneration frequency. These results are in accordance with the previous describing that, OsLEC1 expression was found to be increased in varieties with a high rate of plant regeneration in this study, these results are in accordance with the previous describing that, OsLEC1 gene expressed during somatic embryogenesis maturation in Passiflora species (Leite et al., 2019). Other investigations discovered that the LEC1 gene is a critical regulator of somatic embryogenesis in Arabidopsis thaliana and plays a critical role in somatic embryogenesis (Gaj et al., 2005). It was also discovered that during four weeks of media regeneration, the high regeneration variety of Secale cereale expressed a high level of ScLEC1 (Gruszczyńska and Rakoczy-Trojanowska, 2011). In other words, there is a correlation between morphology and gene expression during somatic embryo development on day 14 in media regeneration.
Furthermore, there were gene differences after the subcultures on day 14. Because embryogenic development in rice takes 14 days, we conducted our research on day 14 after the callus had been differentiated in the second media regeneration subculture. On day 7, RT-PCR revealed no somatic embryogenesis-related gene expression (data not shown).
In addition, OsSERK less influences the regeneration frequency during the development of embryogenic calli after 14 days of media regeneration. Therefore, OsSERK showed lower expression levels than OsLEC1 and OsWOX4 in the varieties that show a higher frequency of plant regeneration, but not for GogoNiti II. Because OsSERK only plays a role in embryo development but not in the maturation process of plant regeneration.
Nevertheless, OsWOX4 indicates that their function is unlikely to correlate in fourteen Javanica rice varieties during the development of embryogenic calli after 14 days of media regeneration. Certain varieties lack or have low expression levels of OsSERK genes but retain a high regeneration frequency. It has been reported that this gene is expressed in somatic embryonic cells that reach the globular stage before transitioning from differentiation to development (Schmidt et al., 1997; Hecht et al., 2001). Additionally, SERK expression increases during the induction of somatic embryogenesis (de Oliveira Santos et al., 2005). As a result, OsSERK is required for embryo development but not for the specific process of plant regeneration maturation. Our results confirm this theory that OsSERK less influences the regeneration frequency during the development of embryogenic calli after 14 days of media regeneration.
However, somatic embryogenesis may be affected by other regulatory genes. Interestingly, several embryogenesis-related genes, ABI3, AGL15, FUS3, BBM, and LEA, are involved in somatic embryo development (Ikeda et al., 2006). Among other varieties that showed high plant regeneration frequency, only Gogoniti II showed a positive response with high number of plantlets. This is because Gogoniti II had very high OsSERK accumulation followed by OsWOX4 and OsLEC1.
In gene expression, which helped Gogoniti II respond well compared to other varieties like MS Pendek, Pandan Ungu and Genjah Nganjuk during process embryogenesis until becoming plantlet. However, SERK also influences some varieties like GogoNiti II, which showed high OsSERK expression and high plant regeneration frequency. This is because Gogoniti II had very high OsSERK accumulation followed by OsWox4 and OsLEC1 in gene expression, which helped Gogoniti II respond well compared to other varieties like MS Pendek, Pandan Ungu and Genjah Nganjuk during process embryogenesis until becoming plantlet. Previously it was shown that SERK gene may perform specific and redundant functions in plant development (Singla et al., 2009).
According to a previous study, the expression of Coffee canephora, which is WOX4, gradually decreased during embryo maturation but remains detectable. Thus, it could be related to the high plant regeneration frequency associated with low OsWOX4 expression, but the high expression associated with low plant regeneration frequency. It was shown that OsWOX4 is required to promote and maintain vascular procambium (Nic-Can et al., 2013).
Our first hypothesis was that pigmentation content in seeds causes a difference in plant regeneration frequency. This is because pigmented rice is high in antioxidants like phenolics, anthocyanin, and gamma-oryzanols (Pradipta et al., 2020), which is important during callus induction until plant regeneration. However, this did not prove to be the case. Instead, we found that all varieties had different responses and showed no specific pigmentation in performance. Furthermore, we conclude characteristics of pigmentation properties have no effect on tissue culture traits and gene expression.
Our data show that pigmented rice (MS Pendek and GogoNiti II) and non-pigmented rice (Pandan Ungu) have a high percentage of plant regeneration correlated to OsLEC1 accumulation after 14 days because somatic embryo requires OsLEC1 expression for further develop. Genjah Nganjuk (non-pigmented rice), on the other hand, has a high percentage of plant regeneration but low expression of OsLEC1. Thus, it can be concluded that other regulatory genes are involved in molecular regulation of somatic embryogenesis. Among 14 Javanica rice, genetic background of rice had a significant influence on how well the plantlet performs in tissue culture. Moreover, pigmentation and non-pigmentation both for tissue culture traits and gene expression are not affected by the characteristics of pigmentation properties.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Authors’ Contribution: The authors confirm contribution to the paper as follows: study conception and design: Kyung-Min Kim, Mohammad Ubaidillah, Siti Nabilah; data collection: Siti Nabilah; analysis and interpretation of results: Mohammad Ubaidillah, Tri Handoyo, Siti Nabilah; draft manuscript preparation: Siti Nabilah. All authors reviewed the results and approved the final version of the manuscript.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Chen R, Xu N, Yu B, Wu Q, Li X, Wang G, Huang J (2020). The WUSCHEL-related homeobox transcription factor OsWOX4 controls the primary root elongation by activating OsAUX1 in rice. Plant Science 298: 110575. DOI 10.1016/j.plantsci.2020.110575. [Google Scholar] [CrossRef]
de Oliveira Santos M, Romano E, Yotoko KS, Tinoco ML, Dias BB, Aragao FJ (2005). Characterisation of the cacao somatic embryogenesis receptor-like kinase (SERK) gene expressed during somatic embryogenesis. Plant Science 168: 723–729. DOI 10.1016/j.plantsci.2004.10.004. [Google Scholar] [CrossRef]
Gaj MD, Zhang S, Harada JJ, Lemaux PG (2005). Leafy Cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222: 977–988. DOI 10.1007/s00425-005-0041-y. [Google Scholar] [CrossRef]
Gruszczyńska A, Rakoczy-Trojanowska M (2011). Expression analysis of somatic embryogenesis-related SERK, LEC1, VP1 and NiR ortologues in rye (Secale cereale L.). Journal of Applied Genetics 52: 1–8. DOI 10.1007/s13353-010-0015-z. [Google Scholar] [CrossRef]
Gulzar B, Mujib A, Malik MQ, Sayeed R, Mamgain J, Ejaz B (2020). Genes, proteins and other networks regulating somatic embryogenesis in plants. Journal of Genetic Engineering and Biotechnology 18: 1–15. DOI 10.1186/s43141-020-00047-5. [Google Scholar] [CrossRef]
Heang D, Sassa H (2012). Overexpression of a basic helix-loop-helix gene Antagonist of PGL1 (APG) decreases grain length of rice. Plant Biotechnology 29: 65–69. DOI 10.5511/plantbiotechnology.12.0117a. [Google Scholar] [CrossRef]
Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt ED, Boutilier K, Grossniklaus U, de Vries SC (2001). The arabidopsis somatic embryogenesis receptor kinase 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiology 127: 803–816. DOI 10.1104/pp.010324. [Google Scholar] [CrossRef]
Hoque KM, Azdi ZA, Prodhan SH (2013). Development of callus initiation and regeneration system of different indigenous indica rice varieties. Journal of Biology 1: 46–51. [Google Scholar]
Ikeda M, Umehara M, Kamada H (2006). Embryogenesis-related genes; its expression and roles during somatic and zygotic embryogenesis in carrot and Arabidopsis. Plant Biotechnology 23: 153–161. DOI 10.5511/plantbiotechnology.23.153. [Google Scholar] [CrossRef]
Kumar V, van Staden J (2017). New insights into plant somatic embryogenesis: An epigenetic view. Acta Physiologiae Plantarum 39: 1–17. DOI 10.1007/s11738-017-2487-5. [Google Scholar] [CrossRef]
Leite CT, Ferreira DA, Vieira AT, Praça-Fontes MM, Ferreira A, Carvalho CR, Clarindo WR (2019). In vitro responses in Passiflora species with different chromosome numbers, ploidy levels and nuclear 2C values: Revisiting and providing new insights. Plant Cell, Tissue and Organ Culture 136: 549–560. DOI 10.1007/s11240-018-01536-9. [Google Scholar] [CrossRef]
Mahdavi-Darvari F, Noor NM, Ismanizan I (2015). Epigenetic regulation and gene markers as signals of early somatic embryogenesis. Plant Cell, Tissue and Organ Culture 120: 407–422. DOI 10.1007/s11240-014-0615-0. [Google Scholar] [CrossRef]
Nic-Can GI, López-Torres A, Barredo-Pool F, Wrobel K, Loyola-Vargas VM, Rojas-Herrera R, De-la-Peña C (2013). New insights into somatic embryogenesis: Leafy cotyledon1, Baby Boom1 and WUSCHEL-Related Homeobox4 are epigenetically regulated in Coffea canephora. PLoS One 8: e72160. DOI 10.1371/journal.pone.0072160. [Google Scholar] [CrossRef]
Park HS, Ryu HY, Kim BH, Kim SY, Yoon IS, Nam KH (2011). A subset of OsSERK genes, including OsBAK1, affects normal growth and leaf development of rice. Molecules and Cells 32: 561–569. DOI 10.1007/s10059-011-0178-4. [Google Scholar] [CrossRef]
Pradipta S, Ubaidillah M, Siswoyo TA (2020). Physicochemical, functional and antioxidant properties of pigmented rice. Current Research in Nutrition and Food Science Journal 8: 837–851. DOI 10.12944/CRNFSJ.8.3.15. [Google Scholar] [CrossRef]
Rupps A, Raschke J, Rümmler M, Linke B, Zoglauer K (2016). Identification of putative homologs of Larix decidua to Babyboom (BBMLeafy cotyledon1 (LEC1WUSCHEL-related Homeobox2 (WOX2) and Somatic Embryogenesis Receptor-like Kinase (SERK) during somatic embryogenesis. Planta 243: 473–488. DOI 10.1007/s00425-015-2409-y. [Google Scholar] [CrossRef]
Salaün C, Lepiniec L, Dubreucq B (2021). Genetic and molecular control of somatic embryogenesis. Plants 10: 1467. DOI 10.3390/plants10071467. [Google Scholar] [CrossRef]
Schmidt ED, Guzzo F, Toonen MA, de Vries SC (1997). A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124: 2049–2062. DOI 10.1242/dev.124.10.2049. [Google Scholar] [CrossRef]
Singla B, Khurana JP, Khurana P (2009). Structural characterization and expression analysis of the SERK/SERL gene family in rice (Oryza sativa). International Journal of Plant Genomics 2009: 1–8. DOI 10.1155/2009/539402. [Google Scholar] [CrossRef]
Suraiya M, Alina W (2018). Efficient callus induction and regeneration in selected indica rice. Agronomy 8: 77. DOI 10.3390/agronomy8050077. [Google Scholar] [CrossRef]
Supplementary Materials


 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |