DOI:10.32604/biocell.2022.018078

| BIOCELL DOI:10.32604/biocell.2022.018078 |  |
| Article |
Identification of key long noncoding RNAs and their biological functions in hepatocellular carcinoma
1Medical College, Soochow University, Suzhou, 215006, China
2Department of Ultrasound, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou, 121001, China
3Department of General Hepatobiliary Surgery, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou, 121001, China
*Address correspondence to: Yuhong Li, 20194155082@stu.suda.edu.cn; Liang Wang, liangsurgery@163.com
Received: 28 June 2021; Accepted: 18 August 2021
Abstract: Long noncoding RNAs (lncRNAs) are vital regulators in tumorigenesis and metastasis. However, the pathological role of lncRNAs in hepatocellular carcinoma (HCC) is still unclear. In this study, we filtered out three lncRNAs from The Cancer Genome Atlas (TCGA) data that were screened for basic expression and clinical research. We selected lncRNA-NEAT1 for further study to explore its function in HCC progression and its regulatory mechanism. We identified three differentially expressed lncRNAs (DElncRNAs) in tumor and adjacent normal tissues from the TCGA library using data mining methods: lncRNA-NEAT1, lncRNA-MAGI2-AS3 and lncRNA-HCG11. Their basic expression levels were detected by qPCR. Then, we selected lncRNA-NEAT1 as a potentially important lncRNA to verity its biological function and mechanism in HCC cell lines. lncRNA-NEAT1, lncRNA-MAGI2-AS3 and lncRNA-HCG11 were overexpressed in liver cancer tissues and cell lines. We found that silencing NEAT1 in vitro can inhibit the proliferation of HuH-7 and Li-7 cells, inhibit cell migration, and induce apoptosis as well as significantly increase the level of miR-16-5p. We also confirmed that miR-16-5p has a significant correlation with Bcl-2. When NEAT1 is silenced, the expression of Bcl-2 decreases. Inhibiting miR-16-5p can restore Bcl-2 to its original level. We conclude that miR-16-5p1/lncRNA NEAT1 plays a crucial role in regulating the delivery of Bcl-2 in HCC. Overall, the miR-16-5p/lncRNA-NEAT1/Bcl-2 signaling axis may be a promising target for HCC treatment.
Keywords: Long noncoding RNAs; Hepatocellular carcinoma; NEAT1; miR-16-5p; Bcl-2
Hepatocellular carcinoma (HCC) is one of the most common and fatal cancers worldwide. It is the third leading cause of cancer-related death and is responsible for approximately 90% of all primary liver cancer cases (Craig et al., 2020; Sahu et al., 2019). It is a highly prevalent and difficult-to-treat malignant tumor. It has a multifaceted molecular pathogenesis. China has the largest number of liver cancer cases in the world, accounting for 55% of newly diagnosed liver cancer cases globally, and approximately 45% of deaths (Gunsar, 2017). Several major diseases, such as hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, and metabolic diseases and alcohol intake promote hepatocarcinogenesis (Singal and El-Serag, 2015). Because of the high morbidity, mortality and various risk factors related to the liver, at present, the focus of liver cancer research is exploring its early detection markers, prognostic biomarkers, and new therapeutic targets. To identify potential targets for diagnosis and therapeutic intervention, it is necessary to further understand the molecular mechanisms behind the development of HCC. Medical researchers have improved their comprehensive understanding of HCC through the discovery of many noncoding RNAs (ncRNAs), such as long noncoding RNAs (lncRNAs) and miRNAs.
lncRNAs are noncoding RNA transcripts with a length of more than 200 nucleotides. lncRNAs are involved in the regulation of multiple cellular processes, such as chromatin remodeling, epigenetic modification, transcriptional and post-transcriptional expression levels, and protein expression, and have certain effects on the pathological and physiological processes of the body (Abbastabar et al., 2018). The latest progress shows the pathophysiological process of lncRNA dysregulation and development, including development, cancer invasion and metastasis (Qu et al., 2016). Abnormal lncRNAs can promote the occurrence and metastasis of HCC, which is related to poor prognosis of HCC. Some lncRNAs have been reported to regulate the development of HCC, and their targets include remote HOXA transcription; maternal expression of MEG3, which is highly upregulated in liver cancer; and ZNFX1 antisense RNA 1 (Zhang et al., 2019).
Chang et al. (2016) documented that the inhibitory effect of lncRNA GAS5, demonstrated by promoting HCC cell growth and invasion, was mediated by regulating vimentin. Also, the increased level of lncRNA GAS5 was correlated with a better prognosis. lncRNA metastasis-related lung adenocarcinoma transcript 1 and HCC upregulate the expression of lncRNA in HCC tissues, which may be a good prognostic biomarker for therapeutically resected HCC (Leti et al., 2017). In this study, The Cancer Genome Atlas (TCGA) data were used to investigate the lncRNA and mRNA expression data of many HCC patients, and three lncRNAs were screened for basic expression and clinical sample research. In addition, this study selected a representative lncRNA among the three lncRNAs, NEAT1, to verify its biological functions. We hypothesize that the regulatory effect of lncRNA-NEAT1 in the growth and apoptosis of HCC cells is related to its influence on the expression of miR16-5p.
In summary, our study aims to explore the role of lncRNA-NEAT1 in the occurrence and development of HCC and its possible mechanism, as well as provide new clues for potential targets for clinical diagnosis and treatment of HCC.
Gene expression profile of hepatocellular carcinoma
The mature miRNA RNA sequencing (RNA-seq) [log2 (RPM + 1)] and exon RNA-seq [log2 (RPKM + 1)] data and the corresponding clinical data were recorded on TCGA (https://genome-cancer.ucsc.edu/). Samples with log2 (rPKM + 1) = 0 were screened out. The differentially expressed profiles of mRNA, lncRNA and miRNA in paired adjacent normal and tumor samples were obtained.
Screening of differentially expressed lncRNAs
In the TCGA database, the classical Bayesian method provided by the limma R package in R/Bioconductor (Smyth GK) was introduced. A linear model of microarray data and R & Bioconductor 2011 bioinformatics and computational biology solutions (version 3.10.3, http://www.bioconductor.org/packageges/2.9/bioc/html/limma.html) were used. We analyzed differentially expressed lncRNA (DElncRNA) in paired adjacent normal and tumor tissues using the Benjamin and Hochberg method. We obtained the P-value through multiple inspections and corrections. P < 0.05 and a logarithmic change >0.585 may be a marker of HCC.
Protein–protein interaction network integration
The STRING database (http://string-db.org/) is an online search tool used to identify interactions between DElncRNAs. Interaction pairs with scores greater than 0.4 are considered significant. The protein-protein interaction (PPI) network is constructed and visualized using Cytoscape software (version 3.4.0; http://www.cytoscape.org/), which is a bioinformatics platform used for construction and visualization of a molecular interaction network. Networking is the main purpose of Cytoscape software. Genes, proteins, or molecules can be represented by each node. The interactions between these biomolecules are represented by the connections between nodes, thus presenting interactions and pathway relationships between proteins encoded by DElncRNAs in NPCs. Corresponding central nodal proteins can be core regulators and exert important physiological functions.
Functional annotation of differentially expressed lncRNAs and modular lncRNAs
A P-value of < 0.05 and an enrichment count of at least five were important criteria for determining significance when using the David database (version 6.8, https://DAVID-d.ncifcrf.gov/) for DElncRNAs. For the lncRNA module, P < 0.05 and at least three enrichment counts were determined as the threshold for determining significant enrichment in the process of liver cancer.
This study recruited 40 patients with HCC. All patients were diagnosed with HCC from January 2011 to January 2014 at the First Affiliated Hospital of Jinzhou Medical University. The research study was carried out in accordance with the ethical guidelines of the Declaration of Helsinki and was approved by the Research Ethics Committee of the Research Institute of Jinzhou Medical University (No. 2018-03). All patients signed an informed consent form.
The Li-7, HuH-7, SNU-387, RBE and Hep3b cell lines were purchased from the National Cell Line Resource Infrastructure. The THLE-2 cell line was purchased from the American Type Culture Collection (ATCC). HuH-7 cells were incubated in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Grand Island, NY) containing 10% fetal bovine serum (FBS) (Gibco, Rockville, Maryland, USA) and penicillin (100 μg/mL) and streptomycin (0.1 mg/mL). Li-7, SNU-38 and RBE cells were cultured in RPMI-1640 (GIBCO, 31800022) supplemented with 10% FBS and 0.1% penicillin/streptomycin. Hep3b cells were cultured in the minimum essential medium (Eagle’s) containing Earle’s balanced salt and 10% FBS. Cells were incubated at 37°C and 5% CO2.
Cell grouping and transfection
HuH-7 and Li-7 cells were placed into six groups: sh-control (transfected with NEAT1, MAGI2-AS3 and HCG11 negative control shRNA), sh-NEAT1 (lentivirus transfected with short hairpin RNA against NEAT1), sh-MAGI2-AS3 (lentivirus transfected with short hairpin RNA against MAGI2-AS3), sh-HCG11 (lentivirus transfected with short hairpin RNA against HCG11), sh-NEAT1+miR inhibitor (transfected with short lentivirus-containing lentiviral anti-NEAT1 hairpin RNA and treated with miR inhibitor), and sh-control + miR inhibitor (NEAT1, MAGI2-AS3 and HCG11 negative control shRNA transfection and treatment with an miR inhibitor). After sequencing, lipofectamine 2000 (Invitrogen, USA) was used to transfect sh-NEAT1, sh-MAGI2-AS3, sh-HCG11 or sh-control into HuH-7 and Li-7 cells. Cells were incubated in the culture medium with 0.5 mg/mL G418 (Sigma-Aldrich) for about 4 weeks. Lipofectamine 2000 was also used to transfect miR-16-5p inhibitors into HuH-7 and Li-7 cells.
Transwell assay for cell migration detection
During logarithmic growth, Huh-7 and Li-7 cells were starved for 24 h, separated the next day, and then centrifuged and resuspended to adjust the cell concentration to 2 × 105 cells/mL. After 24 h of incubation, the transwell chamber was removed. Before air drying, the liquid in the upper chamber was discarded, and 4% paraformaldehyde was added to fix the cells for 15 min. Next, 0.1% crystal violet was employed to stain cells for about 20 min. Then, cells were washed to remove residual crystal violet and dried. Finally, we observed the cells and took photos under an inverted microscope and randomly selected the field of view (×100) to determine the number of migrated cells.
Total RNA was isolated from tissues or cells using TRIzol reagent (Invitrogen, CA). cDNA was synthesized with a reverse transcription kit (Vazyme, China) according to the manufacturer’s protocol. The expression of selected genes was determined using the ABI StepOnePlus real-time PCR system (Applied Biosystems, Foster City, USA) with GAPDH (for protein expression) and U6 (for RNA expression) as internal controls. The primers used in this study are listed in Table 1.
The Cell Counting Kit 8 (CCK-8; Beyotime, China) was used to determine the growth of Li-7 and Huh-7 cells. In general, cells were seeded into 96-well plates at a density of 5 × 103. On days 1, 2, 3 and 4, CCK-8 solution was added and incubated for the indicated time to determine the absorbance values of each well. The optical density (OD) values between the control and treated groups, which were examined at 450 nm with a microplate reader, were used to detect cell proliferation.
The FITC/Apoptosis Kit (Invitrogen) was used to assess cell apoptosis. Huh-7 and Li-7 cells were plated into 6-well plates at a density of 1 × 105/well. After 24 h of incubation, PBS buffer was used to wash cells, followed by suspending the cells in annexin binding buffer. Five microliters of Annexin V and 0.1 μg of propidium iodide (PI) were used to stain cells to determine the percentage of apoptosis using flow cytometry (BD, USA).
Dual-luciferase reporter gene assay
The fragment containing the wild-type or mutant binding site for NEAT1 was inserted into a pGL3-basic luciferase reporter plasmid. Next, Huh-7 cells and Li-7 cells were plated in a 24-well plate and co-transfected with luciferase reporter plasmids and hsa_NEAT1 WT and NEAT1 Mut (1 μg). After 48 h of culture, the Dual-Luciferase Reporter Assay system (Promega) was used to assess cellular luciferase activity, which was calculated by the ratio of firefly luciferase activity to Renilla luciferase activity.
Statistical analysis was performed using SPSS 23.0 software (IBM, Armonk, NY, USA). Each experiment was conducted three times independently. The measured data were presented as the mean ± standard deviation. The significant difference between two groups or between multiple groups was examined by using the t-test or one-way analysis of variance (ANOVA), respectively. The least significant differences (LSD) method was used to compare the variances between groups. The Pearson correlation coefficient was used to analyze the correlation between NEAT1 and miR-16-5p. Kaplan–Meier analysis was used to measure survival data, and the log-rank test was used for comparison. A P-value less than 0.05 (two-tailed) was considered statistically significant.
Expression profiling of genes from the TCGA HCC dataset compared with normal liver tissue
The TCGA database was used to examine the expression of the hub gene and DElncRNAs with prognostic significance. Consistent with previous results, significant overexpression of all prognostic DElncRNAs and hub genes was found in HCC tumor tissues compared with adjacent non-cancerous tissues (Figs. 1A and 1B).
Reconstruction of the lncRNA-associated ceRNA network
A new competing endogenous (ceRNA) network was reconstructed, using newly screened hub genes and DElncRNAs. There were 14 hub genes, including 7 miRNAs (hsa-miR-107, hsa-miR-1297, hsa-miR-24-3p, hsa-miR-216b-5p, hsa-miR-217, hsa-miR-338-3p, and hsa-miR-107) as well as four lncRNAs (FAM182B, SNHG6, SNHG1, and SNHG3) in total. The reestablished network was consistent with 10 lncRNA-miRNA-mRNA pathways, including hsa-miR-374a-5p, hsa-miR-590-3p, hsa-miR-98-5p, lncRNA NEAT1, lncRNA MAGI2-AS3 and lncRNA HCG11, of which lncRNA HCG11 had the highest connection rate with the hub gene.
Functional annotation and pathway enrichment analysis of DElncRNAs
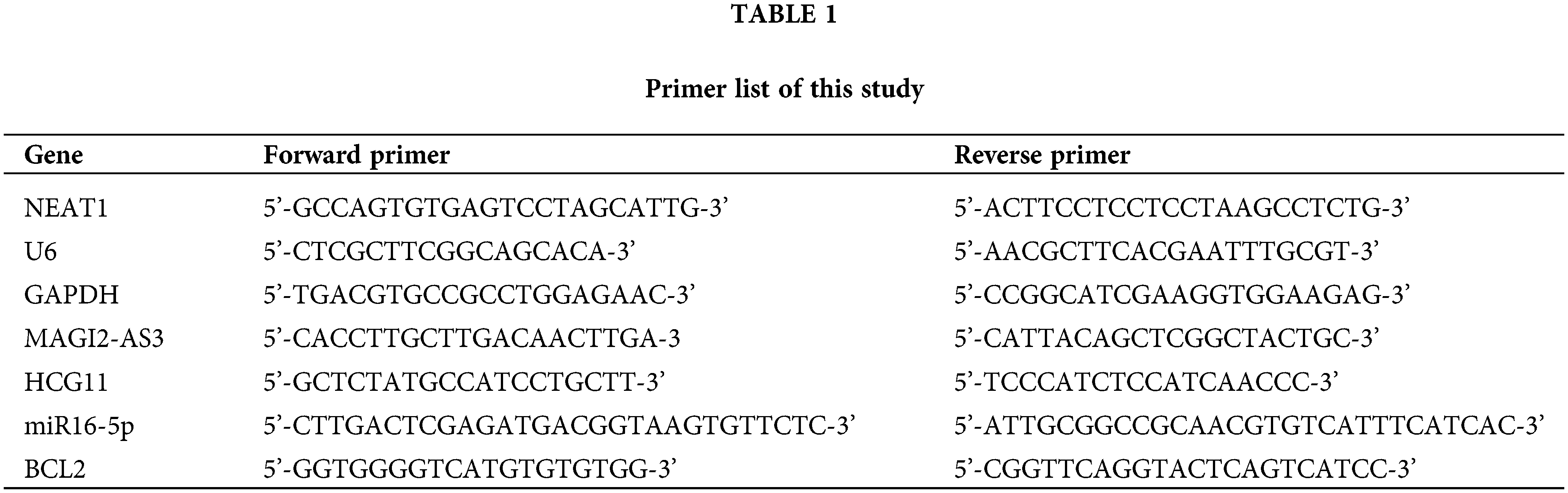
To further illustrate the characteristics and roles of the 126 DEGs in HCC progression, analysis of functional annotation and pathway enrichment for the above DEGs in the ceRNA network was performed, which showed the following: (1) for biological pathways (BP), the DEGs of the ceRNA network were enriched in muscle contraction, GO:2000027~regulation of organ morphogenesis, and striated muscle cell differentiation; (2) for cellular components (CC), the DEGs were obviously enriched in extracellular matrix, contractile fiber, and sarcolemma; (3) for molecular functions (MF), the DEGs were enriched in glycosaminoglycan binding, channel activity and passive transmembrane transporter activity (Figs. 1C and 1D). They were enrolled in an interaction network (Fig. 1E).
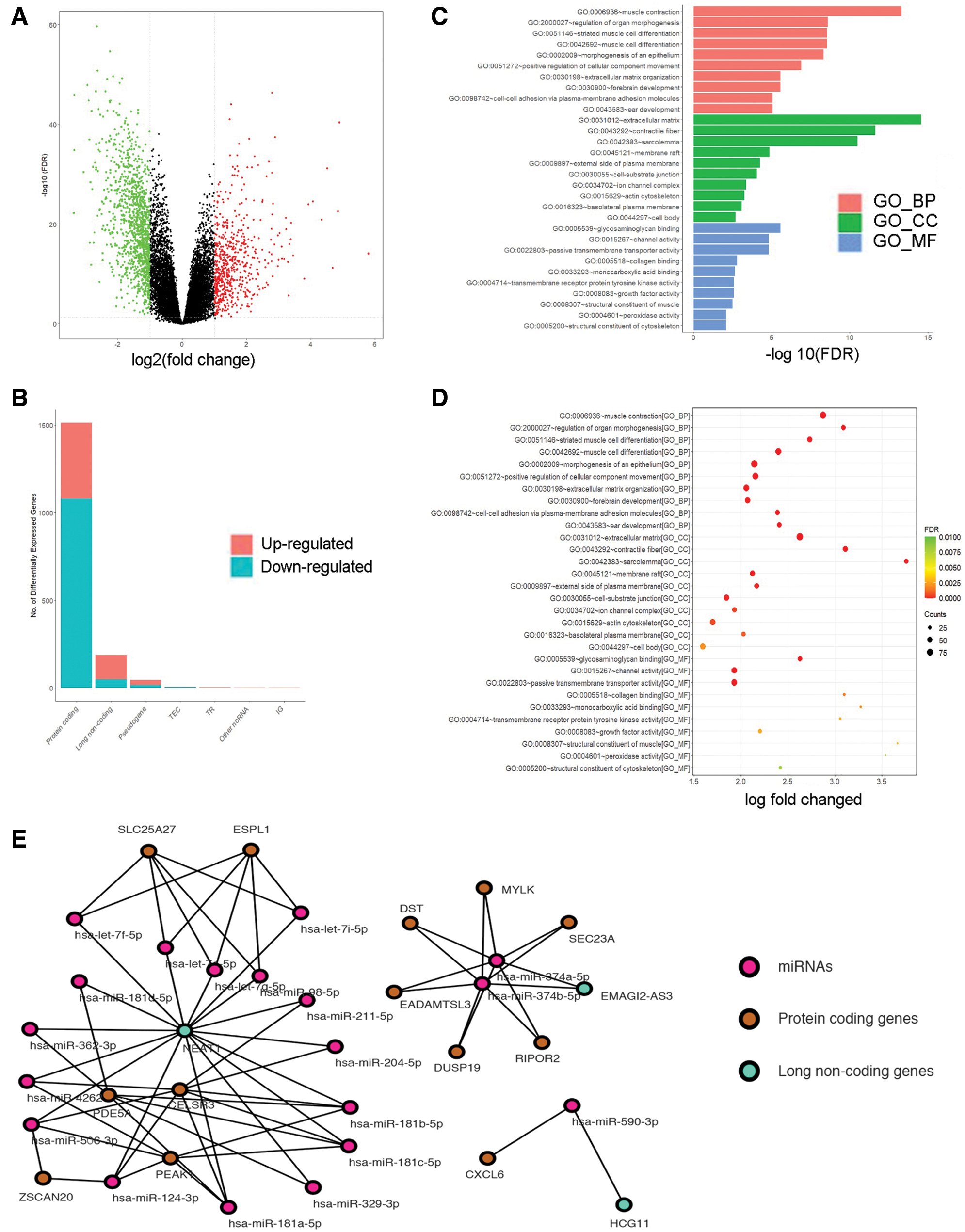
Figure 1: Screening the DEGs in hepatocellular carcinoma (HCC) in the TCGA database and GO and PPI network analysis with the DEGs. (A, B) Analyzing the expression of RNA in HCC. Red squares represent upregulated genes, and the green squares represent upregulated genes. (C, D) Gene Ontology (GO) enrichment significance of different functional groups DElncRNA. GO analysis divided the DElncRNAs into three functional groups: cellular components, molecular functions, and biological processes. (E) Construction of lncRNA-mRNA networks for prognostic lncRNAs. The blue nodes represent interactions between DElncRNAs. Only the 90 DElncRNAs that interacted with other DElncRNAs are shown in the network.
NEAT1, MAGI2-AS3 and HCG11 are abnormally expressed in HCC tumor tissues and cells
To confirm the functions of NEAT1, MAGI2-AS3 and HCG11 in HCC progression, RT-qPCR was employed to assess the expression of NEAT1, MAGI2-AS3 and HCG11 in 40 pairs of HCC para-cancerous and tumor tissues. As shown (Figs. 2A, 2D, and 2G), cancer tissue had higher NEAT1, MAGI2-AS3 and HCG11 expression than that of para-cancer tissue (P < 0.05). According to the median values, the expression of NEAT1, MAGI2-AS3 and HCG11 was classified as the top 50% or the bottom 50%. The Kaplan–Meier survival curves showed that the survival time in patients with expression levels in the bottom 50% for NEAT1 (Fig. 2B), MAGI2-AS3 (Fig. 2E), or HCG11 (Fig. 2H) was significantly longer than in patients with expression levels in the top 50% NEAT1, MAGI2-AS3 or HCG11. HCC cells (HuH-7, Hep3b, Li-7, SNU-387 and RBE cells) exhibited increased expression of NEAT1, MAGI2-AS3 and HCG11 compared to THLE-2 cells. Moreover, the most significantly increased expression level was found in HuH-7 and Li-7 cells (Figs. 2C, 2F, and 2I, all P < 0.05). As a result, these two cell lines were used for later assays.
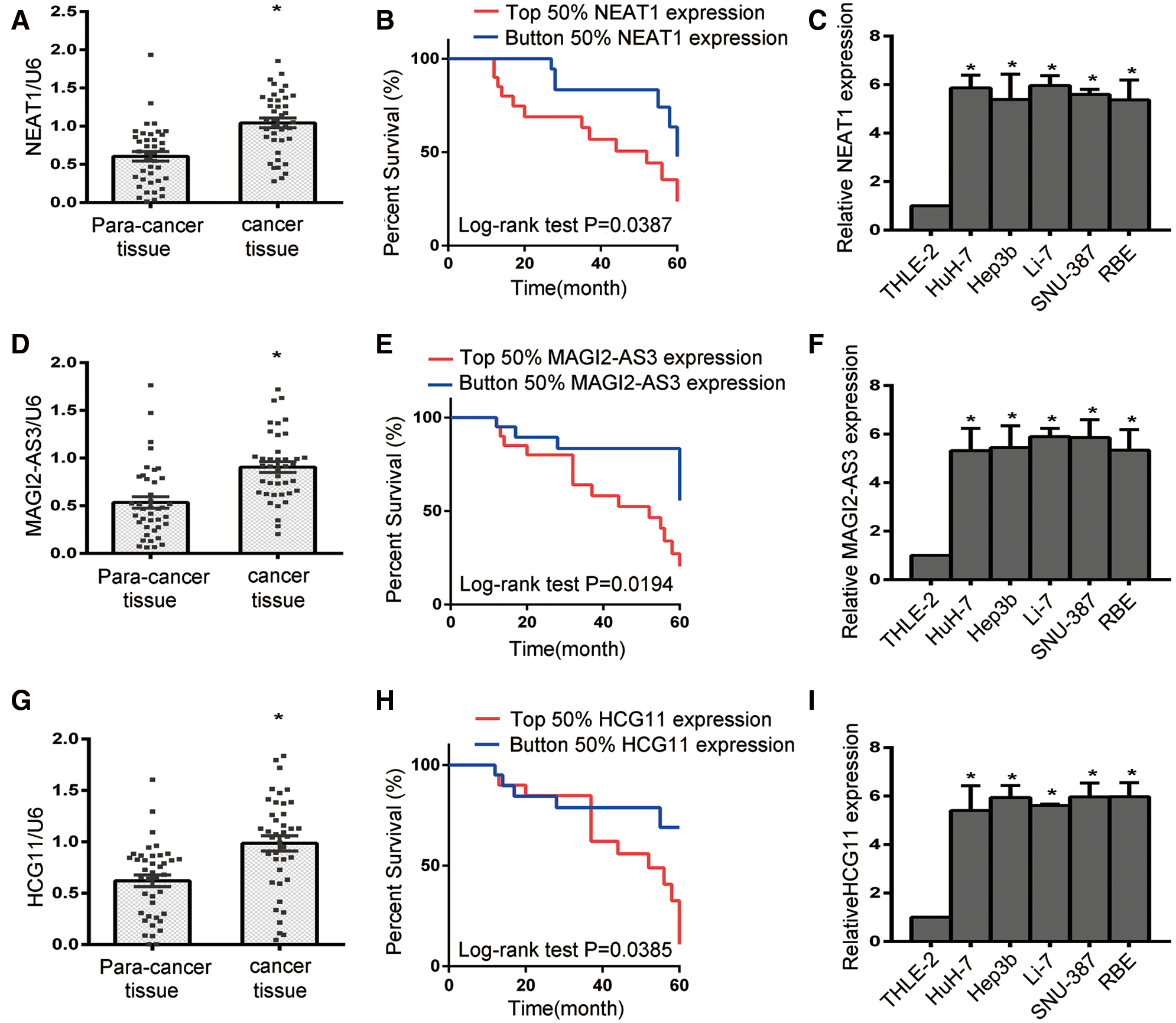
Figure 2: The expression of lncRNA NEAT1, MAGI2-AS3 and HCG11 in para-cancer tissue, cancer tissue and liver cancer cell lines. (A, D, G) The expression levels of NEAT1, MAGI2-AS3 and HCG11 in liver cancer tissue were higher than those in para-cancer tissue (N = 40). (B, E, H) The survival curves for HCC patients for NEAT1 (B), MAGI2-AS3 (E) and HCG11 (H). (C, F, I) NEAT1 (C), MAGI2-AS3 (F) and HCG11 (I) expression in HCC cells (HuH-7, Hep3b, Li-7, SNU-387 and RBE) and normal human liver epithelial cells (THLE-2).
NEAT1 silencing led to a reduced cell proliferation and migration and induced cell apoptosis in vitro
In order to further document the functions of NEAT1, MAGI2-AS3 and HCG11 in HCC progression, HuH-7 and Li-7 cells were transfected with sh-NEAT1, sh-MAGI2-AS3 and sh-HCG11 vectors respectively. First, the knockdown efficiency of NEAT1, MAGI2-AS3 and HCG11 was assessed by qRT-PCR (Figs. 3A–3C, all P < 0.05). Compared to the NC group, the sh-NEAT1, sh-MAGI2-AS3 and sh-HCG11 groups had decreased NEAT1, MAGI2-AS3 and HCG11 expression.
Next, the proliferation ability of the cells was detected by CCK-8 assay. As shown in Figs. 3D and 3E, silencing of NEAT1, MAGI2-AS3 and HCG11 resulted in reduced cellular growth. Moreover, the transwell assay was used to determine the migration ability of HCC cells (Figs. 3F and 3G, P < 0.05). Although there was no significant change in the sh-MAGI2-AS3 group (P < 0.05), knockdown of NEAT1 and HCG1 led to a reduced number of transitional cells, thus indicating that NEAT1 and HCG1 could act as an oncogene in HCC progression. Furthermore, flow cytometry was used to detect cell apoptosis (Figs. 3H and 3I, P < 0.05). Compared with the control group, both the sh-NEAT1 and sh-MAGI2-AS3 groups had obviously increased numbers of apoptotic cells, while the apoptotic cell numbers in the sh-HCG11 group did not significantly change (P < 0.05). These results showed that NEAT1 has a more obvious effect on HuH-7 and Li-7 cells than MAGI2-AS3 and HCG11. Overall, our studies suggest that NEAT1 could act as a key regulator in HCC progression by promoting cell proliferation, supporting cell migration, and inhibiting cell apoptosis.
Figure 3: Silencing NEAT1 could inhibit cell proliferation, suppress cell migration, and induce cell apoptosis of HCC cells in vitro. (A) Expression of NEAT1 in Huh-7 and Li-7 cells. (B) Expression of MAGE-AS3 in Huh-7 and Li-7 cells. (C) Expression of HCG11 in Huh-7 and Li-7 cells. (D, E) The CCK-8 assay was used to determine the growth of Huh-7 (D) and Li-7 (E) cells. (F) Comparison of Huh-7 and Li-7 cell migration in the sh-NEAT1, sh-Magig2-AS3, sh-HCG11 and sh-control groups. *P < 0.05 compared with the control group. (G) Cell migration evaluated by transwell assays in Huh-7 and Li-7 cells. (H) Comparison of Huh-7 and Li-7 cell apoptosis in the sh-NEAT1, sh-Magig2-AS3, sh-HCG11 and sh-control groups. *P < 0.05 compared with the control group. (I) Bar graph of apoptosis rates in Huh-7 and Li-7 cells. *Compared with the sh-control group, P < 0.05.
miR-16-5p was a target of NEAT1
To further illustrate how NEAT1 functions as an oncogene, its potential target was predicted using bioinformatic websites. A binding sequence between NEAT1 and miR-16-5p was identified (Fig. 4A).
The results of the luciferase assay showed that miR-16-5p could decrease the activity of the luciferase reporter vector on HuH-7 and Li-7 cells that contained wild-type NEAT1 fragments, but it could not decrease the luciferase activity of the vector that contained the mutant sequence (Figs. 4B and 4C, P < 0.05). In addition, NEAT1 silencing induced the expression of miR-16-5p in both HuH-7 and Li-7 cells. Moreover, the above induced expression of miR-16-5p was diminished after treatment with miR inhibitor. Also, compared with the control group, miR inhibitor treatment led to a lower level of miR-16-5p expression in the sh-control group (Fig. 4D, P < 0.05). The apoptosis assay on HuH-7 and Li-7 cells showed that the proliferative ability in sh-NEAT1 group cells was significantly decreased (Figs. 3D and 3E), but the apoptosis rate in sh-NEAT1 group cells was increased when compared with the sh-control group. After treatment with miR inhibitor, the apoptosis rate in the sh-NEAT group was reduced to the level of that in the sh-control group. The apoptosis rate in the sh-control + miR inhibitor group was lower than that of the sh-control group (Figs. 4E and 4F, P < 0.05). The results indicate that NEAT1 may inhibit the apoptosis of HuH-7 and Li-7 cells by binding to miR-16-5p.
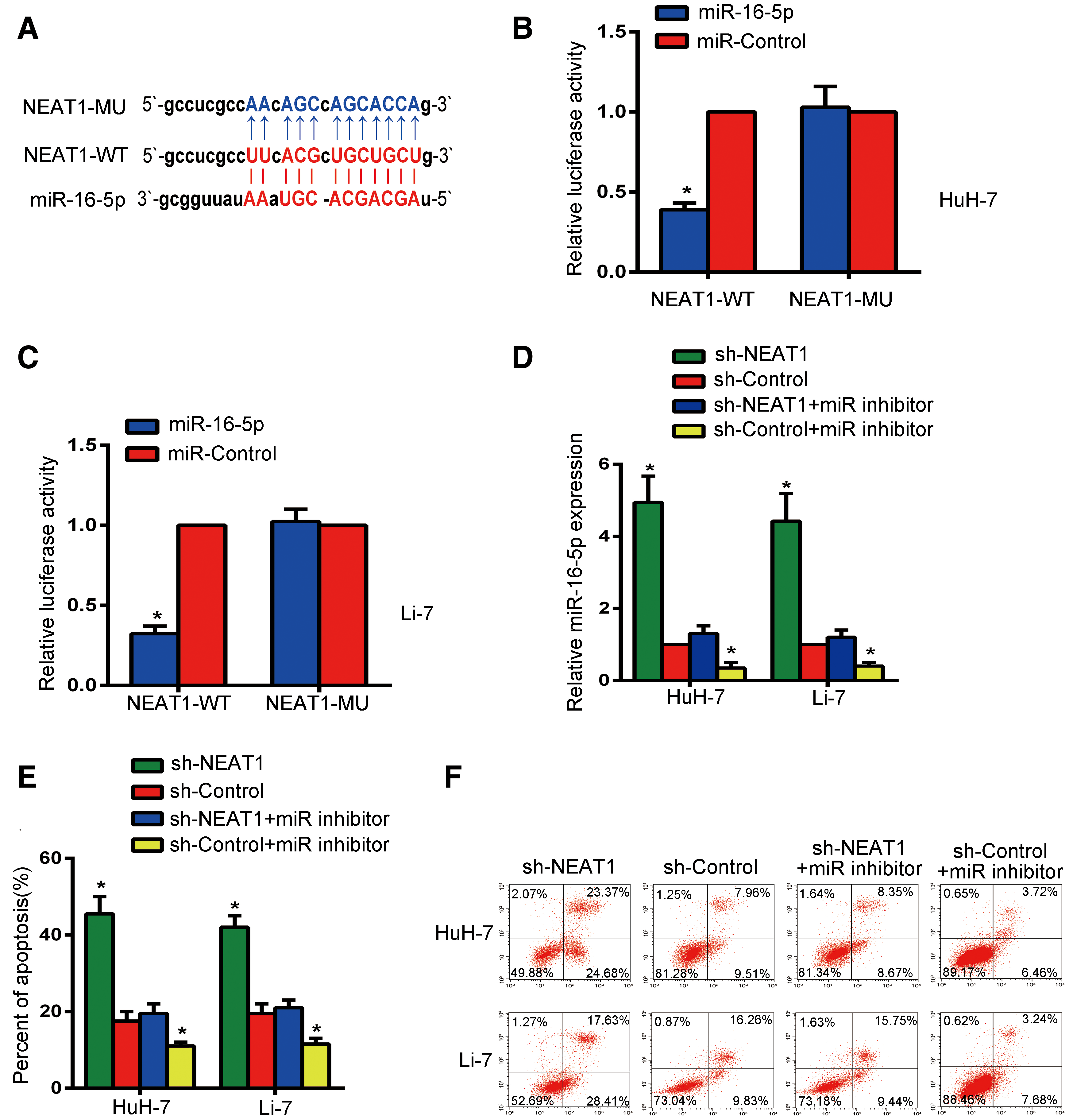
Figure 4: NEAT1 may inhibit the apoptosis of HuH-7 cells and Li-7 cells by binding mir16-5p. (A) Bioinformatics analysis predicted the existence of a binding sequence between miR-16-5p and NEAT1. (B, C) The wild-type NEAT1 (NEAT1-WT) reporter gene and mutated NEAT1 (NEAT1-MU) reporter gene were transfected into Huh-7 cells (B) and Li-7 cells (C) to detect miR-16-5p-regulated luciferase activity. (D) The level of miR-16-5p was examined by qRT-PCR in Huh-7 and Li-7 cells after NEAT1 knockout and NEAT1 combined with miR inhibitor treatment. (E) Bar graph of apoptosis rates of Huh-7 and Li-7 cells. (F) The apoptosis rate of Li-7 and Huh-7 cells in the sh-NEAT1, sh-control, sh-NEAT1 + miR inhibitor and sh-control + miR inhibitor groups were detected by flow cytometry. *Means P < 0.05, compared with the sh-control or miR-control groups.
Bcl-2 was a target gene of miR-16-5p
The biological prediction website also confirmed a binding sequence between Bcl-2 and miR-16-5p (Fig. 5B). The expression of Bcl-2 in HuH-7 and Li-7 cells was quantified by qRT-PCR (Fig. 5A). Compared with the sh-control group, the expression of Bcl-2 was decreased after transfection of NEAT1 shRNA. However, the sh-control + miR inhibitor group had a higher Bcl-2 expression than the other groups, while the sh-NEAT1 + miR inhibitor group had no significant changes in Bcl-2 expression compared to the sh-control group (P < 0.05). Co-transfection with miR-16-5p and Bcl-2-WT decreased the luciferase activity of HuH-7 and Li-7 cells (P < 0.05), as indicated by the dual-luciferase reporter gene assay, while there was no obvious difference in luciferase activity after co-transfection with Bcl-2-MUT (P < 0.05, Figs. 5C and 5D).
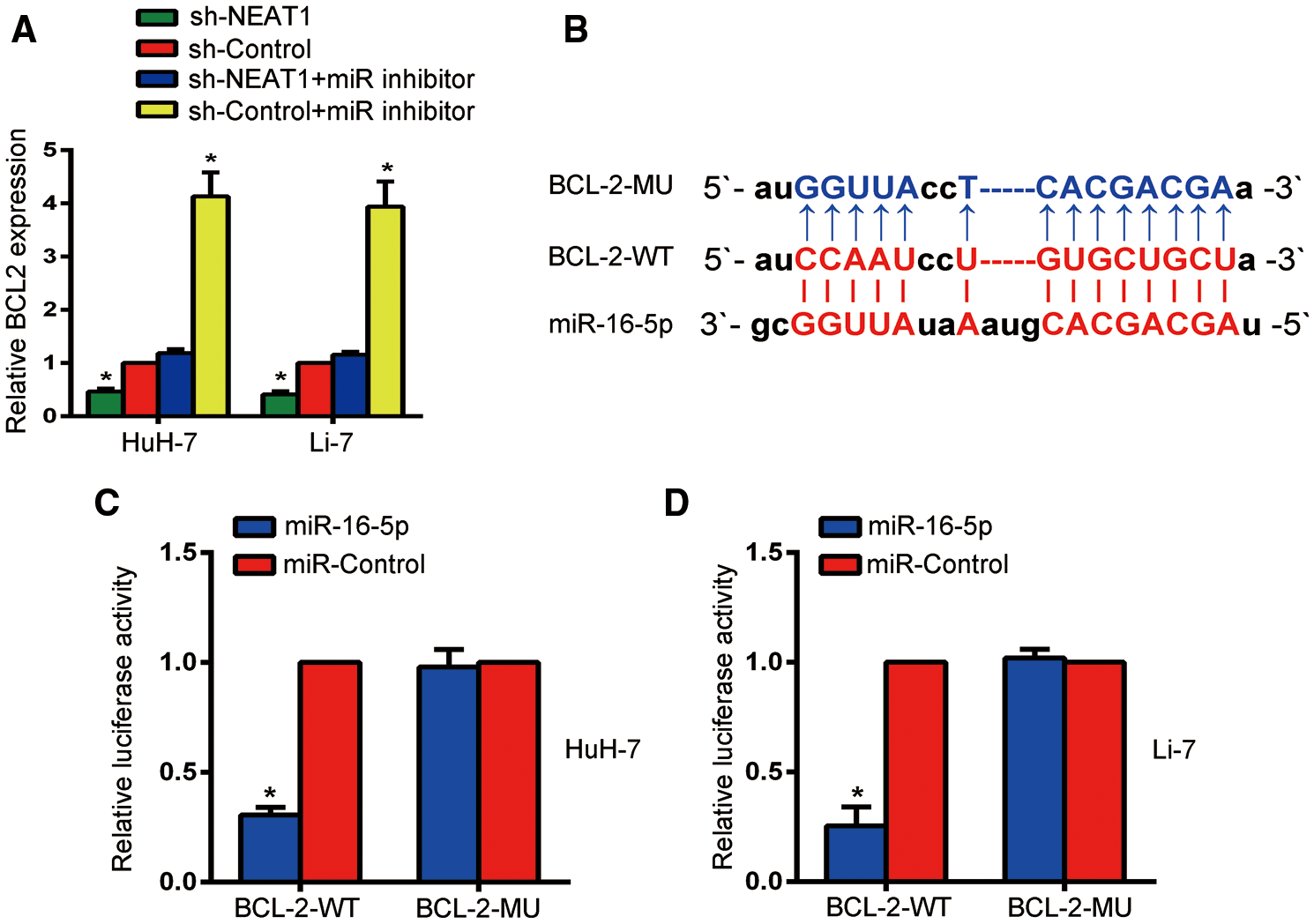
Figure 5: miR-16-5p may regulate Bcl-2. (A) RT-qPCR was used to determine the expression level of Bcl-2 in Huh-7 and Li-7 cells. (B) Bioinformatics analysis predicted a potential binding sequence between and Bcl-2 and miR-16-5p. (C, D) Relative luciferase activity was inhibited in Huh-7 cells (C) and Li-7 cells (D) transfected with miR-16-5p and WT Bcl-2, but not in MUT Bcl-2. *Means P < 0.05, compared with the sh-control or miR-control groups.
In this study, we screened three lncRNAs, lncRNA NEAT1, lncRNA MAG12-AS3 and lncRNA HCG11, in HCC using integrated bioinformatics analyses. These three lncRNAs were overexpressed in HCC tissues and cells. In addition, the Kaplan–Meier curve also demonstrated that they could act as promising prognostic markers in HCC. Patients were divided into higher or lower expression groups according to the median expression values of NEAT1, MAG12-AS3 and HCG11. Kaplan–Meier analysis suggested that patients with higher levels of NEAT1, MAG12-AS3 and HCG11 had significantly shorter survival times than patients with lower expression. In HCC, the contribution of NEAT1, MAG12-AS3 and HCG11 to HCC progression has also been discovered by recent research studies (Li et al., 2019; Niu et al., 2020; Pu et al., 2019), which further confirm the correlation of NEAT1, MAG12-AS3 and HCG11 to the tumor cell epithelial-to-mesenchymal transition (EMT) and the migration and invasion capabilities of HCC. The effects of these three lncRNAs were further verified by cell transfection experiments in vitro, which showed that silencing NEAT1, MAG12-AS3 and HCG11 could significantly reduce the proliferation rate of HCC cells. In addition, we also observed cell migration and apoptosis after silencing the three lncRNAs. We found that, after silencing NEAT1 and HCG11, the cell migration ability was reduced and the migration ability of the sh-MAG12-AS3 group was not affected. After silencing NEAT1 and MAG12-AS3, cell apoptosis was observed, but no obvious apoptosis was observed in the sh-HCG11 group. Therefore, we selected NEAT1 for additional studies to explore its biological function in HCC.
Our results revealed that an increased level of NEAT1 was found in both HCC tissues and cells. NEATI silencing promoted HCC apoptosis and suppressed cell growth and migration. There are many studies related to NEAT1 in HCC. Ren et al. (2017) showed that NEAT1 promotes deterioration of HCC (Mang et al., 2017). In addition, silencing of NEAT1_2 has been shown to exhibit a strengthening role in the radiosensitivity of HCC cells. In addition, miR-101-3p directly regulates WEE1. Thus, the mechanism underlying how lncRNA NEAT1_2 radiosensitizes HCC cells is that the miR-101-3p/WEE1 axis could led to a reduced level of lncRNA NEAT1_2 (Chen and Zhang, 2019). Recently, studies have shown that lncRNAs can be used as ceRNA to regulate the level of its target genes through spongy miRNA, thus playing a regulatory role in tumor development (Lou et al., 2020). Notably, NEAT1, as a ceRNA, can sponge miRNA-22-3p to promote proliferation and invasion of HCC and inhibit apoptosis (Zhou et al., 2019). NEAT1 may target miR-199a-3p to regulate the UCK2 pathway, thus contributing to HCC progression under hypoxic conditions. All in all, NEAT1 might be considered as a new promising therapeutic approach for HCC treatment (Zhang et al., 2020). In addition, Yu et al. (2019) reported that NEAT1 promotes colorectal cancer by secreting mir-193a-3p. Our studies have shown a negative correlation between NEAT1 and miR-16-5p, as shown by an increased level of miR-16-5p caused by NEAT1 silencing. When inhibiting miR-16-5p, its expression level in the sh-NEAT1 group decreased to the original level. Previous studies have found that silencing NEAT1 could increases the rate of apoptosis. Interestingly, we found that silencing NEAT1 also inhibited miR-16-5p, and the apoptosis rate decreased. Therefore, we believe that NEAT1, as a ceRNA, can sponge miR-16-5p to enhance cell growth. Briefly, after silencing NEAT1, more miR-16-5p was released, which led to an increase in apoptosis of HCC cells. More than that, the suppressive role of miR-16-5p in HCC progression was mediated by regulating IGF1R.
Additionally, IGF1R silencing is responsible for the suppressive function of miR-16-5p on the proliferation and migration of SMMC-7721 cells (Cheng et al., 2019). The inhibitory function of miR-16-5p in tumor development was further confirmed by Ruan and Qian (2019). They reported that the level of miR-16-5p was low in breast cancer tissue and its inhibitory effect is achieved by inhibiting the AKT3 level and suppressing the NF-κB pathway.
Emerging reports have indicated Bcl-2 protein family function is a key regulator in cell apoptosis. Moreover, Bax acts as a master proapoptotic regulator in the family (Chen et al., 2019; Sharief et al., 2002). Bcl-2 has been documented to prevent Bax from releasing cyt-C, thus inhibiting activation of downstream apoptotic proteins, and ultimately inhibiting the process of apoptosis (Wang et al., 2016). Based on the study by Zhuang et al. (2020), the inhibitory function of miR-202 in the development of HCC was mediated by suppressing Bcl-2 expression, thus demonstrating that miR-202 could be a promising target for HCC treatment. Chaudhuri et al. (2018) reported that NTKR3 and its downstream effector Bcl-2 could be regulated by miR-125a-5p and miR-16-5p. In addition, a decreased level of miR-16-5p was discovered in breast cancer, and the upregulated expression of miR-16-5p was related to the negative regulation of target gene Bcl-2, leading to cell growth inhibition and apoptosis enhancement (Mavrogiannis et al., 2018).
In conclusion, we cannot rule out that NEAT1 can act as a ceRNA and competitively bind to miR-16-5p, thereby inhibiting the interaction of miR-16-5p and Bcl-2. Gradually increasing the level of Bcl-2 suppresses apoptosis, and the proliferation of HCC increased. Our study found that a decreased level of Bcl-2 resulted from interference of NEAT1. Moreover, miR-16-5p silencing caused the Bcl-2 level to rebound, thus confirming a significant correlation between miR-16-5p and Bcl-2.
Taken together, our results demonstrated that miR-16-5p could promote cell apoptosis by targeting lncRNA NEAT1. lncRNA NEAT1 contributes to the growth and metastasis of HCC cells and suppresses apoptosis of HCC cells via the competitive sponging of miR-16-5p to inhibit its expression. Furthermore, the lncRNA NEAT1/miR-16-5p axis could act as a key player in regulating the level of Bcl-2 in HCC, thus suggesting the lncRNA NEAT1/miR-16-5p/Bcl-2 signaling axis as a new promising therapeutic approach in HCC treatment.
Finally, further studies are needed to determine the function of NEAT1 and its underlying mechanism, which would provide a novel method to treat human cancer.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Authors’ Contribution: The authors confirm contribution to the paper as follows: study conception and design: YHL; data collection: FC; analysis and interpretation of results: FC and LW; draft manuscript preparation: YHL and LW. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: The research study was carried out in accordance with the ethical guidelines of the Declaration of Helsinki and was approved by the Research Ethics Committee of the Research Institute of Jinzhou Medical University (No. 2018-03) at March 07, 2018. All patients signed an informed consent form.
Funding Statement: This work was supported by (1) 2019 Xing Liao Ying Cai Plan of Liaoning Province (XLYC1802049). (2) 2019 Natural Science Foundation Project of Liaoning Province (2019-ZD-0803). (3) 2020 Natural Science Foundation Project of Liaoning Province (2020-MS-297).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Abbastabar M, Sarfi M, Golestani A, Khalili E (2018). lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI Journal 17: 900. [Google Scholar]
Craig AJ, von Felden J, Garcia-Lezana T, Sarcognato S, Villanueva A (2020). Tumour evolution in hepatocellular carcinoma. Nature Reviews Gastroenterology & Hepatology 17: 139–152. DOI 10.1038/s41575-019-0229-4. [Google Scholar] [CrossRef]
Chang L, Li C, Lan T, Wu L, Yuan Y et al. (2016). Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Molecular Medicine Reports 13: 1541–1550. DOI 10.3892/mmr.2015.4716. [Google Scholar] [CrossRef]
Chaudhuri AD, Dastgheyb RM, Yoo S-W, Trout A, Talbot CCJr et al. (2018). TNFα and IL-1β modify the miRNA cargo of astrocyte shed extracellular vesicles to regulate neurotrophic signaling in neurons. Cell Death & Disease 9: 363. DOI 10.1038/s41419-018-0369-4. [Google Scholar] [CrossRef]
Chen X, Zhang N (2019). Downregulation of lncRNA NEAT1_2 radiosensitizes hepatocellular carcinoma cells through regulation of miR-101-3p/WEE1 axis. Cell Biology International 43: 44–55. DOI 10.1002/cbin.11077. [Google Scholar] [CrossRef]
Chen Y, Zhang L, Liu S, Yao B, Zhang H et al. (2019). Sam68 mediates high glucose-induced podocyte apoptosis through modulation of Bax/Bcl-2. Molecular Medicine Reports 20: 3728–3734. DOI 10.3892/mmr.2019.10601. [Google Scholar] [CrossRef]
Cheng B, Ding F, Huang C, Xiao H, Fei F, Li J (2019). Role of miR-16-5p in the proliferation and metastasis of hepatocellular carcinoma. European Review for Medical and Pharmacological Sciences 23: 137–145. [Google Scholar]
Gunsar F (2017). Liver transplantation for hepatocellular carcinoma beyond the Milan criteria. Experimental and Clinical Transplantation: Official Journal of the Middle East Society for Organ Transplantation 15: 59–64. [Google Scholar]
Leti F, Legendre C, Still CD, Chu X, Petrick A et al. (2017). Altered expression of MALAT1 lncRNA in nonalcoholic steatohepatitis fibrosis regulates CXCL5 in hepatic stellate cells. Translational Research 190: 25–39. DOI 10.1016/j.trsl.2017.09.001. [Google Scholar] [CrossRef]
Li M, Zhang Y, Ma L (2019). LncRNA HCG11 accelerates the progression of hepatocellular carcinoma via miR-26a-5p/ATG12 axis. European Review for Medical and Pharmacological Sciences 23: 10708–10720. [Google Scholar]
Lou W, Ding B, Fu P (2020). Pseudogene-derived lncRNAs and their miRNA sponging mechanism in human cancer. Frontiers in Cell and Developmental Biology 8: 85. DOI 10.3389/fcell.2020.00085. [Google Scholar] [CrossRef]
Mang Y, Li L, Ran J, Zhang S, Liu J et al. (2017). Long noncoding RNA NEAT1 promotes cell proliferation and invasion by regulating hnRNP A2 expression in hepatocellular carcinoma cells. OncoTargets and Therapy 10: 1003. DOI 10.2147/OTT. [Google Scholar] [CrossRef]
Mavrogiannis AV, Kokkinopoulou I, Kontos CK, Sideris DC (2018). Effect of vinca alkaloids on the expression levels of microRNAs targeting apoptosis-related genes in breast cancer cell lines. Current Pharmaceutical Biotechnology 19: 1076–1086. DOI 10.2174/1389201019666181112103204. [Google Scholar] [CrossRef]
Niu Y, Tang G, Wu X, Wu C (2020). LncRNA NEAT1 modulates sorafenib resistance in hepatocellular carcinoma through regulating the miR-149-5p/AKT1 axis. Saudi Journal of Gastroenterology: Official Journal of the Saudi Gastroenterology Association 26: 194. DOI 10.4103/sjg.SJG_4_20. [Google Scholar] [CrossRef]
Pu J, Wang J, Wei H, Lu T, Wu X et al. (2019). lncRNA MAGI2-AS3 prevents the development of HCC via recruiting KDM1A and promoting H3K4me2 demethylation of the RACGAP1 promoter. Molecular Therapy-Nucleic Acids 18: 351–362. DOI 10.1016/j.omtn.2019.08.020. [Google Scholar] [CrossRef]
Qu Z, Yuan CH, Yin CQ, Guan Q, Chen H, Wang FB (2016). Meta-analysis of the prognostic value of abnormally expressed lncRNAs in hepatocellular carcinoma. OncoTargets and Therapy 9: 5143. DOI 10.2147/OTT. [Google Scholar] [CrossRef]
Ruan L, Qian X (2019). MiR-16-5p inhibits breast cancer by reducing AKT3 to restrain NF-κB pathway. Bioscience Reports 39: BSR20191611. DOI 10.1042/BSR20191611. [Google Scholar] [CrossRef]
Sahu SK, Chawla YK, Dhiman RK, Singh V, Duseja A et al. (2019). Rupture of hepatocellular carcinoma: A review of literature. Journal of Clinical and Experimental Hepatology 9: 245–256. DOI 10.1016/j.jceh.2018.04.002. [Google Scholar] [CrossRef]
Sharief M, Douglas M, Noori M, Semra Y (2002). The expression of pro-and anti-apoptosis Bcl-2 family proteins in lymphocytes from patients with multiple sclerosis. Journal of Neuroimmunology 125: 155–162. DOI 10.1016/S0165-5728(02)00024-3. [Google Scholar] [CrossRef]
Singal AG, El-Serag HB (2015). Hepatocellular carcinoma from epidemiology to prevention: Translating knowledge into practice. Clinical Gastroenterology and Hepatology 13: 2140–2151. DOI 10.1016/j.cgh.2015.08.014. [Google Scholar] [CrossRef]
Wang Q, Zhang L, Yuan X, Ou Y, Zhu X et al. (2016). The relationship between the Bcl-2/Bax proteins and the mitochondria-mediated apoptosis pathway in the differentiation of adipose-derived stromal cells into neurons. PLoS One 11: e0163327. DOI 10.1371/journal.pone.0163327. [Google Scholar] [CrossRef]
Yu H-M, Wang C, Yuan Z, Chen GL, Ye T, Yang BW (2019). LncRNA NEAT1 promotes the tumorigenesis of colorectal cancer by sponging miR-193a-3p. Cell Proliferation 52: e12526. DOI 10.1111/cpr.12526. [Google Scholar] [CrossRef]
Zhang F, Ding L, Cui L, Barber R, Deng B (2019). Identification of long non-coding RNA-related and-coexpressed mRNA biomarkers for hepatocellular carcinoma. BMC Medical Genomics 12: 15–24. DOI 10.1186/s12920-019-0472-0. [Google Scholar] [CrossRef]
Zhang Q, Cheng Q, Xia M, Huang X, He X, Liao J (2020). Hypoxia-induced lncRNA-NEAT1 sustains the growth of hepatocellular carcinoma via regulation of miR-199a-3p/UCK2. Frontiers in Oncology 10: 998. DOI 10.3389/fonc.2020.00998. [Google Scholar] [CrossRef]
Zhou X, Wang X, Zhou Y, Cheng L, Zhang Y, Zhang Y (2019). Long noncoding RNA NEAT1 promotes cell proliferation and invasion and suppresses apoptosis in hepatocellular carcinoma by regulating miRNA-22-3p/akt2 in vitro and in vivo. OncoTargets and Therapy 12: 8991. DOI 10.2147/OTT. [Google Scholar] [CrossRef]
Zhuang D, Liang L, Zhang H, Feng X (2020). MiR-202 suppresses hepatocellular carcinoma progression via downregulating BCL2 expression. Oncology Research 28: 399–408. DOI 10.3727/096504020X15864296270581. [Google Scholar] [CrossRef]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |