DOI:10.32604/biocell.2022.019806

| BIOCELL DOI:10.32604/biocell.2022.019806 |  |
| Article |
Genome-wide identification of NAC gene family and expression analysis under abiotic stresses in Salvia miltiorrhiza
1Zhejiang Province Key Laboratory of Plant Secondary Metabolism and Regulation, College of Life Sciences and Medicine, Zhejiang Sci-Tech University, Hangzhou, 310018, China
2Institute of Crop Science, Ministry of Agriculture and Rural Affairs Laboratory of Spectroscopy Sensing, Zhejiang University, Hangzhou, 310058, China
*Address correspondence to: Ling Xu, lxu@zstu.edu.cn
Received: 16 October 2021; Accepted: 27 December 2021
Abstract: NAC (NAM, ATAF, CUC) is a class of transcription factors involved in plant growth regulation, abiotic stress responses, morphogenesis and metabolism. Salvia miltiorrhiza is an important Chinese medicinal herb, but the characterization of NAC genes in this species is limited. In this study, based on the Salvia miltiorrhiza genomic databases, 82 NAC transcription factors were identified, which were divided into 14 groups. Meanwhile, phylogenetic analysis, gene structure, chromosomal localization and potential role of SmNACs in abiotic stress conditions were also studied. The results revealed that some SmNACs had different structures than others, which advised that these genes may have multiple/distinct functions. Real-time quantitative polymerase chain reaction (RT-qPCR) analysis showed that SmNACs exhibited differential expression patterns under salt and drought stress. The NaCl induced salinity treatments modulated the expression of several SmNAC genes more in roots compared with leaves. Conversely, under drought stress conditions, more genes were upregulated in leaves compared with roots. These results will be useful for the further study involved in the functional characteristics of SmNAC genes, especially in response to salt and drought stresses, thereby may facilitate genetic breeding in Salvia miltiorrhiza.
Keywords: Salvia miltiorrhiza; Arabidopsis thaliana; NAC transcription factors; Genome-wide analysis; Abiotic stress
Salvia miltiorrhiza (Lamiaceae), as an important traditional Chinese medicinal herb that is widely used to treat cardiovascular diseases (CVD) (Hou et al., 2020; Wang et al., 2017). Salvia miltiorrhiza contains water-soluble phenolic compounds, such as salvianolic acid A, salvianolic acid B and fat-soluble substances, including tanshinone I, tanshinone IIA, dihydrotanshinone, etc (Jung et al., 2020). Modern pharmacology investigations have shown that these bioactive compounds have diverse pharmacological effects that were mediated through antiapoptotic, antioxidative, and anti-inflammatory responses (Wu et al., 2016; Zhou et al., 2011).
The NAC family is a largest class of plant transcriptional factors, whereas, term NAC is derived from closely related three protein families, namely no apical meristem (NAM), Arabidopsis transcription activation factor (ATAF)1/2) and cup-shaped cotyledon (CUC2) (Olsen et al., 2005). A typical NAC proteins contain a N-terminal that is highly homologous to the DNA-binding region of NAC domain, composed of 150 amino acids (Nakashima et al., 2012). The NAC domain consists of five conserved regions, and are named as A to E. The binding of NAC protein to its target is controlled by subdomains C and D (Mathew et al., 2016). The C-terminal interacts with DNA or other transcription factors and has a highly variable transcriptional regulatory region (Shen et al., 2019). Like other TF families such as MYB, WRKY and AP2/ERF, NAC is also involved in regulating various physiological and developmental processes in plants. The NAC gene family have been identified in many plants, for example, around 71 NAC TF were in Solanum muricatum (Yang et al., 2021), 102 in Theobroma cacao (Shen et al., 2019), 106 in Prunus dulcis (Zafar et al., 2021), 142 in Actinidia (Jia et al., 2021) and 151 in Oryza sativa (Nuruzzaman et al., 2010). However, the diversity and numbers of NAC genes in Salvia miltiorrhiza are rarely reported.
The chromosome numbers of Salvia miltiorrhiza are 2n = 2x = 16, and it is considered as a tetraploids (Zhao et al., 2006). Sequencing of the whole genome of Salvia miltiorrhiza was completed in 2020 (Song et al., 2020). Its genome size is about 594,750,066 base pairs. In the present study, genome-wide identification and expression analysis of NAC genes under salt and drought conditions were studied. The genomic structure, phylogenetic relationships and chromosome location of a NAC protein in Salvia miltiorrhiza were identified and discussed. A total of 82 NAC genes were identified and their expression patterns were analyzed under salinity and drought stress using real-time quantitative (RT-qPCR) PCR. This study is to lay a foundation for understanding the regulatory mechanism of NAC genes and may help to elucidate the molecular mechanisms underlying salt/drought stress responses, and to design optimal genetic improvement strategies in Salvia miltiorrhiza.
Database search and sequence analysis of NAC genes in Salvia miltiorrhiza
The Salvia miltiorrhiza genome sequence data was obtained from the China National Center for Bioinformation (https://bigd.big.ac.cn). The nucleotide sequences and amino acid data of 96 AtNAC genes were downloaded from The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/index.jsp). The Hidden Markov Model (HMM, http://hmmer.org/) search was performed against the Salvia miltiorrhiza protein database using the NAM domain (PF02365, e-value ≤ 1.1e-10). The candidate sequences were submitted in the Pfam database (http://pfam.xfam.org) and NCBI Conserved Domains Database for verification (https://www.ncbi.nlm.nih.gov/cdd). The results showed that 82 NAC proteins exist the Salvia miltiorrhiza genomes (Table 1). The isoelectric point and protein molecular weight of SmNAC proteins were predicted using the pI/Mw tool on the online software ExPASy (http://web.expasy.org/protparam).
Multiple sequence alignment, and phylogenetic analysis
The ClustalW analysis was used to analyze the multiple sequence alignment on the NAC domain sequences of Arabidopsis and Salvia miltiorrhiza NAC genes. The MEGA 7.0 software and Maximum Likelihood method (ML, bootstrap = 1000) were used to construct a phylogenetic tree (Sudhir et al., 2018). The sequences for phylogenetic trees are shown in Table S1.
The Perl script was used to extract mRNA location information from the Salvia miltiorrhiza GFF file to determine the SmNAC location, and TBtools 1.007 software was used to map 9 chromosomes in the Salvia miltiorrhiza genome.
Protein properties and sequence analysis
MEME online software (http://meme-suite.org/tools/meme) was used to analyze the conversed motif prediction of 82 SmNAC with the following conditions: zoops, 10 motifs with an optimum motif width between 10 and 50 residues, and any number of repetitions. The CDS and UTR location information in the mRNA of SmNACs was selected from the GFF file of Salvia miltiorrhiza genomes for complete gene structure identification.
Plant materials and abiotic stress treatments
Salvia miltiorrhiza seeds were obtained in Shanxi Province, China. Healthy seeds were grown in potting soil composed of peat, perlite and vermiculite (4:3:1). When Salvia miltiorrhiza plants were five months old, uniform plants were selected for abiotic stress treatments. The plants were carefully uprooted and were directly transformed into the pots containing Hoagland solution with desired levels of salinity and drought treatments. For salt stress treatments, 100 mM, and 200 mM of NaCl induced salinity, and for drought 5%, and 10% polyethylene glycol (PEG6000) induced drought stress treatments were selected, whereas control treatment was without addition of any salt (NaCl) or PEG additions. Leaves and roots were harvested after 24 h of stress exposure, and were frozen in liquid nitrogen and stored at –80°C for total RNA extraction. All experiments were repeated with three biological replicates.
RNA extraction and cDNA synthesis
RNA was extracted using RNAprep Pure Plant Plus Kit (TIANGEN Kit, DP441) from the leaves and roots in Salvia miltiorrhiza for tissue-specific expression. Determination of RNA concentration and purity (OD260/OD280 = 1.8–2.1) was measured using Nucleic acid micrometer (Thermo, NANODROP 2000). The RNA was reversely transcribed into cDNA using the TaKaRa reverse transcription kit (Dalian, China) for qRT-PCR analysis.
Gene expression analyses by real-time quantitative PCR (RT-qPCR)
RT-qPCRs were performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme, China). Primers were designed using online tool: https://www.genscript.com/tools/real-time-pcr-taqman-primer-design-tool. The length of amplicons is between 150 bp and 200 bp. SmActin was used as the reference gene (Han et al., 2020). The gene expression was calculated using 2−ΔΔCT method. Initially, eighty-two genes were selected for RT-qPCR assay with three biological replicates. However, expression levels of thirty-two genes were low, and therefore fifty genes were selected for further analysis.
The gene expression data were presented as mean values of three biological replicates with standard error (SE). The fisher least significant difference (LSD) test to determine significant differences (P ≤ 0.05). Heat map was performed using the OmicStudio tools at https://www.omicstudio.cn/tool.
Identification of NAC gene family in Salvia miltiorrhiza
A sequencing data of 96 NAC genes were downloaded from TAIR. A total of 82 NAC proteins were identified from the Salvia miltiorrhiza genome after comparison with the Hidden Markov Model (HMM) search using the NAM domain (PF02365). For convenience, the 82 NAC genes were successively named SmNAC1-SmNAC82. The characteristic parameters of the predicted protein encoded by SmNAC are shown in Table 1. The approximate length of NAC protein ranged from 120 amino acid (SmNAC36) to 608 (SmNAC25) amino acids, with an average length of 311aa. The isoelectric point (pIs) of NAC proteins ranged from 4.53 (SmNAC43) to 9.73 (SmNAC26), and 55 pIs < 7.00, 27 pIs > 7.00. The molecular weights (MWs) ranged from 14014.88 Da (SmNAC26) to 68033.50 Da (SmNAC2).
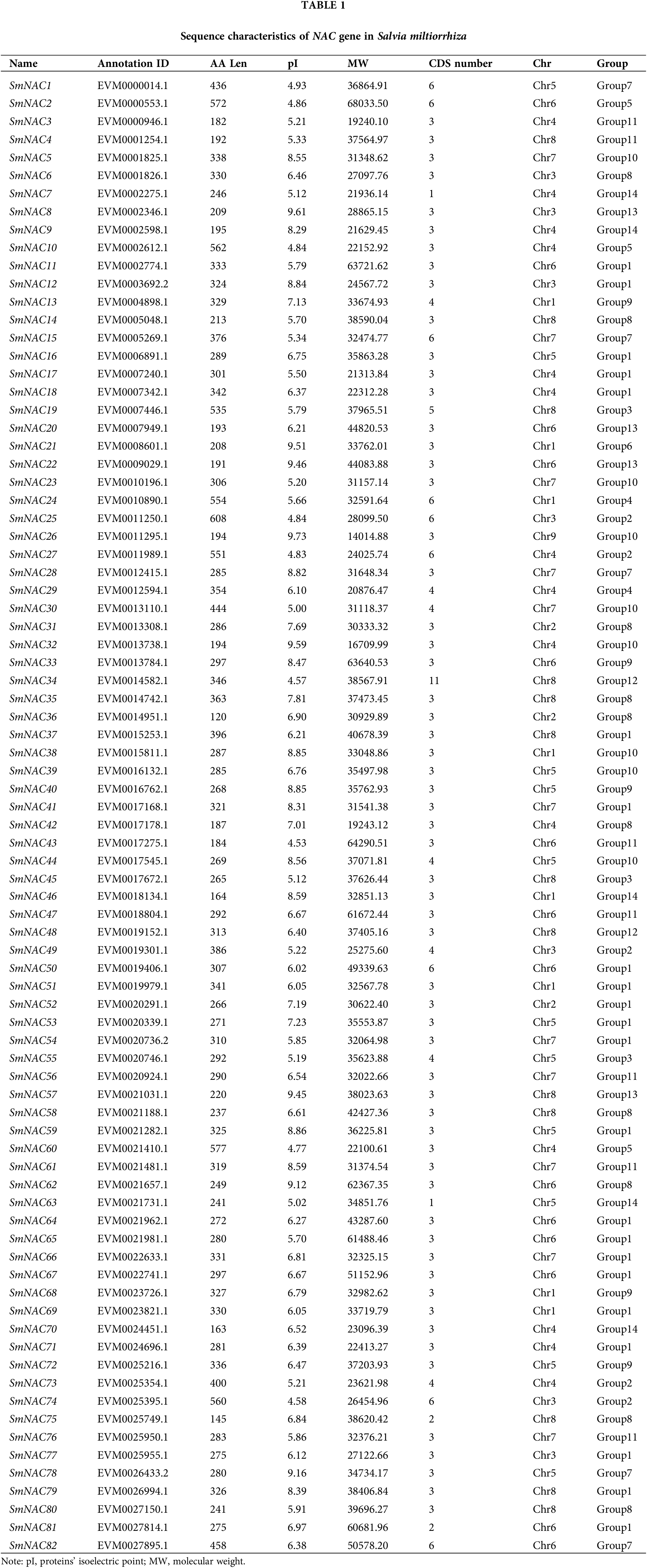
SmNAC gene structure and conserved sequence analysis
The structural diagrams of SmNAC genes were established by the results of MEME motif analysis. We divided SmNAC into 14 groups, as G1-G14. Among them, motif 1, motif 2, motif 3, motif 4, motif 5, and motif 6 are the most widely distributed (Fig. 1b).
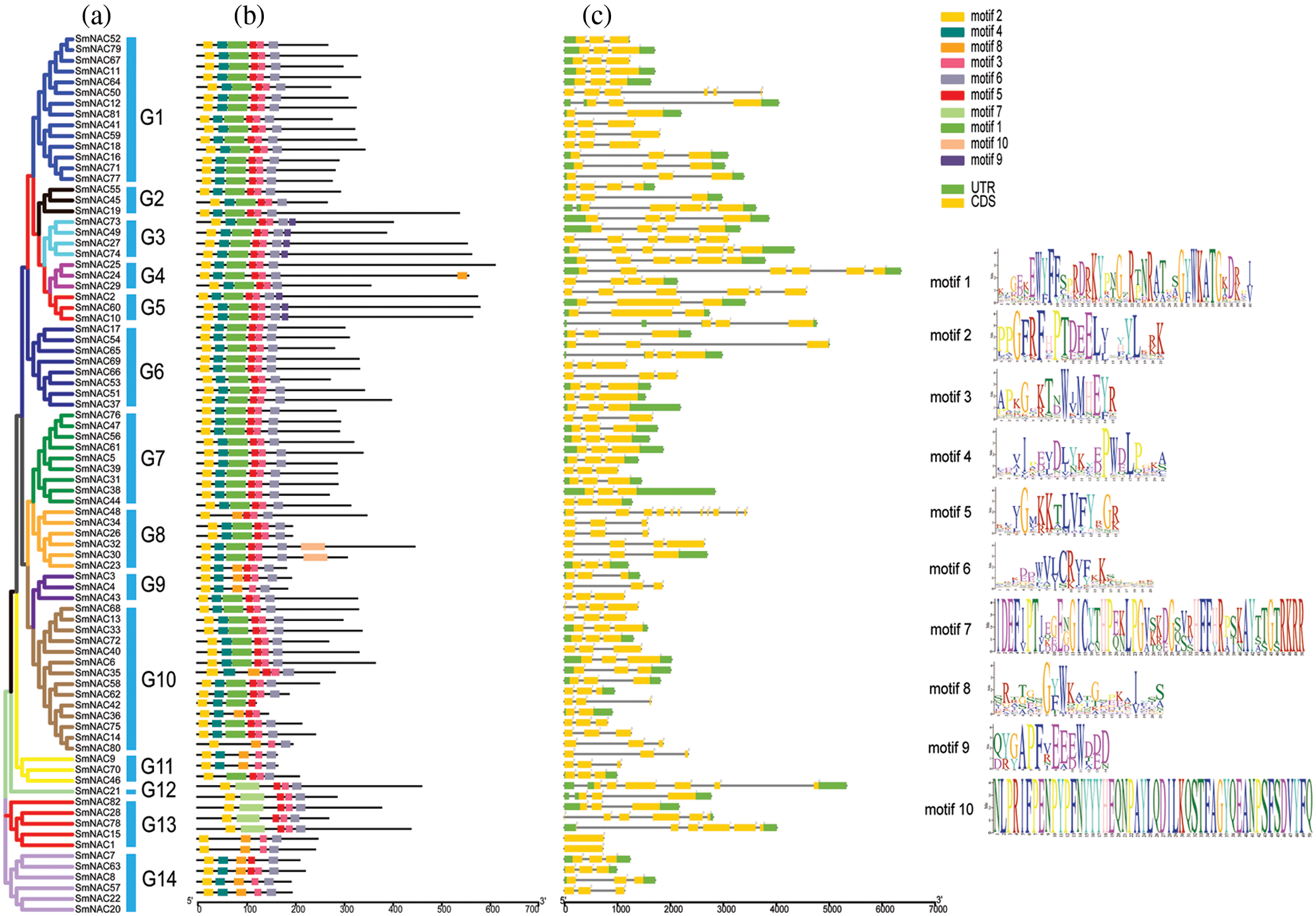
Figure 1: Phylogenetic relationship, conserved protein motif structure and gene structure of SmNAC gene in Salvia miltiorrhiza. (a) Phylogenetic tree was constructed using Mega 7.0 software based on the domains of Salvia miltiorrhiza NAC proteins, with branching details shown in different colors. (b) The motif composition of Salvia miltiorrhiza NAC proteins. (c) The motifs, numbers 1 to 10, are displayed in different colored boxes.
The results have shown that the motif composition across different members of each group was similar, this represents that the gene structure is conserved in a specific subfamily. In addition, each SmNAC has its own specificity, with motif 10 specific to SmNAC23 and SmNAC30 this may be related to the important function of NAC transcription factors. The function of most conserved motifs remains to be elucidated. Exon identification was performed on the identified SmNAC members to further understand the diversity among NAC family members in Salvia miltiorrhiza (Fig. 1c). All SmNAC genes had one to 11 protein-coding DNA sequence (CDS).
Phylogenetic tree analysis of SmNAC gene family
Phylogenetic analysis of Arabidopsis thaliana and Salvia miltiorrhiza NAC genes was carried out, in order to further understand the evolutionary relationship. A total of 261 NAC’s, of which 179 were from Arabidopsis thaliana and 82 were from Salvia miltiorrhiza, were used to construct the phylogenetic tree (Fig. 2). They have divided them into 14 subgroups (G1 to G14) according to their evolutionary branches. The results also showed that the distribution of SmNAC members are uneven among all subgroups. Group1 (G1) contained the highest (22) SmNAC members, followed by Group8 with 10 members, and Group6 with only one member.
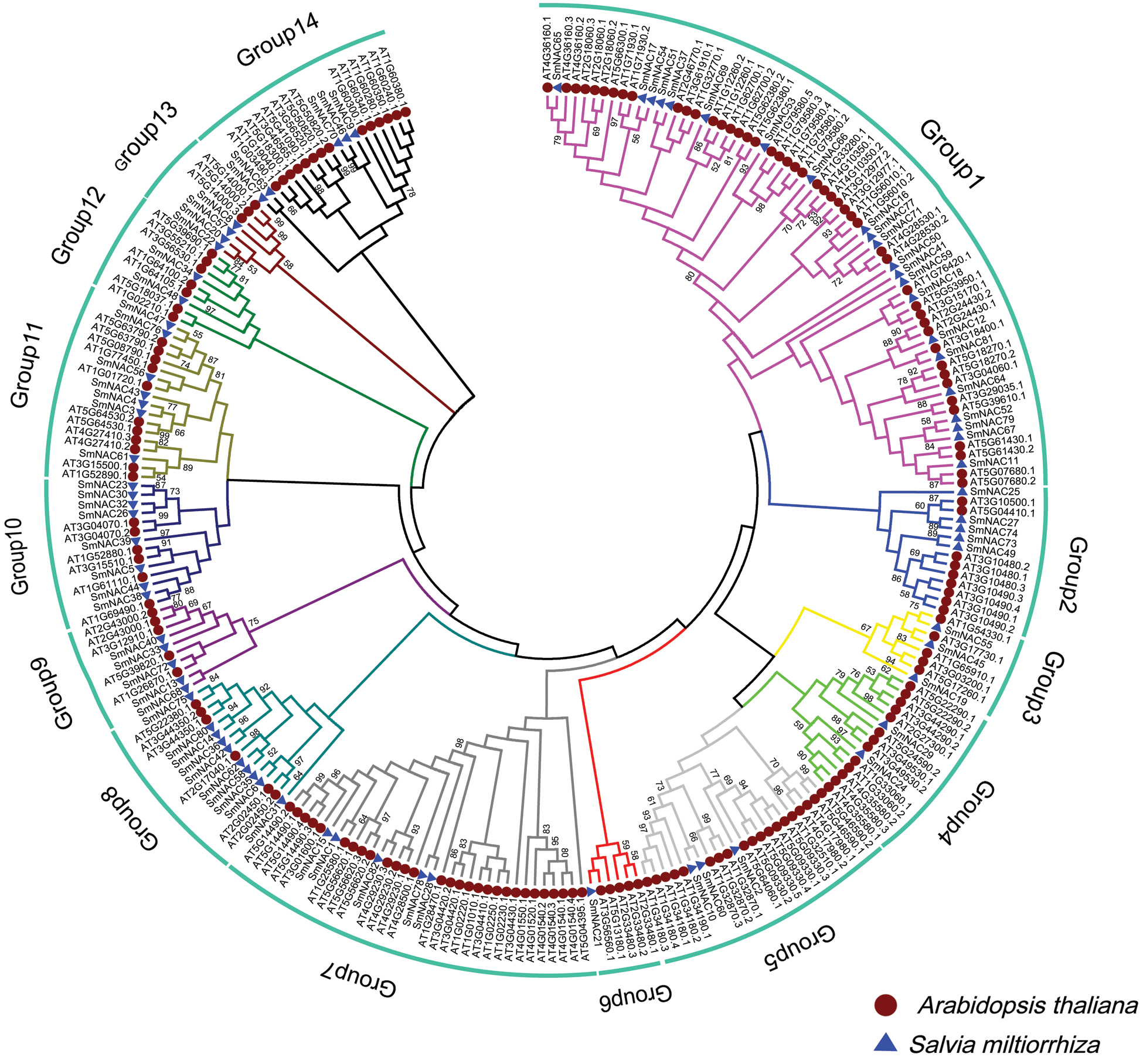
Figure 2: Phylogenetic analysis of NAC in Arabidopsis thaliana and Salvia miltiorrhiza. The tree amplified a total of 1000 Bootstrap replicates using the maximum likelihood (ML) method using MEGA 7.0. The tree divided these NAC proteins into 14 groups, named Group1 to Group14. NAC protein members of A. thaliana and Salvia miltiorrhiza are labeled with red circles and blue triangles, respectively, with different colored branches representing different group.
Chromosomal localization and distribution of SmNAC genes
The TB tools software was used to visualize the location of the acquired genes on the chromosomes. We mapped 82 SmNAC genes to GWHA0SJ00000006, GWHA0SJ00000010, GWHA0SJ00000013, GWHA0SJ00000020, GWHA0SJ00000023, GWHA0SJ00000040, GWHA0SJ00000057, GWHA0SJ00000084, and GWHA0SJ00000114 were all on nine chromosomes, (Fig. 3). A total of 14 genes were located at Chr4 and Chr6, 13 genes on Chr8, 11 gens on Chr5 and Chr7, and so on (Fig. 3).
Figure 3: Distribution of SmNAC genes in chromosomes (Chrs). The vertical bars represent the chromosomes in the Salvia miltiorrhiza genome, with the Chr number at the top.
Differential expressions of SmNAC genes under salt and drought stress
We selected 44 SmNAC genes out of 82 genes to verify the effect of abiotic stresses on SmNAC genes of Salvia miltiorrhiza. The transcription levels of these genes were relatively high in different tissues (Fig. 4). We divided 44 SmNAC genes into six groups. In groups I–II, the expression patterns of 18 SmNAC genes were significantly modulated due to abiotic stresses in different tissues of Salvia miltiorrhiza. On the one hand, under NaCl stress, 10 SmNAC genes were Upregulated in roots and eight SmNAC genes were Upregulated in leaves. The results also showed that the expression of SmNAC genes in roots of S. miltiorrhiae was more upregulated than that in leaves under NaCl stress. In addition, under drought stress, 15 SmNAC genes were upregulated in leaves and six SmNAC genes were upregulated in roots.
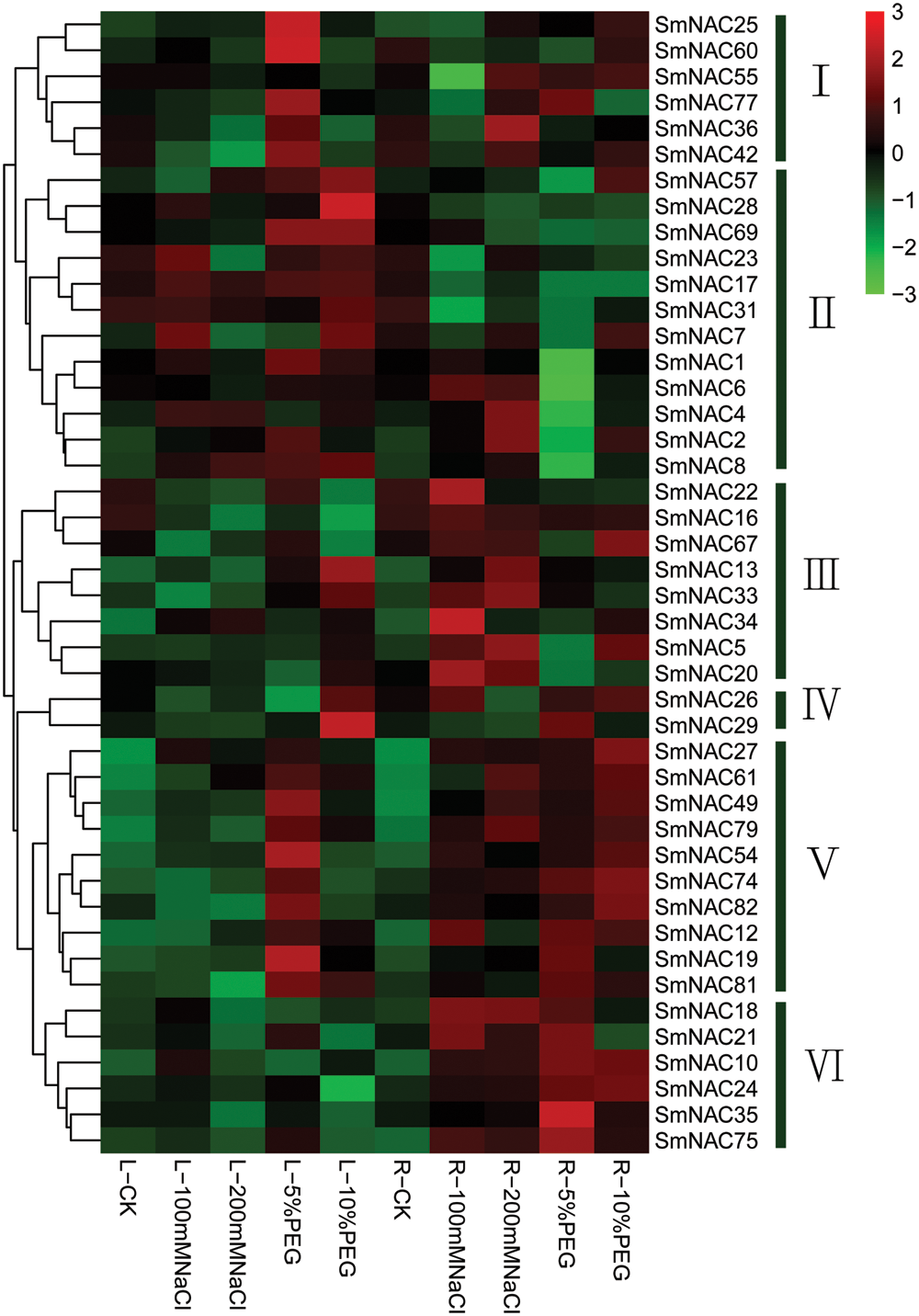
Figure 4: Expression patterns of 44 SmNAC genes in leaves and roots of Salvia miltiorrhiza. L/R-CK (control leaf/root), L/R-100 mM NaCl (Salvia miltiorrhiza leaves/roots under 100 mmol L–1 NaCl treatment), L/R-200 mM NaCl (Salvia miltiorrhiza leaves/roots under 200 mmol L–1 NaCl treatment), L/R-5% PEG (Salvia miltiorrhiza leaves/roots under 5% PEG treatment), L/R-10% PEG (Salvia miltiorrhiza leaves/roots under 10% PEG treatment), Colored blocks represent low/low expression (green), high/up expression (red), and no expression/no change (black).
In groups III–V, most of the SmNAC genes were upregulated in only roots. Among them, 22 SmNAC genes were up-regulated under salt stress, and 20 SmNAC genes were up-regulated under drought stress conditions. The results showed that in abiotic stress environment (salt or drought), NAC gene transcripts were mainly induced in roots. With the increase of Na+ induced salinity and the increased drought level, the expression of some SmNAC genes in roots were abundantly expressed. For example, SmNAC8 and SmNAC13 (Fig. 5b) were significantly up-regulated under salt conditions. The genes SmNAC24, SmNAC25, and SmNAC54 (Fig. 5d) were significantly up-regulated under drought conditions, indicating that NAC genes play an important role in coping with abiotic stress environment. In addition, with the increase of stress intensity, the expression of some genes was down-regulated or inhibited by high concentration of NaCl and drought stress. In this study, 18 genes were significantly expressed in different tissues under abiotic stresses (salt/drought). SmNAC22 gene was significantly up-regulated by 5.83 times in roots under 100 mM NaCl stress (Fig. 5b). Under 200 mM NaCl stress, SmNAC8 gene was up to 13.05 times significantly up-regulated in leaves (Fig. 5a). Moreover, under 5% PEG stress, SmNAC8 gene was up 16.65 times significantly in leaves (Fig. 5c), while under 10% PEG stress, SmNAC8 gene was up 22.53 times significantly in roots (Fig. 5d). SmNAC8 gene expression was gradually induced, by the increase of Na+ concentration (Figs. 5a and 5b), while SmNAC25, SmNAC54 gene expression was gradually induced by the increase of PEG concentration (Fig. 5d). In addition, with the increase of Na+ concentration, the expression levels of SmNAC1 in leaves and SmNAC22 and SmNAC34 in roots were significantly down-regulated. Similarly, with the increase of PEG concentration, SmNAC1, SmNAC2, SmNAC19, SmNAC25, SmNAC54, SmNAC60 in leaves and SmNAC35, SmNAC81 in roots were also significantly down-regulated (Figs. 5a and 5c). Conversely, SmNAC57 expression in leaves was significantly down-regulated. These results indicated that SmNAC gene had different regulatory mechanisms when Salvia miltiorrhiae responded to abiotic stresses.
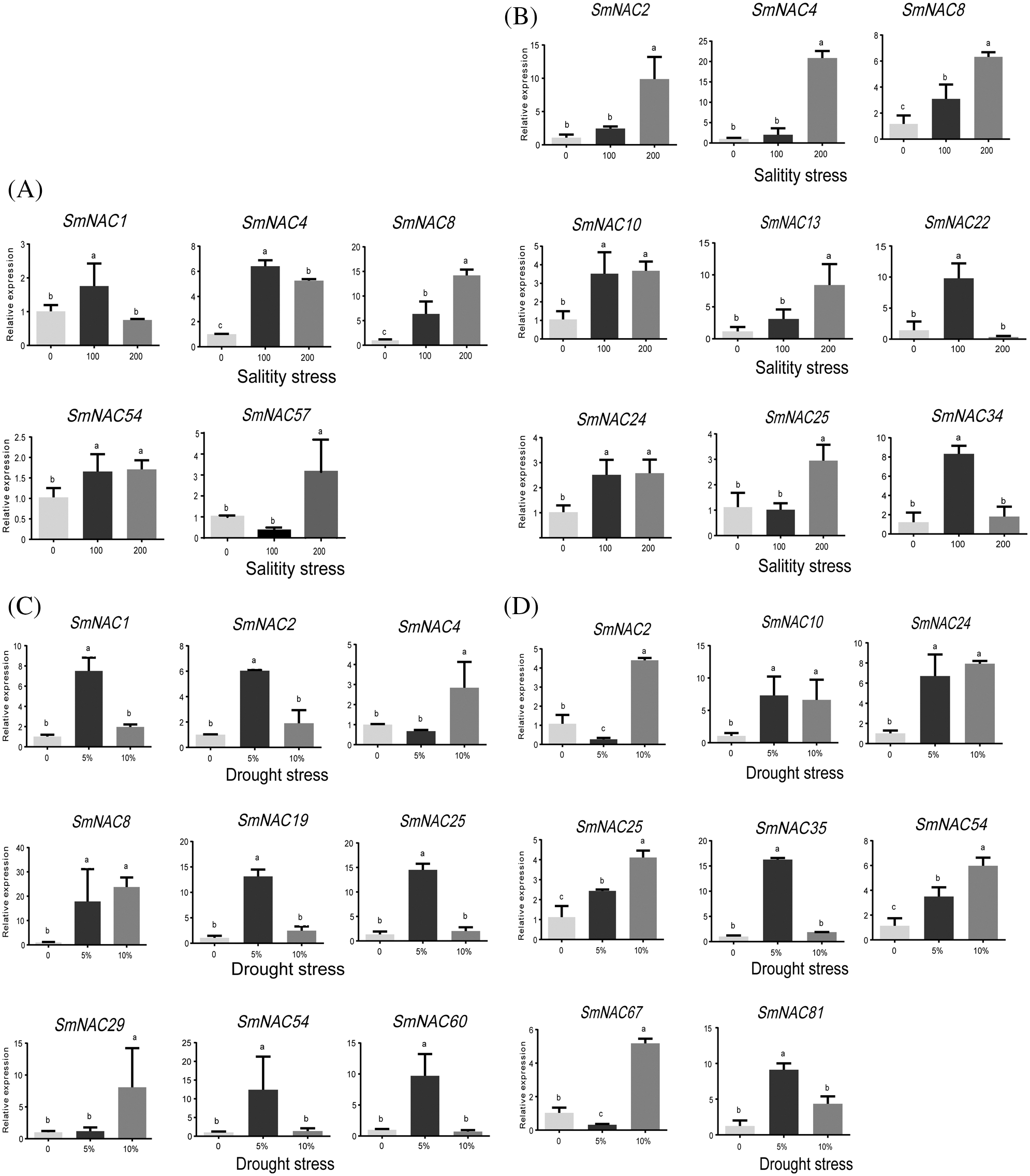
Figure 5: Expression pattern of SmNAC gene responding to NaCl and PEG in different tissues of Salvia miltiorrhiza. (a) Expression pattern of some genes in leaves under salt stress. (b) Expression patterns of some genes in roots of under salt stress. (c) Expression patterns of some genes in leaves under drought stress. (d) Expression patterns of some genes in roots under drought stress.
In the present study, we identified and characterized 82 SmNACs genes in Salvia miltiorrhiza plants (Fig. 2). Gene replication can increase the number of genes, and it is speculated that gene replication (WGD) events occur in different lineages (Prince and Bryan, 2002). For example, four large repeating events have been found in the Arabidopsis thaliana genome (Vision et al., 2000; Blanc et al., 2003), and approximately 83–123 Mai of GWD events have been occurred in tomatoes (Tomato Genome Consortium, 2012). Therefore, the decrease in the number of SmNAC genes may be due to the lack of response to gene replication (WGD) events during the evolutionary process of Salvia miltiorrhiza.
Based on the protein motif and gene structure analysis of Salvia miltiorrhiza, the SmNAC family was divided into 14 groups. The motif and CDS analysis showed that the most closely related members of the phylogenetic tree shared a common motif composition (Fig. 1a). Despite the length, molecular weight and PI of SmNAC genes varied greatly, the gene structure was relatively conserved. Most SmNAC have similar motifs (motif 1, motif 2, motif 3, motif 4, motif 5, and motif 6). Motif 2 is common in all SmNAC genes, suggesting that this motif is necessary for NAC protein functioning. At the same time, there are also changes in genetic structure in a given cluster of genes. For example, the motif structure of SmNAC23, SmNAC30, SmNAC34 in G8 and SmNAC43 in G9 is different, indicating that SmNACs have diversity in the evolutionary process. In previous studies, a similar phenomenon was found in tomato (Jin et al., 2020). In addition, we found that SmNAC21 was isolated in a single group, and its protein function needs to be further explored. The SmNAC gene has three CDS (Fig. 1c), and the number of CDS ranged from 1 to 11, indicating that the NAC proteins had functional similarities. At the same time, phylogenetic tree analysis revealed the separation of different types of NAC transcription factors in Salvia miltiorrhiza, and we divided the SmNAC gene family into 14 groups (Fig. 2). The NAC family members of Arabidopsis thaliana and Salvia miltiorrhiza are not only homologous, but they may be evolved from the same ancestor. However, these genes were unevenly distributed on the chromosomes of Salvia miltiorrhiza, with the number of each chromosome varying from 1 to 14 (Fig. 3), indicating that there was no positive correlation between chromosome length and the number of NAC genes, a situation similar to that of peanut (Yuan et al., 2020).
Generally, gene expression patterns provide clues to gene function. In previous studies, AgNaAC63 and AgNaAC47 in celery can be highly expressed in leaves under adverse conditions (heat, cold, drought and salt) (Duan et al., 2020). We performed a phylogenetic analysis of Salvia miltiorrhiza using Arabidopsis thaliana to study evolutionary relationships and predict drought or salt-responsive genes. We determined the expression levels of 82 SmNAC genes in the leaves and root tissues of Salvia miltiorrhiza. As shown in Fig. 4, 44 SmNACs genes were detected in leaves and roots showing tissue and development-specific expression patterns. These genes may play an important role in the growth and development of Salvia miltiorrhiza, and their exact functions still need to be clarified in future studies. In previous studies, expressions of ANAC55 (At3G15500) and ANAC72 (At4G27410) in Arabidopsis thaliana were also induced by drought and high salinity (Tran et al., 2004). Interestingly, we found that SmNAC4 was in Group11 with ANAC55 (At3G15500) and ANAC72 (At4G27410). Meanwhile, SmNAC4 was detected to be highly expressed in different tissues. We speculated that SmNAC4, as a key gene, plays a critical role in response to abiotic stress. In addition, SmNAC2, SmNAC8 and SmNAC25 were also significantly expressed in different tissues (Fig. 5), suggesting that they may also be involved in specific activities in the growth and development of Salvia miltiorrhiza under adverse stress conditions. In the future, more studies are needed to examine the specific function of the SmNAC family gene in Salvia miltiorrhiza to explore their dynamic role in plant growth and development under normal and environmentally stressed conditions.
Availability of Data and Materials: All data generated or analysed during this study are included in this article.
Authors’ Contribution: Ling Xu and Xin Li conceived the study. Xin Li, Juanjuan Li and Jianmin Pan conducted the experiment. Xin Li and Jianmin Pan analyzed the data. Xin Li and Juanjuan Li wrote the manuscript. Faisal Islam and Ling Xu revised the manuscript. Ling Xu, Zhuoni Hou and Zongqi Yang supported and supervised the experiment. All authors revised and approved the manuscript.
Ethics Approval: Not applicable.
Funding Statement: This work was supported by the National Natural Science Foundation of China (31871694, 31800255), the Fundamental Research Funds of Zhejiang Sci-Tech University (14042216-Y).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Blanc G, Hokamp K, Wolfe KH (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Research 13: 137–144. DOI 13.doi 10.1101/gr.751803. [Google Scholar]
Duan AQ, Yang X, Feng K, Liu JX, Xu ZS, Xiong AS (2020). Genome-wide analysis of NAC transcription factors and their response to abiotic stress in celery (Apium graveolens L). Computational Biology and Chemistry 84: 107186. DOI 10.1016/j.compbiolchem.2019.107186. [Google Scholar] [CrossRef]
Han LM, Hua WP, Cao XY, Yan JA, Chen C, Wang ZZ (2020). Genome-wide identification and expression analysis of the superoxide dismutase (SOD) gene family in S. miltiorrhiza. Gene 742: 144603. DOI 10.1016/j.gene.2020.144603. [Google Scholar] [CrossRef]
Hou Z, Li Y, Su F, Chen J, Zhang X, Xu L, Yang D, Liang Z (2020). Application of 1H-NMR combined with qRT-PCR technology in the exploration of rosmarinic acid biosynthesis in hair roots of Salvia miltiorrhiza Bunge and Salvia castanea f. tomentosa Stib. Planta 253: 2. DOI 10.1007/s00425-020-03506-y. [Google Scholar] [CrossRef]
Jia D, Jiang Z, Fu H, Chen L, Liao G, He Y, Huang C, Xu X (2021). Genome-wide identification and comprehensive analysis of NAC family genes involved in fruit development in kiwifruit (Actinidia). BMC Plant Biology 21: 44. DOI 10.1186/s12870-020-02798-2. [Google Scholar] [CrossRef]
Jin JF, Wang ZQ, He QY, Wang JY, Li PF, Xu JM, Zheng SJ, Fan W, Yang JL (2020). Genome-wide identification and expression analysis of the NAC transcription factor family in tomato (Solanum lycopersicum) during aluminum stress. BMC Genomics 21: 288. DOI 10.1186/s12864-020-6689-7. [Google Scholar] [CrossRef]
Jung I, Kim H, Moon S, Lee H, Kim B (2020). Overview of Salvia miltiorrhiza as a potential therapeutic agent for various diseases: An update on efficacy and mechanisms of action. Antioxidants (Basel) 9: 857. DOI 10.3390/antiox9090857. [Google Scholar] [CrossRef]
Mathew IE, Das S, Mahto A, Agarwal P (2016). Three rice NAC transcription factors heteromerize and are associated with seed size. Front Plant Science 7: 1638. DOI 10.3389/fpls.2016.01638. [Google Scholar] [CrossRef]
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012). NAC transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta 1819: 97–103. DOI 10.1016/j.bbagrm.2011.10.005. [Google Scholar] [CrossRef]
Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene 465: 30–44. DOI 10.1016/j.gene.2010.06.008. [Google Scholar] [CrossRef]
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005). NAC transcription factors: Structurally distinct, functionally diverse. Trends in Plant Science 10: 79–87. DOI 10.1016/j.tplants.2004.12.010. [Google Scholar] [CrossRef]
Prince VE, Bryan PF (2002). Splitting pairs: The diverging fates of duplicated genes. Nature Reviews Genetics 3: 827–837. DOI 10.1038/nrg928. [Google Scholar] [CrossRef]
Shen S, Zhang Q, Shi Y, Sun Z, Zhang Q, Hou S, Wu R, Jiang L, Zhao X, Guo Y (2019). Genome-wide analysis of the NAC domain transcription factor gene family in Theobroma cacao. Genes 11: 35. DOI 10.3390/genes11010035. [Google Scholar] [CrossRef]
Song Z, Lin C, Xing P, Fen Y, Jin H, Zhou C, Gu YQ, Wang J, Li X (2020). A high-quality reference genome sequence of Salvia miltiorrhiza provides insights into tanshinone synthesis in its red rhizomes. Plant Genome 13: e20041. DOI 10.1002/tpg2.20041. [Google Scholar] [CrossRef]
Sudhir K, Glen S, Michael CK, Koichiro T (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. DOI 10.1093/molbev/msy096. [Google Scholar] [CrossRef]
Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. DOI 10.1038/nature11119. [Google Scholar] [CrossRef]
Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498. DOI 10.1105/tpc.104.022699. [Google Scholar] [CrossRef]
Vision TJ, Brown DG, Tanksley SD (2000). The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117. DOI 10.1126/science.290.5499.2114. [Google Scholar] [CrossRef]
Wang L, Ma R, Liu C, Liu H, Zhu R et al. (2017). Salvia miltiorrhiza: A potential red light to the development of cardiovascular diseases. Current Pharmaceutical Design 23: 1077–1097. DOI 10.2174/1381612822666161010105242. [Google Scholar] [CrossRef]
Wu CF, Bohnert S, Thines E, Efferth T (2016). Cytotoxicity of Salvia miltiorrhiza against multidrug-resistant cancer cells. The American Journal of Chinese Medicine 44: 871–894. DOI 10.1142/S0192415X16500488. [Google Scholar] [CrossRef]
Yang S, Zhu H, Huang L, Zhang G, Wang L, Jiang X, Zhong Q (2021). Transcriptome-wide and expression analysis of the NAC gene family in pepino (Solanum muricatum) during drought stress. PeerJ 9: e10966. DOI 10.7717/peerj.10966. [Google Scholar] [CrossRef]
Yuan C, Li C, Lu X, Zhao X, Yan C, Wang J, Sun Q, Shan S (2020). Comprehensive genomic characterization of NAC transcription factor family and their response to salt and drought stress in peanut. BMC Plant Biology 20: 454. DOI 10.1186/s12870-020-02678-9. [Google Scholar] [CrossRef]
Zafar Z, Fatima S, Bhatti MF (2021). Comprehensive analyses of NAC transcription factor family in almond (Prunus dulcis) and their differential gene expression during fruit development. Plants 10: 2200. DOI 10.3390/plants10102200. [Google Scholar] [CrossRef]
Zhao HX, Zhang L, Fan X, Yang RW, Ding CB, Zhou YH (2006). Studies on chromosome numbers of Salvia miltiorrhiza, S. flava and S. evansiana. Zhongguo Zhong Yao Za Zhi 31: 1847–1849. [Google Scholar]
Zhou Y, Li W, Xu L, Chen L (2011). In Salvia miltiorrhiza, phenolic acids possess protective properties against amyloid β-induced cytotoxicity, and tanshinones act as acetylcholinesterase inhibitors. Environmental Toxicology and Pharmacology 31: 443–452. DOI 10.1016/j.etap.2011.02.006. [Google Scholar] [CrossRef]
Supplementary Materials

 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |