DOI:10.32604/cmc.2021.014158

| Computers, Materials & Continua DOI:10.32604/cmc.2021.014158 |  |
| Article |
Brain Tumor Classification Based on Fine-Tuned Models and the Ensemble Method
1Department of Information Technology, Malaysia University of Science and Technology, Selangor, 47810, Malaysia
2Department of Information and Engineering Le2i Laboratory, University of Burgundy Dijon, 21078, France
3Department of Information Technology Faculty of Computing and Information Technology King Abdulaziz, University Jeddah, 21589, Saudi Arabia
*Corresponding Author: Neelum Noreen. Email: neelum.noreen@pg.must.edu.my
Received: 02 September 2020; Accepted: 06 December 2020
Abstract: Brain tumors are life-threatening for adults and children. However, accurate and timely detection can save lives. This study focuses on three different types of brain tumors: Glioma, meningioma, and pituitary tumors. Many studies describe the analysis and classification of brain tumors, but few have looked at the problem of feature engineering. Methods are needed to overcome the drawbacks of manual diagnosis and conventional feature-engineering techniques. An automatic diagnostic system is thus necessary to extract features and classify brain tumors accurately. While progress continues to be made, the automatic diagnoses of brain tumors still face challenges of low accuracy and high false-positive results. The model presented in this study, which offers improvements for feature extraction and classification, uses deep learning and machine learning for the assessment of brain tumors. Deep learning is used for feature extraction and encompasses the application of different models such as fine-tuned Inception-v3 and fine-tuned Xception. The classification of brain tumors is explored through deep- and machine-learning algorithms including softmax, Random Forest, Support Vector Machine, K-Nearest Neighbors, and the ensemble technique. The results of these approaches are compared with existing methods. The Inception-v3 model has a test accuracy of 94.34% and achieves the highest performance compared with other recently reported methods. This improvement may be sufficient to support a significant role in clinical applications for brain tumor analysis. Furthermore, this type of approach can be used as an effective decision-support tool for radiologists in medical diagnostics as a second opinion based on the magnetic resonance imaging (MRI) analysis. It may also save valuable time for radiologists who have to manually review numerous MRI images of patients.
Keywords: Inception; Xception; classification; deep learning; feature extraction; MRI
The early detection of a brain tumor is necessary for proper diagnosis, and magnetic resonance imaging (MRI) is widely used for this purpose. Prompt intervention is also important to prevent the tumor from affecting other tissues of the brain. Early diagnosis is a challenging task because of the difficulty of correctly classifying normal and abnormal tissues. In this context, automatic diagnostic systems can help to overcome the limitations of manual diagnosis and conventional techniques.
Computer-Aided Diagnosis (CAD) has been used in various ways by neuro-oncologists to assist in early diagnosis of disease. CAD applications in neuro-oncology comprise tumor grading, classification, and detection [1]. Detection is the primary problem in brain tumor analysis and treatment. The accurate detection and classification of brain tumors need to be improved, and the number of false-positive results needs to be reduced.
The automatic detection of brain tumors is necessary not only to make accurate assessments and timely diagnoses but also to reduce cost and save time for radiologists. The feature engineering techniques based on handcrafted interventions are not sufficient for the accurate detection of tumor types. Therefore, researchers have looked for ways to automatically extract features and classify tumors. This study aims to classify brain tumors on the basis of deep- and machine-learning approaches. Deep learning is used for feature extraction by the application of different models such as fine-tuned versions of Inception-v3 and Xception. The classification of brain tumors is explored using deep- and machine-learning techniques such as softmax, Support Vector Machine (SVM), Random Forest (RF), K-Nearest Neighbors (KNN), and the ensemble technique.
In this study, two main approaches are proposed for feature extraction and classification. In the first approach, the features are extracted from MRI images using Inception-v3. Next, these features are given to deep classifiers such as softmax and machine learning, which include SVM, RF, KNN, and ensemble technique. In the second approach, the Xception model is applied for feature extraction and classification of brain tumors from MRI images. Furthermore, the extracted features are provided to other machine learning classifiers such as SVM, RF, KNN, and ensemble. In both cases, the ensemble model that is based on the extracted features from fine-tuned Inception-v3 and Xception outperforms the other models.
The rest of this paper is organized into various sections. Section 2 deals with the work related to brain tumor feature extraction and classification. Section 3 describes the dataset used for experiments. Section 4 explains the model based on deep and machine learning. Section 5 and Section 6 show and discuss the results of the proposed models, respectively. Section 7 concludes the work and provides a direction for future work.
Various methods have been developed in the past few years to recognize brain tumors in MRI images. These methods range from classical image processing to neural network-based machine-learning approaches.
Sajjad et al. [2] suggested a framework based on deep learning to categorize brain tumors into multiple classes and grades. A transfer learning approach was used to classify tumors at different stages. The data were passed to a convolutional neural network (CNN) model to predict the grade of the tumor. Two different datasets, namely Radiopaedia (before and after augmentation) and brain tumor, were used for this analysis. The proposed system delivered 90.67% accuracy with augmented data and 87.38% without augmented data.
Patel et al. [3] described a technique to determine intracranial hemorrhage (ICH) using three-dimensional computed tomography without contrast. This method combined a repetitive two-way neural network that used long short-term memory (LSTM) and CNN to determine ICH at the image level. The identification of intangible cultural heritage was carried out through the formation of convolutional neural networks.
Shakeel et al. [4] applied the Multilayer Perceptron Backpropagation Neural Network (MLBPNN) for brain tumor classification. An infrared sensor was used for MLBPNN analysis. NN, SVM, and grayscale backpropagation images were used to determine the statistical characteristics. First, MRI images were selected using several types of filters. Second, multifractals were employed. A two-level approach (Adaboost classifier and NN backward propagation) was used to classify the brain tumor images.
Minz et al. [5] suggested an effective method for MRI that should be used as an Adaboost machine-learning algorithm. This system consists of three phases: feature extraction, classification, and preprocessing. The preprocessing stage was used to clear the noise in the basic data. This stage changes the red, green, and blue image to gray levels and uses an intermediate filter and a threshold division. A gray-level co-occurrence matrix (GLCM) technology was used in the extraction step for the features, and 22 characteristics were recovered by MRI. The reinforcement technology (ADA boost) was applied for classification. This approach had an accuracy of 89.90%, thereby improving brain tumor classification.
Singh et al. [6] used a new method that included the normalization and division of K-Means. The first process eliminates unwanted noise or signals from the input image. Multiple filters are used in an MRI image to reduce noise, normalize the previously prepared image sketch, and complete an MRI classification. Finally, the K-Means algorithm is used to divide the image and remove the tumor from the MRI. Naive Bayes and SVMs are employed to classify the magnetic resonance and provide clear estimates. The naive compilations of Bayes and SVM delivered 87.23% and 91.49% accuracy, respectively.
A model that used direct holistic 3D reconstruction of an image was proposed by Zhou et al. [7]. First, the 3D holistic image was converted into two-dimensional slices, and these images were subsequently given to the DenseNet for feature extraction from the two-dimensional slices. The recurrent neural network model was trained and tested to perform classification of the images. Two different datasets, one proprietary and the other public, were used for testing. The DenseNet uses CNN as a convolutional auto-encoder for sequence learning. DenseNet CNN and DenseNet short-term memory were also used for tumor screening and classification. This approach produced 92.13% accuracy with DenseNet based on LSTM.
In another study by Cheng et al. [8], tumor classification was developed in two stages by creating an offline database and restoring it from the Internet. In the offline database stage, brain tumor images were created sequentially. This procedure includes tumor extraction, metric distance learning, and segmentation. In the online learning stage, a similar process was performed on the images inserted into the brain. The extracted function was then compared with the distance learning metrics stored in an online database. This process showed 94.68% accuracy in classification.
An alternative method was introduced for brain tumor classification by combining discrete wavelet transform (DWT) and 2D Gabor filter techniques [9]. A figshare dataset, which contained 3,064 MRI slices, was used for the analysis and comprised three types of brain tumors, namely meningioma, glioma, and pituitary. Features were formed on the basis of the statistical feature set. This approach produced 91.9% accuracy in the classification of meningioma, glioma, and pituitary brain tumors.
Abiwinanda et al. [9] applied a CNN to diagnose gliomas, meningiomas, and tumors of the pituitary gland. Their approach involved applying a simple CNN structure that consisted of a laminated layer, a maximum mass, and a straight layer, which was completely connected to the next hidden layer. The CNN was formed from a set of brain tumor data containing 3064 T-1 images of contrast-enhanced MRI.
Cheng developed a brain tumor dataset [10], which was also used by many other researchers to look at the problems of brain tumor classification and feature extraction. This tumor dataset has become a benchmark to evaluate the proposed models and is used in this study.
Afshar et al. [11] proposed a model based on modified capsule neural network (CapsNet) architecture to classify brain tumors. The authors claimed that their model had two advantages. It eliminated the need for the exact annotation of the tumor and the CapsNet on the main area, and simultaneously showed the relationship with surrounding tissues. Their proposed model achieved 90.89% accuracy, which was an improvement in classification accuracy compared with previous CapsNets and CNNs.
This research used the figshare dataset, which is also used by many researchers as a benchmark for the classification of brain tumors [12–17]. Some researchers only consider the MRI tumor region in their studies, whereas other investigators extract important features from the tumor region of MRI images [18–20]. Many researchers have transformed this database prior to classification [21–23].
A deep wavelet autoencoder-based deep neural network for high-level feature extraction from the images of brain tumors has been described [24]. Various parts of the images were transmitted into the deep wavelet autoencoder after segmentation of the images. Next, the brain tumor images were encoded by a deep wavelet autoencoder, and the images were subsequently processed by the deep CNN. A comparison between the deep autoencoder classifier and different classifiers was performed at the end of the extraction. The deep autoencoder classifier was more reliable and efficient than other non-deep neural network classifiers and had an accuracy of 96%. In a further study [25], a deep neural network with auto-encoding module was applied to classify brain tumors. The image was segmented before performing the DNN layer processing, and features were extracted from the image. Intensity-based images and textures were extracted using a gray interference matrix (GLCM) and DWT. In the last step, the DNN layers consisted of two automatic code machines, and a softmax layer was implemented for classification.
Talo et al. [26] proposed a pre-trained CNN model (ResNet34) to classify the abnormal and normal conditions of the brain using the MRI dataset. This type of technique provided a clear image of the abnormal brain tumor. A radiologist analyzed the abnormal brain of the patient in a daily routine. However, the analysis of the many MRI images within a limited time was extremely difficult. Computer-aided diagnostic tool provided an accurate and fast analysis of the MRI images. The proposed model had an accuracy of 100% using a fivefold cross-validation based on 613 magnetic resonance images.
The literature survey reveals that some techniques have been developed to enable classification, while other techniques are designed to realize feature extraction based on machine and deep learning. Machine learning is suitable for small data and has limited tuning capabilities. In contrast, deep learning is suitable for massive data and can be tuned in various ways. A trade-off exists in accuracy depending on the selection of one of these learning techniques. One is suitable for classification, and the other performs better in feature transformation. Therefore, our study described here selects deep and machine-learning models for brain tumor detection using MRI images. The results support the use of the proposed method for brain tumor detection based on MRI images.
An MRI dataset containing 3,064 brain MRI slices was used for training and testing of the proposed models. This dataset was obtained from 233 different patients. The dataset comprises three different and prominent classes of brain tumors: Meningioma, glioma, and pituitary (Fig. 1). Specifically, it has 930 images of pituitary tumors, 708 images of glioma, and 1426 images of glioma tumors. The brain tumor dataset was provided in matfile and was first converted into a PNG image format for processing. All deep learning models require a dataset in the batch format. Therefore, this dataset was converted into a tensor format from the PNG image files with different batch sizes for use in the proposed models. Min–max normalization was used within the range of 0–1 for the pixel intensity.
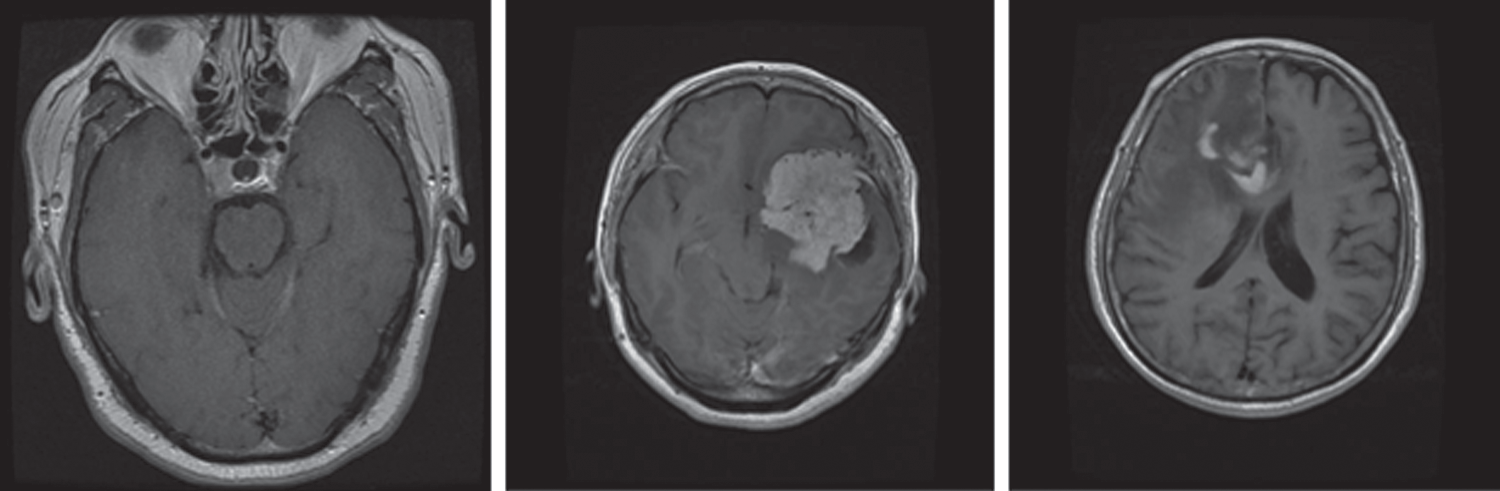
Figure 1: MRI sample images of different brain tumors
This section describes the proposed model based on deep learning and machine learning. Several deep and machine learning models are available, but Inception-v3, Xception, SVM, RF, and KNN were selected here because of their proven capability for detection and classification as demonstrated in different research domains.
Inception-v3 is a broadly used image identification model available to increase the accuracy of ImageNet datasets. It can also use low-level functions for fine tuning. Previously, fine-tuning ImageNet weights have been moved to a CNN to determine Inception-v3. All units were fine-tuned for use with Inception-v3, and then the fully connected layer was changed. The initial weights for Inception were kept constant during training and focused solely on training the top layer. However, all weights can be trained at a lower learning rate. The proposed model for fine-tuned Inception-v3 is shown in Fig. 2. The features are extracted from the fully connected layer and passed to softmax for classification.
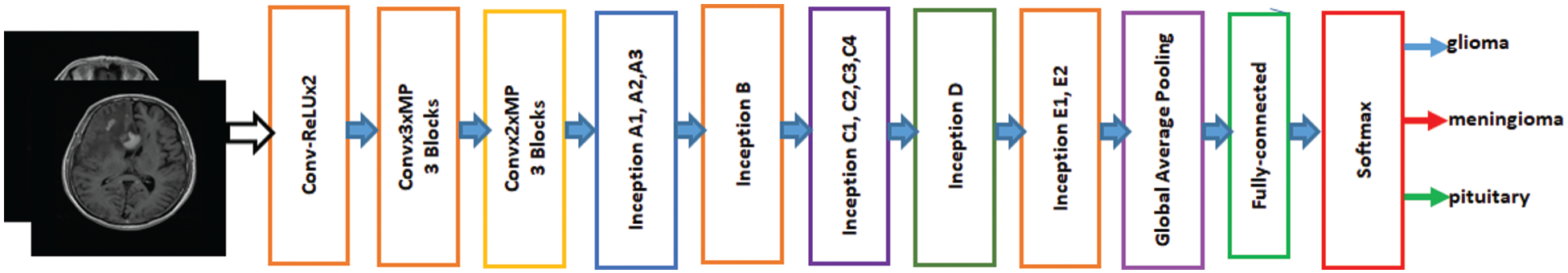
Figure 2: Fine-tuned Inception-V3 model
Francois Chollet proposed the Xception Model, which has been used in image detection [27]. Xception is an addition to the inception model. It replaces the standard inception modules with depth-wise separable convolutions. Xception has been trained on the ImageNet database with more than one million images and has the advantage that it has learned the comprehensive features of various images. The Xception model is fine-tuned on the brain tumor dataset to classify brain tumors into three different types. The proposed model of the fine-tuned Xception model is shown in Fig. 3. The features are extracted from the last layer that is fully connected and provided to the softmax classifier.
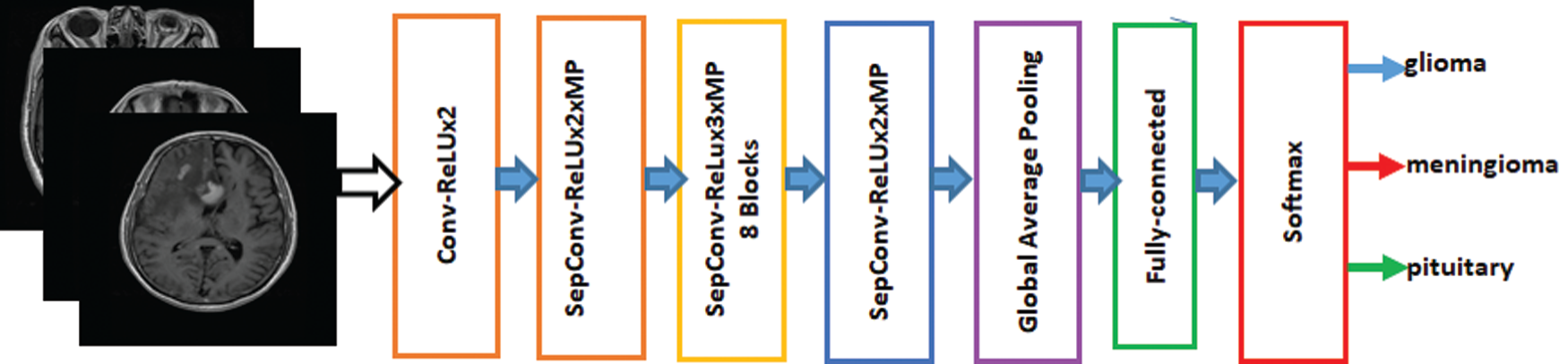
Figure 3: Fine-Tuned Xception model
SVM, which uses statistics, optimization, and functional analysis techniques, was proposed in 1992 by Vladimir Wapnik of AT&T Bell Labs and his team [23]. SVM is used for regression and classification tasks. However, it is widely used in text classification, writing recognition, medicine, pattern recognition, and biological analysis [28]. Therefore, it is used here to classify different categories of brain tumors for improved performance compared with other classifications. SVM separates the hyperplane between training instances and increases margins and reduces classification errors [29].
Leo Breiman proposed RF, which has many applications in image detection and analysis [30]. RF also creates an ensemble of trees that vote for the most popular categories. The ensemble classification approach has demonstrated its high accuracy and superiority. Consequently, RF has received considerable attention from researchers because of its heightened performance in image classification [31,32]. Typically, RF classifiers produce more accurate results when the number of trees in the forest is greater.
Thomas Cover developed KNN, and it has been used effectively for regression and classification in different domains. The KNN approach is a simple, non-parametric, and easy-to-use algorithm for supervised machine learning. Moreover, it has been used in statistical applications since the early 1970s [31]. This technique is unique compared with the earlier-mentioned techniques in that it only works by storing the training data provided. When a new query or instance is started, an identical set of related instances or neighbors is retrieved from the memory and used to classify the new instance [33].
Ensemble is a machine-learning technique that combines several basic models to create an optimized predictive model [34]. Many ensemble methods are identified in the literature, but the majority of the votes method is selected for this work. The majority method is generally used in classification problems. This method uses several models to make predictions for each data point. Predictions for each model are considered a “vote” [35]. Predictions for most models are used as final predictions. The ensemble model based on Inception-v3 and Xception is shown in Figs. 4 and 5. The features are extracted from the last fully connected layer of the fine-tuned Inception-v3 and given to different classifiers as follows: KNN, SVM, and RF. These classifiers are fused on the basis of majority voting. Similarly, the Xception model is the ensemble model based on KNN, SVM, and RF.
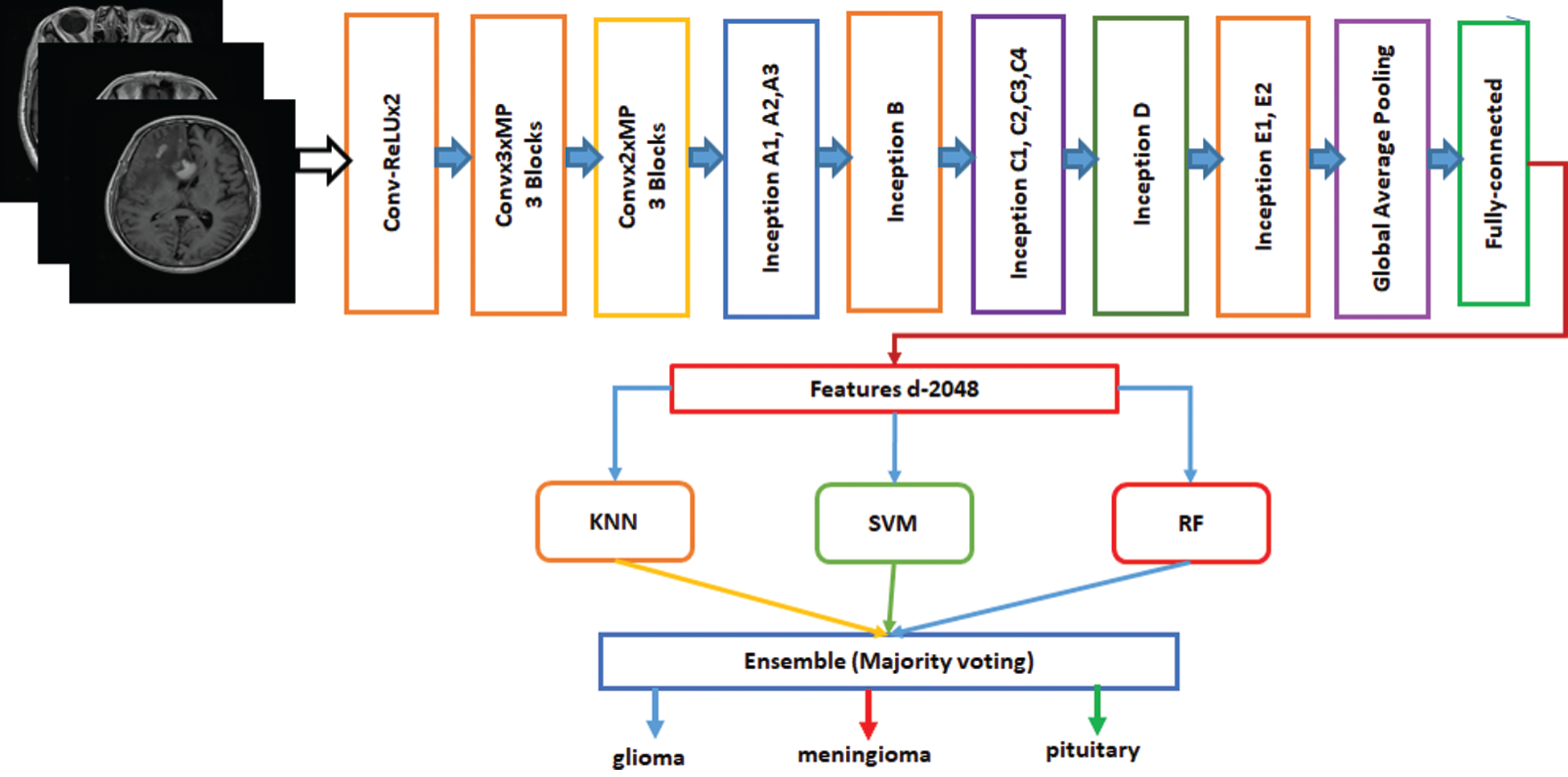
Figure 4: Fine-tuned Inception-v3 model with ensemble method based on machine learning algorithms
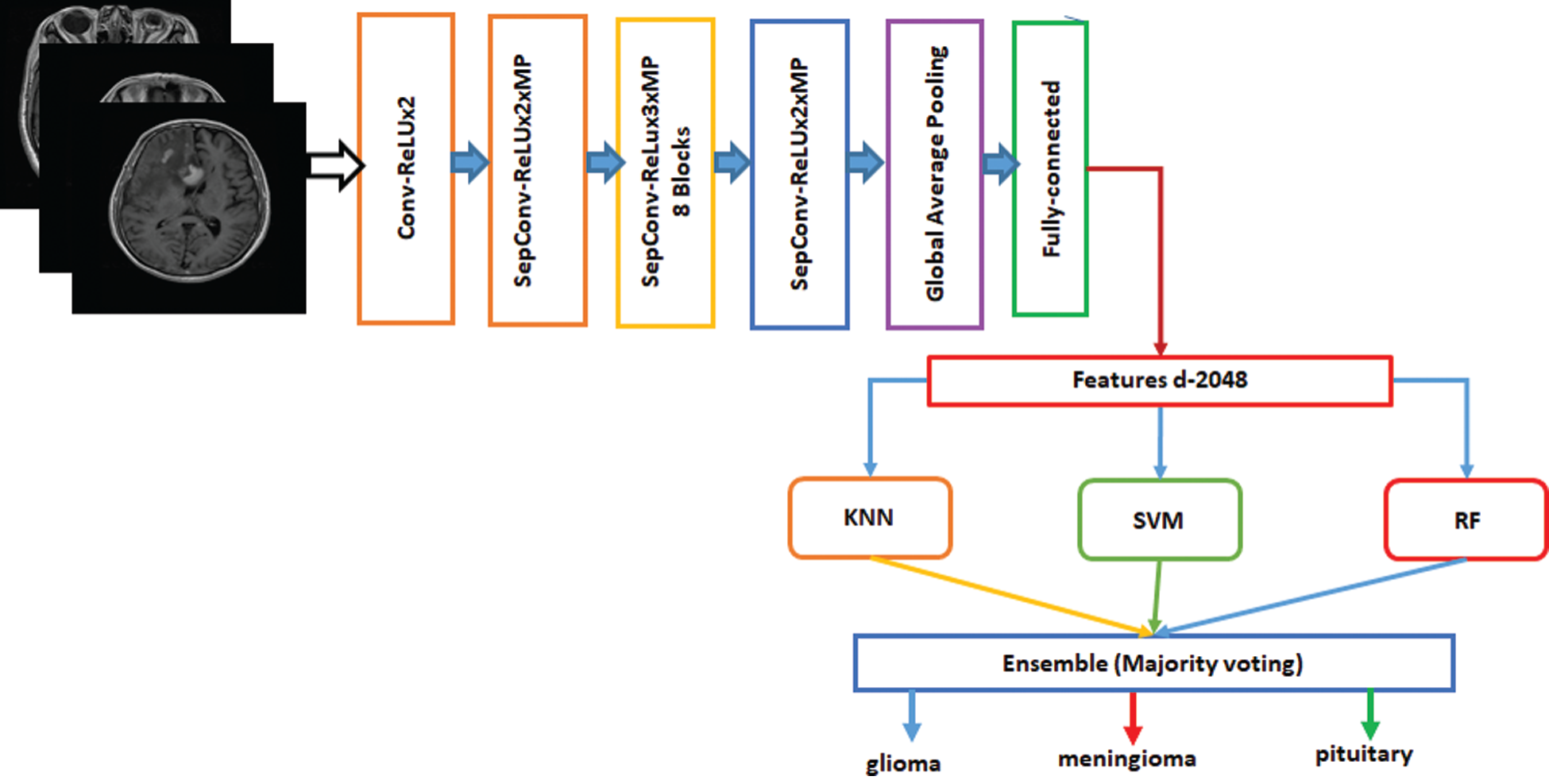
Figure 5: Fine-tuned Xception model with ensemble method based on machine learning algorithms
The proposed models including Inception-v3, Xception, KNN, SVM, and RF were trained on the training dataset, which represents 80% of the total dataset. Next, the trained models were tested on 20% of the total dataset. The proposed models use different hyperparameters during the training phase. These parameters were further optimized using the loss function and Adam optimizers. In addition, 100 epochs, a batch size of 20, a categorical cross-entropy loss function, a 0.0001 learning rate, and an Adam optimizer were used to train the proposed models. The softmax classifier was used for fine-tuned Inception-v3 and Xception. Furthermore, the Keras and tensor flow library were used to train the deep-learning models. The scikit-learn was used for the machine-learning classifiers.
In this study, validation was performed with accuracy, precision, recall, and F1 score. These parameters were measured as true positive (TP), true negative (TN), false positive (FP), and false negative (FN) values. Each of these performance measurements is represented by Eqs. (1)–(5):
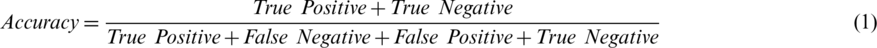




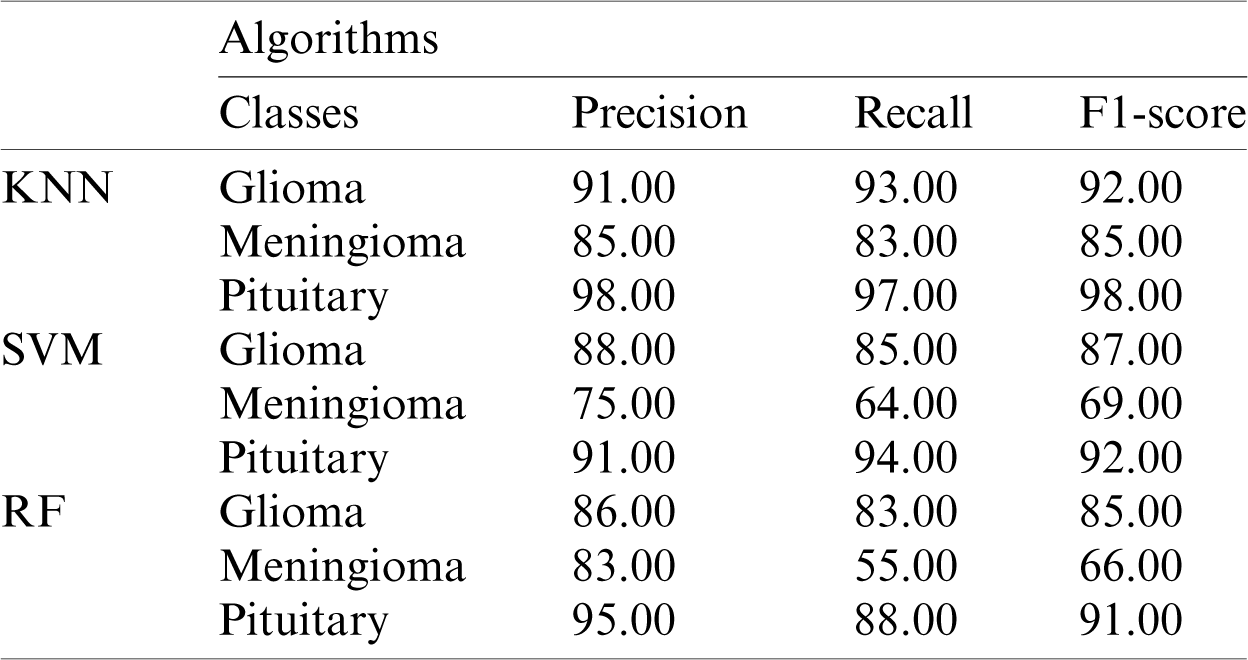
The performance metric score for Inception-v3 based on machine-learning algorithms is shown in Tab. 1.
Table 1: Performance metrics based on Inception-v3 model features using KNN, SVM, and RF
The receiver operating characteristic (ROC) curve for the proposed Inception-v3 model is shown in Fig. 6. The proposed ensemble model uses machine-learning classifiers, which outperforms the other models.
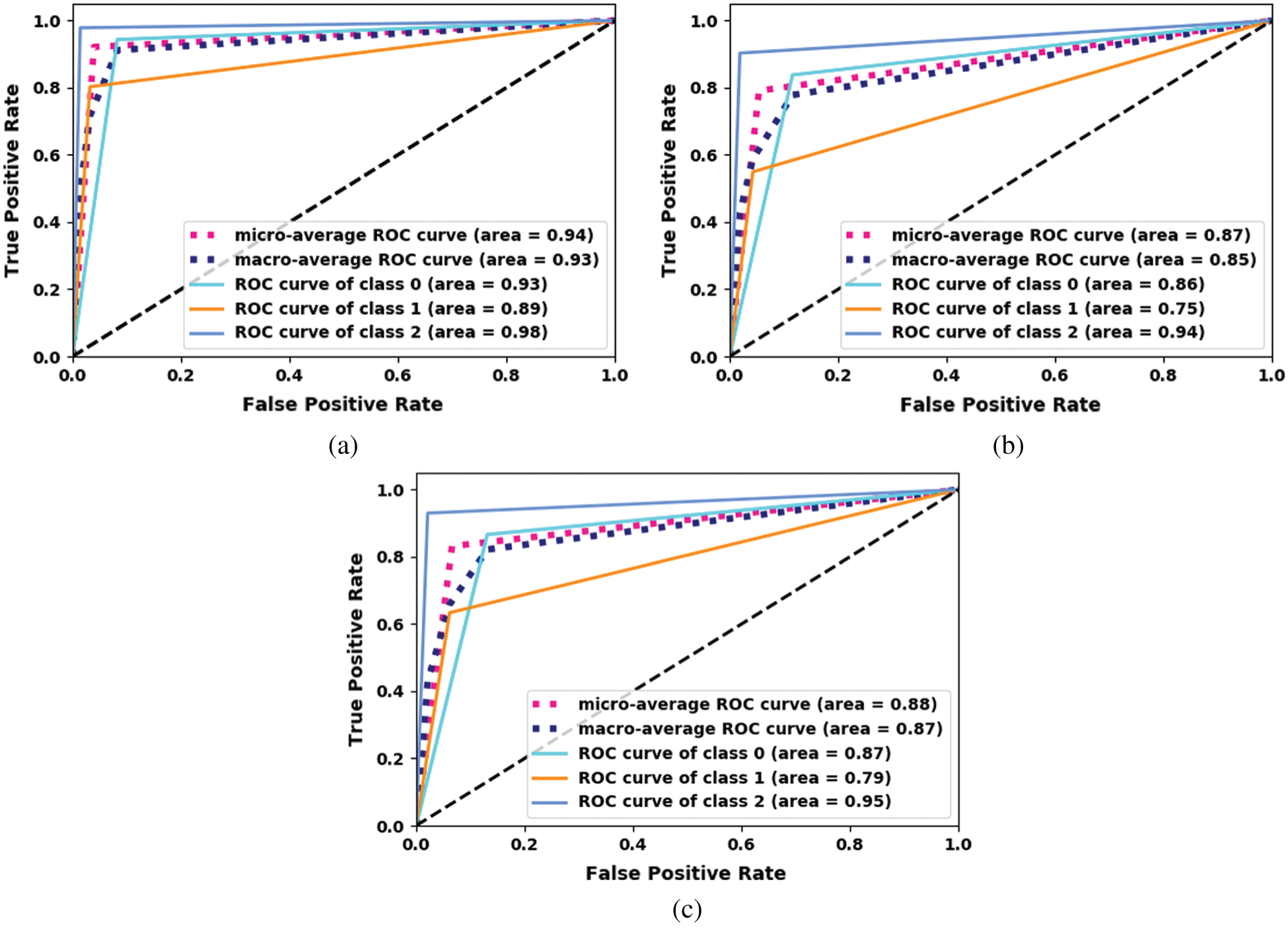
Figure 6: ROC curve for the proposed Inception-v3. (a) Inception-V3-KNN (b) Inception-V3-RF (c) Inception-V3-SVM
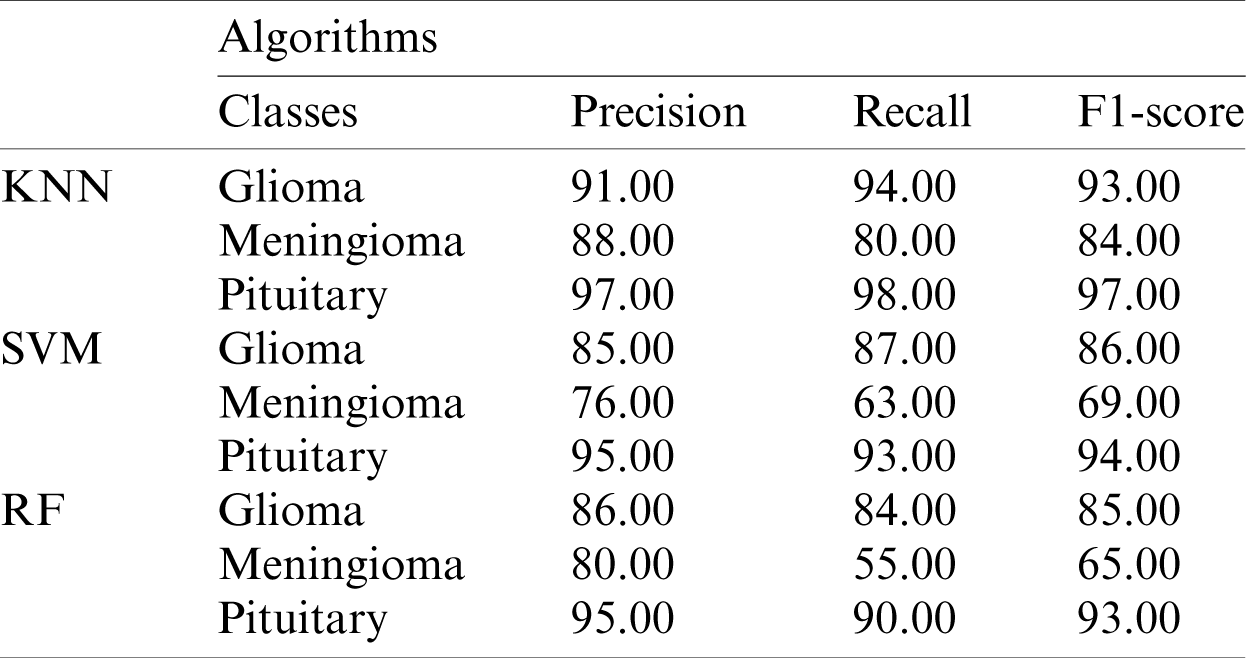
The performance metrics based on the Xception model using machine-learning algorithms for each class are shown in Tab. 2.
Table 2: Performance metrics based on the Xception model features using KNN, SVM, and RF
The ROC curve for the proposed fine-tuned Xception model is shown in Fig. 7. The proposed ensemble model using machine-learning classifiers outperforms the other models.
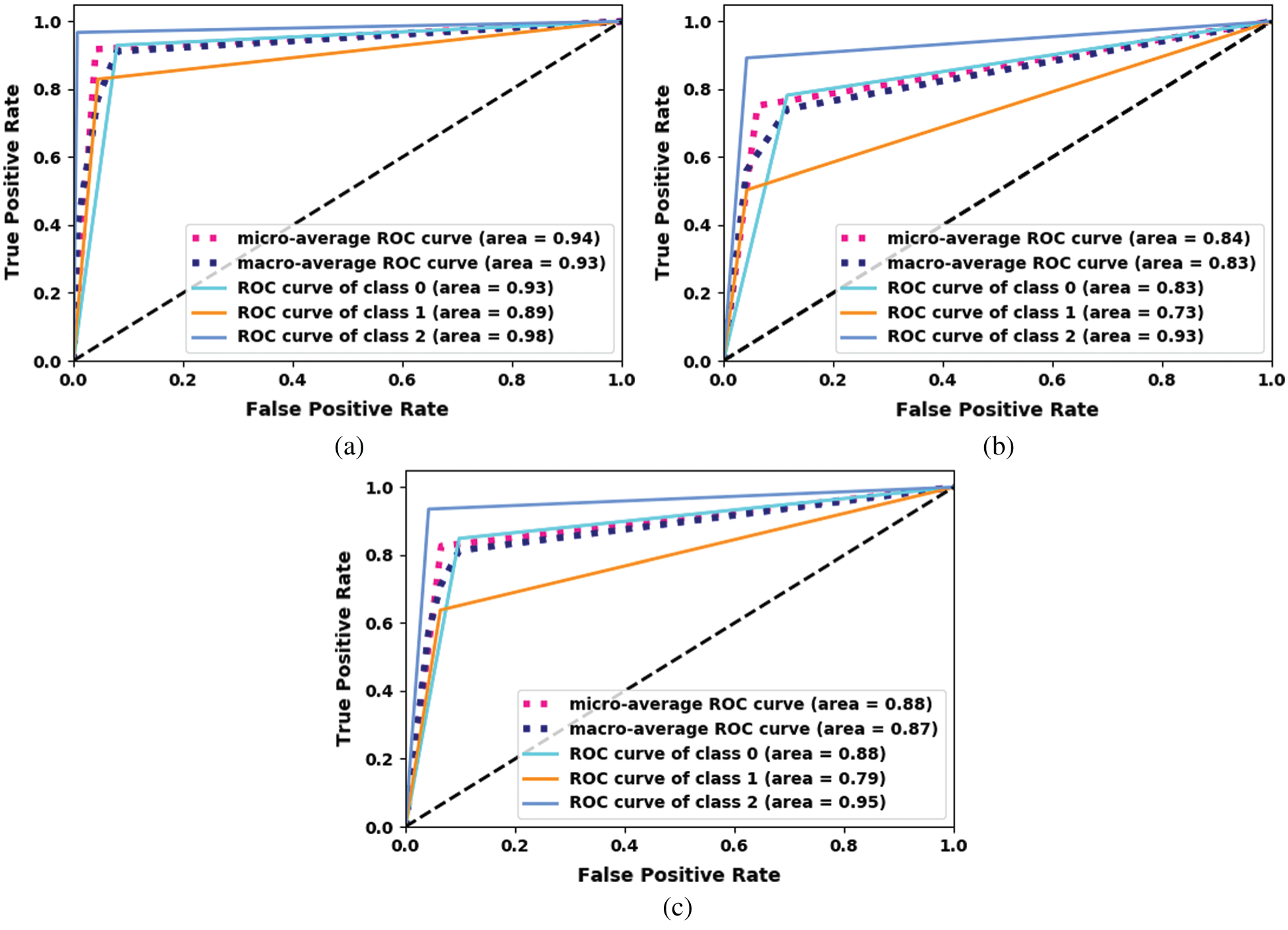
Figure 7: ROC curve for the proposed Xception. (a) Xception-KNN (b) Xception-RF (c) Xception-SVM
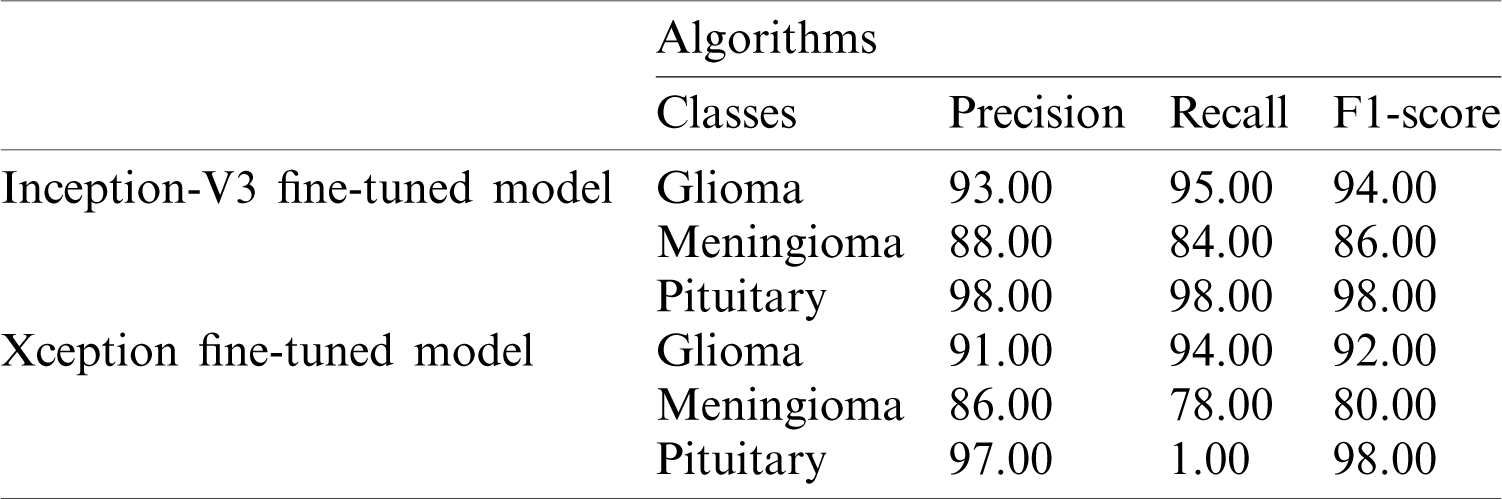
The performance metrics based on the Inception-v3 and Xception models using softmax classifiers are shown in Tab. 3.
Table 3: Performance metrics based on the Inception-v3 and Xception models using softmax classifiers
A comparison of the accuracy of the proposed models is shown in Tab. 4.
Table 4: Accuracy comparison among the proposed models
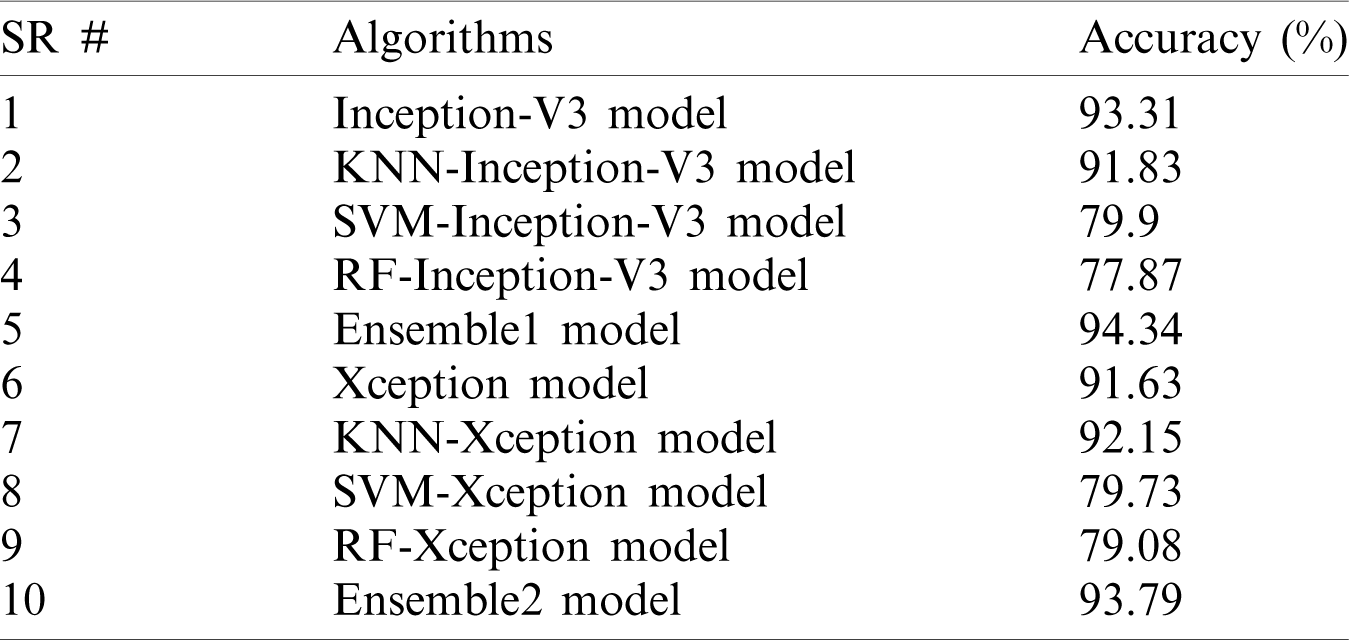
Fig. 8 shows the feature map of the proposed model at different convolutional layers. This map is mostly used for the reconstruction of the original brain tumor images. This also helps to understand the features that our model detects. Furthermore, this map helps to observe features at the early and later layers of the deep network. Low-level (including colors, edges) and high-level features (shapes and objects) are presented at the early and later layers of the model. Sixteen feature maps are used in convolutional layer1; 32 feature maps are used in convolutional layer2; 64 feature maps are used in convolutional layer3; and 128 feature maps are used in convolutional layer4.
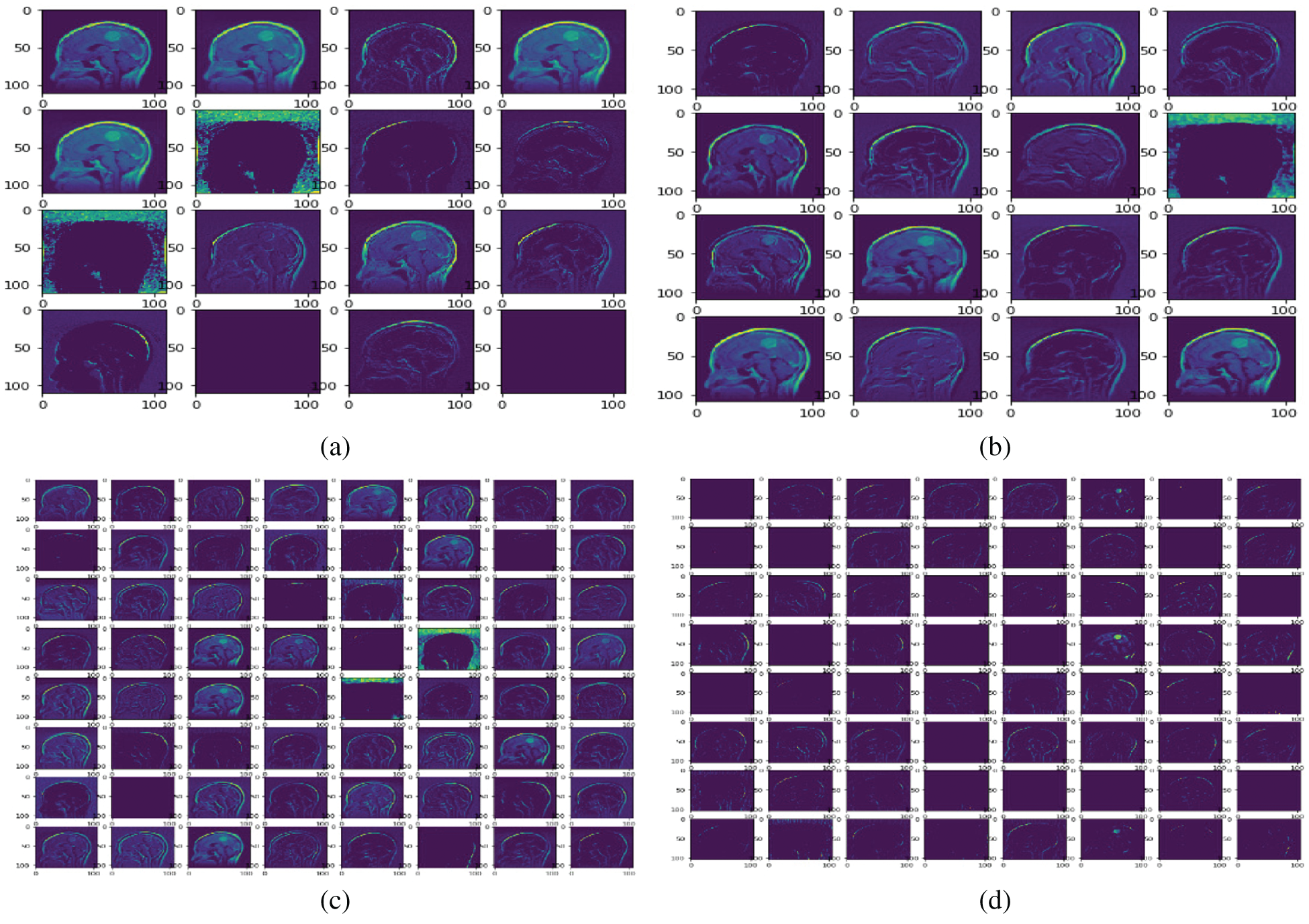
Figure 8: Sample feature maps for convolutional layer1 (FM-C1), convolutional layer2 (FM-C2), convolution layer3 (FM-C3), and convolutional layer4 (FM-C4) for the proposed model. (a) FM-C1 (b) FM-C2 (c) FM-C3 (d) FM-C4
The K-fold cross-validation based on training and testing samples has been evaluated to determine the robustness of the proposed model. The ensemble classifier accuracy for a 10-fold cross-validation based on the proposed Inception-V3 and Xception models is shown in Figs. 9 and 10.
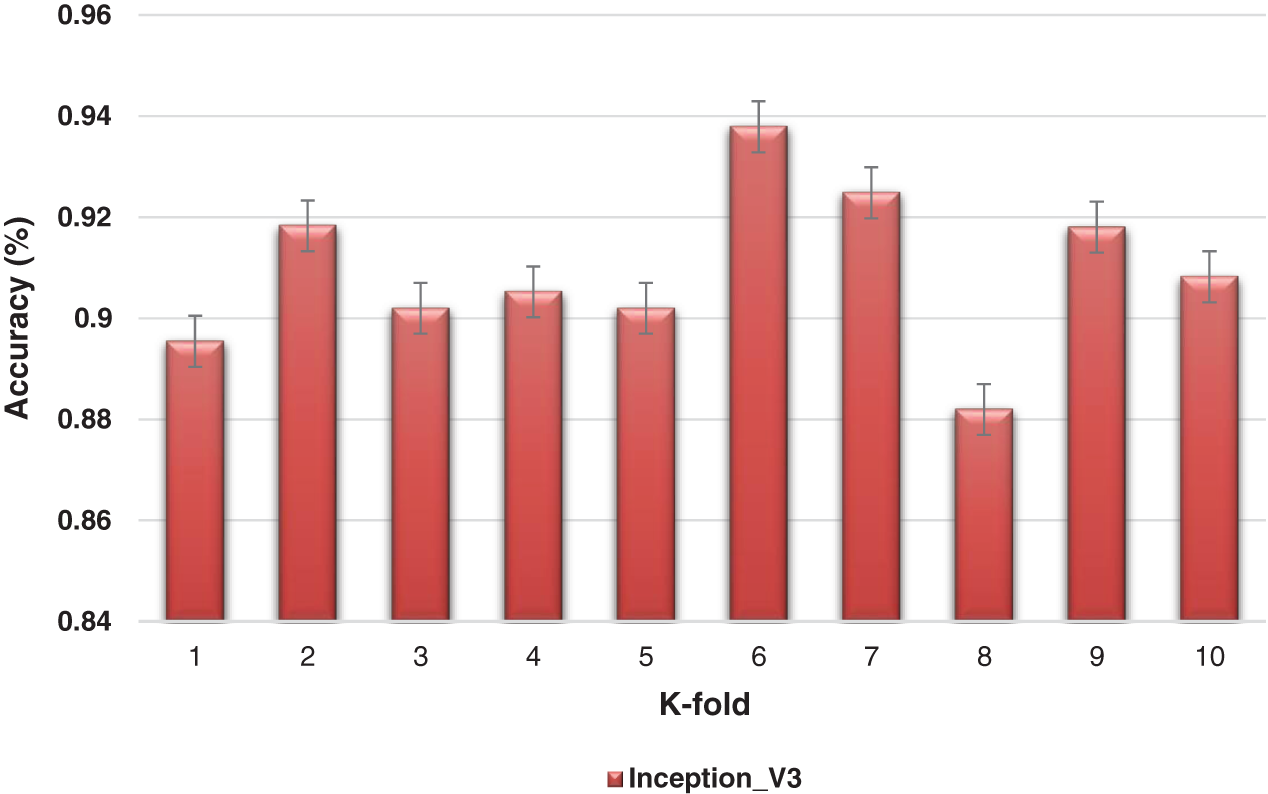
Figure 9: K-fold cross-validation of the ensemble models based on Inception-v3
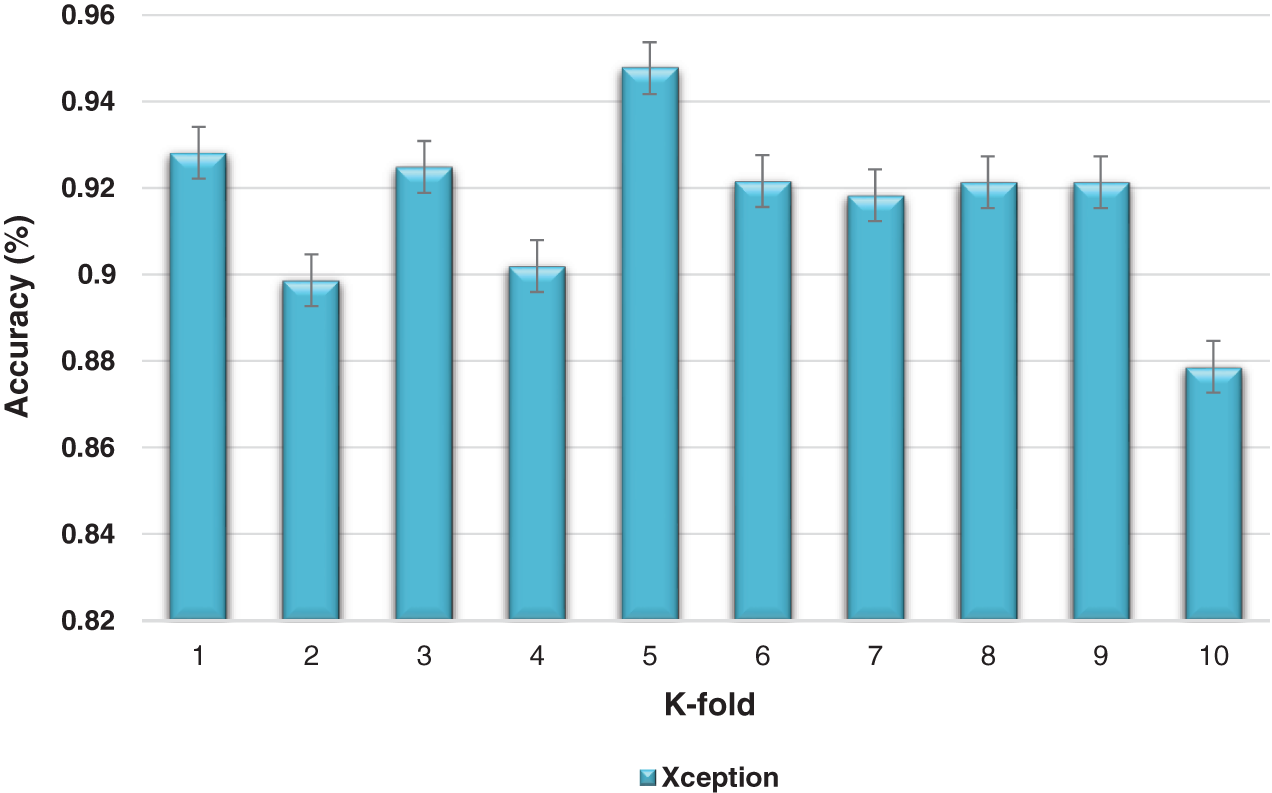
Figure 10: Kfold cross-validation of the ensemble models based on Xception
Recent developments and enhancements of medical image tools have bestowed convenience and innovation to health practitioners. Such advances are helping to improve many areas of medicine including the identification of diseases, their treatments, and the timely decision-making for clinical applications.
Hospitals produce substantial medical data on a daily basis. Expert clinical support systems are vital to help healthcare practitioners make the right decisions. Research in the medical informatics domain supports clinicians and other professionals in their quest to have the best options available to make good use of these burgeoning amounts of data.
Early identification and effective treatment options are necessary to deal effectively with brain tumor diseases. The treatment options depend on the tumor stage, the pathological type of disease, and the grade of tumor at the time of diagnosis. Conventional identification systems at the early phase of relevant feature extraction use some rudimentary machine-learning-oriented algorithms that extract only low- and high-level features.
Here we have presented a unique combination of deep- and machine-learning methods for feature extraction and classification. The ensemble method outperformed in the classification of brain tumors. The proposed model was compared with recent techniques and experiments and showed remarkable performance and accuracy versus other deep-learning methods as indicated in Tab. 5.
Table 5: Comparison between the proposed and existing state-of-the-art models
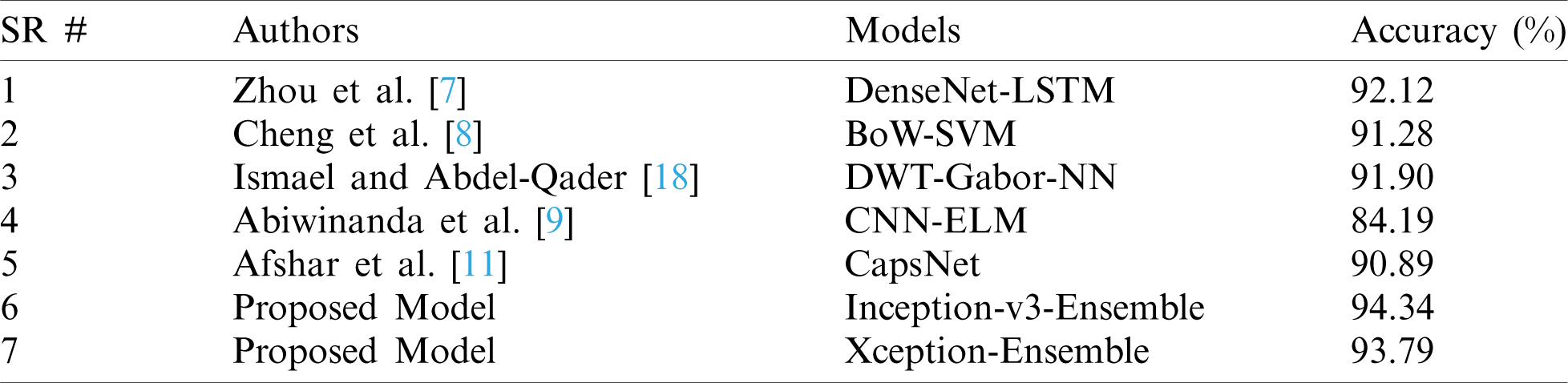
Fine tuning of a few layers in the model gives remarkable predictive results. However, precise and accurate identification is not possible without full consideration of the overall prediction model. Therefore, solutions based on machine learning and deep learning are important to improve the performance of existing systems versus conventional systems. The proposed model in our study outperforms the existing models. Furthermore, the model based on the ensemble approach performed the best among other proposed models.
In this paper, a model is proposed for feature extraction and classification of brain tumors using deep- and machine-learning models. Inception-v3 and Xception were used for feature extraction because of their benefits. Classification was performed using softmax, SVM, RF, KNN, and ensemble. The Inception-v3 model with softmax was combined with other models such as Inception-v3-SVM, Inception-v3-RF, Inception-v3-KNN, and ensemble for the purpose of using machine learning for clinical applications related to brain tumors. Similarly, the Xception model and its combination with other models such as Xception-SVM, Xception-RF, Xception-KNN, and the ensemble method were explored.
The performance of the proposed models was addressed and compared with the existing models in brain tumor classification. The models based on the ensemble method produce highest testing accuracy and outperformed the existing models. This work could be important for the clinical applications of brain tumor analysis. In the future, other techniques based on the deep and machine learning model will be investigated.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Y. Gu, X. Lu, L. Yang, B. Zhang, D. Yu et al. (2018). , “Automatic lung nodule detection using a 3D deep convolutional neural network combined with a multi-scale prediction strategy in chest CTs,” Computers in Biology and Medicine, vol. 103, pp. 220–231. [Google Scholar]
2. M. Sajjad, S. Khan, K. Muhammad, W. Wu, A. Ullah et al. (2019). , “Multi-grade brain tumor classification using deep CNN with extensive data augmentation,” Journal of Computational Science, vol. 30, pp. 174–182. [Google Scholar]
3. A. Patel, S. C. Van De Leemput, M. Prokop, B. Van Ginneken and R. Manniesing. (2019). “Image level training and prediction: Intracranial hemorrhage identification in 3D non-contrast CT,” IEEE Access, vol. 7, pp. 92355–92364. [Google Scholar]
4. P. M. Shakeel, T. E. E. Tobely, H. Al-Feel, G. Manogaran and S. Baskar. (2019). “Neural network based brain tumor detection using wireless infrared imaging sensor,” IEEE Access, vol. 7, pp. 5577–5588. [Google Scholar]
5. A. Minz and C. Mahobiya. (2017). “MR image classification using adaboost for brain tumor type,” in 2017 IEEE 7th Int. Advance Computing Conf., Hyderabad, India, pp. 701–705. [Google Scholar]
6. G. Singh and M. Ansari. (2016). “Efficient detection of brain tumor from MRIs using K-means segmentation and normalized histogram,” in 2016 1st India Int. Conf. on Information Processing, Delhi, India, pp. 1–6. [Google Scholar]
7. Y. Zhou, Z. Li, H. Zhu, C. Chen, M. Gao et al. (2018). , “Holistic brain tumor screening and classification based on densenet and recurrent neural network,” in International MICCAI Brainlesion Workshop, Granada, Spain, pp. 208–217. [Google Scholar]
8. J. Cheng, W. Huang, S. Cao, R. Yang, W. Yang et al. (2015). , “Correction: Enhanced performance of brain tumor classification via tumor region augmentation and partition,” PloS One, vol. 10, pp. e0144479. [Google Scholar]
9. N. Abiwinanda, M. Hanif, S. T. Hesaputra, A. Handayani and T. R. Mengko. (2018). “Brain tumor classification using convolutional neural network,” in World Congress on Medical Physics and Biomedical Engineering. Singapore: Springer, pp. 183–189. [Google Scholar]
10. J. Cheng. (2017). “Brain magnetic resonance imaging tumor dataset,” Figshare MRI Dataset Version 5, . [Online]. Available: https://doi.org/10.6084/m9.figshare.1512427.v5. [Google Scholar]
11. P. Afshar, A. Mohammadi, K. N. Plataniotis, A. Oikonomou and H. Benali. (2019). “From handcrafted to deep-learning-based cancer radiomics: Challenges and opportunities,” IEEE Signal Processing Magazine, vol. 36, pp. 132–160. [Google Scholar]
12. H. H. Sultan, N. M. Salem and W. Al-Atabany. (2019). “Multi-classification of brain tumor images using deep neural network,” IEEE Access, vol. 7, pp. 69215–69225. [Google Scholar]
13. H. Kutlu and E. Avcı. (2019). “A novel method for classifying liver and brain tumors using convolutional neural networks, discrete wavelet transform and long short-term memory networks,” Sensors (Basel), vol. 19, pp. 1–19. [Google Scholar]
14. Z. N. K. Swati, Q. Zhao, M. Kabir, F. Ali, Z. Ali et al. (2019). , “Brain tumor classification for MR images using transfer learning and fine-tuning,” Computerized Medical Imaging and Graphics, vol. 75, pp. 34–46. [Google Scholar]
15. M. Shaikh, V. A. Kollerathu and G. Krishnamurthi. (2019). “Recurrent attention mechanism networks for enhanced classification of biomedical images,” in 2019 IEEE 16th Int. Sym. on Biomedical Imaging, Venice, Italy, pp. 1260–1264. [Google Scholar]
16. A. Rehman, S. Naz, M. I. Razzak, F. Akram and M. Imran. (2020). “A deep learning-based framework for automatic brain tumors classification using transfer learning,” Circuits, Systems, and Signal Processing, vol. 39, no. 2, pp. 757–775. [Google Scholar]
17. P. C. Tripathi and S. Bag. (2020). “Non-invasively grading of brain tumor through noise robust textural and intensity based features,” in Computational Intelligence in Pattern Recognition. vol. 999. Singapore: Springer, pp. 531–539. [Google Scholar]
18. M. R. Ismael and I. Abdel-Qader. (2018). “Brain tumor classification via statistical features and back-propagation neural network,” in IEEE Int. Conf. on Electro/Information Technology, Rochester, MI, USA, pp. 252–257. [Google Scholar]
19. J. Kotia, A. Kotwal and R. Bharti. (2019). “Risk susceptibility of brain tumor classification to adversarial attacks,” in Advances in Intelligent Systems and Computing. vol. 1061. Cham: Springer, pp. 181–187. [Google Scholar]
20. A. K. Anaraki, M. Ayati and F. Kazemi. (2019). “Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms,” Biocybernetics and Biomedical Engineering, vol. 39, no. 1, pp. 63–74. [Google Scholar]
21. H. N. T. K. Kaldera, S. R. Gunasekara and M. B. Dissanayake. (2019). “Brain tumor Classification and Segmentation using Faster R-CNN,” in Advances in Science and Engineering Technology Int. Conf., Dubai, United Arab Emirates, pp. 1–6. [Google Scholar]
22. A. Kharrat and N. E. J. I. Mahmoud. (2019). “Feature selection based on hybrid optimization for magnetic resonance imaging brain tumor classification and segmentation,” Applied Medical Informatics, vol. 41, no. 1, pp. 9–23. [Google Scholar]
23. C. Cortes and V. Vapnik. (1995). “Support-vector networks,” Machine Learning, vol. 20, pp. 273–297. [Google Scholar]
24. P. Kumar Mallick, S. H. Ryu, S. K. Satapathy, S. Mishra, G. N. Nguyen et al. (2019). , “Brain MRI image classification for cancer detection using deep wavelet autoencoder-based deep neural network,” IEEE Access, vol. 7, pp. 46278–46287. [Google Scholar]
25. S. S. Gawande and V. Mendre. (2017). “Brain tumor diagnosis using deep neural network (DNN),” International Journal of Advanced Research in Electrical, Electronics and Instrumentation Engineering, vol. 5, no. 5, pp. 10196–10203. [Google Scholar]
26. M. Talo, U. B. Baloglu, O. Yıldırım and U. R. Acharya. (2019). “Application of deep transfer learning for automated brain abnormality classification using MR images,” Cognitive Systems Research, vol. 54, pp. 176–188. [Google Scholar]
27. F. Chollet. (2017). “Xception: Deep learning with depthwise separable convolutions,” in 2017 IEEE Conf. on Computer Vision and Pattern Recognition, Honolulu, HI, pp. 1800–1807. [Google Scholar]
28. Y. Ma and G. Guo. (2014). “Multi-class support vector machine—Chapter no. 2,” in Support Vector Machines Applications, vol. 2. New York, NY, USA: Springer, pp. 23–48. [Google Scholar]
29. R. Gandhi. (2018). Support Vector Machine-Introduction to Machine Learning Algorithms, Canada: Towards Data Science, . https://towardsdatascience.com/support-vector-machine-introduction-to-machine-learning-algorithms-934a444fca47. [Google Scholar]
30. L. Breiman. (2001). “Random forests,” Machine Learning, vol. 45, pp. 5–32. [Google Scholar]
31. K. Fawagreh, M. M. Gaber and E. Elyan. (2014). “Random forests: From early developments to recent advancements,” Systems Science & Control Engineering: An Open Access Journal, vol. 2, pp. 602–609. [Google Scholar]
32. P. Thanh Noi and M. Kappas. (2018). “Comparison of random forest, k-nearest neighbor, and support vector machine classifiers for land cover classification using sentinel-2 imagery,” Sensors, vol. 18, pp. 18. [Google Scholar]
33. Z. Omary. (2008). “Development of a workflow for the comparison of classification techniques,” M.S. thesis, School of Computing, Technological Univ, Dublin, Ireland. [Google Scholar]
34. S. Singh. (2018). “A Comprehensive guide to ensemble learning (with Python codes),” Analytics Vidhya, . [Online]. Available: https://www.analyticsvidhya.com. [Google Scholar]
35. P. Hong, L. Chengde, L. Linkai and Z. Qifeng. (2007). “Accuracy of classifier combining based on majority voting,” in 2007 IEEE Int. Conf. on Control and Automation, Guangzhou, China, pp. 2654–2658. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |