DOI:10.32604/cmc.2021.015436

| Computers, Materials & Continua DOI:10.32604/cmc.2021.015436 |  |
| Article |
Hep-Pred: Hepatitis C Staging Prediction Using Fine Gaussian SVM
1Center for Cyber Security, Faculty of Information Science and Technology, Universiti Kebansaan Malaysia (UKM), Bangi, 43600, Selangor, Malaysia
2School of Information Technology, Skyline University College, University City Sharjah, Sharjah, 1797, UAE
3Department of Computer Science, Government College University, Faisalabad, 38000, Pakistan
4Department of Computer Science, School of Systems and Technology, University of Management and Technology, Lahore, 54000, Pakistan
5Department of Computer Science, Lahore Garrison University, Lahore, 54000, Pakistan
6School of Computer Science, National College of Business Administration & Economics, Lahore, 54000, Pakistan
7CCSIS, Institute of Business Management, Karachi, 75190, Sindh, Pakistan
*Corresponding Author: Muzammil Hussain. Email: muzammil.hussain@umt.edu.pk
Received: 21 November 2020; Accepted: 26 March 2021
Abstract: Hepatitis C is a contagious blood-borne infection, and it is mostly asymptomatic during the initial stages. Therefore, it is difficult to diagnose and treat patients in the early stages of infection. The disease’s progression to its last stages makes diagnosis and treatment more difficult. In this study, an AI system based on machine learning algorithms is presented to help healthcare professionals with an early diagnosis of hepatitis C. The dataset used for our Hep-Pred model is based on a literature study, and includes the records of 1385 patients infected with the hepatitis C virus. Patients in this dataset received treatment dosages for the hepatitis C virus for about 18 months. A former study divided the disease into four main stages. These stages have proven helpful for doctors to analyze the liver’s condition. The traditional way to check the staging is the biopsy, which is a painful and time-consuming process. This article aims to provide an effective and efficient approach to predict hepatitis C staging. For this purpose, the proposed technique uses a fine Gaussian SVM learning algorithm, providing 97.9% accurate results.
Keywords: Hepatitis C; artificial intelligence; Hep-Pred; support vector machine; machine learning; hepatitis staging
Hepatitis C is a disease affecting the human population on a global level. It is a blood-borne infection that can spread through direct contact with an infected person’s blood or body fluids containing blood. Almost 71 million people are chronically ill because of this disease worldwide, and an estimated 399,000 people died of this disease in 2016 [1].
According to the WHO (World Health Organization), hepatitis C is a global disease. The WHO report also mentioned that 3–4 million people get a new infection of this virus every year. Poor developing countries of Asia and Africa show the highest prevalence of this infection when compared to developed countries in Europe and North America. Furthermore, in countries like Pakistan, China, and Egypt, the number of people with chronic diseases is higher [2–4].
The hepatitis C virus shows symptoms only at the later stages. Around 80% of infected people do not suffer any symptoms after getting an infection at the initial stages, leading to more liver damage with increased mortality rates. There is no proper vaccine available for the hepatitis C virus. Therefore, finding out the degree to which the liver of the affected patient is damaged could help guide clinicians in the diagnosis and treatment of chronic infection, and aid them in managing it properly. Proper management is crucial in the control of disease by preventing the transmission of the virus among people [1,5–7].
Advancements in artificial intelligence (AI) help clinicians with the timely diagnosis and more efficient treatment of patients. Research has been performed comparing AI to human efficiency in the diagnosis of diseases. This research showed that AI was equally comparable to humans in diagnosis, and actually outperformed human efficiency when compared to less experienced doctors [8–12]. Herein, we develop an AI technique using previously available data collected from 1385 patients who got treatment dosages for the hepatitis C virus for about 18 months in Egyptian patients [13].
The 29 attributes used in this study are provided by [13]. These are age, gender, BMI, fever, nausea/vomiting, headache, diarrhea, fatigue, jaundice, epigastric pain, WBC, RBC, HGB, platelets, AST 1, ALT 1, ALT 4, ALT 12, ALT 24, ALT 36, ALT 48, ALT after 24 w, RNA Base, RNA 4, RNA 12, RNA EOT, RNA EF, baseline staging, and staging.
In 75% to 80% of hepatitis C cases, the hepatitis C infection’s progression into its last stages happens as early diagnosis is not possible because of the lack of symptoms. Moreover, sometimes patients with chronic conditions can take years to show symptoms. Then, at the last stage, the liver’s functionality has been destroyed completely, making treatment difficult [14]. As Lok states in an article cited in Michigan Medicine [15], the best treatment with the highest potential of recovery is only possible when the disease is diagnosed at earlier stages. Lok further says that patients infected with hepatitis C mostly show symptoms when they develop liver cirrhosis, increasing the risk for such patients to develop liver cancer. Therefore, a useful and novel diagnostic system for hepatitis C addresses the need for early diagnosis that could help clinicians offer timely treatments to the infected patients. Also, early diagnosis will reduce the chances of transmission of the virus to other people [16].
AI-based disease diagnostics and prediction techniques could help the timely diagnosis of acute infections and chronic diseases. Keltch, Lin, and Bayrak (2014) implemented four different types of AI techniques to the data available publicly for 424 hepatitis C patients. Their proposed model helps to predict the stage of fibrosis by comparing results of standard serum markers to the results taken from biopsies. Keltch et al. (2014) proposed novel approaches and other AI techniques that could help predict hepatitis B and hepatitis C in millions of people worldwide without performing biopsies, which could benefit the overall healthcare system. The authors of [17] also suggest in their study that AI techniques could be applied to different kinds of structured and unstructured healthcare datasets. The most popular artificial intelligence systems include machine learning methods for structured datasets.
Pietrangelo (2018) and Lok (2016) both insist that, in order to avoid complications due to disease progression, the best results of treatment are only possible with the early detection of the infectious stage. Therefore, treatment should be started as soon as possible. Lok and her team are developing a novel system with the help of machine learning methods that could incorporate many datasets to create more accuracy in predicting the risk of developing fibrosis and its progression from mild to moderate [18–22].
In [23], 29 algorithmic parameters that are the symptoms of hepatitis C infection are used to develop an AI technique for detecting human beings’ disease. A dataset has been made publicly available by Kamal et al. (2019) from Egyptian patients who underwent treatment for the hepatitis C virus for 18 months, including 29 symptom attributes. With the proposed diagnostic method, researchers, scientists, and health practitioners will also predict the infection stages without requiring that patients go through liver biopsies.
Research presented in [24] developed an artificial neural network (ANN) system using a data mining approach on a large socio-medical dataset. This system can make a successful predictive diagnosis of the patients who can potentially get hepatitis C virus infection.
Research conducted at the University of Michigan [25] stated that it is a big challenge to reduce the disease management expenses for hepatitis C patients. Therefore, the scientists of Michigan University developed a system using a predictive analytics algorithm to identify the patients having a high risk. For further complications, the authors suggest that their algorithm could help high-risk patients through immediate and effective treatment. The authors claim that their system provides more accuracy when compared with former studies.
Further, authors in [26] identified how factors like gender and obesity could affect hepatitis C infection prevalence among different populations. The study emphasizes the importance of all the mentioned parameters in developing any system, whether manual or through AI, to get highly accurate results and to acquire healthcare professionals’ most efficient treatment strategies.
In similar research, a significant difference is observed in patients for contributing factors like sex, body mass index (BMI), bilirubin, alanine aminotransferase ALT, and other parameters. It is found that the mean BMI value for male patients older than 60 years is lower than the female patients that are younger than 60 years. Furthermore, it is concluded that higher BMI values mean a high risk of early onset of hepatitis C complications in hepatitis C virus patients [27].
A comprehensive analysis of three data mining techniques is presented by [28], i.e., decision trees, naïve Bayes, and neural networks, to predict hepatitis C virus infection.
The progress in the development of machine learning artificial intelligence techniques for predicting esophageal varices (a complication of hepatitis C virus) in chronic hepatitis C patients is discussed in [29]. Researchers of this study also mentioned that 9 variables among 24 are found to be the most significant for analysis through their developed system.
Another research study [30] emphasizes the importance of decision tree learning algorithms to achieve high accuracy in predicting hepatitis C virus infected patients, especially those at high risk of developing advanced liver fibrosis because of hepatitis C virus infection. It can ultimately decrease or even replace liver biopsy, an invasive method that has drawbacks. Several research studies have been presented on hepatitis C, its prediction, and a detailed analysis thereof [31–37]. Similar examples can be found in [38–41]. These examples show that hepatitis C has a very severe impact and demands more research be done in order to succeed in combating this particular disease. Therefore, this article presents a prediction model, Hep-Pred, to be used against hepatitis C.
The proposed method of this research article is the novel model Hep-Pred. The complete layout diagram of the proposed model is shown in Fig. 1.
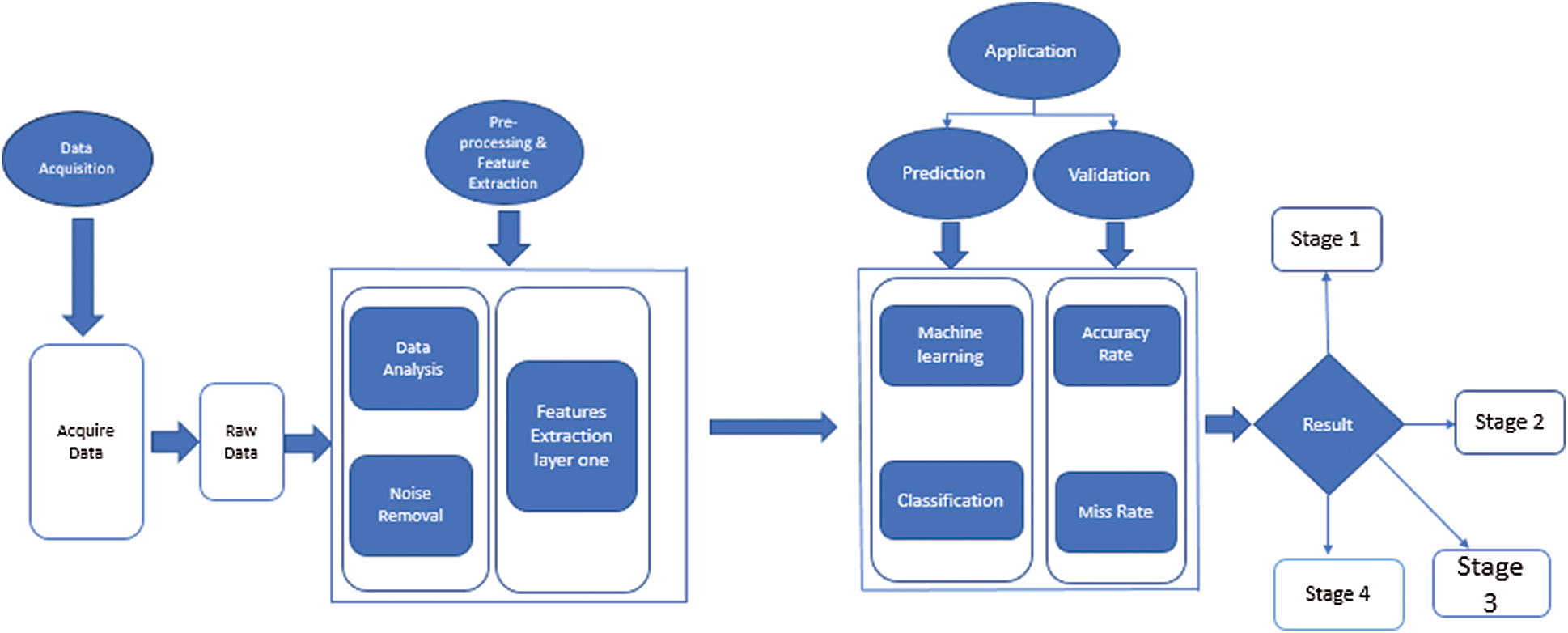
Figure 1: Proposed for hepatitis c stage prediction
Fig. 1 shows that the proposed model is divided into four phases. In the first, the system acquires a raw dataset from the UCI repository. The data contain many noise and garbage values, which can lead the system toward the wrong prediction. So, at the second step (preprocessing), the system cleans the dataset and then finds the dataset feature to perform classification. For classification, the system uses the fine Gaussian support vector machine (SVM) algorithm. After training, the system arrives at a decision.
The raw dataset contains a lot of noise, outliers, and garbage values. At the preprocessing and feature extraction stage, the system removes these values under medical experts’ supervision. After that, the proposed system finds the min and max value of selected features and saves them for further processing.
Following preprocessing, the benchmark data is given to a fine Gaussian SVM for future training. SVM is a supervised training algorithm and works on the hyperplane. The complete mathematical description of SVM is discussed below.
The equation of the line is
where f is a slope of the line, and z is the intersect; therefore
Let
Eq. (2) is the hyperplane equation and derived from 2-dimensional vectors, but it can be used for n number of dimensions.
The direction of a vector
where
As we know that
Eq. (3) can also be written as
The dot product can be computed as the above equation for n-dimensional vectors.
Let us suppose
If sign (L) > 0 then x is correctly classified and if sign (L) < 0 then x is incorrectly classified. Let suppose M is the given a dataset, and the system computes L on training data, then
Then Q is called the functional margin of the dataset.
In the comparison of hyperplanes, the hyperplane with the largest Q will be selected. Where Q is called the geometric margin of the dataset, the main objective is to find an optimal hyperplane, which means that the system needs to find the values of
The Lagrangian function is
From the above two Eqs. (5) and (6), we get
After substituting the Lagrangian function
subject to
Because the constraints are inequalities, we extend the Lagrangian multipliers method to the Karush-Kuhn-Tucker (KKT) conditions. The complementary condition of KKT states that
where
Hence,
These are called support vectors, which are the closest points to the hyperplane. According to the above Eq. (10)
To compute the value of b, we get
Multiplying both sides by y in Eq. (12), then we get
where
Then,
E is the number of support vectors. Once we have the hyperplane, we can then use the hyperplane to make predictions. Where the hypothesis function is
The SVM algorithm’s primary goal is to find a hyperplane that could separate the data and find the optimal hyperplane.
For the results and simulation, Matlab R2018 was used. Different kinds of algorithms were applied to the benchmark dataset for training, and the results of these algorithms are shown in Tab. 1.
Table 1: Accuracy of different algorithms on a benchmark dataset

Tab. 1 shows that the fine Gaussian SVM gives the best results with a 97.9% accuracy rate on 5 cross-validations. The confusion matrix and ROC graph of the proposed method are shown in Figs. 2–6.
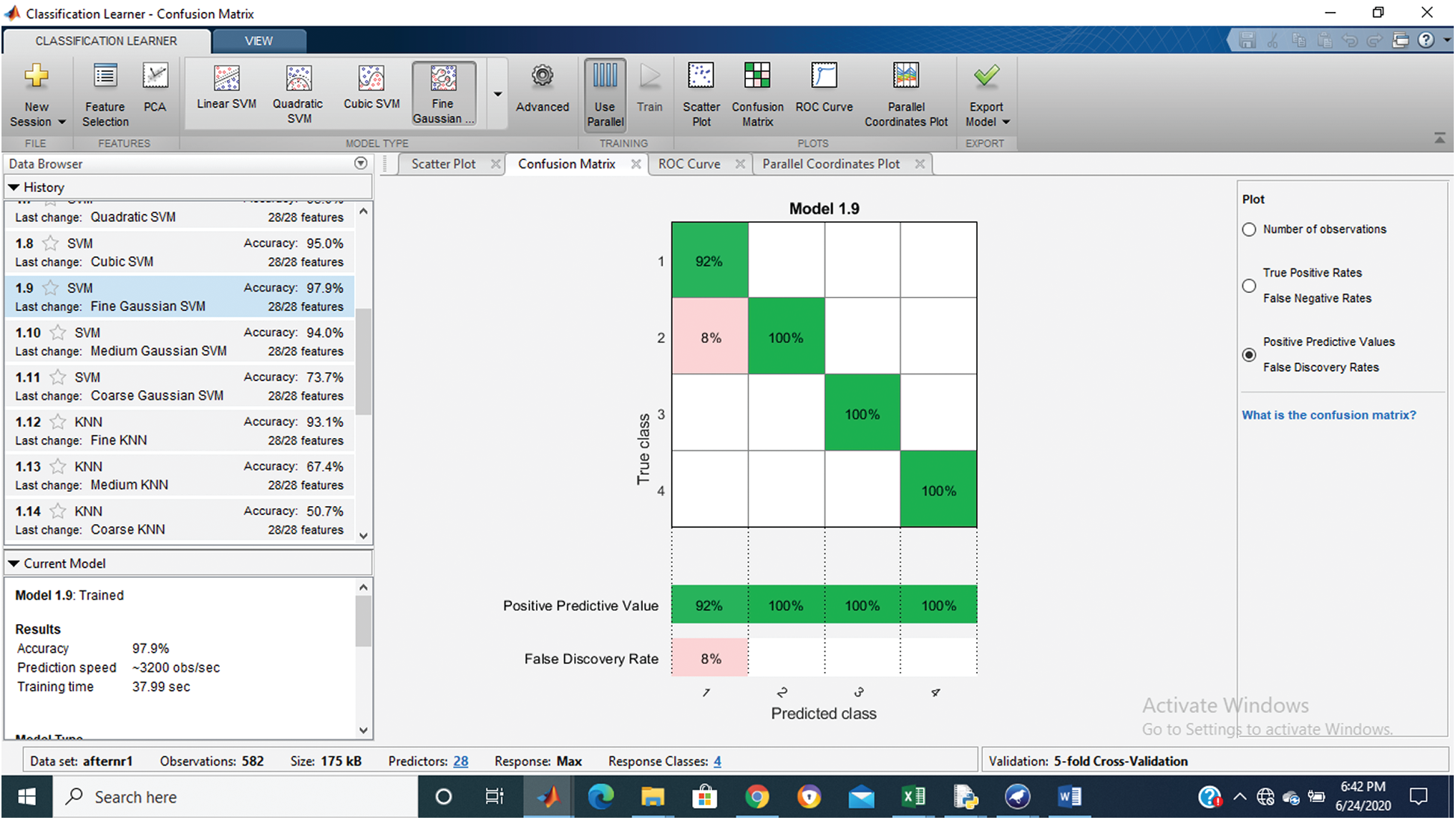
Figure 2: Confusion matrix of proposed Hep-Pred
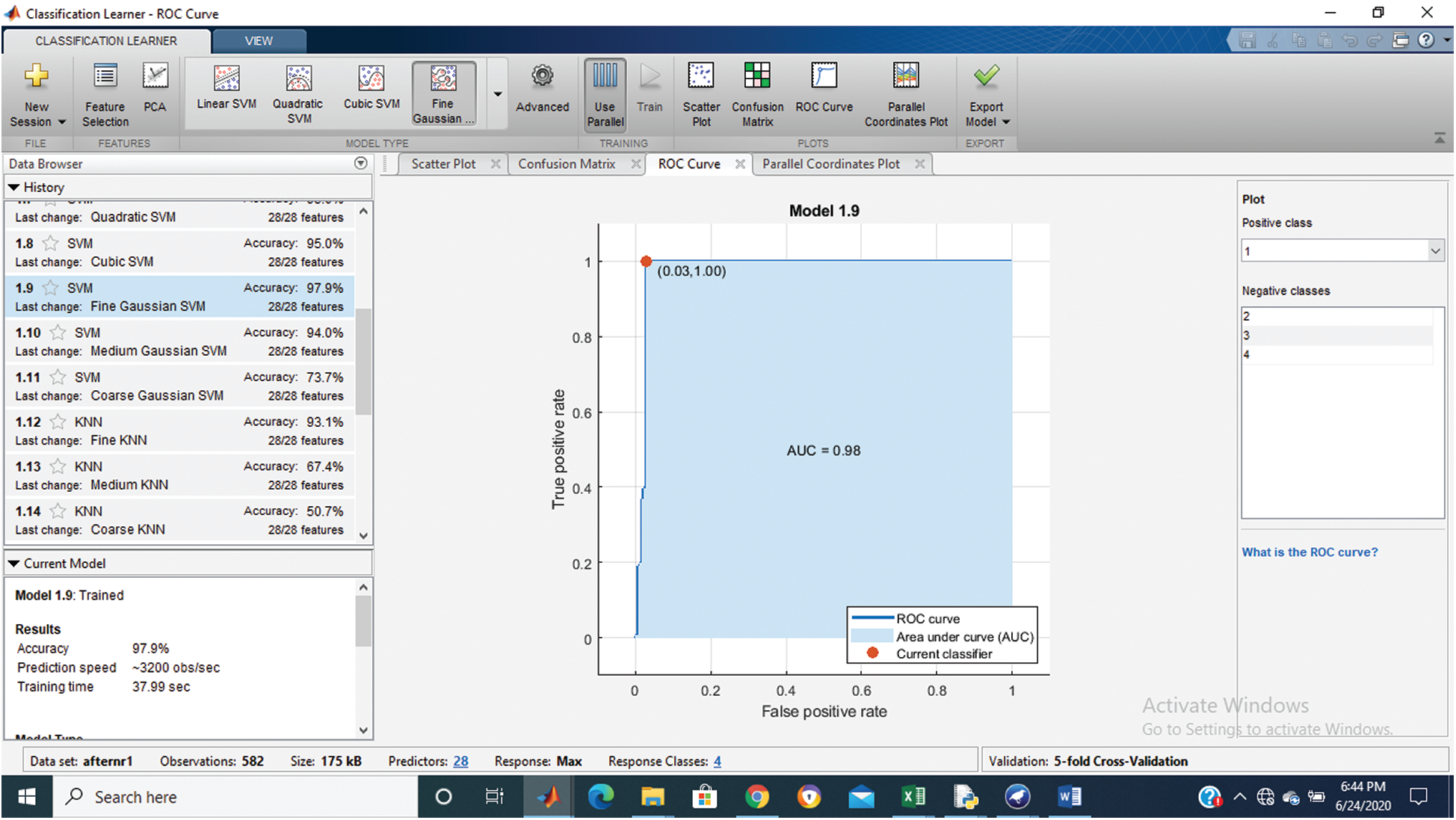
Figure 3: Class stage 1 roc of proposed Hep-Pred
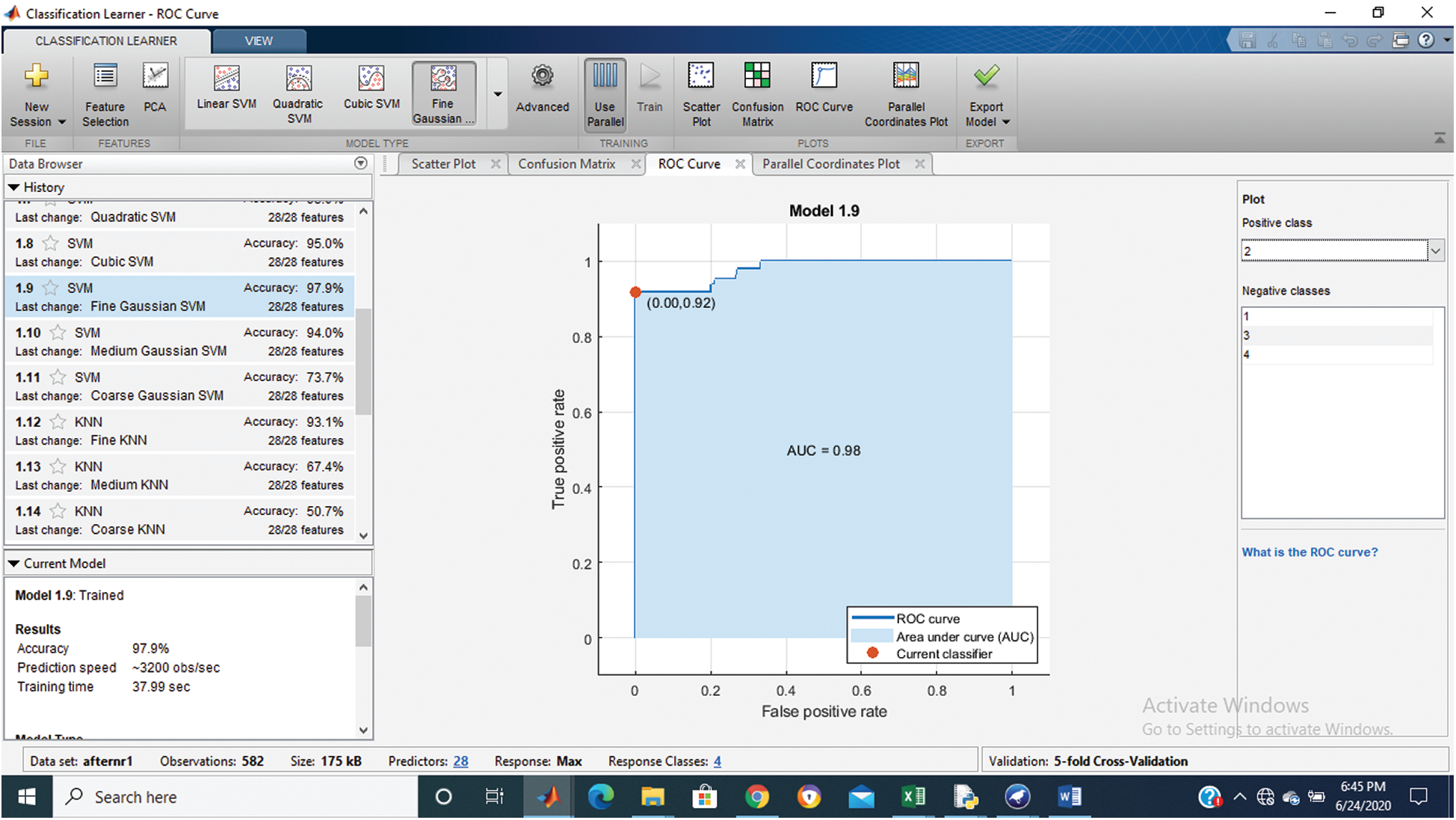
Figure 4: Class stage 2 roc of proposed Hep-Pred
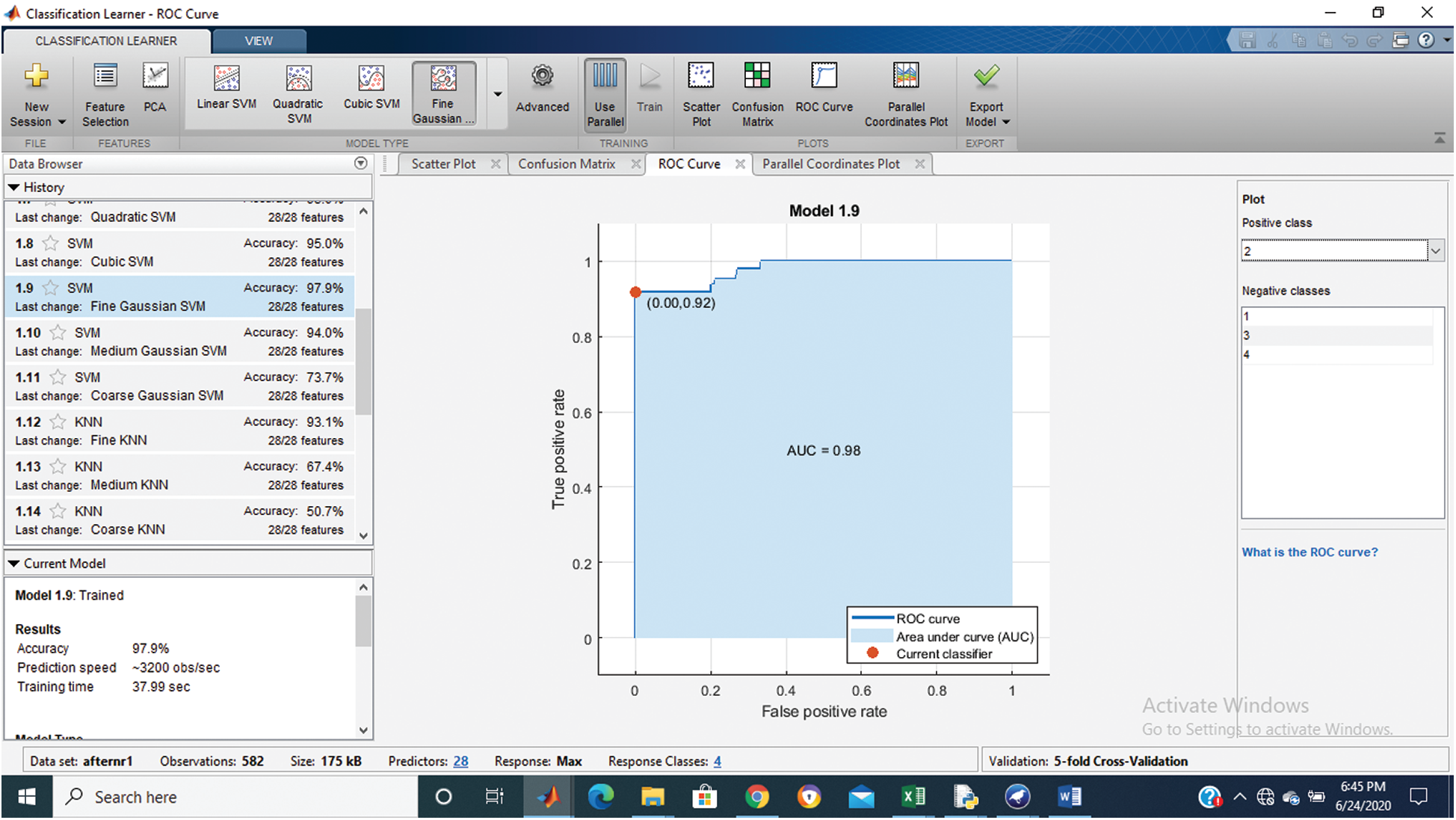
Figure 5: Class stage 3 roc of proposed Hep-Pred
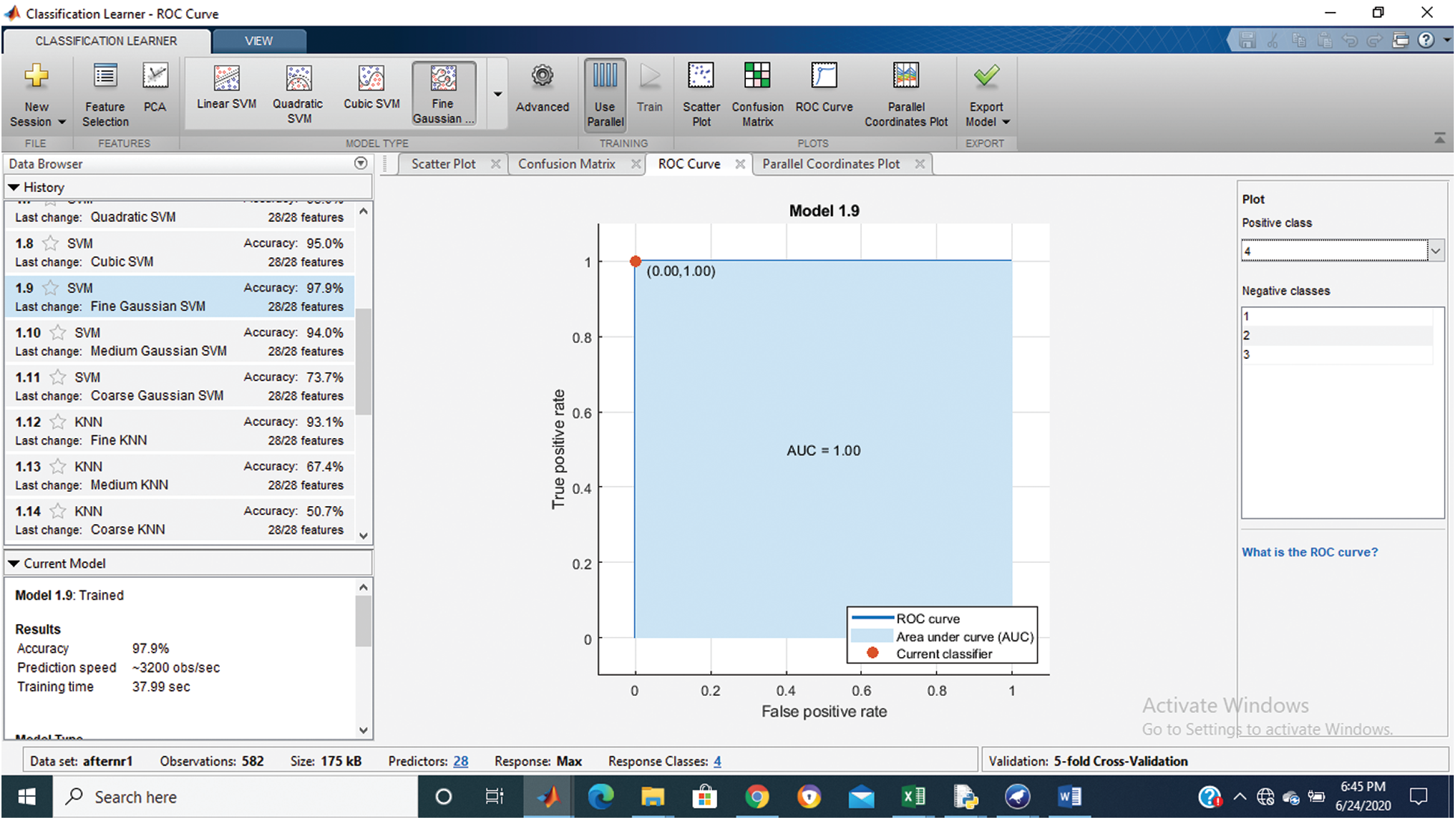
Figure 6: Class stage 4 roc of proposed Hep-Pred
Fig. 2 shows the confusion matrix of the proposed method, and Figs. 3–6 shows the roc graph of each class.
The traditional way to check the hepatitis staining is liver biopsy, a painful and time-consuming procedure, so researchers endeavor to find an easy and accurate alternative. The comparison of the proposed method with other researchers’ work is shown in Tab. 2. Tab. 2 shows that the accuracy of an alternative proposed method [30] is less than Hep-Pred.
Table 2: The comparison of the proposed method with other

The liver is a vital organ of the human body, actively participating in the filtering of blood coming from the digestive system, and is responsible for removing toxins. The proposed Hep-Pred model will help doctors, and other paramedical staff, check a person’s liver health. The dataset for simulation was collected from a UCI repository donated in 2019. A fine Gaussian SVM learning algorithm is used for training the model. The model gives us 97.9% accurate results in 5 cross-validations. In the future, we can enhance the model efficiency by adding more patients’ data. Further, other liver diseases can also be considered for future research.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. World Health Organization, “Hepatitis C [Hepatitis C],” WHO, 2020. [Online]. Available: https://www. who.int/news-room/fact-sheets/detail/hepatitis-c#:~:text=Key facts,major cause of liver cancer [Accessed: 30-Aug-2020]. [Google Scholar]
2. R. Stoean, C. Stoean, M. Lupsor, H. Stefanescu and R. Badea, “Evolutionary-driven support vector machines for determining the degree of liver fibrosis in chronic hepatitis C,” Artificial Intelligence in Medicine, vol. 51, no. 1, pp. 53–65, 2011. [Google Scholar]
3. A. A. Mohamed, T. A. Elbedewy, M. El-Serafy, N. El-Toukhy, W. Ahmed et al., “Hepatitis C virus: A global view,” World Journal of Hepatology, vol. 7, no. 26, pp. 2676–2680, 2015. [Google Scholar]
4. R. Huang, H. Rao, M. Yang, Y. Gao, J. Wang et al., “Noninvasive measurements predict liver fibrosis well in hepatitis C virus patients after direct-acting antiviral therapy,” Digestive Diseases and Sciences, vol. 65, no. 5, pp. 1491–1500, 2020. [Google Scholar]
5. Z. Cheng, Y. Zhang and C. Zhou, “QSAR models for phosphoramidate prodrugs of 2’-methylcytidine as inhibitors of hepatitis C virus based on PSO boosting,” Chemical Biology & Drug Design, vol. 78, no. 6, pp. 948–959, 2011. [Google Scholar]
6. L. Singh, R. R. Janghel and S. P. Sahu, “Classification of hepatic disease using machine learning algorithms,” in Advances in Biomedical Engineering and Technology. Berlin, Germany: Springer, pp. 161–173, 2021. [Google Scholar]
7. J. Vergniol, J. Foucher, E. Terrebonne, P. Bernard, B. le Bail et al., “Noninvasive tests for fibrosis and liver stiffness predict 5-year outcomes of patients with chronic hepatitis C,” Gastroenterology, vol. 140, no. 7, pp. 1970–1979, 2011. [Google Scholar]
8. J. Shen, C. J. P. Zhang, B. Jiang, J. Chen, J. Song et al., “Artificial intelligence versus clinicians in disease diagnosis: Systematic review,” JMIR Medical Informatics, vol. 21, no. 8, pp. 1–15, 2019. [Google Scholar]
9. Y. Murawaki, Y. Ikuta, K. Okamoto, M. Koda and H. Kawasaki, “Diagnostic value of serum markers of connective tissue turnover for predicting histological staging and grading in patients with chronic hepatitis C,” Journal of Gastroenterology, vol. 36, no. 6, pp. 399–406, 2001. [Google Scholar]
10. C. Lackner, G. Struber, B. Liegl, S. Leibl, P. Ofner et al., “Comparison and validation of simple noninvasive tests for prediction of fibrosis in chronic hepatitis C,” Hepatology, vol. 41, no. 6, pp. 1376–1382, 2005. [Google Scholar]
11. C.-T. Wai, J. K. Greenson, R. J. Fontana, J. D. Kalbfleisch, J. A. Marrero et al., “A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C,” Hepatology, vol. 38, no. 2, pp. 518–526, 2003. [Google Scholar]
12. P. Halfon, M. Bourlière, G. Pénaranda, R. Deydier, C. Renou et al., “Accuracy of hyaluronic acid level for predicting liver fibrosis stages in patients with hepatitis C virus,” Comparative Hepatology, vol. 4, no. 1, pp. 6, 2005. [Google Scholar]
13. M. Nasr, K. El-Bahnasy, M. Hamdy and S. M. Kamal, “A novel model based on non invasive methods for prediction of liver fibrosis,” in 2017 13th Int. Computer Engineering Conf., Cairo, Egypt, vol. 2018-January, pp. 276–281, 2017. [Google Scholar]
14. Healthline, “What are the stages of hepatitis C?-HepatitisC.net,” Healthline Media, 2018. [Online]. Available: https://hepatitisc.net/living/what-are-the-stages-of-hepatitis-c/ [Accessed: 15-Aug-2020]. [Google Scholar]
15. MHealth Lab, “Early detection, early treatment for hepatitis C,” MHealth Lab, 2016. [Online]. Available: https://labblog.uofmhealth.org/rounds/early-detection-early-treatment-for-hepatitis-c [Accessed: 20-Aug-2020]. [Google Scholar]
16. E. Gupta, M. Bajpai and A. Choudhary, “Hepatitis C virus: Screening, diagnosis, and interpretation of laboratory assays,” Asian Journal of Transfusion Science, vol. 8, no. 1, pp. 19–25, 2014. [Google Scholar]
17. F. Jiang, Y. Jiang, H. Zhi, Y. Dong, H. Li et al., “Artificial intelligence in healthcare: Past, present and future,” Stroke and Vascular Neurology, vol. 2, no. 4, pp. 230–243, 2017. [Google Scholar]
18. M. A. Konerman, L. A. Beste, T. Van, B. Liu, X. Zhang et al., “Machine learning models to predict disease progression among veterans with hepatitis C virus,” PLoS One, vol. 14, no. 1, pp. e0208141, 2019. [Google Scholar]
19. N. Papadopoulos, S. Vasileiadi, M. Papavdi, E. Sveroni, P. Antonakaki et al., “Liver fibrosis staging with combination of APRI and FIB-4 scoring systems in chronic hepatitis C as an alternative to transient elastography,” Annals of Gastroenterology, vol. 32, no. 5, pp. 498, 2019. [Google Scholar]
20. Y. Zhao, P. H. Thurairajah, R. Kumar, J. Tan, E. K. Teo et al., “Novel non-invasive score to predict cirrhosis in the era of hepatitis C elimination: A population study of ex-substance users in Singapore,” Hepatobiliary & Pancreatic Diseases International, vol. 18, no. 2, pp. 143–148, 2019. [Google Scholar]
21. J. Tani, A. Morishita, T. Sakamoto, K. Takuma, M. Nakahara et al., “Simple scoring system for prediction of hepatocellular carcinoma occurrence after hepatitis C virus eradication by direct‐acting antiviral treatment: All kagawa liver disease group study,” Oncology Letters, vol. 19, no. 3, pp. 2205–2212, 2020. [Google Scholar]
22. S. C. R. Nandipati, C. XinYing and K. K. Wah, “Hepatitis C virus (HCV) prediction by machine learning techniques,” Applications of Modelling and Simulation, vol. 4, pp. 89–100, 2020. [Google Scholar]
23. T. M. K. Motawi, N. A. H. Sadik, D. Sabry, N. N. Shahin and A. S., “Fahim, rs2267531, a promoter SNP within glypican-3 gene in the X chromosome, is associated with hepatocellular carcinoma in Egyptians,” Scientific Reports, vol. 9, no. 1, pp. 1–10, 2019. [Google Scholar]
24. M. Reiser, B. Wiebner and J. Hirsch, “Neural-network analysis of socio-medical data to identify predictors of undiagnosed hepatitis C virus infections in Germany (DETECT),” Journal of Translational Medicine, vol. 17, no. 1, pp. 1–7, 2019. [Google Scholar]
25. J. Bresnick, “Predictive analytics identify high risk Hepatitis C patients,” Health IT Analytics, 2015. [Online]. Available: https://healthitanalytics.com/news/predictive-analytics-identify-high-risk-hepatitis-c- patients/ [Accessed: 15-Oct-2020]. [Google Scholar]
26. Y. C. Tsao, J. Y. Chen, W. C. Yeh, Y. S. Peng and W. C. Li, “Association between visceral obesity and hepatitis C infection stratified by gender: A cross-sectional study in Taiwan,” BMJ Open, vol. 7, no. 11, pp. e017117, 2017. [Google Scholar]
27. T. Akiyama, T. Mizuta, S. Kawazoe, Y. Eguchi, Y. Kawaguchi et al., “Body mass index is associated with age-at-onset of HCV infected hepatocellular carcinoma patients,” World Journal of Gastroenterology, vol. 17, no. 7, pp. 914–921, 2011. [Google Scholar]
28. A. A. A. Radwan and H. Mamdouh, “An analysis of hepatitis C virus prediction using different data mining techniques,” International Journal of Computer Science, Engineering and Information Technology, vol. 3, no. 4, pp. 209–220, 2013. [Google Scholar]
29. S. M. Abd El-Salam, M. M. Ezz, S. Hashem, W. Elakel, R. Salama et al., “Performance of machine learning approaches on prediction of esophageal varices for Egyptian chronic hepatitis C patients,” Informatics in Medicine Unlocked, vol. 17, no. September, pp. 100267, 2019. [Google Scholar]
30. S. Hashem, G. Esmat, W. Elakel, S. Habashy, S. Abdel Raouf et al., “Accurate prediction of advanced liver fibrosis using the decision tree learning algorithm in chronic hepatitis c Egyptian patients,” Gastroenterology Research and Practice, vol. 2016, pp. 1–7, 2016. [Google Scholar]
31. E. W. Abd El-Wahab, H. A. Ayoub, A. A. Shorbila, A. Mikheal, M. Fadl et al., “Noninvasive biomarkers predict improvement in liver fibrosis after successful generic DAAs based therapy of chronic hepatitis C in Egypt,” Clinical Epidemiology and Global Health, vol. 8, no. 4, pp. 1177–1188, 2020. [Google Scholar]
32. J.-P. Zarski, S. David-Tchouda, C. Trocme, J. Margier, A. Vilotitch et al., “Non-invasive fibrosis tests to predict complications in compensated post-hepatitis C cirrhosis,” Clinics and Research in Hepatology and Gastroenterology, vol. 44, no. 4, pp. 524–531, 2020. [Google Scholar]
33. K. Fujita, K. Oura, H. Yoneyama, T. Shi, K. Takuma et al., “Albumin-bilirubin score indicates liver fibrosis staging and prognosis in patients with chronic hepatitis C,” Hepatology Research, vol. 49, no. 7, pp. 731–742, 2019. [Google Scholar]
34. S. Hashem, M. ElHefnawi, S. Habashy, M. El-Adawy, G. Esmat et al., “Machine learning prediction models for diagnosing hepatocellular carcinoma with HCV-related chronic liver disease,” Computer Methods and Programs in Biomedicine, vol. 196, pp. 105551, 2020. [Google Scholar]
35. H. M. Fayed, H. S. Mahmoud and A. E. M. Ali, “The utility of retinol-binding protein 4 in predicting liver fibrosis in chronic hepatitis C patients in response to direct-acting antivirals,” Clinical and Experimental Gastroenterology, vol. 13, pp. 53, 2020. [Google Scholar]
36. O. Hegazy, M. Allam, A. Sabry, M. A. S. Kohla, W. Abogharbia et al., “Liver stiffness measurement by transient elastography can predict outcome after hepatic resection for hepatitis C virus-induced hepatocellular carcinoma,” The Egyptian Journal of Surgery, vol. 38, no. 2, pp. 313, 2019. [Google Scholar]
37. X. Li, H. Xu and P. Gao, “Fibrosis index based on 4 factors (fib-4) predicts liver cirrhosis and hepatocellular carcinoma in chronic hepatitis C virus (HCV) patients,” Medical Science Monitor: International Medical Journal of Experimental and Clinical Research, vol. 25, pp. 7243, 2019. [Google Scholar]
38. M. W. Nadeem, M. A. Al Ghamdi, M. Hussain, M. A. Khan, K. M. Khan et al., “Brain tumor analysis empowered with deep learning: A review, taxonomy, and future challenges,” Brain Sciences, vol. 10, no. 2, pp. 118, 2020. [Google Scholar]
39. H. Malik, M. S. Farooq, A. Khelifi, A. Abid, J. N. Qureshi et al., “A comparison of transfer learning performance versus health experts in disease diagnosis from medical imaging,” IEEE Access, vol. 8, pp. 139367–139386, 2020. [Google Scholar]
40. M. W. Nadeem, H. G. Goh, A. Ali, M. Hussain and M. A. Khan, “Bone age assessment empowered with deep learning: A survey, open research challenges and future directions,” Diagnostics, vol. 10, no. 10, pp. 781, 2020. [Google Scholar]
41. H. Khalid, M. Hussain, M. A. Al Ghamdi, T. Khalid, K. Khalid et al., “A comparative systematic literature review on knee bone reports from MRI, X-rays and CT scans using deep learning and machine learning methodologies,” Diagnostics, vol. 10, no. 8, pp. 518, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |