DOI:10.32604/cmc.2022.018742

| Computers, Materials & Continua DOI:10.32604/cmc.2022.018742 |  |
| Article |
Epilepsy Radiology Reports Classification Using Deep Learning Networks
1Department of Computer Engineering, Halic University, Istanbul, 34445, Turkey
2Department of Computer Engineering, Istanbul University – Cerrahpasa, Istanbul, 34320, Turkey
3Department of Computer Engineering, Sivas Cumhuriyet University, Sivas, 58140, Turkey
*Corresponding Author: Eylem Yucel. Email: eylem@iuc.edu.tr
Received: 19 March 2021; Accepted: 14 June 2021
Abstract: The automatic and accurate classification of Magnetic Resonance Imaging (MRI) radiology report is essential for the analysis and interpretation epilepsy and non-epilepsy. Since the majority of MRI radiology reports are unstructured, the manual information extraction is time-consuming and requires specific expertise. In this paper, a comprehensive method is proposed to classify epilepsy and non-epilepsy real brain MRI radiology text reports automatically. This method combines the Natural Language Processing technique and statistical Machine Learning methods. 122 real MRI radiology text reports (97 epilepsy, 25 non-epilepsy) are studied by our proposed method which consists of the following steps: (i) for a given text report our systems first cleans HTML/XML tags, tokenize, erase punctuation, normalize text, (ii) then it converts into MRI text reports numeric sequences by using index-based word encoding, (iii) then we applied the deep learning models that are uni-directional long short-term memory (LSTM) network, bidirectional long short-term memory (BiLSTM) network and convolutional neural network (CNN) for the classifying comparison of the data, (iv) finally, we used 70% of used for training, 15% for validation, and 15% for test observations. Unlike previous methods, this study encompasses the following objectives: (a) to extract significant text features from radiologic reports of epilepsy disease; (b) to ensure successful classifying accuracy performance to enhance epilepsy data attributes. Therefore, our study is a comprehensive comparative study with the epilepsy dataset obtained from numeric sequences by using index-based word encoding method applied for the deep learning models. The traditional method is numeric sequences by using index-based word encoding which has been made for the first time in the literature, is successful feature descriptor in the epilepsy data set. The BiLSTM network has shown a promising performance regarding the accuracy rates. We show that the larger sized medical text reports can be analyzed by our proposed method.
Keywords: Epilepsy; radiology text report analysis; natural language processing; feature engineering; index-based word encoding; deep learning networks-based text classification
The systems used in transferring the MRI radiology reports to the electronic medical records systems are being updated continuously and integrated leading to potential researches and applications in the area of radiology [1–3]. Because the majority of MRI radiology reports are unstructured and free form language, extracting information manually is a time-consuming, and unmanageable task. And it is prone to human error, labour intensive and requires specific expertise [4]. Natural Language Processing (NLP) is widely used in the analysis of unstructured text data [2]. NLP techniques are rule-based and statistical Machine Learning (ML). The rule-based NLP techniques are widely used in the clinical tasks, such as recordings for the incidence findings in radiology reports and employed as string mapping using a set of predefined keywords by the experts. The ML-based techniques learn the lexical and clinical characteristics of pre-labeled report content to achieve classification [5].
In the recent literature, various studies regarding the radiology and clinical reports have been classified and identified. A new model to identify acute lung injury [3] using the chest X-ray report in two corpora that was modeled by the 6-gram and maximum entropy methods. The F-measure accuracy was 91%. In [6], the authors applied the rule based and conditional random fields methods on the radiology and pathology reports for the hepatocellular carcinoma classification. The F-score accuracy was obtained greater than 80%. In [7], a model was introduced to extract radiology reports BoW features by the Multinomial Naïve Bayes algorithm for the six compartments patellar cartilage. The accuracy rate based on BoW features were obtained by 88.76%. In [8], deep CNN-based, LSTM-based, BiLSTM-based classification models were applied on pulmonary nodular findings radiology reports by the word embedding features. The classification accuracies were based on F1-score which were 89.69%, 88.36%, 89.44%, respectively. Furthermore, the clinical notes for the smoking status and proximal femur were classified by the CNN model with 0.92 according to F1 score and 0.97 according to fracture classification in [9]. In the [10], brain tumor status was investigated from MRI reports based on statistical-SVM and rule based-SVM. The brain masses (metastasis, meningioma, gliomas grade ii, gliomas grade iii, glioblastomas) were classified by the SVM modelling in the [11]. The classification accuracy was obtained by 85%. The clinical examination unigram and bigram features were modeled by the TF-IDF and SVM algorithms in the [12]. The accuracy based on n-gram feature was 90.6%. The brain MRI ischemic stroke documents frequency matrix features were classified by the single decision tree in the [13] by the 98% classification accuracy. EHRs notes pertaining to diabetes BoW features were identified by the SVM in the [14]. The clinical reports for the breast pathology were identified by the rule based and boosting methods in the [15] by the 97% classification accuracy rate. The pathology reports were analyzed for the keyword extraction by the fine-tuning and deep learning approaches in the [16]. The bidirectional encoder representations from transformers model was successful model for the precision and recall values. The head computed tomography reports were identified by the general labeling that were combined Word2Vec word embedding, LSTM-attention methods in the [17] by the 97% classification accuracy rate. The pulmonary embolism reports were classified for the presence and chronicity with the CNN and RNN attention approaches in the [18]. The RNN precision and recall values were more successful than CNN model. Pulmonary nodules were diagnosed by the CNN and LSTM algorithms obtained the same precision, recall and F1 score values which were 91%, 90%, 90%, respectively in the [19]. However, the number of NLP studies to classify epilepsy and non-epilepsy from radiology reports of brain MRI has found to be limited.
The goal of this study is to classify epilepsy patients automatically with the implementation of LSTM, BiLSTM, and CNN networks based on the real free-text brain MRI reports.
In our proposed method, firstly the 122 real MRI radiology text reports (97 epilepsy, 25 non-epilepsy) are converted into the numerical sequences by using index-based word encoding. Secondly, the networks of the LSTM, BiLSTM, and CNN are created and trained by the word embedding layer. Finally, the testing and validation text data are trained with the proposed networks.
Contrarily the previous studies, our proposed method adopt the following contributions: 1) The clever way present to information extraction from the MRI radiology reports, 2) The suggestion a clinical support system to classify epilepsy and non-epilepsy by automatically, 3) It brings together the traditional NLP technique that is index-based word encoding method, and modern deep learning models that are LSTM, BiLSTM, and CNN for the comparing classification accuracies.
This paper is organized as follows: A brief description of the real MRI radiology report, text analysis for text feature extraction are presented in Section 2. The experimental results obtained by the proposed training approach with the deep learning RNN and CNN networks are given in Section 3. The conclusion is addressed in Section 4.
In this study, the performed MRI sampling is a single-center retrospective case-control study. The study protocol has been approved by the Ethics Committee of Halic University, with a waiver of informed consent. The Avicenna Hospital in Turkey stores the entire medical records in a clinical data warehouse on MS-SQL, which has allowed us to screen all brain MRI reports recorded between June 30, 2006 and March 19, 2020. The brain MRI has been examined by the radiologists and their descriptions and findings have been stored on the MS-SQL server. The formats of the reports are the rich text files (.rtf) which were queried data by MS-SQL server with the reports having the size of 5 KB in Turkish. The MRI reports selected from the database were inquired to determine whether they indicate an epilepsy or non-epilepsy, resulting in the fact that 97 patients are epilepsy and 25 patients are non-epilepsy. The detailed results of the adult individuals are given in Tab. 1.
An MRI report is a text source consisting of a sequence of words. A piece of text is a sequence of words which can be correlated. Before the text preprocessing the LSTM, BiLSTM and CNN will first learn the correlated words and then classify the sequence data depending on the degree of the correlations dependencies [20–29]. In this study, “Text Analytics” (The Mathworks ©) has been used, which classify texts using NLP algorithms [22]. The full-text brain MRI reading sentences have been;
• cleaned from the HTML/XML entities,
• parsed into “tokens,” such as numbers, punctuations, symbols and hyphens. These tokens have been removed from the text data.
• used by the lowercase letterings
• removed from connector words (e.g., “e{g}er (if)”, “ve (and)”, “i{c}in (for)”, “gibi (as)”, etc.) which have trivial lexical meaning. Before training LSTM, BiLSTM, and CNN, it is necessary to extract of the meaningful text the features. The number of the words in the report of each patients are approximately 4500 before text preprocessing. After text preprocessing, the number of the words in the report of each patients are approximately 1500 words which form the feature vectors obtained by using the index-based word encoding approach.
Word encoding techniques [30–33] transform words into numbers and texts into number vectors. The index-based word encoding is the most frequently used encoding technique which is the first step to create a dictionary that matches words to indexes. Based on this dictionary, each document consists of a sequence of indexes that encode a word. The index-based word encoding establishes the document vectors of different lengths. Fig. 1, represents an example of the index-based word encoding for preprocessed non-epilepsy MRI radiology text reports.

Figure 1: An example of index-based word encoding
It is the most suitable deep learning model for the long sequences in extracting sequential information from the text data. The sequence of words is essential for the network's meaning. The RNN networks have two functions: (1) extracting important meaning from the sequential of the words, (2) it builds up the memory that summarizes previous calculations [34–36]. The RNN general formula is defined by Eq. (1).
The current hidden state
The features obtained from index-based word encoding have been trained by a deep neural network for the purpose of classification. Three one-layer architecture is considered for the uni-directional LSTM word encoder: The learned features are fed into two fully connected layers where a softmax function is used as the activation in the last layer. Given that N samples and
BiLSTM structure allows the network to receive both backward and forward information about the sequence at each step. It works in two ways, one from the past to the future and the other from the future to the past. With this approach, what sets it apart from being unidirectional is protecting information from the future in LSTM working backwards. It can preserve information from both the past and the future at any point in time [42,43]. The LSTM and BiLSTM networks general structures can be seen in Figs. 2a, and 2b respectively.
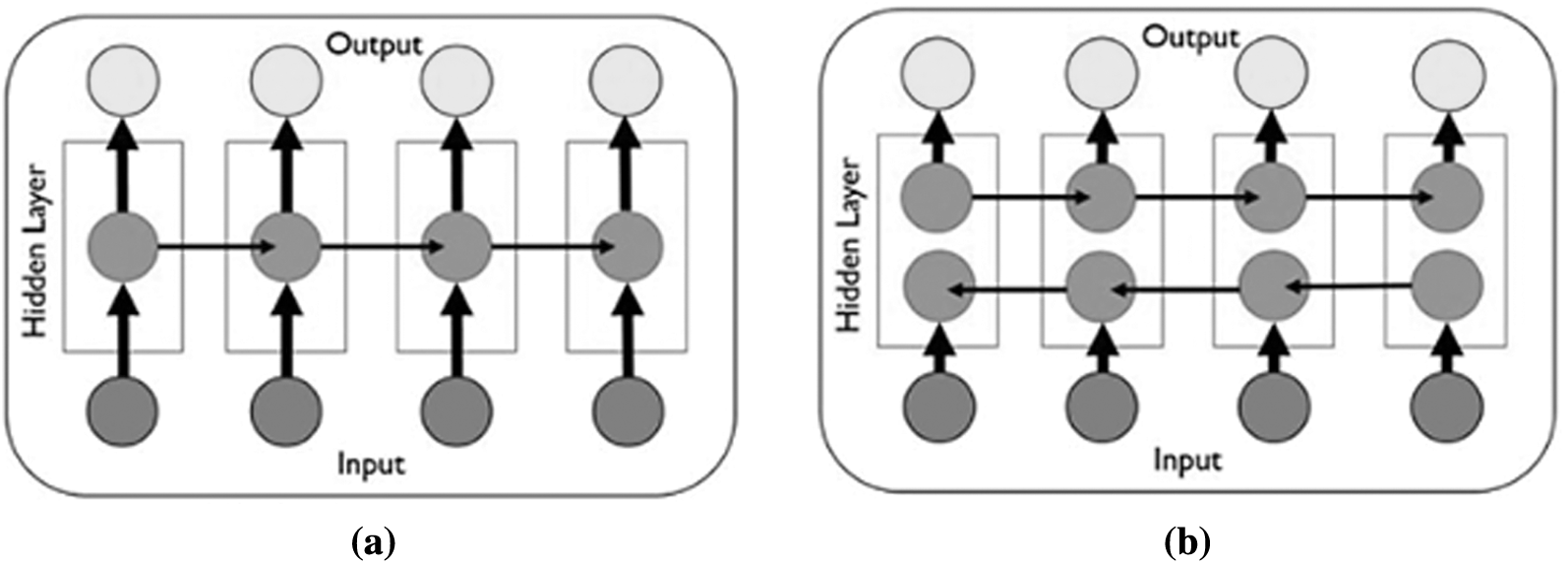
Figure 2: The general structure of RNN networks (a) The LSTM network general structure [41] (b) The BiLSTM network general structure [43]
CNN has three layers which are convolution, activation (Rectified Lineer Unit-ReLU), and pooling in Eqs. (4)–(6), respectively. The convolution layer activates the data features. The ReLU provides a faster and more effective training process by equating negative values to zero and preserving positive values. The pooling layer reduces the number of parameters that the network must learn. The model learns the weight and bias values in the training process and updates it continuously with each new training example. After learning the properties in layers, the architecture of a CNN switches to classification. The next layer is a fully connected layer that outputs a vector of K dimensions where K is the number of classes that the network can predict. The last layer of CNN architecture uses a classification layer like softmax to provide classification output [37,44–47].
In this study, the dataset is 1D vector, so that the sum of the dot product and filter can be presented as Eq. (4).
According to Eq. (5), the floor function of
The CNN algorithm is the most suitable for the repetitive data such as images, signals, audio. CNN is also applied to the repeating text data. The input is collection of sentences which is obtained word vectors. The network is based on the convolutianal filter that calculates as small matrices of the weights. The filter slides over the rows in the text and dot products calculates in the linear unit between weights and words [37,47]. In this study,
The evaluations have been performed on the hold-out validation. The positive class has been defined as epilepsy, and the negative class was defined as the non-epilepsy. Three measures have been sensitivity, specificity, and accuracy are described as in Tab. 2.

Sensitivity, specificity, accuracy, precision, recall, F-Measure, and Geometric Mean (G-Mean) formulations are given in Eqs. (7)–(13) [40,48,49].
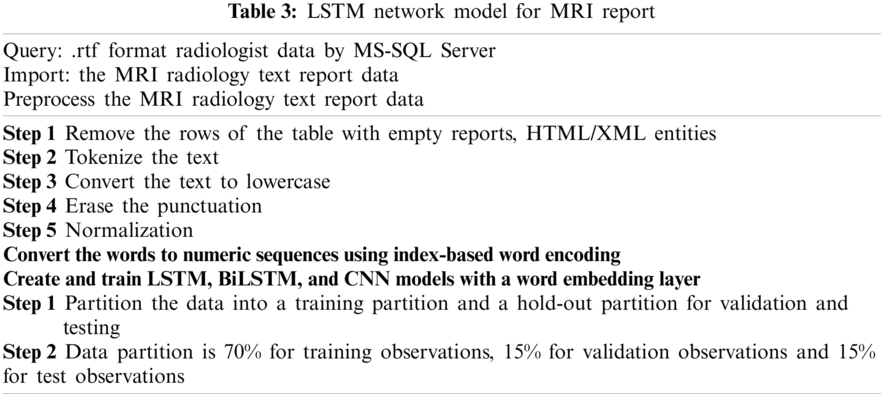
In this subsection we give the algorithm for the proposed method, which is given in Fig. 3 and the flowchart of this algorithm in Tab. 3.
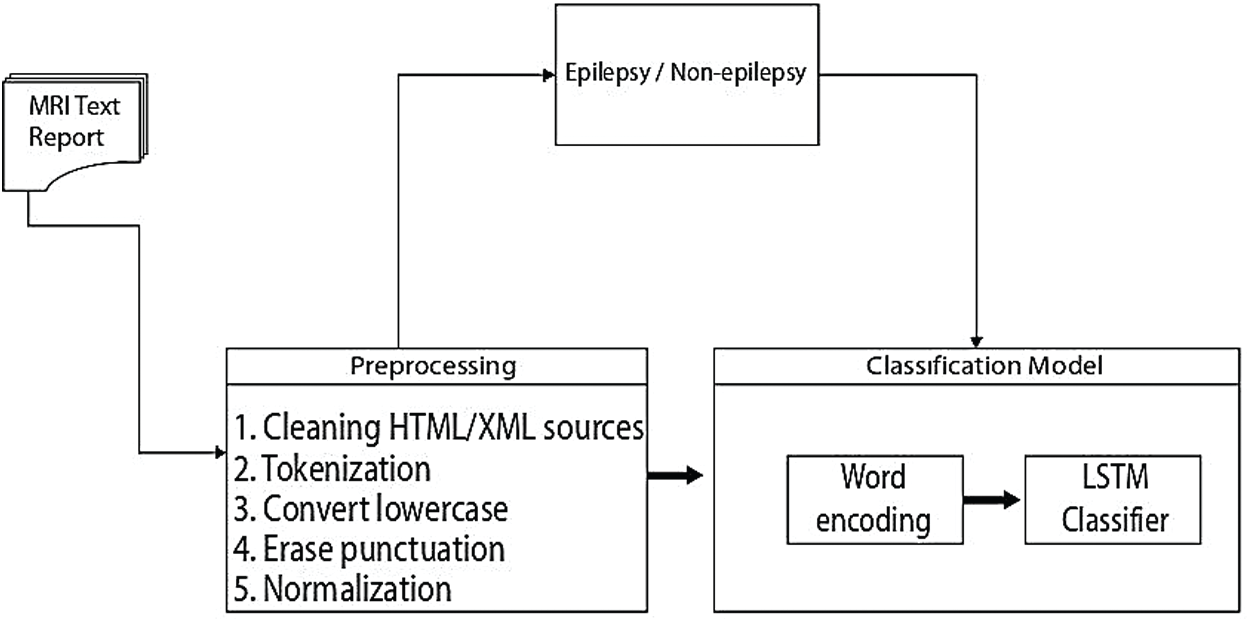
Figure 3: An overview of the proposed epilepsy/non-epilepsy classification framework
3.2 Word Cloud for Training/Validation/Testing Data
The word cloud indicates the size of the word that appears on the document data set is obtained from MRI radiology text reports. The words have been grouped in clusters according to how frequently they are mentioned for training, validation, and testing. The data obtained from MRI radiology text reports have been accurately transferred to the word cloud model. The word cloud for training is given in Fig. 4a, for validation is given in Fig. 4b and for testing is given in Fig. 4c.
Figure 4: Word cloud for the MRI dataset (a) Training data (b) Validation data (c) Test data
3.3 Sequences of Numeric Indices
The documents have been entered into an LSTM, BiLSTM, and CNN networks for training, validation, and testing. A word encoding has been used to convert the documents into the numerical index sequences as shown in Fig. 5.
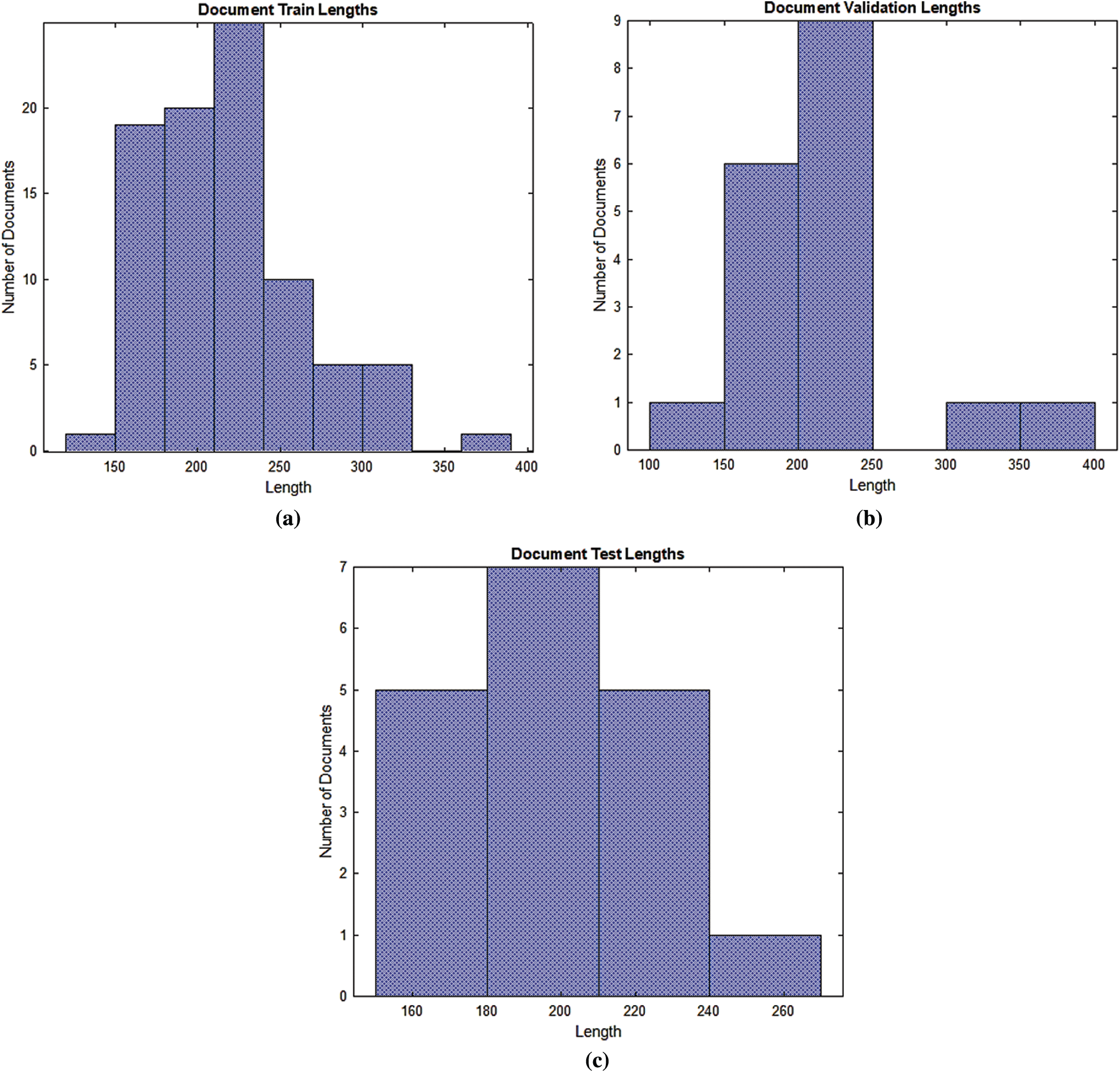
Figure 5: Documents into the numerical index sequences (a) Training document (b) Validation document (c) Test document
According to Fig. 5, most of the training, validation and testing documents have been less than 150 tokens. The setting of the training, validation and testing MRI radiology text report data are shown in Tab. 4. The optimum values have been selected for the best accurate LSTM, BiLSTM, CNN automatic classification of epilepsy and non-epilepsy MRI radiology reports with the highest accuracy rates.
3.4 The Training of the LSTM and BiLSTM Networks
The general LSTM and BiLSTM networks structures have been given in Figs. 6a and 6b. According to Fig. 6, the proposed LSTM and BiLSTM networks options have been set to be as follows:
Step 1: sequence input layer and input size are set to 1,
Step 2: the dimension of the word embedding layer and the word encoding layer are set to 100 dimensions and 1126 real words for training, 677 words for validation, 540 words for testing are used.
Step 3: the number of hidden units is set to 180,
Step 4: the fully connected layers are used for the classification, which are softmax layer and classification layer.
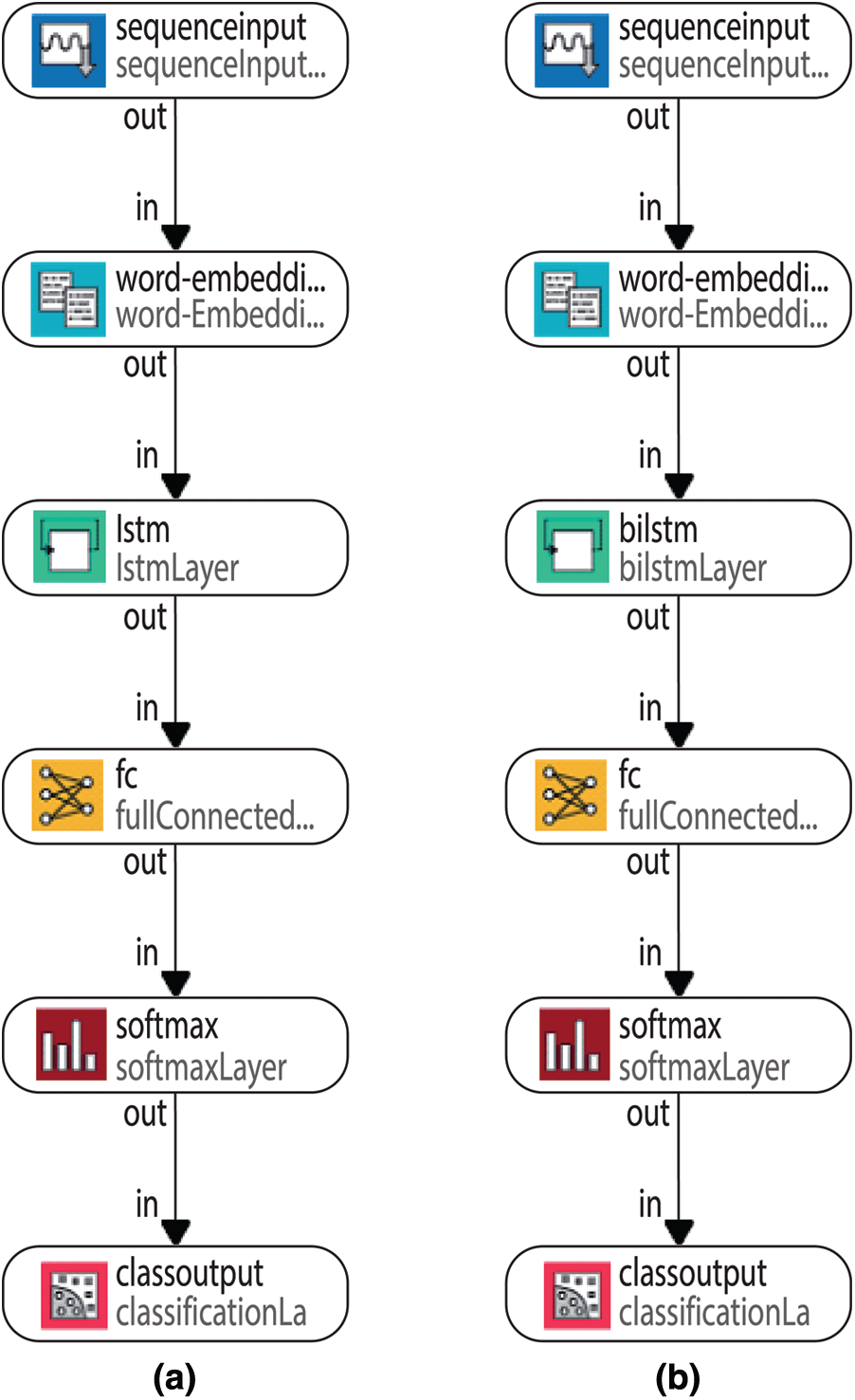
Figure 6: General LSTM and BiLSTM networks structures (a) LSTM network (b) BiLSTM layer
When the solver in the systems have been set to ‘adam’, the data are trained for 100 epochs, the gradient thresholds have been set to 1 and the initial learning rates have been set to 0.0001 and 0.001 for the LSTM and the BiLSTM networks, respectively. By default, the training network models have been used as a Graphics Processing Unit (GPU). The same parameters have been used for testing and validation which used for the training of LSTM and BiLSTM networks. In order to make a prediction of epilepsy and non-epilepsy, the preprocessed test and validation data are used. The training process for the LSTM network has been given in the Fig. 7a. The confusion matrices of the LSTM network have been given in the Fig. 7b.
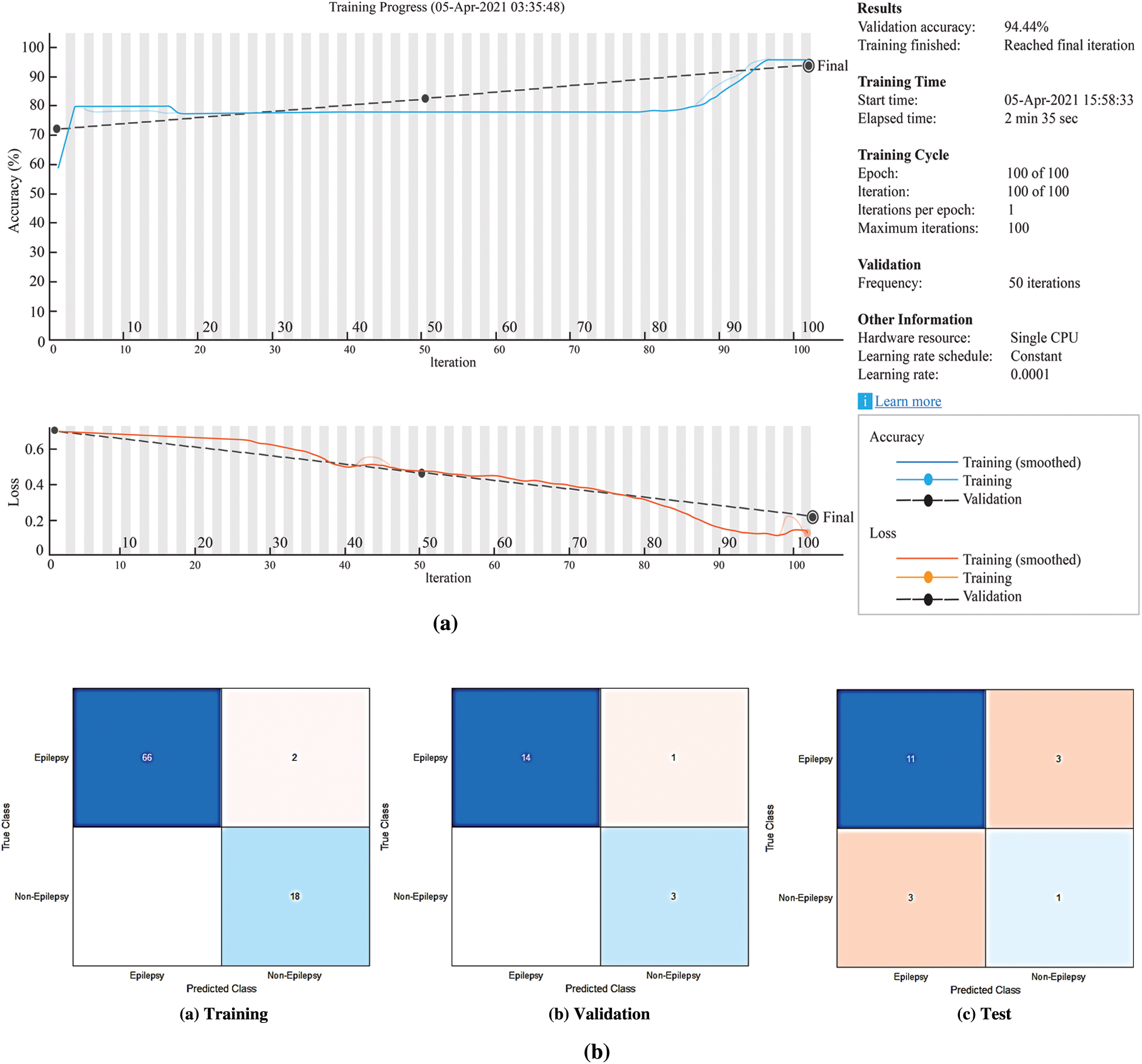
Figure 7: The experimental results for the proposed LSTM network (a) The LSTM network training process (b) Confusion matrices of the LSTM network
According to Fig. 7b, the accuracy rates are the proportion of the labels that the accuracy rates for training, validation and testing are 97.67%, 94.44% and 66.67%, respectively.
The training process for the BiLSTM has been given in the Fig. 8a. The confusion matrices of the BiLSTM network have been given in the Fig. 8b.
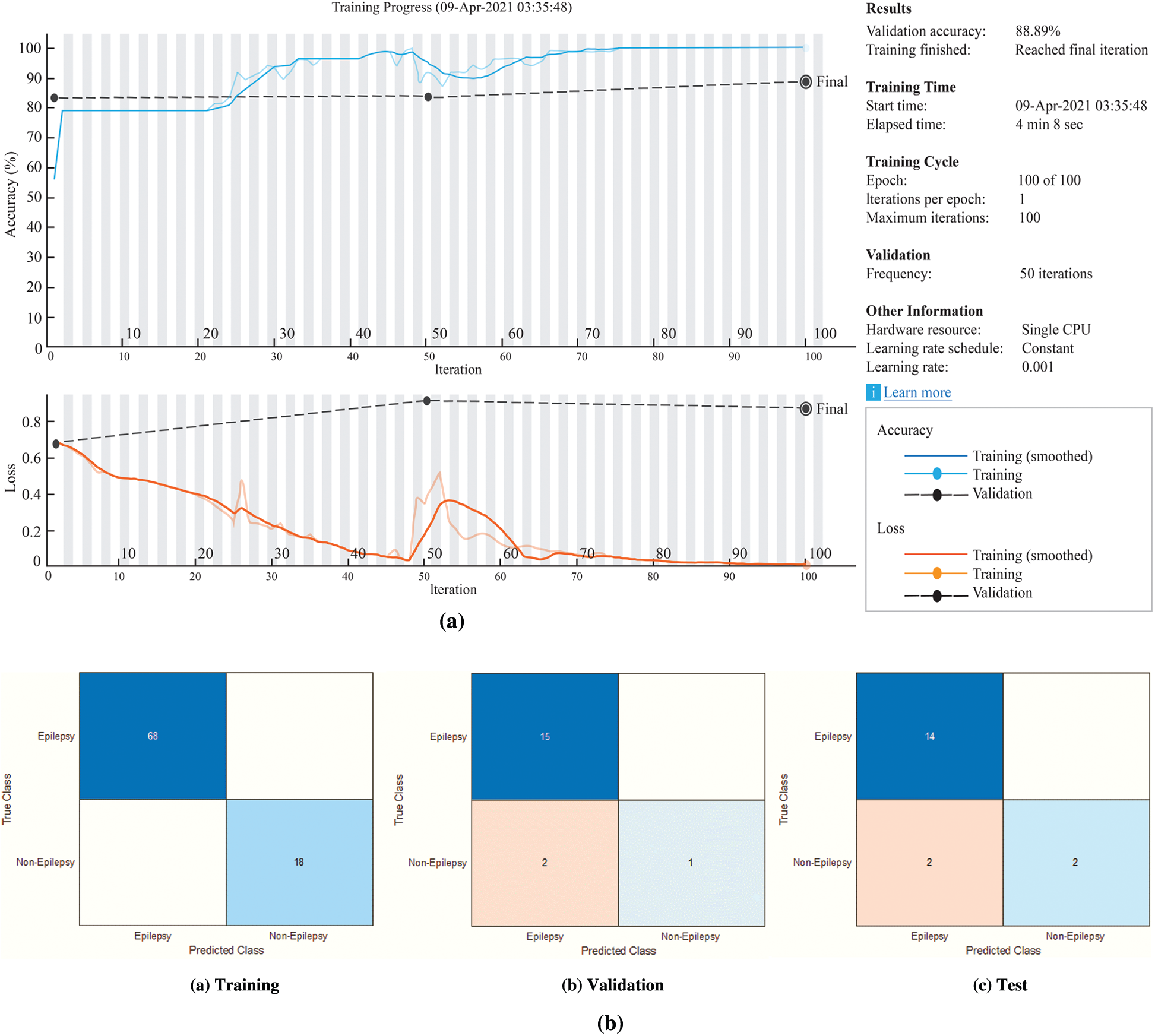
Figure 8: The experimental results for the proposed BiLSTM network (a) The BiLSTM network training process (b) Confusion matrices of the BiLSTM network
According to Fig. 8b, the accuracy rates are the proportion of the labels that the accuracy rates for training, validation and testing are 100%, 88.89% and 88.89%, respectively.
In this study, 1-D CNN model were constructed for the MRI text reports classification has been as in Fig. 9.
To train the CNN, the input size value has been set to 1. The mini-batch size has been used 15 with 150 epochs. 1-D CNN has been trained with the scattering sequences. The scattering sequences have been set 1-by-3, where 3 is the number of time steps and 5 is the number of scatter paths. To use this in a 1-D convolutional network, transfer and reshape the scattering sequences by 3-by-1-by-1-by-to training samples. The results of the loss, test loss, and accuracy have been obtained 36.85%, 51.94%, and 77.90%, respectively, at the end of 770th epoch. The training loss and validation loss plot for our proposed 1-D CNN have been shown in Fig. 10. According to Fig. 10, as the training loss decreases, the validation loss increases.
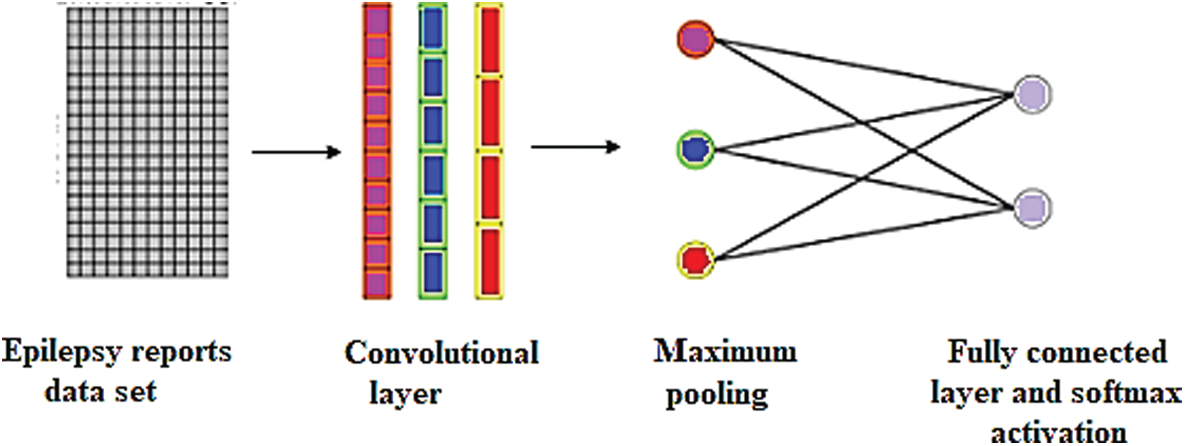
Figure 9: Single-layer CNN for the MRI text reports classification
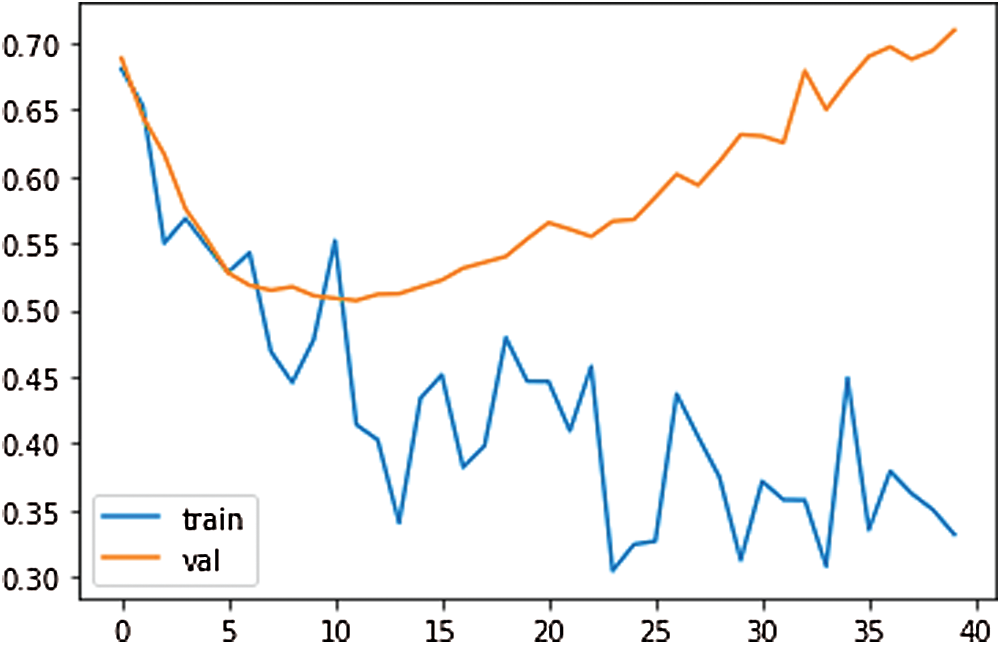
Figure 10: The training loss and validation loss plot for proposed 1-D CNN
It was carried out by calculating the hypothesis test for the p-value. The p-value, as it is useful to interpret the null hypothesis statistical test results is considered less than the level of significance (usually 0.05) and it is statistically significant. In this study, the test analysis of variance (ANOVA) was used. When the p-value is lower the significance, level rejects the null hypothesis and alternative hypothesis of the data is supported. The only way ANOVA design was used to detect differences in results, accuracy rate, sensitivity, specificity, precision, recall, F-measure, and G-Mean [49]. Each statistical analysis was used for the p-value which was 0.05 (significance level). The columns of the matrix represent 7-performance metrics (accuracy rate, sensitivity, specificity, precision, recall, F-measure, and G-Mean) for the LSTM, BiLSTM and CNN models. Fig. 11 showed the 7 performance evaluation metrics. In Fig. 11, modelling epileptic MRG text reports with the LSTM, BiLSTM, and CNN algorithms presented that were efficient models for classification. The p-value was obtained as 2.70521e-06 indicates that the models were not the same.
According to Fig. 11, the statistical testing rejected the null hypothesis on the proposed models which were LSTM, BiLSTM, and CNN. All the results showed that the proposed methods had significantly improved the performance as p-values in all cases were our statistical threshold of 0.05.
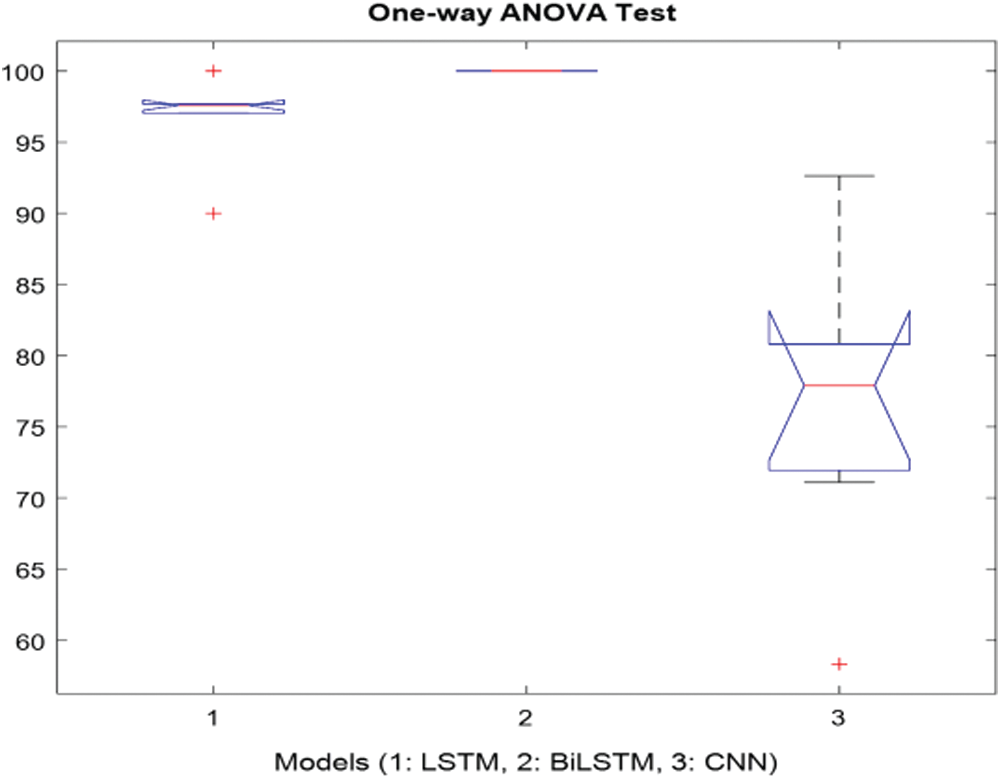
Figure 11: One-way ANOVA test for the proposed models
3.7 The Evaluation of the Experimental Results
Fig. 12 presents the elapsed time for the LSTM, BiLSTM, and CNN. According to Fig. 12, the CNN is the most time-consuming algorithm for the training process and the LSTM network is the least time-consuming algorithm for the training process.
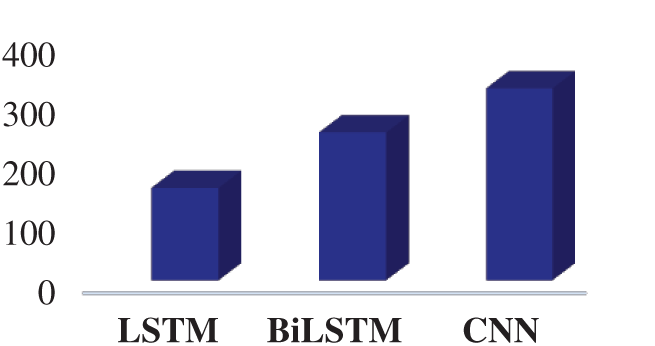
Figure 12: The elapsed time for the proposed networks
Significant Artificial Intelligence (AI) applications have been developed for the various fields. One of the notable developments with AI is NLP applications. The NLP methods process medical text data obtained from various sources. Meaningful information could be extracted about unstructured data including EHRs/EMRs, clinical notes and clinical records, and others, physician note, nurse note, triage note, radiology note, patient schedule, discharge summary, laboratory record, incident report, imaging report. The pathology report, patient narrative, drug label and information provided by the patient, text message, online discussion, state license list, and medical publication allow disease follow-up [50–52]. Although there are various studies with the MRI reports in the literature, there are almost no studies on epilepsy patients. In the literature, the models were developed with text-based medical data were given in Tab. 5.
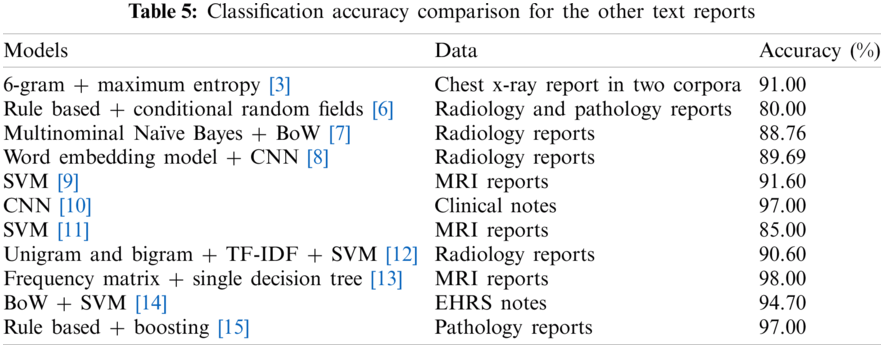
The results in Tab. 6 were implemented by Matlab, and Phyton. The classification accuracy rates were obtained from LSTM, BiLSTM, and CNN networks applied to the epilepsy MRI dataset. The LSTM, BiLSTM, and CNN networks classification sensitivities, specificities, and accuracy rates results for the training were presented on Tab. 6. The BiLSTM training network can protect input data by operating it in two ways, from past to future and from future to past, by using it bi-directionally. Therefore, in this study, BiLSTM modeling provided the best accuracy for epileptic and non-epileptic classification compared to LSTM and CNN models. Also, both RNN algorithms have more successful classification accuracy than the CNN algorithm. In literature, very limited number of studies exist on classification modellings of the MRI radiology reports belonging with the epileptic individuals. In this study, three different mathematical models were developed comparatively for the MRI report data of individuals with epilepsy, written by a radiologist and diagnosed by a neurologist with epilepsy. Classification of MRI radiology reports by processing them with the LSTM, BiLSTM and CNN algorithms are completely new approach.

This study has some limitations. Initially, the algorithm was developed using MRI radiology reports of healthy individuals, 97 with epilepsy and 25 non-epilepsy. MRG report of individuals in the data set is increasing day by day. However, we are constantly working to adapt our algorithm to the growth of the data set. Thus, in the future it can help physicians in the faster and more effective identification of epilepsy more information will be inferred. Secondly, the models have been made with LSTM, BiLSTM and CNN, which are among the deep learning models. While BiLSTM has been given the most successful classification score during the training process, the classification accuracy has been obtained as 88.89% for the testing and validation processes. There is a need for new models where test and validation performance can be increased. Thirdly, classification of epilepsy, more effective modeling should be developed by adding data set such as EEG and laboratory results as well as MRI reports to the data set.
Transferring MRI radiology reports to electronic health record systems, which are constantly updated, integrated, and shared data, has led to the potential for advancement in radiology research and practice. Because the majority of MRI radiology reports are unstructured and free-form language, extracting information manually is time-consuming, often unmanageable, and prone to human error, so it requires special expertise. This study was developed as a decision support system to assist medical professionals in the diagnosis of epilepsy.
The main contribution of this study, to the real MRI reports of individuals suffering from epilepsy in Turkey by applying the traditional methods of numeric sequences by using index-based word encoding have been obtained data sets precedent with epilepsy. Modeling of the epilepsy dataset with the LSTM, BiLSTM, and CNN models, which are popular deep learning models, are presented comparatively. When this study is compared to other works [6–19], no study like this study was found for the actual MRI data set of patients with the epilepsy. Among our proposed methods, due to the BiLSTM network's ability to operate both backward and forward information, it has been proven to give the best classification result. The following objectives can be considered as the future works: Firstly, the size of the samples can be increased. Secondly, classification of epilepsy, to make fine judgment, more effective modeling can be developed by adding data set such as EEG and laboratory results as well as MRI reports to the data set. Thirdly, our proposed LSTM, BiLSTM, and CNN networks can be used for the other medical text reports datasets. Fourthly, the features can be extracted from the other NLP techniques such as n-grams, BoW. Finally, the new classification and mathematical modeling will be applied to compare with our proposed models.
Acknowledgement: The authors are grateful to deputy chief physician Dr. Lutfi Hocaoglu of Avicenna Hospital in Turkey for their support and understanding during this research.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. T. Rüber, B. David and C. E. Elger, “MRI in epilepsy: Clinical standard and evolution,” Current Opinion in Neurology, vol. 31, no. 2, pp. 223–231, 2018. [Google Scholar]
2. M. Woolfe, D. Prime, L. Gillinder, D. Rowlands, S. O'keefe et al., “Automatic detection of the epileptogenic zone: An application of the fingerprint of epilepsy,” Journal Neuroscience Methods, vol. 325, no. 2019, pp. 1–20, 2019. [Google Scholar]
3. I. Solti, C. R. Cooke, F. Xia and M. M. Wurfel, “Automated classification of radiology reports for acute lung injury: Comparison of keyword and machine learning based natural language processing approaches,” in Proc. IEEE Int. Conf. Bioinformatics and Biomedicine Workshop, BIBMW, Washington, United States, pp. 314–319, 2009. [Google Scholar]
4. K. Krabbe, P. Gideon, P. Wagn, U. Hansen, C. Thomsen et al., “MR diffusion imaging of human intracranial tumours,” Neuroradiology, vol. 39, no. 7, pp. 483–489, 1997. [Google Scholar]
5. J. M. Provenzale, S. Mukundan and D. P. Barboriak, “Diffusion-weighted and perfusion MR imaging for brain tumor characterization and assessment of treatment response,” Radiology, vol. 239, no. 3, pp. 632–649, 2006. [Google Scholar]
6. L. Chen, L. Song, Y. Shao, D. Li and K. Ding, “Using natural language processing to extract clinically useful information from Chinese electronic medical records,” International Journal of Medical Informatics, vol. 124, pp. 6–12, 2019. [Google Scholar]
7. L. Chen, R. Shah, T. Link, M. Bucknor, S. Majumdar et al., “Text sentiment analysis for cartilage abnormalities detection from radiology reports,” Osteoarthritis Cartilage, vol. 7, no. 1, pp. 400–401, 2019. [Google Scholar]
8. J. Yuan, H. Zhu and A. Tahmasebi, “Classification of pulmonary nodular findings based on characterization of change using radiology reports,” in Proc. Joint Summits Translational Science Proc., AMIA, Bethesda, USA, pp. 285–294, 2019. [Google Scholar]
9. L. T. E. Cheng, J. Zheng, G. K. Savova and B. J. Erickson, “Discerning tumor status from unstructured MRI reports-completeness of information in existing reports and utility of automated natural language processing,” Journal of Digital Imaging, vol. 23, no. 2, pp. 119–132, 2010. [Google Scholar]
10. Y. Wang, S. Sohn, S. Liu, F. Shen, L. Wang et al., “A clinical text classification paradigm using weak supervision and deep representation,” BMC Medical Informatics and Decision Making, vol. 19, no. 1, pp. 1–13, 2019. [Google Scholar]
11. E. I. Zacharaki, S. Wang, S. Chawla, D. S. Yoo, R. Wolf et al., “Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme,” Magnetic Resonance in Medicine, vol. 62, no. 6, pp. 1609–1618, 2009. [Google Scholar]
12. P. H. Chen, H. Zafar, M. Galperin-Aizenberg and T. Cook, “Integrating natural language processing and machine learning algorithms to categorize oncologic response in radiology reports,” Journal of Digital Imaging, vol. 31, no. 2, pp. 178–184, 2018. [Google Scholar]
13. C. Kim, V. Zhu, J. Obeid and L. Lenert, “Natural language processing and machine learning algorithm to identify brain MRI reports with acute ischemic stroke,” PLOS One, vol. 14, no. 2, pp. 1–13, 2019. [Google Scholar]
14. A. Wright, A. B. McCoy, S. Henkin, A. Kale and D. F. Sittig, “Use of a support vector machine for categorizing free-text notes: Assessment of accuracy across two institutions,” Journal of the American Medical Informatics Association, vol. 20, pp. 887–890, 2013. [Google Scholar]
15. A. Yala, R. Barzilay, L. Salama, M. Griffin, G. Sollender et al., “Using machine learning to parse breast pathology reports,” Breast Cancer Research and Treatment, vol. 161, no. 2, pp. 203–211, 2017. [Google Scholar]
16. Y. Kim, J. H. Lee, S. Choi, J. M. Lee, J. H. Kim et al., “Validation of deep learning natural language processing algorithm for keyword extraction from pathology reports in electronic health records,” Scientific Reports, vol. 10, no. 1, pp. 1–9, 2020. [Google Scholar]
17. Y. Barash, G. Guralnik, N. Tau, S. Soffer, T. Levy et al., “Comparison of deep learning models for natural language processing-based classification of non-English head CT reports,” Neuroradiology, vol. 62, no. 10, pp. 1247–1256, 2020. [Google Scholar]
18. B. Imon, Y. Ling, M. C. Chen, S. A. Hasan, C. P. Langlotz et al., “Comparative effectiveness of convolutional neural network (CNN) and recurrent neural network (RNN) architectures for radiology text report classification,” Artificial Intelligence in Medicine, vol. 97, pp. 79–88, 2019. [Google Scholar]
19. Y. Jianbo, H. Zhu and A. Tahmasebi, “Classification of pulmonary nodular findings based on characterization of change using radiology reports,” AMIA Summits on Translational Science Proc., vol. 2019, pp. 285–294, 2019. [Google Scholar]
20. M. Berry and J. Kogan, “Text mining: Applications and theory,” in Text Extraction, Classification, and Clustering, 1st ed., United Kingdom: John Wiley & Sons Press, pp. 1–35, 2010. [Google Scholar]
21. H. Takci and T. Gungor, “A high performance centroid-based classification approach for language identification,” Pattern Recognition Letters, vol. 33, no. 16, pp. 2077–2084, 2012. [Google Scholar]
22. X. Zhang, W. Lu, F. Li, X. Peng and R. Zhang, “Deep feature fusion model for sentence semantic matching,” Computers, Materials & Continua, vol. 61, no. 2, pp. 601–616, 2019. [Google Scholar]
23. G. Mujtaba, L. Shuib, N. Idris, W. L. Hoo, R. G. Raj et al., “Clinical text classification research trends: Systematic literature review and open issues,” Expert Systems with Applications, vol. 116, pp. 494–520, 2019. [Google Scholar]
24. J. T. Oliva, H. D. Lee, N. Spolaôr, W. S. R. Takaki, C. S. R. Coy et al., “A computational system based on ontologies to automate the mapping process of medical reports into structured databases,” Expert Systems with Applications, vol. 115, pp. 37–56, 2019. [Google Scholar]
25. S. Albahli, A. Algsham, S. Aeraj, M. Alsaeed, M. Alrashed et al., “Covid-19 public sentiment insights: A text mining approach to the gulf countries,” Computers, Materials & Continua, vol. 67, no. 2, pp. 1613–1627, 2021. [Google Scholar]
26. W. L. Martinez and A. R. Martinez, “Introduction to exploratory data analysis,” in Exploratory Data Analysis with MATLAB, 3rd ed., London: Taylor & Francis Group, pp. 3–27, 2017. [Google Scholar]
27. S. P. Conlon, B. J. Reithel, M. W. Aiken and A. I. Shirani, “A natural language processing based group decision support system,” Decision Support Systems, vol. 12, no. 3, pp. 181–188, 1994. [Google Scholar]
28. B. M. Decker, C. E. Hill, S. N. Baldassano and P. Khankhanian, “Can antiepileptic efficacy and epilepsy variables be studied from electronic health records? A review of current approaches,” Seizure, vol. 85, pp. 138–144, 2021. [Google Scholar]
29. C. I. Chesñevar, M. Sabaté-Carrové and A. G. Maguitman, “An argument-based decision support system for assessing natural language usage on the basis of the web corpus,” International Journal of Intelligent Systems, vol. 21, no. 11, pp. 1151–1180, 2006. [Google Scholar]
30. A. E. Omolara, A. Jantan, O. I. Abiodun and H. E. Poston, “A novel approach for the adaptation of honey encryption to support natural language message,” in Proc. of the Int. Multi Conf. of Engineers and Computer Scientists, Hong Kong, pp. 1–7, 2018. [Google Scholar]
31. S. Mankad, H. S. Han, J. Goh and S. Gavirneni, “Understanding online hotel reviews through automated text analysis,” Service Science, vol. 8, no. 2, pp. 124–138, 2016. [Google Scholar]
32. D. D. Wickens, “Some characteristics of word encoding,” Memory & Cognition, vol. 1, pp. 485–490, 1973. [Google Scholar]
33. M. Khayyat, L. Lam and C. Y. Suen, “Arabic handwritten word spotting using language models,” in Proc.: 13th Int. Conf. on Frontiers in Handwriting Recognition, Bari, pp. 1–7, 2012. [Google Scholar]
34. K. Shaukat, S. Luo, V. Varadharajan, I. A. Hameed and M. Xu, “A survey on machine learning techniques for cyber security in the last decade,” IEEE Access, vol. 8, pp. 222310–222354, 2020. [Google Scholar]
35. X. Wei, L. Zhou, Z. Zhang, Z. Chen and Y. Zhou, “Early prediction of epileptic seizures using a long-term recurrent convolutional network,” Journal of Neuroscience Methods, vol. 32, pp. 1–10, 2019. [Google Scholar]
36. H. Eskandari, M. Imani and M. P. Moghaddam, “Convolutional and recurrent neural network based model for short-term load forecasting,” Electric Power Systems Research, vol. 195, no. 107173, pp. 1–14, 2021. [Google Scholar]
37. B. Zhong, X. Xing, P. Love, X. Wang and H. Luo, “Convolutional neural network: Deep learning-based classification of building quality problems,” Advanced Engineering Informatics, vol. 40, pp. 46–57, 2019. [Google Scholar]
38. C. Zhang, K. Qiao, L. Wang, L. Tong, G. Hu et al., “A visual encoding model based on deep neural networks and transfer learning for brain activity measured by functional magnetic resonance imaging,” Journal of Neuroscience Methods, vol. 325, pp. 1–9, 2019. [Google Scholar]
39. R. Samli, S. Senan, E. Yucel and Z. Orman, “Some generalized global stability criteria for delayed cohen–Grossberg neural networks of neutral-type,” Neural Networks, vol. 116, pp. 198–207, 2019. [Google Scholar]
40. I. Soltesz and K. Staley, “Validating models of epilepsy,” in Computational Neuroscience in Epilepsy, 1st ed., USA: Academic Press, pp. 3–17, 2008. [Google Scholar]
41. S. Hochreiter and J. Schmidhuber, “Long short-term memory,” Neural Computation, vol. 9, no. 8, pp. 1735–1780, 1997. [Google Scholar]
42. K. Shaukat, S. Luo, V. Varadharajan, I. A. Hameed, S. Chen et al., “Performance comparison and current challenges of using machine learning techniques in cybersecurity,” Energies, vol. 13, no. 2509, pp. 1–27, 2020. [Google Scholar]
43. A. Graves and J. Schmidhuber, “Framewise phoneme classification with bidirectional LSTM and other neural network architectures,” Neural Networks, vol. 18, no. 5–6, pp. 602–610, 2005. [Google Scholar]
44. V. Sorin, Y. Barash, E. Konen and E. Klang, “Deep learning for natural language processing in radiology—Fundamentals and a systematic review,” Journal of the American College of Radiology, vol. 17, no. 5, pp. 639–648, 2020. [Google Scholar]
45. K. S. Dar, A. B. Shafat and M. U. Hassan, “An efficient stop word elimination algorithm for urdu language,” in 14th Int. Conf. on Electrical Engineering/Electronics, Computer, Telecommunications and Information Technology, Phuket, Thailand, pp. 911–914, 2017. [Google Scholar]
46. S. Kiranyaz, O. Avci, O. Abdeljaber, T. Ince, M. Gabbouj et al., “1D convolutional neural networks and applications: A survey,” Mechanical Systems and Signal Processing, vol. 151, no. 107398, pp. 1–21, 2021. [Google Scholar]
47. P. Li and K. Mao, “Knowledge-oriented convolutional neural network for causal relation extraction from natural language texts,” Expert Systems with Applications, vol. 115, pp. 512–523, 2019. [Google Scholar]
48. S. Bayrak, E. Yucel and H. Takci, “Classification of extracranial and intracranial EEG signals by using finite impulse response filter through ensemble learning,” in Proc. of the Twenty Seventh Signals Processing and Communications Applications Conf., Sivas, Turkey, pp. 1–4, 2019. [Google Scholar]
49. T. M. Alam, K. Shaukat, I. A. Hameed, S. Luo, M. U. Sarwar et al., “An investigation of credit card default prediction in the imbalanced datasets,” IEEE Access, vol. 8, pp. 201173–201198, 2020. [Google Scholar]
50. Z. Shen, “Natural language processing (NLP) applications in patient care: A systematic analysis,” Quarterly Review of Business Disciplines, vol. 7, no. 3, pp. 223–244, 2020. [Google Scholar]
51. K. Shaukat, M. U. Hassan, N. Masood and A. B. Shafat, “Stop words elimination in urdu language using finite state automaton,” International Journal of Asian Language Processing, vol. 27, no. 1, pp. 21–32, 2017. [Google Scholar]
52. K. Shaukat, F. Iqbal, T. M. Alam, G. K. Aujla, L. Devnath et al., “The impact of artificial intelligence and robotics on the future employment opportunities,” Trends in Computer Science and Information Technology, vol. 5, no. 1, pp. 050–054, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |