DOI:10.32604/cmc.2022.019529

| Computers, Materials & Continua DOI:10.32604/cmc.2022.019529 |  |
| Article |
Optimized Convolutional Neural Network Models for Skin Lesion Classification
1Department of Electronics and Automation, Universidad Autónoma de Manizales, Manizales, 170001, Colombia
2SEDMATEC, Corporación Universitaria Autónoma de Nariño, Pasto, 520002, Colombia
3Department of Computer Science, Faculty of Computers and Information, South Valley University, Qena, 83523, Egypt
4Department of Computer Science, Universidad Autónoma de Manizales, Manizales, 170001, Colombia
5Department of Systems and Informatics, Universidad de Caldas, Manizales, 170001, Colombia
*Corresponding Author: Reinel Tabares-Soto. Email: rtabares@autonoma.edu.co
Received: 16 April 2021; Accepted: 14 June 2021
Abstract: Skin cancer is one of the most severe diseases, and medical imaging is among the main tools for cancer diagnosis. The images provide information on the evolutionary stage, size, and location of tumor lesions. This paper focuses on the classification of skin lesion images considering a framework of four experiments to analyze the classification performance of Convolutional Neural Networks (CNNs) in distinguishing different skin lesions. The CNNs are based on transfer learning, taking advantage of ImageNet weights. Accordingly, in each experiment, different workflow stages are tested, including data augmentation and fine-tuning optimization. Three CNN models based on DenseNet-201, Inception-ResNet-V2, and Inception-V3 are proposed and compared using the HAM10000 dataset. The results obtained by the three models demonstrate accuracies of 98%, 97%, and 96%, respectively. Finally, the best model is tested on the ISIC 2019 dataset showing an accuracy of 93%. The proposed methodology using CNN represents a helpful tool to accurately diagnose skin cancer disease.
Keywords: Deep learning; skin lesion; convolutional neural network; data augmentation; transfer learning
Cancer is one of the most important diseases because it is a leading cause of death before the age of 70 years in 112 of 183 countries, reducing life expectancy in every country [1]. In 2020, 19.3 million people worldwide were diagnosed with cancer and almost 10.0 million deaths occurred. These numbers are expected to increase to 24.6 million diseased people and 12.9 million deaths [2]. Skin cancer is the most diagnosed type of cancer; one out of every three diagnostics is skin cancer. There two main categories, namely, melanoma and nonmelanoma, which account for 324,635 and 1.2 million cases, respectively. Nonetheless, skin cancer could be prevented or successfully treated if countries develop efficient cancer prevention and early detection programs [3]. Even melanoma and keratinocyte carcinoma, the most aggressive skin cancers, including basal cell carcinoma and squamous cell carcinoma, can have a 5-year survival rate of 98% if diagnosed at an early stage [4,5]. Therefore, early diagnosis and treatment of skin cancer can minimize the damage caused. Nonetheless, its accurate recognition through skin lesion images is complex due to the similarity between different skin lesions and the limited number of dermatologists with professional knowledge [6,7]. Therefore, skin cancer identification based on image processing techniques and artificial intelligence applied to skin lesion images has become a serious scientific challenge.
Diagnosis and skin cancer detection based on image analysis have been traditionally performed by screening and visual inspection. However, these approaches depend on the dermatologist’s expertise and, consequently, the results are time-consuming, complex, subjective, and error-prone [8]. This fact is caused by the complex nature of the skin lesion images due to two factors: the characteristics of the lesions including texture, size, color, shape, and location; and the presence of multiple artifacts in the images, such as hair, veins, and charts of color calibration and ruler marks [9–11]. Furthermore, the skin cancer diagnostics’ paradigm is far from a binary problem (i.e., melanoma or nonmelanoma). Nowadays, the challenge includes multiple skin lesions that restrict specialized analyses, including Melanoma (Mel), Melanocytic nevus (Nv), Basal cell carcinoma (Bcc), Actinic Keratosis (Ak), Benign keratosis lesion (Bkl), Dermatofibroma (Df), Vascular lesion (Vasc), and Squamous cell carcinoma (Scc) [12]. Consequently, computer-aided diagnosis (CAD) systems become necessary for the preliminary diagnosis of different lesions.
CAD systems have employed traditional machine learning techniques for skin lesion image processing, following the conventional medical image analysis pipeline. This pipeline includes image preprocessing, image segmentation, feature extraction, and classification, where multiple techniques and approaches have been tested without achieving a successful performance [6,8]. The limited generalization capacity of current approaches can be attributed to the selection of a preprocessing approach, the high complexity of segmentation of the region of interest, the requirement of specific expertise to extract useful features related to physical skin lesion characteristics, and the low accuracy rate of the classical classifiers [6,9,10,13,14]. For these reasons, CAD systems are still human-dependent.
Recently, as a natural next step, computers have learned the features that optimally represent image characteristics, leading to the development of a new machine learning branch called deep learning [15,16]. The deep learning method can automatically mine the deep-seated nonlinear relationship in target images and does not need to establish feature estimation and extraction that are required in the traditional image recognition methods [17,18]. The first kind of deep learning model used for skin lesion image processing was the convolutional neural network (CNN). This architecture was demonstrated to exceed a dermatologist’s performance in distinguishing melanoma from non-melanoma [9,19,20]. Li et al. [6] provided an extensive review of CNN deep learning models and compared the most popular architectures such as AlexNet, VGG, GoogleNet, Inception, ResNet, DenseNet, and others. The most accurate model is based on residual learning and separable convolution, with an accuracy of 99.5% [21]. However, this accuracy was achieved for a binary problem.
When the skin lesion classification problem is treated as a multi-class problem, the CNN models require additional steps, either data augmentation [10,12,22–26] or an ensemble of classifiers [22,27–29], to reach an accuracy above 80%. Another deep learning technique named transfer learning improves the performance by taking advantage of previously trained architectures to fix former layers into the new deep learning model. Commonly, the model weights are obtained from ImageNet [30] and the transfer learning approach is used for two purposes: feature extraction [11,31] (while another approach performs the classification step); or direct classification [10,13,32,33]. Since transfer learning provides a base model setup, it provides the possibility to incorporate the optimization and fine-tuning process [34,35], which is an open issue. Hence, new models could improve the performance in skin cancer diagnostics.
In this paper, an optimization process of transfer learning models is proposed for multiple skin lesion classification. This was successfully addressed by using DenseNet-201, Inception-V3, and Inception-ResNet-V2 architectures and pre-training the weights of each model with ImageNet. Two datasets of skin lesion images, HAM10000 and ISIC 2019, are used to compare model performances. Since the datasets present class imbalance, we conducted four experiments: pre-trained models without data augmentation, without optimization, with data augmentation, and the proposed optimization. As a result, the convolutional layers added to the former transfer model improved the overall performance.
The paper is organized as follows. Section 2 presents the datasets and the proposed methodology. The experimental results are provided in Section 3, followed by a discussion of the results in Section 4. Finally, the paper is concluded in Section 5.
The methodology illustrated in Fig. 1 and developed for skin lesion image classification comprises the following stages: first, the number of images is increased by data augmentation; then, transfer learning is used for each tested model; and last, the model’s accuracy is obtained. The methodology consists of four different processing experiments, in which the feature space is treated as follows: i) the unbalanced dataset is composed of the raw images; ii) the dataset is balanced; iii) the transfer learning models are optimized for the unbalanced dataset, and iv) the parameter optimization stage and the balanced dataset were tested.
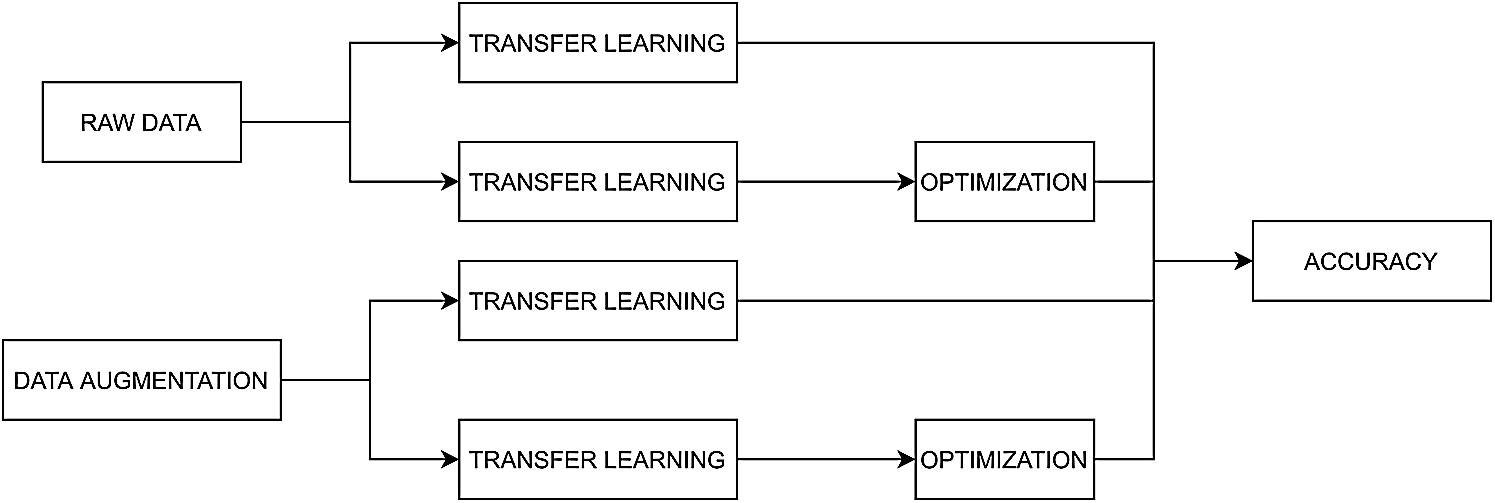
Figure 1: Diagram of the methods developed for skin lesion images classification
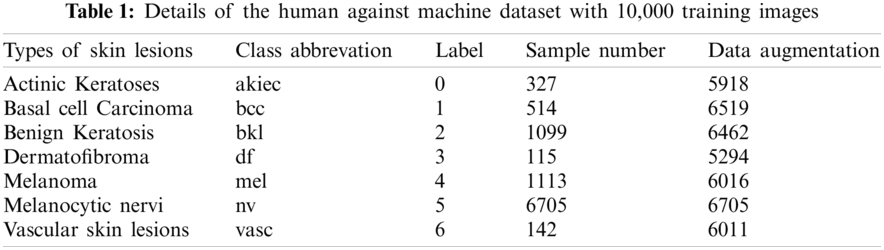
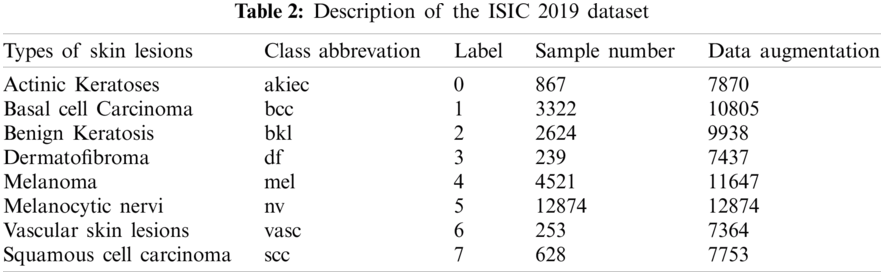
One of the used datasets was the Human Against Machine (HAM10000), an excellent series of multi-source dermatoscopic images of common pigmented skin lesions. This dataset is collected from different populations and stored by other modalities. It consists of 10015 dermatoscopic images released as a training set for academic machine learning purposes and is publicly available through the ISIC archive [36,37]. HAM10000 includes a representative collection of all essential diagnostic categories in the realm of pigmented lesions. As shown in Tab. 1, it includes seven skin lesion types, where label 5 (nv) is the most abundant, indicating that the labels are unbalanced. The other dataset was ISIC 2019 that contains the HAM10000 dataset. It comprises 25328 dermatoscopic images from many sites, applying different preprocessing methods. It contains images of the classes actinic keratosis (akiec), basal cell carcinoma (bcc), benign keratosis (bkl), dermatofibroma (df), melanoma (mel), melanocytic nevus (nv), vascular lesion (vasc), and squamous cell carcinoma (scc) [29,38]. This dataset has one more class than HAM10000, for a total of eight classes. Tab. 2 shows the number of images for each class in the ISIC 2019 dataset.
The images were preprocessed before the authors published them. All images were manually cropped based on the center of the lesion, and manual histogram corrections were applied to enhance visual contrast and color reproduction. Additionally, the authors manually selected the images with correct labeling and filtered them to eliminate disturbances such as jewelry and bubbles that alter the skin injury, as described in [36]. Several image samples for different skin lesion classes are shown in Fig. 2.
To ensure that the learning model is not affected by the class with the most images (nv), we increased the classes with fewer images for all datasets. For this purpose, we performed an analysis on each label and applied translations, random rotates, and other transformations using the Data generator Keras function to generate different images. For the HAM10000 dataset, we increased the dataset to a total of 42925. The image input size was transformed from 450 × 600 × 3 to 224 × 224 × 3 since it improves the transfer learning model design [24]. Additionally, the dataset was split into ≈ 75% training (32201 images), ≈ 17% validation (7150), and ≈ 8% testing (3574 images). For ISIC 2019, the dataset was augmented to 75688 images and split into ≈ 75% training (56774 images), ≈ 17% validation (12611 images), and ≈ 8% testing (6303 images). Furthermore, the image size was changed to 150 × 150 × 3 for the design of the transfer learning model since the number of images was higher and the size of each image was initially 767 × 1022 × 3. Image size reduction is used to decrease the training time, considering that previously the ImageNet weights were previously fixed.
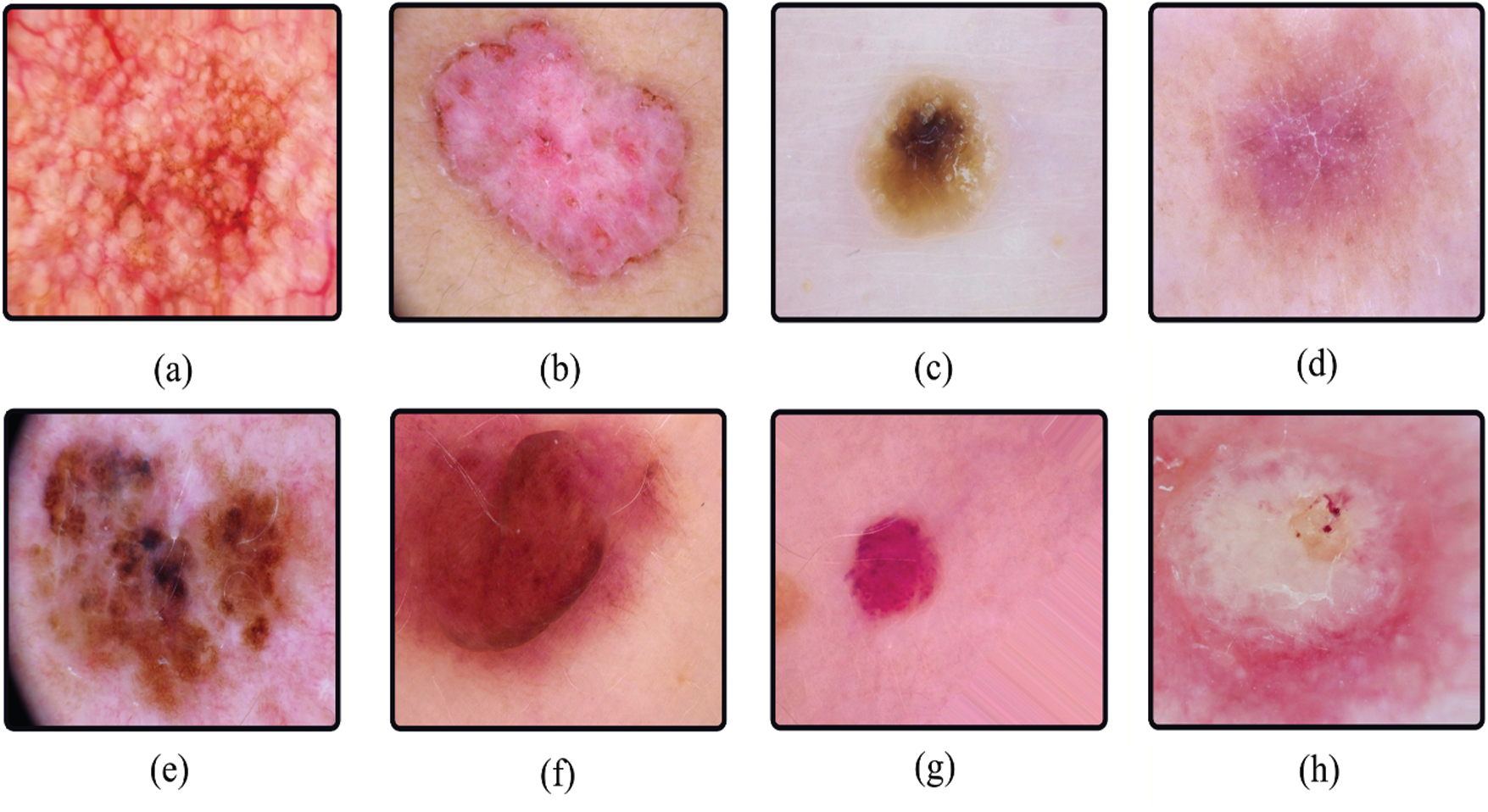
Figure 2: Sample images of each class in HAM10000 (a–g) and ISIC 2019 (a–h). (a) akiec, (b) bcc, (c) bkl, (d) df, (e) mel, (f) nv, (g) vasc, (h) sec
2.3 Transfer Learning and Architectures
Transfer learning is prevalent and beneficial because it considers a pre-trained architecture with weights. For this, datasets with millions of images are used. References [16,39] demonstrated that a pre-trained architecture is the best alternative for medical image analysis containing a priori information on the images. We used fine-tuning to modify some of the feature extraction layers by changing some network weights; the base weights were taken from ImageNet [30], then a new configuration of the dense layers was proposed. The fully connected layers are outlined in purple in Figs. 3–5.
The architectures of DenseNet-201, Inception-V3, and Inception-ResNet-V2 are used for transfer learning and displayed in (Figs. 3–5). There are different block colors in each figure that indicate the fine-tuned layers in red, the non-modified in blue, and the dense layers in purple. DenseNet was proposed to resolve the vanishing gradient problem since it preserves information through additive identity transformations, increasing its complexity. DenseNet uses layer-to-layer connectivity and connects each previous layer to the incoming layer in a feed-forward fashion. It utilizes dense blocks, and feature maps of all the last layers are used as inputs into all subsequent layers [8]. The best result provided by Keras on ImageNet is 0.936 using a specific DenseNet-201.
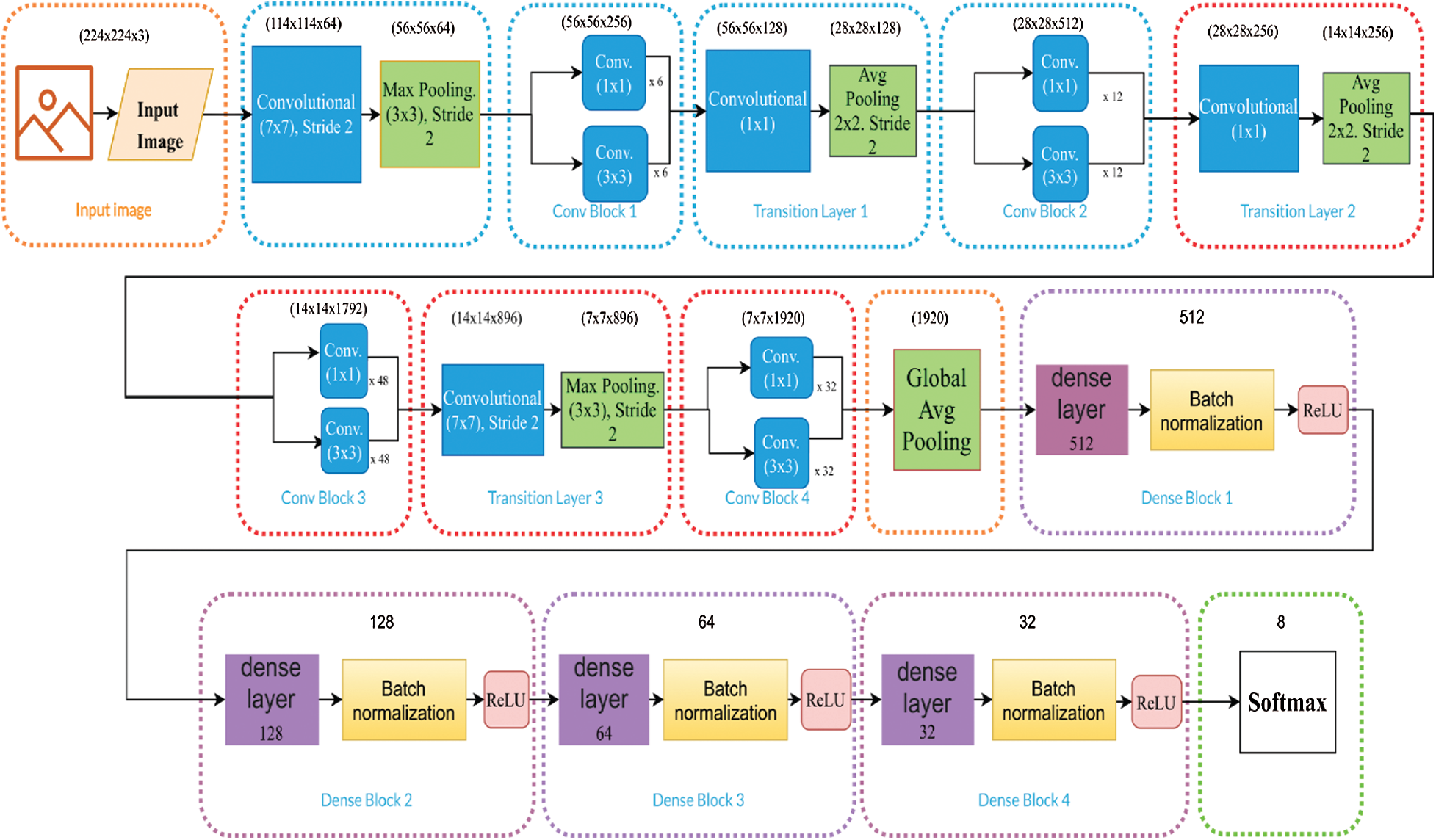
Figure 3: DenseNet-201 model optimization, where the number of hidden layers are 512, 128, 64, 32, and the ReLU function and a Softmax activation function are used

Figure 4: Inception-ResNet-V2 model optimization, where the number of hidden layers are 512, 128, 64, 32, and the ReLU function and a Softmax activation function are used

Figure 5: Inception-V3 model optimization, where the number of hidden layers are 512, 128, 64, 32, and the ReLU function and a Softmax activation function are used
Inception-ResNet-V2 uses a sophisticated architecture to retrieve essential features from the images. The initial layers of the network consist of standard convolutional layers followed by a maximum accumulation layer. The next stage simultaneously convolutes an entry using different filter sizes for each convolution and merges them. The following parts of the network repeat inceptions and residual 10 or 20 times, where the network uses desertion layers to make the filter values equal to 0 to prevent overfitting [40]. The best result provided by Keras on ImageNet is 0.953.
Inception-V3 CNN architecture is based on inception modules. A series of parallel convolutions with different kernel sizes are used for feature extraction. The input image is projected onto a sequence of convolutional and pooling layers; then, inception modules for feature extraction are stacked [22]. The activation function of all convolutions is ReLU. The classifier is developed with a dropout layer and softmax output layers to reduce overfitting. The best result provided by Keras on ImageNet is 0.937.
Optimization focused on fine-tuning each model to modify the network weights and selecting the convolutions to be modified. Then, an exhaustive search was performed to determine the appropriate number of hidden layers for the model's classification layers. Figs. 3–5 for DenseNet-201, Inception-ResNet-V2, and Inception-V3, respectively, show that the number of fully connected hidden layers are 512, 128, 64, 32. Here, we use the ReLU as a nonlinear activation function. We initially normalized the dataset by applying a division by 255 and converting it to float, which allows better learning of the model. Furthermore, we used batch normalization layers in the model's dense blocks between dense layers and the activation function, as outlined in the purple blocks in Figs. 3–5. This type of normalization modified its variance between 0 and 1, allowing deeper networks to converge more easily. Also, we determined the optimizer for the three chosen models, which consisted of analyzing optimizers such as Adam, Adamax, Adamgrad, SGD, among others provided by Keras. The output layer was set to the number of classes contained in each dataset. The classification blocks of each model, with a Softmax activation function, are outlined in purple in Figs. 3–5. The Learning Rate Scheduler function was employed to decrease the rate of change across time so the model learns slower and we avoid overfitting. The model was trained for an initial rate of 0.001 and 30 epochs.
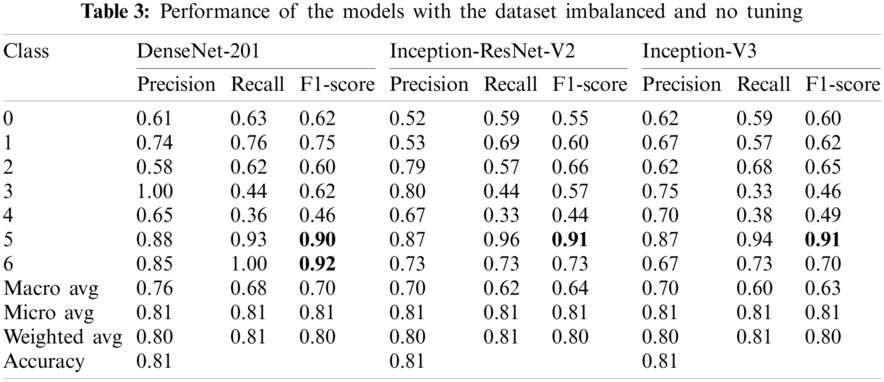
In this work, the Google Collaboratory platform based on Python was used, which is available to work with GPU in the cloud. For all experiments, three commonly used transfer learning models were selected to solve the classification task of skin lesion images: DenseNet-201 [11], Inception-ResNet-V2 [41], Inception-V3 [32]. As explained in Section 2, we applied models without tuning the raw images, and their model performance is shown in Tab. 3. The models with the balanced dataset have a high performance compared to previous experiments, as shown in Tab. 4. Parameter tuning was performed as described in Section 2.4, and the learning rates and classification accuracy are shown in Tab. 5. We found that the best optimizer is Adamax for all models; therefore, this configuration was used for experiments 3 and 4. Experiment 3 tested the models with the tuned parameters and the imbalanced dataset (see Tab. 6), while experiment 4 used both a balanced dataset and tuned transfer learning models (Tab. 7). As shown in Tab. 7, it is possible to identify each specific label accurately, and there were cases in which the model ultimately distinguished a label from the others.




The models are stable and error-free during the training and validation stages. Fig. 6 shows the evolution of accuracy and loss of the training and validation processes for experiment 4. The confusion matrix shown in Fig. 7 helps to observe the errors that the models must identify in some classes. The ROC curve represents true positives and false positives, allowing us to distinguish the success rate achieved by our model for the dataset of seven classes as shown in Fig. 8.

Figure 6: Evolution of training and validation for (a) accuracy and (b) loss of the proposed models
To validate the high performance obtained on the HAM10000 dataset, we used the optimized DenseNet-201 transfer learning model to classify the eight classes of the ISIC 2019 dataset, including data augmentation (see Tab. 8).
Fig. 9 shows the evolution of accuracy and loss of training and validation tasks using the ISIC2019 dataset. Furthermore, it displays the confusion matrix and ROC curves with confidence intervals at 95% of confidentiality.
In this work, four experiments were performed to demonstrate the capacity of CNNs to classify different skin cancer images from two relevant datasets. In these experiments, a robust study was conducted to show the reliability of the results obtained based on multiple metrics such as ROC curves with AUC by class with confidence intervals, Precision, Recall, F1-Score, Accuracy, confusion matrices, and training curves of accuracy and loss. The best results were achieved by DenseNet-201, obtaining 98% accuracy on the 2018 dataset and 93% on the 2019 dataset. Concerning the first experiment (Tab. 3), using models without tuning, the performance varied according to the number of images per class. The only exception was class 6 (vasc) on model DenseNet-201, which showed the highest F1 score. Class 5 (nv) contained the highest number of images and showed a high and similar performance across all models in this experiment.
The second experiment (Tab. 4) corresponded to the use of non-optimized transfer learning models with the balanced dataset. A data augmentation stage in essential to achieve high performance. As shown in this experiment, the number of augmented images depends on the difference between each class and class 5 (nv). Classes 0 (akiec), 1 (bcc), 3 (df), and 6 (vasc) initially contained the fewest images (Tab. 1), and displayed a remarkable increase in precision, recall, and F1 score, compared to the results shown in Tab. 3. Moreover, class 5 (nv) showed a minor increase in performance and was the only class without added images. The third experiment tested the models using the tuned parameters and the imbalanced dataset. In comparison to the results shown in Tab. 4, the performance for each model increased by one percentage point with the tuned the parameters as shown in Tab. 6. Finally, the fourth experiment used a balanced dataset and tuned transfer learning models (Tab. 7). For this experiment, label 6 (vasc) obtained a percentage of accuracy, recall, and F1-Score of 100%. Overall, promising results were obtained, which may help to diagnose the skin lesion correctly. Fig. 6 shows that the DenseNet-201 model obtains the lowest losses in its validation stage and this is reflected by its accuracy since it is the best performing. Both Inception-ResNet-V2 and Inception-V3 have similar behaviors for accuracy and loss.
In general, class 4 (mel) is the lowest-performing, followed by 2 (bkl) and 5 (nv). Moreover, the classifier tends to confuse classes 2, 4, and 5, as shown in Fig. 7. As a result, an average accuracy of 98% is reached for all classes using DenseNet-201 (the best performing model). Furthermore, Inception-ResNet-V2, with 97% average accuracy, and Inception-V3, with 96%, present the lowest-performance for classes 2 (bkl) and 4 (mel) as mentioned above in Fig. 8. Tab. 8 shows the different metric values for the best model, indicating that the proposed method achieves high performance for all classes with an overall accuracy of 93%. Moreover, class 3 (df) is the lowest-performing, and classes 2 (bkl), 6 (vasc) are the highest-performing. DenseNet-201 model obtains low loss in its validation stage and high performance (see Fig. 9). However, this model achieves a maximum accuracy and lowest loss for the training set in very few epochs. The confusion matrix shows that class 3 (df) is the most difficult to classify, followed by class 1 (bcc) (Fig. 9). However, with HAM10000, the classification of class 3 showed one of the highest accuracies. The ROC curves show the proposed model's high performance for each of the classes, as seen in Fig. 9 using 95% of confidentiality. The AUC obtained shows the capacity of the methodological approach to classify skin lesions using deep learning models based on transfer learning and hyper-parameters optimization.

Figure 7: Confusion matrices of the proposed transfer learning models, where classes 0 to 6 represent ‘akiec’, ‘bcc’, ‘bkl’, ‘df’, ‘mel’, ‘nv’ and ’vasc’, respectively
A comparison with state-of-the-art models that were applied to HAM10000 is depicted in Tab. 9. The best accuracy results are obtained with our methodology, “optimized DenseNet-201 including data augmentation”. The improvements differ between 3% to 8% from other models of different authors. Concerning the ISIC 2019 dataset (see Tab. 10), our results do not outperform the state-of-the-art methods. The best model reported is “1D fractal and DenseNet-201 that uses features for ensemble classifiers like k-NN and SVMs.” The network proposed here is the best obtained for the HAM10000 dataset (DenseNet-201 optimized). However, for the ISIC 2019 dataset, different techniques, such ensembles of CNNs, 1D fractal can be applied and classification with traditional machine learning models, such as k-NN and SVM can be performed.

Figure 8: ROC Curves of the proposed transfer learning models with a confidence interval of 95%. Note that classes 0 to 6 represent ‘akiec’, ‘bcc’, ‘bkl’, ‘df’, ‘mel’, ‘nv’, and ‘vasc’, respectively


Figure 9: Evolution of training and validation for (a) accuracy, (b) loss, and (c) confusion matrix and (d) ROC curves of the proposed DenseNet-201 model on dataset ISIC 2019. Confidence intervals are presented at 95%


In this work, a total of four experiments for the classification of skin lesions were described. We showed the use of transfer learning, hyper-parameter optimization, and data augmentation to improve the ability to identify skin lesions. Three novel models in transfer learning were used namely, DenseNet-201, Inception-ResNet-V2, and Inception-V3. The first model achieved the best performance under different metrics. The last experiment incorporated all the techniques and allowed obtaining accuracy values up to 98% on the HAM10000 dataset and 93% on the ISIC 2019 dataset. In future studies, we aim to work more in-depth on the ISIC 2019 dataset by testing more complex CNNs architectures. Additionally, we propose to design new architectures and computational elements of CNNs to detect skin lesions, using preprocessing techniques such as contrast enhancement and segmentation to provide better information as input to the CNNs to improve detection accuracy.
Funding Statement: This research is supported by the Universidad Autónoma de Manizales, Manizales, Colombia under project No. 589-089.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram et al., “Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries,” CA: A Cancer Journal for Clinicians, vol. 71, no. 3, pp. 209–249, 2021. [Google Scholar]
2. C. Ferlay, J. Colombet, M. Soerjomataram, I. Mathers, D. Parkin et al., “Global cancer observatory: Cancer tomorrow. Lyon, France: International Agency for Research on Cancer, 2020. [Online]. Available: https://gco.iarc.fr/tomorrow [Accessed: 30-May-2021]. [Google Scholar]
3. M. Tougaçar, Z. Cömert and B. Ergen, “Intelligent skin cancer detection applying autoencoder, MobileNetV2 and spiking neural networks,” Chaos, Solitons & Fractals, vol. 144, pp. 110714, 2021. [Google Scholar]
4. P. Fontanillas, B. Alipanahi, N. A. Furlotte, M. Johnson, C. H. Wilson et al., “Disease risk scores for skin cancers,” Nature Communications, vol. 12, no. 160, pp. 1–13, 2021. [Google Scholar]
5. K. Urban, S. Mehrmal, P. Uppal, R. L. Giesey and G. R. Delost, “The global burden of skin cancer: A longitudinal analysis from the global burden of disease study, 1990–2017,” JAAD International, vol. 2, no. 1, pp. 98–108, 2021. [Google Scholar]
6. L.-F. Li, X. Wang, W. J. Hu, N. N. Xiong, Y. X. Du et al., “Deep learning in skin disease image recognition: A review,” IEEE Access, vol. 8, no. 1, pp. 208264–208280, 2020. [Google Scholar]
7. L. Faes, S. K. Wagner, D. J. Fu, X. Liu, E. Korot et al., “Automated deep learning design for medical image classification by health-care professionals with no coding experience: A feasibility study,” The Lancet Digital Health, vol. 1, no. 5, pp. 232–242, 2019. [Google Scholar]
8. A. Adegun and S. Viriri, “Deep learning techniques for skin lesion analysis and melanoma cancer detection: A survey of state-of-the-art,” Artificial Intelligence Review, vol. 54, no. 1, pp. 811–841, 2020. [Google Scholar]
9. A. Esteva, B. Kuprel, R. A. Novoa, J. Ko, S. M. Swetter et al., “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, no. 7639, pp. 115–118, 2017. [Google Scholar]
10. K. M. Hosny, M. A. Kassem and M. M. Foaud, “Classification of skin lesions using transfer learning and augmentation with Alex-net,” PLOS One, vol. 14, no. 5, pp. e0217293, 2019. [Google Scholar]
11. E. O. Molina, S. Solorza and J. Álvarez, “Classification of dermoscopy skin lesion color-images using fractal-deep learning features,” Applied Sciences, vol. 10, no. 17, pp. 5954, 2020. [Google Scholar]
12. I. Iqbal, M. Younus, K. Walayat, M. U. Kakar and J. Ma, “Automated multi-class classification of skin lesions through deep convolutional neural network with dermoscopic images,” Computerized Medical Imaging and Graphics, vol. 88, no. 1, pp. 101843, 2021. [Google Scholar]
13. K. Thurnhofer and E. Dominguez, “A convolutional neural network framework for accurate skin cancer detection,” Neural Processing Letters, vol. 1, no. 1, pp. 1–21, 2020. [Google Scholar]
14. R. Tabares-Soto, S. Orozco-Arias, V. Romero-Cano, V. S. Bucheli, J. L. Rodríguez-Sotelo et al., “A comparative study of machine learning and deep learning algorithms to classify cancer types based on microarray gene expression data,” PeerJ Computer Science, vol. 2020, no. 4, pp. 1–22, 2020. [Google Scholar]
15. G. Litjens, T. Kooi, B. E. Bejnordi, A. A. A. Setio, F. Ciompi et al., “A survey on deep learning in medical image analysis,” Medical Image Analysis, vol. 42, no. December 2012, pp. 60–88, 2017. [Google Scholar]
16. M. A. Bravo, H. B. Arteaga, R. Tabares, J. I. Padilla and S. Orozco, “Cervical cancer classification using convolutional neural networks, transfer learning and data augmentation,” EIA Magazine, vol. 18, no. 35, pp. 1–12, 2021. [Google Scholar]
17. M. Hassaballah and A. I. Awad, Deep Learning in Computer Vision: Principles and Applications, 1st ed., Boca Raton, FL: CRC Press, 2020. [Google Scholar]
18. V. Sze, Y. H. Chen, T. J. Yang and J. S. Emer, “Efficient processing of deep neural networks,” Synthesis Lectures on Computer Architecture, vol. 15, no. 2, pp. 1–341, 2020. [Google Scholar]
19. V. Dick, C. Sinz, M. Mittlböck, H. Kittler and P. Tschandl, “Accuracy of computer-aided diagnosis of melanoma a meta-analysis,” JAMA Dermatology, vol. 155, no. 11, pp. 1291–1299, 2019. [Google Scholar]
20. R. C. Maron, M. Weichenthal, J. S. Utikal, A. Hekler, C. Berking et al., “Systematic outperformance of 112 dermatologists in multiclass skin cancer image classification by convolutional neural networks,” European Journal of Cancer, vol. 119, no. 1, pp. 57–65, 2019. [Google Scholar]
21. R. Sarkar, C. C. Chatterjee and A. Hazra, “Diagnosis of melanoma from dermoscopic images using a deep depthwise separable residual convolutional network,” IET Image Processing, vol. 13, no. 12, pp. 2130–2142, 2019. [Google Scholar]
22. A. H. Shahin, A. Kamal and M. A. Elattar, “Deep ensemble learning for skin lesion classification from dermoscopic images,” in Proc. CIBEC, Cairo, Egypt, pp. 150–153, 2018. [Google Scholar]
23. S. Kaymak, P. Esmaili and A. Serener, “Deep learning for two-step clas-sification of malignant pigmented skin lesions,” in 14th IEEE Sym. on Neural Networks and Applications, Serbia and Montenegro (S&Mpp. 1–6, 2018. [Google Scholar]
24. E. H. Mohamed and W. H. Behaidy, “Enhanced skin lesions classification using deep convolutional networks,” in Proc. ICICIS, Cairo, Egypt, pp. 180–188, 2019. [Google Scholar]
25. H. Nahata and S. P. Singh, “Deep learning solutions for skin cancer detection and diagnosis,” Machine Learning with Health Care Perspective, vol. 13, no. 1, pp. 159–182, 2020. [Google Scholar]
26. N. Gessert, T. Sentker, F. Madesta, R. Schmitz, H. Kniep et al., “Skin lesion diagnosis using ensembles, unscaled multi-crop evaluation and loss weighting,” arXiv, vol. 1, no. 1, pp. 1–5, 2018. [Google Scholar]
27. A. G. C. Pacheco, A. R. Ali and T. Trappenberg, “Skin cancer detection based on deep learning and entropy to detect outlier samples,” arXiv, vol. 1, no. 1, pp. 1–6, 2019. [Google Scholar]
28. S. A. A. Ahmed, B. Yanikouglu, Ö. Göksu and E. Aptoula, “Skin lesion classification with deep CNN ensembles,” in Proc. SIU, Gaziantep, Turkey1, pp. 1–4, 2020. [Google Scholar]
29. N. Gessert, M. Nielsen, M. Shaikh, R. Werner and A. Schlaefer, “Skin lesion classification using ensembles of multi-resolution EfficientNets with meta data,” MethodsX, vol. 7, no. 1, pp. 100864, 2020. [Google Scholar]
30. O. Russakovsky, J. Deng, H. Su, J. Krause, S. Satheesh et al., “ImageNet large scale visual recognition challenge,” International Journal of Computer Vision, vol. 115, no. 3, pp. 211–252, 2015. [Google Scholar]
31. M. A. Khan, M. Y. Javed, M. Sharif, T. Saba and A. Rehman, “Multi-model deep neural network based features extraction and optimal selection approach for skin lesion classification,” in Proc. ICCIS, Sakaka, Saudi Arabia, pp. 1–7, 2019. [Google Scholar]
32. V. Sai, N. R. G. Bhavya, M. Ramya, S. Y. Sujana and T. Anuradha, “Classification of skin cancer images using TensorFlow and inception v3,” International Journal of Engineering & Technology, vol. 7, pp. 717–721, 2018. [Google Scholar]
33. D. Moldovan, “Transfer learning based method for two-step skin cancer images classification,” in Proc. EHB, Iasi, Romania, pp. 1–4, 2019. [Google Scholar]
34. A. M. Alqudah, H. Alquraan and I. A. Qasmieh, “Segmented and non-segmented skin lesions classification using transfer learning and adaptive moment learning rate technique using pretrained convolutional neural network,” Journal of Biomimetics, Biomaterials and Biomedical Engineering, vol. 42, no. 1, pp. 67–78, 2019. [Google Scholar]
35. M. A. Kassem, K. M. Hosny and M. M. Fouad, “Skin lesions classification into eight classes for ISIC, 2019 using deep convolutional neural network and transfer learning,” IEEE Access, vol. 8, no. 1, pp. 114822–114832, 2020. [Google Scholar]
36. P. Tschandl, C. Rosendahl and H. Kittler, “The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions,” Scientific Data, vol. 5, no. 1, pp. 180161, 2018. [Google Scholar]
37. N. Codella, V. Rotemberg, P. Tschandl, M. E. Celebi, S. Dusza et al., “Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration,” arXiv, pp. 1–12, 2019. [Google Scholar]
38. T. D. Tô, D. T. Lan, T. T. H. Nguyen, T. T. N. Nguyen, H. P. Nguyen et al., “Ensembled skin cancer classification (ISIC 2019 challenge submission),” HAL Archives-Ouvertes, no. 1, pp. 1–7, 2019. [Google Scholar]
39. N. Tajbakhsh, J. Y. Shin, S. R. Gurudu, R. T. Hurst, C. B. Kendall et al., “Convolutional neural networks for medical image analysis: Full training or fine tuning,” IEEE Transactions on Medical Imaging, vol. 35, no. 5, pp. 1299–1312, 2016. [Google Scholar]
40. Y. Bhatia, A. Bajpayee, D. Raghuvanshi and H. Mittal, “Image captioning using google’s inception-resnet-v2 and recurrent neural network,” in 2019 Twelth Int. Conf. on Contemporary Computing, Noida, India, pp. 1–6, 2019. [Google Scholar]
41. A. Rezvantalab, H. Safigholi and S. Karimijeshni, “Dermatologist level dermoscopy skin cancer classification using different deep learning convolutional neural networks algorithms,” arXiv, vol. 1, no. 1, pp. 1–15, 2018. [Google Scholar]
42. H. Benbrahim, H. Hachimi and A. Amine, “Deep convolutional neural network with TensorFlow and Keras to classify skin cancer images,” Scalable Computing: Practice and Experience, vol. 21, no. 3, pp. 379–390, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |