DOI:10.32604/cmc.2022.019076

| Computers, Materials & Continua DOI:10.32604/cmc.2022.019076 |  |
| Article |
Ensembles of Deep Learning Framework for Stomach Abnormalities Classification
Department of Artificial Intelligence, Faculty of Computer Science and Information Technology, Universiti Malaya, 50603 Kuala Lumpur, Malaysia
*Corresponding Author: Chu Kiong Loo. Email: ckloo.um@um.edu.my
Received: 01 April 2021; Accepted: 18 June 2021
Abstract: Abnormalities of the gastrointestinal tract are widespread worldwide today. Generally, an effective way to diagnose these life-threatening diseases is based on endoscopy, which comprises a vast number of images. However, the main challenge in this area is that the process is time-consuming and fatiguing for a gastroenterologist to examine every image in the set. Thus, this led to the rise of studies on designing AI-based systems to assist physicians in the diagnosis. In several medical imaging tasks, deep learning methods, especially convolutional neural networks (CNNs), have contributed to the state-of-the-art outcomes, where the complicated nonlinear relation between target classes and data can be learned and not limit to hand-crafted features. On the other hand, hyperparameters are commonly set manually, which may take a long time and leave the risk of non-optimal hyperparameters for classification. An effective tool for tuning optimal hyperparameters of deep CNN is Bayesian optimization. However, due to the complexity of the CNN, the network can be regarded as a black-box model where the information stored within it is hard to interpret. Hence, Explainable Artificial Intelligence (XAI) techniques are applied to overcome this issue by interpreting the decisions of the CNNs in such wise the physicians can trust. To play an essential role in real-time medical diagnosis, CNN-based models need to be accurate and interpretable, while the uncertainty must be handled. Therefore, a novel method comprising of three phases is proposed to classify these life-threatening diseases. At first, hyperparameter tuning is performed using Bayesian optimization for two state-of-the-art deep CNNs, and then Darknet53 and InceptionV3 features are extracted from these fine-tunned models. Secondly, XAI techniques are used to interpret which part of the images CNN takes for feature extraction. At last, the features are fused, and uncertainties are handled by selecting entropy-based features. The experimental results show that the proposed method outperforms existing methods by achieving an accuracy of 97% based on a Bayesian optimized Support Vector Machine classifier.
Keywords: Gastrointestinal tract; deep learning; Bayesian optimization hyperparameters; explainable AI; uncertainty handling; feature fusion
Gastrointestinal tract (GIT) diseases are nowadays becoming a common disease worldwide, and approximately 2.8 million new cases have been diagnosed in recent years [1,2]. Even worse, 1.8 million deaths occur every year due to esophageal, colorectal, and stomach cancer [3]. Since 2017, there have been 765,000 people dying of stomach cancer, and a global analysis reveals that colon cancer has caused 525,000 deaths, making it one of the most prevalent forms of cancer in the United States of America [4]. To start treating numerous GIT diseases, endoscopy and wireless capsule endoscopy are the basis for diagnosing stomach cancer, as shown in Fig. 1. In some cases, the infected region of the GIT may be captured only in one or two frames, and sometimes it may be overlooked by the doctors resulting in an incorrect diagnosis [5]. Moreover, some abnormalities may be too small to be easily detected by the naked eye. Furthermore, different physicians may have distinct findings when they analyze the same image [6]. In the early detection of stomach disorders, computerized automated detection systems for stomach infections from endoscopic images were developed by several researchers. By detecting these life-threatening gastric infections early on, the mortality rate of patients as shown in Fig. 2 can be decreased. Advances in technology, especially computer vision techniques, make artificial intelligence (AI)-based systems more feasible [7] to aid gastroenterologists with their diagnosis [8]. Over the decades, research on artificial intelligence has continued to reveal its efficiency in specific fields of medical imaging [9]. Thus, in the domain of medical image processing and analysis, the application of deep learning is a hot topic nowadays [10,11].
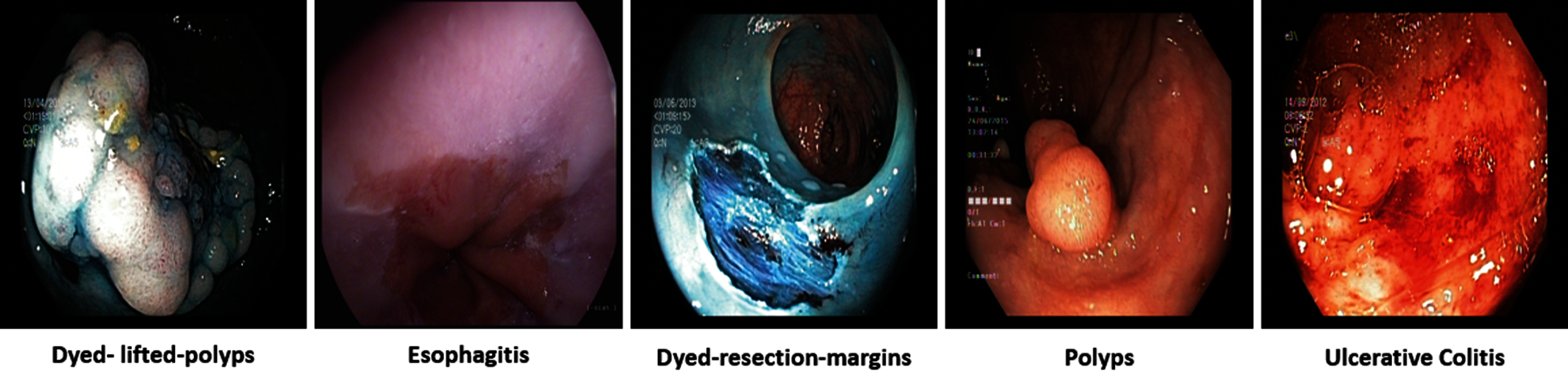
Figure 1: GI tract abnormalities
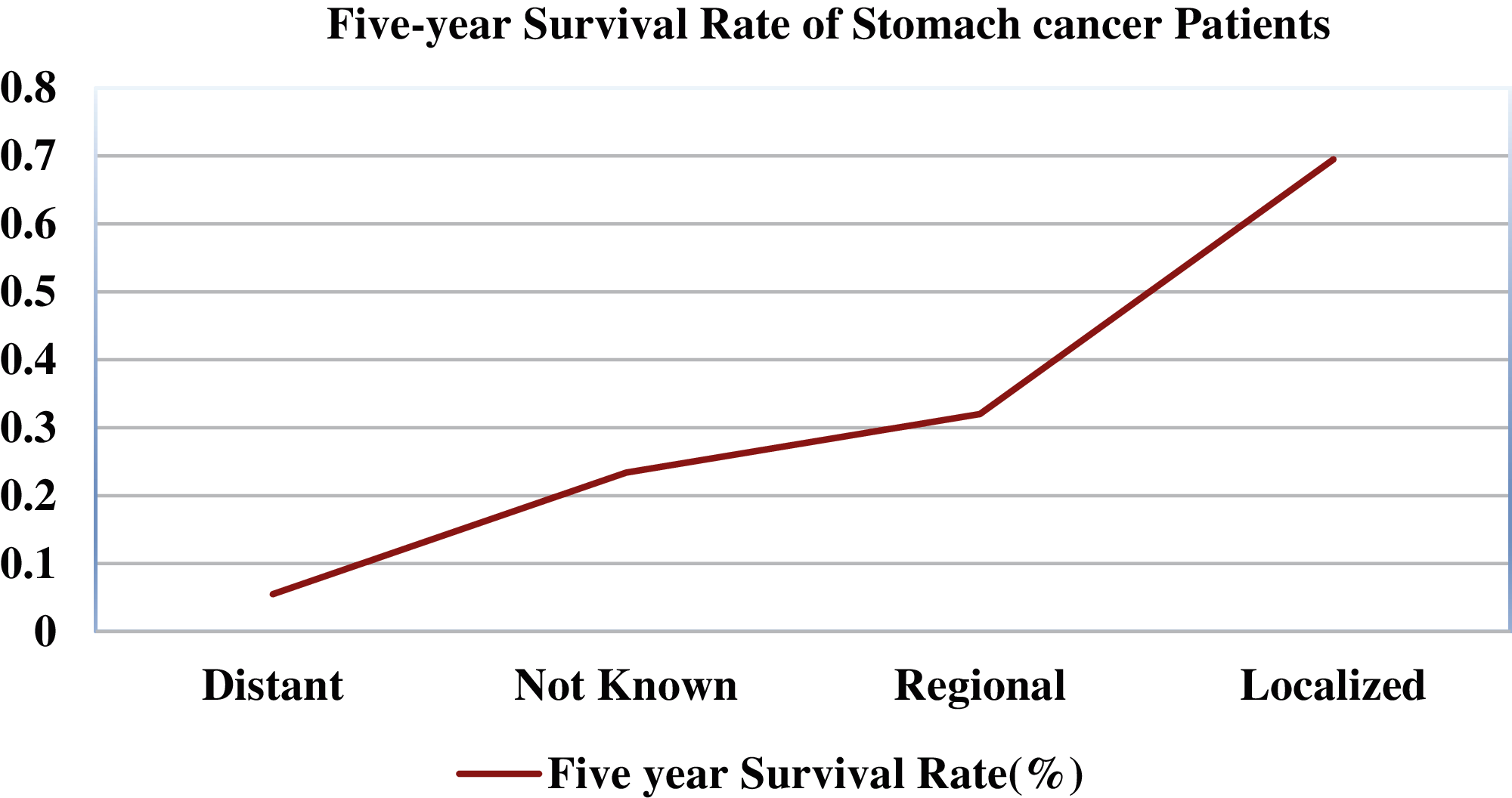
Figure 2: Five-year survival rate of stomach cancer patients relating to cancer stage
As a vigorous technique of artificial intelligence, deep learning can be used for medical disease classification, due to its potential to enhance the skills of health practitioners in the early detection of these diseases. As a form of deep learning, Convolutional Neural Network (CNN) can be applied to extract deep high-level features from a single image. CNN can convert input data into images, signals, multi-dimensional data, and videos, among other formats. Input, convolutional, activation, pooling, fully-connected (FC), and output layers make up a basic CNN model, where low-level features are extracted from the former layers in CNN and high-level features are extracted from deeper layers, and then features are vectorized in the FC layer for final classification. Several deep CNN models are pretrained on thousands of images from ImageNet and can be extended to medical early detection, such as stomach abnormalities and other diseases, through transfer learning. Meanwhile, hyperparameters have a key role in influencing the training behavior of deep CNN. An effective approach to decrease the challenge of hyperparameter tuning is Bayesian optimization, which can be utilized for the global optimization of black-box functions [12]. Generally, CNN acts like a black box, where the internal mechanisms and the result generation process remain elusive in terms of how they produce output data predictions.
Furthermore, deep learning methods have obtained human-like outcomes on several tasks in medical image analysis and diagnosis [13,14]. Explainable AI (XAI) has also been the subject of several studies in general, but it has yet to be extensively explored in the field of medical imaging. The importance of interpretability in the medical field is discussed by [15]. Deep learning approaches have mainly two kinds of uncertainties: aleatoric uncertainty and epistemic uncertainty. The former measures the noise inherent during data generation, while the latter measures the uncertainty in the parameters of the model [16]. Fine-tuning the hundreds of thousands of CNN's trainable parameters requires vast amounts of data, which may arise epistemic uncertainty. Handling the information on uncertainty can improve the trust in deep-learning approaches that are frequently viewed as ‘black-box’ methods, making them more suitable for medical diagnosis. And estimates of uncertainty can increase the speed of the analysis process because, as opposed to reviewing the whole images for diseases not identified by the CNN, medical professionals may spend their time evaluating the most uncertain regions. Finally, both medical imaging functions and other safety-critical activities can be addressed by uncertainty handling approaches. When monitoring the experiment result, the uncertainty information can be measured via entropy, whose framework has been successfully applied in multiple areas, including statistics, mathematics, and information theory. Also, the selection of entropy-based features approach is presented in [17]. To allow the users to individually review the basis of their decisions, the United States Food and Drug Administration needs the application to justify its clinical opinions because these systems are lacking interpretability and difficult to trust their reliability.
Besides, the hyperparameter optimization problem requires recognizing a hyperparameter that produces a precise methodology in a timely manner. However, the evaluation of CNNs takes a long time and requires tremendous computing power [18]. Furthermore, researchers have limited insight into the impact of each hyperparameter on the model's performance, which results in extremely vast boundaries and a much larger space than required. Hence, testing each possible hyperparameter set of a CNN model can be computationally expensive [19]. In the field of medical imaging, it is necessary to explain the decisions of deep learning when validating the results of a model and to handle uncertainties associated with deep features when incorporating AI-based medical diagnosis systems. Meanwhile, deep learning models have millions of neurons, whose structure is too complex to explain how CNN regards certain images as different diseases, which may generate the assumption that the models are untrustworthy. In addition, it may cause a variety of concerns because it is hard to tell whether the justifications behind the findings are ill-formatted or even inaccurate.
It should be noted that in previous research, the existing CNN techniques of GI tract abnormality classification have not involved hyperparameter optimization in training or XAI in CNN decision interpretation. Also, they did not focus on uncertainty handling. We are the first to integrate them into GIT abnormality classification. In this paper, we propose a novel deep CNN-based methodology to solve these challenges, which can reliably infer the multiple GI tract diseases, and our key contributions are as follows:
i. A novel deep CNN-based Bayesian optimized and Explainable Gastrointestinal Net (BX-GI Net) is proposed to classify GI tract diseases by hyperparameter optimization, human-understandable visual explanations, and uncertainty handling.
ii. After Bayesian optimization of hyperparameters, deep features are extracted from two pretrained deep CNN models, i.e., Darknet53 [20] and Inception-V3 [21], and then XAI methods, i.e., LIME [22] and Grad-CAM [23], are applied to interpret how CNN extracts deep features to gain the trust of medical professionals.
iii. By selecting entropy-based optimized features from the pool of features, the uncertainty associated with extracted deep features of both CNN models is conducted to provide a black-box-free deep learning-based diseases diagnosis system.
In previous literature, researchers of the computer vision domain presented many techniques for detecting and classifying GIT diseases. More recently, deep learning has increased the performance of many fields like medical imaging, object classification, and many others. Akram et al. [24] proposed a fully automated method for stomach disease classification from Wireless Capsule Endoscopy (WCE) images based on DenseNet, where a color-based saliency method was developed for ulcer detection, with the top 50% features selected by Tsallis entropy-based features optimization heuristically. These selected features were classified through a multi-layer neural network and attain an accuracy of 99.5%. Fan et al. [25] employed a deep learning-based method for erosion and ulcer detection from WCE images, which includes thousands of ulcer images. An Alexnet model was utilized to direct extract features from original images instead of preprocessing data and segmentation. After that, an evaluation of the WCE database was performed, and an accuracy of 95.16% was obtained. Diamantis et al. [26] presented a fully connected framework for detecting various gastric modalities, such as polyps, bleeding, and ulcers, from WCE images. In [27], two CNN models were utilized for feature extraction, and then these features were fused through the Euclidean fisher vector and reduced through the entropy-based approach. In the end, the geometric features were combined with reduced features for final classification. Alaskar et al. [28] proposed a deep learning-based method for classification ulcers from WCE images. They combined the feature information of two CNN models, namely AlexNet and GoogleNet, for the classification of ulcer and non-ulcer regions.
In recent years, the application of CNN for automated disease detection has grown significantly in images and videos of the GI tract. Manually tuning hyperparameters can increase the possibility of improving the classification performance with the best configurations. However, it is time-consuming. Therefore, it is necessary to have an automatic hyperparameter strategy to improve outcomes without manual intervention. A CNN-based method [29] is proposed using hyperparameters tuning based on Bayesian optimization for the diagnosis of COVID-19 in X-ray images. In [30], hyperparameters of machine learning models like Support Vector Machine (SVM) and k-nearest neighbors (KNN) are also optimized by Bayesian optimization to classify COVID-19 images. In a number of areas, including image recognition, object detection, etc., deep CNNs have proven very effective, but the drawback is that due to their black-box nature, it is difficult for human beings, particularly a layman, to understand how deep CNN decides. Several methods have been proposed to explain CNN's decisions and determine what features induce the network to produce a specific prediction by visualizing the network's inner layers. To increase trustworthiness in the decision-making process, uncertainty is also important to be included as an external insight into point prediction. Whereas the softmax regularly found at the end of a CNN is often interpreted as model confidence, which is usually not an ideal example. For the uncertainty estimation of deep learning, a test time dropout approach [31] is presented by Monte Carlo (MC) samples of the prediction. The MC sample variance is evaluated for image-based diabetic retinopathy diagnosis [32] and demonstrates this context is beneficial. Besides, epistemic uncertainty in COVID-19 classification from CT and X-ray images using deep learning features is calculated by entropy. Several studies have suggested approaches to resolve the lack of transparency in deep learning models. Similarly, these techniques need to be applied in the deep learning-based diagnosis of GI tract diseases [33].
In this research, a novel human-understandable automated classification system for GI tract diseases is proposed based on endoscopic images. The proposed method validates the feature extracted portion of the image by comprising the Bayesian optimization of CNN hyperparameters, the deep features extraction after fine-tuning, and the visualization of CNN's layers by XAI techniques. The uncertainty handling of deep features is conducted by selecting entropy-based best features from the pool of fused features. Selected features are then fed to a Bayesian optimized classifier for the classification of GI tract diseases. Thus, the name of this proposed CNN-based architecture is BX-GI Net: Bayesian optimized and Explainable Gastrointestinal Net. A detailed illustration of the proposed method is shown in Fig. 3, and each step is explained as follows.
In our experiment, endoscopic images were acquired to validate the proposed methodology described above. For this purpose, a benchmark dataset Kvasir [34] with multiclass gastrointestinal tract diseases was utilized, consisting of eight classes named esophagitis, dyed lifted polyps, dyed resection margins, polyps, ulcerative colitis, normal cecum, normal-pylorus, and normal-z-line.
3.2 The Bayesian Optimization of CNN Hyperparameters
The optimization of hyperparameters is a critical challenge, especially in the context of medical diagnosis using artificial intelligence. Hyperparameter tuning is intended to achieve the best results by searching for the best response of hyperparameters for a deep learning algorithm, and automatically tunning of CNN hyperparameters is referred to as a black box because its objective function is unknown. A plethora of hyperparameter tunning techniques exists in the previous literature, where Bayesian optimization, as an outstanding approach in machine learning [35], tackles an unknown objective function and involves an estimation using existing prior knowledge just the same as posterior probability. A probability model is set up to represent the performance to obtain certain values. Then the most suitable values of hyperparameter are chosen based on these probabilities to determine the objective function. In mathematical form, it can be represented as:
where N is considered search space,
In this research, the initial learning rate's most important hyperparameter of two CNN models, i.e., Darknet53 and Inception-V3, is optimized using Bayesian optimization to gain the benefits from transfer learning.
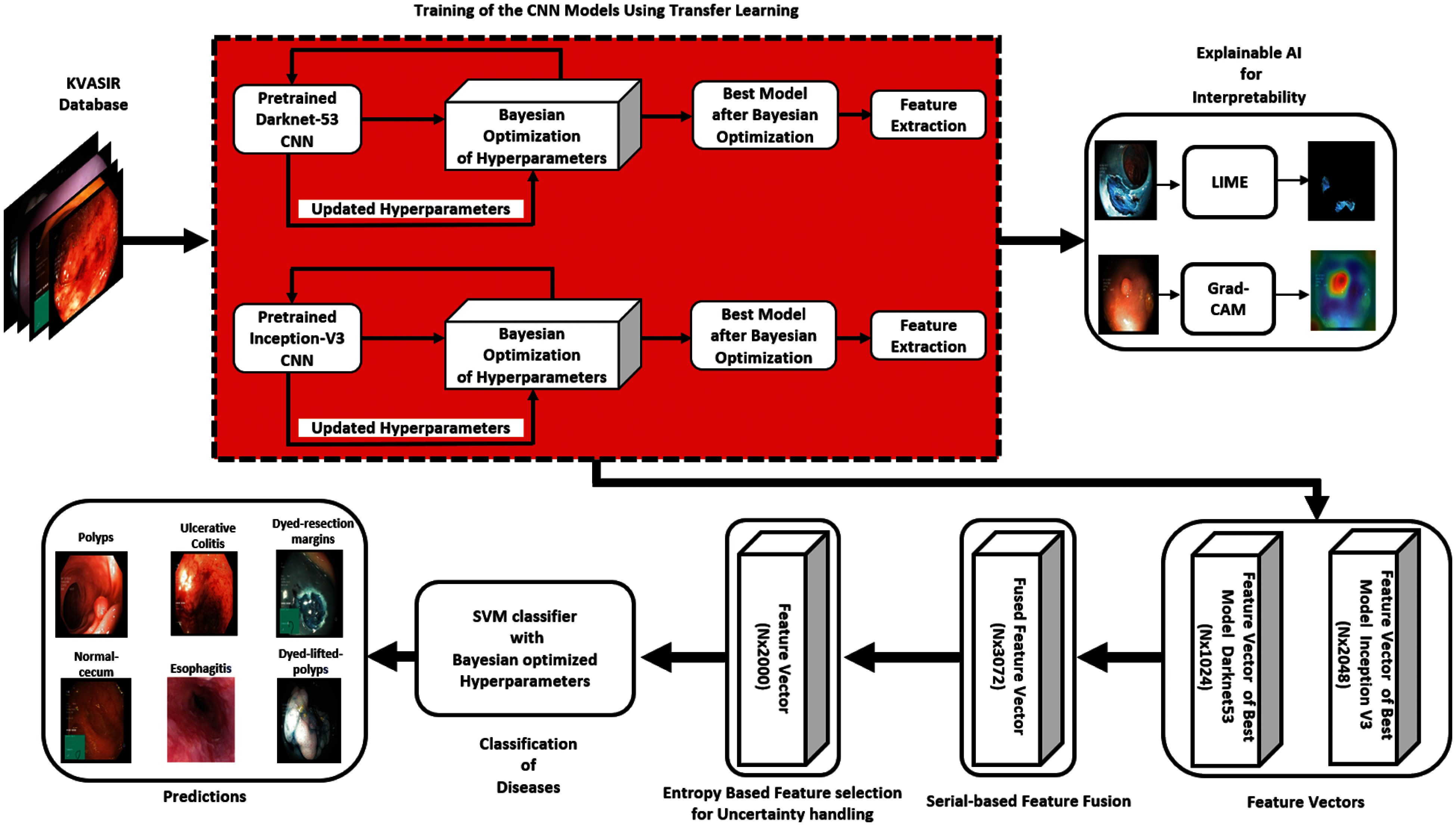
Figure 3: Proposed architecture diagram
In the last few years, significant development has been demonstrated in deep CNNs. Generally, along with a classifier, a deep model is the integration of low-level, mid-level, and high-level elements. The levels of features are enriched by multiple stacked layers in deep learning models. Two pretrained deep CNN models, Darknet53 and Inception-V3, we fine-tuned on the above-mentioned dataset using Bayesian optimization of hyperparameters, then deep-feature extraction took place at the last Global Average Pooling layers of both nets.
In the health care sector, artificial intelligence is generally used for dealing with the issue of transparency and explainability of CNN's decisions to answer questions like why physicians should trust this model's prediction. It is important for medical staff to fully believe that deep learning tools can assist them with the early diagnosis because it is able to produce an explanation justifying why the algorithm gives a certain prediction or regardless of how precise a model is. Different from other models, such as decision trees, which are explainable, the existing state-of-the-art AI models in healthcare applications are based on neural networks, which are black boxes and lack the explainability for their predictions, especially in high-risk circumstances, such as medical diagnosis.
3.4.1 Local Interpretable Model-Agnostic Explanations (LIME)
LIME generates explanations by splitting an image into superpixels. The clusters of pixels can provide local contextual details of different parts from the image with similar features, textures, and colors. A distribution of perturbed images is created by selectively hiding superpixels. An impact of perturbations on the correct probabilities of class prediction is calculated, and a linear model with this data is trained. Each superpixel is then provided the relevance against classification with weight values, where positive values indicate the effect on accurate classification, but rather negative values are within the opposite direction. As mentioned in [36], the arithmetically LIME can be explained as:
The model
3.4.2 Gradient Weighted Class Activation Mapping (Grad-CAM)
Grad-CAM is an XAI technique that can be implemented to any Deep CNN without any network adjustment. In comparison, it is a generalized form of class activation mapping (CAM) that involves a deep CNN network consisting of a stack of convolution layers, accompanied by softmax, activation, and global average pooling. It is applied to visualize the attention of each layer to understand the layer-wise feature map relevance. When the input and class are given, it generates a heat map of the activation class, whose color represents a class activation map related to the specified class image. It uses gradient knowledge flows from CNN's last convolution layer to allocate relevance to every neuron for a specific prediction. As explained by [23], to obtain the Grad-CAM of any class c,
When backpropagating gradients of activations, an actual calculation
3.5 Uncertainty Handling Using Entropy
Entropy is a measure of a system's amount of uncertainty. When monitoring the result of an experiment, the information of uncertainty based on entropy can be measured, whose framework can be used in multiple areas, including statistics, mathematics, and information theory.
At first, a serial-based fusion is conducted from extracted feature vectors of Darknet53 and Inception-V3. Assume the
To handle the uncertainty associated with extracted deep features, the entropy-based feature selection is performed on the fused vector with the dimension of
At last, the final fused vector of
The detailed experimental results of the proposed methodology are discussed in this section. A benchmark dataset Kvasir of GI tract endoscopy is utilized for experiments collected by endoscopic instruments from Vestre Viken Health Trust (VV), Norway. The dataset comprises images, annotations, and the diagnosis confirmed by health professionals experienced endoscopists, including several classes showing pathological findings, endoscopic procedures, and anatomical landmarks in the GI tract, and each class has thousands of images. The number of images is adequate for various tasks like deep learning, machine learning, image retrieval, etc. The Bayesian optimization of hyperparameters is performed at the state-of-the-art deep CNN models InceptionV3 and Darknet-53 using transfer learning, and the ratio of training and testing is 50:50 for both deep CNN models. The 10-fold cross-validation is carried out in the training and the testing of classifiers. Multiple classifiers like Fine KNN, Fine Gaussian SVM, Fine Tree, and Medium KNN are used to equate the findings with a Bayesian optimized SVM classifier. The following metrics are considered for performance evaluation: recall, precision, F1 score, the area under the curve (AUC), false positive rate (FPR), false negative rate (FNR), time, and accuracy. For simulation purposes, MATLAB R2020b Deep Learning Toolbox is used, and the hardware comprises Core-i5 desktop, 6 GB NVIDIA GPU, and 16 GB of RAM.
4.1 Experimental Results and XAI Visualizations
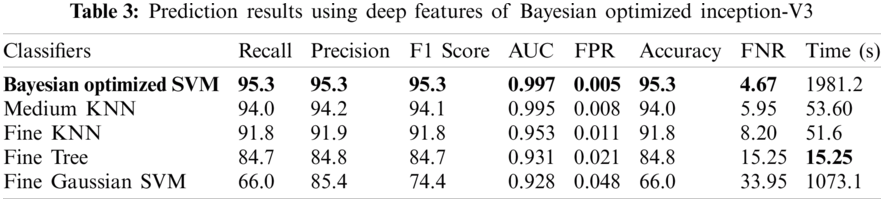
Numerical results for all experiments are discussed in this section in detail. At first, the hyperparameter initial learning rate of both pretrained deep CNNs Darknet53 and Inception V3 is optimized using Bayesian optimization. After the optimization process, the best models for both deep CNNs are selected. The objective function for optimization Darknet-53 achieves after 20 iterations. After that, the Bayesian optimized learning rate of 0.00028129 is considered the best. Meanwhile, for inception-V3, it achieves in 17 iterations with a learning rate of 0.0010421 as the best. And as mentioned earlier, transfer learning is utilized for both the deep CNNs, so the number of epochs is set to 6. The optimizer utilized in this work is Stochastic Gradient Descent with momentum (SGDM) while training both CNNs. The best model parameters are used to validate the proposed methodology, and parameters attained during Bayesian optimization of Inception-V3 are shown in Tab. 1. Deep features are extracted using the Bayesian optimized best Darknet-53 model and then fed to different classifiers, including Bayesian optimized SVM and others, as mentioned in Tab. 2. Compared to others, the highest accuracy achieves 94% by the Bayesian optimized SVM classifier, and the false negative rate (FNR) is 6.05, the area under the curve (AUC) is 0.997, recall, precision rate, F1 score are 93.9, and the execution time is 10149 s. But in terms of execution time, the Fine Tree classifier outperforms over all other classifiers, as shown in Tab. 2. Fig. 4 shows the visual results of XAI technique LIME and indicates which part of the image is considered by Darknet-53 for feature extraction.
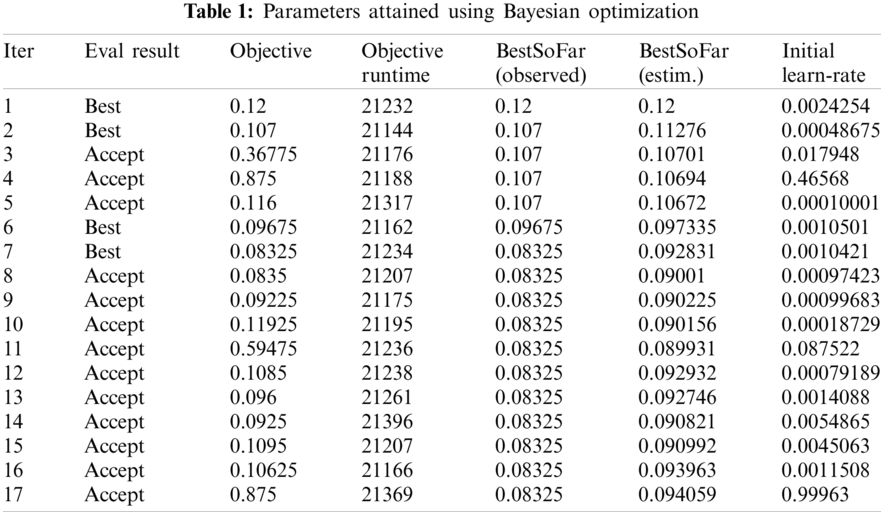
The results in Tab. 2 are taken from the classification of GI tract diseases using Bayesian optimized Inception-V3 deep features. Again, the Bayesian optimized SVM classifier outperforms in the classification by securing the accuracy of 95.3%, FNR, AUC, recall, precision rate, F1 score, and execution time are presented in the table. From the prescriptive of execution time, the Fine Tree achieves the minimum time to classify. Grad-CAM explanations for Inception V3 are shown in Fig. 5, making the proposed methodology trustful for the medical practitioners to incorporate AI-based disease diagnoses system in practical medical applications.
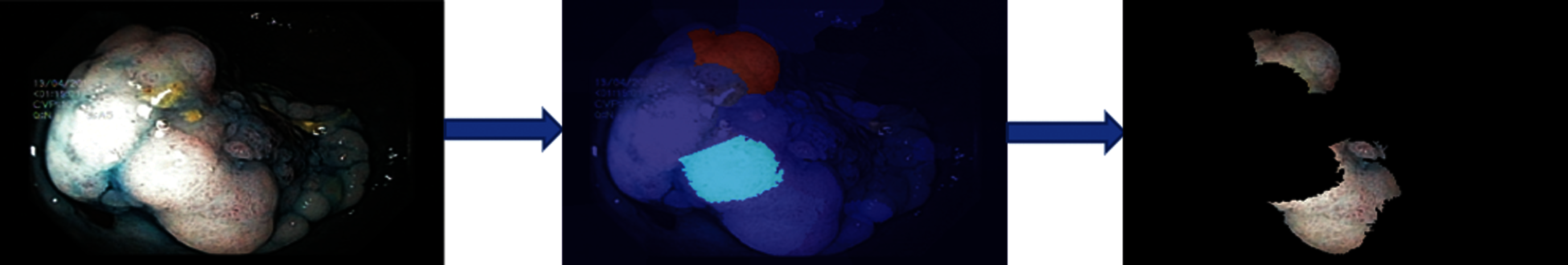
Figure 4: Showing explanations of XAI technique LIME on Bayesian optimized Darknet-53
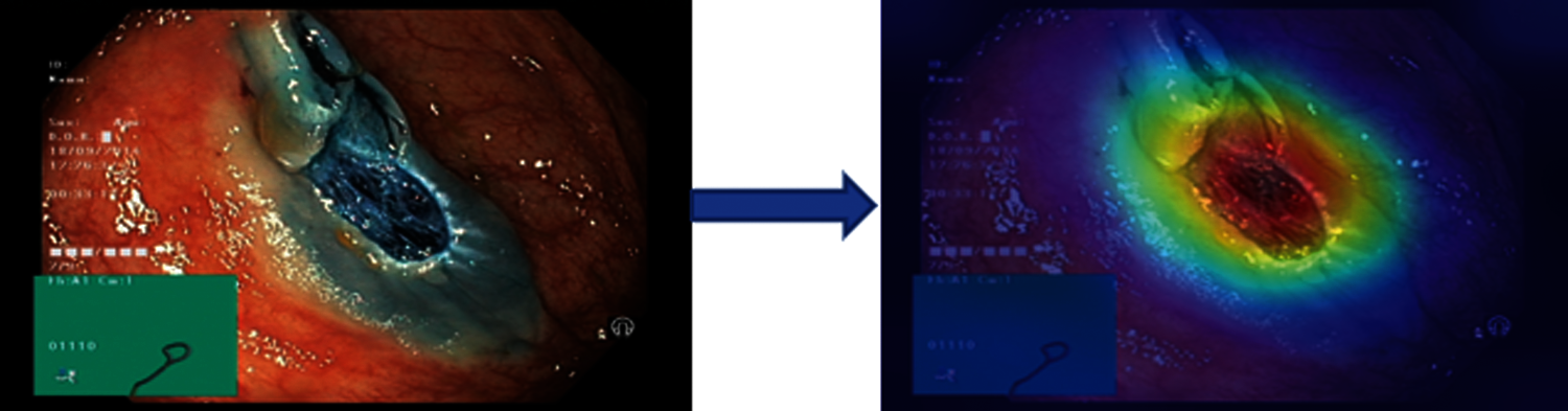
Figure 5: Showing explanations of XAI technique grad-CAM technique on inception-V3
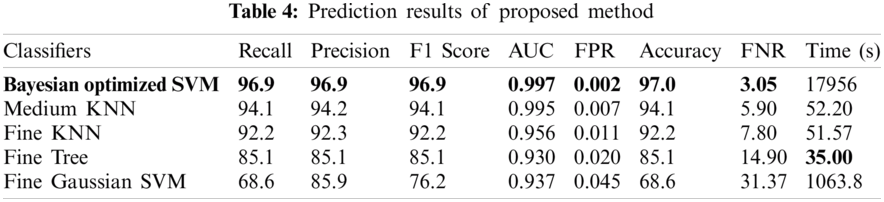
In Tab. 4, the results of the proposed methodology are presented, obtained by the fusion of both vectors, and then the uncertainty associated with deep features is handled by the reduction of uncertain features. Two thousand best-optimized entropy-based features are selected and then fed into classifiers for classification. The maximum accuracy of 97.0% is achieved by Bayesian optimized SVM, which shows the increase in accuracy as compared to both Bayesian optimized Inception-V3 and Darknet-53. During the optimization process, the following hyperparameters of SVM are considered for optimization: multiclass method, box constraint level, kernel scale, kernel function, and standardize data. The minimum classification error plot and the best hyperparameters values are shown in Fig. 5. In the proposed work, the FNR for Bayesian optimized SVM is just 3.05%, and the highest one is 31.37% for Fine Gaussian SVM. From the prescriptive time, the Fine Tree's result is only in 35.0 s, but the prediction accuracy is 85.1%, which is low as compared to Bayesian optimized SVM. The confusion matrix of the proposed work for cubic SVM is presented in Fig. 6, which depicts that the class normal-pylorus has achieved a higher correct prediction accuracy of 99.4%. Based on these findings, it is obvious that the Bayesian optimized SVM provides better results than other classifiers.
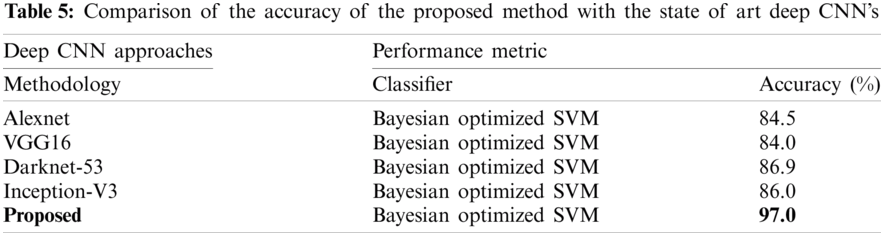
The proposed method is discussed in detail in this section. The proposed architecture contains several key modules, and each module needs to be thoroughly evaluated. At first, the results are obtained for Bayesian optimized Inception-V3 and Darknet53 features, but the accuracy is not sufficient to compare with existing methodologies. Therefore, to remove the uncertainty associated with the extracted features and optimized results, the fusion of both optimized feature vectors, and then the entropy-based feature selection is proposed. As shown in Tab. 4, the accuracy increases to 97.0%. The accuracy given in this table instead of that in Tabs. 2 and 3 has increased. Furthermore, the comparison of the proposed method with the state-of-the-art deep CNNs, like Inception-V3, Alexnet, VGG16, etc., is conducted, and results are shown in Tab. 5. This experiment's aim is to determine the reliability of this proposed novel BX-GI Net for the classification of GI tract diseases, Tab. 5 depicts that Alexnet achieves the accuracy of 84.5%, VGG16 achieves 84.0%, Darknet53 achieves 86.9%, and Inception-V3 achieves 86.0%, but the proposed framework achieves the accuracy of 97.0%. This clearly indicates that the proposed approach outperforms the above-mentioned deep CNNs.
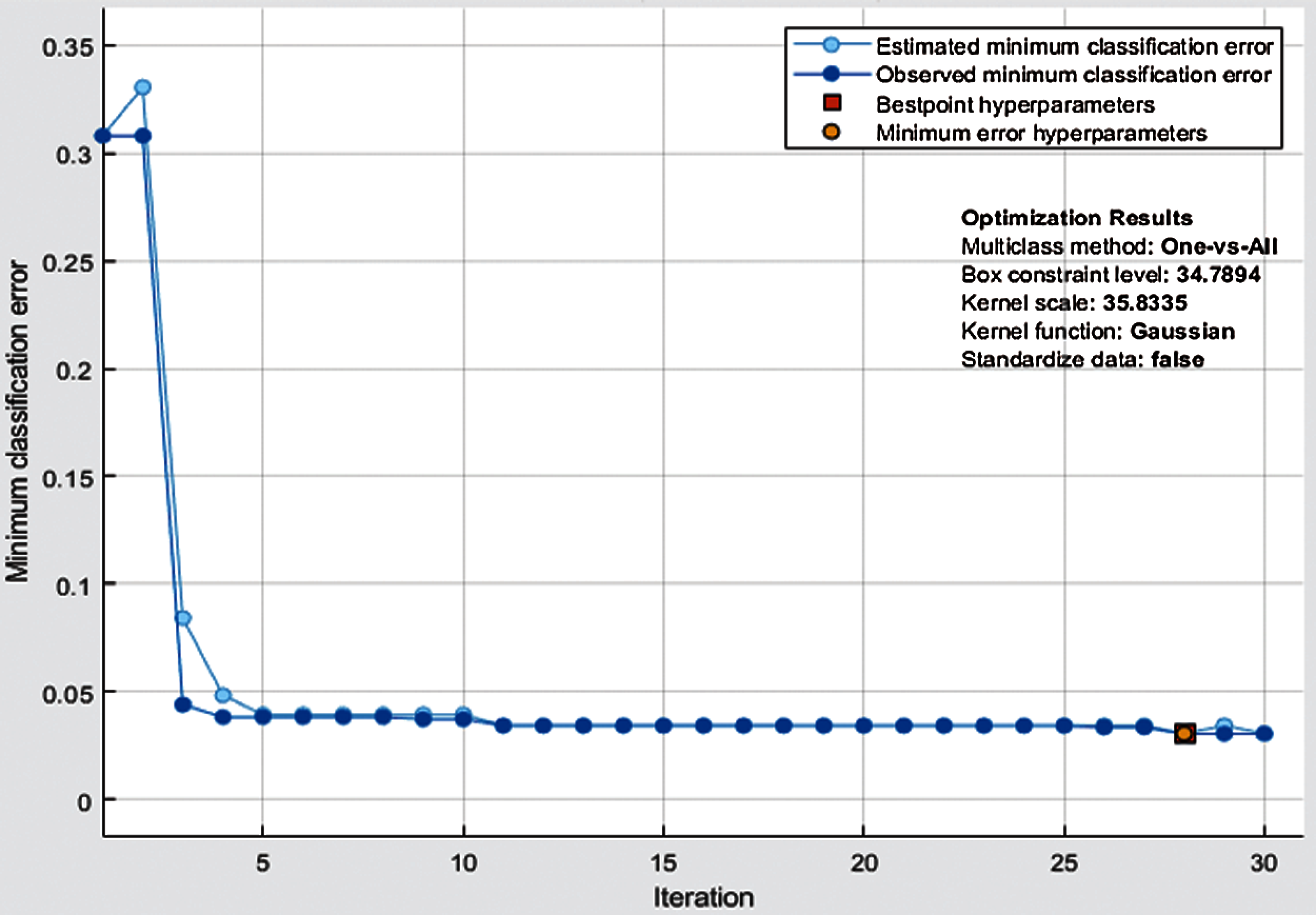
Figure 6: Minimum classification error plot of Bayesian-optimized SVM
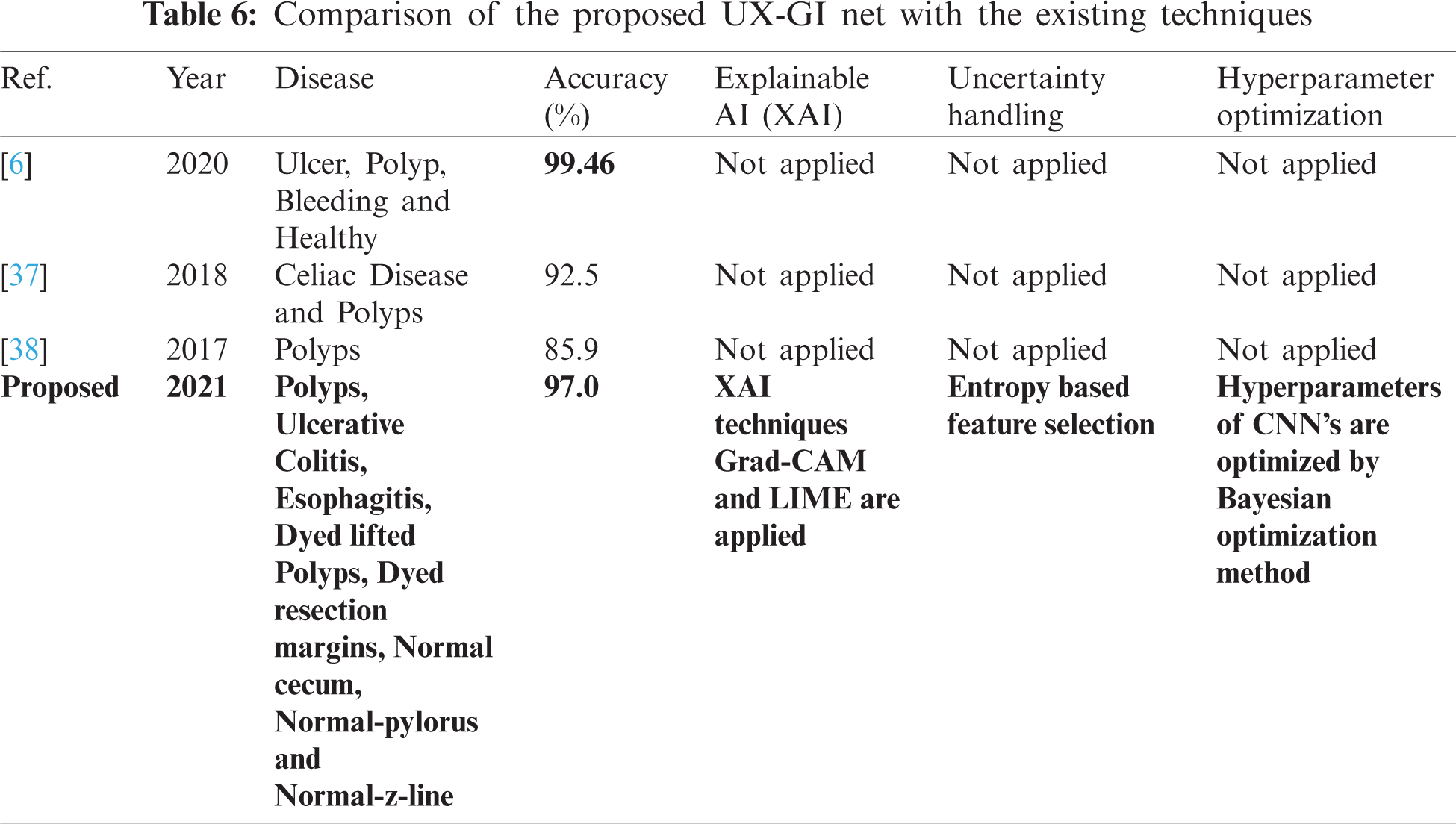
A few related published techniques are compared in Tab. 6, based on accuracy, Bayesian optimization of hyperparameters, XAI, and uncertainty handling. In [6], the author proposed a deep CNN features-based technique and afterward selected optimal features through DE evolutionary algorithm. It enhanced the crow search ECSA algorithm then fused the features using Max Correlation, yet the accuracy achieved higher than the proposed method. But there are no XAI techniques used to validate which part of images the Deep CNN is taking for feature extraction. Furthermore, hyperparameters of CNN are not optimized. In addition, uncertainty associated with characteristics, which is an integral part of any medical disease detection method for effective diagnosis to avoid life-threatening conditions, is also not discussed. But in terms of XAI and uncertainty handling, our proposed approach considers this analysis. Researchers in [37,38] also presented related deep CNN approaches without any hyperparameter optimization technique or uncertainty handling, so the accuracy is less than our proposed method. It shows that our proposed method proves to be more effective as compared to these published approaches.
In this research, a novel framework, namely BX-GI Net, based on deep CNN is proposed to classify GI tract diseases. The research demonstrates that a deep CNN-based system can achieve optimizable results using hyperparameter optimization. And their decisions can be interpretable and explainable by using XAI. Two state-of-the-art pretrained CNN models’ performance is improved by tuning the hyperparameters with Bayesian optimization. XAI techniques, i.e., Grad-CAM and LIME, are implemented to visualize which part of images is considered by deep CNNs for feature extraction. The uncertainty associated with deep features is also handled using entropy-based feature selection to avoid uncertain predictions. The proposed method achieves an accuracy of 97.0% for the classification of GI tract diseases from endoscopic images, which outperforms existing methods. Our results conclude that Bayesian optimization is the best method to select optimal hyperparameters for dealing with deep Convolutional Neural Networks (deep CNNs). Also, entropy is one of the useful techniques for handling uncertainty associated with deep features. The main limitation of this work is the complexity of the model. Also, the imbalanced datasets make the training process more difficult.
Acknowledgement: We would like to thank TopEdit (https://www.topeditsci.com) for its linguistic assistance during the preparation of this manuscript.
Funding Statement: This research was supported by the Universiti Malaya Impact-oriented Interdisciplinary Research Grant Programme (IIRG)-IIRG002C-19HWB, Universiti Malaya Covid-19 Related Special Research Grant (UMCSRG) CSRG008-2020ST and Partnership Grant (RK012-2019) from University of Malaya.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. S. Kadry, M. Alhaisoni, Y. Nam, Y. Zhang, V. Rajinikanth et al., “Computer-aided gastrointestinal diseases analysis from wireless capsule endoscopy: A framework of best features selection,” IEEE Access, vol. 8, pp. 132850–132859, 2020.
2. A. Liaqat, M. Sharif, M. Mittal, T. Saba, K. S. Manic et al., “Gastric tract infections detection and classification from wireless capsule endoscopy using computer vision techniques: A review,” Current Medical Imaging, vol. 4, pp. 1–31, 2020.
3. A. Majid, N. Hussain, M. Alhaisoni, Y. D. Zhang, S. Kadry et al., “Multiclass stomach diseases classification using deep learning features optimization,” Computers, Materials and Continua, vol. 67, pp. 3381–3399, 2021.
4. R. L. Siegel, K. D. Miller and A. Jemal, “Cancer statistics, 2019,” CA: A Cancer Journal for Clinicians, vol. 69, pp. 7–34, 2019.
5. A. Majid, M. Yasmin, A. Rehman, A. Yousafzai and U. Tariq, “Classification of stomach infections: A paradigm of convolutional neural network along with classical features fusion and selection,” Microscopy Research and Technique, vol. 83, pp. 562–576, 2020.
6. M. S. Sarfraz, M. Alhaisoni, A. A. Albesher, S. Wang and I. Ashraf, “Stomachnet: Optimal deep learning features fusion for stomach abnormalities classification,” IEEE Access, vol. 8, pp. 197969–197981, 2020.
7. M. A. Khan, F. Ahmed, M. Mittal, L. M. Goyal, D. J. Hemanth et al., “Gastrointestinal diseases segmentation and classification based on duo-deep architectures,” Pattern Recognition Letters, vol. 131, pp. 193–204, 2020.
8. J. Naz, M. Sharif, M. Yasmin, M. Raza and M. A. Khan, “Detection and classification of gastrointestinal diseases using machine learning,” Current Medical Imaging, vol. 1, pp. 1–27, 2020.
9. I. M. Nasir, M. Sharif, M. Alhaisoni, S. Kadry, S. A. C. Bukhari et al., “A blockchain based framework for stomach abnormalities recognition,” Computers, Materials and Continua, vol. 67, pp. 141–158, 2021.
10. N. Hussain, A. Majid, M. Alhaisoni, S. A. C. Bukhari, S. Kadry et al., “Classification of positive COVID-19 CT scans using deep learning,” Computers, Materials and Continua, vol. 66, pp. 1–16, 2021.
11. T. Akram and Y. D. Zhang, “Attributes based skin lesion detection and recognition: A mask RCNN and transfer learning-based deep learning framework,” Pattern Recognition Letters, vol. 143, pp. 58–66, 2021.
12. G. Litjens, T. Kooi, B. E. Bejnordi, A. A. A. Setio, F. Ciompi et al., “A survey on deep learning in medical image analysis,” Medical Image Analysis, vol. 42, pp. 60–88, 2017.
13. I. Ashraf, M. Alhaisoni, R. Damaševičius, R. Scherer, A. Rehman et al., “Multimodal brain tumor classification using deep learning and robust feature selection: A machine learning application for radiologists,” Diagnostics, vol. 10, pp. 565, 2020.
14. A. Rehman, T. Saba, Z. Mehmood, U. Tariq and N. Ayesha, “Microscopic brain tumor detection and classification using 3D CNN and feature selection architecture,” Microscopy Research and Technique, vol. 84, pp. 133–149, 2021.
15. A. Adadi and M. Berrada, “Peeking inside the black-box: A survey on explainable artificial intelligence (XAI),” IEEE Access, vol. 6, pp. 52138–52160, 2018.
16. P. Natekar, A. Kori and G. Krishnamurthi, “Demystifying brain tumor segmentation networks: Interpretability and uncertainty analysis,” Frontiers in Computational Neuroscience, vol. 14, pp. 6, 2020.
17. T. W. Chang, Y. P. Huang and F. E. Sandnes, “Efficient entropy-based features selection for image retrieval,” in 2009 IEEE Int. Conf. on Systems, Man and Cybernetics, Las Vegas, USA, pp. 2941–2946, 2009.
18. M. I. Sharif, M. A. Khan, M. Alhussein et al., “A decision support system for multimodal brain tumor classification using deep learning,” Complex & Intelligent Systems, 2021. https://doi.org/10.1007/s40747-021-00321-0.
19. E. Bochinski, T. Senst and T. Sikora, “Hyper-parameter optimization for convolutional neural network committees based on evolutionary algorithms,” in 2017 IEEE Int. Conf. on Image Processing, NY, USA, pp. 3924–3928, 2017.
20. J. Redmon, S. Divvala, R. Girshick and A. Farhadi, “You only look once: Unified, real-time object detection,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, NY, USA, pp. 779–788, 2016.
21. C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens and Z. Wojna, “Rethinking the inception architecture for computer vision,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Las Vegas, USA, pp. 2818–2826, 2017.
22. M. T. Ribeiro, S. Singh and C. Guestrin, ““Why should i trust you?” explaining the predictions of any classifier,” in Proc. of the 22nd ACM SIGKDD Int. Conf. on Knowledge Discovery and Data Mining, NY, USA, pp. 1135–1144, 2016.
23. R. R. Selvaraju, M. Cogswell, A. Das and D. Batra, “Grad-cam: Visual explanations from deep networks via gradient-based localization,” in Proc. of the IEEE Int. Conf. on Computer Vision, Venice, Italy, pp. 618–626, 2017.
24. T. Akram, M. Yasmin and R. S. Nayak, “Stomach deformities recognition using rank-based deep features selection,” Journal of Medical Systems, vol. 43, pp. 1–15, 2019.
25. S. Fan, L. Xu, Y. Fan, K. Wei and L. Li, “Computer-aided detection of small intestinal ulcer and erosion in wireless capsule endoscopy images,” Physics in Medicine and Biology, vol. 63, pp. 165001, 2018.
26. D. E. Diamantis, D. K. Iakovidis and A. Koulaouzidis, “Look-behind fully convolutional neural network for computer-aided endoscopy,” Biomedical Signal Processing and Control, vol. 49, pp. 192–201, 2019.
27. M. Rashid, M. Yasmin and U. J. Tanik, “Deep CNN and geometric features-based gastrointestinal tract diseases detection and classification from wireless capsule endoscopy images,” Journal of Experimental and Theoretical Artificial Intelligence, vol. 6, pp. 1–23, 2019.
28. H. Alaskar, A. Hussain, N. Al-Aseem, P. Liatsis and D. Al-Jumeily, “Application of convolutional neural networks for automated ulcer detection in wireless capsule endoscopy images,” Sensors, vol. 19, pp. 1265, 2019.
29. F. Ucar and D. Korkmaz, “COVIDiagnosis-net: Deep Bayes-squeezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images,” Medical Hypotheses, vol. 140, pp. 109761, 2020.
30. M. Nour, Z. Cömert and K. Polat, “A novel medical diagnosis model for COVID-19 infection detection based on deep features and Bayesian optimization,” Applied Soft Computing, vol. 97, pp. 106580, 2020.
31. Y. Gal and Z. Ghahramani, “Dropout as a Bayesian approximation: Representing model uncertainty in deep learning,” in Int. Conf. on Machine Learning, Leicester, UK, pp. 1050–1059, 2016.
32. C. Leibig, V. Allken, M. S. Ayhan, P. Berens and S. Wahl, “Leveraging uncertainty information from deep neural networks for disease detection,” Scientific Reports, vol. 7, pp. 1–14, 2017.
33. S. Bach, A. Binder, G. Montavon, F. Klauschen and W. Samek, “On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation,” PloS One, vol. 10, pp. e0130140, 2015.
34. P. K. Randel, K. R. Griwodz, C. Eskeland, S. L. Lange, D. Johansen et al., “Kvasir: A multi-class image dataset for computer aided gastrointestinal disease detection,” in Proc. of the 8th ACM on Multimedia Systems Conf., NY, USA, pp. 164–169, 2009.
35. J. Wu, X. Y. Chen, H. Zhang, L. D. Xiong and S. H. Deng, “Hyperparameter optimization for machine learning models based on Bayesian optimization,” Journal of Electronic Science and Technology, vol. 17, pp. 26–40, 2019.
36. M. T. Ribeiro, S. Singh and C. Guestrin, “Why should I trust you?”: Explaining the predictions of any classifier,” in Proc. of the 22nd ACM SIGKDD Int. Conf. on Knowledge Discovery and Data Mining, San Francisco, California, USA, pp. 1–6, 2016.
37. G. Wimmer, A. Vécsei, M. Häfner and A. Uhl, “Fisher encoding of convolutional neural network features for endoscopic image classification,” Journal of Medical Imaging, vol. 5, pp. 034504, 2018.
38. R. Zhang, Y. Zheng, T. W. C. Mak, R. Yu, J. Y. Lau et al., “Automatic detection and classification of colorectal polyps by transferring low-level CNN features from nonmedical domain,” IEEE Journal of Biomedical and Health Informatics, vol. 21, pp. 41–47, 2016.
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |