DOI:10.32604/cmc.2022.021995

| Computers, Materials & Continua DOI:10.32604/cmc.2022.021995 |  |
| Article |
Intelligent Biomedical Electrocardiogram Signal Processing for Cardiovascular Disease Diagnosis
1Department of Electronics and Communication Engineering, University College of Engineering Ariyalur, Ariyalur, 621704, India
2School of Computing, SRM Institute of Science and Technology, Chennai, 603203, India
3Department of Electronics and Communication Engineering, K. Ramakrishnan College of Engineering, Tiruchirappalli, 621112, India
4Department of Computer Science, Factually of Science & Art at Mahayil, King Khalid University, Saudi Arabia
5Faculty of Computer and IT, Sana'a University, Yemen
6Department of Biomedical Engineering, College of Engineering, Princess Nourah Bint Abdulrahman University, Saudi Arabia
7Department of Computer and Self Development, Preparatory Year Deanship, Prince Sattam Bin Abdulaziz University, Alkharj, Saudi Arabia
*Corresponding Author: R. Krishnaswamy. Email: krishnaspecial@gmail.com
Received: 22 July 2021; Accepted: 23 August 2021
Abstract: Automatic biomedical signal recognition is an important process for several disease diagnoses. Particularly, Electrocardiogram (ECG) is commonly used to identify cardiovascular diseases. The professionals can determine the existence of cardiovascular diseases using the morphological patterns of the ECG signals. In order to raise the diagnostic accuracy and reduce the diagnostic time, automated computer aided diagnosis model is necessary. With the advancements of artificial intelligence (AI) techniques, large quantity of biomedical datasets can be easily examined for decision making. In this aspect, this paper presents an intelligent biomedical ECG signal processing (IBECG-SP) technique for CVD diagnosis. The proposed IBECG-SP technique examines the ECG signals for decision making. In addition, gated recurrent unit (GRU) model is used for the feature extraction of the ECG signals. Moreover, earthworm optimization (EWO) algorithm is utilized to optimally tune the hyperparameters of the GRU model. Lastly, softmax classifier is employed to allot appropriate class labels to the applied ECG signals. For examining the enhanced outcomes of the proposed IBECG-SP technique, an extensive simulation analysis take place on the PTB-XL database. The experimental results portrayed the supremacy of the IBECG-SP technique over the recent state of art techniques.
Keywords: Biomedical signals; ecg; disease diagnosis; artificial intelligence; parameter tuning; gru model
Cardiovascular diseases (CVD) is a worldwide healthcare problem since they contribute around 30% of overall death and 10% burden of the overall disease [1]. In 2005, around 58 million mortalities occur throughout the world, 17 million deaths existed because of CVD [2]. Based on stroke statistics and heart disease, the adapted population is attributed to the CVD death consist of poor eating habits, blood pressure, etc [3]. Specifically, CVD is connected to a poor standard of livings, like smoking, physical inactivity, alcohol consumption, and increased intake of fat. As the occurrence of obesity has dramatically augmented in recent years and now it has attained a pandemic proportion [4]. Further, in America, 16.7% of women, and 21.3% of men above eighteen years, continue to smoke. While the several persons with CVD are retreating in many higher income nations, middle and low income countries have increased the prevalence, in which 82% of CVD death occurs in both male and female. In disease occurrence research, the WHO determined CVD from enzymes, symptoms, and ECG abnormalities [5]. Among these diagnoses approaches, ECG can potentially deliver cost saving, since the signals attainment is non invasive needs only medium qualified labor. But, ECG alone is frequently inadequate for diagnosing CVDs, like acute myocardial infarction/acute coronary syndrome. It is significant for improving the accuracy of ECG based diagnoses since it decreases the requirement for high cost diagnoses tools. Further, a high accuracy permits us for detecting earlier CVD. Conventionally, enhancing the diagnoses accuracy needs further training to screen physicians. But, training is a substantial recurring cost factor. CAD system gives us hope for decreasing cost and simultaneously improve the accuracy of ECG diagnoses. The lessening of cost rises from human labor is substituted by machines.
Computerized detection of ECG abnormalities is regularly utilized for classifying cardiologist's long term ECG records. Feature extraction method includes Hermite function, wave shape function, statistical feature, and wavelet based feature. Approaches for classifying this extracted feature include k-nearest neighbour (KNN) rules, SVMs, ANNs, linear discriminants, and DTs. Advanced automatic ECG detection system is always based on a pattern matching architecture which signifies the ECG signal as a series of stochastic patterns. They need complicated feature extraction approaches and higher sampling rate and hence it takes time [6]. For real world execution in the hospital at moderate cost, this system should utilize a low sampling rate and a simple set of features. A constraint of various processes is utilized for automate classification of ECG is the incapability of handling huge intraclass variants. They are heavily based on supervised training datasets and carry out poorly while processing huge amounts of novel ECG records [7]. Additionally, classification method doesn't execute if there are wider interpatient variants in ECG signal. Therefore, unreliable competence creates classification methods inconsistent in the medical practice. The DL is a novel ML method i.e., becomes conventional for pattern detection [8]. It was effectively utilized to image verification, object recognition, speech recognition, and classification. DL methods are well-enhanced in the accuracy of detection tools. They are utilized for creating deep, multistage architectures for recognition systems and unsupervised learning.
This paper introduces an intelligent biomedical ECG signal processing (IBECG-SP) technique for CVD diagnosis. The proposed IBECG-SP technique inspects the ECG signals for decision making. It also uses gated recurrent unit (GRU) model for the feature extraction of the ECG signals. Besides, earthworm optimization (EWO) algorithm is utilized to optimally tune the hyperparameters of the GRU model. The design of EWO algorithm to tune the GRU model for ECG signal recognition shows the novelty of the work. Finally, softmax classifier is employed to allot correct class labels to the ECG signals. To investigate the betterment of the proposed IBECG-SP technique, a comprehensive set of experimentation is achieved on the benchmark PTB-XL dataset. In short, the key contributions of the study are given as follows.
• Designs a new IBECG-SP technique to inspect the ECG signals for decision making
• Propose a GRU model to extract the features of the ECG signals
• Employ EWO algorithm to adjust the parameters of the GRU model
• Present a softmax classifier for allocating class labels to the ECG signals
• Validate the results of the IBECG-SP technique on the PTB-XL dataset
Zhang et al. [9] employed a DL approach for building a model for automatic classification and detection of ECG signal. First, they trained CNNs for detecting CVD in ECG signals by a training dataset of 259,789 ECG signals gathered from the cardiac functions room. In Vijayarangan et al. [10], a new application of the Unet integrated by Inception and Residual block is projected to extract the R-peak from the ECG. Further, the problem formulation strongly handles the problems of sparsity and variability of ECG R-peaks. The projected network was trained by a database having ECG events which contain CVDs and was validated with 3 conventional ECG detectors on a valid set. Using objectives for achieving an optimal detection of HD, proper DL and ML methods were detected [11] and the essential classification methods were tested and developed. The GAN method is preferred using objectives for handling imbalanced data by creating and with further false data for detecting purposes. Furthermore, a cooperative method utilizing GAN and LSTM is advanced in this study determines high efficiency than single DL approach. Rath et al. [12] proposed an effective diagnose of CHD from the ECG recording of the subject that employs a robust and simple LSTM network approach of detecting CHD. The conventional PTB diagnostic database version 1.0.0 PhysioNet containing ECG signal recording of 268 subjects is utilized in this approach. The 3 phases of LSTM network using 64, 256, and 128 models in all stages and with 20% arbitrary dropouts of weights among models are applied for developing the detection method.
Khan et al. [13] proposed a generalized method for processing each format of ECG. The SSD MobileNet v2-based DNN framework is utilized for detecting CVD. The research concentrated on identifying the 4 main cardiac abnormalities with 98% accuracy outcomes have been computed. Liang et al. [14] aimed to enhance accuracy by integrating the CNN and BiLSTM. As we know, this method hasn't been explored to date. In this research, dataset-1 using individual lead ECG and dataset-2 using twelve-lead ECG have been utilized for exploring a viable and practical heartbeat event classifier. An evolution neural (method I) and a DL (method II) which integrates CNN and BiLSTM model has been evaluated and compared in processed heartbeat event classification.
Tyagi et al. [15] utilized the pre-processing and FS for appropriate heart diseases classification, in which DWT is utilized for reducing noise and segmenting ECG signal and GOA is utilized to select R-peaks features from the extracted feature set based on R-R intervals and R-peaks which helps to achieve improved classification accuracy. In order to test and train of proposed HCM, the conventional MIT-BIH arrhythmia dataset is used for hybrid CNN framework. The variety of accurate R-R intervals and R-peaks are an important aspect and due to the lack of apposite pre-processing stages such as signal decomposition, noise removal, filtering, and smoothing, the exclusivity of extracted feature is lesser.
Hasan et al. [16] presented a technique for classifying different kinds of heart diseases via 1D DCNN. First, Every ECG signal undergo decomposition via EMD and high order IMF is integrated for creating an adapted ECG signal. It is considered that the usage of EMD will give a wide ranging of data and denoising efficiency. Such processing signals are fed to the CNN framework which categorizes the record based on the CVD by a softmax regressor in the network. It is noted that the CNN framework learnt the intrinsic features of the adapted ECG signal better than raw ECG signal. Hammad et al. [17] presented a DNN approach for ameliorating the above-mentioned challenges. This approach comprises learning phase in which classification accuracy is enhanced by a strong feature extraction protocol. Then, utilizing GA procedure for aggregating an optimal integration of classification and feature extraction. Though several methods are available in the literature, the classification method doesn't execute if there are wider interpatient variants in ECG signal. Therefore, unreliable competence creates classification methods inconsistent in the medical practice. The DL is a novel ML method i.e., becomes conventional for pattern detection which is effectively utilized to image verification, object recognition, speech recognition, and classification.
3 The Proposed Biomedical Signal Processing Technique
Fig. 1 demonstrates the overall working process of IBECG-SP model. This study has focused on the design of IBECG-SP technique for ECG signal classification. The proposed IBECG-SP technique encompasses four levels namely data preprocessing, feature extraction, parameter optimization, and data classification. The detailed working of these modules is discussed in the succeeding sections.
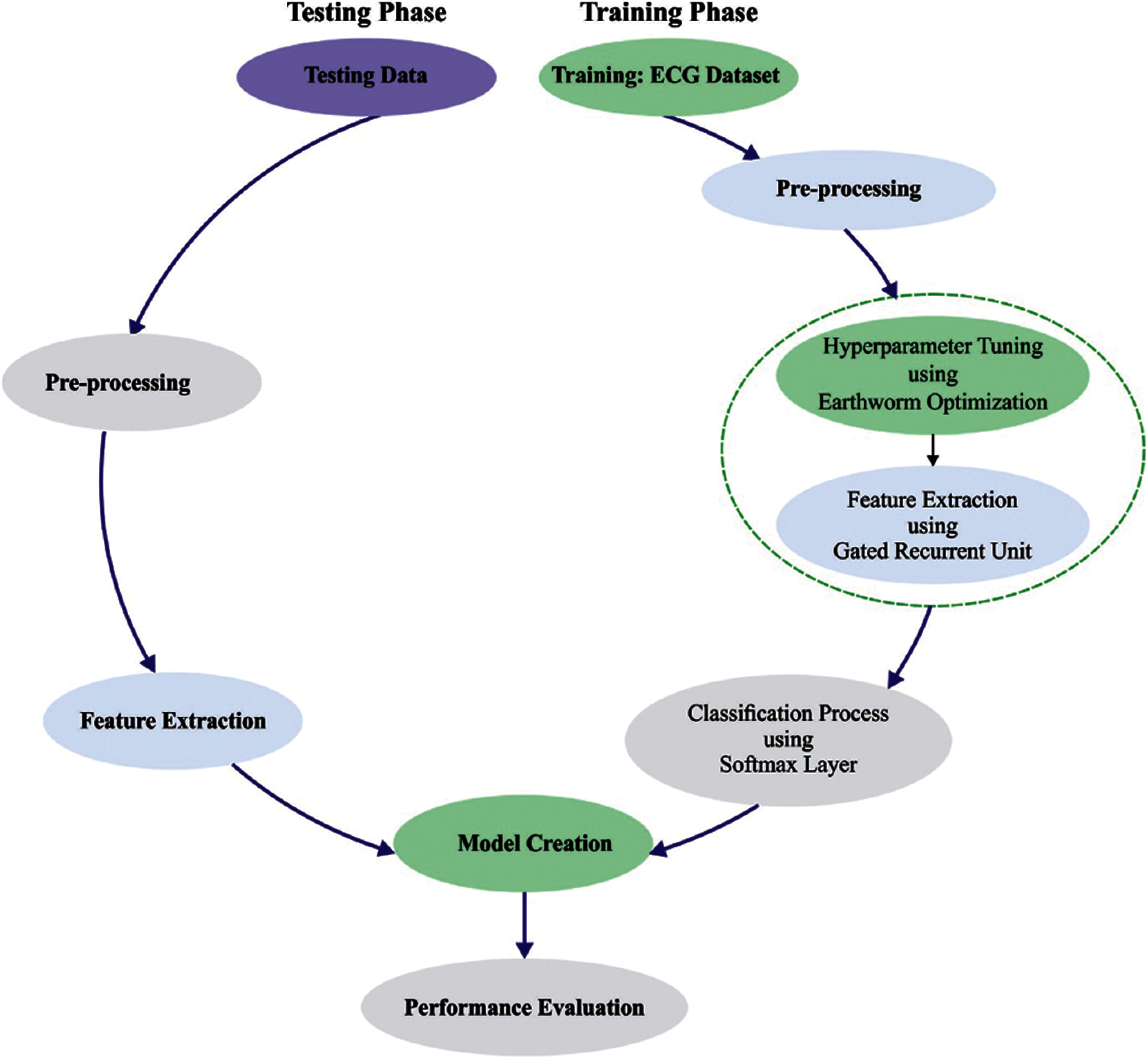
Figure 1: Overall process of IBECG-SP method
3.1 Level I: Data Pre-Processing
At the first stage, the ECG signals are pre-processed to transform them into compatible format. Here, a set of 3000 ECG signals are employed to validate the presented technique. Among them, 35 signals encompass NULL classes which are rejected from the database. Then, the rest of the 2965 ECG signals are used for experimentation. Moreover, a sampling rate of 100 is preferred amongst the two sampling rates of 100 and 500 from the database.
3.2 Level II: Feature Extraction
Next to the ECG signal pre-processing stage, the resultant ECG signals are fed into the GRU model to derive useful features. Deep neural networks (DNN) resolve the restrictions of shallow network and has the robust capability of non-linear fitting. But the classical DNN models do not consider the temporal relationships among the classified instances leading to loss of data during classification. The RNN models are developed to resolve the issue of time dependencies. The RNN creates a feedback link amongst the hidden layers and therefore the network can sustain the learned data to the present moment and compute the end outcomes of the network with the input of present moment. The efficiency of the RNN to solve timing related issues are evaluated in different areas of applications. But it suffers from the vanishing gradient process resulting in poor convergence of the network and fails to overcome the effect of long-term dependency.
Several models to improve the RNN performance are presented and a widely employed network is the Long Short Term Memory (LSTM) [18]. The LSTM includes
where
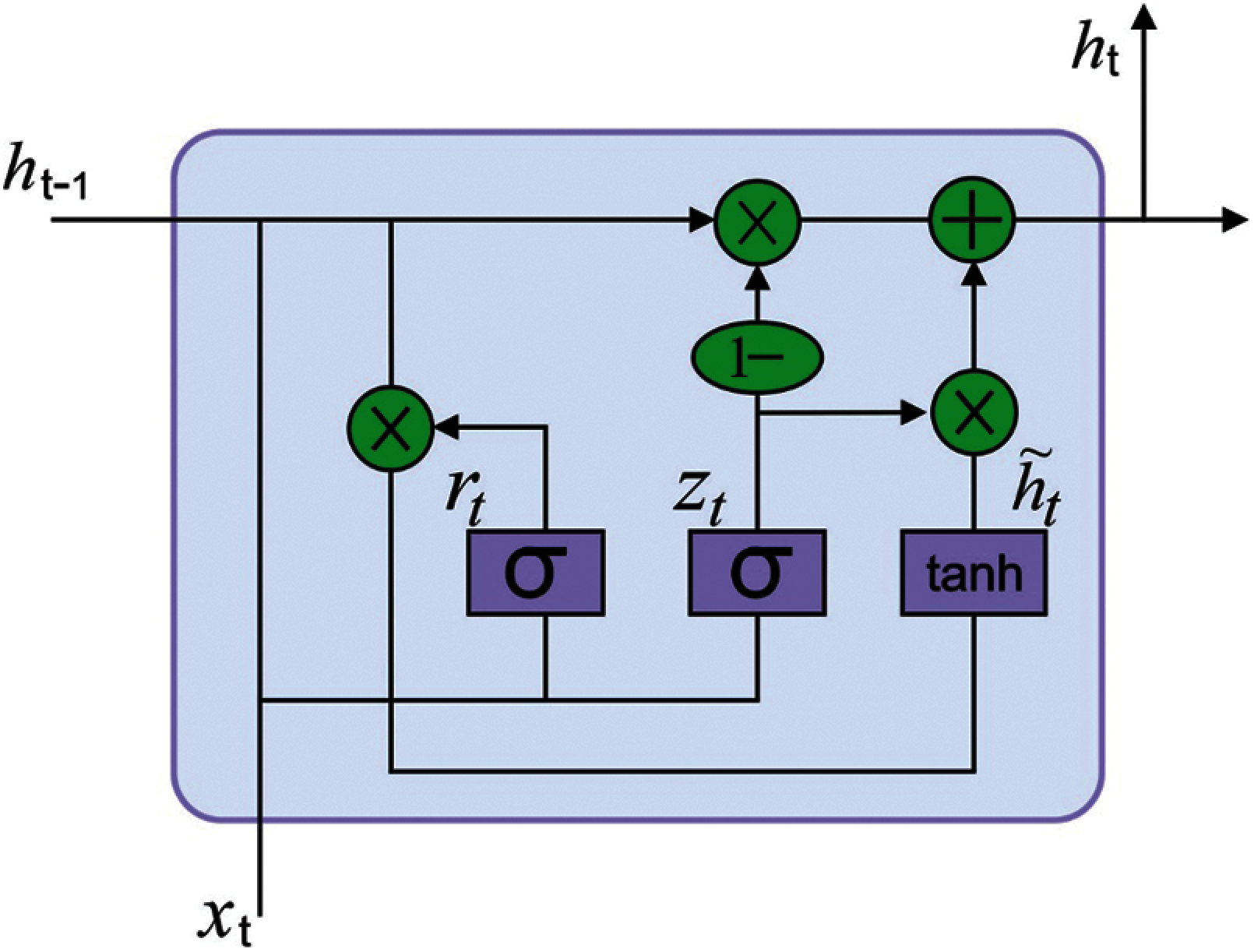
Figure 2: Structure of GRU
3.3 Level III: Parameter Optimization
In order to improve the ECG recognition outcome of the GRU model, the EWO algorithm is used which is inspired from the reproductivity process of earthworms (EW) for solving optimization issues [19]. The EWO was dependent upon few fundamental rules as follow: (A) All EWs in the population is imitating offsprings by 2 and only 2 types of reproduction. (B) The genes limited as child EW is s similar length as that parent EW. (C) A few EW individuals of preceding generation containing optimum are performed straightaway to the subsequent iteration with no modification.
The EW is a kind of hermaphrodite. It implies every one of them performs male as well as female sex organs. Thus, the sole parent EW creates a child EW by itself. The reproduction_1 is defined as follows:
The above formula explains the process of creating
The Reproduction_2 utilizes an enhanced type of crossover operator. Assume, M be the amount of child EWs and it can be 1, 2, or 3 in one of the belongings. The amount of parent EWs (N) is some integer which is more than 1. During this work, uniform crossover is executed with
Initially, 2 offsprings
If
Otherwise,
Finally, the created EW
Afterward, the creating EWs
where
where
where,
At this point,
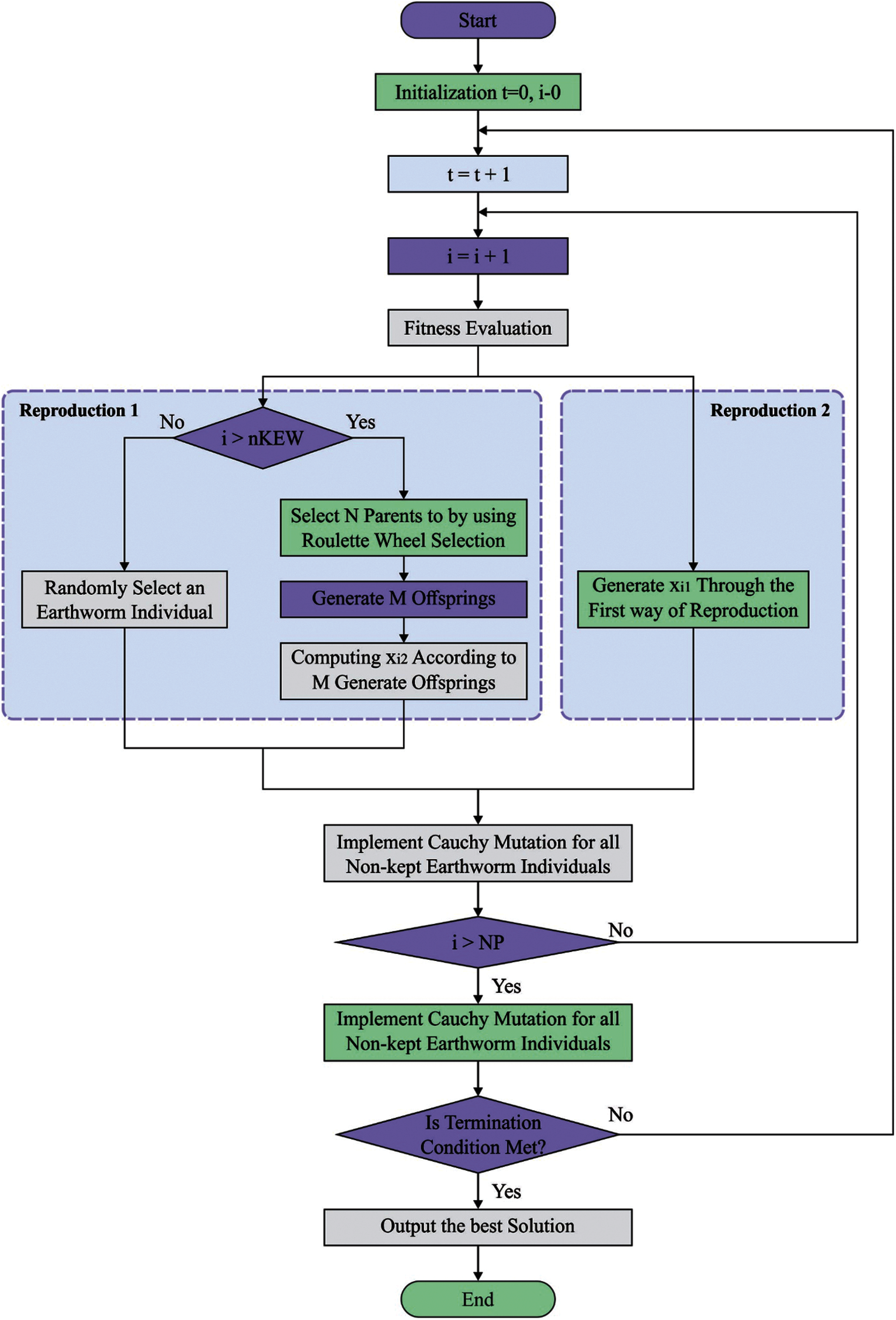
Figure 3: Flowchart of EWO

3.4 Level IV: Data Classification
At the final stage, the softmax (SM) classifier is employed to categorize the ECG signal using the features from the previous process. Owing to the multi-objective classification process of ECG recognition, SM classifier is employed as the last outcome layer of the GRU:
where
This study employs PTB-XL dataset [21] that includes 21837 ECG signals of 10s duration from 18885 persons in which 52% of persons are male and the residual 48% of the persons are female. ECG data utilized to annotation follow the SCP-ECG typical and assigned to 3 non-mutually select classes namely diagnostic, form, and rhythm. Entirely, 71 different records have existed that decomposed as to 44 diagnostics, 12 rhythm, and 19 form statements. Also, the PTB-XL data includes 5 class labels namely normal ECG (NORM), conduction disturbance (CD), myocardial infarction (MI), hypertrophy (HYP), and: ST/T changes (STTC). Besides, an entire 24 subclass labels are also given.
Fig. 4 inspects the ECG recognition performance of the proposed IBECG-SP technique under the execution run-1. The IBECG-SP technique has classified the samples into ‘CD’ class with the maximum accuracy of 88.270%. Besides, the samples under ‘HYP’ class are identified by the IBECG-SP technique with an accuracy of 95.190%. In line with, the IBECG-SP technique classified the samples under ‘MI’ class with an accuracy of 89.010%. Additionally, the IBECG-SP technique has identified the ‘NORM’ class samples with an accuracy of 77.210%. Furthermore, the IBECG-SP technique has classified the samples into ‘STTC’ class with an accuracy of 86.101%. At last, the IBECG-SP technique has categorized the dataset by accomplishing a maximum average accuracy of 87.138%.
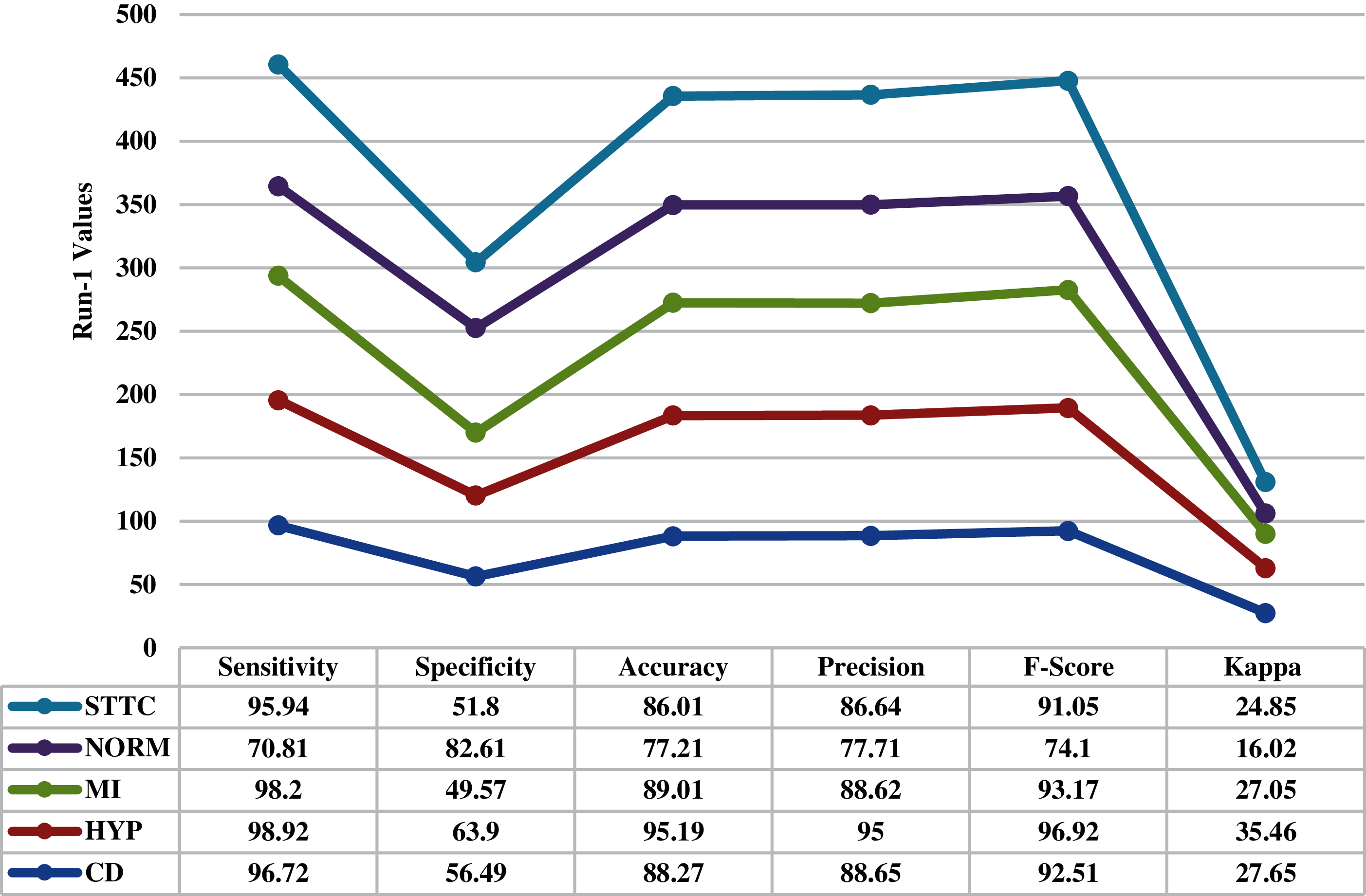
Figure 4: Result analysis of IBECG-SP technique under execution run-1
Fig. 5 examines the ECG recognition efficiency of the presented IBECG-SP approach under the execution run-2. The IBECG-SP method has classified the instances as to ‘CD’ class with the maximal accuracy of 89.050%. Along with that, the samples under ‘HYP’ class are recognized by the IBECG-SP technique with an accuracy of 94.920%. In addition, the IBECG-SP methodology classified the samples under ‘MI’ class with an accuracy of 88.410%. Moreover, the IBECG-SP manner has identified the ‘NORM’ class instances with an accuracy of 77.410%. Likewise, the IBECG-SP approach has classified the samples into ‘STTC’ class with an accuracy of 86.380%. Finally, the IBECG-SP algorithm has classified the dataset with a superior average accuracy of 87.234%.
Fig. 6 reviews the ECG recognition performance of the projected IBECG-SP method under the execution run-3. The IBECG-SP manner has classified the samples as to ‘CD’ class with the superior accuracy of 89.250%. In line with, the samples under ‘HYP’ class are identified by the IBECG-SP manner with an accuracy of 94.780%. Followed by, the IBECG-SP algorithm classified the instances under ‘MI’ class with an accuracy of 89.910%. Moreover, the IBECG-SP method has recognized the ‘NORM’ class samples with an accuracy of 77.680%. Next, the IBECG-SP algorithm has classified the samples into ‘STTC’ class with an accuracy of 86.750%. Eventually, the IBECG-SP approach has resulted to an increased average accuracy of 87.474%.

Figure 5: Result analysis of IBECG-SP technique under execution run-2

Figure 6: Result analysis of IBECG-SP technique under execution run-3
An average results analysis of the IBECG-SP technique under three different runs takes place in Tab. 1. The results ensured that the IBECG-SP technique has gained effective outcomes under all three runs. For instance, under run-1, the IBECG-SP technique results to an average sensitivity of 92.118%, specificity of 60.874%, accuracy of 87.138%, precision of 87.324%, F-score of 89.550%, and kappa of 26.206%. Eventually, under run-2, the IBECG-SP approach provided an average sensitivity of 92.368%, specificity of 59.770%, accuracy of 87.234%, precision of 87.280%, F-score of 89.630%, and kappa of 26.064%. Meanwhile, under run-3, the IBECG-SP methodology has obtainable average sensitivity of 92.660%, specificity of 60.400%, accuracy of 87.474%, precision of 87.388%, F-score of 89.852%, and kappa of 26.452%.
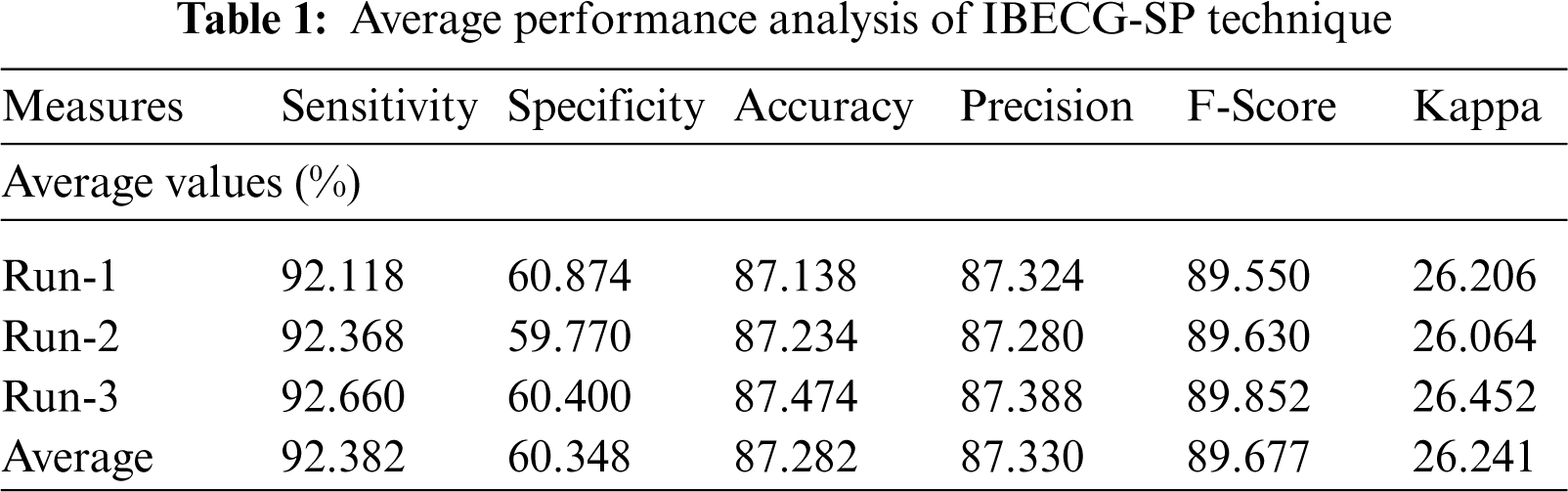
For examining the better performance of the proposed IBECG-SP technique, a comparison study interms of accuracy is made in Tab. 2 and Fig. 7. The results depicted that the LR and DT models have accomplished lower accuracy values of 37.380% and 27.900% respectively. At the same time, the KNC technique has gained slightly reduced outcomes with a moderate accuracy of 66.890%. Followed by, the RF and One Dim. DCNN techniques have reached a reasonable accuracy of 79.830% and 73% respectively. Moreover, the DLECG-PTBXL and GBT models have provided a competitive accuracy of 87.280% and 84.700% respectively. However, the proposed IBECG-SP technique has exhibited improved outcomes a maximum accuracy of 87.280%.
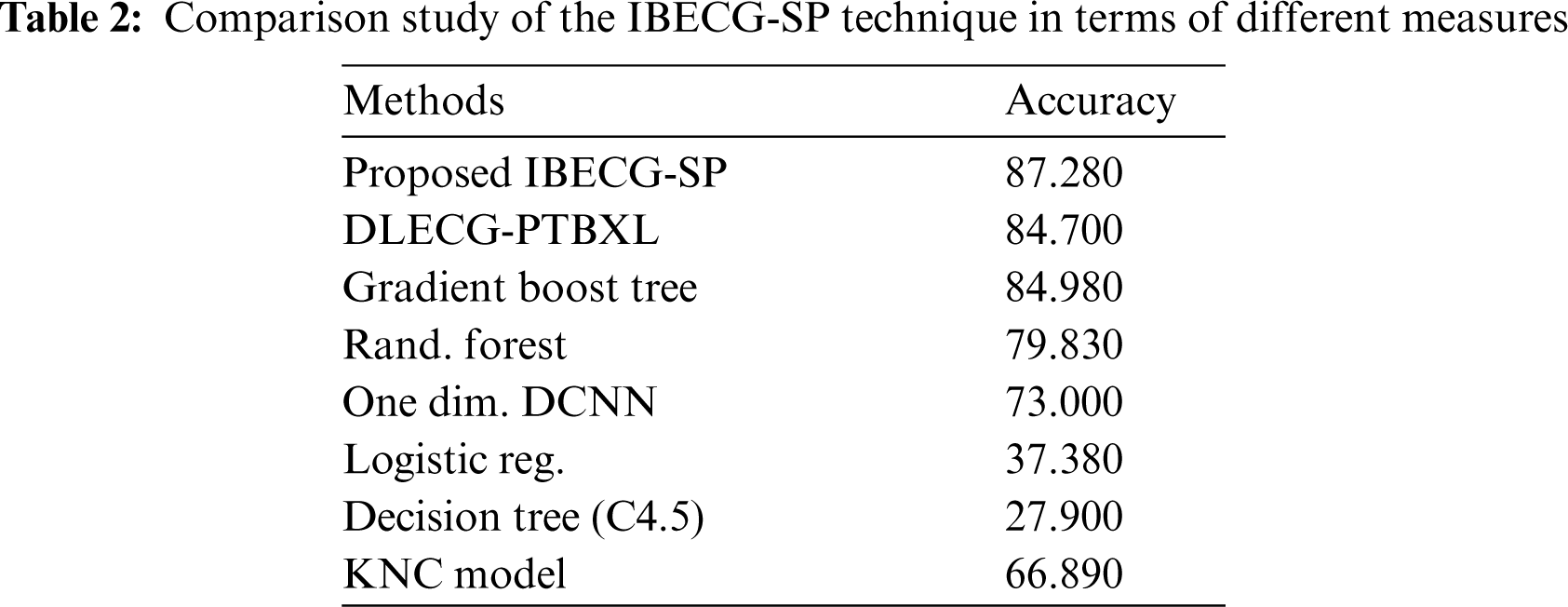

Figure 7: Accuracy analysis of IBECG-SP model with existing techniques
By looking into the aforementioned result analysis, it is obvious that the IBECG-SP technique has the ability to recognize ECG signals and thereby diagnose CVD effectively.
This study has focused on the design of IBECG-SP technique for ECG signal classification. The proposed IBECG-SP technique mainly focused to examine the ECG signals for decision making. The proposed IBECG-SP technique encompasses four levels namely data pre-processing, GRU based feature extraction, EWO based parameter optimization, and SM based data classification. The design of EWO algorithm to tune the GRU model for ECG signal recognition resulted to enhanced outcomes. To investigate the betterment of the proposed IBECG-SP technique, a comprehensive set of experimentations take place. The experimental results portrayed the supremacy of the proposed IBECG-SP technique over the recent techniques. In future, the diagnostic performance of the IBECG-SP technique can be boosted by advanced DL architectures.
Funding Statement: The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work under Grant Number (RGP 2/209/42). This research was funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-Track Path of Research Funding Program.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. O. Faust and E. Y. K. Ng, “Computer aided diagnosis for cardiovascular diseases based on ECG signals: A survey,” Journal of Mechanics in Medicine and Biology, vol. 16, no. 1, pp. 1640001, 2016. [Google Scholar]
2. R. Bharathi, T. Abirami, S. Dhanasekaran, D. Gupta, A. Khanna et al., “Energy efficient clustering with disease diagnosis model for IoT based sustainable healthcare systems,” Sustainable Computing: Informatics and Systems, vol. 28, pp. 100453, 2020. [Google Scholar]
3. A. S. Go, D. Mozaffarian, V. L. Roger, E. J. Benjamin, J. D. Berry et al., “Executive summary: Heart disease and stroke statistics—2013 update: A report from the american heart association,” Circulation, vol. 127, no. 1, pp. 143–152, 2013. [Google Scholar]
4. M. Elhoseny, K. Shankar and J. Uthayakumar, “Intelligent diagnostic prediction and classification system for chronic kidney disease,” Scientific Reports, vol. 9, no. 1, pp. 9583, 2019. [Google Scholar]
5. K. Shankar, S. K. Lakshmanaprabu, D. Gupta, A. Maseleno and V. H. C. de Albuquerque, “Optimal feature-based multi-kernel SVM approach for thyroid disease classification,” The Journal of Supercomputing, vol. 76, no. 2, pp. 1128–1143, 2020. [Google Scholar]
6. S. M. Mathews, C. Kambhamettu and K. E. Barner, “A novel application of deep learning for single-lead ECG classification,” Computers in Biology and Medicine, vol. 99, pp. 53–62, 2018. [Google Scholar]
7. D.-N. Le, V. S. Parvathy, D. Gupta, A. Khanna, J. J. P. C. Rodrigues et al., “IoT enabled depthwise separable convolution neural network with deep support vector machine for COVID-19 diagnosis and classification,” International Journal of Machine Learning and Cybernetics, 2021.https://doi.org/10.1007/s13042-020-01248-7 [Google Scholar]
8. A. Rajagopal, A. Ramachandran, K. Shankar, M. Khari, S. Jha et al., “Optimal routing strategy based on extreme learning machine with beetle antennae search algorithm for low earth orbit satellite communication networks,” International Journal of Satellite Communications and Networking, vol. 39, no. 3, pp. 305–317, 2021. [Google Scholar]
9. X. Zhang, K. Gu, S. Miao, X. Zhang, Y. Yin et al., “Automated detection of cardiovascular disease by electrocardiogram signal analysis: A deep learning system,” Cardiovascular Diagnosis and Therapy, vol. 10, no. 2, pp. 227–235, 2020. [Google Scholar]
10. S. Vijayarangan, V. R. B. Murugesan, P. S. P. J. Joseph and M. Sivaprakasam, “RPnet: A deep learning approach for robust r peak detection in noisy ecg,” in 2020 42nd Annual Int. Conf. of the IEEE Engineering in Medicine & Biology Society (EMBCMontreal, QC, Canada, pp. 345–348, 2020. [Google Scholar]
11. A. Rath, D. Mishra, G. Panda and S. C. Satapathy, “Heart disease detection using deep learning methods from imbalanced ECG samples,” Biomedical Signal Processing and Control, vol. 68, pp. 102820, 2021. [Google Scholar]
12. A. Rath, D. Mishra and G. Panda, “LSTM-Based cardiovascular disease detection using ecg signal,” Cognitive Informatics and Soft Computing, vol. 1317, pp. 133–142, 2021. [Google Scholar]
13. A. H. Khan, M. Hussain and M. K. Malik, “Cardiac disorder classification by electrocardiogram sensing using deep neural network,” Complexity, vol. 2021, pp. 1–8, 2021. [Google Scholar]
14. Y. Liang, S. Yin, Q. Tang, Z. Zheng, M. Elgendi et al., “Deep learning algorithm classifies heartbeat events based on electrocardiogram signals,” Frontiers in Physiology, vol. 11, pp. 569050, 2020. [Google Scholar]
15. A. Tyagi and R. Mehra, “Intellectual heartbeats classification model for diagnosis of heart disease from ECG signal using hybrid convolutional neural network with GOA,” SN Applied Sciences, vol. 3, no. 2, pp. 265, 2021. [Google Scholar]
16. N. I. Hasan and A. Bhattacharjee, “Deep learning approach to cardiovascular disease classification employing modified ecg signal from empirical mode decomposition,” Biomedical Signal Processing and Control, vol. 52, pp. 128–140, 2019. [Google Scholar]
17. M. Hammad, A. M. Iliyasu, A. Subasi, E. S. L. Ho and A. A. A. El-Latif, “A multitier deep learning model for arrhythmia detection,” IEEE Transactions on Instrumentation and Measurement, vol. 70, pp. 1–9, 2021. [Google Scholar]
18. B. Yan and G. Han, “LA-GRU: Building combined intrusion detection model based on imbalanced learning and gated recurrent unit neural network,” Security and Communication Networks, vol. 2018, pp. 1–13, 2018. [Google Scholar]
19. G. G. Wang, S. Deb and L. D. S. Coelho, “Earthworm optimisation algorithm: A bio-inspired metaheuristic algorithm for global optimisation problems,” International Journal of Bio-Inspired Computation, vol. 12, no. 1, pp. 1, 2018. [Google Scholar]
20. I. Ghosh and P. K. Roy, “Application of earthworm optimization algorithm for solution of optimal power flow,” in 2019 Int. Conf. on Opto-Electronics and Applied Optics (OptronixKolkata, India, pp. 1–6, 2019. [Google Scholar]
21. P. Wagner, N. Strodthoff, R. D. Bousseljot, D. Kreiseler, F. I. Lunze et al., “PTB-XL, a large publicly available electrocardiography dataset,” Scientific Data, vol. 7, no. 1, pp. 154, 2020. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |